Abstract
L-Theanine content has tissues and cultivars specificity in tea plant (Camellia sinensis L.), the correlations of theanine metabolic related genes expression profiles with theanine contents were explored in this study. L-theanine contents in the bud and 1st leaf, 2nd leaf, 3rd leaf, old leaf, stem, and lateral root were determined by HPLC from three C. sinensis cultivars, namely ‘Huangjinya’, ‘Anjibaicha’, and ‘Yingshuang’, respectively. The theanine contents in leaves and root of ‘Huangjinya’ were the highest, followed by ‘Anjibaicha’, and ‘Yingshuang’. The theanine contents in the leaves reduced as the leaf mature gradually, and in stem were the least. Seventeen genes encoding enzymes involved in theanine metabolism were identified from GenBank and our tea transcriptome database, including CsTS1, CsTS2, CsGS1, CsGS2, CsGOGAT-Fe, CsGOGAT-NAD(P)H, CsGDH1, CsGDH2, CsALT, CsSAMDC, CsADC, CsCuAO, CsPAO, CsNiR, CsNR, CsGGT1, and CsGGT3. The transcript profiles of those seventeen genes in the different tissues of three tea plant cultivars were analyzed comparatively. Among the different cultivars, the transcript levels of most selected genes in ‘Huangjinya’ were significantly higher than that in the ‘Anjibaicha’ and ‘Yingshuang’. Among the different tissues, the transcript levels of CsTS2, CsGS1, and CsGDH2 almost showed positive correlation with the theanine contents, while the other genes showed negative correlation with the theanine contents in most cases. The theanine contents showed correlations with related genes expression levels among cultivars and tissues of tea plant, and were determined by the integrated effect of the metabolic related genes.
Keywords: theanine, biosynthesis, content identification, gene expression profiling, Camellia sinensis L.
Introduction
L-theanine, or called as γ-glutamyl-L-ethylamide or γ-ethylamino-L-glutamic acid, is a unique non-protein amino acid in tea plant (Camellia sinensis (L.) O. Kuntze) (Deng et al., 2008). All theanine that naturally exists in tea plant belongs to L-theanine, D-theanine can only be produced industrially. L-theanine is an important indicator in the quality evaluation of green tea because of the particular umami taste that largely influences the flavor of tea (Yamaguchi and Ninomiya, 2000). The functions of theanine related to food science and human nutrition have been extensively studied since theanine was first discovered in tea plant leaves. Previous studies have shown that theanine can improve memory and learning ability by activating relative central neurotransmitters (Haskell et al., 2008). To a certain extent, theanine can reduce blood-pressure, maintain stability, and promote relaxation and concentration by inhibiting the negative effects of caffeine (Yokogoshi et al., 1995; Kakuda et al., 2000). In addition, theanine performs positive functions in anti-diseases action, including enhancing anti-tumor activity, preventing vascular diseases, and neuroprotection (Rogers et al., 2008; Liu et al., 2009; Zhao and Zhao, 2014). Numerous physiological functions make L-theanine as one of the hot spots in the development and utilization of functional components in tea.
As an important and highly abundant free amino acid in tea plants, L-theanine, almost can be detected in all tissues (Deng et al., 2009). However, L-theanine content in tea plants considerably varies among different cultivars and tissues. For example, the tea plant cultivars with small leaf contain higher amino acid contents than the cultivars with large leaf (Jiang, 2013). The small-leaf tea cultivars are suitable for producing the green tea, and the large-leaf cultivars are used to process the black tea (Wu et al., 2014). In general, the theanine content of the young leaves is the highest, and followed by the old leaves, root, and stem (Selvendran and Selvendran, 1973). Within a year, the theanine content is the highest in spring, and then decreases gradually, after which it rises during autumn to a certain degree. In winter (dormancy period), the theanine in tea plant is accumulated and stored in the root. Until to the March and April of the next year (germination period), theanine shifts to shoots and remains at a relative low level in roots until to August (Takeo, 1979). Moreover, L-theanine content is influenced by natural environmental conditions, such as light intensity and concentration of ammonia (Ashihara, 2015).
The theanine metabolism pathway, including synthesis, transportation, and hydrolysis, has been studied for more than sixty years since theanine was discovered by Sakato (Deng et al., 2010). Numerous pivotal enzymes participate in the metabolism pathway of theanine. The gene expression levels of theanine metabolic enzymes largely influence the content, distribution, and metabolism rules of theanine in tea plants (Shi et al., 2011). In theanine biosynthesis, numerous structural genes encode enzymes that directly catalyze reaction steps that lead to the formation of theanine. The genes encoding TS and GS have been identified and cloned from tea plant, and the sequences of TS and GS genes present high consistency (Okada et al., 2006; Rana et al., 2008a). The partial coding sequences of GOGAT and GDH have been logged in GenBank. The genes in tea plant encoding to ALT and two tea-specific enzymes, AIDA, and ThYD, are not identified gene sequences (Tsushida and Takeo, 1985; Shi et al., 2011). However, SAMDC and ADC, which are both pivotal enzymes in polyamine metabolism, play similar roles with AIDA.
Ethylamine, one of the hydrolyzates of theanine, is further oxidized to acetaldehyde by AO, which is another precursor of catechin phloroglucinol that participates in catechin biosynthesis (Kito et al., 1968). In addition, NiR, which provides NH4+ for GS/ GOGAT cycle and α-ketoglutarate reductive amination reaction, performs important functions in theanine synthesis. Interestingly, similar to GS in bacteria, γ-GGT can also catalyze theanine synthesis with glutamine and ethylamine as substrate (Mu et al., 2015).
To obtain novel insight into L-theanine metabolism in tea plant, the genes encoding the enzymes involved in the theanine metabolism pathway are identified based on GenBank and our tea plant transcriptome (Wu et al., 2014). The expression patterns of seventeen metabolic-related genes and L-theanine content in the bud and 1st leaf, 2nd leaf, 3rd leaf, old leaf, stem, and root of the three selected tea cultivars were detected and analyzed by RP-HPLC method. The three C. sinensis cultivars, ‘Huangjinya’, ‘Anjibaicha’, and ‘Yingshuang’, were used in this experiment (Figure 1). Based on the obvious difference of geographic and climate characteristics in these three tea production areas, the morphology and physiology of the three tea plants were different. The theanine contents and the ability of adaptation to a single or multiple stresses were different among the three tea plant cultivars. The mainly characteristics of the three tea plant cultivars were listed in Table 1. This work is aimed to predict the relationship between structural genes and L-theanine content and the distribution at transcript level, and to provide reference for the further study of regulation mechanisms of theanine metabolism.
FIGURE 1.

Three Camellia sinensis cultivars ‘Huangjinya’(HJY), ‘Anjibaicha’(AJBC), and ‘Yingshuang’(YS) (1) bud, (2) 1st leaf, (3) 2nd leaf, (4) 3rd leaf, (5) old leaf, (6) stem, (7) root.
Table 1.
The characteristics description of the three Camellia sinensis cultivars, ‘Yingshuang’, ‘Anjibaicha’, and ‘Huangjinya’.
| C. sinensis cultivar | Breeding method | Variation | Shoot color | Characteristics | Resistance |
|---|---|---|---|---|---|
| Yingshuang | Artificial breeding | Non | Green | Lower ratio of polyphenols and amino acids | Strong cold resistance (Zhang et al., 2014) |
| Anjibaicha | Natural variation | Low temperature- induced albino | White (below 20°C) | High levels of amino acids, low levels of chlorophylls, catechins, and caffeine (Li et al., 2011; Feng et al., 2014) | Sensitive to low temperature (Li et al., 2015a) |
| Huangjinya | Natural variation | Light-induced albino | Yellow (1.5∼6∗104 lux) | High levels of amino acids (Wang et al., 2012) | Sensitive to low temperature and glare |
Materials and Methods
Plant Material Cultivation and Sampling
Two-year-old cutting seedlings of three C. sinensis cultivars (‘Huangjinya’, ‘Anjibaicha’, and ‘Yingshuang’) were selected for this study (Figure 1). The tea plants were grown in a matrix composed of peat, vermiculite and perlite (volume ratio = 3:2:1) under the natural environment. On April 2016, the samples of the bud and 1st leaf, 2nd leaf, 3rd leaf, old leaf, new stem (no lignification), and lateral root of three tea cultivars were harvested, immediately frozen in liquid nitrogen, and stored at -80°C.
Determination of L-Theanine Content
The bud and 1st leaf, 2nd leaf, 3rd, old leaf, new stem, and lateral root of tea plants from three cultivars were dried until constant weight in an oven at 80°C for 24 h. According to the method of GB/T 23193-2008 (China), L-theanine was extracted from different tissues separately, and then theanine content was determined by Reverse-Phase High-Performance Liquid Chromatography (RP-HPLC). The Agilent 1200 Series (Agilent Technologies Co., CA, USA), a Zorbax Eclipse XDB-C18 analytical column (250 mm × 4.6 mm inner diameter, 5 μm nominal particle size), and G1314A UV detector were used for chromatographic separation and detection. The volume ratio of mobile phase A (20 mmol⋅L-1 ammonium acetate) and mobile phase B (20 mmol⋅L-1 ammonium acetate: methanol: acetonitrile = 1: 2: 2, volume ratio) ratio was modified to 3:2 to better separate the peak in stems and roots.
RNA Isolation and cDNA Synthesis
RNA was extracted from the buds, leaves, stems, and roots of tea plants according to the instruction manual of the Quick RNA isolation Kit (Huayueyang Biotech Co., Ltd., Beijing, China). RNA concentration and quality were detected by NanoDrop spectrophotometer and agarose gel electrophoresis (12 g⋅L-1). One microgram of each RNA sample (ng⋅μL-1) was reverse transcribed into the first strand cDNA, after which gDNA was removed by using the PrimeScriptTM RT reagent Kit (TaKaRa Biotech Co., Ltd., Dalian, China). Synthesized cDNA was stored at –20°C for quantitative real-time PCR (qRT-PCR) analysis.
Gene Identification and Primer Design
Nucleotide sequences of genes were searched in the GenBank of NCBI. Genes that are not accessed from NCBI were searched through our transcriptome databases of tea plants (Wu et al., 2014). Genes were further identified by BLAST in NCBI by using the deduced amino acid sequences. Then reads per kilobase per million mapped reads (RPKM) of identified genes in three tea plant cultivars (C. sinensis ‘Chawansanhao’, ‘Ruchengmaoyecha’, ‘Anjibaicha’) were searched from transcriptome and were log2 transformed.
Sequences possessing a highly conserved domain and high similarity with other species were selected for qRT-PCR. The primer pairs of identified genes for qRT-PCR were designed by Primer Premier 5. Principles of primer design were as follows: temperature range of 55°C to 65°C, lengths in the range of 18–24 bp, and GC content of 45 to 60 %.
qRT-PCR Analysis
qRT-PCR was performed using an SYBR Premix Ex Taq kit (TaKaRa Biotech Co., Ltd., Dalian, China), iQTM5 software, and iQTM5 qRT-PCR system. The cycling profile is as follows: denaturation at 95°C for 5 min; 40 cycles at 95°C for 5 s, and 60°C for 30 s; and 61 cycles of melt curve analysis at 65°C for 10 s. The 20 μL total volume of the reaction system contained 10 μL of SYBR Premix Ex Taq, 7.2 μL of double-distilled H2O, 0.4 μL each of primer, and 2 μL of diluted cDNA. Each independent RNA and corresponding cDNA sample was analyzed in triplicate for each tea plant cultivar and each tissue. Each reaction was technically repeated for three times to ensure the accuracy.
Statistical Analysis
The mean values and standard deviation (SD) of theanine contents and gene mRNA levels were calculated based on three independent biological replicates. The gene expression levels were calculated relative to the CsTBP gene by the 2-ΔΔCT method (Livak and Schmittgen, 2001). The gene mRNA level of bud and 1st leaf in ‘Yingshuang’ was defined as 1. Significant differences in theanine contents and gene mRNA levels were detected by Duncan’s multiple-range test at the 5 % level with SPSS 17.0 software. The column chart was performed with Origin 6.0 software.
Results
Tea Plants of the Three C. sinensis Cultivars
Candidate tissues, including the bud, 1st leaf, 2nd leaf, 3rd leaf, old leaf, stem, and lateral root from three C. sinensis cultivars, ‘Huangjinya’, ‘Anjibaicha’, and ‘Yingshuang’, were selected to detect of the L-theanine contents, respectively (Supplementary Figure S1). The three tea plant cultivars are shown in Figure 1, and the shoots of ‘Anjibaicha’ turned into the early re-greening phase from the albescent phase.
L-Theanine Contents in Different Tissues among the Three C. sinensis Cultivars
The HPLC profiles of theanine in different tissues among the three C. sinensis cultivars were shown in Figure 2. The profiles of leaves, stem, and root were different, the retention time were all almost at 4.1 min. In addition, the height of profiles showed positive correlation with the area, except the 3rd leaf of ‘Anjibaicha’. The detailed information of HPLC profiles and profiles of standard samples were listed in Supplementary Table S1 and Figure S2.
FIGURE 2.
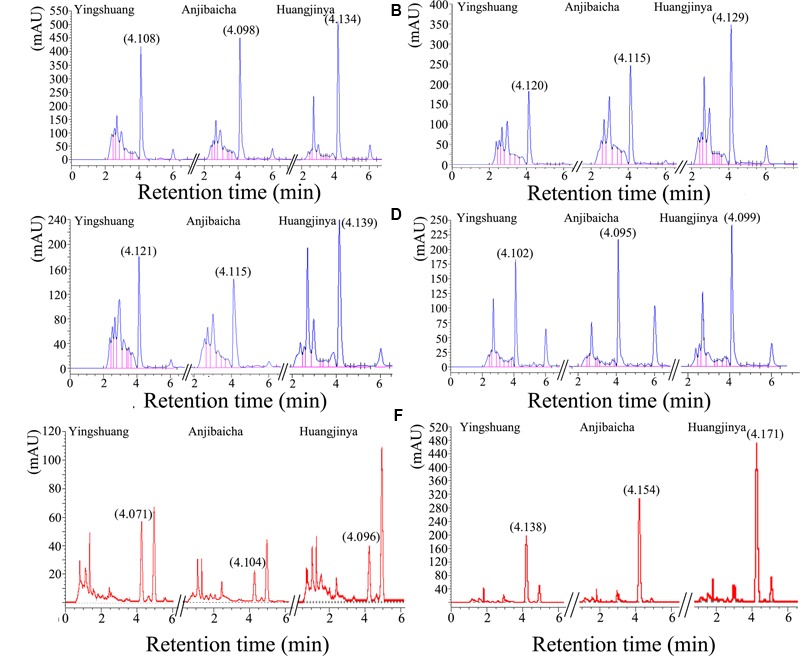
The high-performance liquid chromatography (HPLC) profiles of different tissues in three C. sinensis cultivars. (A) The bud and 1st leaf, (B) 2nd leaf, (C) 3rd leaf, (D) old leaf, (E) stem, (F) root.
The L-theanine contents show evident difference among the three C. sinensis cultivars (Figure 3). Among the three tea plant cultivars, ‘Huangjinya’ showed the highest theanine content in the leaves and roots, whereas ‘Yingshuang’ contained the lowest content. The theanine contents of the bud and 1st leaf, 2nd leaf, 3rd leaf, mature leaf, and roots in ‘Huangjinya’ were 1.5-, 1.6-, 1.4-, 1.4-, and 2.6-folds of ‘Yingshuang’, respectively. However, the maximum theanine content in stems was in ‘Yingshuang’, whereas the minimum content was found in ‘Anjibaicha’.
FIGURE 3.
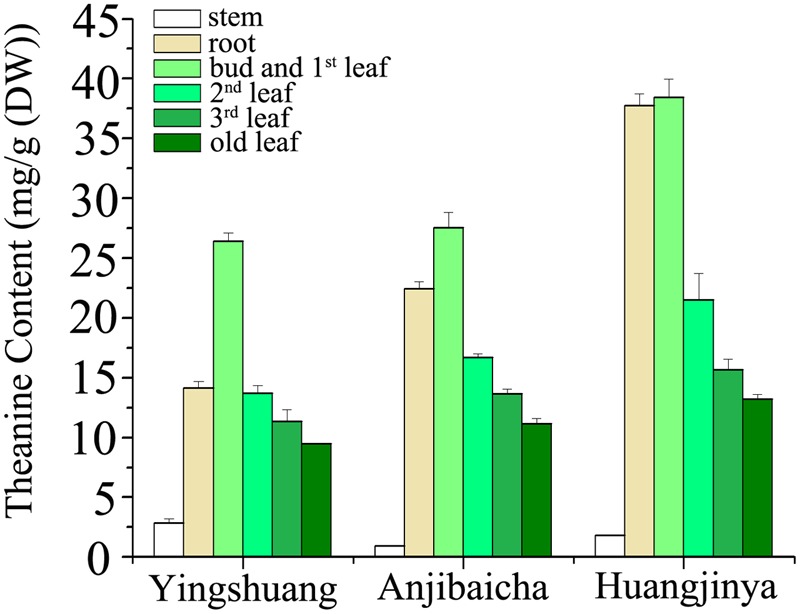
L-theanine content in various tissues of three C. sinensis cultivars. Values are the means of three independent experiments and are calculated as mg theanine equivalents per 1 g DW (mg/g).
The theanine content of stem was considerably below that of leaves and roots (Figure 3). Comparatively from bud to root, the bud and 1st leaf presented the highest theanine amount among the tissues, and the stem showed the lowest. In ‘Huangjinya’ and ‘Anjibaicha’, the theanine content in bud and 1st leaf was approximate to that in root. Theanine content in leaves decreased gradually with the position of leaves moving downward among all three tea cultivars. In general, the theanine distribution among the different parts of the three tea cultivars was similar extremely.
Gene Encoding the Enzymes Involved in L-Theanine Metabolism in Tea Plants
Genes encoding the metabolic enzymes involved in catalysis of theanine synthesis and hydrolysis were identified and analyzed in tea plant. CsTS, CsGS, CsAIDA, CsGOGAT, CsGDH, and CsALT as structural genes encode the enzymes implicated in the synthesis of theanine. CsThYD and CsAO genes encode enzymes that hydrolyze the synthesized theanine (Figure 4).
FIGURE 4.
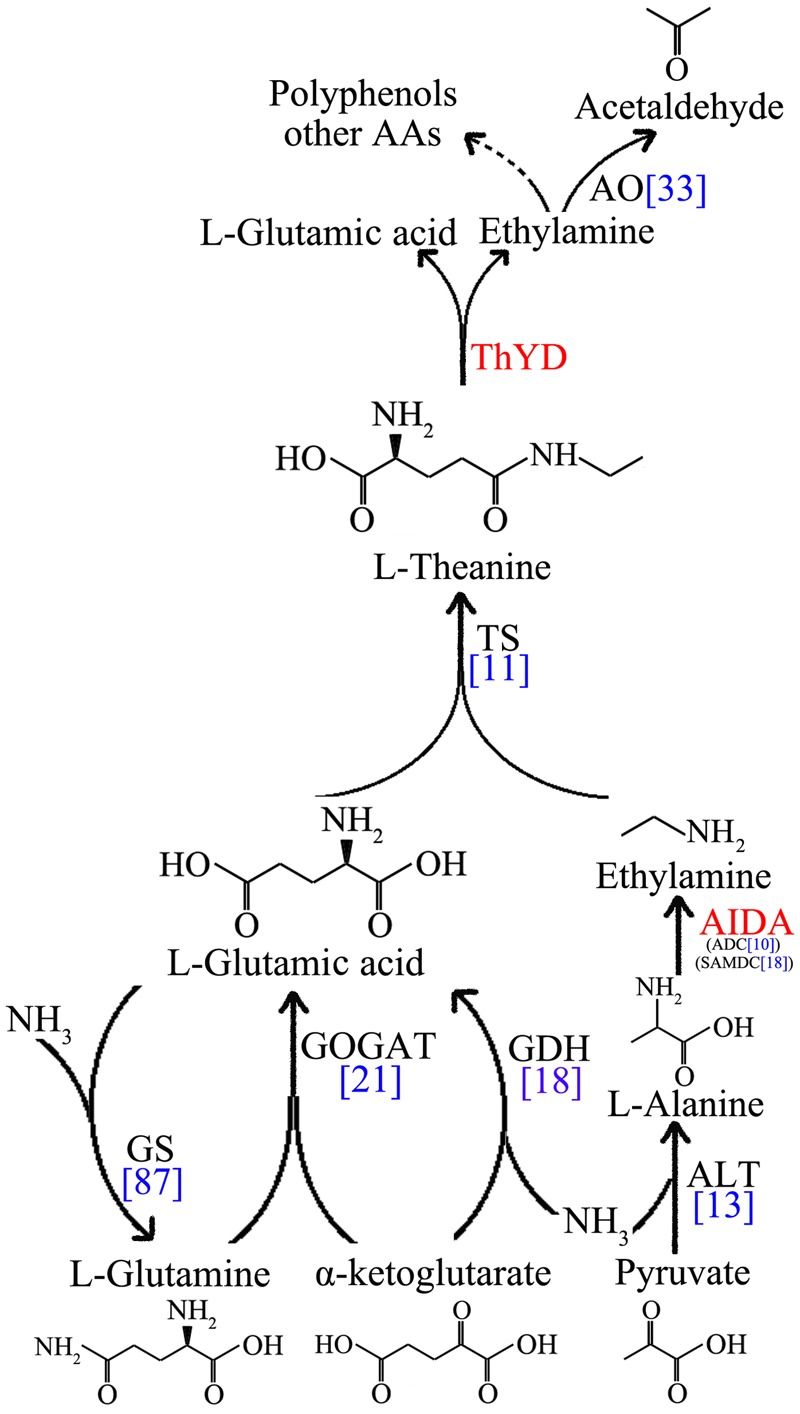
Schematics of the L-theanine metabolism pathway. Enzymes that are not identified in tea plants are marked in red. The unigenes number of theanine metabolic enzymes are marked in blue.
However, CsAIDA and CsThYD genes code specific enzymes in tea plant with no orthologs from other species at present (Shi et al., 2011). Thus, the genes were not annotated and identified in our database. Here, the transcripts of CsSAMDC and CsADC genes, sharing similar domains with CsAIDA, were found in the transcriptome (Zhang et al., 2015). In addition, NR and NiR, which oxidize the nitrate to ammonium by collective effect, are also required for theanine synthesis. Ammonium is indispensable to the GS/GOGAT cycle and in the reductive amination of α-ketoglutarate. GGT, which was used to synthesize theanine in bacteria, also exists in tea plants.
In this study, a total of 11, 87, 9, 12, 18, 13, 33, 18, 10, 9, 1, and 3 putative transcripts for CsTS, CsGS, CsGOGAT-Fe, CsGOGAT-NAD(P)H, CsGDH, CsALT, CsAO, CsSAMDC, CsADC, CsNR, CsNiR, and CsGGT were identified from the transcriptome (Table 2 and Supplementary Table S2). Seventeen genes were identified for further study by BLASTP, including CsTS1 (GenBank ID: DD410895.1) and CsTS2 (GenBank ID: DD410896.1) two genes searched from the GenBank, and fifteen genes (CsGS1, CsGS2, CsGOGAT-Fe, CsGOGAT-NAD(P)H, CsGDH1, CsGDH2, CsALT, CsSAMDC, CsADC, CsCuAO, CsPAO, CsNiR, CsNR, CsGGT1, and CsGGT3) verified from the transcriptome (Table 3 and Supplementary File S1, Table S3). The nucleotide and amino acid sequence alignments of those fifteen theanine metabolic related genes among tea plant and other species were listed in Supplementary Table S4.
Table 2.
Numbers of unigenes involved in the theanine biosynthesis pathway.
| Enzymes | Number of unigenes |
|---|---|
| SAMDC | 18 |
| ADC | 10 |
| ALT | 13 |
| GDH | 18 |
| AO | 33 |
| NR | 9 |
| NiR | 1 |
| GOGAT-Fe | 9 |
| GOGAT-NAD(P)H | 12 |
| GGT | 3 |
| TS | 11 |
| GS | 87 |
Table 3.
L-theanine metabolic related genes in tea plant.
| Gene family | Annotation | GenBank ID/Unigene ID | Sequence length (bp) | Encoded enzyme number |
|---|---|---|---|---|
| TS | TS1 | DD410895.1 | 1071 | EC 6.3.1.6 |
| TS2 | DD410896.1 | 1071 | ||
| GS | GS1 | T4_ BMK.28374 | 1068 | EC 6.3.1.2 |
| GS2 | T2_ BMK.52923 | 1296 | ||
| GOGAT | GOGAT-Fe | T2_ BMK.52873 | 4881 | EC 1.4.1.14 |
| GOGAT- NAD(P)H | T4_ BMK.68558 | 5583 | ||
| NR | NR | T1_ BMK.80048 | 2160 | EC 1.7.1.1 |
| NiR | NiR | T4_ BMK.66088 | 1761 | EC 1.7.7.1 |
| GGT | GGT3 | T2_ BMK.38431 | 1884 | EC 2.3.2.2 |
| GGT1 | T4_ BMK.61745 | 1824 | ||
| GDH | GDH2 | T2_ BMK.12564 | 1239 | EC 1.4.1.3 |
| GDH1 | T1_ BMK.76899 | 1233 | ||
| ALT | ALT | T4_ BMK.66138 | 1623 | EC 2.6.1.2 |
| AO | CuAO | T1_ BMK.69752 | 2355 | EC 1.4.3.6 |
| PAO | T1_ BMK.39464 | 2037 | ||
| SAMDC | SAMDC | T2_ BMK.35350 | 1077 | EC 4.1.1.50 |
| ADC | ADC | T4_ BMK.63973 | 2169 | EC:4.1.1.19 |
Then the unigene expression changes in different tea plant cultivars after quantification using the RPKM values (Figure 5 and Supplementary Table S5). Among the selected 17 unigenes, except the CsGDH1, CsGGT1, CsGGT3, and CsNR, most of them expresses lower level in the C. sinensis ‘Ruchengmaoyecha’ (large-leaf cultivar) than that in the other two cultivars C. sinensis ‘Chawansanhao’ and ‘Anjibaicha’ (small-leaf cultivar). It may be deduced that the expression of metabolic genes involved in theanine metabolism were correlated with theanine content to a large extent.
FIGURE 5.
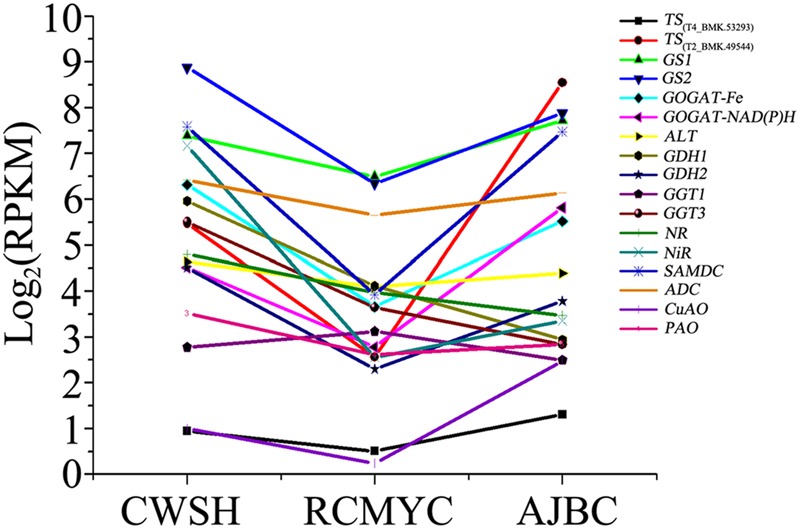
Expression levels of L-theanine metabolism pathway genes among C. sinensis cultivars ‘Chawansanhao’, ‘Ruchengmaoyecha’, and ‘Anjibaicha’.
Expression Profiles of Genes Involved in L-Theanine Metabolism in Different Tissues of Three C. sinensis Cultivars
To further validate the correlation between gene expression level and theanine content, the expression profiles of the genes encoding to the enzymes involved in theanine metabolism were detected in different tissues of three C. sinensis cultivars by using qRT-PCR. The expression profiles of the theanine metabolic genes among different tissues and different cultivars are shown in Figure 6. Nucleotide sequences of primer pairs used for qRT-PCR that are specific to each theanine metabolic related gene are given in Table 4.
FIGURE 6.
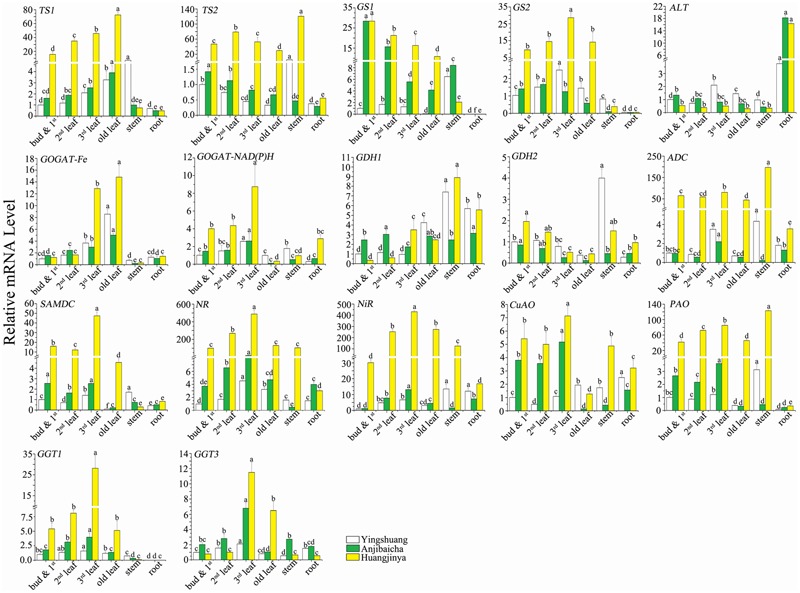
Expression profiles of L-theanine metabolism related genes among various tissues of C. sinensis cultivars ‘Huangjinya’, ‘Anjibaicha’, and ‘Yingshuang’. The gene mRNA level of bud and 1st leaf in ‘Yingshuang’ is defined as 1. The different lowercase letters on the bar graph indicate significant differences at P < 0.05.
Table 4.
Nucleotide sequences of primers specific to L-theanine metabolic related genes and CsTBP gene used for qRT-PCR.
| Gene | Forward primer 5′–3′ | Reverse primer 5′–3′ |
|---|---|---|
| TS1 | AGACCGCCGACATCAACAC | ATGGCTTCCACAGCAGAGT |
| TS2 | CCTAAACCTATTGAGGG TGACTG | TCCTGTAAGCCGACGCTCATT |
| GS1 | ATCAGTTGTGGATGGCTCG | CACTTCGCATGGACTTGGTAC |
| GS2 | GTGGCACCAACGGAGAAGT | CAAGGATGTATCTAGCGCACC |
| NiR | GGACAGGCTGCCCAAATAG | TCACTCCCAATCCTCCCTC |
| NR | TTGATGCTTGGGCTGACA | ACGGACCAGGGATGTGCT |
| GGT1 | GATGAATCTTGGTGATCCTGAT | TTCCACCGTCCACCATAGT |
| GGT3 | GGAGTCAGCTTCAAGATCACG | CAGTAGGCGTCGAGAAGTCAC |
| GDH1 | GAGCTGAAGACATACATGACCA | GCACGAGCAACACGATTAA |
| GDH2 | ATGTGGGACGAAGAGAAGGTG | GCAACACGATTCACTCCCAG |
| ALT | CGAGTCCTACGAGTCTTAT TATGC | GGAGGCGTTGACAATAGAATG |
| CuAO | CAGGTTGTTGAGGTGAATGTTA | AATCCAGTGGCGAGCAGA |
| PAO | GTCGGGGTGACGATACCTTAG | CACCACTTAGCGTCGACATTAT |
| GOGAT-Fe | TGCTGGTATGACTGGAGGTT | CAACTGCCAGAATAGCGGTA |
| GOGAT- NAD(P)H | GCAGCGAGGAGATGATTGA | CACCTTCCACATTGGTTGAG |
| SAMDC | TTCCAGCCAAGCGAGTTC | AACCTCCCTCCTTGCCGA |
| ADC | GGGCTTATGAGGAGGCAC | GCAAGAGGGTCCTGGCAT |
| CsTBP | GGCGGATCAAGTGTTGGAA GGGAG | ACGCTTGGGATTGTATTCGG CATTA |
Expression Profiles in Different Tissues
With the maturity of leaf increases, the transcript abundance of CsTS1 and CsGOGAT-Fe were increased, but that of the CsTS2, CsGS1, and CsGDH2 decreased. The transcript levels of CsGS2, CsGOGAT-NAD(P)H, CsNR, CsNiR, CsPAO, CsGGT1, and CsGGT3 were gradually increased in the bud and 1st leaf, 2nd leaf, and 3rd leaf, but later declined in old leaf. The expression level of CsADC, CsSAMDC, and CsCuAO in 3rd leaf reached to maximum, then followed in bud and 1st leaf, 2nd leaf, old leaf. The mRNA level of ALT presented low and no significant difference in leaves of different parts.
Among the selected genes, most of them showed low transcript abundance in stem and root. The CsTS1, CsTS2, CsGS1, CsGS2, CsPAO, and CsGGT1 showed the lowest transcript abundance in root, but the CsALT showed the highest. The transcript levels of CsGS1, CsGS2, CsGGT1 were close to zero. The stem transcript levels of CsTS2, CsGDH1, CsADC, and CsPAO reached the highest among the tissues in ‘Yingshuang’ and ‘Huangjinya’. In addition, in stem of ‘Yingshuang’, the expression levels of CsTS1, CsGDH2, CsSAMDC, and CsNiR were significantly higher than that in other parts.
The genes belonging to same family, such as the CsGOGAT-Fe and CsGOGAT-NAD(P)H, CsGGT1 and CsGGT3, CsPAO and CsCuAO, CsNR, and CsNiR, the transcript levels showed similar trend from bud to root. However, the tendency of expression levels of CsTS1 and CsTS2, CsGS1 and CsGS2, CsGDH1 and CsGDH2 was almost opposite among different tissues.
Expression Profiles in Three Tea Plant Cultivars
Among cultivars, ‘Huangjinya’ showed the highest leaves mRNA levels of most theanine metabolic related genes, moreover the lowest levels were usually observed in ‘Yingshuang’. The gene mRNA levels in ‘Huangjinya’ exhibited tens and even hundreds of times of the lowest levels. However, the leaves expression level of CsALT, the bud and 1st leaf and 2nd leaf expression levels of CsGDH1 and CsGGT1 in ‘Huangjinya’ were the lowest among three tea cultivars.
Structural genes directly involved in theanine synthesis, CsTS2, CsGDH1, CsGDH2, CsGS2, CsGOGAT-Fe, CsGOGAT-NAD(P)H, and CsADC, expressed the lowest stem transcript levels in ‘Anjibaicha’. The highest root mRNA levels of CsTS2, CsGDH2, CsGOGAT-Fe, CsGOGAT-NAD(P)H, CsADC, CsSAMDC, and CsNiR were detected in ‘Huangjinya’. Furthermore, the root mRNA level of CsALT in ‘Anjibaicha’ and ‘Huangjinya’ was notably higher than that in ‘Yingshuang’. The expression patterns of the genes from bud to root among different tea cultivars were highly similar.
Discussion
The L-theanine metabolism pathway involves in the unique accumulation and translocation pathway of nitrogen in tea plant, which belongs to one of perennial economic crops with ammonium resistance (Konishi et al., 1969). At present, a few studies about the enzymes involved in the theanine metabolism pathway at gene level have been reported (Shi et al., 2011; Li et al., 2015b). To better understand the theanine metabolic mechanism at molecular level and provide references for identification and breeding of high theanine tea plant resources, it is essential to explore the correlations between theanine metabolic related genes and contents.
In this study, different tissues of three tea plant cultivars ‘Huangjinya’, ‘Anjibaicha’, and ‘Yingshuang’ were selected, including buds and 1st leaf, 2nd leaf, 3rd leaf, old leaf, stems, and roots. Among different tissues, the stem shows the lowest theanine content, thereby indicating that stems may only act as the transport part of theanine (Oh et al., 2008). In the ‘Huangjinya’ and ‘Anjibaicha’, root theanine content was close to the content of bud and 1st leaf, and higher than previously reported. This finding may be due to variety specificity and weakened transportation of theanine from root to leaves and bud at the re-greening phase of albino tea (Hahlbrock et al., 1971). Among the different cultivars, they presented highly similar distribution tendencies of theanine contents from bud to root. The theanine content in ‘Huangjinya’, ‘Anjibaicha’, and ‘Yingshuang’ decreased by degrees among the tissues except stems, thereby it confirmed that albino tea plants contained higher theanine concentration than non-albino varieties (Li et al., 2011; Feng et al., 2014). Particularly, the high theanine content of ‘Huangjinya’ explained why it is provided with high quality and high popularity.
In qRT-PCR analysis, the high expression levels of most theanine metabolic genes in ‘Huangjinya’ with high theanine concentration. It was basically consistent with the prediction of transcriptome by RPKM values, that is the higher transcripts levels of related genes in small-leaf tea cultivars (‘Chawansanhao’ and ‘Anjibaicha’, higher theanine content) than that in large-leaf cultivar (‘Ruchengmaoyecha’, lower theanine content). In ‘Huangjinya’, the expression levels of CsNR and CsNiR reached to several hundreds of times higher than the other two cultivars. While the NR and NiR play key role in providing the source of ammonium in theanine synthesis (Yang et al., 2013). In addition, the high expression levels of CsTS1, CsTS2, CsGS2, CsADC, and CsSAMDC in ‘Huangjinya’ also offered the possibility for synthesis of abundant theanine.
However, the expression patterns of the selected genes among tissues exist difference at different degrees, even though the genes belong to a family. The CsTS1 and CsTS2, CsGS1, and CsGS2 showed almost opposite correlations with the theanine content in leaves. The transcript levels of CsTS1 and CsTS2 in root were lower than in leaves, but in shoot were less than and more than in mature leaves, respectively. Those comparison results among shoot, mature leaves, and root were identical with the results of Deng (Deng et al., 2008). The negative correlation between mRNA levels of CsGS1 and theanine concentration in tea plant was comparable to that, which was previously reported for rice (Oryza sativa L.) and tobacco (Nicotiana tabacum L.) transfected CsGS gene. In rice and tobacco, the overexpression of transfected CsGS gene caused the decrease in free amino acids, including glutamic acid and glutamine (Migge et al., 2000; Cai et al., 2009). The positive correlation between mRNA level of CsGDH2 and theanine content, possibly because GDH is strongly activated by abundant amide compounds, such as glutamine and theanine, to assist GS/GOGAT (Rana et al., 2008b). In addition, most genes showed lower expression levels in root than that in other tissues. However, the expression level of CsALT was up-regulated in root and much higher than that in stem and leaves. The CsALT was the key gene in the process from ammonia converted to ethylamine. The ethylamine showed high conversion rate to synthesize theanine in root (Deng et al., 2009). Then the theanine might act as an easily transported nitrogenous compound were transported to other tea plant tissues (Deng et al., 2012). The high expression level of CsALT in root was consistent with the high synthesis rate of theanine, and promoted indirectly the transportation of theanine.
In this study, some genes expression level showed positive correlations with theanine contents among different tea plant cultivars, which explained the potential reason of the tea plant cultivar contains high amount of theanine at molecular level. However, some other theanine metabolic related genes exhibited negative correlations among different tissues. Ashihara mentioned that the root of tea plant played important roles in theanine synthesis. The synthetic theanine is preferentially transported and accumulated in the tender tissues (Ashihara, 2015). In addition, the theanine in mature leaves were probably degraded more quickly than in young leaves. Tsushida and Takeo also demonstrated that the degradation products of theanine as substrates were used for the synthesis of other polyphenols and other amino acids (Tsushida and Takeo, 1985).
In addition, we can see the high similarity of expression profiles of CsGOGAT-Fe and CsGOGAT-NAD(P)H, CsADC and CsSAMDC, CsNR and CsNiR, CsPAO and CsCuAO, CsGGT1 and CsGGT3 among different parts. It may be deduced that they play synergistic roles in regulation of theanine metabolism. Therefore, the theanine metabolism is co-regulated through many related genes, which may apply synergistic or antagonistic effect. The tissue, cultivar, developmental stages, and growth environment also can influence the expression profiles of theanine metabolic related genes, the genes regulation of theanine metabolism became complicated. This study revealed the correlation between theanine content and genes expression to a certain extent, and provide support for the high theanine tea plant germplasm at gene level, as well as molecular theory foundation for quality improvement of tea plants.
Author Contributions
JZ and ZL initiated and designed the research. ZL and JZ performed the experiments. ZL, ZW, HL, YW, and JZ analyzed the data. JZ contributed reagents, materials, analysis tools. ZL wrote the paper. JZ and ZL revised the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The research was supported by the National Natural Science Foundation of China (31570691).
Abbreviations
- ADC
Arginine decarboxylase
- AIDA
L-alanine decarboxylase
- ALT
alanine transaminase
- AO
amine oxidase
- CuAO
copper methylamine oxidase
- DW
dry weight
- GDH
glutamate dehydrogenase
- γ-GGT
γ-glutamyl transpeptidase
- GOGAT
glutamine-2-Oxoglutarate synthetase
- GOGAT-Fe
ferredoxin-dependent glutamate synthase
- GOGAT-NAD(P)H
NAD(P)H-dependent glutamate synthase
- GS
glutamine synthetase
- NiR
nitrite reductase
- NR
nitrate reductase
- PAO
primary amine oxidase
- RP-HPLC
Reverse phase high performance liquid chromatography
- SAMDC
S-adenosylmethionine decarboxylase
- ThYD
L-theanine hydrolase
- TS
L-theanine synthetase
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00498/full#supplementary-material
The bud and 1st leaf, 2nd leaf, 3rd leaf, and old leaf from three C. sinensis cultivars, ‘Yingshuang’, ‘Anjibaicha’, and ‘Huangjinya’.
The chromatogram of theanine standard samples. (A) 0.01 mg/mL; (B) 0.02 mg/mL; (C) 0.05 mg/mL; (D) 0.1 mg/mL; (E) 0.2 mg/mL.
The domains of selected theanine metabolic related genes.
The original data of HPLC profiles.
The annotations of theanine metabolic related genes in tea plant transcriptome.
The annotations of selected theanine metabolic related genes for qRT-PCR.
The nucleotide and amino acid sequence alignments of theanine metabolic related genes among tea plant and other species.
The RPKM and Log2(RPKM) values of the selected theanine metabolic related genes.
References
- Ashihara H. (2015). Occurrence, biosynthesis and metabolism of theanine (gamma-Glutamyl-L-ethylamide) in plants: a comprehensive review. Nat. Prod. Commun. 10 803–810. [PubMed] [Google Scholar]
- Cai H. M., Zhou Y., Xiao J. H., Li X. H., Zhang Q. F., Lian X. M. (2009). Overexpressed glutamine synthetase gene modifies nitrogen metabolism and abiotic stress responses in rice. Plant Cell Rep. 28 527–537. 10.1007/s00299-008-0665-z [DOI] [PubMed] [Google Scholar]
- Deng W. W., Ogita S., Ashihara H. (2008). Biosynthesis of theanine (gamma-ethylamino-L-glutamic acid) in seedlings of Camellia sinensis. Phytochem. Lett. 1 115–119. 10.1016/j.phytol.2008.06.002 [DOI] [Google Scholar]
- Deng W. W., Ogita S., Ashihara H. (2009). Ethylamine content and theanine biosynthesis in different organs of Camellia sinensis seedlings. Z. Naturforsch. C 64 387–390. 10.1515/znc-2009-5-614 [DOI] [PubMed] [Google Scholar]
- Deng W. W., Ogita S., Ashihara H. (2010). Distribution and biosynthesis of theanine in Theaceae plants. Plant Physiol. Biochem. 48 70–72. 10.1016/j.plaphy.2009.09.009 [DOI] [PubMed] [Google Scholar]
- Deng W. W., Ogita S., Chen Q., Zhang Z. Z., Hu X. Y. (2012). Effect of salt treatment on theanine biosynthesis in Camellia sinensis seedlings. Plant Physiol. Biochem. 56 35–40. 10.1016/j.plaphy.2012.04.003 [DOI] [PubMed] [Google Scholar]
- Feng L., Gao M. J., Hou R. Y., Hu X. Y., Zhang L., Wan X. C., et al. (2014). Determination of quality constituents in the young leaves of albino tea cultivars. Food Chem. 155 98–104. 10.1016/j.foodchem.2014.01.044 [DOI] [PubMed] [Google Scholar]
- Hahlbrock K., Ebel J., Ortmann R., Sutter A., Wellmann E., Grisebach H. (1971). Regulation of enzyme activities related to the biosynthesis of flavone glycosides in cell suspension culture of parsley (Petroselinum hortense). Biochim. Biophys. Acta 244 7–15. 10.1016/0304-4165(71)90114-0 [DOI] [PubMed] [Google Scholar]
- Haskell C. F., Kennedy D. O., Milne A. L., Wesnes K. A., Scholey A. B. (2008). The effects of L-theanine, caffeine and their combination on cognition and mood. Biol. Psychol. 77 113–122. 10.1016/j.biopsycho.2007.09.008 [DOI] [PubMed] [Google Scholar]
- Jiang H.-B. (2013). Diversity of tea landraces based on agronomic and quality traits in yunnan province. J. Plant Genet. Res. 14 634–640. [Google Scholar]
- Kakuda T., Nozawa A., Unno T., Okamura N., Okai O. (2000). Inhibiting effects of theanine on caffeine stimulation evaluated by EEG in the rat. Biosci. Biotechnol. Biochem. 64 287–293. 10.1271/bbb.64.287 [DOI] [PubMed] [Google Scholar]
- Kito M., Kokura H., Izaki J., Sasaoka K. (1968). Theanine, a precursor of the phloroglucinol nucleus of catechins in tea plants. Phytochemistry 7 599–603. 10.1016/S0031-9422(00)88234-5 [DOI] [Google Scholar]
- Konishi S., Matsuda T., Takahashi E. (1969). Synthesis of theanine and L-glutamic acid γ-methylamide in Thea sinensis, Camellia sasanqua, and Oryza sativa. V. Metabolism and regulation of theanine and related compounds in the tea plant. Nippon Dojo Hiryo. Zasshi 40 107–112. [Google Scholar]
- Li C. F., Yao M. Z., Ma C. L., Ma J. Q., Jin J. Q., Chen L. (2015a). Differential metabolic profiles during the albescent stages of ‘Anji Baicha’ (Camellia sinensis). PLoS ONE 10:e0139996 10.1371/journal.pone.0139996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C. F., Zhu Y., Yu Y., Zhao Q. Y., Wang S. J., Wang X. C., et al. (2015b). Global transcriptome and gene regulation network for secondary metabolite biosynthesis of tea plant (Camellia sinensis). BMC Genomics 16:560 10.1186/s12864-015-1773-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Huang J., Liu S., Li J., Yang X., Liu Y., et al. (2011). Proteomic analysis of young leaves at three developmental stages in an albino tea cultivar. Proteome Sci. 9:44 10.1186/1477-5956-9-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Duan H. Y., Luan J. L., Yagasaki K., Zhang G. Y. (2009). Effects of theanine on growth of human lung cancer and leukemia cells as well as migration and invasion of human lung cancer cells. Cytotechnology 59 211–217. 10.1007/s10616-009-9223-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2- ΔΔCT method. Methods 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Migge A., Carrayol E., Hirel B., Becker T. W. (2000). Leaf-specific overexpression of plastidic glutamine synthetase stimulates the growth of transgenic tobacco seedlings. Planta 210 252–260. 10.1007/PL00008132 [DOI] [PubMed] [Google Scholar]
- Mu W. M., Zhang T., Jiang B. (2015). An overview of biological production of L-theanine. Biotechnol. Adv. 33 335–342. 10.1016/j.biotechadv.2015.04.004 [DOI] [PubMed] [Google Scholar]
- Oh K., Kato T., Xu H. (2008). Transport of nitrogen assimilation in xylem vessels of green tea plants fed with NH 4-N and NO 3-N. Pedosphere 18 222–226. 10.1016/S1002-0160(08)60010-7 [DOI] [Google Scholar]
- Okada Y., Koseki M., Chu M. (2006). Protein and cDNA Sequences of Two Theanine Synthetases from Camellia sinensis. Japan Patent No. 2006254780. Chiyoda-ku Tokyo: Japan Patent Office. [Google Scholar]
- Rana N. K., Mohanpuria P., Yadav S. K. (2008a). Cloning and characterization of a cytosolic glutamine synthetase from Camellia sinensis (L.) O. Kuntze that is upregulated by ABA, SA, and H2O2. Mol. Biotechnol. 39 49–56. [DOI] [PubMed] [Google Scholar]
- Rana N. K., Mohanpuria P., Yadav S. K. (2008b). Expression of tea cytosolic glutamine synthetase is tissue specific and induced by cadmium and salt stress. Biol. Plant. 52 361–364. 10.1007/s10535-008-0075-7 [DOI] [Google Scholar]
- Rogers P. J., Smith J. E., Heatherley S. V., Pleydell-Pearce C. W. (2008). Time for tea: mood, blood pressure and cognitive performance effects of caffeine and theanine administered alone and together. Psychopharmacology 195 569–577. 10.1007/s00213-007-0938-1 [DOI] [PubMed] [Google Scholar]
- Selvendran R., Selvendran S. (1973). The distribution of some nitrogenous constituents in the tea plant. J. Sci. Food Agric. 24 161–166. 10.1002/jsfa.2740240208 [DOI] [PubMed] [Google Scholar]
- Shi C. Y., Yang H., Wei C. L., Yu O., Zhang Z. Z., Jiang C. J., et al. (2011). Deep sequencing of the Camellia sinensis transcriptome revealed candidate genes for major metabolic pathways of tea-specific compounds. BMC Genomics 12:131 10.1186/1471-2164-12-131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeo T. (1979). Formation of amino acids induced by ammonia application and seasonal level fluctuation of amino acids contents in tea plant. Chagyo Kenkyu 56 70. [Google Scholar]
- Tsushida T., Takeo T. (1985). An enzyme hydrolyzing L-theanine in tea leaves. Agric. Biol. Chem. 49 2913–2917. 10.1271/bbb1961.49.2913 [DOI] [Google Scholar]
- Wang K., Li N., Lu J., Zheng X., Liang Y., Han Z., et al. (2012). Effect of shading on gene expression profile in leaf of tea cultivar “Huangjinya”. J. Tea 4:016. [Google Scholar]
- Wu Z.-J., Li X.-H., Liu Z.-W., Xu Z.-S., Zhuang J. (2014). De novo assembly and transcriptome characterization: novel insights into catechins biosynthesis in Camellia sinensis. BMC Plant Biol. 14:277 10.1186/s12870-014-0277-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S., Ninomiya K. (2000). Umami and food palatability. J. Nutr. 130 921s–926s. [DOI] [PubMed] [Google Scholar]
- Yang Y., Li X., Ratcliffe R., Ruan J. (2013). Characterization of ammonium and nitrate uptake and assimilation in roots of tea plants. Russ. J. Plant Physiol. 60 91–99. 10.1134/S1021443712060180 [DOI] [Google Scholar]
- Yokogoshi H., Kato Y., Sagesaka Y. M., Takiharamatsuura T., Kakuda T., Takeuchi N. (1995). Reduction effect of theanine on blood-pressure and brain 5-hydroxyindoles in spontaneously hypertensive rats. Biosci. Biotechnol. Biochem. 59 615–618. 10.1271/bbb.59.615 [DOI] [PubMed] [Google Scholar]
- Zhang H. B., Xia E. H., Huang H., Jiang J. J., Liu B. Y., Gao L. Z. (2015). De novo transcriptome assembly of the wild relative of tea tree (Camellia taliensis) and comparative analysis with tea transcriptome identified putative genes associated with tea quality and stress response. BMC Genomics 16:298 10.1186/s12864-015-1494-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zhu X., Chen X., Song C., Zou Z., Wang Y., et al. (2014). Identification and characterization of cold-responsive microRNAs in tea plant (Camellia sinensis) and their targets using high-throughput sequencing and degradome analysis. BMC Plant Biol. 14:271 10.1186/s12870-014-0271-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Zhao B. L. (2014). The neuroprotective effect of L-Theanine and its inhibition on nicotine dependence. Chin. Sci. Bull. 59 4014–4019. 10.1007/s11434-014-0529-6 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The bud and 1st leaf, 2nd leaf, 3rd leaf, and old leaf from three C. sinensis cultivars, ‘Yingshuang’, ‘Anjibaicha’, and ‘Huangjinya’.
The chromatogram of theanine standard samples. (A) 0.01 mg/mL; (B) 0.02 mg/mL; (C) 0.05 mg/mL; (D) 0.1 mg/mL; (E) 0.2 mg/mL.
The domains of selected theanine metabolic related genes.
The original data of HPLC profiles.
The annotations of theanine metabolic related genes in tea plant transcriptome.
The annotations of selected theanine metabolic related genes for qRT-PCR.
The nucleotide and amino acid sequence alignments of theanine metabolic related genes among tea plant and other species.
The RPKM and Log2(RPKM) values of the selected theanine metabolic related genes.


