Abstract
Brain cancers demonstrate a complex metabolic behavior so as to adapt the external hypoxic environment and internal stress generated by reactive oxygen species. To survive in these stringent conditions, glioblastoma cells develop an antagonistic metabolic phenotype as compared to their predecessors, the astrocytes, thereby quenching the resources expected for nourishing the neurons. The complexity and cumulative effect of the large scale metabolic functioning of glioblastoma is mostly unexplored. In this study, we reconstruct a metabolic network comprising of pathways that are known to be deregulated in glioblastoma cells as compared to the astrocytes. The network, consisted of 147 genes encoding for enzymes performing 247 reactions distributed across five distinct model compartments, was then studied using constrained-based modeling approach by recreating the scenarios for astrocytes and glioblastoma, and validated with available experimental evidences. From our analysis, we predict that glycine requirement of the astrocytes are mostly fulfilled by the internal glycine–serine metabolism, whereas glioblastoma cells demand an external uptake of glycine to utilize it for glutathione production. Also, cystine and glucose were identified to be the major contributors to glioblastoma growth. We also proposed an extensive set of single and double lethal reaction knockouts, which were further perturbed to ascertain their role as probable chemotherapeutic targets. These simulation results suggested that, apart from targeting the reactions of central carbon metabolism, knockout of reactions belonging to the glycine–serine metabolism effectively reduce glioblastoma growth. The combinatorial targeting of glycine transporter with any other reaction belonging to glycine–serine metabolism proved lethal to glioblastoma growth.
Electronic supplementary material
The online version of this article (doi:10.1007/s11693-015-9183-9) contains supplementary material, which is available to authorized users.
Keywords: Astrocyte, Glioblastoma, Metabolic demand reaction, Mitochondrial ATP synthesis, Glycine, Cystine
Introduction
Human brain, in order to ensure its proper functioning, has to account for a highly perplexing conduct which is maintained by the interplay between its distinctive cell sorts. An emerging area of interest in the last decade, pertaining to brain metabolism, has been the study of behavioral aspects of astrocytes (Bouzier-Sore and Pellerin 2013) and their cancerous counterpart, glioblastoma (Wolf et al. 2010). Several studies have been performed to understand the metabolic alterations incurred within the astrocytes, which lead to their phenotypic manifestation as glioblastoma (Chinnaiyan et al. 2012). However, the cumulative effect of a large scale metabolism on the metabolic functioning of glioblastoma still remains unaddressed. The effect of the mutual connectivity of the individual pathways within its metabolic network and the difference in response they show in the astrocytic and glioblastoma scenarios is also largely unknown. Glioblastoma cells can exhibit diversified response to same stimulus and show a great metabolic heterogeneity, which enable them to thrive even in a glucose starved condition (Griguer et al. 2005). A few of the metabolic phenomena, such as a higher accumulation of glycine in the glioblastoma cells (Hattingen et al. 2009) and the disruption of primary brain tumor growth with the inhibition of cystine (Chung et al. 2005), are known, but the reason to such behavior is still not understood properly. Knowledge about the alternative metabolites, which help the glioblastoma cells to thrive with altered metabolism, also remains largely unexplored.
One of the pioneering work in cancer biology was the discovery of Warburg effect in 1924 (Warburg 1956), which suggested that cancer cells might adapt to a primitive glycolytic pattern of embryonic cells and mitochondrial injury as well as metabolic switching of glycolysis to aerobic glycolysis might be essential for cancer development. Several studies have been carried out to delineate the advantage of such a modification in the tumorous cells. These phenomena are also observable in glioblastoma, enabling them to suffice their rapacious requirements (Zhou et al. 2011). In addition to this remarkable discovery, several other experimental and statistical analyses have been conducted to delineate the phenomenal changes in the properties of glioblastoma as an effect of metabolic alterations in different enzymes belonging to different pathways like tryptophan metabolism (Sahm et al. 2013), cysteine metabolism (Ye et al. 1999), glutamine and glutamate metabolism (Wise et al. 2008). Properties of these individual metabolic pathways have been studied in both astrocytes and glioblastoma, but the difference in their response as a part of a large metabolic network, in the two scenarios, is yet to be studied. A new arena of in silico studies have also been employed in the past decade to obtain a large-scale network understanding of glioblastoma. Different types of dynamic modeling approaches, such as spatiotemporal modeling (Burgess et al. 1997; Tracqui et al. 1995), partial differential equation modeling (Swanson et al. 2003), ordinary differential equations, have been used to detect the growth and invasion of glioblastoma cells (Mandonnet et al. 2008). These studies, however, are only a partial picture to the unaddressed questions, and hence, further studies are required to address the same. Further, the aforementioned studies have been largely limited to understand the metabolism of glioblastoma in parts, and have not been focused to identify or predict feasible drug targets on a network scale. Moreover, these studies have also overlooked the context-dependent understanding of glioblastoma metabolism and its role in achieving specific biological goals (Banerji 2013).
Varieties of chemotherapeutic agents are available commercially to treat cancer, possessing a high degree of target specificity and better clinical manifestation. Gleevec (imatinib), Iressa (gefitinib), Herceptin (trastuzumab), rituximab are a few examples of presently available therapeutics. However, due to multiple genetic and epigenetic alterations, the progression and disease manifestation of cancer turns out to be a complex phenomenon to understand. The malignant cancer cell populations become heterogeneous even within a specific cancer type containing diverse genetic changes, which further alters over time due to genetic instability (Pelicano et al. 2006). A multiple targeting approach in this scenario is favored over single targets to effectively deal with the random mutations generated in a cancer population. The effectiveness of the available therapeutics also has to be monitored, as many of the existing therapeutics are potentially harmful to the normal tissues too and are neurotoxic in nature.
In the present work, to understand the complex differences in the metabolic behavior of astrocyte and glioblastoma, we develop a context-specific constraint-based model for astrocyte/glioblastoma metabolism, and analyze it using flux balance analysis (FBA). For specific comparison between the two scenarios, we have considered those pathways which are known to get deregulated in glioblastoma when compared to the normal astrocyte. Our model accommodates a total of 13 pathways, the abnormal functioning of which have been reported in glioblastoma literature. The model has 247 reactions, with 39 exchange reactions and 69 transport reactions associated with 147 genes. Analyzing this large network using flux balance analysis, the differences in the individual pathway response as a part of large metabolic network in astrocyte and glioblastoma scenarios were delineated. It further aims to capture the properties of the included pathways and metabolites in glioblastoma cells (which help in its growth), to understand the differences in the uptake and utilization of metabolites (which can be categorized as essential and non-essential) and release of overflow metabolites in the two scenarios, and to predict probable chemotherapeutic targets through in silico single and double reaction knockout analyses of the glioblastoma model.
The results generated from both astrocytic and glioblastoma scenarios corroborated qualitatively with the experimentally available information further validating its feasibility in predicting biologically reasonable phenomena. By analyzing the steady state flux profiles generated by flux balance analysis of the model, the fate of a few metabolites, their essentiality in glioblastoma growth and the path followed by them to contribute to the optimization of objective functions (mitochondrial ATP synthesis and glioblastoma growth) were interpreted. Single and double reaction knockout analyses were performed, to determine the essentiality of the reactions involved in the metabolic network, in governing the growth properties of glioblastoma. Potential drug targets were identified from those set of essential reactions. To determine the extent of regulation that could be imposed on those drug targets and to analyze them quantitatively, they were further simulated for chemotherapeutic intervention scenarios with the motive of either reducing the glioblastoma growth to zero, or to reduce it to the growth rate of a normal astrocyte.
Thus, our simple but extensive modeling approach provides a deep insight into the consequences of glioblastoma metabolism substantiating the strength of suitable in silico approaches in understanding metabolic networks and predicting biologically reasonable disease scenarios. Additionally, the classification of reaction knockouts combined with simulation of chemotherapeutic interventions could largely predict reaction pairs as feasible drug targets, further supporting the large-scale applicability of constraint-based models in predicting reasonable chemotherapeutic target combinations.
Materials and methods
Model reconstruction
Information regarding the association of enzymes to crucial metabolic reactions, their appropriate subcellular locations, transports and exchanges were compiled using a variety of data sources. The basis of this reconstruction was to identify the gene–protein–reaction (GPR) network along with appropriate transports and exchanges. The GPR was reconstructed considering reactions that contribute to ATP synthesis and glioblastoma growth.
The reactions considered in the model and their corresponding Enzyme Commission Numbers (EC Numbers) were curated from Expasy Enzyme (Bairoch 2000) and KEGG (Kanehisa et al. 2014). The genes to the enzymatic reactions considered in the model were obtained from NCBI Gene (Edgar et al. 2002). Molecular function of these reactions and their biological process was obtained from UniProt (Consortium 2014), KEGG (Kanehisa et al. 2014) and through literature survey. Information regarding the subcellular localization of the reactions was compiled through extensive literature search and those reactions, for which literature support for subcellular localization was limited or not available; cytosol was taken to be the default compartment of the reaction. A list of reactions, their corresponding genes, enzymes, UniProt ID and KEGG ID was compiled with appropriate literature support to gather evidences related to biological significance and subcellular localization of the reactions (Online Resource 1). Most of the internal reactions along with 12 transport reactions were associated with their corresponding genes, which accounted for 147 genes in the model. All the metabolites and the corresponding reactions in which they were involved were distributed into five different compartments: Extracellular space, Cytoplasm, Mitochondria, Mitochondrial intermembrane space and Nucleus. All these information were organized in the rBioNet toolbox, a MATLAB extension of the COBRA Toolbox (Schellenberger et al. 2011), to reconstruct the constraint-based metabolic model. The reconstructed metabolic network consisted of 13 pathways that are significantly affected during the transformation from astrocyte to glioblastoma (see Table S1 of Online Resource 2). The detailed pathway diagram has been drawn in CellDesigner version 4.3 and has been provided in Fig. S1 of Online Resource 2.
Flux balance analysis (FBA)
Flux balance analysis is a mathematical approach to analyze the flow of metabolites through a metabolic network, where the metabolic reactions are represented in a tabulated form of reaction matrix, of stoichiometric coefficients of each reaction. In our metabolic network, this relationship was established between the metabolites and the reactions in the form of an S-matrix which comprised of 159 metabolites and 247 reactions, building up the S-matrix of dimension ‘159 × 247’. The score assigned to each element of the S-matrix, Sxy, represented the stoichiometry of the metabolite ‘x’ in the reaction ‘y’. A positive score signified the production of the metabolite and a negative score implied its consumption in the reaction. The column vector v had 247 fluxes, including 39 exchange reactions and 69 transport reactions. FBA formalizes the flux distribution through the whole metabolic network as the dot product of the S-matrix with the vector v. All the reactions in the model were organized in the rBioNet toolbox, where their fluxes were constrained between a lower bound v lb and an upper bound v ub. All the reversible reactions were bounded between v lb = −1000 and v ub = 1000. The irreversible reactions in the model were bounded either from 0 to 1000 or −1000 to 0 with respect to the substrate and products defined for that reaction as per available information from literature. The bounds to the exchange reactions were fixed as per the requirement of the system for uptake or release of the exchange metabolites. Those exchanges which were known to be taken in were bounded between [−1000 to 0] and those which were known to be released out were bounded between [0 to 1000]. Rest of the exchanges was bounded between [−1000 to 1000] to analyze their role in the metabolism by simulating the model using FBA.
Selection of objective function
The metabolic requirement of the cancerous cells (glioblastoma, in the present case) is not completely sufficed by diverting flux towards production of ATP through Oxidative Phosphorylation, which necessitates the requirement of an altered metabolism which can fulfill both the energy and metabolic requirement for the growth of the cells. Therefore, in our study, we defined two objective functions:
- ATP synthesis through oxidative phosphorylation (ATPSyn)
1 - a metabolic demand reaction that will dually fulfill the requirements of growth and ATP (GBM_BM). To define the metabolic requirement of the model ribose-5-phosphate, r5p(c), oxaloacetate, oaa(m), succinate, succ(m) and glutathione glt(c) were included as components of the objective function, selected on the basis of their contribution as (a) precursor to the nucleotide biosynthesis and synthesis of amino acids like valine, lysine, methionine, threonine, etc. (Lee et al. 2006), (b) intermediates for maintaining redox balance in different cellular compartments and biosynthesis of other cellular components required for cell growth (Covert et al. 2001; Pistollato et al. 2010), (c) preventing damage to cellular components caused by reactive oxygen species produced due to hypoxia or other cellular stress (Chung et al. 2005):
2
Creation and validation of astrocytic and glioblastoma scenario
Astrocytic brain tumors, commonly known as glioblastoma, are the most frequent human brain tumors, encompassing 50 % of the cases (Jellinger 1977). These emerge as manifestations of multiple alterations in the metabolic (Wolf et al. 2010) and signaling pathways (Kleihues and Ohgaki 2000) of astrocytes. Hence, in the model, selected pathways which were known to be deregulated in the astrocyte-derived glioblastoma (see Table S1 of Online Resource 2) were considered to define the metabolic differences between astrocyte and glioblastoma scenarios. Bounds to the flux through a few enzymes which defined the differences between the two scenarios were assigned on the basis of literature support. Both the objective functions were optimized for the two scenarios. Limited bounds were assigned to a few reactions to create the astrocyte scenario. The rest of the reactions fluxes were allowed to vary between a wide range of [−1000 to 1000] or [0 to 1000] or [−1000 to 0] as per the reversibility or irreversibility of the reactions. The model was then simulated to obtain results that were in accordance with the experimentally available data defining the features of astrocyte (Mangia et al. 2009; Marrif and Juurlink 1999; Pellerin and Magistretti 1994). Bounds to the mitochondrial reactions—‘glutaminase’ [−50, 50], ‘glutamate dehydrogenase’ [−150, 150], ‘mitochondrial pyruvate carboxylase’ [−10, 10] and cytoplasmic reactions—‘acetyl-CoA carboxylase’ [0, 100], ‘L-carnitine O-palmitoyltransferase’ [0, 20], and ‘cytoplasmic malate dehydrogenase’ [−50, 50], were fixed and the model was analyzed using FBA to create the astrocytic scenario.
Perturbations were performed to the same astrocytic model by varying the lower and upper bounds to a few reactions that were experimentally found to be deregulated in glioblastoma, and then the model was simulated to create the glioblastoma scenario. Bounds were released to a few reactions, which were imposed in the astrocytic scenario: ‘glutaminase’ [−1000, 1000] and ‘acetyl-CoA carboxylase’ [0, 1000]. New bounds were assigned to another set of reactions to generate the glioblastoma scenario: ‘glutamate dehydrogenase’ [−200, 200], ‘Cytochrome c Oxidase (complex IV)’ [−10, 10], ‘Trans_Glutamate (ATP)’ [−90, 90] and ‘glycine exchange’ [−500, 500]. This model was analyzed using both ‘ATPSyn’ and ‘GBM_BM’ as objective function. This model was again validated with experimental data available for glioblastoma (Hertz and Zielke 2004; Wise et al. 2008; Ye et al. 1999). The effectiveness of GBM_BM in determining the growth properties of glioblastoma was verified with experimental evidence. The result has been provided in Fig. S2 of Online Resource 2.
In-silico prediction of minimal essential metabolite for glioblastoma growth
Glioblastoma cells are grown in commercially available MEM or DMEM media (Anton and Glod 2014; Guessous et al. 2013; Ye et al. 1999). However, due to lack of sufficient literature that report the essential metabolites required for glioblastoma growth even at glucose starved conditions, an in silico simulation was performed to check the fate of some key metabolites that contribute to the growth in the glioblastoma. The entry of each carbon source was considered in the model, one at a time and the corresponding solution of the GBM_BM objective function (growth) was computed. Also, the fate of the most essential metabolite with another input carbon source within the model was checked and the optimal solution of the GBM_BM objective was calculated. This was performed to identify the most important carbon sources required for enhancing glioblastoma growth.
Single and double reaction knockouts in glioblastoma
A reaction knockout strategy was chosen, instead of gene knockout approach, to completely nullify the functional effect of the reaction in the network. Reaction knockout predictions allowed the identification of reactions that could be targeted for either completely inhibiting or reducing the glioblastoma growth. Each of the 247 reactions in the metabolic network was knocked down individually to predict the mutations that could be lethal to the glioblastoma growth. For performing the knockout, flux through each reaction in the network was constrained to zero and solution of the GBM_BM objective function was computed for each knockout. Double reaction knockouts were also performed, with a combination of two reactions to be knocked down simultaneously. The single and double knockouts were classified on the basis of percentage reduction of flux through the objective function, GBM_BM, from its optimal value. The optimal value of the objective function for the astrocytic scenario in the model corresponded to the normal growth rate.
Simulation of chemotherapeutic interventions
Further, we filtered the results of double reaction knockouts for identifying feasible chemotherapeutic combinations to target glioblastoma growth. For performing in silico chemotherapeutic interventions, we divided the reactions into three groups as per their essentiality. We predicted the putative feasible ranges for each of these reaction combinations, in which chemotherapeutics can effectively target glioblastoma growth either for its complete inhibition or bring it back to the normal astrocytic growth rate.
Results
Properties of the model
The present context-specific model for glioblastoma metabolism has a total of 247 reactions, with 39 exchange reactions and 69 transport reactions. Most of the internal reactions along with a few transport reactions have been associated with their corresponding genes, which accounts for 147 genes in the model.
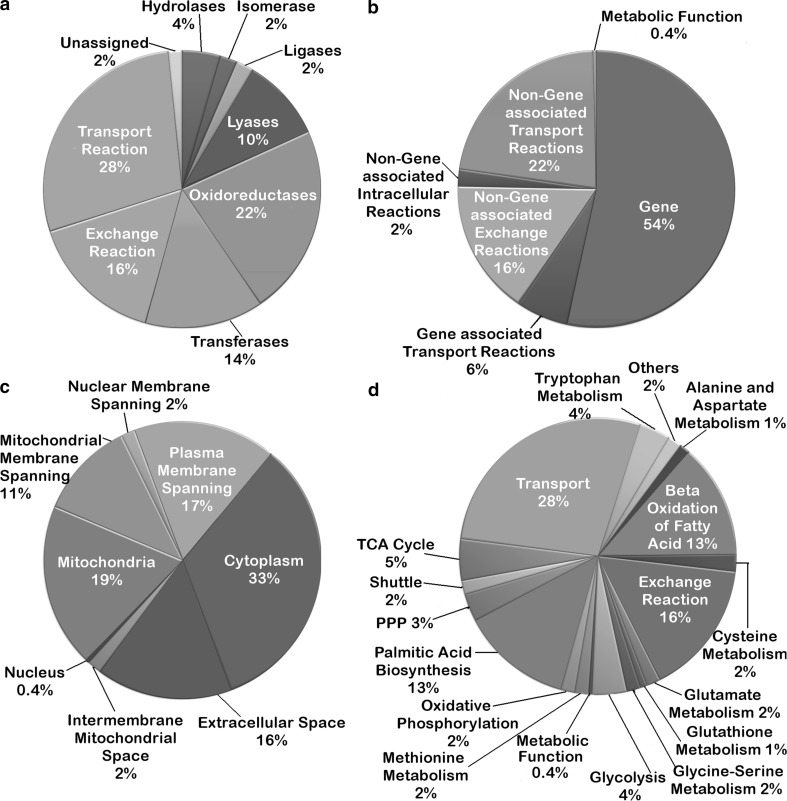
The present model for glioblastoma metabolism can be classified on the basis of the following four categories: (1) enzyme commission number, (2) gene non-gene association, (3) sub-cellular locations, and (4) metabolic processes (Fig. 1). A large number of the reactions in the model belonged to the class 1 category of enzyme classification i.e., the oxidoreductases (22 %). These set of enzymes catalyze the oxidation of one chemical species and the simultaneous reduction of the other by transfer of electrons from one species to another. The other classes of enzymes in this classification scheme were the transferases (14 %) followed by lyases (10 %), hydrolases (4 %), isomerases (2 %), and ligases (2 %). Another 28 % of the reactions belonged to transport reactions and 16 % to extracellular exchange reactions, which occurred spontaneously in the system (Fig. 1a).
Fig. 1.
Classification of the properties of reconstructed metabolic model. The model reconstruction has been classified on the basis of a enzyme commission number or E.C. number, b gene-non gene association, c cellular compartments, and d metabolic processes, respectively
The reactions can also be classified on the basis of their association with genes to understand gene reaction associations (Fig. 1b). 60 % of the model reactions were gene-associated, out of which 6 % were transport reactions. The rest of the reactions were classified as: Non-Gene associated Exchange Reactions (16 %), Non-Gene associated Intracellular Reactions (2 %) and Non-Gene associated Transport Reactions (22 %).
In the classification shown in Fig. 1c, the cytosolic and mitochondrial reactions contributed to 54 % of the total reactions in the model. 2 % of the reactions belonged to the mitochondrial intermembrane space model compartment that specifically accounted for oxidative phosphorylation. The transport reactions were categorized according to the membrane to which it is associated. Transports accounted for 30 % of the total reactions: Mitochondrial membrane spanning (11 %), Nuclear membrane spanning (2 %) and Plasma Membrane spanning (17 %).
With reference to the metabolic processes, 23 % of the reactions belonged to fatty acid metabolism inclusive of both biosynthesis and beta oxidation of palmitic acid. The rest of the pathways contributed to 30 % of the total count of which 14 % belonged to Glycolytic, PPP, TCA cycle and Oxidative phosphorylation pathway and 2 % were contributed each by Glycine–Serine Metabolism, Cysteine Metabolism, Methionine Metabolism and Glutamate Metabolism, without taking into account the transport and exchange reactions. Another set of reactions, namely, cytosolic ATPase (ATPS), cytoplasmic malate dehydrogenase (MDH(Cyto)), Phosphoenolpyruvate carboxykinase (GTP) (PEP_CarbK_1), mitochondrial pyruvate carboxylase (Pyr_Carbm) which could not be assigned strictly under any particular pathway, were categorized as ‘Others’ which contributed 2 % of reactions to the (Fig. 1d).
Validation of astrocytic and glioblastoma model
The reconstructed metabolic model was validated for both the astrocytic and the glioblastoma scenarios, using mitochondrial ATP synthesis as the objective function. The astrocytic scenario was created first, by fixing bounds of a few reactions. Few known perturbations from experiments were introduced to the astrocytic scenario so as to create the glioblastoma scenario (See “Materials and methods” section for details).
Astrocyte
Required changes were made to the bounds of certain reactions during simulation of the astrocyte model and the optimal range of bounds within which it showed the properties of astrocyte was estimated (see “Materials and methods” section). The model astrocyte scenario was analyzed and validated, using mitochondrial ATP synthesis (ATPSyn) as the objective function. The astrocyte scenario was validated for a number of experimental observations, such as pyruvate recycling, lactate production and effect of glutamate.
Astrocytes prefer a glucose-dependent metabolism where glucose is catabolized to pyruvate that enters the TCA cycle thereby leading to ATP synthesis (Mangia et al. 2009) and partly to the formation of lactate so as to suffice the neuronal requirement. This property was examined in the model astrocytic scenario by performing a robustness analysis of glucose uptake with increasing oxygen uptake. The default flux balance analysis (FBA) of model astrocytic scenario suggested an optimal flux of 160 for oxygen uptake from the environment. The uptake of oxygen was thus, varied up to its optimal flux and its effect on glucose uptake was observed. Increase in oxygen uptake led to linear but proportional increase in glucose uptake (Fig. 2a). This inferred the utilization of glucose to produce lactate by the astrocytes without affecting the mitochondrial respiratory chain. Further, a slight dip in the glucose uptake rate was observed when the flux through the oxygen uptake was more than 130. But simultaneously, flux through mitochondrial ATP synthesis continued to increase, signifying that the decrease in glucose uptake did not affect the ATP synthesis. The probable reason for this may be the recycling of pyruvate from the TCA cycle intermediates. Reports also suggest that one of the TCA cycle intermediates, citrate, may produce oxaloacetate, which is subsequently converted to pyruvate through the activity of malic enzyme or by the combined activity of PEP carboxykinase and pyruvate kinase (Sonnewald et al. 1996).
Fig. 2.
Validation of astrocyte scenario. Properties of astrocyte: a increase in glucose uptake driven towards mitochondrial ATP synthesis and lactate production, b increase in the activity of lactate dehydrogenase and pyruvate kinase in hypoxia conditions, c increase in glucose utilization and lactate production with increasing glutamate uptake
Similar to this, model simulations suggested recycling of pyruvate by utilization of TCA produced oxaloacetate through PEP carboxykinase and pyruvate kinase reactions. This resulted in a reduced dependence of pyruvate production on glucose uptake. The pyruvate so formed was catabolized into the TCA cycle and compensated for maintaining ATP production proportional to oxygen uptake.
The activity of lactate dehydrogenase and pyruvate kinase increased during anoxic conditions as compared to normoxic conditions in astrocytes (Marrif and Juurlink 1999). To verify this property, normoxic and hypoxic conditions were created in the model by constraining the oxygen uptakes at the optimum (flux value = −120) and low (flux value = −2) values and ensuring sufficient glucose uptake in the model. It was observed qualitatively that the model is capable of capturing this feature of astrocytes (Fig. 2b). Although the actual experimental result was generated by incubating the astrocyte cells in a completely oxygen deprived anoxic condition for 6 h, creating such a situation in the in silico analysis would lead to zero ATP synthesis (objective function considered for validation) in the model due to its dependence on oxygen. Hence, the property was verified for hypoxic conditions only. Similar pathway-based down-regulation in activity of certain proteins under hypoxic stress has been reported earlier (Maity et al. 2000) but for the signaling pathways.
In astrocytes, the uptake of glucose increases with increase in glutamate uptake thus leading to increased lactate production (Pellerin and Magistretti 1994). This situation was created in the model by regulating the exchanges of glucose, glutamate, glutamine and oxygen. By varying the glutamate uptake from 0 to 450, a corresponding increase in glucose uptake and hence, lactate production was observed during model simulations. Further, it was observed that highest lactate production was at a glutamate uptake flux of 450 (Fig. 2c).
Glioblastoma
In the model, the astrocytic scenario was further perturbed to create the glioblastoma scenario. To validate the glioblastoma scenario, steady state fluxes of certain reactions obtained by simulating for glioblastoma scenario was compared with that of the astrocyte scenario, while keeping all the nine inputs open to the system. The “Warburg effect”, which states a reduction of ATP production through mitochondrial respiration and an increase in glucose utilization to increase the flux towards aerobic glycolysis (Zhou et al. 2011), was observed in the glioblastoma scenario (Fig. 3a).
Fig. 3.
Validation of glioblastoma scenario. Properties of glioblastoma: a reduced mitochondrial ATP synthesis and increased glucose utilization in glioblastoma scenario; b reversal in glutamate and glutamine utilization and increase in cystine uptake in glioblastoma scenario
For further validation of this scenario, few experimental observations were replicated from the model. Glutaminolysis is a known property of glioblastoma cells, where the uptake and utilization of glutamine is favored over glutamate for compensating the loss of glutamate through cysteine–glutamate antiporter (Wise et al. 2008), which is a property that is exactly opposite to that of astrocytes (Hertz and Zielke 2004). Also, uptake of cystine increases in the glioblastoma cells due to enhanced activity of cystine–glutamate antiporter (Ye et al. 1999). All these differences in the exchange properties of glioblastoma could be observed through our model, when the entry of all the input metabolites was allowed (Fig. 3b).
Validation of glioblastoma metabolic demand reaction (GBM_BM)
A separate metabolic demand reaction was also introduced in the model glioblastoma scenario so as to understand the influence of different metabolites on glioblastoma growth. Considering this reaction as the cellular objective, the glioblastoma scenario was further studied for its metabolic properties. All the further analyses have been performed keeping the GBM_BM metabolic demand reaction as the objective function. For the verification of the objective function—‘GBM_BM’ in representing the properties of glioblastoma, a qualitative analysis was performed to compare the activity of certain reported reactions in the astrocytic and glioblastoma scenario. The fold change in activity from astrocytic to glioblastoma scenario as predicted from the model was compared to existing proteome data extracted from young glioblastoma patients (Deighton et al. 2014). The results of this comparison are listed in Table 1. Data was available as fold change in expression for eight reactions of the model. Out of the eight reactions, predicted activity for five reactions was qualitatively found to be in correspondence with the experimental observations.
Table 1.
Comparison of model prediction with the data available for enzyme expression in young patients
| Uniprot ID | Reaction name | Model abbreviation | Fold change | Model prediction | Gene abbreviation | Fold change | Experimental prediction |
|---|---|---|---|---|---|---|---|
| O43175 | D-3-phosphoglycerate dehydrogenase | PGDH | 0.9313 | D | PHGDH | 0.55 | D |
| P04075 | Fructose-bisphosphate aldolase A | FBA | 0.9175 | D | ALDOA | 0.71 | D |
| P50213 | Isocitrate dehydrogenase [NAD] subunit alpha, mitochondrial | IDH | 0.0000 | D | IDH3A | 0.48 | D |
| P18669 | Phosphoglycerate mutase 1 | PGM | 2.4046 | U | PGAM1 | 1.6 | U |
| Q9Y617 | Phosphoserine aminotransferase | PST | 0.9313 | D | PSAT1 | 0.53 | D |
| P00367 | Glutamate dehydrogenase, mitochondrial | GlutDH | 0.0000 | D | GLUD1 | 1.4 | U |
| P60174 | Triosephosphate isomerase | TPI | 0.7401 | D | TPI1 | 2.1 | U |
| P17174 | Aspartate aminotransferase, cytoplasmic | ASPTc | 1.0732 | U | GOT1 | 0.53 | D |
N.B.: Regulation in enzymatic expression (up-regulation or ‘U’ and down-regulation or ‘D’) for eight reactions present in our model could be related to the enzymatic profile available for young glioblastoma patients
Opposing roles of glycine and glutamate uptake in astrocytes
Evidences state that glycine content of neuronal cells was higher than that of glial cells (Roux and Supplisson 2000). Also, most of the CNS tissues sufficed their glycine requirement via the internal glycine–serine metabolism pathway derived from glucose via 3-phosphoglycerate (Nicklas and Browning 1978), even though astrocyte cultures fed with glycine were capable of utilizing it by maintaining intracellular levels of glutathione, serine and creatine (Dringen et al. 1998). Further, high uptake of glycine in astrocytes was observed to be tightly coupled with high secretion of Na+ and Cl− (Zafra and Gimenez 1986). In the model, we have assumed the uptake of glycine to be a normal uptake devoid of its coupling with Na+ and Cl−. To simulate the effect of variation in extracellular Na+ and Cl− ions on glycine uptake, the bounds on glycine uptake in the astrocytic scenario was varied. Further, bounds on uptake of all the aforementioned input metabolites were released so as to allow their utilization within the model. It was observed that glycine uptake had a dominant influence on glutamate/glutamate uptake cycle of astrocytes. Glutamate taken in by the cell is catabolized through TCA, glutamine production and release, and release of glutamate through cystine–glutamate antiporter. Under low glycine uptake (flux value of glycine uptake <−60), most of the glutamate taken into the cell, forms glutamine and is released from the cell ensuring lower uptake of cystine and hence, less glutamate release through cystine–glutamate antiporter (Fig. 4). Whereas, increased glycine uptake is driven towards increased synthesis of glutathione. Simultaneously, an equal increase in flux through cystine uptake (cystine–glutamate antiporter) also takes place. Cystine is then provided for cysteine biosynthesis, which combines with glycine for glutathione production. For increased uptake of cystine, an equal efflux of glutamate through the antiporter is also required. Glutamate required for this efflux is provided by its uptake through glutamate-ATP transporter. To compensate for the amount of glutamate lost through efflux, glutamine uptake increases and this glutamine leads to glutamate synthesis. This result suggests that the astrocyte prefers higher glycine uptake so as to combat oxidative stress whereas a lower uptake of glycine is preferred when there is a higher requirement of glutamine by the surrounding neurons.
Fig. 4.
Effect of glycine uptake on glutamate utilization by astrocyte. Change in the uptake and release of glutamate, glutamine and cystine–glutamate antiporter with increasing uptake of glycine in the astrocyte scenario. The uptake of glutamate reduces with increasing uptake of glycine and the glutamine exchange reverses its direction of flow of flux
Difference in pathway response between the astrocytic and glioblastoma scenarios
Cells tend to either maximize ATP synthesis or optimally use metabolites from the environment to satisfy their cellular demand for optimum growth. The choice of an objective function that can be used to capture actual biological scenarios is a primary requirement for performing FBA. To understand the roles of the aforementioned cellular objectives, the model was simulated in both the astrocytic and glioblastoma scenarios for the two objective functions: mitochondrial ATP synthesis and GBM_BM metabolic demand reaction separately (See “Materials and methods” section).
Maximization of mitochondrial ATP synthesis
FBA simulations for maximization of ATP synthesis revealed a number of metabolic features of the glioblastoma scenario.
Increase in glycolytic flux in glioblastoma Simulations for ATP synthesis as the objective function demonstrated a significant increase in the flux through the glycolytic and pentose phosphate pathways in the glioblastoma scenario as compared to the astrocyte but a corresponding decrease in ATP synthesis (Fig. 5a, b). To create the glioblastoma scenario, a reduced activity of Complex IV of the electron transport chain was assumed (Chatterjee et al. 2006). ATP synthesis is largely dependent on Complex IV for redox balance. Hence, decreased ATP synthesis is observed. Under the reduced activity of Complex IV, the deficiency of electrons for ATP synthesis is partly met through Complex I and III of electron transport chain. This led to an increased synthesis of oxaloacetate from phosphoenolpyruvate through the PEP carboxykinase and aspartate aminotransferase reactions. Hence, flux through glycolysis is largely increased in the glioblastoma scenario for the provision of phosphoenolpyruvate. The glycolytic dependence of ATP synthesis is a unique feature of glioblastoma cells (when compared to astrocytes) that could be captured from the model (Vander Heiden et al. 2009). A comparison of the steady state flux profiles of astrocyte versus glioblastoma with ATPSyn as objective has been provided in Table S2 of Online Resource 2.
Increased cystine uptake in glioblastoma Simulations for ATP synthesis as the objective function also demonstrated an increased uptake of cystine (Fig. 5c). The total flux of cystine is distributed into cysteine biosynthesis which is then distributed towards a relatively low amount of glutathione biosynthesis (Fig. 5f) and largely towards production of pyruvate through the cysteine dioxygenase (CD), cysteine sulfinate transaminase (CST), and the spontaneous 3snpyr (SPON1) reactions. This pyruvate is utilized for acetyl coA synthesis and hence, the biosynthesis of fatty acids which are further released in the extracellular environment.
Increased catabolism of glutamine in glioblastoma Reactions belonging to glutamate metabolism showed a higher activity which was due to higher glutaminolysis in glioblastoma scenario (Wise et al. 2008). This was due to the uptake of glutamine by the glioblastoma cells, from external medium, which was converted to glutamate within the cell. The glutamate that was formed was mostly used by the cystine–glutamate antiporter (anti_cystine_glut) in order to uptake cystine. Cystine then is utilized in the cysteine metabolism pathway for pyruvate synthesis that enters TCA cycle (Fig. 5d).
Decreased glycine–serine biosynthesis in glioblastoma Simulations of the model for ATP synthesis as the objective function also demonstrated an increased uptake of glycine (Fig. 5e). It could be observed that in glioblastoma, glycine was preferred to be taken inside the cell whereas in contrary it is being synthesized within the astrocyte. This was because the glycolytic flux was completely utilized into TCA cycle for maximizing ATP production instead of being distributed into the mitochondrial TCA cycle and glycine–serine metabolism.
Fig. 5.
Pathway response with maximization of mitochondrial ATP synthase, ‘ATPSyn’ as the objective. Flow of flux through the different reactions of a glycolysis, b pentose phosphate pathway, c cysteine metabolism, d glutamate metabolism, e glycine–serine metabolism and f glutathione metabolism pathway while maximizing mitochondrial ATP synthesis
Maximization of the metabolic demand
Qualitatively, the same trend of pathway response was observed for the two scenarios while optimizing the model for the metabolic demand reaction ‘GBM_BM’. Although, a few more differences was further observed while considering the GBM_BM demand reaction.
Increased flux through glycolysis and pentose-phosphate pathway in glioblastoma Simulating the model for GBM_BM objective function in both the scenarios suggested an increased flux through the glycolysis and pentose-phosphate pathway (PPP) reactions (Fig. 6a, b). This increased flux is contributed by the glycine uptake through the phosphoglycerate dehydrogenase (PGDH) reactions into glycolysis and hence, PPP so as to provide for ribulose-5-phosphate present in the GBM_BM objective. Further, the lower part of glycolysis was observed to be more active as compared to the upper reactions as reported in a study (Oudard et al. 1996), where a low activity of hexokinase was observed due to the loss of chromosome 10. Apart from this, some amount of glycine is partly distributed through the phosphoenolpyruvate carboxykinase (PEP_CarbK_1) reaction for production of oxaloacetate and succinate which is part of the GBM_BM demand reaction. A comparison of the steady state flux profiles of astrocyte versus glioblastoma with GBM_BM as objective has been provided in Table S3 of Online Resource 2.
Increased cystine uptake in glioblastoma Simulating the model for GBM_BM objective function in both the scenarios further demonstrated a higher increase in cystine uptake and its metabolism as compared to the model simulations using ATP synthesis as objective (Fig. 6c). This was because of the higher requirement of glutathione to meet the metabolic demand of glioblastoma cells to combat oxidative stress.
Reversal of TCA cycle towards production of malate and fumarate in both scenarios A back flux in TCA cycle, from oxaloacetate to fumarate was also observed in experiments, in both cultured astrocytes and in in vivo conditions, which was due to the activity of mitochondrial pyruvate carboxylase (Brekke et al. 2012). Through the model simulation, similar properties in the glioblastoma scenario were observed too (Fig. 6d). The flux through the fumarate hydratase (FUMH) and malate dehydrogenase (MDH) reactions was reversed and enhanced in the glioblastoma scenario. The reason for this reversal was to maximize succinate production through TCA cycle, which was an important component of the metabolic demand reaction.
Fig. 6.
Pathway response with maximization of metabolic function, ‘GBM_BM’ as the objective. Flow of flux through the different reactions of a glycolysis, b pentose phosphate pathway, c cysteine metabolism, d TCA cycle; while maximizing the metabolic function, GBM_BM, for glioblastoma growth. A positive flux shows progression of reaction in forward direction and a negative flux implies flow of flux in reverse direction
Essentiality of metabolites and reactions in glioblastoma growth
The important metabolites that contributed significantly to glioblastoma growth were identified. Single and double reaction knockouts analyses were performed to identify the sets of reactions, which could significantly reduce glioblastoma growth. The analyses were performed using the GBM_BM objective function.
Determination of metabolites essential for glioblastoma growth
Glioblastoma cell lines can show extremely long survival under glucose starved conditions by undergoing physiological adaptations to utilize alternatives and thus, combat nutrient deprivation (Griguer et al. 2005). In order to determine those metabolites which essentially contributed to glioblastoma survival, even at glucose starved conditions, the metabolic fate of eight carbon sources namely, glucose, cystine, methionine, tryptophan, palmitate, glutamate, glutamine, and glycine through the network, was investigated. From the simulation results, cystine was found to be an essential metabolite for glioblastoma growth, although glucose was largely required for satisfying the metabolic demand and increasing glioblastoma growth rate. A complete deprivation of glucose did not lead to zero growth although growth rate was largely reduced. Previous experimental findings have also suggested a large reduction in cell growth due to glycolytic blockade by glucose starvation (Griguer et al. 2005). Also, effect of glucose in combination with cystine was more pronounced in glioblastoma growth, instead of cystine alone as input (Fig. 7). The simultaneous uptake and utilization of cystine and glucose served as the minimal metabolite requirement that could drive all those pathways which lead to synthesis of objective function components [Eq. (2)]. The essential role of cystine is to produce glutathione that would be required to combat oxidative stress. This observation was in agreement with the available experimental evidences, where cystine uptake was proven to be essential (Chung et al. 2005) and cysteine metabolism was identified to be a novel pathway in the glioblastoma growth (Prabhu et al. 2014). And the role of glucose was to produce ribulose-5-phosphate, oxaloacetate and succinate through PPP and TCA cycle. Consequently, this minimal combination resulted in a solution higher than any other combination, thereby accounting for optimal glioblastoma growth. Restricting the uptake of either of these metabolites led to either zero growth or a highly reduced growth rate (<20 % of the optimal value).
Fig. 7.
Essentiality of metabolites in glioblastoma growth. Flux through different pathways and metabolic function in different input conditions to the glioblastoma scenario. Here flux through the different pathways and the metabolic function is maximum for cystine and glucose as input. The metabolites have been abbreviated as: Cys cystine, Glut glutamate, Glu glucose, Gln glutamine, Gly glycine, Met methionine, Try tryptophan, PA palmitic acid
Predictions from single and double reaction knockouts
From the observations made in the previous section, cystine was found to be an important metabolite for glioblastoma survival. To further analyze the essentiality of the reactions involved in the metabolism of cystine, and also to find the other important reactions in the model, which could be targeted for reducing glioblastoma growth, single and double reaction knockout analyses were performed (Table 2 and Table S4 and Table S5 of Online Resource 2). All the single and double reaction knockout results were categorized as cases of lethal, trivial and non-trivial lethal and non-trivial solutions.
Table 2.
Total number of single and double lethal reaction knockouts
| Deletion | Lethal | Trivial lethal | Non-trivial lethal | Non-trivial total | Total cases |
|---|---|---|---|---|---|
| Single | 6 | NA | 6 | 208 | 208 |
| Double | 1268 | 1227 | 41 | 20,301 | 21,528 |
N.B.: The lethal double reactions knockouts are categorized as trivial and non-trivial lethal. Those knockout combinations, of which at least one is involved in single lethal reaction knockout, are considered to be “trivial”. Those combinations in which neither of the reactions is involved in single lethal reaction knockout are considered to be “non-trivial”
NA not applicable
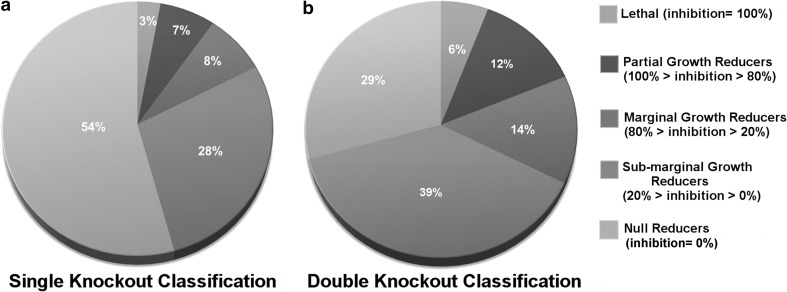
Glioblastoma cells can thrive on different metabolic pathways for survival and show great metabolic heterogeneity (Griguer et al. 2005). Similarly, it was observed that around 3 % (6 reactions) of the total single knockouts (208 reactions) and 6 % (1268 reactions) of the total double knockouts (21,528) were lethal to the glioblastoma scenario. A low number of lethal single knockouts suggested the robustness of metabolism in sustenance of the glioblastoma cells through alternative routes. Knockout analysis was performed on the network using GBM_BM as the objective function.
Knockout analysis identified ribulose phosphate isomerase (RPI), a part of the pentose phosphate pathway to govern a lethal phenotype. In many type of cancers, it has been experimentally observed that PPP drives the glycolytic flux for production of ribose-5-phosphate and NADPH that can be used by cancers cells for detoxification of reactive oxygen species (Boada et al. 2000). RPI represents a rate limiting-step for ribose-5-phosphate production in the PPP pathway. As ribose-5-phosphate is an essential component to meet the cellular metabolic demand, RPI was predicted to govern a lethal phenotype in the glioblastoma scenario. Also, in different types of cancers, high level of glutathione contents have been experimentally observed to combat with the oxidative stress experienced by the cancer cells (Ogunrinu and Sontheimer 2010). Glutamate–cysteine ligase (GCL), rate-limiting step for production of glutathione was predicted to govern a lethal phenotype as it is the penultimate step for glutathione production. Similarly, glutathione synthase (GS), the ultimate step of glutathione synthesis from glutamate and cysteine was also predicted to govern a lethal phenotype. The cystine–glutamate antiporter (Anti_cystine_glut) and cystine reductase (CystRed) reactions are involved in production of cysteine. In the previous results, it was demonstrated that cystine was sufficient for production of the components of the GBM_BM objective. Hence, both the reactions were predicted to demonstrate lethality when knocked out.
Of the 1268 lethal double knockout reactions, 41 were non-trivial, which included reactions from glycolytic, pentose phosphate, TCA cycle and glycine–serine metabolism pathway and a few transport reactions. The most typical observation of glioblastoma metabolism through experiments was the increased flux through glycolysis for a high production of ATP and corresponding reduction in glioblastoma growth under glucose starvation, even though their survival was maintained (Griguer et al. 2005). A combinatorial targeting of the glycolytic pathway with the PPP, TCA cycle and glycine–serine metabolic pathways was hence, found to be more effective in combating glioblastoma growth. Thus, the knockdown of a glycolytic pathway reaction in combination to a pentose phosphate pathway reaction or a TCA cycle reaction hindered the production of r5p or oaa or succ. Consequently, the double knockouts proved to be lethal to the glioblastoma growth. The in silico results also yielded reactions belonging to glycine–serine metabolism as good targets in combination with each other. Glycine was necessarily required for glutathione production. When the availability of glycine was blocked through the knockdown of both the internal glycine–serine metabolism and the external source of glycine uptake, this paired knockout led to the inhibition of glutathione production, and hence proved lethal. Consequently, dual targeting the reactions of this pathway was effective in reducing the growth.
The knockouts reaction results were further classified as lethal, growth reducers and null reducers on the basis of percentage inhibition in the metabolic demand reaction rate in the glioblastoma scenario (Fig. 8). Knockouts which led to 100 % inhibition of metabolic demand reaction were considered to be “Lethal”. Reaction knockouts which caused a flux reduction of >80 % of the flux through the metabolic demand were considered to be “Partial growth reducers”. Those set of reaction knockouts which inhibited the flux of metabolic demand within 20–80 % of the default value, were considered as “Marginal growth reducers”. The class of ‘sub-marginal growth reducers’ was considered for those set of knockouts which could not bring effective reduction (0–20 % inhibition) through the objective function. Analysis of the double knockout showed that 48 % of the partial growth reducers belonged to the glycolytic pathway. The rest of the 52 % were mostly constituted by the reactions of TCA cycle, PPP, Oxidative phosphorylation and Glycine–serine metabolism. The larger fraction of both single and double reaction knockouts which belonged to sub-marginal growth reducers and null reducers which were indicative of the robust and redundant reactions of the glioblastoma metabolic network.
Fig. 8.
Single and double reaction knockout predictions. The results of single and double reaction knockout predictions are summarized. a, b Single and double knockout reaction classifications, respectively. The predictions are classified in five categories: (1) lethal (inhibition = 100 %), (2) partial growth reducers (100 % < inhibition < 80 %), (3) marginal growth reducers (80 % < inhibition < 20 %), (4) sub-marginal growth reducers (20 % < growth < 0 %), (5) null reducers (inhibition = 0 %), with the percentage inhibition imposed by them being indicated in the parentheses
Chemotherapeutic interventions in glioblastoma scenario
The reaction knockout analyses could predict a subset of reactions, which were crucial in glioblastoma growth. To identify the feasibility of targeting these reactions and their effectiveness, these reactions were simulated for their effect as chemotherapeutics for either inhibiting, or bringing down the growth rate of the glioblastoma cells to a normal range. For this analysis, the previously identified growth reducer reactions leading to reduced growth (0 < GBM_BM solution < glioblastoma optimum) were chosen.
From the simulation studies it was observed that in order to completely reduce the flux through the metabolic function, targeting the lethal single knockout reactions required a complete reduction of flux through them. Whereas targeting the lethal double knockout reactions were more effective, as partial reduction of flux through those combinations brought a complete reduction of flux through the metabolic demand reaction. Recent experimental approaches also suggest that targeting a combination of proteins instead of each individual protein of the combination, could bring about a more pronounced anti-tumorigenic effect (Oliveira-Ferrer et al. 2013). As such, combinations from non-trivial lethal knockout reactions were simulated which could be targeted most effectively for efficient growth reduction.
Of the 41 non-trivial lethal double knockout predictions, 36 combinations were chosen for testing their chemotherapeutic intervening properties, which excluded a few transport reactions. Each reaction combination was simulated by varying the flux through individual reactions of the combination simultaneously, to obtain the effective reduction of flux through both of these reactions which reduced glioblastoma growth completely and to obtain a feasible flux range through both the reactions for which the growth rate was reduced to the normal level. The effective reduction of flux was depicted in percentage, which was defined as percentage reduction of flux through any particular reaction. The simulation results for the 10 most effective combinations have been shown as contour plots in Fig. 9. A list of all the 36 chemotherapeutic combinations has been provided in Table S6 of Online Resource 2. The percentage reduction of flux value for complete reduction of growth and for Normal growth, for each reaction of the combinations has been listed in Table S7 of Online Resource 2.
Fig. 9.
Chemotherapeutic intervention scenarios and effective combination of target reactions. Percentage reduction of flux through combination of essential double knockout reactions a hexokinase (HEX) and fructose-1,6-bisphoasphate aldolase (FBA), b ribulose phosphate-3 epimerase (RPE) and 6-phosphogluconolactonase (6PGLase), c fumarate hydratase (FUMH) and alpha ketoglutarate dehydrogenase (AKGDH), d glycine transport (Trans_glycine) and Phosphoglycerate dehydrogenase (PGDH), e hexokinase (HEX) and triose phosphate isomerase (TPI), f glucose transport (Trans_glucose) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), g phosphofructokinase (PFK) and Hexokinase (HEX), h succinyl-CoA synthetase (SCS) and fumarate hydratase (FUMH), i ribulose phosphate-3 epimerase (RPE) and glucose-6-phosphate dehydrogenase (G6PDH) and j glucose transport (Trans_glucose) and phosphoglycerate kinase (PGK), and its effect on the flux through the metabolic function, GBM_BM (different regions in the contour plots)
Inhibitors to a few of these target reactions were already available (see Table S8 of Online Resource 1). In-silico study on the core metabolism in cancer cells showed that reactions of glycolytic, TCA cycle, oxidative phosphorylation and pentose phosphate pathway could be good targets to check cancer cell progression (Resendis-Antonio et al. 2010). Interestingly, our context-specific constraint based metabolic model specific to glioblastoma could identify reactions belonging to cysteine metabolism and reaction combinations of glycine–serine pathway to be potential targets for controlling glioblastoma growth.
These potent reaction pairs of the glycine–serine metabolism give way to discovery/formulation of combinatorial drugs that can inhibit them. Therapeutic agents to target the glycine receptors are already known. Inhibitors like Picrotoxin targeted the neuronal γ-aminobutyric acid and homomeric glycine receptors (Wang et al. 2006), whereas strychnine hydrochloride was found to be a potent antagonist specific to the glycine receptor (García-Colunga and Miledi 1999). These could be employed beneficially to understand the activity of the glycine transporters in glioblastoma too, as evidences state a correlation between the glycine transporter activities with the distribution of its receptors (Jursky and Nelson 1995). In recent years, many pharmaceutical companies have also developed potent and selective inhibitors for glycine transporters. SSR 504734 and SSR 103800, a series of N-(2-aryl-cyclohexyl) substituted spiropiperidines and ORG 25935 are a few compounds, which showed promising results as inhibitors of glycine transporters (Hashimoto 2010).
Discussion
In the present work, a context specific metabolic network for astrocyte/glioblastoma has been developed, that studies the adaptation of complex metabolic reactions within the network under scenario-specific conditions to achieve specific biological goals like growth and maximization of ATP synthesis. Through this model, we tried to investigate the cumulative effect of a large scale metabolism on the metabolic functioning of glioblastoma, the effect of the mutual connectivity of the individual pathways within the metabolic network and the difference in response they show in the astrocytic and glioblastoma scenarios. Also, through the in silico approach, we tried to gain some insight into the alternative metabolic routes and metabolites which contributed to the metabolic heterogeneity of glioblastoma. The present model was capable of yielding results which were in correspondence to the experimentally proved phenomena of both astrocytes and glioblastoma (Deighton et al. 2014; Mangia et al. 2009; Marrif and Juurlink 1999; Pellerin and Magistretti 1994; Wise et al. 2008; Zhou et al. 2011).
From our study, specific pathways were observed to demonstrate a co-operative effect in the astrocyte and glioblastoma scenarios. In the model astrocyte scenario, dependence of glycolysis for mitochondrial ATP synthesis reduced with increased uptake and utilization of glutamate as energy source, due to coupling of both glutamate and glucose catabolism via TCA cycle (Fig. 2c). A similar phenomenon was reported in a previous experimental study (Pellerin and Magistretti 1994). Similarly, in glioblastoma scenario, conversion of glutamine to glutamate and its extracellular release was coupled with cystine uptake and its catabolism to produce glutathione. Previously, through experiments, the cooperative effect of glutamate and cysteine metabolism was speculated and briefly studied in glioblastoma (Ye et al. 1999). Evidences stated that glycine content of neuronal cells was higher than that of glial cells (Roux and Supplisson 2000), and most of the CNS tissues sufficed their glycine requirement via the internal glycine–serine metabolism pathway derived from glucose via 3-phosphoglycerate (Nicklas and Browning 1978), even though astrocyte cultures fed with glycine were capable of utilizing it by maintaining intracellular levels of glutathione, serine and creatine (Dringen et al. 1998). Based on these literature evidences and predictions from the model, it was inferred that astrocytes take up glycine from external sources at a lower level and most of its requirement in the astrocytes was fulfilled by the internal glycine–serine metabolic pathway. Further, a higher uptake of glycine by astrocytes from external source resulted in a reduction in its glutamate uptake rate and an excess of glycine caused glutaminolysis in the astrocytes. On the contrary, glioblastoma showed an increased glycine uptake which was driven towards increased synthesis of glutathione. This increased production of glutathione was simultaneously accompanied with the increased uptake of cystine, which is taken in by the cystine–glutamate antiporter. Hence, a higher amount of glutamate was lost through efflux. This loss was compensated by glutamine uptake which is subsequently converted into glutamate within the cell.
Glioblastoma cell lines can show extremely long survival under glucose starved conditions, which are indicative to the fact that these cells undergo physiological adaptations to overcome nutrient deprivation (Griguer et al. 2005). From the model, it was predicted that cystine could be one essential metabolite that could serve to glioblastoma survival and growth, even at glucose starved conditions. The utilization of glucose was found to be coupled to the cystine uptake in the model. This was perhaps because of the choice of objective function. The objective function included ribulose-5-phosphate, oxaloacetate, succinate and glutathione, which were generated by glycolysis and cysteine metabolism pathways. Consequently, a minimal combination of cystine and glucose could drive a considerable amount of flux through the objective function, as compared to any other minimal combination.
In-silico study on the core metabolism in cancer cells showed that reactions of glycolytic, TCA cycle, oxidative phosphorylation and pentose phosphate pathway are essential for cancer cell progression (Resendis-Antonio et al. 2010) suggesting that central and highly connected proteins (enzymes) can prove to be useful drug targets (Jeong et al. 2001). These enzymes belong to central carbon and energy metabolism that essentially meet the energy requirements of the cell and hence are important. However, the centrality of enzymes does not always imply lethality in complex protein networks (Raman et al. 2014). Similar to this observation, in our metabolic network, we identified some unique enzymatic reactions that did not belong to the aforementioned central metabolic pathways but still were deemed to be absolutely essential for glioblastoma growth. Pertaining to this, targeting the reactions belonging to cysteine metabolism (‘Cystine glutamate antiporter’, Anti_cystine_glut, and ‘cystine reductase’, CystRed) and dual targeting of the reactions belonging to glycine–serine metabolism along with glycine transporter could potentially suppress glioblastoma growth.
Thus to summarize the present study, the designed model could predict—that most of the astrocytic glycine requirement was fulfilled by the internal glycine–serine metabolism pathway, and excess glycine in the environment of growing astrocytes might have an effect on its glutamate metabolism. Further, it was observed that cystine and glucose were two vital metabolites, which could significantly contribute to glioblastoma growth. From model analysis for chemotherapeutic interventions, it was observed that reactions of cysteine metabolism and dual targeting of reactions belonging to glycine–serine metabolism could be potential chemotherapeutic targets for effective inhibition of glioblastoma growth.
The present metabolic model has both its advantages and limitations. The steady state assumption is a limitation to the model, which does not allow us to consider the intermediary dynamic changes in the flux profiles of the reactions. Even though, the present context-specific model suffices to achieve the biological goal of glioblastoma growth, considering only a subset of the whole genomic network might lead to results which deviate from the real biological scenario. In order to overcome such limitations, our predictions can further be validated through experiments, so as to help the chemists and biologists to discover small molecule inhibitors against brain cancer. An in vitro experimental approach, by treating the astrocyte-derived glioblastoma cell lines like LN-229 and U87MG with combinations of inhibitors/chemotherapeutic agents mentioned in Table S8 of Online Resource 2 for ensuring their synergy and selectivity to inhibit cell growth and proliferation (Lehár et al. 2009), can further validate the model predictions.
Therefore, our study not only contributes in understanding the complexities, differences and consequences of glioblastoma metabolism for predicting biologically reasonable disease scenarios, but also provides a deep insight into identification of important targets as well as chemotherapeutic interventions.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We thank Council of Scientific and Industrial Research, XII Five Year Plan Project “GENESIS” (BSC0121) and Department of Biotechnology, Government of India (Project Code: BT/PR13689/BID/07/363/2010) for providing financial support to perform this work. Abhishek Subramanian acknowledges the research fellowship provided by DBT-BINC fellowship program.
References
- Anton K, Glod J. An orchestrated response to tumor signals by macrophages and mesenchymal stem cells potentiates interleukin-6 secretion in glioblastoma. Cell Death Ther. 2014;1:2353–7817. doi: 10.2478/cdth-2014-0001. [DOI] [Google Scholar]
- Bairoch A. The ENZYME database in 2000. Nucleic Acids Res. 2000;28:304–305. doi: 10.1093/nar/28.1.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerji A. An attempt to construct a (general) mathematical framework to model biological “context-dependence”. Syst Synth Biol. 2013;7:221–227. doi: 10.1007/s11693-013-9122-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boada J, Roig T, Perez X, Gamez A, Bartrons R, Cascante M, Bermúdez J. Cells overexpressing fructose-2, 6-bisphosphatase showed enhanced pentose phosphate pathway flux and resistance to oxidative stress. FEBS Lett. 2000;480:261–264. doi: 10.1016/S0014-5793(00)01950-5. [DOI] [PubMed] [Google Scholar]
- Bouzier-Sore AK, Pellerin L. Unraveling the complex metabolic nature of astrocytes. Front Cell Neurosci. 2013;7:179. doi: 10.3389/fncel.2013.00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brekke E, Walls AB, Norfeldt L, Schousboe A, Waagepetersen HS, Sonnewald U. Direct measurement of backflux between oxaloacetate and fumarate following pyruvate carboxylation. Glia. 2012;60:147–158. doi: 10.1002/glia.21265. [DOI] [PubMed] [Google Scholar]
- Burgess PK, Kulesa PM, Murray JD, Alvord EC., Jr The interaction of growth rates and diffusion coefficients in a three-dimensional mathematical model of gliomas. J Neuropathol Exp Neurol. 1997;56:704–713. doi: 10.1097/00005072-199706000-00008. [DOI] [PubMed] [Google Scholar]
- Chatterjee A, Mambo E, Sidransky D. Mitochondrial DNA mutations in human cancer. Oncogene. 2006;25:4663–4674. doi: 10.1038/sj.onc.1209604. [DOI] [PubMed] [Google Scholar]
- Chinnaiyan P, et al. The metabolomic signature of malignant glioma reflects accelerated anabolic metabolism. Cancer Res. 2012;72:5878–5888. doi: 10.1158/0008-5472.CAN-12-1572-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WJ, Lyons SA, Nelson GM, Hamza H, Gladson CL, Gillespie GY, Sontheimer H. Inhibition of cystine uptake disrupts the growth of primary brain tumors. J Neurosci. 2005;25:7101–7110. doi: 10.1523/JNEUROSCI.5258-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium U Activities at the universal protein resource (UniProt) Nucleic Acids Res. 2014;42:D191–D198. doi: 10.1093/nar/gkt1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covert MW, Schilling CH, Palsson B. Regulation of gene expression in flux balance models of metabolism. J Theor Biol. 2001;213:73–88. doi: 10.1006/jtbi.2001.2405. [DOI] [PubMed] [Google Scholar]
- Deighton RF, et al. The proteomic response in glioblastoma in young patients. J Neurooncol. 2014;119:79–89. doi: 10.1007/s11060-014-1474-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dringen R, Verleysdonk S, Hamprecht B, Willker W, Leibfritz D, Brand A. Metabolism of glycine in primary astroglial cells: synthesis of creatine, serine, and glutathione. J Neurochem. 1998;70:835–840. doi: 10.1046/j.1471-4159.1998.70020835.x. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Colunga J, Miledi R. Modulation of nicotinic acetylcholine receptors by strychnine. Proc Natl Acad Sci. 1999;96:4113–4118. doi: 10.1073/pnas.96.7.4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griguer CE, Oliva CR, Gillespie GY. Glucose metabolism heterogeneity in human and mouse malignant glioma cell lines. J Neurooncol. 2005;74:123–133. doi: 10.1007/s11060-004-6404-6. [DOI] [PubMed] [Google Scholar]
- Guessous F, et al. Oncogenic effects of miR-10b in glioblastoma stem cells. J Neurooncol. 2013;112:153–163. doi: 10.1007/s11060-013-1047-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto K. Glycine transport inhibitors for the treatment of schizophrenia. Open Med Chem. 2010;4:10–19. doi: 10.2174/1874104501004010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattingen E, Lanfermann H, Quick J, Franz K, Zanella FE, Pilatus U. 1H MR spectroscopic imaging with short and long echo time to discriminate glycine in glial tumours. Magn Reson Mater Phys, Biol Med. 2009;22:33–41. doi: 10.1007/s10334-008-0145-z. [DOI] [PubMed] [Google Scholar]
- Hertz L, Zielke HR. Astrocytic control of glutamatergic activity: astrocytes as stars of the show. Trends Neurosci. 2004;27:735–743. doi: 10.1016/j.tins.2004.10.008. [DOI] [PubMed] [Google Scholar]
- Jellinger K. Glioblastoma multiforme: morphology and biology. Acta Neurochir. 1977;42:5–32. doi: 10.1007/BF01406628. [DOI] [PubMed] [Google Scholar]
- Jeong H, Mason SP, Barabási A-L, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- Jursky F, Nelson N. Localization of glycine neurotransmitter transporter (GLYT2) reveals correlation with the distribution of glycine receptor. J Neurochem. 1995;64:1026–1033. doi: 10.1046/j.1471-4159.1995.64031026.x. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014;42:D199–D205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleihues P, Ohgaki H. Phenotype vs genotype in the evolution of astrocytic brain tumors. Toxicol Pathol. 2000;28:164–170. doi: 10.1177/019262330002800121. [DOI] [PubMed] [Google Scholar]
- Lee JM, Gianchandani EP, Papin JA. Flux balance analysis in the era of metabolomics. Brief Bioinform. 2006;7:140–150. doi: 10.1093/bib/bbl007. [DOI] [PubMed] [Google Scholar]
- Lehár J, Krueger AS, Avery W, Heilbut AM, Johansen LM, Price ER, Rickles RJ, Short GF, 3rd, Staunton JE, Jin X, Lee MS, Zimmermann GR, Borisy AA. Synergistic drug combinations tend to improve therapeutically relevant selectivity. Nat Biotechnol. 2009;27:659–666. doi: 10.1038/nbt.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maity A, Pore N, Lee J, Solomon D, O’Rourke DM. Epidermal growth factor receptor transcriptionally up-regulates vascular endothelial growth factor expression in human glioblastoma cells via a pathway involving phosphatidylinositol 3′-kinase and distinct from that induced by hypoxia. Cancer Res. 2000;60:5879–5886. [PubMed] [Google Scholar]
- Mandonnet E, Pallud J, Clatz O, Taillandier L, Konukoglu E, Duffau H, Capelle L. Computational modeling of the WHO grade II glioma dynamics: principles and applications to management paradigm. Neurosurg Rev. 2008;31:263–269. doi: 10.1007/s10143-008-0128-6. [DOI] [PubMed] [Google Scholar]
- Mangia S, Simpson IA, Vannucci SJ, Carruthers A. The in vivo neuron-to-astrocyte lactate shuttle in human brain: evidence from modeling of measured lactate levels during visual stimulation. J Neurochem. 2009;109(Suppl 1):55–62. doi: 10.1111/j.1471-4159.2009.06003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrif H, Juurlink BH. Astrocytes respond to hypoxia by increasing glycolytic capacity. J Neurosci Res. 1999;57:255–260. doi: 10.1002/(SICI)1097-4547(19990715)57:2<255::AID-JNR11>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Nicklas WJ, Browning ET. Amino acid metabolism in glial cells: homeostatic regulation of intra-and extracellular milieu by C-6 glioma cells. J Neurochem. 1978;30:955–963. doi: 10.1111/j.1471-4159.1978.tb12387.x. [DOI] [PubMed] [Google Scholar]
- Ogunrinu TA, Sontheimer H. Hypoxia increases the dependence of glioma cells on glutathione. J Biol Chem. 2010;285:37716–37724. doi: 10.1074/jbc.M110.161190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira-Ferrer L, Wellbrock J, Bartsch U, Penas EM, Hauschild J, Klokow M, Bokemeyer C, Fiedler W, Schuch G. Combination therapy targeting integrins reduces glioblastoma tumor growth through antiangiogenic and direct antitumor activity and leads to activation of the pro-proliferative prolactin pathway. Mol Cancer. 2013;12:1–14. doi: 10.1186/1476-4598-12-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudard S, et al. High glycolysis in gliomas despite low hexokinase transcription and activity correlated to chromosome 10 loss. Br J Cancer. 1996;74:839–845. doi: 10.1038/bjc.1996.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelicano H, Martin D, Xu R, Huang P. Glycolysis inhibition for anticancer treatment. Oncogene. 2006;25:4633–4646. doi: 10.1038/sj.onc.1209597. [DOI] [PubMed] [Google Scholar]
- Pellerin L, Magistretti PJ. Glutamate uptake into astrocytes stimulates aerobic glycolysis: a mechanism coupling neuronal activity to glucose utilization. Proc Natl Acad Sci. 1994;91:10625–10629. doi: 10.1073/pnas.91.22.10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pistollato F, et al. Hypoxia and succinate antagonize 2-deoxyglucose effects on glioblastoma. Biochem Pharmacol. 2010;80:1517–1527. doi: 10.1016/j.bcp.2010.08.003. [DOI] [PubMed] [Google Scholar]
- Prabhu A, Sarcar B, Kahali S, Yuan Z, Johnson JJ, Adam KP, Kensicki E, Chinnaiyan P. Cysteine catabolism: a novel metabolic pathway contributing to glioblastoma growth. Cancer Res. 2014;74:787–796. doi: 10.1158/0008-5472.CAN-13-1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman K, Damaraju N, Joshi GK. The organisational structure of protein networks: revisiting the centrality–lethality hypothesis. Syst Synth Biol. 2014;8:73–81. doi: 10.1007/s11693-013-9123-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resendis-Antonio O, Checa A, Encarnación S. Modeling core metabolism in cancer cells: surveying the topology underlying the Warburg effect. PLoS ONE. 2010;5:e12383. doi: 10.1371/journal.pone.0012383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux MJ, Supplisson S. Neuronal and glial glycine transporters have different stoichiometries. Neuron. 2000;25:373–383. doi: 10.1016/S0896-6273(00)80901-0. [DOI] [PubMed] [Google Scholar]
- Sahm F, et al. The endogenous tryptophan metabolite and NAD + precursor quinolinic acid confers resistance of gliomas to oxidative stress. Cancer Res. 2013;73:3225–3234. doi: 10.1158/0008-5472.CAN-12-3831. [DOI] [PubMed] [Google Scholar]
- Schellenberger J, et al. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox v2. 0. Nat Protoc. 2011;6:1290–1307. doi: 10.1038/nprot.2011.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnewald U, Westergaard N, Jones P, Taylor A, Bachelard H, Schousboe A. Metabolism of [U-13C5] glutamine in cultured astrocytes studied by NMR spectroscopy: first evidence of astrocytic pyruvate recycling. J Neurochem. 1996;67:2566–2572. doi: 10.1046/j.1471-4159.1996.67062566.x. [DOI] [PubMed] [Google Scholar]
- Swanson KR, Bridge C, Murray J, Alvord EC. Virtual and real brain tumors: using mathematical modeling to quantify glioma growth and invasion. J Neurol Sci. 2003;216:1–10. doi: 10.1016/j.jns.2003.06.001. [DOI] [PubMed] [Google Scholar]
- Tracqui P, Cruywagen G, Woodward D, Bartoo G, Murray J, Alvord E. A mathematical model of glioma growth: the effect of chemotherapy on spatio-temporal growth. Cell Prolif. 1995;28:17–31. doi: 10.1111/j.1365-2184.1995.tb00036.x. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1103. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D-S, Mangin J-M, Moonen G, Rigo J-M, Legendre P. Mechanisms for picrotoxin block of α2 homomeric glycine receptors. J Biol Chem. 2006;281:3841–3855. doi: 10.1074/jbc.M511022200. [DOI] [PubMed] [Google Scholar]
- Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Wise DR, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci USA. 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf A, Agnihotri S, Guha A. Targeting metabolic remodeling in glioblastoma multiforme. Oncotarget. 2010;1:552–577. doi: 10.18632/oncotarget.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z-C, Rothstein JD, Sontheimer H. Compromised glutamate transport in human glioma cells: reduction–mislocalization of sodium-dependent glutamate transporters and enhanced activity of cystine–glutamate exchange. J Neurosci. 1999;19:10767–10777. doi: 10.1523/JNEUROSCI.19-24-10767.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zafra F, Gimenez C. Characterization of glycine uptake in plasma membrane vesicles isolated from cultured glioblastoma cells. Brain Res. 1986;397:108–116. doi: 10.1016/0006-8993(86)91374-0. [DOI] [PubMed] [Google Scholar]
- Zhou Y, et al. Metabolic alterations in highly tumorigenic glioblastoma cells: preference for hypoxia and high dependency on glycolysis. J Biol Chem. 2011;286:32843–32853. doi: 10.1074/jbc.M111.260935. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.