Abstract
Hypertension is a major risk factor of cardiovascular diseases and a most important health problem in developed countries. Investigations on pathophysiology of hypertension have been based on gene products from coding region that occupies only about 1% of total genome region. On the other hand, non-coding region that occupies almost 99% of human genome has been regarded as “junk” for a long time and went unnoticed until these days. But recently, it turned out that non-coding region is extensively transcribed to non-coding RNAs and has various functions. This review highlights recent updates on the significance of non-coding RNAs such as micro RNAs and long non-coding RNAs (lncRNAs) on the pathogenesis of hypertension, also providing an introduction to basic biology of non-coding RNAs. For example, microRNAs are associated with hypertension via neuro-fumoral factor, sympathetic nerve activity, ion transporters in kidneys, endothelial function, vascular smooth muscle phenotype transformation, or communication between cells. Although reports of lncRNAs on pathogenesis of hypertension are scarce at the moment, new lncRNAs in relation to hypertension are being discovered at a rapid pace owing to novel techniques such as microarray or next-generation sequencing. In the clinical settings, clinical use of non-coding RNAs in identifying cardiovascular risks or developing novel tools for treating hypertension such as molecular decoy or mimicks is promising, although improvement in chemical modification or drug delivery system is necessary.
Keywords: Biomarker, cardiovascular disease, hypertension, lncRNA, long non-coding RNA, microRNA, miRNA, therapy
1. Introduction
Hypertension is a common but one of the most important health problems, because it is a major risk factor for many cardiovascular diseases. So it is very important to prevent, diagnose early and treat hypertension and its complications. Hypertension is a multifactorial disease involving complex interplay of genetic and environmental factors. As a pathogenesis of hypertension, neuro-fumoral factors, such as renin-angiotensin-aldosteron system, sympathetic nervous system, or ion transporters in kidneys and other tissues, have been extensively investigated for a long time. A genetic mutations and abnormal synthesis of transcription factors, regulating synthesis of protein factors mentioned above according to extracellular environmental changes, have drawn attention of researchers.
But it has been recognized from early days that only 1.5% of human genome is coding sequence [1] (exon), and other non-protein-coding DNA sequences are considered as “junk”. But a large part of these non-protein-coding DNA sequences has turned out to be subjected to transcription into introns, retrotransposons, and non-coding RNAs (ncRNAs) that lead to degradation of messenger RNAs (mRNAs) and/or inhibition of translation [2, 3] or other types of regulations of gene expressions. In addition, it has also recently become evident that these transcripts without being translated into proteins are overwhelming messenger RNAs in quantity and quality, owing to innovative techniques such as microarray or next-generation sequencing. Recent studies report the possibility that these functional sequences encompass a larger proportion of the human genome than previously thought, and RNA transcripts from these functional sequences are reported to be detected from 75% of the genome according to the recent study [4].
In the small cosmos of the cell of our body, contrary to “visible” proteins, “invisible” ncRNAs, so to speak, are often referred to as the “Dark Matter” of the genome [5]. It also plays an important role in the quantity and quality in regulating many gene expressions as fine tuner [6-8]. So it is very important to know the regulating mechanism on neuro-fumoral factors or genetic predisposition to hypertension by the “Dark Matter” from vast majority of the genome.
The relevance of the ncRNAs to various human diseases has been reported recently [9-12]. Of these reports on the relation between ncRNAs and cardiovascular diseases or hypertension, most of the reports were on microRNAs (miRNAs), but reports on long ncRNAs (lncRNAs), another member of ncRNAs, in relation to hypertension or cardiovascular diseases are limited in number at present [13-15]. So in this review, miRNAs that may have relation with hypertension or cardiovascular diseases will be reviewed mainly on recent interesting updates, besides the lncRNAs (including transcript of retrotransposon family) that may take part in the pathogenesis or progression of complications in hypertensive patients, because they might increase importance hereafter in understanding the pathogenesis or target organ damage of hypertension. Further we will discuss their potential of becoming a tool for identifying cardiovascular risk or novel therapeutic method in the near future.
1.1. Classification of ncRNAs
Non-coding RNAs can be classified into three types according to their size (nucleotides, nt); small (about 20 nt), intermediate-sized (20-200 nt) and long (>200 nt) (Table 1). Conventionally, ncRNAs other than long ncRNAs are considered small ncRNAs [16]. Small ncRNAs are widely studied, which contain small interfering RNAs (siRNAs), micro RNAs (miRNAs), PIWI-interacting RNAs (piRNAs), and others. Generally speaking, siRNAs are made from exogenous double strand RNAs by virus infection, artificial transfection, etc. and piRNAs are specifically expressed in reproductive cells. Intermediate-sized ncRNAs contain small nucleolar RNAs (snoRNAs) that modify ribosome RNA (rRNA) and small nuclear RNAs (snRNAs) that take part in splicing in the classical protein synthesis pathway. Long non-coding RNAs (lncRNAs) are gaining more interest and studies seem to progress rapidly. LncRNAs contain homeobox transcript antisense RNA (HOTAIR) that takes part in epigenetic regulation of transcription [17], and X-inactivation specific transcript (XIST) that inactivates X-chromosome. Expression of XIST is also regulated by TSIX (reverse spelling of Xist), antisense lncRNA transcribed from antisense strand of XIST [18].
Table 1.
Representative non-coding RNAs and their functions.
| Size | Name | Functions |
|---|---|---|
| Small (~20 nt) | miRNAs siRNA piRNAs |
Degradation or translation repression of target mRNAs Degradation of target mRNAs Repression of reproductive cell-specific gene expression |
| Intermediate (20~200 nt) | snoRNAs snRNAs |
rRNA modification RNA splicing |
| Long (200 nt~) | HOTAIR XIST TSIX |
Maintaining inactive chromatin as scaffold by repressing genes including HOXD genes X-chromosome inactivation Repression of XIST (antisense transcript of XIST) |
1.2. Hypertension and miRNAs
1.2.1. The Biology of microRNAs
It is considered that about 2000 human microRNAs (miRNAs) exist and control over 1/3 of entire genes including development, differetiation, cell growth, neoplastic transformation and so on.
Mature miRNAs are small non-coding single stranded RNAs of 18-24 nt, transcribed from intronic or exonic regions of protein-coding gene or from intergenic regions by promorters, and regulate protein-coding gene expressions. As shown in Fig. (1), miRNAs are transcribed by RNA polymerase II in the nucleus into primary miRNAs (pri-miRNAs). The pri-miRNAs are folded into a hairpin shape, then capped, polyadenylated, and cleaved by the enzyme complex made of RNase III endonuclease Drosha and dsRNA-binding protein Pasha into preccursor miRNAs (pre-miRNAs). Subsequently, they are transported into cytoplasm by the mediation of Ran-GTP and exportin-5, and processed by another RNase III, Dicer, into a short (25-25 nt) transient double strand RNA. Then, it is included in a protein complex called RISC (RNA-induced silencing complex), where passenger strand of the miRNA is degraded by the Argonaute protein that forms the main part of RISC. This mature miRNA binds to complementary 3’ untranslated region (3’-UTR) in the target mRNA and blocks target gene expression.
Fig. (1).
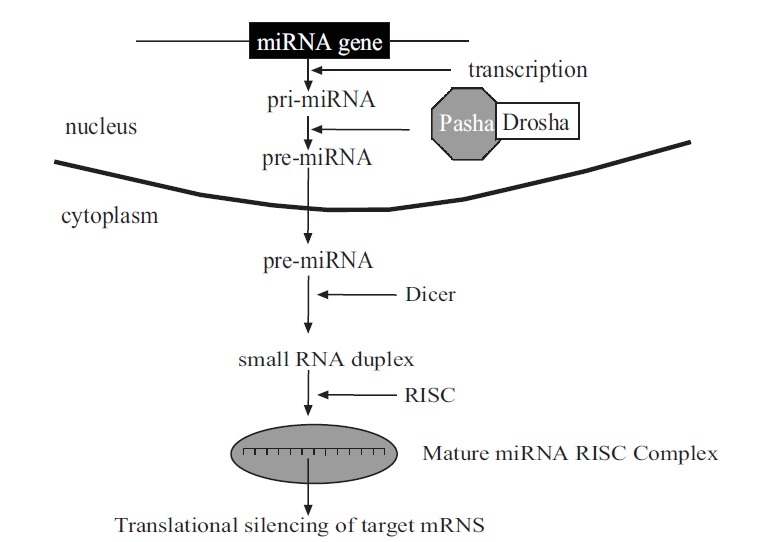
Biogenesis and mechanism of action of miRNAs. Pri-miRNA: primary miRNA. Pre-miRNA: precursor miRNA. Drosha: RNAase III endonuclease. Pasha: dsRNA binding protein. Dicer: RNAase III. RISC: RNA-induced silencing complex.
1.2.2. MicroRNAs and Complications of Hypertension
Abnormal miRNAs expression and function have been under intense investigation in relation to pathogenesis or target organ damages of hypertension, and we would like to refer details to reviews that report their relationship with hypertension [19, 20], cardiovascular diseases [21-27] and vascular diseases [28-31]. Other clinical and experimental studies report the miRNAs’ participation in the complications of hypertension such as, atherosclerosis [32-35], acute myocardial infarction [36-40] or predicting major adverse cardiac events after PCI [41], cardiac hypertrophy [42-44] or fibrosis [45-48], heart failure [49-51], atrial fibrillation [52] or other arrhythmias [51], stroke [53-59], intracranial aneurysm [60], aortic aneurysm [61], PAD [62-65], renal dysfunction [66-74], metabolic syndrome [75, 76], and target organ damage [77, 78].
1.2.3. MicroRNAs and Renin-Angiotensin-Aldosterone System
MicroRNAs are also related with the pathogenesis of hypertension. Renin-angiotensin-aldosterone system has an important role in blood pressure control. SNPs in miRNA (miR-765) binding site of RAAS genes are reported to relate with abnormal blood pressure control [79]. SNP in miRNA binding site of arginine vasopressin 1A receptor gene (AVPR1A SNP rs11174811) was associated with increased blood pressure, whereas SNPs in bradykinin 2 gene (BDKRB2 SNPs rs5225 and rs2069591) and thromboxane A2 receptor gene (TBXA2R SNP rs13306046) were associated with decreased blood pressure. It is strange that increased expression of thromboxan A2 receptor caused by the SNP in miRNA binding site lowered the blood pressure. The authors have explained this phenomenon by statistical artifact due to the low minor allele frequency of this SNP and resulting low number of homozygous carrier of the minor T-allele. Interestingly, they also reported that increased expression of mineralcorticoid receptor (NR3C2) by SNP in miRNA binding site increased plasma levels of VWF and FVIII and associated with increased risk of myocardial infarction in the carriers of the G-allele of the SNP rs5534. However, this SNP was not associated with an increase in blood pressure. Association between angiotensin II (AngII) type-1 receptor gene (AGTR1) A1166C polymorphysm and miR-155 has also been reported in young untreated hypertensives [80]. AGTR1 protein expression was positively correlated with systolic and diastolic blood pressure and negatively correlated with miR-155 expression level. According to their report, TGF-β1 was negatively correlated with miR-155 and positively with AGTR1 protein expression, suggesting the modulator role of TGF-β1 in the interplay between miR-155 and AGTR1 protein expression. But in another report, although AGTR1 protein expression was negatively correlated with miR-155 expression level, relation between miR-155 and TGF-β1 was not identified [81]. Contribution of enhanced renin synthesis induced by lower miRNA-181a in sympathetically mediated activation of the intrarenal renin-angiotensin-aldosteron system and blood pressure elevation has been reported in genetically hypertensive mice [82]. In another study with kidneys of white European subjects, miRNA, has-miR-181a and has-miR-663 (Homo Sapiens) were reported to be able to explain the elevation in intrarenal renin mRNA [83]. Angiotensinogen and angiotensin converting enzyme 1 gene expression, following AngII type-1 receptor activation by AngII, was reported to be negatively regulated by miR-483-3p in vascular sooth muscles of humans and rodents. It is suggested that AT1R-regulated miR-483-3p has the potential role as a negative regulator of RAS components [84]. MiR-132 and miR-212 are reported to be increased in AngII-induced hypertension in human samples [85]. This may be regulated through Gαq-ERK1/2 activation because another activator of Gαq-ERK1/2 signaling, endothelin-1, has also upregulated these miRNAs. In another report, AngII also upregulated the levels of miR-132 and other miRNAs in rat cardiac fibroblasts [86]. MMP9 was the target for miR-132, MMP16 for miR-146b, and TIMP3 for miR-181b respectively, and potential role of these AngII-induced miRNAs in cardiac fibrosis was suggested. Association between increased blood pressure with SNP located in 3’-UTR in human angiotensinogen gene has also been reported, suggesting the role of miRNAs in blood pressure control through angiotensinogen gene expression regulation [87]. Up-regulation of miRNA-21 expression by ANGII was observed, and up-regulation of miRNA-21 intracellular levels increased aldosterone secretion but not cortisol in the human adrenocortical cell line [88]. Authors are speculating that abnormal aldosterone secretion by altered miR-21 expression may lead to hypertension and primary aldosteronism. It was reported that miR-24 can dicer-dependently bind to the 3' untranslated region of CYP11B1 (11β-hydroxylase) and CYP11B2 (aldosterone synthase) genes and can post-transcriptionally regulate the expression of these two genes to modulate cortisol and aldosterone production [89]. Expression of above-mentioed mineral corticoid receptor gene, NR3C2, is also reported to be supressed by miR-124 and miR-135a, suggesting that these miRNAs could participate in the regulation of renin-angiotensin-aldosterone system and blood pressure regulation [90].
1.2.4. MicroRNAs and other Humoral Factors
Increased release of catecholamine from adrenal medulla leads to blood pressure elevation. Chromogranin A (CHGA) is a precursor of the catecholamine release-inhibitor catestatin. Has-miR-637 was reported to cause altered formation and an incorrect trafficking of the chromaffin granules through impaired acidification of intracellular vesicle such as catecholamine storage vesicles of the adrenal medulla and impaired CHGA processing [91]. As mentioned above, cortisol, another important steroid hormone other than aldosterone from adrenal cortex in regulating blood pressure, is regulated by miRNA [89]. To protect intracellular mineral corticoid receptor from excessive activation of cortisol of high intracellular concentration, intracellular cortisol is degraded by 11β-hydroxysteroid dehydrogenase type 2 (11β-HSD2) into inactive cortisone. A decreased activity of 11β-HSD2 causes salt-sensitive hypertension. Different expressions of miRNAs were revealed in kidneys of 2 models between each high and low 11β-HSD2 activity: Splague-Dawley rats with low and Wister rats with high 11β-HSD2 activity, and with and without uninephrectomy [92]. The association of variant (rs5068) in the 3’-UTR of NPPA, gene encoding atrial natriuretic peptide (ANP), with blood pressure has been reported by the population study [93]. The same group reported that mir-425 silences NPPA mRNA and regulates ANP production on the basis of that rs5068 genotype (AG vs. AA) [94]. ANP levels were up to 50% higher in AG individuals than in AA individuals, but B-type natriuretic peptide levels did not differ by rs5068 genotype. They showed miR-425 to bind for the A but not the G allele of rs5068 and miR-425 silenced NPPA mRNA in an allele-specific manner. MiR-21 was shown to contract human vascular smooth muscle cells (HVSMC) and ANP blocked miR-21-induced HVSMC contraction [95]. The authors reported that targets of miR-21 include cofilin-2 (inhibitor of actin polimerization) and myosin phosphatase and Rho interacting protein (M-RIP).
1.2.5. MicroRNAs and Endothelial Dysfunction
Endothelial dysfunction contributes to the pathogenesis of hypertension such as impaired vasodilatation, release of inflammatory and procoagulant mediators, and defective angiogenesis (capillary rarefaction). Endothelial nitric oxide synthase (eNOS) expression and NO production were reported to be decreased by miR-155, and inflammatory cytokines such as TNF-α inreased miR-155 expression [96]. Interestingly, this TNF-α-induced up-regulation of miR-155 was attenuated by simvastatin through interfering mevalonate-geranylgeranyl-pyrophosphate-RhoA signaling pathway. Tumor suppressive miRNA, miR-505, targets FGF18, a proangiogenetic factor, and impairs the migration and tube formation of endothelial cells [97]. Aerobic exercise training (ET) may prevent microvascular abnormalities in hypertension. ET repaired capillary rarefaction, restored eNOS, VEGF (target of miR16), Bcl-2 (target of miR21) levels, and reduced miR-16, -21, but increased miR-126 (represses miR-16, -21) [98]. Recently, importance of increasing endothelial tip cell formation and arteriolar branching by miR-30a expression and downregulation of endothelial DLL4, a target of miR-30a and an inhibitor of tip cell formation, regarding the microvascular rarefaction process in hypertension were reported [99]. Networks of miRNAs in endothelium control the response to various atherosclerotic stimuli such as low shear stress, oxidized LDL, and AngII by modulating of inflammatory transcription factors or apoptosis of endothelial cells [100]. Interestingly, miR-126 is transferred from apoptotic endothelial cells via microvesicles, which allows cell-to-cell communication and reduces atherosclerosis and suppresses chemokine CXCL12.
1.2.6. MicroRNAs and Vascular Smooth Muscle Cells (VSMCs)
Resistance in small arteries determines peripheral vascular resistance and also regulates tissue perfusion and systemic blood pressure. Sustained hypertension causes vascular remodeling through proliferation and phenotype conversion of VSMCs. So, factors influencing contractility, proliferation or phenotype of VSMCs are important in understanding pathogenesis and therapy of hypertension. Involvement of miRNAs in arterial remodeling is reviewed [101]. Phenotype conversion of VSMCs from contractile type to synthetic type plays an important role in arterial remodeling. Phenotype-specific gene expression in synthetic and contrctile VSMCs is also mediated and controlled by miRNAs network [100-107]. Nebivolol, a β1 blocker andβ3 activator, may prevent arterial dysfunction and remodeling by inhibiting miR-320 that targets insulin growth factor-1 receptor (IGR1R), and by overexpressing miR-26b and miR-21 that target phosphatase and tensin homolog on chromosome ten (PTEN) in Dahl Salt Sensitive (DSS) hypertensive rat model [103]. As mentioned above, nebivolol has also been reported to attenuate cardiac remodeling, hypertrophy and fibrosis through inducing miR-27a, -29a targeting Sp1 and miR–133a targeting Cdc42 in DSS hypertensive rat model [47]. The role of PTEN as a target of miR-21 in arterial remodeling is depicted in other reports (miR-143-FRA-1-miR-21 axis) [102]. In addition, endothelial cells and VSMCs may communicate, mediated by extracellular vesicle with miR-143 and -145 to control VSMC phenotypes [35], and forced expression of mir-145 may prove to be a promising strategy for stabilizing atheroscrerotic plaques [108].
1.2.7. MicroRNAs and other Pathophysiological Factors in Hypertension
Abnormal function of ion transporters in renal tubules is involved in pathophysiological processes of hypertension. With-no-lysine kinase-4 (WNK4) is a regulator of sodium reabsorption. WNK4 transcription decrease by epigenetic modulation following β2 adrenergic receptor stimulation is reported [109]. Suppression of human WNK4 expression by miR-296 is reported [110]. Endothelin-1 (ET-1) expression may be suppressed by miR-125a-5p and miR-125b-5p that target prepro-ET-1 mRNA [111]. Association of hypertension with oxidative stress [112], circadian clock [113], and cytomegalovirus infection via miRNAs [114] is also reported.
Relationship between vascular inflammation and hypertension has been discussed for a long time, and regulation of NF-κB signaling by miRNAs (negative feedback by miR-146a, m-181b, -10a and -155?, enhanced by miR-92a) is summarized by Cheng et al. [115].
1.2.8. MicroRNAs as Biomarkers and Therapeutic Modalities
Last but not the least, we would like to review the potential of miRNAs as marker of target organ damage and therapy by miRNAs, based on the pathophysiology of hypertension.
As mentioned above, endothelial cells and VSMCs seem to communicate by extracellular vesicle with miRNA [35]. Surprisingly, systemic circulating miRNAs are also discovered in plasma and many blood cells. These circulating miRNAs are surprisingly stable under various harsh conditions, because they are packaged in microparticles such as exosomes, microvesicles, and apoptotic bodies in complex with RNA-binding proteins such as Argonaute2 or with HDL to resist from degradation. Detailed role and mechanism of miRNAs in cell-to-cell communication are summarized in another review article [116]. These circulating miRNAs function as mediators of cell-to-cell communication and regulate gene expressions of remote target cells. Although many miRNAs are reported to associate with various cardiovascular diseases including complications related to hypertension and with pathophysiology of hypertension, reports on association between essential hypertension and circulating miRNA in human are scarce. Li et al. identified three miRNAs, hcmv-miR-UL112, let-7c, and mir-296-5p, in 127 hypertensive patients and 67 control subjects [114]. Intriguingly, one of these miRNAs was human cytome- galovirus (HCMV)-encoded miRNA (hcmv-miR-UL112), suggesting the possibility of involvement of HCMV in the pathogenesis of essential hypertension. They also reported elevation of HCMV titers in hypertensives. But the possibility of these miRNA as biomarkers of hypertension is uncertain because of interpatient variation and others.
Various miRNAs are involved in the pathogenesis of hypertension, so clinical application of miRNAs, miRNA-targeted or miRNA replacement therapies, is promising as a modality for treating hypertension. Several possible strategies have been introduced to suppress endogenous miRNAs effectively. Various chemically modified oligonucleotides, antagomirs or anti-miRs, have been introduced such as 2’-O-Methyl, 2’-Fluoro (F), 2’-O-MethoxyEthyl (MOE), Locked Nucleic Acid (LNA). Another progress has been made in the stability and delivery to target tissues of miRNAs by introducing cholesterol-conjugated (Chol) antagomirs. But, Thum et al. have reported a difference in the repression activity of between 3 antagomirs (anti-miR-21s) different in size and chemical modifications (22-mer Chol/2’-O-Methyl, 22-mer F/MOE, and 8-mer LNA) using the model of pressure overload-induced cardiac hypertrophy (TAC) [117]. The 8-mer had weaker repressing activity compared to other anti-miR-21s. Another strategy is to replace the decreased miRNA. In this case, miRNA mimicks are used by introducing synthetic double-stranded precursor-miRNA molecules. In vivo administration of antagomir to miRNA targeting Chromogranin A (CHGA) has been reported. CHGA is a protein crucial for biogenesis and trafficking of the chromaffin granules in neurons and neuroendocrine tissues such as adrenal medulla, and influences catecholamine secretion or blood pressure homeostasis centrally and peripherally [91] as mentioned above. Friese et al. reported that CHGA protein is increased in the adrenal gland and plasma but decreased in the brainstem of spontaneously hypertensive rats (SHR), compared to Wister-Kyoto rats [118]. They also demonstrated that in vivo i.p. administration of miR-22 antagomir (LNA oligonucleotide) to SHR caused blood pressure reduction (~18 mmHg in both SBP and DBP). Rhee et al. showed modified miR-30a-based short hairpin RNA (shRNAmir) driven by the cytomegalovirus effectively silenced vascular smooth muscle-specific noncardiac form of the L-type calcium channel [119].
Another problem in applying miRNAs to clinical use is that multiple miRNAs may target one gene and one miRNA may target multiple genes and they constitute complicated miRNA networks. And miRNA may suppress or increase target gene(s) expression. These facts may cause unexpected side effects, but on the other hand, may cover multiple etiologies of hypertension by modulation of single miRNA and unexpected favorable effect. Therefore, careful observation and long-term follow up of side effects are needed in putting them into clinical use.
Some reviews are available on the therapy with miRNAs regarding other hypertention-related cardiovascular diseases [19, 120].
1.3. Hypertension and Long Non-Coding RNAs
Reports on lncRNAs that studied the relation with hypertension are scarce at present, but explosive increase of interest in that field can be expected.
1.3.1. The Biology of Long Non-Coding RNAs
LncRNAs are greater than 200 nt, mostly transcribed by RNA polymerase II, are 3’ polyadenylated, 5’ capped, and spliced but not translated into proteins. LncRNAs are classified according to their relative genomic locations with coding regions: intergenic, intronic, sense, and antisence lncRNAs (Fig. 2). Intergenic lncRNAs are also called as lincRNAs (large intergenic/intervening non-coding RNAs). LncRNAs encompass numerous biological functions such as transcription, translation, splicing, imprinting, cell cycle, development, heat shock response, etc. LncRNAs regulate transcription positively and negatively, but molecular mechanism is not fully understood. Wang et al. classified molecular mechanisms into four archetypes: ArchetypeI: Signals, ArchetypeII: Decoys, ArchetypeIII: Guides, ArchetypeIV: Scaffolds [121]. Combination of these archetypes also can be seen.
Fig. (2).
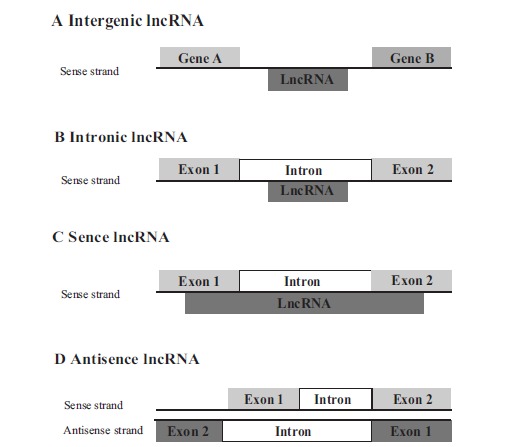
Classification of lncRNAs in relation to the location of protein-coding genes. (A) Intergenic lncRNA may be transcribed from both coding and template strands by promotors. (B) Intronic lncRNAs are transcribed from introns of protein-coding genes. (C) Sense lncRNAs are transcribed from the sense strand of protein coding genes, overlapping with exon, intron or entire sequence of protein coding genes. (D) Antisense lncRNAs are transcribed from the antisense strand of the protein-coding genes, overlapping with exon, intron or entire sequence of protein coding genes.
1.3.2. Hypertension and Long Non-Coding RNAs
Gopalakrishnan et al. showed differentially expressed lncRNAs between Dahl salt-sensitive (S) versus Dahl salt-resistant (R) and Dahl salt-sensitive versus spontaneously hypertensive rat (SHR) comparisons, respectively [122]. They identified 3273 transcripts as rat lncRNAs, and detected differential expression of 273 lncRNAs between S versus R rats. 20 mRNAs were identified as associated mRNAs to 21 differetially expressed lncRNAs of these differentially expressed lncRNAs in comparison of Dahl S versus R rats. They suggested 4 target lncRNA-mRNA-associated genes, Ankyrin Repeat and SOCS Box-Containing 3 (Asb3), cation transport regulator homolog 2 (Chac2), peroxisomal membrane 11B (Pex11b), and Sp5 transcription factor (Sp5) as candidate determinants of blood pressure in the comparison of S versus R rats. In the S (versus R comparison), lncRNAs, targeting Asb3, Chac2, and Pex11b genes respectively, were increased, and protein products of these target genes were decreased. On the contrary, lncRNA that targets Sp5 gene was decreased. All the protein expression patterns were inversely correlated to the lncRNA expression patterns. Little is known about Asb3, but Chung et al. reported that Asb3 is a negative regulator of cellular responses to TNF-α by ubiquitination and proteasome-mediated degradation of tumor necrosis factor receptor II (TNF-R2) [123]. So S rat may have increased insulin resistant and inflammation state due to decreased TNF-R2 degradation. Chac protein is a proapoptic protein induced during endoplasmic reticulum stress. Chac proteins cleave glutathione and overpexpression of Chac proteins is reported to lead to glutathione depletion and enhanced apoptosis [124]. Significance of decrease of Chac proteins in S rat is unclear but may have relation to atherosclerotic or proliferative tendency of VSMC. The Pex11b, family of peroxisomal membrane proteins (PMPs), regulates peroxisome size and number in both higher and lower eukaryotes [125]. Peroxisomes play an important role in a number of essential metabolic pathways, including in humans, the biosynthesis of ether phospholipids and bile acids, β-oxidation of fatty acids, and the detoxification of glyoxylate and of reactive oxygen species [126, 127]. Pex11b is also reported to correlate with fat metabolism, insulin, and blood sugar-related traits in SHR [128]. Decrease of Pex11b and increase of Sp5 protein may be related to metabolic futures of S rat [122]. Nagase et al. reported the transcript of retrotransposon family, REPT1, markedly expressed in proximal tubules of Dahl salt-sensitive rats compared with salt-resistant rats, and this is transcribed by polymerase II and has polyA tail. They also reported that this transcript contained a sequence highly homologous to the 3’-noncoding region of the nicotinic acetylcholine receptor α7 gene without any coding region of it. They also reported that REPT1 contained an element whose expression was modulated by nerve growth factor (NGF). They suggested that REPT1 transcript might interfere with the function of Na transporters in the proximal tubules [129].
Leung et al. analyzed and identified all protein-coding and lncRNAs regulated by AngII in rat VSMCs using transcriptome and epigenome profiling [130]. They showed that lncRNAs serve as promoters for distally neighboring miRNAs implicated in VSMCs proliferation by the fact that Lnc-Ang362, one of the AngII-upregulated lncRNAs, is located in the neighboring proximal position to miR-221 and miR-222, implicated in cell proliferation, and these 2 miRNAs are cotranscribed as part of the host transcript of Lnc-Ang362 enhanced by AngII. They also reported that knockdown of Lnc-Ang362 produced reduction in the expression of Mcm7, member of the minichromosome maitenance protein complex, which contains DNA helicase activity necessary for initiation of DNA replication and cell cycle progression.
Regulation of eNOS by overlapping antisense RNA transcript is also reported. Robb et al. showed that expression of eNOS is regulated post-transcriptionally by antisense mRNA (sONE) derived from a transcription unit (NOS3AS) on the opposite DNA strand from that of eNOS transcription (NOS3) at human chromosome 7q36 [131]. Ho et al. of their group recently reported that post-transcriptional inhibition of NOS3 expression by sONE is protected by heterogenous nuclear ribonucleoprotein E1 (hnRNP E1) [132].
Regulation of nuclear receptor (NR) function by lncRNAs seems promising and is reviewed [133]. Lanz et al. reported that the steroid receptor RNA activator (SRA) binds and activates several NRs [134], and the secondary structure of total SRA transcript was determined by Novikova et al. [135]. SRA may act as a scaffold for other co-regulatory proteins. Growth arrest-specific 5 (GAS5), another lncRNA regulating NRs activities, binds glucocorticoid receptor (GR) and negatively regulates its transcriptional activity, acting as a molecular decoy [136]. As GR has similar responsive element with mineral corticoid receptor (MR), GAS5 may regulate MR. Modulation of expression of NRs by these lncRNAs seems attractive but clinical implication of these lncRNAs is limited to tumors such as breast and prostate cancers so far.
Investigations on the effect of lncRNAs on NF-κB in the context of blood vessel inflammation are just beginning, but several lncRNAs are reported regarding this issue, such as lncRNA-Cox2, PACER, Lethe, etc. [115]. As for these lncRNAs, association between lncRNAs and blood pressure has not been investigated so far and is open for further study.
Clinically, Vausort et al. showed that expression of lncRNAs in peripheral blood cells is regulated after myocardial infarction, and is correlated with cardiovascular risk factors and prognosis [137]. They reported that lncRNAs, cyclin-dependent kinase inhibitor 2B antisense RNA 1 (ANRIL), potassium voltage-gated channel, KQT-like subfamily, member 1 opposite strand/antisense transcript 1 (KCNQ1OT1), myocardial infarction associated transcript (MIAT), and metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) were significant univariable predictors of left ventricular dysfunction, and ANRIL and KCNQ1OT1 improved the prediction of left ventricular dysfunction in multivariable analyses. They also reported that levels of ANRIL were associated with hypertension.
2. FUTURE PERSPECTIVES
We are just beginning to study the new frontier of the role of ncRNAs, especially lncRNAs, in hypertension pathogenesis, and unknown mechanism can be added in the near future. Thus, ncRNAs may represent new and promising biomarkers and therapeutic targets in diagnosing, treating and preventing hypertension and its complications. But ncRNA-based therapy may have advantages and disadvantages because these RNAs target multiple genes.
Another problem is that investigations showing relation with blood pressure are limited in number. As a biomarker, circulating miRNAs are promising because of their stability but further investigations are expected such as combination of miRNAs. As a therapeutic method, improvements in delivery and modification in antagomirs or mimicks for replacing repressed miRNAs. For understanding mechanism of hypertension, further investigations into networks of ncRNAs or cell-to-cell communication of miRNAs by exosomes or microvesicles may be useful. Studies on lncRNAs and hypertension are only beginning but explosive increase of new findings in this field can be expected. As a new therapeutic method, getting aptamer by systematic evolution of ligands by exponential enrichment (SELEX) method or decoy nucleic acid may be of use.
ACKNOWLEDGEMENTS
Declared none.
CONFLICT OF INTEREST
No conflicts of interest, financial or otherwise, are declared by the author.
REFERENCES
- 1.Lander E.S., Linton L.M., Birren B., et al. International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Carninci P., Hayashizaki Y. Noncoding RNA transcription beyond annotated genes. Curr. Opin. Genet. Dev. 2007;17(2):139–144. doi: 10.1016/j.gde.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 3.Claverie J.M. Fewer genes, more noncoding RNA. Science. 2005;309(5740):1529–1530. doi: 10.1126/science.1116800. [DOI] [PubMed] [Google Scholar]
- 4.Kellis M., Wold B., Snyder M.P., et al. Defining functional DNA elements in the human genome. Proc. Natl. Acad. Sci. USA. 2014;111(17):6131–6138. doi: 10.1073/pnas.1318948111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ponting C.P., Belgard T.G. Transcribed dark matter: meaning or myth? Hum. Mol. Genet. 2010;19(R2):R162–R168. doi: 10.1093/hmg/ddq362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mercer T.R., Dinger M.E., Mattick J.S. Long non-coding RNAs:insights into functions. Nat. Rev. Genet. 2009;10(3):155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 7.Guttman M., Rinn J.L. Modular regulatory principles 0f large ono-coding RNAs. Nature. 2012;482(7385):339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krishnan J., Mishra R.K. Emerging trends of non-coding RNAs in gene activation. FEBS J. 2014;281(1):34–45. doi: 10.1111/febs.12578. [DOI] [PubMed] [Google Scholar]
- 9.Esteller M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011;12(12):861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 10.Wapinski O., Chang H.Y. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21(6):354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 11.Sun M., Kraus W.L. Long noncoding RNAs: New “Links” between gene expression and cellular outcomes in endocrinology. Mol. Endocrinol. 2013;27(9):1390–1402. doi: 10.1210/me.2013-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mattick J.S. The central role of RNA in human development and cognition. FEBS Lett. 2011;585(11):1600–1616. doi: 10.1016/j.febslet.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 13.Peters T., Schroen B. Missing links in cardiology: long non-coding RNAs enter the arena. Pflugers Arch. 2014;466(6):1177–1187. doi: 10.1007/s00424-014-1479-1. [DOI] [PubMed] [Google Scholar]
- 14.Papait R., Kunderfranco P., Stirparo G.G., Latronico M.V., Condorelli G. Long non-coding RNA: A new player of heart failure? J. Cardiovasc. Transl. Res. 2013;6(6):876–883. doi: 10.1007/s12265-013-9488-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schonrock N., Harvey R.P., Mattick J.S. Long non-coding RNAs in cardiac development and pathophysiology. Circ. Res. 2012;111(10):1349–1362. doi: 10.1161/CIRCRESAHA.112.268953. [DOI] [PubMed] [Google Scholar]
- 16.Ma L., Bajic V.B., Zang Z. On the classification of long non-coding RNAs. RNA Biol. 2013;10(6):925–933. doi: 10.4161/rna.24604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rinn J.L., Kertesz M., Wang J.K., et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao J., Sun B.K., Erwin J.A., Song J.J., Lee J.T. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322(5902):750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bátkai S., Thum T. MicroRNAs in hypertension: mechanisms and therapeutic targets. Curr. Hypertens. Rep. 2012;14(1):79–87. doi: 10.1007/s11906-011-0235-6. [DOI] [PubMed] [Google Scholar]
- 20.Synetos A., Toutouzas K., Stathogiannis K., et al. MicroRNAs in arterial hypertension. Curr. Top. Med. Chem. 2013;13(13):1527–1532. doi: 10.2174/15680266113139990101. [DOI] [PubMed] [Google Scholar]
- 21.Zhang C. MicroRNAs: role in cardiovascular biology and disease. Clin. Sci. 2008;114(12):699–706. doi: 10.1042/CS20070211. [DOI] [PubMed] [Google Scholar]
- 22.Schroen B., Heymans S. MicroRNAs and beyond: the heart reveals its treasures. Hypertension. 2009;54(6):1189–1194. doi: 10.1161/HYPERTENSIONAHA.109.133942. [DOI] [PubMed] [Google Scholar]
- 23.Xu J., Zhao J., Evan G., Xiao C., Cheng Y., Xiao J. Circulating microRNAs: novel biomarkers for cardiovascular diseases. J Mol Med (Berl) 2012;90(8):865–875. doi: 10.1007/s00109-011-0840-5. [DOI] [PubMed] [Google Scholar]
- 24.Creemers E.E., Tijsen A.J., Pinto Y.M. Circulating microRNAs: novel biomarkers and extracellular communicators in cardiovascular disease? Circ. Res. 2012;110(3):483–495. doi: 10.1161/CIRCRESAHA.111.247452. [DOI] [PubMed] [Google Scholar]
- 25.Tijsen A.J., Pinto Y.M., Creemers E.E. Circulating microRNAs as diagnostic biomarkers for cardiovascular diseases. Am. J. Physiol. Heart Circ. Physiol. 2012;303(9):H1085–H1095. doi: 10.1152/ajpheart.00191.2012. [DOI] [PubMed] [Google Scholar]
- 26.van Empel V.P., De Windt L.J., da Costa Martins P.A. Circulating miRNAs: reflecting or affecting cardiovascular disease? Curr. Hypertens. Rep. 2012;14(6):498–509. doi: 10.1007/s11906-012-0310-7. [DOI] [PubMed] [Google Scholar]
- 27.Quiat D., Olson E.N. MicroRNAs in cardiovascular disease: from pathogenesis to prevention and treatment. J. Clin. Invest. 2013;123(1):11–18. doi: 10.1172/JCI62876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sen C.K., Gordillo G.M., Khanna S., Roy S. Micromanaging vascular biology: tiny microRNAs play big band. J. Vasc. Res. 2009;46(6):527–540. doi: 10.1159/000226221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang C. MicroRNAs in vascular biology and vascular disease. J. Cardiovasc. Transl. Res. 2010;3(3):235–240. doi: 10.1007/s12265-010-9164-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qin S., Zhang C. MicroRNAs in vascular disease. J. Cardiovasc. Pharmacol. 2011;57(1):8–12. doi: 10.1097/FJC.0b013e318203759b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jamaluddin M.S., Weakley S.M., Zhang L., et al. miRNAs: roles and clinical applications in vascular disease. Expert Rev. Mol. Diagn. 2011;11(1):79–89. doi: 10.1586/erm.10.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Santovito D., Mezzetti A., Cipollone F. MicroRNAs and atherosclerosis: new actors for an old movie. Nutr. Metab. Cardiovasc. Dis. 2012;22(11):937–943. doi: 10.1016/j.numecd.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 33.Santovito D., Mandolini C., Marcantonio P., et al. Overexpression of microRNA-145 in atherosclerotic plaques from hypertensive patients. Expert Opin. Ther. Targets. 2013;17(3):217–223. doi: 10.1517/14728222.2013.745512. [DOI] [PubMed] [Google Scholar]
- 34.Sun X., Belkin N., Feinberg M.W. Endothelial microRNAs and atherosclerosis. Curr. Atheroscler. Rep. 2013;15(12):372. doi: 10.1007/s11883-013-0372-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hergenreider E., Heydt S., Tréguer K., et al. Athero-protective communication between endothelial cells and smooth muscle cells through miRNAs. Nat. Cell Biol. 2012;14(3):249–256. doi: 10.1038/ncb2441. [DOI] [PubMed] [Google Scholar]
- 36.Wang G.K., Zhu J.Q., Zhang J.T., et al. Circulating microRNA: a novel potential biomarker for early diagnosis of acute myocardial infarction in humans. Eur. Heart J. 2010;31(6):659–666. doi: 10.1093/eurheartj/ehq013. [DOI] [PubMed] [Google Scholar]
- 37.Kuwabara Y., Ono K., Horie T., et al. Increased microRNA-1 and microRNA-133a levels in serum of patients with cardiovascular disease indicate myocardial damage. Circ Cardiovasc Genet. 2011;4(4):446–454. doi: 10.1161/CIRCGENETICS.110.958975. [DOI] [PubMed] [Google Scholar]
- 38.Long G., Wang F., Duan Q., et al. Human circulating microRNA-1 and microRNA-126 as potential novel indicators for acute myocardial infarction. Int. J. Biol. Sci. 2012;8(6):811–818. doi: 10.7150/ijbs.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Long G., Wang F., Duan Q., et al. Circulating miR-30a, miR-195 and let-7b associated with acute myocardial infarction. PLoS One. 2012;7(12):e50926. doi: 10.1371/journal.pone.0050926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang F., Long G., Zhao C., et al. Plasma microRNA-133a is a new marker for both acute myocardial infarction and underlying coronary artery stenosis. J. Transl. Med. 2013;11:222. doi: 10.1186/1479-5876-11-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu X.Y., Chen J.Y., Zheng Z.W., et al. Plasma miR-126 as a potential marker predicting major adverse cardiac events in dual antiplatelet-treated patients after percutaneous coronary intervention. EuroIntervention. 2013;9(5):546–554. doi: 10.4244/EIJV9I5A90. [DOI] [PubMed] [Google Scholar]
- 42.Cheng Y., Ji R., Yue J., et al. MicroRNAs are aberrantly expressed in hypertrophic heart: do they play a role in cardiac hypertrophy? Am. J. Pathol. 2007;170(6):1831–1840. doi: 10.2353/ajpath.2007.061170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dong D.L., Chen C., Huo R., et al. Reciprocal repression between microRNA-133 and calcineurin regulates cardiac hypertrophy: a novel mechanism for progressive cardiac hypertrophy. Hypertension. 2010;55(4):946–952. doi: 10.1161/HYPERTENSIONAHA.109.139519. [DOI] [PubMed] [Google Scholar]
- 44.Heymans S., Corsten M.F., Verhesen W., et al. Macrophage microRNA-155 promotes cardiac hypertrophy and failure. Circulation. 2013;128(13):1420–1432. doi: 10.1161/CIRCULATIONAHA.112.001357. [DOI] [PubMed] [Google Scholar]
- 45.Bauersachs J. Regulation of myocardial fibrosis by MicroRNAs. J. Cardiovasc. Pharmacol. 2010;56(5):454–459. doi: 10.1097/FJC.0b013e3181ee81df. [DOI] [PubMed] [Google Scholar]
- 46.Castoldi G., Di Gioia C.R., Bombardi C., et al. MiR-133a regulates collagen 1A1: potential role of miR-133a in myocardial fibrosis in angiotensin II-dependent hypertension. J. Cell. Physiol. 2012;227(2):850–856. doi: 10.1002/jcp.22939. [DOI] [PubMed] [Google Scholar]
- 47.Ye H., Ling S., Castillo A.C., et al. Nebivolol induces distinct changes in profibrosis microRNA expression compared with atenolol, in salt-sensitive hypertensive rats. Hypertension. 2013;61(5):1008–1013. doi: 10.1161/HYPERTENSIONAHA.111.00892. [DOI] [PubMed] [Google Scholar]
- 48.Guo T.S., Zhang J., Mu J.J., et al. High-salt intake suppressed microRNA-133a expression in Dahl SS rat myocardium. Int. J. Mol. Sci. 2014;15(6):10794–10805. doi: 10.3390/ijms150610794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Montgomery R.L., Hullinger T.G., Semus H.M., et al. Therapeutic inhibition of miR-208a improves cardiac function and survival during heart failure. Circulation. 2011;124(14):1537–1547. doi: 10.1161/CIRCULATIONAHA.111.030932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dickinson B.A., Semus H.M., Montgomery R.L., et al. Plasma microRNAs serve as biomarkers of therapeutic efficacy and disease progression in hypertension-induced heart failure. Eur. J. Heart Fail. 2013;15(6):650–659. doi: 10.1093/eurjhf/hft018. [DOI] [PubMed] [Google Scholar]
- 51.Duygu B., Poels E.M., da Costa Martins P.A. Genetics and epigenetics of arrhythmia and heart failure. Front. Genet. 2013;4:219. doi: 10.3389/fgene.2013.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McManus D.D., Lin H., Tanriverdi K., et al. Relations between circulating microRNAs and atrial fibrillation: data from the Framingham Offspring Study. Heart Rhythm. 2014;11(4):663–669. doi: 10.1016/j.hrthm.2014.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rink C., Khanna S. MicroRNA in ischemic stroke etiology and pathology. Physiol. Genomics. 2011;43(10):521–528. doi: 10.1152/physiolgenomics.00158.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tan J.R., Koo Y.X., Kaur P., et al. microRNAs in stroke pathogenesis. Curr. Mol. Med. 2011;11(2):76–92. doi: 10.2174/156652411794859232. [DOI] [PubMed] [Google Scholar]
- 55.Long G., Wang F., Li H., et al. Circulating miR-30a, miR-126 and let-7b as biomarker for ischemic stroke in humans. BMC Neurol. 2013;13:178. doi: 10.1186/1471-2377-13-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koutsis G., Siasos G., Spengos K. The emerging role of microRNA in stroke. Curr. Top. Med. Chem. 2013;13(13):1573–1588. doi: 10.2174/15680266113139990106. [DOI] [PubMed] [Google Scholar]
- 57.Tan J.R., Tan K.S., Koo Y.X., et al. Blood microRNAs in Low or No Risk Ischemic Stroke Patients. Int. J. Mol. Sci. 2013;14(1):2072–2084. doi: 10.3390/ijms14012072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sepramaniam S., Tan J.R., Tan K.S., et al. Circulating microRNAs as biomarkers of acute stroke. Int. J. Mol. Sci. 2014;15(1):1418–1432. doi: 10.3390/ijms15011418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li W.Y., Jin J., Chen J., Guo Y., Tang J., Tan S. Circulating microRNAs as potential non-invasive biomarkers for the early detection of hypertension-related stroke. J. Hum. Hypertens. 2014;28(5):288–291. doi: 10.1038/jhh.2013.94. [DOI] [PubMed] [Google Scholar]
- 60.Li P., Zhang Q., Wu X., et al. Circulating microRNAs serve as novel biological markers for intracranial aneurysms. J. Am. Heart Assoc. 2014;3(5):e000972. doi: 10.1161/JAHA.114.000972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Albinsson S., Swärd K. Targeting smooth muscle microRNAs for therapeutic benefit in vascular disease. Pharmacol. Res. 2013;75:28–36. doi: 10.1016/j.phrs.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 62.Zhou X., Yuan P., He Y. Role of microRNAs in peripheral artery disease. Mol. Med. Rep. 2012;6(4):695–700. doi: 10.3892/mmr.2012.978. [review]. [DOI] [PubMed] [Google Scholar]
- 63.Imanishi T., Akasaka T. MicroRNAs in peripheral artery disease. Curr. Top. Med. Chem. 2013;13(13):1589–1595. doi: 10.2174/15680266113139990107. [DOI] [PubMed] [Google Scholar]
- 64.Kloos W., Vogel B., Blessing E. MiRNAs in peripheral artery disease - something gripping this way comes. Vasa. 2014;43(3):163–170. doi: 10.1024/0301-1526/a000345. [DOI] [PubMed] [Google Scholar]
- 65.Stather P.W., Sylvius N., Wild J.B., Choke E., Sayers R.D., Bown M.J. Differential microRNA expression profiles in peripheral arterial disease. Circ Cardiovasc Genet. 2013;6(5):490–497. doi: 10.1161/circgenetics.111.000053. [DOI] [PubMed] [Google Scholar]
- 66.Liang M., Liu Y., Mladinov D., et al. MicroRNA: a new frontier in kidney and blood pressure research. Am. J. Physiol. Renal Physiol. 2009;297(3):F553–F558. doi: 10.1152/ajprenal.00045.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang G., Kwan B.C., Lai F.M., et al. Intrarenal expression of miRNAs in patients with hypertensive nephrosclerosis. Am. J. Hypertens. 2010;23(1):78–84. doi: 10.1038/ajh.2009.208. [DOI] [PubMed] [Google Scholar]
- 68.Liu Y., Taylor N.E., Lu L., et al. Renal medullary microRNAs in Dahl salt-sensitive rats: miR-29b regulates several collagens and related genes. Hypertension. 2010;55(4):974–982. doi: 10.1161/HYPERTENSIONAHA.109.144428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Denby L., Ramdas V., McBride M.W., et al. miR-21 and miR-214 are consistently modulated during renal injury in rodent models. Am. J. Pathol. 2011;179(2):661–672. doi: 10.1016/j.ajpath.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Marques F.Z., Campain A.E., Tomaszewski M., et al. Gene expression profiling reveals renin mRNA overexpression in human hypertensive kidneys and a role for microRNAs. Hypertension. 2011;58(6):1093–1098. doi: 10.1161/HYPERTENSIONAHA.111.180729. [DOI] [PubMed] [Google Scholar]
- 71.Chandrasekaran K., Karolina D.S., Sepramaniam S., et al. Role of microRNAs in kidney homeostasis and disease. Kidney Int. 2012;81(7):617–627. doi: 10.1038/ki.2011.448. [DOI] [PubMed] [Google Scholar]
- 72.Kriegel A.J., Mladinov D., Liang M. Translational study of microRNAs and its application in kidney disease and hypertension research. Clin. Sci. (Lond.) 2012;122(10):439–447. doi: 10.1042/CS20110159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wing M.R., Ramezani A., Gill H.S., Devaney J.M., Raj D.S. Epigenetics of progression of chronic kidney disease: fact or fantasy? Semin. Nephrol. 2013;33(4):363–374. doi: 10.1016/j.semnephrol.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Khella H.W., Bakhet M., Lichner Z., Romaschin A.D., Jewett M.A., Yousef G.M. MicroRNAs in kidney disease: an emerging understanding. Am. J. Kidney Dis. 2013;61(5):798–808. doi: 10.1053/j.ajkd.2012.09.018. [DOI] [PubMed] [Google Scholar]
- 75.Karolina D.S., Tavintharan S., Armugam A., et al. Circulating miRNA profiles in patients with metabolic syndrome. J. Clin. Endocrinol. Metab. 2012;97(12):E2271–E2276. doi: 10.1210/jc.2012-1996. [DOI] [PubMed] [Google Scholar]
- 76.Patella F., Rainaldi G. MicroRNAs mediate metabolic stresses and angiogenesis. Cell. Mol. Life Sci. 2012;69(7):1049–1065. doi: 10.1007/s00018-011-0775-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Heggermont W.A., Heymans S. MicroRNAs are involved in end-organ damage during hypertension. Hypertension. 2012;60(5):1088–1093. doi: 10.1161/HYPERTENSIONAHA.111.187104. [DOI] [PubMed] [Google Scholar]
- 78.Kontaraki J.E., Marketou M.E., Zacharis E.A., Parthenakis F.I., Vardas P.E. MicroRNA-9 and microRNA-126 expression levels in patients with essential hypertension: potential markers of target-organ damage. J. Am. Soc. Hypertens. 2014;8(6):368–375. doi: 10.1016/j.jash.2014.03.324. [DOI] [PubMed] [Google Scholar]
- 79.Hansen JL, Doggen C, Quax PH, Sheikh SP, Rosendaal FR, Nossent AY1 SNPs in microRNA binding sites in 3'-UTRs of RAAS genes influence arterial blood pressure and risk of myocardial infarction. Am. J. Hypertens. 2011;24(9):999–1006. doi: 10.1038/ajh.2011.92. [DOI] [PubMed] [Google Scholar]
- 80.Ceolotto G., Papparella I., Bortoluzzi A., et al. Interplay between miR-155, AT1R A1166C polymorphism, and AT1R expression in young untreated hypertensives. Am. J. Hypertens. 2011;24(2):241–246. doi: 10.1038/ajh.2010.211. [DOI] [PubMed] [Google Scholar]
- 81.Zheng L., Xu C.C., Chen W.D., et al. MicroRNA-155 regulates angiotensin II type 1 receptor expression and phenotypic differentiation in vascular adventitial fibroblasts. Biochem. Biophys. Res. Commun. 2010;400(4):483–488. doi: 10.1016/j.bbrc.2010.08.067. [DOI] [PubMed] [Google Scholar]
- 82.Jackson K.L., Marques F.Z., Watson A.M., et al. A novel interaction between sympathetic overactivity and aberrant regulation of renin by miR-181a in BPH/2J genetically hypertensive mice. Hypertension. 2013;62(4):775–781. doi: 10.1161/HYPERTENSIONAHA.113.01701. [DOI] [PubMed] [Google Scholar]
- 83.Marques F.Z., Campain A.E., Tomaszewski M., et al. Gene expression profiling reveals renin mRNA overexpression in human hypertensive kidneys and a role for microRNAs. Hypertension. 2011;58(6):1093–1098. doi: 10.1161/HYPERTENSIONAHA.111.180729. [DOI] [PubMed] [Google Scholar]
- 84.Kemp J.R., Unal H., Desnoyer R., Yue H., Bhatnagar A., Karnik S.S. Angiotensin II-regulated microRNA 483-3p directly targets multiple components of the renin-angiotensin system. J. Mol. Cell. Cardiol. 2014;75:25–39. doi: 10.1016/j.yjmcc.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Eskildsen T.V., Jeppesen P.L., Schneider M., et al. Angiotensin II Regulates microRNA-132/-212 in Hypertensive Rats and Humans. Int. J. Mol. Sci. 2013;14(6):11190–11207. doi: 10.3390/ijms140611190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jiang X., Ning Q., Wang J. Angiotensin II induced differentially expressed microRNAs in adult rat cardiac fibroblasts. J. Physiol. Sci. 2013;63(1):31–38. doi: 10.1007/s12576-012-0230-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mopidevi B., Ponnala M., Kumar A. Human angiotensinogen +11525 C/A polymorphism modulates its gene expression through microRNA binding. Physiol. Genomics. 2013;45(19):901–906. doi: 10.1152/physiolgenomics.00056.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Romero D.G., Plonczynski M.W., Carvajal C.A., Gomez-Sanchez E.P., Gomez-Sanchez C.E. Microribonucleic acid-21 increases aldosterone secretion and proliferation in H295R human adrenocortical cells. Endocrinology. 2008;149(5):2477–2483. doi: 10.1210/en.2007-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Robertson S., MacKenzie S.M., Alvarez-Madrazo S., et al. MicroRNA-24 is a novel regulator of aldosterone and cortisol production in the human adrenal cortex. Hypertension. 2013;62(3):572–578. doi: 10.1161/HYPERTENSIONAHA.113.01102. [DOI] [PubMed] [Google Scholar]
- 90.Sõber S., Laan M., Annilo T. MicroRNAs miR-124 and miR-135a are potential regulators of the mineralocorticoid receptor gene (NR3C2) expression. Biochem. Biophys. Res. Commun. 2010;391(1):727–732. doi: 10.1016/j.bbrc.2009.11.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wei Z., Biswas N., Wang L., et al. A Common Genetic Variant in the 3'-UTR of Vacuolar H+-ATPase ATP6V0A1 Creates a Micro-RNA Motif to Alter Chromogranin A Processing and Hypertension Risk. Circ Cardiovasc Genet. 2011;4(4):381–389. doi: 10.1161/CIRCGENETICS.111.959767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rezaei M, Andreu T, Neuenschwander S, et al. Regulation of 11β-hydroxysteroid dehydrogenase type 2 by microRNA. Hypertension . 2014;64(4):860–6. doi: 10.1161/HYPERTENSIONAHA.114.00002. [DOI] [PubMed] [Google Scholar]
- 93.Newton-Cheh C., Larson M.G., Vasan R.S., et al. Association of common variants in NPPA and NPPB with circulating natriuretic peptides and blood pressure. Nat. Genet. 2009;41(3):348–353. doi: 10.1038/ng.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Arora P., Wu C., Khan A.M., et al. Atrial natriuretic peptide is negatively regulated by microRNA-425. Clin. Investig. (Lond.) 2013;123(8):3378–3382. doi: 10.1172/JCI67383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kotlo K.U., Hesabi B., Danziger R.S. Implication of microRNAs in atrial natriuretic peptide and nitric oxide signaling in vascular smooth muscle cells. Am. J. Physiol. Cell Physiol. 2011;301(4):C929–C937. doi: 10.1152/ajpcell.00088.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sun H.X., Zeng D.Y., Li R.T., et al. Essential role of microRNA-155 in regulating endothelium-dependent vasorelaxation by targeting endothelial nitric oxide synthase. Hypertension. 2012;60(6):1407–1414. doi: 10.1161/HYPERTENSIONAHA.112.197301. [DOI] [PubMed] [Google Scholar]
- 97.Yang Q., Jia C., Wang P., et al. MicroRNA-505 identified from patients with essential hypertension impairs endothelial cell migration and tube formation. Int. J. Cardiol. 2014;177(3):925–934. doi: 10.1016/j.ijcard.2014.09.204. [DOI] [PubMed] [Google Scholar]
- 98.Fernandes T., Magalhães F.C., Roque F.R., Phillips M.I., Oliveira E.M. Exercise training prevents the microvascular rarefaction in hypertension balancing angiogenic and apoptotic factors: role of microRNAs-16, -21, and -126. Hypertension. 2012;59(2):513–520. doi: 10.1161/HYPERTENSIONAHA.111.185801. [DOI] [PubMed] [Google Scholar]
- 99.Jiang Q., Lagos-Quintana M., Liu D., et al. miR-30a regulates endothelial tip cell formation and arteriolar branching. Hypertension. 2013;62(3):592–598. doi: 10.1161/HYPERTENSIONAHA.113.01767. [DOI] [PubMed] [Google Scholar]
- 100.Nazari-Jahantigh M., Wei Y., Schober A. The role of microRNAs in arterial remodelling. Thromb. Haemost. 2012;107(4):611–618. doi: 10.1160/TH11-12-0826. [DOI] [PubMed] [Google Scholar]
- 101.Quintavalle M., Condorelli G., Elia L. Arterial remodeling and atherosclerosis: miRNAs involvement. Vascul. Pharmacol. 2011;55(4):106–110. doi: 10.1016/j.vph.2011.08.216. [DOI] [PubMed] [Google Scholar]
- 102.Horita H.N., Simpson P.A., Ostriker A., et al. Serum response factor regulates expression of phosphatase and tensin homolog through a microRNA network in vascular smooth muscle cells. Arterioscler. Thromb. Vasc. Biol. 2011;31(12):2909–2919. doi: 10.1161/ATVBAHA.111.233585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ling S., Nanhwan M., Qian J., et al. Modulation of microRNAs in hypertension-induced arterial remodeling through the β1 and β3-adrenoreceptor pathways. J. Mol. Cell. Cardiol. 2013;65:127–136. doi: 10.1016/j.yjmcc.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 104.Joshi S.R., Comer B.S., McLendon J.M., Gerthoffer W.T. MicroRNA Regulation of Smooth Muscle Phenotype. Mol. Cell. Pharmacol. 2012;4(1):1–16. [PMC free article] [PubMed] [Google Scholar]
- 105.Parmacek M.S. MicroRNA-modulated targeting of vascular smooth muscle cells. J. Clin. Invest. 2009;119(9):2526–2528. doi: 10.1172/JCI40503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wu W.H., Hu C.P., Chen X.P., et al. MicroRNA-130a mediates proliferation of vascular smooth muscle cells in hypertension. Am. J. Hypertens. 2011;24(10):1087–1093. doi: 10.1038/ajh.2011.116. [DOI] [PubMed] [Google Scholar]
- 107.Kontaraki J.E., Marketou M.E., Zacharis E.A., Parthenakis F.I., Vardas P.E. Differential expression of vascular smooth muscle-modulating microRNAs in human peripheral blood mononuclear cells: novel targets in essential hypertension. J. Hum. Hypertens. 2014;28(8):510–516. doi: 10.1038/jhh.2013.117. [DOI] [PubMed] [Google Scholar]
- 108.Albinsson S., Swärd K. Targeting smooth muscle microRNAs for therapeutic benefit in vascular disease. Pharmacol. Res. 2013;75:28–36. doi: 10.1016/j.phrs.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 109.Mu S., Shimosawa T., Ogura S., et al. Epigenetic modulation of the renal β-adrenergic-WNK4 pathway in salt-sensitive hypertension. Nat. Med. 2011;17(5):573–580. doi: 10.1038/nm.2337. [DOI] [PubMed] [Google Scholar]
- 110.Mao J., Li C., Zhang Y., Li Y., Zhao Y. Human with-no-lysine kinase-4 3'-UTR acting as the enhancer and being targeted by miR-296. Int. J. Biochem. Cell Biol. 2010;42(9):1536–1543. doi: 10.1016/j.biocel.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 111.Li D., Yang P., Xiong Q. MicroRNA-125a/b-5p inhibits endoyhelin-1 expression in vascular endothelial cells. J. Hypertens. 2010;28(8):1646–1654. doi: 10.1097/HJH.0b013e32833a4922. [DOI] [PubMed] [Google Scholar]
- 112.Magenta A., Greco S., Gaetano C., Martelli F. Oxidative stress and microRNAs in vascular diseases. Int. J. Mol. Sci. 2013;14(9):17319–17346. doi: 10.3390/ijms140917319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hansen K.F., Sakamoto K., Obrietan K. MicroRNAs: a potential interface between the circadian clock and human health. Genome Med. 2011;3(2):10. doi: 10.1186/gm224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Li S., Zhu J., Zhang W., et al. Signature microRNA expression profile of essential hypertension and its novel link to human cytomegalovirus infection. Circulation. 2011;124(2):175–184. doi: 10.1161/CIRCULATIONAHA.110.012237. [DOI] [PubMed] [Google Scholar]
- 115.Cheng H.S., Njock M.S., Khyzha N., Dang L.T., Fish J.E. Noncoding RNAs regulate NF-κB signaling to modulate blood vessel inflammation. Front. Genet. 2014;5:422. doi: 10.3389/fgene.2014.00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Creemers E.E., Tijsen A.J., Pinto Y.M. Circulating microRNAs: novel biomarkers and extracellular communicators in cardiovascular disease? Circ. Res. 2012;110(3):483–495. doi: 10.1161/CIRCRESAHA.111.247452. [DOI] [PubMed] [Google Scholar]
- 117.Thum T., Chau N., Bhat B., et al. Comparison of different miR-21 inhibitor chemistries in a cardiac disease model. J. Clin. Invest. 2011;121(2):461–462. doi: 10.1172/JCI45938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Friese R.S., Altshuler A.E., Zhang K., et al. MicroRNA-22 and promoter motif polymorphisms at the Chga locus in genetic hypertension: functional and therapeutic implications for gene expression and the pathogenesis of hypertension. Hum. Mol. Genet. 2013;22(18):3624–3640. doi: 10.1093/hmg/ddt213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rhee S.W., Stimers J.R., Wang W., Pang L. Vascular smooth muscle-specific knockdown of the noncardiac form of the L-type calcium channel by microRNA-based short hairpin RNA as a potential antihypertensive therapy. J. Pharmacol. Exp. Ther. 2009;329(2):775–782. doi: 10.1124/jpet.108.148866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Siasos G., Tousoulis D., Tourikis P., et al. MicroRNAs in cardiovascular therapeutics. Curr. Top. Med. Chem. 2013;13(13):1605–1618. doi: 10.2174/15680266113139990109. [DOI] [PubMed] [Google Scholar]
- 121.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;16;43(6):904–14. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gopalakrishnan K., Kumarasamy S., Mell B., Joe B. Genome-wide identification of long noncoding RNAs in rat models of cardiovascular and renal disease. Hypertension. 2015;65(1):200–210. doi: 10.1161/HYPERTENSIONAHA.114.04498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chung A.S., Guan Y.J., Yuan Z.L., Albina J.E., Chin Y.E. Ankyrin repeat and SOCS box 3 (ASB3) mediates ubiquitination and degradation of tumor necrosis factor receptor II. Mol. Cell. Biol. 2005;25(11):4716–4726. doi: 10.1128/MCB.25.11.4716-4726.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kumar A., Tikoo S., Maity S., et al. Mammalian proapoptotic factor ChaC1 and its homologues function as γ-glutamyl cyclotransferases acting specifically on glutathione. EMBO Rep. 2012;13(12):1095–1101. doi: 10.1038/embor.2012.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Knoblach B., Rachubinski R.A. Phosphorylation-dependent activation of peroxisome proliferator protein PEX11 controls peroxisome abundance. J. Biol. Chem. 2010;285(9):6670–6680. doi: 10.1074/jbc.M109.094805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Waterham H.R., Ebberink M.S. Genetics and molecular basis of human peroxisome biogenesis disorders. Biochim. Biophys. Acta. 2012;1822(9):1430–1441. doi: 10.1016/j.bbadis.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 127.van Roermund C.W., Tabak H.F., van Den Berg M., Wanders R.J., Hettema E.H. Pex11p plays a primary role in medium-chain fatty acid oxidation, a process that affects peroxisome number and size in Saccharomyces cerevisiae. J. Cell Biol. 2000;150(3):489–498. doi: 10.1083/jcb.150.3.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Morrissey C., Grieve I.C., Heinig M., et al. Integrated genomic approaches to identification of candidate genes underlying metabolic and cardiovascular phenotypes in the spontaneously hypertensive rat. Physiol. Genomics. 2011;43(21):1207–1218. doi: 10.1152/physiolgenomics.00210.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Nagase M., Kato A., Ono T., Suzuki Y., Hirose S., Fujita T. Retrotransposons transcribed preferentially in proximal tubules of salt-hypertensive rats. Kidney Int. 1999;55(3):995–1004. doi: 10.1046/j.1523-1755.1999.055003995.x. [DOI] [PubMed] [Google Scholar]
- 130.Leung A., Trac C., Jin W., et al. Novel long noncoding RNAs are regulated by angiotensin II in vascular smooth muscle cells. Circ. Res. 2013;113(3):266–278. doi: 10.1161/CIRCRESAHA.112.300849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Robb G.B., Carson A.R., Tai S.C., et al. Post-transcriptional regulation of endothelial nitric-oxide synthase by an overlapping antisense mRNA transcript. Biol. Chem. 2004;279(36):37982–37996. doi: 10.1074/jbc.M400271200. [DOI] [PubMed] [Google Scholar]
- 132.Ho J.J., Robb G.B., Tai S.C., et al. Active stabilization of human endothelial nitric oxide synthase mRNA by hnRNP E1 protects against antisense RNA and microRNAs. Mol. Cell. Biol. 2013;33(10):2029–2046. doi: 10.1128/MCB.01257-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ottaviani S., de Giorgio A., Harding V., Stebbing J., Castellano L. Noncoding RNAs and the control of hormonal signaling via nuclear receptor regulation. J. Mol. Endocrinol. 2014;53(2):R61–R70. doi: 10.1530/JME-14-0134. [DOI] [PubMed] [Google Scholar]
- 134.Lanz R.B., McKenna N.J., Onate S.A., et al. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell. 1999;97(1):17–27. doi: 10.1016/s0092-8674(00)80711-4. [DOI] [PubMed] [Google Scholar]
- 135.Novikova I.V., Hennelly S.P., Sanbonmatsu K.Y. Structural architecture of the human long non-coding RNA, steroid receptor RNA activator. Nucleic Acids Res. 2012;40(11):5034–5051. doi: 10.1093/nar/gks071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kino T., Hurt D.E., Ichijo T., Nader N., Chrousos G.P. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 2010;3(107):ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Vausort M., Wagner D.R., Devaux Y. Long noncoding RNAs in patients with acute myocardial infarction. Circ. Res. 2014;115(7):668–677. doi: 10.1161/CIRCRESAHA.115.303836. [DOI] [PubMed] [Google Scholar]


