ABSTRACT
On a broader scale, T cell density and localization in colorectal cancer liver metastases have prognostic and predictive implications. As T cell distribution at higher resolutions has not been fully investigated, a detailed resolution analysis of T cell distribution was performed. Patient tissues were divided into 10 µm distance classes between the tumor border and adjacent normal liver. Thereby, distinct density patterns of T cell localization in relation to the malignant tissue could be detected. At a distance of 20 to 30 µm to the tumor, a decrease of CD3 T cells is common. Within this area, cytotoxic Granzyme B and CD8+ T cells were found to be significantly reduced as well as CD163 macrophages were increased and identified to be in close contact with T cells. Our data suggests a physical or functional border within this region. Survival analysis revealed improved overall survival in patients with high T cells numbers at the direct tumor border. Interestingly, the decreased T cells in the 20 to 30 µm region were also found to be significantly associated with improved survival. Consequently, the detailed localization of T cells, despite blockade, could be associated with improved clinical outcome. The high-resolution analysis represents new insights into relevant heterogenous T cell distributions especially related to clinical responses. As the paradoxical observation of localization-dependent prognostic relevance of T cell densities is only detectable by detailed spatial analyses, this investigation of spatial profiles at higher resolutions is suggested as a new biomarker for survival and response to therapies.
KEYWORDS: Colorectal cancer liver metastases, immunosuppression, prognosis, tumor immunology, tumor microenvironment, T cell distribution
Abbreviations
- CCL5
C–C chemokine ligand 5
- CCR5
C–C chemokine receptor 5
- CRC
colorectal cancer
- GranB
Granzyme B
- LM
liver metastasis
- MSI
microsatellite instability
- OS
overall survival
- PD-L1
programmed death-ligand 1
- TAM
tumor-associated macrophages
- TIL
tumor-infiltrating lymphocytes
Introduction
Metastatic colorectal cancer (CRC) accounts for more than 55.000 deaths per year in the United States alone.1 Around 15% of the patients with liver metastases (LM) can be cured by surgical resection of the metastases.2 Immune system interactions with the primary tumor and the metastases have an important influence on the clinical course of patients. Immune cells infiltrating or surrounding colorectal primary tumors3 and especially tumor-infiltrating lymphocytes (TILs) represent a major prognostic factor.4,5 On a broader scale, high densities of TILs were shown to be correlated with an improved survival in patients with primary CRC as well as LM.6-14 Nevertheless, tumor heterogeneity in many cancers represents an obstacle in detailed analyses of T cell distributions, whereas colorectal cancer liver metastases (CRC-LM) are rather homogenous with a clear border between liver and tumor tissue allowing more precise analyses of spatial distributions.15,16 Especially, T cells and their relation to tumor tissue are gaining increased attention, as the majority of the T cells can be found in the stromal part of primary CRC without direct contact to tumor epithelium.17 Evaluation of LM revealed heterogeneous immune cell infiltration into the metastasis center dependent of histopathological tumor conditions like necrosis or fibrosis. Previous analyses, however, did not reveal a relationship of these lymphocytic infiltrates and the clinical course.17 In contrast, in the invasive margin, immune cell infiltrations showed remarkably strong variations between different patients and allowed prediction of responses to chemotherapy.13,17 While the only few T cells in contact with the tumor or within the epithelial compartment are generally regarded as being engaged in direct antitumor activity, more distant T cells might be blocked by immunosuppressive factors and exploited by the tumor.18-20 It is therefore of high interest to analyze differences in this distribution. The spatial distribution of T cells at the invasive margin of colorectal cancer LM has not been investigated in detail. The aim of this study was to explore the pattern of detailed T cell distribution in the invasive margin of CRC-LM patients using automated high-throughput image quantification analyses on whole slide sections samples. This quantitative approach allows the identification of distribution patterns in 10 µm steps within 100 µm distance from the tumor border and represents a high-resolution map of tumor infiltrating cells. By using serial sections, T cell distance analysis not only to the tumor tissue but also to the existing barriers like immunosuppressive cells is possible. Surface tissue areas of 36 patients were analyzed, allowing robust statistical evaluation. Clustering analyses revealed distinct distribution patterns that could be associated with improved overall survival (OS).
Results
The classification of the 10 µm distance classes within the 100 µm area between the tumor and adjacent liver as outlined in Fig. 1 and the following cell quantification analyses could be performed successfully for all 36 patients. Thereby, bar plots for all distance classes were generated. This revealed distinct patterns that were detectable within each patient. For CD3, T cell quantification representative results of different patients are shown in Fig. S1. Apparently, CD3 T cells are located in all distance classes with maximum densities at different distances.
Figure 1.
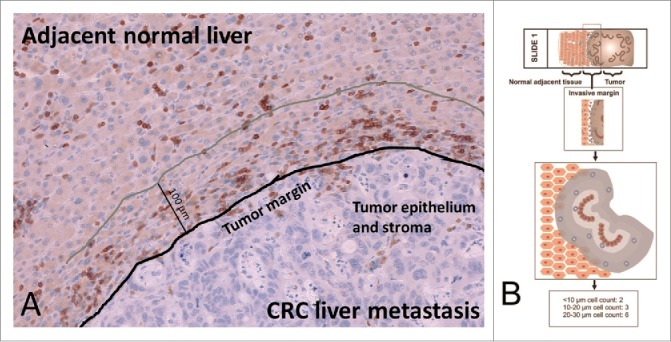
(A) Representative invasive margin of a CRC liver metastasis with annotations (CD3 positive lymphocytes appear dark brown). (B) The invasive margin of liver metastases is analyzed separately, encompassing 100 µm into normal adjacent liver from the tumor epithelium. Shaded areas highlight distinct distance classes of 10 µm in relation to the tumor epithelium.
As shown in Fig. 2, the distribution of CD3 cell densities obviously varied between the different distance classes (Kruskal–Wallis test, p = 0.0001). The statistically significant differences between adjacent distance classes are marked by asterisks (post-hoc Mann–Whitney U tests). It was observed that in the 20–30 µm distance class median T cell densities significantly decreased below 100 cells/mm2, whereas in the distance classes closer than 20 µm and further away than 30 µm from the tumor epithelium median densities higher than 150 cells/mm2 were found. To identify patterns of T cell densities, unsupervised clustering was performed. To avoid possible bias due to different lengths of the invasive margin between different patients, we randomly selected one single field from each patient for further pattern analysis. The clustering analysis revealed distinct groups as shown in Fig. 3 (non-informative distance classes were removed stepwise, informative distance classes shown). The adjacent distance classes 10–20 µm and 20–30 µm clustered, whereas the most proximal distance classes (< 10 µm) and the classes with the highest distance from the tumor epithelium (90–100 µm) were found to be least related to the 20–30 µm distance classes. Furthermore, clustering of patients according to their T cell densities within single distance classes was observed. Survival analysis comparing patients with high and low CD3 T cell densities within each distance class resulted in a significant prolongation of OS for patients with high CD3 cell numbers in the most proximal distance class (< 10 µm), while in the 20–30 µm region a significant prolongation of survival for patients with low T cell numbers was observed (Fig. 4). No significant differences between high and low T cell infiltrates were found in all other distance classes.
Figure 2.
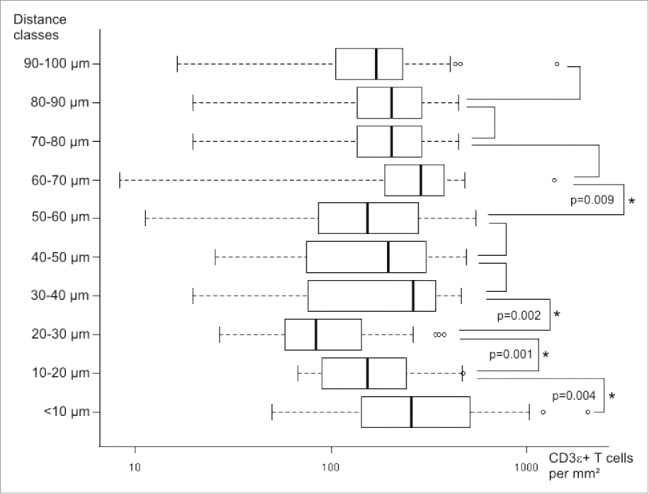
Boxplots of identified CD3 T cell densities in different distance classes of 36 patients. Statistically significant differences between adjacent distance classes are marked with asterisks (p values given for Kruskal–Wallis with post-hoc Mann–Whitney U tests).
Figure 3.
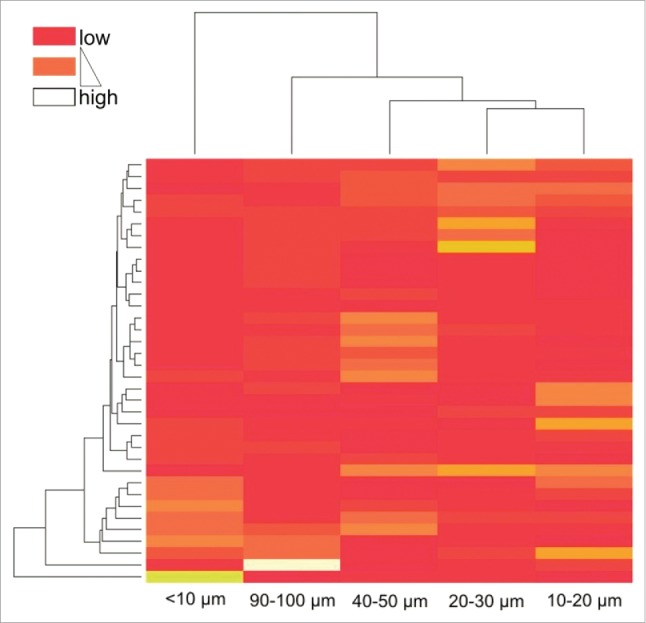
Heatmap based on unsupervised clustering encompassing randomly selected single fields from all 36 patients. Color code indicates clustering of patients according to their relative T cell densities within single distance classes.
Figure 4.
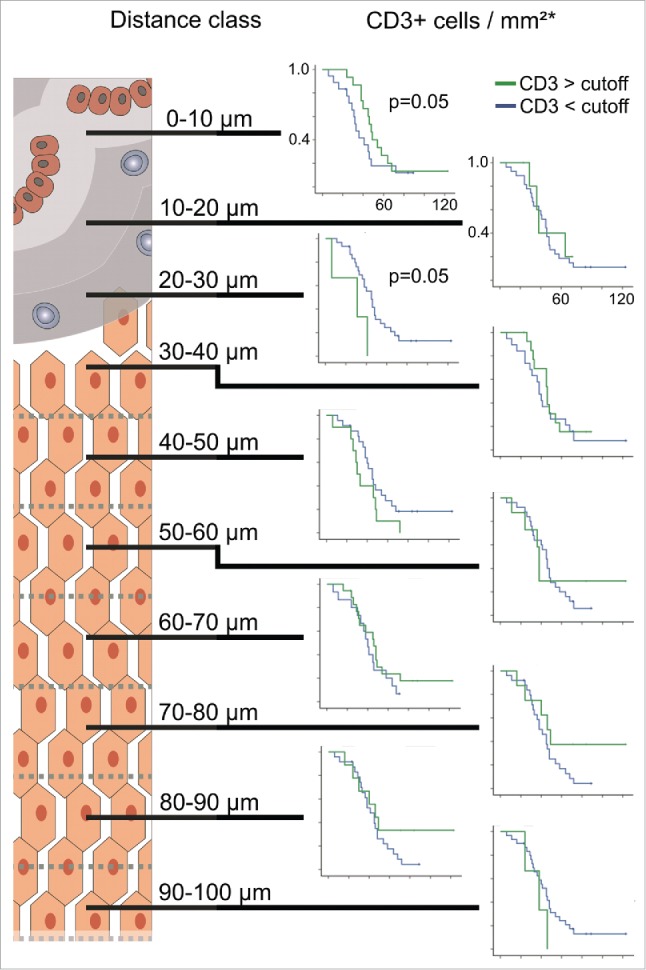
Kaplan–Meier plots for estimated overall survival probabilities of CD3 T cell high and CD3 T cell low patient groups within all distance classes. Graphs indicate cumulative survival (y-axis) and survival in months (x-axis). p values for statistically significant differences between groups are shown (Breslow test). Cutoff values are taken from previous works.6,13
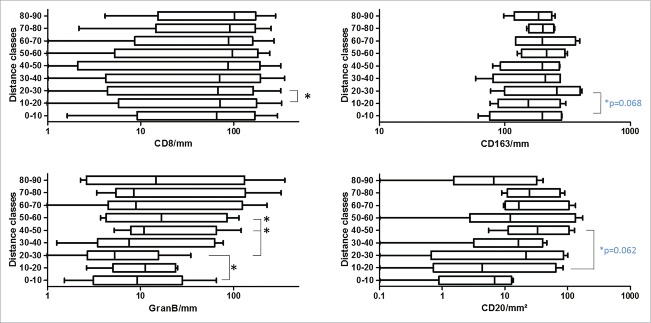
To get a more detailed picture of the CD3 cell distribution pattern, we did a similar distribution and survival analysis (only CD8+) in representative patient samples for CD8+ (n = 10), Granzyme B (n = 6), CD163 (n = 4) and CD20 (n = 4) as shown in Fig. 5 (and Fig. S2). Especially, GranB was found to significantly “decrease” at the distance of 20–30 µm (Wilcoxon signed-rank test, p = 0.03) following a significant increase further away from the tumor (p = 0.03). In line with this, also CD8+ cells slightly but significantly decreased in the 20–30 µm region (p = 0.049). CD8+ cells, however, were rather evenly distributed among the whole area analyzed with a median cell number of ∼87 cells/mm2. Furthermore, survival analysis showed that high CD8+ infiltrates in the >10 µm region significantly elongated OS similar to CD3 (Fig. 6). Nevertheless, no significant differences between CD8+ high and low infiltration were observed in the other distance classes. In contrast to the T cells, CD163 macrophages showed a trend to increase in the 20–30 µm distance class (Fig. 5). While in the other distance classes CD163 cells were rather evenly distributed with a median of ∼200 cells/mm2, the distribution of CD20 cells was found to be more heterogenous with least cells at the tumor border, and slightly increasing densities further away (Fig. 5 and Fig. S2). From all cell types analyzed, GranB and CD20 cells appeared to be the smallest populations present in the tissues with median cell counts of 9 and 16 cells/mm2, respectively. In Figs. S3 and 4, representative images of CD3, CD8+, GranB, CD163 and CD20 immunostainings are shown.
Figure 5.
Boxplots of identified CD8+, GranB, CD163 and CD20 cell densities in different distance classes. Statistically significant differences between adjacent distance classes are marked with asterisks (Wilcoxon signed-rank test; blue p values given for paired t-tests; patient numbers for CD8+ n = 10, GranB n = 6, CD163 n = 4 and CD20 n = 4).
Figure 6.
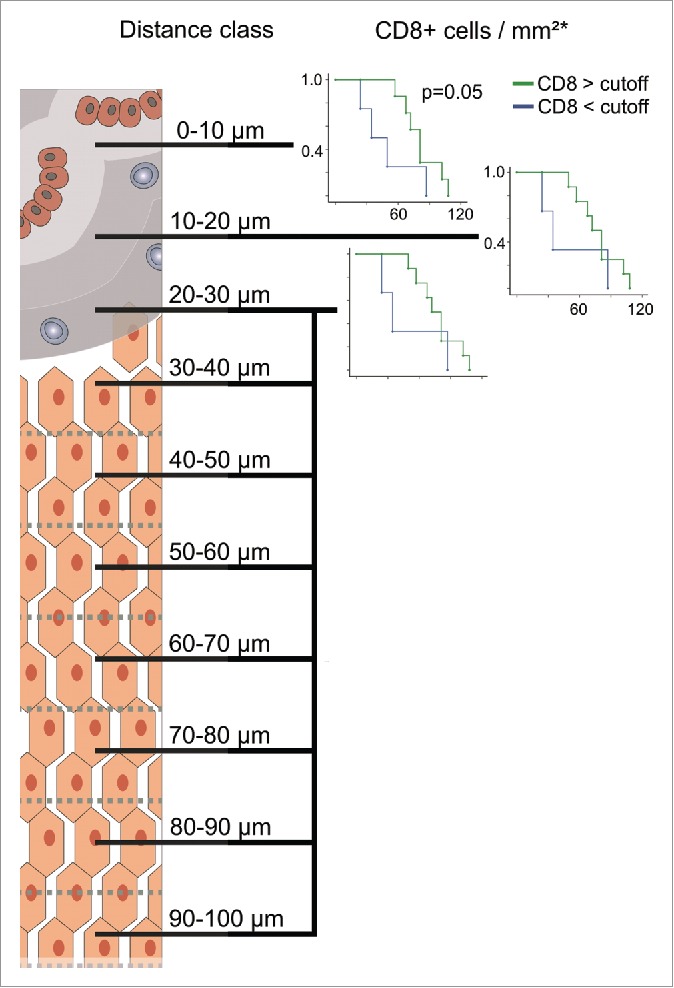
Kaplan–Meier plots for estimated overall survival probabilities of CD8+ high and low patient groups within the first three distance classes. Graphs indicate cumulative survival (y-axis) and survival in months (x-axis). p values for statistically significant differences between groups are shown (Breslow test). Cutoff values are taken from previous works.6,13
In the course of this analysis, we further wanted to understand whether CD163 cells achieve immunosuppressive effects against T cells. We hypothesized that both cell types might be in close contact with each other and therefore analyzed the localization of T cells in relation to CD163 macrophages using serial section analysis (Fig. 7). We identified that 52–81% (median 68%) of CD3 lymphocytes within the tumor microenvironment were in close contact (< 10 µm) with CD163 macrophages. In addition, immunofluorescent double staining of CD163 and programmed death-ligand 1 (PD-L1) showed that the majority of CD163 macrophages were PD-L1 positive (Fig. 8).
Figure 7.
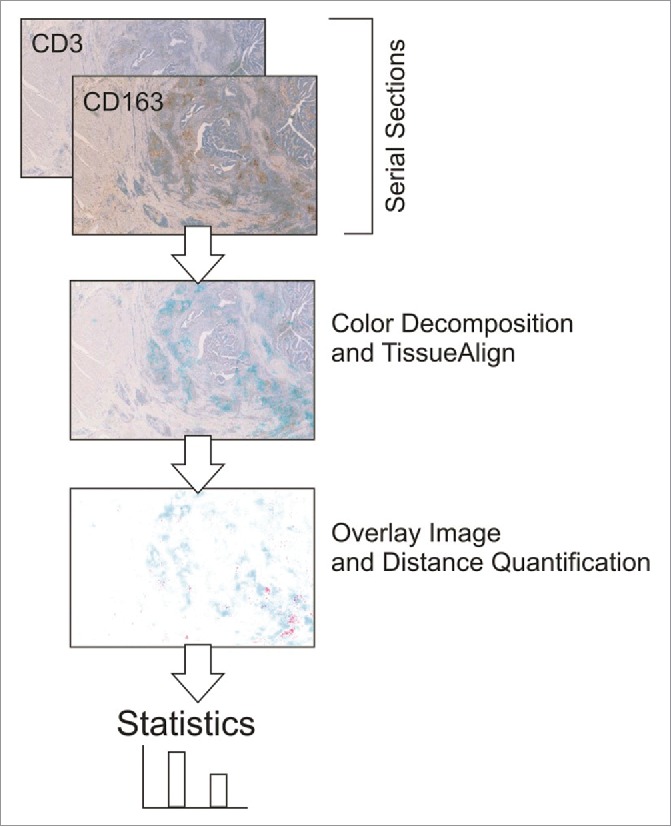
Processing of serial section images for the quantification of distances between CD3 T cells and CD163 macrophages.
Figure 8.
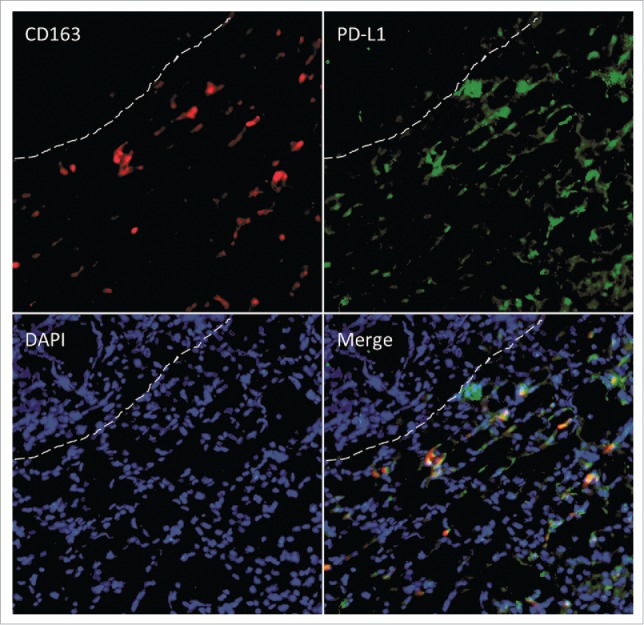
Immunofluorescent double staining for CD163 (red) and PD-L1 (green) at the invasive margin. Dashed lines separate the tumor (top left) from the adjacent liver. DAPI was used as counterstain.
Discussion
Image analysis is a powerful tool to analyze spatial distributions as well as patterns of cells in the tumor microenvironment25,26 and allows in combination with virtual microscopy the automated analysis of large tissue areas. Among the inflammatory cells in the composition of the tumor stroma and epithelium, T cells play a crucial role in regulating tumor growth and progression. It was shown that the invasive margin and its immune cell composition are predictive for the clinical course of cancer,6,13,17,27 and that immunophenotypes and distribution of those can change under therapy.28 Whereas the sheer quantity of T cells is associated with clinical outcome,6-9,13,29 it has not yet been established whether there are any significant differences in the spatial distribution of T cells at the invasive margin. For this reason, we have investigated the detailed localization of CD3ε+ T cells in the invasive margin of colorectal cancer LM. CD3ε is a pan-T cell marker and shows great heterogeneity in distribution intratumorally and at the invasive margin.17 The interface between metastatic liver lesions and adjacent liver allows the precise evaluation of T cell localization, while in the primary tumor the heterogeneity is much more pronounced and is therefore difficult for this analysis.15
The analyzed regions within 100 µm distance from the tumor show a significant “drop” of T cell density in the region of 20–30 µm from tumor epithelium, whereas higher T cell densities are found in close proximity and more far away from the tumor as schematically shown in Fig. 9. Similar examination of CD8+ cells revealed that this CD3 T cells subset is likewise affected by showing a slight but significant “drop” at 20–30 µm from the tumor. The ratio between CD8+ and CD4+ cells varied across distance classes. In general, the CD8+ cell numbers observed among the complete analyzed regions from 0 to 100 µm were ∼45% (87 cells/mm2) of the CD3 cell numbers (195 cells/mm2) leading to the assumption that the other major CD3 cell subtype (CD4+) is evenly present in this area. This is, however, not the case at the location of the T cell “drop” in the 20–30 µm region. Here, a median of ∼90 CD3 cells/mm2 and ∼67 CD8+ cells/mm2 was counted, meaning that CD4+ cells are more affected to decrease in this region. Interestingly, when examining spatial cytotoxic activity by GranB immunostaining it was likewise found to be significantly decreased at the 20–30 µm distance. As natural killer cells have been shown to be very rare in CRC-LM, cytotoxic CD8+ T cells are supposed to be the main source of GranB.30 These findings strikingly confirm not only a reduction of cell quantities but also a reduction of cytotoxic T cell function at the 20–30 µm region.
Figure 9.
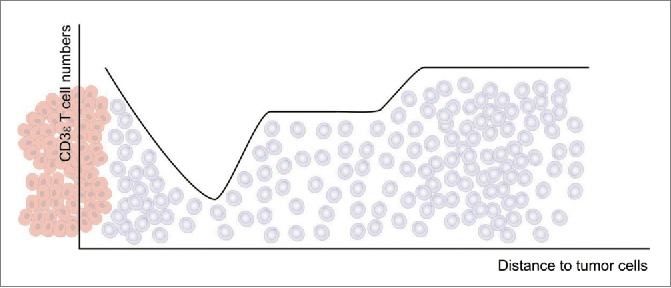
Overview of the observed T cell densities (gray cells) in relation to the distance to the tumor epithelium (red cells).
Those new observations prompted us to further investigate the possible factors involved in this heterogenous T cell distribution patterns. As regulatory T cell numbers are generally low in CRC-LM (3.5 cells/mm2),13 we decided to repeat our spatial distribution analysis to examine the localization of CD20 B cells and CD163 macrophages. This latter tumor-associated macrophage (TAM) population, which is characterized by a tumor-promoting and immunosuppressive phenotype and associated with poor clinical outcome in many different tumors,31-33 we found highly enriched in CRC-LM tissues (∼200 cells/mm2). In our exploratory analyses, we noticed an increase (p = 0.068) at the 20–30 µm region, whereas no relevant differences between the other examined distances classes were observed. This opposite distribution to the T cells leads to the assumption that CD163 cells fill the gap between the tumor distant and tumor proximal T cells. Our data suggests that CD163 macrophages are forming a physical barrier which only a specific T cell subset can overcome. Even more important, this barrier could have functional impact, because it also marks the border, where cytotoxic GranB significantly decreased at the tumor margin. Indeed, TAM have been shown to directly suppress T cell responses through immune inhibitory checkpoint molecules like PD-L1.34,35 Interestingly, these PD-L1 positive TAM were also found to be in close contact with T cells. This data goes in line with our own results showing a high percentage (68%) of T cells being in direct contact with CD163 macrophages, and furthermore, the majority of CD163 cells in the invasive margin were found to express PD-L1. Our data, therefore, suggests itself that TAM directly suppress T cells and consequently GranB production via immune checkpoints. The fact that in the invasive margin of CRC-LM 98% of all CD3 T cells express the checkpoint receptor PD-1 further encourages this assumption.19 In contrast to CD163, the distribution of CD20 B cells is very heterogenous among the distance classes with lower cell densities at the tumor border and increased densities further away (p = 0.062). The role of tumor-infiltrating CD20 B cells is controversial. Some studies report about a protective role against tumors,36-38 whereas others connect B cells with enhanced tumor growth and suppression of antitumor immunity.36,39,40 Notably, B cells can polarize macrophages toward immunosuppressive phenotypes.41 Nevertheless, B cell numbers appeared to be very low with ∼16 cells/mm2 among the complete tissues analyzed and especially compared with CD163 (∼200 cells/mm2) a minor role for CD20 B cells is presumed. Besides immunosuppressive cells, the distribution of the T cells in the tissues could also be accounted for other factors. Chemotactic fields, which might be generated by tumor, endothelial cells or even macrophages, could induce the observed T cell distribution patterns.19,42 Another explanation could be cell-matrix interactions.43,44 Physical properties of the extracellular matrix can influence directional cell movement and might govern the positioning of T cells in the direct vicinity of tumor cells. In addition, CD31, which is a marker for angiogenesis and expressed on endothelial cells, also has a functional role in leukocyte migration.45,46 We analyzed two CRC-LM patient tissues for detailed distribution of CD31 (Fig. S5). Nevertheless, CD31 area was generally very high and showed strong heterogeneity among patients (∼700–8,000 mm2). In addition to that, CD31 can be expressed by leukocytes including T cells and thereby even having inhibitory effects on T cell activation46,47 underscoring its highly diverse role in tumors. In this study, we performed a high-resolution analysis of different immune cell subsets. Consequently, the detailed functional mechanisms and cell interactions involved in the detected spatial T cell distribution patterns need to be investigated in future studies. The clear limitation of our study is the low sample number for some markers that limited our ability to identify statistically significant differences between the distance classes.
Whereas we could not see distinct patterns limited to single patients (data not shown), the clustering analysis revealed that specific groups of patients show patterns of CD3 T cell aggregates, which were limited to a given region (e.g., 0–10 µm or 20–30 µm, Fig. 3). Indeed, OS analysis showed that patients with high T cell densities (CD3 as well as CD8+) within the >10 µm distance class had a survival benefit compared with patients with low T cells in the aforementioned distance (Figs. 4–6). Interestingly, the opposite effect was found at the 20–30 µm distance from the tumor. Here, low CD3 T cell numbers had positive effects on patient survival. Consequently, despite the physical or functional T cells barrier, the observed pattern of T cell distribution at 20–30 µm distance obviously is beneficial for patients (under palliative therapy). When considering this observation from the other side, high T cell numbers unexpectedly might have a tumor promoting effect. Our own previous work illustrated the exploitation of CCL5-producing T cells in the invasive margin of CRC-LM patients by supporting CCL5 receptor (CCR5)-expressing tumor cells.19 Furthermore, blockade of CCR5 led to repolarization of TAM resulting in beneficial clinical responses. These findings might explain, why especially in patients with low T cells in the 20–30 µm region a better survival is observed than in patients with high T cell numbers, and furthermore, it shows that reeducated macrophages, which were found to be increased at the 20–30 µm distance and which are in fact generally high in the invasive margin, can render antitumor effects under therapy. In addition to that, our results indicate that T cell induced antitumor effects can be conducted by T cells that have engaged the tumor margin at the 0–10 µm distance class, which is indeed also true for the cytotoxic CD8+ subtype, while T cells further away remain blocked. The contrary outcome of our detailed distribution analysis that both localization-dependent high as well as low T cell numbers have clinical benefit might explain startling observations for similar tumors, where in one study high48 and in another study low T cell densities49 indicate prognostic relevance. Our finding might further elucidate related controversial observations, where studies identify high densities of immunosuppressive cell populations being associated with favorable prognosis.31,50,51 The reasons for the controversial observation of local T cell decrease being associated with favorable prognosis and if the observed patterns change under therapy are currently investigated. The paradoxical observation of localization-dependent prognostic relevance of T cell densities is only decipherable by detailed spatial analyses. Therefore, the analysis of spatial distribution patterns at higher resolution, and especially the distance classes <10 and 20–30 µm alone, could serve as more precise prognostic markers for responses to chemotherapy and OS compared with cell densities measured on a broader scale.
In summary, we present here for the first time detailed data on the local spatial distribution of immune cells in colorectal cancer LM. Thereby, distinct patterns of immune cells reveal a physical or functional T cell barrier at 20–30 µm distance from the tumor epithelium with decreased T cell and cytotoxic Gran B densities as well as increased CD163 macrophages. These specific distribution patterns with either high T cells at the direct tumor border or low T cells at the 20–30 µm distance are associated with improved OS. This detailed analysis of spatial profiles within the microenvironment, therefore, represents new insights into T cell distribution, function as well as influence of T cell localization on clinical responses and introduces possible spatial immunosuppressive hurdles that need to be overcome by successful immunotherapies. We suggest the analysis of spatial profiles within the < 10 and 20–30 µm distance from the tumor epithelium as new predictive biomarker for survival and response to therapies.
Materials and methods
Tissue samples
The study was approved by the Medical Ethics Committee of the University of Heidelberg. Tumor samples were used after receiving a signed informed consent from all 36 patients. Patients were included in this study when diagnosed with stage UICC IV CRC after metastatic disease in a surgically resected liver metastasis was histologically proven. None of the patients received neoadjuvant treatment. Samples had to contain (a) metastasis, (b) invasive margin and (c) adjacent liver (Fig. 1A). Patients received standard of care palliative therapies and follow-up was obtained as described previously.13 All samples were screened for microsatellite instability (MSI) using established protocols.21 No MSI sample was included in this analysis. Clinical data as well as detailed pathological reports showed no evidence for the presence of ethyltoxic liver damage, chronic inflammation due to ongoing infections (e.g., hepatitis) or other chronic inflammatory conditions.
Immunohistochemistry and immunofluorescence staining
Tissue sections (4 µm) were prepared from formalin-fixed, paraffin-embedded material for sequent immunohistochemically detection of T cells (CD3+, CD8+), Granzyme B (GranB+), B cells (CD20+) and TAM (CD163+). All processing steps were performed with a fully automated staining system (Leica BOND-MAX™, Leica Microsystems, Germany) to achieve maximum reproducibility. Briefly, after deparaffinization and rehydration, slides were boiled for 20 min in either 10 mM citrate buffer (pH 6) for CD3, CD8+ and CD20 or 1 mM EDTA buffer (pH8) for CD163 and GranB to retrieve the antigens. The endogenous peroxidase activity was blocked by incubation with 3–4% (v/v) hydrogen peroxide for 10 min. The sections were blocked with 10% normal goat serum. Mouse monoclonal antibodies recognizing human CD3ε (1:50 dilution, clone PS1, Acris #DM112–05), CD8+ (1:50 dilution, clone 4B11, Novocastra #NCL-CD8–4B11), CD20 (1:50 dilution, clone L26, Novocastra #NCL-L-CD20-L26), GranB (1:20 dilution, clone 11F1, Novocastra #NCL-Gran-B) and human CD163 (1:500 dilution, clone EDHu-1, AbDSerotec #MCA1853) were applied as primary antibodies for 30 min at room temperature. Slides were then incubated with the secondary antibodies and subsequent visualization was performed according to the manufacturer's instructions (Bond Polymer Refine Detection Kit, Leica #DS9800). Antigen detection was performed by a color reaction with 3,3-di-amino-benzidine (DAB). The sections were counterstained with hematoxylin and mounted with Aquatex (VWR #1085620050). Controls without primary antibody and isotype controls were used for all antibodies. Positive controls for the presence of T cells and macrophages consisted of adjacent normal tissue.
For immunofluorescence double staining of CD163 (1:1000 dilution, clone EPR19518, abcam #ab182422) and PD-L1 (1:100 dilution, clone 29E.2A3, BioLegend #329710) deparaffinization and rehydration of slides was performed following 20 min in 1 mM EDTA buffer (pH8) to retrieve the antigens. After incubation of the first primary antibodies overnight at 4 °C, secondary antibodies (Alexa Fluor 594, Life Technologies #A21207; Alexa Fluor 488, Life Technologies #A-11029) were applied for 1 h. DAPI (AppliChem #1001) was used for counterstain and sections were mounted using Vectashield (Vector #H-1000).
Evaluation of immunostainings and tissue classification
Using the NDP Nanozoomer system (Hamamatsu Photonics, Japan) complete microscopic images of full tissue sections were automatically obtained. Cell quantification and distance analysis were performed with the VisioMorph software (Visiopharm, Denmark). Image segmentation and subsequent classification according to color and morphology was established on a separate training set with manually counted cell numbers previously.22 This approach in combination with serial cuts of tissue blocks allows robust and reproducible large scale histological evaluations with high precision across complete (adjacent) sections.
1 mm2 regions of the invasive margin (Fig. 1A) were generated in an automated process, visually controlled and adapted correspondingly. Then, the positions of the T cells in relation to the tumor epithelium were identified in consecutive 10 µm steps (Fig. 1B). Distances to tumor epithelium were grouped into distinct classes (distance classes), e.g., “below 10 µm,” “10–20 µm,” “20–30 µm” and so on. So for each tissue section, T cell quantities in 10 distance classes within 100 µm from the tumor epithelium into the normal adjacent liver were established. Immune cells and other markers were quantified in this process by spectral and morphological criteria as described before15,22 in combination with aggregation of data from the spatial registry.
To evaluate the distance between CD163 macrophages and T cells within the tumor microenvironment, serial sections were stained with the respective markers and after scanning of the sections, these were computationally analyzed using VisioMorph software (Fig. 7).
Statistical analyses
Statistical analyses were performed using the SPSS 18.0 and 22 packages (IBM) as well as GraphPad Prism version 5. To discriminate CD3 cell counts between distance classes Kruskal–Wallis tests with post-hoc Mann–Whitney U tests were performed. Results with two-tailed p values <0.01 were judged to be statistically significant. Boxplots depict the minimum, first quartile, median, third quartile and maximum. Measurements with values between 1.5 and 3 lengths of the box away from the upper and lower border of the box were classified as outliers (white circles). The length of the box equals the interquartile range. To discriminate cell counts between single distance classes for CD8+ and GranB Wilcoxon signed-rank and for CD20 and CD163 paired t-tests were performed. Results with two-tailed p values <0.05 were judged to be statistically significant. Unsupervised hierarchical clustering analysis using Euclidean distance was performed with the R software package.23 OS curves were computed using the Kaplan–Meier estimator (Breslow test) and significance was assumed for p values of 0.05 or less. Cutoff values were adapted (quantile-quantile class) from previous work,13 which is in line with findings from primary CRC tumors.24
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors thank the NCT tissue bank for their support.
Funding
This work was funded by the Helmholtz Alliance on Immunotherapy of Cancer (NH) and through a grant from the Cancer Research Institute (DJ). The study sponsors had not role in the study design, in the collection, analysis and interpretation of data; in the writing of the manuscript; and in the decision to submit the manuscript for publication.
References
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Smigal C, Thun MJ. Cancer Statistics, 2006. CA Cancer J Clin 2006; 56:106-30; PMID:16514137; http://dx.doi.org/ 10.3322/canjclin.56.2.106 [DOI] [PubMed] [Google Scholar]
- 2.O'Connell JB, Maggard MA, Ko CY. Colon cancer survival rates with the new American Joint Committee on Cancer sixth edition staging. J Natl Cancer Inst 2004; 96:1420-5; PMID:15467030; http://dx.doi.org/ 10.1093/jnci/djh275 [DOI] [PubMed] [Google Scholar]
- 3.Pihl E, Nairn R, Milne B, Cuthbertson A, Hughes E, Rollo A. Lymphoid hyperplasia: a major prognostic feature in 519 cases of colorectal carcinoma. Am J Pathol 1980; 100:469-80; PMID:7406021 [PMC free article] [PubMed] [Google Scholar]
- 4.Ohtani H. Focus on TILs: Prognostic significance of tumor infiltrating lymphocytes in human colorectal cancer. Cancer Immun 2007; 7:4; PMID:17311363 [PMC free article] [PubMed] [Google Scholar]
- 5.Ropponen KM, Eskelinen MJ, Lipponen PK, Alhava E, Kosma VM. Prognostic value of tumour-infiltrating lymphocytes (TILs) in colorectal cancer. J Pathol 1997; 182(3):318-24; PMID:9349235; http://dx.doi.org/ [DOI] [PubMed] [Google Scholar]
- 6.Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pagès C, Tosolini M, Camus M, Berger A, Wind P et al.. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science (80) 2006; 313:1960-4; PMID:17008531; http://dx.doi.org/16371631 10.1126/science.1129139 [DOI] [PubMed] [Google Scholar]
- 7.Pagès F, Berger A, Camus M, Sanchez-Cabo F, Costes A, Molidor R, Mlecnik B, Kirilovsky A, Nilsson M, Damotte D et al.. Effector memory T cells, early metastasis, and survival in colorectal cancer. N Engl J Med 2005; 353:2654-66; PMID:16371631; http://dx.doi.org/ 10.1056/NEJMoa051424 [DOI] [PubMed] [Google Scholar]
- 8.Naito Y, Saito K, Shiiba K, Ohuchi A, Saigenji K, Nagura H, Ontani1 H. CD8+ T cells infiltrated within cancer cell nests as a prognostic factor in human colorectal cancer. Cancer Res 1998; 58:3491-4; PMID:9721846 [PubMed] [Google Scholar]
- 9.Prall F, Duehrkop T, Weirich V, Ostwald C, Lenz P, Nizze H, Barten M. Prognostic role of CD8+ tumor-infiltrating lymphocytes in stage III colorectal cancer with and without microsatellite instability. Hum Pathol 2004; 35(7):808-16; PMID:15257543; http://dx.doi.org/ 10.1016/j.humpath.2004.01.022 [DOI] [PubMed] [Google Scholar]
- 10.Diederichsen ACP, Hjelmborg JVB, Christensen PB, Zeuthen J, Fenge C. Prognostic value of the CD4+/CD8+ ratio of tumour infiltrating lymphocytes in colorectal cancer and HLA-DR expression on tumour cells. Cancer Immunol Immunother 2003; 52:423-8; PMID:12695859; http://dx.doi.org/ 10.1007/s00262-003-0388-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Menon AG, Janssen-van Rhijn CM, Morreau H, Putter H, Tollenaar RA, van de Velde CJH, Fleuren GJ, Kuppen PJK. Immune system and prognosis in colorectal cancer: a detailed immunohistochemical analysis. Lab Invest 2004; 84:493-501; PMID:14968119; http://dx.doi.org/ 10.1038/labinvest.3700055 [DOI] [PubMed] [Google Scholar]
- 12.Salama P, Phillips M, Grieu F, Morris M, Zeps N, Joseph D, Platell C, Iacopetta B. Tumor-infiltrating FOXP3+ T regulatory cells show strong prognostic significance in colorectal cancer. J Clin Oncol 2009; 27:186-92; PMID:19064967; http://dx.doi.org/ 10.1200/JCO.2008.18.7229 [DOI] [PubMed] [Google Scholar]
- 13.Halama N, Michel S, Kloor M, Zoernig I, Benner A, Spille A, Pommerencke T, Von Knebel Doeberitz M, Folprecht G, Luber B et al.. Localization and density of immune cells in the invasive margin of human colorectal cancer liver metastases are prognostic for response to chemotherapy. Cancer Res 2011; 71:5670-7; PMID:21846824; http://dx.doi.org/ 10.1158/0008-5472.CAN-11-0268 [DOI] [PubMed] [Google Scholar]
- 14.Emile J, Mauer M, Tanis E, Julie C, Nordlinger B, Aust D, Roth A, Lutz MP, Gruenberger T, Wrba F et al.. Prognostic impact of immune response in resectable colorectal liver metastases treated by surgery alone or surgery with perioperative FOLFOX in the randomised EORTC study 40983. Eur J Cancer 2015; 51(17):2708-17; PMID:26342674; http://dx.doi.org/ 10.1016/j.ejca.2015.08.014 [DOI] [PubMed] [Google Scholar]
- 15.Halama N, Zoernig I, Spille A, Michel S, Kloor M, Grauling-Halama S, Westphal K, Schirmacher P, Jäger D, Grabe N. Quantification of prognostic immune cell markers in colorectal cancer using whole slide imaging tumor maps. Anal Quant Cytol Histol 2010; 32:333-40; PMID:21456345 [PubMed] [Google Scholar]
- 16.Halama N, Spille A, Lerchl T, Brand K, Herpe E, Welte S, Keim S, Lahrmann B, Klupp F, Kahlert C et al.. Hepatic metastases of colorectal cancer are rather homogeneous but differ from primary lesions in terms of immune cell infiltration. Oncoimmunology 2013; 2:e24116; PMID:23734335; http://dx.doi.org/ 10.4161/onci.24116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Halama N, Michel S, Kloor M, Zoernig I, Pommerencke T, Von M, Doeberitz K, Schirmacher P, Weitz J, Grabe N et al.. The localization and density of immune cells in primary tumors of human metastatic colorectal cancer shows an association with response to chemotherapy. Cancer Immun 2009; 9:1; PMID:19226101 [PMC free article] [PubMed] [Google Scholar]
- 18.Wu AA, Drake V, Huang H-S, Chiu S, Zheng L. Reprogramming the tumor microenvironment: tumor-induced immunosuppressive factors paralyze T cells. Oncoimmunology 2015; 4:e1016700; PMID:26140242; http://dx.doi.org/ 10.1080/2162402X.2015.1016700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Halama N, Zoernig I, Berthel A, Kahlert C, Klupp F, Suarez-Carmona M, Suetterlin T, Brand K, Krauss J, Lasitschka F et al.. Tumoral immune cell exploitation in colorectal cancer metastases can be targeted effectively by Anti-CCR5 therapy in cancer patients. Cancer Cell 2016; 29:587-601; PMID:27070705; http://dx.doi.org/ 10.1016/j.ccell.2016.03.005 [DOI] [PubMed] [Google Scholar]
- 20.Coussens LM, Zitvogel L, Palucka AK. Neutralizing tumor-promoting chronic inflammation: a magic bullet? Science 2013; 339:286-91; PMID:23329041; http://dx.doi.org/ 10.1126/science.1232227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Findeisen P, Kloor M, Merx S, Sutter C, Woerner SM, Dostmann N, Benner A, Dondog B, Pawlita M, Dippold W et al.. T25 repeat in the 3´untranslated region of the CASP2 gene: A sensitive and specific marker for microsatellite instability in colorectal cancer. Cancer Res 2005; 65:8072-8; PMID:16166278; http://dx.doi.org/ 10.1158/0008-5472.CAN-04-4146 [DOI] [PubMed] [Google Scholar]
- 22.Halama N, Zoernig I, Spille A, Westphal K, Schirmacher P, Jaeger D, Grabe N. Estimation of immune cell densities in immune cell conglomerates: An approach for high-throughput quantification. PLoS One 2009; 4:e7847; PMID:19924291; http://dx.doi.org/ 10.1371/journal.pone.0007847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.R Development Core Team R: A Language and Environment for Statistical Computing [Internet]. R Found Stat Comput. Vienna Austria: 2015; Available from: http://www.r-project.org/; PMID:26321800 [Google Scholar]
- 24.Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pagès C, Tosolini M, Camus M, Berger A, Wind P et al.. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Sci (New York, NY) 2006; 313:1960-4; PMID:17008531; http://dx.doi.org/18774635 10.1126/science.1129139 [DOI] [PubMed] [Google Scholar]
- 25.Guidolin D, Nico B, Crivellato E, Marzullo A, Vacca A, Ribatti D. Tumoral mast cells exhibit a common spatial distribution. Cancer Lett 2009; 273(1):80-5; PMID:18774635; http://dx.doi.org/ 10.1016/j.canlet.2008.07.032 [DOI] [PubMed] [Google Scholar]
- 26.Valous NA, Lahrmann B, Halama N, Bergmann F, Jäger D, Grabe N. Spatial intratumoral heterogeneity of proliferation in immunohistochemical images of solid tumors. Med Phys 2016; 43:2936; PMID:27277043; http://dx.doi.org/ 10.1118/1.4949003 [DOI] [PubMed] [Google Scholar]
- 27.Zitvogel L, Tesniere A, Kroemer G. Cancer despite immunosurveillance: immunoselection and immunosubversion. Nat Rev Immunol 2006; 6:715-27; PMID:16977338; http://dx.doi.org/ 10.1038/nri1936 [DOI] [PubMed] [Google Scholar]
- 28.Keim S, Zoernig I, Spille A, Lahrmann B, Brand K, Herpel E, Grabe N, Jäger D, Halama N. Sequential metastases of colorectal cancer immunophenotypes and spatial distributions of infiltrating immune cells in relation to time and treatments. Oncoimmunology 2012; 1:593-9; PMID:22934251; http://dx.doi.org/ 10.4161/onci.20179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herman Fridman W, Pagès F, Sautès-Fridman C, Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat Publ Gr 2012; 12:298-306; PMID:22419253; http://dx.doi.org/21325295 10.1038/nrc3245 [DOI] [PubMed] [Google Scholar]
- 30.Halama N, Braun M, Kahlert C, Spille A, Quack C, Rahbari N, Koch M, Weitz J, Kloor M, Zoernig I et al.. Natural killer cells are scarce in colorectal carcinoma tissue despite high levels of chemokines and cytokines. Clin Cancer Res 2011; 17:678-89; PMID:21325295; http://dx.doi.org/ 10.1158/1078-0432.CCR-10-2173 [DOI] [PubMed] [Google Scholar]
- 31.Ruffell B, Coussens LM. Macrophages and therapeutic resistance in cancer. Cancer Cell 2015; 27:462-72; PMID:25858805; http://dx.doi.org/ 10.1016/j.ccell.2015.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Komohara Y, Jinushi M, Takeya M. Clinical significance of macrophage heterogeneity in human malignant tumors. Cancer Sci 2014; 105:1-8; PMID:24168081; http://dx.doi.org/ 10.1111/cas.12314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol 2008; 8:958-69; PMID:19029990; http://dx.doi.org/ 10.1038/nri2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuang DM, Zhao Q, Peng C, Xu J, Zhang J-P, Wu C, Zheng L. Activated monocytes in peritumoral stroma of hepatocellular carcinoma foster immune privilege and disease progression through PD-L1. J Exp Med 2009; 206:1327-37; PMID:19451266; http://dx.doi.org/ 10.1084/jem.20082173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kryczek I, Zou L, Rodriguez P, Zhu G, Wei S, Mottram P, Brumlik M, Cheng P, Curiel T, Myers L et al.. B7-H4 expression identifies a novel suppressive macrophage population in human ovarian carcinoma. J Exp Med 2006; 203:871-81; PMID:16606666; http://dx.doi.org/ 10.1084/jem.20050930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwartz M, Zhang Y, Rosenblatt JD. B cell regulation of the anti-tumor response and role in carcinogenesis. J Immunother Cancer 2016; 4(40):1–15; PMID:27437104; http://dx.doi.org/ 10.1186/s40425-016-0145-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berntsson J, Nodin B, Eberhard J, Micke P, Jirström K. Prognostic impact of tumour-infiltrating B cells and plasma cells in colorectal cancer. Int J Cancer 2016; 139:1129-39; PMID:27074317; http://dx.doi.org/ 10.1002/ijc.30138 [DOI] [PubMed] [Google Scholar]
- 38.Nielsen JS, Sahota RA, Milne K, Kost SE, Nesslinger NJ, Watson PH, Nelson BH. CD20+ tumor-infiltrating lymphocytes have an atypical CD27 - memory phenotype and together with CD8+ T cells promote favorable prognosis in ovarian cancer. Clin Cancer Res 2012; 18:3281-92; PMID:22553348; http://dx.doi.org/ 10.1158/1078-0432.CCR-12-0234 [DOI] [PubMed] [Google Scholar]
- 39.Shimabukuro-Vornhagen A, Schlößer HA, Gryschok L, Malcher J, Wennhold K, Garcia-Marquez M, Herbold T, Neuhaus LS, Becker HJ, Fiedler A et al.. Characterization of tumor-associated B-cell subsets in patients with colorectal cancer. Oncotarget 2014; 5:4651-64; PMID:25026291; http://dx.doi.org/ 10.18632/oncotarget.1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lundgren S, Berntsson J, Nodin B, Micke P, Jirström K. Prognostic impact of tumour-associated B cells and plasma cells in epithelial ovarian cancer. J Ovarian Res 2016; 9:21; PMID:27048364; http://dx.doi.org/ 10.1186/s13048-016-0232-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gunderson AJ, Kaneda MM, Tsujikawa T, Nguyen AV, Affara NI, Ruffell B, Gorjestani S, Liudahl SM, Truit M, Olson P et al.. Bruton tyrosine kinase–Dependent immune cell cross-talk drives pancreas cancer. Cancer Discov 2016; 6:270-85; PMID:26715645; http://dx.doi.org/ 10.1158/2159-8290.CD-15-0827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bromley SK, Peterson DA, Gunn MD, Dustin ML. Cutting edge: hierarchy of chemokine receptor and TCR signals regulating T cell migration and proliferation. J Immunol 2000; 165:15-9; PMID:10861029; http://dx.doi.org/ 10.4049/jimmunol.165.1.15 [DOI] [PubMed] [Google Scholar]
- 43.Geiger B, Bershadsky A, Pankov R, Yamada KM, Correspondence BG. Transmembrane extracellular matrix– cytoskeleton crosstalk. Nat Rev Mol Cell Biol 2001; 2:793-805; PMID:11715046; http://dx.doi.org/ 10.1038/35099066 [DOI] [PubMed] [Google Scholar]
- 44.Xia N, Thodeti CK, Hunt TP, Xu Q, Ho M, Whitesides GM, Westervelt R, Ingber DE. Directional control of cell motility through focal adhesion positioning and spatial control of Rac activation. FASEB J 2008; 22(6):1649-59; PMID:18180334; http://dx.doi.org/12417630 10.1096/fj.07-090571 [DOI] [PubMed] [Google Scholar]
- 45.Dangerfield J, Larbi KY, Huang MT, Dewar A, Nourshargh S. PECAM-1 (CD31) homophilic interaction up-regulates alpha6beta1 on transmigrated neutrophils in vivo and plays a functional role in the ability of alpha6 integrins to mediate leukocyte migration through the perivascular basement membrane. J Exp Med 2002; 196:1201-11; PMID:12417630; http://dx.doi.org/ 10.1084/jem.20020324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feng Y, Chen X, Zhang X. Roles of PECAM-1 in cell function and disease progression. Eur Rev Med Pharmacol Sci 2016; 20(19):4082-8; PMID:27775789 20400708 [PubMed] [Google Scholar]
- 47.Fornasa G, Groyer E, Clement M, Dimitrov J, Compain C, Gaston A-T, Varthaman A, Khallou-Laschet J, Newman DK, Graff-Dubois S et al.. TCR stimulation drives cleavage and shedding of the ITIM receptor CD31. J Immunol 2010; 184:5485-92; PMID:20400708; http://dx.doi.org/ 10.4049/jimmunol.0902219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Houdt IS, Sluijter BJR, Moesbergen LM, Vos WM, De Gruijl TD, Molenkamp BG, Van Den Eertwegh AJM, Hooijberg E, Van Leeuwen PAM, Meijer CJLM et al.. Favorable outcome in clinically stage II melanoma patients is associated with the presence of activated tumor infiltrating T-lymphocytes and preserved MHC class I antigen expression. Int J Cancer 2008; 123:609-15; PMID:18498132; http://dx.doi.org/ 10.1002/ijc.23543 [DOI] [PubMed] [Google Scholar]
- 49.Hillen F, Baeten CIM, Van De Winkel A, Creytens D, Van Der Schaft DWJ, Winnepenninckx V, Griffioen AW. Leukocyte infiltration and tumor cell plasticity are parameters of aggressiveness in primary cutaneous melanoma. Cancer Immunol Immunother 2008; 57:97-106; PMID:17602225; http://dx.doi.org/ 10.1007/s00262-007-0353-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Q, Liu L, Gong C, Shi H, Zeng Y, Wang X. Prognostic significance of tumor-associated macrophages in solid tumor: A meta-analysis of the literature. PLoS One 2012; 7:e50946; PMID:23284651; http://dx.doi.org/21644034 10.1371/journal.pone.0050946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ladoire S, Martin F, Ghiringhelli F. Prognostic role of FOXP3+ regulatory T cells infiltrating human carcinomas: The paradox of colorectal cancer. Cancer Immunol Immunother 2011; 60:909-18; PMID:21644034; http://dx.doi.org/ 10.1007/s00262-011-1046-y [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.