Abstract
Pyroptosis is a lytic form of cell death distinguished from apoptosis, ferroptosis, necrosis, necroptosis, NETosis, oncosis, pyronecrosis and autophagy. Proinflammatory caspases cleave a gasdermin D (GSDMD) protein to generate a 31 kDa N-terminal domain. The cleavage relieves the intramolecular inhibition on the gasdermin-N domain, which then moves to the plasma membrane to exhibit pore-forming activity. Thus, GSDMD acts as the final and direct executor of pyroptotic cell death. Owing to the selective targeting of the inner leaflet of the plasma membrane with the pore-forming that determines pyroptotic cell death, GSDMD could be a potential target to control cell death or extracellular bacterial infections. Intriguingly, other gasdermin family members also share similar N-terminal domains, but they present different cell death programs. Herein, we summarize features and functions of the novel player proteins in cell death, including GSDMD triggering pyroptosis, Gsdma3/GSDMA initiating autophagy/apoptosis and DFNA5 inducing apoptosis/secondary necrosis. The gasdermin N terminus appears to be a novel pore-forming protein. This provides novel insight into the underlying roles and mechanisms of lytic or nonlytic forms of programmed cell death, as well as their potential applications in inflammation-associated diseases.
Facts
Cell death is a fundamental feature of life and functions in many important biological processes, including development, homeostasis and pathogenesis.
The inflammasome-activated gasdermin-N domain of GSDMD drives pyroptotic cell death through selective targeting of the inner leaflet of the plasma membrane with the pore-forming activity.
Gsdma3 and its human homolog GSDMA induce autophagy/apoptosis via different signaling pathways.
DFNA5 cleavage or DFNA5 mutation releases the autoinhibition activity of the C-terminal domain, contributing to cell apoptosis/secondary necrosis.
Open questions
What is the detailed mechanistic process by which the N-terminal cytotoxic activity of GSDMD drives pyroptotic cell death?
Could the N terminal of DFNA5 serve as an apoptotic trigger to initiate the secondary necrosis in response to pathogen infection?
How could critical target molecules in the activated signaling pathways by which GSDMD, GSDMA and DFNA5 mediate cell death be identified to pave therapeutic approaches of inflammasome-associated inflammatory and metabolic disorders?
Pyroptosis is an inflammatory form of programmed cell death that is critical for amplifying protective immunity during infection.1 A dedicated set of inflammatory caspases (caspase-1, -4, -5 and -11) in the canonical and non-canonical inflammasome signaling pathways leads to inflammatory responses, especially in pyroptosis.2, 3, 4, 5, 6 But, how these caspases initiate this cell-death program remains unclear.
This picture depicting a pyroptotic process has not been changed markedly until 2015 when two independent studies led by Dixit and Shao identified cleavage of the protein GSDMD by inflammatory caspases determining pyroptotic cell death.7, 8 They demonstrated that inflammatory caspases cleaved GSDMD to generate a 31 kDa N-terminal fragment and a 22 kDa C-terminal fragment, and the N-terminal GSDMD fragment alone induced pyroptosis when expressed ectopically. GSDMD is just one member of the evolutionarily and structurally conserved gasdermin gene family. Intriguingly, all gasdermins (GSDMA, GSDMB, GSDMC, and GSDMD in humans; Gsdma1–3, Gsdmc1–4, and Gsdmd in mice) are composed of a similar N-terminal domain, which is also shared by the extended gasdermin family members DFNA5 and DFNB59 (deafness, autosomal-dominant 5/autosomal-recessive 59). This gasdermin-N domain in GSDMD, Gsdma3, GSDMA, GSDMB, GSDMC or DFNA5 has been proved to induce extensive cell death in human 293T cells.7, 9 Given the sequence similarity between gasdermin N-terminal domains, it is not surprising that all gasdermins induce membrane permeability pores and subsequent cell death.9, 10
The identification of GSDMD as a critical mediator of pyroptosis has hinted possible functions of other gasdermin family members in programmed cell death. However, a comprehensive summary of mechanism by which gasdermin family members are involved in the programmed cell death has not been undertaken until now. In this review, we summarize current literatures on possible functions of gasdermin family members in the programmed cell death, specifically including GSDMD triggering pyroptosis, GSDMA/Gsdma3 causing autophagy or apoptosis, and DFNA5 inducing apoptosis. In particular, gasdermin-N appears to represent a novel pore-forming protein (PFP) to insert into cell membranes, determining pyroptotic cell death.
Whole Picture of Gasdermin Family
Gasdermins belong to a recently discovered novel gene family. Besides GSDMD, this family also includes GSDMA, GSDMB, GSDMC, DFNA5 and DFNB59. Both GSDMA and GSDMB are located at human chromosome 17q21.1, but GSDMC and GSDMD at chromosome 8q24. Mice lack GSDMB, but encode three Gsdma (Gsdma1–3) and four Gsdmc (Gsdmc1–4). Paralogous genes Gsdma, Gsdma2 and Gsdma3 are clustered on mouse chromosome 11, but Gsdmc, Gsdmc2, Gsdmc3 and Gsdmc4 on chromosome 15. Gasdermin family genes have distinctive expressional patterns. Each is tissue- and differentiation stage-specific and expressed in epithelium of skin and gastrointestinal tract.11 GSDMA, GSDMC and GSDMD are all reported to have tumor suppressor activity, but GSDMB is considered to be an oncogene, associated with immune response diseases, such as childhood-onset asthma.12, 13, 14, 15, 16, 17, 18
The gasdermin family shares ~45% overall sequence homology. Structures of the gasdermins are composed of two domains, gasdermin-N and -C domains. Of note, the N-terminal and C-terminal domains are well conserved among the Gsdm/GSDM family proteins. However, the middle GSDM protein sequence shows no similarities among the family members. There are nine conserved sequence regions and all members encode a leucine-rich protein.11 It was presumed that the GSDM proteins may function as transcriptional factors because two potential leucine zippers that might serve as DNA-binding domains located in their C terminus. However, there is no nuclear localization signal sequence and DNA-binding motifs in the GSDM proteins, and GSDMA is assayed only in the cytoplasm, but not in the nucleus after expressed in 293T cells in vitro. For this reason, there is no strong evidence to support this hypothesis.
Pore-Forming Activity of GSDMD Determines Pyroptotic Cell Death
Pyroptosis is a lytic and highly inflammatory type of the programmed cell death, initiated by inflammasome (or pyroptosome) formation in response to microbial infection or other danger signals. This type of cell death is characteristic of cell swelling, lysis, and release of cytoplasmic content, thus restricting the replication of intracellular pathogens and attracting effector cells of the immune system.1 However, excessive pyroptosis is detrimental to normal tissue and cells. To avoid damage to the host organism, pyroptosis is tightly regulated by the activation of inflammatory caspases, such as caspase-1, murine caspase-11, and its human homologs caspases-4 and -5. They are activated within the inflammasome, a multi-protein complex that is assembled by cytosolic pattern-recognition receptors upon recognition of various cytosolic danger- or pathogen-associated molecular patterns. Although the basis of inflammasome assembly and caspase activation has well been established, it is still unknown how these caspases initiate this cell-death program.
Identification of GSDMD as a critical mediator of pyroptosis
In 2015, several landmark studies independently identified GSDMD as a central mediator of pyroptotic cell death caused by downstream of both caspase-1 and caspase-11, and showed that it was essential for IL-1β secretion.7, 8, 19 These studies demonstrated that GSDMD is cleaved by caspases into a 31 kDa N-terminal fragment and a 22 kDa C-terminal fragment, and that the N terminus itself, but not full-length or C-terminal GSDMD, is capable of triggering pyroptosis when expressed aberrantly, suggesting that the GSDMD C terminus has an autoinhibitory effect on the intrinsic activity of the pyroptosis-inducing GSDMD N terminus. Since then, a series of reports has now shown that the cytotoxicity of this N-terminal fragment is because of its ability to target and insert cellular membrane lipids, and permeabilize the membrane, therefore representing a novel class of PFPs.9, 19, 20, 21, 22, 23, 24
Oligomerized gasdermin-N forms membrane pores during a pyroptotic process
Shao and colleague9 further revealed the molecular mechanism of gasdermin-N domain-induced pyroptosis. They showed that the gasdermin-N domains of human GSDMD, GSDMA, GSDMB, GSDMC, and DFNA5 and of mouse Gsdma3 were cytotoxic to the mammalian cells. A possible mechanism is that the gasdermin-N domains directly disrupt cellular membrane to cause pyroptosis. To confirm this possibility, the engineered forms of GSDMD, GSDMA and Gsdma3 were used, in which the N and C domains remained non-covalently bound together after cleavage to avoid the toxic effects of the N domain while the localization of these domains was able to be examined. Their results demonstrate that only the N-terminal domains of gasdermins can specifically bind to phosphoinositide and cardiolipin on the plasma membrane, which are an important component of the inner mitochondrial membranes and some bacterial membranes.9
How do the gasdermin-N domains translocate to the plasma membrane? After cleavage, full-length GSDMD was located in the cytosol, while GSDMD-C also remained in the cytosol.22 As a general strategy applied in many signaling pathways, GSDMD-N also experienced monomer-to-oligomer transition prior to its migration to the plasma membrane, which is initiated by caspase-mediated removal of C-terminal inhibitory domain.20 It seems that there are multiple steps in the oligomerization process before the formation of a higher-order complex. The high-resolution crystal structure analysis of mouse Gsdma3 indicates the structural basis for the autoinhibition function of its α-helical C terminal. That is, it can hinder the activity of the Gsdma3 N terminal conserved most likely in the gasdermin family members. Human GSDMD shares ~70% homology with Gsdma3. As the first 15 N-terminal amino acids are highly conserved in gasdermins, GSDMD can also form α-helix structure.9 As the mutation of five well conserved and positively charged residues in GSDMD-NT (N terminal) may prevent GSDMD-NT oligomerization and GSDMD-N-HBD (hormone-binding domain)-mediated cell death, the N-terminal α-helix should be indispensible for GSDMD-mediated pyroptosis.20 Furthermore, Shao and colleague9 also proved that L192 on the α4 helix and E15 on helix α1 take part probably in oligomerization and insert into the membrane, resulting in the GSDMD pore formation.
Gasdermin-N domain represents a new PFP
Notably, GSDMD-N induced pyroptosis only when delivered to the cytosol, but not when added extracellularly. This is fully consistent with the localization of phosphoinositides in the cytoplasmic leaflet of the plasma membrane. The gasdermin-N could lead to leakage of the liposomes consisting of phosphoinositide or cardiolipin, but did not impact the ones composed of cholesterol or phosphatidylethanolamine.9 The liposome-floating assay revealed that GSDMD P30 was strongly colocated with mitochondrial- or plasma membrane-like liposomes.20 This finding indicates that just like MLKL protein, which is involved in necroptosis, gasdermins also kill cells from the cytosol. In addition, the gasdermin-N shows no notable similarity in structural homology to other PFPs. Therefore, the gasdermin-N domain appears to represent a new type of PFPs. Further study showed that items with diameters of 10 nm or less could pass through the GSDMD-N-induced pores. Indeed, under a negative electron microscope the pores could be visualized to have an inner diameter of 10–15 nm in nearly all cardiolipin-, phosphoinositide-, or natural lipid-containing liposomes that had been co-cultured with GSDMD-N.9, 21, 24 Of note, these pores are wide enough for the passage of mature IL-1β (4.5 nm) and caspase-1 (7.5 nm).9 Consistent with the above finding that GSDMD-N forms the pores in the plasma membrane, the latest study from Han's group further demonstrated that GSDMD-N-based pores are non-selective to different ions. A large number of GSDMD-N-formed pores could conceivably destruct the plasma membrane, thereby promoting the formation of the apoptotic body-like cell protrusions, and finally inducing plasma membrane disruption.20 However, because GSDMD-N-based pores exhibit ion non-selectivity, there should not be a substantial increase in intracellular osmolarity, which explains why pyroptotic cells appear less swelling and bursting in comparison with necroptotic cells, whereas the latter has ion selection.20 Taken together, these breakthrough findings support that the GSDMD-N domain forms the pores on the membrane driving pyroptosis (Figure 1).
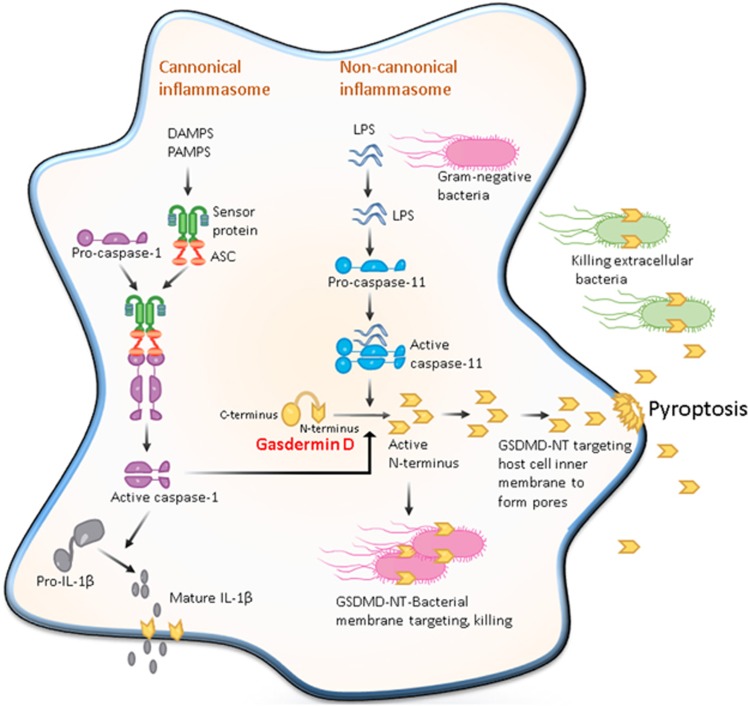
Figure 1.
Pore-forming activity of GSDMD determines pyroptotic cell death. The canonical inflammasome pathway is triggered by different cytoplasmic sensor proteins recognizing multiple pathogens and inflammatory agents. These recruit procaspase-1 monomers through the adaptor protein ASC and activate the caspase by dimerization. Caspase-1 can also initiate pyroptosis by cleaving gasdermin D although other pyroptosis-inducing caspase-1 substrates may exist. Caspase-1 also processes the proinflammatory cytokine pro-IL-1β to generate mature IL-1β, which is presumably released by cell lysis during pyroptosis. The non-canonical inflammasome pathway is triggered by bacterial lipopolysaccharide (LPS) molecules in the cytoplasm of infected cells. Direct binding of LPS to the protein procaspase-11 causes the protein dimerization to become active caspase-11. In human cells, caspase-4 and -5 perform the function of mouse caspase-11. These caspases cleave the protein gasdermin D, releasing its amino-terminal domain from inhibition by its carboxy-terminal domain. The cytotoxic N-terminal fragment of gasdermin D is then released and targets phospholipids on the host cell membrane and assembled into the pores. Processed GSDMD-NT, via its affinity for cardiolipin and phosphatidylserine, may also target and kill intracellular bacteria. GSDMD-NT pore formation in the host cell leads to pyroptosis to produce apoptotic body-like cell protrusions. IL-1β is released from the cell upon membrane rupture
Unanswered questions about N-terminal proteolytic fragment of GSDMD triggering pyroptosis
Although the strong evidence supports that GSDMD-NT pore formation in eukaryotic cells drives pyroptosis, some questions remain to be answered. First, given gasdermin-C is a soluble protein, whereas gasdermin-N is insoluble. When GSDMD is cleaved in the absence of a target membrane,22 we cannot help raise a question that gasdermin-C does act not only as an inhibitory domain, but also simultaneously as a solubility tag for the N-terminal domain, may be by shielding certain hydrophobic or amphipathic segments required for membrane insertion. Thus, further work is needed to show the precise degradation mechanism of gasdermin-N after cytolysis. Second, although it has already been confirmed that GSDMD is essential for pyroptosis induced by caspase-4, -5 or -11,7, 8, 19 the data from Kayagaki et al.8 diverge on the requirement of GSDMD for pyroptosis resulting from caspase-1 activation. Now, the jury is still out on whether GSDMD is an essential mediator of caspase-1-induced pyroptosis. If not, other pyroptosis-inducing caspase-1 substrates must await discovery. Third, could gasdermin-derived peptides be potentially developed as antimicrobial agents to treat extracellular bacterial infections? GSDMD-N could drive pyroptosis in eukaryotic cells, but also is bactericidal, being able to kill different species of bacteria, including Escherichia coli, Staphylococcus aureus and Bacillus megaterium protoplasts. GSDMD also reduces intracellular numbers of Listeria monocytogenes, although it is not clear whether it is able to kill intracellular bacteria directly, or whether this is a consequence of host cell death.24 GSDMD shares similar features with antimicrobial peptides (AMPs), some of which are expressed by macrophages, and can interact with membranes, finally penetrating them.25 The GSDMD effective dose 50 (ED50), which means a dose of compound repressing 50% of bacterial growth, is ~100 nM for both E. coli and S. aureus,24 comparable to the AMPs LL-37 and HBD-3.26 As GSDMD kills host cells via its selectively targeting to the inner leaflet of the plasma membrane, its N terminal, a gasdermin-derived peptide that is analogous to antimicrobial peptides, might be developed as AMPs to treat bacterial infections.
Evidence of Gsdma3/GSDMA Involving Autophagy or Apoptosis
Gsdma3 mutation induces alopecia and excoriation
The Gsdma3 gene is a key member of gasdermin gene family, located at mouse chromosome 11. It is expressed in mouse skin keratinocytes, associated with skin differentiation and inflammation. At present, nine spontaneous and ENU (N-ethyl-N-nitrosourea)-induced mutations of Gsdma3 are listed in public databases (Mouse Genome Informatics database, http://www.informatics.jax.org). Although it is, to date, reported that all Gsdma3 mutations causing alopecia and excoriation are located on the well conserved C- terminal domain, but not on the N terminal or the middle part of this protein, the precise regulation of Gsdma3 inducing an alopecia phenotype remains a mystery.
Gsdma3 is required for a TNF-α-induced apoptotic signaling in keratinocytes
Previous reports suggest that Gsdma3 is associated with tumor necrosis factor TNF-α-induced apoptosis.18, 27, 28 In morphology, Lei et al.29 showed that Gsdma3 gene mutation gave rise to abnormal catagen with unshortened length and unshrunk structure of the hair follicle so that the development of catagen phase was inhibited. By testing the expression of caspase-3, it was confirmed that the Gsdma3 gene mutation suppressed the catagen-associated apoptosis of hair follicle keratinocytes. In 2012, further study demonstrated that Gsdma3 was a key mediator in the TNF-α-induced apoptosis pathway.28 After in vivo TNF-α induction, Gsdma3 expression was significantly augmented in keratinocytes, suggesting that TNF-α acts as a new mediator of Gsdma3 expression. As a result, Gsdma3 causes the catagen-associated apoptosis of hair follicle keratinocytes by directly enhancing the caspase-3 expression (Figure 2a).
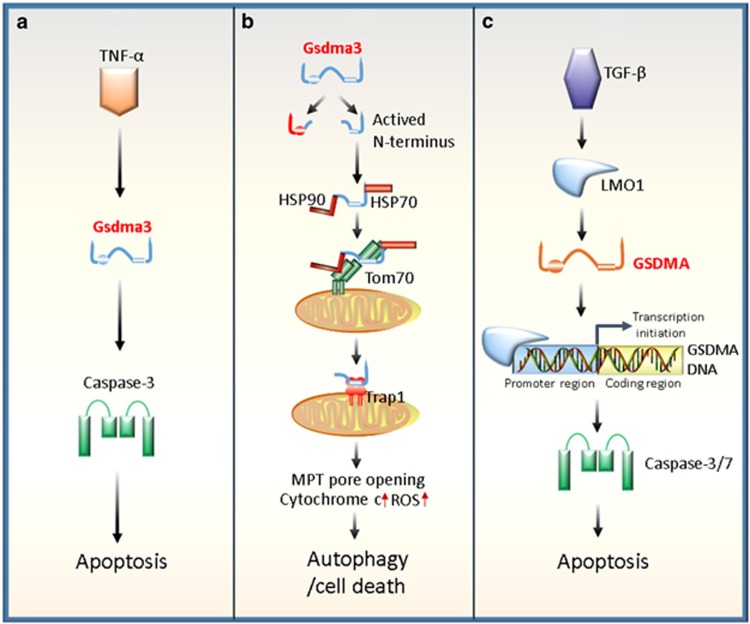
Figure 2.
Regulatory roles of Gsdma3/GSDMA in autophagy or apoptosis. (a) Gsdma3 acts as a key mediator in the TNF-α-induced apoptosis pathway. TNF-α can upregulate Gsdma3, whereas Gsdma3 causes the catagen-associated apoptosis of hair follicle keratinocytes by directly enhancing the caspase-3 expression. (b) Gsdma3 causes autophagy through a mitochondria-dependent pathway. Mutant Gsdma3 loses its whole C-terminal domain, consequently releasing the intrinsic pro-autophagic activity of the N-terminal domain. Then, the unmasked N-terminal domain of Gsdma3 is associated with Hsp90 to be delivered to mitochondria through mitochondrial importer receptor Tom70, where it interacts with the mitochondrial chaperone Trap1 and induces mitochondrial permeability transition (MPT) pore opening, mitochondrial reactive oxygen species (ROS) production, cytochrome c releasing, resulting finally in cell death. (c) Human GSDMA involves transforming growth factor (TGF)-β-induced apoptosis. TGF-β upregulates GSDMA expression by LIM domain only 1 (LMO1) induction through a sequence to which LMO1 binds, in a GSDMA promoter region. Ultimately, the increased GSDMA induces apoptosis of the pit cells of human gastric epithelium
Loss of Gsdma3 self-regulation causes autophagy via targeting mitochondria
In 2015, a study led by Shi et al.10 provided another answer to this question. The mutations of the Gsdma3 C-terminal domain could induce autophagy. Using an in vitro assay system, they found that the wild-type (WT) N-terminal domain displayed a pro-autophagic activity, which could be reversed by co-expression of the C-terminal domain. These data indicate that the pro-autophagic activity of the Gsdma3 is relieved through an intramolecular autoinhibition. Further studies reveal that the expressions of class-II microtubule-associated protein light chain 3 (LC3), an autophagy marker, and mouse orthologue of a key autophagic protein of Schizosaccharomyces pombe Atg8 (autophagy-related protein 8), were significantly enhanced in the AE mutant skin, indicating that autophagy is activated here.10 Consistent with the pro-autophagic activity of the N-terminal domain, the mutation or loss of the C-terminal domain of Gsdma3 binds directly to mitochondria to induce a significant decline in mitochondrial activity and an increase in the ROS level, resulting ultimately in autophagy.10 However, the detailed process by which the N-terminal domain is recruited to mitochondria remains unclear. Lin et al. showed that the unmasked N terminus of Gsdma3 interacts with a Hsp90/Hsp70 machinery complex for targeting mitochondria via Tom70, a mitochondrial importer receptor. Here, it has a further interaction with the mitochondrial chaperone Trap1 so as to induce mitochondrial permeability transition pore opening, mitochondrial ROS production and cytochrome c releasing, resulting finally in cell death (Figure 2b).30 Utilizing a tandem affinity purification method, Zanon et al.31 reported that Parkin might interact directly with GSDMA. As human GSDMA and rat Gsdma are orthologs of mouse Gsdma, their structure and function are conserved as well.32 When mitochondria are damaged, Parkin binds to PTEN-induced putative kinase 1 (PINK1), a mitochondrial targeting signal, and is recruited to mitochondrial outer membrane to initiate the mitophagy process,33, 34 while PINK1 is imported into the mitochondria by a unique pathway that also requires Tom70.35 Hence, it will be crucial to test whether Gsdma3 can regulate this process. Because all the Gsdma3 mutants are deficient in the whole C-terminal domain, which releases the intrinsic activity of the N-terminal pro-autophagy, they possibly induce a common alopecia phenotype by a mitochondria-dependent pathway. However, the Gsdma3 null mice did not show visible skin phenotypes, suggesting that the autophagy ‘activation' induced by the N-terminal domain may require additionally physiological or pathological signals or that there are other regulatory mechanisms.10
Crystal structure analyses of Gsdma3 reveal the molecular foundation of autoinhibited cell death
Regarding a pore-forming mechanism of GSDMD, Shao's laboratory demonstrated a 1.90 Å crystal structure of Gsdma3.9 On the basis of this crystal structure, the autoinhibitory action of the Gsdma3 C-terminal domain can be well explained. Among nine reported Gsdma3 mutant alleles, 259RDW (insertion after residue 259 with three mistranslated residues RDW) and 366stop (a premature stop at residue 366) encode truncated Gsdma3 devoid of inter-domain contacts. Y344C, Y344H and A348T directly contact Gsdma3-N. T278P and L343P are near the direct-contacting residues. In addition, 412EA (duplication of E411A412) disrupts the Gasdermin-N-binding surface. They all weaken inter-domain interactions, making the Gsdma3 constitutively activated. The Gsdma3 itself is not cleaved by inflammatory caspases, though. Five of its mutants (T278P, L343P, Y344C, A348T and 412EA) were found capable of instructing 293T cells to undergo apoptosis or autophagy. Thus, Gsdma3 is also called an autoinhibited cell death-inducing factor.
Human GSDMA is a target of LMO1 in a TGF-β-dependent apoptotic signaling
The exactly expressional regulation mechanism of Gsdma3 is still unclear. However, human GSDMA, which acts as a potential suppressor in gastric cancer, was found to act as a target of LIM domain only 1 (LMO1) and involved in transforming growth factor (TGF)-β-induced apoptosis.18 TGF-β upregulates GSDMA expression by LMO1 induction through a sequence to which LMO1 binds in a GSDMA promoter region (Figure 2c). Given the above described evidence that Gsdma3 gene mutation results in the TNF-α-induced apoptosis, that loss of the Gsdma3 self-regulation contributes to autophagy, or that GSDMA involves the TGF-β-dependent apoptotic signaling, it will be of great significance to search for precise endogenous signaling molecules that may open-up the Gsdma3 protein and activate its function in future studies.
DFNA5 is Implicated in an Apoptotic Pathway
DFNA5 was originally identified as a gene responsible for autosomal-dominant non-syndromic sensorineural hearing loss (HL).36 At least four DFNA5 mutations associated with HL have, to date, been recognized.37, 38, 39, 40, 41 They all lead to exon 8 skipping during splicing, contributing to a frame shift and a premature termination of the protein. These findings support that DFNA5-associated HL is caused by exon 8 skipping, whereas mutations in other parts of this gene may not result in HL at all. In additional, although there are less published papers implicating DFNA5, almost all have a possible involvement in cancer biology. Hence, it is considered as a tumor suppressor.42, 43, 44, 45 The precise physiological function of the DFNA5 correlated to its tumor suppressive characteristics remains to be elucidated. On the other hand, various functional studies hinted that DFNA5 harbors a proapoptotic function.46, 47
Apoptosis-inducing activity of DFNA5-N domain
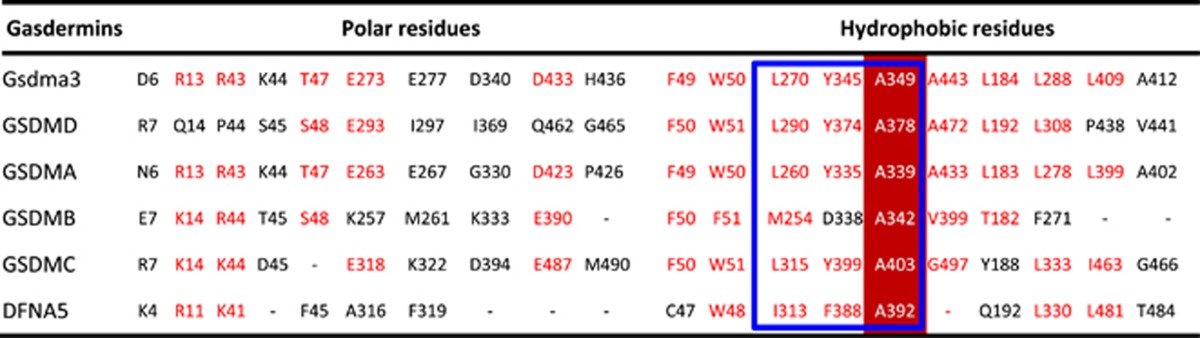
Transfecting the cell death-causing DFNA5 mutant into yeast48 or mammalian cells49 showed a toxic effect of the mutant protein, which was not coming after the transfection of its wild-type. Other transfection studies in mammalian cell lines have further strengthened the evidence, that is, the critical role of DFNA5 in apoptotic pathways.50 The transfection of its mutant resulted in cell death, whereas the transfection of its WT did not. This effect was observed not only in human cell lines, including HEK293T, COS7, MCF7, and HELA cells, but also in the yeast model Saccharomyces cerevisiae.51 Morphological analysis in combination with fluorescent labeling identified apoptosis as the cause of the observed cell death.50 Interestingly, the apoptosis-inducing domain is shared by WT and mutant DFNA5, leading to a hypothesis that the apoptotic induction is an intrinsic feature of DFNA5. Further experimental evidence for this hypothesis was provided by hydrophobic cluster analysis of DFNA5, revealing that it contains two globular domains connected by a hinge region. The N-terminal domain located in exon 2 and exon 6 displays a proapoptotic activity, whereas the C-terminal domain consisting of the amino acid residues 282–496 (exon 8 included) may fold back to mask the apoptosis-inducing function of the N-terminal domain (Figure 3). Using the yeast system and human HEK293T cells further confirms that the N-terminal but not the C-terminal domain leads to the apoptotic cell death. On the basis of the crystal structure of Gsdma3, we can find that the residues in gasdermin-C that make the hydrophobic core largely determining the autoinhibition are highly conserved in the gasdermin family (Table 1). Interestingly, all DFNA5 mutations which skip exon 8 are devoid of the critical residues (I313, F388 and A392) that make the hydrophobic core of DFNA5 (Table 1). Thus, the mutation of DFNA5 releases the autoinhibition activity of the C-terminal domain, resulting in the activation of the N-terminal function (Figure 3). Given that PJVK shows a high N-terminal similarity to DFNA5, it was used to test whether the transfection of the N-terminal is also responsible for apoptotic cell death, but it was not the case.50 Therefore, the apoptosis-inducing role may be specific for DFNA5.
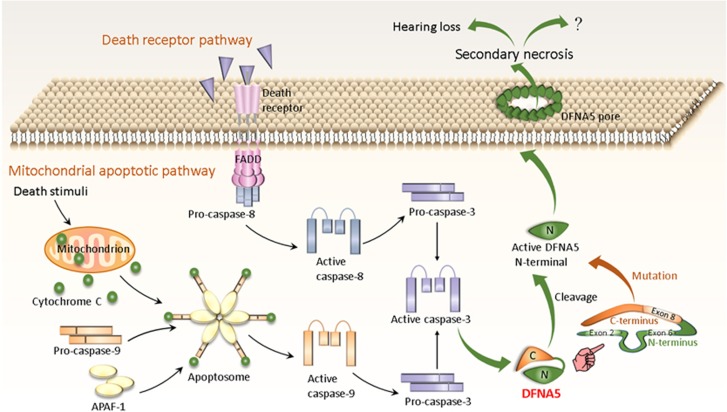
Figure 3.
Potential mechanistic process of apoptosis/secondary necrosis mediated by DFNA5. Intrinsic apoptosis is initiated by various death stimuli. They lead to permeabilization of the outer mitochondrial membrane, causing the release of cytochrome c. The latter binds to Apaf-1, followed successively by apoptosome formation, procaspase-9 recruitment/activation and activation of procaspase-3. Caspase-3 can also be activated by extrinsic or receptor-mediated apoptosis, and directly by active caspase-8. DFNA5 consists of two globular domains. The N-terminal domain of the apoptosis-inducing region located in exon 2 and exon 6 displays a proapoptotic activity, whereas the C-terminal domain, including exon 8, may serve as a regulator that folds back to mask the apoptosis-inducing N-terminal domain. Mutant DFNA5 skipping of exon 8 can change and shortens its C-terminal domain, thereby releasing the autoinhibition activity of the C-terminal domain with the activation of the N-terminal function. In other respect, active caspase-3 also can cleave DFNA5 to generate active DFNA5-N terminal that targets the plasma membrane and permeabilizes it by forming DFNA5 pores to induce secondary necrosis cell death, ultimately leading to HL
Table 1. The residues are involved in the autoinhibitory interactions in the gasdermin family.
Identical residues are highlighted by dark red background and conserved residues are red-colored. Residues in gasdermin-C that make the hydrophobic core are highly conserved in the gasdermin family, which are marked by a blue box
Cleavage of DFNA5 mediates progression of apoptotic cells into secondary necrosis
Consistent with the above findings of the apoptosis-inducing ability of DFNA5, a recent study shows that mutant DFNA5 significantly increases the level of ROS and upregulates several cytochrome c oxidase (COX) genes involved in the cellular oxidative stress in yeast Saccharomyces cerevisiae as well as human embryonic kidney 293T cells.51 In 2015, the DFNA5 mutant was further found co-localized with fragmented mitochondria and four different mitochondrion-related proteins, including Fis1 (a mitochondrial fission protein), Por1 (a voltage-dependent anion channel protein), Aac1 (a mitochondrial adenine nucleotide translocator 1) and Aac3, suggesting that they implicate this event. Moreover, through a microarray study the significance of mitochondria is confirmed in the DFNA5-induced cell death in both yeast and human cells.52 In addition, the gene ontology analysis suggests that the DFNA5 mutant is closely correlated to mitochondria, MAPK pathways and the endoplasmic reticulum.52
An exciting study further reveals that gasdermin-N domain of DFNA5 is involved in secondary necrosis.53 The latter is regarded as a terminal phase following the completion of the apoptotic program.54 It displays membrane permeabilization, swelling and lysis, which looks like the cellular phenotypes of pyroptosis or necroptosis.55, 56, 57, 58 Alnemri et al. proved that DFNA5 is cleaved by apoptotic caspase-3 with the generation of a necrotic DFNA5-N fragment. This fragment directly targets the plasma membrane to permeabilize it. Therefore, DFNA5 functions to regulate apoptotic cells disassembly to progress into the secondary necrosis (Figure 3).53 However, could the N terminal of DFNA5 serve as an apoptotic trigger to initiate the secondary necrosis in response to pathogen infection? Does DFNA5 mutant-induced HL result from the secondary necrosis of hearing cells? Elucidating precise mechanisms of DFNA5-mediated cytotoxic activity will pave the therapeutic approaches for DFNA5 mutant-induced HL or other related diseases.
Concluding Remarks
On the basis of GSDMD emerging as a novel member of cell death inducers, there has been gradually increasing interest in shedding light on possible functions of other gasdermin family members in programmed cell death. As described here, the breakthrough progress regarding GSDMD, Gsdma3/GSDMA or DFNA5 that are closely associated with pyroptotosis, apoptosis or autophagy has been made in our deep understanding of the gasdermin N-terminal-inducing cell death. Especially, the GSDMD-N domain forms the pores on cellular membrane, thereby driving pyroptosis. Therefore, gasdermin-N terminus may be a novel type of PFP. It can make pores on membranes through inserting and binding membrane lipids, consequently mediating cell death. However, detailed molecular events of the gasdermin-N terminus-initiating cell death remain unclear. Thus, elucidating the gasdermin N-terminal's cytotoxic activity may lead to identification of novel crucial molecular targets in the process of cell and bacterial death so as to pave therapeutic approaches for inflammasome-associated inflammatory and metabolic disorders.
Acknowledgments
This project was supported by the National Natural Science Foundation of China (Grant Numbers 81172824, 30971465) to F Xing.
Footnotes
Edited by JM Hardwick
The authors declare no conflict of interest.
References
- Jorgensen I, Miao EA. Pyroptotic cell death defends against intracellular pathogens. Immunol Rev 2015; 265: 130–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayagaki N, Warming S, Lamkanfi M, Vande Walle L, Louie S, Dong J et al. Non-canonical inflammasome activation targets caspase-11. Nature 2011; 479: 117–121. [DOI] [PubMed] [Google Scholar]
- Kayagaki N, Wong MT, Stowe IB, Ramani SR, Gonzalez LC, Akashi-Takamura S et al. Noncanonical inflammasome activation by intracellular LPS independent of TLR4. Science 2013; 341: 1246–1249. [DOI] [PubMed] [Google Scholar]
- Hagar JA, Powell DA, Aachoui Y, Ernst RK, Miao EA. Cytoplasmic LPS activates caspase-11: implications in TLR4-independent endotoxic shock. Science 2013; 341: 1250–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Zhao Y, Wang Y, Gao W, Ding J, Li P et al. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature 2014; 514: 187–192. [DOI] [PubMed] [Google Scholar]
- Broz P, Dixit VM. Inflammasomes: mechanism of assembly, regulation and signalling. Nat Rev Immunol 2016; 16: 407–420. [DOI] [PubMed] [Google Scholar]
- Shi J, Zhao Y, Wang K, Shi X, Wang Y, Huang H et al. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 2015; 526: 660–665. [DOI] [PubMed] [Google Scholar]
- Kayagaki N, Stowe IB, Lee BL, O'Rourke K, Anderson K, Warming S et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature 2015; 526: 666–671. [DOI] [PubMed] [Google Scholar]
- Ding J, Wang K, Liu W, She Y, Sun Q, Shi J et al. Pore-forming activity and structural autoinhibition of the gasdermin family. Nature 2016; 535: 111–116. [DOI] [PubMed] [Google Scholar]
- Shi P, Tang A, Xian L, Hou S, Zou D, Lv Y et al. Loss of conserved Gsdma3 self-regulation causes autophagy and cell death. Biochem J 2015; 468: 325–336. [DOI] [PubMed] [Google Scholar]
- Tamura M, Tanaka S, Fujii T, Aoki A, Komiyama H, Ezawa K et al. Members of a novel gene family, Gsdm, are expressed exclusively in the epithelium of the skin and gastrointestinal tract in a highly tissue-specific manner. Genomics 2007; 89: 618–629. [DOI] [PubMed] [Google Scholar]
- Zihlif M, Obeidat NM, Zihlif N, Mahafza T, Froukh T, Ghanim MT et al. Association between gasdermin A and gasdermin B polymorphisms and susceptibility to adult and childhood asthma among Jordanians. Genet Test Mol Biomarkers 2016; 20: 143–148. [DOI] [PubMed] [Google Scholar]
- Acevedo N, Reinius LE, Greco D, Gref A, Orsmark-Pietras C, Persson H et al. Risk of childhood asthma is associated with CpG-site polymorphisms, regional DNA methylation and mRNA levels at the GSDMB/ORMDL3 locus. Hum Mol Genet 2015; 24: 875–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Kang MJ, Kim BJ, Kwon JW, Song YH, Choi WA et al. Polymorphisms in GSDMA and GSDMB are associated with asthma susceptibility, atopy and BHR. Pediatr Pulmonol 2011; 46: 701–708. [DOI] [PubMed] [Google Scholar]
- Lluis A, Schedel M, Liu J, Illi S, Depner M, von Mutius E et al. Asthma-associated polymorphisms in 17q21 influence cord blood ORMDL3 and GSDMA gene expression and IL-17 secretion. J Allergy Clin Immunol 2011; 127: 1587–1594 e6. [DOI] [PubMed] [Google Scholar]
- Komiyama H, Aoki A, Tanaka S, Maekawa H, Kato Y, Wada R et al. Alu-derived cis-element regulates tumorigenesis-dependent gastric expression of GASDERMIN B (GSDMB). Genes Genet Syst 2010; 85: 75–83. [DOI] [PubMed] [Google Scholar]
- Saeki N, Usui T, Aoyagi K, Kim DH, Sato M, Mabuchi T et al. Distinctive expression and function of four GSDM family genes (GSDMA-D) in normal and malignant upper gastrointestinal epithelium. Genes Chromosomes Cancer 2009; 48: 261–271. [DOI] [PubMed] [Google Scholar]
- Saeki N, Kim DH, Usui T, Aoyagi K, Tatsuta T, Aoki K et al. GASDERMIN, suppressed frequently in gastric cancer, is a target of LMO1 in TGF-beta-dependent apoptotic signalling. Oncogene 2007; 26: 6488–6498. [DOI] [PubMed] [Google Scholar]
- He WT, Wan H, Hu L, Chen P, Wang X, Huang Z et al. Gasdermin D is an executor of pyroptosis and required for interleukin-1beta secretion. Cell Res 2015; 25: 1285–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, He WT, Hu L, Li J, Fang Y, Wang X et al. Pyroptosis is driven by non-selective gasdermin-D pore and its morphology is different from MLKL channel-mediated necroptosis. Cell Res 2016; 26: 1007–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aglietti RA, Estevez A, Gupta A, Ramirez MG, Liu PS, Kayagaki N et al. GsdmD p30 elicited by caspase-11 during pyroptosis forms pores in membranes. Proc Natl Acad Sci USA 2016; 113: 7858–7863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sborgi L, Ruhl S, Mulvihill E. GSDMD membrane pore formation constitutes the mechanism of pyroptotic cell death. EMBO J 2016; 35: 1766–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo HM, Rathkey J, Boyd-Tressler A. Active caspase-1 induces plasma membrane pores that precede pyroptotic lysis and are blocked by lanthanides. J Immunol 2016; 197: 1353–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zhang Z, Ruan J, Pan Y, Magupalli VG, Wu H et al. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature 2016; 535: 153–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour SC, Pena OM, Hancock RE. Host defense peptides: front-line immunomodulators. Trends Immunol 2014; 35: 443–450. [DOI] [PubMed] [Google Scholar]
- Chen X, Niyonsaba F, Ushio H, Okuda D, Nagaoka I, Ikeda S et al. Synergistic effect of antibacterial agents human beta-defensins, cathelicidin LL-37 and lysozyme against Staphylococcus aureus and Escherichia coli. J Dermatol Sci 2005; 40: 123–132. [DOI] [PubMed] [Google Scholar]
- Foitzik K, Lindner G, Mueller-Roever S, Maurer M, Botchkareva N, Botchkarev V et al. Control of murine hair follicle regression (catagen) by TGF-beta1 in vivo. FASEB J 2000; 14: 752–760. [DOI] [PubMed] [Google Scholar]
- Lei M, Bai X, Yang T, Lai X, Qiu W, Yang L et al. Gsdma3 is a new factor needed for TNF-alpha-mediated apoptosis signal pathway in mouse skin keratinocytes. Histochem Cell Biol 2012; 138: 385–396. [DOI] [PubMed] [Google Scholar]
- Lei M, Gao X, Yang L, Yang T, Lian X. Gsdma3 gene is needed for the induction of apoptosis-driven catagen during mouse hair follicle cycle. Histochem Cell Biol 2011; 136: 335–343. [DOI] [PubMed] [Google Scholar]
- Lin PH, Lin HY, Kuo CC, Yang LT. N-terminal functional domain of Gasdermin A3 regulates mitochondrial homeostasis via mitochondrial targeting. J Biomed Sci 2015; 22: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanon A, Rakovic A, Blankenburg H, Doncheva NT, Schwienbacher C, Serafin A et al. Profiling of Parkin-binding partners using tandem affinity purification. PLoS ONE 2013; 8: e78648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Mizushina Y, Kato Y, Tamura M, Shiroishi T. Functional conservation of Gsdma cluster genes specifically duplicated in the mouse genome. G3 2013; 3: 1843–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiyama A, Okamoto K. PINK1/Parkin-mediated mitophagy in mammalian cells. Curr Opin Cell Biol 2015; 33: 95–101. [DOI] [PubMed] [Google Scholar]
- Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol 2011; 12: 9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Lu Q, Rapaport D, Kozjak-Pavlovic V. Tom70 is essential for PINK1 import into mitochondria. PLoS ONE 2013; 8: e58435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Laer L, Huizing EH, Verstreken M, van Zuijlen D, Wauters JG, Bossuyt PJ et al. Nonsyndromic hearing impairment is associated with a mutation in DFNA5. Nat Genet 1998; 20: 194–197. [DOI] [PubMed] [Google Scholar]
- Chai Y, Chen D, Wang X, Wu H, Yang T. A novel splice site mutation in DFNA5 causes late-onset progressive non-syndromic hearing loss in a Chinese family. Int J Pediatr Otorhinolaryngol 2014; 78: 1265–1268. [DOI] [PubMed] [Google Scholar]
- Park HJ, Cho HJ, Baek JI, Ben-Yosef T, Kwon TJ, Griffith AJ et al. Evidence for a founder mutation causing DFNA5 hearing loss in East Asians. J Hum Genet 2010; 55: 59–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff AM, Luijendijk MW, Huygen PL, van Duijnhoven G, De Leenheer EM, Oudesluijs GG et al. A novel mutation identified in the DFNA5 gene in a Dutch family: a clinical and genetic evaluation. Audiol Neuro-otol 2004; 9: 34–46. [DOI] [PubMed] [Google Scholar]
- Yu C, Meng X, Zhang S, Zhao G, Hu L, Kong X. A 3-nucleotide deletion in the polypyrimidine tract of intron 7 of the DFNA5 gene causes nonsyndromic hearing impairment in a Chinese family. Genomics 2003; 82: 575–579. [DOI] [PubMed] [Google Scholar]
- Cheng J, Han DY, Dai P, Sun HJ, Tao R, Sun Q et al. A novel DFNA5 mutation, IVS8+4A>G, in the splice donor site of intron 8 causes late-onset non-syndromic hearing loss in a Chinese family. Clin Genet 2007; 72: 471–477. [DOI] [PubMed] [Google Scholar]
- Akino K, Toyota M, Suzuki H, Imai T, Maruyama R, Kusano M et al. Identification of DFNA5 as a target of epigenetic inactivation in gastric cancer. Cancer Sci 2007; 98: 88–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Chang X, Yamashita K, Nagpal JK, Baek JH, Wu G et al. Aberrant promoter methylation and tumor suppressive activity of the DFNA5 gene in colorectal carcinoma. Oncogene 2008; 27: 3624–3634. [DOI] [PubMed] [Google Scholar]
- Kim MS, Lebron C, Nagpal JK, Chae YK, Chang X, Huang Y et al. Methylation of the DFNA5 increases risk of lymph node metastasis in human breast cancer. Biochem Biophys Res Commun 2008; 370: 38–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CJ, Tang L, Shen DW, Wang C, Yuan QY, Gao W et al. The expression and regulation of DFNA5 in human hepatocellular carcinoma DFNA5 in hepatocellular carcinoma. Mol Biol Rep 2013; 40: 6525–6531. [DOI] [PubMed] [Google Scholar]
- Lage H, Helmbach H, Grottke C, Dietel M, Schadendorf D. DFNA5 (ICERE-1) contributes to acquired etoposide resistance in melanoma cells. FEBS Lett 2001; 494: 54–59. [DOI] [PubMed] [Google Scholar]
- Masuda Y, Futamura M, Kamino H, Nakamura Y, Kitamura N, Ohnishi S et al. The potential role of DFNA5, a hearing impairment gene, in p53-mediated cellular response to DNA damage. J Hum Genet 2006; 51: 652–664. [DOI] [PubMed] [Google Scholar]
- Gregan J, Van Laer L, Lieto LD, Van Camp G, Kearsey SE. A yeast model for the study of human DFNA5, a gene mutated in nonsyndromic hearing impairment. Biochim Biophys Acta 2003; 1638: 179–186. [DOI] [PubMed] [Google Scholar]
- Van Laer L, Vrijens K, Thys S, Van Tendeloo VF, Smith RJ, Van Bockstaele DR et al. DFNA5: hearing impairment exon instead of hearing impairment gene? J Med Genet 2004; 41: 401–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Op de Beeck K, Van Camp G, Thys S, Cools N, Callebaut I, Vrijens K et al. The DFNA5 gene, responsible for hearing loss and involved in cancer, encodes a novel apoptosis-inducing protein. Eur J Hum Genet 2011; 19: 965–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Rossom S, Op de Beeck K, Franssens V, Swinnen E, Schepers A, Ghillebert R et al. The splicing mutant of the human tumor suppressor protein DFNA5 induces programmed cell death when expressed in the yeast Saccharomyces cerevisiae. Front Oncol 2012; 2: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Rossom S, Op de Beeck K, Hristovska V, Winderickx J, Van Camp G. The deafness gene DFNA5 induces programmed cell death through mitochondria and MAPK-related pathways. Front Cell Neurosci 2015; 9: 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers C, Fernandes-Alnemri T, Mayes L, Alnemri D, Cingolani G, Alnemri ES. Cleavage of DFNA5 by caspase-3 during apoptosis mediates progression to secondary necrotic/pyroptotic cell death. Nat Commun 2017; 8: 14128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva MT. Secondary necrosis: the natural outcome of the complete apoptotic program. FEBS Lett 2010; 584: 4491–4499. [DOI] [PubMed] [Google Scholar]
- Silva MT, do Vale A, dos Santos NM. Secondary necrosis in multicellular animals: an outcome of apoptosis with pathogenic implications. Apoptosis 2008; 13: 463–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanden Berghe T, Vanlangenakker N, Parthoens E, Deckers W, Devos M, Festjens N et al. Necroptosis, necrosis and secondary necrosis converge on similar cellular disintegration features. Cell Death Differ 2010; 17: 922–930. [DOI] [PubMed] [Google Scholar]
- Bergsbaken T, Fink SL, Cookson BT. Pyroptosis: host cell death and inflammation. Nat Rev Microbiol 2009; 7: 99–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink SL, Cookson BT. Caspase-1-dependent pore formation during pyroptosis leads to osmotic lysis of infected host macrophages. Cell Microbiol 2006; 8: 1812–1825. [DOI] [PubMed] [Google Scholar]