Abstract
Breast cancer subtypes display distinct biological traits that influence their clinical behavior and response to therapy. Recent studies have highlighted the importance of chromatin structure regulators in tumorigenesis. The RNF20-RNF40 E3 ubiquitin ligase complex monoubiquitylates histone H2B to generate H2Bub1, while the deubiquitinase (DUB) USP44 can remove this modification. We found that RNF20 and RNF40 expression and global H2Bub1 are relatively low, and USP44 expression is relatively high, in basal-like breast tumors compared with luminal tumors. Consistent with a tumor-suppressive role, silencing of RNF20 in basal-like breast cancer cells increased their proliferation and migration, and their tumorigenicity and metastatic capacity, partly through upregulation of inflammatory cytokines. In contrast, in luminal breast cancer cells, RNF20 silencing reduced proliferation, migration and tumorigenic and metastatic capacity, and compromised estrogen receptor transcriptional activity, indicating a tumor-promoting role. Notably, the effects of USP44 silencing on proliferation and migration in both cancer subtypes were opposite to those of RNF20 silencing. Hence, RNF20 and H2Bub1 have contrasting roles in distinct breast cancer subtypes, through differential regulation of key transcriptional programs underpinning the distinctive traits of each subtype.
Ductal carcinomas of the breast are grouped into several main subtypes, each displaying distinct phenotypic features and clinical behavior.1, 2, 3 Estrogen receptor-positive (ER+) tumors of the Luminal A and B subtypes are often responsive to anti-estrogenic treatment and display favorable clinical outcomes. In contrast, triple-negative (TN) tumors of the basal-like subtype are typically poorly differentiated and highly aggressive.2, 4 Although the mutation profiles associated with these subtypes have been characterized extensively,1, 5 the underlying molecular programs dictating their distinct traits are still not fully understood.
Post-translational histone modifications impinge on all aspects of chromatin function, and are extensively implicated in cancer development.6 Monoubiquitylation of histone H2B (H2Bub1) on Lys120 is executed mainly by the E3 ubiquitin ligase complex comprised of RNF20 and RNF40.7, 8 Conversely, this modification can be erased by deubiquitylating enzymes (DUBs). Numerous DUBs have been reported to deubiquitylate H2Bub1,9 most notably USP22,10 which is often upregulated in cancer and is part of a cancer gene signature associated with stem cell-like features and bad prognosis,11 and USP44,12 reported to contribute to breast cancer aggressiveness.13 Modulation of H2Bub1 levels can upregulate the expression of specific gene sets in a cell type-specific manner, while repressing other gene sets.12, 14 Regulation of transcription through H2Bub1 can occur through recruitment of SWI/SNF complexes, interactions with TFIIS, and, potentially, additional mechanisms.15
Reduced global levels of H2Bub1 and of RNF20 and RNF40 mRNA, relative to corresponding non-cancerous tissue, were observed in various tumor types, including breast cancer,16, 17, 18, 19, 20, 21, 22 suggesting that this reduction provides an advantage for tumor growth. Such a reduction could be driven by a variety of mechanisms, including RNF20 promoter hypermethylation14 or increased expression of pertinent DUBs.9, 13, 23, 24 We previously reported that RNF20 silencing promotes the migration of non-transformed human mammary epithelial MCF10A cells, and facilitates transformation of mouse cells, consistent with a tumor-suppressive role.14 Moreover, reduced levels of RNF20 and H2Bub1, observed in colitis and colorectal cancer in mice and humans, can increase the expression of cytokines via the NF-κB pathway, promoting chronic inflammation and colitis-associated colorectal cancer.22 Together, these findings suggest a role for H2Bub1 in cancer prevention and for RNF20 as a tumor suppressor.
Other studies, however, suggested pro-tumorigenic activities of RNF20. Thus, it was shown that the SMURF2 E3 ubiquitin ligase, a direct negative regulator of RNF20 and thereby of H2Bub1, is downregulated in many breast tumors, while RNF20 protein is upregulated.25 Likewise, a positive contribution of H2Bub1 to cancer was demonstrated in leukemias involving MLL gene rearrangements.26
In the present study, we sought to resolve the potential discrepancy between findings indicating a tumor-suppressive role of RNF20 and H2Bub1 in breast cancer17, 21, 27 and those suggesting a tumor-promoting role.25, 28 We report that the effects of RNF20 on mammary tumorigenesis are subtype-dependent: whereas RNF20 exhibits tumor-suppressive features in basal-like breast cancer cells, it supports the tumorigenicity of luminal breast cancer cells. In basal-like cancers cells, RNF20 suppresses the NF-κB-dependent expression of cytokines, known to contribute to the growth of this tumor subtype. In contrast, RNF20 enhances the expression of targets of the estrogen receptor, the main pathway driving luminal breast cancer growth. In agreement, H2Bub1 levels tend to be higher in luminal tumors as compared to basal-like tumors. Hence, the opposing effects of RNF20, and most probably H2Bub1, on the different breast cancer subtypes are mediated, at least in part, by differential regulation of genes whose function is closely associated with subtype identity.
Results
Levels of H2Bub1 and its regulators differ between breast cancer subtypes
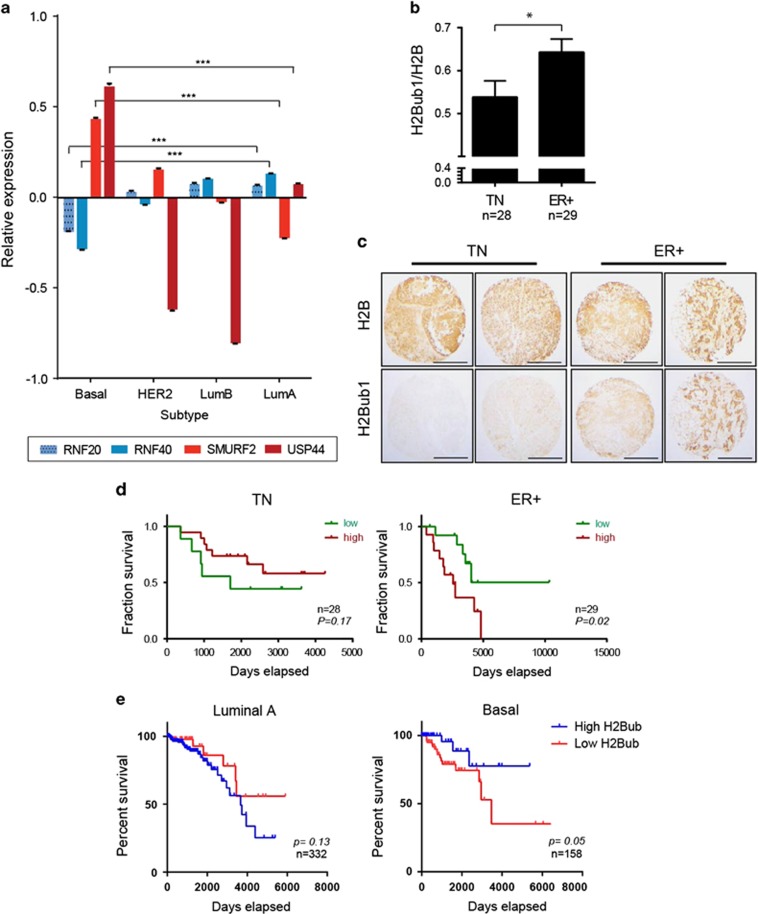
To obtain a detailed view of the expression of H2Bub1 regulators in breast cancer, we examined the mRNA levels of RNF20, RNF40, SMURF2, USP22 and USP44 in published gene-expression profiles of large cohorts of breast cancers that include the major subtypes of the disease.1 We found that RNF20 and RNF40 mRNA levels were significantly lower in basal-like tumors relative to tumors of the luminal A and B subtypes (Figure 1a). In contrast, SMURF2 and USP44, predicted to downregulate H2Bub1, displayed the opposite pattern, showing increased levels in basal-like tumors relative to luminal tumors. USP22 expression was comparable in the different subtypes (data not shown).
Figure 1.
Levels of H2Bub1 and its regulators differ between breast cancer subtypes. (a) mRNA expression levels of the H2Bub1 pathway genes RNF20, RNF40 (positive regulators) and SMURF2, USP44 (negative regulators) in 837 breast tumors included in the TCGA database and stratified according to tumor subtype. Values are shown relative to mean across all samples (0.0). Error bars, S.E. ***P<0.001. (b) Mean ratios of intensity of H2Bub1 staining relative to total H2B staining in a breast cancer tissue microarray (TMA), segregated into triple-negative (TN) and estrogen receptor-positive (ER+) tumors. Error bars, S.E. *, P<0.05. (c) Examples of tumor specimens from the TMA in b, stained for total H2B or H2Bub1. Scale bar, 500 μm. (d) Cancer-related survival rates of the patients carrying the tumors analyzed in b, comparing patients with a high H2Bub1/H2B ratio (red) to those with a low ratio (green), across TN or ER+ tumors. P values were calculated using Gehan–Breslow–Wilcoxon test. (e) Survival of patients with luminal A (left) or basal-like (right) tumors in the TCGA data set, predicted to have either high (blue) or low (red) H2Bub1 scores. Low H2Bub1 score was considered when either RNF20 or RNF40 display low expression (Z-score < –1) and when SMURF2 was highly expressed (Z-score > 1) within a given sample; high H2Bub1 pathway was considered when the opposite criteria were met
The combination of low RNF20/RNF40 and high SMURF2/USP44 expression in basal-like tumors suggested that H2Bub1 levels may be lower in these tumors than in luminal tumors. Indeed, analysis of a breast cancer tissue microarray stained with antibodies against H2Bub1 and total H2B, confirmed that TN cancers, which include basal-like tumors,4 display, on average, weaker H2Bub1 staining than ER-positive tumors, representing the luminal subtypes (Figures 1b and c).
Interestingly, H2Bub1 levels showed an opposite association with clinical outcome in patients carrying tumors of the different subtypes. Among patients carrying ER+ tumors, those with high H2Bub1 staining had a shorter mean survival time than those with low H2Bub1, while an opposite trend was observed in patients with TN tumors (Figure 1d). To further support these distinct associations, we interrogated the TCGA breast cancer mRNA database, defining an H2Bub1 level-predictive score based on the relative levels of RNF20, RNF40 and SMURF2 mRNA in each tumor. This analysis, which allowed a better definition of the different subtypes, revealed a significant association of the H2Bub1-high scoring basal-like tumors with improved patient survival, whereas luminal A tumors displayed an opposite trend (Figure 1e and Supplementary Table 1). A similar trend, albeit weaker, was observed in the METABRIC data set (Supplementary Figure S1 and Supplementary Table 1).
Together, these results indicate that basal-like tumors tend, on average, to have lower H2Bub1 levels than other subtypes, and that within these tumors low H2Bub1 is associated with poor prognosis. In contrast, luminal tumors tend to have more H2Bub1, and within these tumors it is high H2Bub1 that is associated with poorer outcome. These distinct associations might account for the apparent discrepancies in previous reports, which did not segregate the analyzed tumors into molecular subtypes.
H2Bub1 modulation exerts opposing effects on the proliferation and motility of basal-like versus luminal breast cancer cells
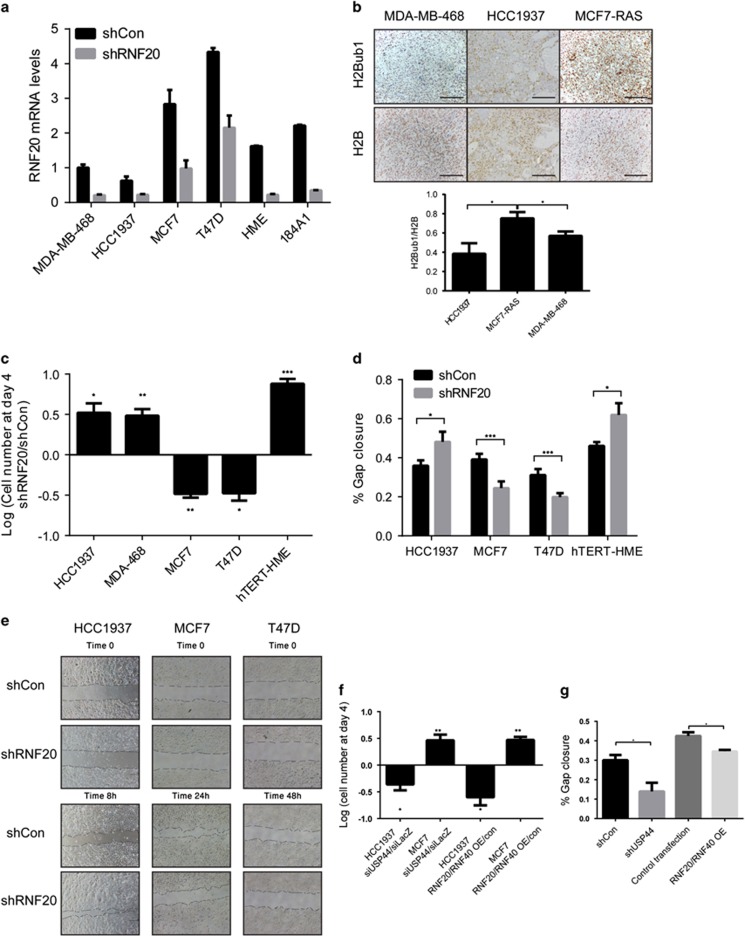
To experimentally address the possibility that H2Bub1 exerts opposing effects in the different breast cancer subtypes, we investigated the effects of RNF20 and H2Bub1 manipulation on the properties of cells representing these subtypes. Consistent with our observations in human tumors, basal-like cell lines, including HCC1937 and MDA-MB-468, expressed lower levels of RNF20 and RNF40 mRNA (shCon, Figure 2a and Supplementary Figure S2A) and protein (Supplementary Figure S2B) than luminal lines, including T47D and MCF7. The immortalized normal mammary epithelial cell lines hTERT-HME and 184A1 expressed intermediate levels of these transcripts (Figure 2a). Consistent with this, tumors that developed upon transplantation of basal-like cancer cell lines into mouse mammary glands showed weaker H2Bub1 staining than luminal cancer-derived tumors (Figure 2b).
Figure 2.
RNF20 silencing augments proliferation and migration of basal-like breast cancer cells but decreases these abilities in luminal breast cancer cells, while USP44 silencing causes opposite effects. (a) qRT-PCR analysis of RNF20 mRNA in HCC1937, MDA-MB-468, T47D, MCF7, hTERT-HME and 184A1 cells, stably expressing RNF20 shRNA (shRNF20) or control shRNA (shCon). Values were normalized to GAPDH mRNA. Error bars, S.E. (b) Sections of tumor xenografts, formed by orthotopic injection of MCF7-RAS, MDA-MB-468 or HCC1937 cells, stained for H2Bub1 and H2B. Scale bar, 100 μm. (lower panel) Quantification of staining. *P<0.05. Error bars, S.E. (c) Relative numbers of HCC1937, MDA-MB-468, MCF7, T47D and hTERT-HME cells stably expressing RNF20 shRNA (shRNF20) or control shRNA (shCon), 4 days after plating equal numbers of cells. Values were calculated by comparing numbers of shRNF20 cells to those of the corresponding shCon cells. *P<0.05; **P<0.01, ***P<0.001. Error bars, S.E. (d) Percentage of gap closure in cultures of MCF7, T47D, HCC1937 and hTERT-HME cells stably expressing RNF20 shRNA (shRNF20) or a control shRNA (shCon) in a 'scratch' experiment end point, calculated by analysis of images exemplified in e. Values indicate means of closure±S.E. in three assay replicates. *P<0.05; ***P<0.001. (e) Representative images of MCF7, T47D and HCC1937 cells stably expressing RNF20 shRNA (shRNF20) or control shRNA (shCon) at the start and end points of a scratch assay; end points varied according to the relative migratory capacity of each cell line. (f) Relative numbers of HCC1937 and MCF7 cells transfected with USP44 siRNA or siLacZ as control, or with RNF20 and RNF40 expression plasmids, measured as described in c. *P<0.05; **P<0.01. Error bars, S.E. (g) Percentage gap closure of HCC1937 cells transfected with either USP44 siRNA or siLacZ as control, or with a combination of RNF20 and RNF40 expression plasmids. Values indicate means of closure±S.E. in three assay replicates. *P<0.05
Next, we silenced RNF20 in representative luminal and basal-like cancer lines by stable infection with a lentivirus expressing RNF20 shRNA (Figure 2a and Supplementary Figure S2B). RNF20 silencing reduced H2Bub1 levels in all tested lines (Supplementary Figure S2B). Importantly, it increased the proliferation rates of the basal-like HCC1937 and MDA-MB-468 cells (Figure 2c). In contrast, the proliferation of MCF7 and T47D cells was decreased upon RNF20 silencing, indicating that it supports the proliferation of these luminal cancer cells. Furthermore, while the migration rate (assessed by a gap closure 'scratch' assay) of RNF20-silenced HCC1937 cells was increased, MCF7 and T47D cells displayed reduced migration (Figures 2d and e).
In agreement, overexpression of RNF20 together with RNF40 had the opposite effect in both cell types: it suppressed the proliferation and migration of HCC1937 cells, yet enhanced MCF7 cell proliferation (Figures 2f and g). Furthermore, silencing of the H2Bub1 DUB USP44,12 which increased H2Bub1 levels (Supplementary Figure S2C), inhibited HCC1937 cell proliferation and migration, but promoted MCF7 cell proliferation (Figures 2f and g). This further implies that the effects of RNF20 manipulation are due to altered H2Bub1 levels. Thus, RNF20 and H2Bub1 support the proliferation and motility of cultured luminal breast cancer cells, but restrict these traits in basal-like cancer cells.
Interestingly, in normal mammary epithelium-derived hTERT-HME cells, silencing of RNF20 increased both the proliferation and gap closure rates (Figures 2c and d), in agreement with our previous findings in non-transformed MCF10A cells,14 and resembling its effects in the basal-like cancer lines. To further assess the impact of RNF20 depletion on the features of non-transformed human mammary epithelial cells, we transiently depleted RNF20 mRNA from 184A1 cells and tested their ability to resume DNA synthesis after EGF starvation.29 Strikingly, in this experimental setting, RNF20 silencing bypassed the requirement for EGF in order for the cells to enter S-phase effectively (Supplementary Figure S2D), implying that RNF20 depletion reduces the dependence of those cells on growth factor signaling.
RNF20 promotes the growth of luminal tumors but limits basal-like tumor growth
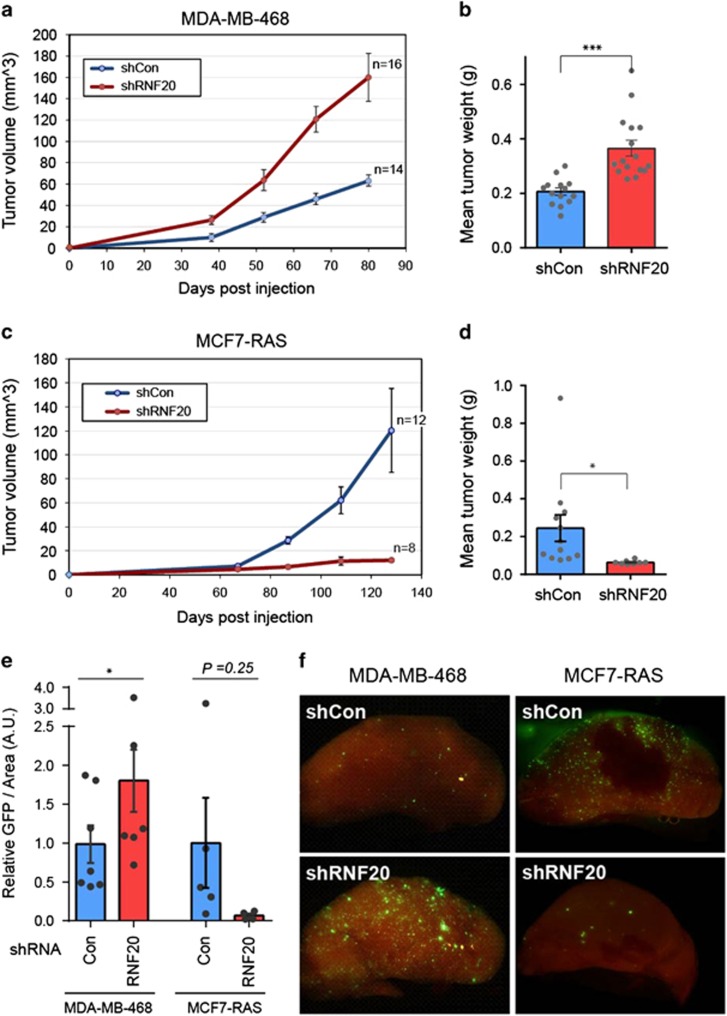
To further explore the impact of RNF20 on basal-like versus luminal cancer, we compared the tumorigenic capacity of control and RNF20-silenced cells, injected into the mammary glands of immunocompromised NSG mice. Remarkably, relative to control cells, RNF20-silenced basal-like MDA-MB-468 cells gave rise to faster growing mammary tumors, which developed at an earlier onset (Figures 3a and b). As expected, the RNF20-silenced tumors displayed reduced H2Bub1 (Supplementary Figure S3A and B).
Figure 3.
RNF20 silencing augments the tumorigenicity and metastatic seeding of basal-like breast cancer cells but decreases them in luminal breast cancer cells. (a) Growth curves of tumors formed by MDA-MB-468 cells stably expressing RNF20 shRNA (shRNF20) or control shRNA (shCon), injected into the mammary glands of NSG mice. Values indicate average tumor volume±S.E. n=number of tumors included in each group. (b) Final weights of tumors shown in a after excision. Dots indicate individual tumors. ***P<0.001. Error bars, S.E. (c) Growth curves of tumors formed by MCF7-RAS cells stably expressing RNF20 shRNA or control shRNA, measured as in a. (d) Final weights of excised tumors in c. *P<0.05. Error bars, S.E. (e) Lung disseminated cells in mice injected with either MDA-MB-468 or MCF7-RAS cells, quantified by image analysis of GFP fluorescence in lungs (A.U., arbitrary units). *P<0.05. Error bars, S.E. (f) Representative images of metastatic cells (green) in lungs of tumor bearing mice, analyzed in e
Since MCF7 cells display very low orthotopic tumorigenicity, we infected them with a retrovirus expressing oncogenic mutant H-RAS immediately before injection. In contrast to MDA-MB-468 cells, RNF20-silenced MCF7-RAS cells yielded tumors that grew more slowly than controls, and in some cases gave rise to only residual growth (Figures 3c and d). Concordantly, the abundance of lung disseminating cells, indicative of metastatic potential, changed upon RNF20 silencing in a manner corresponding to primary tumor size (Figures 3e and f). These findings further support the conjecture that RNF20 is tumor suppressive in basal-like cancer, but pro-tumorigenic in luminal cancer.
RNF20 silencing augments the expression of cytokine genes in basal-like breast cancer cells but reduces the expression of ER-target genes in luminal cancer cells
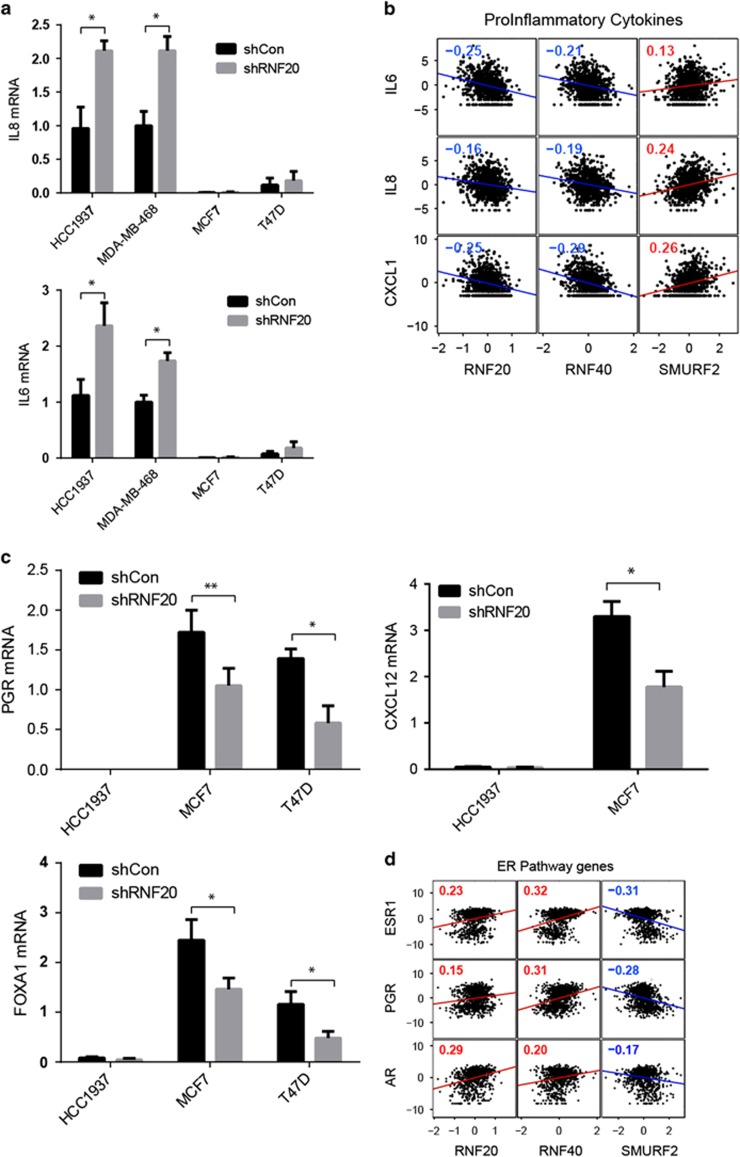
Pro-inflammatory cytokines can enhance the growth and migration of many types of cancer cells. Notably, the relative expression of many cytokine-encoding genes is higher in basal-like tumors than in luminal tumors30 (Supplementary Figure S4A). Indeed, the basal-like cell lines expressed far more IL8 and IL6 mRNA than the luminal lines (Figure 4a). Notably, RNF20 silencing further increased IL8 and IL6 expression in the basal-like cells (Figure 4a). Moreover, in agreement with earlier observations,22 RNF20 silencing decreased the repressive chromatin modification H3K9me3 on the promoters of these genes (Supplementary Figure S4B). Consistent with this, we found that the levels of IL6, IL8 and CXLC1 mRNA in individual breast tumors are inversely correlated with RNF20 and RNF40 mRNA levels, but positively correlated with SMURF2 mRNA (Figure 4b). RNF20 silencing augmented cytokine gene expression also in luminal cancer cells; however, the relative levels of these transcripts, even after RNF20 silencing, were much lower than in the basal-like cells (Figure 4a), probably rendering them inconsequential. Hence in basal-like breast cancer cells, RNF20, and presumably H2Bub1, repress cytokine-encoding genes.
Figure 4.
Regulation of cytokine genes and estrogen receptor target genes by RNF20 in basal-like and luminal breast cancer cells. (a) IL8 (top) and IL6 (bottom) mRNA levels in HCC1937, MDA-MB-468, MCF7 and T47D cells, stably expressing RNF20 shRNA or control shRNA, measured by qRT-PCR. Values were normalized to GAPDH mRNA. *P<0.05. Error bars, S.E. (b) Pearson correlation between expression levels of H2Bub1-related genes (RNF20, RNF40 and SMURF2) and pro-inflammatory cytokine genes across 837 tumor samples (TCGA database). Numbers indicate correlation R-values, P<0.05 in all panels. (c) Relative PGR (top), FOXA1 (bottom) and CXCL12 (right) mRNA levels in the indicated cell lines, determined as in a. *P<0.05; **P<0.01. Error bars, S.E. (d) Correlation between expression levels of indicated H2Bub1-regulating genes and ER pathway genes in the TCGA samples described in b
Luminal tumors and cell lines typically express the estrogen receptor-α (encoded by the ESR1 gene) and its transcriptional targets, such as the progesterone receptor (PGR) gene (Supplementary Figure S4A), and are dependent on estrogen receptor (ER) signaling for proliferation, survival and tumor progression. H2Bub1 was previously shown to promote ER target gene expression,21, 27 in conjunction with the histone chaperone SUPT6H.21 In agreement, RNF20 silencing in luminal cancer cells decreased the expression of the ER targets PGR and CXCL12 (Figure 4c), which can drive breast cancer progression, proliferation and migration,31, 32 as well as of FOXA1, encoding the pioneer transcription factor regulating ER function.33 The same genes were downregulated also in RNF20-silenced MCF7-RAS cells (Supplementary Figure S5A). Furthermore, in human breast cancers, expression of ER target genes correlates positively with RNF20 and RNF40 mRNA, but negatively with SMURF2 mRNA (Figure 4d). Thus, in luminal breast cancer cells, RNF20 and H2Bub1 promote ER target gene activation.
Overall, these results suggest that RNF20 and H2Bub1 affect the expression of genes that execute subtype-specific functions, possibly underpinning the opposite effects of this pathway on proliferation and tumorigenicity in the different subtypes.
We also observed differential effects of RNF20 silencing on several additional genes. Interestingly, RNF20 silencing upregulated the mRNA of the Polycomb factor and oncogene EZH2 (refs 34, 35) in HCC1937 cells, but decreased it in MCF7 cells (Supplementary Figure S5B), consistent with their respective changes in proliferation and motility. EZH2 may thus also contribute to the differential effects of shRNF20 on the proliferation and tumorigenicity of these cells. In addition, RNF20 knockdown elevated fibronectin (FN1) mRNA levels in HCC1937 but not MCF7 cells (Supplementary Figure S5C), suggesting that some of the HCC1937 cell population might have transitioned towards a more mesenchymal-like differentiation state.
RNF20 restricts NF-κB activity in basal-like breast cancer cells and promotes ER pathway activity in luminal cancer cells
To further elucidate the opposite impact of RNF20 on basal-like versus luminal breast cancer features, we investigated the links between RNF20 and key signaling pathways in both tumor-derived cell types.
NF-κB is a major regulator of cytokine gene transcription. Recently, we showed that RNF20 and H2Bub1 decrease NF-κB activity, restricting inflammatory cytokine secretion.22 We therefore asked whether changes in NF-κB activity might mediate the anti-proliferative effects of RNF20 in basal-like cells.
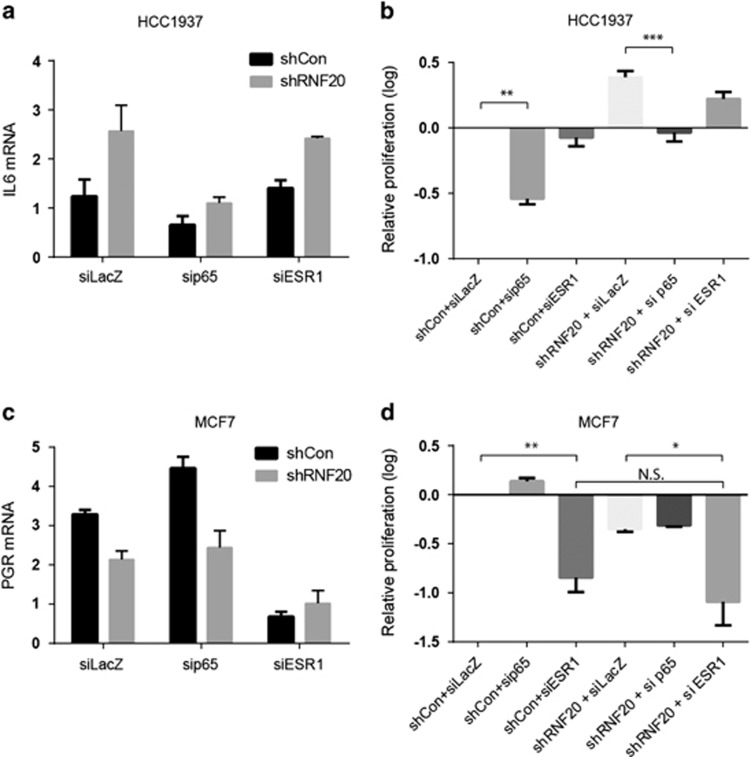
Silencing of the p65 subunit of NF-κB (Supplementary Figure S6A) downregulated IL6 mRNA in HCC1937 cells, and compromised the increase in IL6 mRNA observed upon RNF20 silencing (Figure 5a). Importantly, p65 silencing markedly attenuated the proliferation of these cells, and combined silencing of RNF20 and p65 failed to stimulate their proliferation beyond that observed in control cells (shCon+siLacZ) (Figure 5b). Hence, the positive effects of RNF20 depletion in these basal-like cancer cells rely, at least in part, on NF-κB.
Figure 5.
RNF20 promotes ER pathway activity in luminal breast cancer cells and restricts NF-kB activity in basal-like cells. (a) Relative expression of IL6 mRNA assessed by qRT-PCR in HCC1937 cells stably expressing RNF20 shRNA (shRNF20) or control shRNA (shCon) and transiently transfected with siRNA oligos targeting either p65, ESR1 or LacZ as control. Expression was normalized to GAPDH mRNA. Error bars, S.E. (b) Relative numbers of HCC1937 cells transfected as in a, counted 4 days post seeding and normalized to the shCon + siLacZ culture. **P<0.01; ***P<0.001. Error bars, S.E. (c) Relative expression of PGR mRNA, assessed by qRT-PCR in MCF7 cells stably expressing RNF20 shRNA (shRNF20) or control shRNA (shCon), transfected as in a. Expression was normalized to GAPDH mRNA. Error bars, S.E. (d) Relative numbers of MCF7 cells transfected as in a, counted 4 days post seeding and normalized to the shCon + siLacZ culture. *P<0.05; **P<0.01. N.S., non-significant. Error bars, S.E.
As expected, transient silencing of ESR1 (Supplementary Figure S6B) strongly decreased PGR expression in MCF7 cells (Figure 5c). However, no additional decrease in PGR mRNA was seen when both ESR1 and RNF20 were silenced (Figure 5c), indicating that the transcriptional effect of RNF20 on PGR is dependent on ER activity. Concordantly, the inhibitory effect of ESR1 silencing on MCF7 cell proliferation was not significantly augmented by simultaneous silencing of RNF20 (Figure 5d). These findings support the hypothesis that RNF20 promotes luminal breast cancer cell proliferation, at least partly, by enhancing ER activity.
In sum, our findings suggest that H2Bub1 can possess tumor-suppressive features in basal-like breast cancer, in part by restricting the activity of NF-κB and the expression of pro-inflammatory cytokines, but might exert tumor-promoting effects in luminal cancers, presumably by augmenting estrogen receptor transcriptional signaling.
Discussion
Earlier studies have provided apparently conflicting answers to the question whether H2Bub1 suppresses or promotes breast cancer.17, 21, 25, 27 We now report that both answers are correct, in a manner dictated by the particular subtype of breast cancer.
In TN/basal-like tumors, RNF20, RNF40 and H2Bub1 levels tend to be low and USP44 expression is relatively high, similar to what has been observed in several other types of cancer and consistent with a tumor-suppressive role. Concordantly, the prognosis of basal-like breast cancer patients with lower H2Bub1 tends to be worse than that of patients with high H2Bub1, indicating that in this type of disease H2Bub1 may indeed restrict tumor progression. In contrast, luminal tumors tend to display relatively higher levels of RNF20, RNF40 and H2Bub1, consistent with the tumor-promoting effects observed in this subtype. Indeed, luminal breast cancer patients with low H2Bub1 have a better prognosis than those with higher levels. Notably, a recent study, published while this paper was under revision, also supports a cancer-promoting role of RNF20 and RNF40 in luminal breast cancer.28
Our findings imply that the differential effects of RNF20 on the growth and metastatic potential of luminal and basal-like tumors are dictated by the differences in the key molecular mechanisms driving each tumor subtype. Although in luminal breast cancer estrogen receptor activity is a central driver, in basal-like/TN breast cancer NF-κB and cytokines are important contributors.36 In agreement with previous work,27 we found that RNF20 supports the efficient transcription of ER target genes. In contrast, H2Bub1 downregulation augments NF-κB activity and elevates the expression of pro-inflammatory cytokines, as recently observed also in an in vivo model of intestinal inflammation.22 In basal-like cancers, cytokines such as IL6 and IL8 are abundantly expressed, and promote tumor aggressiveness in both humans and mice.4, 37, 38, 39 Moreover, in these tumors, NF-κB and inflammatory cytokines can enhance tumor growth by eliciting stem cell-like features and increasing the fraction of tumor-initiating cells within the population.40, 41 Our observation that EZH2 mRNA increases in RNF20-silenced basal-like cells is also consistent with their enhanced proliferation, and suggests that their differentiation state may be partly altered. However, the specific roles played by EZH2 in this context will require further analysis. Likewise, downregulation of CXCL12 mRNA upon RNF20 silencing in luminal breast cancer cells, may contribute to their attenuated proliferation and migration.
One potential implication of these findings is that luminal tumors with low H2Bub1 might express lower levels of ER target genes, and instead might recruit alternative signaling pathways to drive tumor progression. If indeed so, anti-estrogenic therapy might be less effective in such cases than in patients with higher H2Bub1. On the other hand, basal-like and triple-negative breast cancers with low H2Bub1 could depend on the production of inflammatory cytokines more than those with high H2Bub1, and may thus respond better to anti-inflammatory treatments or inhibition of cytokine signaling (e.g., STAT3 inhibitors).
Altogether, our study demonstrates that the impact of H2Bub1 on cancer is context-dependent, and greatly depends on the particular molecular pathways that drive a given cancer. H2Bub1 is required for double-strand DNA break repair,42, 43 and its partial loss promotes genomic instability;18, 44 this might suggest that H2Bub1 should be a tumor suppressor by default. Likewise, the ability of H2Bub1/RNF20 to selectively support the expression of cancer-inhibitory genes such as TP53, while repressing proto-oncogenes such as c-MYC and c-FOS,14 is also consistent with a generic tumor suppressor function of this chromatin modification. Indeed, in many cancer types, in which H2Bub1 has been analyzed by immunohistochemistry, advanced disease is associated with decreased H2Bub1. Our observation that RNF20 silencing promotes the proliferation and migration of non-transformed mammary cells is consistent with this. Nevertheless, in cancer types where H2Bub1 supports the efficient execution of transcriptional programs that underpin the cancerous behavior, such as luminal breast cancer and leukemias with MLL gene rearrangements,25, 26 the generic tumor suppressor features of H2Bub1 may be overridden by the stronger tumor-specific selective advantages offered by H2Bub1, resulting in a drive to retain high H2Bub1 levels.
Materials and methods
Breast cancer gene and protein expression and survival analyses
Breast cancer mRNA expression data was downloaded from the TCGA portal (http://cancergenome.nih.gov/). A list of included samples is presented in Supplementary Table 1. Expression of each gene was normalized to the mean across samples and log2 transformed. Breast cancer subtypes were predicted as previously described, using the PAM50 gene predictor.45 Gene-expression correlations were calculated using Matlab employing the Pearson coefficient. Breast tumor microarrays were obtained from the Fox Chase Cancer Center (Philadelphia, PA, USA). Consecutive sections were subjected to immunohistochemical staining for H2Bub1 and H2B and quantified as previously described.22 To generate Kaplan–Meier survival plots, tumors were divided into H2Bub1-high and -low groups based on the ratio between H2Bub1 and H2B across samples, samples with ratio higher then the median were considered ‘H2Bub1 high', and those below ‘H2Bub1 low'. P-value was calculated using Gehan–Breslow–Wilcoxon test. To generate Kaplan–Meier plots from TCGA expression data, mRNA level of key H2Bub1 regulator has examined in individual samples. Those that displayed low expression (Z-score <–1) of RNF20 or RNF40 and highly expressed (Z-score >1) SMURF2 were considered low H2Bub1 pathway; samples that did not meet this criteria were considered H2Bub1 high.
Cell culture and gene perturbation
MCF7 cells were cultured at 37 °C in complete DMEM medium (Biological Industries, Beit-Haemek, Israel) supplemented with penicillin, streptomycin and 5% heat-inactivated fetal calf serum. HCC1937 cells were cultured at 37 °C in complete RPMI medium (Biological Industries) supplemented with penicillin and streptomycin and 5% heat-inactivated fetal calf serum. T47D cells were cultured at 37 °C in complete RPMI medium (Biological Industries) supplemented with penicillin and streptomycin, as well as 5 μg/ml insulin and 5% heat-inactivated fetal calf serum. MDA-MB-468 cells were cultured at 37 °C in Leibovitz L15 (Biological Industries) containing 10% FBS and supplemented with penicillin and streptomycin. Cell lines were obtained from ATCC (Manassas, VA, USA), and authenticated by STR profiling. HME primary mammary epithelial cells expressing the hTERT gene,46 were obtained from Robert Weinberg. Normal 184A1 mammary epithelial cells29 and HME-hTERT cells were grown in were grown in MEGM media (Lonza, Basel, Switzerland) mixed 1:1 with DMEM:F12, supplemented with insulin, EGF and hydrocortisone. siRNA transfection, RNA extraction, qRT-PCR western blot analysis and ChIP assay were performed as previously described.22 For stable RNA silencing, the pLKO.1-puro lentiviral vector was used. We employed standard virus generation and infection procedures, packaging with the pHRΔ8.2 and pCMV-VSV-G vectors. BrdU incorporation analysis in cells exposed to EGF pulses was as previously described.29
Proliferation and “scratch” assays
For proliferation assays, cells were seeded at 1 × 106 cells per 10 cm plate. Four days later, cells were harvested and counted. For scratch assays, cells were seeded in a culture insert (Ibidi, Martinsried, Germany). The insert was removed 24 h following seeding, and migration of cells was monitored at the indicated times.
Tumor xenografts
For xenograft implantation, 1 × 106 viable GFP-labeled cells were injected in 20 μl culture medium containing 25% Matrigel (BD-Biosciences, San Jose, CA, USA) into both #4 mammary glands of 6-week-old female NSG mice. Tumor growth during the experiment was measured by palpation, and at the end of the experiment excised tumors were weighed. Tumors were formalin-fixed and paraffin-embedded for immunohistochemistry. To assess metastatic burden, lungs were pictured and the GFP-positive area was quantified and normalized to the total lung area. All experiments involving animals were performed under approval of the Hebrew University Animal Use and Care Committee.
Immunohistochemistry
For IHC analysis, the following antibodies were used: anti-histone H2B (ab52484, Abcam, Cambridge, UK, and 07-371, Millipore, Billerica, MA, USA) and anti-H2Bub1 (56, 05-1312, Millipore). IHC was performed on paraffin-embedded tissues, employing heat-mediated antigen retrieval in 10mM citric acid (pH6.0). The UltraVision LP Detection System was used (#TL-060-HD, Thermo Scientific, Waltham, MA, USA, Scientific, Bioanalytica, Greece) according to the manufacturer's instructions. Evaluation was performed by measuring the H2Bub1/H2B ratio as previously described.22
Antibodies
The following primary antibodies were used for western blot analysis and ChIP: anti-H2B (07-371, Millipore); anti-RNF20 (ab32639, Abcam); anti-GAPDH (MAB374, Millipore); anti-H3K9me3 (ab8898, Abcam); and anti H3 (ab1791, Abcam). The anti-H2Bub1 antibody was previously described.47
Primers
For mRNA amplification the following primers were used:
IL-8: Fw 5′GGCAGCCTTCCTGATTTCTG3′
Rev 5′CTTGGCAAAACTGCACCTTCA3′
RNF20: Fw 5′GAACAGCGACTCAACCGACA3′
Rev 5′GGAATTCACCCGTTCTAGGACTT3′
GAPDH: Fw 5′AGCCTCAAGATCATCAGCAATG3′
Rev 5′CACGATACCAAAGTTGTCATGGAT3′
IL-6: Fw 5′AGCCCTGAGAAAGGAGACATGTA3′
Rev 5′TCTGCCAGTGCCTCTTTGCT3′
p65: Fw 5′CTCCGCGGGCAGCAT3′
Rev 5′TCCTGTGTAGCCATTGATCTTGAT3′
PGR: Fw 5′GTCCTTACCTGTGGGAGCTG3′
Rev 5′CGATGCAGTCATTTCTTCCA3′
ESR1: Fw 5′AGCTACTGTTTGCTCCTAACTTGCT3′
Rev 5′CCACCATGCCCTCTACACATT3′
EZH2: Fw 5′GCGCGGGACGAAGAATAATCAT3′
Rev 5′TACACGCTTCCGCCAACAAACT3′
Cxcl12: Fw 5′CGTCAAGCATCTCAAAATTCTCA3′
Rev 5′CAGCCGGGCTACAATCTGA3′
FN1: Fw 5′GACTGGGACGTTTTATCAAATTGG3′
Rev 5′TTGAGCTTGGATAGGTCTGTAAAGG3′
For ChIP analysis:
IL8 κB site: Fw5′AAACTTTCGTCATACTCCGTATTTGA3′
Rev 5′TCTCTTGGCAGCCTTCCTGA3′
IL6 kB site: Fw5′AGCCTCAATGACGACCTAAGCT3′
Rev 5′CGTCCTTTAGCATGGCAAGAC3′
Acknowledgments
This study was supported in part by grant 293438 (RUBICAN) from the European Research Council (MO), the Dr Miriam and Sheldon G Adelson Medical Research Foundation (MO), the Israel Cancer Research Fund (IB) and a Jacob and Lena Joels Memorial Foundation Senior Lectureship for Excellence in the Life and Medical Sciences (IB). MO is incumbent of the Andre Lwoff chair in molecular biology.
Author contributions
OT and RG designed and performed the experiments, and wrote the manuscript. IP and VG performed and analyzed histological studies. HM aided with the cell culture experiments. EP and BM conducted pathological analysis and provided suggestions. YZ and YY performed the analysis in Supplementary Figure S2D. MO and IB supervised the work and wrote the manuscript.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by G Melino
The authors declare no conflict of interest.
Supplementary Material
References
- TCGA,. Comprehensive molecular portraits of human breast tumours. Nature 2012; 490: 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakha EA, El-Sayed ME, Reis-Filho J, Ellis IO. Patho-biological aspects of basal-like breast cancer. Breast Cancer Res Treat 2009; 113: 411–422. [DOI] [PubMed] [Google Scholar]
- Prat A, Perou CM. Deconstructing the molecular portraits of breast cancer. Mol Oncol 2011; 5: 5–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prat A, Adamo B, Cheang MC, Anders CK, Carey LA, Perou CM. Molecular characterization of basal-like and non-basal-like triple-negative breast cancer. Oncologist 2013; 18: 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis MJ, Perou CM. The genomic landscape of breast cancer as a therapeutic roadmap. Cancer Discov 2013; 3: 27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos EI, Reinberg D. Histones: annotating chromatin. Annu Rev Genet 2009; 43: 559–599. [DOI] [PubMed] [Google Scholar]
- Osley MA. Regulation of histone H2A and H2B ubiquitylation. Brief Funct Genomic Proteomic 2006; 5: 179–189. [DOI] [PubMed] [Google Scholar]
- Kim J, Guermah M, McGinty RK, Lee JS, Tang Z, Milne TA et al. RAD6-Mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell 2009; 137: 459–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs G, Oren M. Writing and reading H2B monoubiquitylation. Biochim Biophys Acta 2014; 1839: 694–701. [DOI] [PubMed] [Google Scholar]
- Zhang XY, Pfeiffer HK, Thorne AW, McMahon SB. USP22, an hSAGA subunit and potential cancer stem cell marker, reverses the polycomb-catalyzed ubiquitylation of histone H2A. Cell Cycle 2008; 7: 1522–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glinsky GV, Glinskii AB, Berezovskaya O, Smith BA, Jiang P, Li XM et al. Dual-color-coded imaging of viable circulating prostate carcinoma cells reveals genetic exchange between tumor cells in vivo, contributing to highly metastatic phenotypes. Cell Cycle 2006; 5: 191–197. [DOI] [PubMed] [Google Scholar]
- Fuchs G, Shema E, Vesterman R, Kotler E, Wolchinsky Z, Wilder S et al. RNF20 and USP44 regulate stem cell differentiation by modulating H2B monoubiquitylation. Mol Cell 2012; 46: 662–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Sun B, Zhao X, Li Y, Liu Y, Yao Z et al. USP44+ cancer stem cell subclones contribute to breast cancer aggressiveness by promoting vasculogenic mimicry. Mol Cancer Therapeut 2015; 14: 2121–2131. [DOI] [PubMed] [Google Scholar]
- Shema E, Tirosh I, Aylon Y, Huang J, Ye C, Moskovits N et al. The histone H2B-specific ubiquitin ligase RNF20/hBRE1 acts as a putative tumor suppressor through selective regulation of gene expression. Genes Dev 2008; 22: 2664–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shema E, Kim J, Roeder Robert G, Oren M. RNF20 inhibits TFIIS-facilitated transcriptional elongation to suppress pro-oncogenic gene expression. Mol Cell 2011; 42: 477–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MA, Dickson KA, Jackson S, Clarkson A, Gill AJ, Marsh DJ. The tumor suppressor CDC73 interacts with the ring finger proteins RNF20 and RNF40 and is required for the maintenance of histone 2B monoubiquitination. Hum Mol Genet 2012; 21: 559–568. [DOI] [PubMed] [Google Scholar]
- Urasaki Y, Heath L, Xu CW. Coupling of glucose deprivation with impaired histone H2B monoubiquitination in tumors. PLoS One 2012; 7: e36775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernikova SB, Razorenova OV, Higgins JP, Sishc BJ, Nicolau M, Dorth JA et al. Deficiency in mammalian histone H2B ubiquitin ligase Bre1 (Rnf20/Rnf40) leads to replication stress and chromosomal instability. Cancer Res 2012; 72: 2111–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson LL, Guppy BJ, Sawchuk L, Davie JR, McManus KJ. Regulation of chromatin structure via histone post-translational modification and the link to carcinogenesis. Cancer Metastasis Rev 2013; 32: 363–376. [DOI] [PubMed] [Google Scholar]
- Wang ZJ, Yang JL, Wang YP, Lou JY, Chen J, Liu C et al. Decreased histone H2B monoubiquitination in malignant gastric carcinoma. World J Gastroenterol 2013; 19: 8099–8107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedi U, Scheel AH, Hennion M, Begus-Nahrmann Y, Ruschoff J, Johnsen SA. SUPT6H controls estrogen receptor activity and cellular differentiation by multiple epigenomic mechanisms. Oncogene 2014; 34: 465–473. [DOI] [PubMed] [Google Scholar]
- Tarcic O, Pateras IS, Cooks T, Shema E, Kanterman J, Ashkenazi H et al. RNF20 links histone H2B ubiquitylation with inflammation and inflammation-associated cancer. Cell Rep 2016; 14: 1462–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman RT, Stanek TJ, Esteso P, Gearhart JD, Knudsen KE, McMahon SB. The epigenetic modifier ubiquitin-specific protease 22 (USP22) regulates embryonic stem cell differentiation via transcriptional repression of sex-determining region Y-box 2 (SOX2). J Biol Chem 2013; 288: 24234–24246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zhu L, Guo T, Wang Y, Yang J. Decreased H2B monoubiquitination and overexpression of ubiquitin-specific protease enzyme 22 in malignant colon carcinoma. Hum Pathol 2015; 46: 1006–1014. [DOI] [PubMed] [Google Scholar]
- Blank M, Tang Y, Yamashita M, Burkett SS, Cheng SY, Zhang YE. A tumor suppressor function of Smurf2 associated with controlling chromatin landscape and genome stability through RNF20. Nat Med 2012; 18: 227–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E, Kawaoka S, Yu M, Shi J, Ni T, Yang W et al. Histone H2B ubiquitin ligase RNF20 is required for MLL-rearranged leukemia. Proc Natl Acad Sci USA 2013; 110: 3901–3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prenzel T, Begus-Nahrmann Y, Kramer F, Hennion M, Hsu C, Gorsler T et al. Estrogen-dependent gene transcription in human breast cancer cells relies upon proteasome-dependent monoubiquitination of histone H2B. Cancer Res 2011; 71: 5739–5753. [DOI] [PubMed] [Google Scholar]
- Duan Y, Huo D, Gao J, Wu H, Ye Z, Liu Z et al. Ubiquitin ligase RNF20/40 facilitates spindle assembly and promotes breast carcinogenesis through stabilizing motor protein Eg5. Nat Commun 2016; 7: 12648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwang Y, Sas-Chen A, Drier Y, Shay T, Avraham R, Lauriola M et al. Two phases of mitogenic signaling unveil roles for p53 and EGR1 in elimination of inconsistent growth signals. Mol Cell 2011; 42: 524–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman ZC, Poage GM, den Hollander P, Tsimelzon A, Hill J, Panupinthu N et al. Growth of triple-negative breast cancer cells relies upon coordinate autocrine expression of the proinflammatory cytokines IL-6 and IL-8. Cancer Res 2013; 73: 3470–3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos EA, Grochoski M, Braun-Prado K, Seniski GG, Cavalli IJ, Ribeiro EM et al. Epigenetic changes of CXCR4 and its ligand CXCL12 as prognostic factors for sporadic breast cancer. PLoS One 2011; 6: e29461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasmin R, Siraj S, Hassan A, Khan AR, Abbasi R, Ahmad N. Epigenetic regulation of inflammatory cytokines and associated genes in human malignancies. Mediators Inflamm 2015; 2015: 201703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 2005; 122: 33–43. [DOI] [PubMed] [Google Scholar]
- Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci USA 2003; 100: 11606–11611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietersen AM, Horlings HM, Hauptmann M, Langerod A, Ajouaou A, Cornelissen-Steijger P et al. EZH2 and BMI1 inversely correlate with prognosis and TP53 mutation in breast cancer. Breast Cancer Res 2008; 10: R109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Taguchi Y, Ito-Kureha T, Semba K, Yamaguchi N, Inoue J. NF-κB non-cell-autonomously regulates cancer stem cell populations in the basal-like breast cancer subtype. Nat Commun 2013; 4: 2299. [DOI] [PubMed] [Google Scholar]
- Barbie TU, Alexe G, Aref AR, Li S, Zhu Z, Zhang X et al. Targeting an IKBKE cytokine network impairs triple-negative breast cancer growth. J Clin Invest 2014; 124: 5411–5423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim G, Ouzounova M, Quraishi AA, Davis A, Tawakkol N, Clouthier SG et al. SOCS3-mediated regulation of inflammatory cytokines in PTEN and p53 inactivated triple negative breast cancer model. Oncogene 2015; 34: 671–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aceto N, Duss S, MacDonald G, Meyer DS, Roloff TC, Hynes NE et al. Co-expression of HER2 and HER3 receptor tyrosine kinases enhances invasion of breast cells via stimulation of interleukin-8 autocrine secretion. Breast Cancer Res 2012; 14: R131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendellen MF, Bradford JW, Lawrence CL, Clark KS, Baldwin AS. Canonical and non-canonical NF-kappaB signaling promotes breast cancer tumor-initiating cells. Oncogene 2014; 33: 1297–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T, Yang SJ, Hwang D, Song J, Kim M, Kyum Kim S et al. A basal-like breast cancer-specific role for SRF-IL6 in YAP-induced cancer stemness. Nat Commun 2015; 6: 10186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyal L, Lerenthal Y, Gana-Weisz M, Mass G, So S, Wang S-Y et al. Requirement of ATM-dependent monoubiquitylation of histone H2B for timely repair of DNA double-strand breaks. Mol Cell 2011; 41: 529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Kato A, Kobayashi J, Yanagihara H, Sakamoto S, Oliveira Douglas VNP et al. Regulation of homologous recombination by RNF20-dependent H2B ubiquitination. Mol Cell 2011; 41: 515–528. [DOI] [PubMed] [Google Scholar]
- Chernikova SB, Dorth JA, Razorenova OV, Game JC, Brown JM. Deficiency in Bre1 impairs homologous recombination repair and cell cycle checkpoint response to radiation damage in mammalian cells. Radiat Res 2010; 174: 558–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol 2009; 27: 1160–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elenbaas B, Spirio L, Koerner F, Fleming MD, Zimonjic DB, Donaher JL et al. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev 2001; 15: 50–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minsky N, Shema E, Field Y, Schuster M, Segal E, Oren M. Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nat Cell Biol 2008; 10: 483–488. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.