Abstract
Arginine methylation is a common posttranslational modification found on nuclear and cytoplasmic proteins that has roles in transcriptional regulation, RNA metabolism and DNA repair. The protozoan parasite Toxoplasma gondii has a complex life cycle requiring transcriptional plasticity and has unique transcriptional regulatory pathways. Arginine methylation may play an important part in transcriptional regulation and splicing biology in this organism. The T. gondii genome contains five putative protein arginine methyltransferases (PRMTs), of which PRMT1 is important for cell division and growth. In order to better understand the function(s) of the posttranslational modification monomethyl arginine (MMA) in T. gondii, we performed a proteomic analysis of MMA proteins using affinity purification employing anti-MMA specific antibodies followed by mass spectrometry. The arginine monomethylome of T. gondii contains a large number of RNA binding proteins and multiple ApiAP2 transcription factors, suggesting a role for arginine methylation in RNA biology and transcriptional regulation. Surprisingly, 90% of proteins that are arginine monomethylated were detected as being phosphorylated in a previous phosphoproteomics study which raises the possibility of interplay between MMA and phosphorylation in this organism. Supporting this, a number of kinases are also arginine methylated. Because PRMT1 is thought to be a major PRMT in T. gondii, an organism which lacks a MMA-specific PRMT, we applied comparative proteomics to understand how PRMT1 might contribute to the MMA proteome in T. gondii. We identified numerous putative PRMT1 substrates, which include RNA binding proteins, transcriptional regulators (e.g. AP2 transcription factors), and kinases. Together, these data highlight the importance of MMA and PRMT1 in arginine methylation in T. gondii, as a potential regulator of a large number of processes including RNA biology and transcription.
Arginine methylation occurs on cytoplasmic and nuclear proteins and has important functions in many pathways including epigenetic and transcriptional regulation, RNA splicing and the DNA damage response (1). At the molecular level, methylation of arginine does not alter charge, but changes protein or nucleic acid binding affinity by increasing hydrophobicity and steric hindrance, preventing hydrogen bonding (1, 2). In instances involving a methyl transfer reaction with S-adenosyl methionine, arginine methylation increases hydrogen bonding capacity (3). Although the focus of many arginine methylation studies has been arginine methylation of histones (4), this post-translational modification (PTM) also occurs on a large number of non-histone proteins that have diverse functions. For example, arginine methylation of transcription factors can inhibit their degradation by preventing phosphorylation events required for ubiquitin-mediated destruction (5). The largest family of nonhistone arginine methylated proteins are the RNA binding proteins (RBP); in some cases, arginine methylation negatively regulates binding of RBP to RNA (6) and in others, enhances binding (7). The overrepresentation of RBP in arginine methylated proteins is thought to be related to the high frequency of glycine-arginine (GAR) motifs, which are targeted by the enzymes that catalyze arginine methylation (8). However, this preference for GAR motifs was recently challenged by an exhaustive study of arginine methylation in humans, in which only one third of the (mostly novel) 8030 methylation sites were found within GAR motifs (7), suggesting that arginine monomethylation does not occur in a sequence context dependent manner.
Arginine methylation is mediated by a family of protein arginine methyltransferases (PRMT) 1, which are classified Types I to IV by the type of arginine methylation they catalyze (supplemental Table S1). Individual PRMT family members differ significantly in terms of their biochemical properties and substrate specificities (9), suggesting that they have non-redundant functions. The four types of arginine methylation each have a potentially different function. Although MMA is an intermediate step preceding possible dimethylation (omega-NG-dimethylarginine) and can be catalyzed by all PRMTs, the importance of MMA as a terminal PTM has been debated (10). Type I and II PRMTs can transfer a second methyl to either the same or another nitrogen on arginine, forming omega NG-NG-asymmetric dimethylarginine (ADMA) or omega NG-NG-symmetric dimethylarginine (SDMA) respectively. Few type III PRMT have been identified; PRMT7 is found in humans (11), C. elegans (12), kinetoplastidae (13), choanoflagellates, and trypanosomatids (14). Of these only trypanosomatid PRMT7 harbors exclusively terminal MMA methyltransferase activity. Type IV PRMTs, which catalyze the addition of a methyl group to the guanidine nitrogen of arginine, are rare but present in fungi and plants (15, 16).
Across a large number of tissues and cell types, the ratios of the different types of modification are estimated to be roughly 3:2:1 for ADMA/MMA/SDMA (17, 18). Although MMA is often considered a transitory modification, robust site-specific regulation of MMA (19) and the restriction of some PRMTs to MMA addition (14) suggests that MMA is biologically relevant on its own. Importantly, there is dynamic interplay between the different types of methyl modifications, with ADMA capable of blocking SDMA and MMA on the same substrate (20). However, although PRMT type determines the class of methyl modification, a proteome-wide study of MMA in humans demonstrated that specific PRMTs determine the function of a substrate, as seen in the change of the binding capacity of HNRNPUL1 with the knockdown of PRMT4 and PRMT1 but not with PRMT5 (7). Together, these findings extend the regulatory capacity of the methylation machinery, indicating that distinct PRMT enzymes are required to regulate separate biological functions of the same substrate.
Until recently, arginine methylation was regarded as a permanent modification; however, recent studies suggest that there is dynamic regulation of this PTM. For example, under actinomycin D-induced transcriptional arrest, monomethylated sites decrease, whereas corresponding dimethyl and protein expression levels do not (19). In addition, reversible methylation of arginine on tumor necrosis factor receptor-associated factor 6, mediated by jumonji domain-containing protein JMJD6, is important for TOLL-like receptor signaling (21). JMJD6 also demethylates histone 3 (H3R2me2) and histone 4 (H4R3me2) (22), providing a mechanism for dynamic histone methylation (23). Recent work has implicated peptidyl arginine deiminases (PADIs) as putative demethylases that function by deiminating arginines to citrulline thus preventing methylation [reviewed in (24)]. These findings suggest that arginine methylation PTMs are more dynamic than previously thought.
PRMT enzymes are conserved in many kingdoms of life with extended sets of PRMTs present in protozoa compared with other simple eukaryotes (25). The assortment of PRMTs present in each protozoan organism differs, suggesting that PRMTs probably have unique roles in the biology of different parasites. Toxoplasma gondii is an important human and veterinary pathogen that has a complicated life cycle, with multiple rounds of infection of different hosts and cell types. In host tissues, it reversibly differentiates between a rapidly replicating tachyzoite stage and a slow-growing, cyst-forming bradyzoite form, both of which are important in the pathogenesis of this infection. Changes in transcription occur during life cycle transitions (26) and PTMs are important regulators of the T. gondii cell cycle (27). In previous work, we identified multiple monomethylated arginine residues on T. gondii histones (28, 29). T. gondii, encodes five PRMTs, four of which are predicted to be type I PRMTs and one that is predicted to be a type II enzyme, based on sequence similarity to human homologues (supplemental Table S1). Two of the type I PRMTs, TgPRMT1 and TgPRMT4, possess protein arginine methyltransferase activity and modify histones (29, 30). TgPRMT4 localizes to the nucleus of the parasite where it has been implicated in gene regulation and parasite development (30). Furthermore, inhibition of TgPRMT4 induces differentiation to bradyzoites, thus supporting a role for arginine methylation in regulation of life cycle transitions. TgPRMT2 is a noncanonical TgPRMT, reported to have weak homology to PRMT6, a type I PRMT (25).
TgPRMT1, on the other hand, localizes primarily to the cytosol and pericentriolar regions and ensures correct segregation of daughter cells during parasite replication (29). Like human PRMT1 (20), TgPRMT1 is not essential to viability, however deletion of TgPRMT1 results in loss of synchronous replication and a disrupted cell cycle, along with changes in gene expression (29). In mammalian cells, PRMT1 is the major arginine methyltransferase, responsible for over 90% of ADMA deposition (31). TgPRMT1 is also thought to be a major contributor to the arginine methylome in T. gondii and appears to negatively regulate histone H3 monomethylation (29). The effects of ablation of TgPRMT1 on the arginine methylome of T. gondii are unknown; loss of PRMT1 in mammalian cells leads to an increase in MMA and SDMA, caused by substrate scavenging by other PRMTs (20). This suggests that PRMT1 also plays a role in regulating the substrate specificity of other PRMTs and that there is interplay between the three types of arginine methylation. It is unknown whether this also occurs in T. gondii. In the absence of a Type III PRMT7 homolog in T. gondii, it is plausible that MMA is catalyzed by PRMTs of other types, as either an intermediate or terminal modification.
To expand our understanding of arginine methylation, the T. gondii MMA proteome was mapped and compared with the MMA proteome of PRMT1 KO parasites that have previously been phenotypically characterized (29). TgPRMT1 is a suitable candidate for mediating monomethylation in T. gondii. The T. gondii arginine monomethylome is enriched in nuclear and cytoplasmic proteins and proteins that bind nucleic acids, such as RNA binding proteins, are abundantly represented. Surprisingly, almost 90% of MMA proteins were previously shown to be targets of phosphorylation by phosphoproteomics (32). In the PRMT1 KO, a significant decrease in MMA proteins was observed, suggesting that TgPRMT1 is responsible for a considerable proportion of MMA. Putative TgPRMT1 monomethylarginine substrates were also identified, which included kinases and RNA binding proteins. Together, these findings implicate MMA as an important regulator of nuclear activity and suggest that TgPRMT1 is a major regulator of MMA in T. gondii. In addition, crosstalk between phosphorylation and arginine methylation likely plays a role in cell cycle checkpoint control in this organism.
EXPERIMENTAL PROCEDURES
Cell Culture
Fifteen 150 cm2 plates containing human foreskin fibroblasts (HFF) were grown to confluency in Dulbecco's Modified Eagles Medium (Gibco - Life Technologies, Grand Island, NY), containing 10% fetal bovine serum (Gibco - Life Technologies), 1% l-glutamine (Gibco - Life Technologies), and 1% Penicillin/Streptomycin (Gibco - Life Technologies). HFF were infected with 2.5 × 108 freshly lysed T. gondii tachyzoites [strains: RHΔhxgprt, RHΔhxgprtΔku80, RHΔhxgprtΔprmt1 (PRMT1KO), RHΔhxgprtΔprmt1::PRMT1RFP (PRMT1COMP)] (29). For harvesting of intracellular tachyzoites, infected cells were incubated at 37 °C with 5% CO2 and harvested before parasite egress, at ∼36 h postinfection. Extracellular tachyzoites were removed by aspirating the media and washing with 10 ml of phosphate buffered saline (PBS). Infected cells were harvested using a cell scraper and collected in a 500-ml beaker. The suspension was passed through a 27G needle three times, using a manual press to mechanically break infected cells, following which the parasite suspension was vacuum filtered through a 3-micron filter (GE Water & Process Technologies, Trevose, PA) to remove host cell debris. The filtrate was evenly divided into 50 ml conical tubes and centrifuged at 3000 rpm for 20 min at 4 °C. The supernatant was aspirated and multiple pellets were resuspended in 22 ml of 1× PBS. The suspension was then centrifuged at 3000 rpm for 20 min at 4 °C. The supernatant was removed by aspiration and the dry pellet was stored at −80 °C preceding lysis and protein extraction. To harvest extracellular tachyzoites, cells were infected as above and floating parasites were harvested by centrifugation at ∼48 h post infection. The pellet was resuspended in PBS and vacuum filtered as above, before storing at −80 °C.
Preparation of Lysates and Peptides
Parasite pellets were solubilized in 10 ml of urea lysis buffer (20 mm HEPES pH 8.0, 9.0 m urea, 1 mm sodium orthovanadate (activated), 2.5 mm sodium pyrophosphate, 1 mm β-glycerol-phosphate). The suspension was then cooled on ice for 1 min before sonicating for 30 s at 35% amplitude on Fisher Scientific Sonic Dismembrator model 500 (Thermo Fisher Scientific, Waltham, MA). This was repeated three times. Samples were then centrifuged at 20,000 × g at 4 °C for 15 min. The cleared supernatant was transferred to a new 50 ml conical tube. The capped tube was placed in a dry ice/ethanol bath for 30 min or until the protein extract was completely frozen. Samples were stored at −80 °C prior to immunoaffinity enrichment.
Sample Preparation
Samples were prepared according to Guo et al. (33). Briefly, samples were reduced, alkylated, trypsin digested, lyophilized and stored at −80 °C. Peptide immunoaffinity purification was performed using protein A agarose beads and methylation motif specific antibodies (Fig. 1A). The antibodies used for immunoprecipitation are commercially available: Me-R4–100 (CST #8015, Cell Signaling Technology, Danvers, MA) and R*GG (D5A12) (CST #8711, Cell Signaling Technology) and were generated from New Zealand White rabbits immunized with the following antigen libraries: (XXXXXXXR*XXXXXX), where X represents a mixture of all naturally occurring amino acids with the exceptions of tryptophan (W), cysteine (C), tyrosine (Y) and a second library (XXXXXXXR*GGXXX) in order to reflect the arginine-glycine rich background in which monomethylation occurs (33). The eluted peptides were analyzed by mass spectrometry. Monomethyl motif-enriched peptides were separated on a reversed-phase high pressure liquid chromatography column after which an Orbitrap mass spectrometer was used to collect tandem mass spectra. Sample preparation and LC-MS/MS was performed according to (33).
Fig. 1.
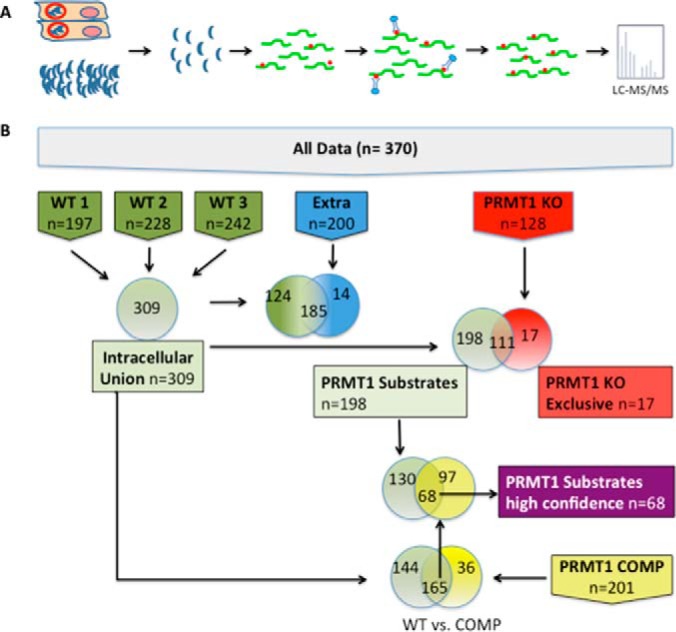
Affinity Purification of MMA peptides and data sets. A, Diagram of affinity purification strategy. T. gondii tachyzoites were harvested either from infected cells (top) or as free floating extracellular parasites (bottom) and were filtered to remove host cell debris. Parasites were lysed and digested with trypsin to release peptides (green lines). MMA-modified peptides (red dots) were enriched by immunoaffinity purification using a mixture of two monoclonal antibodies raised against arginine monomethylation at R* and R*GG motifs (* = site of arginine methylation). Purified peptides were identified by LC-MS/MS and database search using the parameters described in the materials and methods. B, Diagram of data sets. “All data” represents all 370 MMA proteins that were identified within this study. Six biological samples were analyzed in two technical replicates (merged data for both replicates are presented). Three data sets are composed of proteins detected in wild type intracellular tachyzoites: two biological replicates from wild type (WT) intracellular RHΔhxgprt tachyzoites (WT1, WT2), one data set from intracellular RHΔhxgprtΔku80 tachyzoites (WT3). Three data sets consist of proteins detected in: (1) Extracellular wild-type parasites (EXTRA); (2) TgPRMT1 knockout (PRMT1KO) parasites; and (3) TgPRMT1 knockout parasites genetically complemented with PRMT1mRFP (PRMT1COMP) (29). All proteins present in any of the three biological replicates of intracellular wild-type parasites are included in “Intracellular Union”. The other data sets shown are derived from comparative analysis of the initial data sets and manual filtering of the protein lists. The candidate “TgPRMT1 substrates” data set represents the proteins present in the WT Intracellular Union data set but not in the PRMT1KO data set. The “high confidence TgPRMT1 substrate” data set consists of those proteins in the TgPRMT1 substrate data set (i.e. not present in the PRMT1KO) for which methylation was restored in PRMT1COMP. The 'PRMT1KO exclusive' data set consists of the proteins present in the PRMT1KO and not present in the WT Intracellular Union data set.
Experimental Design and Statistical Rationale
In this study the following six biological samples were prepared and analyzed: (1, 2) RHΔhxgprt intracellular (n = 2, technical replicates for each of 2 biological replicates (WT1 and WT2)), (3) RHΔprmt1Δhxgprt (n = 2, technical replicates), (4) RHΔhxgprtΔprmt1::PRMT1RFP (n = 2, technical replicates), (5) RHΔhxgprtΔku80 intracellular (WT3) (n = 2, technical replicates) and (6) RHΔhxgprtΔku80 extracellular (n = 2, technical replicates). The technical replicates of each biological sample were combined and analyzed as a single sample, as presented in the results section of this manuscript. RHΔprmt1Δhxgprt is a strain in which TgPRMT1 is knocked out (PRMT1KO) and RHΔhxgprtΔprmt1::PRMT1RFP (PRMT1COMP) is the complemented PRMT1 knockout parasite in which TgPRMT1 has been genetically restored (29). A cartoon depicting the biological samples and composite data sets made after comparative analysis of the initial data sets is shown (Fig. 1B). The RHΔhxgprt intracellular biological samples were analyzed as a combined data set. RHΔhxgprtΔku80 extracellular parasites were analyzed in order to study changes in MMA proteins and parasite biology because of previously observed cell cycle arrest in a G0-like state (34, 35) and changes in PTM proteomes (27, 32, 36).
RHΔhxgprtΔku80 intracellular and RHΔhxgprtΔku80 extracellular parasite samples were prepared from the same lysates as those prepared for the study of the ubiquitin proteome of T. gondii (27). Specifically, MMA peptides were purified from the flow-through samples that had been depleted of ubiquitinated peptides. A similar yield of MMA peptides was obtained with RHΔhxgprtΔku80 intracellular parasite flow-through as from the RHΔhxgprt samples obtained from whole cell lysates (Fig. 1B). In addition, in error tolerant searches of the ubiquitin data sets (Mascot from Matrix Science, version 2.5.1) we did not detect any MMA sites, indicating that few, if any, MMA peptides were lost in the preceding ubiquitin enrichment step. There is also little overlap between proteins targeted by MMA and ubiquitin (27).
Database Searches
Raw mass spectra from LC-MS/MS were converted to Mascot Generic Format (MGF) files using Proteome Discoverer 1.2 (Thermo Fisher Scientific) software and then searched against a combined database (entries = 27608) of Homo sapiens (downloaded from Uniprot.org, Feb 25th, 2015) proteins and Toxoplasma gondii ME49 (downloaded from ToxoDB.org, version 12) proteins using an in-house Mascot search engine (Matrix Science, version 2.5.1, Boston, MA) and the Mascot default decoy database to obtain protein and peptide %FDRs. The following search parameters were used: trypsin, three missed cleavages; fixed modification of carbamidomethylation (Cys); variable modifications of oxidation (Met) and Methyl (Arg); monoisotopic masses; peptide mass tolerance of 5.0 ppm; product ion mass tolerance of 0.4 Da. Error tolerant searches were performed using either the same parameters or including the following variable modifications: methylation (R), oxidation (M), succinyl (K), phospho (STY). DAT files obtained from these searches were uploaded to Scaffold Q+ (Proteome Software, version 4.3.2, Proteome Software, Inc, Portland, Oregon). The following filters were used for protein and peptide validation: 95% minimum protein probability, minimum number peptides of 1 and 95% peptide probability. Contaminant proteins from human fibroblasts were excluded. As the immunoprecipitation technique results in selective purification of peptides with an MMA modification and only a single peptide in a protein may have this PTM, we included single peptide identifications of proteins in our data sets. These settings were based on previous experience with this technique (Cell Signaling Inc.) and our previous proteomic studies using similar immunoaffinity approaches (27). These files were exported to Scaffold PTM (Proteome Software, version 2.1.2.1). The Ascore algorithm (37) was used to localize MMA sites and a cutoff of 95% localization confidence was applied. The protein decoy FDR for this data was 7.8% and the peptide decoy FDR was 1.1%. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD004083 and 10.6019/PXD004083.
Bioinformatics Analysis of Protein and Peptide Hits
MMA proteins were categorized into subcellular compartments and functional groups using information obtained from the following databases: PFAM (http://pfam.xfam.org), http://tdrtargets.org, UNIPROT (http://uniprot.org), SUPFAM (http://supfam.org), http://prosite.expasy.org, TOXODB (htttp://www.toxodb.org) and literature searches (Dec. 2015). Gene Ontology (GO) and KEGG pathway analysis were both performed in TOXODB (http://www.toxodb.org). Enrichment analysis was performed using a custom R script for hypergeometric testing of enrichment of MMA proteins in gene sets that were defined previously (27, 35, 38). The p values of enrichment were adjusted for multiple hypothesis testing using the Bonferroni correction method and to control for random enrichment, 1000 random gene sets were generated and a p value of random enrichment was obtained. These values were used to generate a normalized p value, the “adjusted” p value, by dividing the experimental p value by the random enrichment p value. Motif logos of the six amino acid residues surrounding the R methyl sites were obtained from Scaffold PTM software. Heatmaps depicting amino acid residue enrichment and depletion were created with iceLogo software (http://iomics.ugent.be/icelogoserver/main.html).
RESULTS
General Features of the Arginine Monomethylome of Intracellular Tachyzoites
To study the arginine monomethylome of Toxoplasma gondii, we infected human foreskin fibroblasts with two wild type Type I T. gondii strains, RHΔhxgprt or RHΔhxgprtΔku80. All of the parasites analyzed in this study were tachyzoites grown under standard culture conditions (pH 7, 5% CO2) in vitro. Intracellular parasites were selectively harvested because this stage is highly transcriptionally active and arginine methylation is typically abundant on proteins involved in transcription (1).
Parasite lysates were prepared and MMA peptides were purified by affinity purification preceding LC-MS/MS (Fig. 1). From intracellular tachyzoites, 470 MMA sites were identified on 309 unique MMA proteins (>95% protein confidence, 7.7% protein decoy FDR). Sites were localized using the AScore algorithm (37) and a cutoff of 95% confidence of localization was applied. Three biological replicates (each consisting of two technical replicates) of intracellular wild type parasites were analyzed. Two were strain RHΔhxgprt and the other from strain RHΔhxgprtΔku80, strains that are highly similar in growth and virulence and commonly used for generation of T. gondii genetic mutants. MMA peptides from RHΔhxgprtΔku80 were purified using the flow-through of an enrichment experiment for ubiquitinated peptides from T. gondii (27) as input. Overall, 139 proteins and 346 MMA sites were common to all intracellular data sets (protein confidence >95%, 7.8% protein decoy FDR).
In surveys of arginine methylomes (MMA, ADMA, SDMA) in other organisms, 1970 arginine methylation sites (ADMA and MMA) were detected on 910 proteins in human HCT116 cells (33), and 1332 methylarginines on 676 proteins were detected in the arginine methylome of Trypanosoma brucei, another protozoan parasite (39). The present study focused solely on monomethylation; assuming the ratio of ADMA: MMA: SDMA in T. gondii is similar to that of humans (17, 18), we detected comparable protein numbers and MMA sites in T. gondii to those observed in human cells (33).
Immunoaffinity purification experiments may be biased for high abundance proteins. The coverage of proteins within our data set was compared with gene sets generated from transcript expression data as representing proteins whose mRNA are found from 0 to 100% expression percentiles (27). MMA proteins are enriched in proteins whose transcript expression levels are between 0 and 5 and 80% expression percentiles (supplemental Fig. S1A), indicating that both highly and lowly expressed proteins are present in the arginine methylome.
Arginine methylation typically occurs in GAR regions, and therefore to immunoprecipitate MMA peptides, antibodies specific to R*GG and R* (* = methyl site) were used. Consistent with existing literature and validating our approach, the amino environment surrounding MMA sites in T. gondii is rich in glycine and arginine residues (supplemental Fig. S2). This motif strongly resembles that of human MMA proteins (supplemental Fig. S2A; adapted from (33)) suggesting similar substrate specificities between human and T. gondii arginine methyltransferases. Most of the MMA sites detected occurred on a RGG, RG or XRX substrate motif (supplemental Fig. S2) with only few proteins in the intracellular wild-type data set matching to other motifs (supplemental Fig. S3). A heat map of amino acid residues surrounding arginine methylation sites in comparison to the entire T. gondii proteome is shown in supplemental Fig. S2B. Alanine, serine and proline are enriched in regions flanking the central arginine but are excluded from the positions immediately surrounding the methylated arginine residue. Most charged and hydrophobic residues are depleted in MMA peptides and isoleucine, lysine and glutamic acid are depleted at all positions within the regions examined (supplemental Fig. S2B).
Many MMA Proteins Localize to the Nucleus and Bind Nucleic Acids
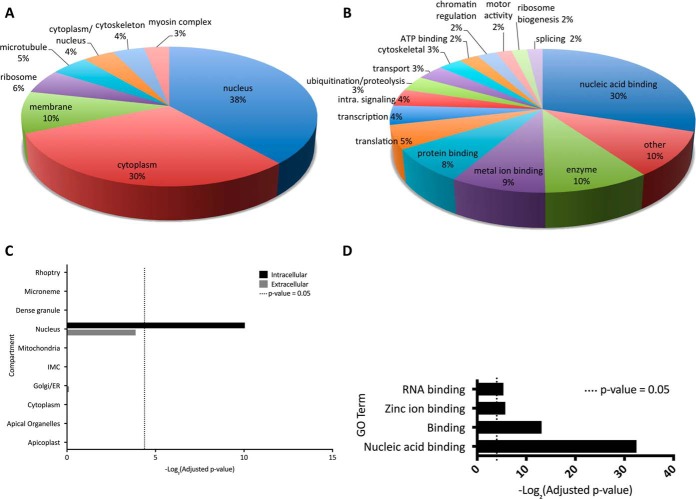
Arginine methylated proteins were manually annotated with predicted (based on GO terms) or known localization and function derived from the literature. Almost 30% of identified intracellular parasite MMA proteins are hypothetical proteins. T. gondii arginine methylated proteins are concentrated in the nucleus (38%) and cytoplasm (30%) (Fig. 2A; proteins missing biological function and cellular compartment GO term annotation were excluded from these statistics) In human cells, MMA is roughly equally distributed in the cytoplasm and nuclear compartment (7). Compared with a background of all T. gondii proteins, the MMA proteome is statistically enriched for nuclear proteins (Fig. 2C). Several cytoskeletal proteins were modified, including myosins J, F, E, and myosin heavy chain. β-tubulin, which our group has previously demonstrated to be methylated at the C terminus, was also modified (40). Few MMA proteins were detected in the mitochondria or the parasite apicoplast organelle, although it should be noted that relatively few proteins of the entire predicted proteome have been located to these compartments in T. gondii.
Fig. 2.
Inferred cellular compartments and functions of MMA proteins. A, Pie chart depicting 125 proteins of the arginine monomethylome categorized by subcellular compartment. Proteins of unknown localization are excluded (184 proteins; 64% hypothetical proteins). B, Pie chart depicting 207 of the MMA proteins categorized by function (molecular and biological) excluding unknown proteins (102 proteins; 93% hypothetical proteins). The unknown proteins were excluded from the manual annotations. Hypothetical proteins constitute 29% of the methylome and include proteins for which there was not enough information available to assign them to a cell compartment. C, Enrichment analysis of methylome proteins using cellular compartment gene sets as defined by (27) demonstrates that MMA proteins are enriched in proteins that localize to the nucleus; -log2(adjusted p value) is displayed with the dotted line indicating significant enrichment (adjusted p value = 0.05). D, Molecular function GO terms significantly (p value < 0.05) enriched in the MMA proteome of intracellular wild type parasites demonstrates that MMA proteins are enriched for GO terms associated with nucleic acid binding. GO terms were obtained from toxodb.org; -log2 (adjusted p value) is displayed, dotted line indicates adjusted p value of 0.05.
MMA proteins have a wide range of functions (Fig. 2B) but are enriched in DNA and RNA binding proteins (Fig. 2D). Many of these DNA and RNA binding proteins are highly modified by MMA, with up to 7 MMA sites (supplemental Table S2). Proteins containing an RNA recognition motif are the most abundantly MMA-modified nucleic acid binding proteins, constituting 29% of nucleic acid binding proteins and 6% of the total T. gondii arginine monomethylome. Of the 82 proteins predicted to contain an RNA Recognition Motif (RRM) (identified using PFAM domain searches), 23 are arginine monomethylated. Splicing factors were arginine monomethylated, including TGME49_319530, important for alternative splicing in T. gondii (41), as reported for its homologue in human (7). In addition, we detected arginine methylation on a large number of splicing factors and DEAD box helicases. These findings suggest that MMA has an important role in RNA biology in T. gondii.
Apetela 2 (ApiAP2) are conserved Apicomplexan transcription factors that regulate developmental stages of the Apicomplexan life cycle (42). Ten ApiAP2 were detected as being arginine methylated (AP2VIIa-4, AP2VIIa-5, AP2VIIa-7, AP2VIII-2, AP2VIII-4, AP2X-1, AP2XI-5, AP2XII-1, AP2XII-5, APVIIb-1). Notably, the arginine methylation sites never fall within an AP2 domain (supplemental Table S3), suggesting any functional regulation is allosteric. A number of other candidate regulators of transcription were also detected as being MMA-modified, such as general transcription factor E and a transcription elongation factor (SPT6).
Abundance of Stage-specific Proteins in the Arginine Monomethylome
Arginine methylation by TgPRMT4 has been implicated as a negative regulator of T. gondii differentiation (30). To determine whether the arginine monomethylome is enriched for stage specific proteins, we calculated the enrichment of stage-specific gene sets (35) in the MMA proteome. Surprisingly, MMA proteins are statistically enriched in gene sets that are either specifically up-regulated in bradyzoites or in tachyzoites (supplemental Fig. 1B).
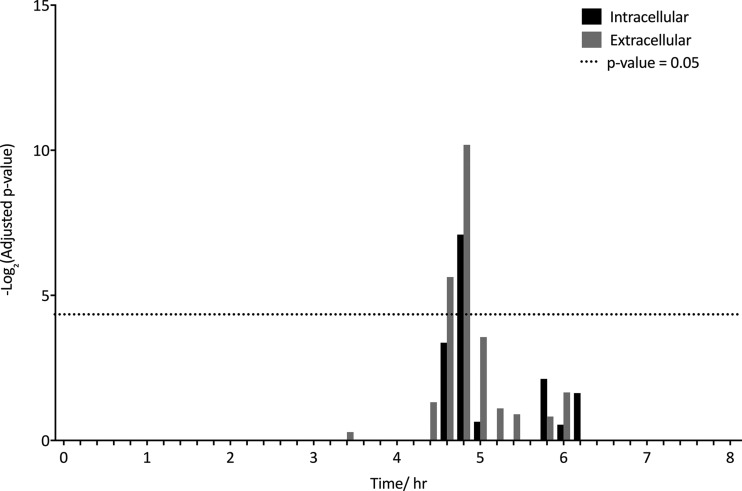
Deletion of TgPRMT1 in T. gondii causes cell cycle defects (29) and thus arginine methylation may be involved in cell cycle dynamics. T. gondii has an 8 h cell cycle consisting of distinct G1 and S/M regulated subtranscriptomes (43). Gene sets consisting of genes up-regulated at different time points in G1 and S/M phase (35) were used to determine whether MMA proteins are enriched for cell cycle regulated genes. Unlike other posttranslational modifications (27), arginine methylated proteins are not significantly enriched in S/M regulated genes and were only enriched in proteins whose genes were up-regulated in mid-G1 phase at 4.8 h (Fig. 3).
Fig. 3.
MMA proteins are enriched in mid-G1 up-regulated genes. MMA proteins were tested for enrichment of G1 regulated genes using predefined gene sets corresponding to 12 min time points during 8 h cell cycle (35). -log2 (p value) is displayed with the dotted line indicating significant enrichment (adjusted p value = 0.05). No significant enrichment of S/M genes was found.
Interplay Between MMA and Phosphorylation in T. gondii
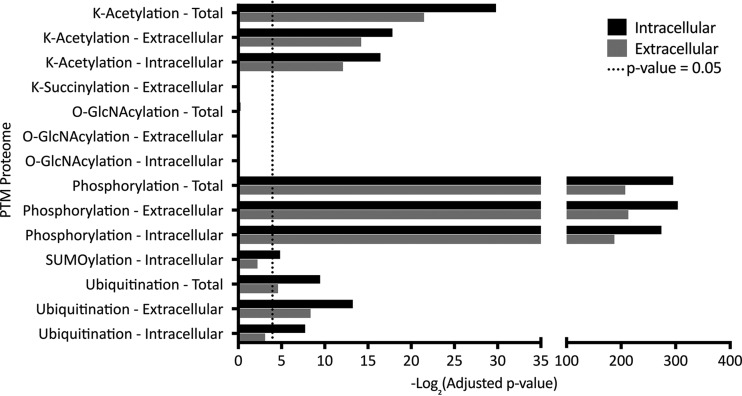
Crosstalk between PTM occurs in many organisms including T. gondii (27). PTM can promote or inhibit the occurrence of another PTM, or act in a combinatorial manner. In addition, adjacent posttranslational modifications can prevent arginine methylation by masking methylation sites or via steric hindrance (44). To explore cross-talk between arginine methylation and other PTM, we analyzed the significance of overlaps between the arginine monomethylome and proteins detected in previously published proteome-wide PTM data sets surveying phosphorylation, lysine acetylation, lysine succinylation, sumoylation, and ubiquitination (27, 32, 45–48) and the O-GlcNAc proteome (Silmon de Monerri & Kim, unpublished). The results of this analysis are shown in Fig. 4. The arginine methylome is significantly enriched in phosphorylated (-log2 p value 305.1), ubiquitinated (-log2 p value 13.5), and acetylated proteins (-log2 p value 33.9). Notably, we observed the greatest interaction to be with phosphoproteins in the arginine monomethylome, representing 89% of MMA proteins.
Fig. 4.
Crosstalk between MMA and other PTM. Interactions between arginine methylation and other PTMs including acetylation, O-GlcNAcylation, ubiquitination and phosphorylation were assessed by testing whether the arginine methylated proteins were significantly enriched for proteins detected in other PTM proteomes. -log2 adjusted p values are plotted, and the dotted line indicates statistically significant (adjusted p value = 0.05) enrichment.
MMA sites were detected on seven protein kinases of various types and three protein phosphatases. T. gondii possesses a large family of calcium dependent protein kinases (CDPKs) that have roles in signaling, host cell invasion and cell division (49). Of these, CDPK2A and CDPK7 are MMA modified on two sites in the N-terminal region, which is important for functional regulation (50). MMA sites were also detected on two cell cycle-associated kinases, a cyclin-dependent protein kinase (CDK) and serine-arginine protein kinase (SRPK).
Validation of Arginine Methylation Sites in GCN5b Complex
The lysine acetyltransferase GCN5b is a master regulator of transcription in T. gondii (51). Because arginine methylation is often involved in gene regulation, occurring on transcription factors and other nuclear proteins, GCN5b is a candidate for regulation by arginine methylation. Wang et al. immunoprecipitated GCN5b and its interaction partners and identified 20 proteins including RRM-containing proteins (51). Upon re-examining this data for PTMs, we detected arginine monomethylation on five of the 20 immunoprecipitated proteins: myosin F, ADA2-A transcriptional coactivator SAGA component, transcription elongation factor SPT6, an RNA recognition motif (RRM) domain containing protein (TGME49_262620) and a hypothetical protein of unknown function (TGME49_280590). In addition, several other proteins identified as being arginine monomethylated in this study were independently validated by database searches for MMA modifications in the GCN5b immunoprecipitation mass spectrometry data, including AP2VIII-4 (TGME49_272710) and beta tubulin (TGME49_266960) (supplemental Table S4).
Changes in the Arginine Monomethylome of Extracellular Parasites
One reason for mapping the MMA proteome of T. gondii is to determine whether MMA is dynamically regulated in extracellular tachyzoites. During transcriptional arrest, arginine methylation changes (19), indicating that it is dynamic. In extracellular tachyzoites, cell cycle is arrested in a G0-like state (34, 35) and changes in PTM proteomes are observed (27, 32, 36). To determine whether these changes are accompanied by altered MMA, we surveyed the arginine monomethylome of extracellular parasites. Extracellular T. gondii RHΔhxgprtΔku80 that had spontaneously egressed were harvested, lysed and subjected to immunoaffinity purification using anti-MMA antibodies. This yielded 198 arginine methylated proteins (7.7% protein decoy FDR) and 288 MMA sites. Of these proteins, 185 overlap with the 309 MMA proteins detected in intracellular parasites and 14 proteins are uniquely modified in extracellular tachyzoites. There are 181 fewer MMA sites in extracellular tachyzoites. This suggests that there is an overall decrease in MMA in extracellular tachyzoites, consistent with reduced MMA during the G1 cell cycle arrest that has been reported to occur in extracellular parasites (34).
Overall, the MMA proteome of extracellular tachyzoites shares many features with that of intracellular tachyzoites. A large number of MMA proteins identified in extracellular tachyzoites are nuclear, though this enrichment was no longer statistically significant (Fig. 2C). MMA proteins from extracellular tachyzoites are enriched for genes up-regulated at 4.8 h in G1 phase but not S/M phase (Fig. 3), and are enriched in tachyzoite and bradyzoite gene sets (supplemental Fig. S1B). The same highly significant enrichment of phosphoproteins was also observed (Fig. 4). In addition, there is no significant change in the amino acid environment of MMA sites in extracellular tachyzoites (supplemental Fig. S2A).
The 14 proteins that were detected as being MMA modified in only extracellular tachyzoites consist of three metabolic enzymes (phosphatidylinositol 3- and 4-kinase, phosphoglycerate mutase family protein, serine esterase (DUF676) protein) and several hypothetical proteins. Proteins that were not detected as being arginine monomethylated in extracellular tachyzoites, but were in intracellular tachyzoites, were examined by GO term analysis and this did not demonstrate an enrichment of these proteins for any particular function. Some of the ApiAP2 transcription factors that were arginine methylated in intracellular tachyzoites (AP2VIIa-7, AP2XI-5, AP2XII-1) and other transcriptional regulators such as SWI2/SNF2 chromatin remodeling complex proteins were absent in the extracellular arginine monomethylome (52).
Perturbation of the MMA Proteome by Disruption of TgPRMT1
MMA is the initial step in methylation and its addition can theoretically be catalyzed by any type of PRMT. In mammalian cells, on ablation of PRMT1, an increase in MMA and SDMA is observed (20) suggesting substrate scavenging by other PRMT. In T. gondii, few TgPRMTs have been studied and it is not known whether the same effect occurs on deletion of TgPRMT1. T. gondii and other unicellular eukaryotes, aside from Trypanosomatids, lack a type III PRMT7 homologue, thus the enzyme that performs MMA addition in these organisms is unknown. Recent work suggests that TgPRMT1 is the major PRMT in T. gondii (29, 30) and it is possible that TgPRMT1 is able to catalyze MMA addition in T. gondii. Thus, to understand the contribution of TgPRMT1 to global arginine methylation and dissect the underlying mechanism of the TgPRMT1 knockout phenotype, we surveyed the MMA proteome of intracellular TgPRMT1 knockout tachyzoites (PRMT1KO) and a genetically complemented strain (PRMT1COMP) (29), harvested at 36 h postinfection. The PRMT1KO and PRMT1COMP strains were generated in the RHΔhxgprt strain background (29) and therefore the MMA proteome of the RHΔhxgprt strain was used as the wild type strain for comparison. Analyzed data sets are summarized in Fig. 1.
Of the MMA proteins identified in all three replicates of wild type tachyzoites (139 proteins), 50% were not detected in PRMT1KO parasites. These proteins may be composed of both TgPRMT1 substrates and proteins or peptides whose abundances are close to the detection threshold of this study. By comparing the MMA proteomes of the wild type, knockout and complemented strains, proteins that were more likely to be TgPRMT1 substrates were identified. Proteins that were detected in PRMT1COMP and wild type parasites but not PRMT1KO parasites were considered highly probable TgPRMT1 substrates.
Using these stringent criteria, 68 high confidence TgPRMT1 candidate substrates were identified and are listed in supplemental Table S5. PRMTs often exhibit variation in substrate specificity. The amino acid environment surrounding MMA sites of candidate TgPRMT1 substrates was analyzed and they are similar to the global MMA sequence preferences, i.e. no TgPRMT1 specific motif was evident in this data set, although on average proline was less frequently seen flanking the modified arginine residue in the PRMT1 substrates.
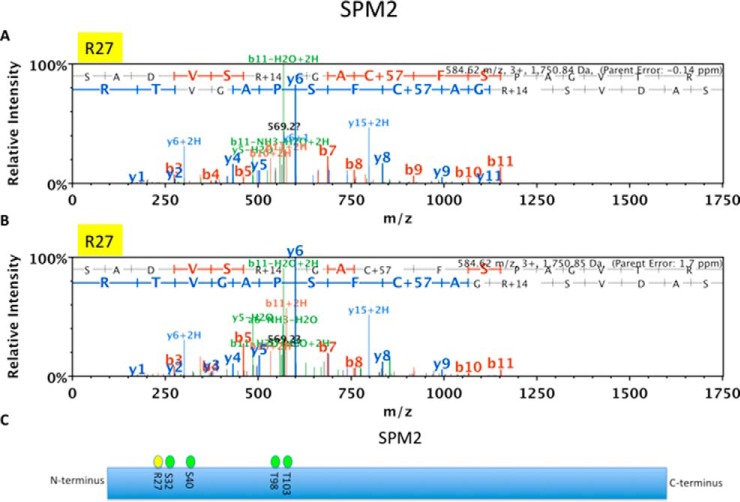
Like global MMA proteins, candidate TgPRMT1 substrates have a variety of functions. GO analysis did not reveal any particular pathways enriched within TgPRMT1 substrates, suggesting that TgPRMT1 has multiple functions or that it is a master regulator. Most TgPRMT1 substrates localize to the nucleus, which is unexpected considering the localization of TgPRMT1 to the cytosol with a concentration in pericentriolar regions (29). Two cytoskeletal proteins of interest that are likely TgPRMT1 substrates are myosin E, whose function is unknown, and SPM2 (Fig. 5), a component of the subpellicular microtubules, some of which regulate division (53). Of the nuclear substrates, a SET domain containing histone lysine methyltransferase was identified, with a single MMA site in the protein N terminus at R287. Two AP2 transcription factors, AP2VIII-2 (Fig. 6) and AP2XII-1 (supplemental Fig. S4), are highly confident TgPRMT1 substrates, as well as several RNA binding proteins.
Fig. 5.
Spectra of microtubule-associated Protein SPM2 and its MMA sites in intracellular and PRMT1COMP. LC-MS/MS spectrum of microtubule-associated protein SPM2 derived peptide (SADVSR*GACFSPAGVTR) showing MMA sites on (A) Arginine R27 in WT2 with an error of −0.14 ppm and (B) R27 in PRMT1 COMP with an error of 1.7 ppm. C, Schematic of microtubule-associated protein SPM2 with MMA sites (yellow) and previously mapped serine and threonine phosphorylation sites (green) (32).
Fig. 6.
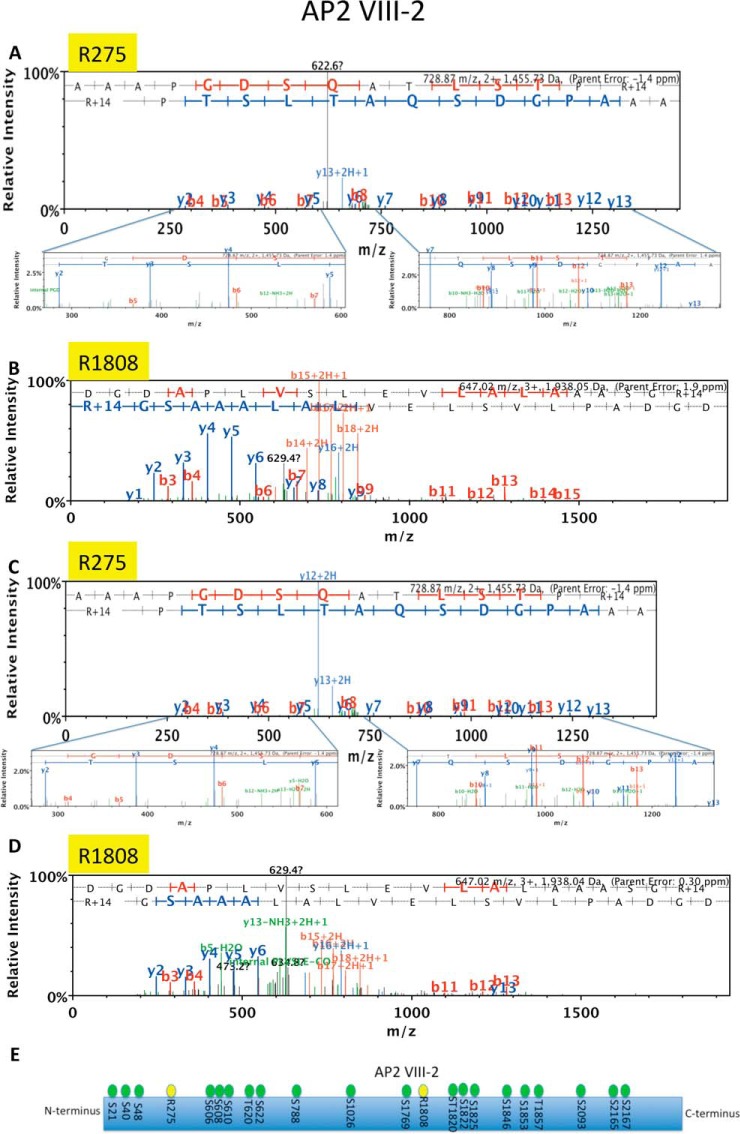
Spectra of AP2 VIII-2 and its two MMA sites in intracellular and PRMT1 COMP. LC-MS/MS spectra of AP2VIII-2 derived peptides (AAAPGDSQATLSTPR* (A,C) and DGDAPLVSLEVLALAAASGR* (B,D)) showing MMA sites: R275 in WT3 with an error of −1.4 ppm (A), R1808 in WT3 with an error of 1.9 (B), R275 in PRMT1COMP with an error of −1.4 ppm (C) and R1808 in PRMT1COMP with an error of 0.30 ppm (D); none of these sites were detected in PRMT1KO parasites. E. Schematic of AP2VIII-2 with MMA sites (yellow) and previously mapped serine and threonine phosphorylation sites (green) (32).
Following the same trend as the entire arginine monomethylome, 48 of the candidate TgPRMT1 substrate proteins are also targets of phosphorylation (32). Of the kinases identified in intracellular tachyzoites, calcium dependent protein kinase 7 (CDPK7) was identified as a putative TgPRMT1 substrate. Two MMA sites were detected in intracellular tachyzoites (R463 and R805) (Fig. 7). CDPK7 is involved in cell division and, interestingly, CDPK7 knockout parasites exhibit a similar defect in counting as TgPRMT1 mutant parasites (29, 54). In addition, a single MMA site was detected on TgPRMT1 itself at R17, near the N terminus.
Fig. 7.
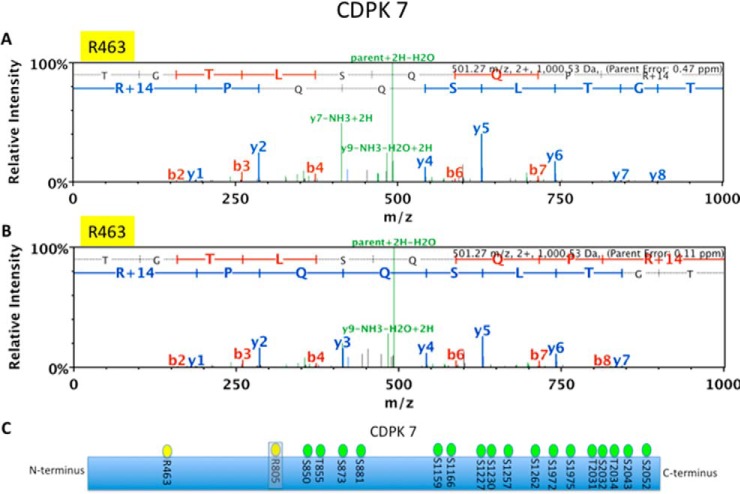
MMA sites detected on CDPK7. LC-MS/MS spectrum of a CDPK7-derived peptide (TGTLSQQPR*) showing MMA at R463 from sample WT2 with an error of 0.47 ppm (A) and in PRMT1COMP strain with an error of 0.11 ppm (B), but not detected in PRMT1KO parasites. C, Schematic of CDPK7 with MMA sites (yellow) and previously mapped serine and threonine phosphorylation sites (green) (32).
Because a global decrease in MMA in the PRMT1 KO was unexpected given prior observations in mammalian cells (20), we performed 2D-PAGE immunoblots (supplemental Fig. S5) using equal amounts of PRMT1KO and PRMT1COMP tachyzoite protein lysates and examined these blots for the presence of MMA, SDMA, and ADMA modifications using methylation specific antibodies (supplemental Methods). SDMA antibodies did not have immunoblot reactivity. The PRMT1COMP parasites had a greater number of MMA and ADMA modified proteins relative to the PRMT1 KO, consistent with PRMT1 being responsible for a significant amount of the observed MMA and ADMA activity in T. gondii.
DISCUSSION
Arginine methylation plays important roles in parasite division and differentiation (29, 30). In this paper we have demonstrated that MMA is highly abundant in T. gondii and that this PTM is found at comparable levels to both ubiquitination and phosphorylation, as was recently shown in humans (7). The MMA proteome of T. gondii likely consists of proteins that are terminally monomethylated as well as those that are transiently monomethylated and later converted to SDMA and ADMA. MMA proteins represent almost 4% of the total T. gondii proteome, demonstrating that MMA is an abundant modification in this organism. This study focused on MMA. Although we cannot evaluate the full extent of ADMA in T. gondii, our 2D-PAGE immunoblots (supplemental Fig. S5B) provide evidence that PRMT1 is important for MMA and ADMA modifications of T. gondii proteins; furthermore, the PRMT1 KO phenotype suggests these modifications have important functions in the biology of this pathogen. In the future, it will be interesting to explore the contributions of each modification (i.e. MMA, ADMA, and SDMA) to global arginine methylation. Comparative proteomics should help identify MMA sites that are found in many Apicomplexa as well as those that are unique to T. gondii and may have specific biological functions in this organism.
Surprisingly, a considerable portion of the MMA proteome is also targeted by phosphorylation in tachyzoites. Although many PTM can regulate the same protein, such significant co-regulation of phosphorylation and arginine methylation has not, to our knowledge, been observed in other organisms. Phosphorylation and arginine methylation are often mutually exclusive [e.g. (55)], but arginine methylation can also promote phosphorylation (56). Proteins detected in the phosphoproteome of tachyzoites are enriched in genes that are up-regulated at several time points during cell cycle, including mid-G1 phase (27). MMA is also enriched in genes upregulated at mid-G1 phase and many of these proteins were detected as being phosphorylated in a previous phosphoproteomic study on intracellular and extracellular tachyzoites (32). This time point likely represents a mid-G1 checkpoint and coincides with a peak in phosphorylation (27). Together, these data suggest interplay between phosphorylation and MMA proteins on genes up-regulated in mid-G1 phase. We also observed a significant enrichment of ubiquitination in the monomethylome. In human cells lysine ubiquitination sites were enriched in regions of unmodified arginine residues, so further studies are needed to evaluate whether crosstalk between the two PTMs is significant (7).
Overall, the arginine monomethylome of T. gondii is enriched for nuclear proteins and proteins that bind nucleic acids such as RNA. RNA binding proteins (RBP) are key targets of arginine methylation in T. gondii and other organisms. Arginine methylation of RBP regulates a large number of RNA processes such as pre-mRNA splicing, RNA stability and translation. Arginine methylated splicing factors that regulate genome-wide alternative splicing (41) were detected in humans and our study, implicating arginine methylation could play a conserved role in regulation of splicing activity. Other nuclear proteins modified by arginine methylation and phosphorylation include several ApiAP2 transcription factors (supplemental Table S3). Cooperation between PTMs is likely to play an essential role in transcriptional control in this organism.
Arginine methylation plays a key role in epigenetic regulation as part of the histone code (1). Arginine residues on histones are monomethylated on several sites in T. gondii (28). In the current study we did not identify any of the histone peptides as being MMA modified, however, in our previous study of histone PTM, methylation of histone H4R3 appeared to be substochiometric (28). Although the affinity purification method is highly sensitive (33), the abundance of MMA on histones may be below the detection limit of this study, given that we did not enrich for histones prior to affinity purification. Alternatively, the failure to detect MMA sites on histones could be because of biases in antibody specificity. A combination of an R-methyl antibody that is not sequence-specific and one that recognizes the R-methyl-GG motif was used with majority of identified sites encoding an RG motif (Supplemental Fig. 2). Histones are highly basic and very few glycine residues are found in their primary sequences.
Arginine methylation typically occurs in GAR motifs, which we confirmed is a feature of arginine methylation sites in T. gondii by analyzing the sequences surrounding identified sites. The importance of GAR motifs in T. gondii is highlighted by a recent study on TgSossB, a single strand DNA and RNA binding protein. Boulila et al. showed that removal of the RGG portion of TgSossB resulted in a severe fitness defect (57). We detected arginine methylation on the C terminus TgSossB, within the GAR domain. These findings highlight a possible role for specific arginine methylation of the GAR domain in processes critical for parasite viability.
RBP interact with RNA through RNA recognition motifs (RRM). Of 86 RRM-containing proteins encoded in T. gondii (58) 19 RRM proteins were arginine methylated at 62 MMA sites. In five of the RRM proteins (TGME49_270880, TGME49_265250, TGME49_291930, TGME49_304760, TGME49_262620), MMA sites fall within RRM domains, suggesting a role for MMA in non-enzymatic regulation of RNA binding. In contrast, MMA appears to play little or no role in the regulation of enzymatic RNA binding domains (RBD), as supported by the lack of MMA sites in any of the four MMA modified DEAD-box or -like RNA helicase domains found in the T. gondii methylome and supported by recent similar findings in humans (7). SF2 (TGME49_319530), however, represents a notable exception to this observation as both of its MMA sites (R88 and R108) were found within its RBD, and in humans MMA is proposed to play a novel regulatory role in SF2 assembly within the nucleus (7).
In other species, MMA is decreased when transcriptional arrest is artificially induced (19). The MMA proteome of extracellular tachyzoites, which are considered to be growth arrested (34, 35), differs from intracellular tachyzoites with 181 fewer MMA sites identified in extracellular tachyzoites; other PTM are known to change in abundance in extracellular tachyzoites (27, 32, 36). Together, PTM are implicated as regulators of cell cycle control in extracellular tachyzoites. To confirm these differences, quantitative proteomics would be required to assess changes in protein abundance.
Arginine methylation is also a potential regulator of parasite differentiation. In this study, MMA proteins were found to be statistically enriched in bradyzoite specific proteins as well as tachyzoite specific proteins, suggesting that arginine methylation plays some role in regulating differentiation. Supporting this, Saksouk and colleagues previously showed that inhibition of the type I TgPRMT4 (referred to as CARM1 by these authors) induces bradyzoite differentiation (30). In T. brucei, patterns of arginine methylation differ between life cycle forms (39); it would be interesting to determine whether global MMA differs between tachyzoites and bradyzoites in T. gondii.
TgPRMT1 is thought to be the major arginine methyltransferase in T. gondii (29, 30). In PRMT1KO parasites, 70 proteins were absent in comparison with the proteins common to all three MMA proteomes of wild type parasites. For a proportion (33 proteins) of these proteins, arginine methylation was restored in genetically complemented parasites. Though quantitative proteomics would provide more definitive answer to what the contribution of TgPRMT1 is to the arginine methylome, this data suggests that TgPRMT1 is a major contributor to the arginine monomethylome. In contrast, loss of PRMT1 in humans and T. brucei results in an increase in MMA (20, 39), attributed to substrate scavenging by other PRMTs in the absence of PRMT1. Although the current study did not survey arginine dimethylation using mass spectrometry, the 2D-PAGE immunoblot analysis provides evidence that TgPRMT1 contributes significantly to ADMA. Further characterization of ADMA and SDMA proteomes of the TgPRMT1 knockout strains could further define the function of TgPRMT1 methylation in the biology of T. gondii.
Though PRMT types have distinct properties (9), there is a degree of redundancy among different PRMTs in humans (25) and T. brucei (59). Whether the five TgPRMTs in T. gondii overlap in function is unclear. TgPRMT4 is essential to parasite viability, suggesting that its function cannot be compensated for by another methyltransferase (30). TgPRMT1 is not essential, although knockout parasites are impaired (29). When TgPRMT expression was assessed by microarray in PRMT1 KO parasites, only a minor increase in TgPRMT4 was observed (1.18-fold change) (29), suggesting that compensatory mRNA up-regulation of redundant TgPRMTs in response to PRMT1KO does not occur. Collectively these data suggest that TgPRMT1 in T. gondii has unique functions that cannot be compensated for by another TgPRMT.
PRMT1 usually functions as a dimer (60). A single MMA site at R17 detected on TgPRMT1 may be important for regulation of the enzyme. Phosphorylation sites have been mapped very close to R17 at T22 and S30 (32) and considering the hypothesized mutually exclusive nature of arginine methylation and phosphorylation, this could suggest two opposing modes of regulation of TgPRMT1 function. Whether this regulation is reflective of automethylation or regulation by another methyltransferase is unclear.
A number of candidate TgPRMT1 substrates were identified in this study. A significant proportion of candidate TgPRMT1 substrates localize to the nucleus. Although TgPRMT1 is primarily a cytosolic enzyme with a role in regulation of the pericentriolar matrix (29), methylation may alter the subcellular localization or protein-protein interactions of substrate. TgPRMT1 catalyzes the formation of ADMA (or even SDMA) at pericentriolar regions. To answer these questions, further proteomics studies assessing ADMA and SDMA in TgPRMT1 knockout parasites will be required.
It has been previously reported that Trypanosoma brucei alpha tubulin is modified by SDMA, beta tubulin modified by MMA and epsilon tubulin modified by SDMA, indicating that methylation may play a role in the regulation of tubulin and processes such as cytoskeletal support and intracellular transport (39). Evidence for the latter comes from arginine methylation found on substrates involved in intra-Golgi transport, vesicular transport proteins and vesicle fusion (39).
Apetela 2 (ApiAP2) are conserved Apicomplexan transcription factors that regulate developmental stages of the Apicomplexan life cycle (42). Arginine methylation of transcription factors can inhibit their degradation by preventing phosphorylation events required for ubiquitin-mediated destruction (5). Interestingly, we observed the greatest interaction to be with phosphoproteins in the arginine monomethylome, representing 89% of MMA proteins, supporting a role for arginine methylation in regulating the phosphoproteome in T. gondii. One possible example of such regulation is AP2XI-5, which is methylated at R647 and has been implicated in transcriptional regulation of virulence factors expressed late in the T. gondii cell cycle (52).
Overall, the data presented here suggest that MMA is an abundant, dynamic PTM in T. gondii that regulates RNA biology and transcription among other functions. Future work should address the contribution of other types of arginine methylation to the arginine monomethylome and the potential role of arginine methylation in differentiation. TgPRMT1 appears to play a role in the regulation of this modification and the identification of TgPRMT1 substrates in this study contributes significantly to our understanding of the function of TgPRMT1 in parasite biology.
Supplementary Material
Acknowledgments
We thank Dr. Kentaro Kato of Obihiro University of Agriculture and Veterinary Medicine, Japan for his kind gift of rabbit anti-TgALD1.
Footnotes
Author contributions: R.R.Y., N.C.S., K.K., and L.M.W. designed research; R.R.Y., N.C.S., and E.N. performed research; R.R.Y., N.C.S., E.N., K.K., and L.M.W. analyzed data; R.R.Y., N.C.S., K.K., and L.M.W. wrote the paper.
 This article contains supplemental Materials.
This article contains supplemental Materials.
1 The abbreviations used are:
- PRMTs
- protein arginine methyltransferases
- ApiAP2
- Apicomplexan Apetala 2 transcription factors
- Arg
- arginine (R)
- cdpk
- calcium dependent protein kinase
- CARM1
- coactivator-associated arginine methyltransferase 1
- Cys
- cysteine (C)
- DUF
- domain of unknown function
- EXTRA
- extracellular
- FDR
- false discovery rate
- GO
- Gene Ontology
- GAR
- glycine-arginine
- H3R2me2
- histone 3 dimethyl arginine 2
- H4R3me2
- histone 4 dimethyl arginine 3
- HFF
- human foreskin fibroblasts
- JMJD6
- jumonji domain-containing protein 6
- KEGG
- Kyoto Encyclopedia of Genes and Genomes
- Lys
- lysine (L)
- MGF
- mascot generic format
- Met
- methionine (M)
- MMA
- monomethyl arginine
- ADMA
- NG-NG-asymmetric dimethylarginine
- SDMA
- omega NG-NG-symmetric dimethylarginine
- PADIs
- peptidyl arginine deiminases
- PBS
- phosphate buffered saline
- PTM
- post translational modification
- PRMT1COMP
- PRMT1 complemented T. gondii strain
- PRMT1KO
- PRMT1 knockout T. gondii strain
- RBD
- RNA binding domain
- RBP
- RNA binding proteins
- RRM
- RNA Recognition Motif
- STY
- serine threonine tyrosine
- SPT6
- suppressor of Ty 6
- Trp
- tryptophan (W)
- Tyr
- tyrosine (Y)
- WT
- wild type.
REFERENCES
- 1. Bedford M. T., and Clarke S. G. (2009) Protein arginine methylation in mammals: who, what, and why. Mol. Cell. 33, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bedford M. T., Frankel A., Yaffe M.B., Clarke S., Leder P., and Richard S. (2000) Arginine methylation inhibits the binding of proline-rich ligands to Src homology 3, but not WW, domains. J. Biol. Chem. 275 16030–16036 [DOI] [PubMed] [Google Scholar]
- 3. Horowitz S., and Trievel R.C. (2012) Carbon-oxygen hydrogen bonding in biological structure. J. Biol. Chem. 287 41576–41582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Molina-Serrano D., Schiza V., and Kirmizis A. (2013) Cross-talk among epigenetic modifications: lessons from histone arginine methylation. Biochem. Soc. Trans. 41, 751–759 [DOI] [PubMed] [Google Scholar]
- 5. Yamagata K., Daitoku H., Takahashi Y., Namiki K., Hisatake K., Kako K., Mukai H., Kasuya Y., and Fukamizu A. (2008) Arginine methylation of FOXO transcription factors inhibits their phosphorylation by Akt. Mol. Cell. 32, 221–231 [DOI] [PubMed] [Google Scholar]
- 6. Wei H. M., Hu H. H., Chang G. Y., Lee Y. J., Li Y. C., Chang H. H., and Li C. (2014) Arginine methylation of the cellular nucleic acid binding protein does not affect its subcellular localization but impedes RNA binding. FEBS Lett. 588, 1542–1548 [DOI] [PubMed] [Google Scholar]
- 7. Larsen S. C., Sylvestersen K. B., Mund A., Lyon D., Mullari M., Madsen M. V., Daniel J. A., Jensen L. J., and Nielsen M. L. (2016) Proteome-wide analysis of arginine monomethylation reveals widespread occurrence in human cells. Sci. Sig. 9, 1–15 [DOI] [PubMed] [Google Scholar]
- 8. Najbauer J., Johnson B. A., Young A. L., and Aswad D. W. (1993) Peptides with sequences similar to glycine, arginine-rich motifs in proteins interacting with RNA are efficiently recognized by methyltransferase(s) modifying arginine in numerous proteins. J. Biol. Chem. 268, 10501–10509 [PubMed] [Google Scholar]
- 9. Herrmann F., Pably P., Eckerich C., Bedford M.T., and Fackelmayer F.O. (2009) Human protein arginine methyltransferases in vivo–distinct properties of eight canonical members of the PRMT family. J.Cell Sci. 122, 667–677 [DOI] [PubMed] [Google Scholar]
- 10. Bachand F. (2007) Protein arginine methyltransferases: From unicellular eukaryotes to humans. Eukaryotic Cell. 6, 889–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zurita-Lopez C. I., Sandberg T., Kelly R., and Clarke S. G. (2012) Human protein arginine methyltransferase 7 (PRMT7) is a type III enzyme forming ω-N G-monomethylated arginine residues. J. Biol. Chem. 287, 7859–7870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Takahashi Y., Daitoku H., Yokoyama A., Nakayama K., Kim J. D., and Fukamizu A. (2011) The C. elegans PRMT-3 possesses a type III protein arginine methyltransferase activity. J. Receptor Signal Transduction Res. 31, 168–172 [DOI] [PubMed] [Google Scholar]
- 13. Ferreira T. R., Alves-Ferreira E. V. C., Defina T. P. A., Walrad P., Papadopoulou B., and Cruz A. K. (2014) Altered expression of an RBP-associated arginine methyltransferase 7 in Leishmania major affects parasite infection. Mol. Microbiol. 94, 1085–1102 [DOI] [PubMed] [Google Scholar]
- 14. Fisk J. C., Sayegh J., Zurita-Lopez C., Menon S., Presnyak V., Clarke S. G., and Read L. K. (2009) A type III protein arginine methyltransferase from the protozoan parasite Trypanosoma brucei. J. Biol. Chem. 284, 11590–11600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McBride A. E., Zurita-Lopez C., Regis A., Blum E., Conboy A., Elf S., and Clarke S. (2007) Protein arginine methylation in Candida albicans: Role in nuclear transport. Eukaryotic Cell 6, 1119–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Niewmierzycka A., and Clarke S. (1999) S-Adenosylmethionine-dependent methylation in Saccharomyces cerevisiae. J. Biol. Chem. 274, 814–824 [DOI] [PubMed] [Google Scholar]
- 17. Paik W. K., and Kim S. (1980) Natural occurrence of various methylated amino acid derivatives. John Wiley & sons: New York [Google Scholar]
- 18. Matsuoka M. (1972) [Epsilon-N-methylated lysine and guanidine-N-methylated arginine of proteins. 3. Presence and distribution in nature and mammals]. Seikagaku 44, 364–370 [PubMed] [Google Scholar]
- 19. Sylvestersen K. B., Horn H., Jungmichel S., Jensen L. J., and Nielsen M. L. (2014) Proteomic analysis of arginine methylation sites in human cells reveals dynamic regulation during transcriptional arrest. Mol. Cell. Proteomics 13, 2072–2088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dhar S., Vemulapalli V., Patananan A. N., Huang G. L., Di Lorenzo A., Richard S., Comb M. J., Guo A., Clarke S. G., and Bedford M. T. (2013) Loss of the major Type I arginine methyltransferase PRMT1 causes substrate scavenging by other PRMTs. Sci. Reports 3, 1311–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tikhanovich I., Kuravi S., Artigues A., Villar M. T., Dorko K., Nawabi A., Roberts B., and Weinman S. A. (2015) Dynamic arginine methylation of TNF receptor associated factor 6 regulates Toll-like receptor signaling. J. Biol. Chem. 290, 10.1074/jbc.M115.653543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cheng D., Côté J., Shaaban S., and Bedford M. T. (2007) The arginine methyltransferase CARM1 regulates the coupling of transcription and mRNA processing. Mol. Cell 25, 71–83 [DOI] [PubMed] [Google Scholar]
- 23. Ng S. S., Yue W. W., Oppermann U., and Klose R. J. (2009) Dynamic protein methylation in chromatin biology. Cell. Mol. Life Sci. 66, 407–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Thompson P. R., and Fast W. (2006) Histone citrullination by protein arginine deiminase: is arginine methylation a green light or a roadblock? ACS Chem. Biol. 1, 433–441 [DOI] [PubMed] [Google Scholar]
- 25. Fisk J. C., and Read L. K. (2011) Protein arginine methylation in parasitic protozoa. Eukaryotic Cell 10, 1013–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Radke J. R., Behnke M. S., Mackey A. J., Radke J. B., Roos D. S., and White M. W. (2005) The transcriptome of Toxoplasma gondii. BMC Biol. 3, 26–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. de Monerri S., Natalie C., Rama R., Yakubu A. L. Chen P. J., Nieves B. E., Weiss L. M., and Kim K. (2015) The ubiquitin proteome of Toxoplasma gondii Reveals roles for protein ubiquitination in cell-cycle transitions. Cell Host Microbe 18, 621–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nardelli S. C., Che F. Y., Silmon de Monerri N. C., Xiao H., Nieves E., Madrid-Aliste C., Angel S. O., Sullivan W. J., Angeletti R. H., Kim K., and Weiss L. M. (2013) The histone code of Toxoplasma gondii comprises conserved and unique posttranslational modifications. mBio. 4, e00922-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. El Bissati K., Suvorova E. S., Xiao H., Lucas O., Upadhya R., Ma Y., Angeletti R. H., White M. W., Weiss L. M., and Kim K. (2016) Toxoplasma gondii arginine methyltransferase 1 (PRMT1) Is Necessary for centrosome dynamics during tachyzoite cell division. MBio. 7, e02094-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Saksouk N., Bhatti M. M., Kieffer S., Aaron T., Musset K., Garin J., Wullivan W. J. Jr, Hakimi M. A., and Smith A. T. (2005) Histone-modifying complexes regulate gene expression pertinent to the differentiation of the protozoan parasite Toxoplasma gondii Mol. Cell. Biol. 25, 10301–10314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tang J., Frankel A., Cook R. J., Kim S., Paik W. K., Williams K. R., Clarke S., and Herschman H. R. (2000) PRMT1 is the predominant type I protein arginine methyltransferase in mammalian cells. J. Biol. Chem. 275, 7723–7730 [DOI] [PubMed] [Google Scholar]
- 32. Treeck M., Sanders J. L., Elias J. E., and Boothroyd J. C. (2011) The phosphoproteomes of Plasmodium falciparum and Toxoplasma gondii reveal unusual adaptations within and beyond the parasites' boundaries. Cell Host Microbe 10, 410–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Guo A., Gu H., Zhou J., Mulhern D., Wang Y., Lee K. A., Yang V., Aguiar M., Kornhauser J., Jia X., Ren J., Beausoleil S. A., Silva J. C., Vemulapalli V., Bedford M. T., and Comb M. J. (2014) Immunoaffinity enrichment and mass spectrometry analysis of protein methylation. Mol. Cell. Proteomics 13, 372–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lescault P.J., Thompson A. B., Patil V., Lirussi D., Burton A., Margarit J., Bond J., and Matrajt M. (2010) Genomic data reveal toxoplasma gondii differentiation mutants are also impaired with respect to switching into a novel extracellular tachyzoite state. PLoS ONE. 5, e14463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Croken M. M., Qiu W., White M. W., and Kim K. (2014) Gene set enrichment analysis (GSEA) of Toxoplasma gondii expression data sets links cell cycle progression and the bradyzoite developmental program. BMC Genomics 15, 515–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Xue B., Jeffers V., Sullivan W. J., and Uversky V. N. (2013) Protein intrinsic disorder in the acetylome of intracellular and extracellular Toxoplasma gondii. Mol. Biosyst. 9, 645–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Beausoleil S. A., Villén J. Gerber S. A., Rush J., and Gygi S. P. (2006) A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol. 24, 1285–1292 [DOI] [PubMed] [Google Scholar]
- 38. Gaji R. Y., Behnke M. S., Lehmann M. M., White M. W., and Carruthers V. B. (2011) Cell cycle-dependent, intercellular transmission of Toxoplasma gondii is accompanied by marked changes in parasite gene expression. Mol. Microbiol. 79, 192–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lott K., Li J. Fisk J. C., Wang H., Aletta J. M., Qu J., and Read L. K. (2013) Global proteomic analysis in trypanosomes reveals unique proteins and conserved cellular processes impacted by arginine methylation. J. Proteomics 91, 210–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xiao H., El Bissati K., Verdier-Pinard P., Burd B., Zhang H., Kim K., Fiser A., Angeletti R. H., and Weiss L. M. (2010) Post-translational modifications to Toxoplasma gondii alpha- and beta-tubulins include novel C-terminal methylation. J. Proteome Res. 9, 359–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yeoh L. M., Goodman C. D., Hall N. E., van Dooren G. G., McFadden G. I., and Ralph S. A. (2015) A serine-arginine-rich (SR) splicing factor modulates alternative splicing of over a thousand genes in Toxoplasma gondii. Nucleic Acids Res. 43, 4661–4675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Balaji S., Madan Babu M., Iyer L. M., and Aravind L. (2005) Discovery of the principal specific transcription factors of Apicomplexa and their implication for the evolution of the AP2-integrase DNA binding domains. Nucleic Acids Res. 33, 3994–4006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Behnke M. S., Wootton J. C., Lehmann M. M., Radke J. B., Lucas O., Nawas J., Sibley L.D., and White M. W. (2010) Coordinated progression through two subtranscriptomes underlies the tachyzoite cycle of Toxoplasma gondii. PLoS ONE. 5, e12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bedford M. T., and Richard S. (2005) Arginine methylation. Mol. Cell 18, 263–272 [DOI] [PubMed] [Google Scholar]
- 45. Foe I. T., Child M. A., Majmudar J. D., Krishnamurthy S., van der Linden W. A., Ward G. E., Martin B. R., and Bogyo M. (2015) Global analysis of palmitoylated proteins in toxoplasma gondii. Cell Host Microbe 18, 501–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li X., Hu X., Wan Y., Xie G., Li X., Chen D., Cheng Z., Yi X., Liang S., and Tan F. (2014) Systematic identification of the lysine succinylation in the protozoan parasite Toxoplasma gondii. J. Proteome Res. 13, 6087–6095 [DOI] [PubMed] [Google Scholar]
- 47. Jeffers V., and Sullivan W. J. (2012) Lysine acetylation is widespread on proteins of diverse function and localization in the protozoan parasite Toxoplasma gondii. Eukaryotic Cell 11, 735–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Braun L., Cannella D., Pinheiro A. M., Kieffer S., Belrhali H., Garin J., and Hakimi M. A. (2009) The small ubiquitin-like modifier (SUMO)-conjugating system of Toxoplasma gondii. Int. J. Parasitol. 39, 81–90 [DOI] [PubMed] [Google Scholar]
- 49. Nagamune K., Moreno S. N., Chini E. N., and Sibley L. D. (2008) Calcium regulation and signaling in apicomplexan parasites. Subcell Biochem. 47, 70–81 [DOI] [PubMed] [Google Scholar]
- 50. Ingram J. R., Knockenhauer K. E., Markus B. M., Mandelbaum J., Ramek A., Shan Y., Shaw D. E., Schwartz T. U., Ploegh H. L., and Lourido S. (2015) Allosteric activation of apicomplexan calcium-dependent protein kinases. Proc. Natl. Acad. Sci. U. S. A. 112, E4975–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang J., Dixon S. E., Ting L. M., Liu T. K., Jeffers V., Croken M. M., Calloway M., Cannella D., Ali Hakimi M., Kim K., and Sullivan W. J. (2014) Lysine Acetyltransferase GCN5b Interacts with AP2 Factors and Is Required for Toxoplasma gondii Proliferation. PLoS Pathogens 10, e1003830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Walker R., Gissot M., Huot L., Alayi T. D., Hot D., Marot G., Schaeffer-Reiss C., Van Dorsselaer A., Kim K., and Tomavo S. (2013) Toxoplasma transcription factor TgAP2XI-5 regulates the expression of genes involved in parasite virulence and host invasion. J. Biol. Chem. 288, 31127–31138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chen C. T., Kelly M., Leon J., Nwagbara B., Ebbert P., Ferguson D.J., Lowery L. A., Morrissette N., and Gubbels M. J. (2015) Compartmentalized Toxoplasma EB1 bundles spindle microtubules to secure accurate chromosome segregation. Mol. Biol. Cell. 26, 4562–4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Morlon-Guyot J., Berry L., Chen C.T., Gubbels M. J., Lebrun M., and Daher W. (2014) The Toxoplasma gondii calcium-dependent protein kinase 7 is involved in early steps of parasite division and is crucial for parasite survival. Cell. Microbiol. 16, 95–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yang J. H., Chiou Y. Y., Fu S. L., Shih I. Y., Weng T. H., Lin W. J., and Lin C. H. (2014) Arginine methylation of hnRNPK negatively modulates apoptosis upon DNA damage through local regulation of phosphorylation. Nucleic Acids Res. 42, 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nakakido M., Deng Z., Suzuki T., Dohmae N., Nakamura Y., and Hamamoto R. (2015) PRMT6 increases cytoplasmic localization of p21CDKN1A in cancer cells through arginine methylation and makes more resistant to cytotoxic agents. Oncotarget 6, 30957–30967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Boulila Y., Tomavo S., and Gissot M. (2014) A RGG motif protein is involved in Toxoplasma gondii stress-mediated response. Mol. Biochem. Parasitol. 196, 1–8 [DOI] [PubMed] [Google Scholar]
- 58. Suvorova E. S., Croken M., Kratzer S., Ting L. M., Conde de Felipe M., Balu B., Markillie M. L., Weiss L. M., Kim K., and White M. W. (2013) Discovery of a splicing regulator required for cell cycle progression. PLoS Gen. 9, e1003305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lott K., Zhu L., Fisk J. C., Tomasello D. L., and Read L. K. (2014) Functional interplay between protein arginine methyltransferases in Trypanosoma brucei. Microbiologyopen 3, 595–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhang X., and Cheng X. (2003) Structure of the predominant protein arginine methyltransferase PRMT1 and analysis of its binding to substrate peptides. Structure 11, 509–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Schwartz D., and Gygi S. P. (2005) An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nature Biotechnol. 23, 1391–1398 [DOI] [PubMed] [Google Scholar]
- 62. Sugi T., Kato K., Kobayashi K., Watanabe S., Kurokawa H., Gong H., Pandey K., Takemae H., and Akashi H. (2010) Use of the kinase inhibitor analog 1NM-PP1 reveals a role for Toxoplasma gondii CDPK1 in the invasion step. Eukaryot Cell 9, 667–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.