Abstract
The CD40 ligand (CD40L/CD154), a member of TNF superfamily, is notably expressed on activated CD4+ T-cells and stimulated platelets. CD40L is linked to a variety of pathologies and to acute transfusion reactions (ATR). Mutations in this gene (CD40LG) lead to X-linked hyper-IgM syndrome. Some CD40LG polymorphisms are associated with variable protein expression. The rationale behind this study is that CD40L protein has been observed to be involved in ATR. We wondered whether genetic polymorphisms are implicated. We investigated genetic diversity in the CD40LG using DHPLC and capillary electrophoresis for screening and genotyping (n = 485 French and Tunisian blood donors). We identified significant difference in the CD40LG linkage pattern between the two populations. Variant minor alleles were significantly over-represented in Tunisian donors (P<0.0001 to 0.0270). We found higher heterogeneity in the Tunisian, including three novel low frequency variants. As there was not a particular pattern of CD40LG in single apheresis donors whose platelet components induced an ATR, we discuss how this information may be useful for future disease association studies on CD40LG.
The CD40 ligand gene (CD40LG), located on the long arm of human X chromosome at position q26.3-q27.1, harbors five exons spanning about 12 kilo base pairs (kb) (http://www.ncbi.nlm.nih.gov/gene/959). The CD40L (CD154), which is a member of the tumor necrosis factor (TNF) superfamily, is a 33 kDa type II membrane glycoprotein. It is mostly expressed on activated CD4+ T-cells and stimulated platelets; presence of this protein is often a characterizing feature of antigen presenting cells1,2,3. CD40L is also expressed on non-leukocyte and non-immune cells such as endothelial cells4. CD40L expression is post-transcriptionally regulated in part through mRNA stability5.
The costimulatory molecule CD40L and its receptor CD40 have essential roles in adaptive immunity; it also plays a role in inflammatory responses, which are important in innate immunity6. The normal interaction between T and B lymphocytes via CD40 and CD40L induces B cell activation, proliferation, differentiation, survival and immunoglobulin isotype switching, thereby regulating B cell commitment to mature plasma or memory B cells3,7. Deficient CD40L expression might induce a reduced antigen response, such as that seen in X-linked hyper-IgM syndrome (HIGM1)8, whereas polymorphisms in this ligand have been associated with various pathologies as autoimmune and infectious diseases9,10. CD40L is cryptic in unstimulated platelets, but is translocated to the cell surface within seconds or minutes after in vitro activation, where it is involved in in vivo thrombus formation11. CD40L expressed on the surface of activated platelets interacts with CD40 to trigger inflammatory responses and expression of tissue factor in endothelial cells12. Approximately 95% of the soluble fragment of CD40L (sCD40L) found in plasma is derived and cleaved from platelets, which are important players in inflammation13, in addition to their roles in hemostasis, thrombosis and platelet regulatory functions14,15,16. The contribution of platelets and their secretory products has been observed in tissue pathology17,18.
Interaction between platelet-derived CD40L and target cells such as a blood transfusion recipient's B-lymphocytes and vascular endothelial cells is considered a highly inflammatory process in transfusions, where it is known to be responsible for adverse events, such as febrile non hemolytic transfusion reactions (FNHTR), atypical allergy and hypotension. Such reactions can be benign or severe signs of inflammation, while others such as ATR and transfusion-related acute lung injury (TRALI) are generally thought to result, at least in part, from elevated sCD40L levels19,20,21,22,23.
However, the vast majority of transfusions, despite inducing high levels of pro-inflammatory molecules, proceed without harm to the patient. This fact gives rise to the hypothesis of genetic susceptibility in the donor population relating to cytokines and/or chemokines and in recipients to the relevant receptor involved (both in physiology and pathophysiology). For this, we wondered whether genetic CD40LG polymorphisms are implicated.
To determine the dispersion of genetic variation in populations that are distinct but have certain common ancestries, we sought to examine CD40LG among populations of volunteer blood donors, in central Tunisia and in metropolitan France (whose platelet component induced or not ATR). We hypothesized that variability within a population would reveal high levels of genetic polymorphism or segregation of particular haplotypes24,25.
Denaturing High Performance Liquid Chromatography (DHPLC) and capillary electrophoresis were performed to analyze polymorphisms in the CD40LG, and to estimate the allele frequencies and CD40LG haplotype structures and patterns of linkage disequilibrium (LD) around a 17 kb region that includes the CD40LG. This study, besides, aimed to provide population genetic data that could be used for future studies on the CD40LG and its association with pathology.
Results
Nucleotide polymorphism analysis
DHPLC analysis found nine SNPs and two variable number tandem repeats (VNTRs) in 10 of the 11 amplicons investigated herein. Homogeneity was tested for males and females in each population (Supplementary Table S1). All genotype frequencies were in Hardy-Weinberg Equilibrium (HWE) in the studied populations for all of the typed SNPs. Table 1 summarizes the allele frequencies of the 11 variants identified.
Table 1. Minor allele frequencies of CD40LG SNPs in the French (n = 211) and Tunisian population (n = 274) identified by DHPLC.
| Minor Allele Frequency (MAF) | Statistic | |||||||
|---|---|---|---|---|---|---|---|---|
| SNP | Marker ID | Position Chr X (Location) | Allele | French population | Tunisian Population | χ2 | p value | PCR Fragment |
| 1 | c.-3525_-3526insCAAACAAAa | 135726882 (5' UTR) | (CAAA)8 | 0.003 | 0.003 | 0.017 | 1b | CD40LG-5'UTR |
| 2 | rs201992677 [-/CAAA] | 135726882 (5' UTR) | (CAAA)7 | 0.175 | 0.364 | 28.141 | <0.0001 | CD40LG-5'UTR |
| 3 | rs3092952 A>G | 135726950 (5'UTR) | G | 0.175 | 0.354 | 25.040 | <0.0001 | CD40LG-5'UTR |
| 4 | rs1126535 T>C | 135730555 (exon 1) | C | 0.154 | 0.224 | 4.998 | 0.027 | CD40LG-E1 |
| 5 | rs147739883 C>T | 135732387 (IVS 1) | T | 0 | 0.003 | 0.828 | 1b | CD40LG-E2 |
| 6 | c.8140A>Ga | 135738547 (exon 4) | G | 0 | 0.003 | 0.828 | 1b | CD40LG-E4 |
| 7 | rs3092923 T>C | 135741185 (IVS 4) | C | 0.116 | 0.326 | 39.424 | <0.0001 | CD40LG-E5A |
| 8 | rs148594123 G>A | 135741443 (exon 5) | A | 0.027 | 0 | 9.793 | 0.0002b | CD40LG-E5B |
| 9 | rs3092921 C>T | 135743000 (3'UTR) | T | 0.113 | 0.314 | 37.392 | <0.0001 | CD40LG-3'UTRB |
| 10 | c.*2367C>Ga | 135743941 (3'UTR) | G | 0 | 0.003 | 0.828 | 1b | CD40LG-3'UTRC |
| 11 | rs3092920 G>T | 135743991 (3'UTR) | T | 0.103 | 0.286 | 33.203 | <0.0001 | CD40LG-3'UTRC |
aNovel mutations identified in this study.
bFisher Exact Test (n<5).
Alleles are denoted as ancestral/derived and are referred to by their dbSNP ID. Polymorphisms in boldface were included in the LD and haplotype analysis.
All the amplicons contained an average of one SNP. However, CD40LG-5′UTR and CD40LG-3′UTR harbored 3 polymorphisms (Table 1). At the genotyping stage, we observed that 11% of the samples from females that displayed one peak at the screening analysis, showed multiple peaks when mixed with the wild homozygous control sample, thus presenting the mutant homozygous state.
Three variants were novel: a non-synonymous c.8140A>G in exon 4, c.*2367C>G in the 3′UTR in the Tunisian population at low frequency (MAF = 0.003 for both), and one insertion in the 5′UTR (c.-3525_-3526insCAAACAAA) in one male from each of the study populations. Seven polymorphisms were transitions, three were A/G and four C/T, while two were transversions (one C/G, one G/T). Eight of the 11 variants were previously reported in the NCBI database. Only three variants were located in the coding region: rs1126535 in exon 1, c.8140 A>G in exon 4 and rs148594123 in exon 5. The others were in the non-coding region with three variants in the 5′UTR, one in IVS4 and three in the 3′UTR (Table 1; Supplementary Figure S1).
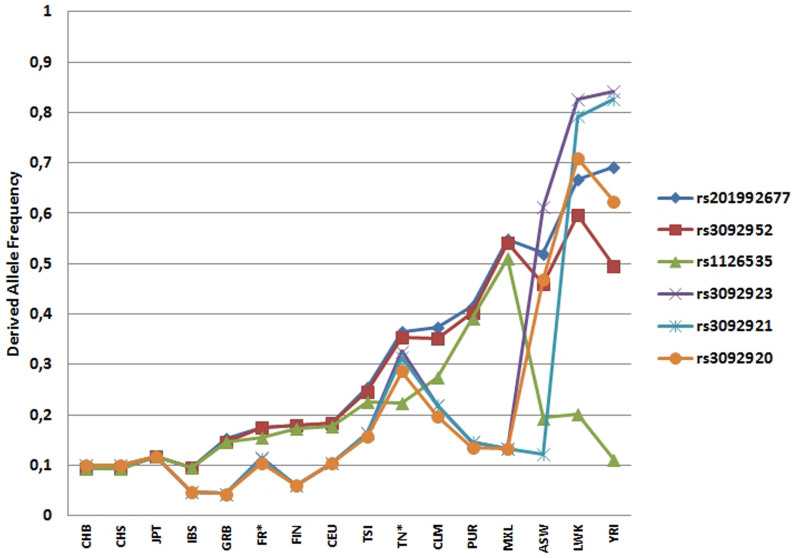
Variant minor alleles with MAF>0.05 were significantly over-represented in the Tunisian individuals (P<0.0001 to 0.027), in comparison with the French individuals (Table 1). In addition, we compared MAFs for the two populations with those extracted from the 1000 genomes project (Figure 1). In the present French population, all the allele frequencies were close to those belonging to the Utah residents with Northern and Western European ancestry (CEU) population (Figure 2). Regarding the Tunisian study, all the allele frequencies of the SNPs fell between those belonging to African ancestry (African Ancestry in Southwest US, ASW; Luhya in Webuye, Kenya, LWK; Africans “Yoruban in Ibadan”, Nigeria, YRI) and those with European origins, the CEU group, British from England and Scotland (GBR) and Tuscan in Italy (TSI), with the exception of rs1126535 for which the frequency is very close to that of TSI.
Figure 1. Derived allele frequencies for six variants with minor allele frequencies in the CD40LG from French (FR) and Tunisian (TN) donors and those extracted from the 1000 genomes database.
African Ancestry in Southwest US (ASW), Han Chinese in Beijing (CHB), Han Chinese South (CHS), Japanese individuals in Tokyo (JPT), Iberian populations in Spain (IBS), British from England and Scotland (GBR), Finnish from Finland (FIN), Utah residents with Northern and Western European ancestry (CEU), Tuscan in Italy (TSI), Colombian in Medellin (CLM), Puerto Rican in Puerto Rico (PUR), Mexican Ancestry in Los Angeles, CA (MXL), African Ancestry in Southwest US (ASW), Luhya in Webuye, Kenya (LWK), Africans “Yoruban in Ibadan”, Nigeria (YRI) taken from the 1000 genomes project. *Present study.
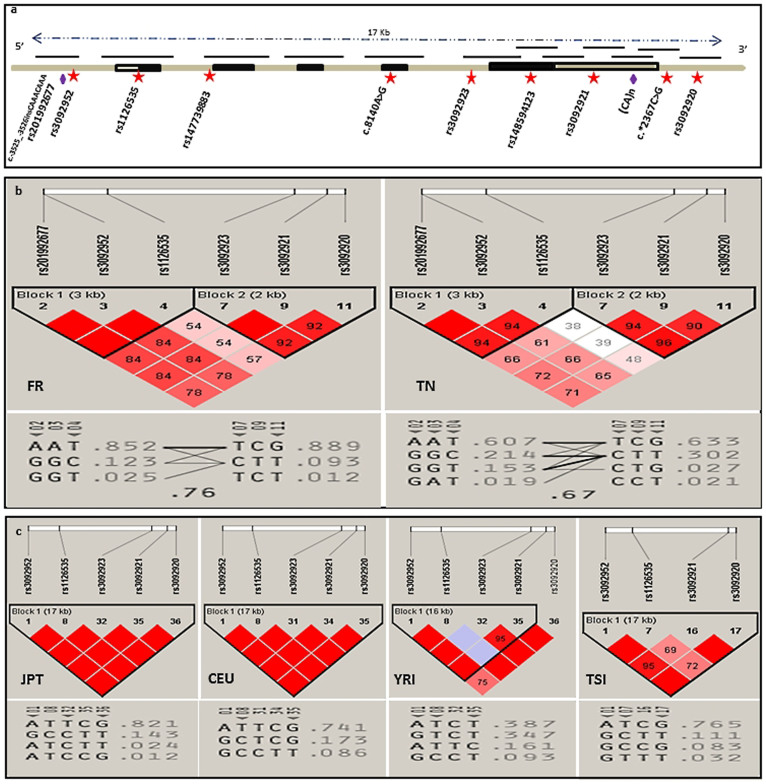
Figure 2. Genomic structure of the CD40LG and the haplotype blocks of the French (FR) and Tunisian (TN) populations (this study) compared with those of JPT, CEU, YRI and TSI (HapMap data).
(a) Genomic structure of the CD40LG gene and the position of the identified polymorphisms. Exons are represented by boxes, with filled boxes denoting translated regions. Upward facing lines indicate amplified fragments, and polymorphisms are shown as red stars for SNPs and purple lozenges for microsatellites. (b) CD40LG LD and haplotype diversity in French and Tunisian populations. (c) CD40LG LD and haplotype diversity in JPT, CEU, YRI and TSI populations. The numbers in boxes represent the pairwise D’ value between adjacent SNPs. For (b) and (c), haplotype frequencies are shown below the LD diagrams. Triangular pointers highlight SNPs which could be tagged for future association analysis.
LD and block haplotype analysis
The haplotype structure of the French and Tunisian populations with YRI, CEU, TSI and Japanese in Tokyo (JPT) populations is shown in Figure 2. There was a marked difference in the pattern of LD between the two populations. Using the solid spine algorithm26 in the Haploview software, two haplotype blocks were identified across the 17 kb region in both cohorts, with evidence of disruption in LD between SNP4 and SNP7. Block 1, spanning 3 kb, encompassed the 5′UTR and the first exon and contained three SNPs (rs201992677, rs3092952, rs1126535), while block 2 encompassed the rest of the gene. In block 1, there were three haplotypes in the French cohort with an AAT predominant one (0.852), which is also the most frequent among the four haplotypes found in the Tunisian cohort (0.607). The second most frequent haplotype was GGC; it was more prevalent in the Tunisian population than in the French one (0.214 versus 0.123). In block 2, there was one additional haplotype in the Tunisian cohort, even though the high-frequency haplotype was TCG for both populations (0.889: French cohort and 0.633: Tunisian cohort). Considering the resulting haplotype blocks, the SNPs defined five and eight core haplotypes in the French and Tunisian cohorts, respectively. The most frequent one was AAT/TCG in both populations (Figure 2).
rs201992677 [-/CAAA], described in the 1000 Genomes data but not in HapMap studies, was in high linkage disequilibrium with rs3092952 with D’ = 1 and r2 = 0.9 in the Tunisian dataset and D’ = 1, r2 = 1 in the French dataset. Surprisingly, all the male donors possessing the ‘G’ minor allele (rs3092952) were also found to carry seven CAAA repeats (rs201992677); in two cases, eight CAAA repeats were identified, which constitute a novel VNTR (c.-3525_-3526insCAAACAAA), instead of the most frequently found six CAAA repeats. Notably, three major ‘A’ type alleles of rs3092952 were unusually associated with seven CAAA repeats (rs201992677) in females originating from Tunisia, but not from France.
(CA)n microsatellite analysis
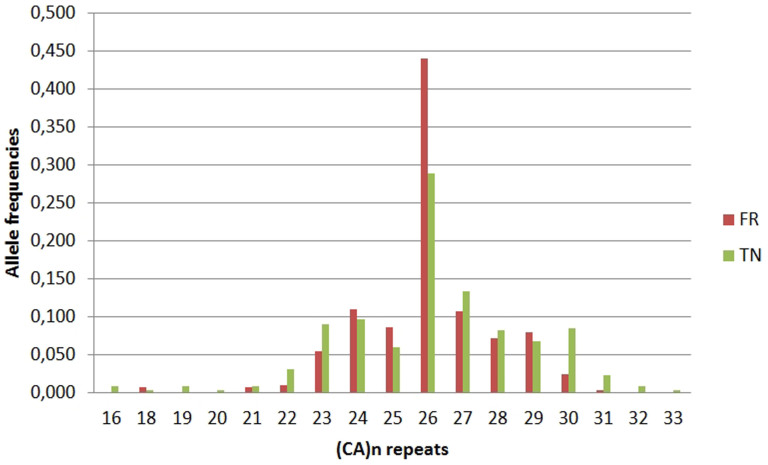
Allele frequencies were calculated in the French and Tunisian donors and the data revealed no deviation from HWE; we had previously verified the different allele frequencies for (CA)n repeats between males and females (P = 0.297 and 0.137 for the French and the Tunisian donors, respectively). Twelve different alleles (from 18 to 31 CA repeats, corresponding to PCR products from 98 to 124 bp) were detected in the French cohort while 17 alleles (with 16 to 33 repeats, corresponding to PCR products from 94 to 128 bp) were observed in the Tunisian cohort. The (CA)26 allele was the central or median allele and the most represented one in both groups, albeit more frequent in French than in Tunisian donors (Figure 3).
Figure 3. Frequency distribution of the number of (CA)n repeats in the CD40LG in the French (FR) and Tunisian (TN) populations.
Allele frequencies correspond to the number of the group of test alleles divided by the total number of alleles.
The allele distribution of the donors carrying 26 CA repeats, and less than or more than 26 CA repeats between the French and the Tunisian cohorts were compared by univariate statistical analysis. The allelic distribution between the French and the Tunisian donors was significantly different for allele groups of repeats as shown in Table 2.
Table 2. Allelic distributions of the CD40LG (CA)n repeat polymorphism in the French and Tunisian cohorts (291 and 353 alleles respectively).
| French cohort | Tunisian cohort | ||
|---|---|---|---|
| (CA)n | Na (frequency) | Na (frequency) | P-valueb |
| <26 CA | 80 (0.275) | 109 (0.309) | 0.00019 |
| 26 CA | 128 (0.440) | 102 (0.289) | |
| >26 CA | 83 (0.285) | 142 (0.402) |
aNumber of alleles.
bBonferroni correction: P-value is significant when <0.016.
Given the large difference between the two studied populations, we conducted a preliminary study in a group of 30 French single apheresis platelet donors whose platelets (in platelet components) induced an ATR. However, we did not observe any allelic or genotypic association with neither SNPs nor CA repeats in 3′UTR, nor haplotype of CD40LG (Supplementary Table S2 and S3).
Discussion
DHPLC is a high-throughput semi-automated method with good accuracy for detecting base pair changes, either transversions or transitions, and can be used as a screening or a genotyping strategy that also allows identification of new polymorphisms, including more than one in the same fragment27. By this strategy, three new polymorphisms, albeit some at a very low frequency, have been identified in the Tunisian and French study groups where there are no population CD40LG reports, yet. We screened and genotyped 11 variants including two VNTRs. Of the three SNPs located in the coding region, the novel c.8140 A>G in exon 4 found in a Tunisian donor resulted in a missense mutation (ATA to GTA) in which isoleucine was substituted for valine in the expressed protein at amino acid position 127 (p.I127V) of the extracellular portion28. This mutation is predicted to be benign by PolyPhen-2 with a score of 0.001 (http://genetics.bwh.harvard.edu/ggi/pph2/). The rare minor allele ‘A’ of the exonic rs148594123 inducing p.G219R was retrieved in exclusively eight healthy French including 4 females at the heterozygous A/G state and 4 ‘A’ hemizygous males, although we did not find the hypomorphic mutation p.G466X in XIAP, the adjacent gene of CD40LG with which cosegregation has been described as resulting in HIGM129. rs147739883, which was found in a Tunisian male, has been previously detected only in the Kenyan population (LWK). In addition, the minor ‘C’ allele of rs1126535 in exon 1 is associated with susceptibility to severe malaria30 and would increase the risk of reduced bone mineral density and osteopenia/osteoporosis (as a medium risk) for this synonymous SNP as it could affect splicing regulation31.
Among the three SNPs located in the 5′UTR, rs3092952 has been shown to provide an active role in CD40LG expression; the minor ‘G’ allele induces increased expression of the membrane and soluble forms of CD40L, thereby leading to enhanced CD40 interactions; this SNP is implicated in myocardial infarction and vascular disorders32. The variant ‘C’ allele of rs3092923, located in IVS4, is described as conferring a marginal reduced risk of follicular lymphoma33. CD40L regulation by the 5′UTR is related to the G allele of rs309295232, which is itself in high LD with the variable number of tandem CAAA repeats located a few bases upstream (rs201992677); therefore, could an increase in expression of the protein also be partially related to the presence of the CAAA repeats? as has been described in transcriptional regulation of the insulin-like growth factor 134.
We defined a core region of 17 kb in CD40LG. There is less than 2 kb in distance between haplotype blocks 1 and 2, whereas Chadha et al. described a distance of 4 kb35. We compared the results obtained for the haplotype structure of the SNPs, and the LD patterns with frequencies for Caucasians (CEU, TSI), Africans (YRI) and Asians (JPT). The LD structure of CD40LG on chromosome X and haplotypes in the corresponding LD blocks differed between the two populations studied. The Tunisian haplotypes were found to be intermediate between the European populations (French in the present study and TSI) and the African populations.
However, considering the HapMap data, the structure of LD in CD40LG and the haplotypes in the CEU European group, were found to be similar to those of the Asian (JPT) population, regardless of the allele frequency differences at each SNP. Our results for the French population were closer to those obtained with a European English cohort by Chadha et al.35, and also from TSI than to those of CEU (Figure 2). As already described, these results do not always correspond with the data (allele frequencies and LD) downloaded from HapMap Phase II and III; this may be explained by local variations within a country and/or by genotyping errors36.
It is worth noting that the French and Tunisian populations, as with any other North African population, do not exist in the HapMap or 1000 genomes datasets. Our results indicate moderate transferability of tagging SNPs on CD40LG from the European-Caucasian population to the French population. For the Tunisian population, this transferability is hardly conceivable. This approach has often been attempted in other populations (e.g. Thai and Spanish), and with other genes, with the aim of genotyping hotSNPs with which to assist future genetic association studies25,37.
The distribution of the frequencies of the CA repeat alleles in the populations studied appears similar to that obtained in others studies, although the major allele identified was 24 to 27 CA repeats. As discussed by Stewart et al, this difference is most likely explained by differences in the systems used for determining the repeat numbers: i.e. denaturing polyacrylamide gel and silver staining38, allele-specific oligonucleotide hybridization of membrane-fixed DNA products39 (major allele with 24 repeats), denaturing polyacrylamide gel and autoradiography40 (26 repeats), capillary electrophoresis using a CEQT 8000 Genetic analysis41 (27 repeats), or a CEQ 2000 genetic analysis system, as in the present study (26 repeats)42.
Different selection pressures may explain the extreme variability of this 3′UTR and its association with immune disorders. For example, polymorphisms of longer (CA)n alleles are seen in autoimmune processes such as seen in patients with systemic lupus erythematosus43, those with a high risk of thromboembolic disease44, or rheumatoid arthritis39,45. However, no association was found in the context of alloimmunity (e.g. rejection of renal transplantation) with either acute graft rejection or patient death, but a strong association was shown with long-term graft failure41. Shorter CA repeats are also linked to rare inherited HIGM1 with an informative usefulness in linkage analysis40. Is there any infectious selection process that could explain the more frequently observed longer alleles in the Tunisian population in relation to the need for enhanced inflammation46? It appears that the number of repeats increases over generations, but would this bear any relationship to ancestral population? This would reinforce the supposed old origins of the Tunisian population47.
CA dinucleotide is the most common repeat sequence in the human genome. Human CD40L is encoded by an unstable mRNA; this instability is conferred by a portion of its 3′UTR including a dual cis-acting element comprising a polypyrimidine-rich region and CA repeats48. It was shown that this CA repeats region is a binding site of nuclear factors and thus could modify the affinity of interaction with CD40LG mRNA and consequently, the translation efficiency48,49,50.
The (CA)24 allele (corresponding to (CA)26 in our study) appears to confer more stability on its mRNA, and longer alleles confer higher expression levels43,45,48.
To the best of our knowledge, ours is the first description of this microsatellite marker in Tunisians. This microsatellite has a distribution characteristic of each population as described in other studies38,39,40. In the present French and Tunisian study, the (CA)26 allele showed a statistically significant difference in frequency between the two populations (0.438 versus 0.291, respectively), while a previous French study found a frequency of 0.319. The frequency of heterozygosity was 0.70 (this study) and 0.79 (DiSanto et al.)40 for the French, and 0.82 for the Tunisian (this study). This result reflects genetic diversity in the Tunisian population, confirmed by the allelic distribution of (CA)n alleles, which has also been reported in the Spanish population, demonstrating that there was also a highly polymorphic marker (of 15 alleles) in Spaniards versus 17 in Tunisians38. The dinucleotide repeat (CA)n is a marker that is highly polymorphic, and therefore useful for genetic studies on the CD40L molecule in relation to immunity.
Both French and North African populations have a tumultuous history with various ancestors but the Tunisian population appears to be significantly more heterogeneous as demonstrated by Y-STR diversity in the Sousse population51; the three novel polymorphisms are also in favor of the extreme heterogeneity in the studied cohort (Figure 1)35,52,53. Heterogeneity in haplotype frequencies is often higher in Africans such as the YRI group, in comparison to the European populations who left the African continent and experienced bottlenecks during their migrations, reducing SNPs diversity (Figure 2). The intermediate Tunisian CD40LG haplotype structure might be explained by its African location and multilayered history53.
As variants repartition is different between the 2 populations, these findings added to the current knowledge base in the studied French and Tunisian populations, may ultimately lead to better investigate the CD40LG associated diseases. Their prevalence were found to be often in support with genetic polymorphisms repartition (Table 1 and 2), e.g. cardiovascular disorders32,54,55, systemic lupus erythematosus43,56,57, rheumatoid arthritis45,58, although exact Tunisian prevalence is difficult to clarify for lack of national epidemiological registers. In addition to CD40L polymorphisms, other genetic markers were found differently associated in the studied populations59 whereas others were similarly associated60. On the other hand, this technical strategy could be used for HIGM1 investigation (we covered 182 of all the 183 mutations in CD40LG that cause HIGM1 (http://www.hgmd.org/ updated in June 2014). It is less expensive than actual used techniques, especially in developing countries such as Tunisia where HIGM syndrome is frequently found46,61,62.
Although several studies have found an association between ATR and high level of sCD40L in the transfused blood component19,20,21,22,23, we did not find significant differences in CD40LG genetic pattern in donors whose platelets induced or not an ATR in the French population. This would be in favor of a CD40L release mechanism not due to a genetic regulation. It is noteworthy that all the previous studies have been performed with small effectives. We will have to extend the CD40LG study to a more important cohort of donors including donors whose donated platelets induced TRALI. It will be also interesting to investigate CD40L receptor (CD40 gene) in the recipients presenting ATR.
In conclusion, we identified polymorphisms known as regulatory within the CD40LG i.e. rs3092952 in the 5′UTR, (CA)n repeats in the 3′UTR, some rare and novel mutations, and polymorphisms involved in several diseases. These polymorphisms could modulate immune responses via interactions between CD40L and its various receptors63. We found a major difference between Maghreb (Tunisian) and European (French) populations in their CD40LG haplotype patterns. Surprisingly, we did not find any difference in CD40LG genetic pattern in apheresis platelet donors whose blood components prepared for transfusion purpose induced or not an ATR. Our findings provide useful information for future disease-association studies on the CD40LG in the context of inflammation and auto-immunity.
Methods
Ethical considerations
Informed and written consent was obtained from all the healthy donors who participated in this study, according to a protocol approved by the Ethics Committees for scientific research at Saint-Etienne (France), F. Bourguiba (Monastir, Tunisia), and F. Hached (Sousse, Tunisia) University Hospitals. All analyses were performed anonymously.
Studied populations
The cohort contained 485 healthy volunteer human blood donors, acting as non-profit donors (211 French donors who presented at the Auvergne-Loire Regional Branch of the French National Blood System EFS collection sites (130 males and 81 females) and 274 Tunisian donors who presented at the Monastir Blood Bank or the Regional Blood Transfusion Centre of Sousse blood collection sites (195 males and 79 females). Blood donors at both locations were unrelated and randomly chosen to enter this study, based on the timing of their donations, and no selection bias was applied to them (Supplementary Table S4). For comparison, the allele and genotype information for other populations were downloaded from HapMap Phase II and III (http://www.hapmap.org, August 2010) and from the 1000 genomes project (http://www.1000genomes.org). Thirty French single apheresis platelet donors whose donated blood components induced an ATR (FNHTR) were also tested (18 males and 12 females) (Supplementary Table S4).
DNA samples
EDTA-treated blood samples were collected from all individuals. DNA was purified from peripheral blood leukocytes using a FlexiGene DNA kit (Qiagen, Paris, France) according to the manufacturer's instructions. DNA concentrations and purity were assessed using the Nanodrop 2000 Spectrophotometer (Thermo Scientific, Wilmington, USA).
SNPs and microsatellite analysis
The nomenclature used herein for reporting sequence alterations abides by the original Human Genome Variation Society guidelines. NC_000023.10 was used as the reference sequence. All information about the SNPs detected was extracted from the public Single Nucleotide Polymorphism database dbSNP built 138 (http://ncbi.nlm.nih.gov/SNP/).
DHPLC analysis: screening and genotyping
To screen for the maximum number of polymorphisms potentially involved in CD40L protein expression or reported to be associated with immune disorders or inflammation32,45,48,64, PCR was performed with eleven primer pairs with corresponding annealing temperatures (Table 3, Supplementary Data S1 for PCR protocols). To promote stable heteroduplex formation, PCR products were denatured for 5 min at 96°C, and then gradually cooled at a rate of 0.5°C per 10 sec over 150 cycles.
Table 3. PCR fragments, primer sequences, annealing temperatures (Ta)°C and conditions of Denaturing High Performance Liquid Chromatography (DHPLC) analysis of CD40LG.
| Primer sequences (5'>3') | DHPLC analysis | |||||
|---|---|---|---|---|---|---|
| Fragment CD40LG- (bp) | Forward primer | Reverse primer | Annealing temperature (°C) | Column temperature (°C)a | Buffer B % | Time shift (min) |
| 5'UTR (285)b | TACTTGGGAGGCTGAGGCAG | TTGACCCCTGCCCACATTT | 62 | [56.9], [57.9] | 63.4 | 0 |
| E1 (362)b | GCGCTCTTAACTAATCCTGAG | TCTTCTATTTGGTTTCTACCATC | 55 | 55.1, 56.1, [57.1] | 64.1 | 0 |
| E2 (283)b | TGCCGTGGAAATGAATGTAG | CCCGATCTAGCAAATGTAGTAAA | 58,5 | 55.6, [56.6] | 62 | 0 |
| E3 (209) | AAACCCCACAGCAGAGCCC | CCTGATGCAACAACACTGGGT | 58 | [56.8], 57.8 | 59 | 0 |
| E4 (310)b | TCAGTGGGAGAGATGTCAGACC | CCCAGGGGAAAAGAGGAGTTTA | 58 | [56.3], 58.3 | 62.5 | 0 |
| E5A (383)b | GAACCATGCTCTGCTTCACCT | CACTTGGCTTGGATCAGTCA | 58 | 58.3, [59.3] | 64.5 | 0 |
| E5B (432)b | GAGAGAATCTTACTCAGAGCTGC | ATGTCTGCATCAGTGGGGTT | 58,5 | 57.2, [60] | 65.4, 63.4 | 0 0.4 |
| E5C (356) | TCATAATACAGCACAGCGGT | TTCCCTTCTGCATCTTCACT | 57,4 | 57.1, [58] | 64 | 0 |
| 3'UTRA (304)b | ATGCAGAAGGGAAATGGGG | GTTAGAAAGGGGGATTGAGAAG | 58 | 58.9, [60.9] | 62.6 | 0 1 |
| 3'UTRB (339)b | GTTGCAGGAGATTGAAAGAAC | TAGTGGCTTGAAGATGCTGC | 55 | [58.5], [58.9] | 63.6 | 0 |
| 3'UTRC (244)b | CCAAGACCTTGTCCCATTCACC | CAGTCATTTTTTACTCCATGAGTGC | 59 | [56.5], [57.3] | 60.6 | 0 |
aColumn temperature selection was based on the melting domains present in the PCR fragments.
The column temperature recommended by the Navigator connection Wadmin software is shown in italics, while the temperature at which a polymorphism was accurately identified is shown within square brackets.
bSNPs were identified in these PCR fragments.
At the screening stage, 5 µL of each PCR product was automatically analyzed by DHPLC using the Wave system (Transgenomic, Ltd. Glasgow, UK)65,66. PCR products were injected into a preheated, fully equilibrated chromatographic column (DNASep Column, Transgenomic Ltd.). A linear gradient of 5% triethylammonium acetate (TEAA) (Buffer A) and 25% acetonitrile-5%TEAA (Buffer B) was generated for each amplicon. Each DHPLC run included a DNA loading step (5% drop for loading Buffer B), a linear separation gradient (2% Buffer B slope per min, 4.5 min), a wash step (75% acetonitrile; Buffer D, 0.5 to 1 min) and an equilibration step (0.9 to 1.2 min). The eluate was detected by an ultraviolet light detector at 260 nm. DHPLC conditions were optimized via fragment melting profile analysis, using the Wave system software Navigator connection WADMIN (Transgenomic, Ltd.).
The melting point (and profile) of each fragment was analyzed according to Fixman and Friere's implementation of Poland's algorithm67, which calculates the probability that a base is in the helical duplex conformation (at 75–90%) or in the non-helical, single-stranded form. Each amplicon was, therefore, assessed at two or three column temperatures (Table 3). The flow speeds, temperatures, retention times and initial Buffer B concentrations of the elution gradient for DHPLC analysis were determined by the software provided. If the sample contains heteroduplex molecules, these will denature at lower concentrations of acetonitrile and will be visualized as peaks (or sometimes a single peak) with shorter retention times than that of a homoduplex. Control samples sequenced previously (homozygous and heterozygous for a known SNP, displaying profiles with single or multiple peaks) were run in parallel.
At the genotyping stage, all the samples from females presenting with one peak at the screening stage were mixed in equal volumes with a reference sample. Such samples were run again using the same DHPLC conditions and the same controls described above. Samples presenting with one peak at the screening stage had the same genotype of the homozygous control if the same single peak that was observed was homozygous for the other allele if multiple peaks were seen. Distorted multiple peak elution profiles differing from the reference profiles indicated the presence of a different polymorphism from the reference one or an additional SNP in the fragment; additional sequencing was applied in these cases.
For the male samples, because the gene of interest is on the X chromosome, a preliminary step was required. This involved mixing the PCR product from each sample with the PCR product of one of the controls (1V/1V) that was previously sequenced and for which no polymorphism for the fragment had been found and was, therefore, automatically genotyped.
DNA sequencing
To identify the type and position of the genetic variants, DNA samples with abnormal DHPLC patterns were re-amplified as described above, and then subjected to direct sequencing in both forward and reverse directions using the same PCR primers. PCR products were purified by Amicon Centrifugal filters (Millipore, Molsheim, France). Sequencing reactions were performed using the GenomeLab DTCS Quick Start mix (Beckman Coulter, Brea, CA). The resulting products were purified with Beckman Coulter's CEQ DTCS kit according to the manufacturer's instructions, subjected to analysis with a CEQ 2000 XL-Beckman Coulter sequencing machine, and assembled using CEQ 8000 Beckman Coulter software (Beckman Coulter, Inc. CA). For each DHPLC profile displayed, 10 samples were sequenced. However, all the heteroduplex profiles for the CD40L-5′UTR fragment were sequenced because of more than one polymorphism.
Capillary electrophoresis: genotyping 3′UTR CA repeats
Samples were resolved by capillary electrophoresis separation, using a CEQ 2000 XL genetic analysis system (Beckman Coulter, Inc. CA, USA). To 0.5 µL of PCR product were added 0.25 µL of a 600 bp standard and 40 µL of sample loading solution (Beckman Coulter). Specifically designed labeled oligonucleotides were excited to fluoresce using a diode laser. The length of the amplified fragment was estimated with reference to the internal ladder and the number of repeats was calculated by analogy with 10 sequenced samples, using the CEQ 8000 Beckman Coulter software provided.
Statistical analysis
Allele and genotype frequencies were calculated and compared using XLSTAT™ (Addinsoft, Paris, France) computer software. The χ2 test was used to compare the allelic distribution between males and females and the allelic frequencies between French and Tunisian populations, using the Bonferroni correction or Fisher's exact test, where appropriate. Statistical significance was taken as P<0.05. For each polymorphism, the quality of the genotype data was assessed by testing for HWE, using the Haploview program http://www.broad.mit.edu/mpg/haploview/26. For analysis of the 3′UTR CA repeats, HWE was tested by comparison of the observed and expected genotype frequencies. Haploview 4.2 was also used to calculate the pairwise linkage disequilibrium (LD) of the polymorphisms for which the minor allele frequency (MAF) was >0.05. LD assessment and CD40LG haplotype definition, was determined using Lewontin's standardized disequilibrium coefficient D’, the squared correlation coefficient r2, while the block definition was based on the solid spine method26. For practical purposes, we introduced rs201992677 [-/CAAA] to the LD analysis, arbitrarily calling ‘A’ the wild type allele with six CAAA repeats, and ‘G’ the variant allele with seven repeats.
Additional information
Accession codes: We submitted the three novel mutations in DDBJ database with following accession numbers
> AB897730 52b053e41553421871002993.CD40LG1
> AB897731 52b053e41553421871002993.CD40LG2
> AB897732 52b053e41553421871002993.CD40LG3
Supplementary Material
Supplementary Information
Acknowledgments
This work was supported by the EFS Auvergne-Loire, Cooperation Région Rhône-Alpes CMIRA, Erasmus Mundus Al-Idrisi, the “Amis de Rémi” Association. We thank the blood donors for taking part in this study.
Footnotes
The authors declare no competing financial interests.
Author Contributions S.L., F.C. and O.G. initiated and completed the project. S.L. and C.A. designed the experiments. C.A. and C.S. performed the experiments. M.H., T.C., S.JY., M.A., C.F., B.P. and C.A. contributed to the recruitment of the blood donors. J.F., V.G.H. and R.T. did the DHPLC analysis. C.A. and A.P. analyzed the data. S.L. drafted the manuscript. All authors reviewed the manuscript.
References
- Horner A. A., Jabara H., Ramesh N. & Geha R. S. gamma/delta T lymphocytes express CD40 ligand and induce isotype switching in B lymphocytes. J Exp Med 181, 1239–44 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grammer A. C. et al. The CD40 ligand expressed by human B cells costimulates B cell responses. J Immunol 154, 4996–5010 (1995). [PubMed] [Google Scholar]
- Van Kooten C. & Banchereau J. CD40-CD40 ligand: a multifunctional receptor-ligand pair. Adv Immunol 61, 1–77 (1996). [DOI] [PubMed] [Google Scholar]
- Wagner A. H., Guldenzoph B., Lienenluke B. & Hecker M. CD154/CD40-mediated expression of CD154 in endothelial cells: consequences for endothelial cell-monocyte interaction. Arterioscler Thromb Vasc Biol 24, 715–20 (2004). [DOI] [PubMed] [Google Scholar]
- Suarez A., Mozo L., Gayo A., Zamorano J. & Gutierrez C. Requirement of a second signal via protein kinase C or protein kinase A for maximal expression of CD40 ligand. Involvement of transcriptional and posttranscriptional mechanisms. Eur J Immunol 27, 2822–9 (1997). [DOI] [PubMed] [Google Scholar]
- Elgueta R. et al. Molecular mechanism and function of CD40/CD40L engagement in the immune system. Immunol Rev 229, 152–72 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foy T. M. et al. Blockade of CD40-CD154 interferes with human T cell engraftment in scid mice. Cell Transplant 7, 25–35 (1998). [DOI] [PubMed] [Google Scholar]
- Korthauer U. et al. Defective expression of T-cell CD40 ligand causes X-linked immunodeficiency with hyper-IgM. Nature 361, 539–41 (1993). [DOI] [PubMed] [Google Scholar]
- MacDonald K. P., Nishioka Y., Lipsky P. E. & Thomas R. Functional CD40 ligand is expressed by T cells in rheumatoid arthritis. J Clin Invest 100, 2404–14 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue S. et al. Enhancement of dendritic cell activation via CD40 ligand-expressing gammadelta T cells is responsible for protective immunity to Plasmodium parasites. Proc Natl Acad Sci U S A 109, 12129–34 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henn V. et al. CD40 ligand on activated platelets triggers an inflammatory reaction of endothelial cells. Nature 391, 591–4 (1998). [DOI] [PubMed] [Google Scholar]
- Lindmark E., Tenno T. & Siegbahn A. Role of platelet P-selectin and CD40 ligand in the induction of monocytic tissue factor expression. Arterioscler Thromb Vasc Biol 20, 2322–8 (2000). [DOI] [PubMed] [Google Scholar]
- Andre P., Nannizzi-Alaimo L., Prasad S. K. & Phillips D. R. Platelet-derived CD40L: the switch-hitting player of cardiovascular disease. Circulation 106, 896–9 (2002). [DOI] [PubMed] [Google Scholar]
- Phipps R. P. et al. The CD40-CD40 ligand system: a potential therapeutic target in atherosclerosis. Curr Opin Investig Drugs 2, 773–7 (2001). [PubMed] [Google Scholar]
- Jin Y. et al. Characterization of soluble CD40 ligand released from human activated platelets. J Med Dent Sci 48, 23–7 (2001). [PubMed] [Google Scholar]
- Inwald D. P., McDowall A., Peters M. J., Callard R. E. & Klein N. J. CD40 is constitutively expressed on platelets and provides a novel mechanism for platelet activation. Circ Res 92, 1041–8 (2003). [DOI] [PubMed] [Google Scholar]
- Langer H. F. & Chavakis T. Platelets and neurovascular inflammation. Thromb Haemost 110, 888–93(2013). [DOI] [PubMed] [Google Scholar]
- Siddiqui T. I., Kumar K. S. & Dikshit D. K. Platelets and atherothrombosis: causes, targets and treatments for thrombosis. Curr Med Chem 20, 2779–97 (2013). [DOI] [PubMed] [Google Scholar]
- Khan S. Y. et al. Soluble CD40 ligand accumulates in stored blood components, primes neutrophils through CD40, and is a potential cofactor in the development of transfusion-related acute lung injury. Blood 108, 2455–62 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumberg N., Gettings K. F., Turner C., Heal J. M. & Phipps R. P. An association of soluble CD40 ligand (CD154) with adverse reactions to platelet transfusions. Transfusion 46, 1813–21 (2006). [DOI] [PubMed] [Google Scholar]
- Cognasse F., Payrat J. M., Corash L., Osselaer J. C. & Garraud O. Platelet components associated with acute transfusion reactions: the role of platelet-derived soluble CD40 ligand. Blood 112, 4779–80; author reply 4780–1 (2008). [DOI] [PubMed] [Google Scholar]
- Hamzeh-Cognasse H. et al. Immune-reactive soluble OX40 ligand, soluble CD40 ligand, and interleukin-27 are simultaneously oversecreted in platelet components associated with acute transfusion reactions. Transfusion 54, 613–25 (2014). [DOI] [PubMed] [Google Scholar]
- Nguyen K. A. et al. A computerized prediction model of hazardous inflammatory platelet transfusion outcomes. PLoS One 9, e97082 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Service S., International Collaborative Group on Isolated, P., Sabatti C. & Freimer N. Tag SNPs chosen from HapMap perform well in several population isolates. Genet Epidemiol 31, 189–94 (2007). [DOI] [PubMed] [Google Scholar]
- Nuchnoi P. et al. Linkage disequilibrium structure of the 5q31-33 region in a Thai population. J Hum Genet 53, 850–6 (2008). [DOI] [PubMed] [Google Scholar]
- Barrett J. C., Fry B., Maller J. & Daly M. J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21, 263–5 (2005). [DOI] [PubMed] [Google Scholar]
- Wagner T. et al. Denaturing high-performance liquid chromatography detects reliably BRCA1 and BRCA2 mutations. Genomics 62, 369–76 (1999). [DOI] [PubMed] [Google Scholar]
- Schonbeck U., Mach F. & Libby P. CD154 (CD40 ligand). Int J Biochem Cell Biol 32, 687–93 (2000). [DOI] [PubMed] [Google Scholar]
- Rigaud S. et al. Human X-linked variable immunodeficiency caused by a hypomorphic mutation in XIAP in association with a rare polymorphism in CD40LG. Blood 118, 252–61 (2011). [DOI] [PubMed] [Google Scholar]
- Toure O. et al. Candidate polymorphisms and severe malaria in a Malian population. PLoS One 7, e43987 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineda B. et al. Gene-gene interaction between CD40 and CD40L reduces bone mineral density and increases osteoporosis risk in women. Osteoporos Int 22, 1451–8 (2011). [DOI] [PubMed] [Google Scholar]
- Malarstig A., Lindahl B., Wallentin L. & Siegbahn A. Soluble CD40L levels are regulated by the -3459 A>G polymorphism and predict myocardial infarction and the efficacy of antithrombotic treatment in non-ST elevation acute coronary syndrome. Arterioscler Thromb Vasc Biol 26, 1667–73 (2006). [DOI] [PubMed] [Google Scholar]
- Skibola C. F. et al. A functional TNFRSF5 gene variant is associated with risk of lymphoma. Blood 111, 4348–54 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. Y. et al. Functional Interaction Between SNPs and Microsatellite in the Transcriptional Regulation of Insulin-Like Growth Factor 1. Hum Mutat 34, 1289–97 (2013). [DOI] [PubMed] [Google Scholar]
- Chadha S. et al. Haplotype structure of TNFRSF5-TNFSF5 (CD40-CD40L) and association analysis in systemic lupus erythematosus. Eur J Hum Genet 13, 669–76 (2005). [DOI] [PubMed] [Google Scholar]
- Montpetit A. et al. An evaluation of the performance of tag SNPs derived from HapMap in a Caucasian population. PLoS Genet 2, e27 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribas G. et al. Evaluating HapMap SNP data transferability in a large-scale genotyping project involving 175 cancer-associated genes. Hum Genet 118, 669–79 (2006). [DOI] [PubMed] [Google Scholar]
- Citores M. J. et al. CD154 polymorphism in Spanish populations. Differences in the allelic distribution between Canary islanders and Peninsulars. Eur J Immunogenet 27, 141–4 (2000). [DOI] [PubMed] [Google Scholar]
- Gomolka M. et al. Immunoprinting: various genes are associated with increased risk to develop rheumatoid arthritis in different groups of adult patients. J Mol Med (Berl) 73, 19–29 (1995). [DOI] [PubMed] [Google Scholar]
- DiSanto J. P. et al. Brief report: prenatal diagnosis of X-linked hyper-IgM syndrome. N Engl J Med 330, 969–73 (1994). [DOI] [PubMed] [Google Scholar]
- Dmitrienko S., Hoar D. I., Balshaw R. & Keown P. A. Immune Response Gene Polymorphisms in Renal Transplant Recipients. Transplantation 80, 1773–1782 (2005). [DOI] [PubMed] [Google Scholar]
- Stewart S., Wickramasinghe D., Dorrance A. E. & Robertson A. E. Comparison of three microsatellite analysis methods for detecting genetic diversity in Phytophthora sojae (Stramenopila: Oomycete). Biotechnol Lett 33, 2217–23 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citores M. J. The dinucleotide repeat polymorphism in the 3′UTR of the CD154 gene has a functional role on protein expression and is associated with systemic lupus erythematosus. Annals of the Rheumatic Diseases 63, 310–317 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugert P. et al. The risk for thromboembolic disease in lupus anticoagulant patients due to pathways involving P-selectin and CD154. Thromb Haemost 97, 573–80 (2007). [PubMed] [Google Scholar]
- Martin-Donaire T. et al. Association of the microsatellite in the 3′ untranslated region of the CD154 gene with rheumatoid arthritis in females from a Spanish cohort: a case-control study. Arthritis Res Ther 9, R89 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejaoui M., Mellouli F., Chouanine R., Dellagi K. & Barbouche M. R. [The hyper-IgM syndrome: 13 observations]. Presse Med 32, 544–9 (2003). [PubMed] [Google Scholar]
- Mirkin S. M. Expandable DNA repeats and human disease. Nature 447, 932–40 (2007). [DOI] [PubMed] [Google Scholar]
- Hamilton B. J. et al. Separate cis-trans pathways post-transcriptionally regulate murine CD154 (CD40 ligand) expression: a novel function for CA repeats in the 3′-untranslated region. J Biol Chem 283, 25606–16 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui J. et al. Intronic CA-repeat and CA-rich elements: a new class of regulators of mammalian alternative splicing. EMBO J 24, 1988–98 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert L. A. et al. A T cell-specific enhancer of the human CD40 ligand gene. J Biol Chem 277, 7386–95 (2002). [DOI] [PubMed] [Google Scholar]
- Fadhlaoui-Zid K. et al. Sousse, Tunisia: tumultuous history and high Y-STR diversity. Electrophoresis 33, 3555–63 (2012). [DOI] [PubMed] [Google Scholar]
- Cherni L. et al. Y-chromosomal STR haplotypes in three ethnic groups and one cosmopolitan population from Tunisia. Forensic Sci Int 152, 95–9 (2005). [DOI] [PubMed] [Google Scholar]
- Henn B. M. et al. Genomic ancestry of North Africans supports back-to-Africa migrations. PLoS Genet 8, e1002397 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andre R. et al. International differences in acute coronary syndrome patients' baseline characteristics, clinical management and outcomes in Western Europe: the EURHOBOP study. Heart 100, 1201–7 (2014). [DOI] [PubMed] [Google Scholar]
- Saidi O. et al. Analyzing recent coronary heart disease mortality trends in Tunisia between 1997 and 2009. PLoS One 8, e63202 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaud L., Fagot J. P., Paita M., Fagot-Campagna A. & Amoura Z. Prevalence and incidence of systemic lupus erythematosus in France: A 2010 nation-wide population-based study. Autoimmun Rev (2014). 10.1016/j.autrev.2014.08.034 [DOI] [PubMed] [Google Scholar]
- Khanfir M. S. et al. TULUP (TUnisian LUPus): a multicentric study of systemic lupus erythematosus in Tunisia. Int J Rheum Dis 16, 539–46 (2013). [DOI] [PubMed] [Google Scholar]
- Guillemin F. et al. Prevalence of rheumatoid arthritis in France: 2001. Ann Rheum Dis 64, 1427–30 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maalej A. et al. Association of IRF5 gene polymorphisms with rheumatoid arthritis in a Tunisian population. Scand J Rheumatol 37, 414–8 (2008). [DOI] [PubMed] [Google Scholar]
- Roulland S. et al. t(14;18) Translocation: A predictive blood biomarker for follicular lymphoma. J Clin Oncol 32, 1347–55 (2014). [DOI] [PubMed] [Google Scholar]
- Grumbt B., Eck S. H., Hinrichsen T. & Hirv K. Diagnostic applications of next generation sequencing in immunogenetics and molecular oncology. Transfus Med Hemother 40, 196–206 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamia S. et al. [Primary immunodeficiency disorders in 51 cases]. Tunis Med 91, 38–43 (2013). [PubMed] [Google Scholar]
- Choi W. S., Jeon O. H. & Kim D. S. CD40 ligand shedding is regulated by interaction between matrix metalloproteinase-2 and platelet integrin alpha(IIb)beta(3). J Thromb Haemost 8, 1364–71 (2010). [DOI] [PubMed] [Google Scholar]
- Teruel M. et al. Analysis of the association between CD40 and CD40 ligand polymorphisms and systemic sclerosis. Arthritis Res Ther 14, R154 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B., Sawyer N. A., Chiu C., Oefner P. J. & Underhill P. A. DNA mutation detection using denaturing high-performance liquid chromatography (DHPLC). Curr Protoc Hum Genet Chapter 7, Unit7 10 (2006). [DOI] [PubMed] [Google Scholar]
- Fackenthal D. L., Chen P. X., Howe T. & Das S. Denaturing high-performance liquid chromatography for mutation detection and genotyping. Methods Mol Biol 1015, 25–54 (2013). [DOI] [PubMed] [Google Scholar]
- Fixman M. & Freire J. J. Theory of DNA melting curves. Biopolymers 16, 2693–704 (1977). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information