Abstract
Lung ischemia-reperfusion (I/R) injury remains one of the most common complications after various cardiopulmonary surgeries. The inflammation response triggered by the released damage-associated molecular patterns (DAMPs) aggravates lung tissue damage. However, little is known about the role of autophagy in the pathogenesis of lung I/R injury. Here, we report that a variety of inflammation-related and autophagy-associated genes are rapidly upregulated, which facilitate the inflammation response in a minipig lung I/R injury model. Left lung I/R injury triggered inflammatory cytokine production and activated the autophagy flux as evidenced in crude lung tissues and alveolar macrophages. This was associated with the release of DAMPs, such as high mobility group protein B1 (HMGB1) and heat shock protein 60 (HSP60). Indeed, treatment with recombinant HMGB1 or HSP60 induced autophagy in alveolar macrophages, whereas autophagy inhibition by knockdown of ATG7 or BECN1 markedly reduced DAMP-triggered production of inflammatory cytokines including IL-1β, TNF and IL12 in alveolar macrophages. This appeared to be because of decreased activation of MAPK and NF-κB signaling. Furthermore, knockdown of ATG7 or BECN1 inhibited Lys63 (K63)-linked ubiquitination of TNF receptor-associated factor 6 (TRAF6) in DAMP-treated alveolar macrophages. Consistently, treatment with 3-MA inhibited K63-linked ubiquitination of TRAF6 in I/R-injured lung tissues in vivo. Collectively, these results indicate that autophagy triggered by DAMPs during lung I/R injury amplifies the inflammatory response through enhancing K63-linked ubiquitination of TRAF6 and activation of the downstream MAPK and NF-κB signaling.
Ischemia-reperfusion (I/R) injury characterized by robust sterile inflammatory responses remains a challenge in diverse clinical situations such as organ transplantation, thromboembolic events and cardiac arrest.1 During I/R injury, acute oxidative or nitrosative stress and consequent necrotic cell death leads to the release of endogenous self-antigens known as damage-associated molecular patterns (DAMPs), such as high mobility group box 1 (HMGB1) and heat shock proteins (HSPs).2 Upon binding to receptors, DAMPs activate mitogen-activated protein kinase (MAPK) and nuclear factor of kappa light polypeptide gene enhancer in B-cells (NF-κB) signaling, which in turn triggers the inflammatory response. The release of inflammatory cytokines such as TNF, IL-1β and IL12 by immune cells aggravates acute tissue damage.3 Owing to its dual blood supply systems and continuous physiologic demand for oxygen uptake and gas exchange, the lung appears especially vulnerable to I/R injury. In addition, the molecular mechanisms of I/R injury in lungs are thought to be more complicated than those in other organs.4 Thus, clarification of key components and better understanding of the process involved in sterile inflammation during lung I/R injury is critical for improving the outcome of patients with acute lung ischemia injury.
Autophagy is an evolutionarily conversed cellular process associated with clearance of damaged proteins and organelles. It is involved in various cellular conditions, such as energy starvation, oxidative stress, and endoplasmic reticulum stress.5 Recent studies have revealed the close association between autophagy and the inflammation response.3, 6 Autophagy induced by pattern recognition receptors (PRRs) can influence the inflammation response through interactions with innate immune signaling pathways.6 As a complex organ that functions primarily in gas exchange, the lung is prone to exposure to various stress-inducing factors, including hypoxia, oxidants, smokes and I/R. This results in constitutive activation of autophagy in cells of lungs as an inducible response to cellular stress.7 Increasing evidence suggests that autophagy participates in the pathogenesis of I/R injury in organs such as the heart, kidney and liver. Dysregulated autophagy either acts as a protective mechanism or contributes to the damaging effect of I/R injury depending on organs and experimental models in question.8, 9, 10 However, the potential relationship between autophagy and the pathogenesis of I/R injury in the context of sterile inflammation during lung I/R injury remains unclear.11
PRRs are crucial for sensing endogenous and exogenous danger signals and initiating innate inflammatory responses.12 Accumulating evidence suggests that Toll-like receptors (TLRs), the primary cellular sensor for pathogen-associated molecular patterns (PAMPs), can regulate autophagy through activation of downstream signaling in macrophages and other types of cells.3, 13 On the other hand, The crosstalk between autophagy and TLRs also modulates inflammatory responses.14, 15 During lung I/R injury, DAMPs are released as danger signals that can be recognized by TLRs. It is therefore of particular interest to explore the role of autophagy in regulating DAMP-mediated sterile inflammation caused by lung I/R injury. Here, we report that a number of inflammation-related and autophagy-associated genes are upregulated or downregulated in a minipig left lung I/R injury model and that DAMPs released after lung I/R injury induce autophagy, which in turn facilitates DAMP-triggered inflammation responses in lung tissues as manifested largely in alveolar macrophages.
Results
Lung I/R injury triggers inflammation responses in lung tissues of minipigs
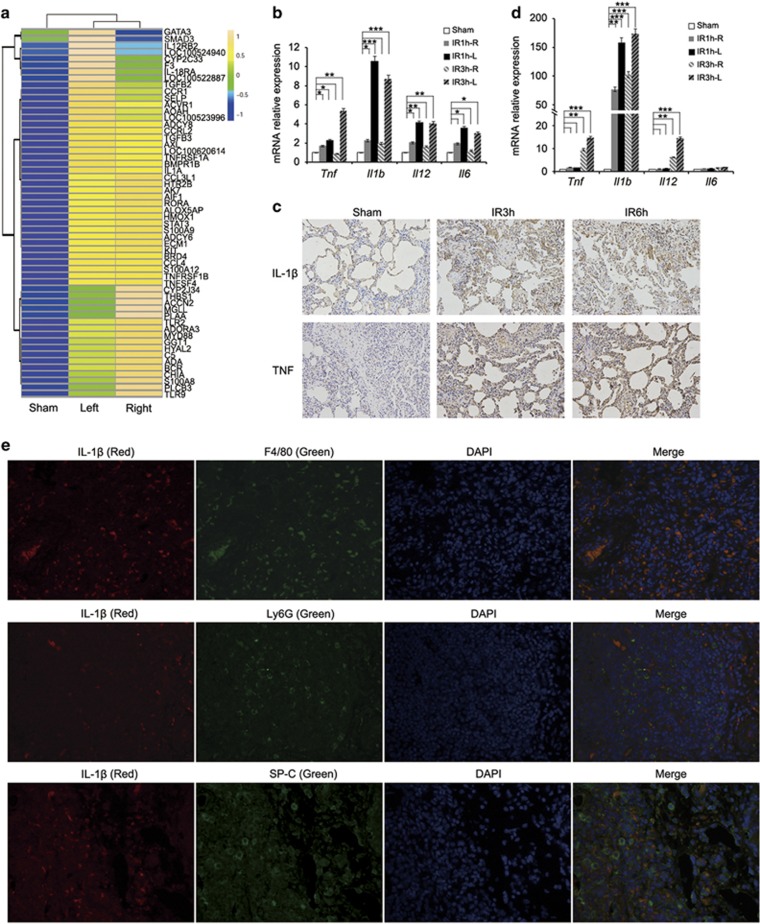
We constructed a left lung I/R injury model of minipigs and monitored the inflammation response in lung tissues. Total RNA from crude left or right lung tissues of minipigs in the sham or I/R group was subjected to microarray analysis. The results showed that 54 of a set of 315 inflammation-related genes were significantly upregulated in tissues of both left and right lungs of minipigs undergoing I/R compared with those in the sham group (Figure 1a, Supplementary Figure S1a and Supplementary Table S1). As shown in Figure 1b, the increase in Tnf, Il1b, Il12 and Il6 mRNA expression was detectable as early as 1 h after the commencement of reperfusion, which is more pronounced in left lungs that had been subjected to I/R injury. Similarly, the expression of IL-1β and TNF at the protein level was also upregulated in left lungs of minipigs in the I/R group (Figure 1c and Supplementary Figure S2). Consistent with the important role of macrophages in the initiation and generation of the early inflammatory response to lung I/R injury,12, 16 alveolar macrophages isolated from bronchoalveolar lavage fluid (BALF) of minipigs with I/R injury expressed elevated levels of Tnf, Il1b and Il12 mRNA (Figure 1d). In support, immunofluorescence analysis showed that alveolar macrophages (F4/80+) but not neutrophils (Ly6G+) or alveolar epithelial cells (SP-C+) were the source of IL-1β production in left lung tissues of minipigs with lung ischemia followed by reperfusion for 1 h (Figure 1e and Supplementary Figure S3). These findings, along with the previous results showing that alveolar macrophage depletion protects lungs from I/R-induced injury and significantly reduces cytokine/chemokine production,16, 17, 18 indicate that lung I/R primarily triggers production of inflammatory cytokines by alveolar macrophages, which in turn have an important role in the pathogenesis of acute lung I/R injury.
Figure 1.
Lung I/R injury triggers the inflammation response in lung tissues of minipigs. Different groups of minipigs underwent 1 h left lung ischemia followed by reperfusion for 1 h (IR1h), 3 h (IR3h) or 6 h (IR6h), or only received sham operations (Sham). (a) Heat maps showing hierarchical clustering of upregulated expressed transcripts of inflammation-related genes in right or left lung tissues from the sham group or minipigs subjected to lung ischemia followed by reperfusion for 1 h. (b) qPCR analysis of Tnf, Il1b, Il12 and Il6 mRNA expression in right (R) or left (L) lung tissues from the sham group or minipigs undergoing lung I/R as indicated (n=3). (c) Immunohistochemical staining of IL-1β and TNF expression in left lung tissues from the sham group or minipigs undergoing lung I/R as indicated. (d) qPCR analysis of Tnf, Il1b, Il12 and Il6 mRNA expression in alveolar macrophages from the sham group or minipigs undergoing lung I/R as indicated (n=3). (e) Immunofluorescence staining of IL-1β, F4/80 (macrophage marker), Ly6G (neutrophil marker) and SP-C (alveolar epithelial cell marker) in left lung tissues from minipigs subjected to lung ischemia followed by reperfusion for 1 h. Original magnification, × 400. Data are representative (c and e), or mean±S.E.M. (b and d), of three individual experiments. *P<0.05, **P<0.01, ***P<0.001
Lung I/R injury induces autophagy in lung tissues of minipigs
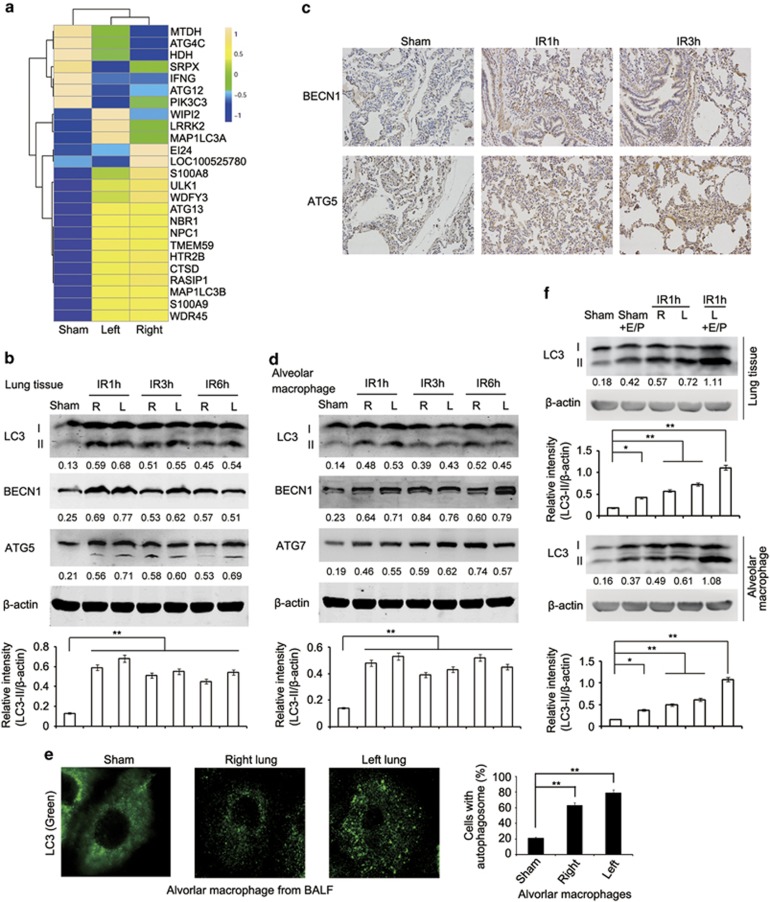
Next, we investigated whether lung I/R injury activates autophagy in lung tissues. Microarray analysis showed that 25 of 70 autophagy related genes were differentially expressed between lungs undergoing I/R injury and those in the sham group (Figure 2a, Supplementary Figures S1b–d and Supplementary Table S2). Induction of autophagy was evidenced by conversion of microtubule-associated protein 1 light chain 3 α/β (MAP1LC3A/B, LC3A/B)-I into LC3-II and the increase in the protein and mRNA expression of Beclin 1 (BECN1) and autophagy related 5 (ATG5) in crude lung tissues from minipigs subjected to I/R (Figures 2b, c and Supplementary Figure S4). Consistently, conversion of LC3, upregulation of BECN1 and autophagy related 7 (ATG7) as well as increased LC3 puncta were also observed in alveolar macrophages isolated from BALF of lungs of both sides from minipigs with I/R (Figures 2d and e). Activation of the autophagy flux was further confirmed using the lysosomal degradation inhibitors E64d and Pepstatin A in vivo (Figure 2f). A pulmonary protective solution with or without the addition of E64d and Pepstatin A was continuously perfused into left lungs of minipigs in the sham or ischemia group for 1 h. As shown in Figure 2f, more pronounced accumulation of LC3-II was observed in crude lung tissues as well as isolated alveolar macrophages of minipigs with ischemia treated with the protective solution in the presence of E64d and Pepstatin A (Figure 2f). Of note, autophagy was not observed in alveolar epithelial cells of minipigs with lung I/R injury (Supplementary Figure S5). Taken together, these results indicate that the autophagy is induced in lungs, and in particular, in alveolar macrophages at relatively early stages of lung I/R injury.
Figure 2.
Lung I/R injury induces autophagy in lung tissues of minipigs. (a) Heat maps showing hierarchical clustering of differentially expressed transcripts of autophagy related genes in right or left lung tissues from the sham group or minipigs subjected to lung ischemia followed by reperfusion for 1 h. (b) Immunoblotting analysis of LC3, BECN1, ATG5 and β-actin (as a loading control) in lysates of right or left lung tissue from the sham group or minipigs subjected to lung I/R as indicated. (c) Immunohistochemical staining of BECN1 and ATG5 in left lung tissue of the sham group or minipigs subjected to lung I/R as indicated. (d) Immunoblotting analysis of LC3, BECN1, ATG7 and β-actin (as a loading control) in lysates of alveolar macrophages from BALF of the sham group or minipigs subjected to lung I/R as indicated. (e) Immunofluorescence analysis of LC3 in alveolar macrophages from BALF of the sham group or minipigs subjected to lung ischemia followed by reperfusion for 1 h. Original magnification, × 630. Quantification of cells with autophagosomes was also shown. (f) Immunoblotting analysis of LC3 and β-actin (as a loading control) in lysates of lung tissues (upper) or alveolar macrophages (lower) from the sham group or minipigs subjected to 1 h left lung ischemia and perfusion with pulmonary protective solution containing E64d (15 μg/ml) and Pepstatin A (15 μg/ml) or DMSO followed by reperfusion for 1 h. The band densitometry was quantified using ImageJ software (National Institutes of Health, Bethesda, MD, USA). Values below lanes represent the relative intensities of the corresponding proteins (LC3-II, BECN1, ATG5 and ATG7) to β-actin in the same lane. The relative band intensities of LC3-II/β-actin were calculated from three independent experiments and shown as mean±S.E.M. Data are representative of three individual experiments (b–f). *P<0.05, **P<0.01
DAMPs are involved in lung I/R injury-triggered autophagy in alveolar macrophages
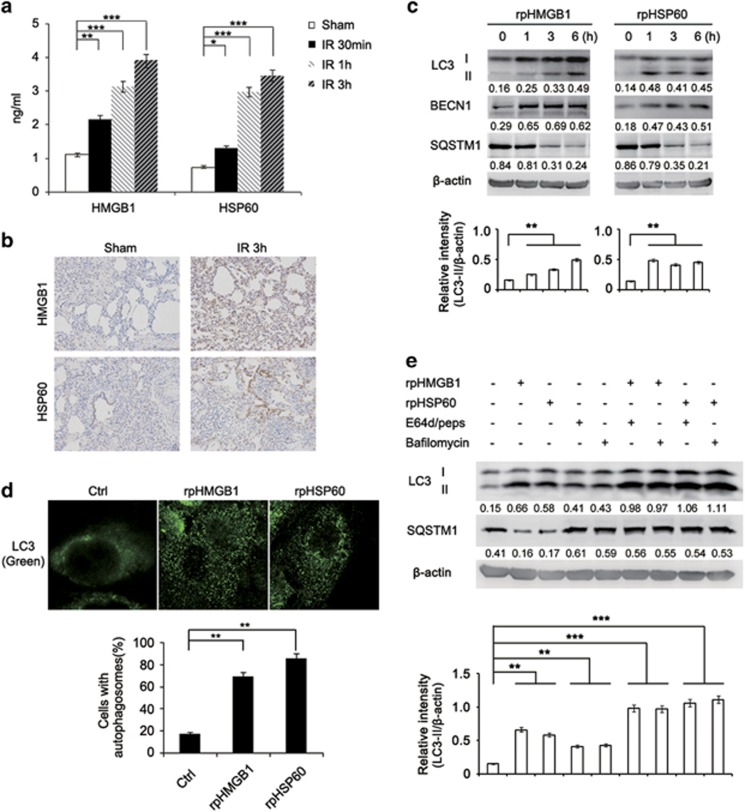
As the significant increase in HMGB1 and HSP60 production was detectable as soon as 30 min after reperfusion in BALF of minipigs with I/R (Figures 3a and b), we examined whether these DAMPs were involved in activation of autophagy. Indeed, treatment with recombinant porcine HMGB1 (rpHMGB1) or HSP60 (rpHSP60) induced autophagy in minipig alveolar macrophages. This was evidenced by conversion of LC3-I into LC3-II, upregulation of BECN1, degradation of sequestosome 1 (SQSTM1) and increased LC3 puncta (Figures 3c and d). The autophagy flux induced by HMGB1 or HSP60 was further confirmed by increased accumulation of LC3-II and inhibition of degradation of SQSTM1 when cells were treated with E64d and Pepstatin A, or the autophagosome–lysosome fusion inhibitor Bafilomycin A1 (Figure 3e). Noticeably, HMGB1 or HSP60 did not trigger apoptosis or necrosis in alveolar macrophages (Supplementary Figure S6). These results suggest that DAMPs released into the extracelluar space after lung I/R contribute to autophagy induction in alveolar macrophages.
Figure 3.
DAMPs are involved in lung I/R injury-triggered autophagy in alveolar macrophages of minipigs. (a) ELISA of HMGB1 and HSP60 production in the BALF of the sham group or minipigs subjected to lung I/R as indicated (n=3). (b) Immunohistochemical staining of HMGB1 and HSP60 in left lung tissues of the sham group or minipigs subjected to lung I/R as indicated. (c) Immunoblotting analysis of LC3, BECN1, SQSTM1 and β-actin (as a loading control) in lysates of alveolar macrophages treated with rpHMGB1 (1 μg/ml) or rpHSP60 (1 μg/ml) for the indicated periods. (d) Immunofluorescence analysis of LC3 in alveolar macrophages stimulated with rpHMGB1 (1 μg/ml) or rpHSP60 (1 μg/ml) for 3 h. Original magnification, × 630. Quantification of cells with autophagosomes is also shown. (e) Immunoblotting analysis of LC3, SQSTM1 and β-actin (as a loading control) in lysates of alveolar macrophages with or without treatment with E64d (15 μg/ml), Pepstatin A (15 μg/ml) or Bafilomycin A1 (30 nM) before stimulation by rpHMGB1 (1 μg/ml) or rpHSP60 (1 μg/ml) for 3 h. Values below lanes represent the relative intensities of the corresponding proteins (LC3-II, BECN1 and SQSTM1) to β-actin in the same lane. The relative band intensities of LC3-II/β-actin were calculated from three independent experiments and shown as mean±S.E.M. Data are mean±S.E.M. (a) or representative (b–e), of three individual experiments. *P<0.05, **P<0.01, ***P<0.001
Autophagy is involved in inflammation responses in lung I/R injury
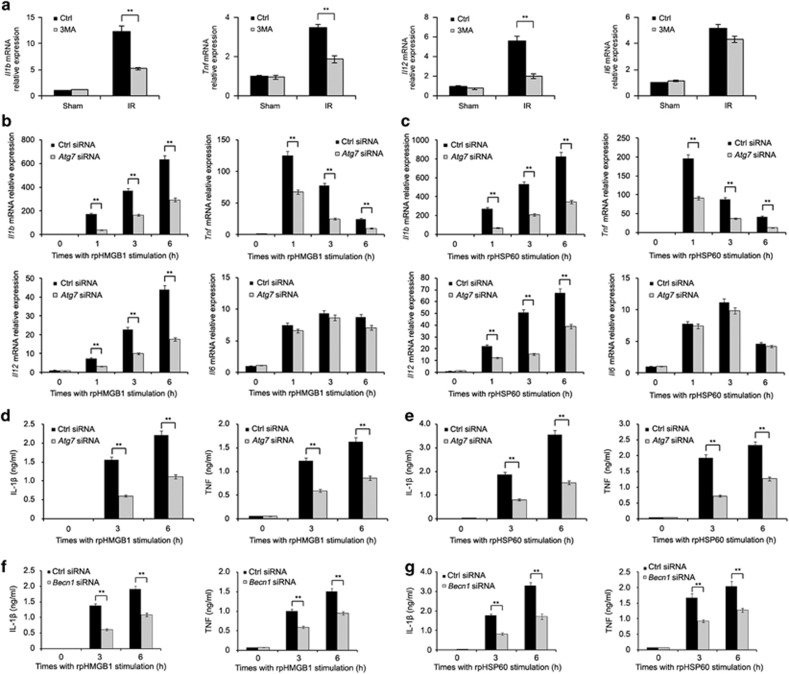
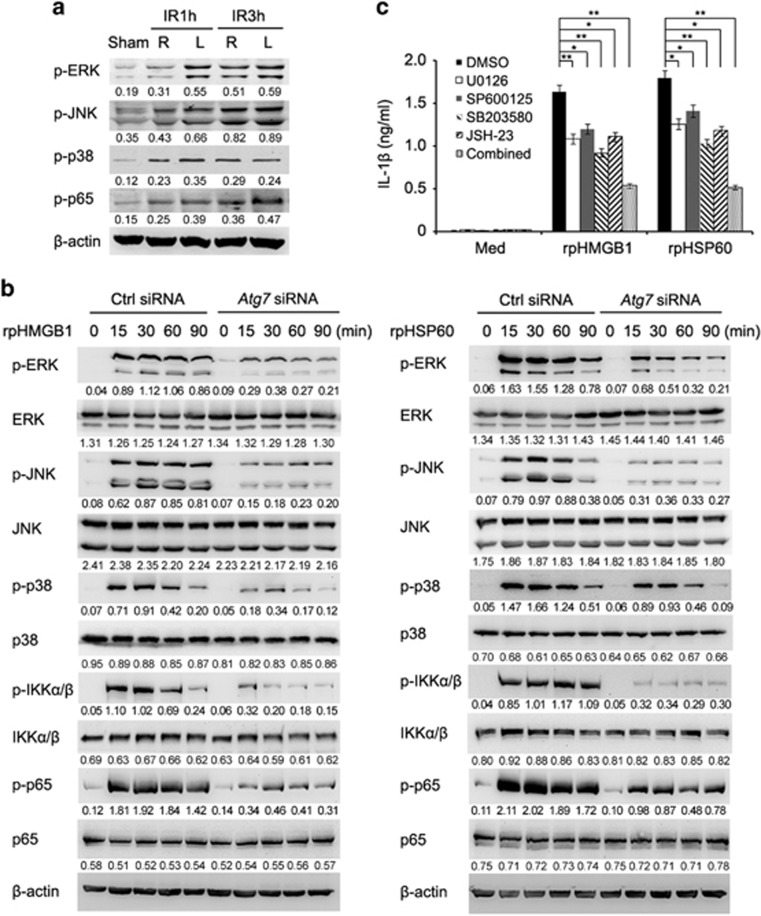
Next, we examined the effect of 3-MA treatment in vivo on inflammatory gene expression in lung tissues. Minipigs were subjected to lung ischemia and perfusion with pulmonary protective solution containing 3-MA or DMSO followed by reperfusion for 1 h. As shown in Figure 4a, 3-MA treatment significantly decreased the levels of Il1b, Tnf and Il12 mRNA, but not the level of Il6 mRNA, in left lung tissues, suggesting that autophagy is involved in the inflammatory response in lung tissues triggered by I/R injury. Furthermore, 3-MA treatment did not affect HMGB1 and HSP60 production in the BALF of minipigs with I/R, which indicates that the decrease of inflammatory cytokine production in 3-MA treatment group is not because of decreased induction and release of DAMPs (Supplementary Figure S7). In minipig alveolar macrophages, stimulation with rpHMGB1 or rpHSP60 significantly increased the expression of Il1b, Tnf, Il12 and Il6 at the mRNA level (Figures 4b and c), suggesting that HMGB1- and HSP60-induced autophagy and the inflammation response may interact with each other. Indeed, knockdown of ATG7 by two individual siRNAs not only inhibited accumulation of LC3-II and SQSTM1 degradation induced by rpHMGB1, but also reduced the production of IL-1β, TNF and IL12 caused by rpHMGB1 or rpHSP60 in alveolar macrophages (Figures 4b–e, Supplementary Figure S8 and S9). Nevertheless, ATG7 knockdown did not significantly affect the expression of Il6 mRNA triggered by rpHMGB1 or rpHSP60 (Figures 4b, c, Supplementary Figures S9b and c). Consistently, ATG7 overexpression increased the production of IL-1β and TNF induced by rpHMGB1 or rpHSP60, suggesting that autophagy promotes DAMP-induced inflammatory cytokine production (Supplementary Figures S10a and b). Overexpression of siRNA-resistant ATG7 mutant with synonymous mutation at siRNA-targeted site could rescue the effect of ATG7 knockdown on inflammatory cytokine production induced by DAMPs in alveolar macrophages (Supplementary Figures S10c and d), suggesting that ATG7 knockdown is specific. What's more, silencing of BECN1, another important autophagy related gene, decreased the production of IL-1β and TNF induced by rpHMGB1 or rpHSP60 in alveolar macrophages (Figures 4f and g). These data indicate that the autophagy activation induced by DAMPs is necessary for DAMP-triggered inflammation responses in alveolar macrophages.
Figure 4.
Treatment with 3-MA or ATG7 knockdown attenuates inflammatory cytokine production in lung tissues or alveolar macrophages respectively. (a) Minipigs were subjected to lung ischemia and perfusion with pulmonary protective solution containing 3-MA (5 mM) or DMSO followed by reperfusion for 1 h or received sham operations (Sham). The levels of Il1b, Tnf, Il12 and Il6 mRNA expression in left lung tissue were detected by qPCR analysis. (b–g) Alveolar macrophages were transfected with the control siRNA, Atg7 siRNA or Becn1 siRNA. Sixty hour later, cells were treated with rpHMGB1 (1 μg/ml) (b, d and f) or rpHSP60 (1 μg/ml) (c, e and g) for the indicated periods. The levels of Il1b, Tnf, Il12 and Il6 mRNA expression were detected by qPCR analysis (b and c), or the levels of IL-1β and TNF in supernatants were measured by ELISA (d–g). Data are mean±S.E.M. of three individual experiments. **P<0.01
As numerous previous studies have shown that endogenous released HMGB1 or HSP60 triggers sterile inflammation mainly through binding TLR4 during I/R injury of different organs including heart, lung and liver,19, 20, 21 we investigated whether HMGB1 or HSP60-induced autophagy activation and inflammation cytokine production via TLR4 in lung I/R injury of minipig. TAK-242 is a novel small molecule that selectively inhibits TLR4 signaling in cells and in vivo models of several species.22 A pulmonary protective solution with or without the addition of TAK-242 was continuously perfused into left lungs of minipigs in the sham or ischemia group for 1 h. Indeed, TAK-242 treatment inhibited conversion of LC3 and decreased the levels of Il1b and Tnf mRNA in left lung tissue of minipig with lung I/R (Supplementary Figures S11a and b). Furthermore, knockdown of TLR4 reduced accumulation of LC3-II and the production of IL-1β and TNF induced by rpHMGB1 or rpHSP60 in alveolar macrophages (Supplementary Figures S11c–e). Thus, our data together with previous studies demonstrate that HMGB1 and HSP60 bind TLR4, then trigger inflammatory cytokine production and activate autophagy, whereas autophagy in turn promotes inflammation response activated by TLR4 signaling during lung I/R.
Autophagy is involved in activation of MAPK and NF-κB signaling induced by DAMPs
We further tested the effect of autophagy on DAMP-activated downstream signal pathways in alveolar macrophages. Lung I/R resulted in increased activation (phosphorylation) of ERK, JNK, p38 and p65 in lung tissues (Figure 5a). Strikingly, ATG7 knockdown substantially attenuated the phosphorylation levels of ERK, JNK, p38, IKKα/β and p65 in alveolar macrophages caused by rpHMGB1 or rpHSP60 (Figure 5b). To clarify the role of MAPK and NF-κB signaling in inflammatory cytokine production triggered by DAMPs, we treated alveolar macrophages with rpHMGB1 or rpHSP60 in the presence of MAPK inhibitors U0126, SP600125, or SB203580 or NF-κB inhibitor JSH-23. As shown in Figure 5c, the increase in IL-1β production triggered by rpHMGB1 or rpHSP60 was reversed, albeit partially, by U0126, SP600125, SB203580 or JSH-23, and more significantly reversed by the combined use of the above MAPK and NF-κB inhibitors. These results indicate that autophagy is involved in DAMP-triggered activation of MAPK and NF-κB signaling that in turn has an important role in the increased inflammatory cytokine production in alveolar macrophages during lung I/R injury.
Figure 5.
Autophagy is involved in activation of MAPK and NF-κB signaling induced by DAMPs in alveolar macrophages of minipigs. (a) Immunoblotting analysis of phosphorylation (p−) levels of ERK, JNK, p38 and p65 in lysates of alveolar macrophages isolated from right or left lungs of minipigs in the sham group or those undergoing lung I/R as indicated. Values below lanes represent the relative intensities of the corresponding proteins to β-actin in the same lane. (b) Immunoblotting analysis of phosphorylation (p−) or total levels of ERK, JNK, p38, IKKα/β and p65 in lysates of alveolar macrophages transfected with the control siRNA or Atg7 siRNA followed by treatment with rpHMGB1 (1 μg/ml) (left) or rpHSP60 (1 μg/ml) (right) for the indicated periods. Values below lanes represent the relative intensities of the phosphorylated proteins to the total proteins or the total proteins to β-actin, respectively, in the same lane. (c) ELISA of IL-1β production in supernatants of alveolar macrophages with or without pretreatment with inhibitors of ERK (U0126), JNK (SP600125), p38 (SB203580) or p65 (JSH-23), or under the combined use of the above inhibitors for 30 min followed by stimulation with rpHMGB1 (1 μg/ml) or rpHSP60 (1 μg/ml) for 3 h. Data are representative (a and b), or mean±S.E.M. (c), of three individual experiments. *P<0.05, **P<0.01
Autophagy is involved in TRAF6 ubiquitination triggered by lung I/R injury
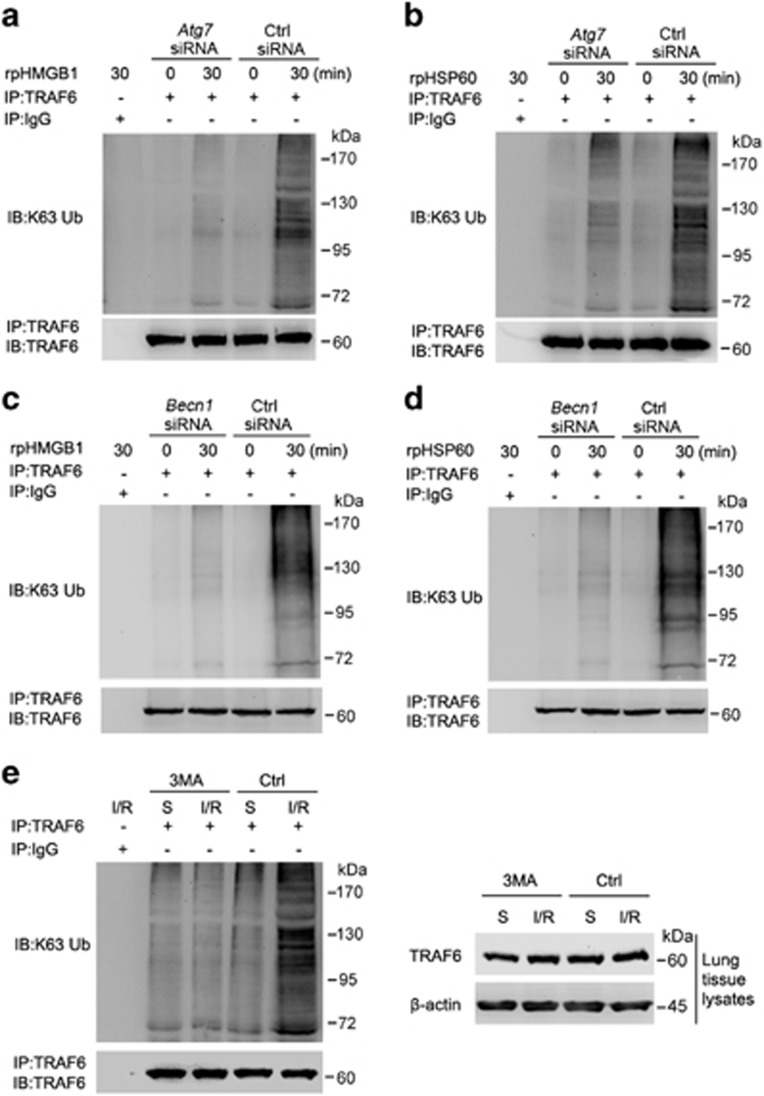
Since HMGB1 and HSP60 is recognized by TLR4, which subsequently induces Lys63 (K63)-linked ubiquitination of TRAF6 leading to activation of downstream MAPK and NFKB signaling and production of inflammatory cytokines,23, 24 we examined whether TRAF6 ubiquitination participates in autophagy-mediated activation of MAPK and NF-κB signaling induced by DAMPs in lungs undergoing I/R injury. Indeed, knockdown of ATG7 or BECN1 markedly reduced K63-linked ubiquitination of TRAF6 triggered by rpHMGB1 or rpHSP60 in alveolar macrophages (Figures 6a–d). In addition, the pulmonary protective solution with addition of the autophagy inhibitor 3-MA substantially attenuated K63-linked ubiquitination of TRAF6 in left lung tissues triggered by I/R injury (Figure 6e). The data from 3-MA treatment suggest that autophagy is involved in K63-linked ubiquitination of TRAF6 in lung tissues triggered by I/R, which has an important role in I/R injury-induced activation of MAPK and NF-κB signaling and the inflammatory response.
Figure 6.
Autophagy is involved in TRAF6 ubiquitination triggered by lung I/R injury. (a–d) Alveolar macrophages were transfected with the control siRNA, Atg7 siRNA (a and b) or Becn1 siRNA (c and d). Sixty hours later, cells were treated with rpHMGB1 (1 μg/ml) (a and c) or rpHSP60 (1 μg/ml) (b and d) for 30 min or left untreated. (e) Minipigs underwent lung ischemia and perfusion with pulmonary protective solution containing 3-MA (5 mM) or DMSO followed by reperfusion for 30 min or received sham operations (Sham). Whole-cell lysates of alveolar macrophages (a–d) or lysates of left lung tissues (e) were subjected to immunoprecipitation with anti-TRAF6 antibody followed by immunoblotting with anti-K63 ubiquitin and anti-TRAF6 antibodies, or immunoblotted with anti-TRAF6 and β-actin antibodies. S: sham group. Data are representative of three individual precipitation experiments
Discussion
In situations of extreme cellular damage, an excessive inflammatory response can result in organ injury and dysfunction, followed by a period of immune hypoactivity that creates an environment in which infectious pathogens thrive.25 I/R injury represents such a potentially maladaptive response of the innate immune system, which is featured by the exacerbated sterile inflammation response triggered by tissue damage.26 However, the molecular mechanism responsible for the pathogenesis of sterile inflammation remains unclear. Endogenous DAMPs are released from cells undergoing necrotic cell death during tissue damage, which activate signaling pathways mediated by PRRs.3 TLRs are well characterized PRRs that detect endogenous or exogenous danger signals. Upon ligation, TLRs activate the downstream MAPK and NF-κB pathways, which eventually lead to the production of inflammatory cytokines.27 Previous studies have implicated TLR4 as a key receptor involved in activation of pro-inflammatory signaling cascades during I/R injury in many organs including lungs, with TLR2 and TLR9 also potentially involved.28 HMGB1, one of the best-characterized DAMPs, is released from immune and non-immune cells in response to stimulation and damage. HMGB1 has been shown to activate TLR4 and contribute to inflammation caused by I/R injury.29, 30 HSPs are another class of pro-inflammatory DAMPs that are released to extracellular milieu under stress conditions.31 In the present study, we constructed a minipig lung I/R model and found that the expression of DAMPs including HMGB1 and HSP60 was significantly increased in BALF and crude lung tissues after I/R. Moreover, stimulation with rpHMGB1 or rpHSP60 activated MAPK and NFKB signaling in alveolar macrophages. As a consequence, the production of inflammatory cytokines such as IL-1β, TNF and IL12 was increased. Collectively, our results reveal that the release of DAMPs is responsible for inflammatory responses during minipig lung I/R injury.
Autophagy is a regulated cellular process of lysosome-dependent turnover of damaged or unnecessary cytoplasmic macromolecules and organelles.32, 33 Apart from its well-acknowledged role in maintenance of normal cellular phenotypes and functions, growing evidence has shown that autophagy is modulated under conditions of cellular stress such as ER stress, oxidative stress, hypoxia and inflammation.34, 35 I/R refers to the arrest and restitution of blood flow that may occur under various clinical conditions, such as cardiac arrest, shock, organ transplantation, respiratory failure.36 The disorder of autophagy activity has been reported in cardiac, renal and hepatic I/R models.35, 37, 38 Because of its rich vascular supply and continuous physiologic demand for gas exchange, the lung is prone to I/R injury.11 Nevertheless, little is known about the role of autophagy in lung I/R injury. Our results in this study have identified a role of autophagy in the pathogenesis of sterile inflammation caused by lung I/R injury. We found that a variety of autophagy related genes were upregulated after lung I/R injury in minipigs, and that the autophagy flux was indeed activated in crude lung tissues and isolated alveolar macrophages after lung I/R injury. Moreover, our results showed that autophagy inhibition in vitro and in vivo attenuated TRAF6 ubiquitination and inhibited the inflammatory cytokine production. Therefore, autophagy is enhanced during lung I/R, which has an important role in the pathogenesis of lung I/R injury.
Recent studies have demonstrated a close link between autophagy and inflammatory signaling.3 Activation of PRRs by PAMPs or DAMPs promotes autophagy activation through a mechanism that has been partially elucidated.6 On the other hand, autophagy is known to regulate the release of DAMPs through undefined mechanisms.39 Moreover, autophagy also participates in the regulation of inflammation mediated by PAMPs or DAMPs.14, 15 K63-linked ubiquitination of TRAF6 that is crucial for activation of NF-κB and MAPK signaling, acts as an initiating signal for TLR-triggered inflammatory responses.23 TRAF6 has been shown to activate autophagy after TLR activation and acts as an E3 ligase to ubiquitinate BECN1 and unc-51 like autophagy activating kinase 1 (ULK1).40, 41 These observations suggest that TRAF6 is also involved in DAMP-triggered autophagy. This, together with our data showing that DAMP-triggered autophagy maintains TRAF6 ubiquitination and enhances DAMP-induced inflammatory injury in lung tissues, suggests that there is a potential positive feedback loop between TRAF6 and autophagy, although the exact molecular mechanism involved remains to be defined.
The exact function of autophagy in I/R injury remains largely uncharacterized. Autophagy may exert both adaptive and maladaptive effects during the pathogenesis of I/R, which may be highly dependent on experimental models and tissue types in question.38, 42, 43 The beneficial role of autophagy is associated with the homeostatic turnover of damaged cellular organelles and proteins, thus promoting recycling of vital metabolic building blocks and regeneration of energy charge.43 However, excessive autophagy may be associated with aberrant degradation of intracellular constituents, leading to autophagic cell death, or alternatively, apoptosis, both of which may contribute to the disease development and progression.44 For instance, in ischemic heart diseases, autophagy is beneficial and adaptive during ischemia, whereas detrimental during reperfusion. Reperfusion injury is the consequence of dysregulated autophagy.45 Consistent with our results, a previous study also found that the autophagy flux was activated during the lung ischemia period, which was further increased during reperfusion in rats.46 Inhibition of autophagy by 3-MA ameliorated lung I/R injury as indicated by a reduced lung wet/dry ratio, myeloperoxidase activity, and malondialdehyde concentrations.46 These findings collectively suggest that increased autophagy may promote sterile inflammation during lung I/R injury. Nevertheless, further studies are needed to reveal the molecular mechanism by which autophagy is initiated. Similarly, how autophagy regulates inflammatory responses during lung I/R injury needs further investigations.
In summary, we have demonstrated that autophagy is induced by DAMPs and contributes to the pathogenesis of the sterile inflammation response during minipig lung I/R injury. Our results indicate that autophagy has important roles in the development of inflammation responses in clinical events involving lung I/R such as lung transplantation.
Materials and methods
The lung I/R injury model of minipigs
Bama minipigs (mean body weight 20 kg) were from Taihe Biotechnology (Taizhou, China). All minipigs were clinically healthy, housed in the animal facility under standard conditions and received humane care. All animal experiments were performed according to the National Institute of Health Guide for the Care and Use of Laboratory Animals, with the approval of the Scientific Investigation Board of Tongji University, Shanghai. The left lung I/R injury model of minipigs was established as reported before.47 Briefly, minipigs were anesthetized and left thoracotomies were performed in the fifth intercostal space. The left pulmonary hilum was dissected, and then the left pulmonary artery, left pulmonary vein and left bronchus from the left pulmonary hilum were clamped individually. Pulmonary protective solution (30 ml/kg body weight) was perfused through the left pulmonary artery and flowed out from the left pulmonary vein. In some experiments, a pulmonary protective solution with the autophagy inhibitor 3-MA (5 mM final concentration) or the lysosomal degradation inhibitor E64d (15 μg/ml) and Pepstatin A (15 μg/ml) was used to perfuse into left lungs. After 60 min of normothermic ischemia and perfusion, the incisions of left pulmonary artery and left pulmonary vein were closed with continuous suture, and left lungs were reperfused. Except for sham operation group, all other minipigs were subjected to left lung ischemia for 1 h, and then randomly divided into different groups to accept reperfusion for 30 min, 1 h, 3 h or 6 h (n=3).
Reagents
E64d (E8640), Pepstatin A (P4265), Bafilomycin A1 (B1793) and 3-MA (M9281) were from Sigma (St. Louis, MO, USA). Recombinant porcine HMGB1 (CSB-EP010553PI) was from Cusabio (Wuhan, China) and recombinant porcine HSP60 was generated in house. Endotoxin level was <1.0 EU per 1 mg of the recombinant HMGB1 or HSP60 protein. Inhibitors of ERK (U0126, 662005), JNK (SP600125, 420119), p38 (SB203580, 559389) and p65 (JSH-23, 481408) were from Merck Millipore (Billerica, MA, USA) and dissolved in DMSO (D8418, Sigma). TAK-242 (tlrl-cli95) was from InvivoGen (San Diego, CA, USA). Antibodies were obtained from the following sources: IL-1β (bs-0812R), TNF (bs-0078R), HSP60 (bs-0191R), Biosynthesis Biotechnology (Beijing, China); BECN1 (3495), ATG5 (8540), ATG7 (8558), ERK (4695), JNK (9252), p38 (9212), IKKα/β (8943), p65 (6956), phospho-ERK (Thr202/Tyr204) (9106), phospho-JNK (Thr183/Tyr185) (9255), phospho-p38 (Thr180/Tyr182) (9215), phospho- IKKα/β (Ser176/180) (2697), phospho-p65 (Ser536) (3033), β-actin (4970), Cell Signaling Technology (Danvers, MA, USA); Lys63-specific ubiquitin (05–1313), Merck Millipore; TRAF6 (sc-7221), SQSTM1 (sc-25575), Santa Cruz (Dallas, TX, USA); HMGB1 (NB100–2322), LC3B (NB600–1384), Novus Biologicals (Littleton, CO, USA).
Microarray analysis
Total RNA from lung tissues of minipigs subjected to 1 h left lung ischemia followed by reperfusion for 1 h or sham group was extracted using the TRIzol Reagent (15596-026, Invitrogen, Carlsbad, CA, USA). Gene expression analysis with Affymetrix GeneChip Porcine Genome Arrays was performed by Shanghai Biotechnology (Shanghai, China). Labeling and hybridization were performed according to the protocols in the Affymetrix GeneChip 3′ IVT Express Kit User Manual (Thermo Fisher Scientific). The slides were scanned by GeneChip Scanner 3000 (Thermo Fisher Scientific, Waltham, MA, USA). The data were analyzed using Genespring Software 11.0 (Agilent Technologies, Santa Clara, CA, USA). Raw data were normalized by MAS5.0s algorithm and expressed in log2 scale. Heatmap, venn and scatter diagrams were conducted using R package (www.r-project.org). Differentially expressed genes (DEGs) were identified with cutoff of absolute fold change 1.5. Then, the DEGs were further biologically interpreted using the Gene ontology (GO) database (geneontology.org) and Kyoto Encyclopedia of Genes and Genomes (KEGG) public pathway resource (www.genome.jp/kegg). To measure the level of significance for biological pathways in the KEGG pathway database represented by DEGs, enrichment analysis was performed by Perl ‘Comparison' package.48 Only pathways that had a P-value ⩽0.05 were considered significant. Array data are available at Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE75560.
Bronchoalveolar lavage and isolation of alveolar macrophages
Bronchoalveolar lavage was performed as described previously.47, 49 Briefly, the BALF was collected by lavaging the left or right lung three times with phosphate-buffered saline (PBS) through a tracheal cannula, which was pooled and centrifuged at 300 × g for 10 min. The acellular supernatant was divided into aliquots and stored at −80 °C until further assay. Cell pellet was washed three times with PBS using centrifugation (300 × g, 5 min) and then resuspended in Dulbecco's Modified Eagle Medium (11965-092, Gibco, Paisley, PA, USA) supplemented with 10% fetal bovine serum (Gibco, 10100147). After 1 h of culture in plates, nonadherent cells were removed by gently washing three times with warm PBS. The adherent alveolar macrophages were subjected to different treatment and analysis.
RT-PCR and qPCR
Total RNA was exacted from cells or tissues using TRIzol regent (Invitrogen, 15596-026) following the manufacturer's instructions. Total RNA (1 μg) was used in a 20 μl reverse-transcription reaction using the First Strand cDNA Synthesis kit (FSK-101, Toyobo, Osaka, Japan), and then cDNA was diluted into 80 μl water as the template of real-time qPCR. qPCR analysis was performed with Applied Biosystems 7500 Real-Time PCR System. Data were normalized by the level of β-actin expression in each sample. The qPCR primers for analysis of cytokine RNA levels were shown in Supplementary Table S3.
Cytokine detection
HMGB1 and HSP60 in BALF, or IL-1β and TNF in cell culture supernatants were measured by ELISA. The kits for HMGB1 (CSB-EL010553PI) and HSP60 (CSB-EL010847PI) were from Cusabio, IL-1β (PLB00B) and TNF (PTA00) were from R&D Systems (Minneapolis, MN, USA).
RNA interference
Small interfering RNA (siRNA) was designed and synthesized by GenePharma (Shanghai, China). The sequences of siRNA specific for minipig Atg7 were 5′-GCTGCTATTTCTGCAATGA-3′ and 5′-GCCGCTGTATGAATTTGAA-3′ (Atg7 siRNA #2). The sequences of siRNA specific for minipig Becn1 and Tlr4 were 5′-CCTGGATCGTGTTACCATT-3′ and 5′-GCATGGAGCTGAATTTCTA-3′, respectively. The sequence of control siRNA was 5′-TTCTCCGAACGTGTCACGT-3′. The siRNA duplexes were transfected into cells using INTERFERin reagent (Polyplus-transfection, 409-10) according to the standard protocol.
Immunoblotting and immunoprecipitation
Cells were washed twice with ice cold PBS and lysed with cell lysis buffer (Cell Signaling Technology, 9803) containing protease inhibitor mixture (Merck Millipore, 539134). Protein concentration of the extracts was measured with BCA assay (23225, Thermo Fisher Scientific, Waltham, MA, USA). Immunoprecipitation and immunoblotting analysis were performed as described previously.50 The detection of LC3 conversion (LC3-I to LC3-II) by immunoblotting was performed as reported previously.51
Immunofluorescence analysis
The detection of LC3 puncta in alveolar macrophages was performed as described previously.51 Cells grown on glass coverslips were fixed with 4% paraformaldehyde and permeabilized with 0.3% Triton X-100. After blocking with BSA, cells were incubated with primary rabbit anti-LC3 antibody (1:800 dilution) at 4 °C overnight followed by incubation with Alexa Fluor 488-conjugated donkey anti-rabbit IgG (Thermo Fisher Scientific, A21206, 1:500 dilution) for 1 h at room temperature. Cells were examined under a confocal laser microscope (Leica TCS SP5II STED, Mannheim, Germany). At least 200 cells on each slide were counted and the percentage of cells with LC3 puncta (autophagosomes) was calculated.
Statistical analysis
The statistical significance of the comparison between two groups was determined with two-tailed student's t-test, whereas comparisons among multiple groups were assessed by analysis of variance (ANOVA). P-values of <0.05 were considered statistically significant.
Acknowledgments
We thank Mr. Junqi Yang for technical assistance. This work is supported by Grants from the National Natural Science Foundation of China (81373146, 81571541, 81422021, 31570871 and 81370433), Basic Research Program of Science and Technology Commission of Shanghai Municipality (14JC1405200), the Foundation for the Author of National Excellent Doctoral Dissertation of China and The national innovative research groups program of the National Natural Science Foundation of China (81521061).
Author contributions
XL, HC, JL, BW and PZ planned, performed and analyzed experiments; XDZ and ZL reviewed the manuscript and provided advice; ZZ and HY designed the overall research, analyzed experiments and wrote the manuscript.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd).
Edited by E Baehrecke
The authors declare no conflict of interest.
Supplementary Material
References
- Eltzschig HK, Bratton DL, Colgan SP. Targeting hypoxia signalling for the treatment of ischaemic and inflammatory diseases. Nat Rev Drug Discov 2014; 13: 852–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Golen RF, Reiniers MJ, Olthof PB, van Gulik TM, Heger M. Sterile inflammation in hepatic ischemia/reperfusion injury: present concepts and potential therapeutics. J Gastroenterol Hepatol 2013; 28: 394–400. [DOI] [PubMed] [Google Scholar]
- Tang D, Kang R, Coyne CB, Zeh HJ, Lotze MT. PAMPs and DAMPs: signal 0 s that spur autophagy and immunity. Immunol Rev 2012; 249: 158–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Hengst WA, Gielis JF, Lin JY, Van Schil PE, De Windt LJ, Moens AL. Lung ischemia-reperfusion injury: a molecular and clinical view on a complex pathophysiological process. Am J Physiol Heart Circ Physiol 2010; 299: H1283–H1299. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature 2008; 451: 1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nat Rev Immunol 2013; 13: 722–7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryter SW, Choi AM. Autophagy in the lung. Proc Am Thorac Soc 2010; 7: 13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb RA, Mentzer RM. Autophagy during cardiac stress: joys and frustrations of autophagy. Annu Rev Physiol 2010; 72: 45–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaja MJ, Ding WX, Donohue TMJr, Friedman SL, Kim JS, Komatsu M et al. Functions of autophagy in normal and diseased liver. Autophagy 2013; 9: 1131–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Hartleben B, Kretz O, Wiech T, Igarashi P, Mizushima N et al. Autophagy plays a critical role in kidney tubule maintenance, aging and ischemia-reperfusion injury. Autophagy 2012; 8: 826–837. [DOI] [PubMed] [Google Scholar]
- Ryter SW, Nakahira K, Haspel JA, Choi AM. Autophagy in pulmonary diseases. Annu Rev Physiol 2012; 74: 377–401. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity 2011; 34: 637–650. [DOI] [PubMed] [Google Scholar]
- Műzes G, Constantinovits M, Fűri I, Tulassay Z, Sipos F. Interaction of autophagy and Toll-like receptors: a regulatory cross-talk—even in cancer cells? Curr Drug Targets 2014; 15: 743–752. [DOI] [PubMed] [Google Scholar]
- Bortoluci KR, Medzhitov R. Control of infection by pyroptosis and autophagy: role of TLR and NLR. Cell Mol Life Sci 2010; 67: 1643–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Into T, Inomata M, Takayama E, Takigawa T. Autophagy in regulation of Toll-like receptor signaling. Cell Signal 2012; 24: 1150–1162. [DOI] [PubMed] [Google Scholar]
- Zhao M, Fernandez LG, Doctor A, Sharma AK, Zarbock A, Tribble CG et al. Alveolar macrophage activation is a key initiation signal for acute lung ischemia-reperfusion injury. Am J Physiol Lung Cell Mol Physiol 2006; 291: L1018–L1026. [DOI] [PubMed] [Google Scholar]
- Tsushima Y, Jang JH, Yamada Y, Schwendener R, Suzuki K, Weder W et al. The depletion of donor macrophages reduces ischaemia-reperfusion injury after mouse lung transplantation. Eur J Cardiothorac Surg 2014; 45: 703–709. [DOI] [PubMed] [Google Scholar]
- Prakash A, Mesa KR, Wilhelmsen K, Xu F, Dodd-o JM, Hellman J. Alveolar macrophages and Toll-like receptor 4 mediate ventilated lung ischemia reperfusion injury in mice. Anesthesiology 2012; 117: 822–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GY, Nuñez G. Sterile inflammation: sensing and reacting to damage. Nat Rev Immunol 2010; 10: 826–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Si R, Feng Y, Chen HH, Zou L, Wang E et al. Myocardial ischemia activates an injurious innate immune signaling via cardiac heat shock protein 60 and Toll-like receptor 4. J Biol Chem 2011; 286: 31308–31319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Deng Y, Su D, Tian J, Gao Y, He Z et al. TLR4 as receptor for HMGB1-mediated acute lung injury after liver ischemia/reperfusion injury. Lab Invest 2013; 93: 792–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha T, Sunamoto M, Kitazaki T, Sato J, Ii M, Iizawa Y. Therapeutic effects of TAK-242, a novel selective Toll-like receptor 4 signal transduction inhibitor, in mouse endotoxin shock model. Eur J Pharmacol 2007; 571: 231–239. [DOI] [PubMed] [Google Scholar]
- Bhoj VG, Chen ZJ. Ubiquitylation in innate and adaptive immunity. Nature 2009; 458: 430–437. [DOI] [PubMed] [Google Scholar]
- Park JS, Svetkauskaite D, He Q, Kim JY, Strassheim D, Ishizaka A et al. Involvement of toll-like receptors 2 and 4 in cellular activation by high mobility group box 1 protein. J Biol Chem 2004; 279: 7370–7377. [DOI] [PubMed] [Google Scholar]
- Rock KL, Latz E, Ontiveros F, Kono H. The sterile inflammatory response. Annu Rev Immunol 2010; 28: 321–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalogeris T, Baines CP, Krenz M, Korthuis RJ. Cell biology of ischemia/reperfusion injury. Int Rev Cell Mol Biol 2012; 298: 229–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill LA. When signaling pathways collide: positive and negative regulation of toll-like receptor signal transduction. Immunity 2008; 29: 12–20. [DOI] [PubMed] [Google Scholar]
- Lin Q, Li M, Fang D, Fang J, Su SB. The essential roles of Toll-like receptor signaling pathways in sterile inflammatory diseases. Int Immunopharmacol 2011; 11: 1422–1432. [DOI] [PubMed] [Google Scholar]
- Tsung A, Tohme S, Billiar TR. High-mobility group box-1 in sterile inflammation. J Intern Med 2014; 276: 425–443. [DOI] [PubMed] [Google Scholar]
- Sims GP, Rowe DC, Rietdijk ST, Herbst R, Coyle AJ. HMGB1 and RAGE in inflammation and cancer. Annu Rev Immunol 2010; 28: 367–388. [DOI] [PubMed] [Google Scholar]
- Jin C, Cleveland JC, Ao L, Li J, Zeng Q, Fullerton DA et al. Human myocardium releases heat shock protein 27 (HSP27) after global ischemia: the proinflammatory effect of extracellular HSP27 through toll-like receptor (TLR)-2 and TLR4. Mol Med 2014; 20: 280–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terman A, Gustafsson B, Brunk UT. Autophagy, organelles and ageing. J Pathol 2007; 211: 134–143. [DOI] [PubMed] [Google Scholar]
- Cecconi F, Levine B. The role of autophagy in mammalian development: cell makeover rather than cell death. Dev Cell 2008; 15: 344–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V, Levine B. Autophagy, immunity, and microbial adaptations. Cell Host Microbe 2009; 5: 527–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell 2008; 132: 27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugam TV, Okun E, Tang SC, Thundyil J, Taylor SM, Woodruff TM. Toll-like receptors in ischemia-reperfusion injury. Shock 2009; 32: 4–16. [DOI] [PubMed] [Google Scholar]
- Ryter SW, Lee SJ, Smith A, Choi AM. Autophagy in vascular disease. Proc Am Thorac Soc 2010; 7: 40–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Wang Y, Chen Y, Cao F. The role of the autophagy in myocardial ischemia/reperfusion injury. Biochim Biophys Acta 2015; 1852: 271–276. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Kang R, Zeh HJ3rd, Lotze MT, Tang D. DAMPs and autophagy: cellular adaptation to injury and unscheduled cell death. Autophagy 2013; 9: 451–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi CS, Kehrl JH. TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy. Sci Signal 2010; 3: ra42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazio F, Strappazzon F, Antonioli M, Bielli P, Cianfanelli V, Bordi M et al. mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat Cell Biol 2013; 15: 406–416. [DOI] [PubMed] [Google Scholar]
- Chen-Scarabelli C, Agrawal PR, Saravolatz L, Abuniat C, Scarabelli G, Stephanou A et al. The role and modulation of autophagy in experimental models of myocardial ischemia-reperfusion injury. J Geriatr Cardiol 2014; 11: 338–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan WJ, Dong HL, Xiong LZ. The protective roles of autophagy in ischemic preconditioning. Acta Pharmacol Sin 2013; 34: 636–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei Y, Thompson MD, Cohen RA, Tong X. Autophagy and oxidative stress in cardiovascular diseases. Biochim Biophys Acta 2015; 1852: 243–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thapalia BA, Zhou Z, Lin X. Autophagy, a process within reperfusion injury: an update. Int J Clin Exp Pathol 2014; 7: 8322–8341. [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Wang JS, Zheng ZK, Tang J, Fan K, Guo H et al. Participation of autophagy in lung ischemia-reperfusion injury in vivo. J Surg Res 2013; 182: e79–e87. [DOI] [PubMed] [Google Scholar]
- Simón Adiego C, González-Casaurrán G, Azcárate Perea L, Isea Viña J, Vara Ameigeiras E, García Martín C et al. Experimental swine lung autotransplant model to study lung ischemia-reperfusion injury. Arch Bronconeumol 2011; 47: 283–289. [DOI] [PubMed] [Google Scholar]
- Audic S, Claverie JM. The significance of digital gene expression profiles. Genome Res 1997; 7: 986–995. [DOI] [PubMed] [Google Scholar]
- Tsai YC, Chang HW, Jeng CR, Lin TL, Lin CM, Wan CH et al. The effect of infection order of porcine circovirus type 2 and porcine reproductive and respiratory syndrome virus on dually infected swine alveolar macrophages. BMC Vet Res 2012; 8: 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zhan Z, Li D, Xu L, Ma F, Zhang P et al. Intracellular MHC class II molecules promote TLR-triggered innate immune responses by maintaining activation of the kinase Btk. Nat Immunol 2011; 12: 416–424. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2016; 12: 1–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.