Abstract
The Rb/E2F network has a critical role in regulating cell cycle progression and cell fate decisions. It is dysfunctional in virtually all human cancers, because of genetic lesions that cause overexpression of activators, inactivation of repressors, or both. Paradoxically, the downstream target of this network, E2F1, is rarely strongly overexpressed in cancer. E2F1 can induce both proliferation and apoptosis but the factors governing these critical cell fate decisions remain unclear. Previous studies have focused on qualitative mechanisms such as differential cofactors, posttranslational modification or state of other signaling pathways as modifiers of the cell fate decisions downstream of E2F1 activation. In contrast, the importance of the expression levels of E2F1 itself in dictating the downstream phenotypes has not been rigorously studied, partly due to the limited resolution of traditional population-level measurements. Here, through single-cell quantitative analysis, we demonstrate that E2F1 expression levels have a critical role in determining the fate of individual cells. Low levels of exogenous E2F1 promote proliferation, moderate levels induce G1, G2 and mitotic cell cycle arrest, and very high levels promote apoptosis. These multiple anti-proliferative mechanisms result in a strong selection pressure leading to rapid elimination of E2F1-overexpressing cells from the population. RNA-sequencing and RT-PCR revealed that low levels of E2F1 are sufficient to induce numerous cell cycle-promoting genes, intermediate levels induce growth arrest genes (i.e., p18, p19 and p27), whereas higher levels are necessary to induce key apoptotic E2F1 targets APAF1, PUMA, HRK and BIM. Finally, treatment of a lung cancer cell line with a proteasome inhibitor, MLN2238, resulted in an E2F1-dependent mitotic arrest and apoptosis, confirming the role of endogenous E2F1 levels in these phenotypes. The strong anti-proliferative activity of moderately overexpressed E2F1 in multiple cancer types suggests that targeting E2F1 for upregulation may represent an attractive therapeutic strategy in cancer.
Studies over the last 30 years have identified the core regulatory network that controls cell cycle entry in mammalian cells.1 In response to growth stimulation, Myc protein rapidly accumulates and contributes to the induction of cyclin D leading to inactivation of the retinoblastoma (Rb) tumor suppressor.2, 3 The subsequent release of E2F transcription factors from Rb-mediated repression leads to transcriptional activation of genes that initiate DNA replication and cell cycle progression and is considered a key step in regulation of mammalian proliferation.4
Despite 30 years of investigation, the role of the founding member of the E2F family, E2F1, in regulating the fates of normal and cancer cells still remains controversial. On one hand, it is generally accepted that activation of E2F1 has a critical role in driving normal cells into the cell cycle.5, 6, 7, 8 On the other hand, it is also well recognized that overexpression of E2F1 promotes apoptosis6, 8, 9, 10 or growth arrest.11, 12, 13
To reconcile these contradictory observations, a common explanation is that the action of E2F1 depends on the cellular context, in terms of the presence or absence of differential cofactors, posttranslational modifications or state of other signaling pathways.14, 15, 16, 17, 18, 19 Another under-appreciated contribution, however, is the quantitative responses of individual cells to E2F1 levels. Expression levels of endogenous E2F1 can exhibit significant cell–cell variability, which is even higher for exogenous transgenes delivered, for example, via adenoviral transduction.20, 21 Depending on the average E2F1 level and the extent of cell–cell variability, one can draw different conclusions from experiments on the same system as E2F1 may trigger opposing effects.
Nevertheless, cell–cell variability represents an opportunity to elucidate the regulatory function of E2F1 in a high-throughput manner if both E2F1 levels and cellular responses can be quantified at a single-cell resolution. To this end, here we have used time-lapse microscopy and flow cytometry to provide the first quantitative analysis of the effects of E2F1 levels on fate decisions in single cells.
Results
Experimental system for studies of E2F1-mediated cell fates at a single-cell resolution
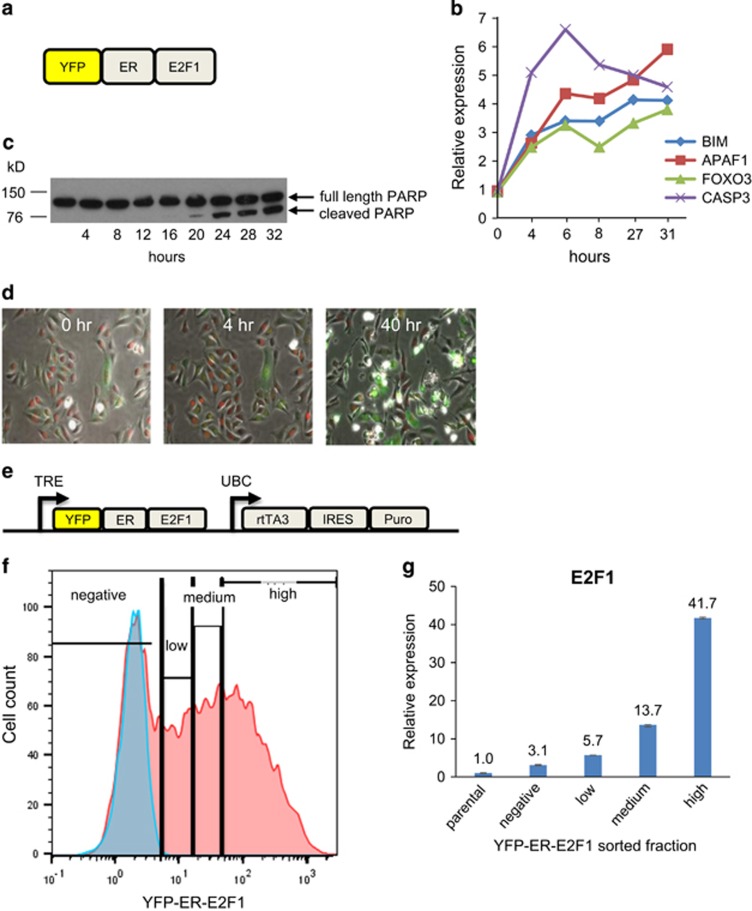
To control and monitor E2F1 activity in single cells, we stably expressed YFP–ER–E2F1 fusion protein in U2OS cells (Figure 1a). The addition of ER ligands, such as tamoxifen and 4-hydroxytamoxifen (OHT), leads to nuclear translocation of the fusion protein and activation of E2F1-mediated transcription. The yellow fluorescent protein (YFP) tag enables real-time monitoring and quantification of nuclear E2F1. Tamoxifen treatment of the engineered cells strongly induced well-characterized apoptotic E2F1 target genes, including BIM, APAF1, CASP3 and FOXO3 (Figure 1b), and induced apoptosis as measured by cleavage of the PARP protein (Figure 1c). These results confirm that the fusion protein is fully functional.
Figure 1.
Experimental system for studies of E2F1-mediated cell fates at a single-cell resolution. (a) Schematic of YFP–ER–E2F1 fusion protein. (b) U2OS cells stably expressing pEYFP-ER-E2F1 were serum starved for 24 h and then treated with 2 μM tamoxifen in starvation medium. Cells were collected at indicated time points and expression of BIM, APAF1, FOXO3 and CASP3 analyzed by real-time qPCR. (c) U2OS cells stably expressing pEYFP-ER-E2F1 were serum starved for 24 h and then treated with 2 μM tamoxifen in starvation medium. Cells were collected at indicated time points and PARP cleavage was analyzed by western blot. (d) Images of U2OS cells expressing YFP-ER-E2F1 (green) and H2B-RFP (red) before (0 h), 4 h and 40 h after exposure to 3 μM tamoxifen. (e) Schematic of pTRIPZ-YFP-ER-E2F1 construct. (f and g) U2OS pTRIPZ-YFP-ER-E2F1 cells were grown in full serum-containing growth medium and treated with 500 ng/ml doxycycline for 48 h followed by addition of 90 nM OHT for an additional 20 h. Cells from the indicated YFP fractions were sorted by flow cytometry and E2F1 expression was analyzed by quantitative RT-PCR. (f) Histogram of YFP expression and sorting gates in induced U2OS pTRIPZ-YFP-ER-E2F1 cells (red) and parental U2OS cells (blue). (g) Real-time quantitative RT-PCR analysis of total E2F1 (endogenous+YFP-ER-E2F1) in sorted fractions. Graph represents the relative E2F1 expression after normalization to GAPDH housekeeping control (mean and range of duplicate PCR reactions). IRES, internal ribosomal entry site; rtTA3, reverse tetracycline-transactivator 3; TRE, tetracycline-responsive element; UBC, ubiquitin C promoter
To facilitate quantification of nuclear YFP-ER-E2F1 in time-lapse experiments, we integrated a gene encoding the histone H2 tagged with a red fluorescent protein (H2B-RFP) into the U2OS cells stably expressing YFP-ER-E2F1 to mark nuclei. As expected, the YFP signal was localized to the cytoplasm before addition of OHT and underwent nuclear translocation following OHT addition (Figure 1d).
We also introduced the YFP–ER–E2F1 fusion protein into a lentiviral doxycycline-inducible vector, which also contains the rTA3 activator (Figure 1e). This integrated design coupled with lentiviral delivery streamlines the creation of multiple inducible cell lines. It also enables modulation of E2F1 activity by dox-mediated transcriptional control and tamoxifen or OHT-mediated localization control minimizing leaky E2F1 activity.
To compare the expression levels of YFP-ER-E2F1 and endogenous E2F1, we sorted the induced U2OS pTRIPZ-YFP-ER-E2F1 into several fraction of increasing YFP intensity and measured total levels of E2F1 mRNA by qPCR. The degree of E2F1 overexpression compared with parental U2OS cells varied from 3-fold to 42-fold in different fractions (Figures 1f and g).
E2F1 expression levels dictate fates of individual cells
We used time-lapse microscopy to analyze the effects of different E2F1 levels on the fates of individual cells. We first performed the experiment in serum-starved conditions to minimize the effects of the endogenous E2F1. We developed a Matlab script to extract the nuclear YFP intensity over time in single cells from time-lapse movies upon OHT induction (Figure 2a). Apoptosis (cell rounding, membrane blebbing and bursting) and mitosis (cell rounding and division to daughter cells) events were detected visually (Supplementary Video). Detection of apoptosis by morphological changes displayed an excellent correlation with 4′,6-diamidino-2-phenylindole (DAPI) staining (Supplementary Figure S1).
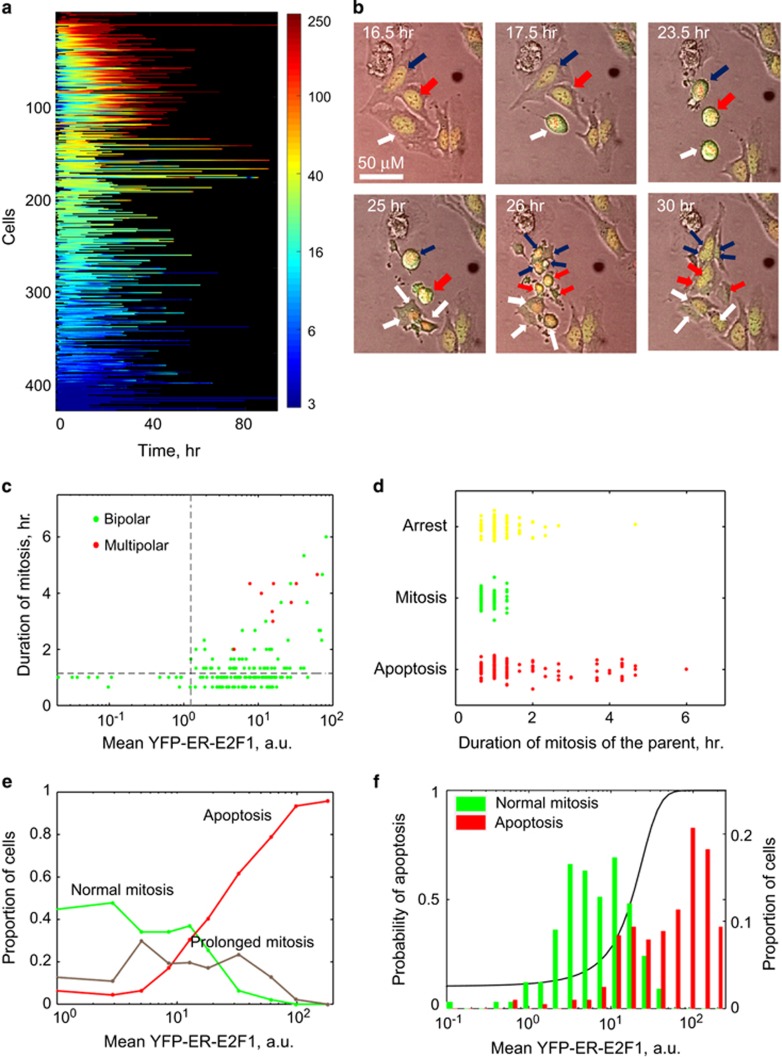
Figure 2.
E2F1 expression levels dictate fates of individual cells. U2OS cells expressing pEYFP-ER-E2F1 and H2B-RFP were serum starved for 24 h, treated with 90 nM OHT and analyzed by time-lapse microscopy for 72 h. Images were taken every 20 min. (a) Single-cell dynamics of nuclear YFP fluorescence for 470 cells. The color represents the intensity of nuclear YFP signal as indicated by the scale bar on the right. (b) Representative images of mitotic arrest and multipolar mitosis. Blue, red and white arrows indicate three cells and their correspondent daughter cells. YFP-ER-E2F1 is shown in green and H2B-RFP in red. (c) Relationship between mean nuclear YFP-ER-E2F1 and duration of mitosis. Multipolar mitosis events are indicated in red. (d) Relationship between length of mitosis and subsequent fate of daughter cells. (e) Cells were grouped into 10 equal bins of increasing mean nuclear YFP-ER-E2F1. Proportions of cells undergoing specified fate (normal mitosis (1 h or less), prolonged mitosis (longer than 1 h) or apoptosis) were calculated for each bin. (f) Histograms of mean nuclear YFP-ER-E2F1 for cells undergoing normal mitosis (mitotic duration of 1 h or less) or apoptosis. Black line indicates the probability of a cell to undergo either normal mitosis or apoptosis for each YFP-ER-E2F1 level as determined by logistic regression. See also Supplementary Figure S1 for ROC curve
YFP-ER-E2F1 expression was highly variable between cells, likely due to differences in copy numbers of the integrated construct (Figure 2a). We examined the relationship between several dynamic characteristics including mean, maximum and integral of nuclear YFP intensity over the life of each cell and cell fates and found that mean nuclear YFP intensity provided the best correlations. Thus, we used this metric in all our analyses.
We found that many cells with high levels of nuclear YFP-ER-E2F1 experienced prolonged mitotic arrest (Figure 2b). For example, the cell marked with a white arrow spent 6 h in mitosis. In contrast, cells with background YFP expression and parental cells not expressing the construct completed mitosis in 1 h or less (Figure 2c). We also observed multiple instances of multipolar mitosis in cells expressing high nuclear YFP-ER-E2F1 (i.e., cells marked with white, blue and red arrows in Figure 2b).
Quantification of mitosis duration as a function of nuclear YFP-ER-E2F1 level revealed a striking threshold effect; whereas all cells with nuclear YFP levels below 1.1 arbitrary unit (a.u.) completed mitosis in 1 h or less, mitotic duration was prolonged in 43% of cells expressing nuclear YFP-ER-E2F1 above this threshold (Figure 2c). The frequency of multipolar mitosis also increased above 1.1 a.u. YFP-ER-E2F1 threshold (Figure 2c). Fate analysis of the daughter cells revealed that almost all daughter cells born from mitosis, which was longer than 2 h, committed apoptosis (Figure 2d).
Single-cell resolution of our methodology enabled us to perform a detailed analysis of the effects of different E2F1 levels on the likelihood to divide normally, arrest in mitosis or commit apoptosis (Figure 2e). We found that moderate increase in nuclear YFP-ER-E2F1 from background levels to ~3 a.u. resulted in a slight increase in the proportion of normally dividing cells without inducing apoptosis. Further increase in nuclear YFP-ER-E2F1 to about 8 a.u. reduced the proportion of normal mitosis because of an increase in the proportion of cells experiencing mitotic arrest. Additional increase in nuclear YFP-ER-E2F1 resulted in a progressively higher proportion of cells undergoing apoptosis, which became the predominant fate for cells with nuclear YFP-ER-E2F1 above 20 a.u.
To test the role of E2F1 levels on cell fates in the presence of growth factors, we repeated the time-lapse experiment in regular serum-containing medium. Decrease in mitosis and increase in apoptosis with increasing E2F1 levels were also observed in these conditions (Supplementary Figure S2).
The level of nuclear YFP-ER-E2F1 was an excellent predictor of a cell's decision to divide normally or to die, as illustrated by area under the curve (AUC=0.901) of the receiver operator characteristic (ROC) curve of a logistic regression predictor (Figure 2f and Supplementary Figure S3A). However, it did not correlate significantly with timing of either mitosis or apoptosis (Supplementary Figure S3B).
In summary, our results show that E2F1 levels have a critical role in the decision whether to die or divide but not when to do it.
E2F1-mediated mitotic arrest is observed in multiple cell lines
To extend our studies to additional cell lines, we created a panel of cell lines stably expressing the inducible pTRIPZ-YFP-ER-E2F1 construct. This panel included colon (HCT116), lung (H1299), bone (U2OS) and breast (MDA-MB-231) cancer cell lines, as well as normal ER-negative hTERT-immortalized mammary epithelial cells HME and non-tumorigenic immortalized rat fibroblast line REF52.
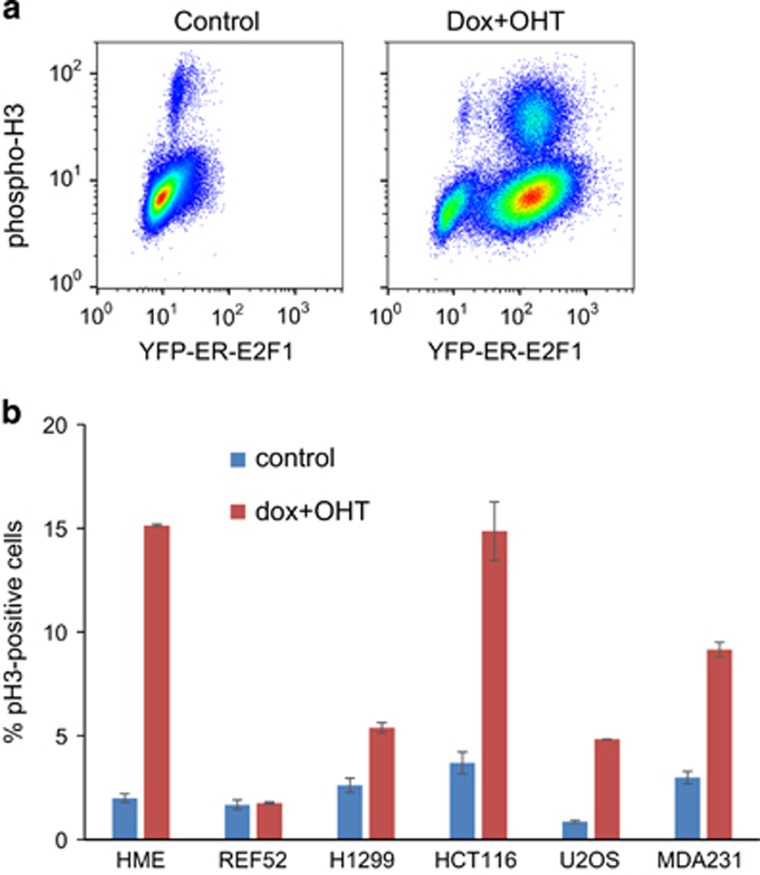
To evaluate the generality and growth factor dependence of E2F1-induced mitotic arrest observed by time-lapse microscopy in U2OS cells, we used phospho-histone H3 (pH3) flow cytometry in normal breast epithelial HME pTRIPZ-YFP-ER-E2F1 cells grown in the presence of growth factors. Histone H3, the core protein of the nucleosome, is phosphorylated at the end of prophase and is an established mitotic marker.22 Phosho-histone H3 staining is used to measure the fraction of cells in mitosis. As the length of mitosis is almost invariable in unstressed proliferating cells, higher proportion of pH3-positive cells corresponds to relatively shorter other cell cycle phases (faster proliferation). pH3 staining is also widely used to assess the effects of perturbations that directly affect mitosis itself (i.e., microtubule poisons such as nocodazole). In this case, higher pH3 staining measures mitotic arrest. E2F1 induction increased the proportion of pH3-positive cells from 1.7 to 13.3% suggesting mitotic arrest (Figure 3b). Mitotic arrest was also confirmed by time-lapse microscopy (data not shown).
Figure 3.
E2F1 induces mitotic arrest in multiple cell lines. (a) HME cells expressing pTRIPZ-YFP-ER-E2F1 in full growth factor containing medium were treated with a combination of 1 μg/ml doxycycline and 90 nM OHT for 24 h. Cells were stained with anti-phospho-histone H3 antibody (mitotic marker) and analyzed by flow cytometry. (b) Six cell lines expressing pTRIPZ-YFP-ER-E2F1 in full serum-containing medium were treated with a combination of 1 μg/ml doxycycline and 90 nM OHT for 18 h. Cells were stained with anti-phospho-histone H3 antibody (mitotic marker) and analyzed by flow cytometry. Data show the percentages of phospho-histone H3-positive cells among YFP-positive cells for the induced samples and total percentage of phospho-histone H3-positive cells for the uninduced controls. Data represent means and ranges of duplicate samples from a representative experiment. See also Supplementary Figure S4 for the raw data
E2F1-mediated mitotic arrest in the presence of growth factors was observed in five out of six cell lines tested (Figure 3b and Supplementary Figure S4). The only cell line that did not show mitotic arrest, REF52, was arrested in G1 and did not enter the cell cycle. Our results suggest that induction of mitotic arrest by deregulated E2F1 is a general novel intrinsic safeguard limiting E2F1-induced proliferation.
Effect of E2F1 on DNA synthesis and DNA content is cell type and dose dependent
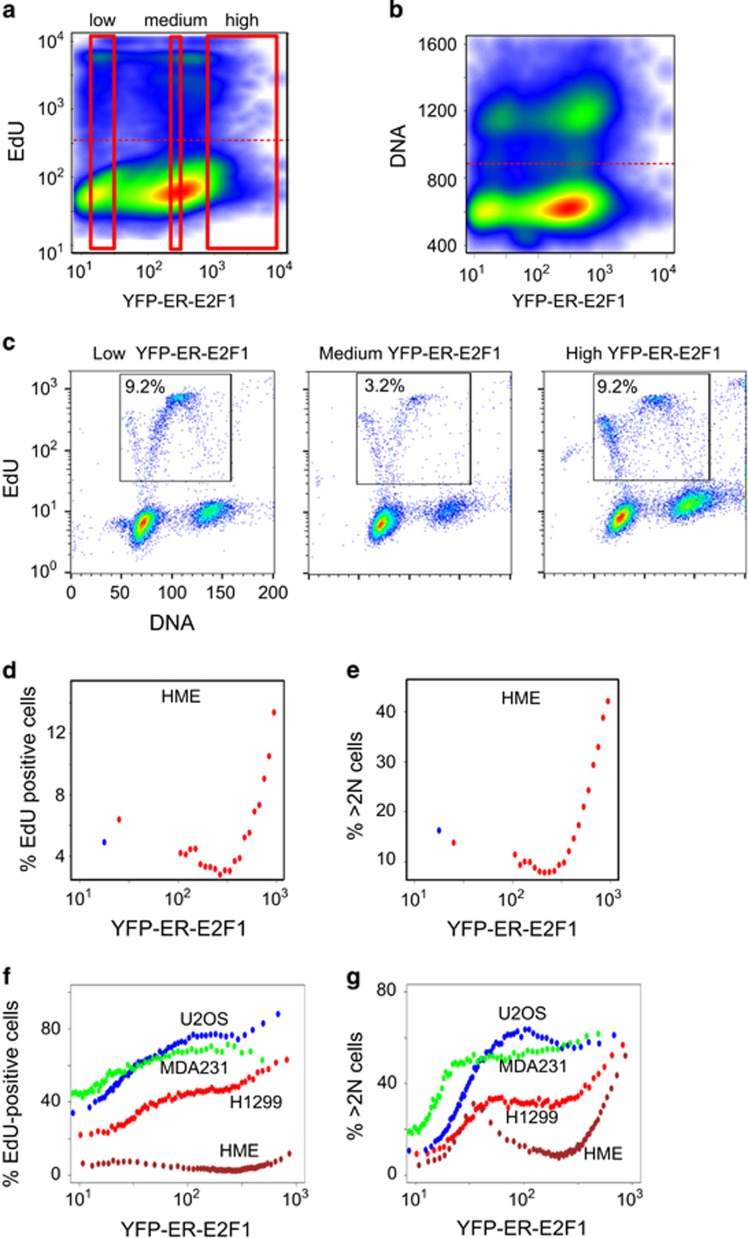
We next used flow cytometry to examine the role of E2F1 levels in regulating other cell cycle transition points. Following growth factor deprivation, pTRIPZ-YFP-ER-E2F1 cell lines were treated with doxycycline and OHT in the presence of a thymidine analog, EdU. Example distributions of EdU and DNA content versus YFP levels in induced untransformed HME cells are shown in Figures 4a and b.
Figure 4.
Dose-dependent effect of E2F1 on DNA synthesis and content. Cells expressing pTRIPZ-YFP-ER-E2F1 were starved of growth factors for 2 days and then treated with a combination of 1 μg/ml doxycycline, 90 nM OHT or control starvation medium in the presence of 10 μM EdU. Cells were fixed and stained for DNA content (DAPI) and DNA synthesis (EdU incorporation) 20 h later. (a) YFP/EdU distribution from induced HME cells. Red horizontal dashed line represents the boundary between EdU-positive and EdU-negative cells. Regions of low, intermediate and high YFP used for plotting DNA/EdU distributions in (c) are marked with red boxes. (b) YFP/DNA distribution from induced HME cells. Red horizontal line represents the boundary between cells with 2 N and >2 N DNA content. (c) DNA/EdU distributions from 25 000 cells with either low, intermediate or high YFP marked with red boxes in a. The percentage of EdU-positive cells is indicated. (d) Data in a were analyzed by bin analysis where cells were divided into 21 bins according to their YFP fluorescence. The first bin contained all cells with YFP fluorescence under 100 a.u. (range of uninduced samples). Red dots represent proportion of EdU-positive cells for each YFP bin. Blue dot represents uninduced control. (e) Bin analysis as in d for DNA content. (f) Analysis as in d for three cancer cell lines and HME normal cells using 60 YFP bins. (g) Analysis as in e for three cancer cell lines and HME normal cells using 60 YFP bins
A dose-response curve is typically constructed from multiple samples treated with a range of concentrations of an effector. In contrast, single-cell measurements using fluorescently labeled effector enable construction of dose-response curves from a single sample, which is more efficient and precise.
We first analyzed the effects of different YFP-ER-E2F1 induction levels on DNA synthesis in normal HME pTRIPZ-YFP-ER-E2F1 cells by calculating the percentage of EdU-positive cells from regions of low, intermediate and high YFP (marked by red boxes in Figure 4a). Surprisingly, we found that intermediate levels of YFP-ER-E2F1 suppressed the basal rate of DNA synthesis, whereas higher levels induced it (Figure 4c). To derive E2F1 dose-response curves, we divided the YFP axis into 21 bins of increasing YFP-ER-E2F1 and calculated the percentage of EdU-positive cells in each bin. This analysis confirmed a biphasic dose response (Figure 4d).
This non-monotonic dose response was consistent with the proportion of cells with >2 N DNA content as a function of the YFP-ER-E2F1 level (Figure 4e). The decrease in the proportion of cells in G2/M by intermediate E2F1 levels below that of the uninduced controls was likely due to inhibition of DNA synthesis. The accumulation of cells with >2 N DNA content at high E2F1 levels was consistent with G2 and/or mitotic arrest.
Similar experiments in cancer cell lines revealed that dose responses had both general and cell type-specific features. Whereas the reduction in DNA synthesis and the fraction of cells with >2 N DNA content by intermediate E2F1 levels was specific to HME cells, the induction of S-phase entry and subsequent G2/M arrest at high levels of E2F1 activity occurred in all tested cell lines (Figures 4f and g).
Cells with elevated E2F1 are eliminated from population
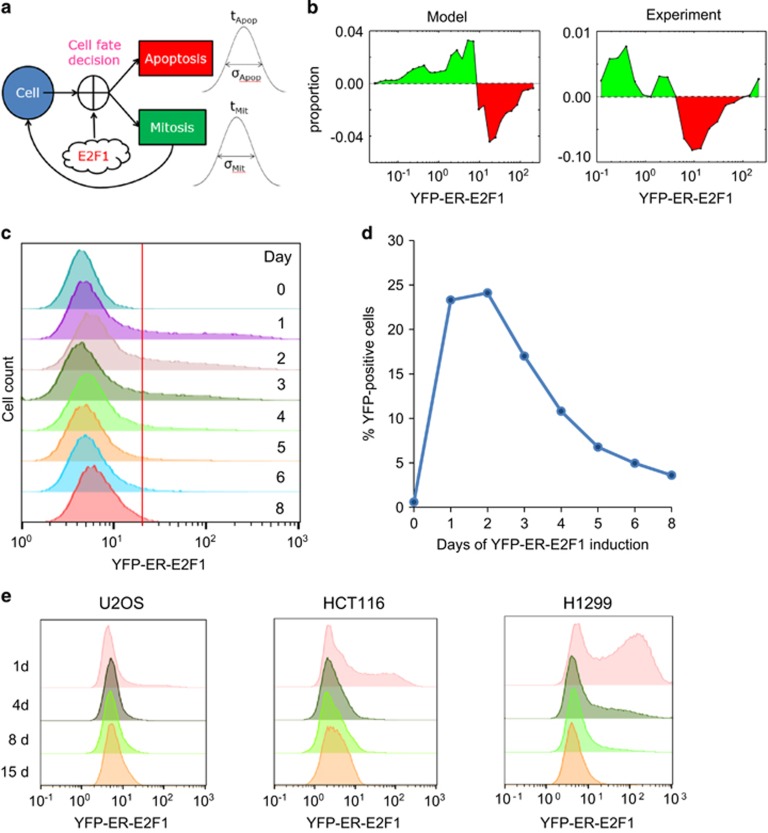
To evaluate the long-term effects of E2F1 levels in a population, we constructed a probabilistic mathematical model that integrates several key parameters directly deduced from microscopy. These include the probabilities of a cell to choose a particular fate, as well as the time to mitosis or apoptosis (Figure 5a and Supplementary Information for detailed model description).
Figure 5.
Cells with elevated E2F1 (remove the empty space between 2 and F) are eliminated from population. (a) Schematics of the computational model. Probability of a single cell to either divide (mitosis) or die (apoptosis) is dependent on E2F1 levels and was experimentally measured by time-lapse microscopy (Figure 2f). In contrast, time to the correspondent event (mitosis or apoptosis) is E2F1 independent and was modeled based on experimentally observed range (Supplementary Figure S3B). (b) Left panel: mean nuclear YFP distribution from 6 to 12 h of the time-lapse experiment described in Figure 2 was used as initial distribution for 25 000 cells. Cell fates were simulated using the model framework in a and final distribution calculated using mean nuclear YFP at 91–96 h. Graphs show the change in the proportion of cells in each YFP bin in the final simulated distribution compared with the initial distribution. Green color indicates enrichment and red color indicates depletion of cell sub-population in a given YFP bin during the experiment as predicted by the simulation. Right panel: same calculation using the experimental distributions. (c) U2OS pTRIPZ-YFP-ER-E2F1 cells were grown in starvation medium in the presence of 1 μg/ml doxycycline and 90 nM OHT. Fresh medium with inducers was changed daily. YFP distributions of the adherent (alive) cells were analyzed by flow cytometry at indicated time points. (d) Dynamics of change in percentages of YFP-positive cells in the population in the experiment in c. (e) Three cancer cell lines expressing pTRIPZ-YFP-ER-E2F1 were treated with a combination of 100 ng/ml doxycycline and 90 nM OHT (D+O) or with control starvation medium. Twenty-one hours before collection of each time point, medium was replaced to fresh starvation medium with 100 ng/ml doxycycline and 90 nM OHT. At the time of collection, one-fourth of the culture was replated in full growth medium to neutralize trypsin and enable attachment. Medium was replaced to starvation medium with or without inducers 24 h later. YFP distributions of adherent (alive) cells were measured at indicated time points by flow cytometry. See also Supplementary Figures S3 for similar analysis in additional cell lines and for RT-PCR analysis
Simulations predicted the relative enrichment of the cell sub-population expressing YFP-ER-E2F1 below a threshold level, and relative depletion of the sub-population expressing YFP-ER-E2F1 above this threshold (Figure 5b). These predictions were confirmed by changes in experimental distributions during our time-lapse experiment (Figure 5b). To evaluate the long-term effects of E2F1 expression levels, we examined detailed population dynamics in U2OS pTRIPZ-YFP-ER-E2F1 by flow cytometry. Following induction of YFP-ER-E2F1, we observed a transient increase in the proportion of YFP-positive cells (Figures 5c and d). However, this sub-population steadily declined after 3 days of induction despite addition of fresh doxycycline and OHT. This rapid decline was observed in three cell lines and by 2 weeks YFP distributions of induced cells were indistinguishable from those of uninduced controls (Figure 5e). Elimination of high YFP expressors from the population over time was confirmed in a similar experiment in six cell lines (Supplementary Figure S5). We also found that cells expressing YFP-ER-E2F1 were eliminated from the population in the presence of serum (Supplementary Figure S6).
To test the possibility that E2F1 expression levels selected during these long-term experiments are below the sensitivity limit of flow cytometry we used RT-PCR. We found that expression levels of E2F1 (YFP-ER-E2F1+endogenous E2F1) and of its target gene CCNE1 were indistinguishable between induced and uninduced samples after 15 days of culture (Supplementary Figure S7). Complete elimination of cells with even slight E2F1 overexpression from the population demonstrates extreme sensitivity of cancer cells to even moderate E2F1 overexpression above the endogenous levels.
Dose-dependent induction of E2F1 target genes
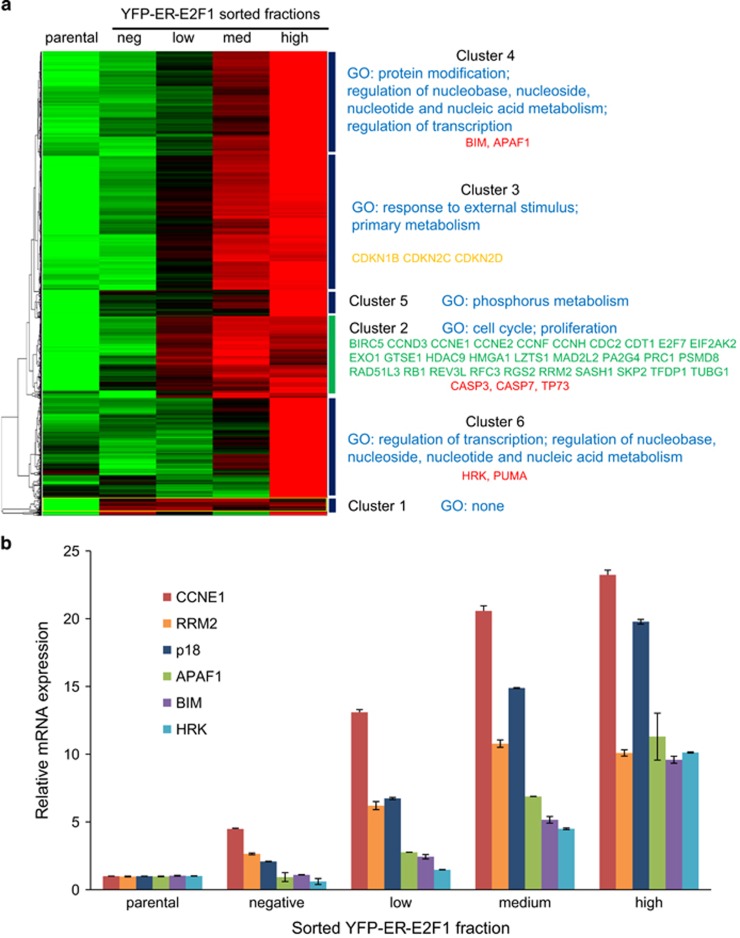
To gain insights into how different E2F1 levels induce different cell fates, we performed a global gene expression RNA-seq analysis in the sorted U2OS pTRIPZ-YFP-ER-E2F1 shown in Figure 1f. Unsupervised hierarchical clustering of 2375 genes induced at least 1.5-fold in the fraction with the highest YFP-ER-E2F1 level compared with the parental cells resulted in six main clusters (Figure 6a). We then analyzed the genes in each cluster for the enrichment of functional categories using GATHER.23
Figure 6.
Dose-dependent induction of E2F1 target genes. U2OS pTRIPZ-YFP-ER-E2F1 cells were grown in full serum-containing growth medium and treated with 500 ng/ml doxycycline for 48 h followed by addition of 90 nM OHT for an additional 20 h. Cells from the indicated YFP fractions were sorted by flow cytometry and expression of E2F1 target genes was analyzed by RNA-seq and quantitative RT-PCR (same samples as in Figures 1f and g). (a) RNA-seq analysis. Unsupervised hierarchical clustering of 2375 genes induced at least 1.5-fold in the fraction with the highest YFP-ER-E2F1 level compared with the parental cells. Red color represents higher expression and green color represents lower expression than the average of the five samples. Statistically significant enriched GO annotations are shown in blue for each cluster. Example cell cycle genes are shown in green, characterized apoptotic E2F1 targets are shown in red, and cell cycle inhibitory CDK inhibitors are shown in yellow. Complete gene lists for each cluster can be found in Supplementary Table S1. (b) Real-time quantitative RT-PCR analysis of indicated E2F1 target genes in sorted fractions. Graph represents the relative E2F1 expression after normalization to GAPDH housekeeping control (mean and range of duplicate PCR reactions)
Cluster 1 is separated from all other clusters and is characterized by higher and E2F1 dose-independent expression in all YFP-ER-E2F1 fractions compared with the parental control cells. No significant enrichment of any functional annotation and no enrichment of E2F1-binding sites were detected in this cluster. Thus, this small cluster likely consists of a heterogeneous collection of genes picked from the transcriptome by our 1.5-fold threshold. In contrast, all other clusters displayed E2F1 dose-dependent increase in gene expression and E2F1-binding sites were highly significantly enriched in the promoters, characteristic of bona fide E2F1 target genes.
Cluster 2 is characterized by strong induction already in the 'YFP-low' fraction. The top overrepresented gene ontology (GO) annotations are 'cell cycle' and 'proliferation'. Many well-characterized E2F1 proliferative targets, including cyclins E1, E2, F, and H, RRM2, and E2F7, belong to this cluster (shown in green in Figure 6a). No significant enrichment of apoptosis-related GO annotations is found in this or in other clusters. Three known E2F1 apoptotic targets caspase 3, caspase 7 and p73 are found in this cluster (shown in red in Figure 6a).
Genes in cluster 3 are weakly induced in the 'YFP-low' fraction and strongly induced in the 'YFP-medium' and 'YFP-high' fractions. This cluster is enriched for genes having a role in 'response to external stimuli' and primary metabolism. Notably, cell cycle inhibitors CDKN1B (p27), CDKN2C (p18) and CDKN2D (p19) (shown in yellow on Figure 6a) also belong to this cluster in correlation with the observed growth arrest phenotype.
Clusters 4, 5 and 6 are characterized by induction requiring higher levels of E2F1. They are enriched with metabolic and transcription-related genes. Importantly, key apoptotic E2F1 targets BIM, APAF1, HRK and PUMA are also found in these clusters. Gene lists of all clusters can be found in Supplementary Table S1.
RT-PCR analysis of selected E2F1 targets confirmed that cyclin E (CCNE1) was induced 4.5-fold in the YFP-negative fraction compared with parental cells, suggesting that E2F1 levels below the detection limit of our flow cytometry were sufficient for the induction of this central regulator of S-phase. Another cell cycle-promoting E2F1 target, ribonucleotide reductase (RRM2), was induced 2.6-fold in the YFP-negative fraction. Notably, a CDK inhibitor, p18 (CDKN2C), was also induced to a similar extent. In contrast, apoptotic E2F1 targets APAF1, BIM and HRK were not induced by these relatively low levels of E2F1 (threefold higher than in parental cells). The induction of CCNE1, RRM2 and p18 was significantly stronger than that of the apoptotic E2F1 targets also in the other sorted fractions.
Taken together, these results show that different target genes have different E2F1 activation thresholds. Numerous proliferation-related target genes are induced already by the lowest E2F1 levels. Intermediate E2F1 levels induce cdk inhibitors, which may be responsible for cell cycle arrest. Finally, although three apoptotic E2F1 targets are induced already by low E2F1 levels, many key apoptotic genes require higher E2F1 levels for induction.
Endogenous E2F1 mediates mitotic arrest and apoptosis in response to proteasome inhibitor, MLN2238
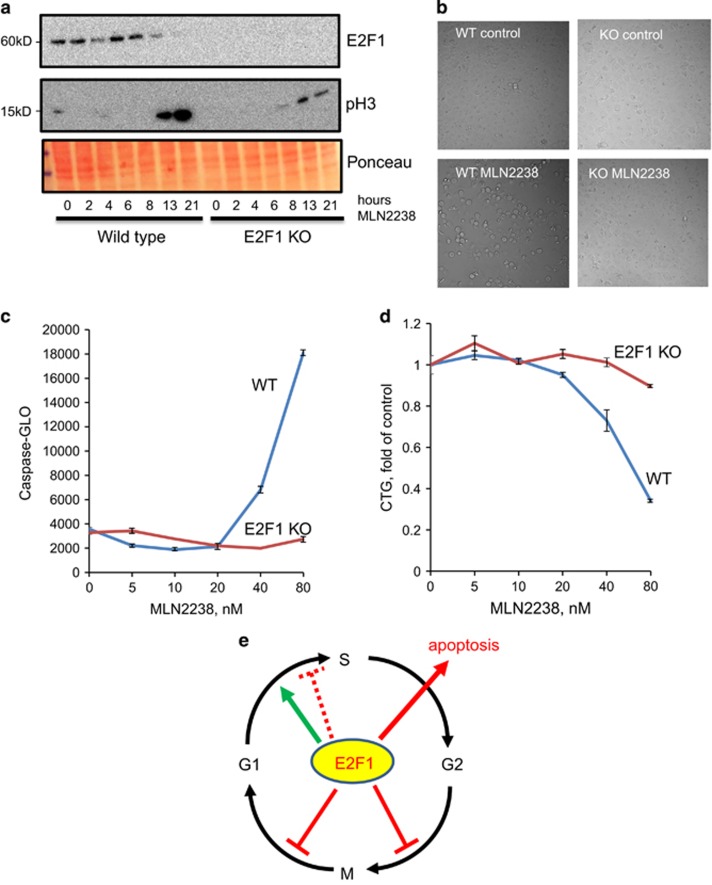
To evaluate the effects of endogenous E2F1 on cell fates, we used CRISPR-mediated knockout of E2F1 in a lung cancer cell line, H1299. We found that knockout of E2F1 did not affect growth rates of cells in unstressed conditions in the presence of serum. These results are in agreement with previous studies that demonstrated dispensability of E2F1 for normal cell growth in the presence of growth factors.24
To examine the role the endogenous E2F1, we treated wild-type and E2F1 KO cells with a proteasome inhibitor, MLN2238. This treatment resulted in a pulse of E2F1 protein induction peaking at 6 h of treatment and declining at later time points (Figure 7a). CRISPR-mediated knockout of E2F1 was very efficient at all time points. We found that MLN2238 induced mitotic arrest as evidenced by cell rounding (Figure 7b) and accumulation of phospho-histone H3 (Figure 7a) in the wild-type cells. This arrest was almost completely abolished in E2F1 KO cells. MLN2238 also induced E2F1-dependent apoptosis as evidenced by a dose-dependent caspase activation (Figure 7c) and reduction in cell number (Figure 7d).
Figure 7.
Endogenous E2F1 mediates mitotic arrest and apoptosis in response to proteasome inhibitor, MLN2238. (a) Western blot of E2F1 and phospho-Histone H3 (mitotic marker) from wild-type or E2F1 KO H1299 cells treated with 80 nM MLN2238 in full growth medium for indicated periods of time. Ponceau staining demonstrates equal loading. (b) Microgaphs (x200) of wild-type or E2F1 KO H1299 cells treated with 80 nM MLN2238 for 21 h in full growth medium. (c) Caspase activity (Caspase-Glo) assays in wild-type or E2F1 KO H1299 cells treated with indicated concentrations of MLN2238 in full growth medium for 21 h (mean and range of duplicate wells). (d) Cell Titer-Glo (CTG) assays in wild-type or E2F1 KO H1299 cells treated with indicated concentrations of MLN2238 in full growth medium for 72 h (mean and range of duplicate wells). (e) Graphical summary of the effects of E2F1 on different cell cycle transitions The green arrow indicates promotion of G1/S transition. The dashed inhibitory arrow indicates inhibition of DNA synthesis (G1 arrest) by intermediate levels of E2F1 in normal HME cells. This inhibition is inactive in the cancer cells we tested. The two thick inhibitory arrows indicate G2 and mitotic arrest by elevated E2F1 levels. High E2F1 levels also promote apoptosis
Collectively, these results show the importance of endogenous E2F1 levels in inducing mitotic arrest and apoptosis during treatment with a proteasome inhibitor.
Discussion
Previous studies have established an important role of qualitative factors, such as posttranslational modifications, differential interactions with cofactors and activity of other signaling pathways, in determining the outcome of E2F1 activation. For example, PI3K pathway selectively prevents E2F1-mediated activation of a subset of E2F target genes, including several apoptotic genes.25 Additional cofactors that cooperate with E2F1 in regulation of specific subsets of E2F1 target genes affecting E2F1-mediated cell fate decisions include Jab1,14 TopBP1,15 RRP1B26 and EPC1.17 Posttranslational modifications affecting target gene specificity and E2F1-mediated apoptosis include acetylation,18 methylation,19 phosphorylation27 and poly(ADP-ribosyl)ation.28
Although these qualitative factors are certainly important in determining the final outcome of E2F1 activation, the quantitative aspect has received much less attention. Our single-cell resolution quantitative measurements of the relationships between E2F1 level and cell fates demonstrate that the levels of E2F1 itself represent an additional critical level of control. Notwithstanding the canonical view of activator E2Fs (E2F1-3) as positive regulators of proliferation, our data demonstrate strong and general growth inhibitory activity of even moderately overexpressed E2F1 in multiple cell lines. Elimination or arrest of cells that express high levels of E2F1 provide an explanation to the fact that unlike Myc or cyclins, E2F1 is very rarely amplified or strongly overexpressed in cancer.
Our results reveal multiple safeguard mechanisms to restrain E2F1-mediated proliferation (Figure 7e). In normal HME cells, inhibition of DNA synthesis by intermediate E2F1 levels suggests activation of the G1/S checkpoint. This checkpoint can be overridden by the further increase in E2F1 levels leading to augmented DNA synthesis. This E2F1-induced G1 arrest is absent in all three tested cancer cell lines, demonstrating frequent inactivation of this checkpoint mechanism in cancer.
The second checkpoint, activated in a progressively higher proportion of cells with increasing E2F1 levels, is G2 arrest. Unlike G1 arrest, which was observed only in non-transformed cells, the G2 arrest checkpoint was operational in all tested cell lines as judged by accumulation of cells with >2 N DNA content. Several studies have described accumulation of cells with 4 N DNA content following E2F1 overexpression but no attempt was made to distinguish G2 arrest from mitotic (spindle) checkpoint activation.11, 29 Here, we show that activation of mitotic arrest represents a third checkpoint mechanism limiting proliferation of cells with deregulated E2F1. Our quantitative time-lapse analysis indicates that this checkpoint is activated by lower E2F1 levels than those triggering massive apoptosis.
Our RNA-seq and RT-PCR dose-response analyses of E2F1 target genes suggest that the mechanistic basis for the induction of different cell fates by increasing E2F1 levels may pertain to differential affinities of the targets. Low levels of E2F1 are sufficient to induce numerous cell cycle-promoting genes, intermediate levels induce growth arrest genes (i.e., p18, p19 and p27), whereas higher levels are necessary to induce key apoptotic E2F1 targets APAF1, PUMA, HRK and BIM. Additional studies are necessary to further test this hypothesis.
The current paradigm of cell cycle targeting antineoplastic drugs aims to inhibit cell cycle progression leading to inhibition of E2F activity. An example of this approach is a cdk4/6 inhibitor palbociclib, recently approved for the treatment of breast cancer. Notably, the efficacy of this drug in slowing tumor growth is limited to a sub-population of patients with intact Rb tumor suppressor.30 Based on our data, we propose that a complementary strategy to consider is to target E2F1 for upregulation. Our results on the crucial role of E2F1 in MLN2238-induced apoptosis provide proof-of-concept for this approach.
Materials and methods
Constructs
pEYFP-ER-E2F1 construct was generated by inserting the NheI digestion fragment containing the ER-E2F1 cassette from pBabePuro-HA.ER.E2F1 (a gift from Dr. Kristian Helin) into the compatible XbaI digestion site of the pEYFP-C1 vector (Clontech, Mountain View, CA, USA). pTRIPZ-YFP-ER-E2F1 construct was generated by inserting the YFP-ER-E2F1 cassette from pEYFP-ER-E2F1 into the pTRIPZ vector (Open Biosystems, Lafayette, CO, USA) between the AgeI and MluI sites replacing the TurboRFP and shRNA cassette. For CRISPR-Cas9-mediated E2F1 knockout, gRNA sequence 5′-GGAGATGATGACGATCTGCG-3′ targeting exon 1 of E2F1 was cloned into LentiCRISPR v.2 (Addgene, Cambridge, MA, USA, #52961).31
Cell lines and cell culture
All cell lines except HME-hTERT were obtained from Duke Cell Culture facility and their identity was authenticated by DNA STR profiling assay.
All cell lines were tested for mycoplasma contamination and found negative. U2OS cells were grown in Dulbecco's modified Eagle's medium with 10% fetal bovine serum (FBS). H1299, HCT116 and MDA-MB-231 cell lines were grown in RPMI with 10% FBS. HME-hTERT from ATCC (Manassas, VA, USA) were grown in MEBM supplemented with MEGM kit (Lonza, Walkersville, MD, USA). Doxycycline and OHT were from Sigma (St. Louis, MO, USA). Stable U2OS YFP-ER-E2F1 cell line was established by transfection of U2OS with pEYFP-ER-E2F1 followed by G418 selection. Cells were subsequently infected with pBabe-puro-H2B-RFP (gift from Dr. Cleveland, Ludwig Institute, San-Diego, CA, USA). YFP/RFP double-positive cells were flow sorted on FACS Diva instrument (BD Biosciences, San Jose, CA, USA).
RNA isolation and RT-PCR
Total RNA was isolated using RNeasy Kit (Qiagen, Valencia, CA, USA) and reverse-transcribed using High-Capacity cDNA Reverse Transcription Kit (ThermoFisher Scientific, Waltham, MA, USA). Real-time PCR was performed on StepOnePlus Real-Time PCR Instrument (ThermoFisher Scientific) using Power SYBR Green Master Mix (ThermoFisher Scientific). Primer sequences could be found in Supplementary Materials and Methods.
RNA-sequencing
RNA-seq was performed at Duke sequencing facility. Detailed protocols of library construction and sequencing can be found in Supplementary Materials and Methods. Data have been deposited to NCBI's Gene Expression Omnibus under the accession number GSE93365.
The raw fastq files were processed with sortmerna32 and trimmomatic33 to filter out the rRNA and adapter sequences, respectively. The output reads were then aligned to the h19 genome assembly using HISAT2.34 Afterward, the read counts were obtained for each gene using HTSeq,35 under the default '-m union' mode. The GTF file used for HTSeq was obtained from the PrimerSeq library36 (http://primerseq.sourceforge.net/gtf.html). The raw counts mapping to the genes were then converted to transcripts per million (TPM) values. For this, the mean fragment length for each condition (S1-S5) were computed using the Picard 'CollectInsertSizeMetrics' module (https://broadinstitute.github.io/picard/).
Genes with TPM>5 in all samples and at least 1.5-fold higher expression in 'YFP-high' fraction than in parental cells were used for clustering. Following logarithmic transformation, gene centering and normalization, unsupervised hierarchical clustering with uncentered correlation and average linkage clustering was performed using Cluster 3 software (http://bonsai.hgc.jp/~mdehoon/software/cluster) and visualized with Java TreeView (http://jtreeview.sourceforge.net/). Enrichment analysis of GO categories for each cluster was performed using GATHER.23
Lentivirus infection
Lentiviral particles were produced using 293T cells co-transfected with pMD2.G (Addgene 12259), and psPAX2 (Addgene 12260) and either pTRIPZ-YFP-ER-E2F1 or LentiCRISPR v.2 targeting construct for E2F1 knockout using Mirus TransIT-LT1 Transfection Reagent. Target cells were infected with lentivirus-containing supernatant and selected with 2.5 μg/ml puromycin.
Western blotting
Cell pellets were lysed with RIPA buffer containing Complete Mini protease inhibitor cocktail (Roche Diagnostics, Indianapolis, IN, USA). Proteins were resolved on 4–20% gradient SDS-PAGE, transferred to PFDF membranes and probed with antibodies for cleaved PARP (Cell Signaling Technologies, Danvers, MA, USA, 9541 S), E2F1 (Santa Cruz Biotechnologies, Dallas, TX, USA, sc-251) or p-Histone H3 (Ser10) (Santa Cruz Biotechnologies, sc-8656- R.)
Time-lapse microscopy
U2OS pEYFP-ER-E2F1/H2B-RFP cells were seeded on chamber slides (ibidi USA, Madison, WI, USA). Time-lapse images were acquired every 20 min on Nikon Eclipse Ti microscope (Nikon Instruments Inc., Melville, NY, USA) equipped with OkoLab environmental system (Pozzuoli, Italy), Spectra-X LED light source (Lumencor Inc., Beaverton, OR, USA), and Andor ZYLA 4.2 camera (Andor Technology Ltd, Belfast, UK). Exposure times were 100 ms for YFP and 50 ms for RFP channels, respectively. Single-cell fluorescence intensity was quantified using a custom Matlab software (MathWorks, Natick, MA, USA). Briefly, nuclei were identified using an adaptive threshold of the H2B-RFP channel and tracked through consecutive time frames using a sum of absolute differences minimization algorithm. Integrated nuclear YFP intensity was then extracted from nuclear masks as defined by the RFP channel and arranged into trajectories using the tracking algorithm.
Flow cytometry
All analytical flow cytometry was performed on MACSQuant VYB (Miltenyi Biotec, San Diego, CA, USA) using fixed parameters in all experiments. For analysis of DNA content, EdU incorporation and YFP intensity cells were incubated with 10 μM EdU for 18 h and then fixed and stained using Click-iT Plus EdU Alexa Fluor 647 Flow Cytometry Assay Kit (ThermoFisher Scientific). Cells were resuspended in 1 mg/ml DAPI and 2 × 105 events were collected from each sample. Analysis was performed in R using flowCore package to import the data.37 Following compensation using single-stained controls, cell aggregates were excluded using area versus width plots of the DAPI channel. Clustering was used for determination of the boundary between 2 N and >2 N DNA content and between EdU-positive and -negative populations. For determination of E2F1 dose responses, cells were divided into the indicated number of bins of increasing YFP fluorescence and percentages of >2 N and of EdU-positive cells were calculated for each YFP bin. For analysis of mitosis, cells were fixed in 1% formaldehyde and stained with anti-phosho-Histone H3 (Ser10) antibody (Santa Cruz Biotechnologies, sc-8656-R) followed by anti-rabbit Alexa 647 (Cell Signaling Technologies #4414 s).
Flow sorting of different YFP fractions for mRNA analysis was performed on FACSDiva (BD Biosciences, San Jose, CA, USA) instrument.
Modeling E2F1 population dynamics
E2F1 population dynamics were simulated using a single-cell stochastic model with parameters derived from time-lapse microscopy. A given cell has an E2F1-dependent probability of undergoing a specific fate, as well as an E2F1-independent fate timing. A batch of 25 000 cells with initial E2F1 distribution equal to that measured in time-lapse microscopy was simulated according to the model framework in Figure 5a. Briefly, for each cell in the batch, a fate is calculated based on its E2F1 expression (from Figure 2f), and fate timing is calculated from the distribution in Supplementary Figure S1B, and the population dynamics are recorded. However, it was observed experimentally that the proportion of cells undergoing mitosis was decreasing with time, and this effect was also included in the finalized model. Full description of modeling can be found in Supplementary Information.
Acknowledgments
We thank Dr. Bernard Mathey-Prevot for critical reading of the manuscript, Drs. Shuqiang Huang and Dong Peng for help with microscopy, Dr. So Young Kim from Duke functional genomics core for help with CRISPR-Cas9-mediated E2F1 knockout, and all members of the You lab for discussions. This research was partially supported by National Institutes of Health (R01 GM106107 to LY, R01 GM117106 to RG), National Science Foundation (DMS1463137 and DMS1418172 to GY), and a David and Lucile Packard Fellowship (LY).
Author contributions
IS and LY conceived and designed the ideas and experiments and wrote the manuscript. IS performed the experiments. IS, MD, AD, DM, RG, GY and LY analyzed the data. CZ, JSK and GY provided reagents. All authors assisted in writing the manuscript.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by H Ichijo
The authors declare no conflict of interest.
Supplementary Material
References
- Bretones G, Delgado MD, Leon J. Myc and cell cycle control. Biochim Biophys Acta 2015; 1849: 506–516. [DOI] [PubMed] [Google Scholar]
- Mateyak MK, Obaya AJ, Sedivy JM. c-Myc regulates cyclin D-Cdk4 and -Cdk6 activity but affects cell cycle progression at multiple independent points. Mol Cell Biol 1999; 19: 4672–4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ. Cancer cell cycles. Science 1996; 274: 1672–1677. [DOI] [PubMed] [Google Scholar]
- van den Heuvel S, Dyson NJ. Conserved functions of the pRB and E2F families. Nat Rev Mol Cell Biol 2008; 9: 713–724. [DOI] [PubMed] [Google Scholar]
- DeGregori J, Kowalik T, Nevins JR. Cellular targets for activation by the E2F1 transcription factor include DNA synthesis- and G1/S-regulatory genes. Mol Cell Biol 1995; 15: 4215–4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalik TF, DeGregori J, Schwarz JK, Nevins JR. E2F1 overexpression in quiescent fibroblasts leads to induction of cellular DNA synthesis and apoptosis. J Virol 1995; 69: 2491–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chellappan SP, Hiebert S, Mudryj M, Horowitz JM, Nevins JR. The E2F transcription factor is a cellular target for the RB protein. Cell 1991; 65: 1053–1061. [DOI] [PubMed] [Google Scholar]
- Shan B, Lee WH. Deregulated expression of E2F-1 induces S-phase entry and leads to apoptosis. Mol Cell Biol 1994; 14: 8166–8173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin XQ, Livingston DM, Kaelin WG Jr, Adams PD. Deregulated transcription factor E2F-1 expression leads to S-phase entry and p53-mediated apoptosis. Proc Natl Acad Sci USA 1994; 91: 10918–10922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg C, Helin K, Sehested M, Karlstrom O. E2F-1-induced p53-independent apoptosis in transgenic mice. Oncogene 1998; 17: 143–155. [DOI] [PubMed] [Google Scholar]
- Elliott MJ, Dong YB, Yang H, McMasters KM. E2F-1 up-regulates c-Myc and p14(ARF) and induces apoptosis in colon cancer cells. Clin Cancer Res 2001; 7: 3590–3597. [PubMed] [Google Scholar]
- Radhakrishnan SK, Feliciano CS, Najmabadi F, Haegebarth A, Kandel ES, Tyner AL et al. Constitutive expression of E2F-1 leads to p21-dependent cell cycle arrest in S phase of the cell cycle. Oncogene 2004; 23: 4173–4176. [DOI] [PubMed] [Google Scholar]
- Lindstrom MS, Wiman KG. Myc and E2F1 induce p53 through p14ARF-independent mechanisms in human fibroblasts. Oncogene 2003; 22: 4993–5005. [DOI] [PubMed] [Google Scholar]
- Hallstrom TC, Nevins JR. Jab1 is a specificity factor for E2F1-induced apoptosis. Genes Dev 2006; 20: 613–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Luo Y, Lin FT, Lin WC. TopBP1 recruits Brg1/Brm to repress E2F1-induced apoptosis, a novel pRb-independent and E2F1-specific control for cell survival. Genes Dev 2004; 18: 673–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shats I, Gatza ML, Liu B, Angus SP, You L, Nevins JR. FOXO transcription factors control E2F1 transcriptional specificity and apoptotic function. Cancer Res 2013; 73: 6056–6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Alla V, Goody D, Gupta SK, Spitschak A, Wolkenhauer O et al. Epigenetic factor EPC1 is a master regulator of DNA damage response by interacting with E2F1 to silence death and activate metastasis-related gene signatures. Nucleic Acids Res 2015; 44: 117–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pediconi N, Ianari A, Costanzo A, Belloni L, Gallo R, Cimino L et al. Differential regulation of E2F1 apoptotic target genes in response to DNA damage. Nat Cell Biol 2003; 5: 552–558. [DOI] [PubMed] [Google Scholar]
- Kontaki H, Talianidis I. Lysine methylation regulates E2F1-induced cell death. Mol Cell 2010; 39: 152–160. [DOI] [PubMed] [Google Scholar]
- Wong JV, Yao G, Nevins JR, You L. Viral-mediated noisy gene expression reveals biphasic E2f1 response to MYC. Mol Cell 2011; 41: 275–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong P, Maddali MV, Srimani JK, Thelot F, Nevins JR, Mathey-Prevot B et al. Division of labour between Myc and G1 cyclins in cell cycle commitment and pace control. Nat Commun 2014; 5: 4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juan G, Traganos F, James WM, Ray JM, Roberge M, Sauve DM et al. Histone H3 phosphorylation and expression of cyclins A and B1 measured in individual cells during their progression through G2 and mitosis. Cytometry 1998; 32: 71–77. [DOI] [PubMed] [Google Scholar]
- Chang JT, Nevins JR. GATHER: a systems approach to interpreting genomic signatures. Bioinformatics 2006; 22: 2926–2933. [DOI] [PubMed] [Google Scholar]
- Chen D, Pacal M, Wenzel P, Knoepfler PS, Leone G, Bremner R. Division and apoptosis of E2f-deficient retinal progenitors. Nature 2009; 462: 925–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallstrom TC, Mori S, Nevins JR. An E2F1-dependent gene expression program that determines the balance between proliferation and cell death. Cancer Cell 2008; 13: 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik JC, Wang B, Liu K, Lue JK, Lin WC. Regulation of E2F1-induced apoptosis by the nucleolar protein RRP1B. J Biol Chem 2010; 285: 6348–6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnevale J, Palander O, Seifried LA, Dick FA. DNA damage signals through differentially modified E2F1 molecules to induce apoptosis. Mol Cell Biol 2012; 32: 900–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari A, Iwasaki T, Pyndiah S, Cassimere EK, Palani CD, Sakamuro D. Regulation of E2F1-induced apoptosis by poly(ADP-ribosyl)ation. Cell Death Differ 2015; 22: 311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atienza C Jr., Elliott MJ, Dong YB, Yang HL, Stilwell A, Liu TJ et al. Adenovirus-mediated E2F-1 gene transfer induces an apoptotic response in human gastric carcinoma cells that is enhanced by cyclin dependent kinase inhibitors. Int J Mol Med 2000; 6: 55–63. [DOI] [PubMed] [Google Scholar]
- Roberts PJ, Bisi JE, Strum JC, Combest AJ, Darr DB, Usary JE et al. Multiple roles of cyclin-dependent kinase 4/6 inhibitors in cancer therapy. J Natl Cancer Inst 2012; 104: 476–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 2014; 11: 783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopylova E, Noe L, Touzet H. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 2012; 28: 3211–3217. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014; 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 2015; 12: 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 2015; 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokheim C, Park JW, Xing Y. PrimerSeq: design and visualization of RT-PCR primers for alternative splicing using RNA-seq data. Genomics Proteomics Bioinformatics 2014; 12: 105–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahne F, LeMeur N, Brinkman RR, Ellis B, Haaland P, Sarkar D et al. flowCore: a Bioconductor package for high throughput flow cytometry. BMC Bioinformatics 2009; 10: 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.