Abstract
Development of hematopoietic populations through the process of differentiation is critical for proper hematopoiesis. The transcription factor CCAAT/enhancer binding protein alpha (C/EBPα) is a master regulator of myeloid differentiation, and the identification of C/EBPα target genes is key to understand this process. Here we identified the Ecotropic Viral Integration Site 2B (EVI2B) gene as a direct target of C/EBPα. We showed that the product of the gene, the transmembrane glycoprotein EVI2B (CD361), is abundantly expressed on the surface of primary hematopoietic cells, the highest levels of expression being reached in mature granulocytes. Using shRNA-mediated downregulation of EVI2B in human and murine cell lines and in primary hematopoietic stem and progenitor cells, we demonstrated impaired myeloid lineage development and altered progenitor functions in EVI2B-silenced cells. We showed that the compromised progenitor functionality in Evi2b-depleted cells can be in part explained by deregulation of cell proliferation and apoptosis. In addition, we generated an Evi2b knockout murine model and demonstrated altered properties of hematopoietic progenitors, as well as impaired G-CSF dependent myeloid colony formation in the knockout cells. Remarkably, we found that EVI2B is significantly downregulated in human acute myeloid leukemia samples characterized by defects in CEBPA. Altogether, our data demonstrate that EVI2B is a downstream target of C/EBPα, which regulates myeloid differentiation and functionality of hematopoietic progenitors.
Regulation of hematopoietic development is essential for proper functioning of the organism and involves tight control of numerous biological processes. This control ensures keeping the balance between different hematopoietic populations, such as hematopoietic stem cells (HSC), progenitors, myeloid, and lymphoid cells. The myeloid lineage is formed by cells at different stages of differentiation, where immature myeloid cells proliferate and differentiate into mature cells, such as granulocytes.1 A block of myeloid differentiation is associated with uncontrolled proliferation of immature myeloid progenitors and is a major characteristic of acute myeloid leukemia (AML).2
CCAAT/enhancer binding proteins (C/EBPs) constitute a family of basic region leucine zipper DNA-binding transcription factors containing six members – α, β, γ, δ, ɛ, and ζ.3 The best characterized C/EBP member is C/EBPα, which plays a critical role in myelopoiesis and controls hematopoietic stem and progenitor cell (HSPC) self-renewal and maintenance.4, 5, 6 Alterations in C/EBPα are related to the development of AML and can be mediated by several mechanisms. Mutations in CEBPA occur in 5–14% of AML cases. Approximately 70% of the CEBPA mutant AMLs carry double mutations.7, 8, 9, 10, 11 Most of these CEBPA double mutant cases carry a C-terminal mutation on one allele (such as the 22C mutation)12 and an N-terminal mutation (like 10N and 22N)12 on the other allele. Besides these common CEBPA mutants some rare mutations, such as mutant 128, have been described.12 The other 30% of CEBPA mutant AMLs carry a single mutation, and those cases are mostly N-terminal mutants. On one hand, mutations in the N-terminal part of CEBPA result in the absence of full-length C/EBPα (42 kDa – p42). These N-terminal mutations lead to increased usage of an alternative start codon and consequently increased expression of a shorter C/EBPα isoform (30 kDa – p30). On the other hand, mutations in the C-terminal basic region leucine zipper affect DNA binding properties of C/EBPα.12 Other mechanisms causing inactivation of C/EBPα are promoter hypermethylation and silencing,13, 14 messenger RNA (mRNA) translational alterations,15 and posttranslational modifications.16, 17
C/EBPα acts through regulation of genes involved in HSPC self-renewal, proliferation, and myeloid development. Therefore, identification of novel C/EBPα target genes is important for a better understanding of hematopoiesis and for developing new therapeutic approaches in AML. Recently, we identified a list of genes upregulated after C/EBPα activation.18 One of the genes identified was Ecotropic Viral Integration site 2B (EVI2B), also known as CD361,19 and originally described as a common viral integration site in retrovirally-induced murine leukemia.20 EVI2B gene codes for a transmembrane glycoprotein expressed in most hematopoietic populations,21 however its function in hematopoiesis remains elusive. Taking into account that EVI2B was identified as a potential C/EBPα target gene and that C/EBPα is a key transcription factor in HSPC self-renewal and myeloid differentiation, we hypothesized that EVI2B might play a role in these processes.
In the present study, we showed that EVI2B is a direct C/EBPα target gene downregulated in a subset of AML samples characterized by defects in C/EBPα. We demonstrated that EVI2B expression is upregulated during neutrophilic differentiation and that EVI2B depletion leads to alterations in myeloid differentiation. Further, we demonstrated that Evi2b-silenced or knockout (KO) HSPC present impaired self-renewal and bone marrow repopulating abilities, which could be caused partially by defected cell cycle progression and cell survival. Altogether, our results demonstrate that EVI2B is an important regulator of HSPC and myeloid differentiation, and that low levels of EVI2B might contribute to the differentiation arrest characteristic of AML.
Results
EVI2B is expressed in mature hematopoietic populations and is upregulated upon C/EBPα activation
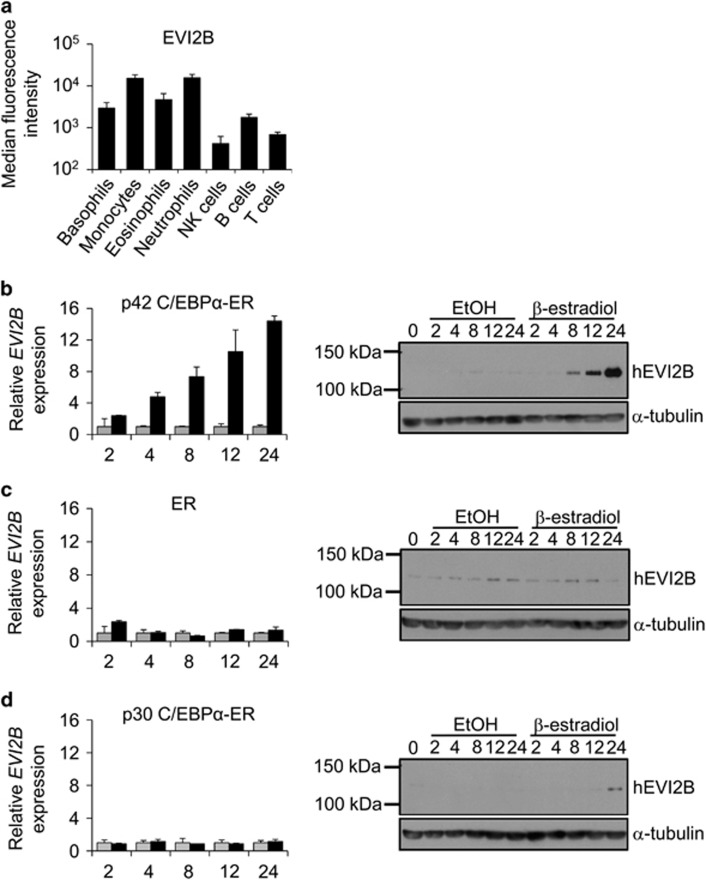
Human buffy coat samples demonstrated highest EVI2B levels on neutrophils and monocytes, followed by eosinophils and basophils (Figure 1a and Supplementary Figure S1). EVI2B was also detected, although at lower levels, on NK, B, and T cells. We investigated whether C/EBPα could regulate expression of EVI2B. We employed K562 cells, which do not express endogenous C/EBPα, stably transfected with a β-estradiol inducible full-length p42 C/EBPα-ER (estrogen receptor).22 We observed that β-estradiol treatment of p42 C/EBPα-ER K562 cells causes increase of EVI2B mRNA and protein level in a time-dependent manner (Figure 1b). Control cells overexpressing ER alone did not show increased EVI2B expression upon β-estradiol stimulation (Figure 1c). Activation of the short form of C/EBPα (p30 C/EBPα-ER) did not result in elevated levels of EVI2B (Figure 1d). Surprisingly, we observed that deletion of C/EBPα in primary murine cells from CebpaloxP/loxP MX1-CRE+ conditional KO mice (lineage−, c-kit+, Sca1+, LKS, and common myeloid progenitors, CMP)6, did not alter Evi2b levels (Supplementary Figure S2). Altogether, these results indicate that EVI2B is strongly expressed in granulocytic cells and that EVI2B expression is upregulated by full-length C/EBPα, but not by the C/EBPα p30 isoform.
Figure 1.
EVI2B is expressed in myeloid cells and upregulated upon p42 C/EBPα-ER activation. (a) Flow cytometric analysis of EVI2B expression in different populations of human buffy coat samples. The y axis is shown in logarithmic scale and represents median fluorescence intensity of EVI2B signal. The x axis shows different hematopoietic populations. Results represent average measurement of four buffy coats. (b–d) Quantitative RT-PCR and western blot analysis of K562 cells overexpressing (b) p42 C/EBPα-ER, (c) ER or (d) p30 C/EBPα-ER treated with 1 μM β-estradiol (black bars) or EtOH vehicle control (gray bars). The y axes represent relative human EVI2B mRNA expression compared to vehicle control treatment. The x axis indicates hours upon stimulation. RT-PCR results represent the average of two independent experiments, each done in duplicate. Western blots: upper panels show murine EVI2B staining and lower panels α-tubulin loading control. Positions of m.w. standards (kDa) are shown on the left
EVI2B is a direct C/EBPα target gene
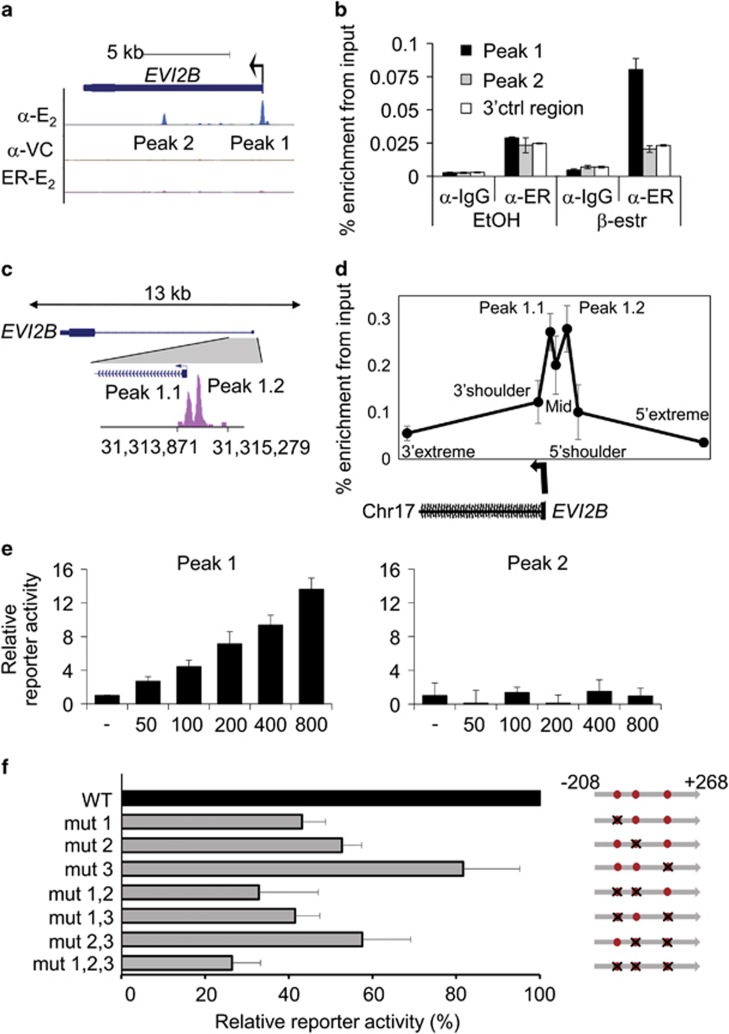
To study whether EVI2B is a direct C/EBPα target gene, we made use of chromatin immunoprecipitation followed by sequencing (ChIP-seq) data acquired in our previous study.18 We identified two potential C/EBPα binding sites in the proximity of EVI2B gene, called peak 1 and 2 (Figure 2a). We validated C/EBPα binding to peak 1 by ChIP-PCR experiments (Figure 2b). To exclude that C/EBPα binding in the promoter of EVI2B (peak 1) was due to C/EBPα overexpression, we re-analyzed ChIP-seq datasets from previous studies (GEO Ids: GSM1187163, GSM1187164 and GSM722424), in which cells endogenously expressing C/EBPα were used. We observed C/EBPα binding in the proximity of EVI2B promoter in U937 cells (Figure 2c) and murine GMP (granulocyte macrophage progenitor) (Supplementary Figure S3), indicating that endogenous C/EBPα binds within the EVI2B gene. ChIP-PCR assays validated the CEBPα binding to peak 1 in U937 cells (Figure 2d).
Figure 2.
EVI2B is a direct C/EBPα target gene. (a) Human C/EBPα ChIP-seq data in K562 C/EBPα-ER expressing cells treated with 1 μM β-estradiol (α-E2) or vehicle control (α-VC), and K562 ER control cells treated with 1 μM β-estradiol (ER-E2) for 6 h. Depicted are potential C/EBPα binding sites (peak 1 and 2). Black arrow indicates transcriptional start site and direction of transcription. (b) ChIP-PCR was performed with K562 C/EBPα-ER expressing cells treated as described in panel A. Immunoprecipitation was performed using antibodies against ER (α-ER) or IgG control antibodies (α-IgG). Q-PCR was performed using oligos in peak 1, peak 2, or a 3'UTR control region (3'ctrl region). The y axis indicates percentage of enrichment from input. (c) ChIP-seq dataset (GSM722424) on U937 human cells assessing the binding of endogenous CEBPα within the EVI2B promoter (peak 1.1 and 1.2). (d) ChIP-PCR assay in U937 cells using seven primer sets covering the promoter region, including the two peaks shown by the U937 ChIP-seq (peak 1.1 and peak 1.2), the region between the peaks (mid) along with the 5' and 3' proximal (shoulders) and distal (extreme) regions flanking the peaks. Error bars represent the standard deviation across the replicates (n=3). CEBPα ChIP enrichment on peak 1.1 and 1.2 regions is significantly higher compared to the distal control regions (P=0.0006 and 0.0013, respectively). (e) Luciferase reporter assay in K562 cells co-transfected with 100 ng EVI2B luciferase reporter vector (peak 1 and 2) and increasing amounts of human CEBPA expression plasmid. The y axis indicates the relative percentage of luciferase activity and it is presented as fold change compared to the empty vector (not containing CEPBA). Results represent the average of two independent experiments, each done in triplicate. (f) Luciferase reporter assays in K562 cells transfected with WT, single mutants, double mutants or a triple mutant of human EVI2B promoter peak 1 luciferase reporter vector and 100 ng CEBPA expression plasmid. The x axis indicates the relative percentage of luciferase activity. Results represent the average of at least three independent experiments. Right part of the Figure graphically depicts human EVI2B promoter peak 1, and 3 C/EBP consensus binding sites (red dots). Black crosses indicate target mutations in C/EBP consensus binding sites from CAAT to CCCT. Numbers on the right side of the panel represent distance from TSS
To assess whether the interactions of these sites with C/EBPα protein are functionally relevant, we carried out luciferase reporter assays. We cloned peak 1 and 2 into a luciferase reporter vector and co-transfected these constructs with increasing concentrations of CEBPA expressing plasmid into K562 cells. We observed that C/EBPα was able to transactivate luciferase reporter in case of peak 1, but not peak 2 (Figure 2e). Next, we identified 3 C/EBP consensus sites within peak 1 conserved through genomes of 8 vertebrate species (Supplementary Figure S4). To investigate which site was essential for transactivation, we performed targeted mutagenesis of these sites individually or in combination. Mutation of all three sites led to 80% ablation of reporter activity (Figure 2f). We observed only partial loss of activity in constructs with 1 or 2 mutations. Altogether, these data indicate that EVI2B is regulated by direct binding of C/EBPα to the promoter region of human EVI2B, and that all 3 C/EBP binding sites are involved. Since all C/EBP family members can bind to CAAT motif, we investigated whether C/EBPβ (ref. 23) and C/EBPγ (ref.24) were able to transactivate EVI2B promoter. As shown in Supplementary Figure S5 neither of these transcription factors could increase luciferase reporter activity.
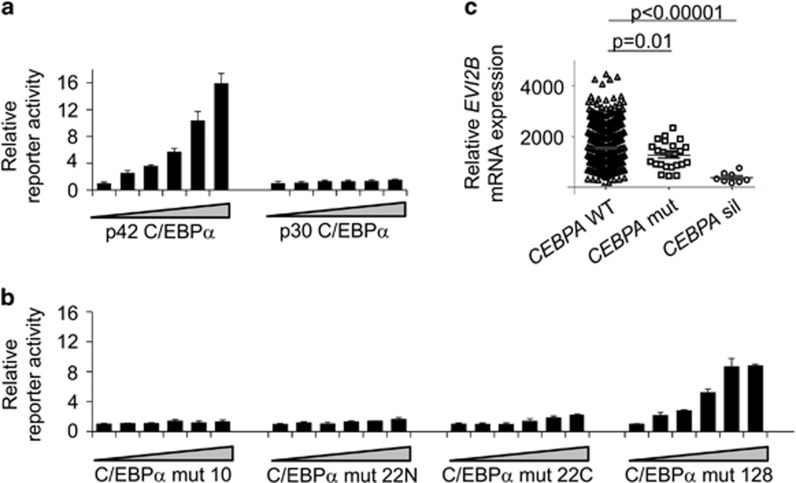
We next assessed whether the p30 C/EBPα isoform could transactivate EVI2B promoter. We observed that p30 C/EBPα was not able to induce EVI2B promoter activity in contrast to p42 C/EBPα isoform (Figure 3a). Further, we investigated whether CEBPA mutant forms identified in human AML patients12 could induce transactivation of the human EVI2B reporter construct. Luciferase assays using EVI2B reporter construct in combination with 3 CEBPA mutants expression plasmids (mut 10, mut 22N, mut 22C) did not result in luciferase activity (Figure 3b). On the contrary, mut 128 was able to induce reporter activity in a dose dependent manner, although transactivation did not reach the level observed in case of the C/EBPα p42 form (Figure 3b). Altogether, these results indicate that C/EBPα p42, and to a lesser extend mut 128, can transactivate human EVI2B promoter, whereas the p30 C/EBPα isoform and other C/EBPα mutant forms were not able to.
Figure 3.
Human EVI2B promoter is transactivated by full-length C/EBPα p42 but not C/EBPα p30 and C/EBPα mutants. (a and b) Luciferase reporter assays assessing transactivation of EVI2B peak 1 by different C/EBPα mutant forms. (a) rat C/EBPα p42 and rat C/EBPα p30, (b) human C/EBPα mut 10, human C/EBPα mut 22N, human C/EBPα mut 22C, and human C/EBPα mut 128. The x axes show increasing concentration of CEBPA construct or its mutant forms (0, 50, 100, 200, 400, 800 ng), the y axes indicate relative reporter activity compared to empty vector control. Results represent the average of three independent experiments. (c) Relative expression of EVI2B mRNA in 493 patient samples with non-affected CEBPA (CEBPA WT), 26 CEBPA mutant cases and 10 CEBPA silenced cases
EVI2B is downregulated in a specific subset of AML patient samples characterized by defects in CEBPA
Since our data indicate that p42 C/EBPα, but not p30 CEBPα nor several C/EBPα mutants, bind and activate human EVI2B promoter, we hypothesized that EVI2B levels would be affected in C/EBPα defective AMLs. We studied EVI2B mRNA expression levels in a cohort of 529 AML samples,25 and observed that EVI2B levels were significantly reduced in 36 patients characterized by defects in CEBPA (Figure 3c). The alterations in CEBPA were caused by either CEBPA promoter hypermethylation and silencing (n=10) or by biallelic mutations in CEBPA (n=26). Interestingly, the reduction of EVI2B was more abrupt in the CEBPA-silenced cases than in the CEBPA-mutated samples, compared to other AML samples which express full-length p42 C/EBPα. In summary, we have identified a subset of AML samples characterized by defects in CEBPA which significantly present decreased levels of EVI2B transcripts.
EVI2B regulates neutrophilic differentiation in vitro
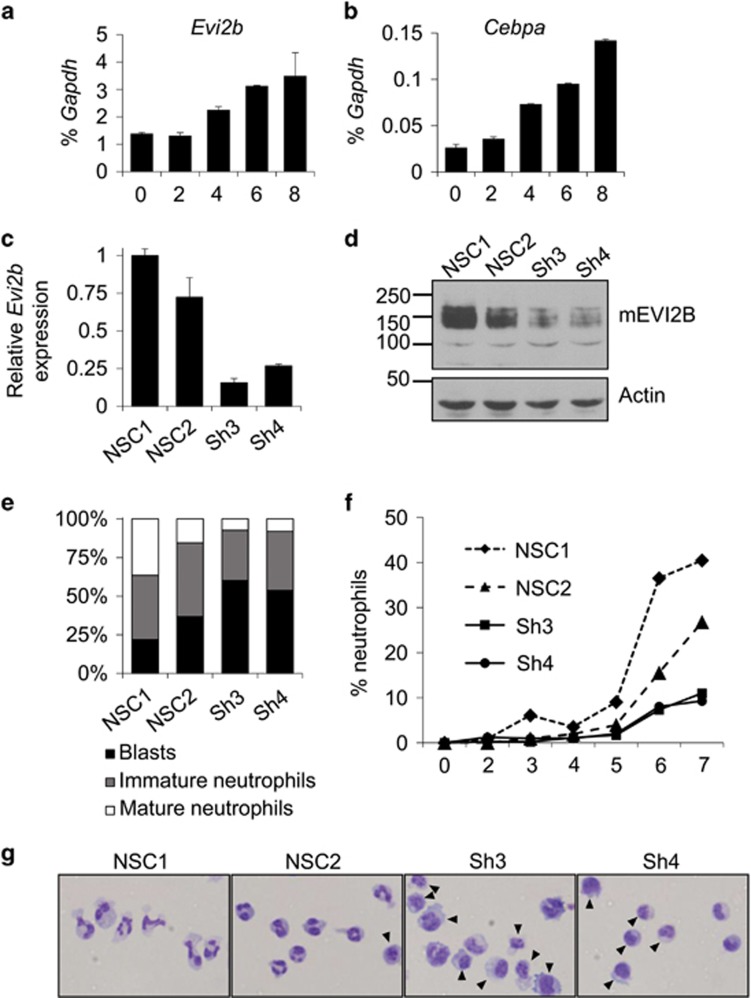
Since EVI2B expression is controlled by C/EBPα and C/EBPα is required for granulocytic differentiation, we hypothesized that EVI2B plays a role in neutrophilic development. We employed the 32D/G-CSF-R cell line as a model of murine granulocytic differentiation.26, 27 We demonstrated that Evi2b mRNA levels are upregulated in 32D/G-CSF-R cells upon treatment with G-CSF in a similar fashion as Cebpa (Figures 4a and b). To test whether depletion of Evi2b affects 32D/G-CSF-R cell differentiation we generated several Evi2b-downregulating shRNA constructs, and cloned them in a lentiviral vector containing a GFP reporter. 32D/G-CSF-R cells were transduced and the infected cells were sorted based on GFP expression. Evi2b shRNA#3 and shRNA#4 were causing 80% Evi2b knockdown in 32D/G-CSF-R cells on both mRNA (Figure 4c) and protein (Figure 4d) level. On the contrary, shRNA#1 was not working efficiently and therefore, along with the non-silencing control (NSC#1), it was used as an additional negative control (further referred as NSC#2) (Figures 4c and d). Of note, murine EVI2B molecular weight determined by SDS-PAGE is ca. 140 kDa (Supplementary Figure S6), however the predicted size of mature EVI2B is 46kDa (421 amino acids). This is probably due to markedly acidic character of this polypeptide (predicted pI 4.6) and multiple N-glycosylation of the extracellular domain. To assess the ability of Evi2b knockdown cells to differentiate towards neutrophils, we transduced 32D/G-CSF-R cells with control and Evi2b-downregulating shRNAs and treated these cells with G-CSF. We observed that the percentage of immature blasts was significantly increased in case of Evi2b knockdown cells (shRNA#3, shRNA#4) compared to controls (NSC#1, NSC#2) (Figures 4e and g). This increase was at the expense of neutrophils, which were significantly reduced in the absence of EVI2B. In addition, we observed that the percentage of neutrophils was decreased during the whole course of differentiation in case of Evi2b-downregulated cells (Figure 4f). These data indicate that Evi2b silencing reduces murine neutrophilic differentiation of 32D/G-CSF-R cells in vitro.
Figure 4.
Evi2b knockdown blocks granulocytic differentiation in mouse 32D/G-CSF-R cells. (a and b) Quantitative RT-PCR in 32D/G-CSF-R cells treated with G-CSF (100 ng/ml) for 8 days. The y axes represent relative expression of (a) Evi2b and (b) Cebpa compared to Gapdh. The x axes indicate days of treatment. Results represent the average of two independent experiments. (c) Quantitative RT-PCR in 32D/G-CSF-R cells transduced with distinct shRNA lentiviral constructs 5 days after infection. The y axis indicates fold change of Evi2b mRNA level after transduction with Evi2b silencing shRNA (Sh3, Sh4) compared to non-silencing controls (NSC1, NSC2). (d) Western blot analysis in 32D/G-CSF-R cells after transduction with the distinct shRNA lentiviral constructs. Upper panel shows murine EVI2B staining and lower panel Actin loading control. Positions of m.w. standards (kDa) are shown on the left. (e) Differential counting of mature neutrophils (white bars), immature neutrophils (gray bars), and blasts cells (black bars) in control and Evi2b-silenced 32D/G-CSF-R cells treated with G-CSF (100 ng/ml) for 7 days in May-Grünwald Giemsa stained cytospins. (f) Percentage of mature neutrophils during the course of G-CSF-induced neutrophilic differentiation of 32D/G-CSF-R cells. Samples were cytospun every day during 7 days of treatment. The x axis demonstrates the time of treatment (days), the y axis represents the percentage of neutrophils. (g) Representative pictures of 32D/G-CSF-R cells treated with G-CSF for 7 days, cytospun and stained with May-Grünwald Giemsa. Black arrows indicate blasts. Original magnification, x40 (e–g) Experiments were performed three independent times and at least 100 cells were counted for each sample
Evi2b silencing impairs HSPC properties in vitro and in vivo
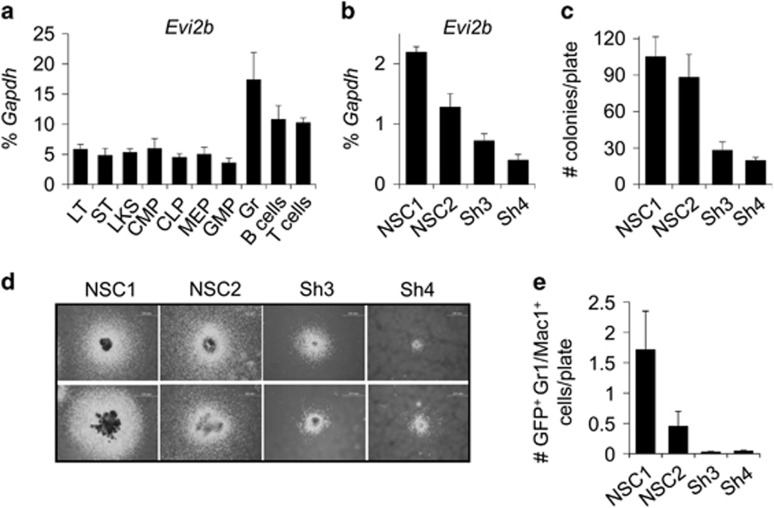
To assess the role of EVI2B in primary myeloid progenitors, we investigated the levels of Evi2b expression in different sorted murine BM populations. We observed that Evi2b mRNA was expressed in stem cells and the LKS fraction, which is enriched for HSPC (Figure 5a). Evi2b transcripts were detected in myeloid and lymphoid progenitors at comparable levels, and increased in mature hematopoietic populations. The highest levels of Evi2b were found in granulocytes (Figure 5a), which is in agreement with our previous observation showing highest levels of EVI2B protein in human mature myeloid cells (Figure 1a).
Figure 5.
Evi2b knockdown impairs murine HSPC granulocytic colony forming ability. (a) Quantitative RT-PCR in different sorted murine BM hematopoietic populations. The y axis represents relative expression of Evi2b compared to Gapdh control. The x axis indicates hematopoietic populations: long-term HSC (LT), short-term HSC (ST), LKS, common myeloid progenitors (CMP), common lymphoid progenitors (CLP), megakaryocyte erythroid progenitors (MEP), granulocyte monocyte progenitors (GMP), granulocytes (Gr), B cells, and T cells. Values are average of 3 independent experiments. (b) Quantitative RT-PCR of sorted LKS cells transduced with controls (NSC#1, NSC#2) and Evi2b-downregulating shRNAs (sh#3, sh#4) 3 days after infection. The y axis represents relative expression of murine Evi2b mRNA compared to Gapdh. (c) Quantification of myeloid colonies formed by LKS cells infected with the shRNA constructs growing in semisolid medium for 14 days. The y axis indicates number of myeloid colonies per plate. (d) Two representative pictures of colonies formed by NSCs or Evi2b-downregulated LKS cells. Original magnification, x4. Colony culture assays were performed at least three times. (e) Number of GFP+ Gr1/Mac1+ granulocytic cells per plate in semisolid cultures 14 days after seeding LKS cells transduced with the distinct shRNA lentiviral constructs
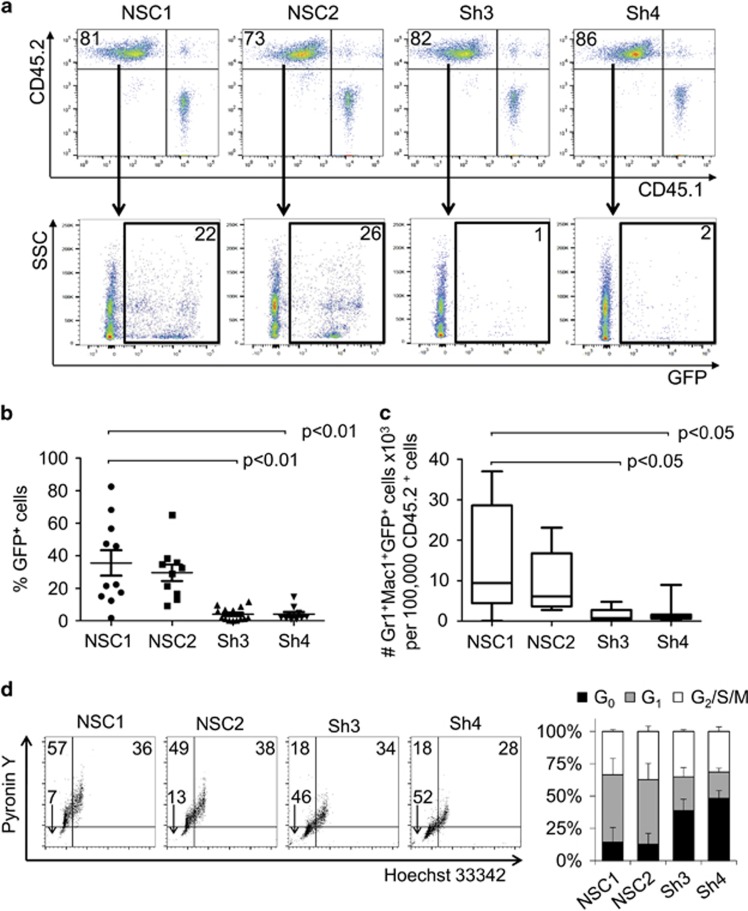
We next investigated whether Evi2b silencing affects myeloid development in murine BM. Using the Evi2b shRNA constructs, we downregulated Evi2b expression in LKS cells (Figure 5b). Infected GFP+ cells were sorted and plated in semisolid medium (Methocult M3434), which allows myeloid colony formation. We observed decreased number and size of myeloid colonies in Evi2b-downregulated cells compared to non-silenced control cells (Figures 5c and d). Consistently, we showed that Evi2b-silenced LKS cells produced significantly less Gr1/Mac1+ granulocytic cells compared to controls (Figure 5e). These results indicate that EVI2B is required for proper murine myeloid development in vitro. To study the effect of Evi2b knockdown on HSPC functions in vivo we transplanted Evi2b-silenced LKS cells (CD45.2+) into lethally irradiated congenic recipient mice (CD45.1+). Since we did not sort GFP+ cells prior to transplantation, the donor-derived CD45.2+ population consists of a mixture of transduced (GFP+) and untransduced (GFP−) cells. GFP positivity was similar in all groups (Supplementary Figure S7). As expected, blood analysis of recipient mice 4 weeks after transplantation did not show changes in the engraftment of total CD45.2+ cells (Figure 6a and Supplementary Figure S8). However, the contribution of GFP+ donor cells to the hematopoietic system of the recipients was strikingly reduced in case of Evi2b-downregulated cells (Figures 6a and b). Consequently, the number of GFP+Gr1/Mac1+ granulocytes originating from the CD45.2+ donor cell population was significantly lower in case of Evi2b-silenced cells compared to controls (Figure 6c). These results demonstrated that Evi2b-downregulated HSPC are functionally impaired, that is possess reduced ability to reconstitute the hematopoietic system of lethally irradiated recipients, and present reduced contribution to the formation of granulocytes in transplanted animals. Altogether, we demonstrated that EVI2B is expressed in immature murine BM cells, detected the highest EVI2B levels in granulocytes, and showed that Evi2b silencing dramatically affects functionality of HSPC in vitro and in vivo.
Figure 6.
Evi2b knockdown impairs repopulating ability of HSPC. (a) Representative dot plot showing engraftment of donor CD45.2+ cells transduced with control (NSC1 and NSC2) or Evi2b-silenced (Sh3 and Sh4) constructs and support CD45.1 spleen cells (upper panels). GFP+ cells within the CD45.2+ gate are shown in the lower panels. Numbers indicate percentage of cells. Blood analysis was performed 4 weeks after transplantation. (b) Quantification of contribution of control or Evi2b-silenced LKS cells to hematopoietic system of the recipient animals 4 weeks after transplantation. The y axis indicates the percentage of GFP+ cells out of CD45.2+ gate. Each group contains at least 10 animals. (c) Number of GFP+ granulocytes derived from the donor (CD45.2+) cells. The Y axis indicates the number of GFP+Gr/Mac1+ × 103 granulocytes per 100.000 Ly5.2+ cells. Each group contains at least five animals. The values on y axes of (b) and (c) were normalized to the original percentage of GFP+ cells measured 2 days after infection. Transplantation experiments were performed three times. (d) Representative contour plots of cell cycle analysis on murine LKS cells transduced with controls NSC1 and NSC2 or Evi2b-downregulating shRNAs Sh3 and Sh4. The x axis indicates Hoechst 33342 staining and the y axis shows Pyronin Y staining. The x and y axes are displayed in linear scale. Numbers indicate the percentage of cells in G0 (lower left quadrant), G1 (upper left quadrant), and G2/S/M phase (upper right quadrant). Right panel indicates quantification of cell cycle analysis in LKS cells transduced with the distinct shRNAs. Black bars indicate percentage of cells in G0, gray bars indicate percentage of cells in G1, and white bars the percentage of cells in G2/S/M. Results are average of three independent experiments
Since Evi2b downregulation in HSPC was associated with reduced number and size of colonies in vitro, as well as with reduced engraftment in vivo, we hypothesized that EVI2B might be involved in regulation of cell proliferation and/or cell death. Cell cycle analysis demonstrated that Evi2b-silenced cells were more quiescent compared to controls (Figure 6d). Annexin V and Cleaved Caspase-3 staining showed a moderate but significant increase in apoptosis of Evi2b-silenced cells compared to control cells (Supplementary Figure S9). Altogether, these data indicate that EVI2B controls cell cycle progression and survival of HSPC thereby maintaining functional properties of these cells.
Evi2b KO mice present reduced G-CSF-induced colony formation, impaired self-renewal and BM reconstitution abilities
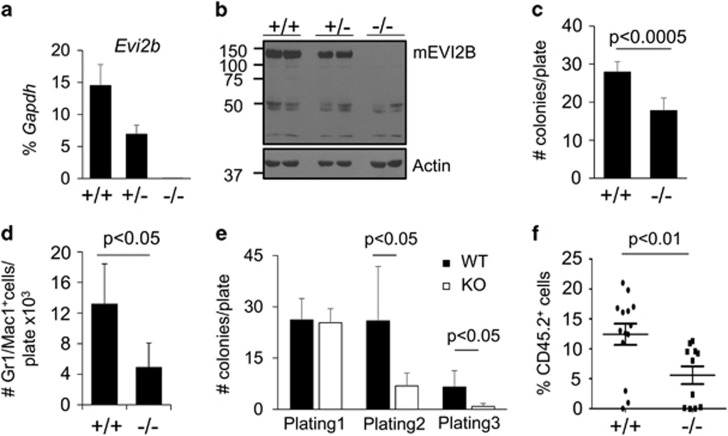
To deepen our understanding of the function of EVI2B in hematopoiesis, we generated Evi2b KO mice (Supplementary Figure S10a). We demonstrated absence of Evi2b mRNA and protein in Evi2b KO BM cells (Figures 7a and b). Since Evi2b gene is located in an intron of NF1 gene (between exons 36 and 37), we demonstrated that NF1 protein levels were not altered by Evi2b deletion (Supplementary Figure S10b). Flow cytometric analysis of steady-state hematopoiesis did not reveal differences between WT and Evi2b KO animals (Supplementary Figure S10c-f and data not shown). Given our previous results, we hypothesized that Evi2b depletion might affect properties of hematopoietic cells under G-CSF-stimulating conditions. We cultured BM cells in semisolid medium (Methocult M3231) supplemented with G-CSF and observed reduced colony formation (Figure 7c) and number of mature granulocytes (Figure 7d) in Evi2b KO compared to WT BM cultures. Since our Evi2b knockdown experiments suggested that EVI2B plays a role in HSPC functionality, we carried out serial re-plating assays (Methocult 3434), which provide an indication of HSPC self-renewal in vitro. Interestingly, the ability of Evi2b KO cells to form colonies at the first plating was not affected (Figure 7e), which is not surprising given the fact that in steady-state conditions the KO mice do not present defects. However, sequential re-plating revealed reduced re-plating capacity in Evi2b KO cells (Figure 7e). Next, we determined whether deletion of Evi2b would impair HSPC functions in vivo. We demonstrated that transplantation of Evi2b KO BM cells into lethally irradiated recipient mice resulted in reduced engraftment (Figure 7f). Altogether, these results demonstrate that Evi2b KO cells, similarly to Evi2b-silenced cells, present impaired HSPC properties, and thereby affected G-CSF-induced differentiation and ability to repopulate lethally irradiated mice.
Figure 7.
Bone marrow cells from Evi2b KO mice present impaired repopulating ability in vivo and impaired plating capacities in vitro. (a) Quantitative RT-PCR in Evi2b WT (+/+), heterozygous (+/−) and KO (−/−) BM nucleated cells. The y axis represents relative Evi2b expression compared to Gapdh control. Each group contains 3 mice. (b) EVI2B expression in EVI2B WT, heterozygous and KO mice assessed on protein level using Western blotting. Equal amounts of cells (0.5 × 106cells) were loaded per lane. Antibodies against murine EVI2B were used to evaluate EVI2B expression in Evi2b KO, heterozygous and WT mice (upper panel). Equal loading was verified by staining the membrane with antibody against Actin (lower panel). Positions of m.w. standards (kDa) are shown on the left. (c) Quantification of colonies formed by 10 000 whole BM cells from WT and Evi2b KO mice plated in semisolid medium M3231 supplemented with G-CSF (100 ng/ml). The y axis represents the number of colonies per plate. Counting was performed 10 days after plating. Results represent the average of 6 animals. (d) Quantification of Gr1/Mac1+ cells in cultures from (c). The y axis represents the number of Gr1/Mac1+ cells per plate × 103. (e) Re-plating assays of WT (black bars) and Evi2b KO (white bars) BM cells using Methocult M3434. Initially 5000 BM nucleated cells were plated per well and 2000 cells were re-plated. Colonies were quantified under microscope and re-plated after 10–14 days in semisolid medium. The y axis indicates the number of colonies per plate. This is a representative experiment out of 2. Results indicate the average of 3 independent cultures. (f) Flow cytometric analysis of blood of lethally irradiated recipient mice transplanted with 100.000 WT or Evi2b KO BM nucleated cells along with 0.5 × 106 BM cells. The y axis indicates the percentage of donor (CD45.2+) cells in the blood of recipients 4 weeks after transplantation. Each group contains at least 11 animals. Results include data from three independent experiments
EVI2B silencing impairs human granulocytic differentiation in cell lines and primary HSPC
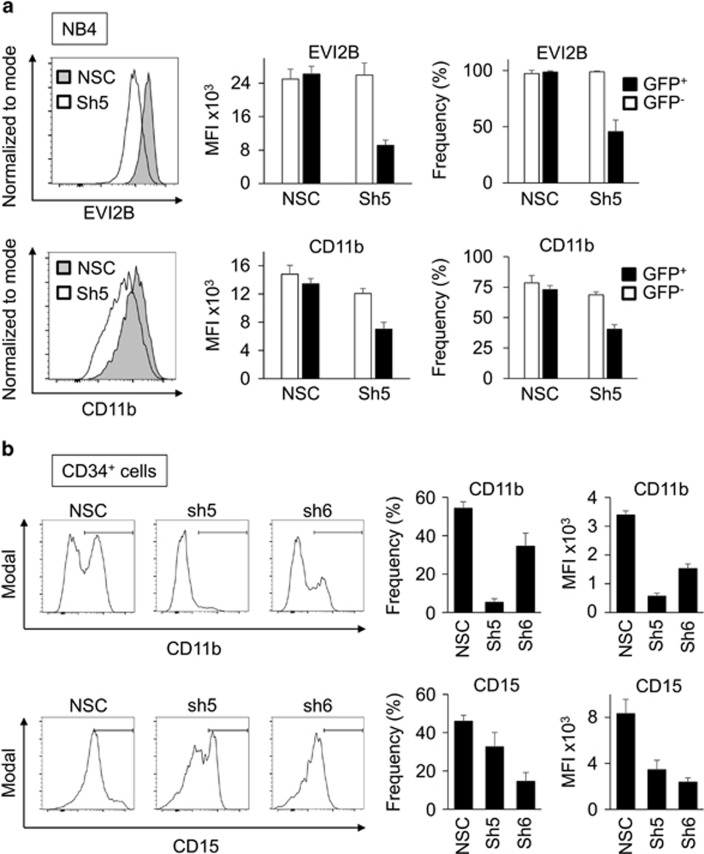
To exclude that the effects of Evi2b silencing were limited to murine cells, we employed a human cell line and primary cells. First, NB4 cells were used as a model of human granulocytic differentiation. We designed shRNA lentiviral constructs downregulating human EVI2B, transduced NB4 cells, and treated them with 1 μM all-trans retinoic acid (ATRA) for 2 days to induce granulocytic differentiation. We observed that downregulation of human EVI2B (shRNA#5) resulted in reduced expression of CD11b in comparison to NSC transduced cells in GFP+, but not in GFP− cells (Figure 8a). In addition, human cord blood CD34+ HSPC were transduced with EVI2B-targeting (shRNA#5 and shRNA#6) or NSC shRNA lentiviral particles. Following transduction, cells were placed in semisolid culture in the presence of cytokines promoting granulocytic differentiation. Due to low EVI2B knockdown efficiency in human CD34+ cells, CD11b and CD15 levels were assessed in EVI2Blow gate in case of EVI2B-downregulating shRNAs and EVI2Btotal gate in case of NSC (see flow cytometry gating in Supplementary Figure S11). We observed that EVI2B-silenced CD34+ cells exhibited lower levels of CD11b and CD15 expression after 10 days in semisolid culture in comparison to NSC control cells (Figure 8b and Supplementary Figure S11). Altogether, these experiments demonstrate that EVI2B silencing impairs human granulocytic differentiation in cell lines and primary HSPC.
Figure 8.
EVI2B silencing blocks granulocytic differentiation in human cell lines and primary HSPC. (a) Representative histogram plots showing EVI2B (upper) and CD11b (lower) levels in NB4 cells transduced with NSC or EVI2B silencing shRNA (Sh5). The y axes indicate cell counts normalized to mode. Right panels indicate mean fluorescence intensity (MFI) and frequency of positive cells in the infected GFP+ and the control GFP− populations. (b) Representative histogram plots showing CD11b and CD15 staining in human CD34+ cells infected with NSC or EVI2B silencing shRNA (Sh5 and Sh6) upon 10 days in culture. The y axes are shown in modal display mode. Right panels show MFI and frequency of positive cells for CD11b and CD15
Discussion
C/EBPα transcription factor, being one of the major hematopoietic regulators, controls stem cell quiescence and is required for production of mature granulocytes.4, 5, 6 Identification of C/EBPα target genes is critical for understanding of hematopoiesis in general and the development of AML. One of the putative C/EBPα target genes we recently identified is EVI2B.18 EVI2B encodes a transmembrane glycoprotein with no similarity to any other gene product, but so far its function remains unknown. The present study provides insight into the biological role of EVI2B, that is, involvement in regulation of myeloid differentiation and HSPC functions.
We showed that the human EVI2B gene is under the control of the full-length C/EBPα p42, which has been demonstrated to be responsible for proper myelopoiesis.4, 5 On the contrary, the differentiation-deficient C/EBPα p30 isoform and the majority of the CEBPA mutant forms originally described in human AML12 were not able to cause EVI2B promoter activity. Consistently, we observed that EVI2B mRNA levels are significantly lower in AML patients with silenced or mutated CEBPA, suggesting that C/EBPα might be an important regulator of EVI2B in human specimens. Intriguingly, we observed that several CEBPA WT AML samples exhibit low EVI2B levels as well. This could be explained by the fact that CEBPA, in addition to mutations and hypermethylation, can be inactivated by other mechanisms, such as protein phosphorylation16 and acetylation.28 It has also been shown that CEBPA can be downregulated by oncogenic fusion proteins such as AML1-ETO.29 AML cases with these types of CEBPA alterations are included in the CEBPA WT group, and this could explain why some AML cases despite having WT CEBPA present very low EVI2B levels.
The fact that ablation of Cebpa in murine bone marrow cells does not affect Evi2b expression levels supports the hypothesis that other transcription factors may regulate Evi2b expression. Thus, we analyzed EVI2B promoter sequence using TFBIND (tfbind.hgc.jp) and observed conserved binding motifs for several transcription factors including PU.1, GFI-1 and other C/EBP proteins. Importantly, these transcription factors are involved in granulocytic differentiation and could regulate EVI2B expression in Cebpa KO cells. Similarly, other transcription factors might be involved in the regulation of EVI2B expression during lymphopoiesis, since EVI2B is abundantly expressed in lymphoid cells which do not express C/EBPα.
In addition to the block in granulocytic differentiation, CEPBA-deficient HSPC present an increased proliferative potential in vitro and in vivo.5, 6 This is partially mimicked by EVI2B-silenced cells, which exhibit reduced potential to produce granulocytes. Nevertheless, EVI2B deficiency did not induce expansion of HSPC but rather induced a proliferation arrest. Results showing affected myeloid lineage development under G-CSF stimulating conditions suggest that the C/EBPα-EVI2B axis functions as an effector of G-CSF during granulocytic differentiation. However, given the different roles of EVI2B and C/EBPα in proliferation of HSPC, we conclude that the C/EBPα-EVI2B axis is not mediating the HSPC expansion. This is not surprising given the fact that C/EBPα, due to its nature, orchestrates expression of a large number of genes and has a broader effect than EVI2B.
Evi2b KO mice did not demonstrate any alterations in steady-state hematopoiesis. A possible explanation may be a compensatory effect due to functional replacement by another so far unidentified protein(s). We evaluated EVI2B expression in healthy human specimens and observed large variability in EVI2B levels, which included some samples with very low EVI2B (data not shown). Therefore, we hypothesize that there could be similar compensatory mechanisms in Evi2b KO mice and EVI2Blow healthy controls that would allow for normal granulocytic differentiation. Bone marrow transplantation experiments demonstrate that EVI2B is required to reconstitute the hematopoietic system of lethally irradiated recipient mice. Therefore, it is tempting to speculate that EVI2B levels in human grafts may be critical to determine engraftment, reconstitution and recovery of human cells in a transplantation setting. Future studies should reveal whether EVI2B might regulate engraftment of human cells during bone marrow transplantation therapy.
In conclusion, our results demonstrate that EVI2B is a novel C/EBPα target gene. The data from our knockdown and KO models and a subset of AML patients characterized by defects in CEBPA, strongly indicate that EVI2B is a player in myeloid differentiation. In addition, our data show that EVI2B regulates the repopulating ability of HSPC. Future studies will aim to elucidate the molecular mechanisms mediating EVI2B function. Ultimately, an interesting question will be whether modulation of EVI2B levels in AML cells could override the granulocytic differentiation arrest characteristic of this disease.
Material and methods
Detailed Materials and Methods are described in Supplementary Materials and Methods.
Primary cells isolation
Four buffy coats of human blood cells were processed by dextran sedimentation for 30 min. and remaining erythrocytes were lysed with NH4Cl-containing lysing solution for 15 min. Mouse BM cells were isolated by crushing the long bones, hips and spine. Murine BM, spleen and blood erythrocytes were lysed using ACK buffer (150 mM NH4Cl, 10 mM KHCO3, 0.1 mM ethylenediaminetetraacetic acid). Hematopoietic CD34+ cells were isolated from umbilical cord blood by separation of mononuclear cells using Ficoll method (PAN Biotech, Aidenbach, Germany) followed by using a CD34+ MicroBead kit (Miltenyi Biotec, Bergisch Gladbach, Germany). Samples were collected with written informed consent in accordance to the Declaration of Helsinki and under Ethics committee approval of the corresponding centers.
Plasmids and targeted mutagenesis
Peak 1 (from −208 bp to +268 bp) and peak 2 (from +5523 bp to +5861 bp) were cloned into pXP2 firefly luciferase reporter vector. All distances are relative to transcription start site (TSS). C/EBP consensus sequence CAAT in EVI2B peak 1 region was mutated to CCCT. Oligos used for targeted mutagenesis can be found in Supplementary Table S1.
Lentiviral transductions
Lentiviral particles were produced as previously described.24 Viral titer in supernatant was 1–5 × 106 TU/ml.
Methylcellulose colony assays
Murine colony culture assays were performed using Methocult M3434 and Methocult M3231 (Stemcell Technologies, Vancouver, BC, Canada). Methocult M3231 was supplemented with 100 ng/ml G-CSF. 500 sorted LKS cells were plated in M3434 and 10 000 whole BM cells were plated in M3231. 2000 human cord blood CD34+ cells were plated into Methocult H4535 (Stemcell Technologies, Vancouver, BC, Canada). Number of colonies was assessed 10 to 14 days after plating. For re-plating assays 5000 BM cells were initially plated, after 10–14 days cells were collected, and 2000 cells re-plated.
Cell cycle and apoptosis analysis
LKS cells were infected with shRNA lentiviral constructs, GFP+ cells sorted 2 days later and plated into semisolid Methocult M3434 medium. Apoptosis and cell cycle were analyzed by flow cytometry after 2 and 4 days in colony culture.
Mice and bone marrow transplantation experiments
CebpaloxP/loxP MX1-CRE+ (Cebpa KO) and control CebpaloxP/loxP MX1-CRE− (WT) mice were treated with pI:pC as previously described.6 Murine primary cells were isolated from C57BL/6NCrl mice. Evi2b KO (B6-Evi2btm1Ciphe) mice were generated by excision of full coding sequence. In bone marrow transplantation assays congenic C57BL/6NCrl mice (CD45.1+) were employed as recipients and lethally irradiated two times using 7.5 Gy with 4 h interval. Approximately 15 000–30 000 shRNA transduced LKS cells (CD45.2+) were transplanted 1 day after infection together with 0.5 × 106 spleen cells as a support (CD45.1+). Two days after transduction a non-transplanted aliquot of transduced cells was analyzed for GFP positivity. In case of competitive transplantation assays using Evi2b KO BM cells (CD45.2+), 100000 cells were transplanted together with 0.5 × 106 whole BM cells (CD45.1+). Recipient mice were bled and analyzed 4 weeks after transplantation. Mice were maintained in the animal facility of the Institute of Molecular Genetics. All experiments were approved by the ethical committee of the Institute.
Statistical analysis
Two-sided, unpaired Student t-test was used to determine the statistical significance of experimental results. P values <0.05 are considered statistically significant. Clustered column charts indicate average plus standard deviation. Scatter dot plots indicate mean with S.E.M. In box and whisker, whiskers indicate minimum and maximum values.
Acknowledgments
We would like to thank Hong Zhang and Min Ye for their technical assistance with Cebpa conditional KO mice, Berta Díez Odriozola and Carlos Lozano Asencio for their contribution. We thank Pavel Draber (IMG, Prague, Czech Republic) for providing us with the TU-01 monoclonal antibody against α-tubulin, and Vladimir Korinek (IMG, Prague, Czech Republic) for providing us with the monoclonal antibody 9E10 against myc. This work was supported by a GACR grant P303/12/0855 to MA-J. TK was supported by Ministry of Health Czech Republic grant 15-26588A and infrastructure support by NPU I no. LO1604, Ministry of Education, Youth and Sports, Czech Republic. This work was supported by the Czech Centre for Phenogenomics (CCP, project no. LM2015040), and OP RDI CZ.1.05/2.1.00/19.0395 (Higher quality and capacity for transgenic models).
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by S Kaufmann
The authors declare no conflict of interest.
Supplementary Material
References
- Orkin SH. Diversification of haematopoietic stem cells to specific lineages. Nat Rev Genet 2000; 1: 57–64. [DOI] [PubMed] [Google Scholar]
- Lowenberg B, Downing JR, Burnett A. Acute myeloid leukemia. N Engl J Med 1999; 341: 1051–1062. [DOI] [PubMed] [Google Scholar]
- Ramji DP, Foka P. CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem J 2002; 365: 561–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radomska HS, Huettner CS, Zhang P, Cheng T, Scadden DT, Tenen DG. CCAAT/enhancer binding protein alpha is a regulatory switch sufficient for induction of granulocytic development from bipotential myeloid progenitors. Mol Cell Biol 1998; 18: 4301–4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Iwasaki-Arai J, Iwasaki H, Fenyus ML, Dayaram T, Owens BM et al. Enhancement of hematopoietic stem cell repopulating capacity and self-renewal in the absence of the transcription factor C/EBP alpha. Immunity 2004; 21: 853–863. [DOI] [PubMed] [Google Scholar]
- Ye M, Zhang H, Amabile G, Yang H, Staber PB, Zhang P et al. C/EBPa controls acquisition and maintenance of adult haematopoietic stem cell quiescence. Nat Cell Biol 2013; 15: 385–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nerlov C. C/EBPalpha mutations in acute myeloid leukaemias. Nat Rev Cancer 2004; 4: 394–400. [DOI] [PubMed] [Google Scholar]
- Wouters BJ, Lowenberg B, Erpelinck-Verschueren CA, van Putten WL, Valk PJ, Delwel R. Double CEBPA mutations, but not single CEBPA mutations, define a subgroup of acute myeloid leukemia with a distinctive gene expression profile that is uniquely associated with a favorable outcome. Blood 2009; 113: 3088–3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy H, Roumier C, Huyghe P, Biggio V, Fenaux P, Preudhomme C. CEBPA point mutations in hematological malignancies. Leukemia 2005; 19: 329–334. [DOI] [PubMed] [Google Scholar]
- Preudhomme C, Sagot C, Boissel N, Cayuela JM, Tigaud I, de Botton S et al. Favorable prognostic significance of CEBPA mutations in patients with de novo acute myeloid leukemia: a study from the Acute Leukemia French Association (ALFA). Blood 2002; 100: 2717–2723. [DOI] [PubMed] [Google Scholar]
- Taskesen E, Bullinger L, Corbacioglu A, Sanders MA, Erpelinck CA, Wouters BJ et al. Prognostic impact, concurrent genetic mutations, and gene expression features of AML with CEBPA mutations in a cohort of 1182 cytogenetically normal AML patients: further evidence for CEBPA double mutant AML as a distinctive disease entity. Blood 2011; 117: 2469–2475. [DOI] [PubMed] [Google Scholar]
- Pabst T, Mueller BU, Zhang P, Radomska HS, Narravula S, Schnittger S et al. Dominant-negative mutations of CEBPA, encoding CCAAT/enhancer binding protein-alpha (C/EBPalpha), in acute myeloid leukemia. Nat Genet 2001; 27: 263–270. [DOI] [PubMed] [Google Scholar]
- Wouters BJ, Jorda MA, Keeshan K, Louwers I, Erpelinck-Verschueren CA, Tielemans D et al. Distinct gene expression profiles of acute myeloid/T-lymphoid leukemia with silenced CEBPA and mutations in NOTCH1. Blood 2007; 110: 3706–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chim CS, Wong AS, Kwong YL. Infrequent hypermethylation of CEBPA promotor in acute myeloid leukaemia. Br J Haematol 2002; 119: 988–990. [DOI] [PubMed] [Google Scholar]
- Perrotti D, Cesi V, Trotta R, Guerzoni C, Santilli G, Campbell K et al. BCR-ABL suppresses C/EBPalpha expression through inhibitory action of hnRNP E2. Nat Genet 2002; 30: 48–58. [DOI] [PubMed] [Google Scholar]
- Ross SE, Radomska HS, Wu B, Zhang P, Winnay JN, Bajnok L et al. Phosphorylation of C/EBPalpha inhibits granulopoiesis. Mol Cell Biol 2004; 24: 675–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radomska HS, Basseres DS, Zheng R, Zhang P, Dayaram T, Yamamoto Y et al. Block of C/EBP alpha function by phosphorylation in acute myeloid leukemia with FLT3 activating mutations. J Exp Med 2006; 203: 371–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liss A, Ooi CH, Zjablovskaja P, Benoukraf T, Radomska HS, Ju C et al. The gene signature in CCAAT-enhancer-binding protein alpha dysfunctional acute myeloid leukemia predicts responsiveness to histone deacetylase inhibitors. Haematologica 2014; 99: 697–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawthon RM, Andersen LB, Buchberg AM, Xu GF, O'Connell P, Viskochil D et al. cDNA sequence and genomic structure of EV12B, a gene lying within an intron of the neurofibromatosis type 1 gene. Genomics 1991; 9: 446–460. [DOI] [PubMed] [Google Scholar]
- Buchberg AM, Bedigian HG, Jenkins NA, Copeland NG. Evi-2, a common integration site involved in murine myeloid leukemogenesis. Mol Cell Biol 1990; 10: 4658–4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matesanz-Isabel J, Sintes J, Llinas L, de Salort J, Lazaro A, Engel P. New B-cell CD molecules. Immunol Lett 2011; 134: 104–112. [DOI] [PubMed] [Google Scholar]
- D'Alo F, Johansen LM, Nelson EA, Radomska HS, Evans EK, Zhang P et al. The amino terminal and E2F interaction domains are critical for C/EBP alpha-mediated induction of granulopoietic development of hematopoietic cells. Blood 2003; 102: 3163–3171. [DOI] [PubMed] [Google Scholar]
- Hirai H, Zhang P, Dayaram T, Hetherington CJ, Mizuno S, Imanishi J et al. C/EBPbeta is required for 'emergency' granulopoiesis. Nat Immunol 2006; 7: 732–739. [DOI] [PubMed] [Google Scholar]
- Alberich-Jorda M, Wouters B, Balastik M, Shapiro-Koss C, Zhang H, Di Ruscio A et al. C/EBPgamma deregulation results in differentiation arrest in acute myeloid leukemia. J Clin investig 2012; 122: 4490–4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaak RG, Wouters BJ, Erpelinck CA, Abbas S, Beverloo HB, Lugthart S et al. Prediction of molecular subtypes in acute myeloid leukemia based on gene expression profiling. Haematologica 2009; 94: 131–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong F, van Buitenen C, Pouwels K, Hoefsloot LH, Lowenberg B, Touw IP. Distinct cytoplasmic regions of the human granulocyte colony-stimulating factor receptor involved in induction of proliferation and maturation. Mol Cell Biol 1993; 13: 7774–7781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorda MA, Lowenberg B, Delwel R. The peripheral cannabinoid receptor Cb2, a novel oncoprotein, induces a reversible block in neutrophilic differentiation. Blood 2003; 101: 1336–1343. [DOI] [PubMed] [Google Scholar]
- Bararia D, Kwok HS, Welner RS, Numata A, Sarosi MB, Yang H et al. Acetylation of C/EBPalpha inhibits its granulopoietic function. Nat Commun 2016; 7: 10968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabst T, Mueller BU, Harakawa N, Schoch C, Haferlach T, Behre G et al. AML1-ETO downregulates the granulocytic differentiation factor C/EBPalpha in t(8;21) myeloid leukemia. Nat Med 2001; 7: 444–451. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.