Abstract
Necroptosis is a form of necrotic cell death that requires the activity of the death domain-containing kinase RIP1 and its family member RIP3. Necroptosis occurs when RIP1 is deubiquitinated to form a complex with RIP3 in cells deficient in the death receptor adapter molecule FADD or caspase-8. Necroptosis may play a role in host defense during viral infection as viruses like vaccinia can induce necroptosis while murine cytomegalovirus encodes a viral inhibitor of necroptosis. To see how general the interplay between viruses and necroptosis is, we surveyed seven different viruses. We found that two of the viruses tested, Sendai virus (SeV) and murine gammaherpesvirus-68 (MHV68), are capable of inducing dramatic necroptosis in the fibrosarcoma L929 cell line. We show that MHV68-induced cell death occurs through the cytosolic STING sensor pathway in a TNF-dependent manner. In contrast, SeV-induced death is mostly independent of TNF. Knockdown of the RNA sensing molecule RIG-I or the RIP1 deubiquitin protein, CYLD, but not STING, rescued cells from SeV-induced necroptosis. Accompanying necroptosis, we also find that wild type but not mutant SeV lacking the viral proteins Y1 and Y2 result in the non-ubiquitinated form of RIP1. Expression of Y1 or Y2 alone can suppress RIP1 ubiquitination but CYLD is dispensable for this process. Instead, we found that Y1 and Y2 can inhibit cIAP1-mediated RIP1 ubiquitination. Interestingly, we also found that SeV infection of B6 RIP3−/− mice results in increased inflammation in the lung and elevated SeV-specific T cells. Collectively, these data identify viruses and pathways that can trigger necroptosis and highlight the dynamic interplay between pathogen-recognition receptors and cell death induction.
Initiation of innate immunity against pathogens occurs upon recognition of pathogen-derived components by pattern-recognition receptors (PRRs). Once engaged, PRR signalling can result in production of inflammatory cytokines to fight pathogens. Infected cells may also be eradicated through cellular death pathways like apoptosis and pyroptosis.1, 2 Signal transduction pathways of cell death and those mediated by PRRs were long considered to be distinct, but recent evidence suggests a dynamic interplay between the two pathways. However, the molecular details of this crosstalk are still not completely clear.
In addition to apoptosis and pyroptosis, an alternate form of caspase-independent programmed cell death termed ‘necroptosis' has been identified.3, 4, 5 Necroptosis can be initiated by TNF or by TLR3/TLR4 ligands, DNA damaging agents and T-cell receptor ligation.6 Signalling in cells deficient of the death receptor adapter protein FADD or caspase-8 leads to deubiquitination of the serine/threonine kinase, RIP1, and its recruitment to family member, RIP3, to form a ‘necrosome' complex. Inhibition of RIP1 kinase activity or deletion of RIP3 prevents necroptosis.7, 8, 9, 10 Necroptosis is inherently inflammatory due to the release of intracellular contents. Thus, it has been suggested that in some cases, recognition of pathogens by PRRs may lead to necroptosis where the ensuing inflammation is crucial in fighting infection.
Given that viruses often encode inhibitors of apoptosis and that disruption of apoptotic machinery may trigger necroptosis, it is not surprising that necroptosis has recently surfaced as a potential antiviral mechanism. For example, vaccinia virus infection can sensitize cells to TNF-induced necroptosis due to expression of the viral caspase inhibitor, B13R.9, 11, 12 Moreover, RIP3-deficient mice die when exposed to vaccinia.9 Mice deficient in the apoptosis inhibitor, cIAP2, succumb to infection by influenza virus due to FasL-dependent necrosis of airway epithelia,13 while the murine cytomegalovirus, MCMV, was found to encode an inhibitor of necroptosis.14 Similarly, the viral protein, ICP6, from herpes simplex virus 1 (HSV-1) directly engages RIP3 to promote necroptotic death.15, 16, 17 Nevertheless, the prevalence of virus-induced necroptosis and molecular mechanisms leading to this form of cellular demise remains to be fully characterized. Here, we report that out of seven viruses (MCMV, lymphocytic choriomeningitis virus – LCMV, Sendai virus – SeV, murine gammaherpesvirus-68 – MHV68, influenza – WSN, Kaposi's sarcoma-associated herpesvirus – KSHV, HSV-1) that were examined, only two, MHV68 and SeV, dramatically induce necroptosis in a necroptotic-sensitive cell line. We found that MHV68-induced necroptosis proceeds through a STING and TNF-dependent pathway, while SeV promotes activation of a novel RIG-I-dependent necroptotic pathway. Furthermore, we have identified two SeV proteins, Y1 and Y2, which are crucial for this process. Moreover, infection of RIP3-deficient mice with SeV resulted in increased pulmonary inflammation as well as SeV-specific T cells. Our findings highlight a novel interaction between PRR sensors and the necroptotic pathway.
Results
Sendai virus and MHV68 induce necroptosis through TNF-independent and dependent manner
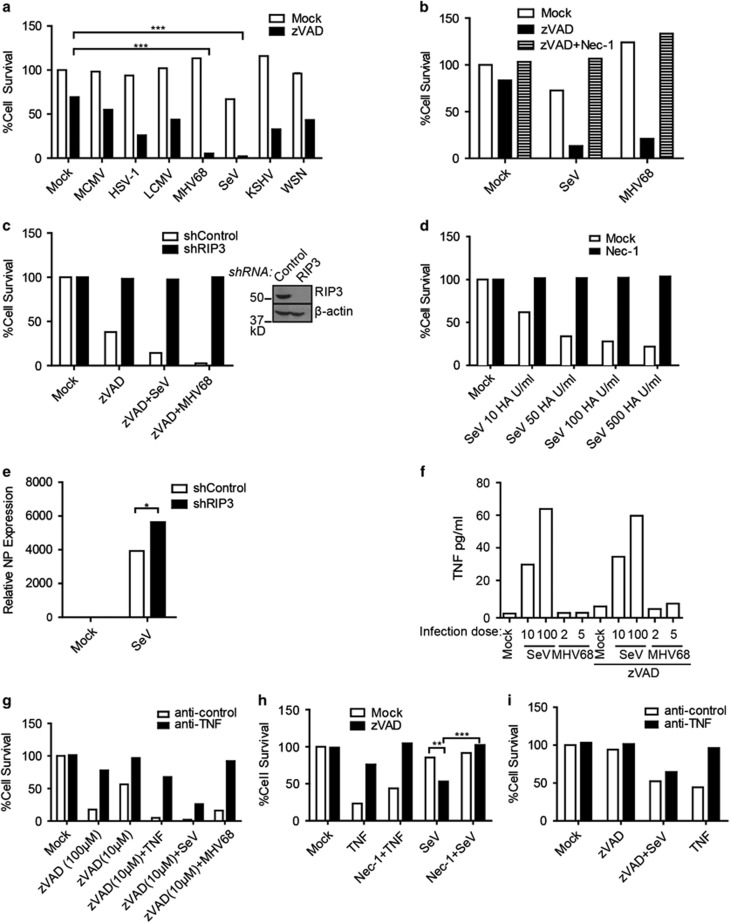
To examine the prevalence of virus-induced necroptosis, we examined several viruses in L929 cells. This murine fibrosarcoma cell line is a prototypic system in which to study necroptosis due to its ability to readily undergo necroptosis in response to zVAD.FMK (zVAD) or TNFα treatment.18 Four different DNA viruses: MCMV, MHV68, HSV-1 and KSHV and three different RNA viruses: SeV, LCMV and WSN were used. Treatment with zVAD alone results in a low level of death at the concentration used. However, additional infection with SeV or MHV68 dramatically enhanced cell death, with less than 5% viable cells remaining (Figure 1a). In contrast, the other viruses examined yielded substantial cell viability. Curiously, neither influenza nor KSHV, two viruses related to Sendai and MHV68, respectively, induced death as dramatically as SeV or MHV68 (Figure 1a). Intriguingly, others have reported necroptotic induction by HSV-1, which we did not observe at our infection condition of MOI 5. Infecting with an MOI of 20, however, did cause increased death of L929 cells (Supplementary Figure S1).15, 16, 17 As the viral titres were not determined in L929 cells, but in standard susceptible cell lines, it is possible that in alternate cell lines and/or other conditions, these other viruses can induce necroptosis. We focused on SeV and MHV68 as they induced the most striking death in L929 cells. We were able to determine that the death induced by these two viruses was necroptosis. Addition of the RIP1 kinase inhibitor Necrostatin-17 rescued SeV and MHV68-induced death (Figure 1b). Moreover, knockdown of RIP3 prevented virally induced cell death (Figure 1c). Infection with SeV alone was able to induce RIP1-dependent necroptosis as well (Figure 1d). Further characterization of virus-induced death revealed necroptosis to occur after 10 h of infection and in a dose-dependent manner (Supplementary Figure S2).
Figure 1.
Sendai virus and MHV68 cause RIP1- and RIP3-dependent necroptosis. (a) L929 cells were treated with zVAD (10 μM) and infected with MCMV (MOI 5), HSV-1 (MOI 5), LCMV (MOI 5), MHV68 (MOI 5), SeV (10 HA U/ml), KSHV (titre giving 70% infection) or WSN (MOI 5) and % cell survival was determined by the CellTiter-Glo assay. (b) Cell death of L929 cells treated with zVAD (10 μM), Necrostatin-1 (20 μM) and infected with SeV (10 HA U/ml) or MHV68 (MOI 5). (c) L929 cells stably expressing mouse shRIP3 or control shRNA were treated with or without zVAD (5 μM) in the absence or presence of SeV (10 HA U/ml) or MHV68 (MOI 5). Right panel: cell lysates of the shRNA retrovirally infected cells were probed by RIP3- or actin-specific antibodies. (d) L929 cells were mock-treated or treated with Necrostatin-1 (Nec-1) at 20 μM and infected with indicated titres of SeV. Cell death was monitored 18–20 h post-infection by CellTiter-Glo. (e) Increased relative expression of viral NP protein from RIP3 shRNA knockdown cells as compared to control shRNA knockdown cells by quantitative RT-PCR. Expression is relative to mock-infected samples after normalization to γ-actin. (f) TNF ELISA from supernatants of L929 cells treated with zVAD and infected with SeV (10 or 100 HA U/ml) or MHV68 (MOI 5 or 10). (g) Death of cells treated with control or anti-TNF neutralizing antibody, zVAD (10 μM and 100 μM), TNF (60 pg/ml), SeV (100 HA U/ml) or MHV68 (MOI 5). (h) LA-4 cells were treated with zVAD (20 μM), TNF (100 ng/ml) or Nec-1 (20 μM) and infected with SeV (100 HA U/ml) or MHV68 (MOI 10). Cell survival was monitored 24 h post-infection. (i) LA-4 cells were treated with anti-control or anti-TNF neutralizing antibodies and treated with zVAD, TNF and infected with SeV (100 HA U/ml) before cell survival was assayed. Experiment repeated twice. Data are averages of triplicates from a single experiment, which is representative of at least three independent experiments (unless otherwise stated). ***P<0.001, **P<0.01, *P<0.05
We found that SeV infection of RIP3 knockdown cells resulted in increased viral loads as measured by RT-PCR of the SeV NP mRNA (Figure 1e). Thus, necroptotic death of the cells may be important in limiting SeV viral load. Given that L929 cells can produce TNF and undergo autocrine-induced necroptosis,18 we next investigated whether TNF production is responsible for the virus-induced necroptosis. ELISA of virally infected supernatants revealed SeV to be a potent inducer of TNF, with up to 60 pg/ml produced upon infection (Figure 1f). In contrast, infection of MHV68 caused production of only 3 pg/ml of TNF. Addition of zVAD did not significantly alter levels of TNF by either virus (Figure 1f). To examine whether TNF was responsible for the ensuing necroptosis, L929 cells were treated with anti-control or anti-TNF neutralizing antibodies prior to addition of zVAD and virus. As a control, we showed that addition of an anti-TNF neutralizing antibody rescued death of L929 cells induced by zVAD and 60 pg/ml of TNF, an amount equivalent to that produced by SeV infection (Figure 1g). In addition, we could rescue death induced by a high dose of zVAD (100 μM) to mimic death induced by endogenous production of TNF (Figure 1g). Inverse to the levels of TNF produced, the same amount of anti-TNF neutralizing antibody rescued MHV68-induced death but not the majority SeV-induced death (Figure 1g). These data suggest that SeV-induced necroptosis occurs through a novel mechanism that is mostly TNF independent, while MHV68 induces necroptosis via TNF, presumably by sensitizing the cells to TNF signalling. In addition to L929 cells, we also found that the lung adenoma cell line, LA-4 cells,19 were sensitive to SeV-induced necroptosis. Treatment of LA-4 cells with TNF alone caused cell death that was primarily apoptotic, as zVAD rescued the majority of it. Infection with SeV and zVAD, however, resulted in necroptosis that was rescued by Nec-1 (Figure 1h). Although not as dramatic as zVAD/SeV-induced necroptosis in L929, zVAD/SeV-induced necroptosis in LA-4 cells was also mostly independent of TNF (Figure 1i). Additionally, we found the lung epithelial type I cell line, LET1,20 was also susceptible to necroptosis when infected with SeV alone (at 500 HA U/ml) or in the presence of zVAD (Supplementary Figure S3).
Necroptosis by SeV is RIG-I dependent while MHV68 is STING dependent
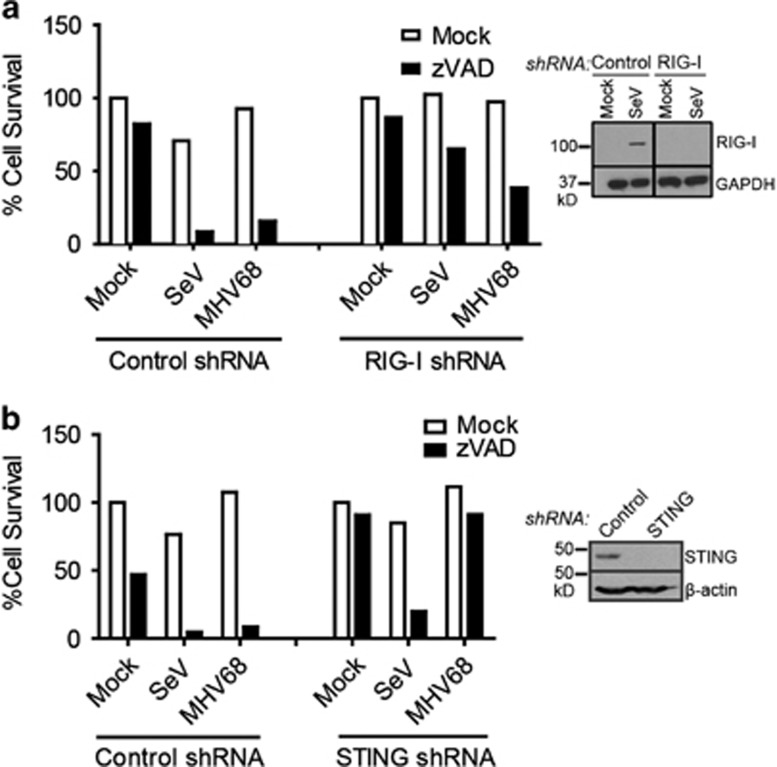
In addition to TNFR family members, activation of certain pattern recognition receptors can elicit necroptotic cell death.21, 22 Innate immune recognition of MHV68 occurs primarily through TLR2 and TLR9.23, 24 However, L929 cells do not express detectable levels of TLR2 or TLR9.25, 26 Thus, induction of necroptosis by MHV68 must occur through an alternate mechanism. SeV, on the other hand, is a potent activator of the cytoplasmic RNA sensor RIG-I.27 The ER-resident molecule Stimulator of IFN Genes (STING) is also important for IFN production upon SeV infection.28 Thus, to determine whether necroptosis induction may be initiated through these pathways, L929 cells stably expressing shRNA against RIG-I or STING were generated (Figures 2a and b). L929 cells express a high level of STING and low basal levels of RIG-I, which are upregulated upon viral infection. We confirmed knockdown of RIG-I in SeV-infected cells (Figure 2a) and knockdown of STING in uninfected cells (Figure 2b). Knockdown with RIG-I but not control shRNA potently rescued SeV-mediated death. Infection with MHV68 yielded minor rescue (Figure 2a). Surprisingly, knockdown of STING was unable to effectively rescue SeV-induced necroptosis, yet drastically rescued MHV68-induced death (Figure 2b). Thus, it appears that SeV initiates necroptosis through activation of RIG-I independently of STING. MHV68, on the other hand, appears to signal through a STING-dependent pathway.
Figure 2.
Sendai virus induces necroptosis through RIG-I while MHV68 signals through STING. (a) L929 cells stably expressing mouse RIG-I shRNA or control shRNA were treated with zVAD (10 μM) and infected with SeV (10 HA U/ml) or MHV68 (MOI 5). Right panel: knockdown was confirmed by western blot of L929 cells infected with SeV to upregulate RIG-I protein expression. (b) L929 cells stably expressing mouse STING shRNA or control shRNA were treated with zVAD (10 μM) and infected with SeV (10 HA U/ml) or MHV68 (MOI 5). Right panel: knockdown was confirmed by western blot. Data are averages of triplicates from a single experiment, which is representative of at least three independent experiments
SeV Y1 and Y2 accessory proteins promote viral necroptosis
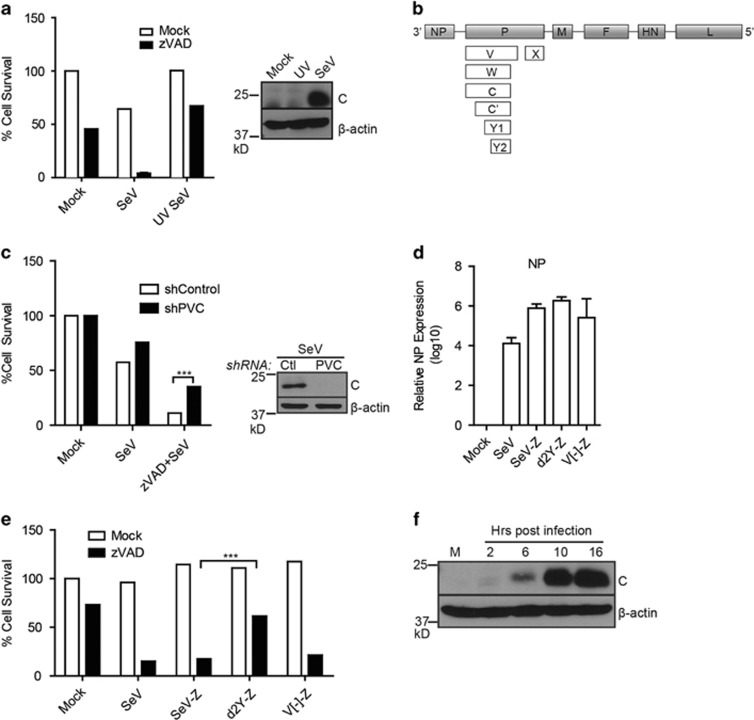
Viruses are known for their ability to modulate host cell responses. To investigate whether expression of viral protein(s) are responsible for eliciting necroptosis of SeV-infected cells, L929 cells were infected with either live or UV-irradiated virus. Irradiation of viruses by ultraviolet rays damages viral genomes to prevent viral gene expression while still allowing viral entry and activation of certain antiviral proteins independent of viral replication. Consistent with this, infection with UV-irradiated SeV still leads to expression of the interferon-stimulated protein, ISG56.29 UV inactivation impaired SeV-induced necroptotic death in L929 and LA-4 cells, suggesting that replication and/or expression of a particular viral protein(s) is required (Figure 3a, Supplementary Figure S4).30 SeV is a small non-segmented, negative, single-stranded RNA virus with six structural genes.31 The P gene of SeV is unique in its ability to produce up to seven different accessory proteins via overlapping reading frames or mRNA editing (Figure 3b). Collectively referred to as ‘C proteins', C, C′, Y1 and Y2 are a set of nested genes initiated at different start codons but which share the same termination site. To study the role of SeV accessory proteins in L929 cells, an shRNA was used to knockdown expression of the P gene (and consequently, the majority of accessory proteins). Immunoblot analysis confirmed knockdown of the C proteins upon infection (Figure 3c). Knockdown partially rescued SeV-induced cell death, suggesting that expression of a particular viral protein(s) is likely required to provoke necroptotic cell death. Accordingly, expression of the C proteins was not seen after infection of L929 cells with UV-irradiated SeV (Figure 3a). As expression of multiple genes was reduced by the PVC shRNA, we then used several mutants previously generated from the SeV Z strain to dissect a role for viral proteins.32, 33, 34 These include a d2Y mutant that lacks Y1 and Y2 proteins from SeV as well as a mutant lacking the V protein (V[-]-Z). Both d2Y and V[-] mutants were reported to behave similarly to WT SeV in vitro.32, 34 Consistent with this, infection with these viral mutants revealed equivalent expression of the SeV NP gene as analysed by RT-PCR, demonstrating a comparable level of infection (Figure 3d). Infection of L929 cells with the wild type (SeV-Z) or V[-]-Z mutant resulted in an equivalent level of necroptotic death as that which is generally seen with the Cantell strain of SeV (Figure 3e; compare SeV to SeV-Z and V[-]-Z). However, the d2Y mutant-infected cells exhibited diminished death, indicating that the Y1/Y2 protein of SeV is at least partially responsible for the necroptotic phenotype observed (Figure 3e). Moreover, we saw expression of ‘C proteins' upregulated 10 h post-infection (Figure 3f), concomitant with the previously determined kinetics of cell death (Supplementary Figure S2). Thus, Y1/Y2 SeV proteins appear to sensitize L929 cells towards necroptotic death.
Figure 3.
Sendai virus Y1/Y2 proteins are required for necroptosis. (a) Infection of L929 cells with ultraviolet-inactivated (UV) SeV rescues cell death as monitored by CellTiter-Glo (experiment repeated three times). Right panel: western blot of L929 cells infected with live or UV-inactivated SeV (10 HA U/ml) after 18 h (performed once) (41). (b) Schematic representation of SeV proteins. Six structural proteins (NP, P, M, F, HN, L) and accessory proteins (V, W, X, C, C', Y1, Y2). (c) L929 cells stably expressing a control (Ctl) shRNA or shRNA against P and accessory proteins (PVC) were treated with zVAD and infected with SeV (10 HA U/ml) and cell survival monitored. Right panel: knockdown was monitored by western blot using antibodies against the C proteins of SeV. (d) L929 cells were infected with Cantell strain of SeV, Z strain of SeV (SeV-Z), or mutants (d2Y-Z, V[-]-Z). 16–18 h post-infection, mRNA was isolated and used for quantitative RT-PCR using oligonucleotides specific for the SeV NP or γ-actin gene. Expression was plotted on a log scale relative to mock-infected samples after normalization to γ-actin. Standard error of the mean was calculated from three independent experiments. (e) Survival of cells infected with Cantell strain of SeV (SeV) or Z strain (SeV-Z, d2Y-Z, V[-]-Z) as measured by CellTiter-Glo assay. (f) Lysates from SeV-infected cells at various time points (M: mock-infected) were probed in a western blot analysis using SeV C protein or β-actin specific antibodies (experiment repeated twice). For 3A, 3C, 3D, 3E, data are averages of triplicates from a single experiment, which is representative of at least three independent experiments. ***P<0.001
zVAD and SeV infection modulate RIP1 ubiquitination
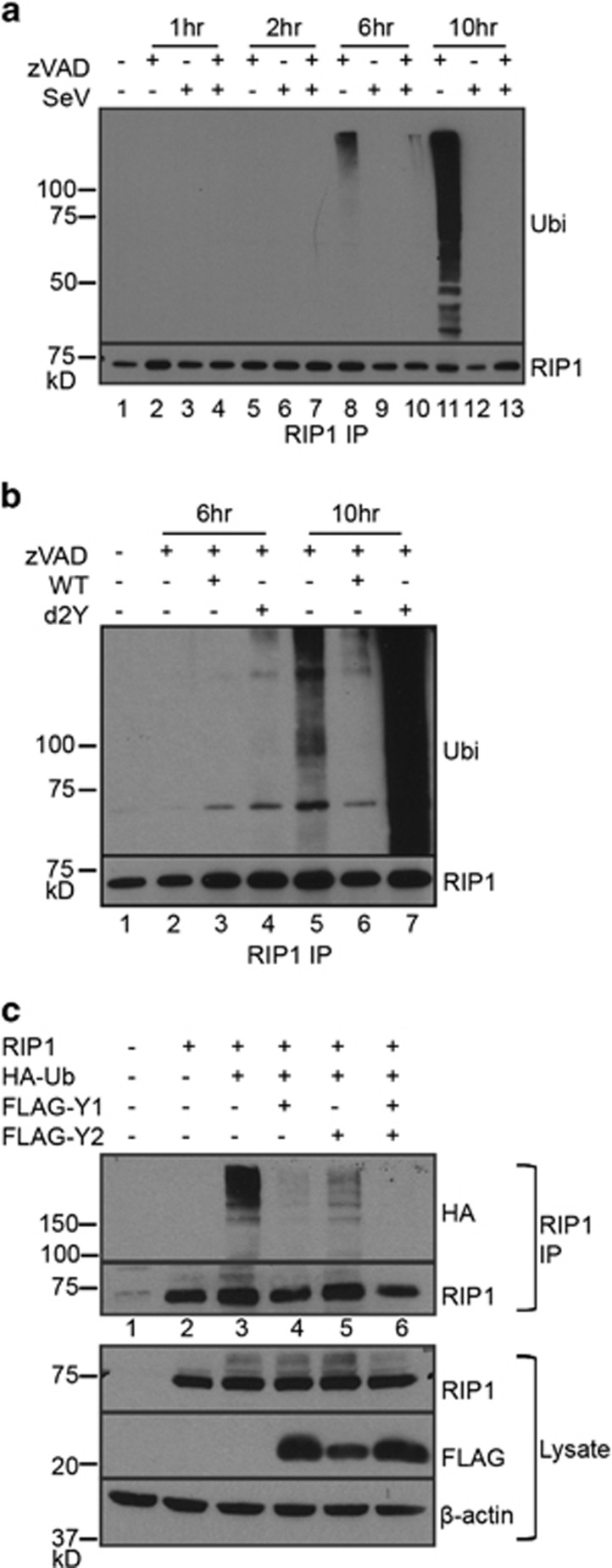
Necroptosis induction has been largely studied downstream of the TNF receptor, where ubiquitination status of RIP1 is a major determinant in the regulation of necroptotic death.3, 4, 5 RIP1 is heavily ubiquitinated when participating in NF-κB signalling. However, upon deubiquitination, RIP1 can associate with either apoptotic or necroptotic signalling machinery.6, 35 Thus, loss of RIP1 ubiquitination can be used to monitor a cell's susceptibility towards cell death. We investigated whether RIP1 ubiquitination status is critical in modulating SeV-induced necroptosis. To examine this, RIP1 was immunoprecipitated from zVAD- and SeV-infected L929 cells and ubiquitination status monitored by immunoblot over time (Figure 4a). Diminished RIP1 ubiquitination was already evident for zVAD and SeV treated cells at 6 h post-infection as compared to cells treated with zVAD alone (Figure 4a). By 10 h post-infection, RIP1 ubiquitination was clearly evident in zVAD treated cells but was completely lost in cells treated with zVAD and SeV (Figure 4a lanes 11 versus 13). The presence of protein species found below the RIP1 band could be due to degradation products or perhaps co-immunoprecipitating RIP3, which others found could persist in the presence of urea.36 Thus, infection with SeV reduces ubiquitination of RIP1, resulting in increased necroptosis.
Figure 4.
Loss of RIP1 ubiquitination determines SeV-induced necroptosis. (a) L929 cells were treated as indicated and cell lysates harvested in 1% SDS lysis buffer. RIP1 immunoprecipitation was carried out from SDS lysates and ubiquitination monitored by immunoblot using ubiquitin-specific antibodies. RIP1-specific antibodies were used to ensure uniform RIP1 immunoprecipitation. (b) Infection with d2Y mutant yields higher RIP1 ubiquitination compared to WT SeV infection. A similar experiment as (a) was carried out but using SeV Z wild type (WT) or d2Y mutant virus. (c) 293T cells were transfected with indicated plasmids and lysed using 1% NP40 lysis buffer. Immunoprecipitated RIP1 was resolved by SDS-PAGE and immunoblotted with anti-HA to determine ubiquitination. RIP1 western blot was performed to ensure uniform RIP1 immunoprecipitation. Bottom panel: lysates of the 293T cells transfected with the indicated constructs were probed for RIP1 or Y1/Y2 expression using RIP1 or FLAG-specific antibodies. All experiments repeated at least three times
To determine whether the Y1/Y2 viral proteins are required for deubiquitination of RIP1, cells infected with the d2Y mutant virus were examined. RIP1 immunoprecipitates revealed that the d2Y mutant virus was no longer able to induce deubiquitination of RIP1 as compared to wild type virus (Figure 4b lanes 6 versus 7). Thus, we implicated the Y1/Y2 protein as promoting deubiquitination of RIP1. In accordance with mutant virus data, overexpression of viral Y1 and Y2 proteins in 293T cells is sufficient to diminish ubiquitination of RIP1 (Figure 4c).
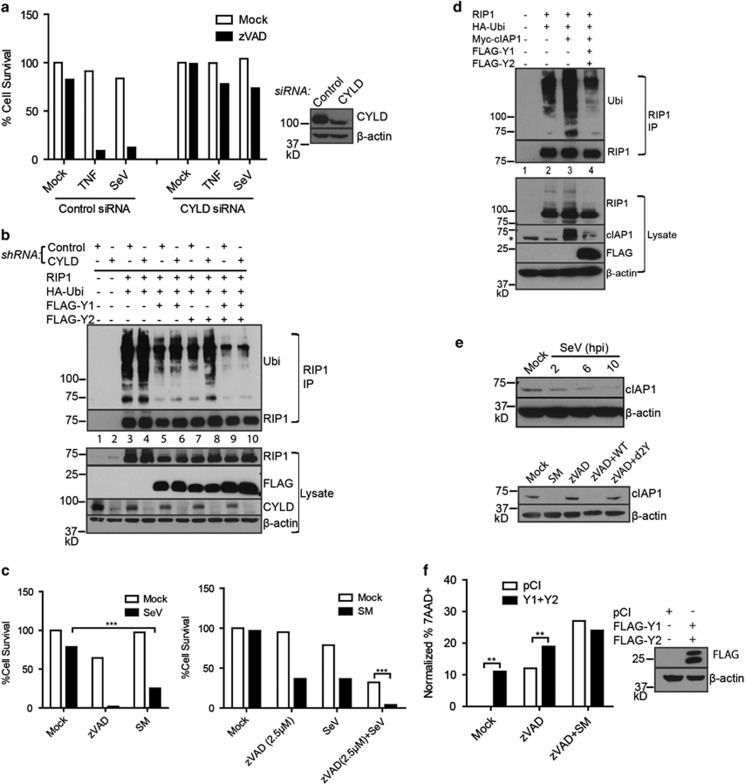
Downstream of TNFR signalling, RIP1 ubiquitination occurs primarily through the E3 ubiquitin ligases cIAP1 and cIAP2 to promote NF-κB activation and cell survival.37, 38, 39 In contrast, the deubiquitinase CYLD is critical in mediating the transition of RIP1 away from NF-κB activation and towards cell death.36 In addition, both CYLD and cIAP proteins act downstream of RIG-I to mediate IFN responses.40, 41 Thus, theoretically, either of these proteins could be involved in RIG-I-dependent SeV-induced death. Y1/Y2 may enhance CYLD's activity to deubiquitinate RIP1 or alternatively, they may inhibit ubiquitination of RIP1. To assess whether CYLD was required for the activity of Y1/Y2, its expression was knocked down from L929 cells using siRNA. Loss of CYLD expression rescued SeV-induced death as expected (Figure 5a). However, knockdown of CYLD did not significantly affect the ability of Y1 or Y2 to downregulate RIP1 ubiquitination (Figure 5b). We next sought to determine whether cIAP-mediated ubiquitination of RIP1 is important in SeV-induced death. L929 cells were treated with an SMAC mimetic to induce degradation of cIAP proteins. Although not as dramatic as zVAD/SeV-induced death, SMAC mimetic and SeV infection also caused significant cell death (Figure 5c, left panel). Moreover, at low concentrations of zVAD, which were not sufficient for dramatic death, addition of SMAC mimetic further sensitized cells to death upon SeV infection (Figure 5c, right panel).
Figure 5.
Sendai virus proteins restrict cIAP1-mediated RIP1 ubiquitination. (a) Knockdown of CYLD protein by siRNA in L929 cells rescues SeV induced necroptosis. Cells transfected with control siRNA or CYLD siRNA were treated with the indicated agent and/or SeV. Right panel: western blot analysis of the control siRNA or CYLD siRNA treated cells using CYLD- or β-actin specific antibodies. (b) Knockdown of CYLD using shRNA in 293T cells overexpressing indicated constructs. RIP1 was immunoprecipitated (RIP1 IP) from 1% NP40 lysates and its ubiquitination was monitored by western blot with ubiquitin-specific antibodies. Lower panel: western blot analysis of lysates with the indicated antibodies. (c) Left panel: treatment of L929 cells with 100 nM SMAC Mimetic (SM) and infection with SeV (10 HA U/ml) causes death of cells. Right panel: SMAC mimetic treatment sensitizes low dose zVAD (2.5 μM) and SeV infection to cell death. (d) RIP1 was immunoprecipitated from 1% NP40 lysates generated from 293T cells transfected with indicated constructs. cIAP1-mediated RIP1 ubiquitination was monitored by western blot. Top panel: RIP1 immunoprecipitates (RIP1 IP) were analysed by western blot analysis using ubiquitin- or RIP1-specific antibodies. Lower panel: lysates from the transfected cells were analysed using the indicated antibodies. Asterisk indicates non-specific band. (e) Top panel: loss of cIAP1 protein expression upon infection with SeV after 10 h post-infection (hpi). Lower panel: lysates from L929 cells treated with indicated agents were resolved by SDS-PAGE and western blot analysis confirmed loss of cIAP1 protein in wild type (WT) SeV-infected samples but not d2Y mutant-infected samples. (f) L929 cells were transfected with pCI or Y1/Y2 constructs and subsequently treated with zVAD (2.5 μM) and SM (100 nM). Cell death was determined by 7AAD+ staining by flow cytometry. For, 5A, 5C, 5F, data are averages of triplicates from a single experiment, which is representative of at least three independent experiments. All experiments repeated at least three times. ***P<0.001, **P<0.01, *P<0.05
To determine whether Y1 and Y2 proteins promote necroptosis through modulation of cIAP proteins, ubiquitination of RIP1 was monitored in 293T cells overexpressing RIP1 and ubiquitin with either cIAP1 alone or a combination of cIAP1, Y1 and Y2 (Figure 5d). RIP1 was immunoprecipitated followed by western blot analysis with ubiquitin-specific antibodies. While RIP1 ubiquitination is increased in the presence of cIAP1, addition of Y1 and Y2 diminished its ubiquitination (Figure 5d). Interestingly, expression of Y1/Y2 proteins also reduces expression of cIAP1, suggesting that they promote cIAP1 protein degradation (Figure 5d). Consistent with this, loss of cIAP1 expression was seen in cells infected with SeV by 10 h post-infection (Figure 5e). Loss of cIAP1 could also be seen from zVAD treated cells infected with wild type SeV, but not with d2Y mutant virus (Figure 5e). To determine if expression of Y1/Y2 was sufficient to induce cell death, L929 cells were transfected with FLAG-tagged Y1 and Y2 constructs and treated with zVAD alone or zVAD plus SMAC mimetic and cell death was monitored by 7AAD. Interestingly, in contrast to addition of SMAC mimetic, expression of Y1/Y2 alone resulted in increased cell death, suggesting that Y1/Y2 might have other biological functions in addition to downregulation of cIAP1 (Figure 5f). Cell death was further enhanced with zVAD treatment and Y1/Y2 expression did not increase death induced by a combination of zVAD and SMAC mimetic treatment. We conclude that the Y1/Y2 proteins from SeV promote necroptotic death by downregulating cIAP1 expression.
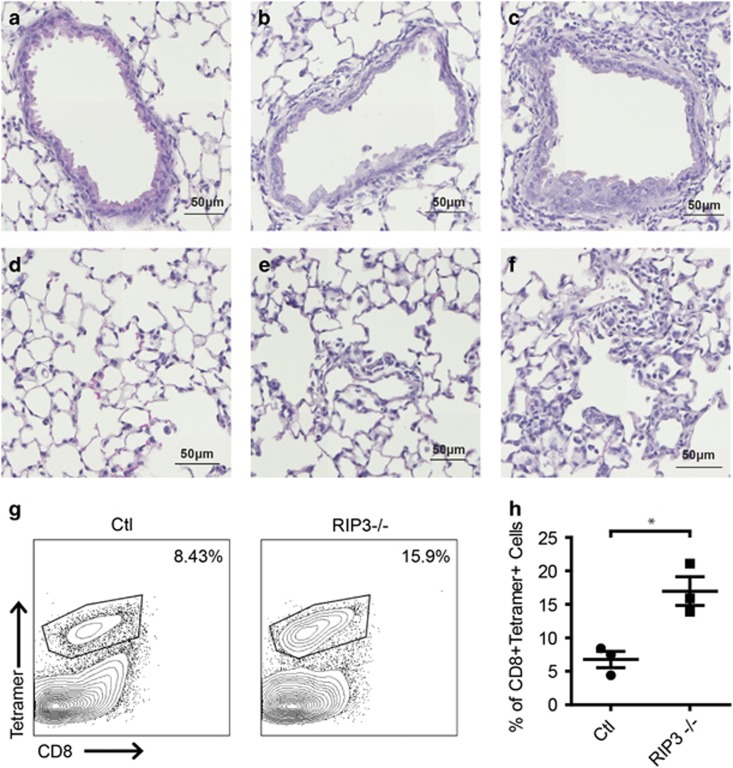
To examine the role of necroptosis in vivo, we infected mice with SeV. SeV causes a pneumonia-like respiratory infection in mice that is often used as a model of human parainfluenza virus infection.42 The severity of infection varies upon mouse strain, with 129/J mice being the most susceptible and C57BL/6 being the most resistant.43 We infected wild type and RIP3-deficient littermate mice (on the C57BL/6 background) with SeV (Cantell strain). Given the resistant nature of the C57BL/6 background, no differences were observed in weight loss upon infection as expected (data not shown). However, histological analysis of H&E stained lung sections exhibited marked cellular infiltration and inflammation as well as thickening of bronchial walls in SeV-infected RIP3-deficient animals (Figures 6c and f) as compared to infected wild-type animals (Figures 6b and e). To examine SeV-specific T cells, we recovered lymphocytes from the lung by bronchoalveolar lavage (BAL) 9 days post-infection. Interestingly, we found that virus-specific CD8+ T cells (revealed by SeV NP324-332/Kb tetramer staining) were increased in RIP3-deficient mice (Figures 6g and h). Moreover, these virus-specific CD8+ T cells exhibited an increased activated phenotype (CD44+Tetramer+) as compared to control mice (data not shown). Although more experiments need to be done, these data illustrate the importance of RIP3 for SeV infection in vivo and suggest that our observation in cell lines also has physiological significance in mice.
Figure 6.
Lung and BAL analysis of SeV-infected wild type and RIP3-deficient mice. (a–f) Representative haematoxylin- and eosin- (h & e) stained lung sections of uninfected wild type (a, d), SeV-infected wild type mice (b, e) and SeV-infected RIP3−/− mice (c, f) 7 days post-infection. (g) Increased frequency of anti-SeV CD8+ T cells found in BAL of SeV-infected RIP3−/− mice as compared to control mice 9 days post-infection. Results are representative of three control (Ctl) and 3 RIP3−/− mice infected with SeV or 1 control and 1 RIP3−/− mice infected with PBS. (h) Quantitation of anti-SeV CD8+ T cells from control and RIP3−/− mice. *P<0.05
Discussion
Necroptosis has been described to occur upon viral infection but we do not have a clear understanding of the prevalence of this phenomenon. In this paper, we find that striking necroptosis was induced by two out of seven virus strains in the L929 cell line under the conditions investigated. We provide evidence that MHV68-induced necroptosis requires activation of the adaptor molecule, STING and proceeds in a TNF-dependent fashion. In contrast to MHV68, SeV-induced necroptosis requires RIG-I activation as well as the presence of virally encoded proteins Y1 and/or Y2. SeV induced ample amounts of TNF; however, necroptosis is surprisingly independent of TNF. The SeV C proteins (including Y1 and Y2) have been suggested to inhibit interferon signalling and apoptosis.44, 45 Our data strongly suggest that Y1/Y2 proteins are involved in necroptosis induction by diminishing cIAP1 protein levels and thus cIAP1-mediated ubiquitination of RIP1. SMAC proteins destabilize cIAP proteins through direct interaction utilizing a conserved, N-terminal, four residue IAP-binding motif (AVPI).46 Examination of Y1/Y2 amino acid sequence from the Cantell and Z strains of SeV reveals a similar motif (ALPI) in Cantell but not in the Z strain (ALLI). Thus, it is unlikely that Y1/Y2 utilize this mechanism to promote necroptosis. In addition, we were unable to detect a direct interaction between Y1/Y2 and cIAP1 (data not shown). The expression of Y1/Y2 alone was able to induce death of L929s while treatment with SMAC mimetic alone did not. These data suggest that Y1/Y2 may have another biological function in addition to their ability to promote cIAP1 degradation. As cIAP proteins themselves are anti-apoptotic, the necroptotic function of Y1/Y2 seems to contradict the speculative role of C proteins as anti-apoptotic. However, closer examination of published data suggests that anti-apoptotic activity reported for C proteins may be specific to the case of interferon-induced apoptosis, as this effect was not seen in interferon-deficient Vero cells.44 Moreover, we find that in addition to destabilizing cIAP1 levels, SeV-induced death still requires CYLD. It is possible that SeV-induced cIAP1 downregulation is itself insufficient to reduce RIP1 ubiquitination to the threshold necessary for its necroptotic function.
Caspase-8 has been shown to cleave RIP1 to downregulate RIG-I signalling.47 In our study, necroptosis induction was most dramatic with addition of the caspase-8 inhibitor zVAD. Thus, basal levels of caspase-8 may be important in restricting RIG-I-dependent necroptosis, therefore yielding canonical IFN production upon RIG-I activation. Caspase-8 expression is variable between tissues and can be inhibited by regulators such as cFLIP.22, 48 In these conditions, the SeV Y1/Y2 proteins, which inhibit cIAP1 activity, likely hinder induction of IFN downstream of RIG-I, instead resulting in necroptosis.
Finally, we have found that infection of RIP3-deficient mice with SeV resulted in increased pulmonary cellular infiltration and pathology as compared to infection with wild type mice. This is consistent with our observation that knockdown of RIP3 in L929 results in increased viral titre, which may in turn lead to increased inflammation in vivo. At 9 days post-infection, however, both wild type and RIP3-deficient mice had cleared the virus (data not shown). Whether there are increased SeV titres at earlier time points await future experiments. RIP3-dependent cellular death is important in viral clearance from influenza-infected mice.49 Interestingly, influenza-infected RIP3−/− mice also exhibited histological abnormalities suggesting that necroptosis might be important for both influenza and SeV viral clearance. However, in contrast to influenza-infected RIP3−/− mice where a decrease of virus-specific T cells was detected,49 we observed an increased frequency of activated SeV-specific CD8+ T cells in RIP3-deficient animals. These data may suggest that SeV have taken advantage of the necroptotic process to decrease viral immunity. In susceptible strains of mice, loss of RIP3 may actually lead to increased resistance to viral pathogenesis. Alternatively, the increase in SeV-specific CD8+ T cells may be in response to the inability of the host to effectively curb viral infection through necroptotic death. As experiments were performed in whole genome RIP3 knockout mice and RIP3 has also been shown to play non-necroptotic roles,50, 51, 52 it would be interesting to see what would happen in mixed lineage kinase domain-like knockout mice and in lung epithelial-, DC- or T cell-specific conditional knockouts. This may help to address further questions of host–pathogen interaction during SeV infection.
Materials and Methods
Cells and reagents
L929 cells (American Type Culture Collection-ATCC; Manassass, VA, USA) and LET1 cells20 were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), penicillin and streptomycin, L-glutamine, and Na pyruvate. LA-4 cells (American Type Culture Collection) were maintained in Ham's F12K media supplemented with 15% FBS, penicillin and streptomycin, and L-glutamine. All cells tested negative for mycoplasma. The following reagents were purchased: Anti-TNF neutralizing antibody (clone MP6-XT22) and anti-rat control (R&D Systems, Minneapolis, MN, USA), zVAD-FMK (Enzo Life Sciences, Farmingdale, NY, USA) and Necrostatin-1 (Sigma-Aldrich, Saint Louis, MO, USA). The following antibodies were purchased: anti-RIP1 (BD Biosciences, San Jose, CA, USA), anti-CYLD (Invitrogen, Waltham, MA, USA), anti-FLAG (Sigma-Aldrich), anti-RIP3 (ProSci Inc., Poway, CA, USA), anti-Ubiquitin (Santa Cruz Biotechnology, Dallas, TX, USA), anti-β-actin, anti-HA (Abcam, Cambridge, UK), anti-RIG-I, anti-STING and anti-GAPDH (Cell Signaling, Danvers, MA, USA), anti-cIAP (R&D Systems). Compound 3 (SMAC mimetic) was a generous gift from Dr Patrick Harran.53 CellTiter-Glo was used to determine cell survival (Promega, Madison, WI, USA).
Mice and viral infection
Mice were used at 6–8 weeks of age. Littermates or age-matched mice were used as controls. RIP3−/− mice were from Dr Xiaodong Wang8 and backcrossed to C57BL/6 for more than 10 generations. Mice were inoculated intranasally with 640 HA U/mouse of SeV (Cantell strain) or an equivalent volume of PBS on day 0, 1 and 2 for BAL collection or on day 0 for histology. For histology, lungs were perfused, fixed in formalin and embedded in paraffin before 5μm sections were taken and stained using haematoxylin and eosin. For BAL collection, after sacrifice, the lungs of each mouse were lavaged with sterile PBS. Collected BAL samples were centrifuged and supernatant removed. Resulting samples were adhered to plastic dishes for 1 h at 37 °C to remove adherent macrophages. Red blood cells were lysed and cells enumerated and stained for flow cytometry. SeV+ T cells were detected using APC-labelled H-2Kb tetramers carrying the SeV NP324–332 peptide (NIH Tetramer Core Facility). Cells were gated on CD45+, CD4− and plotted CD8 versus Tetramer or alternatively gated on CD45+, CD3+, CD8+CD4− and plotted Tetramer versus CD44. Experimental mice were housed in the animal facility at the University of California, Berkeley and all procedures involving animals were approved by the Animal Care and Use Committee.
Viruses
Sendai virus (Cantell strain) was purchased from Charles River (Wilmington, MA, USA). MHV68, WSN, MCMV, HSV-1 and LCMV were purchased from ATCC. SeV Z strain and its mutant were generated in Japan as described.32 MHV68 was propagated in 3T3 cells (from Dr Laurent Coscoy Lab originally from ATCC) as previously described.54 In some experiments, MHV68-GFP from Dr Laurent Coscoy was used. KSHV was a generous gift from Dr Britt Glaunsinger. UV irradiation of virus was carried out as described previously and an equivalent volume of live and UV-irradiated virus was used for infections.55
Virus infection and cell death assay
L929 cells were seeded in 96-well plates overnight. Samples in triplicate were then pretreated with zVAD and/or Necrostatin-1 for 1 h prior to addition of virus. Following 16–18 h of infection, cell survival was monitored by CellTiter-Glo (Promega), as per manufacturer's protocol. Mock samples were set to 100% survival and all other samples were expressed relative to mock. Alternatively, treated cells were harvested and stained using 7AAD and run by flow cytometry. For transfection experiments, low-passage number L929 cells were transfected with pCI or pCI-FLAG-Y1 and pCI-FLAG-Y2 using Lipofectamine2000 (Invitrogen). Eighteen to twenty-four hours post-transfection, cells were treated with indicated reagents and cell death was assayed an additional 10–16 h later. For statistical analysis, the Student's t-test was used to determine significance. ***P<0.001, **P<0.01, *P<0.05.
ELISA and TNFα neutralization assay
To determine TNFα secretion, cells were treated as indicated in the figure legend. Samples of virally infected supernatants were then used to determine the TNFα levels by ELISA (eBioscience, San Diego, CA, USA) as per manufacturer's protocol. For TNF neutralization assays, L929 cells were treated with anti-TNF or anti-rat control antibodies at 4 μg/ml prior to addition of zVAD and infection with SeV or MHV68 with the indicated concentrations. 16–18 hours post-infection, cell survival was measured by CellTiter-Glo ATP Assay (Promega).
shRNA and siRNA knockdown
LMP-RIG-I and LMP-control shRNA retroviral constructs were a gift from Dr Russell Vance. Sequences are as described.56 RIG-I and control retroviruses were produced in Phoenix cells (from Dr Ellen Robey) and L929 cells were subsequently infected. A stably expressing cell line was generated by puromycin (4 μg/ml) selection for 1 week. PTY-STING and PTY-control shRNA lentiviral constructs were a gift from Dr Zhijian Chen.57 293T (from Garry Nolan Lab) cells were used to produce lentivirus. L929 cells were then infected and a stably expressing cell line was generated with puromycin selection. To knockdown CYLD expression, L929 cells were transfected with 75 nM of control or CYLD siRNA (Dharmacon, Lafayette, CO, USA) using the Dharmafect1 Transfection Reagent. After 48 hours, cells were split into 96-well plates and treated the following day with indicated reagents or harvested for western blot analysis. For knockdown of SeV PVC proteins, target sequence 5′-GAAGACCAAGCTGAAGGACTT-3′ was cloned into pSUPER retroviral vector to target SeV PVC. Stable cell lines were selected using puromycin. For knockdown of RIP3 from L929 cells, lentiviral shRNA (pLKO.1-shRIP3) was purchased from Open Biosystems (RMM3981-958994 clone:TRCN0000022535). A stable cell line was selected using puromycin.
qRT-PCR
To confirm that the SeV mutants were able to establish an efficient infection, L929 cells were infected with SeV (Cantell strain) or Z strain: WT, V[−], d2Y mutants. After 16–18 hours of incubation, RNA was harvested by TRIzol (as per manufacturer's protocol) followed by extraction using the RNeasy Mini Kit (Qiagen, Germantown, MD, USA). qRT-PCR was carried out for SeV structural protein NP (Forward oligonucleotide 5′-CAGAGGAGCACAGTCTCAGTGTTC-3′ and reverse oligonucleotide 5′-TCTCTGAGAGTGCTGCTTATCTGTG-3′) and normalized to γ-actin (forward oligonucleotide 5′CTGAGGCTGGCAAGGGTTC 3′ and reverse oligonucleotide 5′CCACCCTGTTAGACTGGCAAG 3′) using SYBR green (Invitrogen). Expression was plotted on a log scale relative to mock-infected samples. To examine viral load, shControl or shRIP3 L929 cells were infected with SeV at 100 HA U/ml. One hour post-infection, cells were washed with PBS and media was replaced. Sixteen to eighteen hours post-infection, cells were collected and RNA harvested using TRIzol followed by extraction using the RNeasy Mini kit. qRT-PCR was carried out using primers specific to SeV NP gene and γ-actin. Expression was normalized to γ-actin and plotted.
Constructs
The following expression plasmids were purchased from Addgene (Cambridge, MA, USA): pDEST-HA-CYLD (#15506) and pCDNA3.1-Myc-cIAP1 (#8311). Murine RIP1 was purchased from Open Biosystems and subcloned into pCI using NotI and EcoRI. SeV Y1 and Y2 was PCR-amplified with the addition of a C-terminal FLAG tag from reverse-transcribed cDNA generated from 293T cells infected with SeV. PCR product was then cloned into pCI vector using EcoRI. pCI-HA-Ubiquitin was a generous gift from Dr Laurent Coscoy.
Western blot and immunoprecipitation
For overexpression studies, plasmids were transfected into 293T cells settled into a six-well plate using Lipofectamine 2000 (following manufacturer's protocol). For western blot analysis, cells were lysed in NP40 lysis buffer (1% Nonidet P-40, 50 mM Tris-Cl at pH 7.6, 150 mM NaCl, 1 mM EDTA, and supplemented with inhibitors (1 mM Na3V04 and NaF, 1 mM phenylmethylsulfonyl fluoride, 1 mM benzamidine, 5 μg/ml pepstatin, 10 μg/ml aprotinin, 10 μg/ml leupeptin and 2 mM n-ethylmaleimide)). Lysates were cleared and 100 μg of protein were resolved by SDS-PAGE.
For endogenous RIP1 immunoprecipitation, L929 cells were seeded into 15-cm dishes and treated with zVAD (20 μM) followed by SeV infection (10 HA U/ml) for the indicated times. Cells were harvested in 1% SDS lysis buffer as previously described.36 Anti-RIP1 (2 μg) antibody and Protein A/G-immobilized beads (Pierce, Waltham, MA, USA) were added to 1 mg of cell lysate and rotated overnight at 4 °C. Immune complexes were washed, boiled and resolved by SDS-PAGE. Alternatively, RIP1 was immunoprecipitated from NP40 lysates of 293T cells transfected in a SIX-well plate.
Acknowledgments
We thank Paul Thomas for LET1 cells, Patrick Harran for the generous gift of Compound 3 (SMAC mimetic) and Russell Vance and Zhijian Chen for retroviral and lentiviral constructs. We also thank Britt Glaunsinger for MHV68 virus and Bin Xue, Neil Sahasrabudhe, Max Stahl and Lin He for help with BAL collection and lung histology. We also thank Holly Aaron and Jen-Yi Lee with the UC Berkeley Imaging Core for slide scanning. We are appreciative of Gagandeep Kumar for helpful comments as well as Sean Sunyoto and Guangzhi Zhang for help on experiments. This work was supported by grants from the National Institutes of Health (R01 AI095299 and T32 AI100829). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by J Silke
The authors declare no conflict of interest.
Supplementary Material
References
- Bergsbaken T, Fink SL, Cookson BT. Pyroptosis: host cell death and inflammation. Nat Rev Microbiol 2009; 7: 99–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blander JM. A long-awaited merger of the pathways mediating host defence and programmed cell death. Nat Rev Immunol 2014; 14: 601–618. [DOI] [PubMed] [Google Scholar]
- Chan FK, Luz NF, Moriwaki K. Programmed necrosis in the cross talk of cell death and inflammation. Annu Rev Immunol 2015; 33: 79–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallach D, Kang TB, Dillon CP, Green DR. Programmed necrosis in inflammation: toward identification of the effector molecules. Science 2016; 352: aaf2154. [DOI] [PubMed] [Google Scholar]
- Christofferson DE, Yuan J. Necroptosis as an alternative form of programmed cell death. Curr Opin Cell Biol 2010; 22: 263–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlangenakker N, Vanden Berghe T, Vandenabeele P. Many stimuli pull the necrotic trigger, an overview. Cell Death Differ 2012; 19: 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degterev A, Hitomi J, Germscheid M, Ch'en IL, Korkina O, Teng X et al. Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat Chem Biol 2008; 4: 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Wang L, Miao L, Wang T, Du F, Zhao L et al. Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-alpha. Cell 2009; 137: 1100–1111. [DOI] [PubMed] [Google Scholar]
- Cho YS, Challa S, Moquin D, Genga R, Ray TD, Guildford M et al. Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell 2009; 137: 1112–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DW, Shao J, Lin J, Zhang N, Lu BJ, Lin SC et al. RIP3, an energy metabolism regulator that switches TNF-induced cell death from apoptosis to necrosis. Science 2009; 325: 332–336. [DOI] [PubMed] [Google Scholar]
- Kaiser WJ, Upton JW, Mocarski ES. Viral modulation of programmed necrosis. Curr Opin Virol 2013; 3: 296–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Beg AA. Induction of necrotic-like cell death by tumor necrosis factor alpha and caspase inhibitors: novel mechanism for killing virus-infected cells. J Virol 2000; 74: 7470–7477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigue-Gervais IG, Labbe K, Dagenais M, Dupaul-Chicoine J, Champagne C, Morizot A et al. Cellular inhibitor of apoptosis protein cIAP2 protects against pulmonary tissue necrosis during influenza virus infection to promote host survival. Cell Host & Microbe 2014; 15: 23–35. [DOI] [PubMed] [Google Scholar]
- Upton JW, Kaiser WJ, Mocarski ES. DAI/ZBP1/DLM-1 complexes with RIP3 to mediate virus-induced programmed necrosis that is targeted by murine cytomegalovirus vIRA. Cell Host & Microbe 2012; 11: 290–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Li Y, Liu S, Yu X, Li L, Shi C et al. Direct activation of RIP3/MLKL-dependent necrosis by herpes simplex virus 1 (HSV-1) protein ICP6 triggers host antiviral defense. Proc Natl Acad Sci USA 2014; 111: 15438–15443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Wu SQ, Liang Y, Zhou X, Chen W, Li L et al. RIP1/RIP3 binding to HSV-1 ICP6 initiates necroptosis to restrict virus propagation in mice. Cell Host & Microbe 2015; 17: 229–242. [DOI] [PubMed] [Google Scholar]
- Guo H, Omoto S, Harris PA, Finger JN, Bertin J, Gough PJ et al. Herpes simplex virus suppresses necroptosis in human cells. Cell Host & Microbe 2015; 17: 243–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YT, Tan HL, Huang Q, Sun XJ, Zhu X, Shen HM. zVAD-induced necroptosis in L929 cells depends on autocrine production of TNFalpha mediated by the PKC-MAPKs-AP-1 pathway. Cell Death Differ 2011; 18: 26–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoner GD, Kikkawa Y, Kniazeff AJ, Miyai K, Wagner RM. Clonal isolation of epithelial cells from mouse lung adenoma. Cancer Res 1975; 35: 2177–2185. [PubMed] [Google Scholar]
- Rosenberger CM, Podyminogin RL, Askovich PS, Navarro G, Kaiser SM, Sanders CJ et al. Characterization of innate responses to influenza virus infection in a novel lung type I epithelial cell model. J Gen Virol 2014; 95: 350–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenabeele P, Galluzzi L, Vanden Berghe T, Kroemer G. Molecular mechanisms of necroptosis: an ordered cellular explosion. Nat Rev Mol Cell Biol 2010; 11: 700–714. [DOI] [PubMed] [Google Scholar]
- Mocarski ES, Guo H, Kaiser WJ. Necroptosis: the Trojan horse in cell autonomous antiviral host defense. Virology 2015; 479-480: 160–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud F, Coulombe F, Gaudreault E, Kriz J, Gosselin J. Involvement of TLR2 in recognition of acute gammaherpesvirus-68 infection. PloS One 2010; 5: e13742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guggemoos S, Hangel D, Hamm S, Heit A, Bauer S, Adler H. TLR9 contributes to antiviral immunity during gammaherpesvirus infection. J Immunol 2008; 180: 438–443. [DOI] [PubMed] [Google Scholar]
- Gaajetaan GR, Geelen TH, Grauls GE, Bruggeman CA, Stassen FR. CpG and poly(I:C) stimulation of dendritic cells and fibroblasts limits herpes simplex virus type 1 infection in an IFNbeta-dependent and -independent way. Antiviral Res 2012; 93: 39–47. [DOI] [PubMed] [Google Scholar]
- Shcheblyakov DV, Logunov DY, Rakovskaya IV, Shmarov MM, Naroditsky BS, Ginzburg AL. Triggering of toll-like receptor-2 in mouse myelomonocytic leukaemia cells WEHI-3B leads to the suppression of apoptosis and promotes tumor progression in vivo. Acta Naturae 2011; 3: 83–93. [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel J, Tan CP, Goubau D, Schulz O, Pichlmair A, Bier K et al. RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell 2010; 140: 397–408. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 2008; 455: 674–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SE, Noyce RS, Mossman KL. Innate cellular response to virus particle entry requires IRF3 but not virus replication. J Virol 2004; 78: 1706–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi K, Komatsu T, Kitagawa Y, Sada K, Gotoh B. Sendai virus C protein plays a role in restricting PKR activation by limiting the generation of intracellular double-stranded RNA. J Virol 2008; 82: 10102–10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faisca P, Desmecht D. Sendai virus, the mouse parainfluenza type 1: a longstanding pathogen that remains up-to-date. Res Vet Sci 2007; 82: 115–125. [DOI] [PubMed] [Google Scholar]
- Irie T, Nagata N, Yoshida T, Sakaguchi T. Paramyxovirus Sendai virus C proteins are essential for maintenance of negative-sense RNA genome in virus particles. Virology 2008; 374: 495–505. [DOI] [PubMed] [Google Scholar]
- Irie T, Kiyotani K, Igarashi T, Yoshida A, Sakaguchi T. Inhibition of interferon regulatory factor 3 activation by paramyxovirus V protein. J Virol 2012; 86: 7136–7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato A, Kiyotani K, Sakai Y, Yoshida T, Nagai Y. The paramyxovirus, Sendai virus, V protein encodes a luxury function required for viral pathogenesis. EMBO J 1997; 16: 578–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitomi J, Christofferson DE, Ng A, Yao J, Degterev A, Xavier RJ et al. Identification of a molecular signaling network that regulates a cellular necrotic cell death pathway. Cell 2008; 135: 1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moquin DM, McQuade T, Chan FK. CYLD deubiquitinates RIP1 in the TNFalpha-induced necrosome to facilitate kinase activation and programmed necrosis. PloS One 2013; 8: e76841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand MJ, Milutinovic S, Dickson KM, Ho WC, Boudreault A, Durkin J et al. cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol Cell 2008; 30: 689–700. [DOI] [PubMed] [Google Scholar]
- Mahoney DJ, Cheung HH, Mrad RL, Plenchette S, Simard C, Enwere E et al. Both cIAP1 and cIAP2 regulate TNFalpha-mediated NF-kappaB activation. Proc Natl Acad Sci USA 2008; 105: 11778–11783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varfolomeev E, Goncharov T, Fedorova AV, Dynek JN, Zobel K, Deshayes K et al. c-IAP1 and c-IAP2 are critical mediators of tumor necrosis factor alpha (TNFalpha)-induced NF-kappaB activation. J Biol Chem 2008; 283: 24295–24299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman CS, O'Donnell MA, Legarda-Addison D, Ng A, Cardenas WB, Yount JS et al. The tumour suppressor CYLD is a negative regulator of RIG-I-mediated antiviral response. EMBO Rep 2008; 9: 930–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao AP, Li S, Zhong B, Li Y, Yan J, Li Q et al. Virus-triggered ubiquitination of TRAF3/6 by cIAP1/2 is essential for induction of interferon-beta (IFN-beta) and cellular antiviral response. J Biol Chem 2010; 285: 9470–9476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skiadopoulos MH, Surman SR, Riggs JM, Elkins WR, St Claire M, Nishio M et al. Sendai virus, a murine parainfluenza virus type 1, replicates to a level similar to human PIV1 in the upper and lower respiratory tract of African green monkeys and chimpanzees. Virology 2002; 297: 153–160. [DOI] [PubMed] [Google Scholar]
- Parker JC, Whiteman MD, Richter CB. Susceptibility of inbred and outbred mouse strains to Sendai virus and prevalence of infection in laboratory rodents. Infect Immun 1978; 19: 123–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama AH, Irie H, Kato A, Nagai Y, Adachi A. Virus multiplication and induction of apoptosis by Sendai virus: role of the C proteins. Microbes Infect 2003; 5: 373–378. [DOI] [PubMed] [Google Scholar]
- Garcin D, Latorre P, Kolakofsky D. Sendai virus C proteins counteract the interferon-mediated induction of an antiviral state. J Virol 1999; 73: 6559–6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Tschopp J, Lin SC. Smac mimetics and TNFalpha: a dangerous liaison? Cell 2007; 131: 655–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajput A, Kovalenko A, Bogdanov K, Yang SH, Kang TB, Kim JC et al. RIG-I RNA helicase activation of IRF3 transcription factor is negatively regulated by caspase-8-mediated cleavage of the RIP1 protein. Immunity 2011; 34: 340–351. [DOI] [PubMed] [Google Scholar]
- Sakamaki K, Tsukumo S, Yonehara S. Molecular cloning and characterization of mouse caspase-8. Eur J Biochem 1998; 253: 399–405. [DOI] [PubMed] [Google Scholar]
- Nogusa S, Thapa RJ, Dillon CP, Liedmann S, Oguin TH3rd, Ingram JP et al. RIPK3 activates parallel pathways of MLKL-driven necroptosis and FADD-mediated apoptosis to protect against Influenza A virus. Cell Host & Microbe 2016; 20: 13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriwaki K, Bertin J, Gough PJ, Chan FK. A RIPK3-caspase 8 complex mediates atypical pro-IL-1beta processing. J Immunol 2015; 194: 1938–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Diaz S, Dillon CP, Lalaoui N, Tanzer MC, Rodriguez DA, Lin A et al. The pseudokinase MLKL and the kinase RIPK3 have distinct roles in autoimmune disease caused by loss of death-receptor-induced apoptosis. Immunity 2016; 45: 513–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Fan C, Zhang H, Zhao Q, Liu Y, Xu C et al. MLKL and FADD are critical for suppressing progressive lymphoproliferative disease and activating the NLRP3 inflammasome. Cell Rep 2016; 16: 3247–3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Thomas RM, Suzuki H, De Brabander JK, Wang X, Harran PG. A small molecule Smac mimic potentiates TRAIL- and TNFalpha-mediated cell death. Science 2004; 305: 1471–1474. [DOI] [PubMed] [Google Scholar]
- Lindquester GJ, Greer KA, Stewart JP, Sample JT. Epstein-Barr virus IL-10 gene expression by a recombinant murine gammaherpesvirus in vivo enhances acute pathogenicity but does not affect latency or reactivation. Herpesviridae 2014; 5: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudreault E, Fiola S, Olivier M, Gosselin J. Epstein-Barr virus induces MCP-1 secretion by human monocytes via TLR2. J Virol 2007; 81: 8016–8024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe KM, McWhirter SM, Vance RE. Identification of host cytosolic sensors and bacterial factors regulating the type I interferon response to Legionella pneumophila. PLoS Pathog 2009; 5: e1000665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Chen ZJ. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci Signal 2012; 5: ra20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.