Abstract
Ubiquitination is a posttranslational modification that regulates many cellular processes including protein degradation, intracellular trafficking, cell signaling, and protein-protein interactions. Deubiquitinating enzymes (DUBs), which reverse the process of ubiquitination, are important regulators of the ubiquitin system. OTUD6B encodes a member of the ovarian tumor domain (OTU)-containing subfamily of deubiquitinating enzymes. Herein, we report biallelic pathogenic variants in OTUD6B in 12 individuals from 6 independent families with an intellectual disability syndrome associated with seizures and dysmorphic features. In subjects with predicted loss-of-function alleles, additional features include global developmental delay, microcephaly, absent speech, hypotonia, growth retardation with prenatal onset, feeding difficulties, structural brain abnormalities, congenital malformations including congenital heart disease, and musculoskeletal features. Homozygous Otud6b knockout mice were subviable, smaller in size, and had congenital heart defects, consistent with the severity of loss-of-function variants in humans. Analysis of peripheral blood mononuclear cells from an affected subject showed reduced incorporation of 19S subunits into 26S proteasomes, decreased chymotrypsin-like activity, and accumulation of ubiquitin-protein conjugates. Our findings suggest a role for OTUD6B in proteasome function, establish that defective OTUD6B function underlies a multisystemic human disorder, and provide additional evidence for the emerging relationship between the ubiquitin system and human disease.
Main Text
Whole-exome sequencing (WES) has proven powerful for discovering new genes associated with genetically heterogeneous disorders, including global developmental delay, intellectual disability, and seizures.1, 2, 3, 4 In our clinical WES cohort of roughly 9,000 unrelated individuals, the majority of whom have neurologic manifestations and belong to the pediatric age group, we identified two unrelated individuals with biallelic variants in OTUD6B (MIM: 612021) with overlapping clinical features. Through GeneMatcher,5 we subsequently identified 10 additional individuals, yielding a total of 12 affected subjects from 6 unrelated families (Figure 1). We obtained written informed consent from all study participants in accordance with protocols approved by institutional review boards at Baylor College of Medicine (USA), University of Kiel (Germany), National Research Center (Egypt), CHU de Nantes (France), and Makassed Islamic Charitable Hospital (Jerusalem).
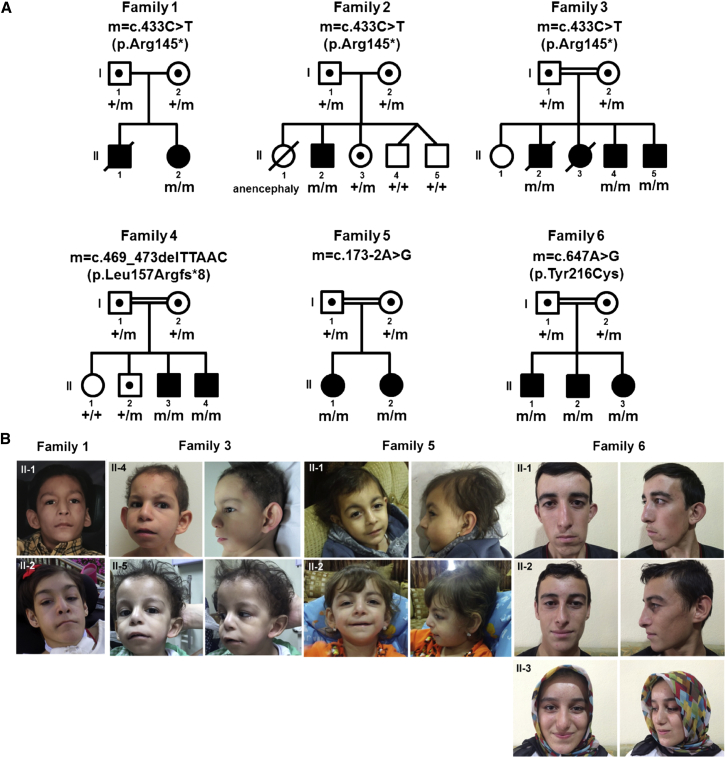
Figure 1.
Pedigrees and Clinical Features of Families with Pathogenic Variants in OTUD6B
(A) Families with biallelic variants in OTUD6B. Allelic status is given below each tested individual. Symbols are as follows: filled, affected; empty, unaffected; dotted, heterozygous carrier; hash, deceased.
(B) Facial photos of affected individuals from families 1, 3, 5, and 6.
A summary of clinical findings is provided in Table 1, detailed case summaries can be found in the Supplemental Note, and pedigrees and photos are available in Figure 1. Of the six families with variants in OTUD6B, two were non-consanguineous and four consanguineous. The cohort comprised eight males and four females, ages 3 to 20 years. All subjects exhibited intellectual disability. Nine subjects (75%) were severely impaired with absent speech and significant global developmental delay, and three subjects had only mild to moderate intellectual disability without delay in language or motor development. All subjects presented with seizures, mostly of the generalized tonic-clonic type, but with varying degrees of severity and frequency (from one episode to daily, refractory epilepsy). Other seizure types noted were absence, myoclonic, and atonic seizures. In nine subjects, particularly those with severe intellectual disability, additional recurrent features included severe microcephaly (Z-scores between −2.8 and −6.5), hypotonia, gross motor delay, and growth retardation. Intrauterine growth restriction (IUGR) was observed in seven pregnancies (58%). Nine subjects had feeding difficulties, which were evident as early as the neonatal period and required gastrostomy tube feeds for three subjects. Six subjects never walked independently. One subject (II-2 in family 1) with congenital spastic quadriplegia is able to walk short distances but is largely wheelchair dependent. One subject (II-2 in family 2) who did attain walking subsequently lost this ability due to severe and progressive spasticity. Among 12 subjects with neuroimaging studies, 6 showed structural brain abnormalities (50%) including cortical changes/white matter volume loss (n = 5), abnormal corpus callosum (n = 3), and dilatation of lateral ventricles (n = 1) (Figure S1).
Table 1.
Clinical Features of Subjects with Biallelic OTUD6B Variants
|
Family 1 |
Family 2 |
Family 3 |
Family 4 |
Family 5 |
Family 6 |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| II-2 | II-2 | II-2 | II-4 | II-5 | II-3 | II-4 | II-1 | II-2 | II-1 | II-2 | II-3 | |
| Gender | female | male | male | male | male | male | male | female | female | male | male | female |
| Ethnic origin | Hispanic | Hispanic/Italy | Egypt | Egypt | Egypt | Syria | Syria | Palestine | Palestine | Turkey | Turkey | Turkey |
| Age at last examination | 18 years, 2 months | 17 years, 2 months | 13 years (died same year) | 8 years, 1 month | 3 years | 9 years, 5 months | 3 years, 11 months | 5 years, 4 months | 3 years, 2 months | 20 years, 7 months | 16 years, 6 months | 14 years, 8 months |
| Intellectual disability | severe | severe | severe | severe | severe | severe | severe | severe | severe | moderate | mild | moderate |
| Speech delay | no words | no words | no words | no words | no words | no words | no words | no words | no words | no delay | no delay | no delay |
| Seizure (onset) | + (12 months) | + (15 months) | + (NA) | + (NA) | + (NA) | + (17 months) | + (3 years) | + (7 months) | + (2 years) | + (18 months) | + (7 years) | + (8 years) |
| IUGR | + | + (poor fetal activity) | + | + (poor fetal activity) | + | + | + | − | − | − | − | − |
| Weight, kg (z-score) | 28 (−8.9) | 47.6 (−2.2) | 15 (−2.2) (at 10 years) | 12 (−4.5) (at 5 years) | 9 (−3.5) | 15.5 (−1.6) (at 5 years 2 months) | 15 (−1) | 15.6 (−1.4) | 10 (−3.4) | 67 (−0.33) | 66 (0.27) | 54 (0.27) |
| Height, cm (z-score) | 129.5 (−5.2) | 137 (−4.9) | 103 (−5) (at 10 years) | 90 (−3.8) (at 5 years) | 80 (−3.7) | 107 (−0.6) (at 5 years 2 months) | 93 (−2.5) | 100 (−2.1) | 87 (−2.0) | 176 (−0.1) | 171 (−0.5) | 159 (−0.4) |
| OFC, cm (z-score) | 47.9 (−4.7) | 52 (−2.8) | 44.5 (−6.5) (at 10 years) | 44 (−5.3) (at 5 years) | 43 (−5.3) | 46.5 (−3.5) (at 5 years 2 months) | 45 (−4.3) | 45 (−4.5) | 44 (−4.3) | 55.7 (−0.35) | 54.5 (−0.59) | 54.3 (0.38) |
| Hypotonia | + | + | + | + | + | + | + | + | + | − | − | − |
| Gross motor delay | congenital quadriplegic, mostly in wheelchair | walked at 3 years 9 months but now non-ambulatory | could sit, non-ambulatory | sits, non-ambulatory | does not sit, non-ambulatory | walked 4 years 6 months | sits, non-ambulatory | crawled at 3 years; stands and walks only with support | does not crawl or pull to stand, non-ambulatory | no delay | no delay | no delay |
| Feeding difficulties | + | + | + | + | + | + | + | + | + | − | − | − |
| Others | FTT, chronic constipation, G-tube feeds | FTT, chronic constipation and diarrhea, G-tube feeds | FTT | FTT | FTT | FTT | FTT, G-tube feeds | |||||
| Additional neurologic features | congenital quadriplegia, ataxia, spasticity | cerebral palsy, ataxia, spastic quadriplegia | autism spectrum disorder | autism spectrum disorder | autism spectrum disorder | − | − | − | − | − | − | − |
| Brain imaging | normal | prominent perivascular spaces, hypoplastic corpus callosum, generalized white matter volume loss, gliosis in bilateral parietal lobes | mild fronto-parietal cortical changes | mild fronto-parietal cortical changes, short corpus callosum | mild fronto-parietal cortical changes | normal | cortical and white matter atrophy | mild dilatation of lateral ventricles mainly occipital horns, deep interhemispheric fissure/hypoplastic corpus callosum | normal | normal | normal | normal |
| Physical Features | ||||||||||||
| CHD | NA | normal echo | PS, ASD | normal echo | PS, ASD, VSD | NA | VSD | NA | TOF, ASD | NA | NA | NA |
| Other congenital malformations | sacral dimple | spina bifida occulta, cryptorchidism | − | cryptorchidism | − | cryptorchidism | sacral dimple cryptorchidism | − | − | − | − | − |
| Head/Neck | displaced posterior hair whorl | brachycephaly, sparse hair | short neck, sloping shoulders | short neck, sloping shoulders | short neck, sloping shoulders | − | − | flat occiput, sparse hair | flat occiput | wide forehead, narrow long asymmetric face | wide forehead, narrow long face | narrow long face |
| Eyes | arched eyebrows, long eyelashes, long palpebral fissures | arched eyebrows, long down-slanting palpebral fissures, ptosis | long palpebral fissures | long palpebral fissures | long palpebral fissures | long palpebral fissures | − | arched eyebrows, deep-set eyes, long eyelashes | deep-set eyes, long eyelashes | down-slanting palpebral fissures | − | − |
| Nose | long nose, hypoplastic alae, high nasal bridge, long philtrum | prominent nasal bridge, short columella, small alae, smooth long philtrum | broad root and prominent nasal bridge, smooth long philtrum | broad root and prominent nasal bridge, smooth long philtrum | broad root and prominent nasal bridge, smooth long philtrum | − | − | slightly long philtrum | slightly long philtrum | tubular nose | − | − |
| Mouth/Chin | high arched palate, cupid bow, dental crowding | thin upper lip, downturned mouth corners, high arched palate, mild retrognathia | very thin upper lip, retrognathia | very thin upper lip, retrognathia | very thin upper lip, retrognathia | − | − | thin upper lip | thin upper lip | high arched palate, dental malocclusion, narrow chin | high arched palate, narrow chin | high arched palate |
| Ears | − | posteriorly rotated ears, bilateral pits | large protruding low-set ears | large protruding low-set ears | large protruding low-set ears | large ears | large ears | large ears | large ears | prominent, dysplastic ears | − | − |
| Scoliosis | + | + | + | − | − | − | + | + | − | − | − | − |
| Extremities | broad thumbs, tapered fingers, long first toes, joint contractures, brachydactyly of other toes, hallux valgus | broad thumbs, bulbous finger tips, forearm asymmetry, feet turned outward, joint contractures overriding toes, planovalgus | broad thumbs, clubbed fingers, clubfoot | broad thumbs, hyperextensibility of interphalangeal joints, overriding toes | broad thumbs, hyperextensibility of interphalangeal joints, overriding toes | broad thumbs and first toes, fetal pads | − | bilateral toe syndactyly | bilateral toe syndactyly | arachnodactyly | hyperextensiblity of elbows | arachnodactyly, hyperextensibility of elbows |
Abbreviations are as follows: IUGR, intrauterine growth restriction; OFC, occipitofrontal circumference; NA, not available; CHD, congenital heart disease; PS, pulmonic stenosis; VSD, ventricular septal defect; ASD, atrioseptal defect; TOF, tetralogy of Fallot; FTT, failure to thrive. For weight and height, data were obtained from last examination, unless otherwise noted. Z-scores were calculated using the CDC charts for children ages 2–20. For head circumference, z-score was calculated based on published data.38
Dysmorphic features were noted in all individuals. Shared facial features included large/protruding ears (n = 8), long philtrum (n = 7), thin upper lip (n = 6), long palpebral fissures (n = 6), high arched palate (n = 5), prominent/high nasal bridge (n = 5), retrognathia (n = 4), and arched eyebrows (n = 3). Abnormalities of fingers and/or toes were also common (n = 10), including broad thumbs (n = 6), overriding toes (n = 3), bulbous finger tips/fetal pads (n = 2), syndactyly (n = 2), arachnodactyly (n = 2), and brachydactyly (n = 1). Other shared features included progressive scoliosis (n = 5), hyperextensible joints (n = 4), joint contractures/clubfoot (n = 3), and sacral dimple/spina bifida occulta (n = 3). Four subjects had congenital heart defects including atrioseptal defect (n = 3), ventricular septal defect (n = 2), pulmonic stenosis (n = 2), and tetralogy of Fallot (n = 1). Four of eight males (50%) had either unilateral or bilateral cryptorchidism.
Not included in the study cohort are two additional deceased affected subjects (II-1 in family 1 and II-3 in family 3) for whom DNA samples were unavailable (Figure 1). Subject II-1 in family 1 had intellectual disability, intractable seizures, poor growth, sacral dimple, progressive scoliosis, and bilateral toe polydactyly. He died at age 12 years due to complications associated with Lennox-Gastaut syndrome. Subject II-3 in family 3 had a congenital heart defect, dysmorphic facial features, and delayed milestones. She died at age 2 years due to recurrent pulmonary infections. Based on the significant overlap of clinical features with those in our study cohort, we suspect that these individuals also carried biallelic pathogenic variants in OTUD6B.
A summary of molecular findings is provided in Figure 1 and Table 2. Four homozygous variants in OTUD6B (GeneBank: NM_016023.3; ClinVar: SCV000492514, SCV000492515, SCV000492516, SCV000492517)—one nonsense, one frameshift, one splice variant, and one missense variant—were identified by WES or whole-genome sequencing (family 6) and subsequently confirmed by Sanger sequencing. Consistent with a recessive mode of inheritance, the unaffected parents in all of the families were heterozygous for the respective familial variants, and all tested unaffected siblings (in families 2 and 4) were either heterozygous for the variant or homozygous for the reference allele. The available affected individuals in families 1, 2, and 3 shared the same homozygous nonsense variant, c.433C>T (p.Arg145∗), located in exon 4 (Figure 2). This variant is annotated in dbSNP 147 (rs368313959) and has a maximum subpopulation allele frequency in the Latino population of 0.0026 (5/1,898 alleles) in the Exome Aggregation Consortium (ExAC) database and 0.00079 (25/31,718) in the Genome Aggregation Database (gnomAD) (Table 2). Therefore, this variant is significantly enriched in our cohort (6 of 12 alleles from 6 families versus 5 of 1,898 reference alleles from the Latino population in ExAC, p = 6.295 × 10−12; Fisher’s exact test). The calculated prevalence of individuals who are homozygous for this variant based on random mating would therefore be approximately 1/145,000 in the Latino population. This variant is not present in the homozygous state in ExAC or gnomAD, but this locus was covered in only 949 and 15,859 Latino individuals in these databases, respectively. Of note, families 1 and 2 were reported to be non-consanguineous, and both are of Hispanic descent. Illumina HumanExome-12v1 (cSNP) array data of the affected subjects from these families showed that OTUD6B lies in a small stretch of absence of heterozygosity (AOH) (5.4 Mb and 7.4 Mb, respectively) in chromosome 8. Also, AOH is seen in multiple chromosomes in these subjects, suggesting that AOH regions likely occurred through identity-by-descent. The affected subjects in families 4, 5, and 6 carried a homozygous frameshift variant (c.469_473delTTAAC [p.Leu157Argfs∗8]), a homozygous splice variant (c.173−2A>G), and a homozygous missense variant (c.647A>G [p.Tyr216Cys]), respectively. These variants were not reported in ExAC or gnomAD.
Table 2.
Variants in OTUD6B
| Families 1,2,3 | Family 4 | Family 5 | Family 6 | |
|---|---|---|---|---|
| Genomic position (hg19) | 8: 92,090,611 | 8: 92,090,647–92,090,651 | 8: 92,083,364 | 8: 92,090,825 |
| Exon/Intron | exon 4 | exon 4 | intron 1 | exon 4 |
| Nucleotide change | c.433C>T | c.469_473delTTAAC | c.173−2A>G | c.647A>G |
| Protein change | p.Arg145∗ | p.Leu157Argfs∗8 | – | p.Tyr216Cys |
| Allele Frequency in ExAC | ||||
| Total | 8/43,688 = 0.00018 | not present | not present | not present |
| Latino | 5/1,898 = 0.0026 | – | – | – |
| African | 1/4,386 = 0.000023 | – | – | – |
| South Asian | 2/8,864 = 0.000023 | – | – | – |
| Number of homozygotes | none | – | – | – |
| Allele Frequency in gnomAD | ||||
| Total | 32/239,902 = 0.00013 | not present | not present | not present |
| Latino | 25/31,718 = 0.00079 | – | – | – |
| African | 1/22,484 = 0.000044 | – | – | – |
| South Asian | 1/25,966 = 0.000039 | – | – | – |
| Number of homozygotes | none | – | – | – |
| SIFT | – | – | – | deleterious |
| PolyPhen2 | – | – | – | probably damaging |
Variant nomenclature is based on GenBank: NM_016023.3. Abbreviations are as follows: ExAC, Exome Aggregation Consortium; gnomAD, Genome Aggregation Database.
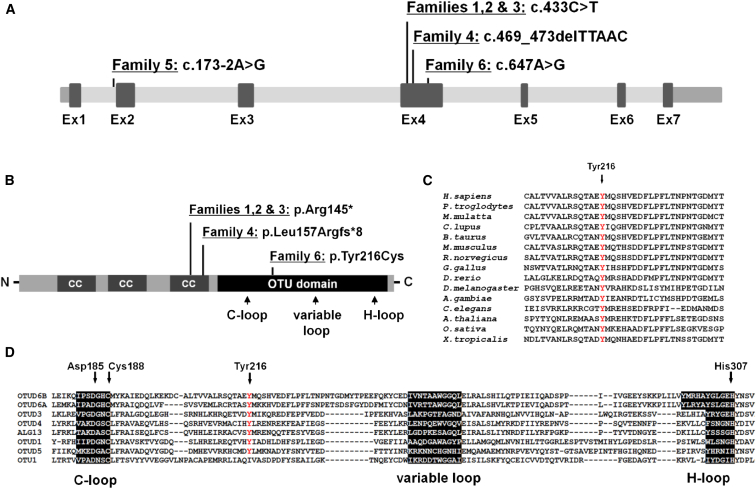
Figure 2.
Location of Variants in OTUD6B and Conservation of the Substituted Tyrosine Residue in Orthologous and Paralogous Proteins
(A) Three of the OTUD6B variants in the described families are located in exon 4 of OTUD6B (GenBank: NM_016023.3, GRCH37/hg19). One is located in a canonical splice acceptor site in intron 1.
(B) Approximate location of amino acid changes relative to key functional domains of the OTUD6B protein (GenBank: NP_057107.3) including three coiled coil domains (CC) and an OTU-like cysteine protease domain. Also shown are the locations of the conserved predicted ubiquitin binding sites within the OTU domain: cysteine loop (C-loop), histidine loop (H-loop), and variable loop.
(C) Clustal Omega alignment of OTUD6B orthologs indicated the substituted tyrosine 216 residue is well conserved across species.
(D) Sequence of the catalytic center of human OTU domain-containing enzymes showing the location of the conserved catalytic residues (Asp185, Cys188, and His307) indicated that tyrosine 216 is also highly conserved across human OTUD enzymes.
OTUD6B maps to chromosome 8q21.3 and encodes a member of the ovarian tumor (OTU) domain-containing subfamily of deubiquitinating enzymes (DUBs). It consists of seven exons that encode a 323-amino acid protein (Figures 2A and 2B). The predicted protein contains several coiled coil (CC) domains and a conserved OTU domain, which is associated with cysteine protease activity. The predicted catalytic residues that function together at the center of the enzyme’s active site are Asp185, Cys188, and His307; the C-loop, variable loop, and H-loop are the predicted ubiquitin-binding regions (Figures 2B and 2D). The location of the variants identified in our cohort in relation to the gene structure is shown in Figure 2. The truncating variants c.433C>T and c.469_473delTTAAC are predicted to lead to nonsense-mediated decay of the mRNA molecule. The c.173−2A>G variant affects an intronic splice acceptor site and is predicted to cause exon 2 skipping and introduction of a premature termination codon. This change is also predicted to result in nonsense-mediated decay of the mRNA molecule. The c.647A>G (p.Tyr216Cys) variant affects a highly evolutionarily conserved tyrosine residue in the OTU domain and is predicted to be deleterious by SIFT and PolyPhen-2 prediction tools (Figure 2C and Table 2). This tyrosine residue is also highly conserved among other OTU domain-containing enzymes and is located between the C-loop and variable loop (Figure 2D). Notably, subjects with predicted loss-of-function alleles (families 1 to 5) have a very severe clinical presentation while subjects with the missense variant (family 6) have a milder phenotype, suggestive of a hypomorphic allele (Table 1).
The International Mouse Phenotyping Consortium (IMPC) is generating a knockout mouse strain for every protein-coding gene in the mouse genome and is employing a standardized, broad-based adult and embryonic phenotyping of the knockout mice to identify gene-to-phenotype associations.6, 7 A C57BL/6N mouse strain harboring a β-galactosidase (lacZ)-tagged, knockout allele of Otud6b, created by deletion of exon 4 (Otud6btm1b(EUCOMM)Wtsi) (Figure 3A), has been produced and phenotyped by the IMPC. Mice homozygous for the Otud6b knockout allele (Otud6btm1b/tm1b) were sub-viable. Out of a total of 97 animals born from heterozygous knockout intercrosses, only 2 homozygous mice (1 male and 1 female) were identified by genotyping tissues collected 1 day after birth, which significantly deviates from the expected Mendelian frequency of 1 wild-type:2 heterozygotes:1 homozygote (p < 1 × 10−5; Table S1). Both homozygotes were found dead at the time of tissue collection, indicating that death occurred on the day of birth. Importantly, the IMPC and others have previously shown that genes causing lethality in mice are enriched for human genes associated with disease.6, 8, 9 Thus, the sub-viability of homozygous mice appears consistent with the severity of OTUD6B loss-of-function variants in humans.
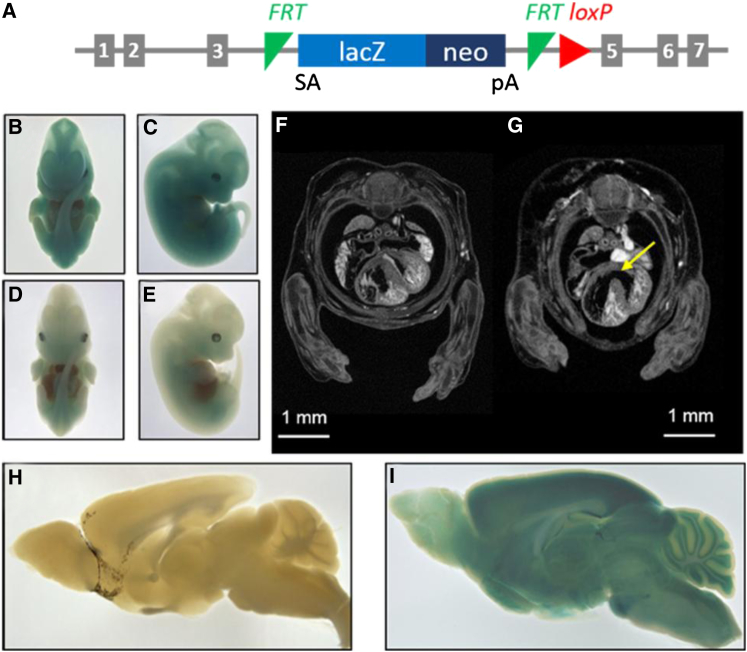
Figure 3.
Phenotypic Effects of Otud6b Deficiency in Mice
(A) Schematic of the Otud6btm1b knockout allele. Exon 4 is deleted and β-galactosidase (lacZ) is expressed following splicing to exon 3 (SA = splice acceptor, pA = polyadenylation signal).
(B–E) LacZ expression, as determined by X-gal staining, from the Otud6btm1b allele is widespread in E12.5 Otud6btm1b/tm1b (B, C) and Otud6btm1b/+ (D, E) embryos.
(F and G) Representative μCT image of E14.5 wild-type (F) and Otud6btm1b/tm1b knockout (G) embryos. The ventricular septal defect in the Otud6btm1b/tm1b knockout embryo is indicated (arrow).
(H and I) Representative X-gal staining images of wild-type (H) and Otud6btm1b/+ (I) adult brains (>50 days of age).
The IMPC performed embryo phenotyping6 to determine when Otud6btm1b/tm1b animals die during embryogenesis, the tissue distribution of lacZ reporter expression in fetal tissue, and the developmental defects that may contribute to sub-viability. Otud6btm1b/tm1b embryos were observed at the expected Mendelian frequencies on embryonic day (E) E18.5 (7 homozygotes in a total of 29 embryos), indicating that Otud6btm1b/tm1b mice die between E18.5 and shortly after birth. Expression of the lacZ reporter was nearly ubiquitous. As reported by the IMPC, lacZ expression was observed in several anatomical locations in Otud6btm1b/tm1b and Otud6btm1b/+ embryos, including the footplate and handplate, oral cavity (mandibular and maxillary processes), liver, lung, somite, tail, ear, eye, brain (forebrain, midbrain, and hindbrain), and heart (Figures 3B–3E and S2A–S2D). Analysis of micro computed tomography (μCT) images indicated that the E18.5 Otud6btm1b/tm1b embryos (n = 2) were smaller (34% reduced total volume, not significant) than wild-type littermates (n = 2) (Figure S3). Interestingly, 7 out of 12 human subjects in our study cohort with biallelic variants in OTUD6B had IUGR (Table 1). Additionally, seven human subjects had failure to thrive, and eight had small stature (Table 1). Furthermore, μCT imaging of Otud6btm1b/tm1b embryos showed ventricular septal defects (VSDs) in 80% of hearts (3 of 3 E14.5 hearts with a VSD; 1 of 2 E18.5 hearts with a VSD) (Figures 3F, 3G, and S4). Blood obscured imaging of the heart in the E18.5 Otud6btm1b/tm1b embryo in which a VSD could not be confirmed. Importantly, VSDs are not a common feature of the C57BL/6N inbred background. Only one VSD has been observed in 150 C57BL/6N embryos analyzed (0.67% incidence). Consistent with the high incidence of VSDs in Otud6btm1b/tm1b mouse embryos, four human subjects in our study cohort had congenital heart defects, two of whom specifically had VSDs (Table 1).
Because homozygotes were sub-viable, heterozygous (Otud6btm1b/+) knockout male and female mice (n = 8 for each sex) were placed into the adult IMPC phenotyping pipeline, and values were compared to C57BL/6N controls. No significant differences in adult phenotypes were reported between Otud6btm1b/+ mice and wild-type controls (using the IMPC suggested p value of 1E-4). However, using the lacZ expression reporter in the Otud6b knockout allele, human disease-relevant expression patterns for Otud6b were identified in Otud6btm1b/+ adult mice. Expression of lacZ was observed in several organ systems including the cardiovascular system (heart and aorta), digestive system (large intestine and stomach), nervous system (spinal cord, peripheral nervous system, pituitary gland, and the brain), and musculoskeletal system (skeletal muscle and cartilage) (data not shown; available on the IMPC website). As reported by the IMPC, within the brain lacZ reporter expression was near ubiquitous, localizing to several anatomical locations including the olfactory bulb, cortex, cerebellum (granular and Purkinje layers with weak LacZ staining in the white matter), brain stem, hypothalamus, midbrain (inferior and superior colliculus), hippocampus (CA1/2/3 and subiculum), striatum, and thalamus (Figures 3H, 3I, and S2E). The widespread expression pattern of Otud6b in adult mice is consistent with a previous study showing Otud6b gene expression in multiple tissues.10 Furthermore, it is consistent with the multiple organ systems affected in the described subjects, including the cardiovascular system (congenital heart defects), nervous system (cognitive disability, seizures, structural brain abnormalities, hypotonia, feeding and digestion issues), musculoskeletal system (scoliosis, abnormalities of the fingers/toes), and digestive system (feeding and digestion issues).
Post-translation modification by ubiquitin is an essential process that occurs in almost all cellular tissues in humans and other eukaryotes. Ubiquitin becomes covalently linked to target proteins by an isopeptide bond involving the ubiquitin C-terminal glycine and ε-amino group of a lysine within the target. Ubiquitination is mediated via the sequential action of a three enzyme conjugation pathway involving an E1-activating enzyme, an E2 conjugating enzyme, and an E3 ligase enzyme.11, 12 Ultimately, the attachment of ubiquitin, or chains of ubiquitin, to a protein substrate creates a signal for the protein’s fate that is primarily associated with degradation via the 26S proteasome but may also result in changes in cellular location, protein activity, and interaction with other proteins.13, 14, 15
Ubiquitination is a reversible process countered by the action of DUBs, which are proteases that specifically cleave ubiquitin isopeptide linkages from protein substrates. Therefore, DUBs can regulate ubiquitin-dependent processes. The human genome encodes approximately 90 DUBs, which are classified into five subfamilies by their sequence diversity: ubiquitin C-terminal hydrolases (UCHs), ubiquitin-specific peptidases (USPs/UBPs), Josephin or Machado-Joseph disease (MJD) proteins, JAMM (Jab1/MPN domain-associated metalloisopeptidase) domain proteins, and lastly, the OTU-domain containing subfamily to which OTUD6B belongs. Several members of the OTU subfamily have been shown to regulate important signaling pathways including NF-κB signaling, interferon signaling, p97-mediated processes, and the DNA damage response.16, 17, 18, 19, 20, 21
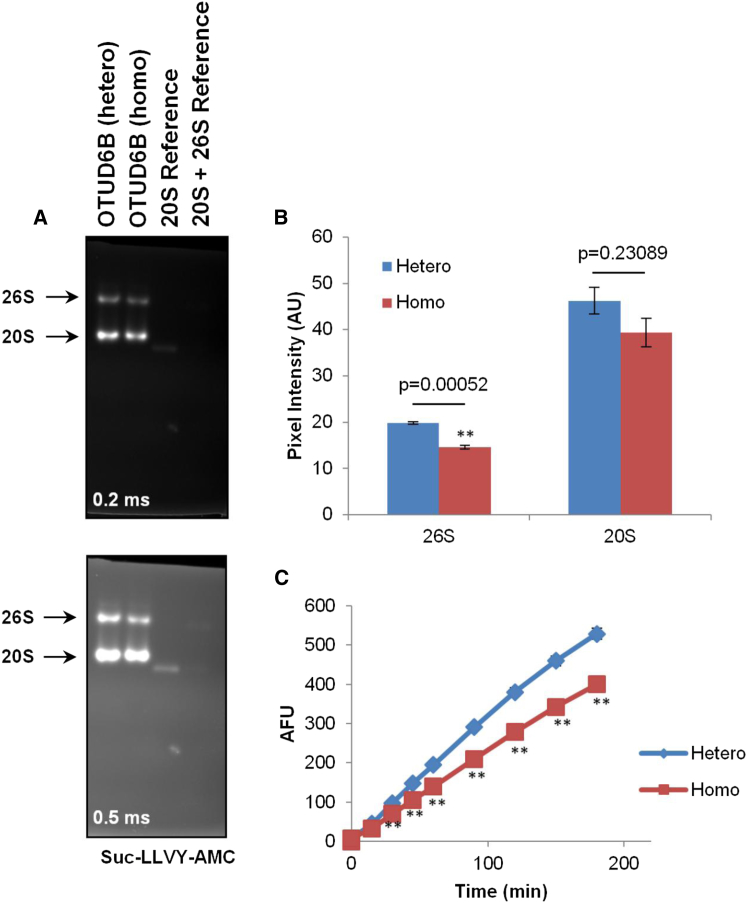
Given the potential role of OTUD6B in removing ubiquitin and/or ubiquitin chains from protein substrates, we reasoned that it may regulate the ubiquitin-proteasome system (UPS). To address the contribution of OTUD6B to the UPS, peripheral blood mononuclear cells (PBMCs) from subjects carrying a heterozygous or a homozygous c.469_473delTTAAC (p.Leu157Argfs∗8) OTUD6B deletion (family 4; subjects I-1 and II-3, respectively) were analyzed for their proteasome content. To this end, proteasome complexes were separated on non-denaturing (native)-PAGE and visualized by their capacity to hydrolyze a Suc-LLVY-AMC fluorogenic substrate. As shown in Figure 4A, both of the 20S and 26S complexes from PBMCs of the homozygous subject exhibited weaker fluorescence intensity than those from the heterozygous subject, indicating that chymotrypsin-like activity was reduced in this sample. Densitometric analysis of the proteasome bands indicated that the intensity of the chymotrypsin-like activity of the 26S proteasome complexes from the homozygous subject was reduced by 20% when compared to those from the heterozygous subject (Figure 4B). Likewise, the activity of the 20S complexes in the homozygous sample was lower than that detected in the heterozygous sample, albeit not significantly. To further confirm these differences, the chymotrypsin-like activity was directly measured in whole-cell extracts by exposing them to 0.1 mM of the Suc-LLVY peptide for various periods of time. As shown in Figure 4C, the 26/20S complexes from the homozygous subject exhibited a decreased capacity to hydrolyze the Suc-LLVY substrate when compared to the same analysis on PBMCs from the heterozygous subject. This difference was statistically significant at all time points. This is in agreement with the data of our above-described, in-gel activity assays and therefore confirms that the homozygous c.469_473delTTAAC OTUD6B deletion compromises the chymotrypsin-like proteasome activity in PBMCs.
Figure 4.
PBMCs from Individual Carrying the Homozygous c.469_473delTTAAC OTUD6B Deletion Exhibit Decreased Chymotrypsin-like Proteasome Activity
(A) Forty micrograms of whole-cell extracts of PBMCs derived from subjects with the heterozygous (hetero) or homozygous (homo) c.469_473delTTAAC OTUD6B deletion were separated by native PAGE, as indicated. Size controls in this experiment consisted of purified blood-derived 20S (20S reference) and a combination of 20S and 26S complexes (reference 20S + 26S) for 600 kDa and 1.2 MDa, respectively. Chymotrypsin-like proteasome activity was examined by incubating the gels with 0.1 mM of Suc-LLVY-AMC for 20 min at 37°C. Two exposure times (0.2 and 0.5 ms) at 360 nm are shown. One representative experiment of three is shown.
(B) Quantification of the 20S and 26S proteasome fluorescent bands from PBMCs with the hetero or homo c.469_473delTTAAC OTUD6B deletion by densitometry using the ImageJ 1.48v software. Shown is the mean pixel intensity calculated from three independent experiments following an exposure time of 0.3 ms. ∗∗p < 0.001 (Student’s t test).
(C) Ten micrograms of whole-cell lysates from PBMCs carrying the hetero or homo OTUD6B deletion were incubated in quadruplicates with 0.1 mM Suc-LLVY in a final volume of 100 μL in non-denaturing conditions on 96-well plates. The fluorescence activity emerging from the release of AMC was measured at 360 nm every 15 min for the first hour, and then every 30 min for the last two hours. ∗∗p < 0.01 (Student’s t test).
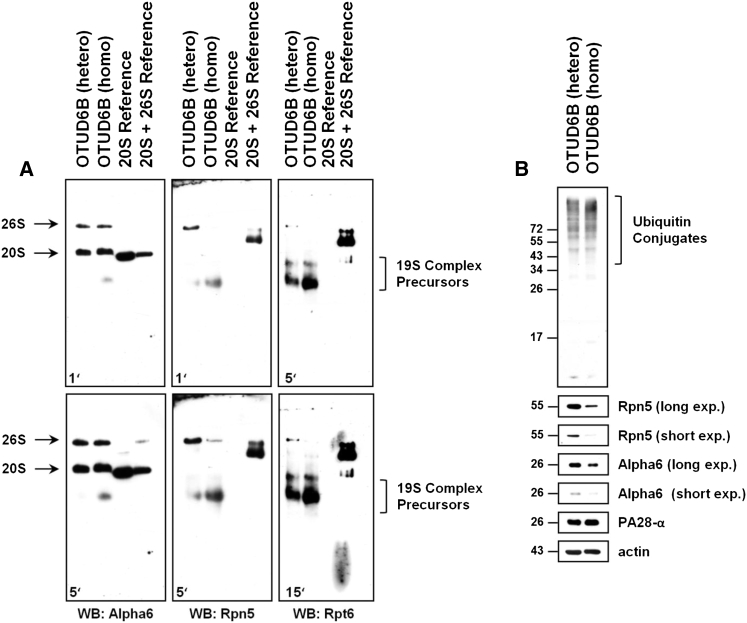
We next tested whether such impaired activity was due to a decreased amount of proteasome complexes in these samples. Protein-cell lysates derived from PBMCs from the heterozygous and homozygous subjects were resolved on native PAGE and subjected to western blotting to detect proteasome subunits: proteasome subunit alpha 6 (PSMA6 or Alpha6), proteasome 26S subunit, non-ATPase 12 (PSMD12 or Rpn5), and proteasome 26S subunit, ATPase 5 (PSMC5 or Rpt6). As illustrated in Figure 5A, probing the membrane with the Alpha6 antibody showed two strongly staining bands in both samples corresponding to the 20S and 26S proteasome complexes in these cells. Strikingly, the incorporation of the Rpn5 and Rpt6 subunits into 26S proteasomes was substantially reduced in the homozygous subject, as shown by decreased intensity of the Rpn5 and Rpt6 bands at the position of the 26S proteasome. These data strongly suggest that the c.469_473delTTAAC OTUD6B homozygous deletion is associated with a 26S proteasome assembly defect. In an attempt to determine whether the c.469_473delTTAAC OTUD6B deletion was a dominant or recessive genetic trait, the 26S proteasome populations from both of the heterozygous and homozygous OTUD6B subjects were then compared to those of OTUD6B wild-type individuals. To this end, proteasome complexes derived from PBMCs of four healthy blood donors were characterized using native-PAGE and western blotting, as described above. As expected, all four wild-type OTUD6B donors exhibit full incorporation of the Rpt6 and Rpn5 subunits in 26S proteasome complexes (Figure S5), confirming that the assembly pathway of the 19S regulatory particle was not compromised in these cells. To enable reliable comparison between wild-type, heterozygous, and homozygous OTUD6B subjects, our 26S reference was used as standard for normalization of the Rpn5 signal intensities emerging from cellular 26S proteasomes between experiments on different gels. Interestingly, our densitometric analysis showed that the relative amounts of Rpn5 in the 26S proteasome complexes was reduced by about 65% and 90% in the OTUD6B heterozygous and homozygous subjects, respectively, when compared with those of the OTUD6B wild-type controls (Figure S5), indicating that impaired proteasome assembly begins with the disruption of either of the two OTUD6B copies. This notion is fully in line with the observation that 19S precursor complexes accumulate in both of the heterozygous and homozygous subjects (Figure 5A) but not in wild-type controls (Figure S5). The lack of visible phenotype in the OTUD6B heterozygous subject suggests the existence of a compensation mechanism.
Figure 5.
The Homozygous c.469_473delTTAAC OTUD6B Deletion Results in Decreased Amounts of 26S Proteasomes and Increased Accumulation of Ubiquitin-Protein Conjugates in PBMCs
(A) Forty micrograms of whole-cell extracts from PBMCs from subjects with the hetero or homo c.469_473delTTAAC OTUD6B deletion were resolved on native PAGE and subjected to western blotting using antibodies to Alpha6 (clone MCP20, Enzo Life Sciences), Rpn5 (clone H-9, Santa Cruz Biotechnology), and Rpt6 (clone p45-110, Enzo Life Sciences), as indicated. Controls consisted of 20S and 20S + 26S references, as described in Figure 4.
(B) Ten micrograms of protein lysates were separated by 15% SDS-PAGE followed by western blotting using antibodies specific for ubiquitin (ref #Z0458, DAKO GmbH), Rpn5 (clone H-9, Santa Cruz), Alpha6 (clone MCP20, Enzo Life Sciences), and PA28-α (K232/1, laboratory stock). Equal protein loading was ensured by probing the membranes with the anti-actin monoclonal antibody.
To further evaluate the functional significance of the c.469_473delTTAAC OTUD6B deletion, we next analyzed the cellular content of the ubiquitin-modified proteins in PBMCs in both of the heterozygous and homozygous subjects by SDS-PAGE. As shown in Figure 5B, the ubiquitin-protein conjugates accumulated much stronger in the homozygous sample than in the heterozygous one. This finding is in agreement with our previous observation that 26S proteasomes from the homozygous subject are defective. Interestingly, our SDS-PAGE analysis further confirmed a decrease in the total amount of the Rpn5 19S subunit and to a lesser extent of the Alpha6 subunit in PBMCs from the homozygous subject. By contrast, the steady-state expression level of the proteasome activator PA28-α remains unaffected in these cells. Interestingly, comparing both of the heterozygous and homozygous subjects with wild-type controls showed that the loss of one OTUD6B copy is sufficient to dramatically affect protein homeostasis, as evidenced by the accumulation of ubiquitin-protein conjugates in the OTUD6B heterozygous subject (Figure S6). Importantly, the increased amount of ubiquitin-modified proteins in subjects with the OTUD6B heterozygous and homozygous variant was paralleled by a substantial reduction of the steady-state expression level of the Rpt6 subunit in both of these samples (Figure S6). However, the expression level of the Rpn5 subunit was decreased in the homozygous OTUD6B subject but not in the heterozygous one, as determined by western blotting. Altogether, these data suggest that the c.469_473delTTAAC OTUD6B deletion results in proteasome malfunction whose severity proportionally increases with the number of affected alleles.
In this work, we identified the homozygous c.469_473delTTAAC OTUD6B deletion as a new member of the growing family of genetic alterations specifically affecting proteasome function.22, 23, 24, 25, 26 To the best of our knowledge, the alteration described in this paper is the first one occurring outside the genes encoding the proteasome maturation protein (POMP) or any of seven alpha and/or beta proteasome subunits. The cellular loss-of-function phenotype of OTUD6B in the homozygous subject was characterized by a substantially reduced incorporation of 19S subunits into 26S proteasomes (Figure 5A) as well as decreased steady-state expression levels of 26S proteasome subunits (Figure 5B). Accordingly, this was accompanied by a drop of the chymotrypsin-like activity (Figure 4) and a concomitant accumulation of ubiquitin-protein conjugates (Figure 5B). It remains unclear whether the defect in 26S proteasome assembly detected in our native PAGE analysis is the cause or the consequence of the overall decreased expression of the proteasome subunits observed in SDS-PAGE. The accumulation of 19S precursor complexes in the homozygous subject suggests that the 19S subunits are not a rate-limiting factor for the formation of 26S proteasome complexes. It is therefore highly likely that the decreased steady-state levels of the proteasome subunits may reflect a higher susceptibility of these subunits to degradation due to their failure to incorporate into 26S proteasomes.
The precise mechanisms by which OTUD6B affects 26S proteasome assembly are ill defined and open unexpected avenues to understanding the regulation of protein homeostasis by the UPS. Given the described role of OTUD6B as a DUB, one can speculate that the formation of 26S complexes is a process regulated by ubiquitin modification. This notion would be in agreement with previous studies reporting the ubiquitination of various proteasome subunits including Rpn10, Rpn13, or Rpt5.27, 28, 29 As such, it is conceivable that any impaired de-ubiquitination of these subunits might impact proteasome assembly and/or function. In this matter, further experiments will attempt to address the capacity of OTUD6B to remove ubiquitin from proteasome subunits.
Interestingly, by transmission electron microscopy, we identified abnormal cytoplasmic inclusions in lymphocytes from subject II-2 in family 2 (Figure S7). We speculate that these inclusions represent accumulation of protein substrates due to an imbalance in ubiquitination/deubiquitination activities in these cells. Although this study was performed in only one subject and only in lymphocytes, it raises the possibility that the inclusions might serve as a biomarker for this syndrome.
Many DUBs have been implicated in human diseases such as neurodegeneration, inflammation, infection, and cancer. For example, pathogenic variants in the ubiquitin-specific protease genes USP9X (MIM: 300072) and USP27X (MIM: 300975) cause X-linked intellectual disability (MIM: 300919, 300968, 300984). De novo loss-of-function variants in USP7 (MIM: 602519) cause a neurodevelopmental disorder.30 Trinucleotide repeat expansion in ATXN3 (MIM: 607047) causes Machado-Joseph disease (MIM: 109150), a neurologic disorder characterized by ataxia, spasticity, and ocular movement abnormalities. Heterozygous truncating variants in A20/TNFAIP3 (MIM: 191163) cause familial Behcet-like auto-inflammatory syndrome (MIM: 616744) and biallelic pathogenic variants in OTULIN (MIM: 615712) cause autoinflammation, panniculitis, and dermatosis syndrome (MIM: 617099). Germline variants in BAP1 (MIM: 603089) and CYLD (MIM: 605018) can cause tumor predisposition syndromes (MIM: 614327, 132700). Maternal germline variants and de novo events in the human OTU domain-containing enzyme ALG13 (MIM: 300776) cause early infantile epileptic encephalopathy 36 (MIM: 300884). Similarly, many ubiquitin ligase genes have been implicated in human diseases.31, 32, 33, 34, 35, 36, 37 For example, defects in the UBR1 (MIM: 605981), MID1 (MIM: 300552), MID2 (MIM: 300204), and CUL4B (MIM: 300304) have been associated with neurodevelopmental disorders.31, 32, 33, 34 Overall, this demonstrates the importance of the ubiquitination/deubiquitination process in human cells and that its disruption can lead to disease.
In summary, we describe a severe intellectual disability syndrome caused by biallelic loss-of-function variants in the deubiquitinating enzyme gene OTUD6B. This syndrome is characterized by cognitive dysfunction with absent speech, seizures, microcephaly, hypotonia, growth retardation, dysmorphism, and other variable features. We also describe a milder phenotype in humans with mild to moderate intellectual disability, seizures, and dysmorphic features caused by a homozygous missense variant in OTUD6B that is presumed to represent a hypomorphic allele. Otud6b knockout mice display several of the features seen in human subjects with OTUD6B loss-of-function variants and demonstrate the power of the International Mouse Phenotyping Consortium as a resource for functional validation of novel disease genes. Finally, in vitro analyses implicate OTUD6B in proteasome function, thereby strengthening the notion that dysregulation of the ubiquitin system is causative of human disease.
Consortia
The members of the Autosomal Recessive working group of the EuroEPINOMICS RES Consortium are Zaid Afawi, Rudi Balling, Nina Barisic, Stéphanie Baulac, Dana Craiu, Peter De Jonghe, Rosa Guerrero-Lopez, Renzo Guerrini, Ingo Helbig, Helle Hjalgrim, Johanna Jähn, Karl Martin Klein, Eric Leguern, Holger Lerche, Carla Marini, Hiltrud Muhle, Felix Rosenow, José Serratosa, Katalin Sterbová, Arvid Suls, Rikke S. Moller, Pasquale Striano, Yvonne Weber, and Federico Zara.
Acknowledgments
We thank the subjects and their families for participating in our research study. This work was supported by NIH K08DK106453 (L.C.B.), NIH U42OD011174 and NIH U54HG006348 (J.D.H., M.E.D., J.R.S., S. Wells, S.J.J., L.T.), NIH T32GM007526-39 (M.J.T.), the Trudy Mandel Louis Charitable Trust (O.E.), and the International Coordination Action grant G0E8614N (S. Weckhuysen, K.H.). The Eurocores program of the European Science Foundation supported the EuroEPINOMICS-RES network (G.A.136.11.N, FWO/ESF-ECRP, and HE5415/3-1). S.H. Caglayan is granted by the TUBITAK project no 110S518 within the Euroepinomics-RES network. The Family Genomics group at the Institute for Systems Biology deserves thanks for their support and project management of the whole-genome sequencing data (family 6). We acknowledge the contribution of Peter De Rijk (University of Antwerp) and the HPC facilities of the University of Luxembourg for computational support. The Department of Molecular and Human Genetics at the Baylor College of Medicine derives revenue from molecular genetic testing offered at the Baylor Miraca Genetics Laboratories. Since mid-October 2015, K.H. is under employment of UCB Pharma (Braine-l’Alleud, Belgium). The company had no part in this study.
Published: March 23, 2017
Footnotes
Supplemental Data include Supplemental Note, seven figures, and one table and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2017.03.001.
Contributor Information
Jason D. Heaney, Email: heaney@bcm.edu.
Magdalena A. Walkiewicz, Email: mwalkiew@bcm.edu.
EuroEPINOMICS RES Consortium Autosomal Recessive working group, S. Hande Caglayan:
Zaid Afawi, Rudi Balling, Nina Barisic, Stéphanie Baulac, Dana Craiu, Peter De Jonghe, Rosa Guerrero-Lopez, Renzo Guerrini, Ingo Helbig, Helle Hjalgrim, Johanna Jähn, Karl Martin Klein, Eric Leguern, Holger Lerche, Carla Marini, Hiltrud Muhle, Felix Rosenow, José Serratosa, Katalin Sterbová, Arvid Suls, Rikke S. Moller, Pasquale Striano, Yvonne Weber, and Federico Zara
Web Resources
Clustal Omega, http://www.ebi.ac.uk/Tools/msa/clustalo/
ExAC Browser, http://exac.broadinstitute.org/
gnomAD Browser, http://gnomad.broadinstitute.org/
Growth Charts, http://www.cdc.gov/growthcharts/clinical_charts.htm
HPC, https://hpc.uni.lu/
International Mouse Phenotyping Consortium, http://www.mousephenotype.org/
OMIM, http://www.omim.org/
UniProt, http://www.uniprot.org/
Supplemental Data
References
- 1.Yang Y., Muzny D.M., Reid J.G., Bainbridge M.N., Willis A., Ward P.A., Braxton A., Beuten J., Xia F., Niu Z. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N. Engl. J. Med. 2013;369:1502–1511. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang Y., Muzny D.M., Xia F., Niu Z., Person R., Ding Y., Ward P., Braxton A., Wang M., Buhay C. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA. 2014;312:1870–1879. doi: 10.1001/jama.2014.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee H., Deignan J.L., Dorrani N., Strom S.P., Kantarci S., Quintero-Rivera F., Das K., Toy T., Harry B., Yourshaw M. Clinical exome sequencing for genetic identification of rare Mendelian disorders. JAMA. 2014;312:1880–1887. doi: 10.1001/jama.2014.14604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sawyer S.L., Hartley T., Dyment D.A., Beaulieu C.L., Schwartzentruber J., Smith A., Bedford H.M., Bernard G., Bernier F.P., Brais B., FORGE Canada Consortium. Care4Rare Canada Consortium Utility of whole-exome sequencing for those near the end of the diagnostic odyssey: time to address gaps in care. Clin. Genet. 2016;89:275–284. doi: 10.1111/cge.12654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dickinson M.E., Flenniken A.M., Ji X., Teboul L., Wong M.D., White J.K., Meehan T.F., Weninger W.J., Westerberg H., Adissu H., International Mouse Phenotyping Consortium. Jackson Laboratory. Infrastructure Nationale PHENOMIN, Institut Clinique de la Souris (ICS) Charles River Laboratories. MRC Harwell. Toronto Centre for Phenogenomics. Wellcome Trust Sanger Institute. RIKEN BioResource Center High-throughput discovery of novel developmental phenotypes. Nature. 2016;537:508–514. doi: 10.1038/nature19356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koscielny G., Yaikhom G., Iyer V., Meehan T.F., Morgan H., Atienza-Herrero J., Blake A., Chen C.K., Easty R., Di Fenza A. The International Mouse Phenotyping Consortium Web Portal, a unified point of access for knockout mice and related phenotyping data. Nucleic Acids Res. 2014;42:D802–D809. doi: 10.1093/nar/gkt977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Georgi B., Voight B.F., Bućan M. From mouse to human: evolutionary genomics analysis of human orthologs of essential genes. PLoS Genet. 2013;9:e1003484. doi: 10.1371/journal.pgen.1003484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dickerson J.E., Zhu A., Robertson D.L., Hentges K.E. Defining the role of essential genes in human disease. PLoS ONE. 2011;6:e27368. doi: 10.1371/journal.pone.0027368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu Z., Zheng Y., Zhu Y., Kong X., Hu L. Evidence for OTUD-6B participation in B lymphocytes cell cycle after cytokine stimulation. PLoS ONE. 2011;6:e14514. doi: 10.1371/journal.pone.0014514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pickart C.M. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 12.Schulman B.A., Harper J.W. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat. Rev. Mol. Cell Biol. 2009;10:319–331. doi: 10.1038/nrm2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glickman M.H., Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 14.Mukhopadhyay D., Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007;315:201–205. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- 15.Schnell J.D., Hicke L. Non-traditional functions of ubiquitin and ubiquitin-binding proteins. J. Biol. Chem. 2003;278:35857–35860. doi: 10.1074/jbc.R300018200. [DOI] [PubMed] [Google Scholar]
- 16.Ernst R., Mueller B., Ploegh H.L., Schlieker C. The otubain YOD1 is a deubiquitinating enzyme that associates with p97 to facilitate protein dislocation from the ER. Mol. Cell. 2009;36:28–38. doi: 10.1016/j.molcel.2009.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu H., Brittain G.C., Chang J.H., Puebla-Osorio N., Jin J., Zal A., Xiao Y., Cheng X., Chang M., Fu Y.X. OTUD7B controls non-canonical NF-κB activation through deubiquitination of TRAF3. Nature. 2013;494:371–374. doi: 10.1038/nature11831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hymowitz S.G., Wertz I.E. A20: from ubiquitin editing to tumour suppression. Nat. Rev. Cancer. 2010;10:332–341. doi: 10.1038/nrc2775. [DOI] [PubMed] [Google Scholar]
- 19.Kayagaki N., Phung Q., Chan S., Chaudhari R., Quan C., O’Rourke K.M., Eby M., Pietras E., Cheng G., Bazan J.F. DUBA: a deubiquitinase that regulates type I interferon production. Science. 2007;318:1628–1632. doi: 10.1126/science.1145918. [DOI] [PubMed] [Google Scholar]
- 20.Keusekotten K., Elliott P.R., Glockner L., Fiil B.K., Damgaard R.B., Kulathu Y., Wauer T., Hospenthal M.K., Gyrd-Hansen M., Krappmann D. OTULIN antagonizes LUBAC signaling by specifically hydrolyzing Met1-linked polyubiquitin. Cell. 2013;153:1312–1326. doi: 10.1016/j.cell.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakada S., Tai I., Panier S., Al-Hakim A., Iemura S., Juang Y.C., O’Donnell L., Kumakubo A., Munro M., Sicheri F. Non-canonical inhibition of DNA damage-dependent ubiquitination by OTUB1. Nature. 2010;466:941–946. doi: 10.1038/nature09297. [DOI] [PubMed] [Google Scholar]
- 22.Brehm A., Liu Y., Sheikh A., Marrero B., Omoyinmi E., Zhou Q., Montealegre G., Biancotto A., Reinhardt A., Almeida de Jesus A. Additive loss-of-function proteasome subunit mutations in CANDLE/PRAAS patients promote type I IFN production. J. Clin. Invest. 2015;125:4196–4211. doi: 10.1172/JCI81260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brehm A., Krüger E. Dysfunction in protein clearance by the proteasome: impact on autoinflammatory diseases. Semin. Immunopathol. 2015;37:323–333. doi: 10.1007/s00281-015-0486-4. [DOI] [PubMed] [Google Scholar]
- 24.Kitamura A., Maekawa Y., Uehara H., Izumi K., Kawachi I., Nishizawa M., Toyoshima Y., Takahashi H., Standley D.M., Tanaka K. A mutation in the immunoproteasome subunit PSMB8 causes autoinflammation and lipodystrophy in humans. J. Clin. Invest. 2011;121:4150–4160. doi: 10.1172/JCI58414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arima K., Kinoshita A., Mishima H., Kanazawa N., Kaneko T., Mizushima T., Ichinose K., Nakamura H., Tsujino A., Kawakami A. Proteasome assembly defect due to a proteasome subunit beta type 8 (PSMB8) mutation causes the autoinflammatory disorder, Nakajo-Nishimura syndrome. Proc. Natl. Acad. Sci. USA. 2011;108:14914–14919. doi: 10.1073/pnas.1106015108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kunimoto K., Kimura A., Uede K., Okuda M., Aoyagi N., Furukawa F., Kanazawa N. A new infant case of Nakajo-Nishimura syndrome with a genetic mutation in the immunoproteasome subunit: an overlapping entity with JMP and CANDLE syndrome related to PSMB8 mutations. Dermatology (Basel) 2013;227:26–30. doi: 10.1159/000351323. [DOI] [PubMed] [Google Scholar]
- 27.Isasa M., Katz E.J., Kim W., Yugo V., González S., Kirkpatrick D.S., Thomson T.M., Finley D., Gygi S.P., Crosas B. Monoubiquitination of RPN10 regulates substrate recruitment to the proteasome. Mol. Cell. 2010;38:733–745. doi: 10.1016/j.molcel.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacobson A.D., MacFadden A., Wu Z., Peng J., Liu C.W. Autoregulation of the 26S proteasome by in situ ubiquitination. Mol. Biol. Cell. 2014;25:1824–1835. doi: 10.1091/mbc.E13-10-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cohen-Kaplan V., Livneh I., Avni N., Fabre B., Ziv T., Kwon Y.T., Ciechanover A. p62- and ubiquitin-dependent stress-induced autophagy of the mammalian 26S proteasome. Proc. Natl. Acad. Sci. USA. 2016;113:E7490–E7499. doi: 10.1073/pnas.1615455113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hao Y.H., Fountain M.D., Jr., Fon Tacer K., Xia F., Bi W., Kang S.H., Patel A., Rosenfeld J.A., Le Caignec C., Isidor B. USP7 acts as a molecular rheostat to promote WASH-dependent endosomal protein recycling and is mutated in a human neurodevelopmental disorder. Mol. Cell. 2015;59:956–969. doi: 10.1016/j.molcel.2015.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geetha T.S., Michealraj K.A., Kabra M., Kaur G., Juyal R.C., Thelma B.K. Targeted deep resequencing identifies MID2 mutation for X-linked intellectual disability with varied disease severity in a large kindred from India. Hum. Mutat. 2014;35:41–44. doi: 10.1002/humu.22453. [DOI] [PubMed] [Google Scholar]
- 32.Quaderi N.A., Schweiger S., Gaudenz K., Franco B., Rugarli E.I., Berger W., Feldman G.J., Volta M., Andolfi G., Gilgenkrantz S. Opitz G/BBB syndrome, a defect of midline development, is due to mutations in a new RING finger gene on Xp22. Nat. Genet. 1997;17:285–291. doi: 10.1038/ng1197-285. [DOI] [PubMed] [Google Scholar]
- 33.Zenker M., Mayerle J., Lerch M.M., Tagariello A., Zerres K., Durie P.R., Beier M., Hülskamp G., Guzman C., Rehder H. Deficiency of UBR1, a ubiquitin ligase of the N-end rule pathway, causes pancreatic dysfunction, malformations and mental retardation (Johanson-Blizzard syndrome) Nat. Genet. 2005;37:1345–1350. doi: 10.1038/ng1681. [DOI] [PubMed] [Google Scholar]
- 34.Zou Y., Liu Q., Chen B., Zhang X., Guo C., Zhou H., Li J., Gao G., Guo Y., Yan C. Mutation in CUL4B, which encodes a member of cullin-RING ubiquitin ligase complex, causes X-linked mental retardation. Am. J. Hum. Genet. 2007;80:561–566. doi: 10.1086/512489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kishino T., Lalande M., Wagstaff J. UBE3A/E6-AP mutations cause Angelman syndrome. Nat. Genet. 1997;15:70–73. doi: 10.1038/ng0197-70. [DOI] [PubMed] [Google Scholar]
- 36.Tomaić V., Banks L. Angelman syndrome-associated ubiquitin ligase UBE3A/E6AP mutants interfere with the proteolytic activity of the proteasome. Cell Death Dis. 2015;6:e1625. doi: 10.1038/cddis.2014.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kitada T., Asakawa S., Hattori N., Matsumine H., Yamamura Y., Minoshima S., Yokochi M., Mizuno Y., Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 38.Rollins J.D., Collins J.S., Holden K.R. United States head circumference growth reference charts: birth to 21 years. J. Pediatr. 2010;156:907–913. doi: 10.1016/j.jpeds.2010.01.009. 913.e1–913.e2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.