Abstract
The platelet-derived growth factor receptor alpha (PDGFRα) exhibits divergent effects in skeletal muscle. At physiological levels, signaling through this receptor promotes muscle development in growing embryos and proper angiogenesis in regenerating adult muscle.1,2 However, both increased PDGF ligand abundance and enhanced PDGFRα pathway activity cause pathological fibrosis.3,4 This excessive collagen deposition, which is seen in aged and diseased muscle,5–7 interferes with proper muscle function and limits the effectiveness of gene- and cell-based therapies for muscle disorders.8,9 Although compelling evidence exists for the role of PDGFRα in fibrosis, little is known about the cells through which this pathway acts. Here we show that PDGFRα signaling regulates a population of muscle-resident fibro/adipogenic progenitors (FAPs) that play a supportive role in muscle regeneration but may also cause fibrosis when aberrantly regulated.10–13 We found that FAPs produce multiple transcriptional variants of PDGFRα with different polyadenylation sites, including an intronic variant that codes for a protein isoform containing a truncated kinase domain. This variant, upregulated during regeneration, acts as a decoy to inhibit PDGF signaling and to prevent FAP over-activation. Moreover, increasing expression of this isoform limits fibrosis in vivo, suggesting both biological relevance and therapeutic potential of modulating polyadenylation patterns in stem cell populations.
Although aberrant PDGFRα signaling is linked to muscle pathology,4,14–17 we sought to test how PDGFRα signaling is regulated during healthy muscle regeneration. PDGFRα is largely unphosphorylated, and hence inactive, in resting muscle but is activated upon injury with levels peaking after five days (Extended Data Fig. 1a and 1b; for gel source data, see Supplementary Fig. 1). Indeed, injured muscle shows a dramatic increase of phospho-PDGFRα positive cells (Extended Data Fig. 1c). Using a mouse containing an H2B-eGFP fusion protein knocked into the PDGFRα locus,2 we found a population of PDGFRα-expressing cells located in interstitial regions (Extended Data Fig. 1d). Fluorescence-activated cell sorting (FACS) analysis revealed that PDGFRα is expressed strongly in over 99% of FAPs and that nearly all PDGFRα-expressing cells are in this population in both uninjured (Extended Data Fig. 1e–1g) as well as and injured muscle (Extended Data Fig. 1h–1m).
To understand how PDGFRα signaling is regulated in FAPs, we performed direct RNA sequencing (DRS) both because its single-molecule approach allows for straightforward quantification and also because it involves sequencing at the polyadenylated tail of RNA, affording the opportunity to evaluate post-transcriptional regulation via polyadenylation.18 A previous study from our lab demonstrated that alternative polyadenylation at the 3′ untranslated region (3′ UTR) of transcripts plays a critical role in modulating stem cell behavior.19
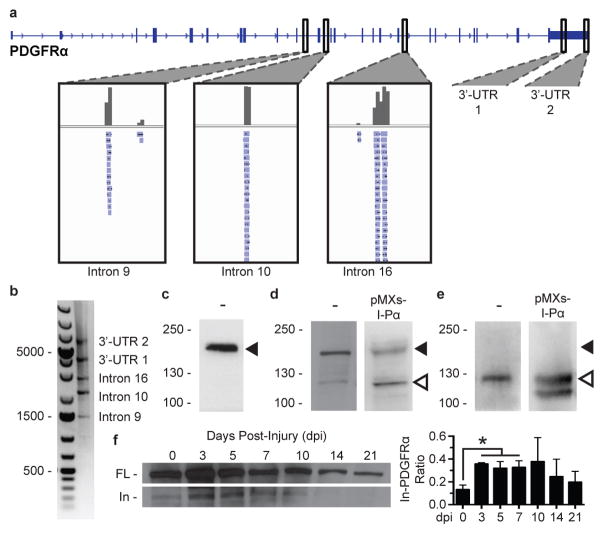
We found that FAPs produce not only transcriptional variants of PDGFRα that harbor different polyadenylation sites within the 3′ UTR but also multiple transcripts that are polyadenylated at introns within the protein coding region (Figure 1a). We confirmed polyadenylation at these sites through 3′ RACE (Figure 1b and Extended Data Fig. 2a). Particularly intriguing was a transcriptional variant (which we term “In-PDGFRα”) that terminated within intron 16 of the PDGFRα. This transcript, the most abundant of the intronic variants, contained the canonical AAUAAA polyadenylation termination motif preceded by an in-frame stop codon sequence, suggesting a protein-coding potential. Such a protein would consist of membrane-bound isoform of PDGFRα containing an intact ligand-binding domain but a truncated kinase domain (Extended Data Fig. 2b and 2c). This type of structural modification was of particular interest as previous studies in cell lines have suggested that protein truncations produced through intronic polyadenylation can significantly impact functionality of the full-length counterpart.20 For example, artificial induction of intronic polyadenylation of tyrosine kinase receptors, such that both the kinase and transmembrane regions were truncated, caused cells to produce soluble decoy receptors that bound ligand or functional receptors, suppressing signaling.21
Figure 1. PDGFRα undergoes intronic polyadenylation in FAPs, resulting in a truncated protein that increases in abundance during regeneration.
(a) Direct RNA sequencing reveals multiple polyadenylation sites for PDGFRα, including two sites within the 3′ UTR and three sites within introns. (b) 3′ RACE, in which the gene-specific primer overlaps with the PDGFRα start codon, produces bands that correspond with the predicted sizes of the polyadenylation variant transcripts. (c–e) Western blot analyses of FAPs untreated (−) or infected with an In-PDGFRα-expressing retrovirus (pMXs-I-Pα). Lysates were probed with antibodies targeting the amino acid sequences at one of the following locations: C-terminus of FL-PDGFRα (c), central region preceding exon 16 (d), C-terminus unique to In-PDGFRα (e). Solid arrowheads denote 180 kDa; open arrowheads denote 120 kDa. Overexpression with In-PDGFRα resulted in a doublet (e); the lower band likely represents an alternative form of In-PDGFRα oftentimes seen for FL-PDGFRα.26 (f) Levels of In-PDGFRα relative to FL-PDGFRα increase during regeneration. Lysates of FAPs collected from uninjured muscle (day 0) or at the indicated times post-injury were probed by Western blot with an antibody targeting a central region of PDGFRα (n = 3 biological replicates of pooled FAPs per time point). Expression level is plotted as a ratio of In-PDGFRα to FL-PDGFRα. Significance calculated using unpaired Student’s t-tests, and error bars represent standard error of the mean (s.e.m). *P < 0.05. For source data, see Supplementary Table 1.
Given that the kinase domain of In-PDGFRα is disrupted, we hypothesized that this protein could act as a decoy to negatively regulate PDGF signaling. We therefore tested whether the In-PDGFRα transcript actually produced functional protein in FAPs. Indeed, while Western blot analysis of FAP lysates using a C-terminal-directed antibody revealed a band at 180 kDa corresponding to the full-length protein (FL-PDGFRα) (Figure 1c), an antibody directed at a central region of PDGFRα detected bands at both 180 kDa and 120 kDa (Figure 1d). Overexpression of In-PDGFRα in FAPs using a retroviral construct resulted in increased levels of a 120 kDa protein (Figure 1d).
Use of the identified stop codon in intron 16 would be expected to result in an extension of stretch of 16 amino acids unique to In-PDGFRα (Extended Data Fig. 2d). We developed a polyclonal antibody directed against this specific 16-amino acid peptide. This antibody detected a band at 120 kDa but not at 180 kDa in FAPs, demonstrating its specificity for In-PDGFRα, and the expected 120 kDa band in cells overexpressing In-PDGFRα (Figure 1e).
To test for specific regulation of In-PDGFRα production, we analyzed FAPs activated in response to muscle injury. Overall levels of FL-PDGFRα increased but there was also a marked increase in the proportion of In-PDGFRα to FL-PDGFRα, peaking three to five days post-injury (Figure 1f), suggesting differential regulation of the intronic variant relative to the full-length form. After three weeks, the relative levels of In-PDGFRα were comparable to those in uninjured muscle (Figure 1f). Using primers that could specifically detect In-PDGFRα and FL-PDGFRα (Extended Data Fig. 2d and 2e), we found a similar temporal pattern at the transcriptional level (Extended Data Fig. 2f).
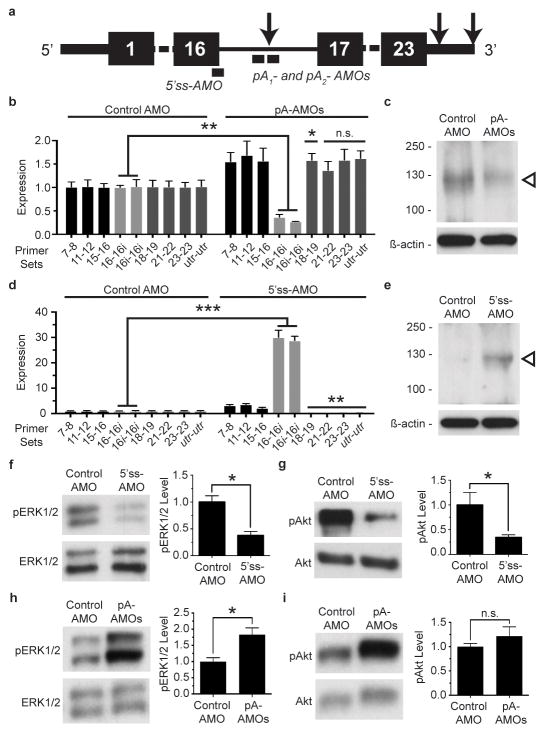
To evaluate the effect of intronic polyadenylation on PDGFRα activity, we used that antisense morpholino oligonucleotides (AMOs), which can alter intronic polyadenylation site usage by blocking splicing and polyadenylation machinery.21 We designed AMOs to target the intronic polyadenylation site (pA-AMOs) and the upstream 5′ splice site (5′ss-AMO) expected to decrease and increase production of In-PDGFRα, respectively (Figure 2a). We measured the impact of these morpholinos on both In-PDGFRα transcript and protein levels. The pA-AMOs did in fact decrease levels of In-PDGFRα without altering FL-PDGFRα levels (Figures 2b and 2c and Extended Data Fig. 2g). Likewise, the 5′ss-AMO increased In-PDGFRα levels, and there was a corresponding reduction of FL-PDGFRα levels (Figures 2d and 2e, and Extended Data Fig. 2g).
Figure 2. Intronic polyadenylation of PDGFRα inhibits signaling through the receptor, while decreasing In-PDGFRα abundance augments signaling.
(a) Morpholino target location along PDGFRα transcript. Black boxes denote exon number. Arrows represent polyadenylation sites. (b–e) Treatment of FAPs with pA-AMOs results in knockdown of the In-PDGFRα transcript (n = 3) (b) and protein (c) while treatment with the 5′ss-AMO upregulates the In-PDGFRα transcript (n = 3) (d) and protein (e). Open arrowheads denote 120 kDa in (c) as well as in (e). At the exposure shown in (e), the expression of the endogenous In-PDGFRα protein is below the level of detection due to the abundance of the protein in the 5′ss-AMO lane. (f–g) Treatment of FAPs with the 5′ss-AMO decreases phosphorylation of ERK1/2 (f) and Akt (g) in response to PDGF-AA. (h–i) On the other hand, treatment of FAPs with the pA-AMOs increases phosphorylation of ERK1/2 (h) with a trend towards enhanced Akt phosphorylation (i) in response to PDGF-AA. pERK1/2 and pAkt levels represent the ratio of phosphorylated protein to total protein. For (b), (d), and (f–i), n = 3 biological replicates of pooled FAPs per condition. In (b) and (d), all conditions were processed together, such that the control in (b) is identical to the control in (d). Significance calculated using unpaired Student’s t-tests in (b) and (d) and using paired ratio t tests in (f–i). Error bars represent s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001. For source data, see Supplementary Table 1.
We then asked whether modulation of the levels of intronic variant could alter PDGFRα signaling. Treatment of cells with PDGF-AA led to phosphorylation of PDGFRα and downstream components including ERK1/2 and Akt (Extended Data Fig. 3a and 3b). If In-PDGFRα serves as a decoy, we would expect decreased PDGFRα signaling with upregulation of the variant. Indeed, pre-treatment of cells with the 5′ss-AMO, which increases In-PDGFRα abundance, led to reduced ERK1/2 and Akt phosphorylation (Figures 2f and 2g), an effect also observed when we overexpressed In-PDGFRα (Extended Data Fig. 3c). Conversely, pre-treatment of cells with the pA-AMOs resulted in increased ERK1/2 and Akt phosphorylation (Figures 2h and 2i).
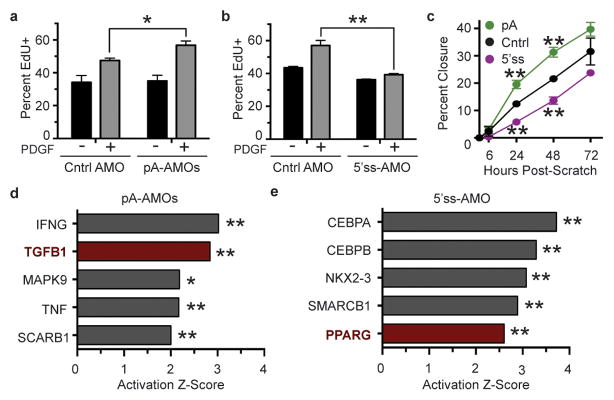
These shifts in signaling were also accompanied by changes in FAP activation. FAPs treated with the pA-AMOs exhibited an enhanced proliferative response to PDGF-AA (Figure 3a and Extended Data Fig. 4a–c), as did cells treated with an In-PDGFRα-specific siRNA (Extended Data Fig. 4d and 5a–d). In contrast, upregulation of In-PDGFRα by 5′ss-AMO pre-treatment blunted PDGF-induced proliferation (Figure 3b and Extended Data Fig. 4e), as did the overexpression of In-PDGFRα (Extended Data Fig. 4f). In independent functional assays, we found that pre-treatment with pA-AMOs enhanced FAP proliferation and migration while 5′ss-AMO pre-treatment resulted in a decrease (Figure 3c).
Figure 3. Intronic polyadenylation of PDGFRα limits activation of FAPs in vitro, and reducing intronic variant levels promotes FAP activation.
(a–b) Treatment of FAPs with pA-AMOs increases FAP proliferation in response to PDGF-AA (a) while 5′ss-AMO treatment blunts proliferation (b). (c) FAPs treated with pA-AMOs show enhanced proliferation and migration while those treated with the 5′ss-AMO displayed delayed proliferation and migration as assessed by scratch assays. (d–e) Ingenuity pathway analysis reveals TGFβ1, a promoter of fibrosis, as a top predicted regulator of gene expression change in FAPs treated with pA-AMOs compared with control treatment (d) while treatment with the 5′ss-AMOs activates genes implicated in reducing fibrosis, including PPARγ (e). For (a–c), n = 3 biological replicates of pooled FAPs per condition or time point. Significance calculated using unpaired Student’s t-tests, and error bars represent s.e.m. For (d–e), two pooled FAP samples per condition were used with the overlap p-value calculated using the Fisher’s Exact test across genes and gene sets. *P < 0.05, **P < 0.01. For source data, see Supplementary Table 1.
We also examined the effects of In-PDGFRα modulation on FAP differentiation. FAPs treated with PDGF-AA displayed an upregulation of key fibrosis markers and an induction of downstream TGF-β signaling (Extended Data Fig. 6a and 6b), consistent with previous studies.12 Interestingly, gene expression of FAPs treated with pA-AMOs, in which In-PDGFRα levels are decreased, revealed a pattern consistent with enhanced activation and fibrotic differentiation. In particular, there was enrichment for DNA replication and cell cycle genes including the MAPK/ERK pathway (Extended Data Fig. 6c–e). Moreover, causal network analysis suggested that TGF-β1 signaling was active in these cells (Figure 3d) with increased expression of associated fibrosis mediators including connective tissue growth factor (CTGF), (data not shown). Conversely, the inhibition of PDGFRα signaling through 5′ss-AMO-mediated In-PDGFRα upregulation did not significantly change gene expression associated with TGF-β1 activation. Instead, there was enrichment for processes related to protein/RNA processing and metabolism (Extended Data Fig. 6f–h). The top predicted regulators included PPARγ (Figure 3e), a gene whose activity is associated with a reduction in fibrosis and TGF-β signaling in a number of tissues.22
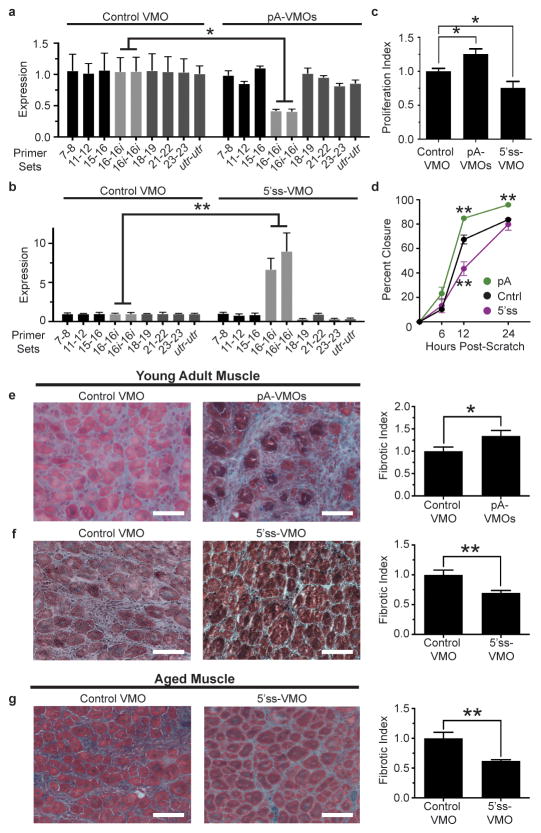
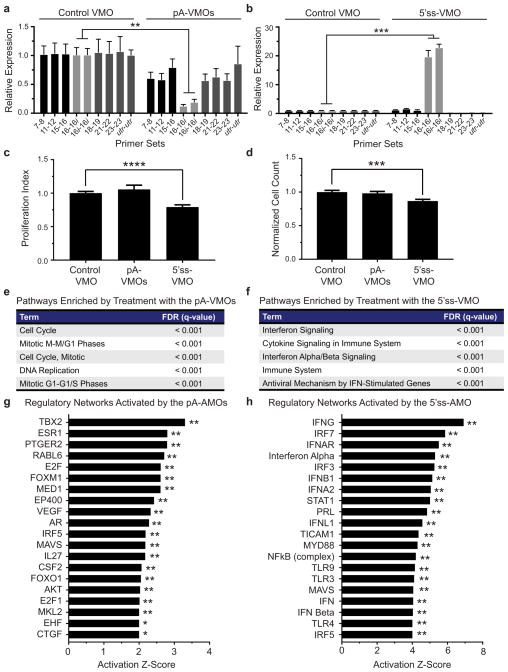
Given these findings and the association of FAPs with fibrosis, we hypothesized that altering PDGF signaling in FAPs by modulating In-PDGFRα levels in vivo would affect muscle fibrosis following injury. Therefore, we designed PDGFRα-specific Vivo-Morpholinos (VMOs), a class of morpholinos that can enter cells directly because of a covalently-bound delivery moiety.23 The VMOs targeting the polyadenylation site (pA-VMOs) and the 5′-splice-site (5′ss-VMO) were designed to contain the same targeting sequences as their AMO counterparts. We tested these VMOs both in vitro and in vivo and observed that their effects on In-PDGFRα and FL-PDGFRα levels mimicked those of their corresponding AMOs (Figures 4a and 4b and Extended Data Fig. 7a and 7b). Using intramuscular injection of VMOs, we found that 5′ss-VMO resulted in a decreased FAP proliferation index and a corresponding decrease in FAP numbers (Extended Data Fig. 7c and 7d). Treatment with pA-VMOs did not enhance FAP proliferation significantly, possibly because cells are already maximally stimulated by the injury response in vivo (Extended Data Fig. 7c and 7d).
Figure 4. Downregulation of In-PDGFRα enhances FAP activation and fibrosis, while enhancing intronic polyadenylation of PDGFRα attenuates fibrosis.
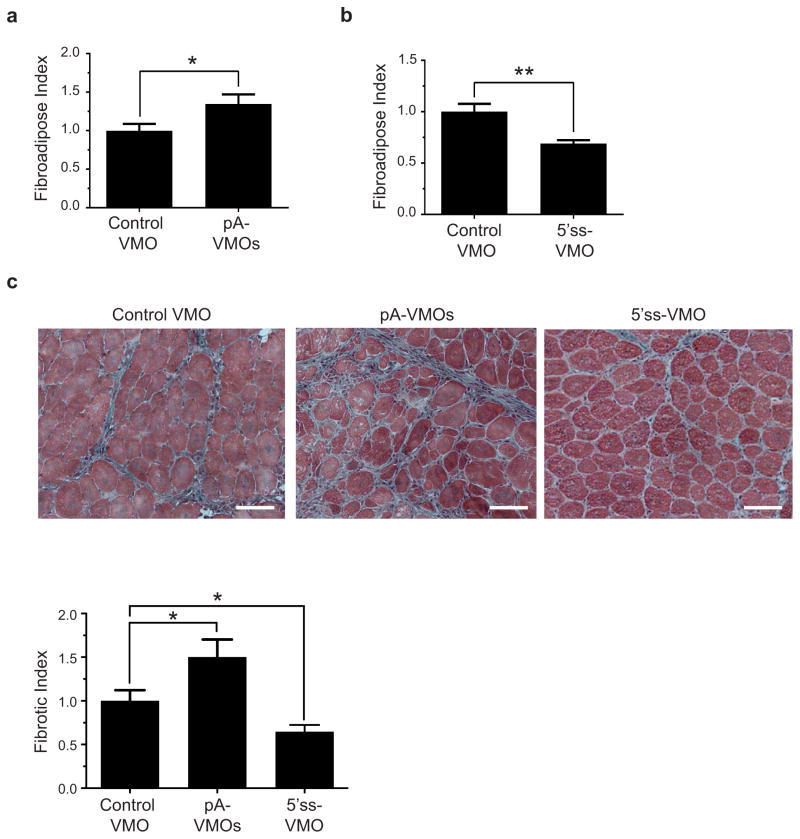
(a–b) FAPs treated in vivo with pA-VMOs show a downregulation of the In-PDGFRα transcript (n = 3) (a), while those treated with the 5′ss-VMO exhibit an upregulation of In-PDGFRα (control: n = 4, 5′ss-VMO: n = 3) (b). (c–d) FAPs treated in vivo with pA-VMOs show enhanced proliferation and migration whereas those treated with the 5′ss-VMO show decreases of both as assessed by EdU incorporation (control: n = 6, pA-VMO: n = 5) (c) and scratch assay (n = 5) (d). (e–f) Representative images of Gomori-trichrome stained cryosections and quantification of fibrosis of glycerol-injured muscles treated with pA-VMOs (control: n = 9, pA-VMO: n = 10) (e) or the 5′ss-VMO (control: n = 9, 5′ss-VMO: n = 10) (f) compared with the control VMO in young adult mice show increased and decreased fibrosis levels, respectively. (g) Reduced fibrosis in glycerol-injured muscle by treatment with the 5′ss-VMO in aged mice (control: n = 9, 5′ss-VMO: n = 10). The fibrotic index is defined as the area of fibrosis divided by total area of muscle, normalized to control-treated muscle. For (a–d) n represents biological replicates of pooled FAPs while for (e–f), n represents individual TA muscles. For histology images, scale bars represent 100μM. Significance calculated using unpaired Student’s t-tests, and error bars represent s.e.m. *P < 0.05, **P < 0.01. For source data, see Supplementary Table 1.
In parallel to our in vitro analyses, FAPs isolated from muscle pretreated with pA-VMOs displayed increased proliferation and enhanced migration (Figures 4c and 4d). Moreover, expression analysis of these cells revealed enrichment for terms related to proliferation including cell cycle and mitosis (Extended Data Fig. 7e). Of note, causal network analysis was significant for a number of proliferation and fibrosis regulators including CTGF, a growth factor that is upregulated by TGF-β and has been shown to increase collagen production by fibroblasts and to promote tissue fibrosis (Extended Data Fig. 7f).8 By contrast, 5′ss-VMO-treated FAPs showed decreased proliferation and migration (Figures 4c and 4d) as well as enrichment for interferon signaling (Extended Data Fig. 7g and 7h), which has been shown to decrease fibroblast activation and reduce fibrosis levels in models of hepatic and renal injury.24
We hypothesized that enhancing FAP activity through inhibition of In-PDGFRα via intramuscular injection of pA-VMOs would enhance fibrosis following injury. Indeed, we found this to be the case in two separate models of muscle injury (Figure 4e and Extended Data Fig. 8). Of course, although FAPs have been implicated as key mediators of fibrosis, the ability of pA-VMOs to promote fibrosis while having a more modest effect on FAP proliferation in vivo (Extended Data Fig. 7c and 7d) could reflect the contribution of other cell types to the fibrotic response. We next tested whether the 5′ss-VMO, because of its potential to block PDGFRα signaling, could decrease fibrosis. Strikingly, not only did injection of the 5′ss-VMO decrease fibrosis following injury in young adult muscle (Figures 4f and Extended Data Fig. 8), but overall levels of fibrosis were also significantly reduced in aged muscle (Figure 4g), where this type of detrimental pathology is more pronounced.5
Our finding that PDGFRα is subject to intronic polyadenylation and that this process influences FAP activation suggests that polyadenylation site choice can serve as a mechanism by which stem cells can fine tune levels of molecular signaling. Moreover, VMOs are particularly attractive candidates for disease treatment because of their sequence specificity, stability, resistance to degradation, and lack of interaction with cellular structures.25 The fact that experimental manipulation of PDGFRα intronic variant levels can reduce tissue fibrosis suggests that modulating polyadenylation site choice could be a viable therapeutic approach to the treatment of fibrotic diseases and disorders.
Materials and Methods
Animals
Wildtype male C57Bl/6 mice and B6.129S4-PDGFRαtm11(EGFP)Sor/J mice (Jackson strain# 007669), which contain an H2B-eGFP fusion protein knocked into the PDGFRα locus, were obtained from Jackson Laboratories. Young adult mice were 6–8 weeks of age; aged mice were 22–24 months of age. Mice were housed and maintained in the Veterinary Medical Unit at Veterans Affairs Palo Alto Health Care Systems. Animal protocols were performed in accordance with the policies of the Administrative Panel on Laboratory Animal Care of Stanford University.
Muscle Injury
Mice were anesthetized using isoflurane. To assess muscle regeneration, 50 μl of a 1.2% barium chloride (BaCl2) solution (Sigma-Aldrich) were injected into tibialis anterior (TA) muscles as described previously.5 To isolate activated FAPs for Western blot analysis and FACS analysis, 50 μl total of 1.2% BaCl2 or 50% v/v glycerol/water were injected throughout the lower hindlimb muscles. For induction of fibrosis, 30 μl of 50% v/v glycerol or 30 μl of a 1.2% BaCl2 solution were injected into TA muscles.
Isolation of FAPs
Muscles were dissected from mice and dissociated mechanically. All hindlimb muscles were used except in experiments where FAPs were isolated from VMOs injected into TA muscles. In this case, only the TA was dissected. The muscle suspension was digested using Collagenase II (760 U/ml; Worthington Biochemical Corporation) in Ham’s F10 (Invitrogen) with 10% horse serum (Invitrogen) for 90 minutes at 37°C with agitation. The suspension was then washed and digested in collagenase II (152 U/ml; Worthington Biochemical Corporation) and dispase (2 U/ml; Invitrogen) for 30 minutes at 37°C with agitation. The resultant mononuclear cells were then stained with the following antibodies: VCAM-1-biotin (clone 429; BioLegend, 105704), CD31-APC (clone MEC 13.3; BioLegend, 102510), CD45-APC (clone 30-F11; BioLegend, 103112) and Sca-1-Pacific Blue (clone D7; BioLegend, 108120) at 1:75. Streptavidin-PE-Cy7 (BioLegend, 405206) at 1:75 was used to amplify the VCAM-1 signal. Fluorescence-activated cell sorting (FACS) was performed using BD-FACS Aria II and BD-FACS Aria III cell sorters equipped with 488 nm, 633 nm and 405 nm lasers. The cell sorters were carefully optimized for purity and viability and sorted cells were subjected to FACS analysis immediately post-sorting to confirm FAP purity.
Direct RNA Sequencing
FAPs were isolated from uninjured C57Bl/6 mice as described above and lysed. RNA was prepared with the RNeasy Mini Kit as per the manufacturer’s instructions (Qiagen). A 3′ blocking reaction was performed using a poly(A) tailing kit (Ambion) and 3′-dATP (Jena Bioscience) and the reaction mixture was incubated at 37°C for 30 minutes. RNA was hybridized to flow cell surfaces for DRS as previously described.18 Raw DRS reads were filtered using the Helicos-developed pypeline, Helisphere, to eliminate reads less than 25 nucleotides long or of low quality. These reads were then mapped to the mouse genome (NCBI37/mm9) using IndexDPgenomic module and reads with a score above 4.3 were allowed. To avoid artifacts from mispriming, reads mapping to regions where more than 4 consecutive adenines were coded genomically immediately 3′ to the mapping were excluded for further analysis. The sequencing data were deposited into the NCBI Sequence Read Archive (Accession SRP079186).
Analysis of Alternative Polyadenylation Sites and Quantitative RT-PCR
Total RNA was extracted from FAPs isolated from uninjured C57Bl/6 mice using TRIzol (Invitrogen) as per the manufacturer’s instructions. To identify the polyadenylation sites, the sample was reverse transcribed using the SMARTer RACE cDNA amplification kit (Clontech) according to the manufacturer’s instructions using the primers listed in Extended Data Table 1. The amplified fragments were subcloned into pGEM-T-Easy (Promega) and sequenced. Sequencing data was visualized with 4Peaks. To assess levels of the intronic variant and UTR variants, primers were designed to span the PDGFRα transcript (Extended Data Table 2). Variant expression was normalized to GAPDH using the comparative CT method (Pfaffl, 2001) and reported relative to the average of control-treated samples.
Retroviral Transfection
A construct corresponding to In-PDGFRα (DNAFORM, AK035501, RIKEN clone 9530057A20) was obtained. This construct was subcloned into the pMXs-IRES-GFP retroviral backbone (Cell BioLabs, Inc.) to generate pMXs-I-Pα. Replication-incompetent retroviral particles were generated by transfection of the 293T human embryonic kidney cell-derived Phoenix helper cell line (gift from Dr. Gary Nolan, Stanford University). Viral supernatant was filtered through 0.45 μm polyethersulfone filters, concentrated using PEG precipitation, and stored at −80°C.
FAPs were plated in 6-well plates and grown in DMEM supplemented with 10% FBS. When cells reached 70% confluency, viral supernatant and polybrene (at a final concentration of 4 μg/ml) were added to the medium. For overexpression experiments, FAPs were incubated with the viral supernatant for 48 hours before analysis. For signaling assays, FAPs were incubated with the viral supernatant for 24 hours. Afterwards, the medium was then changed to serum-free DMEM containing viral supernatant and the cells were incubated for an additional 24 hours. The FAPs were then treated with 1 ng/ml PDGF-AA for 15 minutes, after which the cells were used for Western blot analysis.
Generation of In-PDGFRα-specific Rabbit Polyclonal Antibody
A peptide with the sequence GKSAHAHSGKYDLSVV, which represents the unique carboxy-terminal region of In-PDGFRα protein, was generated (Thermo Scientific Pierce, OE0726). To generate In-PDGFRα rabbit polyclonal antibodies directed against In-PDGFRα, New Zealand White Rabbits that are Specific Pathogen Free were immunized with 0.25 mg of the peptide in Completed Freund’s Adjuvant. The rabbits received three boosters of antigen consisting of 0.10 mg in Incomplete Freund’s Adjuvant at days 14, 42, and 56 post-immunization. Serum was collected at days 70 and 72 (Thermo Scientific Pierce).
Western Blot Analysis
Cells and homogenized tissues were lysed with RIPA lysis buffer supplemented with protease and phosphatase inhibitors (Roche). The lysates were run on Criterion SDS-PAGE gels (Bio-Rad), transferred to nitrocellulose membranes (Fisher Scientific), and analyzed by Western blot using the following rabbit antibodies: PDGFRα polyclonal (1:1000, Cell Signaling, 3174), PDGFRα center (1:100, Abgent, AP14254c), In-PDGFRα custom (1:1000), p-PDGFRα (Tyr 754) polyclonal (1:1000, Cell Signaling, 4547), Akt polyclonal (1:1000, Cell Signaling, 9272), p-Akt polyclonal (1:1000, Cell Signaling, 9271), Plcγ polyclonal (1:1000, Cell Signaling, 5690), p-Plcγ polyclonal (1:1000, Cell Signaling, 2821), Erk polyclonal (1:2000, Cell Signaling, 4695), p-Erk polyclonal (1:2000, Cell Signaling, 4370), Smad2/3 monoclonal (1:1000, Cell Signaling, 8685), and p-Smad2(Ser 465/467)/Smad3(Ser 423/425) monoclonal (1:1000, Cell Signaling, 8828). Membranes were incubated in horseradish peroxidase-labeled secondary antibodies and bands were visualized with enhanced chemiluminescence (Advansta).
siRNA Knockdown of PDGFRα Variants
siRNAs were designed using the Dharmacon siDESIGN center for knockdown of In-PDGFRα and FL-PDGFRα (Supplementary Table 1). To knockdown either In-PDGFRα and FL-PDGFRα in FAPs, approximately 8 × 104 cells were plated in a 12-well plate containing DMEM supplemented with 10% FBS and grown to 70–80% confluence. Cells were incubated in 200 nM of either PDGFRα or control siRNAs using Lipofectamine 2000 (Invitrogen). To assess knockdown, cells were harvested at 24 hours for qPCR analysis. For Western blot analyses, 3 × 105 cells were plated in 6-well plates and incubated in Ham’s F10 (Invitrogen) supplemented with 10% horse serum (Invitrogen) for 24 hours. The medium was then replaced with serum-free Ham’s F10 (Invitrogen) supplemented with 200 nM siRNA and incubated for an additional 24 hours.
Antisense Morpholino Treatment
Morpholinos were designed to target two polyadenylation sites on the intronic variant (pA1: 5′-TGATTACATTATATCTGTCTTTATT-3′ and pA2: 5′-AGCAAAGACCATCATAGCAGAATGA-3′) and the upstream 5′ splice site of the intron (5′ss: 5′-ATGGGCACTTTTACCTAGCATGGAT-3′) (Gene Tools, LLC). For in vitro treatment, cells were grown to 70–80% confluency in DMEM (Invitrogen) supplemented with 10% fetal bovine serum (Atlanta Biologicals). Cells were incubated in 10 μM of the indicated morpholino using the Endo-Porter transfection reagent (Gene Tools, LLC). Cells were harvested at 24 hours for qPCR analysis with RNA isolated using the RNeasy Plus Mini kit with on-column Dnase digestion per manufacturer’s instructions (Qiagen). For Western blot analysis, cells were transfected for 24 hours in Ham’s F10 (Invitrogen) supplemented with 10% horse serum (Invitrogen). The medium was then replaced with serum-free Ham’s F10 (Invitrogen) and incubated for an additional 24 hours. For signaling assays, cells were then incubated for 15 minutes with PDGF-AA (Peprotech) at 0.1 ng/ml or 20 ng/ml for cells treated with pA-AMOs or the 5′ss-AMO, respectively, and lysed for Western blot analysis as described above.
In Vitro EdU Incorporation Assay
For AMO treatment, FAPs were isolated from the uninjured hindlimb muscles of C57Bl/6 mice and seeded at 1 × 105 cells per well in poly-D-lysine coated 8-well chamber slides (BD Biosciences) coated with ECM gel (Sigma-Aldrich). Cells were transfected with 10 μM AMO using Endoporter (Gene Tools) and expanded for two days in Ham’s F10 (Invitrogen) supplemented with 10% horse serum (Invitrogen). The medium was then replaced with Opti-Mem supplemented with 2 ng/ml PDGF-AA ligand and 10 μm EdU (Invitrogen). Cells were fixed in 4% paraformaldehyde (Sigma-Aldrich) after 24 hours.
For siRNA treatment, FAPs were isolated from the uninjured hindlimb muscles of C57Bl/6 mice and seeded at 2 × 105 cells per well in poly-D-lysine coated 8-well chamber slides (BD Biosciences) coated with ECM gel (Sigma-Aldrich). The medium was supplemented with 200 nM siRNA and transfected using Lipofectamine 2000 (Invitrogen). After 24 hours, the medium was replaced with Opti-Mem and the cells were re-transfected with 200 nM siRNA and 50 ng/ml PDGF-AA. In siRNA-treated samples, EdU was not included in this medium. Rather, after 20 hours the medium was replaced with Opti-Mem containing 10 μm EdU (Invitrogen). Cells were fixed 4 hours later.
For retroviral overexpression of In-PDGFRα, FAPs were isolated from uninjured hindlimbs of C57Bl/6 mice and seeded at 2 × 105 cells per well in poly-D-lysine coated 8-well chamber slides (BD Biosciences) coated with ECM gel (Sigma-Aldrich). FAPs were cultured in DMEM supplemented with 10% FBS along with viral supernatant and 4 μg/ml polybrene. After 24 hours, the medium was replaced with serum-free DMEM containing viral supernatant and 20 ng/ml PDGF-AA. Twenty hours later, the medium was replaced with Opti-Mem containing 10 μM EdU. Cells were fixed after 4 hours. For EdU incorporation experiments, cells were stained using the Click-iT EdU Imaging Kit (Invitrogen). Cells were analyzed on a Zeiss Observer Z1 fluorescent microscope (Carl Zeiss) equipped with a Hamamatsu Orca-ER camera (Hamamatsu) and Improvision Volocity software (Perkin Elmer).
Scratch Assay
Cells isolated by FACS from uninjured hindlimb muscles were seeded at a density of 3.5 × 104 cells per well in 96-well plates in Ham’s F10 medium supplemented with 2% horse serum. After 48 hours, cells were nearly confluent and the medium was changed to Ham’s F10 with 2% horse serum and 20 ng/ml PDGF-AA. A wound was made by scratching a 200 μl pipette tip across the monolayer of cells. The initial scratch area was determined immediately and set to 100%. Images were taken at regular intervals and the scratch area at each time point was measured and calculated as a percent of the initial scratch area. Scratch closure is defined as the cell-free area as a percentage of total area.
Microarray Analysis
For in vitro microarray analysis, FAPs were isolated from the uninjured hindlimb muscles of C57Bl/6 mice. Cells were plated at 1 × 106 cells per well in 12-well plates. Cells were grown for 2.5 days in DMEM supplemented with 10% FBS. The medium was switched to Ham’s F10 supplemented with 10% horse serum and transfected with 200 nM siRNA or 10 μM AMO as indicated for 48 hours. For the siRNA-treated cells, the medium was replaced and cells re-transfected after 24 hours. The medium was then replaced with Opti-Mem and cells re-transfected with 200 nM siRNA or 10 μM AMO. After 48 hours, the cells were lysed and RNA prepared with the RNeasy Mini Kit as per the manufacturer’s instructions (Qiagen). For in vivo microarray analysis, TA muscles were each injured with 30 μL of glycerol and injected with the indicated VMO after 3 days. FAPs were then isolated from the muscles 2 days post-VMO injection. Cells were pelleted and RNA prepared from samples as indicated above. The microarray data were obtained using Affymetrix Mouse 1.0 ST. For GSEA, The samples were normalized and processed using Gene Pattern ExpressionFileCreator and PreProcessDataset modules. Expression data were analyzed and visualized with GSEA29 and GENE-E (http://www.broadinstitute.org/cancer/software/GENE-E/). For Ingenuity Pathway Analysis, including causal network analysis, the samples were normalized using Affymetrix Expression Console Software and analyzed for enrichment using IPA (Ingenuity® Systems, www.ingenuity.com). Array data were deposited into Gene Expression Omnibus (Accessions GSE60099 and GSE81744).
Vivo Morpholino Treatment
Vivo-morpholinos were designed to target two polyadenylation sites on the intronic variant (pA1-VMO: 5′-TGATTACATTATATCTGTCTTTATT-3′ and pA2-VMO: 5′-AGCAAAGACCATCATAGCAGAATGA-3′) and the upstream 5′ splice site of the intron (5′ss-VMO: 5′-ATGGGCACTTTTACCTAGCATGGAT-3′) (Gene Tools, LLC). For treatment in vitro, cells were isolated from hindlimb muscles of C57/Bl6 mice and grown to 70–80% confluency in DMEM (Invitrogen) supplemented with 10% fetal bovine serum (Atlanta Biologicals). Cells were incubated in the 10 μM of the indicated morpholino (Gene Tools, LLC). Cells were harvested at 24 hours for qPCR analysis. For in vivo qPCR analysis, TA muscles were injured with glycerol as described above and injected with 250 ng of the indicated VMO at the site of injury 3 days later. FAPs were sorted by FACS 7 days after VMO injection for qPCR analysis.
For ex vivo proliferation and scratch assays, TA muscles were injured with glycerol and injected with 250 ng of the indicated VMO 3 days post-injury. FAPs were isolated 2 days later by FACS. In EdU incorporation studies, cells were seeded at 4 × 104 cells per well in poly-D-lysine coated 8-well chamber slides (BD Biosciences) coated with ECM gel (Sigma-Aldrich). Cells were incubated in 10 ng/ml PDGF-AA (Peprotech) and 10 μM EdU (Invitrogen) for 24 hours. The cells were fixed and stained. In the ex vivo proliferation studies as well as the in vivo proliferation studies described below, the proliferation index was used to denote the percent EdU incorporation normalized to control. In the scratch assays, cells were seeded and treated as described above.
For in vivo proliferation studies, TA muscles were injected with 150 ng of the indicated VMO at 0 and 24 hours. FAPs were isolated at 48 hours via FACS. To assess in vivo proliferation, the cells were exposed to 10 μM EdU immediately after muscle isolation and incubated in 10 μM EdU ex vivo during the collagenase, collagenase/dispase, and antibody incubations as described above. The cells were plated in poly-D-lysine coated 8-well chamber slides (BD Biosciences) coated with ECM gel (Sigma-Aldrich), fixed 1 hour after plating, and stained using the Click-iT EdU Imaging Kit (Invitrogen).
For histological analysis, TA muscles were injured with glycerol or BaCl2 and injected at the site of injury with 250 ng of the indicated VMO. After 7 days, the muscles were snap frozen in isopentane cooled in liquid nitrogen immediately after dissection. Muscles sections were stained with Gomori-Trichrome (Richard-Allan Scientific) per manufacturer’s instructions or oil red O (Sigma-Aldrich) as previously described.30 The Fibrotic Index was calculated as the area of fibrosis divided by total area of muscle normalized to control-treated muscle. The Fibroadipose Index was defined as the area of fibrosis plus the area of adiposis (as detected by Oil Red O staining) divided by total area of muscle, normalized to control.
Statistics and General Methods
Major factors in determining sample size included the level of the effect and the inherent variability in measurements obtained. Animals were excluded from the study only if their health status was compromised, such as occurred when animals had visible wounds from fighting. Samples were not specifically randomized or blinded. However, mouse identifiers were used as possible to blind evaluators to experimental conditions, and all samples within experiments were processed identically for measurement quantification using automated tools as specified.
Extended Data
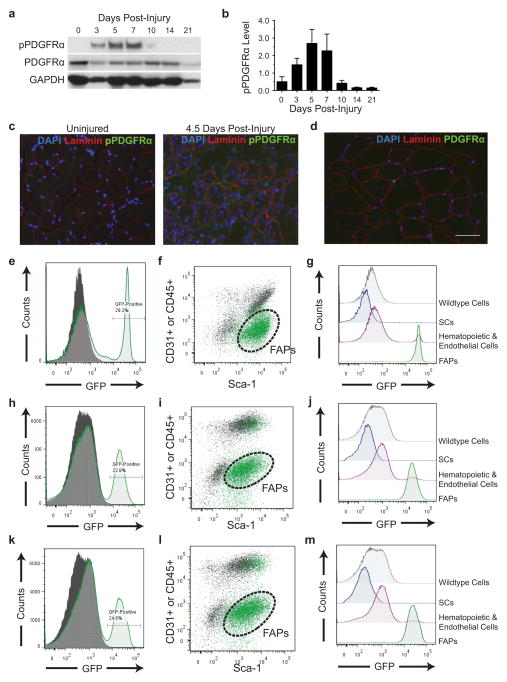
Figure E1. PDGFRα is activated specifically in FAPs during muscle regeneration.
(a) TA muscles were dissected at the indicated days post-injury. The muscle lysates were subjected to Western blot to probe for PDGFRα and phospho-PDGFRα using independent antibodies. (b) Quantification of (a) (n = 3 individual TA muscles per time point). pPDGFRα level refers to expression levels for pPDGFRα normalized for total PDGFRα. Error bars represent s.e.m. (c) Immunofluorescence of an uninjured TA muscle and a muscle 4.5 days post-injury (DAPI = blue, Laminin = red, pPDGFRα = green). (d) Immunofluorescence of an uninjured TA muscle from a PDGFRα-eGFP reporter mouse. (e) FACS histogram analysis of GFP signal in a PDGFRα-eGFP reporter heterozygote (green line) and in a wildtype littermate (gray, solid) in cells from uninjured hindlimb muscles. (f) FACS plot of cells isolated from uninjured hindlimb muscles of PDGFRα-eGFP reporter. GFP-positive cells (shown in green) are overlaid upon all cells (shown in gray). Dashed lines represent the gate for the FAP population. (g) Histograms showing GFP signal in cell populations isolated from uninjured PDGFRα-eGFP reporter mice, as detected by FACS analysis. (h–j) The patterns of expression of (h), (i), and (j), as assessed by FACS analysis of BaCl2-injured muscle were similar to those patterns exemplified in (e), (f), and (g), respectively from uninjured muscle. (k–m) Similarly, the patterns of expression of (k), (l), and (m), as evaluated by FACS analysis of glycerol-injured muscle were also comparable to those patterns exemplified in (e), (f), and (g), respectively, from uninjured muscle. For source data, see Supplementary Table 1.
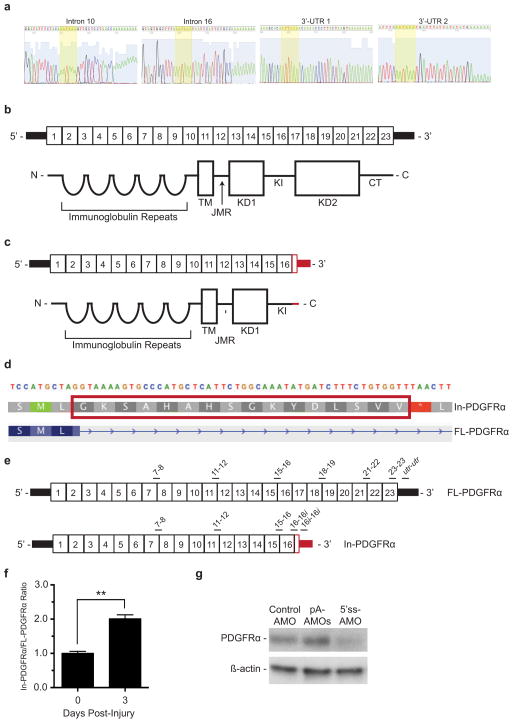
Figure E2. In-PDGFRα transcript and protein structure.
(a) DNA sequencing of 3′ RACE products illustrated in Figure 1b confirms polyadenylation sites of highly expressed variants. (b) The fully-spliced FL-PDGFRα transcript contains 23 exons, as illustrated in the top portion of the figure, that code for the corresponding protein domains pictured below. (TM = transmembrane region, JMR = juxtramembrane region, KD1 = kinase domain 1, KI = interkinase domain, KD2 = kinase domain 2, CT = c-terminal region). (c) The fully spliced In-PDGFRα transcript contains 16 exons, as illustrated in the top portion of the figure, that code for the protein domains shown below. In red are the portions of the transcript and protein that are unique to In-PDGFRα. (d) A close-up view of the genomic sequence that codes for the unique region of In-PDGFRα protein. In FL-PDGFRα, this region is spliced out of the transcript. (e) Map of the locations of amplicons used to assess levels of In-PDGFRα and FL-PDGFRα. Primers amplifying regions of exons 7 to 8 (7–8), exons 11 to 12 (11–12), and exons 15 to 16 (15–16) are common to In-PDGFRα and FL-PDGFRα. Primers designated with 16i target the region canonically referred to as intron 16. In FL-PDGFRα, this region is spliced out. In In-PDGFRα, this region becomes the 3′ UTR. Therefore 16-16i and 16i-16i are specific for In-PDGFRα. Primers amplifying regions of exons 18 to 19 (18–19), exons 21 to 22 (21–22), exon 23 (23–23) and the 3′ UTR (utr-utr) are specific to FL-PDGFRα. (f) Levels of In-PDGFRα transcript relative to FL-PDGFRα transcript increase during regeneration. FAPs were collected from uninjured muscles (day 0) or at 3 days post-injury and RNA levels assessed via qPCR. Expression level is plotted as a ratio of In-PDGFRα to FL-PDGFRα normalized to day 0. For In-PDGFRα primers 16-16i and 16i-16i were averaged, while for FL-PDGFRα, primers 23–23 and utr-utr were averaged. For each time point n = 3 biological replicates of pooled FAPs. Significance calculated using unpaired Student’s t-test, and error bars represent s.e.m. (g) Western blot using the C-terminal PDGFRα antibody shows knockdown of FL-PDGFRα in response to the 5′ss-AMO. For source data, see Supplementary Table 1.
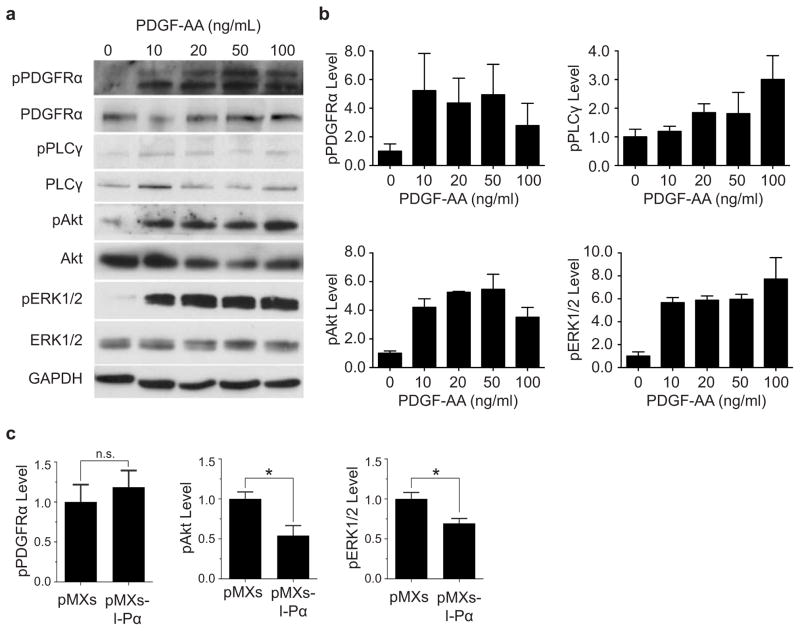
Figure E3. Phosphorylation of PDGFRα and downstream signaling components in FAPs in response to PDGF-AA stimulation is altered by changes in In-PDGFRα expression.
(a) Western blot showing phosphorylation of PDGFRα, PLCγ, Akt, and ERK1/2 in response to PDGF-AA stimulation. (b) Quantification of data in panel (a). (c) Viral overexpression of In-PDGFRα in FAPs results in decreased signaling through Akt and ERK/2 while FL-PDGFRα levels remain constant. For (b) and (c), n = 3 biological replicates of pooled FAPs per condition except for the ERK1/2 condition in (b) where n = 4. Significance calculated using unpaired Student t-tests, and error bars represent s.e.m. *P < 0.05, **P < 0.01. For source data, see Supplementary Table 1.
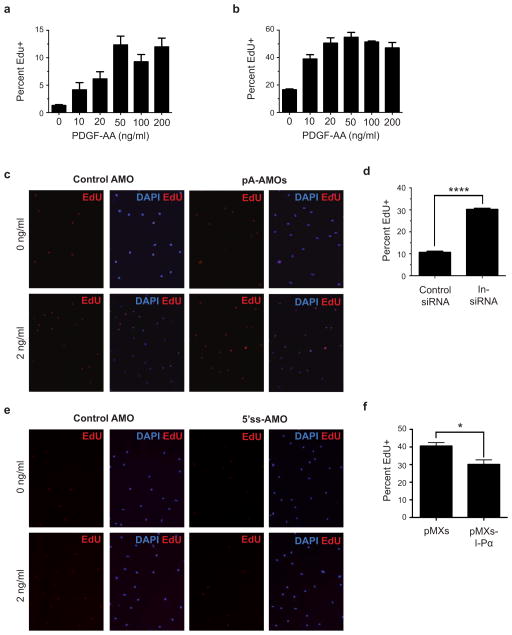
Figure E4. FAPs proliferate in response to PDGF stimulation.
(a) EdU incorporation in FAPs stimulated with indicated amounts of PDGF for 20 hours and incubated in EdU in the final 4 hours. (b) EdU incorporation in FAPs incubated with indicated amounts of PDGF and EdU for 24 hours. (c–f) Knockdown of In-PDGFRα by pre-treatment of FAPs with pA-AMOs (c) or In-siRNA (d) increases FAP proliferation in response to PDGF-AA, whereas In-PDGFRα upregulation by 5′ss-AMO pre-treatment (e) or viral overexpression (f) decreases proliferation. For (a), (b), (d), and (f), n = 3 biological replicates of pooled FAPs per condition. Significance calculated using unpaired Student t-tests, and error bars represent s.e.m. *P < 0.05, ****P < 0.0001. For source data, see Supplementary Table 1.
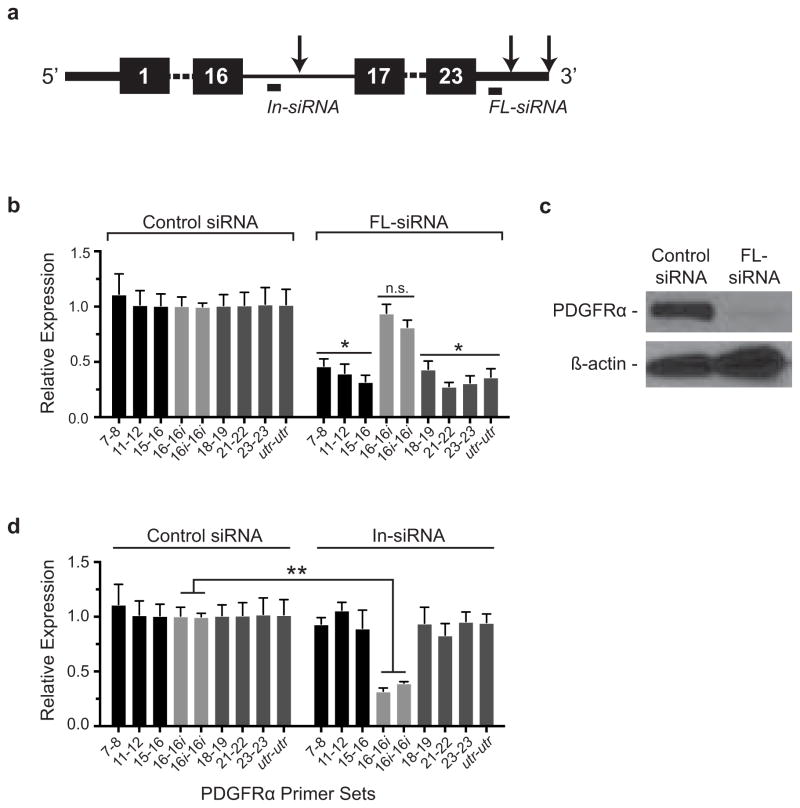
Figure E5. Knockdown of In-PDGFRα and FL-PDGFRα with siRNAs.
(a) Schematic of the FL-PDGFRα transcript showing the location of siRNAs designed to knockdown In-PDGFRα (In-siRNA) and FL-PDGFRα (FL-siRNA). Arrows designate relevant polyadenylation sites. (b–d) FL-siRNA induces transcriptional knockdown of FL-PDGFRα but not In-PDGFRα (b) and protein knockdown of FL-PDGFRα (c). At the same time, In-siRNA induces knockdown of In-PDGFRα (d). For (b) and (d), n = 3 biological replicates of pooled FAPs per condition. In (b) and (d) the samples were all processed together such that the control condition in (b) is identical to the control condition in (d). Significance calculated using unpaired Student t-tests, and error bars represent s.e.m. **P < 0.01. For source data, see Supplementary Table 1.
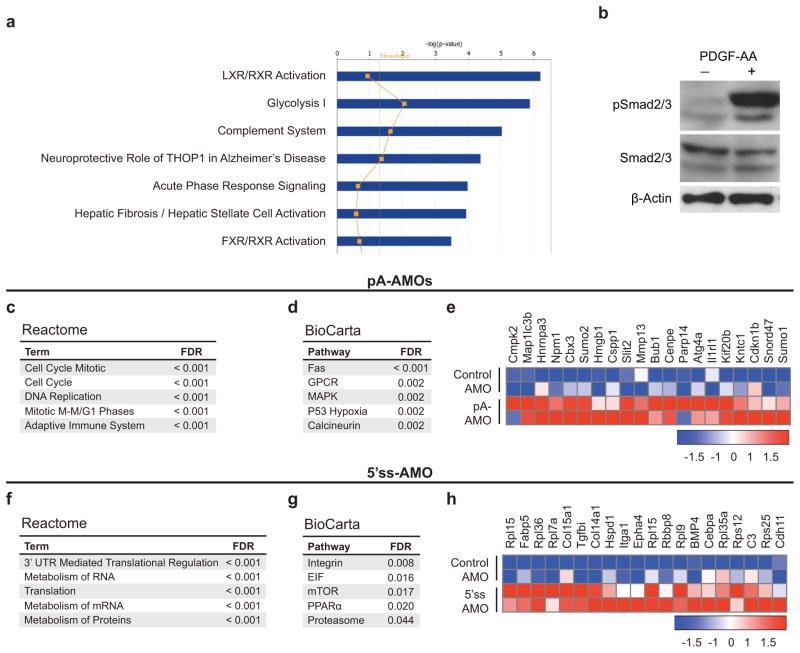
Figure E6. Assessment of molecular pathway changes in response to modulation of In-PDGFRα levels.
(a) Ingenuity pathway analysis of control AMO-treated FAPs stimulated with PDGF-AA (50 ng/ml) show enrichment for genes associated with fibrosis compared with control cells. (b) Western blot showing phosphorylation of Smad2/3 in response to PDGF-AA stimulation (50 ng/ml). (c–d) GSEA of FAPs treated with the pA-AMOs compared with control-treated samples were analyzed for enrichment of pathways in the Reactome (c) and BioCarta (d) databases.27,28 Top sets with a false discovery rate less than 5% are shown. (e) The heat map displays a subset of genes that are upregulated with a false discovery rates less than 0.1% in the pA-AMO- treated FAPs. The colors indicate the Z-score for expression normalized for each gene. Red indicates high expression while blue indicates low expression. (f–g) Similarly, enrichment analyses of FAPs treated with the 5′ss-VMO compared with controls were performed using the Reactome (f) and BioCarta (g) databases.27,28 (h) A heat map displaying top upregulated genes in 5′ss-VMO-treated FAPs with parameters as specified in (e) is shown.
Figure E7. Alteration of In-PDGFRα levels in vivo modulates FAP activity.
(a,b) FAPs treated in vitro with pA-VMOs showed a downregulation of In-PDGFRα (n = 3) (a) whereas those treated with the 5′ss-VMO exhibited an upregulation of In-PDGFRα (n = 3) (b). (c–d) In vivo treatment with 5′ss-VMO decreases FAP proliferation (n = 16) (c) with a corresponding decrease in cell count (n = 24) (d). Treatment with pA-VMOs does not lead to a significant change in proliferation (p = 0.42). (e–f) GSEA of FAPs collected from TAs treated with pA-VMOs (e) or the 5′ss-VMO (f) compared with control-treated samples were analyzed for enrichment of pathways in the Reactome database.27,28 Top sets with a false discovery rate less than 5% are shown. (g–h) Ingenuity pathway analysis of top regulators of gene expression in FAPs treated with pA-AMOs (g) or 5′ss-VMO (h) compared with control treatment. The top hits are shown, excluding those with the “Molecular Type” designated as a chemical or drug. For (a–d), n represents biological replicates of pooled FAPs and p-value is calculated using unpaired Student t-tests. In (g) and (h), two pooled FAP samples per condition were used with the overlap p-value calculated using the Fisher’s Exact test. Error bars represent standard error of the mean. *P < 0.05, **P < 0.01. For source data, see Supplementary Table 1.
Figure E8. Alteration of In-PDGFRα levels in vivo modulates muscle fibrosis.
(a,b) Fibroadipose Index of muscles treated with the pA-VMOs (n = 9) (a) or the 5′ss VMO (control: n = 9, 5′ss-VMO: n = 10) (b). (c) Representative images of Gomori-trichrome staining and quantification of fibrosis of BaCl2-treated muscles from aged mice treated with the Control VMO (n = 10), the pA-VMOs (n = 9), and the 5′ss-VMO (n = 10). The fibrotic index is defined as the area of fibrosis divided by total area of muscle, normalized to control-treated muscle. For (a–c), n represents biological replicates consisting of TA muscles isolated from individual mice. For histology images, scale bars represent 100μM. Significance calculated using unpaired Student t-tests, and error bars represent s.e.m. *P < 0.05, **P < 0.01. For source data, see Supplementary Table 1.
Extended Data Table 1.
PDGFRα gene-specific primers used in 3′ RACE
| Location | Sequence |
|---|---|
| Exon 2 | CTATGGGGACCTCCCACCAGGTCTTTCT |
| Exon 10 | GTCCTTCGCCAAAGTGGAAGAGACCATC |
| Exon 16 | AACGGCGACTACATGGACATGAAGCAAG |
| 3′-UTR (precedes both polyA sites) | CCTCTCCGGACCTCTGAAGAGACCACTC |
Extended Data Table 2.
Primers for PDGFRα variant quantification
| Name | Forward Sequence | Reverse Sequence |
|---|---|---|
| 7–8 | CTCACCGAGATCACCACTGA | CCTTAGCCCGGATCAGCTTT |
| 11–12 | TGTTGGTGCTGTTGGTGATT | CGTGGTTTCTGCTTCCAAAT |
| 15–16 | TGAATCCTGCAGACGAGAGC | ATGTCCATGTAGTCGCCGTT |
| 16-16i | GGCCAGCCTCCTACAAGAAG | TCCCTCTGAGCAGCAAGTTA |
| 16i-16i | AAAAGTGCCCATGCTCATTC | GCTTGGCAGAGCTACCTGTC |
| 18–19 | CTGGCCAGAGACATCATGCA | CATCCACTTCACAGGCAGGA |
| 21–22 | TGTTGCCGGGACAATACAAGA | TATCAGAGTCCACCCGCATG |
| 23–23 | GAGAGAGGACGAGACCATCG | CACCAGGTCCGAGGAATCTA |
| utr-utr | AGAGGACTTGGGTGATGTGG | TCATCCACACAGGCTTACCA |
Supplementary Material
Acknowledgments
We would like to thank members of the Rando lab, especially Antoine de Morree, Marco Quarta, Johnathan Shih, Bryan Yoo, Victor Garcia, and Igor Akimenko for their insightful discussions and experimental assistance as well as Lusijah Rott for guidance with FACS. This work was supported by the Glenn Foundation for Medical Research, by grants from the National Institutes of Health (F30 AG043235) and the California Institute for Regenerative Medicine (TG2-01159) to A.A.M., and by grants from the Department of Veterans Affairs and the NIH (P01 AG036695, R01 AG23806, R01 AR062185, and TR01 AG47820) to T.A.R.
Footnotes
Author Contributions
A.A.M. and T.A.R. conceived the study and were involved in the overall design of experiments. A.A.M. and T.H.C. designed, conducted, and analyzed the microarray and sequencing experiments as well as the FACS analyses. A.A.M. designed the morpholinos and siRNAs and also performed and analyzed the associated experiments to test for their transcriptional effects. A.A.M. and C.T.V.V. designed the viral construct while C.T.V.V. created the construct and performed experiments to analyze expression. A.A.M. and C.T.V.V. designed and performed experiments and data analysis for PDGFRα protein expression and downstream signaling in response to morpholino treatment in vitro and in vivo as well as for muscle fibrosis responses. A.A.M. conducted and analyzed the proliferation assays while C.T.V.V. performed and evaluated the scratch assays. K.F. carried out pilot experiments to assess for PDGFRα signaling in regenerating skeletal muscle and in initial FACS characterization of the FAP population. A.A.M. and T.A.R. wrote the manuscript with helpful input from C.T.V.V. and T.C.
Competing financial interests
Authors declare no conflicts of interest.
Reference List
- 1.Orr-Urtreger A, Bedford MT, Do MS, Eisenbach L, Lonai P. Developmental expression of the alpha receptor for platelet-derived growth factor, which is deleted in the embryonic lethal Patch mutation. Development. 1992;115:289–303. doi: 10.1242/dev.115.1.289. [DOI] [PubMed] [Google Scholar]
- 2.Soriano P. The PDGF alpha receptor is required for neural crest cell development and for normal patterning of the somites. Development. 1997;124:2691–700. doi: 10.1242/dev.124.14.2691. [DOI] [PubMed] [Google Scholar]
- 3.Andrae J, Gallini R, Betsholtz C. Role of platelet-derived growth factors in physiology and medicine. Genes Dev. 2008;22:1276–312. doi: 10.1101/gad.1653708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olson LE, Soriano P. Increased PDGFRalpha activation disrupts connective tissue development and drives systemic fibrosis. Dev Cell. 2009;16:303–13. doi: 10.1016/j.devcel.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brack AS, et al. Increased Wnt signaling during aging alters muscle stem cell fate and increases fibrosis. Science. 2007;317:807–10. doi: 10.1126/science.1144090. [DOI] [PubMed] [Google Scholar]
- 6.Goldspink G, Fernandes K, Williams PE, Wells DJ. Age-related changes in collagen gene expression in the muscles of mdx dystrophic and normal mice. Neuromuscul Disord. 1994;4:183–91. doi: 10.1016/0960-8966(94)90019-1. [DOI] [PubMed] [Google Scholar]
- 7.Conboy IM, Rando TA. Aging, stem cells and tissue regeneration: lessons from muscle. Cell Cycle. 2005;4:407–10. doi: 10.4161/cc.4.3.1518. [DOI] [PubMed] [Google Scholar]
- 8.Mann CJ, et al. Aberrant repair and fibrosis development in skeletal muscle. Skelet Muscle. 2011;1:21. doi: 10.1186/2044-5040-1-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Serrano AL, et al. Cellular and molecular mechanisms regulating fibrosis in skeletal muscle repair and disease. Curr Top Dev Biol. 2011;96:167–201. doi: 10.1016/B978-0-12-385940-2.00007-3. [DOI] [PubMed] [Google Scholar]
- 10.Uezumi A, Fukada S, Yamamoto N, Takeda S, Tsuchida K. Mesenchymal progenitors distinct from satellite cells contribute to ectopic fat cell formation in skeletal muscle. Nat Cell Biol. 2010;12:143–52. doi: 10.1038/ncb2014. [DOI] [PubMed] [Google Scholar]
- 11.Joe AWB, et al. Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nat Cell Biol. 2010;12:153–63. doi: 10.1038/ncb2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uezumi A, et al. Fibrosis and adipogenesis originate from a common mesenchymal progenitor in skeletal muscle. J Cell Sci. 2011;124:3654–64. doi: 10.1242/jcs.086629. [DOI] [PubMed] [Google Scholar]
- 13.Fiore D, et al. Pharmacological blockage of fibro/adipogenic progenitor expansion and suppression of regenerative fibrogenesis is associated with impaired skeletal muscle regeneration. Stem Cell Res. 2016;17 doi: 10.1016/j.scr.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 14.Tidball JG, Spencer MJ, St Pierre BA. PDGF-receptor concentration is elevated in regenerative muscle fibers in dystrophin-deficient muscle. Exp Cell Res. 1992;203:141–9. doi: 10.1016/0014-4827(92)90049-e. [DOI] [PubMed] [Google Scholar]
- 15.Zhao Y, et al. Platelet-derived growth factor and its receptors are related to the progression of human muscular dystrophy: an immunohistochemical study. J Pathol. 2003;201:149–59. doi: 10.1002/path.1414. [DOI] [PubMed] [Google Scholar]
- 16.Ito T, et al. Imatinib attenuates severe mouse dystrophy and inhibits proliferation and fibrosis-marker expression in muscle mesenchymal progenitors. Neuromuscul Disord. 2013;23:349–56. doi: 10.1016/j.nmd.2012.10.025. [DOI] [PubMed] [Google Scholar]
- 17.Huang P, Zhao XS, Fields M, Ransohoff RM, Zhou L. Imatinib attenuates skeletal muscle dystrophy in mdx mice. FASEB J. 2009;23:2539–48. doi: 10.1096/fj.09-129833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ozsolak F, et al. Direct RNA sequencing. Nature. 2009;461:814–8. doi: 10.1038/nature08390. [DOI] [PubMed] [Google Scholar]
- 19.Boutet SC, et al. Alternative Polyadenylation Mediates MicroRNA Regulation of Muscle Stem Cell Function. Cell Stem Cell. 2012;10 doi: 10.1016/j.stem.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mueller AA, Cheung TH, Rando TA. All’s well that ends well: alternative polyadenylation and its implications for stem cell biology. Curr Opin Cell Biol. 2013;25:222–232. doi: 10.1016/j.ceb.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vorlová S, et al. Induction of Antagonistic Soluble Decoy Receptor Tyrosine Kinases by Intronic PolyA Activation. Mol Cell. 2011;43:927–939. doi: 10.1016/j.molcel.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawai T, et al. PPAR-gamma agonist attenuates renal interstitial fibrosis and inflammation through reduction of TGF-beta. Lab Invest. 2009;89:47–58. doi: 10.1038/labinvest.2008.104. [DOI] [PubMed] [Google Scholar]
- 23.Morcos PA, Li Y, Jiang S. Vivo-Morpholinos: a non-peptide transporter delivers Morpholinos into a wide array of mouse tissues. Biotechniques. 2008;45:613–4. 616, 618. doi: 10.2144/000113005. passim. [DOI] [PubMed] [Google Scholar]
- 24.Bansal R, et al. Novel engineered targeted interferon-gamma blocks hepatic fibrogenesis in mice. Hepatology. 2011;54:586–96. doi: 10.1002/hep.24395. [DOI] [PubMed] [Google Scholar]
- 25.Du L, Gatti RA. Potential therapeutic applications of antisense morpholino oligonucleotides in modulation of splicing in primary immunodeficiency diseases. J Immunol Methods. 2011;365:1–7. doi: 10.1016/j.jim.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heinrich MC, et al. PDGFRA activating mutations in gastrointestinal stromal tumors. Science. 2003;299:708–10. doi: 10.1126/science.1079666. [DOI] [PubMed] [Google Scholar]
- 27.Milacic M, et al. Annotating cancer variants and anti-cancer therapeutics in reactome. Cancers (Basel) 2012;4:1180–211. doi: 10.3390/cancers4041180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Croft D, et al. The Reactome pathway knowledgebase. Nucleic Acids Res. 2014;42:D472–7. doi: 10.1093/nar/gkt1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mehlem A, Hagberg CE, Muhl L, Eriksson U, Falkevall A. Imaging of neutral lipids by oil red O for analyzing the metabolic status in health and disease. Nat Protoc. 2013;8:1149–54. doi: 10.1038/nprot.2013.055. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.