Abstract
Legionella pneumophila is an opportunistic bacterial pathogen that causes a severe lung infection termed “Legionnaires' disease.” The pathogen replicates in environmental protozoa as well as in macrophages within a unique membrane-bound compartment, the Legionella-containing-vacuole (LCV). LCV formation requires the bacterial Icm/Dot type IV secretion system, which translocates ca. 300 “effector proteins” into host cells, where they target distinct host factors. The L. pneumophila “pentuple” mutant (Δpentuple) lacks 5 gene clusters (31% of the effector proteins) and replicates in macrophages but not in Dictyostelium discoideum amoeba. To elucidate the host factors defining a replication-permissive compartment, we compare here the proteomes of intact LCVs isolated from D. discoideum or macrophages infected with Δpentuple or the parental strain Lp02. This analysis revealed that the majority of host proteins are shared in D. discoideum or macrophage LCVs containing the mutant or the parental strain, respectively, whereas some proteins preferentially localize to distinct LCVs. The small GTPase Rap1 was identified on D. discoideum LCVs containing strain Lp02 but not the Δpentuple mutant and on macrophage LCVs containing either strain. The localization pattern of active Rap1 on D. discoideum or macrophage LCVs was confirmed by fluorescence microscopy and imaging flow cytometry, and the depletion of Rap1 by RNA interference significantly reduced the intracellular growth of L. pneumophila. Thus, comparative proteomics identified Rap1 as a novel LCV host component implicated in intracellular replication of L. pneumophila.
The causative agent of Legionnaires' disease, Legionella pneumophila, is a ubiquitous environmental bacterium that naturally colonizes complex aquatic biofilms, where the bacteria preferentially parasitize free-living protozoa (1–3). The opportunistic pathogen survives and replicates both in extracellular and intracellular niches. To this end, L. pneumophila adopts a biphasic life style and switches between a transmissive (virulent) and a replicative (nonvirulent) phase (4). The life cycle is regulated by the growth phase (5): bacteria in postexponential phase induce virulence, motility, and stress resistance, whereas exponentially growing bacteria repress these traits and upregulate metabolic pathways (6, 7). Growth phase-specific proteomic analyses of L. pneumophila grown in broth confirmed the up-regulation of virulence and motility traits in postexponential growth phase, but also identified a set of virulence factors (“effector proteins”) exclusively produced in the exponential phase (8, 9).
After uptake by protozoan phagocytes, L. pneumophila establishes a unique intracellular compartment termed “Legionella-containing vacuole” (LCV) 1. The LCV avoids fusion with lysosomes and instead communicates with the host secretory and retrograde trafficking pathways as well as with the endoplasmic reticulum (ER) (10–12). Through an evolutionary conserved mechanism the pathogen also replicates within LCVs in human alveolar macrophages, thus causing the potentially fatal Legionnaires' pneumonia (13). Over 50 Legionella species have been described to date, and more than half of these were found to be linked to human disease, although L. pneumophila accounts for most of the clinical cases (13).
The L. pneumophila Icm/Dot (intracellular multiplication/defective organelle trafficking) T4SS (type IV secretion system) represents the pathogen's major and essential virulence machinery. Using this T4SS, the bacteria translocate more than 300 different so-called “effector proteins” into the host cell, where they subvert cellular processes and govern LCV formation (14, 15). A number of these effectors target components of signal transduction and vesicle trafficking pathways, such as phosphoinositide (PI) lipids (16), Rab family GTPases (17, 18), Ran GTPase (19, 20), or the retromer complex (21).
Proteomic analysis of isolated pathogen compartments has proven instrumental to dissect the intimate and distinct interactions between bacterial pathogens and their host cells (22, 23). Intact pathogen vacuoles harboring L. pneumophila have been isolated from Dictyostelium discoideum amoeba (24, 25), murine RAW 264.7 macrophages (26, 27), bone marrow-derived macrophages (BMM) of A/J mice (28), and human U937 macrophages (29). To purify these LCVs, we established a straight-forward two-step protocol comprising an immuno-magnetic and a density gradient step (30). The immuno-magnetic step exploits the specific LCV membrane localization of the Icm/Dot-secreted effector protein SidC, which binds to the host cell PI lipid PtdIns(4)P (31–33). Using an antibody against SidC and a secondary antibody coupled to magnetic microbeads, LCVs are retained in a magnetic field, washed, eluted and further enriched by standard Histodenz density gradient centrifugation. Thus, the proteome of LCVs purified from D. discoideum (30), RAW 264.7 macrophage-like cells (34) and primary macrophages (28) was determined.
Among the more than 1150 host cell factors identified in the macrophage LCV proteome, 13 small Rab GTPases were detected (34). Their localization to the LCV membrane and their impact on intracellular growth of L. pneumophila was validated by microscopy and RNA interference, respectively. Treatment of BMM with type I (β) or type II (γ) interferon (IFN) substantially modified the LCV composition and induced the production of antimicrobial components such as itaconic acid through IRG1 (immune-responsive gene 1) (28). In contrast, the IFNs apparently neither prevented LCV formation nor promoted endo-lysosomal fusion. Collectively, the LCV proteomics studies provide a rich resource for further hypothesis-driven approaches to elucidate LCV biogenesis. Accordingly, the identification of the small GTPase Ran and its effector RanBP1 on LCVs (30) led to the characterization of the Icm/Dot substrate LegG1 (35, 36) as an RCC1 domain-containing bacterial Ran activator (19).
In this study we investigated the proteome of pathogen vacuoles from D. discoideum amoeba and macrophages infected with either the parental L. pneumophila strain Lp02 or the “pentuple” mutant (37). The pentuple mutant strain (here referred to as “Δpentuple”) is defective for intracellular replication in D. discoideum and other amoeba (Acanthamoeba castellanii, Hartmanella vermiformis), but grows in BMM derived from the A/J mouse strain. The mutant harbors a minimized genome and lacks five gene clusters (Δ2ab, Δ3, Δ4a, Δ6a, Δ7a) comprising 12.7% of the genome (373 genes) and at least 31% of the Icm/Dot-translocated effector proteins (37). The comparative proteomics approach revealed that the presence of active Rap1 on the LCV membrane correlates with intracellular replication of L. pneumophila.
EXPERIMENTAL PROCEDURES
Bacteria, Cells, and Infection
The thymidine auxotroph L. pneumophila strain Lp02 and its mutant derivatives were cultured under aerobic conditions at 37 °C in AYE (ACES yeast extract) broth or grown on CYE (charcoal yeast extract) agar plates containing 0.1 mg ml−1 thymidine. All bacterial strains and plasmids used in this study are listed in Table I. For infections, L. pneumophila Lp02 or Δpentuple harboring plasmid pCR079 (GFP, SidC) or pCR080 (DsRed, SidC) were inoculated at an OD600 of 0.1 in 3 ml AYE medium containing 0.1 mg ml−1 thymidine, 5 μg ml−1 chloramphenicol (Cm) and 1 mm IPTG, and grown to stationary growth phase (ca. 21 h). The infection was synchronized by centrifugation of the bacteria onto host cells (450 × g, 10 min).
Table I. Strains and plasmids used in this study.
| Strain/plasmid | Relevant properties/description a | Reference |
|---|---|---|
| L. pneumophila | ||
| Lp02 | L. pneumophila sg 1 Philadelphia-1, thyA, rpsL, hsdR | (61) |
| YAM10 (Δ2ab) | L. pneumophila Lp02 Δlpg1104-lpg1128, Δlpg1136-lpg1169 | (37) |
| YAS4 (Δ3) | L. pneumophila Lp02 Δlpg1603-lpg1686, tufBC197A | (37) |
| YAM35 (Δ4a) | L. pneumophila Lp02 Δlpg1933-lpg1999 | (37) |
| YAS59 (Δ6a) | L. pneumophila Lp02 Δlpg2369-lpg2465 | (37) |
| YAS64 (Δ7a) | L. pneumophila Lp02 Δlpg2508-lpg2573 | (37) |
| YAM40 (Δpentuple) | L. pneumophila Lp02 Δlpg1104-lpg1128, Δlpg1136-lpg1169, Δlpg1603-lpg1686, tufBC197A, Δlpg1933-lpg1999, Δlpg2369-lpg2465, Δlpg2508-lpg2573 | (37) |
| Lp053 | L. pneumophila Lp02 ΔdotA053 | (62) |
| Plasmids | ||
| pCR079 | pMMB207C-Ptac-RBS-gfp-RBS-MCS-sidC | (21) |
| pCR080 | pMMB207C-Ptac-RBS-dsred-RBS-MCS-sidC | (21) |
| pGFP-Rap1 | GFP-Rap1 (alias RapA) expression construct, pDM317 | (40) |
| pRalGDSRBD-GFP | RalGDSRBD-GFP expression construct, pDM115 | (40) |
| pNT28 | pMMB207C, gfp (constitutive) | (63) |
| pSW001 | pMMB207C, dsRed-express (constitutive) | (64) |
Dictyostelium discoideum Ax3 (lab collection (38)), and strains producing GFP-calnexin (39), GFP-Rap1 (alias RapA) or RalGDSRBD-GFP (40, 41) were grown axenically in HL5 medium at 23 °C with 10 μg ml−1 G418, if necessary. Murine RAW 264.7 macrophages (lab collection, ATCC TIB-71) and A549 lung epithelial carcinoma cells (lab collection) were cultivated in RPMI 1640 medium (Gibco) containing 10% heat-inactivated fetal bovine serum (FBS) and 2 mm glutamine at 37 °C/5% CO2.
Intracellular Replication of L. pneumophila
Exponentially growing D. discoideum amoeba or RAW 264.7 macrophages were suspended in HL5 medium (D. discoideum) or RPMI 1640 medium (macrophages). 2 × 104 cells per well were seeded into 96-well plates and incubated at 23 °C (D. discoideum) or 37 °C/5% CO2 (macrophages) overnight, such that ∼4 × 104 cells per well were present. The HL5 medium was replaced with MB medium (42), and the RPMI 1640 medium was exchanged with fresh medium. The cells were then infected (MOI 1) with L. pneumophila Lp02 or Δpentuple containing plasmid pCR080 (producing DsRed and SidC) or not. The cells were further incubated for 1 h at either 25 °C (D. discoideum) or 37 °C/5% CO2 (macrophages), washed with MB or RPMI 1640 medium, each containing 0.2 mg ml−1 thymidine, and further incubated. At given time points, the cells were lysed with 0.8% saponin, and appropriate dilutions were plated on CYE plates containing thymidine to determine CFU.
Fluorescence Microscopy
To assess the production of SidC in L. pneumophila Lp02 or Δpentuple, bacteria harboring plasmid pCR079 (GFP, SidC) or pCR080 (DsRed, SidC) were grown to stationary growth phase in AYE medium containing 0.1 mg ml−1 thymidine, 5 μg ml−1 Cm and 1 mm IPTG. Bacteria were spun onto poly-l-lysine-coated cover slips, fixed with 4% paraformaldehyde (PFA) for 15 min at room temperature (RT) and mounted on glass slides. The production of GFP or DsRed was analyzed using a Leica TCS SP5 confocal microscope (HCX PLAPO CS, objective 63×/1.4–0.60 oil). The production of SidC was assessed by Western blot using an affinity-purified polyclonal rabbit anti-SidC antiserum (1:1000; (32)).
To visualize SidC during intracellular growth of L. pneumophila, 1 × 105 D. discoideum or RAW 264.7 macrophages were seeded onto poly-l-lysine-coated cover slips in a 24-well plate and incubated overnight. The cells were infected (MOI 50) with L. pneumophila Lp02 or Δpentuple carrying plasmid pCR080 and incubated at 25 °C (D. discoideum) or 37 °C/5% CO2 (macrophages) for 1 h. The infected cells were washed with SorC (Soerensen phosphate buffer containing CaCl2: 2 mm Na2HPO4, 15 mm KH2PO4, 50 μm CaCl2, pH 6.0; D. discoideum) or PBS (macrophages) three times, fixed with 4% PFA and blocked with 3% bovine serum albumin (BSA) in either SorC or PBS for 30 min. The samples were stained for SidC using a polyclonal anti-SidC antibody (1:100; (32)) and a secondary goat anti-rabbit antibody coupled to Cy5 (1:200; Invitrogen: A-10523) and mounted on glass slides using Vectashield mounting medium containing 2 μg ml−1 DAPI.
To investigate the localization of Rap1 on LCVs in D. discoideum, amoeba harboring pGFP-Rap1 (40, 41) were grown to 80% confluency (∼2 × 107 cells per flask) in HL5 medium containing 10 μg ml−1 G418 using three 75 cm2 cell culture flasks per L. pneumophila strain to be tested. Prior to infection, the medium was exchanged to HL5 medium without antibiotics, and the amoeba were infected (MOI 30) with L. pneumophila Lp02 or Δpentuple harboring plasmid pCR080. The infected cells were incubated at 25 °C for 1 h, washed twice with SorC, resuspended in HS homogenization buffer (20 mm HEPES, 250 mm sucrose, 0.5 mm EGTA, pH 7.2) (43) and lysed by nine passages through an ice-cold ball homogenizer with an exclusion size of 8 μm (Isobiotec). The homogenates were spun onto poly-l-lysine-coated cover slips, incubated with blocking solution (3% BSA in SorC, 30 min) and incubated with 30 μl of an anti-SidC antibody (1:100 in blocking solution, 1 h, RT). The cover slips were washed three times with SorC, 30 μl of secondary antibody coupled to Cy5 (1:200 in blocking solution) was added and samples were further incubated for 30 min. The washing steps were repeated, and the samples were mounted on glass slides using Vectashield mounting medium containing 2 μg ml−1 DAPI.
To visualize Rap1 on LCVs in intact RAW 264.7 macrophages, 1 × 105 cells were seeded in 24-well tissue-culture plates on poly-l-lysine coated coverslips and incubated for 24 h. 2 h postinfection (MOI 10) with DsRed-producing L. pneumophila strains (pSW001), cells were fixed with 4% PFA (at 37 °C, 15 min), permeabilized (0.1% Triton-X100, 10 min), incubated with blocking buffer (1% BSA in DPBS) and stained with mouse anti-Rap1 antibody (Abcam: ab175329), and rabbit-anti SidC antibody (32), and samples were mounted on glass slides with ProLong Diamond antifade mounting medium (ThermoFisher Scientific) containing DAPI. Alternatively, macrophages were cultivated for 24 h in up to three T75 flasks per L. pneumophila strain to be tested, and LCVs were isolated 2 h postinfection (see below). The LCV suspensions were centrifuged onto poly-l-lysine-treated sterile cover slips in a 24-well tissue culture plate, washed once with ice-cold DBPS, fixed, and samples were stained with anti-Rap1 and anti SidC antibodies.
Flow Cytometry
The uptake of L. pneumophila by D. discoideum was analyzed by flow cytometry as described (32, 44). In brief, the amoeba were seeded into a 24-well plate (5 × 105 cells/well) and incubated at 25 °C cells for 1–2 h to allow adherence. The cells were then infected (MOI 50) with L. pneumophila Lp02 or Δpentuple harboring plasmid pCR079 (GFP, SidC) and incubated at 25 °C for 30 min. The infected cells were washed three times with SorC to remove extracellular bacteria, detached using a cell scraper and analyzed by flow cytometry using a BD FACS Canto II flow cytometer (BD Biosciences). To quantify uptake, BD Canto II software was used, and an uptake index was defined as the product of the number of cells above the gate threshold and the fluorescence intensity of the cell.
Imaging Flow Cytometry
D. discoideum Ax3 producing GFP-Rap1 or RalGDSRBD-GFP (40) were seeded in 12-well plates (5 × 105 amoeba per well) for 24 h and infected (MOI 5) with L. pneumophila Lp02 or Δpentuple producing DsRed (pSW001). The infected amoeba were detached from the surface at the time points indicated and fixed for 30 min on ice with 4% PFA. Subsequently, 10,000 events were acquired on an imaging flow cytometer (ImageStreamX MkII, Amnis), and after color compensation, analysis was carried out using the IDEAS 6.2 software (Amnis) essentially as described (45). Briefly, GFP-positive in-focus single cells were selected, followed by gating of L. pneumophila-containing cells, using the IDEAS internalization wizard. Only cells containing one bacterium were gated, using the feature [Spot Count_Spot(M04, DsRed, Bright, 8.5, 1, 0)_4], and finally, > 1000 cells per sample were analyzed for colocalization between DsRed and GFP, using the colocalization wizard in IDEAS. This wizard computes the log transformed Pearson's correlation coefficient of the localized bright spots with a radius of 3 pixels or less in two images, providing an imaging flow cytometry (IFC) colocalization score for each cell (mean score for the sample is reported). The IFC data were analyzed by regular two-way ANOVA followed by Bonferroni posthoc tests.
LCV Isolation
Intact LCVs were purified using established protocols for D. discoideum (24) and macrophages (27). Briefly, L. pneumophila Lp02 or Δpentuple harboring plasmid pCR080 (DsRed, SidC) were grown for 21 h at 37 °C in AYE medium supplemented with 0.1 mg ml−1 thymidine and 5 μg ml−1 Cm. D. discoideum amoeba or RAW 264.7 macrophages were grown in 75 cm2 flasks to ∼80% confluency (∼ 2 × 107 cells), infected with L. pneumophila (MOI 50) and incubated at 25 °C (D. discoideum) or 37 °C/5% CO2 (macrophages) for 1 h. All further steps were performed on ice. The infected cells were washed once with SorC (D. discoideum) or PBS (macrophages), resuspended in 3 ml HS homogenization buffer and lysed by nine passages through a ball homogenizer (http://www.isobiotec.com) using an exclusion size of 8 μm. The homogenate was blocked with 3% BSA in SorC or PBS (blocking solution) for 30 min, incubated with an anti-SidC antibody (1:100 in blocking solution; (32)), followed by a secondary anti-rabbit antibody coupled to magnetic beads (20 μl slurry per 0.5 ml homogenate; MACS goat anti-rabbit IgG micro beads; Miltenyi Biotec: 130–048-602). The LCVs were then separated in a magnetic field and further purified by Histodenz density gradient centrifugation as described (27).
Mass Spectrometry and Proteome Analysis
Liquid chromatography (LC) coupled to tandem mass spectrometry (MS/MS) and subsequent proteome analysis were performed as described earlier (34). For each sample, three independent biological replicates were analyzed. To resolve purified LCVs, 1D-SDS-PAGE was used; gel lanes were excised in ten equidistant pieces and subjected to tryptic digestion. Reversed phase column chromatography was then applied to desalt and separate the peptide mixtures. To this end, the samples were loaded onto self-packed columns (Luna 3μ C18(2) 100A, Phenomenex, Aschaffenburg, Germany) in an EASYnLC apparatus (Proxeon, Odense, Denmark) in 0.1% acetic acid/water at a flow rate of 700 nL/min. Peptides were separated in a binary nonlinear gradient of 5–50% acetonitrile in 0.1% acetic acid over 70 min. For MS, an LTQ-Velos Orbitrap mass spectrometer (Thermo Fisher, Bremen, Germany) at a spray voltage of 2.4 kV was used. MS/MS data were recorded for the twenty most intensive precursor ions in the linear ion trap; singly charged ions were not taken into account for MS/MS analysis.
Subsequent database searches were done via Sorcerer with Sequest (version 27, revision 11, SageN, Milptas, CA) without charge state deconvolution and deisotoping, using either a forward-reverse database of combined entries of L. pneumophila Philadelphia-1 (NC002942) and D. discoideum (30739 entries) or a forward-reverse database of combined entries of L. pneumophila Philadelphia-1 and Mus musculus (64993 entries). The L. pneumophila and M. musculus fasta files were downloaded from NCBI on the 19/07/2011 and 06/07/2010, respectively; the D. discoideum fasta files were downloaded from http://dictybase.org on the 19/07/2011. Sequest was used assuming trypsination, a fragment ion mass tolerance of 1.00 Da and a search tolerance of 10 ppm for the overview scans. Furthermore, oxidation of methionine was specified as a variable modification. MS/MS-based peptide and protein identifications were filtered and validated using Scaffold (version 3.5.1, Proteome Software Inc., Portland, OR). Peptides were only accepted when spectra exceeded Xcorr values of 2.2, 3.3 and 3.8 for doubly, triply and quadruply charged peptides with deltaCN values of more than 0.1. Protein identification is based on at least two unique peptides, resulting in a FDR < 0.1%, as calculated by Scaffold. Proteins that contain similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to meet the principle of parsimony. The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (www.ebi.ac.uk/pride/archive/users/profile) (46) with the data set identifier PXD004797. Detailed information on all peptide sequences and protein identification is listed in supplemental Table S9–S16.
RNA Interference
RNA interference experiments were performed using A549 lung epithelial cells. As a first step, siRNA stocks (10 μm) were diluted 1:15 in RNase-free water, and 3 μl of diluted siRNA was added to designated wells in a 96-well plate. Scrambled siRNA or only transfection reagent (mock) were used as negative controls. In a next step, 24.25 μl RPMI 1640 medium without FBS and glutamine and 0.75 μl HiPerFect transfection reagent (Qiagen) were added to each well, mixed and incubated for 10 min at RT. A549 cells were diluted to a concentration of 1.14 × 105 cells ml−1 in RPMI 1640 medium containing 10% FBS/1% glutamine, 175 μl of this dilution (2 × 104 cells) was added to the wells containing the siRNA-HiPerFect transfection complex, and the 96-well plate was incubated for 48 h. The cells were then infected (MOI 50) with L. pneumophila Lp02 harboring plasmid pNT28 (constitutive GFP production), diluted in RPMI 1640 medium and incubated for 1 h at 37 °C/5% CO2. The infected cells were washed three times with RPMI 1640 medium containing 10% FBS/1% glutamine and further incubated for 24 h. Intracellular growth of L. pneumophila was determined by measuring GFP fluorescence in a plate spectrophotometer (FluoStar Optima, BMG Labtech). The depletion efficiency was assessed by Western blot with A549 epithelial cells treated with 4 different siRNAs for 48 h. Rap1 was visualized with a mouse monoclonal anti-Rap1 antibody (1:500; Abcam: ab175329), rabbit polyclonal anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH; 1:1000; Cell Signaling: 2118) served as a loading control, and Qiagen AllStars oligonucleotides were used as a negative control (“scrambled”). The siRNA oligonucleotides used in this study are listed in the supplemental Table S1.
RESULTS
Production of SidC by L. pneumophila Lp02 and ΔPentuple Cluster Deletion Mutant
The aim of this study was to correlate the ability of L. pneumophila to replicate intracellularly in evolutionarily distant phagocytes with the corresponding LCV proteomes. To this end, we used the L. pneumophila pentuple mutant strain (Δpentuple), which lacks 5 gene clusters encoding at least 31% of the Icm/Dot-translocated effector proteins and is defective for intracellular replication in D. discoideum, but grows in BMM derived from the A/J mouse strain (37).
We previously established protocols for the purification and proteomics analysis of intact LCVs from D. discoideum and RAW 264.7 macrophages (27). Enrichment of LCVs from these phagocytes by immuno-affinity separation requires the presence of the bacterial effector protein SidC, which is missing from the Δpentuple mutant (supplemental Table S2). In order to restore the production of SidC in the Δpentuple mutant, we transformed the strain with the plasmids pCR079 or pCR080, producing Ptac-controlled SidC, whereas GFP (pCR079) or DsRed (pCR080) are constitutively produced. The analysis of SidC production by Western blot revealed that Δpentuple harboring pCR080 produced the effector protein in similar amounts as Lp02, regardless of whether the latter contains the plasmid or not (Fig. 1A). Comparable results were obtained for strain Lp02 and the Δpentuple mutant transformed with pCR079 (data not shown). Furthermore, upon growth to stationary phase, similar numbers of Lp02 and Δpentuple bacteria harboring pCR079 or pCR080 produced the fluorescent proteins GFP and DsRed (Fig. 1B). The fluorescence signal for DsRed was even more robust than that for GFP.
Fig. 1.
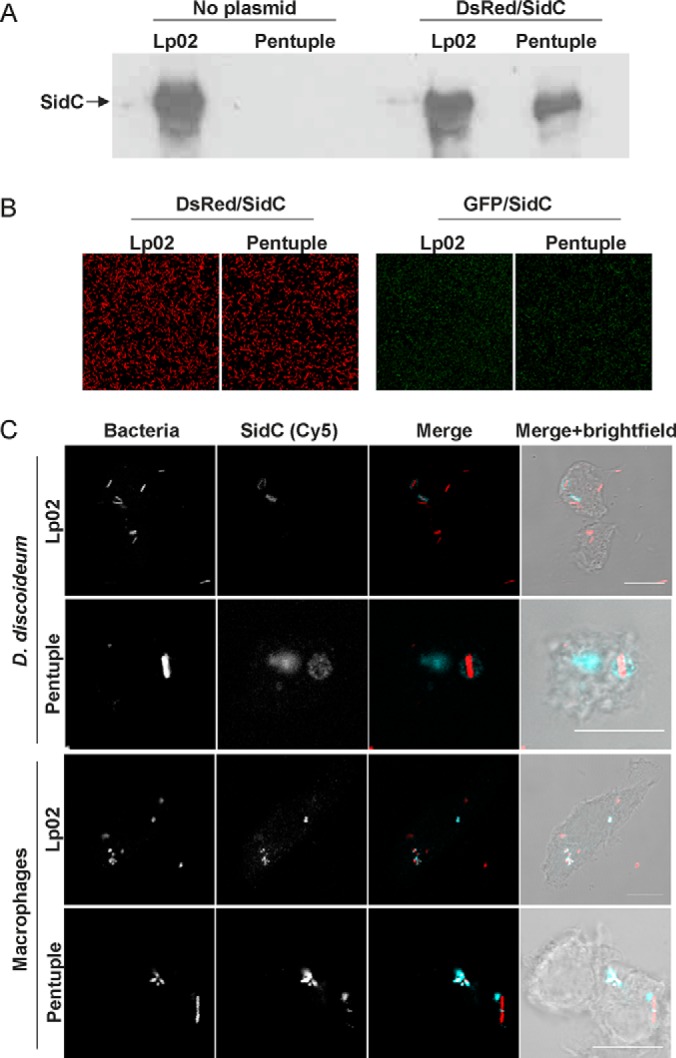
Production of SidC by L. pneumophila Lp02 and Δpentuple deletion mutant. A, The production of the Icm/Dot substrate SidC (106 kDa) was assessed by Western blot in lysates of L. pneumophila Lp02 and the Δpentuple mutant transformed with plasmid pCR080 (DsRed, SidC) or not. B, Fluorescence microscope images of strain Lp02 and Δpentuple harboring plasmid pCR080 (DsRed, SidC; left panels) or pCR079 (GFP, SidC; right panels). C, D. discoideum or RAW 264.7 macrophages were infected (MOI 50, 1 h) with L. pneumophila Lp02 or Δpentuple harboring plasmid pCR080 (DsRed, SidC), fixed and stained for SidC using an anti-SidC antibody and a secondary antibody coupled to Cy5. Images are representatives of three independent experiments. Scale bar, 10 μm.
Next, D. discoideum or RAW 264.7 macrophages infected with strain Lp02 or the Δpentuple mutant harboring pCR080 were stained for SidC and analyzed by fluorescence microscopy (Fig. 1C). These experiments revealed that ectopically produced SidC is translocated and exclusively localizes to the cytoplasm-directed surface of the LCV, as observed for endogenous SidC. In summary, the ectopic expression of sidC in L. pneumophila Lp02 or Δpentuple leads to the production of similar amounts of the effector, which is translocated and decorates the cytoplasmic side of LCVs. These studies establish Lp02 and Δpentuple harboring pCR080 as suitable tools to isolate LCVs using the immuno-magnetic purification protocols.
Intracellular Growth and Uptake of SidC-producing L. pneumophila Lp02 and ΔPentuple
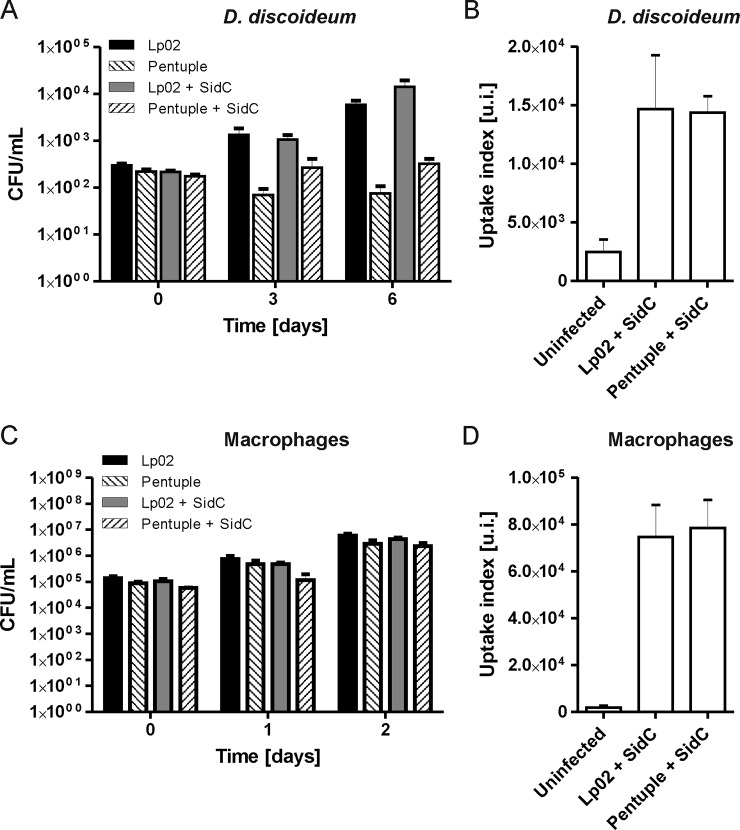
Upon infection of D. discoideum, the parental Lp02 strain produced an increase in CFU of approximately two orders of magnitude in the course of 6 days (Fig. 2A). Intracellular growth was robust and indistinguishable, regardless of whether the strain harbored plasmid pCR080 (DsRed, SidC) or not. In contrast, the Δpentuple mutant did not grow intracellularly in presence or absence of the plasmid, but persisted in the amoeba. Flow cytometry experiments revealed that the uptake of strain Lp02 or Δpentuple by D. discoideum was identical (Fig. 2B). This result indicates that the growth defect of the Δpentuple mutant in amoeba is not due a reduced uptake, and the Icm/Dot-dependent up-regulation of uptake previously described (47) is not caused by the effector proteins lacking in the cluster mutant.
Fig. 2.
Intracellular growth and uptake of SidC-producing L. pneumophila Lp02 and Δpentuple. A, D. discoideum or (C) RAW 264.7 macrophages were infected (MOI 1) with L. pneumophila Lp02 or Δpentuple harboring pCR080 (DsRed, SidC) or not. At given time points the infected cells were lysed, and CFU were determined. Mean and S.D. of triplicates are shown; data are representatives of three independent experiments. B, D. discoideum or (D) RAW 264.7 macrophages were infected (MOI 50, 30 min) with L. pneumophila Lp02 or Δpentuple harboring plasmid pCR079 (GFP, SidC). The infected cells were analyzed by FACS to determine the uptake index (u.i.; product of number of infected cells above the gate threshold and fluorescence intensity). Data represent mean and standard deviation of three independent experiments.
Upon infection of RAW 264.7 macrophages, the strain Lp02 produced an increase in CFU of approximately two orders of magnitude in the course of 2 days (Fig. 2C). The robust intracellular growth was the same in presence or absence of plasmid pCR080 (DsRed, SidC). The Δpentuple mutant also replicated robustly in the macrophages, but reached slightly lower CFU compared with the parental strain, regardless of whether the mutant contained plasmid pCR080 or not. Similar to D. discoideum, the uptake of strain Lp02 and Δpentuple by RAW 264.7 macrophages was identical (Fig. 2D). Overall, the macrophages phagocytosed L. pneumophila considerably more efficiently than the amoeba (Fig. 2B, 2D).
Taken together, the L. pneumophila Δpentuple mutant shows a strong growth defect in D. discoideum amoeba, but replicates like the parental strain in RAW 264.7 macrophages, and thus, the phenotypes of the Δpentuple mutant are similar to the ones previously published (37). Because the ectopic production of SidC does not significantly affect intracellular replication and uptake of Lp02 and Δpentuple, the strains harboring pCR080 are well suited to assess the corresponding LCV proteomes.
Purification and Proteomics of Intact D. discoideum or Macrophage LCVs Harboring Lp02 or ΔPentuple
To isolate intact LCVs harboring L. pneumophila Lp02 or Δpentuple producing SidC, the pathogen vacuoles were enriched by an immuno-affinity step (Fig. 3A), followed by further purification by density centrifugation (Fig. 3B). A high number of intact LCVs were obtained from either D. discoideum or RAW 264.7 macrophages infected with strain Lp02 or the Δpentuple mutant. The yield of LCVs from infected amoeba was higher than the yield from macrophages, as observed previously. Homogenate and pellet (before immuno-magnetic separation), as well as flow-through and eluate (after MACS column separation), were analyzed by immunofluorescence microscopy (Fig. 3A). Enrichment of LCVs was observed for both bacterial strains in both host cells in the pellet after homogenization of the infected cells and in the eluate after immuno-magnetic separation. To purify intact LCVs further, the eluate was then subjected to Histodenz density gradient centrifugation, resulting in a preferential accumulation of intact LCVs in fraction 4 (Fig. 3B).
Fig. 3.
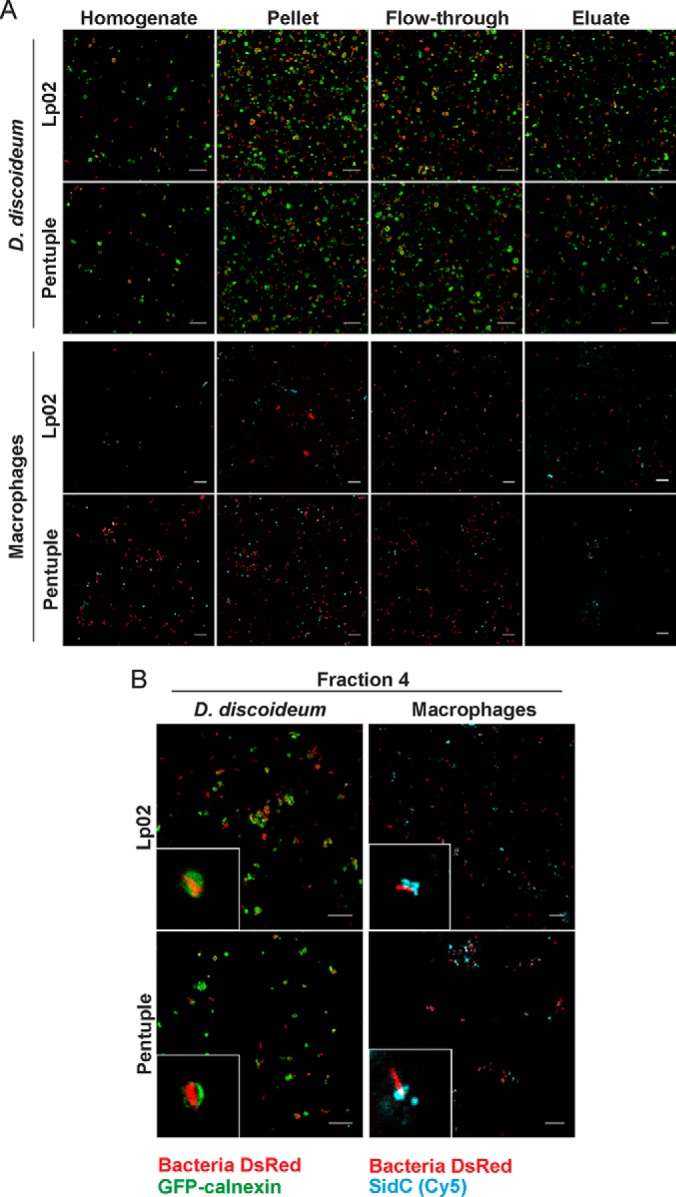
Purification of intact D. discoideum or macrophage LCVs harboring Lp02 or Δpentuple. A, Isolation of LCVs by immuno-affinity separation from D. discoideum producing GFP-calnexin (upper panels) and RAW 264.7 macrophages (lower panels) infected with L. pneumophila Lp02 or Δpentuple harboring pCR080 (DsRed, SidC). Infected cells were lysed using a ball homogenizer (homogenate), followed by centrifugation (pellet) and immuno-magnetic separation using MACS columns, yielding flow-through and eluate. B, LCVs in the eluate were further enriched by Histodenz density gradient centrifugation, yielding the highest amount of intact LCVs in fraction 4. Samples derived from macrophages were stained for SidC using anti-SidC and Cy5-conjugated antibodies (blue). Data representative for three independent experiments are shown. Scale bars, 10 μm.
As fraction 4 after density centrifugation contained the highest number of intact LCVs and also only a neglectable amount of contamination from other host organelles and cell debris, this fraction was analyzed by proteomics. In order to determine soluble and membrane proteins, samples were initially resolved by 1D-SDS-PAGE followed by LC-MS/MS.
Proteome of D. discoideum LCVs Harboring Lp02 or ΔPentuple
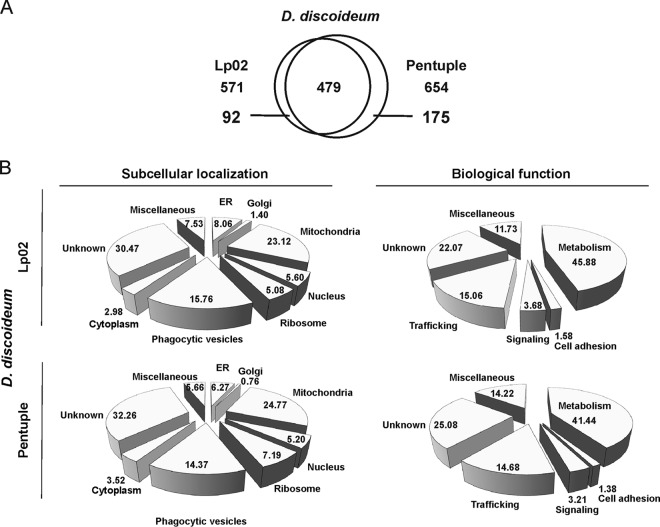
The D. discoideum LCV proteome revealed a total of 571 host proteins from pathogen vacuoles harboring strain Lp02 and 654 proteins from pathogen vacuoles harboring the Δpentuple mutant (Fig. 4A). Comparison of the proteomes indicated that 479 host proteins were commonly present in pathogen vacuoles harboring either Lp02 or Δpentuple, whereas 92 were only present in the Lp02 LCV proteome and 175 only in the Δpentuple LCV proteome.
Fig. 4.
Proteomics of D. discoideum LCVs harboring Lp02 or Δpentuple. A, Venn diagram highlighting the number of host cell proteins identified in the proteome of purified D. discoideum LCVs harboring Lp02 or Δpentuple. B, Pie charts illustrating the subcellular localization and biological function of host proteins identified in the LCV proteomes of D. discoideum amoeba. Data shown is in % indicating the respective theoretical subcellular localization of the different proteins identified.
To define the subcellular localization and biological function of host cell proteins derived from the D. discoideum LCV proteome, we sought to arrange categories and analyze the LC-MS/MS data with the UniProt database (http://www.uniprot.org). Accordingly, host proteins found in the proteome of Lp02 LCVs were classified into the following categories regarding their subcellular location: mitochondria (23.1%), phagocytic vesicles (15.8%), ER (8.1%), nucleus (5.6%), ribosome (5.1%), cytoplasm (3.0%) and Golgi (1.4%) (Fig. 4B). These proteins were then further classified regarding their putative biological functions. Almost half of the identified host cell proteins are implicated in metabolism (45.9%) and a significant number of proteins are involved in trafficking (15.1%). The other identified proteins fall into the categories signaling (3.67%), cell adhesion (1.6%), miscellaneous (not matching any of the other categories; 11.7%), or unknown functions (22.1%).
The localization of host proteins identified in the Δpentuple LCV proteome was comparable to that found in the Lp02 LCV proteome: mitochondria (24.8%), phagocytic vesicles (14.4%), ribosome (7.2%), ER (6.3%), nucleus (5.2%), cytoplasm (3.5%), and Golgi (0.8%) (Fig. 4B). Regarding their biological function, host proteins of the Δpentuple LCV proteome are predicted to be primarily involved in metabolism (41.4%), trafficking (14.7%), signaling (3.2%), cell adhesion (1.4%), miscellaneous (14.2%), or unknown functions (25.1%).
Four hundred seventy-nine host proteins were found in LCVs harboring either the Lp02 strain or the Δpentuple mutant (Fig. 4A). Most of these 479 shared proteins are predicted to be involved in metabolism and transport. For example, several ATPases were identified, including the vacuolar ATPase (VatA, -B, -C, -E, -H, -M), the plasma membrane ATPase PatB and the calcium-transporting ATPase PatA. Furthermore, several host proteins implicated in amino acid metabolism, lipid metabolism or central metabolic pathways were found in the D. discoideum LCV proteome. The second largest group of proteins commonly identified in the D. discoideum Lp02 and Δpentuple LCV proteomes is predicted to be involved in membrane trafficking. Overall, 11 small GTPases, including Rab2A, Rab2B, Rab5A, Rab5B, Rab6, Rab7A, Rab8A, Rab11A, Rab14, and Rab32A, were identified. In addition to these GTPases, NSF attachment proteins, Vamp7B and several host cytoskeletal factors including actin, myosin and comitin were detected. Furthermore, a considerable number of host proteins that are involved in signaling were found, including the small GTPases RasG and RanA, as well as the apoptosis inducing factor Aif. The complete set of shared proteins in the D. discoideum Lp02 and Δpentuple LCV proteomes is listed in supplemental Table S3.
The proteome of LCVs harboring Lp02 comprised 92 host proteins that were exclusively found for this parental strain, and 31 of these are uncharacterized (Table II). Several proteins are implicated in cellular metabolism. Host factors involved in vesicle trafficking include the small GTPase Rab11C, the vacuolar protein Vps45, the interaptin AbpD, the TRAP protein Ssr1 and the vesicle-fusing ATPase NsfA. Other proteins are assigned to the signaling category, including the PI phosphatase Sac1, the Ser/Thr kinase KinX, annexin, polyubiquitin F and the small Ras GTPase Rap1. Finally, a number of cytoskeletal and motor proteins (actin, cortexillin, talin, myosin) as well as the ABC transporter AbcG10 or Mfsd1 were identified specifically in the Lp02 LCV proteome (Table II).
Table II. Selected host proteins identified by MS localizing to purified D. discoideum LCVs harbouring either Lp02 or Δpentuple. Abbreviations: no acronym (N.A.), cytoplasm (CP), plasma membrane (PM), inner membrane (IM), early endosome (EE), late endosome (LE), lysosome (LS), recycling endosome (RE), autophagosome (AP), endoplasmic reticulum (ER), Golgi apparatus (GA), ER-Golgi intermediate compartment (ERGIC), Mitochondria (MC), nucleus (NC). Detection frequency was determined in three independent biological samples, taken 1 h post infection. Complete set of D. discoideum LCV proteins identified by LC-MS/MS is listed in the supplementary Tables S3, S4, and S5.
| Gene | Classification |
Accession number (detection frequency) |
Ref. | ||
|---|---|---|---|---|---|
| Product | Localization | Lp02 | Δpentuple | ||
| Signalling | |||||
| CapB | cAMP-binding protein 2 | DDB_G0277501 (1) | (65) | ||
| Dcd2A | Neutral ceramidase A | DDB_G0293538 (1) | (65) | ||
| FttB | 14-3-3 protein | CP | DDB_G0269138 (3) | (66) | |
| KinX | Probable Ser/Thr-protein kinase KinX | DDB_G0283391 (1) | (65) | ||
| LkhA | Leukotriene hydrolase | CP | DDB_G0269148 (1) | (65, 67) | |
| KsrA-1 | 3-ketodihydro-sphingosine reductase | ER | DDB_G0274015 (2) | (65) | |
| Mdn1 | Midasin | NC | DDB_G0295765 (1) | (65) | |
| N.A. | Regulator complex LAMTOR3 | DDB_G0274833 (1) | (65) | ||
| N.A. | RCC domain protein | DDB_G0292586 (1) | (65) | ||
| Nca2 | Nuclear control of ATPase protein 2 | DDB_G0267648 (1) | (65) | ||
| Nop58 | Nucleolar protein | NC | DDB_G0268098 (1) | (65) | |
| Nup98 | Nuclear pore complex protein Nup98-Nup96 | NC | DDB_G0291390 (1) | (65) | |
| Nup107 | Nucleoporin 107 | NC | DDB_G0285579 (1) | (65) | |
| Nup155 | Nucleoporin 155 | NC | DDB_G0291163 (1) | (65) | |
| NxnA | Annexin A7 | DDB_G0269160 (2) | (65) | ||
| Rap1 | Ras GTPase | PM | DDB_G0291237 (1) | (68, 69) | |
| Rpb3 | RNA polymerase | NC | DDB_G0292244 (3) | (65) | |
| Sac1 | PI phosphatase Sac1 | ER, PM | DDB_G0271630 (2) | (70) | |
| UbqJ | Polyubiquitin J | CP, NC | DDB_G0269458 (3) | (65) | |
| UbqF | Polyubiquitin F | CP, NC | DDB_G0289449 (3) | (65) | |
| Vesicle trafficking | |||||
| AbpD | Interaptin | DDB_G0287291 (1) | (71) | ||
| DlpA | Dynamin-like protein A | CP | DDB_G0268592 (1) | (65) | |
| DymB | Dynamin B GTPase | CP | DDB_G0277851 (1) | (72) | |
| NsfA | Vesicle-fusing ATPase | CP | DDB_G0276153 (1) | (73) | |
| Rab1A | Ras-related protein Rab1A | ER, GA | DDB_G0283757 (3) | (65) | |
| Rab11C | Rab11C | GA, RE | DDB_G0277101 (1) | (65) | |
| Rab21 | Rab21 | EE | DDB_G0286553 (1) | (74, 75) | |
| Rtnlc | Reticulon family protein | ER | DDB_G0293088 (3) | (76, 77) | |
| Ssr1 | TRAP protein | ER, PM | DDB_G0283497 (3) | (78) | |
| Syn5 | Syntaxin 5 t-SNARE | ER, GA | DDB_G0277565 (1) | (65) | |
| Vamp7A | Vamp7A v-SNARE | PM, EE | DDB_G0284951 (1) | (65) | |
| Vps45 | Vacuolar protein | GA, PM | DDB_G0290213 (1) | (65) | |
| Metabolism | |||||
| AlrA | Aldose reductase A | CP | DDB_G0293850 (1) | (65) | |
| Coq4 | Ubiquinone biosynthesis protein Coq4 | DDB_G0292620 (1) | (65) | ||
| Coq9 | Ubiquinone biosynthesis protein Coq9 | DDB_G0274457 (2) | (65) | ||
| Cox11 | Cytochrome c oxidase assembly protein Cox11 | MC | DDB_G0289353 (1) | (65) | |
| CxgE | Cytochrome c oxidase subunit 7e | DDB_G0277837 (1) | (65) | ||
| EnoA | Enolase A | CP | DDB_G0283137 (1) | (65) | |
| FdfT | Squalene synthase | ER, PM | DDB_G0292072 (1) | (65) | |
| Gpi | Glucose 6-P isomerase | CP | DDB_G0283673 (1) | (65) | |
| MasA | Malate synthase | CP | DDB_G0275887 (1) | (65) | |
| Surf1 | SURF1-like protein | DDB_G0274001 (3) | (79) | ||
| Transport | |||||
| AbcG10 | ABC transporter G family | PM | DDB_G0292986 (1) | (80) | |
| McfS | Mitochondrial substrate carrier | CP, MC | DDB_G0290913 (2) | (81) | |
| McsP | Mitochondrial substrate carrier | CP, MC | DDB_G0292034 (1) | (81) | |
| Mfsd1 | Major facilitator superfamily protein 1 | DDB_G0289201 (1) | (65) | ||
| Timm9 | Mitochondrial import IM translocase Tim9 | MC | DDB_G0272931 (1) | (65) | |
| Timm16 | Mitochondrial import IM translocase Tim16 | MC | DDB_G0282195 (1) | (65) | |
| Timm17 | Mitochondrial import IM translocase Tim17 | MC | DDB_G0287627 (2) | (65) | |
| Cytoskeleton/motor proteins | |||||
| AbpC | Gelation factor | CP, PM | DDB_G0269100 (1) | (65) | |
| Act1 | Major actin | CP | DDB_G0274599 (3) | (65) | |
| Act2 | Major actin | CP | DDB_G0274133 (3) | (65) | |
| ArcC | Actin-related protein | CP | DDB_G0283755 (1) | (65) | |
| ArcD | Actin-related protein 2/3 compl. subunit 4 | CP | DDB_G0269102 (2) | (65) | |
| CtxA | Cortexillin | CP, PM | DDB_G0289483 (1) | (82) | |
| LimE | LIM domain protein | CP, PM | DDB_G0279415 (2) | (83) | |
| MlcR | Myosin regulatory light chain | CP | DDB_G0276077 (1) | (65) | |
| TalB | Talin B | CP | DDB_G0287505 (1) | (65) | |
| Miscellaneous | |||||
| Cnrl | SET domain-containing protein | DDB_G0269768 (2) | (65) | ||
| DscA | Discoidin 1 subunit A | CP | DDB_G0273919 (3) | (65) | |
| Kil1 | Sulfotransferase | PM | DDB_G0267630 (1) | (84) | |
| SodA | Superoxide dismutase | MC | DDB_G0267420 (1) | (85) | |
Among the 175 host proteins that were exclusively detected in the Δpentuple LCV proteome, about 70 are uncharacterized. In contrast to the Lp02 LCV, the Δpentuple LCV is associated with RNA polymerase and more ribosomal components (Table II). Among factors involved in cellular metabolism, many components of the respiratory chain (cytochrome C oxidase, Surf1, ubiquinone biosynthesis) were only found in the Δpentuple LCV proteome. Vesicle trafficking pathway factors included dynamin and dynamin-like GTPases, t- (Syn5) and v-SNAREs (Vamp7A) and members of the reticulon family, as well as the small GTPases Rab1 and Rab21. Moreover, signaling pathway components, such as protein 14-3-3, midasin, as well as an RCC domain protein and a cAMP-binding protein were detected. Interestingly, the small GTPase Rap1 was not found in the proteome of Δpentuple LCV proteome but only in the Lp02 LCV proteome (Table II). The complete sets of D. discoideum LCV proteins exclusively found in pathogen vacuoles harboring strain Lp02 or the Δpentuple mutant are listed in supplemental Table S4 and S5.
Proteome of Macrophage LCVs Harboring Lp02 or ΔPentuple
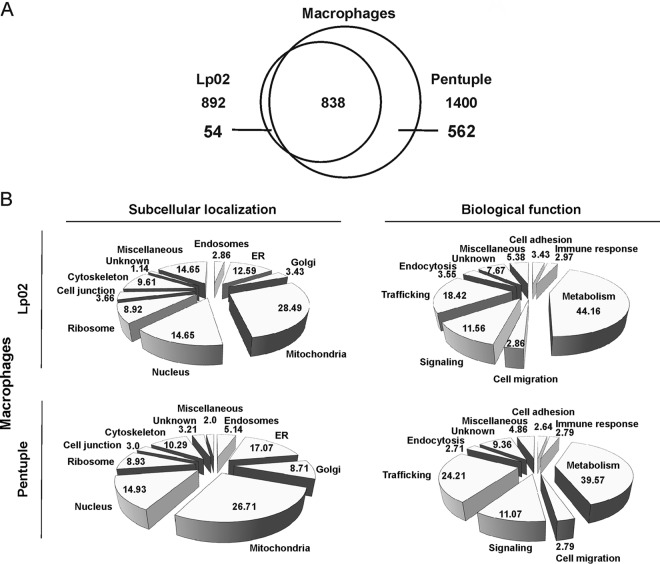
The RAW 264.7 LCV proteome revealed a total of 892 host proteins from pathogen vacuoles harboring strain Lp02 and 1400 host proteins from pathogen vacuoles harboring the Δpentuple mutant (Fig. 5A). Comparison of the proteomes indicated a shared number of 838 host proteins present in pathogen vacuoles harboring either Lp02 or Δpentuple. 54 host proteins accumulated specifically on Lp02-containing LCVs and as many as 562 host proteins were only present on Δpentuple LCVs.
Fig. 5.
Proteomics of RAW 264. 7 macrophage LCVs harboring Lp02 or Δpentuple. A, Venn diagram highlighting the number of host cell proteins identified in the proteome of RAW 264.7 macrophage LCVs harboring Lp02 or Δpentuple. B, Pie charts illustrating the subcellular localization and biological function of host proteins identified in the LCV proteomes of RAW 264.7 macrophages. Data shown is in % indicating the respective theoretical subcellular localization of the different proteins identified.
Data obtained for RAW 264.7 macrophage LCVs were analyzed in the same way as data from D. discoideum. The host proteins were assigned to several categories regarding subcellular localization and biological function (Fig. 5B). Nine categories were defined for the Lp02 LCV proteome regarding cellular localization of identified host proteins: mitochondria (28.5%), nucleus (14.7%) and ER (12.6%). Other proteins are predicted to localize to the cytoskeleton (9.6%), ribosome (8.9%), cell junction (3.7%), Golgi (3.4%), or endosomes (2.9%). 14.7% of all proteins did not fit these categories and were defined as miscellaneous, whereas only 1.1% of the proteins are unknown. The biological function of the identified proteins is predicted to be mainly metabolism (44.1%), trafficking (18.4%), and signaling (11.6%). To a lesser extent, the proteins fell into the categories endocytosis (3.6%), cell adhesion (3.4%), immune response (3.0%), cell migration (2.9%), miscellaneous (5.4%), or unknown functions (7.7%).
Again, the subcellular localization of host proteins found in the Δpentuple LCV proteome was comparable to that of Lp02 LCVs: mitochondria (26.7%), ER (17.1%), nucleus (14.9%), cytoskeleton (10.3%), ribosome (8.9%), Golgi (8.7%), endosome (5.1%), and cell junction (3.0%). Compared with the Lp02 LCV proteome, the Δpentuple LCV proteome comprises of around 560 additional host proteins, most of them being involved in metabolism (39.6%), trafficking (24.2%), and signaling (11.1%). Also, the predicted function of other proteins from the Δpentuple LCV proteome was comparable to the Lp02 LCV proteome: immune response (2.8%), cell migration (2.8%), endocytosis (2.7%), and cell adhesion (2.6%) (Fig. 5B).
Overall, 838 host proteins were commonly identified in the Lp02 and Δpentuple LCV proteomes (Fig. 5A). Similarly to LCVs derived from D. discoideum, the largest group of these shared LCV proteins is implicated in metabolism, including members of fatty acid and lipid metabolism, amino acid metabolism and central metabolic pathways. Moreover, several components of the respiratory chain such as the cytochrome c oxidase Cox, the succinate dehydrogenase and the cytochrome bc1 complex were found. Immune system constituents of the common macrophage LCV proteomes include cell adhesion molecules (CD14, CD44, CD166), β2-microglobulin of the MHC I, as well as components of the complement activating cascade, the Toll signaling pathway and prostaglandin biosynthesis.
Factors influencing vesicle trafficking and membrane dynamics were found in large numbers in the LCV proteome shared by strain Lp02 and the Δpentuple mutant. These include macrophage scavenger receptors, t- and v-SNAREs (syntaxins, Vamps), soluble NSF proteins, as well as synaptogyrin and synaptojanin. Furthermore, we detected a variety of small GTPases such as Arl8, Rab1A, Rab1B, Rab2A, Rab5A, Rab5B, Rab5C, Rab7A, Rab8B, Rab10, Rab11A, Rab14, Rab18, Rab21, and Rab31. Also, many constituents of signaling pathways were identified, including apoptosis mediators (Aifm1, Bcl2l13), the small Ras GTPases Rap1 and KRas, the Rho GTPase Rhot1 and the Rho mediator Arhgdia, the Ran GTPase interactors RanBP2 and RanGAP1, as well as a considerable number of tyrosine and serine/threonine phosphatases. Moreover, a large number of channel and transport proteins were discovered such as the lipid transporter Scp2, vesicle transport proteins (Sec22, Vti1b) and 11 solute carrier (Slc) family transport proteins. The complete set of shared proteins in the macrophage LCV proteomes is listed in supplemental Table S6.
The Lp02 LCV proteome contained 54 specific proteins (Fig. 5A). Among those were the IgG Fc-receptor Fcgr1, T-cell transcription factor Nfat5 and ER aminopeptidase, all associated with the immune system, as well as dynamin 2 and the GTPases Rab6 implicated in membrane dynamics (Table III). Furthermore, signaling pathway components were identified, including the GTPase Rap2C, the Rho guanine exchange factor ArhGEF 2, phosphatidylinositol 4-kinase II (PI4K2α), the Tyr protein kinase Hck, protein phosphatase Ptc7, ubiquitin hydrolase Usp34, and desmoglein (Table III).
Table III. Selected host proteins identified by MS localizing to purified LCVs from RAW 264.7 macrophages harbouring either Lp02 or Δpentuple. Abbreviations: no acronym (N.A.), cytoplasm (CP), plasma membrane (PM), early endosome (EE), late endosome (LE), lysosome (LS), recycling endosome (RE), autophagosome (AP), endoplasmic reticulum (ER), Golgi apparatus (GA), ER-Golgi intermediate compartment (ERGIC), Mitochondria (MC), nucleus (NC). Detection frequency was determined in three independent biological samples, taken 1 h post infection. Complete set of RAW 264.7 macrophage LCV proteins identified by LC-MS/MS is listed in the supplementary Tables S6, S7, and S8.
| Gene | Classification |
Accession number (detection frequency) |
Ref. | ||
|---|---|---|---|---|---|
| Product | Localization | Lp02 | Δpentuple | ||
| Immune response | |||||
| C5ar1 | C5a anaphylatoxin receptor | PM | NP_031603 (1) | (86) | |
| Erap1 | ER aminopeptidase | ER | NP_109636 (3) | (86) | |
| Faf2 | FAS-associated factor 2 | PM | NP_848484 (1) | (86) | |
| Fcgr1 | High affinity IgG Fc-receptor | PM | NP_034316 (2) | (86) | |
| Irgm1 | Immunity-related GTPase family M protein 1 | NP_032352 (1) | (86) | ||
| Itgav | Integrin alpha-V | NP_032428 (1) | (86) | ||
| Lilrb4 | Leukocyte Ig-like receptor subfamily B member 4 | NP_038560 (1) | (86) | ||
| Pbxip1 | Pre-B-cell leukemia transcription factor-interacting protein 1 | NP_666243 (2) | (86) | ||
| Nfat5 | Nuclear factor of activated T-cells 5 | CP, NC | NP_061293 (1) | (86) | |
| Tlr7 | Toll-like receptor 7 | NP_573474 (1) | (86) | ||
| Tnf | Tumor necrosis factor | NP_038721 (2) | (86) | ||
| Signalling | |||||
| Anxa1 | Annexin-A1 | CP, NC | NP_034860 (2) | (86) | |
| Arf4 | ADP-ribosylation factor | ER, PM | NP_031505 (2) | (86) | |
| Arf5 | ADP-ribosylation factor | ER, PM | NP_031506 (1) | (86) | |
| Arf6 | ADP-ribosylation factor | ER, PM | NP_031507 (2) | (86) | |
| Arfrp1 | Arf-related protein 1 isoform 1 | NP_001159464 (1) | (86) | ||
| Arhgdia | Rho GDI 1 | PM | NP_598557 (1) | (87) | |
| Arhgdib | Rho GDI 2 | PM | NP_031512 (1) | (86) | |
| Arhgef2 | Rho GEF 2 | PM | NP_032513 (1) | (88) | |
| Arl2 | ADP-ribosylation factor-like protein 2 | ER, PM | NP_062696 (2) | (86) | |
| Arl6ip1 | Arl6-interacting protein 1 | ER, PM | NP_062292 (1) | (86) | |
| Arl6ip5 | PRA1 family protein 5 | NP_075368 (1) | (86) | ||
| Asah1 | Acid ceramidase | ER, GA | NP_062708 (3) | (89) | |
| Aurkb | Ser/Thr protein kinase 12 | NP_035626 (2) | (86) | ||
| Bak1 | Bcl2 homolog. antagonist | MC | NP_031549 (2) | (90) | |
| Bax | Apoptosis regulator Bax | ER, CP | NP_031553 (3) | (91) | |
| Bri3bp | Bri3-binding protein | ER | NP_084028 (3) | (92) | |
| Cdc42se2 | CDC42 small effector protein 2-like | NP_848741 (1) | (86) | ||
| Dsg1a | Desmoglein | CP, PM | NP_034209 (1) | (86) | |
| Esyt1 | Extended synaptotagmin 1 | NP_035973 (2) | (86) | ||
| Gbf1 | Golgi-specific brefeldin A-resistance GEF 1 | NP_849261 (1) | (86) | ||
| Gnai3 | G protein subunit α-13 | PM, PS | NP_034433 (3) | (93) | |
| Gng12 | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit γ-12 | NP_001171031 (2) | (86) | ||
| Hck | Tyr protein kinase Hck | NP_034537 (1) | (86) | ||
| Icam1 | Intercellular adhesion molecule 1 | PM | NP_034623 (1) | (86) | |
| Itpr2 | Inositol 1,4,5-triphosphate receptor 2 | ER | NP_064307 (1) | (86) | |
| Itpr3 | Inositol 1,4,5-triphosphate receptor 3 | ER | NP_542120 (2) | (94) | |
| Kras | KRas GTPase | NP_067259 (1) | (86) | ||
| Lamtor3 | Mitogen-activated protein | PM | NP_064304 (2) | (86) | |
| Lemd2 | LEM domain-containing protein 2 | NC | NP_666187 (1) | (86) | |
| Lemd3 | LEM domain-containing protein 3 | NC | NP_001074662 (2) | (86) | |
| Lyn | Tyr protein kinase Lyn isoform B | NP_001104566 (2) | (86) | ||
| N.A. | Bcl10-interacting CARD protein | NP_081014 (1) | (86) | ||
| Nup53 | Nucleoporin Nup53 isoform 2 | NP_081367 (3) | (86) | ||
| PI4KIIa | PtdIns(4)-kinase II α | MC, PM | NP_663476 (1) | (95) | |
| Ppp2r1a | Ser/Thr protein phosphatase 2 | CP, NC | NP_058587 (1) | (96) | |
| Pstpip1 | Pro/Ser/Thr phosphatase-interacting protein 1 | NP_035323 (1) | (86) | ||
| Rac1 | Ras-related C3 botulinum toxin substrate 1 | NP_033033 (2) | (86) | ||
| Ralb | Ras-related protein RalB | NP_071722 (1) | (86) | ||
| Rap1A | Ras-related GTPase Rap1A | CP | NP_663516 (1) | NP_663516 (2) | (86) |
| Rap2C | Ras-related GTPase Rap2C | NP_766001 (1) | (86) | ||
| Rhot1 | Mitochondrial Rho GTPase (Miro1) | MC | NP_067511 (2) | (97) | |
| Rhot2 | Mitochondrial Rho GTPase 2 | MC | NP_666111 (1) | (86) | |
| Rsu1 | Ras suppressor protein 1 | NP_033131 (1) | (86) | ||
| Ptgs1 | Prostaglandin G/H synthase 1 | NP_032995 (2) | (86) | ||
| Ptgs2 | Prostaglandin G/H synthase 2 | ER, PM | NP_035328 (1) | (86) | |
| Ptpmt1 | Protein Tyr phosphatase mitochondrial 1 | NP_079852 (2) | (86) | ||
| Pptc7 | Protein phosphatase Ptc7 | MC | NP_796216 (1) | (86) | |
| Sil1 | Nucleotide exchange factor Sil1 | NP_109674 (2) | (86) | ||
| Smpd4 | Sphingomyelin phosphodiesterase 4 | NP_001158083 (2) | (86) | ||
| Usp34 | Ubiquitin hydrolase | CP, NC | NP_001177330 (1) | (86) | |
| Ywhah | 14–3-3 protein subunit eta | CP | NP_035868 (2) | (86) | |
| Ywhaz | 14–3-3 protein subunit zeta/delta | CP | NP_035870 (2) | (98) | |
| Zmpste24 | CAAX prenyl protease 1 | NP_766288 (1) | (86) | ||
| Vesicle trafficking | |||||
| Chmp4b | Charged multivesicular body protein 4B | NP_083638 (1) | (86) | ||
| Dnm2 | Dynamin 2 | NP_001034609 (1) | (86) | ||
| Gf2r | Mannose-6-P receptor | LE, LS | NP_034645 (1) | (21) | |
| Gosr1 | Golgi SNAP receptor complex member 1 | NP_058090 (1) | (86) | ||
| Igf2r | Cation-independent mannose-6-P receptor | NP_034645 (1) | (86) | ||
| Lnp | Lunapark protein isoform a | NP_081409 (1) | (86) | ||
| Napg | Soluble NSF attachment protein gamma | NP_082293 (2) | (86) | ||
| Rab6A | Rab GTPase | RE, GA | NP_001157135 (3) | (86) | |
| Rab8A | Rab GTPase | RE, GA, PM | NP_075615 (1) | (99) | |
| Rab9A | Rab GTPase | RE, GA, PM | NP_062747 (2) | (100) | |
| Rab11B | Rab GTPase | GA, RE | NP_033023 (2) | (86) | |
| Rab27A | Rab GTPase | RE, GA, PM | NP_076124 (1) | (86) | |
| Rab35 | Rab GTPase | RE, GA, PM | NP_937806 (1) | (86) | |
| Rabac1 | Prenylated Rab acceptor protein 1 | NP_034391 (1) | (86) | ||
| Rtn3 | Reticulon-3 isoform 4 | NP_001003934 (1) | (86) | ||
| Scamp 1 | Secretory carrier-associated membrane protein 1 | LE, LS | NP_083429 (2) | (86) | |
| Scamp 2 | Secretory carrier-associated membrane protein 2 | LE, LS | NP_073724 (1) | (86) | |
| Stx3 | Syntaxin-3 t-SNARE | PM | NP_689344 (1) | (86) | |
| Stx4 | Syntaxin-4 t-SNARE | PM | NP_033320 (3) | (101) | |
| Stx5 | Syntaxin-5 t-SNARE | PM | NP_001161271 (2) | (86) | |
| Stx12 | Syntaxin-12 t-SNARE | PM | NP_598648 (2) | (86) | |
| Stxbp1 | Syntaxin-binding protein 1 isoform b | NP_001107041 (1) | (86) | ||
| Syngr1 | Synaptogyrin-1 | PM | NP_033329 (2) | (102) | |
| Trappc3 | Trafficking protein particle complex subunit 3 | ER, GA | NP_038746 (1) | (86) | |
| Vamp7 | Vamp7 v-SNARE | PM, EE | NP_035645 (2) | (103) | |
| VapA | Vamp-associated protein A | ER | NP_038961 (2) | (104) | |
| Vps18 | Vacuolar protein sorting-associated protein 18 | NP_758473 (1) | (86) | ||
| Metabolism | |||||
| Cmc1 | COX assembly mitochondrial protein homolog | NP_080718 (1) | (86) | ||
| Coq6 | Ubiquinone monooxygenase | GA, MC | NP_766170 (1) | (86) | |
| Cox7a2 | Cytochrome c oxidase subunit 7A2 | NP_034075 (3) | (86) | ||
| Cyb5r3 | NADH-cytochrome b5 reductase 3 | NP_084063.1 (3) | (86) | ||
| Cyp51a1 | Lanosterol C14 demethylase | ER, GA | NP_064394 (2) | (105) | |
| Dlat | Dihydrolipoyllysine acetyltransferase | MC | NP_663589 (3) | (86) | |
| Hsd17b7 | 3-Keto-steroid reductase | NP_034606 (2) | (86) | ||
| Lias | Lipoyl synthase | MC | NP_077791 (2) | (86) | |
| Lipa | Lysosomal acid lipase/cholesteryl ester hydrolase | LS | NP_001104570 (1) | (86) | |
| Ndufb3 | NADH dehydrogenase 1 β subcomplex subunit 3 | NP_079873 (3) | (86) | ||
| Nsdhl | Sterol-4-alpha-carboxylate 3-dehydrogenase | NP_035071 (2) | (86) | ||
| Osbpl8 | Oxysterol-binding protein | CP | NP_780698 (2) | (86) | |
| Pfkp | 6-Phosphofructokinase type C | NP_062677 (1) | (86) | ||
| Pgam1 | Phosphoglycerate mutase 1 | MC | NP_075907 (2) | (86) | |
| Uqcc | Ubiquinol-cytochrome c reductase complex | MC | NP_061376 (2) | (86) | |
| Uqcrb | Cytochrome b-c1 complex subunit 7 | MC | NP_080495 (2) | (86) | |
| Transport | |||||
| Abcd3 | ATP-binding cassette subfamily D member 3 | NP_033017 (1) | (86) | ||
| Atp1a3 | Na/K-transport ATPase subunit alpha-3 | PM | NP_659170 (1) | (86) | |
| Atp1b3 | Na/K-transport ATPase subunit beta-3 | PM | NP_031528 (1) | (86) | |
| Atp2b4 | PM Ca-transport ATPase 4 isoform b | PM | NP_998781 (1) | (86) | |
| Atp2c1 | Ca-transport ATPase 2C member 1 | NP_778190 (1) | (86) | ||
| Mmgt1 | Membrane magnesium transport | ER, GA | NP_666346 (2) | (86) | |
| Sec11c | Signal peptidase complex catalytic subunit Sec11c | ER | NP_079744 (2) | (86) | |
| Sec61a1 | Protein transport protein Sec61 | ER, PM | NP_058602 (2) | (86) | |
| Sec62 | Protein transport protein Sec62 | ER, PM | NP_081292 (1) | (86) | |
| Slc2a1 | Solute carrier family 2 (glucose transporter), member 1 | PM | NP_035530 (3) | (86) | |
| Slc2a6 | Solute carrier family 2 (glucose transporter), member 6 | PM | NP_001171098 (1) | (86) | |
| Slc7a5 | Large neutral AA transporter small subunit 1 | PM | NP_035534.2 (2) | (86) | |
| Slc16a1 | Monocarboxylate transporter 1 | PM | NP_033222 (2) | (86) | |
| Slc25a1 | Solute carrier family 25, member 1 | PM | NP_694790 (3) | (86) | |
| Slc25a11 | Mitoch. 2-oxoglutarate/malate transporter | NP_077173 (2) | (86) | ||
| Slc25a33 | Solute carrier family 25, member 33 | NP_081736 (1) | (86) | ||
| Timm17a | Mitochondrial import IM translocase Tim17A | NP_035720 (2) | (86) | ||
| Cytoskeleton/motor proteins | |||||
| Arpc3 | Actin-related protein 2/3 complex subunit 3 | CP | NP_062798 (1) | (86) | |
| Capzb | F-actin-capping protein subunit beta isoform b | CP | NP_033928 (2) | (86) | |
| Coro1A | Coronin 1A | NP_034028 (2) | (86) | ||
| Dynlrb1 | Dynein light chain roadblock-type 1 | CP | NP_080223 (1) | (86) | |
| Fam82b | Regulator of microtubule dynamics | CP | NP_079752 (2) | (86) | |
| Fmnl3 | Formin-like protein 3 | NP_035841 (1) | (86) | ||
| Kif5b | Kinesin-1 heavy chain | NP_032474 (2) | (86) | ||
| Kif11 | Kinesin-like protein Kif11 | NP_034745 (1) | (86) | ||
| Kif23 | Kinesin family member 23 | NP_077207 (2) | (86) | ||
| Kifc1 | Kinesin family member C5B | NP_444403 (1) | (86) | ||
| Map6 | Microtubule-associated protein 6 isoform 2 | CP | NP_001041632 (1) | (86) | |
| Myo5a | Myosin VA | CP | NP_034994 (2) | (86) | |
| Myo19 | Myosin XIX | NP_079690 (1) | (86) | ||
| Tpm1 | Tropomyosin α-1 chain | CP | NP_077745 (1) | (86) | |
| Tpm3 | Tropomyosin α-3 chain | CP | NP_071709 (1) | (86) | |
| Miscellaneous | |||||
| Atad1 | ATPase family AAA domain protein 1 | NP_080763 (2) | (86) | ||
| Calm1 | Calmodulin | NP_033920 (2) | (86) | ||
| CtsB | Cathepsin B | LE, LS | NP_031824 (3) | (86) | |
| Ergic1 | ERGIC protein 1 | ERGIC | NP_080446 (1) | (86) | |
| Fech | Ferrochelatase | NP_032024 (2) | (86) | ||
| Fxn | Frataxin | NP_032070 (2) | (86) | ||
| Golga5 | Golgin-A5 | GA | NP_038775 (3) | (86) | |
| Golm1 | Golgi membrane protein 1 | GA | NP_081583 (2) | (86) | |
| Sod1 | Superoxide dismutase [Cu-Zn] | NP_035564 (2) | (86) | ||
In the Δpentuple LCV proteome more than 560 additional proteins were exclusively found compared with the Lp02 proteome (Fig. 5A). Immune system components include FAS-associated factor 2, TNF, and TLR7, as well as a family M immunity-related GTPase and integrin α5 (Table III). Also, additional ribosomal proteins were identified. Metabolic components include several enzymes implicated in carbohydrate or lipid metabolism, as well as the respiratory chain.
Several vesicle trafficking factors were also only found in the Δpentuple LCV proteome such as Vps18, Rab GTPases (Rab8A, -9A, -11B, -27A, -35), syntaxins (Stx3, -4, -5, -12), synaptogyrin (Syngr1), synaptotagmin (Esyt1), the trafficking protein TRAPP, and (cation-independent) mannose 6-phosphate receptors. Novel LCV-associated proteins, not found in any previous study (30, 34), included the small GTPases Rab9A, Rab27A, and Rab35 as well as the ADP-ribosylation factors Arf4, -5, -6, and Arl2 (Table III). Furthermore, reticulon-3, lunapark, NSF, and the v-SNARE Vamp7 together with Vamp-associated protein A (VapA) were detected. Other host factors involved in signal transduction pathways comprise members of the Rho GTPase family (Rhot1, -2), Rho GDI 1 and 2, Cdc42se2, Rac1, RalB, G proteins and Gbf1, CAAX prenyl protease 1, annexin, Ser/Thr kinase 12, the Tyr kinase Lyn, Ser/Thr phosphatase 2, mitochondrial Tyr phosphatase, nucleotide exchange factor Sil1, inositol receptors (Itpr2 and 3), and members of the 14-3-3 adaptor family. Regulators of apoptosis (Bax, Bak1, Bcl10-interacting CARD protein) were also specifically present in the Δpentuple LCV proteome (Table III).
Moreover, several solute carriers (Slc), ABC transporters, or ion transport ATPases were found in the Δpentuple LCV proteome. Finally, cytoskeletal and motor proteins (F-actin capping protein, regulator of microtubule dynamics, coronin, kinesins, dynein, myosin, tropomyosin) were identified (Table III). The complete sets of proteins exclusively found in the proteome of macrophage LCVs harboring strain Lp02 or the Δpentuple mutant are listed in supplemental Table S7 and S8.
Active Small GTPase Rap1 Localizes to LCVs in D. discoideum
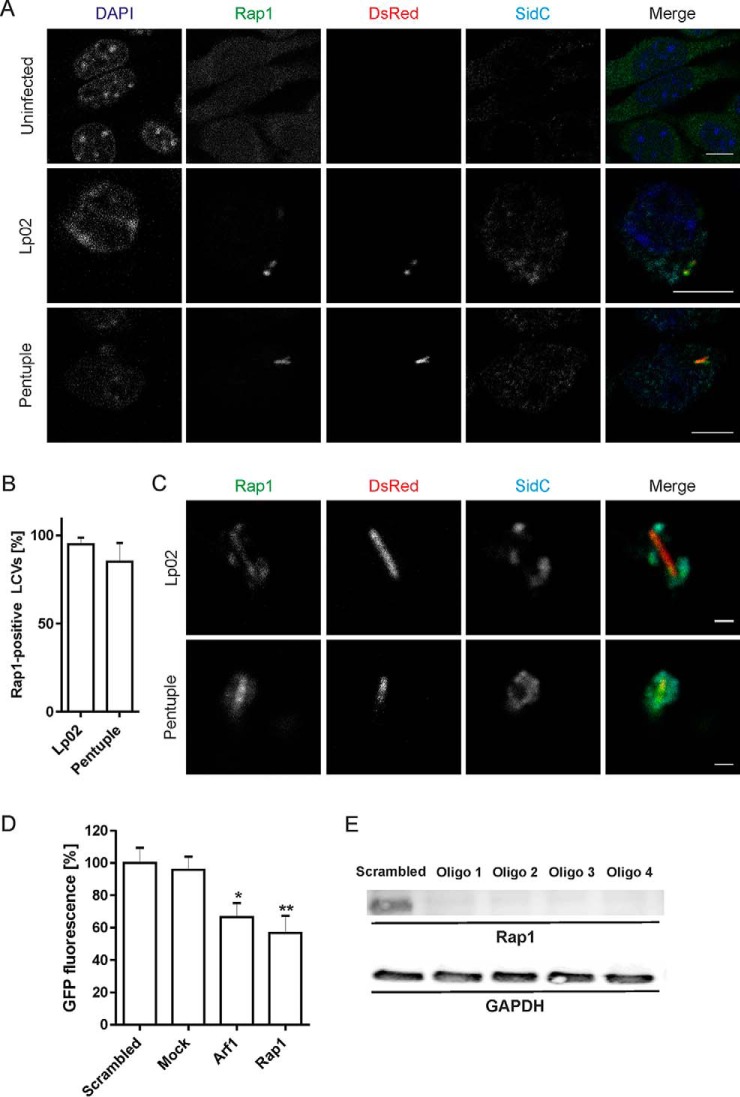
Because the Δpentuple mutant grows in macrophages but not in amoeba (Fig. 2) (37), we searched the LCV proteomes of Lp02 and Δpentuple for differences that might account for the intracellular growth defect. Host factors present on Lp02 and Δpentuple LCVs in macrophages, but only on Lp02 LCVs in D. discoideum might be positively correlated to intracellular bacterial replication. An interesting candidate fulfilling this criterion is Rap1. The small GTPase is conserved in D. discoideum and mammalian cells and controls cell adhesion dynamics and phagocytosis (41, 48–50). Our comparative proteomics approach revealed that Rap1 localizes in D. discoideum to LCVs harboring the parental strain Lp02, but not to LCVs harboring the Δpentuple mutant (Table II), whereas in RAW 264.7 macrophages Rap1 was identified on LCVs harboring either strain (Table III).
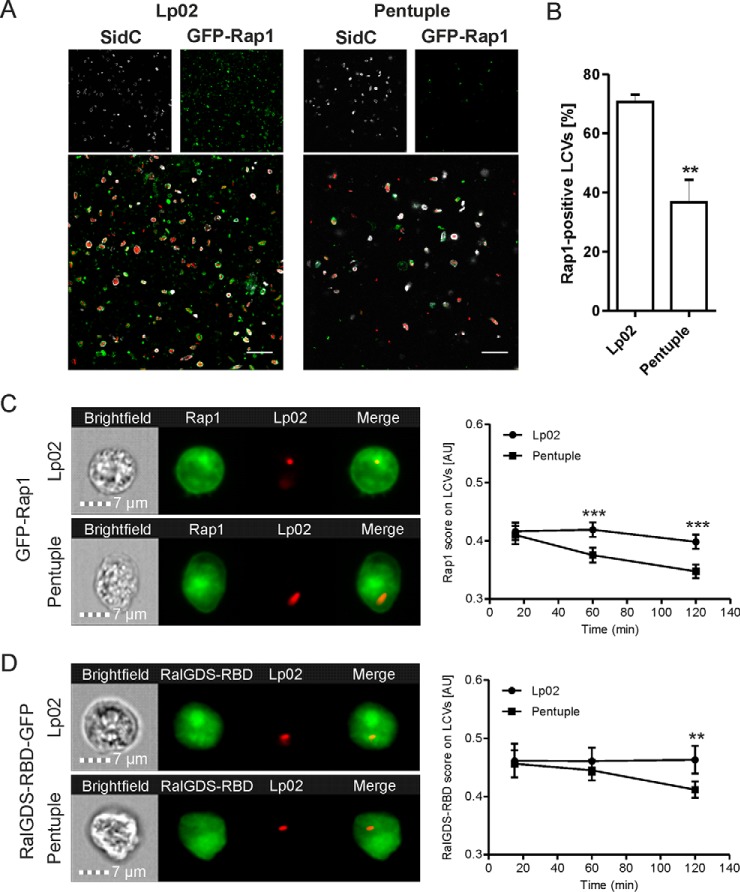
To validate the Rap1 localization pattern identified by proteomics, we used a D. discoideum strain producing GFP-Rap1 (40, 41). The amoeba were infected with L. pneumophila Lp02 or Δpentuple producing SidC and DsRed, homogenized and analyzed by fluorescence microscopy for SidC- and Rap1-positive LCVs (Fig. 6A). Indeed, ∼70% of intact LCVs harboring strain Lp02 stained positive for Rap1, whereas only 35% of LCVs containing the Δpentuple mutant were associated with Rap1 (Fig. 6B). Thus, significantly fewer pathogen vacuoles that do not allow intracellular replication stained positive for Rap1. We also noticed that compared with Δpentuple-infected D. discoideum a higher number of intact LCVs were found in homogenates of Lp02-infected amoeba (Fig. 6A).
Fig. 6.
Active small GTPase Rap1 localizes to LCVs harboring Lp02 in D. discoideum. A, D. discoideum amoeba producing GFP-Rap1 were infected (MOI 30, 1 h) with L. pneumophila Lp02 or Δpentuple harboring plasmid pCR080 (DsRed, SidC). The infected cells were lysed using a ball homogenizer, fixed, stained for SidC and analyzed by fluorescence microscopy. Scale bars, 5 μm. B, Quantification of SidC- and Rap1-positive LCVs harboring Lp02 or Δpentuple. Data show means and standard deviation of at least 150 LCVs per strain counted in three independent experiments (Student's t test: **p < 0.01). Imaging flow cytometry (IFC) of D. discoideum Ax3 producing (C) GFP-Rap1 or (D) RalGDSRBD-GFP, infected (MOI 5) with DsRed-producing L. pneumophila Lp02 or Δpentuple. Representative IFC images from the 2 h time point (left panels). Quantification of IFC colocalization score between GFP and DsRed in > 1′000 cells per sample at the time points indicated (right panels). Data show means and 95% confidence intervals of one representative experiment out of three independent experiments (**p < 0.01, ***p < 0.001).
Next, we assessed the accumulation of Rap1 to pathogen vacuoles by high-throughput imaging flow cytometry (IFC). To this end, D. discoideum producing GFP-Rap1 was infected with DsRed-labeled L. pneumophila Lp02 or Δpentuple, and 10,000 cells each at three different time points were assessed. The IFC colocalization score (see Materials and Methods) for the acquisition of GFP-Rap1 to LCVs was significantly lower for vacuoles containing the Δpentuple mutant compared with the parental strain (Fig. 6C). Analogously, we examined by IFC whether LCV-attached Rap1 is active using a probe for GTP-bound Rap1, RalGDSRBD-GFP (40, 51). Two hours post infection, significantly less active Rap1 localized to vacuoles containing the Δpentuple mutant compared with the parental strain (Fig. 6D) Taken together, active Rap1 preferentially localizes to LCVs containing Lp02 rather than Δpentuple in D. discoideum. This finding is in agreement with the notion that the small GTPase promotes intracellular replication of L. pneumophila.
The Small GTPase Rap1 Localizes to LCVs in Macrophages and Promotes Intracellular Growth of L. pneumophila
Rap1 was identified by proteomics in macrophage LCVs harboring either L. pneumophila Lp02 or Δpentuple (Table III). To validate the presence of Rap1 on the LCVs, RAW 264.7 macrophages were infected with DsRed-producing L. pneumophila, labeled with antibodies against Rap1 or SidC and analyzed by fluorescence microscopy (Fig. 7A). This approach revealed that Rap1 indeed accumulated to nearly 100% on vacuoles harboring either Lp02 or Δpentuple (Fig. 7B). Similarly, Rap1 localized to Lp02- and Δpentuple-containing intact macrophage LCVs, which were isolated using the two-step immuno-affinity protocol (Fig. 7C).
Fig. 7.
The small GTPase Rap1 localizes to LCVs in macrophages and promotes intracellular growth of L. pneumophila. A, RAW 264.7 macrophages were left uninfected or were infected (MOI 10, 2 h) with DsRed-producing L. pneumophila Lp02 or Δpentuple (pSW001), followed by labeling with anti-Rap1 and anti-SidC antibodies as well as DAPI. Bars, 5 μm. B, For quantification of (A), at least 50 infected macrophages were scored each in two independent experiments (means and S.D. are shown). C, RAW 264.7 macrophages were infected as in (A), and intact LCVs were isolated using the two-step immuno-affinity protocol. Isolated LCVs were immuno-labeled as in (A). Bars, 1 μm. D, Human A549 lung epithelial cells were treated for 48 h with siRNAs against Rap1, Arf1, scrambled siRNA or transfection reagent only (“mock”) and infected (MOI 10, 24 h) with GFP-producing L. pneumophila Lp02 (pNT28). Bacterial growth was determined by measuring GFP fluorescence in a plate reader. Data represent mean and standard deviation from three independent experiments (Student's t test: *p < 0.05, **p < 0.01) shown for one oligonucleotide out of four tested. E, A549 cells were treated with 4 different siRNAs (Oligo 1–4) against Rap1 for 48 h. The depletion efficiency was assessed by Western blot using a mouse monoclonal antibody against Rap1 (21 kDa). GAPDH (38 kDa) served as a loading control, and Qiagen AllStars oligonucleotides (“scrambled”) were used as a negative control.
RNA interference was utilized to directly assess a role of Rap1 for intracellular replication of L. pneumophila (Fig. 7D). To this end, human A549 lung epithelial cells were treated with siRNA oligonucleotides targeting Rap1, or—as a positive control—the small GTPase Arf1 (34, 52, 53). Scrambled siRNA or transfection reagent only (mock) served as negative controls. siRNA-treated cells were infected with GFP-producing L. pneumophila Lp02, and intracellular bacterial growth was assessed after 24 h by measuring fluorescence. Interestingly, compared with both negative controls the depletion of Rap1 reduced growth of L. pneumophila Lp02 in A549 cells by almost 50%, which is similar to the growth reduction observed by silencing of Arf1 (Fig. 7D). The treatment of A549 cells with siRNA targeting Rap1 almost completely depleted the small GTPase (Fig. 7E). In summary, Rap1 localizes to macrophage LCVs harboring either the parental strain Lp02 or the Δpentuple mutant, and the small GTPase contributes to intracellular replication of L. pneumophila. Thus, comparative proteomics of Lp02 and Δpentuple LCVs identified a novel host factor, Rap1, localizing preferentially to a replication-permissive pathogen compartment and promoting intracellular replication of L. pneumophila.
DISCUSSION
The aim of this study was to to characterize the host cell proteome defining the membrane-bound, replication-permissive intracellular compartment of L. pneumophila. The rationale was that host factors present on a replication-permissive LCV potentially promote intracellular replication, whereas the absence of a host factor on a nonpermissive LCV might contribute to the replication defect. Alternatively, in a more complex scenario, an inhibitory factor accumulating on LCVs might counteract intracellular replication. This could be the case for the small GTPase Rab21, the depletion of which promotes intracellular replication of L. pneumophila (34). Indeed, Rab21 was identified on Δpentuple LCVs in D. discoideum (Table II), and therefore, the GTPase might act as an inhibitory factor.
The study identified a number of host cell proteins that were enriched on LCVs harboring either Lp02 or Δpentuple in D. discoideum (Table II) or RAW 246.7 macrophages (Table III). The proteomics approach yields semiquantitative data, and thus, we cannot exclude false negative results, i.e. the failure of detecting a given LCV component because of a low(er) abundance. Some of the differentially localizing host factors were identified in three out of three independent biological replicates in LCVs harboring either Lp02 or Δpentuple, but not in both compartments. In this case, the finding is robust and very likely biologically relevant. In other cases, a host factor was identified in one or two out of three independent biological replicates in LCVs harboring either Lp02 or Δpentuple. Even if less robust, these results might still be biologically significant. Despite these technical limitations, the comparative proteomics study presented here allowed the identification of a novel relevant LCV host factor, Rap1, on replication-permissive pathogen vacuoles.
The proteomics analysis was validated by microscopy, which confirmed that in infected D. discoideum activated GTP-bound Rap1 showed stronger accumulation on LCVs harboring Lp02 compared with Δpentuple (Fig. 6). Moreover, Rap1 was identified on Lp02 as well as Δpentuple LCVs in infected RAW 264.7 macrophages, correlating with the finding that both strains replicate in these phagocytes (Fig. 7). Accordingly, the depletion of Rap1 by siRNA resulted in severely reduced intracellular growth of the bacteria.
Rap1 belongs to the Rat sarcoma (Ras) protein superfamily of small GTPases, is conserved in mammalian cells and D. discoideum (50, 54), and essential in the amoeba (41, 55). In mammalian cells, activation of the GTPase (i.e. binding of GTP) is triggered by cell adhesion molecules, cytokines, growth factors like TNFα or IFNγ, or second messengers that are coupled to Rap1 guanine nucleotide exchange factors (GEFs) (54, 56). Rap1 controls cell adhesion dynamics and phagocytosis, especially by mediating the functions of integrins and cadherins (48, 49). Strikingly, several integrins (integrin-α4, -αM, -β1, -β2) were identified in Lp02- and Δpentuple-harboring macrophage LCV proteomes (supplemental Table S6). In this pathway, Rap1 acts upstream of the integrin-associated factor talin and controls the recruitment of the cytoskeletal protein to sites of particle binding and pha-gocytosis (57). Talin was found in the proteome of Lp02- and Δpentuple-harboring macrophage LCVs (supplemental Table S6), and thus, this host factor possibly interacts with Rap1 during uptake of L. pneumophila. The interaction between the D. discoideum homologue of talin and Rap1 is also essential for cellular adhesion of D. discoideum (41). However, the downstream signaling is not yet understood in the amoeba. Interestingly, in D. discoideum talin is exclusively found in the Lp02 LCV proteome and not in the Δpentuple LCV proteome (Table II). Therefore, the presence of Rap1 and talin on LCVs correlates with intracellular replication of L. pneumophila. Because the uptake of the Δpentuple mutant was identical to the parental strain Lp02, Rap1 and talin likely play a role in intracellular bacterial replication after phagosome closure.
In addition to adhesion and phagocytosis, Rap1 also plays a pivotal role for cell growth, proliferation and survival in mammalian cells (54, 58) and is involved in proliferation, differentiation and modulation of the cytoskeleton in D. discoideum (50, 55). This modulatory effect on the cytoskeleton might also account for the decisive role of Rap1 during chemotaxis and cell migration. Here, Rap1 is rapidly activated dependent on cAMP at the leading edge of migrating cells, which leads to the recruitment of downstream effectors like PI 3-kinases (50). Furthermore, Rap1 together with Ras binds to and controls the activity of the target of rapamycin complex 2 (TORC2), a key player in D. discoideum chemotaxis, and might function as a signal relay allowing coordinated cytoskeletal remodeling (59).
Several other interesting candidates for host factors determining the intracellular fate of L. pneumophila in D. discoideum were identified based on their presence on Lp02 LCVs and absence on Δpentuple LCVs (Table II). These candidates include Sac1, an ER-associated PI phosphatase that meta-bolizes PtdIns(4)P on LCVs, thus removing PI-bound effectors (60). Other candidates present on Lp02 but not Δpentuple LCVs are the small GTPase Rab11C, which localizes to LCVs harboring wild-type but not ΔicmT mutant L. pneumophila (34).
Noteworthy, many more host proteins were found to be associated with macrophage LCVs containing the Δpentuple mutant compared with LCVs harboring the parental strain (Table III). Because Δpentuple mutant bacteria replicate in macrophages, some of these factors might promote intracellular growth. Candidate host factors promoting growth of L. pneumophila are several solute carriers (Slc), as well as proteins implicated in vesicle trafficking, membrane dynamics and signaling processes. These include Vps18, Rab GTPases (Rab8A, -9A, -11B, -27A, -35), syntaxins (Stx3, -4, -5, -12), synaptogyrin (Syngr1), synaptotagmin (Esyt1), the trafficking protein TRAPP, and (cation-independent) mannose 6-phosphate receptors. Novel LCV-associated proteins, not found in previous studies (30, 34), comprise the small GTPases Rab9A, Rab27A, and Rab35, as well as the ADP-ribosylation factors Arf4, -5, -6, and Arl2. Furthermore, reticulon-3, lunapark, NSF, and the v-SNARE Vamp7 together with Vamp-associated protein A (VapA) might also promote intracellular replication of L. pneumophila (Table III).
Taken together, our study provides comparative proteomics data for LCVs from two different phagocytic host cells and two different L. pneumophila strains, which show distinct phenotypes regarding intracellular replication. The study reveals a novel small GTPase, Rap1, implicated in intracellular replication of L. pneumophila and provides insights for further hypothesis-driven, mechanistic investigation of the intricate interactions between L. pneumophila and its various eukaryotic host cells.
Supplementary Material
Acknowledgments
We thank Ralph Isberg (Tufts University) for the L. pneumophila Δpentuple and other cluster deletion mutant strains and Arjan Kortholt (University of Groningen) for the D. discoideum GFP-Rap1 and RalGDSRBD-GFP expression constructs, as well as Stephen Weber for help with analyzing D. discoideum.
Footnotes
Author contributions: D.B. and H.H. designed research; J.S., C.M., A.O., C.H., B.S., and A.W. performed research; D.B. contributed new reagents or analytic tools; J.S., C.M., A.O., C.H., B.S., A.W., D.B., and H.H. analyzed data; J.S., C.M., and H.H. wrote the paper.
* This work was supported by the Institute of Medical Microbiology, the University of Zürich (UZH), the UZH Center for Microscopy and Image Analysis, the UZH Flow Cytometry Facility, the Swiss National Science Foundation (SNF; 31003A_153200), the German Research Foundation (DFG; HI 1511/3-1, SPP 1580), the Bundesministerium für Bildung und Forschung (BMBF; 031A410A; Infect-ERA project EUGENPATH) and a “FöFoLe” stipend from the Ludwig-Maximilians University (Faculty of Medicine) awarded to J.S. A.W. was supported by a grant from the Swedish Research Council (2014-396). The group of D.B. was supported by the Bundesministerium für Bildung und Forschung (BMBF; 031A410B; Infect-ERA project EUGENPATH).
 This article contains supplemental Tables.
This article contains supplemental Tables.
1 The abbreviations used are:
- LCV
- Legionella-containing vacuole
- ACES
- N-(2-acetamido)-2-aminoethanesulfonic acid
- AYE
- ACES yeast extract
- BMM
- bone marrow-derived macrophages
- BSA
- bovine serum albumin
- CFU
- colony forming units
- CYE
- charcoal yeast extract
- ER
- endoplasmic reticulum
- FBS
- fetal bovine serum
- GAPDH
- glyceraldehyde 3-phosphate dehydrogenase
- Icm/Dot
- intracellular replication/defective organelle trafficking
- IFC
- imaging flow cytometry
- PYG
- proteose yeast extract glucose
- PFA
- paraformaldehyde
- PI
- phosphoinositide
- Rap1
- Ras-related protein 1
- RT
- room temperature
- T4SS
- type IV secretion system.
REFERENCES
- 1. Declerck P. (2010) Biofilms: the environmental playground of Legionella pneumophila. Environ. Microbiol. 12, 557–566 [DOI] [PubMed] [Google Scholar]
- 2. Abdel-Nour M., Duncan C., Low D. E., and Guyard C. (2013) Biofilms: the stronghold of Legionella pneumophila. Int. J. Mol. Sci. 14, 21660–21675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hoffmann C., Harrison C. F., and Hilbi H. (2014) The natural alternative: protozoa as cellular models for Legionella infection. Cell Microbiol. 16, 15–26 [DOI] [PubMed] [Google Scholar]
- 4. Molofsky A. B., and Swanson M. S. (2004) Differentiate to thrive: lessons from the Legionella pneumophila life cycle. Mol. Microbiol. 53, 29–40 [DOI] [PubMed] [Google Scholar]
- 5. Byrne B., and Swanson M. S. (1998) Expression of Legionella pneumophila virulence traits in response to growth conditions. Infect. Immun. 66, 3029–3034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brüggemann H., Hagman A., Jules M., Sismeiro O., Dillies M. A., Gouyette C., Kunst F., Steinert M., Heuner K., Coppee J. Y., and Buchrieser C. (2006) Virulence strategies for infecting phagocytes deduced from the in vivo transcriptional program of Legionella pneumophila. Cell Microbiol. 8, 1228–1240 [DOI] [PubMed] [Google Scholar]
- 7. Faucher S. P., Mueller C. A., and Shuman H. A. (2011) Legionella pneumophila transcriptome during intracellular multiplication in human macrophages. Front. Microbiol. 2, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hayashi T., Nakamichi M., Naitou H., Ohashi N., Imai Y., and Miyake M. (2010) Proteomic analysis of growth phase-dependent expression of Legionella pneumophila proteins which involves regulation of bacterial virulence traits. PLoS ONE 5, e11718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Aurass P., Gerlach T., Becher D., Voigt B., Karste S., Bernhardt J., Riedel K., Hecker M., and Flieger A. (2016) Life stage-specific proteomes of Legionella pneumophila reveal a highly differential abundance of virulence-associated Dot/Icm effectors. Mol. Cell. Proteomics 15, 177–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hilbi H., Hoffmann C., and Harrison C. F. (2011) Legionella spp. outdoors: colonization, communication and persistence. Environ. Microbiol. Rep. 3, 286–296 [DOI] [PubMed] [Google Scholar]
- 11. Isberg R. R., O'Connor T. J., and Heidtman M. (2009) The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat. Rev. Microbiol. 7, 13–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Personnic N., Bärlocher K., Finsel I., and Hilbi H. (2016) Subversion of retrograde trafficking by translocated pathogen effectors. Trends Microbiol. 24, 450–462 [DOI] [PubMed] [Google Scholar]
- 13. Newton H. J., Ang D. K., van Driel I. R., and Hartland E. L. (2010) Molecular pathogenesis of infections caused by Legionella pneumophila. Clin. Microbiol. Rev. 23, 274–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Finsel I., and Hilbi H. (2015) Formation of a pathogen vacuole according to Legionella pneumophila: how to kill one bird with many stones. Cell. Microbiol. 17, 935–950 [DOI] [PubMed] [Google Scholar]
- 15. Hubber A., and Roy C. R. (2010) Modulation of host cell function by Legionella pneumophila type IV effectors. Ann. Rev. Cell. Dev. Biol. 26, 261–283 [DOI] [PubMed] [Google Scholar]
- 16. Haneburger I., and Hilbi H. (2013) Phosphoinositide lipids and the Legio-nella pathogen vacuole. Curr. Top. Microbiol. Immunol. 376, 155–173 [DOI] [PubMed] [Google Scholar]
- 17. Itzen A., and Goody R. S. (2011) Covalent coercion by Legionella pneumophila. Cell Host Microbe 10, 89–91 [DOI] [PubMed] [Google Scholar]
- 18. Sherwood R. K., and Roy C. R. (2013) A Rab-centric perspective of bacterial pathogen-occupied vacuoles. Cell Host Microbe 14, 256–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rothmeier E., Pfaffinger G., Hoffmann C., Harrison C. F., Grabmayr H., Repnik U., Hannemann M., Wölke S., Bausch A., Griffiths G., Müller-Taubenberger A., Itzen A., and Hilbi H. (2013) Activation of Ran GTPase by a Legionella effector promotes microtubule polymerization, pathogen vacuole motility and infection. PLoS Pathog. 9, e1003598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Simon S., Wagner M. A., Rothmeier E., Müller-Taubenberger A., and Hilbi H. (2014) Icm/Dot-dependent inhibition of phagocyte migration by Legionella is antagonized by a translocated Ran GTPase activator. Cell Microbiol. 16, 977–992 [DOI] [PubMed] [Google Scholar]
- 21. Finsel I., Ragaz C., Hoffmann C., Harrison C. F., Weber S., van Rahden V. A., Johannes L., and Hilbi H. (2013) The Legionella effector RidL inhibits retrograde trafficking to promote intracellular replication. Cell Host Microbe 14, 38–50 [DOI] [PubMed] [Google Scholar]
- 22. Hartlova A., Krocova Z., Cerveny L., and Stulik J. (2011) A proteomic view of the host-pathogen interaction: The host perspective. Proteo-mics 11, 3212–3220 [DOI] [PubMed] [Google Scholar]
- 23. Herweg J. A., Hansmeier N., Otto A., Geffken A. C., Subbarayal P., Prusty B. K., Becher D., Hensel M., Schaible U. E., Rudel T., and Hilbi H. (2015) Purification and proteomics of pathogen-modified vacuoles and membranes. Front. Cell. Infect. Microbiol. 5, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Urwyler S., Finsel I., Ragaz C., and Hilbi H. (2010) Isolation of Legio-nella-containing vacuoles by immuno-magnetic separation. Curr. Protoc. Cell Biol. Chapter 3, Unit 3.34 [DOI] [PubMed] [Google Scholar]
- 25. Finsel I., Hoffmann C., and Hilbi H. (2013) Immunomagnetic purification of fluorescent Legionella-containing vacuoles. Methods Mol. Biol. 983, 431–443 [DOI] [PubMed] [Google Scholar]
- 26. Hoffmann C., Finsel I., and Hilbi H. (2012) Purification of pathogen vacuoles from Legionella-infected phagocytes. J. Vis. Exp. e4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hoffmann C., Finsel I., and Hilbi H. (2013) Pathogen vacuole purification from Legionella-infected amoeba and macrophages. Methods Mol. Biol. 954, 309–321 [DOI] [PubMed] [Google Scholar]
- 28. Naujoks J., Tabeling C., Dill B. D., Hoffmann C., Brown A. S., Kunze M., Kempa S., Peter A., Mollenkopf H. J., Dorhoi A., Kershaw O., Gruber A. D., Sander L. E., Witzenrath M., Herold S., Nerlich A., Hocke A. C., van Driel I., Suttorp N., Bedoui S., Hilbi H., Trost M., and Opitz B. (2016) IFNs modify the proteome of Legionella-containing vacuoles and restrict infection via IRG1-derived itaconic acid. PLoS Pathog. 12, e1005408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bruckert W. M., and Abu Kwaik Y. (2015) Complete and ubiquitinated proteome of the Legionella-containing vacuole within human macrophages. J. Proteom Res. 14, 236–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Urwyler S., Nyfeler Y., Ragaz C., Lee H., Mueller L. N., Aebersold R., and Hilbi H. (2009) Proteome analysis of Legionella vacuoles purified by magnetic immunoseparation reveals secretory and endosomal GTPases. Traffic 10, 76–87 [DOI] [PubMed] [Google Scholar]
- 31. Luo Z. Q., and Isberg R. R. (2004) Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc. Natl. Acad. Sci. U.S.A. 101, 841–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Weber S. S., Ragaz C., Reus K., Nyfeler Y., and Hilbi H. (2006) Legio-nella pneumophila exploits PI(4)P to anchor secreted effector proteins to the replicative vacuole. PLoS Pathog. 2, e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ragaz C., Pietsch H., Urwyler S., Tiaden A., Weber S. S., and Hilbi H. (2008) The Legionella pneumophila phosphatidylinositol-4 phosphate-binding type IV substrate SidC recruits endoplasmic reticulum vesicles to a replication-permissive vacuole. Cell Microbiol. 10, 2416–2433 [DOI] [PubMed] [Google Scholar]
- 34. Hoffmann C., Finsel I., Otto A., Pfaffinger G., Rothmeier E., Hecker M., Becher D., and Hilbi H. (2014) Functional analysis of novel Rab GTPases identified in the proteome of purified Legionella-containing vacuoles from macrophages. Cell Microbiol. 16, 1034–1052 [DOI] [PubMed] [Google Scholar]
- 35. de Felipe K. S., Glover R. T., Charpentier X., Anderson O. R., Reyes M., Pericone C. D., and Shuman H. A. (2008) Legionella eukaryotic-like type IV substrates interfere with organelle trafficking. PLoS Pathog. 4, e1000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ninio S., Celli J., and Roy C. R. (2009) A Legionella pneumophila effector protein encoded in a region of genomic plasticity binds to Dot/Icm-modified vacuoles. PLoS Pathog. 5, e1000278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. O'Connor T. J., Adepoju Y., Boyd D., and Isberg R. R. (2011) Minimization of the Legionella pneumophila genome reveals chromosomal regions involved in host range expansion. Proc. Natl. Acad. Sci. U.S.A. 108, 14733–14740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhou K., Takegawa K., Emr S. D., and Firtel R. A. (1995) A phosphatidylinositol (PI) kinase gene family in Dictyostelium discoideum: biological roles of putative mammalian p110 and yeast Vps34p PI3-kinase homologs during growth and development. Mol. Cell. Biol. 15, 5645–5656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Müller-Taubenberger A., Lupas A. N., Li H., Ecke M., Simmeth E., and Gerisch G. (2001) Calreticulin and calnexin in the endoplasmic reticulum are important for phagocytosis. EMBO J. 20, 6772–6782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kortholt A., Bolourani P., Rehmann H., Keizer-Gunnink I., Weeks G., Wittinghofer A., and Van Haastert P. J. (2010) A Rap/phosphatidylinositol 3-kinase pathway controls pseudopod formation. Mol. Biol. Cell 21, 936–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Plak K., Keizer-Gunnink I., van Haastert P. J., and Kortholt A. (2014) Rap1-dependent pathways coordinate cytokinesis in Dictyostelium. Mol. Biol. Cell 25, 4195–4204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Solomon J. M., Rupper A., Cardelli J. A., and Isberg R. R. (2000) Intracellular growth of Legionella pneumophila in Dictyostelium discoideum, a system for genetic analysis of host-pathogen interactions. Infect. Immun. 68, 2939–2947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Derre I., and Isberg R. R. (2004) Legionella pneumophila replication vacuole formation involves rapid recruitment of proteins of the early secretory system. Infect. Immun. 72, 3048–3053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tiaden A. N., Kessler A., and Hilbi H. (2013) Analysis of Legionella infection by flow cytometry. Methods Mol. Biol. 954, 233–249 [DOI] [PubMed] [Google Scholar]
- 45. Johansson J., Karlsson A., Bylund J., and Welin A. (2015) Phagocyte interactions with Mycobacterium tuberculosis–Simultaneous analysis of phagocytosis, phagosome maturation and intracellular replication by imaging flow cytometry. J. Immunol. Methods 427, 73–84 [DOI] [PubMed] [Google Scholar]
- 46. Vizcaino J. A., Csordas A., del-Toro N., Dianes J. A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., Xu Q. W., Wang R., and Hermjakob H. (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, D447–D456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hilbi H., Segal G., and Shuman H. A. (2001) Icm/Dot-dependent upregulation of phagocytosis by Legionella pneumophila. Mol. Microbiol. 42, 603–617 [DOI] [PubMed] [Google Scholar]
- 48. Retta S. F., Balzac F., and Avolio M. (2006) Rap1: a turnabout for the crosstalk between cadherins and integrins. Eur. J. Cell Biol. 85, 283–293 [DOI] [PubMed] [Google Scholar]
- 49. Chung J., Serezani C. H., Huang S. K., Stern J. N., Keskin D. B., Jagirdar R., Brock T. G., Aronoff D. M., and Peters-Golden M. (2008) Rap1 activation is required for Fc gamma receptor-dependent phagocytosis. J. Immunol. 181, 5501–5509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kortholt A., and van Haastert P. J. (2008) Highlighting the role of Ras and Rap during Dictyostelium chemotaxis. Cell Signal. 20, 1415–1422 [DOI] [PubMed] [Google Scholar]
- 51. Jeon T. J., Lee D. J., Merlot S., Weeks G., and Firtel R. A. (2007) Rap1 controls cell adhesion and cell motility through the regulation of myosin II. J. Cell Biol. 176, 1021–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kagan J. C., and Roy C. R. (2002) Legionella phagosomes intercept vesicular traffic from endoplasmic reticulum exit sites. Nat. Cell Biol. 4, 945–954 [DOI] [PubMed] [Google Scholar]
- 53. Nagai H., Kagan J. C., Zhu X., Kahn R. A., and Roy C. R. (2002) A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science 295, 679–682 [DOI] [PubMed] [Google Scholar]
- 54. Caron E. (2003) Cellular functions of the Rap1 GTP-binding protein: a pattern emerges. J. Cell Sci. 116, 435–440 [DOI] [PubMed] [Google Scholar]
- 55. Kang R., Kae H., Ip H., Spiegelman G. B., and Weeks G. (2002) Evidence for a role for the Dictyostelium Rap1 in cell viability and the response to osmotic stress. J. Cell Sci. 115, 3675–3682 [DOI] [PubMed] [Google Scholar]
- 56. Alsayed Y., Uddin S., Ahmad S., Majchrzak B., Druker B. J., Fish E. N., and Platanias L. C. (2000) IFN-γ activates the C3G/Rap1 signaling pathway. J. Immunol. 164, 1800–1806 [DOI] [PubMed] [Google Scholar]
- 57. Lim J., Dupuy A. G., Critchley D. R., and Caron E. (2010) Rap1 controls activation of the alpha(M)beta(2) integrin in a talin-dependent manner. J. Cell. Biochem. 111, 999–1009 [DOI] [PubMed] [Google Scholar]
- 58. Zhang W., and Liu H. T. (2002) MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. 12, 9–18 [DOI] [PubMed] [Google Scholar]
- 59. Khanna A., Lotfi P., Chavan A. J., Montano N. M., Bolourani P., Weeks G., Shen Z., Briggs S. P., Pots H., Van Haastert P. J., Kortholt A., and Charest P. G. (2016) The small GTPases Ras and Rap1 bind to and control TORC2 activity. Sci. Rep. 6, 25823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hubber A., Arasaki K., Nakatsu F., Hardiman C., Lambright D., De Camilli P., Nagai H., and Roy C. R. (2014) The machinery at endoplasmic reticulum-plasma membrane contact sites contributes to spatial regulation of multiple Legionella effector proteins. PLoS Pathog. 10, e1004222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Berger K. H., and Isberg R. R. (1993) Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol. Microbiol. 7, 7–19 [DOI] [PubMed] [Google Scholar]
- 62. Roy C. R., Berger K. H., and Isberg R. R. (1998) Legionella pneumo-phila DotA protein is required for early phagosome trafficking decisions that occur within minutes of bacterial uptake. Mol. Microbiol. 28, 663–674 [DOI] [PubMed] [Google Scholar]
- 63. Tiaden A., Spirig T., Weber S. S., Brüggemann H., Bosshard R., Buchrieser C., and Hilbi H. (2007) The Legionella pneumophila response regulator LqsR promotes host cell interactions as an element of the virulence regulatory network controlled by RpoS and LetA. Cell Microbiol. 9, 2903–2920 [DOI] [PubMed] [Google Scholar]
- 64. Mampel J., Spirig T., Weber S. S., Haagensen J. A. J., Molin S., and Hilbi H. (2006) Planktonic replication is essential for biofilm formation by Legionella pneumophila in a complex medium under static and dynamic flow conditions. Appl. Environ. Microbiol. 72, 2885–2895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Eichinger L., Pachebat J. A., Glöckner G., Rajandream M. A., Sucgang R., Berriman M., Song J., Olsen R., Szafranski K., Xu Q., Tunggal B., Kummerfeld S., Madera M., Konfortov B. A., Rivero F., Bankier A. T., Lehmann R., Hamlin N., Davies R., Gaudet P., Fey P., Pilcher K., Chen G., Saunders D., Sodergren E., Davis P., Kerhornou A., Nie X., Hall N., Anjard C., Hemphill L., Bason N., Farbrother P., Desany B., Just E., Morio T., Rost R., Churcher C., Cooper J., Haydock S., van Driessche N., Cronin A., Goodhead I., Muzny D., Mourier T., Pain A., Lu M., Harper D., Lindsay R., Hauser H., James K., Quiles M., Madan Babu M., Saito T., Buchrieser C., Wardroper A., Felder M., Thangavelu M., Johnson D., Knights A., Loulseged H., Mungall K., Oliver K., Price C., Quail M. A., Urushihara H., Hernandez J., Rabbinowitsch E., Steffen D., Sanders M., Ma J., Kohara Y., Sharp S., Simmonds M., Spiegler S., Tivey A., Sugano S., White B., Walker D., Woodward J., Winckler T., Tanaka Y., Shaulsky G., Schleicher M., Weinstock G., Rosenthal A., Cox E. C., Chisholm R. L., Gibbs R., Loomis W. F., Platzer M., Kay R. R., Williams J., Dear P. H., Noegel A. A., Barrell B., and Kuspa A. (2005) The genome of the social amoeba Dictyostelium discoideum. Nature 435, 43–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Matto-Yelin M., Aitken A., and Ravid S. (1997) 14-3-3 inhibits the Dictyostelium myosin II heavy-chain-specific protein kinase C activity by a direct interaction: identification of the 14–3-3 binding domain. Mol. Biol. Cell 8, 1889–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Fan D., and Hou L. S. (2015) Novel zinc protease gene isolated from Dictyostelium discoideum is structurally related to mammalian leukotriene A4 hydrolase. Genet. Mol. Res. 14, 16332–16342 [DOI] [PubMed] [Google Scholar]
- 68. Mun H., and Jeon T. J. (2012) Regulation of actin cytoskeleton by Rap1 binding to RacGEF1. Mol. Cells 34, 71–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sasaki A. T., and Firtel R. A. (2009) Spatiotemporal regulation of Ras-GTPases during chemotaxis. Methods Mol. Biol. 571, 333–348 [DOI] [PubMed] [Google Scholar]
- 70. Blagoveshchenskaya A., and Mayinger P. (2009) SAC1 lipid phosphatase and growth control of the secretory pathway. Mol. Biosyst. 5, 36–42 [DOI] [PubMed] [Google Scholar]
- 71. Rivero F., Kuspa A., Brokamp R., Matzner M., and Noegel A. A. (1998) Interaptin, an actin-binding protein of the alpha-actinin superfamily in Dictyostelium discoideum, is developmentally and cAMP-regulated and associates with intracellular membrane compartments. J. Cell Biol. 142, 735–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Rai A., Nothe H., Tzvetkov N., Korenbaum E., and Manstein D. J. (2011) Dictyostelium dynamin B modulates cytoskeletal structures and membranous organelles. Cell Mol. Life Sci. 68, 2751–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Chang L. F., Chen S., Liu C. C., Pan X., Jiang J., Bai X. C., Xie X., Wang H. W., and Sui S. F. (2012) Structural characterization of full-length NSF and 20S particles. Nat. Struct. Mol. Biol. 19, 268–275 [DOI] [PubMed] [Google Scholar]
- 74. Pellinen T., Arjonen A., Vuoriluoto K., Kallio K., Fransen J. A., and Ivaska J. (2006) Small GTPase Rab21 regulates cell adhesion and controls endosomal traffic of beta1-integrins. J. Cell Biol. 173, 767–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Egami Y., and Araki N. (2009) Dynamic changes in the spatiotemporal localization of Rab21 in live RAW264 cells during macropinocytosis. PLoS ONE 4, e6689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Diaz A., and Ahlquist P. (2012) Role of host reticulon proteins in rearranging membranes for positive-strand RNA virus replication. Curr. Opin. Microbiol. 15, 519–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Teng F. Y., and Tang B. L. (2008) Cell autonomous function of Nogo and reticulons: The emerging story at the endoplasmic reticulum. J. Cell. Physiol. 216, 303–308 [DOI] [PubMed] [Google Scholar]
- 78. Journet A., Klein G., Brugiere S., Vandenbrouck Y., Chapel A., Kieffer S., Bruley C., Masselon C., and Aubry L. (2012) Investigating the macropinocytic proteome of Dictyostelium amoebae by high-resolution mass spectrometry. Proteomics 12, 241–245 [DOI] [PubMed] [Google Scholar]
- 79. Poyau A., Buchet K., and Godinot C. (1999) Sequence conservation from human to prokaryotes of Surf1, a protein involved in cytochrome c oxidase assembly, deficient in Leigh syndrome. FEBS Lett. 462, 416–420 [DOI] [PubMed] [Google Scholar]
- 80. Anjard C., Loomis W. F., and Dictyostelium Sequencing Consortium. (2002) Evolutionary analyses of ABC transporters of Dictyostelium discoideum. Eukaryot. Cell 1, 643–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Satre M., Mattei S., Aubry L., Gaudet P., Pelosi L., Brandolin G., and Klein G. (2007) Mitochondrial carrier family: repertoire and peculiarities of the cellular slime mould Dictyostelium discoideum. Biochimie 89, 1058–1069 [DOI] [PubMed] [Google Scholar]
- 82. Faix J., Steinmetz M., Boves H., Kammerer R. A., Lottspeich F., Mintert U., Murphy J., Stock A., Aebi U., and Gerisch G. (1996) Cortexillins, major determinants of cell shape and size, are actin-bundling proteins with a parallel coiled-coil tail. Cell 86, 631–642 [DOI] [PubMed] [Google Scholar]
- 83. Schneider N., Weber I., Faix J., Prassler J., Müller-Taubenberger A., Kohler J., Burghardt E., Gerisch G., and Marriott G. (2003) A Lim protein involved in the progression of cytokinesis and regulation of the mitotic spindle. Cell Motil. Cytoskeleton 56, 130–139 [DOI] [PubMed] [Google Scholar]
- 84. Benghezal M., Fauvarque M. O., Tournebize R., Froquet R., Marchetti A., Bergeret E., Lardy B., Klein G., Sansonetti P., Charette S. J., and Cosson P. (2006) Specific host genes required for the killing of Klebsiella bacteria by phagocytes. Cell Microbiol. 8, 139–148 [DOI] [PubMed] [Google Scholar]
- 85. Akaza Y., Tsuji A., and Yasukawa H. (2002) Analysis of the gene encoding copper/zinc superoxide dismutase homolog in Dictyostelium discoideum. Biol. Pharm. Bull. 25, 1528–1532 [DOI] [PubMed] [Google Scholar]
- 86. Carninci P., Kasukawa T., Katayama S., Gough J., Frith M. C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Kodzius R., Shimokawa K., Bajic V. B., Brenner S. E., Batalov S., Forrest A. R., Zavolan M., Davis M. J., Wilming L. G., Aidinis V., Allen J. E., Ambesi-Impiombato A., Apweiler R., Aturaliya R. N., Bailey T. L., Bansal M., Baxter L., Beisel K. W., Bersano T., Bono H., Chalk A. M., Chiu K. P., Choudhary V., Christoffels A., Clutterbuck D. R., Crowe M. L., Dalla E., Dalrymple B. P., de Bono B., Della Gatta G., di Bernardo D., Down T., Engstrom P., Fagiolini M., Faulkner G., Fletcher C. F., Fukushima T., Furuno M., Futaki S., Gariboldi M., Georgii-Hemming P., Gingeras T. R., Gojobori T., Green R. E., Gustincich S., Harbers M., Hayashi Y., Hensch T. K., Hirokawa N., Hill D., Huminiecki L., Iacono M., Ikeo K., Iwama A., Ishikawa T., Jakt M., Kanapin A., Katoh M., Kawasawa Y., Kelso J., Kitamura H., Kitano H., Kollias G., Krishnan S. P., Kruger A., Kummerfeld S. K., Kurochkin I. V., Lareau L. F., Lazarevic D., Lipovich L., Liu J., Liuni S., McWilliam S., Madan Babu M., Madera M., Marchionni L., Matsuda H., Matsuzawa S., Miki H., Mignone F., Miyake S., Morris K., Mottagui-Tabar S., Mulder N., Nakano N., Nakauchi H., Ng P., Nilsson R., Nishiguchi S., Nishikawa S., Nori F., Ohara O., Okazaki Y., Orlando V., Pang K. C., Pavan W. J., Pavesi G., Pesole G., Petrovsky N., Piazza S., Reed J., Reid J. F., Ring B. Z., Ringwald M., Rost B., Ruan Y., Salzberg S. L., Sandelin A., Schneider C., Schonbach C., Sekiguchi K., Semple C. A., Seno S., Sessa L., Sheng Y., Shibata Y., Shimada H., Shimada K., Silva D., Sinclair B., Sperling S., Stupka E., Sugiura K., Sultana R., Takenaka Y., Taki K., Tammoja K., Tan S. L., Tang S., Taylor M. S., Tegner J., Teichmann S. A., Ueda H. R., van Nimwegen E., Verardo R., Wei C. L., Yagi K., Yamanishi H., Zabarovsky E., Zhu S., Zimmer A., Hide W., Bult C., Grimmond S. M., Teasdale R. D., Liu E. T., Brusic V., Quackenbush J., Wahlestedt C., Mattick J. S., Hume D. A., Kai C., Sasaki D., Tomaru Y., Fukuda S., Kanamori-Katayama M., Suzuki M., Aoki J., Arakawa T., Iida J., Imamura K., Itoh M., Kato T., Kawaji H., Kawagashira N., Kawashima T., Kojima M., Kondo S., Konno H., Nakano K., Ninomiya N., Nishio T., Okada M., Plessy C., Shibata K., Shiraki T., Suzuki S., Tagami M., Waki K., Watahiki A., Okamura-Oho Y., Suzuki H., Kawai J., Hayashizaki Y., Consortium F., Group R. G. E. R., and Genome Science G. (2005) The transcriptional landscape of the mammalian genome. Science 309, 1559–1563 [DOI] [PubMed] [Google Scholar]
- 87. Otera H., Wang C., Cleland M. M., Setoguchi K., Yokota S., Youle R. J., and Mihara K. (2010) Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J. Cell Biol. 191, 1141–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Fukazawa A., Alonso C., Kurachi K., Gupta S., Lesser C. F., McCormick B. A., and Reinecker H. C. (2008) GEF-H1 mediated control of NOD1 dependent NF-kappaB activation by Shigella effectors. PLoS Pathog. 4, e1000228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Jenkins R. W., Clarke C. J., Canals D., Snider A. J., Gault C. R., Heffernan-Stroud L., Wu B. X., Simbari F., Roddy P., Kitatani K., Obeid L. M., and Hannun Y. A. (2011) Regulation of CC ligand 5/RANTES by acid sphingomyelinase and acid ceramidase. J. Biol. Chem. 286, 13292–13303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Fox J., Azad A., Ismail F., and Storey A. (2011) “Licensed to kill”: tyrosine dephosphorylation and Bak activation. Cell Cycle 10, 598–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Jain P., Luo Z. Q., and Blanke S. R. (2011) Helicobacter pylori vacuolating cytotoxin A (VacA) engages the mitochondrial fission machinery to induce host cell death. Proc. Natl. Acad. Sci. U.S.A. 108, 16032–16037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Yamazaki T., Sasaki N., Nishi M., Yamazaki D., Ikeda A., Okuno Y., Komazaki S., and Takeshima H. (2007) Augmentation of drug-induced cell death by ER protein BRI3BP. Biochem. Biophys. Res. Commun. 362, 971–975 [DOI] [PubMed] [Google Scholar]
- 93. Fajardo M., Schleicher M., Noegel A., Bozzaro S., Killinger S., Heuner K., Hacker J., and Steinert M. (2004) Calnexin, calreticulin and cytoskeleton-associated proteins modulate uptake and growth of Legionella pneumophila in Dictyostelium discoideum. Microbiology 150, 2825–2835 [DOI] [PubMed] [Google Scholar]
- 94. Fujimoto T., Machida T., Tsunoda T., Doi K., Ota T., Kuroki M., and Shirasawa S. (2011) Determination of the critical region of KRAS-induced actin-interacting protein for the interaction with inositol 1,4,5-trisphosphate receptor. Biochem. Biophys. Res. Commun. 408, 282–286 [DOI] [PubMed] [Google Scholar]
- 95. Craige B., Salazar G., and Faundez V. (2008) Phosphatidylinositol-4-kinase type II alpha contains an AP-3-sorting motif and a kinase domain that are both required for endosome traffic. Mol. Biol. Cell 19, 1415–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Nanahoshi M., Tsujishita Y., Tokunaga C., Inui S., Sakaguchi N., Hara K., and Yonezawa K. (1999) Alpha4 protein as a common regulator of type 2A-related serine/threonine protein phosphatases. FEBS Lett. 446, 108–112 [DOI] [PubMed] [Google Scholar]
- 97. MacAskill A. F., Brickley K., Stephenson F. A., and Kittler J. T. (2009) GTPase dependent recruitment of Grif-1 by Miro1 regulates mitochondrial trafficking in hippocampal neurons. Mol. Cell. Neurosci. 40, 301–312 [DOI] [PubMed] [Google Scholar]
- 98. Zhou Q., Kee Y. S., Poirier C. C., Jelinek C., Osborne J., Divi S., Surcel A., Will M. E., Eggert U. S., Müller-Taubenberger A., Iglesias P. A., Cotter R. J., and Robinson D. N. (2010) 14–3-3 coordinates microtubules, Rac, and myosin II to control cell mechanics and cytokinesis. Curr. Biol. 20, 1881–1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Buczynski G., Bush J., Zhang L., Rodriguez-Paris J., and Cardelli J. (1997) Evidence for a recycling role for Rab7 in regulating a late step in endocytosis and in retention of lysosomal enzymes in Dictyostelium discoideum. Mol. Biol. Cell 8, 1343–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Cardoso C. M., Jordao L., and Vieira O. V. (2010) Rab10 regulates phagosome maturation and its overexpression rescues Mycobacterium-containing phagosomes maturation. Traffic 11, 221–235 [DOI] [PubMed] [Google Scholar]
- 101. Arasaki K., Toomre D. K., and Roy C. R. (2012) The Legionella pneumophila effector DrrA is sufficient to stimulate SNARE-dependent membrane fusion. Cell Host Microbe 11, 46–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Kedra D., Pan H. Q., Seroussi E., Fransson I., Guilbaud C., Collins J. E., Dunham I., Blennow E., Roe B. A., Piehl F., and Dumanski J. P. (1998) Characterization of the human synaptogyrin gene family. Hum. Genet. 103, 131–141 [DOI] [PubMed] [Google Scholar]
- 103. Pryor P. R., Jackson L., Gray S. R., Edeling M. A., Thompson A., Sanderson C. M., Evans P. R., Owen D. J., and Luzio J. P. (2008) Molecular basis for the sorting of the SNARE VAMP7 into endocytic clathrin-coated vesicles by the ArfGAP Hrb. Cell 134, 817–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Peretti D., Dahan N., Shimoni E., Hirschberg K., and Lev S. (2008) Coordinated lipid transfer between the endoplasmic reticulum and the Golgi complex requires the VAP proteins and is essential for Golgi-mediated transport. Mol. Biol. Cell 19, 3871–3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Homma K., Yoshida Y., and Nakano A. (2000) Evidence for recycling of cytochrome P450 sterol 14-demethylase from the cis-Golgi compartment to the endoplasmic reticulum (ER) upon saturation of the ER-retention mechanism. J. Biochem. 127, 747–754 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.