Abstract
The colonization of Madagascar by Austronesian-speaking people during AD 50–500 represents the most westerly point of the greatest diaspora in prehistory. A range of economically important plants and animals may have accompanied the Austronesians. Domestic chickens (Gallus gallus) are found in Madagascar, but it is unclear how they arrived there. Did they accompany the initial Austronesian-speaking populations that reached Madagascar via the Indian Ocean or were they late arrivals with Arabian and African sea-farers? To address this question, we investigated the mitochondrial DNA control region diversity of modern chickens sampled from around the Indian Ocean rim (Southeast Asia, South Asia, the Arabian Peninsula, East Africa and Madagascar). In contrast to the linguistic and human genetic evidence indicating dual African and Southeast Asian ancestry of the Malagasy people, we find that chickens in Madagascar only share a common ancestor with East Africa, which together are genetically closer to South Asian chickens than to those in Southeast Asia. This suggests that the earliest expansion of Austronesian-speaking people across the Indian Ocean did not successfully introduce chickens to Madagascar. Our results further demonstrate the complexity of the translocation history of introduced domesticates in Madagascar.
Keywords: chicken, human migration, dispersal, Madagascar, mitochondrial DNA
1. Introduction
Beginning in the first few centuries AD, Austronesian speakers from Island Southeast Asia (ISEA) established trade links with India and eventually colonized Madagascar during ca AD 50–500 [1,2]. This makes Madagascar the most westerly point of the great Austronesian expansion. Linguistic and genetic evidence [3–6] suggests a dual ancestry for the indigenous people of Madagascar, involving both African and Southeast Asian origins. For example, Malagasy, the language spoken in Madagascar, is a member of the Austronesian language family related to the Barito and Dayak languages spoken in southeast Kalimantan, Indonesia [7]. However, recent genetic studies indicate that contemporary Malagasy populations are derived from genetic admixture involving Indonesian and African ancestors (i.e. Bantu) [5,6]. In addition to genetic and linguistic evidence, transfers of material culture are also evident in the Austronesian-inherited traditions connecting Madagascar to Indonesia [8].
The prehistoric exchanges between Madagascar and its maritime neighbours around the Indian Ocean rim have influenced the current day distribution of many domestic plants and animals in the region [9–11]. The most notable Madagascan domesticates that have originated from ISEA are the taro (Colocasia esculenta), Asian yam (Dioscorea alata) and banana (Musa sapientum) [10]. Their absence in the intervening regions of India and the Arabian Peninsula makes a translocation via a central Indian Ocean maritime corridor [10] more likely than a coastal route [12]. However, genetic studies of ship-borne commensals found in Madagascar, such as rats (Rattus sp.), the house mouse (Mus musculus) and the shrew (Suncus murinus), suggests India as the point of origin [13–15]. An investigation of diversity in domesticate/commensal species and their translocation patterns around the Indian Ocean rim suggests a deeply entrenched trade and contact network [11]. For instance, exchanges in the Arabian Sea led to the movement of certain cereal crops, such as sorghum (Sorghum bicolor), pearl millet (Pennisetum galucum) and finger millet (Eleusine coracana), from Africa to the Arabian Peninsula and India [16], while Zebu cattle were translocated in the reverse direction from India to the Arabian Peninsula and Africa [17]. Translocations across the Bengal Sea included the movements of mung bean (Vigna raidata) and horsegram (Macrotyloma uniflorum) from India to Southeast Asia [18] and in reverse mango (Mangifera indica) and citron (Citrus medica) from Southeast Asia to India [19]. The protracted connection between India and the Austronesian speakers in Indonesia probably culminated in the development of the Srivijayan Empire in Indonesia and the Malay Peninsula [20]. The Srivijayan religion, culture and even language contains a mixture of influences not only from India but also from Persia [21]. These instances demonstrate that there has been a long history of contact, trade and exchange between geographically distant groups of people around the Indian Ocean rim. However, uncertainty exists as to how chickens became part of this exchange network.
Chickens were domesticated from jungle fowl (Gallus spp.) in East Asia during the Mid–Late Holocene [22–24], with subsequent translocation by humans to other parts of the world, including Africa and Madagascar. Similar to banana, taro and yam, chickens are deeply integrated into the subsistence culture of Africa, indicating a certain level of antiquity [25], with chickens first appearing in Madagascar around the late AD eighth–mid nineth century [26]. Archaeological evidence suggests that chickens were introduced into Africa via trade links with Southeast Asia [27]. Furthermore, on Madagascar, the Malagasy term for chicken and other domesticated animals, is borrowed from Bantu languages (from the east coast of Africa), not Austronesian [9,28], suggesting that chickens were initially established on the east coast of Africa before being introduced into Madagascar.
Previous studies have characterized partial mtDNA control region sequences in African village chickens, which fell into two major mitochondrial lineages, that suggest two origins: (i) in Southeast Asia and (ii) the Indian subcontinent [29–31]. Chickens from Madagascar have been reported to also have a dual geographical origin, from continental Africa and Indonesia, on the basis of the same two mitochondrial lineages [32]. However, Razafindraibe et al. [32] provided no analysis to support their conclusions.
Thus, the geographical origin of Madagascan chickens remains uncertain, with Africa, South Asia and/or ISEA all possible source populations. Here, we assess the genetic relationships and diversity of chicken populations found around the Indian Ocean rim. We aim to address the question of whether indigenous chickens in Madagascar trace their ancestry to Indonesia as reflected in the expansion of the Austronesian culture, or they resulted from a more complex connection with other areas around the Indian Ocean.
2. Material and methods
In total, 3128 chicken sequences from Madagascar, Africa, the Arabian Peninsula, West Asia, East Asia, South Asia, ISEA, Mainland Southeast Asia (MSEA) and the Pacific were used for analyses (figure 1; table 1 and the electronic supplementary material, table S1). Mwacharo et al. [31] sequenced 512 chickens from East Africa (Kenya: 211, Ethiopia: 43, Sudan: 135, Uganda: 123) but only 159 sequences (all from Kenya) were available on GenBank. We generated new sequences from 898 chickens from ISEA and the Pacific and combined these with 2230 existing sequences (electronic supplementary material, table S1). Seventy-nine previously published sequences were available from Madagascar. Two samples have unknown collection locality; the remaining 77 were collected from two regions in central Madagascar. The sampling approach performed in ISEA and the Pacific was approved by the University of Adelaide Animal Ethics Committee (AEC) as part of the study on reconstructing the human colonization of the Pacific (AEC approval number: S-2011-211). Samples from indigenous village chickens consisted of plucked body feathers. Genomic DNA was extracted from feather samples using the salting-out method [48].
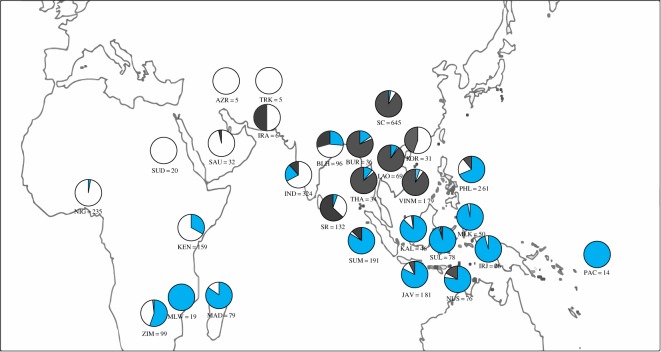
Figure 1.
Frequency distribution of chicken mitochondrial DNA haplogroup in the region under study (blue, haplogroup D; white, haplogroup E; grey, other haplogroups) with geographical location and sample size noted. Sample localities are Azerbaijan (AZR), Bangladesh (BLH), Burma (BUR), India (IND), Iran (IRA), Irian Jaya (IRJ), Java (JAV), Kalimantan (KAL), Kenya (KEN), Korea (KOR), Laos (LAO), Madagascar (MAD), Malawi (MLW), Maluku (MLK), Nigeria (NIG), Nusa Tenggara (NUS), Pacific (PAC; Fiji, Solomon and Vanuatu), Philippines (PHL; Luzon, Visayas and Mindanao), Saudi Arabia (SAU), South China (SC), Sri Lanka (SRI), Sudan (SUD), Sulawesi (SUL), Sumatra (SUM), Thailand (THA), Turkmenistan (TRK), Vietnam (VIE) and Zimbabwe (ZIM).
Table 1.
Details, including collection locality and published source, for 3128 chicken mtDNA control region sequences used in this study. (#Mwacharo et al. [31] sequenced additional chicken samples from East Africa (Ethiopia: 43, Sudan: 135, Uganda: 123) but did not make them available on GenBank. See the electronic supplementary material, table S1 for detailed attributes of the 3128 sequences.)
| region | locality | no. samples | references |
|---|---|---|---|
| Africa# | |||
| Madagascar | 79 | [32]b, [33]a | |
| Malawi | 19 | [29] | |
| Zimbabwe | 99 | [29] | |
| Kenya | 159 | [31] | |
| Nigeria | 235 | [34] | |
| Sudan | 20 | [29] | |
| Arabian Peninsula | |||
| Saudi Arabia | 32 | [35] | |
| West Asia | |||
| Iran | 6 | [22], [33]a | |
| Azerbaijan | 5 | [22] | |
| Turkmenistan | 5 | [22] | |
| South Asia | |||
| India | 324 | [22,23,36] | |
| Sri Lanka | 132 | [37] | |
| Bangladesh | 96 | [38] | |
| East Asia and Mainland Southeast Asia | |||
| China | 645 | [39]a, [40]a, [22,23,41,42]a | |
| Burma | 36 | [22,23,43] | |
| Thailand | 34 | [44], [33]a, [45] | |
| Laos | 69 | [33]a, [23,46] | |
| Vietnam | 179 | [33]a, [45], this study | |
| Korea | 31 | [47] | |
| Island Southeast Asia | |||
| Sumatra | 191 | [44], [33]a, [23], this study | |
| Java | 181 | [44], this study | |
| Kalimantan | 46 | this study | |
| Nusa Tenggara | 76 | [33]a, [22,46], this study | |
| Sulawesi | 78 | this study | |
| Maluku | 50 | this study | |
| Irian Jaya, Papua | 26 | this study | |
| Philippines | 261 | [33]a [46], this study | |
| Pacific | |||
| Solomon | 3 | this study | |
| Vanuatu | 9 | this study | |
| Fiji | 2 | this study | |
aSequences submitted only to GenBank without publication.
bSequences not submitted to any database, sequences obtained directly from the authors.
A 760 base pair (bp) fragment of the mtDNA control region (CR) was PCR-amplified using primers GallP4F (5′-AACTCCCCTACTAAGTGTACCCCC-3′) and GallP4R (5′-TTGACACTGATGCACTTTGGATCG-3′) from position 43 to 802 in the chicken mtDNA genome (GenBank Accession: X52392 [49]). Each PCR (25 µl final volume) contained 10× Hotmaster buffer, 0.25 mM of each dNTP, 0.2 µM of each primer and 0.1 U of Hotmaster Taq DNA polymerase. Thermocycling comprised an initial denaturation and enzyme activation at 94°C for 2 min, followed by 30 cycles of denaturing at 94°C for 20 s, primer annealing at 55°C for 10 s and elongation at 65°C for 60 s, and a final extension at 65°C for 10 m. PCR clean-up and Sanger sequencing were conducted at the Australian Genome Research Facility Ltd. Forward and reverse sequence chromatograms were assembled and manually edited using Geneious 6.5.5 (Biomatters) to obtain a consensus sequence.
DNA sequences, combining newly generated data and existing data, consisted of variable lengths of the mtDNA control region. Sequences were aligned using the MUSCLE algorithm in Geneious v. 6.5.5 and truncated to 349 bp (corresponding to positions 46–394 in the chicken mtDNA genome, GenBank accession no. X52392 [49]), the longest sequence common to all samples. We followed the haplogroup naming system of Miao et al. [23], which differs from that of Mwacharo et al. [31] and Muchadeyi et al. [29]. The number of haplotypes and the number of samples per haplotype were determined by collapsing sequences to unique haplotypes using FaBox 1.40 [50]. Identifying the mitochondrial haplogroup of each unique sequence was done by ordering them into a neighbour-joining tree generated using the Tamura-Nei substitution model and comparing the assignments to those from previously published papers [22,23,51,52].
To assess population genetic differentiation and gene flow among sampling locations, Slatkin's linearized FST was computed using Arlequin v. 3.1 [53] (10 000 permutations). To visualize the relationships between populations, a non-parametric multidimensional scaling plot (MDS) was performed using the Slatkin's linearized FST scores. This was done initially on all chicken mitochondrial haplogroups and then on just haplogroup D, as a substantial proportion of haplotypes found on Madagascar and ISEA belongs to mitochondrial haplogroup D. To further explore geographical structure, an analysis of molecular variance (AMOVA) was also calculated in Arlequin [53] using Indonesia, South Asia, East Africa and Madagascar as groups. These regions were selected for AMOVA because they are the most likely regions involved in the translocation of chickens to Madagascar. Furthermore, the extensive linguistic and human genetic studies mostly involve these regions.
The evolutionary relationships among haplotypes were estimated via a median-joining (MJ) network [54] using Network v. 4.6.1 (fluxus-engineering.com) for haplogroups D and E separately. Pairwise genetic distances between all haplotypes, and again for haplogroup D only, were calculated using Genalex [55] and ordered into principal coordinate analysis (PCoA) plots to further explore the relationships of the lineages and to see how the haplotypes around the Indian Ocean rim are distributed. Intra-population genetic variation and population expansion statistics (i.e. haplotype/nucleotide diversity and Tajima's D/Fu's Fs, respectively) were also calculated at a regional level using Arlequin v. 3.1 [53].
3. Results
3.1. Mitochondrial haplogroup distribution patterns
Chickens in Madagascar cluster into two haplogroups: the majority (85%) of samples belong to haplogroup D and the rest belong to haplogroup E (figure 1). Similarly, in East Africa only haplogroup D and E chickens are observed in publically available sequences, with a positive association between haplogroup D frequency and latitude southwards. By contrast, haplogroup D is not observed in chickens from the Arabian Peninsula and western Asia: the haplogroup composition in these regions is dominated by haplogroup E. Within India, haplogroup E chickens are also observed at a high frequency (67%) with haplogroup D (22%) and all other haplogroups (11%) making up the balance. To the east in MSEA, all haplogroups (A to I) are observed, but the frequency of haplogroup D and E are dramatically lower in comparison to other haplogroups (4.8% and 2.4%, respectively). However, chickens from islands further east (in the Pacific Ocean) have a high proportion of haplogroup D (84%).
3.2. Population genetic structure
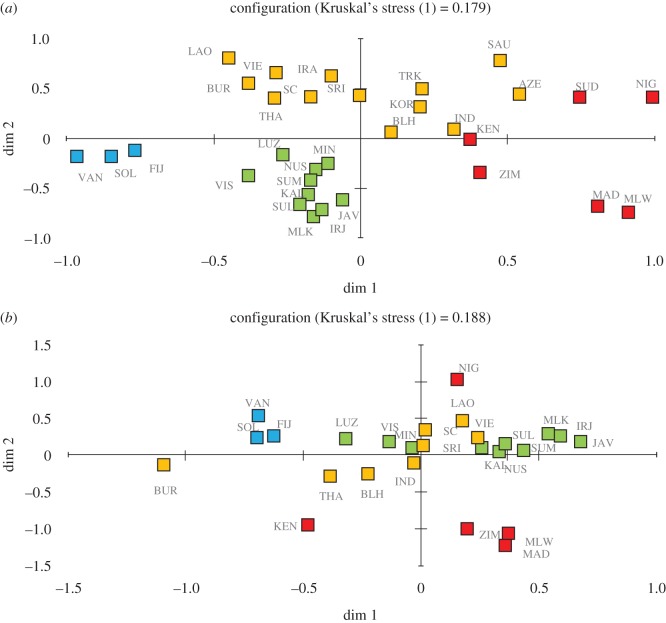
The overall genetic structure in the MDS plot using all haplogroups reveals structuring at a broad geographical scale (figure 2a). Distinctive clustering of populations from ISEA (shown in green) and from the Pacific (in blue) can be seen. African populations, including Madagascar, sit closer to South and West Asian populations, than ISEA populations. Madagascar falls closest to Malawi, which is the closest African population to Madagascar in the dataset. The other continental African populations show broad geographical clines (northern–southern cline follows high–low ‘Dim 2’ values). When only haplogroup D is used, the Madagascan samples form a distinct cluster with the geographically closest East African populations, Malawi and Zimbabwe (figure 2b).
Figure 2.
Multidimensional scaling plots (MDS) for pairwise population Slatkin's linearized FST for (a) 3128 chickens from Asia (orange), Africa (red), ISEA (green) and the Pacific (blue) using all haplogroups. (b) 1081 haplogroup D chickens from the same regions. Azerbaijan, Iran, Korea, Saudi Arabia, Sudan and Turkmenistan are not present in plot B because they do not contain haplogroup D lineages. See figure 1 for locality abbreviations.
The AMOVA performed on all chickens from Indonesia, South Asia, East Africa and Madagascar show population structure (table 2). However, the among-group variance components (regional groupings) are highest when only haplogroup D chickens are used. For haplogroup D samples, the among-group variance component is always significant when Indonesia is separated from Madagascar. The only non-significant among-group variance component is when South Asia was isolated from Indonesia, East Africa and Madagascar as a group. The variance components among the groups (regional grouping) are generally low when using only haplogroup E.
Table 2.
Population genetic structure estimated from the analysis of molecular variance (AMOVA) based on mtDNA control region sequences from relevant regions in the Indian Ocean rim: (A) Indonesia, (B) South Asia, (C) East Africa and (D) Madagascar.
| variance components (%) |
||||||
|---|---|---|---|---|---|---|
| group | n | no. population | no. groups | among groups | among populations within groups | within populations |
| haplogroup D and E combined | ||||||
| no grouping | 1347 | 14 | 1 | … | 31.07 | 68.93 |
| group 1 (A versus B versus C versus D) | 1347 | 14 | 4 | 30.38 | 5.75 | 63.87 |
| group 2 (A, C and D versus B) | 1347 | 14 | 2 | 17.87 | 19.66 | 62.46 |
| group 3 (A versus B, C and D) | 1347 | 14 | 2 | 23.58 | 14.77 | 61.64 |
| group 4 (A, B, versus C, D) | 1347 | 14 | 2 | 11.38 | 24.11 | 64.51 |
| haplogroup D only | ||||||
| no grouping | 845 | 14 | 1 | … | 48.00 | 52.00 |
| group 1 (A versus B versus C versus D) | 845 | 14 | 4 | 51.98 | 6.95 | 41.07 |
| group 2 (A, C, & D versus B) | 845 | 14 | 2 | −1.98 | 49.19 | 52.79 |
| group 3 (A versus B, C and D) | 845 | 14 | 2 | 44.25 | 15.07 | 40.68 |
| group 4 (A, B, versus C, D) | 845 | 14 | 2 | 56.42 | 9.30 | 34.28 |
| haplogroup E only | ||||||
| no grouping | 502 | 10 | 1 | … | 9.86 | 90.14 |
| group 1 (A versus B versus C versus D) | 502 | 10 | 4 | 4.88 | 6.26 | 88.86 |
| group 2 (A, C and D versus B) | 502 | 10 | 2 | 1.80 | 8.65 | 89.55 |
| group 3 (A versus B, C and D) | 502 | 10 | 2 | 4.78 | 8.72 | 86.50 |
| group 4 (A, B, versus C, D) | 502 | 10 | 2 | 0.49 | 9.56 | 89.96 |
3.3. Population dynamics and genetic variability
The genetic differentiation observed from the FST values (table 3) using both haplogroup D and E samples show that the highest level of divergence is between Indonesia and Madagascar (1.33034), whereas it is the lowest between South Asia and Africa (0.14823). When using only haplogroup D samples, the highest level of divergence is still between Indonesia and Madagascar (0.69607) and the lowest is between Africa and Madagascar (0.13317). All population pairwise comparisons were significant at the 5% level.
Table 3.
Matrix of Slatkin's linearized FST between chicken samples from Indonesia, South Asia, East Africa and Madagascar based on mitochondrial control region sequences. (*Significant differences at p < 0.05.)
| population | abbreviation | SA | INDO | AFR | MAD |
|---|---|---|---|---|---|
| haplogroup D and E combined | |||||
| South Asia | SA | 0 | |||
| Indonesia | INDO | 0.61* | 0 | ||
| Africa | AFR | 0.15* | 0.51* | 0 | |
| Madagascar | MAD | 0.80* | 1.33* | 0.29* | 0 |
| haplogroup D | |||||
| South Asia | SA | 0 | |||
| Indonesia | INDO | 0.28* | 0 | ||
| Africa | AFR | 0.38* | 0.67* | 0 | |
| Madagascar | MAD | 0.38* | 0.70* | 0.13* | 0 |
Both Tajima's D and Fu's Fs neutrality statistics indicate that chickens from South Asia, Indonesia, East Africa and Madagascar deviate from neutrality when using only haplogroup D (table 4). However, when using both haplogroup D and E, all consistently deviate from neutrality except Madagascar. These results support a model of demographic expansion of haplogroup D for each of the four regions. Furthermore, the highest level of genetic diversity is observed in South Asia and ISEA. On the other hand, Madagascar has the lowest diversity among the four regional groups being compared, with East Africa next lowest, suggesting a genetic bottleneck effect with every new introduction of chickens into an area.
Table 4.
Genetic diversity measures and historical demographic patterns of chickens from Indonesia, South Asia, East Africa and Madagascar. (N(H), size (no. haplotypes); HD, haplotype diversity; ND, nucleotide diversity; π, mean no. of pairwise difference; SSD, sum of squared differences. *Statistically significant p-values (p < 0.05 for Tajima's D, p < 0.02 for Fu's Fs).)
| molecular diversity indices |
neutrality test |
|||||
|---|---|---|---|---|---|---|
| region | N(H) | HD | ND (SD) | π | Tajima's D | Fu's FS |
| haplogroup D and E combined | ||||||
| Indonesia | 583 (115) | 0.95 | 0.0047 (0.0031) | 1.63 | −2.01* | −27.13* |
| South Asia | 409 (114) | 0.91 | 0.0124 (0.0068) | 4.31 | −1.97* | −25.10* |
| Africa | 276 (42) | 0.91 | 0.0123 (0.0068) | 4.30 | −0.03 | −17.36* |
| Madagascar | 79 (10) | 0.43 | 0.0048 (0.0032) | 1.68 | −1.15 | −1.88 |
| D haplogroup only | ||||||
| Indonesia | 551 (102) | 0.95 | 0.0038 (0.0026) | 1.32 | −2.14* | −28.85* |
| South Asia | 100 (40) | 0.92 | 0.0118 (0.0066) | 4.13 | −1.67* | −25.74* |
| Africa | 127 (19) | 0.78 | 0.0041 (0.0028) | 1.41 | −1.50* | −11.94* |
| Madagascar | 67 (6) | 0.22 | 0.0008 (0.0009) | 0.27 | −1.90* | −5.32* |
3.4. Phylogenetic relationships of Madagascan mtDNA haplotypes in East Africa, South Asia and Indonesia
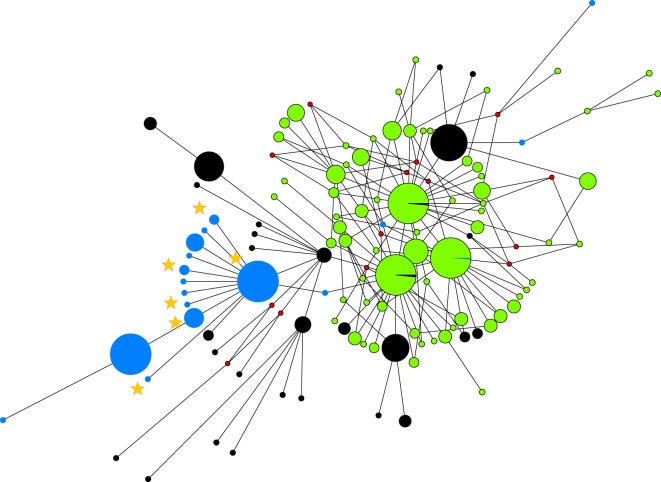
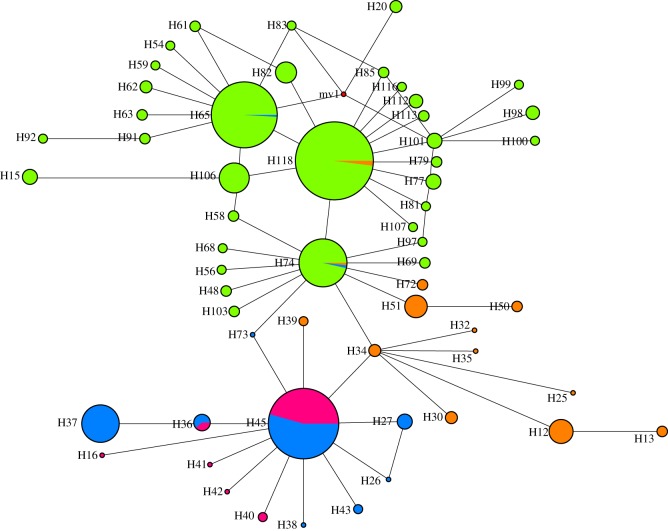
The median-joining phylogenetic network created using only haplogroup D samples show that Madagascar and East Africa share a closely related set of haplotypes and together these two regions have a closer phylogenetic relationship with South Asia than with Indonesia (figure 3). Altogether, there are six Madagascan D haplotypes: H45 and H36 are shared with East Africa and the rest are unique to Madagascar (H16, H40, H41 and H42; figure 4). H45 is the most common D haplotype in East Africa and Madagascar and forms the central node from which the rest of the Madagascan and East African haplotypes radiate. Two predominantly Indonesian haplotypes (H65 and H74) are observed at very low frequencies in continental Africa, but not at all in Madagascar and in fact they are phylogenetically distant to the other Madagascan D haplotypes. The PCoA plot (electronic supplementary material, figure S1) that used the genetic distances between all 112 D haplotypes from Madagascar, East Africa, South Asia and Indonesia also supports the broad separation of regions indicated in the network. Additionally, there are four haplotypes belonging to haplogroup E found on Madagascar. One haplotype is unique to Madagascar, one is shared with East Africa and two are shared with East Africa, South Asia and Indonesia (electronic supplementary material, figure S2).
Figure 3.
Median-joining network depicting the relationship of D haplotypes of chickens from East Africa and Madagascar (blue), South Asia (black) and Indonesia (green) using all observed D haplotypes regardless of frequency. Stars mark the position of the Madagascan samples. Inferred haplotypes are indicated by small red dots.
Figure 4.
Median-joining (MJ) network of mtDNA-CR D haplotypes observed in Madagascar (purple), Africa (blue), South Asia (brown) and Indonesia (green) excluding most haplotypes represented by one sample. The circle sizes are proportional to the haplotype frequencies and the length of the lines corresponds to the number of mutations connecting haplotypes.
4. Discussion
All our population genetic and phylogeographic analyses of the more than 3000 chicken mtDNA sequences from around the Indian Ocean rim strongly support an African origin for Madagascan chickens. The close relationship between Madagascan and East African chickens is seen in the PCoA plots, the phylogeographic patterns in the networks, as well as in the AMOVA and FST analyses. The progression from high to low diversity and stronger to weaker evidence of demographic expansions from South Asia via East Africa to Madagascar also highlights the effect of repeated genetic bottlenecks as each population was introduced to a new landmass. An African origin for Madagascan chickens is consistent with linguistic evidence—Malagasy words for most domestic animals (dog, goat, cow, sheep, donkey, chicken and guinea fowl) are of Bantu (African) origin [28]. Similarly, Malagasy dogs have an African origin [56] and pig tapeworms in Madagascar trace their origins to South Asia and Africa, not ISEA [57]. This suggests that at least two (dog and chicken) and possibly all three (dog, chicken and pig) animal domesticates successfully transported by Austronesians into the Pacific, were either not transported directly to, or failed to establish, in Madagascar, following the first Austronesian voyages across the Indian Ocean.
The presence of only two haplogroups, D and E, in Madagascar and East Africa suggests that these were the only two lineages translocated to these regions that have survived to the present day, with haplogroup D the dominant lineage in both Madagascar and East Africa. Despite a long and complex history of maritime exchange around the Indian Ocean rim a strong phylogeographic signal remains in haplogroup D chickens (figure 3). The phylogeography indicates a shared common ancestry for Madagascan and East African chickens. The star-like radiation stemming from the most common Madagascar haplotype (H45, which is also the most common east African haplotype; figure 3) suggests that the initial chicken populations arrived in Madagascar via the East African coast. What is not known, however, is whether H45 represents the founding lineage, with in situ evolution responsible for the one and two base pair derivations radiating from H45. As the H45 haplotype is currently not observed outside of East Africa/Madagascar and there are no archaeological chicken remains representing the earliest chickens on Madagascar, we cannot tell where H45 originated.
The strong phylogeographic signal within haplogroup D suggests that South Asia, rather than Southeast Asia, is the most likely recent source for East African and Madagascan chickens (figure 3). Therefore, despite the clear linguistic and human genetic associations between Madagascar and Indonesia, the Austronesians do not appear to have successfully translocated chickens directly to Madagascar across the Indian Ocean. However, Austronesians may have transported chickens to Africa indirectly via the Indian subcontinent, rather than via the direct route across the Indian Ocean. Support for this theory can be inferred from the presence of Austronesian speakers in South Asia during the first millennium A.D. [21,58] and the fact that there are no high frequency central nodes in a star-like Indian cluster. Rather, Indian haplotypes belong to two of the high-frequency central nodes in star-like ISEA clusters.
It is interesting that haplogroup D chickens are not observed in the Arabian Peninsula and occur at low frequencies in northeast Africa, suggesting that chickens might have been transported via a direct sea link from India across the Arabian Sea to eastern Africa/Madagascar. Alternatively, Madagascan chickens could also have been transported along a coastal route through the Arabian Peninsula and northeast Africa but with the signal overwritten by subsequent and repeated translocations of haplogroup E chickens.
Haplogroup E is less common in Madagascar compared with haplogroup D. As haplogroup E lacks phylogeographic signature, most probably owing to modern-day translocations (electronic supplementary material, figure S2), it is difficult to derive fine-scale inferences based on haplogroup E other than establishing that it is also most probably South Asian in origin. Furthermore, it is difficult to ascertain whether the arrival of haplogroups D and E was contemporaneous. However, the most parsimonious explanation is that a mixed population of both haplogroup D and E chickens were transported from India to Madagascar via East Africa. Testing this hypothesis is difficult using existing samples. A more deliberate sampling regime, combined with nuclear genetic data and ancient DNA from archaeological samples, would help establish the full history of chicken translocation around the Indian Ocean rim.
5. Conclusion
Mitochondrial DNA data suggest that chickens were introduced into Madagascar from South Asia via East Africa. A scenario whereby chickens arrived in Madagascar along with the expansion of the Austronesian-speaking people directly across the Indian Ocean is not supported. However, it remains a possibility that Austronesian traders and mariners integrated South Asian chickens during their coastal voyages en route to east Africa and Madagascar. Additional sampling of chickens in Madagascar, increased genomic sequencing of existing samples and accessioning of existing mtDNA data from Mwacharo et al. [31] onto publically available databases will help refine our understanding of the ultimate origins of Madagascan chickens.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank Hanta Razafindraibe for making the Madagascan chicken sequences available for this study, and two anonymous reviewers for constructive comments that improved the manuscript.
Ethics
The sampling of chickens in ISEA and the Pacific for this study was approved by the University of Adelaide AEC as part of the study on reconstructing the human colonization of the Pacific (AEC approval number: S-2011-211).
Data accessibility
All mtDNA control region sequences generated during this study are available on Genbank under accession numbers KX642436-KX643333 [59].
Authors' contributions
M.B.H. carried out field and molecular laboratory work, contributed to data analysis and design of the study and drafted the manuscript. V.A.T. contributed to data analysis and design of the study and helped draft the manuscript. J.J.W. contributed to molecular laboratory work and data analysis. P.J.P. helped interpret the results and draft the manuscript. S.S. and A.B.D. carried out field and molecular laboratory work on Indonesian samples. S.K. contributed to molecular laboratory work. J.G. helped arrange collection of samples from Indonesia, and contributed to data analysis and interpretation. J.J.A. contributed to the design of the study, data analysis and interpretation, and helped draft the manuscript. All authors gave their final approval for publication.
Competing interests
The authors declare no competing interests.
Funding
This research was funded by the Australian Research Council (ARC) Discovery Project DP110105187 to J.J.A. and J.G.
References
- 1.Dewar RE, Wright HT. 1993. The culture history of Madagascar. J. World Prehist. 7, 417–466. (doi:10.1007/BF00997802) [Google Scholar]
- 2.Burney DA, et al. 2004. A chronology for late prehistoric Madagascar. J. Hum. Evol. 47, 25–63. (doi:10.1016/j.jhevol.2004.05.005) [DOI] [PubMed] [Google Scholar]
- 3.Beaujard P. 2003. Les arrivées austronésiennes à madagascar: Vagues ou continuum? (partie I). Études Océan Indien. 35–36, 59–147. [Google Scholar]
- 4.Hurles ME, Sykes BC, Jobling MA, Forster P. 2005. The dual origin of the Malagasy in Island Southeast Asia and East Africa: evidence from maternal and paternal lineages. Am. J. Hum. Genet. 76, 894–901. (doi:10.1086/430051) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tofanelli S, Bertoncini S, Castrì L, Luiselli D, Calafell F, Donati G, Paoli G. 2009. On the origins and admixture of Malagasy: new evidence from high-resolution analyses of paternal and maternal lineages. Mol. Biol. Evol. 26, 2109–2124. (doi:10.1093/molbev/msp120) [DOI] [PubMed] [Google Scholar]
- 6.Pierron D, et al. 2014. Genome-wide evidence of Austronesian–Bantu admixture and cultural reversion in a hunter-gatherer group of Madagascar. Proc. Natl Acad. Sci. USA 111, 936–941. (doi:10.1073/pnas.1321860111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dahl OC. 1991. Migration from Kalimantan to Madagascar. Oslo, Norway: Norwegian University Press. [Google Scholar]
- 8.Blench R. 2010. Evidence for the Austronesian voyages in the Indian Ocean. In The global origins and development of seafaring (ed. Anderson A.), pp. 239–248. Cambridge, UK: McDonald Institute Monographs. McDonald Institute for Archaeological Research. [Google Scholar]
- 9.Blench R. 2006. The Austronesians in Madagascar and on the East African coast: surveying the linguistic evidence for domestic and translocated animals. In Tenth Int. Conf. on Austronesian Linguistics, 17–20 January 2006, Palawan, Philippines: Linguistic Society of the Philippines and SIL International. [Google Scholar]
- 10.Fuller DQ, Boivin N, Hoogervorst T, Allaby RG. 2011. Across the Indian Ocean: the prehistoric movement of plants and animals. Antiquity 85, 544–558. (doi:10.1017/S0003598X00067934) [Google Scholar]
- 11.Boivin N, Crowther A, Helm R, Fuller DQ. 2013. East Africa and Madagascar in the Indian Ocean world. J. World Prehist. 26, 213–281. (doi:10.1007/s10963-013-9067-4) [Google Scholar]
- 12.Murdock GP. 1959. Africa: its peoples and their culture history. New York, NY: McGraw-Hill. [Google Scholar]
- 13.Hingston M, Goodman SM, Ganzhorn JU, Sommer S. 2005. Reconstruction of the colonization of southern Madagascar by introduced Rattus rattus. J. Biogeogr. 32, 1549–1559. (doi:10.1111/j.1365-2699.2005.01311.x) [Google Scholar]
- 14.Kurachi M, et al. 2007. Population structure of wild musk shrews (Suncus murinus) in Asia based on mitochondrial DNA variation, with research in Cambodia and Bhutan. Biochem. Genet. 45, 165–183. (doi:10.1007/s10528-006-9051-0) [DOI] [PubMed] [Google Scholar]
- 15.Tollenaere C, et al. 2010. Phylogeography of the introduced species Rattus rattus in the western Indian Ocean, with special emphasis on the colonization history of Madagascar. J. Biogeogr. 37, 398–410. (doi:10.1111/j.1365-2699.2009.02228.x) [Google Scholar]
- 16.Fuller DQ, Boivin N. 2009. Crops, cattle and commensals across the Indian Ocean. Current and potential archaeobiological evidence. Études Océan Indien 42–43, 13–46. (doi:10.4000/oceanindien.698) [Google Scholar]
- 17.Hanotte O, et al. 2002. African pastoralism: genetic imprints of origins and migrations. Science 296, 336–339. (doi:10.1126/science.1069878) [DOI] [PubMed] [Google Scholar]
- 18.Castillo C, Fuller D. 2010. Still too fragmentary and dependent upon chance? Advances in the study of early Southeast Asian archaeobotany. In 50 years of archaeology in southeast Asia (eds Bellina B, Bacus EA, Pryce O, Weissman CJ), pp. 91–111. London, UK: River Books. [Google Scholar]
- 19.Asouti E, Fuller DQ. 2008. Trees and woodlands of South India: archaeological perspectives. Walnut Creek, CA: Left Coast Press. [Google Scholar]
- 20.Munoz PM. 2006. Early kingdoms of the Indonesian Archipelago and the Malay peninsula. Singapore: Editions Didier Millet. [Google Scholar]
- 21.Hall KR. 1977. The coming of Islam to the Archipelago: a re-assessment. In Economic change and cocial interaction in southeast Asia (ed. Hutterer KL.), pp. 213–231. Ann Arbor, MI: Center for South and Southeast Asian Studies, University of Michigan. [Google Scholar]
- 22.Liu Y-P, et al. 2006. Multiple maternal origins of chickens: out of the Asian jungles. Mol. Phylogenet. Evol. 38, 12–19. (doi:10.1016/j.ympev.2005.09.014) [DOI] [PubMed] [Google Scholar]
- 23.Miao YW, et al. 2013. Chicken domestication: an updated perspective based on mitochondrial genomes. Heredity 110, 277–282. (doi:10.1038/hdy.2012.83) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larson G, et al. 2014. Current perspectives and the future of domestication studies. Proc. Natl Acad. Sci. USA 111, 6139–6146. (doi:10.1073/pnas.1323964111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williamson K. 2000. Did chickens go west? In The origins and development of African livestock: archaeology, genetics, linguistics and ethnography (eds Blench R, MacDonald K), pp. 368–448. Abingdon, UK: Routledge. [Google Scholar]
- 26.Mudida N, Horton M. 1996. Subsistence at Shanga: the faunal record. In Shanga: the archaeology of a muslim trading community on the coast of East Africa (ed. Horton M.), pp. 378–393. Nairobi, Kenya: The British Institute in Eastern Africa. [Google Scholar]
- 27.MacDonald KC. 1992. The domestic chicken (Gallus gallus) in sub-Saharan Africa: a background to its introduction and its osteological differentiation from indigenous fowls (Numidinae and Francolinus sp.). J. Archaeol. Sci. 19, 303–318. (doi:10.1016/0305-4403(92)90019-Y) [Google Scholar]
- 28.Adelaar A. 2006. The Indonesian migrations to Madagascar: making sense of the multidisciplinary evidence. In Austronesian diaspora and the ethnogeneses of people in Indonesian archipelago: Proc. Int. Symp (eds Simanjuntak T, Pojoh IHE, Hisyam M), pp. 205–235. Jakarta: Indonesian Institute of Science. [Google Scholar]
- 29.Muchadeyi F, Eding H, Simianer H, Wollny C, Groeneveld E, Weigend S. 2008. Mitochondrial DNA D loop sequences suggest a Southeast Asian and Indian origin of Zimbabwean village chickens. Anim. Genet. 39, 615–622. (doi:10.1111/j.1365-2052.2008.01785.x) [DOI] [PubMed] [Google Scholar]
- 30.Mtileni B, Muchadeyi F, Maiwashe A, Groeneveld E, Groeneveld L, Dzama K, Weigend S. 2011. Genetic diversity and conservation of South African indigenous chicken populations. J. Anim. Breed. Genet. 128, 209–218. (doi:10.1111/j.1439-0388.2010.00891.x) [DOI] [PubMed] [Google Scholar]
- 31.Mwacharo J, Bjørnstad G, Mobegi V, Nomura K, Hanada H, Amano T, Jianlin H, Hanotte O. 2011. Mitochondrial DNA reveals multiple introductions of domestic chicken in East Africa. Mol. Phylogenet. Evol. 58, 374–382. (doi:10.1016/j.ympev.2010.11.027) [DOI] [PubMed] [Google Scholar]
- 32.Razafindraibe H, Mobegi VA, Ommeh SC, Bjørnstad G, Hanotte O, Jianlin H. 2008. Mitochondrial DNA origin of indigenous Malagasy chicken. Ann. N Y Acad. Sci. 1149, 77–79. (doi:10.1196/annals.1428.047) [DOI] [PubMed] [Google Scholar]
- 33.Miyake T. 1997. Gallus gallus mitochondrial DNA for D-loop region. Published only in Genbank. [Google Scholar]
- 34.Adebambo AO, Mobegi VA, Mwacharo JM, Gro B, Jianlin H, Hanotte O. 2010. Lack of phylogeographic structure in Nigerian village chickens revealed by mitochondrial D-loop sequence analysis. Int. J. Poultry Sci. 9, 503–507. (doi:10.3923/ijps.2010.503.507) [Google Scholar]
- 35.Yacoub HA, Fathi MM. 2013. Phylogenetic analysis using D-loop marker of mtDNA of Saudi native chicken strains. Mitochondrial DNA 24, 538–551. (doi:10.3109/19401736.2013.770494) [DOI] [PubMed] [Google Scholar]
- 36.Kanginakudru S, Metta M, Jakati R, Nagaraju J. 2008. Genetic evidence from Indian red jungle fowl corroborates multiple domestication of modern day chicken. BMC Evol. Biol. 8, 174 (doi:10.1186/1471-2148-8-174) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Silva P, Guan X, Ho Shing O, Jones J, Xu J, Hui D, Notter D, Smith E. 2009. Mitochondrial DNA-based analysis of genetic variation and relatedness among Sri Lankan indigenous chickens and the Ceylon junglefowl (Gallus lafayetti). Anim. Genet. 40, 1–9. (doi:10.1111/j.1365-2052.2008.01783.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bhuiyan MSA, Chen S, Faruque S, Bhuiyan AKFH, Beja-Pereira A. 2013. Genetic diversity and maternal origin of Bangladeshi chicken. Mol. Biol. Rep. 40, 4123–4128. (doi:10.1007/s11033-013-2522-6) [DOI] [PubMed] [Google Scholar]
- 39.Fu Y, Niu D, Luo J, Zhang Y-P, Ran H. 1999. Gallus gallus domesticus mitochondrial DNA for D-loop region. Published only in Genbank. [Google Scholar]
- 40.Liu ZG, et al. 2003. Genetic variability of mtDNA sequences in Chinese native chicken breeds Published only in Genbank. [Google Scholar]
- 41.Qu LJ, Yang N, Li XY. 2004. High polymorphism rate found in Chinese game chickens Published only in Genbank. [Google Scholar]
- 42.Tong XM, et al. 2006. Complete sequence and gene organization of the Tibetan chicken mitochondrial genome. Yi Chuan 28, 769–777. [PubMed] [Google Scholar]
- 43.Komiyama T, Ikeo K, Gojobori T. 2003. Where is the origin of the Japanese gamecocks? Gene 317, 195–202. (doi:10.1016/S0378-1119(03)00703-0) [DOI] [PubMed] [Google Scholar]
- 44.Fumihito A, Miyake T, Takada M, Shingu R, Endo T, Gojobori T, Kondo N, Ohno S. 1996. Monophyletic origin and unique dispersal patterns of domestic fowls. Proc. Natl Acad. Sci. USA 93, 6792–6795. (doi:10.1073/pnas.93.13.6792) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berthouly-Salazar C, et al. 2010. Vietnamese chickens: a gate towards Asian genetic diversity. BMC Genet. 11, 53 (doi:10.1186/1471-2156-11-53) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nishibori M, Shimogiri T, Hayashi T, Yasue H. 2005. Molecular evidence for hybridization of species in the genus Gallus except for Gallus varius. Anim. Genet. 36, 367–375. (doi:10.1111/j.1365-2052.2005.01318.x) [DOI] [PubMed] [Google Scholar]
- 47.Lee YJ, et al. 2007. Mitochondrial DNA diversity of Korean Ogol chicken. Asian Aust. J. Anim. Sci. 20, 477 (doi:10.5713/ajas.2007.477) [Google Scholar]
- 48.Nicholls JA, Double MC, Rowell DM, Magrath RD. 2000. The evolution of cooperative and pair breeding in thornbills Acanthiza (Pardalotidae). J. Avian Biol. 31, 165–176. (doi:10.1034/j.1600-048X.2000.310208.x) [Google Scholar]
- 49.Desjardins P, Morais R. 1990. Sequence and gene organization of the chicken mitochondrial genome. A novel gene order in higher vertebrates. J. Mol. Biol. 212, 599–634. (doi:10.1016/0022-2836(90)90225-B) [DOI] [PubMed] [Google Scholar]
- 50.Villesen P. 2007. FaBox: an online toolbox for fasta sequences. Mol. Ecol. Notes 7, 965–968. (doi:10.1111/j.1471-8286.2007.01821.x) [Google Scholar]
- 51.Gongora J, et al. 2008. Indo-European and Asian origins for Chilean and Pacific chickens revealed by mtDNA. Proc. Natl Acad. Sci. USA 105, 10 308–10 313. (doi:10.1073/pnas.0801991105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thomson VA, et al. 2014. Using ancient DNA to study the origins and dispersal of ancestral Polynesian chickens across the Pacific. Proc. Natl Acad. Sci. USA 111, 4826–4831. (doi:10.1073/pnas.1320412111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Excoffier L, Laval G, Schneider S. 2005. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol. Bioinform. 1, 47. [PMC free article] [PubMed] [Google Scholar]
- 54.Bandelt H-J, Forster P, Röhl A. 1999. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48. (doi:10.1093/oxfordjournals.molbev.a026036) [DOI] [PubMed] [Google Scholar]
- 55.Peakall R, Smouse PE. 2006. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 6, 288–295. (doi:10.1111/j.1471-8286.2005.01155.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ardalan A, Oskarsson MCR, van Asch B, Rabakonandriana E, Savolainen P. 2015. African origin for Madagascan dogs revealed by mtDNA analysis. R. Soc. open sci. 2, 140552 (doi:10.1098/rsos.140552) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yanagida T, Carod J-F, Sako Y, Nakao M, Hoberg EP, Ito A, Yao Y-G. 2014. Genetics of the pig tapeworm in Madagascar reveal a history of human dispersal and colonization. PLoS ONE 9, e109002 (doi:10.1371/journal.pone.0109002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mahdi W. 1999. The dispersal of Austronesian boat forms in the Indian Ocean. Archaeol Lang. 3, 144–179. [Google Scholar]
- 59.GenBank. See https://www.ncbi.nlm.nih.gov/genbank/.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- GenBank. See https://www.ncbi.nlm.nih.gov/genbank/.
Supplementary Materials
Data Availability Statement
All mtDNA control region sequences generated during this study are available on Genbank under accession numbers KX642436-KX643333 [59].