Abstract
Background
Acute viral hepatitis E (AVH-E) can often result in acute liver failure (ALF) during pregnancy. microRNAs serve as mediators in drug induced liver failure. We investigated their role as a biomarker in predicting ALF due to HEV (ALF-E).
Methods
We performed next generation sequencing and subsequent validation studies in PBMCs of pregnant (P) self limiting AVH-E, ALF due to HEV (ALF-E) and compared with AVH-E in non-pregnant (NP) females and healthy controls.
Findings
Eleven microRNAs were significantly expressed in response to HEV infection; importantly, miR- 431, 654, 1468 and 4435, were distinctly expressed in pregnant self-limiting AVH-E and healthy females (p = 0.0005), but not in ALF-E. Sixteen exclusive microRNAs differentiated ALF-E from self limiting AVH-E in pregnant females. miR-450b which affects cellular proliferation and metabolic processes through RNF20 and SECB was predominanlty upregulated and correlated with poor outcome (ROC 0.958, p = 0.001).
Interpretation
Our results reveal that a specific microRNA profile can predict fatality in ALF-E in pregnancy. These microRNAs could be exploited as prognostic biomarkers and help in the development of new therapeutic interventions.
Keywords: Health sciences, Virology
1. Introduction
Hepatitis E virus (HEV) infection causes acute viral hepatitis (AVH-E), which is generally a mild and self-limiting disease but can be fatal in the presence of pre-existing liver disease or in pregnancy [1]. The spectrum of infection in pregnancy may vary from asymptomatic transaminitis to severe hepatitis leading to liver failure [2]. During pregnancy, about a quarter of infected women with HEV develop liver failure with high maternal and fetal morbidity and mortality [3]. The cascade of events that precede development of acute liver failure (ALF) in these patients is not well understood. Transcriptional signatures have been used to unravel pathways that dictate immune responses in chronic viral infections and liver failure [4, 5]. Pathogen activated miR expression profiles are crucial for the initiation of pathogenic events and disease outcomes. Similarly, gene expression regulated by specific miRs may vary in response to different infections, in pregnancy and liver failure. microRNA (miRs) profiling has been used in acute hepatitis, drug induced ALF, hepatic encephalopathy etc [6, 7, 8].
microRNAs are a class of highly conserved non-coding single stranded RNA molecules of 18–24 nucleotides that regulate gene expression post-transcriptionally and modulate a range of biological and cellular processes [9, 10]. miR profiles indicate that miR-21, miR-122 and miR-221 are involved in liver regeneration and contribute to spontaneous recovery from ALF [11]. Down regulation of miR-122 has been observed in acute and chronic liver diseases. Its levels inversely correlate with hepatic injury, and may have a bearing in pathogenesis of ALF [12]. However, very few miRs have been investigated In the pathogenesis of ALF [13, 14]. miR-122 has been shown to bind to adjacent sites in the 5′ non-coding region of HCV genomes and regulate HCV RNA accumulation. Anti-miR-122 strategy in HCV-infected chimpanzees showed reduction in HCV RNA levels and liver injury [13]. miR-1, miR-372 and miR-373 have been shown to enhance HBV replication and gene expression [5, 15]. There are no reports of micro-RNA profiles in HEV infection, regulation of infection and hepatic injury, including studies of micro-RNA profile following HEV infection in preganant females.
In this study, we undertook to identify distinctive miR signatures, their gene targets and pathways in a pilot cohort of pregnant and non-pregnant females suffering from acute self-limiting HEV infection or acute liver failure. After the initial discovery cohort, we validated the miR signatures of HEV infection and acute liver failure in a validation cohort of preganant HEV infected females.
2. Methods
2.1. Patients and healthy subjects
In this case control prospective study, thirteen pregnant females with ALF-E, 40 with AVH-E, 20 healthy and 15 non-pregnant females with AVH-E were enrolled between 2012–2014, at the Department of Obstetrics and Gynaecology, Lady Hardinge Medical College, and the Department of Hepatology, Institute of Liver and Biliary Sciences, New Delhi. Most of the AVH-E, ALF-E and healthy pregnant females were in the 26th to 28th week of gestation. Blood samples were collected at the time of hospitalisation. Patients were characterised as described in our earlier study [16] and described briefly as below: Acute viral hepatitis patients had prodromal symptoms, jaundice and elevated serum aminotransferase levels (>5 ULN). Acute liver failure (ALF) was defined by development of any grade of hepatic encephalopathy and coagulopathy (INR > 1.5) within 4 weeks of onset of jaundice. Hepatic encephalopathy was defined and graded using West Haven criteria. Hepatitis E related acute viral hepatitis or ALF was diagnosed on postitivity for anti-HEV IgM and negative for other etiologies (hepatic viruses, drugs, autoimmunity). Healthy females who served as control, had no known past or present history or clinical features of any liver disease and were negative for IgM anti-HAV, HBsAg, Anti-HCV and HIV with normal ALT values. Anti-HEV IgM was determined by micro capture ELISA kit (Wantai biological, Beijing, China). HEV RNA was quantified using RT-PCR method.
2.2. Ethics committee approval
The study protocol was approved by the institutional ethical committee (No. F.8(10)05-06/AC/ILBS/Pt.File/1564) dated 26/5/10 and an informed consent was obtained from each recruited patient or her relatives and healthy subjects.
In this pilot study, peripheral blood mononuclear cells (PBMCs) were isolated from the whole blood in five subjects from each group. To normalise the results, equal number of PBMCs (1–2 × 106) from each subject were pooled. Total RNA was isolated from pooled PBMCs using miRVANA kit (Invitrogen, Bangalore, India) for miR and mRNA analysis.
Subsequently, for the validation of miR and gene expression by QRT-PCR, 1–2 ug of total RNA was isolated from PBMCs of new and independent samples (validation cohort) to make complementary DNA and used for QRT-PCR. Quality and integrity of the total RNA was determined by using Agilent Technologies 2100 Bioanalyzer (Agilent Technologies, Waldbronn, Germany).
2.3. Flow cytometric analysis
Freshly isolated PBMCs were stained with anti-CD14 FITC, anti-CD11b PE for monocytes and macrophages, anti-DC-SIGN-PE/Cy7 for Lin-ve DCs, anti-CD19 Pecy7, anti-CD38 FITC for B cells, anti-CD3 Pecy7, anti-CD4 PE and anti-CD8APC for T cells were used. Intracellular expression of TLRs was analyzed by using anti-TLR3-PE, TLR7-PE, and TLR9-allophycocyanin (BD Pharmingen, San Jose, CA and eBioscience, Inc., San Diego, CA) for flow cytometry staining.
Cells were acquired for flow cytometric analysis using FACSCalibur, Cyan (DAKO) and the results were analyzed using Flow-Jo software8.7.
2.4. Small RNA deep sequencing
Total RNA was size-fractionated on a 15% PAGE gel, and a 16–30 nt fraction was collected. The 5' RNA adapter (5'-GUUCAGAGUUCUACAGUCCGACGAUC-3') was ligated to the RNA pool with T4 RNA ligase. Ligated RNA was size-fractionated on a 15% agarose gel, and a 40–60 nt fraction excised. The 3' RNA adapter (5'-pUCGUAUGCCGUCUUCUGCUUGidT-3'; p, phosphate; idT, inverted deoxythymidine) was subsequently ligated to precipitated RNA using T4 RNA ligase. Ligated RNA was size-fractionated on a 10% agarose gel, and the 70–90 nt fraction (small RNA + adaptors) excised. Small RNAs ligated with adaptors were subjected to RT-PCR (Superscript II reverse transcriptase, 15 cycles of amplification) to produce sequencing libraries. PCR products were purified and small RNA libraries were sequenced using Illumina GAIIx, a massively parallel sequencing technology.
2.5. Microarray analysis
For hybridization, biotin-labeled cRNA samples were prepared according to Illumina's sample labeling procedure. cRNA concentrations were measured using Nanodrop, ND-1000. Labeled, amplified material (750 ng per array) was hybridized to a version3 of the Illumina Human-Ht-12 BeadChip (48 K) according to the Manufacturer's instructions (Illumina, Inc., San Diego, CA).
Methods for RNA sequencing analysis and microarray data analysis, integration of miR and gene expression data, regulatory network modelling and validation of miR and target genes by qRT-PCR.
2.6. Integration of miR and gene expression data
A list of biologically predicted targets of differentially expressed miRs found in our study, was determined by using miRBase. To determine the possible miR − mRNA interactions,the list of targets was compared with the list of differentially expressed genes.
Differentially expressed genes that are negatively regulated by target miR were subjected to biological significance analysis using GOElite tool. Gene ontology categories and pathways that were significantly dysregulated (p < 0.05 and Z score > 2) were considered.
2.7. RNA sequencing and microarray data analysis
Low quality reads were trimmed using NGSQC Tool kit. After elimination of redundancy, sequences ≥18 nt were mapped to the Human genome build HG19. Sequences that perfectly matched the genome along their entire length were considered for subsequent analyses. Genome sequences and annotations of the human genome (Hg19) were downloaded from NCBI. After alignment to the genome, read counts were normalized calculating miRs per million miR alignments. The list of significantly regulated miRs was created using a t-test analysis (p ≤ 0.05) and fold change (≥1.5) cut-off between two conditions (eg, HEV infected vs healthy controls).
For microarray analysis, Intra-array and Inter-array normalization was done by Quantile normalization for each chip/samples and by taking median of all the samples. Volcano plot was used to find out genes that are 2 fold differentially expressed between any 2 conditions (eg.: infected Vs. uninfected) by applying unpaired Student’s t-test for significnce (p < 0.05) and Benjamini Hocheberg based FDR correction. Hierarchical clustering of differentially expressed genes was done by Pearson uncentered algorithm with Average linkage rule to identify up and down regulated gene clusters. The statistical significance of intergroup differences for continuous variables was evaluated by Mann-Whitney U test and for categorical variables with chi-square test.
2.8. miR: mRNA regulatory network modelling
Regulatory network modelling of miR (35 miR) and their target mRNA (172 genes) along with biological categories dysregulated was done using Integromatrix Software from Bionivid Technology Pvt Ltd, Bangalore, India. The resultant file of Integromatrix software was imported into Cytoscape V 8.0. Further Cytoscape V 8.0 was used to model the biological network with emphasis to the connectivity score and p value to understand the synchronisation of miR:mRNA:Pathways in regulation of infection induced regulatory changes.
2.9. Identification of immune cell subsets associated with miRs and their targeted genes
To identify the active set of cells or pathways associated with miRs and their target genes we applied stringent cut-offs in gene data collected from Illumina Human-Ht-12 48 K bead chip. Segmental analysis was done to understand miR mediated gene regulation in pathway specific manner. Further, genes which satisfy a stringent statistical criteria of corrected p value <=0.01, MFE of ≤-7 Kcal/mol and with a 3′ bias were considered for downstream analysis. Thorough review of literature and analysis of pathway databases resulted in identification of genes involved in immune responses, signalling pathways and cellular processes. Experimentally validated targets were considered along with the predicted ones. The list of predicted targets was then compared to our list of differentially expressed genes to conclude possible miR–mRNA interactions.
2.10. Validation of miR and target genes using quantitative RT-PCR (q-RT-PCR)
To validate miR expression data, two step qRT-PCR, cDNA synthesis and real-time PCR amplification was performed. Minimum of 1–2 μg of total RNA was reverse transcribed by using Universal cDNA synthesis kit (Exiqon, USA) and specific miRs primers (Exiqon San Francisco, USA). Further miR PCR Kit (Exiqon, USA) amplification was performed in Roche RT-PCR machine. 5S-rRNA was used as an endogenous control for normalization.
To validate mRNA expression, qRT-PCR was performed in triplicate in ViiA7 real-time PCR machine (ABI, Whitefield, Bangalore, India) using Syber Green (Sigma, St. Louis, MO, USA) with specific primers for genes (Table 1). 18sRNA served as endogenous control for normalization. Relative quantification of miR and each gene was determined by calculating the Log RQ of each sample’s Ct value.
Table 1.
Primers for Genes which were used for qRT-PCR.
| Gene | Primer Sequence |
|---|---|
| BIRC3 | 5′-GTTTCTGTGGCTTGCCTTCA-3′ 3′-TTTCCTCCCCTCACTTGGTC-5′ |
| GDPD1 | 5′-AGAAGCAGCGATTCCTCAGT-3′ 3′-CAAGGTAAGGTGGGAGCTCA-5′ |
| MAPK1 | 5′-CTAACGTTCTGCACCGTGAC-3′ 3′-AGAATGCAGCCTACAGACCA-5′ |
| NFIB | 5′-CCACCAAGCAGCAAAAGAC-3′ 3′-TGGAGATGCAGAGCTGAACA-5′ |
| PDCD4 | 5′-TGGATTAACTGTGCCAACCA-3′ 3′-TCTCAAATGCCCTTTCATCC-5′ |
| RNF20 | 5′-GAGCTCTTATCCCGGAAGCT-3′ 3′-TCCCACTGCAGGTCATCAAT-5′ |
| Sptlc1 | 5′-AGTGGGTTCTGGTGGAGATG-3′ 3′-CAAGAGGTTCTGGTTGCCAC-5′ |
| TNFSF15 | 5′-GCTGAGAAGGAATGCGACAG-3′ 3′-AAGCACATGGGGTTTGTCAC-5′ |
2.11. Statistical analysis
The analysis was done using SPSS version 22 (IBM Corp: Armonk NY) and the clinical parameters were represented either as median (range) or in percentage as appropriate. Derangements in expression of miRs and genes were analysed using principal component analysis. The comparison between groups was done using one way ANOVA or Kruskal Wallis test, wherever necessary. The post-hoc comparison was done by using LSD method or by probability adjustment. Besides this, Student’s t-test and Chi-square test were used for comparison. The correlation between the different miRs in ALF was evaluated by using Spearman's rank correlation coefficient. The diagnostic test was also applied to find out the discriminatory power of mir450 using ROC. The log transformation was applied and the results are represented as log fold. A p value of <0.05 was considered to be significant.
2.12. Role of the funding source
The funder of the study had no role in study design, data collection, data analysis, data interpretation, or writing this article. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication.
3. Results
The clinical characteristics of the patients and healthy controls enrolled in this study are shown in Table 2.
Table 2.
Clinical Characteristics of HEV-related patients and healthy control subjects.
| Parameters Median (Range) | ALF-E(P) (N = 12) | AVH-E(P) (N = 44) | AVH-E(NP) (N = 10) | HC(P) (N = 20) | P Value |
|---|---|---|---|---|---|
| Median Age (years; range) | 25 | 24 | 25 | 24 | NS |
| (21–31) | (18–31) | (20–36) | (19–30) | ||
| Median ALT (IU/L; range) | 132 | 117 | 99 | 28* | P < 0.001 |
| (115–772) | (55–999) | (58–233) | (20–32) | ||
| Median AST (IU/L; range) | 147 | 134 | 112 | 24.5* | P < 0.001 |
| (125–1583) | (60–2639) | (52–1538) | (20–28) | ||
| Median Bilirubin (mg/dl; range) | 7.5 | 7 | 10 | 0.3* | P < 0.001 |
| (3.9–22) | (2–20) | (2–32) | (0.1–0.7) | ||
|
Median INR (range) |
3 | 1.2% | 1.3% | 1* | * and % P < 0.001 |
| (2.5–4) | (1.1–1.3) | (0.9–1.4) | (0.9–1.3) | ||
| Mortality | 5/12 | – | – | – | – |
| Median HEV RNA (Copies/mL) | |||||
| High Viral Load = 2.64 x 105(2.33 x 105–1.6 x 106) | – | 20% | 16% | – | – |
| Low Viral Load = 3.2 x 103(3.25 x 103–2.65 x 104) | 82% | 80% | 34% | ||
| Undetectable (less than 100 copies/ml) | 18% | 50% | |||
| Serum Albumin (g/dl) | 3.05 | – | – | – | – |
| (2.8–3.35) | |||||
| Arterial Ammonia (μgm/dl) | 155 | – | – | – | – |
| (149–724) | |||||
| Grade of Encephalopathy | – | – | – | – | |
| I–II | 7(60%) | ||||
| III–IV | 4(40%) | ||||
| Hemoglobin (g/dl) | 11 | – | – | – | – |
| (8.8–11.5) | |||||
| Platelets (10 ∼ 9/l) | 170 | – | – | – | – |
| (94–330) |
3.1. Derangements in expression of miRs and gene expression during pregnancy
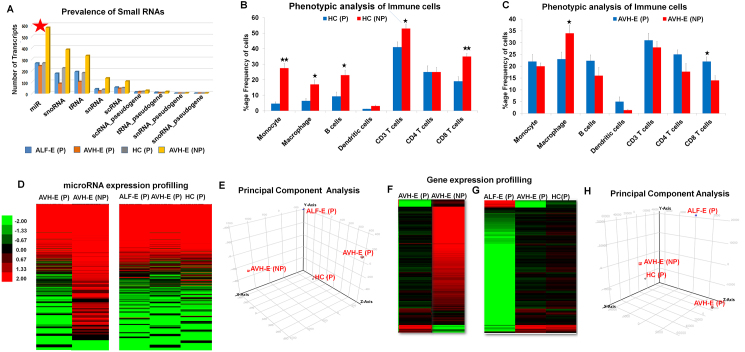
Small RNAs play a major role in post-transcriptional regulation of gene expression and particularly regulate anti-viral defence mechanisms. First, we assessed the total number of differentially expressed small RNAs in ALF-E(P), AVH-E(P), HC(P) and AVH-E(NP) from total PBMCs using Illumina GAIIx genome analyzer deep sequencing platform. For each group, deep sequencing generated an average of 30 million reads of 50 bp length. Expression of total small RNAs was masked in pregnant females than non pregnant, although, expression of miRs waspredominant in all groups (Fig. 1A). Low expression of miRs may be related with overall decline in immune cells during pregnancy (Fig. 1B).
Fig. 1.
(A) Global analysis of small RNAs identified using deep sequencing. (B-C) Frequency of Immune cells in peripheral blood of pregnant and non-pregnant healthy controls(HC) and acute HEV patients. (D) Averaged expression signature of miRs in pooled PBMCs isolated from pregnant AVH-E, ALF-E and HC and non pregnant AVH-E. Pregnant AVH-E and non pregnant AVH-E was compared and also pregnant ALF-E and AVH-E was compared with HC. Each lane represents the average signal log intensity of five patients in each group as independent technical replicates. Expression was displayed in red and green for increased and decreased expression levels and black for no changed expression levels. Color intensity was calibrated to expression level as illustrated at the side of heat map. (E) miR expression data from different groups were applied to principal component analysis. The use of this technical analysis allowed us to visualise the observed variance between groups. (F) Averaged gene expression signature showing decreased global gene expression in AVH-E(P) compared to AVH-E(NP). (G) Heat maps of gene expression in pregnant ALF-E, AVH-E and HC. (H) Principal component analysis (PCA) visualise observed variance in groups.
During HEV infection, there was an increase in the number of all immune cells, significantly only for CD8 + T-cells. The macrophage numbers showed a decline (Fig. 1C).
Low quality reads were trimmed using NGSQC Tool kit. Read counts were normalized by calculating miRs per million miR alignments. The list of significantly regulated miRs was created using a t-test analysis (p ≤ 0.05) and fold change (≥1.5) cut-off between two conditions (eg, HEV infected versus healthy controls).
A total of 415 miR probes were detectable in one or more groups, however, expression of miRs was reduced to 60% in AVH-E(P) comparison to AVH-E(NP) (Fig. 1D). There was no significant difference in number of miRs expressed in ALF-E, AVH-E and HC during pregnancy.
Principal component analysis was used to evaluate the overall variance between four groups and PCA separated pregnant (P) from non-pregnant (NP) groups (Fig. 1E). Further, regression analysis showed a linear correlation in pregnant AVH-E and ALF-E (R2 = 0.957), ALF-E and healthy controls (R2 = 0.928) and AVH-E and controls (R2 = 0.984) (Fig. 2A).
Fig. 2.
(A-B) Scatterplot representation of comparative global miR and gene expression profiles between AVH-E(P), ALF-E(P) and HC(P) patient groups. X and Y axis shows expression levels of miRs and genes between two groups. R2 value, a measure of goodness-of-fit of linear regression indicates the degree of correlation between the subjects.
In order to find differentially expressed genes, the same samples were processed for gene expression analysis. Like the miR expression, there was a reduction in the number of genes expressed in pregnant than non-pregnant females (Fig. 1F). A total of 2,295 genes were expressed in all groups and unsupervised hierarchical clustering was done to understand their expression pattern across the groups (Fig. 1G). It was interesting to note that using their global expression profile, principal component analysis could distinguish self limiting AVH-E(P) from ALF-E(P) (Fig. 1H).
In addition, scatter plot analysis, distinction could be made between pregnant AVH-E and ALF-E (R2 = 0.06) and AVH-E and HC (R2 = 0.01) (Fig. 2B).
3.2. Distinct miR signatures associated with HEV infection
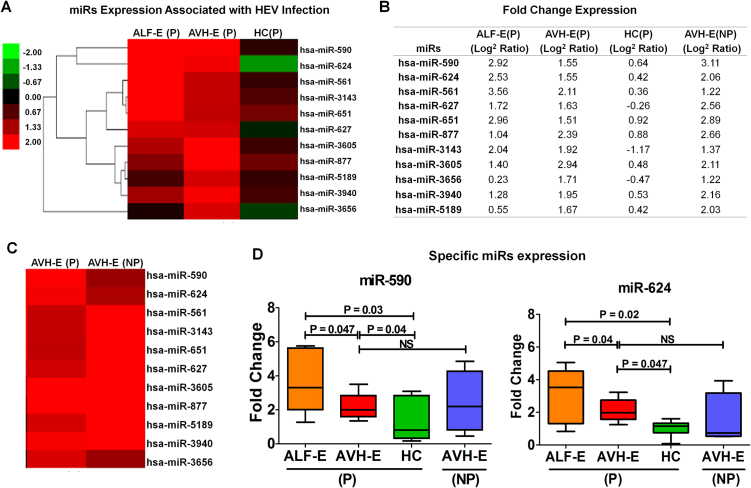
To determine the impact of HEV infection on miR expression profile, unsupervised hierarchical clustering analysis was done in pregnant hepatitis patients and compared with controls. It revealed 11 distinct miRs; viz. miRs-561, 590, 624, 627, 651, 877, 3143, 3605, 3656, 3940, and 5189 with <2 fold expression (p < 0.003). as indicators of HEV Infection (Fig. 3A-B). The miR profile in AVH-E(P) and AVH-E(NP) (Fig. 3C) was comparable.
Fig. 3.
(A) Distinct miR expression in pregnant HEV infection compared to HC. (B) Difference in fold change (log2 ratio) expression of various miRs in three groups. (C) No difference in miR profiling in AVH-E(P) and AVH-E(NP) (D) Fold change expression of miR-590 and miR-624 of various subjects. Data are expressed as box plots in which the horizontal lines indicate the 25th, 50th, and 75th percentiles of the fold change measured by qRT-PCR.
The expression of miR-590 and miR-624 was analysed by qRT-PCR in the validation cohort and found significant increase in pregnant hepatitis patients than controls (Fig. 3D).
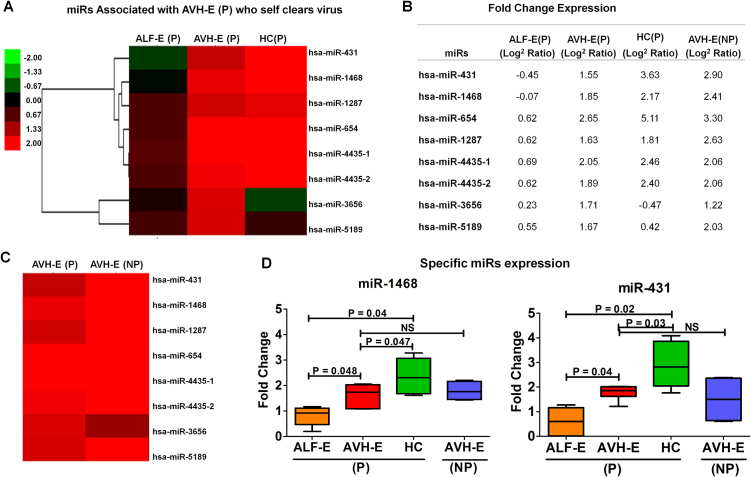
3.3. miR signatures associated with AVH-E patients who self-cleared the virus
Majority of pregnant females have self-limiting HEV infection without developing ALF. It is not known, which miRs are associated with HEV clearance during pregnancy. Supervised hierarchical clustering analysis identified miRs-431, 654, 1468 and 4435, which were over expressed in pregnant AVH-E patients with spontaneous resolution of HEV infection (Fig. 4A-B). These miRs are signatures of natural, self-limiting hepatitis E infection. These miRs were equally expressed in AVH-E(NP) and HC(P) (Fig. 4C).
Fig. 4.
(A) miRs expressed as indicators of virus self clearance. Hierarchical clustering showing high expression of specific miRs in AVH-E(P) and decrease in ALF-E (P) patients. (B) Fold change (log2 ratio) expression of specific miRs. (C) miR profiling in AVH-E(P) and AVH-E(NP) does not show any difference. (D) Box plots showed fold change expression of miR-431 and 1468 by qRT-PCR.
On the other hand, expression of miRs-431, 654, 1468 and 4435 was reduced in ALF-E patients (Fig. 4A-B). There is a possibility that dim expression of these miRs is pathogenetic for the development of acute liver failure. The expression of miR-431 and 1468 was analysed by qRT-PCR in validation cohort of all groups (Fig. 4D).
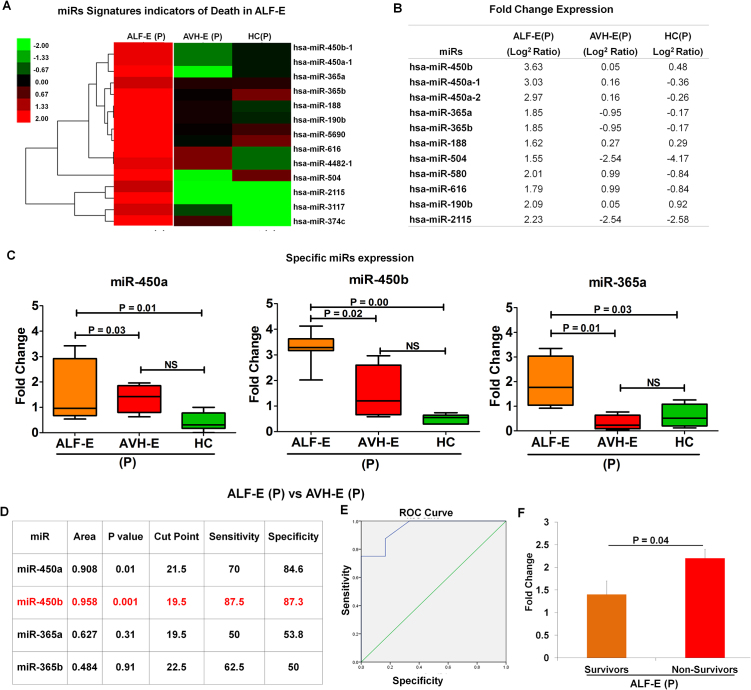
3.4. Distinct miRs associated with HEV induced acute liver failure
HEV infection varies from a clinically self-limiting condition to one associated with ALF. To understand the molecular patterns associated with HEV induced ALF during pregnancy, hyper geometric analysis of miRs was conducted, which revealed that 16 miRs were exclusively expressed >2 fold change (p < 0.002) in pregnant ALF-E patients (Fig. 5A); namely miR 188, 190b, 374c, 365a, 365b, 450a-1, 450a-2, 450b, 504, 580, 616, 2115, 3117, 4482, 4772 and 5690. These miRs were considered as signature miRs of HEV induced acute liver failure (Fig. 5B). We validated miR-365a-3p, 450a1-5p and 450b-5p, in a large cohort of pregnant AVH-E, ALF-E and HC (Fig. 5C), AVH-E (NP) [data not shown].
Fig. 5.
(A) Heat map showed comprehensive miR expression linked with ALF related to HEV infection as indicators of death signatures. (B) Fold change (log2 ratio) expression of specific miRs. (C) Box plots showing qRT-PCR data of higher fold change expression of miR-450a, 450b and miR-365a in ALF-E(P). (D-E) ROC curve analysis of miR-450b between ALF-E (P) and AVH-E (P) showed a cut off of ΔCt ≤ 19.5 (Sensitivity = 87.5, specificity = 87.3; p = 0.001) in ALF in pregnancy. (F) Higher expression of miR-450b in ALF non-survivors than ALF patients who survived.
3.5. miR-450b is a strong predictor for mortality in ALF-E patients
In ALF-E, out of 12 patients, seven died. To determine the association of miRs with the occurrence of ALF-E, ROC curves of four miRs were analysed. The area under the ROC curves showed that only miR-450b is a strong predictor (AUC 95.8, p = 0.001) of ALF in HEV infected pregnant females (Fig. 5D-E). A cut-off of ΔCt ≤ 19.5 (Sensitivity = 87.5%, specificity = 87.3%) in ALF-E versus AVH-E was a strong predictor of development of ALF (p = 0.001). qRT-PCR data showed that greater expression of miR-450b was observed in ALF-E non-survivors compared to survivers (Fig. 5F).
3.6. Identification of immune cell subsets associated with differentially expressed miRs and their targeted genes
Total of 2,295 genes were found to be expressed in our patients, 642 of these are associated with cellular immunity and are targeted mainly by miR-504-3p, miR-877-3p, miR-877-5p, miR-590-3p and miR-616-5p. To identify the active set of cells or pathways associated with miRs and their target genes, we applied stringent cut-offs in the gene data collected from Illumina Human-Ht-12 48 K bead chip. Segmental analysis was done to understand miR mediated gene regulation in pathway specific manner.
Genes related to miRs expressed in patients with self limiting HEV infection were associated mainly with T cells, B cells and plasmacytoid DCs and interferon signalling pathway (Fig. 6). However, in ALF patients, most of the genes expressed were related to neutrophils, monocytes, macrophages, eosinophils and TGF-β, TNF-α, IL-6 cytokine signalling pathways (Fig. 6).
Fig. 6.
Differential gene expression data represented by bar graphs showes various number of genes related to both innate and adaptive immune cells as well as Type 1 and Type 2 immune signalling pathways expressed in self limiting AVH-E(P) and ALF-E(P) groups.
Over all, cell proliferation and cell cycle pathway genes were expressed in patients with self limiting AVH-E but apoptosis, inflammation and cell death processes were associated with ALF (Fig. 6).
3.7. Pathways linked to miRs and their targeted genes in HEV infection
Total of 643 genes were associated with miRs-561, 590, 624, 627, 651, 877, 3143, 3605, 3656, 3940, and 5189 expressed with HEV infection.
Mitarbase data analysed specific target genes like ATXN2, BIRC3, TNFSF1β, PEG3, XAF1, AHR, PDCD4, CREB1, MCM8, NF1B, TDRD1 were associated with miRs 561-3p, 561-5p, 590-5p, 624-3p and 877-5p. Network analysis using cytoscope tool analyzed that these miR-target genes regulate mainly, cell death and DNA metabolic processes (Fig. 7A). We have quantified mRNA expression of BIRC3, PDCD4 and NF1B genes in validation cohort of pregnant HEV infected and healthy controls and observed significant upregulation of BIRC3, PDCD4 and NF1B in HEV infected patients (Fig. 7B-D).
Fig. 7.
Network analyses for pathways associated with miRs and target genes. Predicted targets of differentially expressed miRs was determined by using mirtarBase data. To determine the possible miR − mRNA interactions, the list of targets was generated using Integromatrix Software (Proprietary of Bionivid Technology Pvt Ltd, Bangalore, India). Red lines indicated miRs inhibiting the expression of target genes, and regulating different processes. (A) miRs and targeted genes related to cell death and DNA metabolic processes in HEV infection. (B-D) mRNA expression levels of targeted genes by qRT-PCR. (E) miRs and genes regulated homeostasis processes in patients who self cleared the virus. (F-H) Bar graphs showed mRNA expression of specific genes by qRT-PCR.
3.8. Pathways linked to miRs and their targeted genes in self-limiting HEV infection
microRNAs 431, 1468 and 4435 were specifically related to gene targets GDPD1, TNFSF15, SPTLC1 and IL17RD regulating the cellular homeostasis processes and metal ion binding (Fig. 7E). qRT-PCR of TNFSF15, SPTLC1 and GDPD1 validated the mRNA expression of in all subjects (Fig. 7F-H).
3.9. Pathways linked to miR- targeted genes in acute liver failure
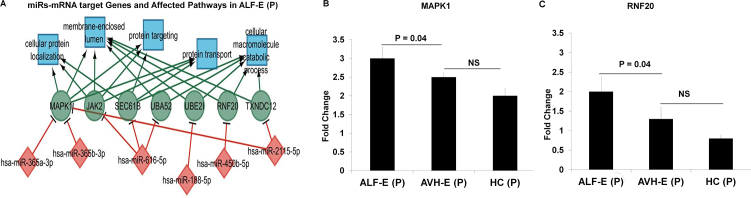
Network analysis revealed that out of the 16 miRs expressed in ALF-365a-3p, 365b-3p, 616-5p, 188-5p, 450b, and 2115-5c regulate MAPK1, JAK2 SEC61B, UBA52, UBE21, RNF20 and TXNDC12 genes which are primarily involved in protein localization or metabolic processes, protein targeting and transport, proliferation, innate immunity, tumor suppression and cellular homeostasis (Fig. 8A). Target genes MAPK1 related to miR365a and 365b and RNF20 related miR450b were specifically validated in all hepatitis E patients and healthy controls (Fig. 8B-C). Increased expression of these miRs can tweak or deregulate the cellular processes in ALF.
Fig. 8.
(A) miR signatures and their targeted genes in pregnant patients with acute liver failure. (B-C) Bar Diagram showing specific gene expression by qRT-PCR.
As miR-450b was indicated as strong predictor of liver failure and death with more than 87% sensitivity and specificity. We have specifically focused on miR-450b regulated cells and cellular processes.
miR450b particularly involved with WASF2, PPMIF, SP110, IMP4, SERBP1, CREB3L2, LDLR and many genes which influence the cells including CD8, granulocyte, monocyte, dendritic cells, eosinophils and regulate the Interferon, MYD88, cell proliferation, cell death pathways (Fig. 9).
Fig. 9.
miR-450b interaction with immune cells showing involvement of various pathways and genes.
4. Discussion
Our data demonstrates for the first time the speciifc imprints of transcriptional signatures asosciated with HEV infection in self- limiting disease and acute liver failure in pregnancy. In our study, all HEV infected patients belonged to genotype 1, hence, ruled out influence of genotype on transcriptional signatures.
Furthermore, significant effect of pregnancy was observed in lowering more than 50% miRs and genes compared to non-pregnants. Lower expression of miRs and genes may be due to suppressed immunity or non-functional immune cells in pregnancy [16, 17]. Expression of miRs regulate considerable number of genes, acting not only in a particular module but also cross linking the individual modules. In our study, distinctly identified miRs were seen to preferentially target the gene expression profiles of NK cells, neutrophils, monocytes, macrophages, T cells, B cells, plasmacytoid DCs and eosinophils.
Hepatitis E is mostly a neglected disease with little information on molecular mechanisms associated with disease pathogenesis. Till date there is no data on the miRNAs expressed during HEV infection. Our miRNA profiling results showed specific expression of miRs (561, 590, 624, 627, 651, 877, 3143, 3605, 3656, 3940 and 5189) in HEV infection during pregnancy. These miRs were maily related to cell death and DNA metabolic processes. None of these miRs have been reported earlier either in HEV infection or in other viral hepatitis. In fact, miRs-, 3605, 3656, 3940, 5189 are novel miRs which have not been reported in any physiological conditions. However, miRs-561 and 590 were reported in acetaminophen drug induced hepatotoxicity andin HCCs by direct targeting TGF-β RII [18]. Additionally, miR- 627, 651 and 877 reglaute cancer cell proliferation by modulating processes like apoptosis and cell cycle progression [19, 20, 21]. miRs related to HEV infection significantly lead to the upregulation of BIRC3 and PDCD4 genes, which have also been implicated in cell death in hepatitis B virus infection [22, 23].
Most of the AVH-E(P) females, who cleared the HEV infection spontaneously, were asscociated with expression of miRs-431, 654, 1468 and 4435. miR-431 and 654 were reported so far in cellular regeneration [24], proliferation, apoptosis of cancer cells [25] and miR-1468 in resolving pathogenic insults by recruiting neutrophils [26]. Hence, these miRs reflect protective effect in self-clearance of HEV infection. miR-4435 is a novel miR and has not been reported in any physiological condition till date.
Our data revelaed that in self-limiting AVH-E, miRs targeted most of the genes which were linked to cell proliferation and cell cycle pathway and were associated with Nk cells, T cells, B-cells, plasmacytoid DCs, toll like receptors (TLRs), IFN-γ.
In other reports also, during recovery phase of HEV infection, expression of TLR3 was reported to be increased to restore the IFN-γ response [27]. Altered expression of NK cells was also observed in acute HEV infections which play major role in direct killing of virus-infected cells in AVH-E [26].
Around, 25% pregnant females with HEV infection develop acute liver failure with high maternal and fetal morbidity and mortality. It is not clear, why such a rapid liver failure develops following HEV infection in pregnancy. It is important to note that there may be inability to induce protective miRs as in self limited HEV infection, resuling in severe and progressive loss of functional liver cell mass leading to ALF.
In HEV induced ALF, we identified 16 uniquely expressed miRs. Most of these miRs have not been earlier reported in context of acute liver failure, but have been reported in hepatocellular carcinoma [28], insulin resistance [29], leukemia [30], breast cancer [31], and prostate cancer. Only, two miR-504 and miR-374c had been reported to be upregulated in mice with acute liver failure-induced hepatic encephalopathy [32] and HBV related acute on chronic liver failure [23]. In animal models of drug induced ALF (drug/DGalN/LPS) different miRs from our study like miR-15b, 16, 197, 122 and 221 were expressed [8, 9, 11]. In alcohol induced ACLF patients miR-16 was also reported to be involved [29]. However, in our study, these miRs were not expressed.
In HEV induced ALF-E, genes related to cell death, apoptosis and inflammation processes involving neutrophils, monocytes and especially macrophages appeared to be more in numbers than self-limiting HEV infection. Previously, also we have demonstrated the increased number of macrophages in HEV induced ALF with severely compromised TLR downstream signalling [16].
It has been observed that in acetaminophen-induced ALF, activated monocytes and macrophages secreted TNF-α caused hepatocellular injury [33]. But in fulminant autoimmune hepatitis and HBV induced acute on chronic liver failure, liver damage is driven by CD4, CD8 T-cells secreted TNF-α and IFN-γ [34, 35, 36].
Although we have observed, overexpression of T regulatory suppressor genes in HEV induced ALF, dominant upregualtion of miR-450b in ALF-E pregnant patients was shown to regulate several other genes like PPMIF, WASF2, CALM1, ETS1, Bid, IMP4 and TNFSF1β involved in the process of cellular inflammation and apoptosis [37, 38, 39]. It is well known that directly or indirectly TNF-α pathway is known for hepatocyte apoptosis and liver injury.
Thus, we surmise that HEV induced miR-450b negatively regulates cell survival and thereby apoptotic cell death contributed more in ongoing inflammation and injury leading to liver failure.
In HEV induced ALF, genes also related to eosinophils were more expressed, a possible reflection of an attempt by failing liver to regenerate [40]. Which was similar to the observations of fulminant hepatic failure where eosinophils with high IL-6 expression were reported to be involved in liver damage [41].
In summary, we were able to demonstrate that the mechanism for diverse outcome of HEV infection could be due to the interaction of distinct miRs in specific immune pathways leading to inflammatory response, liver failure or death. The cellular homeostasis was associated with self limiting HEV infection and was regulated by miR-431, 1468 and 4435. Severeliver injury regulated by apoptosis and cell death in HEV induced ALF was dominantly associated to miR- 450b. Therefore, these novel miR signatures may help in developing therapeutic interventions in HEV infected pregnant females who can develop liver failure.
Declarations
Author contribution statement
Nirupma Trehanpati: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Rashi Sehgal, Ashish Vyas, Ritu Khosla, Arshi Khanam: Performed the experiments.
Guresh Kumar, Rakhi Maiwall, Sharda Patra, Gayatri Ramakrishna: Contributed reagents, materials, analysis tools or data.
Madavan Vasudevan: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Shyam Kottilil: Contributed reagents, materials, analysis tools or data; Wrote the paper.
Shiv Kumar Sarin: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This work was supported by HEV grant NO. VIR/17/2009/ECD-1 from Indian Council of Medical Research, Govt. of India, New Delhi.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
Dr. Ekta Gupta from department of virology, ILBS helped us in detecting viral loads and genotyping in HEV infected samples. Corresponding author is responsible for submitting this paper for publication in this journal.
Contributor Information
Nirupma Trehanpati, Email: trehanpati@gmail.com.
Shiv Kumar Sarin, Email: shivsarin@gmail.com.
References
- 1.Navaneethan U., Al Mohajer M., Shata M.T. Hepatitis E and pregnancy: understanding the pathogenesis. Liver Int. 2008;28(9):1190–1199. doi: 10.1111/j.1478-3231.2008.01840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khuroo M.S., Kamili S. Aetiology, clinical course and outcome of sporadic acute viral hepatitis in pregnancy. J. Viral Hepat. 2003;10(1):61–69. doi: 10.1046/j.1365-2893.2003.00398.x. [DOI] [PubMed] [Google Scholar]
- 3.Patra S., Kumar A., Trivedi S.S., Puri M., Sarin S.K. Maternal and fetal outcomes in pregnant women with acute hepatitis E virus infection. Ann. Intern. Med. 2007;147(1):28–33. doi: 10.7326/0003-4819-147-1-200707030-00005. [DOI] [PubMed] [Google Scholar]
- 4.TrehanPati N., Geffers R., Sukriti, Hissar S., Riese P., Toepfer T. Gene expression signatures of peripheral CD4+ T cells clearly discriminate between patients with acute and chronic hepatitis B infection. Hepatology. 2009;49(3):781–790. doi: 10.1002/hep.22696. [DOI] [PubMed] [Google Scholar]
- 5.John K., Hadem J., Krech T., Wahl K., Manns M.P., Dooley S. MicroRNAs play a role in spontaneous recovery from acute liver failure. Hepatology. 2014;60(4):1346–1355. doi: 10.1002/hep.27250. [DOI] [PubMed] [Google Scholar]
- 6.Mi S., Zhang J., Zhang W., Huang R.S. Circulating microRNAs as biomarkers for inflammatory diseases. Microrna. 2013;2(1):63–71. doi: 10.2174/2211536611302010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Starkey Lewis P.J., Dear J., Platt V., Simpson K.J., Craig D.G., Antoine D.J. Circulating microRNAs as potential markers of human drug-induced liver injury. Hepatology. 2011;54(5):1767–1776. doi: 10.1002/hep.24538. [DOI] [PubMed] [Google Scholar]
- 8.An F.M., Yu D.S., Xie Q., Gong B.D., Wang H., Guo Q. [The role of miRNA-122 expression during the acute liver failure in mice induced by D-GalN/LPS] Zhonghua Gan Zang Bing Za Zhi. 2010;18(7):527–532. doi: 10.3760/cma.j.issn.1007-3418.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 9.Kumar R., Shalimar, Sharma H., Goyal R., Kumar A., Khanal S. Prospective derivation and validation of early dynamic model for predicting outcome in patients with acute liver failure. Gut. 2012;61(7):1068–1075. doi: 10.1136/gutjnl-2011-301762. [DOI] [PubMed] [Google Scholar]
- 10.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 11.Sharma A.D., Narain N., Handel E.M., Iken M., Singhal N., Cathomen T. MicroRNA-221 regulates FAS-induced fulminant liver failure. Hepatology. 2011;53(5):1651–1661. doi: 10.1002/hep.24243. [DOI] [PubMed] [Google Scholar]
- 12.Dubin P.H., Yuan H., Devine R.K., Hynan L.S., Jain M.K., Lee W.M. Micro-RNA-122 levels in acute liver failure and chronic hepatitis C. J. Med. Virol. 2014;86(9):1507–1514. doi: 10.1002/jmv.23987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jopling C.L., Yi M., Lancaster A.M., Lemon S.M., Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309(5740):1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 14.Wu X.J., Li Y., Liu D., Zhao L.D., Bai B., Xue M.H. miR-27a as an oncogenic microRNA of hepatitis B virus- related hepatocellular carcinoma. Asian Pac. J. Cancer Prev. 2013;14(2):885–889. doi: 10.7314/apjcp.2013.14.2.885. [DOI] [PubMed] [Google Scholar]
- 15.Antoine D.J., Dear J.W., Goldring C.E., Park B.K. Understanding the pathophysiological regulatory role of microRNAs in acute liver failure. Hepatology. 2015;61(4):1439–1440. doi: 10.1002/hep.27349. [DOI] [PubMed] [Google Scholar]
- 16.Sehgal R., Patra S., David P., Vyas A., Khanam A., Hissar S. Impaired monocyte-macrophage functions and defective Toll-like receptor signaling in hepatitis E virus-infected pregnant women with acute liver failure. Hepatology. 2015;62(6):1683–1696. doi: 10.1002/hep.28143. [DOI] [PubMed] [Google Scholar]
- 17.Warning J.C., McCracken S.A., Morris J.M. A balancing act: mechanisms by which the fetus avoids rejection by the maternal immune system. Reproduction. 2011;141(6):715–724. doi: 10.1530/REP-10-0360. [DOI] [PubMed] [Google Scholar]
- 18.Jiang X., Xiang G., Wang Y., Zhang L., Yang X., Cao L. MicroRNA-590-5p regulates proliferation and invasion in human hepatocellular carcinoma cells by targeting TGF-beta RII. Mol. Cells. 2012;33(6):545–551. doi: 10.1007/s10059-012-2267-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gragnani L., Piluso A., Fognani E., Zignego A.L. MicroRNA expression in hepatitis C virus-related malignancies: A brief review. World J. Gastroenter. 2015;21(28):8562–8568. doi: 10.3748/wjg.v21.i28.8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Padi S.K., Zhang Q., Rustum Y.M., Morrison C., Guo B. MicroRNA-627 mediates the epigenetic mechanisms of vitamin D to suppress proliferation of human colorectal cancer cells and growth of xenograft tumors in mice. Gastroenterology. 2013;145(2):437–446. doi: 10.1053/j.gastro.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang X., Qin J., Lu S. Up-regulation of miR-877 induced by paclitaxel inhibits hepatocellular carcinoma cell proliferation though targeting FOXM1. Int. J. Clin. Exp. Pathol. 2015;8(2):1515–1524. [PMC free article] [PubMed] [Google Scholar]
- 22.Weng L., Du J., Zhou Q., Cheng B., Li J., Zhang D. Identification of cyclin B1 and Sec62 as biomarkers for recurrence in patients with HBV-related hepatocellular carcinoma after surgical resection. Mol. Cancer. 2012;11:39. doi: 10.1186/1476-4598-11-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Damania P., Sen B., Dar S.B., Kumar S., Kumari A., Gupta E. Hepatitis B virus induces cell proliferation via HBx-induced microRNA-21 in hepatocellular carcinoma by targeting programmed cell death protein4 (PDCD4) and phosphatase and tensin homologue (PTEN) PloS One. 2014;9(3) doi: 10.1371/journal.pone.0091745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu D., Murashov A.K. MicroRNA-431 regulates axon regeneration in mature sensory neurons by targeting the Wnt antagonist Kremen1. Front. Mol. Neurosci. 2013;6:35. doi: 10.3389/fnmol.2013.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Formosa A., Markert E.K., Lena A.M., Italiano D., Finazzi-Agro E., Levine A.J. MicroRNAs, miR-154, miR-299-5p, miR-376a, miR-376c, miR-377, miR-381, miR-487b, miR-485-3p, miR-495 and miR-654-3p, mapped to the 14q32.31 locus, regulate proliferation, apoptosis, migration and invasion in metastatic prostate cancer cells. Oncogene. 2014;33(44):5173–5182. doi: 10.1038/onc.2013.451. [DOI] [PubMed] [Google Scholar]
- 26.Mellett M., Atzei P., Horgan A., Hams E., Floss T., Wurst W. Orphan receptor IL-17RD tunes IL-17A signalling and is required for neutrophilia. Nat. Commun. 2012;3:1119. doi: 10.1038/ncomms2127. [DOI] [PubMed] [Google Scholar]
- 27.Majumdar M., Ratho R.K., Chawla Y., Singh M.P. Role of TLR gene expression and cytokine profiling in the immunopathogenesis of viral hepatitis E. J. Clin. Virol. 2015;73:8–13. doi: 10.1016/j.jcv.2015.09.011. [DOI] [PubMed] [Google Scholar]
- 28.Chen Z., Huang Z., Ye Q., Ming Y., Zhang S., Zhao Y. Prognostic significance and anti-proliferation effect of microRNA-365 in hepatocellular carcinoma. Int. J. Clin. Exp. Pathol. 2015;8(2):1705–1711. [PMC free article] [PubMed] [Google Scholar]
- 29.Hung T.M., Ho C.M., Liu Y.C., Lee J.L., Liao Y.R., Wu Y.M. Up-regulation of microRNA-190b plays a role for decreased IGF-1 that induces insulin resistance in human hepatocellular carcinoma. PloS One. 2014;9(2) doi: 10.1371/journal.pone.0089446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jinlong S., Lin F., Yonghui L., Li Y., Weidong W. Identification of let-7a-2-3p or/and miR-188-5p as prognostic biomarkers in cytogenetically normal acute myeloid leukemia. PloS One. 2015;10(2) doi: 10.1371/journal.pone.0118099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cizeron-Clairac G., Lallemand F., Vacher S., Lidereau R., Bieche I., Callens C. MiR-190b, the highest up-regulated miRNA in ERalpha-positive compared to ERalpha-negative breast tumors, a new biomarker in breast cancers? BMC Cancer. 2015;15:499. doi: 10.1186/s12885-015-1505-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vemuganti R., Silva V.R., Mehta S.L., Hazell A.S. Acute liver failure-induced hepatic encephalopathy s associated with changes in microRNA expression rofiles in cerebral cortex of the mouse [corrected] Metab. Brain Dis. 2014;29(4):891–899. doi: 10.1007/s11011-014-9545-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Possamai L.A., Antoniades C.G., Anstee Q.M., Quaglia A., Vergani D., Thursz M. Role of monocytes and macrophages in experimental and human acute liver failure. World J. Gastroenterol. 2010;16(15):1811–1819. doi: 10.3748/wjg.v16.i15.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Streetz K., Leifeld L., Grundmann D., Ramakers J., Eckert K., Spengler U. Tumor necrosis factor alpha in the pathogenesis of human and murine fulminant hepatic failure. Gastroenterology. 2000;119(2):446–460. doi: 10.1053/gast.2000.9364. [DOI] [PubMed] [Google Scholar]
- 35.Robinson R.T., Wang J., Cripps J.G., Milks M.W., English K.A., Pearson T.A. End-organ damage in a mouse model of fulminant liver inflammation requires CD4+ T cell production of IFN-gamma but is independent of Fas. J. Immunol. 2009;182(5):3278–3284. doi: 10.4049/jimmunol.0803417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anderson A.E., Sayers B.L., Haniffa M.A., Swan D.J., Diboll J., Wang X.N. Differential regulation of naive and memory CD4+ T cells by alternatively activated dendritic cells. J. Leukoc. Biol. 2008;84(1):124–133. doi: 10.1189/jlb.1107744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jurmeister S., Baumann M., Balwierz A., Keklikoglou I., Ward A., Uhlmann S. MicroRNA-200c represses migration and invasion of breast cancer cells by targeting actin-regulatory proteins FHOD1 and PPM1F. Mol. Cell. Biol. 2012;32(3):633–651. doi: 10.1128/MCB.06212-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee S.J., Baserga S.J. Imp3p and Imp4p, two specific components of the U3 small nucleolar ribonucleoprotein that are essential for pre-18S rRNA processing. Mol. Cell. Biol. 1999;19(8):5441–5452. doi: 10.1128/mcb.19.8.5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu D.S., An F.M., Gong B.D., Xiang X.G., Lin L.Y., Wang H. The regulatory role of microRNA-1187 in TNF-alpha-mediated hepatocyte apoptosis in acute liver failure. Int. J. Mol. Med. 2012;29(4):663–668. doi: 10.3892/ijmm.2012.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goh Y.P., Henderson N.C., Heredia J.E., Red Eagle A., Odegaard J.I., Lehwald N. Eosinophils secrete IL-4 to facilitate liver regeneration. Proc. Natl. Acad. Sci. USA. 2013;110(24):9914–9919. doi: 10.1073/pnas.1304046110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.dos Santos D.C., da Silva Gomes Martinho J.M., Pacheco-Moreira L.F., Carvalho Viana de Araujo C., Caroli-Bottino A., Pannain V.L. Eosinophils involved in fulminant hepatic failure are associated with high interleukin-6 expression and absence of interleukin-5 in liver and peripheral blood. Liver Int. 2009;29(4):544–551. doi: 10.1111/j.1478-3231.2008.01872.x. [DOI] [PubMed] [Google Scholar]