Abstract
We report the development, characterization, and validation of a peptide specific for the extracellular domain of HER2. This probe chemistry was developed for molecular imaging by using a structural model to select an optimal combination of amino acids that maximize the likelihood for unique hydrophobic and hydrophilic interactions with HER2 domain 3. The sequence KSPNPRF was identified and conjugated with either FITC or Cy5.5 via a GGGSK linker using Fmoc-mediated solid-phase synthesis to demonstrate fiexibility for this chemical structure to be labeled with different fluorophores. A scrambled sequence was developed for control by altering the conformationally rigid spacer and moving both hydrophobic and hydrophilic amino acids on the C-terminus. We validated peptide specificity for HER2 in knockdown and competition experiments using human colorectal cancer cells in vitro, and measured a binding affinity of kd = 21 nM and time constant of k = 0.14 min−1 (7.14 min). We used this peptide with either topical or intravenous administration in a preclinical model of colorectal cancer to demonstrate specific uptake in spontaneous adenomas and to show feasibility for real time in vivo imaging with near-infrared fluorescence. We used this peptide in immunofluorescence studies of human proximal colon specimens to evaluate specificity for sessile serrated and sporadic adenomas. Improved visualization can be used endoscopically to guide tissue biopsy and detect premalignant lesions that would otherwise be missed. Our peptide design for specificity to HER2 is promising for clinical translation in molecular imaging methods for early cancer detection.
Graphical abstract
INTRODUCTION
Antibodies, enzyme-activated probes, and lectins are being developed for use as molecular probes to improve detection specificity with molecular imaging.1-3 These targeting moieties can be labeled with bright fluorescent dyes to achieve high contrast and produce near-infrared (NIR) emission for deep tissue imaging.4 Compared with conventional whole body imaging methods, these properties may improve cancer staging. However, clinical usefulness of some probes has been limited by slow binding onset, long circulation times, and increased background.5,6 Molecular expression can also be visualized in precancerous lesions well before gross architectural changes of cancer become apparent, and may be useful for early detection. Recently, targeted imaging with peptides has been demonstrated as a diagnostic tool in clinical studies to guide tissue biopsy in the digestive tract.7,8 Peptides are well suited for in vivo imaging because of high diversity, small size, labeling fiexibility, and minimal immunogenicity, and are well-suited for clinical use because of rapid binding kinetics, deep tissue penetration, lack of toxicity, and affordable cost.9
Human epidermal growth factor receptor 2 (HER2) is a member of the tyrosine kinase family that also includes HER1 (EGFR), HER3, and HER4, and is located on chromosome 17q21.10-12 It encodes a 185 kDa transmembrane protein that lacks a natural ligand and functions as a coreceptor to form homo and heterodimers with other HER family members. Dimerization results in activation of signaling cascades that include the MAPK and PI3K/AKT pathways that are essential for cell proliferation and differentiation.13,14 There is evidence that HER2 is highly overexpressed in many tumors. Amplification and/or over expression of this gene has been associated with mitogenesis, malignant transformation, increased motility, invasion, and metastasis.15-17 HER2 over-expression may also predict cancer prognosis, correlate with tumor size, and reflect stage. These findings motivate the development of HER2 as an imaging target to help select patient populations that are more likely to benefit from therapy and avoid unnecessary treatment, reduce side effects, and decrease cost.
Colorectal cancer (CRC) is a leading cause of cancer death worldwide that can be prevented with improved methods for early detection.18,19 Conventional white light colonoscopy is the standard method for CRC surveillance. However, evidence is mounting that mortality benefit is conferred primarily to the distal rather than proximal colon.20-24 Proximal lesions are more likely to be flat in appearance and difficult to visualize.25 These lesions can be more aggressive than visible polyps, and 5 times more likely to harbor carcinoma in some patient populations.26 Thus, molecular imaging methods may improve detection and hence prevention of CRC. Previously, specific imaging agents for EGFR, c-Met, and mucin-1 have been demonstrated in preclinical and clinical studies.27-29 Previous immunohistochemistry studies have shown a wide range of 10–83% for HER2 overexpression in CRC.12,17,30 This variability may result from sampling error, small sample size, limited study populations, differences in technique, and nonstandard scoring systems.31-34 To date, little is known about HER2 overexpression in proximal versus distal colon.
Here, we aim to design and validate a NIR-labeled peptide specific for the extracellular domain of HER2 for use with in vivo molecular imaging applications. We will illustrate potential for early detection of CRC by demonstrating specific binding to adenomas in mouse colon and characterizing overexpression of membranous HER2 in precancerous lesions from proximal human colon.
RESULTS
Peptide Selection using Structural Model
We identified peptides that bind specifically to the extracellular domain (ECD) of HER2 (2A91) using a structural model (Figure 1A).35 The ECD was targeted because it is exposed and accessible to imaging. We generated ~1000 candidates using mimotopes36 by considering Arg or Lys amino acid residues at the N-terminus to form hydrophilic interactions with HER2-ECD.37,38 Hydrophobic residues such as Phe, Trp, Val, Met, Ile, and Leu were appended at the C-terminus to increase the likelihood of hydrophobic/hydrophilic interactions. Other amino acids such as Ser, His, Arg, Tyr, Thr, Asp, and Asn were used to increase peptide diversity.39 Either the N- or C-terminus of the peptides were connected by a conformationally rigid spacer group, such as PFP, PNP, PYP, and PWP, in the middle region. We evaluated binding interactions of these candidates to HER2-ECD using Pepsite-2.40 We synthesized the 5 leading candidates based on P-value, attached a FITC label for imaging with Flo1-HER2 and Q-hTERT cells (control) using confocal microscopy. A Western blot shows differences in HER2 expression for these cells (Figure 1B). We selected KSPNPRF based on the highest mean fluorescence intensity from binding to Flo1-HER2 cells (Figure 1C). From our model, this peptide binds to HER2 domain 3. The peptide (black) was then labeled with Cy5.5 (red) via a GGGSK linker (blue) on the C-terminus, hereafter KSP*-Cy5.5 (Figure 1D). Cy5.5 was chosen for improved quantum yield, photostability, and imaging depth. The linker prevents steric hindrance by spatially separating the fluorophore from the peptide. We developed a scrambled sequence PPSNFKR for use as control by altering the conformationally rigid spacer PNP and moving both hydrophobic and hydrophilic amino acids at the C-terminus. We also linked this control peptide to Cy5.5 via GGGSK, hereafter PPS*-Cy5.5 (Figure 1E). The absorbance spectra of KSP*-Cy5.5 and PPS*-Cy5.5 at 5 μM in PBS shows a maximum at 680 nm (Figure 1F). The fluorescence emission peak occurs at 708 nm in the NIR spectrum. For both peptides, we achieved >98% purity with HPLC, and measured an experimental mass-to-charge (m/z) ratio on mass spectrometry of 1794.98, which agrees with expected values (Figure S1A,B).
Figure 1.
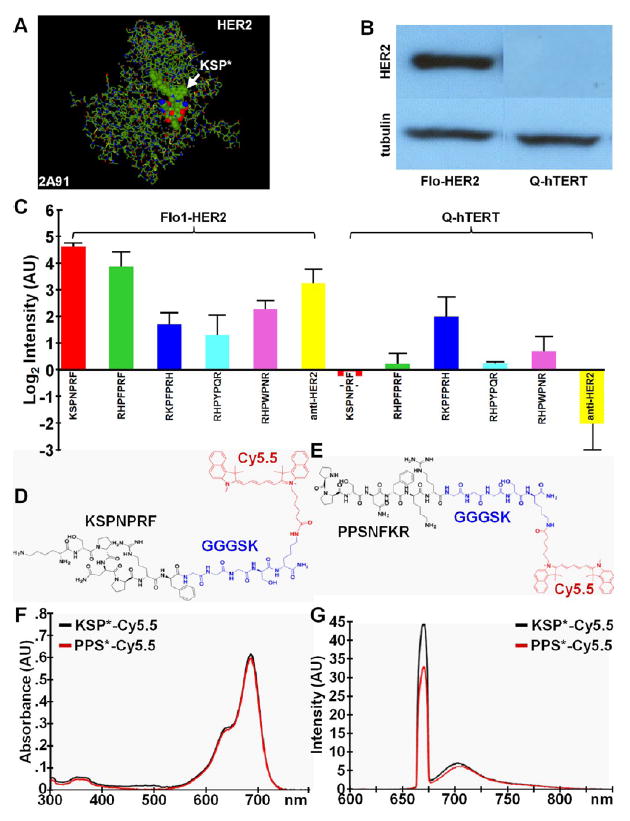
Selection of peptide specific for HER2. (A) Structural model (2A91) for HER2-ECD was used to evaluate binding interactions between target and candidate peptides. (B) Western blot of Flo1-HER2 and Q-hTERT (control) cells show difference in HER2 expression. (C) Top 5 peptide sequences were FITC-labeled and evaluated with confocal microscopy with cells in vitro. KSPNPRF showed the highest mean fluorescence intensity compared with the other peptides. Data was transformed to log base 2. (D) Chemical structure of KSPNPRF peptide (black) labeled with Cy5.5 fluorophore (red) via a GGGSK linker (blue); KSP*-Cy5.5. (E) Scrambled control peptide PPSNFKR; PPS*-Cy5.5. (F) Peak absorption occurs at λex = 680 nm and (G) maximum fluorescence emission is seen at 708 nm for both peptides.
Confocal Fluorescence Microscopy
We performed confocal microscopy to demonstrate peptide binding to the plasma membrane (arrows), and observed significantly greater fluorescence intensity for KSP*-Cy5.5 than PPS*-Cy5.5 to HT29 and SW480 cells (Figure 2A-D). An AF488-labeled anti-HER2 antibody also showed significantly greater signal than the control peptide (Figure 2E,F). Quantified results are shown (Figure 2G). Western blot shows difference in expression of HER2 for HT29 and SW480 (Figure 2H).
Figure 2.
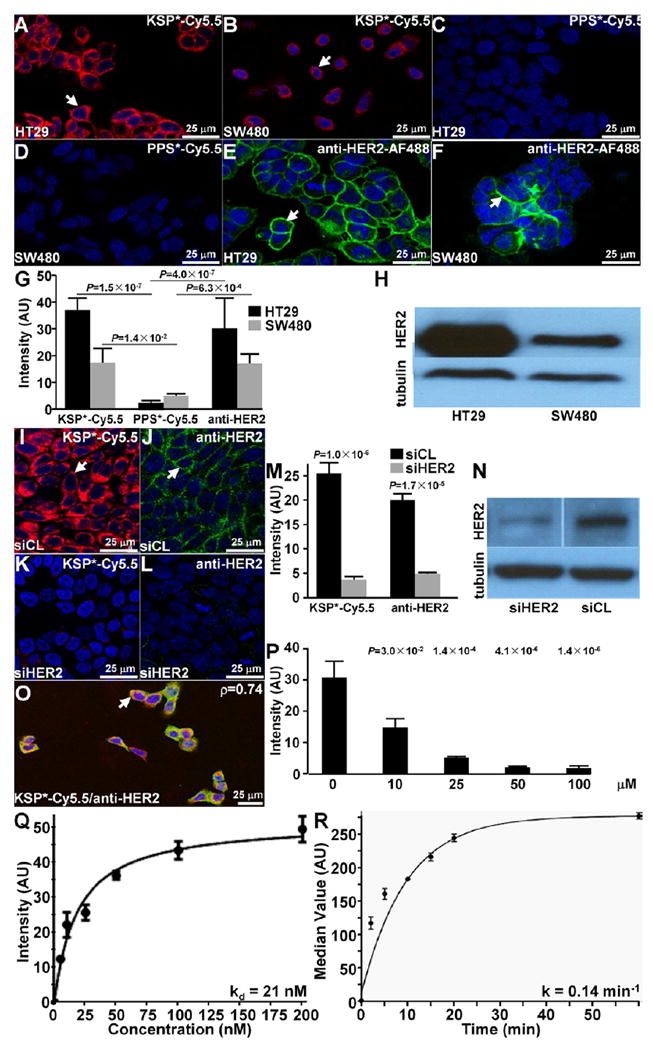
Validation of specific peptide binding to HER2 expressed on cell surface. On confocal microscopy, (A,B) KSP*-Cy5.5 (red) binds to the surface (arrows) of HT29 and SW480 cells, while (C,D) PPS*-Cy5.5 (negative control) shows minimal binding. (E,F) Anti-HER2-AF488 (green) is used as a positive control. The mean fluorescence intensity for KSP*-Cy5.5 was significantly greater than that for PPS*-Cy5.5 for both HT29 and SW480 cells, P = 1.5 × 10−7 and 1.4 × 10−2, respectively, by paired t test. Similarly, the results for anti-HER2-AF488 were greater than that for PPS*-Cy5.5, P = 4.0 × 10−7 and 6.3 × 10−4, respectively, by paired t test. (G) Each result is an average of 3 images collected independently. (H) Western blot shows differences in HER2 expression between HT29 and SW480 cells. (I,J) KSP*-Cy5.5 (red) and anti-HER2-AF488 (green) binds significantly greater to the surface (arrows) of siCL treated control HT29 cells compared to (K,L) siHER2 knockdown cells, P = 1.0 × 10−4 and 1.7 × 10−4, respectively, by paired t test. (M) Results are an average of 3 images collected independently. (N) Western blot shows effective knockdown of HER2 (siHER2) compared to control (siCL). (O) Binding of KSP*-Cy5.5 and anti-HER2-AF488 colocalizes to surface of HT29 cells, Pearson’s coefficient p = 0.74. (P) On competition, we found the fluorescence intensities to decrease in a concentration dependent manner, P-values for intensity at each time point compared to that at 0 min are shown above data points. Each result is an average of 3 independent measurements. We measured (Q) an apparent dissociation constant (binding affinity) of kd = 21 nM, R2 = 0.98 for KSP*-Cy5.5 to HT29 cells, and (R) an apparent association time constant k = 0.14 min−1 (7.1 min); R2 = 0.92. Results for each measurement are representative of 3 independent experiments.
siRNA Knockdown of HER2 and Colocalization
We performed siRNA knockdown experiments with HT29 cells to validate specific binding of KSP*-Cy5.5 to HER2. On confocal microscopy, KSP*-Cy5.5 (red) and AF488-labeled anti-HER2 antibody (green) bind strongly to the surface (arrow) of control HT29 cells (transfected with siCL, nontargeting siRNA) (Figure 2I,J). Significantly reduced fluorescence intensities were observed for HT29 knockdown cells (transfected with siHER2, targeting siRNA) (Figure 2K,L). Quantified results are shown (Figure 2M). Western blot shows effective HER2 knockdown (Figure 2N). Also, we observed colocalization of peptide and antibody binding (arrow) on merged images (Pearson’s coefficient p = 0.743) (Figure 2O).
Competition for Peptide Binding
We performed competition studies to demonstrate that binding occurs with the peptide rather than the fluorophore by adding unlabeled KSP* to compete with KSP*-Cy5.5 to HT29 cells. We found the fluorescence intensities to decrease significantly in a concentration dependent manner (Figure 2P).
Characterization of Peptide Binding
We measured the binding parameters of KSP*-Cy5.5 to HT29 cells using flow cytometry, and found an apparent dissociation constant of kd = 21 nM, R2 = 0.98 (Figure 2Q), and an apparent association time constant of k = 0.14 min−1 (7.14 min), R2 = 0.92, to support rapid binding with topical administration (Figure 2R).
Effect of Peptide on Cell Signaling Pathways
We evaluated the effect of the KSP* peptide binding on HER2-mediated signaling in SKBR3 cells. On Western blot, we observed no change in phosphorylation of either HER2 (p-HER2) or of downstream AKT (p-AKT) and ERK (p-ERK) with addition of KSP* at 5 and 100 μM (Figure S2). By comparison, the addition of lapatinib, a tyrosine kinase inhibitor known to interrupt HER2 signaling in solid tumors, showed reduced expression of p-HER2, p-AKT, and p-ERK in a concentration dependent manner. Cells treated with 1% DMSO and untreated cells showed no suppression of HER mediated signaling.
In Vivo Imaging of HER2 Overexpressed in Mouse Colonic Neoplasia
We evaluated specific binding of KSP*-Cy5.5 to colonic adenomas that overexpress HER2 in CPC;Apc mice. This mouse was genetically engineered to somatically delete an Apc allele under Cre regulation, and develops both polypoid and flat adenomas spontaneously.41 This model is representative of human disease because Apc mutations are found in >80% of sporadic colorectal cancers.42 Imaging was performed in vivo using a small animal endoscope that is sensitive to NIR fluorescence.27 A white light image of a polypoid adenoma (arrow) was first identified (Figure 3A). KSP*-Cy5.5 was then topically administered, and allowed to incubate. After ~7 min the unbound peptides were rinsed away, and the fluorescence image shows increased intensity (arrow) at the polyp (Figure 3B). After this signal went away 2–3 days later, endoscopy was repeated using PPS*-Cy5.5 (control) from the same area, and minimal signal was observed (Figure 3C). A flat adenoma (arrow) with subtle appearance on white light is shown (Figure 3D). The corresponding fluorescence image shows increased intensity with visible crypt-like structures (Figure 3E). Images collected several days later with PPS*-Cy5.5 show minimal signal (Figure 3F). In all fluorescence images, surrounding normal colonic mucosa produced minimal background. Quantified results show significantly greater fluorescence intensity for adenomas compared to normal (Figure 3G).
Figure 3.
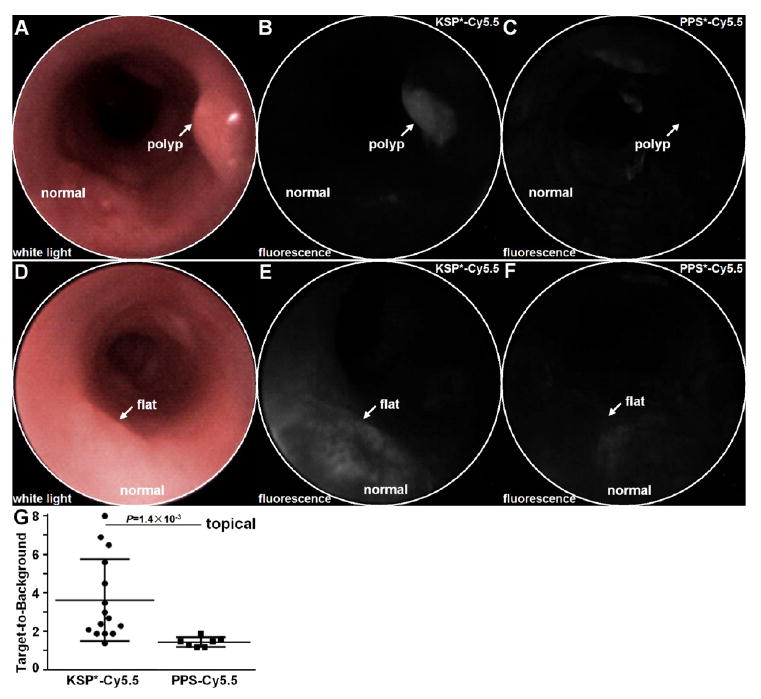
In vivo imaging of mouse colonic dysplasia with topical HER2 peptide. (A) Endoscopic image with white light illumination shows a spontaneous polyp (arrow). (B) NIR fluorescence image after topical administration of KSP*-Cy5.5 shows increased intensity from polyp (arrow). (C) Image of same region with PPS*-Cy5.5 control several days later shows significantly reduced signal. (D) White light and NIR fluorescence images with (E) KSP*-Cy5.5 and (F) PPS*-Cy5.5 show similar results with a flat adenoma (arrow). (G) From n = 7 mice, topically administered KSP*-Cy5.5 (n = 15 adenomas) resulted in a higher mean (±std) T/B ratio than PPS*-Cy5.5 (n = 7 adenomas), 3.64 ± 2.13 and 1.46 ± 0.25, respectively, P = 1.4 × 10−3 by unpaired t test.
KSP*-Cy5.5 was also administered systemically via tail vein prior to endoscopy (Figure 4A). NIR fluorescence images were collected over time in each mouse, and a representative image of a polypoid adenoma at 100 min after i.v. peptide injection is shown (Figure 4B). In n = 4 mice, the mean intensity was found to peak at 100 min (Figure 4C).
Figure 4.
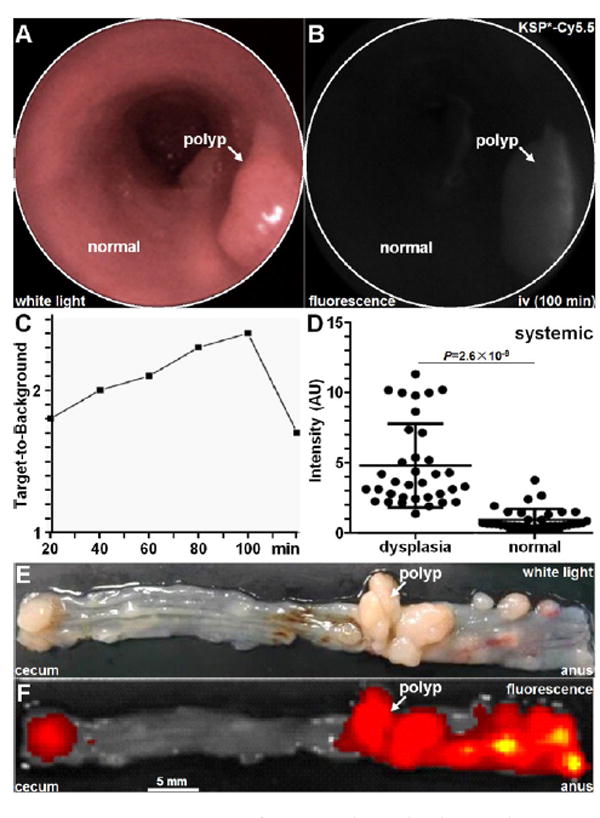
In vivo imaging of mouse colonic dysplasia with systemic HER2 peptide. (A) Representative white light and (B) fluorescence image from polyp collect 100 min after intravenous administration of KSP*-Cy5.5 shows region of increased intensity (arrow). (C) Peak T/B ratio from colonic adenomas in n = 4 mice was observed at 100 min. (D) The mean fluorescence intensity from adenomas was 5.03-fold higher than that from uninvolved surrounding normal mucosa, P = 2.6 × 10−9 by paired t test. (E) Representative white light image of exposed mucosal surface of excised mouse colon shows locations of numerous polyps. (F) Corresponding fluorescence image show increased fluorescence intensities at site of polyps.
Ex Vivo Macroscopic Validation of HER2 Overexpressed in Mouse Colonic Neoplasia
After imaging, the animals were euthanized, and the colon was excised and divided longitudinally to expose the mucosal surface for collection of macroscopic white light and fluorescence images (Figure 4E,F). The tissues were sectioned along planes parallel to the surface, and the pathologist (SRO) identified regions of dysplasia and normal on histology while blinded to the imaging results. In n = 4 mice, we found significantly greater mean fluorescence intensities for dysplasia compared to normal (Figure 4D).
Ex Vivo Microscopic Validation of HER2 Overexpressed in Mouse Colonic Neoplasia
We also sectioned the specimens for collection of confocal images to perform microscopic validation of peptide binding. We found increased cell surface staining of KSP*-Cy5.5 compared with PPS*-Cy5.5 to dysplastic crypts (arrow) (Figure 5A,B). Minimal staining was observed for either peptide with normal mouse colonic mucosa (Figure 5D,E). Corresponding histology (H&E) is shown (Figure 5C,F).
Figure 5.
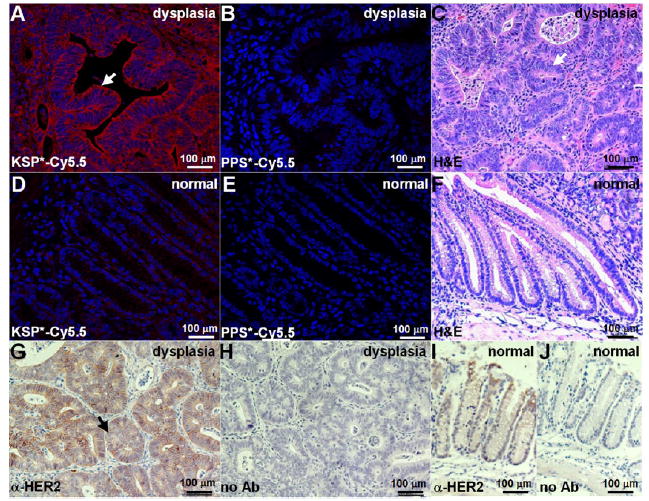
Validation of specific peptide binding to HER2 overexpressed by mouse colonic dysplasia. On confocal microscopy, we found intense staining of (A) KSP*-Cy5.5 compared to (B) PPS*-Cy5.5 to sections of dysplasia. (C) Histology (H&E) shows features of low-grade dysplasia (arrows). Minimal staining was observed with either (D) KSP*-Cy5.5 or (E) PPS*-Cy5.5 to normal colon. (F) Histology (H&E) for normal. (G) On immunohistochemistry with a known antibody, we confirmed overexpression of HER2 in dysplasia. (H) No antibody (control) with dysplasia. Normal colonic mucosa (I) with and (J) without antibody (control).
We performed immunohistochemistry with a known antibody to validate overexpression of HER2 in mouse colonic dysplasia (Figure 5G). A serial section with no primary antibody used (control) shows minimal reactivity (Figure 5H). By comparison, normal mouse colonic mucosa showed minimal reactivity either with or without anti-HER2 (Figure 5I,J).
Binding of HER2 Peptide to Human Proximal Colonic Neoplasia
We demonstrate potential for clinical translation of the HER2 peptide by examining specific binding to overexpressed HER2 on formalin-fixed, paraffin-embedded (FFPE) specimens of human proximal colon. On confocal microscopy, we observed greater binding of both KSP*-Cy5.5 (red) and AF488-labeled anti-HER2 antibody (green) to dysplastic versus normal crypts on adjacent sections (Figure 6A,B, respectively). Corresponding histology (H&E) is shown (Figure 6C). At higher magnification, colocalization of peptide and antibody binding to the surface (arrow) of dysplastic colonocytes can be seen (Figure 6D). Also, differences in fluorescence intensities between dysplasia and normal from the dashed red and green boxes in Figure 6A with KSP*-Cy5.5 (red) can be appreciated (Figure 6E,F). Differences in fluorescence intensities between dysplasia and normal from the dashed red and green boxes in Figure 6B with AF488-labeled anti-HER2 antibody (green) can be seen (Figure 6G,H). Quantified fluorescence intensities (mean ± std) for normal (n = 30), hyperplastic polyps (n = 12), sessile serrated adenomas (n = 13), and sporadic adenomas (n = 30) are shown (Figure 6I). The mean result for adenoma was significantly greater than that for normal, hyperplasia, and SSA, while the difference between SSA and normal was nonsignificant.
Figure 6.
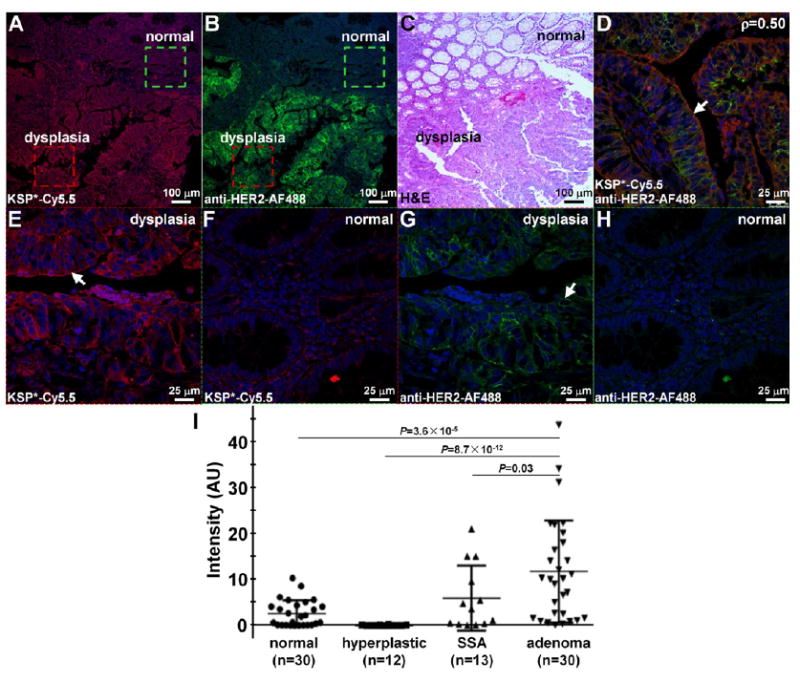
Validation of specific peptide binding to HER2 overexpressed by human proximal colonic neoplasia. Increased fluorescence intensity was observed for staining of (A) KSP*-Cy5.5 (red) and (B) anti-HER2-AF488 antibody (green) to human colonic dysplasia. (C) Corresponding histology (H&E) from adjacent section. (D) Co-localization of binding (arrow) by peptide (red) and antibody (green) in dysplasia is shown, Pearson’s correlation coefficient p = 0.50. High magnification image of dashed red and green boxes in (A) show increased staining of KSP*-Cy5.5 peptide (red) to (E) cell surface in dysplasia (arrow) versus (F) normal. High magnification image of dashed red and green boxes in (B) show increased staining of anti-HER2-AF488 antibody (green) to (G) dysplasia versus (H) normal. (I) Quantified results showed mean (±sem) intensities to be 2.72 ± 0.31, 0.12 ± 0.08, 6.07 ± 0.73, and 11.9 ± 0.61 for normal (n = 30), hyperplastic polyps (n = 12), sessile serrated adenomas (n = 13), and sporadic adenomas (n = 30), respectively. The P-values for differences in the mean results for sporadic adenomas and that for normal, hyperplastic polyps, and sessile serrated adenomas were P = 3.6 × 10−5, P = 8.7 × 10−12, and P = 0.03, respectively, by Tukey’s multiple comparisons. The difference between the mean intensities for SSA and normal colon was P = 0.20.
Overexpression of HER2 in Human Proximal Colon
We performed immunohistochemistry with a known antibody to validate overexpression of HER2 in human colonic dysplasia. We observed weak staining (0+/1+) of HER2 either on the membrane or in the cytoplasm of cells in the epithelium of normal colon and hyperplastic polyps (Figure 7A,B). By comparison, we found various intensities of staining in dysplastic crypts for both sessile serrated adenomas (SSA) and sporadic adenomas (Figure 7C,D). Contrast in reactivity between dysplasia and normal can appreciated for both polypoid and flat adenomas (Figure 7E,G). Differences in HER2 expression between neoplasia and normal crypts in the same field-of-view support HER2 expression specific to dysplasia. Magnified views show intense staining on the membrane of dysplastic colonocytes (Figure 7F,H). The control experiment was performed without use of primary antibody (Figure 7I). An expert GI pathologist (SRO) using a standard IHC scoring system revealed overexpression, defined by either 2+ or 3+ staining, in 0% (0/30) of normal, 25% (3/12) of hyperplastic polyps, 14% (2/14) of SSA, and 41% (12/29) of adenomas (Figure 7J). These results support HER2 as a promising early target for detection of proximal colon cancers.
Figure 7.
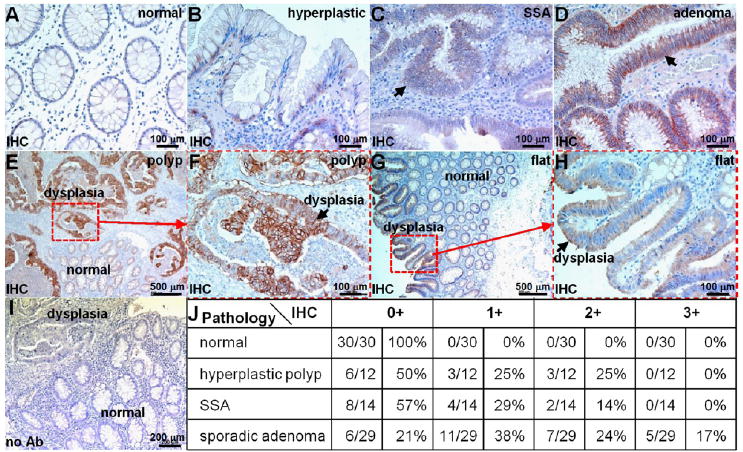
Overexpression of HER2 in human proximal colonic neoplasia. On immunohistochemistry (IHC) of archived specimens, minimal staining was observed from all sections of (A) normal colon and most sections of (B) hyperplastic polyps. Intense cell surface staining was seen from some (C) sessile serrated adenomas (SSA) and (D) many sporadic adenomas. (E) Differences in HER2 expression between dysplastic and normal crypts from a polypoid adenoma can be seen. (F) Magnified view from dashed red box in (E) shows intense staining (arrow) from dysplastic colonocytes. (G) Difference in staining between dysplastic and normal crypts is shown for flat adenoma. (H) Magnified view from dashed red box in (G). (I) No antibody (control). (J) Consensus between 2 expert gastroenterology pathologists using a standard IHC scoring system revealed overexpression, defined by either 2+ or 3+ staining, in 0% (0/30) of normal, 25% (3/12) of hyperplastic polyps, 14% (2/14) of SSA, and 41% (12/29) of adenomas.
DISCUSSION
Here, we used a structural model to identify a peptide that binds specifically to the extracellular domain of HER2 by selecting amino acids that increase the likelihood of hydrophobic/hydrophilic interactions with the target. We found KSP* to bind HER2 domain 3 with high affinity and rapid kinetics, validated specific binding on knockdown and competition experiments in cells, and demonstrated specific binding to either polypoid or flat adenomas in mice in vivo using either topical or intravenous administration. We also found HER2 to be overexpressed in precancerous lesions in human proximal colon, the most common region for missed CRC, and showed greater peptide binding to adenomas compared with normal mucosa and hyperplastic polyps. We measured a binding affinity of kd = 21 nM for the KSP*-Cy5.5 peptide. Previously reported HER2 peptides have shown a range in kd from 30 nM for optical imaging39 to ~200–300 nM for SPECT imaging.43,44 The SPECT result has limitations for in vivo detection of low HER2 receptor densities, requiring highly sensitive imaging systems. KSP* was found to bind rapidly with topical administration, k = 0.14 min−1 (7.1 min), and is compatible with the high volume work flow of endoscopy. We found this peptide to have no effect on HER2 signaling, an advantage for diagnostic imaging. By comparison, antibodies can stimulate the HER2 pathway and potentially accelerate cell proliferation and tumorigenesis.45-50
Although increased acceptance of colonoscopy has resulted in reduced incidence and mortality from CRC, the results are much less than expected. Cases of CRC diagnosed after colonoscopy are common, and as many as 1 in 10 CRCs are found in patients who have completed this procedure.51-53 Although interval cancers may occur for a variety of reasons, most tumors are thought to arise from prevalent lesions that were missed because of inadequate visualization. While efforts to improve quality have focused on increased instrument withdrawal time, adenoma detection rate, and bowel prep quality,54 reports of interval cancers in patients undergoing careful endoscopic examination show that conventional colonoscopy can be ineffective even under optimal conditions.55,56 Other advanced endoscopic techniques, such as narrowband imaging (NBI)57-59 and chromoendoscopy,60-62 have been investigated in efforts to improve polyp visualization; however, these technologies do not offer much improvement in the adenoma detection rate or in patient outcomes.63,64
Molecular imaging detects overexpressed targets, and has the potential to improve CRC surveillance. Target specific imaging agents provide a biological basis for detecting disease, and can also establish prognosis, guide therapy, and monitor treatment. A more comprehensive picture of HER2 expression in tumors that are highly heterogeneous may overcome limitations of the small size of biopsy specimens used for evaluation with IHC and FISH.65-68 Other HER2 specific targeting moieties, such as antibodies and antibody fragments, can achieve the high binding affinities required for therapy, but are limited for diagnostics by slow binding kinetics, long half-lives, and increased background.69-75 Compared with peptides, they are much more difficult and expensive to mass manufacture.76,77 The KSP* peptide may also be clinically useful as an adjunct to conventional white light colonoscopy to guide tissue biopsy. On immunohistochemistry, we found HER2 to be overexpressed in sessile serrated and sporadic adenomas from human proximal colon. These lesions are precursors of CRC; thus our results support HER2 as an early target. The added time and cost for use of an exogenous imaging agent can be justified in high risk patients, such as those with Lynch Syndrome, inflammatory bowel disease, or history of multiple polyps, if the frequency of surveillance can be reduced.78,79 Clinical use of peptides with endoscopes that are sensitive to fluorescence has recently been demonstrated in the clinic.80
Future development of this peptide will include clinical validation studies. We have previously studied the peptide VRPMPLQ in patients with confocal endomicroscopy. This sequence was identified using phage display from human colonic polyps obtained via biopsy.7 This peptide was labeled with FITC, and specific binding was validated with topical application in vivo on dysplastic polyps. Because this peptide was selected empirically, the target is unknown and its clinical use may not be widely generalizable. The use of a NIR fluorophore, such as Cy5.5, instead of FITC provides greater photostability, less background, and deeper tissue penetration. With topical administration, risk for toxicity is minimal and undesired probe biodistribution to nontarget tissues is avoided, a major concern with systemic delivery. A high concentration can be delivered to saturate target binding and maximize signal, albeit to localized regions of the tissue only. With systemic administration, peptides can be delivered to the entire colon28 with better target access and greater imaging depth, but rigorous pharmacology/toxicity studies are needed to establish safety.
While we found promising results with this peptide alone, disease heterogeneity in a broad patient population may require imaging of multiple targets using multiplexed imaging methods.81 We labeled this peptide with either FITC or Cy5.5 to demonstrate fiexibility for use of a broad range of fluorophores. Our HER2 specific peptide was optimized based on a known target and validated on premalignant human proximal lesions. Furthermore, this peptide can be used in a multimer configuration to detect multiple targets concurrently and potentially detect disease at lower levels of molecular expression.82
EXPERIMENTAL PROCEDURES
Cells, Chemicals, and Materials
Human HT29, SW480, Q-hTERT, and SKBR3 cells were obtained from the ATCC. We used McCoy’s Medium for HT29 and SKBR3, Dulbecco’s Modified Eagle Medium (DMEM) for SW480, and Keratinocyte-SFM media (Gibco) for Q-hTERT cells. Flo1-HER2 cells were maintained in DMEM supplemented with G418. All cells were cultured at 37 °C in 5% CO2, and were supplemented with 10% fetal bovine serum (FBS) with 1% penicillin/streptomycin. Penicillin/streptomycin was omitted for the siRNA knockdown studies. FBS was omitted for Keratinocyte-SFM media. The cells were passaged using 0.25% EDTA containing trypsin (Mediatech Inc.). The cell numbers were counted with a hemocytometer. Peptide synthesis reagents were obtained from either Anaspec or AAPPTEC and were of the highest grade available (>99% purity) and used without further purification. Solvents and other chemical reagents were purchased from Sigma-Aldrich, unless otherwise mentioned.
Peptide Selection Using Structural Model
The crystal structure (ID#2A91) of HER2 extracellular domain (ECD) was obtained from the Protein Data Bank (PDB).83 A peptide library with 7 random amino acid residues was aligned with 2A91 using PepSite-2.40 Approximately 1000 peptide sequences were generated using mimotopes.36 We evaluated binding of the candidate peptides with the 2A91 protein structure specified by chain A. The software comprehensively evaluated all possible combinations of predicted binding motifs for each of the 7 amino acids, and generated a 3D model of binding interactions between the candidate peptides and the HER2-ECD target. For each peptide, the raw score was fitted to a Gumbel distribution and then ordered by P-value.
Flo-1-HER2 and Q-hTERT cells were seeded with ~7500 cells in 12-well plates with 1.5-mm-thick glass cover slides, and grown to ~70% confluence. The cells were gently washed with cold PBS before adding peptide. 10 μM of FITC-labeled peptide was added to each well and incubated at 4 °C for 1 h. After incubation, the cells were washed 3× with PBS, fixed in ice-cold 4% PFA for 10 min, and washed 1× with PBS. Subsequently, the cells were embedded on microscope slides with ProLong Gold reagent containing DAPI (Invitrogen), and the slides where analyzed with a Leica TCS SP5 confocal microscope (Leica Microsystems) using a 63× oil immersion objective. The FITC-labeled peptides and anti-HER2 antibody were tested in quadruplicate.
Peptide Synthesis
We synthesized and labeled the peptides with either FITC or Cy5.5- using standard Fmoc-mediated solid-phase chemical synthesis.84 We used Fmoc and Boc protected L-amino acids, and synthesis was assembled on rink amide MBHA resin. The peptides were synthesized on a PS3 automatic synthesizer (Protein Technologies Inc.). The C-terminus lysine was incorporated as Fmoc-Lys (ivDde)–OH, and the N-terminus amino acid was incorporated with Boc protection to avoid unwanted Fmoc removal during deprotection of the ivDde moiety prior to fluorophore labeling. Upon complete assembly of the peptide, the resin was transferred to a reaction vessel for manual labeling with dye. The ivDde side chain protecting group was removed with 5% hydrazine in DMF (3× for 20 min) with continuous shaking at room temperature (RT). The resin was washed with dimethylforma-mide (DMF) and dichloromethane (DCM) 3× each for 1 min. The protected resin-bound peptide was incubated overnight with either FITC or Cy5.5-NHS ester (Lumiprobe LLC) in the presence of DIEA, and the completion of the reaction was monitored by a qualitative Ninhydrin test. Upon completion of labeling, the peptide was cleaved from the resin using TFA:TIS:H2O (95:2.5:2.5 v/v/v) for 4 h with shaking in the dark at RT. After separating the peptide from the resin, the filtrate was evaporated with N2 gas followed by precipitation with chilled diethyl ether and stored overnight at −20 °C. The precipitate was centrifuged at 3000 rpm for 5 min and washed with diethyl ether 3× and centrifuged in between each washing step. The crude peptides were dissolved in 1:1 acetonitrile/H2O (v/v) and purified by prep-HPLC with a C18 column (Waters Inc.) using a water (0.1% TFA)–acetonitrile (0.1% TFA) gradient. The final purity of the peptides was confirmed by analytical C18-column. Further characterization was performed with either ESI (Waters Inc.) or Q-TOF (Agilent Technologies) mass spectrometry. The scrambled (control) peptide PPS*-Cy5.5 was synthesized, labeled, and purified similarly.
Spectral Measurements
The peptide absorbance spectra were collected using a spectrophotometer (NanoDrop 2000, Thermo Scientific). Fluorescence emission from a 5 μM peptide solution diluted in PBS was collected with a fiber coupled spectrophotometer (Ocean Optics) using a diode-pumped solid state laser (Technica Laser Inc.) with λex = 671 nm. The spectra were plotted with Origin 6.1 software (OriginLab Corp).
Confocal Fluorescence Microscopy
Cells were washed with PBS and incubated with 5 μM of KSP*-Cy5.5 and PPS*-Cy5.5 for 30 min at 4 °C. The cells were then washed 3× in PBS, fixed with ice cold 4% paraformaldehyde (PFA) for 10 min, washed 1× with PBS, and then mounted on glass slides with ProLong Gold reagent containing DAPI (Invitrogen). Confocal fluorescence images were collected with Cy5.5, and DAPI filters. For antibody staining, the cells were prefixed with cold methanol for 10 min at −20 °C and blocked with 2% BSA for 30 min at RT. Cells were incubated with 1:450 dilution of anti HER2 antibody overnight at 4 °C. The cells were washed 3× with PBS and processed for secondary staining. Goat-anti-rabbit Alexa-Fluor 488 (AF488) was added to the cells and incubated for 1 h at RT. Cells were further washed 3× with PBS and mounted onto glass coverslips. Fluorescence intensities from 3 independent images were quantified using custom Matlab (Mathworks) software.
siRNA Knockdown of HER2 and Co-Localization
We examined HER2 knockdown in HT29 cells using ON-TARGETplus human ERBB2 siRNA (#L-003126-00-0005), ON-TARGETplus Nontargeting pool (#D-001810-10-05), and DharmaFECT transfection reagents (Thermo Scientific). Briefly, cells were seeded in 6-well culture plates at 30% confluence in McCoy’s 5A medium supplemented with 10% fetal bovine serum without antibiotics. The next day, cells were transfected with siRNA at a final concentration of 5 μM/L using oligofectamine (Thermo Scientific). Knockdown of HER2 was confirmed by Western blot. Cells were first washed in PBS and then lysed in RIPA buffer containing 1% Nonidet P40, 0.5% sodium deoxycholate, 0.1% SDS, 10 mg/mL phenylmethylsulfonylfluoride, and 1 mM sodium orthovanadate. Aliquots were placed on ice for 30 min and centrifuged at 14 000 rpm for 10 min. Protein aliquots were denatured in loading buffer at 95 °C for 5 min, separated on SDS-polyacrylamide gels (SDS-PAGE), and transferred onto PVDF membranes. The membrane was blocked with blocking buffer (5% skim milk in 0.1% PBST) for 30 min. The membranes were incubated with anti-HER2 primary antibody at 4 °C overnight. After washing 5× with PBST and 5× with PBS, the membrane was incubated for 1 h in peroxidase-conjugated secondary antibody (1:5000 dilution; GE Healthcare), and were developed using the Western blot chemiluminescent substrate (GE Healthcare). The luminescent signal was detected by exposure to X-ray film (Denville Scientific).
We evaluated co-localization of binding by KSP*-Cy5.5 and anti-HER2 antibody to the surface of HT29 cells. KSP*-Cy5.5 at 5 μM concentration was incubated for 1 h at 4 °C. The cells were washed and fixed with 4% PFA for 5 min, and then incubated with primary anti-HER2 and secondary AF488-labeled antibody. We administered the peptide first because of its lower affinity, and used a low concentration (<10 μM) to minimize interference with antibody binding.
Competition for Peptide Binding
Specific binding of KSP*-Cy5.5 to HT29 cells was validated on competitive inhibition with unlabeled KSP* peptide. Approximately 7500 HT29 cells were grown to ~70% confluence on coverslips in triplicate. Unlabeled KSP* peptide at 0, 10, 25, 50, and 100 μM was added and incubated with the cells for 30 min at 4 °C. The cells were washed 3× with PBS, and further incubated with 2 μM of KSP*-Cy5.5 for another 30 min at 4 °C. The cells were washed 3× with PBS and fixed with 4% PFA for 10 min. The cells were washed with PBS and mounted with ProLong Gold reagent containing DAPI (Invitrogen). Confocal fluorescence images were collected at each concentration, and intensities from 3 independent images were quantified using custom Matlab (Mathworks) software.
Measurement of Peptide Binding Affinity and Time Constant
We measured the apparent dissociation constant of the HER2 peptide to HT29 cells as an assessment of binding affinity. KSP*-Cy5.5 was serially diluted in PBS at concentrations of 0, 5, 10, 25, 50, 100, and 200 nM. Approximately 105 HT29 cells were incubated with KSP*-Cy5.5 at 4 °C for 1 h and washed with cold PBS. The mean fluorescence intensities were measured with flow cytometry (BD LSRII, BD Biosciences). The equilibrium dissociation constant kd = 1/ka was calculated by performing a least-squares fit of the data to the nonlinear equation I = (I0 + Imaxka[X])/(I0 + ka[X]). I0 and Imax are the initial and maximum fluorescence intensities, corresponding to no peptide and at saturation, respectively, and [X] represents the concentration of the bound peptide.85 Graphpad prism analysis software (Graphpad Software Inc.) was used to calculate kd and ka.
We measured the apparent association time constant of the peptide to HT29 cells to assess binding kinetics. HT29 cells were grown to ~80% confluence in 10 cm dishes, and detached with PBS-based cell dissociation buffer (Invitrogen). Approximately 105 cells were incubated with 5 μM KSP*-Cy5.5 at 4 °C for various time intervals ranging from 0 to 60 min. The cells were centrifuged, washed with cold PBS, and fixed with 4% PFA. Flow cytometry was performed, and the median fluorescence intensity (y) was ratioed with that of HT29 cells without addition of peptide at different time points (t) using Flowjo software. The rate constant k was calculated by fitting the data to a first-order kinetics model, y(t) = Imax[1 –exp(–kt)], where Imax = maximum value,80 using Graphpad Prism 5.0 software.
Effect of HER2 Peptide on Cell Signaling
SKBR3 cells were grown in 6 well plates to ~60% confluence. Unlabeled KSP* peptide was added to separate wells at concentrations of 5 and 100 μM. Lapatinib, a tyrosine kinase inhibitor, was used as a positive control and was added in concentrations of 1, 25, 100, and 1000 nM. Lapatinib stock solution starting at 1 mg/mL was diluted in DMSO and PBS. 1% DMSO treated cells and untreated cells were used as control. After 48 h following treatment, the cells were harvested for Western blot analysis. Anti-HER2 antibody (Cell Signaling, #2165), phospho-HER2/ErbB2 (Tyr1248) antibody (Cell Signaling, #2247), anti-AKT (Cell Signaling, #4691P), anti-ERK1/2 (Cell Signaling, #4695P), antiphospho-AKT (pS473), (Cell Signaling, #4060P), anti-phospho-ERK1/2 (Cell Signaling, #4370P), and anti-tubulin (Invitrogen, #32–2600) were used per manufacturer’s instructions.
In Vivo Imaging of Mouse Colonic Dysplasia with Topical HER2 Peptide
Mouse imaging studies were performed with approval of the University of Michigan Committee on the Use and Care of Animals (UCUCA). The mice were housed in pathogen-free conditions and supplied water ad libitum under controlled conditions of humidity (50 ± 10%), light (12/12 h light/dark cycle), and temperature (25 °C). Anesthesia was induced and maintained via a nose cone with inhaled isoflurane mixed with oxygen at a concentration of 2–4% at a flow rate of ~0.5 L/min. The colon was first rinsed with tap water to remove mucous and debris. White light illumination was used first to identify anatomic landmarks, including polyps, mucosal folds, colonic segment, and distance of endoscope tip from anus, to register the in vivo images with histology. The Cy5.5-labeled peptides were locally delivered at a concentration of 100 μM in a volume of 1.5 mL through the instrument channel (3 Fr). After 5 min for incubation, the colon was rinsed 3× with a tap water to remove the unbound peptides. Imaging was performed using a small animal endoscope (Karl Storz Veterinary Endoscopy).27 During the imaging, we recorded the distance between the endoscope tip and the anus and the clockwise location of each region of high intensity. Endoscopy was repeated 2–3 days later to confirm disappearance of any residual signal from KSP*-Cy5.5, and the mice were imaged with the PPS*-Cy5.5 control.
White light and fluorescence videos were exported in avi format with 24 (RGB) and 8 (grayscale) bit digital resolution for the color and fluorescence images, respectively. Streams that showed minimum motion artifact and absence of debris (stool, mucus) were selected for quantification. Individual frames were exported using custom Matlab software. On the fluorescence images, 3 regions of interest (ROI) with dimensions of 25 × 25 pixels were picked to represent a range of staining for diseased (target) and normal (background) tissues and averaged. The mean intensities within these ROI were determined to calculate the target to-background (T/B) ratio.
In Vivo Imaging of Mouse Colonic Dysplasia with Systemic HER2 Peptide
Mice were anaesthetized using isoflurane, administered 200 μL of KSP*-Cy5.5 peptide at 300 μM concentration via the tail vein. The imaging was performed prior to peptide injection to assess baseline fluorescence and after injection until the maximum signal was observed. The ratio of fluorescence intensity in the adenoma to that in the normal region was calculated by drawing regions of interest at each time point.
Ex Vivo Macroscopic Validation of HER2 Over Expressed in Mouse Colonic Neoplasia
After imaging, the colon was excised, flushed with PBS, and opened longitudinally for imaging with the IVIS 200 system using a Cy5.5 filter (Caliper Life Sciences, Hopkinton, MA). NIR fluorescence images were collected using 675 nm excitation and 720 nm emission with 1 s exposure. A ruler was placed next to the specimen to determine the distance from the anus for registration with the endoscopy and histology images. The specimen was then processed for histology by cutting sections in the plane parallel to the mucosal surface. Digital images were collected with a Zeiss Axiovision microscope (Thornwood, NY) using 5× magnification, and stitched together using Image Composite Editor (Microsoft, Redmond, WA). An expert gastrointestinal pathologist (SRO) who was blinded to the imaging results reviewed the histology, and identified regions of dysplasia and normal colon. Fluorescence intensities were measured from ellipsoid regions of interest (ROI) around the adenoma and an annulus of equal area around adjacent normal using Living Image 4.0 software (PerkinElmer; Hopkinton, MA). The intensity was defined by the sum of the radiance from each pixel inside the ROI/number of pixels (photons/s/cm2/sr).
Peptide Immunofluorescence on Mouse Colonic Adenomas
Specimens of mouse colonic adenoma were formalin fixed and processed, as described previously. Serial 5 μm sections were incubated with 2 μM of either KSP*-Cy5.5 or PPS*-Cy5.5 for 10 min at RT. The sections were washed 3× with PBS, and mounted with Prolong Gold reagent containing DAPI (Invitrogen). Adjacent sections were processed for histology (H&E).
Binding of HER2 Peptide to Human Proximal Colonic Neoplasia
Formalin-fixed, paraffin-embedded (FFPE) specimens of human proximal colon were obtained from the archived tissue bank in the Department of Pathology. 5-μm-thick sections were cut, and mounted onto glass slides (Superfrost Plus, Fischer Scientific). The tissues were deparaffinized, and antigen retrieval was performed, as describe below. The sections were blocked with protein serum for 15 min at RT followed by rinsing with PBS. The sections were then stained with KSP*-Cy5.5 at 5 μM concentration for 10 min at RT. The sections were then washed 3× for 3 min with PBS and further incubated with anti-HER2 antibody overnight at 4 °C. Sections were washed 3× with PBST and mounted with Prolong Gold reagent containing DAPI (Invitrogen) using #1 cover glass (1.5 μm thickness). We placed 3 boxes with dimensions of 20 × 20 μm2 completely within colonic epithelium in each image, and measured the mean fluorescence intensities using custom Matlab software. Regions of saturated intensities were avoided.
Over Expression of HER2 in Human Proximal Colon
Immunohistochemistry (IHC) was performed using 1:450 dilution of anti-HER2/neu (#29D8; rabbit monoclonal antibody #2165, Cell Signaling Inc.). Vectastain Elite kit (Vector Laboratories Inc.) was used per manufacturer’s instructions. Briefly, formalin-fixed specimens were deparaffinized using a standard dehydration/rehydration protocol and antigen unmasking was performed by boiling the slides in 10 mM sodium citrate buffer with 0.05% Tween at pH 6.0, and then maintaining at a sub-boiling temperature for 15 min. The slides were cooled for 30 min. The sections were washed 3× with dH2O for 3 min, and then incubated in 3% H2O2 in methanol for 10 min. The sections were washed 3× in dH2O for 2 min and in PBST for 5 min. Blocking was performed with protein blocking agent (#X0909, Dako) for 15 min at RT. The blocking solution was washed 1× with PBS. The sections were incubated with anti-HER2/neu antibody overnight at 4 °C in a humidified chamber and then washed 3× in PBST for 5 min. A 1:200 dilution of biotinylated secondary antibody (goat anti-rabbit IgG) was added to each section and incubated for 30 min at RT, and then removed by washing 3× with PBST for 5 min. Premixed Elite Vectastain ABC reagent from Vector Laboratories was added to each section and incubated for 30 min at RT. The sections were washed 3× in PBS for 5 min, and developed with 3,3’-diaminobenzidine (DAB) substrate. The reaction was monitored for up to 5 min, and then quenched by immersing the slides in dH2O. Hematoxylin was added as a counterstain for ~20 s, and the sections were dehydrated in increasing concentrations of ethyl alcohol (2× each at 70%, 80%, 95%, and 100%). Coverslips were mounted using permount mounting medium (#SP15–100, Fisher) in xylene. Serial sections were processed for routine histology (H&E).
IHC scoring was performed independently by expert gastroenterology pathologists (SRO, HA) without prior knowledge of clinical information or imaging results. The scoring was performed according to the DAKO HercepTest guidelines as follows: 0+, no reactivity or membrane staining in ≥10% of tumor cells; 1+, faint/barely perceptible partial membrane staining in ≥10% of tumor cells; 2+, weak-to-moderate complete or basolateral membrane staining in ≥10% of tumor cells; 3+, moderate-to-strong complete membrane staining in ≥10% of tumor cells. IHC scoring with 2+ and 3+ were considered to be positive for HER2 overexpression.
Statistical Analysis
Fluorescence intensities were transformed in base-2 log if needed to improve normality and stabilize variance. The fold-change between classification pairs was estimated using the antilog of the difference in the log-transformed data. We fit the human data with 4 histological classifications, including normal, hyperplastic polyp, sessile serrated adenoma, and sporadic adenoma, with a one-way ANOVA model and used Tukey’s multiple comparisons. Co-localization of peptide and antibody binding was evaluated on Pearson’s correlation coefficient.
Supplementary Material
Acknowledgments
Funding was provided in part by National Institutes of Health U54 CA163059, U01 CA189291, R01 CA142750, R01 CA200007, and R01 EB020644 (T.D.W.) and by Mary L. Petrovich.
ABBREVIATIONS
- AU
arbitrary units
- NIR
near-infrared
- ROI
regions of interest
- RT
room temperature
- T/B
target-to-background
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.bioconjchem.5b00565.
Additional figures (PDF)
Author Contributions
Designed research − BPJ, TDW. Performed research − BPJ, AP, JZ, XD, TDW. Contributed new reagents or analytic tools − BPJ, XD, QZ, TDW. Analyzed data − BPJ, AP, RK, SRO, HA, TDW. Wrote manuscript − BPJ, TDW.
Notes
The authors declare the following competing financial interest(s): BPJ and TDW are co-inventors on a patent disclosure submitted to the University of Michigan on the peptide.
References
- 1.Liu J, Zuo X, Li C, Yu T, Gu X, Zhou C, Li Z, Goetz M, Kiesslich R, Li Y. In vivo molecular imaging of epidermal growth factor receptor in patients with colorectal neoplasia using confocal laser endomicroscopy. Cancer Lett. 2013;330:200–7. doi: 10.1016/j.canlet.2012.11.044. [DOI] [PubMed] [Google Scholar]
- 2.Mitsunaga M, Kosaka N, Choyke PL, Young MR, Dextras CR, Saud SM, Colburn NH, Sakabe M, Nagano T, Asanuma D, Urano Y, Kobayashi H. Fluorescence endoscopic detection of murine colitis-associated colon cancer by topically applied enzymatically rapid-activatable probe. Gut. 2013;62:1179–86. doi: 10.1136/gutjnl-2011-301795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bird-Lieberman EL, Neves AA, Lao-Sirieix P, O’Donovan M, Novelli M, Lovat LB, Eng WS, Mahal LK, Brindle KM, Fitzgerald RC. Molecular imaging using fluorescent lectins permits rapid endoscopic identification of dysplasia in Barrett’s esophagus. Nat Med. 2012;18:315–21. doi: 10.1038/nm.2616. [DOI] [PubMed] [Google Scholar]
- 4.Luo S, Zhang E, Su Y, Cheng T, Shi C. A review of NIR dyes in cancer targeting and imaging. Biomaterials. 2011;32:7127–38. doi: 10.1016/j.biomaterials.2011.06.024. [DOI] [PubMed] [Google Scholar]
- 5.Thurber GM, Schmidt MM, Wittrup KD. Antibody tumor penetration: transport opposed by systemic and antigen-mediated clearance. Adv Drug Delivery Rev. 2008;60:1421–34. doi: 10.1016/j.addr.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chauhan VP, Stylianopoulos T, Boucher Y, Jain RK. Delivery of molecular and nanoscale medicine to tumors: transport barriers and strategies. Annu Rev Chem Biomol Eng. 2011;2:281–98. doi: 10.1146/annurev-chembioeng-061010-114300. [DOI] [PubMed] [Google Scholar]
- 7.Hsiung PL, Hardy J, Friedland S, Soetikno R, Du CB, Wu AP, Sahbaie P, Crawford JM, Lowe AW, Contag CH, Wang TD. Detection of colonic dysplasia in vivo using a targeted heptapeptide and confocal microendoscopy. Nat Med. 2008;14:454–8. doi: 10.1038/nm1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sturm MB, Joshi BP, Lu S, Piraka C, Khondee S, Elmunzer BJ, Kwon RS, Beer DG, Appelman HD, Turgeon DK, Wang TD. Targeted imaging of esophageal neoplasia with a fluorescently labeled peptide: first-in-human results. Sci Transl Med. 2013;5:184ra61. doi: 10.1126/scitranslmed.3004733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee S, Xie J, Chen X. Peptides and peptide hormones for molecular imaging and disease diagnosis. Chem Rev. 2010;110:3087–111. doi: 10.1021/cr900361p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brandt-Rauf PW, Pincus MR, Carney WP. The c-erbB-2 protein in oncogenesis: molecular structure to molecular epidemiology. Crit Rev Oncog. 1994;5:313–329. doi: 10.1615/critrevoncog.v5.i2-3.100. [DOI] [PubMed] [Google Scholar]
- 11.Tao RH, Maruyama IN. All EGF (ErbB) receptors have preformed homo- and heterodimeric structures in living cells. J Cell Sci. 2008;121:3207–17. doi: 10.1242/jcs.033399. [DOI] [PubMed] [Google Scholar]
- 12.Baselga J, Swain SM. Novel anticancer targets: revisiting ERBB2 and discovering ERBB3. Nat Rev Cancer. 2009;9:463–475. doi: 10.1038/nrc2656. [DOI] [PubMed] [Google Scholar]
- 13.Katz M, Amit I, Yarden Y. Regulation of MAPKs by growth factors and receptor tyrosine kinases. Biochim Biophys Acta Mol Cell Res. 2007;1773:1161–76. doi: 10.1016/j.bbamcr.2007.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fornaro L, Lucchesi M, Caparello C, Vasile E, Caponi S, Ginocchi L, Masi G, Falcone A. Anti-HER agents in gastric cancer: from bench to bedside. Nat Rev Gastroenterol Hepatol. 2011;8:369–383. doi: 10.1038/nrgastro.2011.81. [DOI] [PubMed] [Google Scholar]
- 15.Ross JS. The HER-2/neu oncogene in tumors of the gastrointestinal tract. Cancer Invest. 2001;19:554–68. doi: 10.1081/cnv-100103852. [DOI] [PubMed] [Google Scholar]
- 16.Seo AN, Kwak Y, Kim DW, Kang S-B, Choe G, Kim WH, Lee HS. HER2 Status in Colorectal Cancer: Its Clinical Significance and the Relationship between HER2 Gene Amplification and Expression. PLoS One. 2014;9:e98528. doi: 10.1371/journal.pone.0098528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khelwatty SA, Essapen S, Bagwan I, Green M, Seddon AM, Modjtahedi H. Co-Expression of HER Family Members in Patients with Dukes’ C and D Colon Cancer and Their Impacts on Patient Prognosis and Survival. PLoS One. 2014;9:e91139. doi: 10.1371/journal.pone.0091139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferlay J, Soerjomataram I, Ervik M, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. IARC CancerBase No 11 [Internet] International Agency for Research on Cancer; Lyon, France, 2013: 2012. GLOBOCAN v 1.0, Cancer Incidence and Mortality Worldwide. [Google Scholar]
- 19.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008. Int J Cancer. 2010;127:2893–917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 20.Nishihara R, Wu K, Lochhead P, Morikawa T, Liao X, Qian ZR, Inamura K, Kim SA, Kuchiba A, Yamauchi M, et al. Long-term colorectal-cancer incidence and mortality after lower endoscopy. N Engl J Med. 2013;369:1095–105. doi: 10.1056/NEJMoa1301969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh H, Nugent Z, Demers AA, Kliewer EV, Mahmud SM, Bernstein CN. The reduction in colorectal cancer mortality after colonoscopy varies by site of the cancer. Gastroenterology. 2010;139:1128–37. doi: 10.1053/j.gastro.2010.06.052. [DOI] [PubMed] [Google Scholar]
- 22.Baxter NN, Goldwasser MA, Paszat LF, Saskin R, Urbach DR, Rabeneck L. Association of colonoscopy and death from colorectal cancer. Ann Intern Med. 2009;150:1–8. doi: 10.7326/0003-4819-150-1-200901060-00306. [DOI] [PubMed] [Google Scholar]
- 23.Lakoff J, Paszat LF, Saskin R, Rabeneck L. Risk of developing proximal versus distal colorectal cancer after a negative colonoscopy: a population-based study. Clin Gastroenterol Hepatol. 2008;6:1117–21. doi: 10.1016/j.cgh.2008.05.016. [DOI] [PubMed] [Google Scholar]
- 24.Bressler B, Paszat LF, Vinden C, Li C, He J, Rabeneck L. Colonoscopic miss rates for right-sided colon cancer: a population-based analysis. Gastroenterology. 2004;127:452–6. doi: 10.1053/j.gastro.2004.05.032. [DOI] [PubMed] [Google Scholar]
- 25.Rondagh EJ, Bouwens MW, Riedl RG, Winkens B, de Ridder R, Kaltenbach T, Soetikno RM, Masclee AA, Sanduleanu S. Endoscopic appearance of proximal colorectal neoplasms and potential implications for colonoscopy in cancer prevention. Gastrointest Endosc. 2012;75:1218–25. doi: 10.1016/j.gie.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 26.Soetikno RM, Kaltenbach T, Rouse RV, Park W, Maheshwari A, Sato T, Matsui S, Friedland S. Prevalence of nonpolypoid (flat and depressed) colorectal neoplasms in asymptomatic and symptomatic adults. JAMA. 2008;299:1027–35. doi: 10.1001/jama.299.9.1027. [DOI] [PubMed] [Google Scholar]
- 27.Zhou J, Joshi BP, Duan X, Pant A, Qiu Z, Kuick R, Owens SR, Wang TD. EGFR Overexpressed in Colonic Neoplasia Can be Detected on Wide-Field Endoscopic Imaging. Clin Transl Gastroenterol. 2015;6:e101. doi: 10.1038/ctg.2015.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burggraaf J, Kamerling IM, Gordon PB, Schrier L, de Kam ML, Kales AJ, Bendiksen R, Indrevoll B, Bjerke RM, Moestue SA, et al. Detection of colorectal polyps in humans using an intravenously administered fluorescent peptide targeted against c-Met. Nat Med. 2015;21:955–961. doi: 10.1038/nm.3641. [DOI] [PubMed] [Google Scholar]
- 29.Bando T, Muguruma N, Ito S, Musashi Y, Inayama K, Kusaka Y, Tadatsu M, Ii K, Irimura T, Shibamura S, Takesako K. Basic Studies on a Labeled Anti-mucin Antibody Detectable by Infrared-fluorescence Endoscopy. J Gastroenterol. 2002;37:260–269. doi: 10.1007/s005350200033. [DOI] [PubMed] [Google Scholar]
- 30.Yao YL, Shao J, Zhang C, Wu JH, Zhang QH, et al. Proliferation of Colorectal Cancer Is Promoted by Two Signaling Transduction Expression Patterns: ErbB2/ErbB3/AKT and MET/ErbB3/MAPK. PLoS One. 2013;8:e78086. doi: 10.1371/journal.pone.0078086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gill MK, Manjari M, Jain K. Expression of Her-2/neu in colon carcinoma and its correlation with the histological grades and the lymph nodes status. J Clin Diagn Res. 2011;5:1564–8. [Google Scholar]
- 32.Half E, Broaddus R, Danenberg KD, Danenberg PV, Ayers GD, Sinicrope FA. HER-2 receptor expression, localization, and activation in colorectal cancer cell lines and human tumors. Int J Cancer. 2004;108:540–8. doi: 10.1002/ijc.11599. [DOI] [PubMed] [Google Scholar]
- 33.Kavanagh D, Chambers G, O’ Grady L, et al. Is overexpression of HER-2 a predictor of prognosis in colorectal cancer? BMC Cancer. 2009;9:1. doi: 10.1186/1471-2407-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nathanson DR, Culliford AT, Shia J, et al. HER 2/neu expression and gene amplification in colon cancer. Int J Cancer. 2003;105:796–802. doi: 10.1002/ijc.11137. [DOI] [PubMed] [Google Scholar]
- 35.Garrett TP, McKern NM, Lou M, Elleman TC, Adams TE, Lovrecz GO, Kofler M, Jorissen RN, Nice EC, Burgess AW, Ward CW. The crystal structure of a truncated ErbB2 ectodomain reveals an active conformation, poised to interact with other ErbB receptors. Mol Cell. 2003;11:495–505. doi: 10.1016/s1097-2765(03)00048-0. [DOI] [PubMed] [Google Scholar]
- 36.http://www.mimotopes.com.
- 37.Lemmon MA. Ligand-induced ErbB receptor dimerization. Exp Cell Res. 2009;315:638–648. doi: 10.1016/j.yexcr.2008.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Franklin MC, Carey KD, Vajdos FF, Leahy DJ, de Vos AM, Sliwkowski MX. Insights into ErbB signaling from the structure of the ErbB2-pertuzumab complex. Cancer Cell. 2004;5:317–328. doi: 10.1016/s1535-6108(04)00083-2. [DOI] [PubMed] [Google Scholar]
- 39.Wang Z, Wang W, Bu X, Wei Z, Geng L, Wu Y, Dong C, Li L, Zhang D, Yang S, et al. Microarray Based Screening of Peptide Nano Probes for HER2 Positive Tumor. Anal Chem. 2015;18:8367–72. doi: 10.1021/acs.analchem.5b01588. [DOI] [PubMed] [Google Scholar]
- 40.Trabuco LG, Lise S, Petsalaki E, Russell RB. Nucleic Acids Res. 2012;40:W423–426. doi: 10.1093/nar/gks398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hinoi T, Akyol A, Theisen BK, Ferguson DO, Greenson JK, Williams BO, Cho KR, Fearon ER. Mouse model of colonic adenoma-carcinoma progression based on somatic Apc inactivation. Cancer Res. 2007;67:9721–30. doi: 10.1158/0008-5472.CAN-07-2735. [DOI] [PubMed] [Google Scholar]
- 42.Rowan AJ, Lamlum H, Ilyas M, Wheeler J, Straub J, Papadopoulou A, Bicknell D, Bodmer WF, Tomlinson IP. APC mutations in sporadic colorectal tumors: A mutational “hotspot” and interdependence of the “two hits”. Proc Natl Acad Sci U S A. 2000;97:3352–7. doi: 10.1073/pnas.97.7.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Larimer BM, Thomas WD, Smith GP, Deutscher SL. Affinity maturation of an ERBB2-targeted SPECT imaging peptide by in vivo phage display. Mol Imaging Biol. 2014;16:449–58. doi: 10.1007/s11307-014-0724-5. [DOI] [PubMed] [Google Scholar]
- 44.Geng L, Wang Z, Yang X, Li D, Lian W, Xiang W, Wang W, Bu X, Lai W, Hu Z, Fang Q. Structure-based Design of Peptides with High Affinity and Specificity to HER2 Positive Tumors. Theranostics. 2015;5:1154–1165. doi: 10.7150/thno.12398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Banappagari S, Alecia M, Call, Krystal F, Vicente GM, Gujar A, Satyanarayanajois S. Design, synthesis and characterization of peptidomimetic conjugate of BODIPY targeting HER2 protein extracellular domain. Eur J Med Chem. 2013;65:60–69. doi: 10.1016/j.ejmech.2013.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Banappagari S, Ronald S, Satyanarayanajois SD. A Conformationally Constrained Peptidomimetic Binds to the Extracellular Region of HER2 Protein. J Biomol Struct Dyn. 2010;28:289–308. doi: 10.1080/07391102.2010.10507360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jamalan M, Zeinali M, Asadabadi EB. Design of Peptidomimetics for Inhibition of HER2 Receptor Dimerization by a Combination of Virtual Screening, MD Simulations, and QSAR In Silico Methods. Chem Biol Drug Des. 2013;81:455–462. doi: 10.1111/cbdd.12062. [DOI] [PubMed] [Google Scholar]
- 48.Nakajima H, Mizuta N, Sakaguchi K, Fujiwara I, Yoshimori A, Takahashi S, Takasawa R, Tanuma S. Development of HER2-antagonistic peptides as novel anti-breast cancer drugs by in silico methods. Breast Cancer. 2008;15:65–72. doi: 10.1007/s12282-007-0018-8. [DOI] [PubMed] [Google Scholar]
- 49.Schroeder RL, Stevens CL, Sridhar J. Small molecule tyrosine kinase inhibitors of ErbB2/HER2/Neu in the treatment of aggressive breast cancer. Molecules. 2014;19:15196–212. doi: 10.3390/molecules190915196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tian C, Ding P, Yuan Z, Li H, Zhao Y, Sun L, Guo Q, Wang Z, Sun L, Zhang L, Jiang Z. A novel dual EGFR/HER2 inhibitor KU004 induces cell cycle arrest and apoptosis in HER2-overexpressing cancer cells. Apoptosis. 2015;20:1599. doi: 10.1007/s10495-015-1164-7. [DOI] [PubMed] [Google Scholar]
- 51.Sanduleanu S, Masclee AM, Meijer GA. Interval cancers after colonoscopy-insights and recommendations. Nat Rev Gastroenterol Hepatol. 2012;9:550–4. doi: 10.1038/nrgastro.2012.136. [DOI] [PubMed] [Google Scholar]
- 52.Robertson DJ, Lieberman DA, Winawer SJ, Ahnen DJ, Baron JA, Schatzkin A, Cross AJ, Zauber AG, Church TR, Lance P, et al. Colorectal cancers soon after colonoscopy: a pooled multicohort analysis. Gut. 2014;63:949–56. doi: 10.1136/gutjnl-2012-303796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Farrar WD, Sawhney MS, Nelson DB, Lederle FA, Bond JH. Colorectal cancers found after a complete colonoscopy. Clin Gastroenterol Hepatol. 2006;4:1259–64. doi: 10.1016/j.cgh.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 54.Rex DK, Schoenfeld PS, Cohen J, Pike IM, Adler DG, Fennerty MB, Lieb JG, Park WG, Rizk MK, Sawhney MS, et al. Quality indicators for colonoscopy. Am J Gastroenterol. 2015;110:72–90. doi: 10.1038/ajg.2014.385. [DOI] [PubMed] [Google Scholar]
- 55.Shergill AK, Conners EE, McQuaid KR, Epstein S, Ryan JC, Shah JN, Inadomi J, Somsouk M. Protective association of colonoscopy against proximal and distal colon cancer and patterns in interval cancer. Gastrointest Endosc. 2015;82:529–37. doi: 10.1016/j.gie.2015.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.le Clercq CM, Winkens B, Bakker CM, Keulen ET, Beets GL, Masclee AA, Sanduleanu S. Metachronous colorectal cancers result from missed lesions and non-compliance with surveillance. Gastrointest Endosc. 2015;82:325–33. doi: 10.1016/j.gie.2014.12.052. [DOI] [PubMed] [Google Scholar]
- 57.Adler A, Aschenbeck J, Yenerim T, Mayr M, Aminalai A, Drossel R, Schröder A, Scheel M, Wiedenmann B, Rösch T. Narrow-band versus white-light high definition television endoscopic imaging for screening colonoscopy: a prospective randomized trial. Gastroenterology. 2009;136:410–6. doi: 10.1053/j.gastro.2008.10.022. [DOI] [PubMed] [Google Scholar]
- 58.East JE, Ignjatovic A, Suzuki N, Guenther T, Bassett P, Tekkis PP, Saunders BP. A randomized, controlled trial of narrow-band imaging vs high-definition white light for adenoma detection in patients at high risk of adenomas. Colorectal Dis. 2012;14:e771–8. doi: 10.1111/codi.12014. [DOI] [PubMed] [Google Scholar]
- 59.Pasha SF, Leighton JA, Das A, Harrison ME, Gurudu SR, Ramirez FC, Fleischer DE, Sharma VK. Comparison of the yield and miss rate of narrow band imaging and white light endoscopy in patients undergoing screening or surveillance colonoscopy: a meta-analysis. Am J Gastroenterol. 2012;107:363–70. doi: 10.1038/ajg.2011.436. [DOI] [PubMed] [Google Scholar]
- 60.Kahi CJ, Anderson JC, Waxman I, Kessler WR, Imperiale TF, Li X, Rex DK. High-definition chromocolonoscopy vs. high-definition white light colonoscopy for average-risk colorectal cancer screening. Am J Gastroenterol. 2010;105:1301–7. doi: 10.1038/ajg.2010.51. [DOI] [PubMed] [Google Scholar]
- 61.Lapalus MG, Helbert T, Napoleon B, Rey JF, Houcke P, Ponchon T Société Française d’Endoscopie Digestive. Does chromoendoscopy with structure enhancement improve the colonoscopic adenoma detection rate? Endoscopy. 2006;38:444–8. doi: 10.1055/s-2006-925265. [DOI] [PubMed] [Google Scholar]
- 62.Le Rhun M, Coron E, Parlier D, Nguyen JM, Canard JM, Alamdari A, Sautereau D, Chaussade S, Galmiche JP. High resolution colonoscopy with chromoscopy versus standard colonoscopy for the detection of colonic neoplasia: a randomized study. Clin Gastroenterol Hepatol. 2006;4:349–54. doi: 10.1016/j.cgh.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 63.Dinesen L, Chua TJ, Kaffes AJ. Meta-analysis of narrow-band imaging versus conventional colonoscopy for adenoma detection. Gastrointestinal Endoscopy. 2012;75:604–11. doi: 10.1016/j.gie.2011.10.017. [DOI] [PubMed] [Google Scholar]
- 64.Rex DK, Clodfelter R, Rahmani F, Fatima H, James-Stevenson TN, Tang JC, Kim HN, McHenry L, Kahi CJ, Rogers NA, et al. Narrow-band imaging versus white light for the detection of proximal colon serrated lesions: a randomized, controlled trial. Gastrointestinal Endoscopy. 2015 doi: 10.1016/j.gie.2015.03.1915. [DOI] [PubMed] [Google Scholar]
- 65.Gill MK, Manjari M, Jain K. Expression of Her-2/neu in colon carcinoma and its correlation with the histological grades and the lymph nodes status. JCDR. 2011;5:1564–8. [Google Scholar]
- 66.Half E, Broaddus R, Danenberg KD, Danenberg PV, Ayers GD, Sinicrope FA. HER-2 receptor expression, localization, and activation in colorectal cancer cell lines and human tumors. Int J Cancer. 2004;108:540–8. doi: 10.1002/ijc.11599. [DOI] [PubMed] [Google Scholar]
- 67.Kavanagh D, Chambers G, O’ Grady L, et al. Is overexpression of HER-2 a predictor of prognosis in colorectal cancer? BMC Cancer. 2009;9:1. doi: 10.1186/1471-2407-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nathanson DR, Culliford AT, Shia J, et al. HER 2/neu expression and gene amplification in colon cancer. Int J Cancer. 2003;105:796–802. doi: 10.1002/ijc.11137. [DOI] [PubMed] [Google Scholar]
- 69.Choi WI, Lee JH, Kim JY, Heo SU, Jeong YY, Kim YH, Tae G. Targeted Antitumor Efficacy and Imaging via Multifunctional Nano-Carrier Conjugated with Anti-HER2 trastuzumab. Nanomedicine. 2014;14:00532–2. doi: 10.1016/j.nano.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 70.McCormack DR, Walsh AJ, Sit W, Arteaga CL, Chen J, Cook RS, Skala MC. In vivo hyperspectral imaging of microvessel response to trastuzumab treatment in breast cancer xenografts. Biomed Opt Express. 2014;5:2247–61. doi: 10.1364/BOE.5.002247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Miyake KK, Nakamoto Y, Kanao S, Tanaka S, Sugie T, Mikami Y, Toi M, Togashi K. Journal Club: Diagnostic value of (18)F-FDG PET/CT and MRI in predicting the clinicopathologic subtypes of invasive breast cancer. AJR, Am J Roentgenol. 2014;203:272–9. doi: 10.2214/AJR.13.11971. [DOI] [PubMed] [Google Scholar]
- 72.Kim HY, Wang X, Wahlberg B, Edwards WB. Discovery of hapten-specific scFv from a phage display library and applications for HER2-positive tumor imaging. Bioconjugate Chem. 2014;25:1311–22. doi: 10.1021/bc500173f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sörensen J, Sandberg D, Sandström M, Wennborg A, Feldwisch J, Tolmachev V, Åström G, Lubberink M, Garske-Román U, Carlsson J, Lindman H. First-in-human molecular imaging of HER2 expression in breast cancer metastases using the 111In-ABY-025 affibody molecule. J Nucl Med. 2014;55:730–5. doi: 10.2967/jnumed.113.131243. [DOI] [PubMed] [Google Scholar]
- 74.Rizvi SB, Rouhi S, Taniguchi S, Yang SY, Green M, Keshtgar M, Seifalian AM. Near-infrared quantum dots for HER2 localization and imaging of cancer cells. Int J Nanomed. 2014;9:1323–37. doi: 10.2147/IJN.S51535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Larimer BM, Thomas WD, Smith GP, Deutscher SL. Affinity maturation of an ERBB2-targeted SPECT imaging peptide by in vivo phage display. Mol Imaging Biol. 2014;16:449–58. doi: 10.1007/s11307-014-0724-5. [DOI] [PubMed] [Google Scholar]
- 76.Zábrady M, Hrdinová V, Müller B, Conrad U, Hejátko J, Janda L. Targeted In Vivo Inhibition of Specific Protein-Protein Interactions Using Recombinant Antibodies. PLoS One. 2014;9:e109875. doi: 10.1371/journal.pone.0109875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ryu JH, Shin M, Kim SA, Lee S, Kim H. In vivo fluorescence imaging for cancer diagnosis using receptor-targeted epidermal growth factor-based nanoprobe. Biomaterials. 2013;34:9149–59. doi: 10.1016/j.biomaterials.2013.08.026. [DOI] [PubMed] [Google Scholar]
- 78.ASGE Standards of Practice Committee. Shergill AK, Lightdale JR, Bruining DH, Acosta RD, Chandrasekhara V, Chathadi KV, Decker GA, Early DS, Evans JA, et al. The role of endoscopy in inflammatory bowel disease. Gastrointestinal Endoscopy. 2015;81:1101–1121. doi: 10.1016/j.gie.2014.10.030. [DOI] [PubMed] [Google Scholar]
- 79.Kamiński MF, Hassan C, Bisschops R, Pohl J, Pellisé M, Dekker E, Ignjatovic-Wilson A, Hoffman A, Longcroft-Wheaton G, Heresbach D, Dumonceau JM, East JE. Advanced imaging for detection and differentiation of colorectal neoplasia: European Society of Gastrointestinal Endoscopy (ESGE) Guideline. Endoscopy. 2014;46:435–49. doi: 10.1055/s-0034-1365348. [DOI] [PubMed] [Google Scholar]
- 80.Joshi BP, Duan X, Kwon RS, Piraka C, Elmunzer BJ, Lu S, Rabinsky E, Beer DG, Appelman HD, Owens SR, Kuick R, Doguchi N, Turgeon DK, Wang TD. Multimodal endoscope can quantify wide-field fluorescence detection of Barrett’s neoplasia. Endoscopy. 2015;48 doi: 10.1055/s-0034-1392803. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Joshi BP, Miller SJ, Lee CM, Seibel EJ, Wang TD. Multispectral endoscopic imaging of colorectal dysplasia in vivo. Gastroenterology. 2012;143:1435–7. doi: 10.1053/j.gastro.2012.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Joshi BP, Liu Z, Elahi SF, Appelman HD, Wang TD. Near-infrared-labeled peptide multimer functions as phage-mimic for high affinity, specific targeting of colonic adenomas in vivo. Gastrointestinal Endoscopy. 2012;76:1197–206. doi: 10.1016/j.gie.2012.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.http://www.rcsb.org/pdb.
- 84.Fields GB, Noble RL. Solid phase peptide synthesis utilizing 9-fluorenylmethoxycarbonyl amino acids. Int J Pept Protein Res. 1990;35:161–214. doi: 10.1111/j.1399-3011.1990.tb00939.x. [DOI] [PubMed] [Google Scholar]
- 85.Thomas R, Chen J, Roudier MM, Vessella RL, Lantry LE, Nunn AD. In vitro binding evaluation of 177Lu-AMBA, a novel 177Lu-labeled GRP-R agonist for systemic radio-therapy in human tissues. Clin Exp Metastasis. 2009;26:105–19. doi: 10.1007/s10585-008-9220-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


