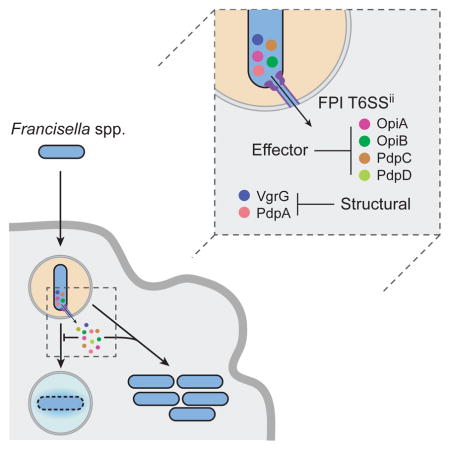
Summary
The intracellular bacterial pathogen Francisella tularensis causes tularemia, a zoonosis that can be fatal. The type VI secretion system (T6SS) encoded by the Francisella pathogenicity island (FPI) is critical for the virulence of this organism. Existing studies suggest that the complete repertoire of T6SS effectors delivered to host cells is encoded by the FPI. Using a proteome-wide approach, we discovered that the FPI-encoded T6SS exports at least three effectors encoded outside of the island. These proteins share features with virulence determinants of other pathogens and we provide evidence that they can contribute to intramacrophage growth. The remaining proteins we identified are encoded within the FPI. Two of these FPI-encoded proteins constitute effectors, whereas the others form a unique complex required for core function of the T6SS apparatus. The discovery of secreted effectors mediating interactions between Francisella and its host significantly advances our understanding of the pathogenesis of this organism.
eTOC blurb
The Francisella pathogenicity island (FPI) encodes a unique type VI secretion system. Eshraghi et al. discover several effector proteins secreted by this system, some of which are encoded by genes unlinked to the FPI. Effectors encoded within and outside the FPI cooperate to enhance the growth of Francisella in macrophages.
Introduction
The Francisella are host-adapted Gram-negative γ-proteobacteria endemic to much of the northern hemisphere (Kingry and Petersen, 2014). Francisella tularensis, the etiologic agent of tularemia, is a category A biodefense agent that is transmitted by a wide range of animals, including ticks. A signature feature of Francisella genomes is the presence of a gene cluster referred to as the Francisella Pathogenicity Island (FPI) (Nano and Schmerk, 2007). This region of the genome is approximately 25 kilobases, has extraordinarily low G+C content, and is essential for intramacrophage growth and virulence. Within F. tularensis, subspecies novicida (F. novicida) is unique by virtue of its single FPI; the genomes of subsp. tularensis, holartica, and mediasiatica contain two nearly identical FPI copies. Together with its relatively low virulence in humans, this feature of F. novicida has made it a valuable model for the study of FPI function.
The FPI is thought to encode a protein secretion system with structural and functional similarity to the bacterial type VI secretion system (T6SS) (Nano and Schmerk, 2007; Russell et al., 2014a). Indeed, based on gene content and phylogeny, the FPI-encoded secretion system was recently proposed to represent a unique T6SS subtype, T6SSii (Russell et al., 2014b). Representatives of T6SSi are widespread among Proteobacteria, whereas T6SSiii have so far been identified only in Bacteroidetes. Studies have revealed that T6SSi and T6SSiii predominantly function as intercellular protein delivery pathways that mediate antagonism between bacteria; however, a small number of T6SSi have been shown to target effector proteins to host cells (Hood et al., 2010; Schwarz et al., 2010).
In addition to informatic-based predictions, there are several experimental observations that support a functional relatedness between the FPI-encoded secretion system and canonical T6SSs. For instance, the major proteins exported by the systems, IglC and Hcp, respectively, adopt a structure that is closely related to gpV, the tube protein of lambda bacteriophage (de Bruin et al., 2011). The systems also appear to share a dynamic filament composed of two proteins, TssB–TssC and IglA–IglB in T6SSi,iii and T6SSii, respectively (Basler et al., 2012; Clemens et al., 2015). Energy released by contraction of this complex is postulated to drive T6S-dependent effector delivery.
Although the similarities between the FPI-encoded T6SSii and more thoroughly characterized T6S pathways are extensive, profound differences also exist between the systems. Currently, these differences prohibit the application of a common framework toward understanding FPI function. Secretion by T6SSi and T6SSiii pathways depends on the disassembly of contracted TssB–TssC filaments by a conserved AAA+-family ATPase, ClpV (Cianfanelli et al., 2016). Neither a ClpV homolog nor a functional substitute has been identified in association with the FPI-encoded secretion system. Other notable differences between the systems relate to effector recognition and export. T6SSi pathways secrete a multi-domain valine-glycine repeat protein, VgrG, that closely resembles the bacteriophage T4 tailspike, consisting of proteins gp27 and gp5. Recent studies show that a subset of effector proteins bind VgrG proteins through PAAR adaptor domains in order to be recognized and transported into recipient cells (Shneider et al., 2013; Whitney et al., 2015). The FPI does not encode a canonical PAAR domain protein and the VgrG protein associated with the FPI lacks the larger gp27-related domain (Rigard et al., 2016).
Substrate identification remains a major hurdle in understanding how the FPI promotes pathogenesis. Published attempts have employed indirect measures of export, such as the fusion of substrates to reporter enzymes. A strength of such methods is that they permit in vivo measurements; however, as implemented for the study of T6SSii, they have been limited to candidate substrates encoded within the FPI. Furthermore, if similar mechanisms underlie T6SSi and T6SSii substrate recognition and export, effector–reporter fusions are unlikely to be tolerated and could produce spurious results. Supporting this notion, there is little agreement among published studies pertaining to T6SSii substrates. Using C-terminal fusions of select FPI-encoded proteins to adenylate cyclase, Klose and colleagues concluded that VgrG and IglI, but not the Hcp homolog IglC, reach the cytoplasm of J774 macrophage cells infected by F. novicida (Barker et al., 2009). A later study by Broms et al., which analyzed the translocation of C-terminal fusions of the same proteins to β-lactamase, found IglC, but not VgrG and IglI access the cytoplasm of J774 cells infected by F. novicida (Broms et al., 2012). Here we used a proteome-wide approach to directly measure the secreted structural components and effectors of the FPI-encoded T6SSii pathway. This led to the unexpected finding that a subset of T6SSii-secreted virulence factors of F. novicida are encoded outside of the FPI and that export via T6SSii requires a unique secreted protein complex.
Results
An Improved Allelic Exchange System for F. novicida
One barrier to progress in the Francisella field is the lack of a facile method for generating precise genomic deletions and insertions in F. novicida. To address this, we constructed an allelic exchange vector that uses p-chlorophenylalanine – a conditionally toxic amino acid analog – for counterselection, as first implemented in Escherichia coli (Kast, 1994). In this system, expression of a phenylalanyl-tRNA synthetase (pheS) variant with relaxed substrate specificity confers sensitivity to p-chlorophenylalanine, which becomes incorporated into proteins. Sequence alignment of E. coli and F. novicida pheS genes enabled us to design an F. novicida pheS allele that confers sensitivity to p-chlorophenylalanine (pheS A305G). This allele was placed under control of the F. novicida promoter driving bacterioferritin expression and cloned into the parent vector pEX18km, generating the plasmid pEX18-pheS-km (Figure S1) (Charity et al., 2007). Implementation of this counterselection strategy to drive allelic exchange improved mutation efficiency and allowed for the construction of all strains generated in this study.
Identification of F. novicida T6SSii Substrates
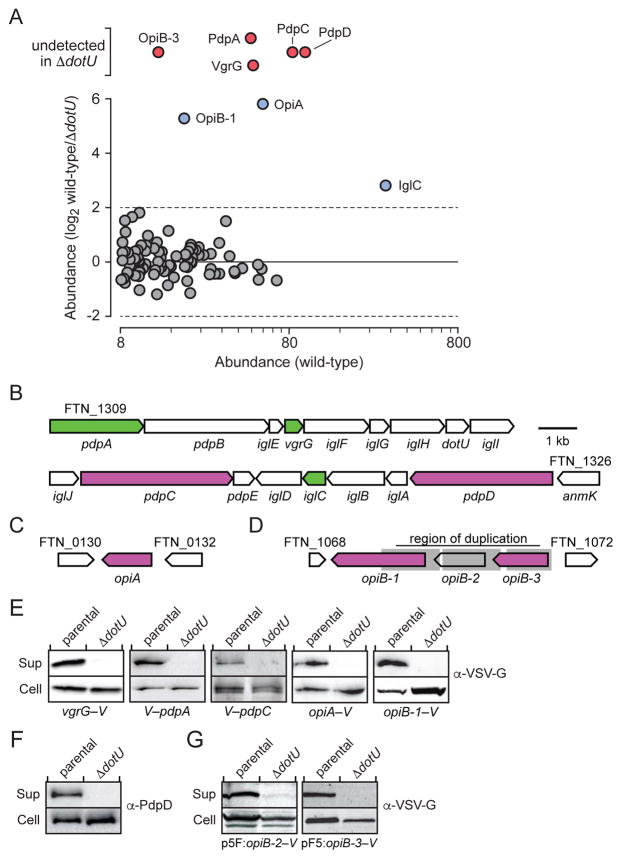
To our knowledge, a proteome-wide, direct biochemical approach for defining T6SSii substrates has not been reported. We reasoned that this was likely due to repression of the system in vitro, a property observed in many previously characterized T6SSi pathways (Silverman et al., 2012). Recently, it was shown that the addition of excess potassium to rich media triggers increased T6SSii substrate export by F. novicida (Clemens et al., 2015). Rich media that contain proteolytic digests are not optimal for extracellular proteome studies; however, we found that in defined media more suitable for mass spectrometry (MS) analyses, high levels of potassium promoted F. novicida lysis. To circumvent these problems, we subjected stabilized F. novicida supernatant samples derived from rich media to extensive dialysis prior to precipitation. This treatment diminished the concentration of media-derived peptides sufficiently to permit the detection of 95 F. novicida proteins following stringent filtering criteria of duplicate samples (Figure 1A and Table S1). As is frequently observed in extracellular proteomes, abundant, stable cytoplasmic proteins were detected among an enriched set of predicted secreted proteins. Nonetheless, reflecting the high level of T6SSii activity achieved by potassium treatment, the three proteins we identified with greatest spectral counts are each encoded within the FPI.
Figure 1. The T6SSii System Facilitates the Export of at Least Eight Proteins, Including Three Encoded Outside of the FPI.
(A) Quantitative mass spectrometric-based comparison of the extracellular proteomes of F. novicida wild-type and ΔdotU. Proteins highlighted in red and blue are absent or detected in significantly lower abundance in ΔdotU relative to the wild-type, respectively.
(B–D) Regions of the F. novicida genome encoding T6SSii substrates: the FPI (B) and two distant genomic loci (C and D). Functions assigned in this study are indicated by fill color; secreted structural components are green and effectors are purple. Grey shading denotes the region of duplication overlapping opiB loci.
(E–G) Western blot analysis probing candidate T6SSii substrate levels in the supernatant (Sup) and cell fractions of F. novicida wild-type and ΔdotU. Genetic backgrounds are denoted below the western blots. Substrates were detected by antibodies against N- (V–) or C-terminal (–V) vesicular stomatitis virus glycoprotein (VSV-G) epitope fusions encoded at native chromosomal loci (E), protein-specific antibodies (F), or via VSV-G fusions produced ectopically (G).
To define the T6SSii-dependent secretome, we compared the wild-type extracellular proteome to that of a T6SSii-inactive reference strain. For this purpose, we employed an F. novicida strain bearing an in-frame deletion in dotU. Consistent with previous studies demonstrating the requirement for DotU (TssL) in T6SSi function, we found that secretion of the two known T6SSii substrates, VgrG and IglC (Hcp), is compromised in F. novicida ΔdotU (Durand et al., 2015). VgrG was not detected in the ΔdotU-derived samples and IglC levels were substantially lower relative to the wild-type (Figure 1A). In addition to these known T6SSii substrates, six proteins in wild-type samples were undetected or displayed significantly diminished abundance in those derived from ΔdotU (Figure 1A, Table S1). Three of these, PdpA, PdpC, and PdpD, are encoded within the FPI (Figure 1B). Interestingly, the remaining proteins are encoded by open reading frames located outside of the FPI (FTN_0131, FTN_1069, FTN_1071) (Figures 1C and 1D). Henceforth we refer to these proteins as outside pathogenicity island A (OpiA), OpiB-1 and OpiB-3, respectively. The genes encoding OpiB-1 and OpiB-3 flank a third open reading frame, which we refer to as opiB-2 (Figure 1D). These genes appear to have arisen from multiple duplication events, as they share a high degree of identity (93%) that encompasses the full length of opiB-2 and opiB-3 and extends to intragenic regions.
To validate our MS results, we sought to visualize the export of each of the putative T6SSii substrates by Western blot. Although we did not detect unique OpiB-2 peptides in the extracellular proteome of F. novicida, based on homology and linkage between the opiB genes, we included this protein in our study. Western analysis comparing cell and supernatant fractions prepared from wild-type and ΔdotU backgrounds confirmed T6SSii-dependent export of each candidate substrate (Figure 1E–G). Based on these data, we conclude that the T6SSii pathway of F. novicida facilitates the export of at least eight proteins, including three encoded outside of the FPI (OpiA, OpiB.1 and OpiB.3).
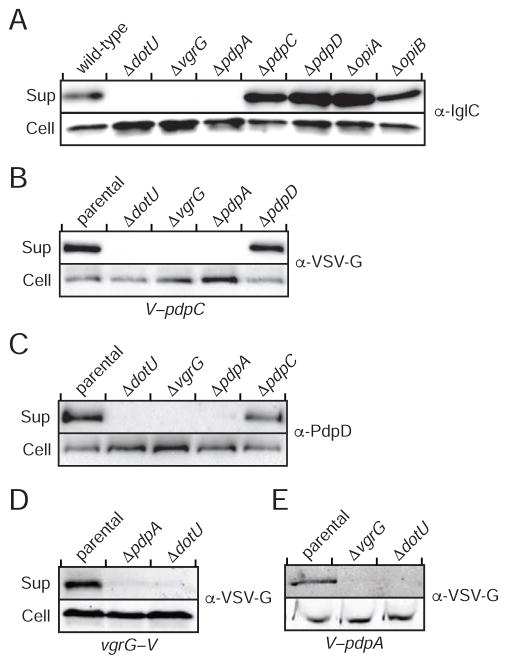
T6SSii Substrates Constitute Two Functional Classes
Type VI secretion substrates can be structural components, effectors, or proteins that serve both functions. We sought to determine which of these categories each of the T6SSii-secreted proteins we identified by MS belong to. A common measure of basal T6SS function is the export of an Hcp-related protein; thus, we examined IglC export in F. novicida strains bearing in-frame deletions in the genes encoding each substrate. We found that OpiA and the OpiB proteins are not required for IglC export, which taken together with the fact that both are encoded outside of the FPI and share properties with known effectors (see below), led us to conclude that these proteins likely function as effectors (Figure 2A). Among the FPI-encoded substrates, we observed that IglC secretion occurs independently of PdpC and PdpD, but requires VgrG and PdpA.
Figure 2. T6SSii Substrates Distribute into Two Phenotypic Classes.
(A–E) Analysis of supernatant and cell fractions from the indicated F. novicida strains. (A) Core function of the T6SSii apparatus as determined by IglC secretion in strains lacking substrate-encoding genes. Sequence duplication within the opiB loci prohibited their individual manipulation, thus ΔopiB corresponds to a deletion of opiB-1-opiB-3. (BE) Western blot analysis of supernatant (Sup) and cell fractions of wild-type F. novicida (C) or strains expressing chromosomally-encoded VSV-G fusions to pdpC (V–pdpC) (B), vgrG (vgrG–V) (D), pdpA (V–pdpA) (E).
In the T6SSi pathway, VgrG proteins serve critical roles in effector export. PdpA is unrelated to T6SSi proteins, but its requirement for IglC secretion suggested that both proteins might contribute to the export of other substrates. To test this prediction, and to further investigate the functional diversity among FPI-encoded T6S substrates, we examined the genetic determinants of their secretion. Our data showed that the export of PdpC and PdpD requires VgrG and PdpA (Figures 2B and 2C), and that the latter two proteins are uniquely co-dependent for secretion (Figures 2D and 2E). From these data, we conclude that the F. novicida T6SSii pathway exports at least two classes of substrates, those that facilitate core function of the apparatus (VgrG and PdpA) and those that serve no apparent function for the apparatus itself (PdpC, PdpD, OpiA, OpiB-1 and OpiB-3). We postulated that the latter represent effector proteins.
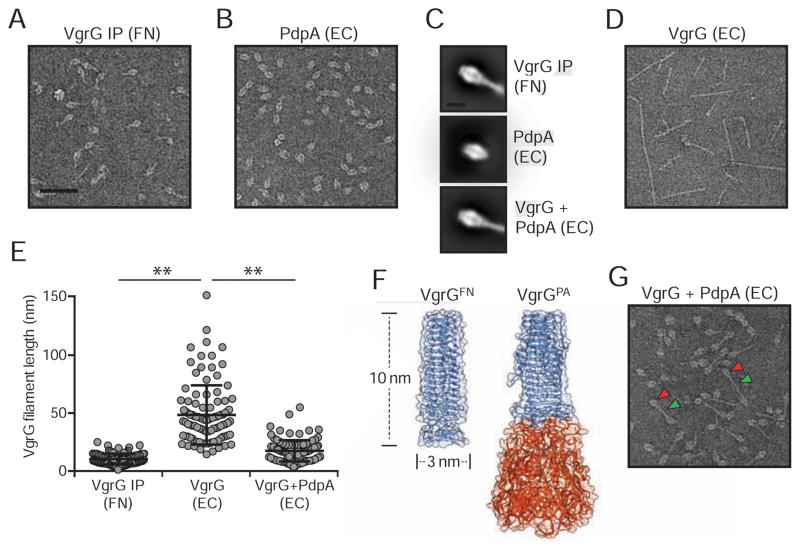
VgrG Forms Filaments and Interacts Directly with PdpA
The mutual requirement of VgrG and PdpA for IglC export suggested that these proteins might participate in a common complex essential for T6SSii function. Indeed, we readily detected PdpA in association with VgrG by co-immunoprecipitation (Figure 3A). F. novicida VgrG is distinct from its relatives associated with the T6SSi pathway. For instance, F. novicida VgrG is predicted to encompass only the gp5-related domain of T6SSi VgrG proteins. In the T6SSi pathway, this region associates with PAAR domains, which serve as adaptors that recruit cognate effector proteins (Shneider et al., 2013). Previous work by our group identified a domain (DUF4280) encoded within the FPI gene iglG that is predicted to adopt the distinctive pyramidal fold of the PAAR domain (Russell et al., 2014b). However, we found that although IglG is required for core T6SSii function, it does not mediate the interaction of VgrG and PdpA (Figures 3B and C). Moreover, IglG precipitated with the uncharacterized FPI-encoded protein IglF but not the VgrG–PdpA complex (Figure 3D). A concurrent study identified an interaction between IglG and IglF using a bacterial two-hybrid approach, suggesting that the proteins associate directly (Rigard et al., 2016).
Figure 3. VgrG Interacts with PdpA Independent of the Predicted PAAR-like Protein IglG.
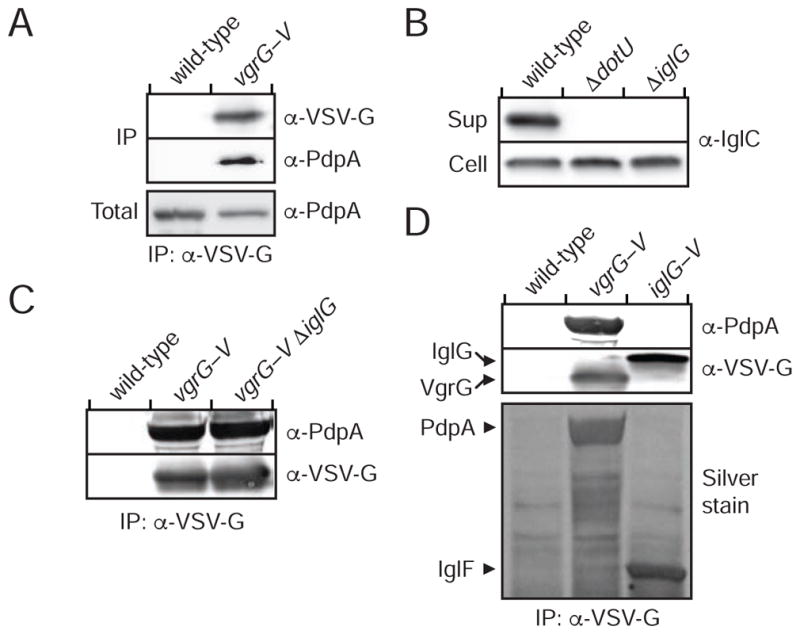
(A, C and D) Western blot analysis of samples derived from the indicated F. novicida strains prior to (Total) or following (IP) anti-VSV-G immunoprecipitation. The band denoted IglF in (D) was identified via mass spectrometry.
(B) anti-IglC western blot analysis of supernatant and cell fractions from the indicated strains.
The unique attributes of F. novicida VgrG motivated us to probe its structure. We began by examining the eluate of VgrG immunoprecipitate from F. novicida by electron microscopy (EM). Particles obtained by this method generally exhibited a distinctive racket-like structure (Figures 4A and S2A). Given our finding that VgrG interacts with PdpA, we speculated that these structures are minimally composed of these two proteins; however, we could not ascertain their molecular organization based solely on these constraints. To determine the placement of PdpA within the structures, we heterologously expressed and purified the protein from E. coli. Inspection of raw images revealed a clear structural resemblance between isolated PdpA and the head region of the racket-like structure (Figures 4B and S2B). Class averages generated from >65,000 particles from each sample provided further support to this apparent structural correlation (Figure 4C).
Figure 4. PdpA Interacts Directly with VgrG and Limits Filament Length.
(A, B, D, G) Negatively stained electron micrographs of (A) VgrG-V immunoprecipitated from F. novicida (FN) or (B) PdpA, (D) VgrG, or (G) VrgG and PdpA heterologously expressed and purified from E. coli (EC). Arrowheads indicate particle regions corresponding PdpA (red) and VgrG (green). Scale bar represents 50 nm. Representative negative stain electron micrographs are presented in Figure S2.
(C) Class averages of PdpA–VgrG-VSV-G complexes immunoprecipitated from F. novicida (top), or PdpA (middle) or PdpA–VgrG (bottom) expressed and purified from E. coli. Scale bar represents 10nm.
(E) Filament lengths of VgrG purified from F. novicida, or E. coli (EC) heterologously expressing VgrG or co-expressing PdpA and VgrG (n=100/group). Horizontal lines represent mean and standard deviation for each group. Asterisks denote statistically significant differences (** p ≤ 0.001).
(F) Structural model of F. novicida VgrG (VgrGFN) adjacent to the crystal structure of VgrG1 from P. aeruginosa (VgrGPA; PDB ID, 4MTK) (Spinola-Amilibia et al., 2016). Regions bearing structural similarity to gp5 and gp27 from T4 bacteriophage are colored blue and orange, respectively.
Our placement of PdpA led us to hypothesize that the remaining, handle portion of the structure is comprised of VgrG. Heterologously expressed and purified F. novicida VgrG forms narrow filaments of width similar to the handle region; however, the lengths of these filaments greatly exceed those observed in particles purified from F. novicida (Figures 4D, 4E and S2C). Although the structure of VgrG has not been determined, it is predicted to resemble the gp5-like α-helix domain of T6SSi VgrG proteins. We used this information to generate a homology model of VgrG, which suggested that the protein could form a trimeric interlaced α-helix with dimensions closely matching those of the racket handle (Figure 4F). If this model is correct, it would imply that the VgrG filaments observed upon over-expression in E. coli are formed from end-to-end stacking of VgrG trimers. Consistent with this, we found that VgrG bearing a short (four amino acid) linker to GFP at its N-terminus formed filaments resembling those of the wild-type protein, but decorated with GFP at their periphery (Figures S3A and S3B). Additionally, VgrG bearing a single non-conservative amino acid substitution at the C-terminus of the protein (G164R) – localized to the predicted contact point between trimers – disrupted polymerization (Figures S3C–F). Analysis of an F. novicida strain expressing VgrG (G164R) from the native vgrG locus indicated that this substitution does not grossly disrupt VgrG stability or folding, as the variant protein accumulates to wild-type levels and maintains interaction with PdpA (Figure S3G).
The handle region of the structures we purified from F. novicida resembles a single predicted VgrG trimer, leading us to question the functional relevance filaments formed upon over-expression in E. coli. To investigate this, we measured T6SS function in F. novicida expressing vgrG (G164R), which is defective in filamentation. Secretion of IglC and PdpA are maintained in this strain, suggesting that large VgrG polymers are not a functionally relevant form of the protein in vivo (Figure S3H).
Together, our data suggest that the racket-shaped particles we isolated from F. novicida contain a single trimer of VgrG capped by PdpA, and furthermore that the association of these proteins blocks formation of VgrG polymers. To interrogate this model, we co-expressed VgrG–His6 with native PdpA and isolated the resulting complexes using Ni2+-affinity chromatography. PdpA co-purified with VgrG, consistent with a direct interaction between these proteins. Analysis of this material by EM revealed a predominant species with morphology closely resembling that found in F. novicida VgrG immunoprecipitate (Figures 4G and S2D). One difference between VgrG–PdpA complexes is that the handle region of those produced in E. coli is on average two-fold longer than those produced in F. novicida (Figure 4E). Thus, upon over-expression in E. coli, the presence of PdpA attenuates, but does not completely abrogate VgrG polymerization. In summary, these data reveal the topology of a unique VgrG-containing complex with components that are required for effector secretion via the T6SSii.
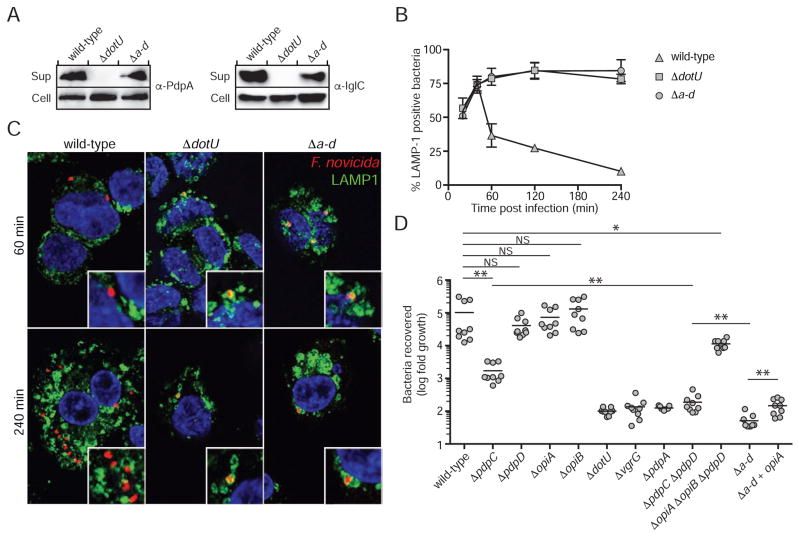
Effectors Encoded Within and Outside of the FPI Contribute to F. novicida Intramacrophage Growth
It is likely that each of the effectors we identified plays a specialized role in the complex intracellular lifestyle of Francisella. However, we hypothesized that if these proteins represent most or all of T6SSii effectors, their cumulative action should approximate that of the FPI apparatus. Testing of this hypothesis requires a means of phenotypically separating the activity of the known effectors from the apparatus they transit. In the T6SSi pathway, this has been difficult to accomplish because effector proteins often contain adaptor domains that are required for apparatus assembly, and thus the secretion of additional effectors (Cianfanelli et al., 2016). Analogous domains are not apparent in the effector proteins we identified, and we found that a strain bearing in-frame deletions in pdpC pdpD opiA and opiB (Δa-d) maintains core apparatus function, as observed by the export of IglC and PdpA (Figure 5A).
Figure 5. T6SSii effectors act in concert to facilitate phagosomal escape and intramacrophage growth.
(A) Core T6SSii function is maintained in a strain lacking all effectors identified in this study. Western blot analysis of secreted structural proteins in the cell and supernatant fractions of the indicated strains.
(B) Phagosomal escape of wild type and mutant F. novicida strains in human macrophage-like THP-1 cells. THP-1 cells were infected as described in Materials & Methods, processed for immunofluorescence staining of bacteria (red), LAMP1 (green) and nuclei (blue) and localization of bacteria in LAMP1-positive phagosomes was scored over a 4h time course. Data represent mean ± stdev of three independent experiments.
(C) Representative confocal micrographs of 60 and 240 min time points of experiment described in (B). Arrows indicate areas magnified in insets. Scale bars, 5 and 1 μm.
(D) Growth of the indicated F. novicida strains in PMA-differentiated THP-1 cells 24 hours post infection. Data from each technical replicate was normalized to the number of intracellular bacteria immediately post infection and data from each of three biological replicates was normalized to ΔdotU. Asterisks denote statistically significant differences cited in the text (* p ≤ 0.05, ** P ≤ 0.001).
While the contribution of T6SSii activity to cytoplasmic replication of Francisella remains controversial, there is general agreement that the activity of the system is critical for phagosomal escape (Celli and Zahrt, 2013). Phagosomes containing F. novicida strains that lack T6SSii activity are not disrupted and eventually fuse with lysosomal compartments, where bacteria within them are rapidly killed. To investigate the contribution of the effectors we identified to the process of phagosomal escape, we compared the co-localization of wild-type, ΔdotU, and Δa-d strains with LAMP1, which marks late endosomal and lysosomal membranes. Our analysis showed that compared to wild-type bacteria, Δa-d and ΔdotU strains are equally impaired in phagosomal escape, suggesting that although Δa-d contains a basally functional T6SSii pathway, the factors required to escape the phagosome are inactivated in this strain (Figure 5B and 5C).
Though our study defines OpiA and OpiB as T6SSii substrates, prior studies have uncovered regulatory links between the corresponding genes and the FPI. MglA is an RNA polymerase-associated protein that positively regulates FPI gene expression (Charity et al., 2007; Lauriano et al., 2004). In a transciptome-wide analysis of F. novicida, Monack and colleagues identified opiA and opiB as additional MglA targets that are coordinately regulated with FPI genes (Brotcke et al., 2006). This is reminiscent of the coordinate regulation of primary T6SSi genes clusters with unlinked genes that encode accessory elements (Hood et al., 2010). In another study, Wehrly et al. found that similar to genes within the FPI, opiA expression is activated during infection of bone marrow-derived macrophages (Wehrly et al., 2009). Despite the potential functional implications for OpiA and OpiB garnered from these studies, the authors found that neither are required to support intramacrophage growth of F. novicida. We tested F. novicida ΔopiA and ΔopiB strains for growth in human macrophage-like (THP-1) and lung epithelial-like (A549) cells, and obtained similar findings (Figures 5D and S4A). This was not specific to F. novicida; deletion of opiA and opiB orthologs from F. tularensis subsp. holarctica LVS also did not measurably impact intracellular growth (Figure S4B). The intracellular growth characteristics of Francisella strains lacking PdpC and PdpD have also been reported (Chou et al., 2013). Consistent with these earlier findings, we found that ΔpdpC is attenuated in intramacrophage growth, whereas ΔpdpD proliferation is equivalent to wild-type (Figure 5D).
It is not uncommon for bacterial effectors to serve redundant functions (Galan, 2009; Isaac and Isberg, 2014; van Schaik et al., 2013). Taking this into account, we generated an F. novicida strain lacking the three effectors that did not affect intracellular growth when inactivated individually (ΔopiA ΔopiB ΔpdpD). Surprisingly, this strain showed only a small attenuation in intracellular growth (p = 0.04) (Figure 5D). This result suggested that either these proteins only minimally contribute to F. novicida intracellular growth or that under the conditions of our assay, their role is masked by PdpC. To distinguish these possibilities, we measured the intracellular growth of Δa-d. The degree of attenuation of the resulting strain exceeded that of ΔpdpC. Indeed, we found that this strain is less able to proliferate intracellularly than is ΔdotU or strains lacking either of the other secreted structural proteins we identified (Figures 5D and S4A). These data indicate that effectors identified in this study – beyond PdpC – contribute to intracellular growth. Furthermore, our results suggest that in the absence of a functional secretion system, intracellular lysis of F. novicida may contribute to effector release.
To begin to assess the contribution of individual effectors, we compared the intracellular growth phenotype of ΔpdpC ΔpdpD to that of ΔpdpC and Δa-d. Our results showed that PdpD enhances intracellular growth in the absence of PdpC, and additionally suggested that the Opi proteins also play a significant role in this background (Figure 5D). Restoration of opiA in F. novicida Δa-d returned growth to the level of ΔpdpC ΔpdpD, while our attempts to restore intracellular growth through opiB complementation were unsuccessful. These data demonstrate that effectors encoded within and outside of the FPI contribute to the intracellular growth of F. novicida.
F. novicida Effectors are Conserved Amongst Francisella spp. Pathogenic to Mammals
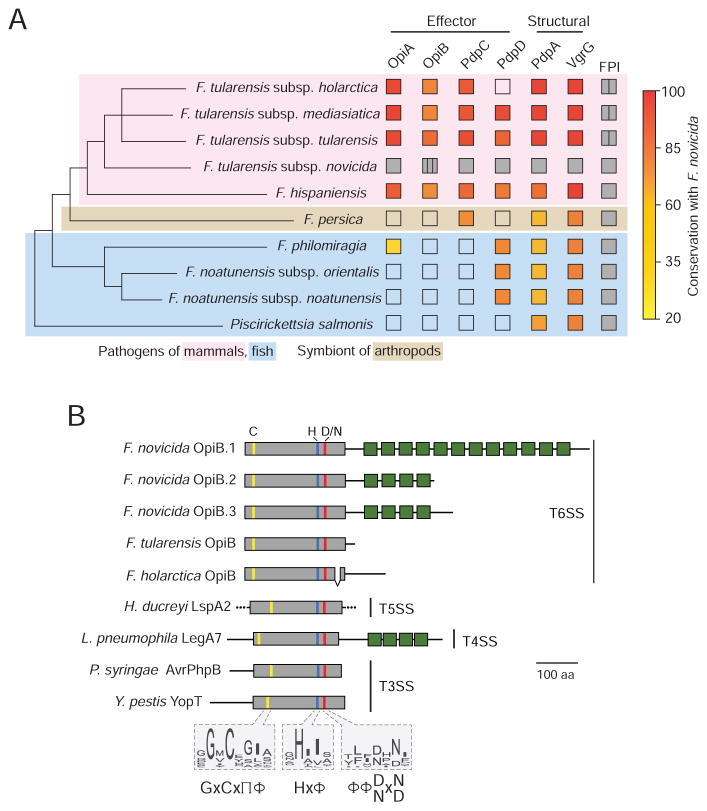
The Francisella are host-associated organisms thought to derive from a common ancestor that occupied marine habitats. Phylogenetic analyses conducted by Sjodin et al. suggest that one clade of Francisella remain in this habitat as fish pathogens, and that a second clade changed their host range to terrestrial organisms including mammals and arthropods (Sjodin et al., 2012). We found that genes located both inside and outside of the FPI encode F. novicida effector proteins. As a first step toward understanding the broader role that these proteins might play in pathogenesis, we examined their distribution among Francisella and closely related organisms that possess the FPI. Interestingly, we found that with the exception of a distant opiA homolog found in F. philomiragia, opiA and opiB are restricted to Francisella spp. associated with pathogenesis in mammals (Figure 6A). PdpC displays a similar distribution, whereas the distribution of pdpD is sporadic.
Figure 6. OpiA and OpiB are Virulence-associated Proteins.
(A) Phylogenetic tree of Francisella and closely related organisms that possess the FPI (see methods) indicating the presence (filled) or absence (open) of secreted F. novicida T6SSii effectors and structural proteins identified in this study. If present, the degree of conservation with the F. novicida ortholog is indicated (fill color). The tree is adapted from Sjodin et al.; branch lengths do not represent evolutionary distance (Sjodin et al., 2012). Vertical lines within boxes indicate multiple copies of a given element and background colors denote host-specificities as indicated.
(B) Overview of OpiB domain organization and conservation of its predicted N-terminal protease domain among diverse effector proteins. All proteins are shown to scale. The location of predicted or experimentally determined catalytic residues are indicated by colored lines, and box fill denotes protease domain (grey) and ankyrin motifs (green). Conserved catalytic motifs based on the alignment of a broader group of representative orthologs are shown as sequence logos. Amino acid composition at each position is shown according to (Aasland et al., 2002). The experimentally determined or predicted export pathway of each protein is provided at right. Proteins shown are: OpiB-1 A0Q6U1, OpiB-2 A0Q6U2, OpiB-3 A0Q6U3, OpiBFT Q5NH59, OpiBLVS Q2A3V2, LspA2 Q9ZHL3, LegA7 Q5ZYG9, AvrPhpB Q9F3T4, YopT O68703.
In contrast, PdpA and VgrG, the secreted structural components we identified in this study, are conserved within each of the bacteria we analyzed. These findings suggest that bacteria with the FPI utilize a variable complement of effectors, and furthermore that these effectors could reflect, or even underlie, host tropism. For instance, the acquisition of opiA, opiB and pdpC may have been an important innovation en route to pathogenesis in mammals.
OpiB-family Effectors are Exported by Diverse Secretion Systems
Homologs of OpiA were not found outside of Francisella and our analyses failed to identify characterized domains or motifs within the protein. Conversely, we readily detected homology between ankryin repeat domains and the C-terminus of F. novicida OpiB proteins (Figure 6B). Ankyrin repeats mediate protein–protein interactions and are most often found in eukaryotic proteins. When found in bacteria, proteins with these motifs are strongly associated with the translocated effectors of intracellular pathogens and symbionts (Al-Khodor et al., 2010). The number of ankyrin repeats varies among F. novicida paralogs and they are degenerate or absent from orthologs in other F. tularenesis subspecies. In contrast, the N-terminus of OpiB is conserved among Francisella orthologs (61% identity) and displays no overall sequence similarity to characterized proteins. We thus turned to hidden markov model-based iterative search algorithms to define more distant homologs of the Francisella OpiB proteins. This method revealed motifs and secondary structural features within OpiB consistent with cysteine proteases in the CA clan (Barrett and Rawlings, 2001). Within the CA clan, we found that OpiB belongs to a distant and previously unrecognized branch of the C58 family, defined by Shao et al., that comprises known type III secretion effectors from Yersinia and Pseudomonas spp. (Figure S5) (Shao et al., 2002). Remarkably, our analyses showed that members of the OpiB branch include type IV secretion (T4S)-translocated effectors of unknown function in Legionella (e.g. LegA7) and type V (two partner)-exported virulence factors of Haemophilus (Habyarimana et al., 2010; Huang et al., 2011; Janowicz et al., 2004) (Figure 6B). LegA7 also contains C-terminal ankryin repeat motifs, which lend an overall topology similar to F. novicida OpiB proteins. These findings suggest that the N-terminal domain of OpiB constitutes an evolutionarily plastic cysteine protease and they point to previously unrecognized similarities between effectors of the T4 and T6 secretion systems.
Discussion
Our findings provide biochemical and genetic evidence supporting the assignment of OpiA, OpiB, PdpC and PdpD as secreted effectors of the Francisella T6SSii pathway. While we cannot rule out the possibility that VgrG and PdpA also act as effectors, our data reveal that evaluating the direct contribution of these proteins to host interactions will be confounded by their requirement for the secretion of other substrates. A study measuring the translocation of TEM-1 reporter fusions by F. tularensis LVS suggested that several additional FPI-encoded proteins are T6SSii substrates, including IglE, IglF, IglI, IglJ, and PdpE (Broms et al., 2012). It is possible that these proteins are bona fide T6SSii substrates, but escaped detection in our study. This could be due to low abundance, differences in T6SSii activity in vitro versus in vivo, or our use of F. novicida rather than F. tularensis LVS. The latter is unlikely given the high level of sequence conservation and gene content between the FPIs of these strains. We speculate that using reporter fusions for identifying secreted proteins yields false-positive results, potentially explaining the assignment of at least a subset of these purported substrates. Indeed, Barker et al. detected translocation of VgrG–CyaA into macrophage cytoplasm from a F. novicida strain lacking the FPI (ΔpdpA-pdpD) (Barker et al., 2009). A subsequent study, and our findings herein, clearly demonstrate that VgrG export is dependent on multiple FPI genes (Clemens et al., 2015).
Many questions remain regarding the mechanism by which the F. novicida T6SSii translocates effectors. In the T6SSi pathway, VgrG and Hcp proteins play critical roles in effector recognition and translocation. VgrG proteins use adaptor domains such as PAAR to recognize specific cognate effectors, whereas Hcp-family proteins can directly interact with multiple effector proteins (Shneider et al., 2013; Silverman et al., 2013). There is not a prototypical PAAR domain protein encoded by the FPI and we uncovered an interaction between F. novicida VgrG and the secreted protein PdpA. How this protein complex mediates effector export is not known. If export is mediated by transient or exclusively intracellular interactions with the VgrG–PdpA complex, these may have escaped detection in the co-immunoprecipitation assays we employed. Hcp proteins of the T6SSi pathway bind effectors within the lumen of their hexameric ring-shaped structure (Silverman et al., 2013). It is notable that while IglC adopts a fold similar to the Hcp monomer, it has not been shown to form higher order structures (de Bruin et al., 2011). Like its bacteriophage equivalent lambda gpV, IglC may adopt a hexameric structure only in the context of an assembled tube, or this protein may function in a manner unique to the T6SSii pathway (Pell et al., 2009). Whether or not Hcp oligomerizes, the small volume within its interior may preclude the relatively large T6SSii effectors (approximately 50–150 kDa) we identified from this mode of recognition and transport. Indeed, Hcp-associated effectors of the T6SSi pathway are without known exception less than 50 kDa, whereas VgrG-associated effectors of this pathway, such as the Rhs family of proteins, can exceed 200 kDa (Koskiniemi et al., 2013; Whitney et al., 2014).
We discovered that one of the effectors secreted by the F. novicida T6SSii, OpiB, is related to group of effectors translocated by the specialized Dot/Icm T4SS of Legionella. In light of this finding, it is noteworthy that these systems share two key structural subunits, IcmF (PdpB) and DotU and that amongst the three classes of T6SSs, the FPI-encoded pathway shares the fewest components (Russell et al., 2014b). Recognizing that T6SSii is an amalgam of the T4 and T6 secretion pathways has mechanistic implications and could help direct future investigations of this unique virulence factor. Furthermore, the close phylogenetic relatedness of Francisella and Legionella raises the possibility that an ancestral system containing IcmF and DotU facilitated evolution of an intracellular lifestyle and then became specialized within the two lineages through acquisition of T6 or T4 components, respectively.
The literature offers little insight into the molecular mechanism by which the effectors we identified contribute to Francisella pathogenesis. Only one, OpiB, contains conserved domains detected by our bioinformatic analyses. The N-terminus of the protein consists of a putative cysteine protease domain that is related to the YopT (C58) family of proteins defined by Shao and Dixon (Shao et al., 2002). While this distant similarity is intriguing, it does not specify the target of OpiB, since substrate diversity is observed even between closely related members of the family (Zhu et al., 2004). Ankyrin domains, localized to the C-terminus of OpiB, mediate protein–protein interaction and, similarly, have diverse targets. For example, the ankyrin repeats of L. pneumophila AnkB target the protein to membranes, while those of AnkX are involved in disrupting microtubule-dependent vesicle trafficking (Ge and Shao, 2011; Pan et al., 2008; Price et al., 2010).
F. novicida must actively commandeer the host in order to avoid cellular defenses. A wealth of data indicate that T6SSii is required for endosomal escape of Francisella; however, there are conflicting data pertaining to its requirement for establishing a replicative cytoplasmic niche (Meyer et al., 2015; Wu et al., 2015). We found that a strain lacking each of the five effectors identified in our secretome analysis retains core apparatus function, but displays phagosomal escape and intracellular growth defects equal to or greater than strains containing an inactive T6SSii apparatus. This implies that F. novicida may require a relatively small number of proteins to facilitate its intracellular lifestyle. By comparison, L. pneumophila uses hundreds of effector proteins, albeit these often serve redundant functions (Isaac and Isberg, 2014). We cannot rule out that the T6SSii of Francisella secretes additional effectors that were undetected because they fell below the detection limit, nor can we exclude the possibility that proteins released by alternative secretion machineries directly mediate host interactions for this organism. In either case, defining the mechanism by which the effectors we discovered act on the host stands to significantly advance our understanding of Francisella pathogenesis.
Experimental procedures
Strains and Culture Conditions
Francisella strains used in this study are listed in Table S2. Conditions for maintaining mammalian cell lines and bacterial strains can be found in Supplemental Experimental Procedures.
Secretion Assays
Overnight cultures of F. novicida were washed, diluted to an optical density at 600 nm (OD600) of 0.04 and grown in TSBC supplemented with 5% potassium chloride (TSBC+KCl) until they reached OD600 1.4. The sequence identity between the opiB genes prohibited the construction of chromosomally-encoded epitope fusions to opiB-2 and opiB-3; therefore, to assess secretion of these proteins we expressed OpiB-2 and OpiB-3 fused to VSV-G on pF5 in the presence of 5 μg/mL of kanamycin. The cell and supernatant fractions were then collected and processed as previously described (Hood et al., 2010).
Preparation of Extracellular Proteome
After growth in TSBC+KCl, the cells were separated from secreted proteins by centrifugation and filtration. Protease inhibitors (1 mM AEBSF, 10 μM leupeptin, 1 μM pepstatin) were added to the filtered supernatants before dialysis with a 10 kDa molecular weight cut off against phosphate buffered saline (PBS) containing 1 mM MgCl2 and 1 mM CaCl2 at 4°C. The retained proteins were precipitated, resuspended in 100 μL of 6 M urea and 50 mM NH4HCO3, reduced and alkylated with dithiotreitol and iodoactamide, respectively, and digested with trypsin (50:1 protein:trypsin ratio). The resulting peptides were desalted with C18 spin columns (Pierce) following the manufacturer’s protocol and mass spectrometry was performed as described in Supplemental Experimental Procedures.
Negative Staining Electron Microscopy
Protein samples purified from E. coli were diluted to a concentration of 0.04 mg/mL and then adsorbed to glow-discharged carbon-coated copper mesh grids. Proteins isolated from F. novicida were purified as described in the immunoprecipitation section above, eluted with 100 μg/mL VSVG peptide and directly adsorbed without further dilution. Samples were incubated on the grids for 30 seconds prior to staining with 2% uranyl formate. Micrographs were acquired using the Leginon software (Suloway et al., 2005) on a 100kV FEI Tecnai G2 Spirit with a Gatan Ultrascan 4000 4k × 4k CCD camera at 52,000 nominal magnification (pixel size of 2.07 Å at the specimen level). The defocus ranged from 1.0 to 2.0 μm. The details of image analysis can be found in Supplemental Experimental Procedures.
Modeling of the F. novicida VgrG Protein
The amino acid sequence for F. novicida VgrG was submitted to the protein homology/analogy recognition engine (Phyre2) server for structure prediction (Kelley et al., 2015). The output model from Phyre was comprised of a single VgrG polypeptide that was built using the gp5-like portion of P. aeruginosa VgrG1 (PDB ID, 4MTK) as a template. To generate the trimeric arrangement found in the template structure, three copies of F. novicida VgrG were aligned to each of the subunits of VgrG1 using the secondary-structure matching alignment tool in COOT (Emsley et al., 2010). The resulting model was used for all structure figures, which were generated using UCSF Chimera (Pettersen et al., 2004).
Infection Assays
Infection assays were performed similar to previous reports with some modifications outlined in Supplemental Experimental Procedures (Clemens et al., 2015).
Immunofluorescence of F. novicida Infection
At each time point after infection, THP-1 cells were gently washed three times with PBS and fixed for 10 min in 3% paraformaldehyde in PBS at 37°C. Immunofluorescence staining was carried out as described previously using mouse anti-human LAMP1 (clone H4A3) and goat anti-Francisella (BEI Resources) primary antibodies, followed by Alexa Fluor™ 488-conjugated donkey anti-mouse and Alexa Fluor™ 568-conjugated donkey anti-goat (1:500, Invitrogen) secondary antibodies (Starr et al., 2008). Further details can be found in Supplemental Experimental Procedures.
Supplementary Material
Highlights.
Proteome-wide analysis identifies diverse Francisella T6SS substrates
T6SS effectors are encoded within and outside the Francisella pathogenicity island (FPI)
FPI-encoded substrates VgrG and PdpA facilitate core function of the T6SS apparatus
Effectors encoded within and outside the FPI contribute to intramacrophage growth
Acknowledgments
The authors would like to thank Tamir Gonen and Hongjin Zheng for preliminary EM analyses, Dan Stetson and Brad Cookson for sharing reagents, Josh Woodward for critical review of the manuscript, and members of the Mougous Lab for helpful discussions. J.C.W. was supported by a postdoctoral research fellowship from CIHR, training grants from the NIH supported A.E. and H.E.L. (AI055396), A.C.W. (GM008268), and K.M.R. (HD055148) and J.D.M. holds an Investigator in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund. The work was supported by The University of Maryland Baltimore School of Pharmacy Mass Spectrometry Center (SOP1841-IQB2014 to Y.A.G. and D.R.G.) and National Institutes of Health grants AI081693 (to S.L.D.) and AI080609 (to J.D.M.).
Footnotes
Author contributions
Conceptualization, A.E. and J.D.M.; Investigation, A.E., J.K., A.C.W., H.E.L., C.M., K.M.R., J.C.W., M.C.R., B.R.R., B.Q.T., Y.A.G., D.V. and J.D.M.; Writing-original draft, A.E., A.C.W., H.E.L., M.C.R., S.B.P., Y.A.G., D.V. and J.D.M.; Writing-reviewing and editing, A.E., J.K., A.C.W., H.E.L., C.M., K.M.R., J.C.W., M.C.R., S.B.P., Y.A.G., D.R.G., S.L.D., J.C., D.V. and J.D.M.; Project administration, J.D.M.; Funding acquisition, D.R.G., S.L.D., J.C., D.V. and J.D.M.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aasland R, Abrams C, Ampe C, Ball LJ, Bedford MT, Cesareni G, Gimona M, Hurley JH, Jarchau T, Lehto VP, et al. Normalization of nomenclature for peptide motifs as ligands of modular protein domains. FEBS letters. 2002;513:141–144. doi: 10.1016/s0014-5793(01)03295-1. [DOI] [PubMed] [Google Scholar]
- Al-Khodor S, Price CT, Kalia A, Abu Kwaik Y. Functional diversity of ankyrin repeats in microbial proteins. Trends Microbiol. 2010;18:132–139. doi: 10.1016/j.tim.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker JR, Chong A, Wehrly TD, Yu JJ, Rodriguez SA, Liu J, Celli J, Arulanandam BP, Klose KE. The Francisella tularensis pathogenicity island encodes a secretion system that is required for phagosome escape and virulence. Mol Microbiol. 2009;74:1459–1470. doi: 10.1111/j.1365-2958.2009.06947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett AJ, Rawlings ND. Evolutionary lines of cysteine peptidases. Biol Chem. 2001;382:727–733. doi: 10.1515/BC.2001.088. [DOI] [PubMed] [Google Scholar]
- Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature. 2012;483:182–186. doi: 10.1038/nature10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broms JE, Meyer L, Sun K, Lavander M, Sjostedt A. Unique substrates secreted by the type VI secretion system of Francisella tularensis during intramacrophage infection. PLoS One. 2012;7:e50473. doi: 10.1371/journal.pone.0050473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brotcke A, Weiss DS, Kim CC, Chain P, Malfatti S, Garcia E, Monack DM. Identification of MglA-regulated genes reveals novel virulence factors in Francisella tularensis. Infection and immunity. 2006;74:6642–6655. doi: 10.1128/IAI.01250-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celli J, Zahrt TC. Mechanisms of Francisella tularensis intracellular pathogenesis. Cold Spring Harb Perspect Med. 2013;3:a010314. doi: 10.1101/cshperspect.a010314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charity JC, Costante-Hamm MM, Balon EL, Boyd DH, Rubin EJ, Dove SL. Twin RNA polymerase-associated proteins control virulence gene expression in Francisella tularensis. PLoS Pathog. 2007;3:e84. doi: 10.1371/journal.ppat.0030084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou AY, Kennett NJ, Nix EB, Schmerk CL, Nano FE, Elkins KL. Generation of protection against Francisella novicida in mice depends on the pathogenicity protein PdpA, but not PdpC or PdpD. Microbes Infect. 2013;15:816–827. doi: 10.1016/j.micinf.2013.07.005. [DOI] [PubMed] [Google Scholar]
- Cianfanelli FR, Monlezun L, Coulthurst SJ. Aim, Load, Fire: The Type VI Secretion System, a Bacterial Nanoweapon. Trends Microbiol. 2016;24:51–62. doi: 10.1016/j.tim.2015.10.005. [DOI] [PubMed] [Google Scholar]
- Clemens DL, Ge P, Lee BY, Horwitz MA, Zhou ZH. Atomic structure of T6SS reveals interlaced array essential to function. Cell. 2015;160:940–951. doi: 10.1016/j.cell.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin OM, Duplantis BN, Ludu JS, Hare RF, Nix EB, Schmerk CL, Robb CS, Boraston AB, Hueffer K, Nano FE. The biochemical properties of the Francisella pathogenicity island (FPI)-encoded proteins IglA, IglB, IglC, PdpB and DotU suggest roles in type VI secretion. Microbiology (Reading, England) 2011;157:3483–3491. doi: 10.1099/mic.0.052308-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand E, Nguyen VS, Zoued A, Logger L, Pehau-Arnaudet G, Aschtgen MS, Spinelli S, Desmyter A, Bardiaux B, Dujeancourt A, et al. Biogenesis and structure of a type VI secretion membrane core complex. Nature. 2015;523:555–560. doi: 10.1038/nature14667. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan JE. Common themes in the design and function of bacterial effectors. Cell host & microbe. 2009;5:571–579. doi: 10.1016/j.chom.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge J, Shao F. Manipulation of host vesicular trafficking and innate immune defence by Legionella Dot/Icm effectors. Cellular microbiology. 2011;13:1870–1880. doi: 10.1111/j.1462-5822.2011.01710.x. [DOI] [PubMed] [Google Scholar]
- Habyarimana F, Price CT, Santic M, Al-Khodor S, Kwaik YA. Molecular characterization of the Dot/Icm-translocated AnkH and AnkJ eukaryotic-like effectors of Legionella pneumophila. Infection and immunity. 2010;78:1123–1134. doi: 10.1128/IAI.00913-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood RD, Singh P, Hsu F, Guvener T, Carl MA, Trinidad RR, Silverman JM, Ohlson BB, Hicks KG, Plemel RL, et al. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell host & microbe. 2010;7:25–37. doi: 10.1016/j.chom.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Boyd D, Amyot WM, Hempstead AD, Luo ZQ, O’Connor TJ, Chen C, Machner M, Montminy T, Isberg RR. The E Block motif is associated with Legionella pneumophila translocated substrates. Cellular microbiology. 2011;13:227–245. doi: 10.1111/j.1462-5822.2010.01531.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaac DT, Isberg R. Master manipulators: an update on Legionella pneumophila Icm/Dot translocated substrates and their host targets. Future microbiology. 2014;9:343–359. doi: 10.2217/fmb.13.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janowicz DM, Fortney KR, Katz BP, Latimer JL, Deng K, Hansen EJ, Spinola SM. Expression of the LspA1 and LspA2 proteins by Haemophilus ducreyi is required for virulence in human volunteers. Infection and immunity. 2004;72:4528–4533. doi: 10.1128/IAI.72.8.4528-4533.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kast P. pKSS--a second-generation general purpose cloning vector for efficient positive selection of recombinant clones. Gene. 1994;138:109–114. doi: 10.1016/0378-1119(94)90790-0. [DOI] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nature protocols. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingry LC, Petersen JM. Comparative review of Francisella tularensis and Francisella novicida. Front Cell Infect Microbiol. 2014;4:35. doi: 10.3389/fcimb.2014.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskiniemi S, Lamoureux JG, Nikolakakis KC, t’Kint de Roodenbeke C, Kaplan MD, Low DA, Hayes CS. Rhs proteins from diverse bacteria mediate intercellular competition. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:7032–7037. doi: 10.1073/pnas.1300627110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauriano CM, Barker JR, Yoon SS, Nano FE, Arulanandam BP, Hassett DJ, Klose KE. MglA regulates transcription of virulence factors necessary for Francisella tularensis intraamoebae and intramacrophage survival. Proc Natl Acad Sci U S A. 2004;101:4246–4249. doi: 10.1073/pnas.0307690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer L, Broms JE, Liu X, Rottenberg ME, Sjostedt A. Microinjection of Francisella tularensis and Listeria monocytogenes reveals the importance of bacterial and host factors for successful replication. Infection and immunity. 2015;83:3233–3242. doi: 10.1128/IAI.00416-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nano FE, Schmerk C. The Francisella pathogenicity island. Ann N Y Acad Sci. 2007;1105:122–137. doi: 10.1196/annals.1409.000. [DOI] [PubMed] [Google Scholar]
- Pan X, Luhrmann A, Satoh A, Laskowski-Arce MA, Roy CR. Ankyrin repeat proteins comprise a diverse family of bacterial type IV effectors. Science. 2008;320:1651–1654. doi: 10.1126/science.1158160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pell LG, Kanelis V, Donaldson LW, Howell PL, Davidson AR. The phage lambda major tail protein structure reveals a common evolution for long-tailed phages and the type VI bacterial secretion system. Proc Natl Acad Sci U S A. 2009;106:4160–4165. doi: 10.1073/pnas.0900044106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Price CT, Al-Khodor S, Al-Quadan T, Abu Kwaik Y. Indispensable role for the eukaryotic-like ankyrin domains of the ankyrin B effector of Legionella pneumophila within macrophages and amoebae. Infection and immunity. 2010;78:2079–2088. doi: 10.1128/IAI.01450-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigard M, Broms JE, Mosnier A, Hologne M, Martin A, Lindgren L, Punginelli C, Lays C, Walker O, Charbit A, et al. Francisella tularensis IglG Belongs to a Novel Family of PAAR-Like T6SS Proteins and Harbors a Unique N-terminal Extension Required for Virulence. PLoS Pathog. 2016;12:e1005821. doi: 10.1371/journal.ppat.1005821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell AB, Peterson SB, Mougous JD. Type VI secretion system effectors: poisons with a purpose. Nature reviews Microbiology. 2014a;12:137–148. doi: 10.1038/nrmicro3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell AB, Wexler AG, Harding BN, Whitney JC, Bohn AJ, Goo YA, Tran BQ, Barry NA, Zheng H, Peterson SB, et al. A type VI secretion-related pathway in Bacteroidetes mediates interbacterial antagonism. Cell host & microbe. 2014b;16:227–236. doi: 10.1016/j.chom.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz S, Hood RD, Mougous JD. What is type VI secretion doing in all those bugs? Trends Microbiol. 2010;18:531–537. doi: 10.1016/j.tim.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao F, Merritt PM, Bao Z, Innes RW, Dixon JE. A Yersinia effector and a Pseudomonas avirulence protein define a family of cysteine proteases functioning in bacterial pathogenesis. Cell. 2002;109:575–588. doi: 10.1016/s0092-8674(02)00766-3. [DOI] [PubMed] [Google Scholar]
- Shneider MM, Buth SA, Ho BT, Basler M, Mekalanos JJ, Leiman PG. PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature. 2013;500:350–353. doi: 10.1038/nature12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman JM, Agnello DM, Zheng H, Andrews BT, Li M, Catalano CE, Gonen T, Mougous JD. Haemolysin Coregulated Protein Is an Exported Receptor and Chaperone of Type VI Secretion Substrates. Molecular cell. 2013;51:584–593. doi: 10.1016/j.molcel.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman JM, Brunet YR, Cascales E, Mougous JD. Structure and Regulation of the Type VI Secretion System. Annual review of microbiology. 2012;66:453–473. doi: 10.1146/annurev-micro-121809-151619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjodin A, Svensson K, Ohrman C, Ahlinder J, Lindgren P, Duodu S, Johansson A, Colquhoun DJ, Larsson P, Forsman M. Genome characterisation of the genus Francisella reveals insight into similar evolutionary paths in pathogens of mammals and fish. BMC genomics. 2012;13:268. doi: 10.1186/1471-2164-13-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinola-Amilibia M, Davo-Siguero I, Ruiz FM, Santillana E, Medrano FJ, Romero A. The structure of VgrG1 from Pseudomonas aeruginosa, the needle tip of the bacterial type VI secretion system. Acta Crystallogr D Struct Biol. 2016;72:22–33. doi: 10.1107/S2059798315021142. [DOI] [PubMed] [Google Scholar]
- Starr T, Ng TW, Wehrly TD, Knodler LA, Celli J. Brucella intracellular replication requires trafficking through the late endosomal/lysosomal compartment. Traffic (Copenhagen, Denmark) 2008;9:678–694. doi: 10.1111/j.1600-0854.2008.00718.x. [DOI] [PubMed] [Google Scholar]
- Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. Automated molecular microscopy: the new Leginon system. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- van Schaik EJ, Chen C, Mertens K, Weber MM, Samuel JE. Molecular pathogenesis of the obligate intracellular bacterium Coxiella burnetii. Nat Rev Microbiol. 2013;11:561–573. doi: 10.1038/nrmicro3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrly TD, Chong A, Virtaneva K, Sturdevant DE, Child R, Edwards JA, Brouwer D, Nair V, Fischer ER, Wicke L, et al. Intracellular biology and virulence determinants of Francisella tularensis revealed by transcriptional profiling inside macrophages. Cellular microbiology. 2009;11:1128–1150. doi: 10.1111/j.1462-5822.2009.01316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney JC, Beck CM, Goo YA, Russell AB, Harding BN, De Leon JA, Cunningham DA, Tran BQ, Low DA, Goodlett DR, et al. Genetically distinct pathways guide effector export through the type VI secretion system. Molecular microbiology. 2014;92:529–542. doi: 10.1111/mmi.12571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney JC, Quentin D, Sawai S, LeRoux M, Harding BN, Ledvina HE, Tran BQ, Robinson H, Goo YA, Goodlett DR, et al. An interbacterial NAD(P)(+) glycohydrolase toxin requires elongation factor Tu for delivery to target cells. Cell. 2015;163:607–619. doi: 10.1016/j.cell.2015.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YC, Wu TH, Clemens DL, Lee BY, Wen X, Horwitz MA, Teitell MA, Chiou PY. Massively parallel delivery of large cargo into mammalian cells with light pulses. Nature methods. 2015;12:439–444. doi: 10.1038/nmeth.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M, Shao F, Innes RW, Dixon JE, Xu Z. The crystal structure of Pseudomonas avirulence protein AvrPphB: a papain-like fold with a distinct substrate-binding site. Proc Natl Acad Sci U S A. 2004;101:302–307. doi: 10.1073/pnas.2036536100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.