Abstract
The expression, functional reconstitution and first NMR characterization of the human growth hormone secretagogue (GHS) receptor reconstituted into either DMPC or POPC membranes is described. The receptor was expressed in E. coli. refolded, and reconstituted into bilayer membranes. The molecule was characterized by 15N and 13C solid-state NMR spectroscopy in the absence and in the presence of its natural agonist ghrelin or an inverse agonist. Static 15N NMR spectra of the uniformly labeled receptor are indicative of axially symmetric rotational diffusion of the G protein-coupled receptor in the membrane. In addition, about 25% of the 15N sites undergo large amplitude motions giving rise to very narrow spectral components. For an initial quantitative assessment of the receptor mobility, 1H-13C dipolar coupling values, which are scaled by molecular motions, were determined quantitatively. From these values, average order parameters, reporting the motional amplitudes of the individual receptor segments can be derived. Average backbone order parameters were determined with values between 0.56 and 0.69, corresponding to average motional amplitudes of 40–50° of these segments. Differences between the receptor dynamics in DMPC or POPC membranes were within experimental error. Furthermore, agonist or inverse agonist binding only insignificantly influenced the average molecular dynamics of the receptor.
G protein-coupled receptors (GPCRs) belong to the largest class of mammalian membrane proteins and have received significant attention due to their high pharmacological relevance. In spite of extensive attempts to crystallize GPCRs, a successful strategy was only developed about a decade ago. Crystallization succeeded after intracellular loop 3 that was found to be most responsible for helix movement, was rigidified by introduction of T4 lysozyme, truncations, thermostabilization, or by anti- or nanobody binding1,2,3. Such receptor engineering proved to be a very successful strategy and led to a number of crystal structures solved for various GPCRs4. But proteins are dynamic molecules that do not possess a unique and rigid conformation, but are best described as dynamic ensembles of more or less different structures5,6. This particularly applies to membrane proteins, which reside in liquid-crystalline bilayers that are also highly dynamic7.
In spite of the success of crystallography, the need to study native GPCRs in their relevant membrane environment remains as GPCRs are highly dynamic molecules that undergo various conformational changes upon activation by an extracellular ligand and subsequent G-protein interaction8. At this point, GPCR dynamics have to be addressed explicitly. The most important dynamic structural alteration has been observed for helix 6 of the heptahelical membrane proteins. Specific labeling of the protein with fluorescence9,10, EPR11,12, or NMR13,14,15,16,17,18,19 probes have highlighted the dynamical alterations of the molecule upon inverse agonist or agonist binding as well as G-protein interaction. The most complete set of data that describes the interesting structural dynamics of GPCRs is available for the β2-adrenergic receptor (β2AR). Unliganded receptor and inverse agonist bound β2AR seem to exist in two inactive conformations that exchange rapidly with a frequency in the kilohertz range18. Agonist binding shifts the equilibrium towards a conformation that can bind cytoplasmatic G-protein, this process is incomplete resulting in an even increased conformational heterogeneity, which includes the active, possibly several intermediate, and the inactive states. Only G-protein binding results in the complete transition to the active conformation, which essentially abolishes the aforementioned dynamics18. This and other data suggests that the assumption of just one active receptor structure may represent a too simplistic representation of the function of GPCRs9.
Energy landscapes are a useful tool to integrate GPCR structure, dynamics, and function8,20 and represent a general means to describe the dynamic character of proteins6. For GPCRs, models of the energy landscapes of the molecules upon activation and G-protein binding show pronounced minima for the ground state and the active state, locked by the G-protein21. However, there are also several local minima, separated by low energy barriers that are populated upon binding of agonists and inverse agonists as demonstrated for the β2AR18.
So far, explicit data on the molecular dynamics of GPCRs is rather limited. In addition to the aforementioned data on the dynamic activation of GPCRs, one study has addressed the molecular dynamics of the CXCR1 receptor reconstituted in bilayer membranes22. This investigation came to the conclusion that the receptor undergoes axially symmetric rotational diffusion in the membrane and about 10% of the residues are subject to larger amplitude motions. In contrast, the molecular dynamics of the human neuropeptide Y receptor type 2 has been systematically studied revealing that the individual segments of the molecule are subject to large amplitude motions, expressed by average order parameters between 0.55 and 0.67 in the protein backbone, which corresponds to amplitudes of the segmental motions up to about 50° 23,24. Here, we report results on the molecular dynamics of the human growth hormone secretagogue (GHS) receptor, which plays a key role in the regulation of appetite and food intake and, therefore, represents an interesting target for intervening with eating disorders25. The ligand of the GHS receptor is the octanoylated growth hormone ghrelin consisting of 28 amino acids26,27,28. We used solid-state NMR spectroscopy to study the molecular dynamics of the GHS receptor reconstituted into DMPC or POPC membranes.
Results
Pharmacological characterization of the recombinant GHS receptor
Prior to biophysical investigation of the GHS receptor, a detailed pharmacological characterization was carried out using homogenous fluorescence assays as described in the literature29. Figure 1A shows the dose-dependent atto520-ghrelin ligand binding plots of the GHS receptor.
Figure 1.
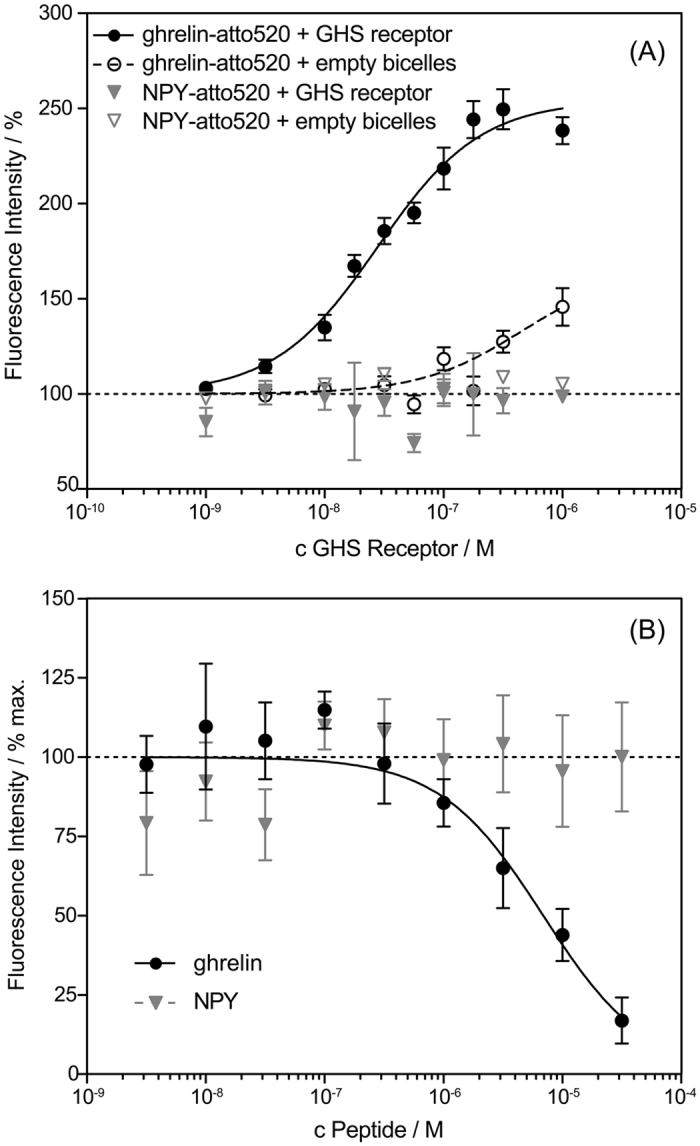
(A) Saturation binding of atto520-labeled ghrelin (c = 100 nM) to increasing amounts of GHS receptor-containing bicelles or empty bicelles. As a control, atto520-labeled NPY was used, which did not display enhanced fluorescence in the presence of GHS receptor-loaded or empty bicelles. Data reflect fluorescence enhancement upon binding. The inflection point (EC50 = 28 nM) for GHS receptor binding is approximately at the limit of the assay of Ltotal/2 = 50 nM; demonstrating high functionality of the system. (B) Displacement of atto520-ghrelin binding to the GHS receptor by unlabeled ghrelin. In contrast, neuropeptide Y (NPY) was not able to displace the bound atto520-ghrelin ligand. Results represent mean +/− SEM of three independent assays each performed in triplicates.
The inflection point for GHS receptor binding was determined at EC50 = 28 nM, which is approximately at the limit of the assay; demonstrating high functionality of the system. The assay limit is dictated by the total concentration of the labeled ligand (Ltotal). Fluorescence enhancement will be maximal when all labeled ligand molecules are bound by receptor. Assuming a 1:1 stoichiometry, half-maximal signal is reached at receptor concentrations equivalent to Ltotal/2 + Kd30, leading to an assay limit of Ltotal/2 for high-affinity systems. Unfortunately, the use of lower tracer concentrations closer to the reported sub-nanomolar Kd was obviated by the sensitivity of the fluorescence reader. Ghrelin also binds to empty phosphatidylcholine bicelles, but at much higher concentration as determined before26. As an additional control, we used fluorescently labeled neuropeptide Y (atto520-NPY), which should neither bind to the GHS receptor nor to empty phosphatidylcholine bicelles. Accordingly, no change in fluorescence was recorded.
Next, displacement experiments were carried out at a tracer peptide concentration far above the Kd, resulting in a large Cheng-Prusoff shift and further distortion by ligand depletion31,32. Applying these correction factors and using the Kd of 0.15 nM as reported by Els et al.33, a Ki of 10 nM can be calculated from the IC50 of 6870 nM. This corresponds well to values observed previously both in vivo and in vitro34,35. The specificity of displacement was further underlined by using NPY as a control, which did not result in displacement of the atto520-ghrelin ligand (Fig. 1B).
Static 15N CP spectra of the GHS receptor in DMPC
A standard method to characterize membrane proteins is the analysis of static 15N CP NMR spectra of uniformly 15N labeled membrane protein36. Fully rigid membrane proteins that do not undergo axially symmetric rotational diffusion show NMR spectra, which are characterized by broad anisotropic lineshapes with an asymmetry parameter η ≠ 0. Axially symmetric rotational diffusion of the membrane protein is manifested by anisotropic lineshapes indicative of axially symmetric chemical shift anisotropy tensors (η = 0). Superimposed to these broad lineshapes, large amplitude motions give rise to very narrow lines detected at the isotropic chemical shift of the respective resonances22,24. However, as static 15N NMR spectra are almost always acquired using cross polarization (CP), the recorded spectra show a strong bias towards rigid structures and dynamic features are only detected if sufficiently long CP contact times are used24.
Figure 2 shows static 15N NMR spectra of the GHS receptor reconstituted into DMPC membranes recorded with CP contact times of 70 μs (A), 2 ms (B), and 8 ms (C) at a temperature of 37 °C. All spectra show anisotropic lineshapes that can be simulated using an axially symmetric CSA tensor with a span of Δσ = 128–138 ppm. This is an indication that the receptor is in a monomeric form. As longer CP contact times are used, also narrow 15N NMR lines are detected, which are indicative of receptor segments undergoing large amplitude motions. These can be assigned to the Lys and Arg side chains resonating at higher magnetic field, but also to amide signals of the receptor backbone at chemical shifts of ~108, 116, and 124 ppm. This observation indicates that in addition to the axially symmetric rotational diffusion of the GHS receptor in the membrane, significant portions of the molecule also undergo large amplitude motions. We have previously quantified the proportion of highly mobile segments of the Y2 receptor by acquiring static 15N NMR spectra as a function of CP contact time as well as directly excited 15N NMR spectra24. It could be shown that static 15N CP spectra recorded at a CP contact time of 8 ms can be readily quantified to provide the relative proportion of relatively rigid and highly mobile residues24. Following this analysis, we can estimate that about 25% of the residues of the GHS receptor undergo large amplitude motions in DMPC membranes, which is significantly smaller than what has been determined for the Y2 receptor, for which about 40% of the entire spectral intensity of the static 15N NMR spectra was attributed to the isotropic signals.
Figure 2. Static 15N CP NMR spectra of the uniformly 15N-labeled GHS receptor reconstituted into DMPC membranes at a temperature of 37 °C.
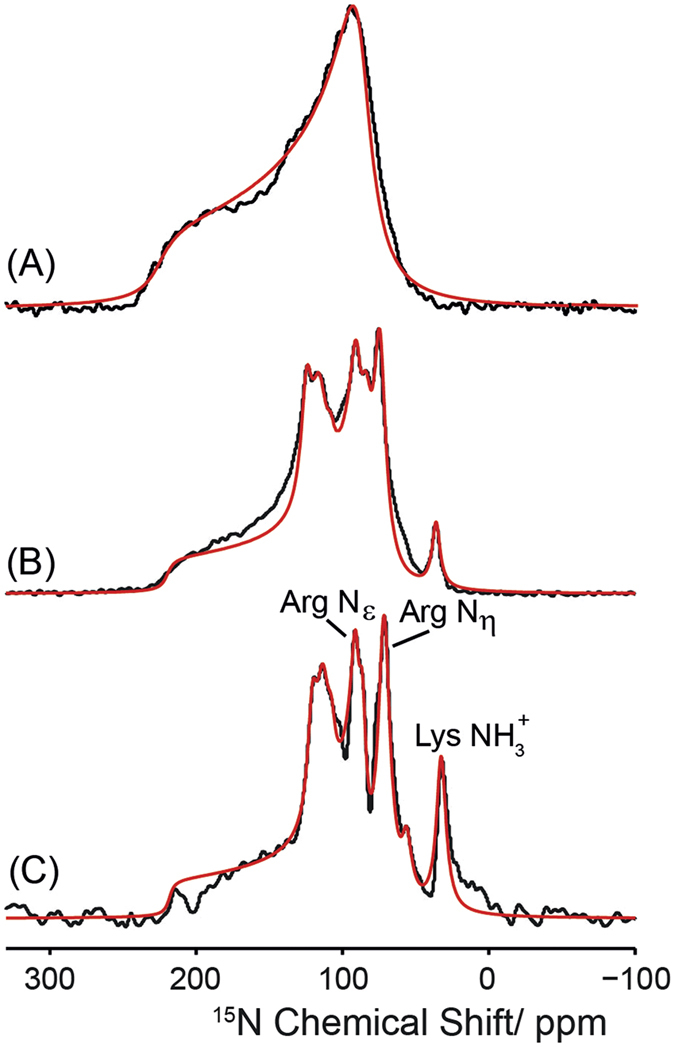
NMR spectra were acquired using CP with contact times of 70 μs (A), 2 ms (B), and 8 ms (C). Assignment of the prominent side chains is given in the NMR spectrum acquired at the longest CP contact time. Superimposed with the experimental data, best fit numerical simulations of the spectral line shapes are shown.
13C MAS NMR studies of the GHS receptor in membranes
13C NMR experiments recorded under MAS conditions provide a more direct way to investigate the molecular dynamics of membrane proteins. Provided that high resolution can be achieved either by selective labeling37 or quasi-crystalline preparation of the membrane protein of interest38,39,40, even side-specific dynamics information such as segmental order parameters can be determined. In the case of G protein-coupled receptors, spectral resolution is limited and only a single case has been reported in the literature where a large amount of sequential assignments have been achieved41. In our hands, the GHS receptor as well as the Y2 receptor reconstituted into DMPC or POPC membranes23,24,42 yielded relatively broad NMR lines under MAS at moderate frequencies rendering sequential assignments impossible so far.
Figure 3 shows typical 13C MAS NMR spectra of the uniformly 13C labeled GHS receptor reconstituted into DMPC-d54 membranes with perdeuterated lipid chains. In both 13C CPMAS (A) and directly excited 13C MAS NMR spectra (B), the NMR lines of the receptor are relatively broad and dispersed in a narrow chemical shift region as typical for α-helical membrane proteins41. Directly excited 13C MAS NMR spectra (B) show several narrow NMR lines, indicative of highly mobile sites especially for the 13CO resonances and the aromatic rings. A narrow signal from the Arg 13Cε side chain can also be well resolved. Some narrow lines can also be observed in the 13C INEPT spectrum of the GHS receptor in particular in the side chain region (C). However, the INEPT spectrum in the backbone region is dominated by lipid signals, which means that the fraction of receptor segments that undergo large amplitude motions is relatively small. Again, this is somewhat in contrast to our previous investigations on the molecular dynamics of the Y2 receptor23, where 13C INEPT spectra showed a larger proportion of narrow lines. Qualitatively, the 13C MAS NMR spectra of the GHS receptor reconstituted into POPC membranes are similar (data not shown), however, due to the lack of deuteration, more lipid signals are detected in the directly excited and INEPT NMR spectra.
Figure 3. 13C MAS NMR spectra of the uniformly 13C-labeled GHS receptor reconstituted into chain perdeuterated DMPC-d54 membranes using different 13C excitation pulse sequences.
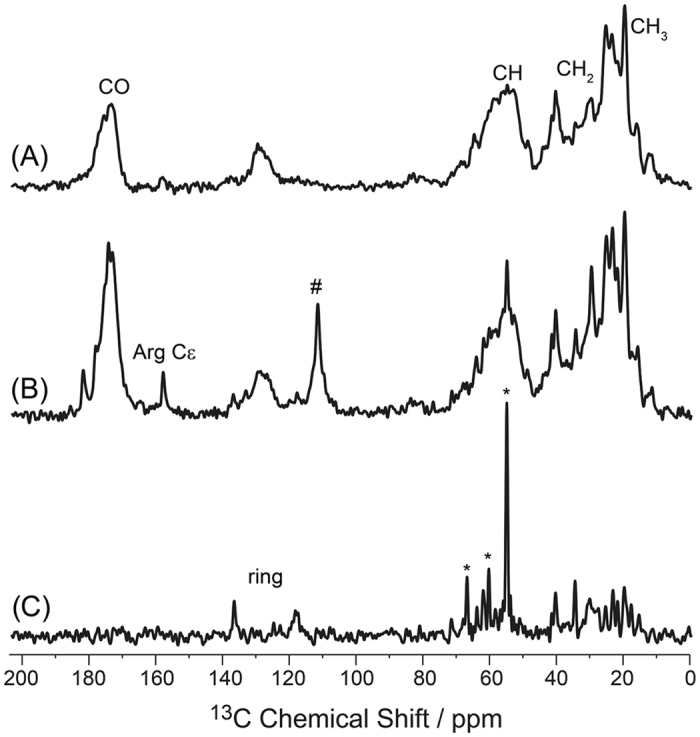
(A) 13C CPMAS spectrum using a CP contact time of 20 μs, (B) directly excited 13C MAS NMR spectrum. (C) 13C INEPT NMR spectrum. All NMR spectra were recorded at 37 °C at a MAS frequency of 7 kHz using Spinal64 decoupling with a radio frequency amplitude of ~65 kHz. The asterisk denotes lipid signals from the DMPC headgroup and glycerol backbone and the hashtag indicates the Teflon signal from the probe that is detected in directly excited 13C MAS NMR spectra.
Motional amplitudes of the GHS receptor in POPC and DMPC membranes
For a more quantitative discussion of the dynamics of the GHS receptor, we carried out 1H-13C dipolar coupling measurements using the DipShift pulse sequence43, which is a separated local field experiment44. Molecular motions of the C-H bond vector with a given amplitude scale down the dipolar couplings resulting in a reduced coupling constant provided these motions are faster than ~40 μs. Typical dipolar dephasing curves for two independent preparations and measurements are shown in Fig. S1. From this motional averaging, a molecular order parameter can be derived, which describes the amplitude of the motions of the respective segment. Typically, dipolar dephasing curves obtained under MAS conditions are measured, from which the exact value for the dipolar coupling is extracted in a simulation. All preparations and measurements were done at least twice; error bars represent the difference between two independent preparations.
The average order parameters of the resolved segments of the GHS receptor in POPC and DMPC membranes are reported in Fig. 4. Similar to the static 15N measurements, 1H-13C DipShift experiments that use a cross polarization transfer step are biased by the presence of motions in the molecule. At short CP contact time, only the rigid segments are efficiently excited and contribute to the spectral intensity, which most likely involve the transmembrane helices of the receptor. DipShift experiments acquired at longer CP contact times also excite the more mobile parts of the receptor, which are constituted by larger loops and tail regions. All 13C sites are equally excited if directly excited DipShift experiments are carried out. As a consequence, the measured order parameters are largest for short CP contact times and decrease upon increase of the contact times. Lowest order parameters are always determined for the directly excited DipShift experiments. In addition to the contact time dependence of the order parameters, we also observe order parameter differences for the individual positions in the protein backbone and sidechain. Highest order parameters are observed for the Cα and Gly Cα sites, followed by CH2 groups in the side chain and lowest order parameters are observed for the CH3 groups. Order parameters of the GHS receptor in POPC membranes are typically slightly higher than those in DMPC bilayers but the difference is much smaller as determined for the Y2 receptor before23,24. Without the motional bias of the CP experiment, lowest order parameters determined for the protein backbone were 0.60–0.69 for POPC and 0.56–0.65 for DMPC membranes. These values are slightly higher than what has been observed for the Y2 receptor before23,24. Order parameters values of the GHS receptor in DMPC and POPC membranes are reported in Tables S1 and S2 in the Supporting Information.
Figure 4. 1H-13C NMR order parameters for backbone and side-chain carbons of the uniformly 13C labeled GHS receptor reconstituted into POPC (filled symbols) and DMPC membranes (open symbols) at varying CP contact times and a temperature of 37 °C.
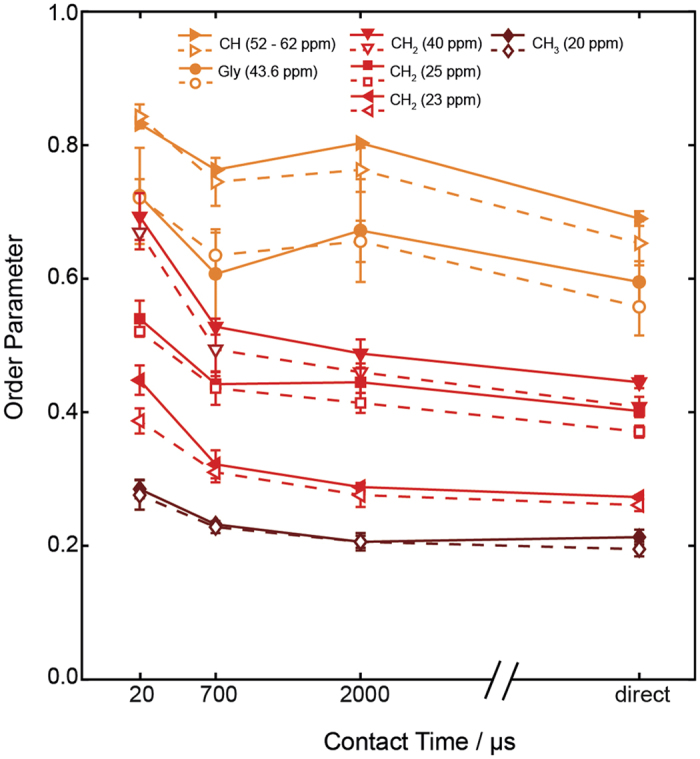
C-H dipolar coupling constants were measured using DipShift experiments and averaged over several signals in the same 13C chemical shift region or for specific peaks.
As INEPT-based polarization transfer also provided some receptor signals, we measured the motionally averaged dipolar couplings of receptor segments that could be excited by INEPT using the separated local field experiment r-PDLF45. Residues that can be detected by INEPT-based pulse sequences have to be highly mobile, which is the case especially for the termini of the receptor22,41. Figure S2 shows experimental r-PDLF spectra of the GHS receptor in the presence of its ligand ghrelin as well as numerical simulations to determine the motionally averaged dipolar coupling for the signals detected at 23 and 40 ppm, respectively. Such chemical shifts are typical for the aliphatic side chains and could correspond to Cγ sites of amino acids such as Lys, Thr, or Val and the Cβ site of Leu, respectively. The determined order parameters are 0.09 for the peak at 23 ppm and 0.05 for the peak at 40 ppm.
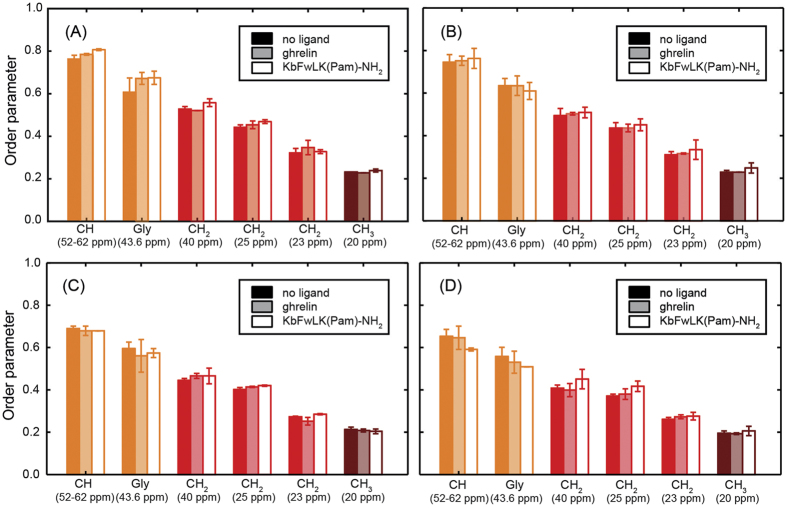
Order parameters of the GHS receptor in the presence of the agonist and an inverse agonist
Agonists and inverse agonists shift the equilibrium of active and inactive G protein-coupled receptor towards the active or inactive conformation, respectively. As the GHS receptor has been reported to display a ~50% constitutive activity35,46, studying the molecular dynamics of the receptor in the presence of either agonist or inverse agonist represents an interesting topic8. Figure 5 shows the order parameters of the GHS receptor in the absence as well as in the presence of either the agonist ghrelin or the inverse agonist KbFwLK(Pam)-NH2 recorded in DipShift experiments that were excited by CP at a contact time of 700 μs (A, B) or by direct 13C excitation (C, D) both in POPC (A, C) and DMPC membranes (B, D). Order parameters determined by CP at a contact time of 700 μs are generally higher than those obtained from directly excited DipShift experiments irrespective of the presence or absence of any ligand. Order parameters determined from the CP based DipShift experiments at 700 μs contact time, which preferentially detects the more rigid structures show a tendency to be slightly higher for the ligand bound GHS receptor. However, this is just a trend, which is not consistent for all receptor segments studied and only in a few cases outside of the error margin of the experiment, which was determined from preparations and measurements carried out in duplicate. Order parameters determined by direct 13C excitation, which do not have a bias by molecular motions with varying amplitudes are identical within the experimental error for all three studied cases. No systematic differences between receptor preparations in saturated DMPC versus monounsaturated POPC membranes were observed. All order parameters are reported in Tables S1 and S2.
Figure 5. Summary of the order parameter measurements of the GHS receptor in the absence of ligand (filled bars) and in the presence of the agonist ghrelin (light filled bars) and the inverse agonist KbFwLK(Pam)-NH2 (empty bars) performed this study.
Order parameters were determined from DipShift experiments conducted using CP excitation at a contact time of 700 μs (A,B) as well as by direct excitation (C,D). The GHS receptor was reconstituted into POPC (A,C) or DMPC membranes (B,D) and investigated at a temperature of 37 °C.
Discussion
We describe the expression, functional reconstitution in lipid membranes, and preliminary NMR investigation of the human GHS receptor from E. coli culture. The receptor could be labeled with stable 13C and 15N isotopes allowing for an initial characterization by solid-state NMR. At the current preliminary state, unfortunately, the spectral resolution does not allow for a site-specific assignment of the NMR lines and the parameters that have been determined from the solid-state NMR measurements are averaged over the entire molecule. This situation lags behind the capabilities of modern NMR spectroscopy as demonstrated for other GPCRs41 or membrane proteins of similar size38,47. We suspect that the mobility of the molecule in fluid membranes is the reason for the limited spectral resolution. For a more site-specific approach, expression in auxotrophic E. coli strains or cell free expression might improve the situation. Although with the current initial results, only information averaged over the entire molecule can be obtained, there are some important conclusion that can be drawn with regard to the biological meaning of the results of this study.
The explicit studies on the GHR receptor dynamics confirm that GPCRs are highly dynamic membrane proteins
Our data of the ground state dynamics of the GHS receptor in membranes indicate segmental fluctuations and rapid conformational transitions of the molecule on a submicrosecond timescale. This correlation time window includes local dynamics, mobility of secondary structure elements as well as more collective motions. In addition to the axially symmetric rotational diffusion of the protein in the membrane, we detected fluctuations in the backbone of the receptor described by relatively low order parameters. Although no site resolution has been achieved, the data allow some conclusions about domain-specific dynamics. At the very short CP contact time of 20 μs, only the rigid segments of the receptor such as the central parts of the transmembrane α-helices are excited, these segments show relatively low conformational freedom expressed by a high order parameter of 0.72–0.84. However, this is clearly smaller than what has been measured for the transmembrane segments of Anabaena sensory rhodopsin, where site-specific order parameters of 0.9 and higher have been reported for the transmembrane helices (vide infra)38. In contrast, we can also excite very mobile receptor domains using the INEPT polarization transfer scheme, which revealed that signals with order parameters lower than 0.1 can be detected, which likely belong to the highly mobile termini. This domain-specific data demonstrates that the conformational flexibility of GPCRs is broadly distributed between the individual segments from highly mobile to very restricted.
These dynamic properties represent a consequence of the energy landscapes, on which these molecules exist20, which is described as a rugged surface that allows for a multitude of conformational states, separated by low energy barriers on the order of the thermal energy that can be overcome by fluctuations of various amplitudes6. This translates into an inherent flexibility of the individual segments of the GPCR at equilibrium. Such fluctuations have also been observed in recent molecular dynamics simulations of the neurotensin receptor48. This receptor shows quite significant root mean square fluctuations in the backbone in particular for the loop and tail structures. Interestingly, these molecular dynamics simulations show that thermostabilization of the neurotensin receptor leads to a significant reduction of these fluctuations48. Also, crystallographic B factors of this and other GPCRs are relatively high and crystal structures often lack electron density for loop and tail structures, which suggests that these segments may undergo large amplitude motions or are statically disordered11,49.
The GHS receptor is more mobile than other membrane proteins of comparable size
Compared to other membrane proteins of similar size38,50,51, the order parameters of the GHS receptor are significantly lower. For the backbone, the measured order parameters translate into average fluctuations with amplitudes on the order of 40–50°. Compared to the neuropeptide Y2 receptor using the same set of experiments23,24 the GHS receptor shows slightly higher order parameters. An obvious difference between the GHS and the Y2 receptor is the distribution between the lengths of the loops and tails on the one and transmembrane α-helical segments on the other hand. In the GHS receptor, about 56% of the residues constitute loops and tails, while this number increases to 61% for the Y2 receptor. In particular, the termini of the Y2 receptor are significantly longer (in total 107 vs. 80 residues). Assuming that loops and termini are more flexible than the helical segments, the difference in the dynamics between the two molecules can be well accounted for.
It is obvious that β-barrel membrane proteins are intrinsically more stabilized than molecules of predominantly α-helical structure. Intrastrand hydrogen bonds lead to high structural homogeneity of β-barrels, which are expressed in high backbone order parameters on the order of 0.9 as observed for OmpA from different sources47,51. On the secondary structure level, isolated α-helices are more flexible. In particular, the helix ends are subject to dynamic processes that have been seen both experimentally52 and in molecular dynamics simulations53. Furthermore, transmembrane α-helices are also known to show some interesting structural plasticity, which includes dynamic transitions between α-helical and β-sheet structures54,55. The α-helical protein colicin Ia in the membrane bound state showed order parameters between 0.88 and 0.93 for the backbone at a CP contact time of 0.7 ms50, which is much higher than what has been observed for the GHS receptor under these conditions here (order parameters for the backbone at a CP contact time of 0.7 ms were between 0.61 and 0.81).
The general heptahelical architecture of a membrane protein is not the determining factor for its molecular dynamics in fluid lipid membranes
GPCRs appear to be specifically more flexible than heptahelical microbial rhodopsins such as bacteriorhodopsin56,57, proteorhodopsin39, or Anabaena sensory rhodopsin38, for which more ordered structures with higher order parameters have been reported. This may in part be due to the 2D crystalline nature of the host membrane, but also membrane reconstituted microbial rhodopsins in the lamellar liquid-crystalline lipid phase showed high order39. Although Gly-rich motifs (GxxG and GxxxG) are discussed to be responsible for structural plasticity of fusion peptides58 the transmembrane regions of the GHS receptor have fewer Gly residues than the respective segments of both proteorhodopsin or Anabaena sensory rhodopsin. Therefore, the only obvious difference in the topology of the GPCRs and the microbial rhodopsins are the significantly longer loops that connect the transmembrane segments, which likely provides the molecular origin for the enhanced mobility of the interesting GPCRs.
Agonist or inverse agonist binding does not alter the average dynamics of the GHS receptor
Another obvious difference between individual members of the GPCR family is the constitutive activity of the GHS receptor, which is on the order of 50%35,46. As well investigated for the β2 adrenergic receptor, ligand binding alters the shape of the energy landscape and the receptor populates different conformations8,20. However, agonist bound β2 adrenergic receptor was found to be also highly dynamic and to interconvert between inactive, intermediate, and active conformations18. To further investigate the dynamics of the GHS receptor in the activated state as well as at lower efficacy, we studied the dynamics of the molecule in the presence of the agonist ghrelin as well as an inverse agonist. By and large, we did not see a conclusive trend of order parameter changes in response to agonist or inverse agonist binding. The bulk measurement as conducted here is likely not sensitive to detect the influence of the few residues that might change their dynamics in response to ligand binding. This suggests that although the energy landscape of the receptor is changed upon ligand binding, the molecular mobility remains on a relatively high level. This agrees with fluorescence studies on the GHS receptor8 and the results on the β2 adrenergic receptor, which show that the agonist-bound molecule is highly dynamic and interconverts between inactive, intermediate, and active conformations with varying timescales18.
The membrane environment only slightly influences the molecular dynamics of the GHS receptor
Although GPCRs are characteristic for specific tissues and are found in lipid membranes of highly specific compositions, we note that the molecular dynamics of the GHS receptor is also sensitive to the membrane environment. Although unsaturated lipid membranes (such as POPC) are in general more mobile that saturated ones (i.e. DMPC), the molecular mobility of the GHS receptor in DMPC was a bit higher than in POPC59. This can be related to the hydrophobic thickness of either membrane, which is higher in POPC than in DMPC, which leads to a slight increase in the α-helix length of the receptor, which rigidifies these residues to decrease the overall order parameters of the molecule24,60. This underlines the dynamic adaptivity and the structural plasticity of the GHS receptor and possibly of many members of the GPCR family. Such high flexibility increasingly appears as a general property of many GPCRs23,24 required for the biological function of these molecules.
In summary, although in this preliminary state of the project, no site-specific dynamics information was achieved, the experiments show that the GHS receptor is highly mobile in liquid-crystalline lipid membranes. Given the current resolution of the NMR spectra of the GHS receptor, no site-specific assignments were possible, restricting the conclusions to an average picture, which nevertheless underlines the outstanding position of the physiologically highly important GPCRs among other membrane proteins. In situations, where spectral resolution limits the assignability of the NMR spectra, specific labeling by cell-free synthesis or usage of auxotrophic E. coli strains may provide further insights into the molecular dynamics of GPCRs going beyond the initial characterization provided here.
Methods
Materials
The lipids 1-palmityl-2-oleyl-sn-glycero-3-phospocholine (POPC) or 1,2-dimyristol-sn-glycero-3-phosphocholine (DMPC) along with the chain deuterated analog DMPC-d54 as well as 1,2-dihexanoyl-sn-glycero-3-phosphocholine (DHPC) were purchased from Avanti Polar Lipids, Inc. (Alabaster, USA). Chemicals for fermentation were purchased from Sigma-Aldrich (Taufkirchen, Germany) and isotopically labeled ammonium salts and glucose were purchased from Cambridge Isotopes, USA.
Peptide Synthesis
Neuropeptide Y (NPY), ghrelin and the inverse agonist KbFwLK(Pam)-NH2 (b = β-(3-benzothienyl)-D-alanine) were synthesized by Fmoc/tert-butyl strategy as described before28,34. Fluorescent tracer [Dpr3-Oct; Dpr16-atto520]-ghrelin (Dpr, diaminopropionic acid; Oct, octanoic acid) for in vitro functionality assay was prepared similarly as described for a fluorescein-conjugated ghrelin analogue34. [Boc-G1; Dpr3(Mtt); Dpr16(Dde)]-ghrelin was synthesized automatically on R-Wang resin. Mtt (4-methyltrityl) protection group was then cleaved by repeated treatment with 94/5/1 dichloromethane/triisopropylsilane/trifluoroacetic acid (v/v/v) (15 × 1 min), and resin was washed with 97.5/2.5 (v/v) dichloromethane/N,N-diisopropylethylamine (2 × 10 min). Octanoic acid was coupled using 1-hydroxybenzotriazole/diisopropylcarbodiimide (5 eq. each) in dimethylformamide overnight. For coupling of atto520 dye, Dde (2-acetyldimedone) protection group was cleaved by repeated treatment with 98/2 (v/v) dimethylformamide/hydrazine (10 × 10 min), and 1 eq. of atto520 (free acid; Sigma-Aldrich, Taufkirchen, Germany) was coupled using 1-hydroxybenzotriazole/diisopropycarbodiimide in dimethylformamide overnight. The same protocol was applied for the preparation of [Dpr22-atto520]-NPY as control.
Expression of the GHS receptor
The human wild type GHSR1a including an N-terminal eightfold His-tag was overexpressed in Escherichia coli in defined mineral salt medium using a fed-batch fermentation process as described before42,61. For uniform 15N labeling, 15NH4Cl and (15NH4)2SO4 (Cambridge Isotope Lab., USA) was added as sole nitrogen source during fermentation. Furthermore, uniform 13C labeling was achieved by applying U-13C6 glucose (Cambridge Isotope Lab., USA) during the feed phase 30 minutes prior to the induction of expression through addition of ITPG. Expression times varied from 3 to 4 hours. Preparation of inclusion bodies, followed by the solubilization and the purification of the receptor was conducted as previously recorded62.
Receptor folding and NMR sample preparation
To induce the formation of the disulfide bridge in the GHS receptor, the purified receptor was dialyzed against a folding buffer containing 1 mM oxidized (GSSG) and 2 mM (GSH) reduced glutathione at pH 8, as previously described24. For receptor reconstitution POPC or DMPC was suspended in 50 mM NaP (pH 8) reaching a lipid concentration of 10 mg/ml. Subsequently, lipid dispersions were merged with a sixfold (in case of POPC) or fourfold (for DMPC) molar excess of the detergent DHPC by heating to 40 °C. For POPC/DHPC solutions, three cycles of temperature jumps between 0 °C and 40 °C were completed. The receptor dispersion was incubated at a molar ratio of 1:200 (receptor: lipid) and three additional heating/cooling cycles were conducted. Removal of the detergent was accomplished by application of BioBeads SM2 (Bio-Rad Lab., Germany), where 50 mg/ml were added twice. To examine the influence of ligands, after bicelle preparation, either the natural agonist ghrelin or an inverse agonist KbFwLK(Pam)-NH2 were added in a molar ratio of 1:2 (receptor:ligand). Preparations were completed by ultracentrifugation of the sample at 86,000 rpm for ~12 hours. Final MAS samples contained ~6 mg of receptor. Samples used for static 15N NMR spectra contained ~12 mg of receptor. Chain deuterated DMPC-d54 instead of DMPC was used to suppress aliphatic lipid signals.
In vitro binding assay
Functionality of GHS receptor preparations was tested by a homogeneous fluorescence binding assay using [Dpr3-Oct; Dpr16-atto520]-ghrelin (ghrelin-atto520), detecting fluorescence enhancement upon receptor binding29. Functionality of the fluorescent tracer ghrelin-atto520 was verified in 3H-inositol phosphate accumulation assays as described before displaying an EC50 value of 1.7 nM (pEC50 = 8.8 ± 0.10; ghrelin: pEC50 = 9.0 ± 0.15).
For saturation binding analysis, 25 nM or 100 nM of ghrelin-atto520 was incubated with increasing concentration of GHS receptor containing bicelles in binding buffer (Tris 50 mM pH 7, 10 mM CaCl2, 1 mM MgCl2, 0.01% TritonX-100) or bicelles prepared in the absence of receptor protein for 2 h under gentle agitation in opaque 96 well plates. For displacement assays, 3.16 μM GHS receptor in buffer was incubated with 100 nM ghrelin-atto 520 and increasing concentrations of unlabeled peptides. Fluorescence intensity was measured in a plate reader (FlexStation 3, Molecular Devices, Sunnyvale, USA); using linear polarized fluorescence light, an excitation wavelength of 520 nm, an emission wavelength of 540 nm (beam splitter 530 nm), PMT medium, 90° detection angle. Experiments were carried out three times independently in triplicates. Data were normalized to fluorescence in unbound state (100%), and combined into a single curve.
Data were fitted to the Hill equation to determine the inflection point. For displacement experiments, Goldstein-Barrett correction31 was applied to determine Ki values, using the KD of 0.15 nM reported previously33.
NMR experiments
Static 15N CP NMR spectra were acquired on a Bruker Avance I 750 MHz NMR spectrometer (Bruker Biospin, Rheinstetten) at a temperature of 37 °C. Standard CP spectra with contact times of either 70 μs, 2 ms, or 8 ms were recorded using Hahn echo detection. TPPM decoupling at a radio field strength of ~62 kHz was applied. Typical 90° pulse lengths were 7 μs and 6 μs on the 1H and 15N channel, respectively. Spectra line shapes were simulated using MathCad, considering anisotropic and isotropic contributions.
13C magic-angle spinning (MAS) NMR experiments were carried out on a Bruker Avance III 600 MHz NMR spectrometer using MAS probes equipped with 3.2 or 4 mm spinning modules at a temperature of 37 °C. The 90° pulses were usually set to 4 μs for both, 1H and 13C channels. The heteronuclear decoupling sequence Spinal64 was used at a field strength of ~65 kHz. One dimensional 13C NMR experiments (either CP MAS, directly excited and INEPT experiments) were conducted at a MAS frequency of 7 kHz. For the 13C CP MAS NMR spectra, a contact time of 700 μs was used. The 1H-13C dipolar couplings were measured using the DipShift pulse sequence43 at a MAS frequency of 5 kHz. For homonuclear 1H-1H decoupling, the FSLG sequence was used63. Excitation of 13C nuclei was either accomplished by direct excitation or applying CP with contact times ranging from 20 μs to 2 ms. While using a short contact time of 20 μs the RODEO DipShift experiment64 was used to avoid distortion effects in the dephasing curves. Dephasing signals were acquired over one rotor period for all resolved isotropic chemical shift signals. Averaged dipolar coupling strengths were determined by best fit of experimental dephasing curves as described in the literature50. Order parameters were calculated as the ratio of the motionally averaged dipolar coupling strength and the rigid limit value of the dipolar coupling as determined experimentally50,65. R-PDLF experiments using a R187145 pulse scheme were carried out at 5 kHz MAS frequency. Polarization transfer was accomplished by an INEPT step. For 1H and 13C pulse lengths of 5 μs were used. The decoupling field strength by use of Spinal 64 was set to 20 kHz to allow acquisition of a larger number of increments in the indirect dimension.
Additional Information
How to cite this article: Schrottke, S. et al. Expression, Functional Characterization, and Solid-State NMR Investigation of the G Protein-Coupled GHS Receptor in Bilayer Membranes. Sci. Rep. 7, 46128; doi: 10.1038/srep46128 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We would like to acknowledge Prof. Dr. Kay Saalwächer and Dr. Tiago Mendes Ferreira for useful discussions and help in implementing the r-PDLF experiment. Parts of this study were supported by the EU and the Free State of Saxony (ESF 100148835 and 100227414).
Footnotes
The authors declare no competing financial interests.
Author Contributions D.H. conceived the project. S.S., A.K., H.A.S., M.B., S.E.-H., P.S., A.B.-S., and D.H. designed the experiments. A.K., S.E.-H. and A.B.-S. contributed reagents. S.S., A.K., G.V., H.A.S., D.W., M.B., P.S. and D.H. performed the experiments and analyzed results. S.S., and D.H. wrote the manuscript. D.H. coordinated the project and supervised the research. All authors contributed through scientific discussions.
References
- Rosenbaum D. M. et al. GPCR engineering yields high-resolution structural insights into beta(2)-adrenergic receptor function. Science 318, 1266–1273 (2007). [DOI] [PubMed] [Google Scholar]
- Warne T. et al. Structure of a beta(1)-adrenergic G-protein-coupled receptor. Nature 454, 486–491 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steyaert J. & Kobilka B. K. Nanobody stabilization of G protein-coupled receptor conformational states. Curr. Opin. Struct. Biol. 21, 567–572 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatakrishnan A. J. et al. Molecular signatures of G-protein-coupled receptors. Nature. 494, 185–194 (2013). [DOI] [PubMed] [Google Scholar]
- Frauenfelder H., Sligar S. G. & Wolynes P. G. The energy landscapes and motions of proteins. Science. 254, 1598–1603 (1991). [DOI] [PubMed] [Google Scholar]
- Henzler-Wildman K. & Kern D. Dynamic personalities of proteins. Nature. 450, 964–972 (2007). [DOI] [PubMed] [Google Scholar]
- Huster D. Investigations of the structure and dynamics of membrane-associated peptides by magic angle spinning NMR. Prog. Nucl. Magn. Reson. Spectrosc. 46, 79–107 (2005). [Google Scholar]
- Mary S. et al. Ligands and signaling proteins govern the conformational landscape explored by a G protein-coupled receptor. Proc. Natl. Acad. Sci. USA 109, 8304–8309 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay J. F. & Farrens D. L. Structural dynamics and energetics underlying allosteric inactivation of the cannabinoid receptor CB1. Proc. Natl. Acad. Sci. USA 112, 8469–8474 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X. et al. Coupling ligand structure to specific conformational switches in the beta2-adrenoceptor. Nat. Chem. Biol. 2, 417–422 (2006). [DOI] [PubMed] [Google Scholar]
- Hubbell W. L., Altenbach C., Hubbell C. M. & Khorana H. G. Rhodopsin structure, dynamics, and activation: a perspective from crystallography, site-directed spin labeling, sulfhydryl reactivity, and disulfide cross-linking. Adv. Protein Chem. 63, 243–290 (2003). [DOI] [PubMed] [Google Scholar]
- Columbus L. & Hubbell W. L. A new spin on protein dynamics. Trends Biochem. Sci. 27, 288–295 (2002). [DOI] [PubMed] [Google Scholar]
- Liu J. J. et al. Biased signaling pathways in beta2-adrenergic receptor characterized by 19F NMR. Science 335, 1106–1110 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okude J. et al. Identification of a conformational equilibrium that determines the efficacy and functional selectivity of the mu-opioid receptor. Angew. Chem. Int. Ed. 54, 15771–15776 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T. H. et al. The role of ligands on the equilibria between functional states of a G protein-coupled receptor. J. Am. Chem. Soc. 135, 9465–9474 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horst R., Liu J. J., Stevens R. C. & Wuthrich K. Beta-adrenergic receptor activation by agonists studied with 19F NMR spectroscopy. Angew. Chem. Int. Ed. 52, 10762–10765 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofuku Y. et al. Efficacy of the beta(2)-adrenergic receptor is determined by conformational equilibrium in the transmembrane region. Nat. Commun. 3, 1045 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manglik A. et al. Structural insights into the dynamic process of beta2-adrenergic receptor signaling. Cell. 161, 1101–1111 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sounier R. et al. Propagation of conformational changes during mu-opioid receptor activation. Nature. 524, 375–378 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deupi X. & Kobilka B. K. Energy landscapes as a tool to integrate GPCR structure, dynamics, and function. Physiology. 25, 293–303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manglik A. & Kobilka B. The role of protein dynamics in GPCR function: insights from the beta2AR and rhodopsin. Curr. Opin. Cell Biol. 27, 136–143 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S. H. et al. Local and global dynamics of the G protein-coupled receptor CXCR1. Biochemistry. 50, 2371–2380 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt P. et al. The G protein-coupled neuropeptide Y receptor type 2 is highly dynamic in lipid membranes as revealed by solid-state NMR spectroscopy. Chemistry 20, 4986–4992 (2014). [DOI] [PubMed] [Google Scholar]
- Thomas L. et al. The molecular dynamics of the G protein-coupled neuropeptide Y2 receptor in monounsaturated membranes investigated by solid-state NMR spectroscopy. J. Biomol. NMR 61, 347–359 (2015). [DOI] [PubMed] [Google Scholar]
- Wiedmer P. et al. Ghrelin, obesity and diabetes. Nat. Clin. Pract. Endocrinol. Metab. 3, 705–712 (2007). [DOI] [PubMed] [Google Scholar]
- Vortmeier G. et al. Integrating solid-state NMR and computational modeling to investigate the structure and dynamics of membrane-associated ghrelin. PLoS. One. 10, e0122444 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima M. et al. Ghrelin is a growth-hormone-releasing acylated peptide from stomach. Nature. 402, 656–660 (1999). [DOI] [PubMed] [Google Scholar]
- Kostelnik K. B. et al. High metabolic in vivo stability and bioavailability of a palmitoylated ghrelin receptor ligand assessed by mass spectrometry. Bioorg. Med. Chem. 23, 3925–3932 (2015). [DOI] [PubMed] [Google Scholar]
- Casiraghi M. et al. Functional modulation of a G protein-coupled receptor conformational landscape in a lipid bilayer. J. Am. Chem. Soc. 138, 11170–11175 (2016). [DOI] [PubMed] [Google Scholar]
- Hulme E. C. & Trevethick M. A. Ligand binding assays at equilibrium: validation and interpretation. Br. J. Pharmacol. 161, 1219–1237 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein A. & Barrett R. W. Ligand dissociation constants from competition binding assays: errors associated with ligand depletion. Mol. Pharmacol. 31, 603–609 (1987). [PubMed] [Google Scholar]
- Carter C. M., Leighton-Davies J. R. & Charlton S. J. Miniaturized receptor binding assays: complications arising from ligand depletion. J. Biomol. Screen. 12, 255–266 (2007). [DOI] [PubMed] [Google Scholar]
- Els S. et al. An aromatic region to induce a switch between agonism and inverse agonism at the ghrelin receptor. J. Med. Chem. 55, 7437–7449 (2012). [DOI] [PubMed] [Google Scholar]
- Park M. et al. Bioorthogonal Labeling of Ghrelin Receptor to Facilitate Studies of Ligand-Dependent Conformational Dynamics. Chem. Biol. 22, 1431–1436 (2015). [DOI] [PubMed] [Google Scholar]
- Damian M. et al. Ghrelin receptor conformational dynamics regulate the transition from a preassembled to an active receptor:Gq complex. Proc. Natl. Acad. Sci. USA 112, 1601–1606 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opella S. J. Protein dynamics by solid state nuclear magnetic resonance. Methods Enzymol. 131, 327–361 (1986). [DOI] [PubMed] [Google Scholar]
- Reuther G. et al. The lipidated membrane anchor of the N-ras protein shows an extensive dynamics as revealed by solid-state NMR. J. Am. Chem. Soc. 128, 13840–13846 (2006). [DOI] [PubMed] [Google Scholar]
- Good D. B. et al. Conformational dynamics of a seven transmembrane helical protein Anabaena Sensory Rhodopsin probed by solid-state NMR. J. Am. Chem. Soc. 136, 2833–2842 (2014). [DOI] [PubMed] [Google Scholar]
- Yang J., Aslimovska L. & Glaubitz C. Molecular dynamics of proteorhodopsin in lipid bilayers by solid-state NMR. J. Am. Chem. Soc. 133, 4874–4881 (2011). [DOI] [PubMed] [Google Scholar]
- Lorieau J. L. & McDermott A. E. Conformational flexibility of a microcrystalline globular protein: Order parameters by solid-state NMR spectroscopy. J. Am. Chem. Soc. 128, 11505–11512 (2006). [DOI] [PubMed] [Google Scholar]
- Park S. H. et al. Structure of the chemokine receptor CXCR1 in phospholipid bilayers. Nature. 491, 779–783 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser A. et al. Unwinding of the C-Terminal Residues of Neuropeptide Y is critical for Y(2) Receptor Binding and Activation. Angew. Chem. Int. Ed. 54, 7446–7449 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munowitz M. G., Griffin R. G., Bodenhausen G. & Huang T. H. Two-dimensional rotational spin-echo nuclear magnetic resonance in solids: correlation of chemical shift and dipolar interactions. J. Am. Chem. Soc. 103, 2529–2533 (1981). [Google Scholar]
- Waugh J. S. Uncoupling of local field spectra in nuclear magnetic resonance: determination of atomic positions in solids. Proc. Natl. Acad. Sci. USA 73, 1394–1397 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvinskikh S. V., Zimmermann H., Maliniak A. & Sandstrom D. Measurements of motionally averaged heteronuclear dipolar couplings in MAS NMR using R-type recoupling. J. Magn Reson. 168, 194–201 (2004). [DOI] [PubMed] [Google Scholar]
- Damian M. et al. High constitutive activity is an intrinsic feature of ghrelin receptor protein: a study with a functional monomeric GHS-R1a receptor reconstituted in lipid discs. J. Biol. Chem. 287, 3630–3641 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurel O. et al. Local and Global Dynamics in Klebsiella pneumoniae Outer Membrane Protein a in Lipid Bilayers Probed at Atomic Resolution. J. Am. Chem. Soc. 139, 1590–1597 (2017). [DOI] [PubMed] [Google Scholar]
- Lee S. et al. Structural dynamics and thermostabilization of neurotensin receptor 1. J. Phys. Chem. B. 119, 4917–4928 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X., Zhao X. & Watts A. G-protein-coupled receptor structure, ligand binding and activation as studied by solid-state NMR spectroscopy. Biochem. J. 450, 443–457 (2013). [DOI] [PubMed] [Google Scholar]
- Huster D., Xiao L. & Hong M. Solid-state NMR investigation of the dynamics of soluble and membrane-bound colicin Ia channel-forming domain. Biochemistry 40, 7662–7674 (2001). [DOI] [PubMed] [Google Scholar]
- Liang B., Arora A. & Tamm L. K. Fast-time scale dynamics of outer membrane protein A by extended model-free analysis of NMR relaxation data. Biochim. Biophys. Acta. 1798, 68–76 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel A., Scheidt H. A. & Huster D. The distribution of lipid attached EPR probes in bilayers. Application to membrane protein topology. Biophys. J. 85, 1691–1701 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quint S. et al. Residue-specific side-chain packing determines the backbone dynamics of transmembrane model helices. Biophys. J. 99, 2541–2549 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H. & Hong M. Membrane-dependent conformation, dynamics, and lipid interactions of the fusion peptide of the paramyxovirus PIV5 from solid-state NMR. J. Mol. Biol. 425, 563–576 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J. X., Yau W. M. & Tycko R. Evidence from solid-state NMR for nonhelical conformations in the transmembrane domain of the amyloid precursor protein. Biophys. J. 100, 711–719 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barré P., Yamaguchi S., Saito H. & Huster D. Backbone dynamics of bacteriorhodopsin as studied by 13C solid-state NMR spectroscopy. Eur. Biophys. J. 32, 578–584 (2003). [DOI] [PubMed] [Google Scholar]
- Saito H. Dynamic pictures of membrane proteins in two-dimensional crystal, lipid bilayer and detergent as revealed by site-directed solid-state C-13 NMR. Chem. Phys. Lipids 132, 101–112 (2004). [DOI] [PubMed] [Google Scholar]
- Yao H. & Hong M. Conformation and lipid interaction of the fusion peptide of the paramyxovirus PIV5 in anionic and negative-curvature membranes from solid-state NMR. J. Am. Chem. Soc. 136, 2611–2624 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huster D., Arnold K. & Gawrisch K. Influence of docosahexaenoic acid and cholesterol on lateral lipid organization in phospholipid membranes. Biochemistry 37, 17299–17308 (1998). [DOI] [PubMed] [Google Scholar]
- Soubias O., Niu S. L., Mitchell D. C. & Gawrisch K. Lipid-rhodopsin hydrophobic mismatch alters rhodopsin helical content. J. Am. Chem. Soc. 130, 12465–12471 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger C., Montag C., Berndt S. & Huster D. Optimization of Escherichia coli cultivation methods for high yield neuropeptide Y receptor type 2 production. Protein Expr. Purif. 76, 25–35 (2011). [DOI] [PubMed] [Google Scholar]
- Schmidt P. et al. A reconstitution protocol for the in vitro folded human G protein-coupled Y2 receptor into lipid environment. Biophys. Chem. 150, 29–36 (2010). [DOI] [PubMed] [Google Scholar]
- Bielecki A., Kolbert A. C. & Levitt M. H. Frequency-switched pulse sequences: homonuclear decoupling and dilute spin NMR in solids. Chem. Phys. Lett. 155, 341–345 (1989). [Google Scholar]
- Kurz R. et al. Avoiding bias effects in NMR experiments for heteronuclear dipole-dipole coupling determinations: principles and application to organic semiconductor materials. Chemphyschem 14, 3146–3155 (2013). [DOI] [PubMed] [Google Scholar]
- Barré P., Zschörnig O., Arnold K. & Huster D. Structural and dynamical changes of the bindin B18 peptide upon binding to lipid membranes. A solid-state NMR study. Biochemistry 42, 8377–8386 (2003). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.