Abstract
The acquired enamel pellicle is an oral, fluid-derived protein layer that forms on the tooth surface. It is a biologically and clinically important integument that protects teeth against enamel demineralization, and abrasion. Tooth surfaces are exposed to different proteinaceous microenvironments depending on the enamel location. For instance, tooth surfaces close to the gingival sulcus contact serum proteins that emanate via this sulcus, which may impact pellicle composition locally. The aims of this study were to define the major salivary and serum components that adsorb to hydroxyapatite, to study competition among them, and to obtain preliminary evidence in an in vivo saliva/serum pellicle model. Hydroxyapatite powder was incubated with saliva and serum, and the proteins that adsorbed were identified by mass spectrometry. To study competition, saliva and serum proteins were labeled with CyDyes, mixed in various proportions, and incubated with hydroxyapatite. In vivo competition was assessed using a split-mouth design, with half the buccal tooth surfaces coated with serum and the other half with saliva. After exposure to the oral environment for 0 min, 30 min and 2 h, the pellicles were analyzed by SDS-PAGE. In pure saliva- or serum-derived pellicles, 82 and 84 proteins were identified, respectively. When present concomitantly, salivary protein adsorbers effectively competed with serum protein adsorbers for the hydroxyapatite surface. Specifically, acidic proline-rich protein, cystatin, statherin and protein S100-A9 proteins competed off apolipoproteins, complement C4-A, haptoglobin, transthyretin and serotransferrin. In vivo evidence further supported the replacement of serum proteins by salivary proteins. In conclusion, although significant numbers of serum proteins emanate from the gingival sulcus, their ability to participate in dental pellicle formation is likely reduced in the presence of strong salivary protein adsorbers. The functional properties of the acquired enamel pellicle will therefore be mostly dictated by the salivary component.
Keywords: dental pellicle, gingival crevicular fluid, gingivitis, proteomics, dental enamel, biofilms
Introduction
Teeth exposed to proteinaceous oral environments form a tooth integument that is composed almost entirely of oral, fluid-derived proteins known as acquired enamel pellicles (Dawes et al. 1963). The major biological importance of this tooth integument is its protective effect of tooth surfaces against acid insults and abrasion, and it plays a role in guiding the attachment of early microbial colonizers (Kolenbrander and London 1993). One of the oral fluid constituents that might contribute to the formation of the acquired enamel pellicle is gingival crevicular fluid, which emanates from the gingival sulcus. Gingival fluid represents a serumnal transudate under gingivally healthy conditions (Lamster and Ahlo 2007), that can transform into an exudate with the onset of gingivitis (Loe and Holm-Pedersen 1965). As this fluid exits the sulcular space, it mixes with saliva (Oppenheim 1970). Consequently, the cervical portions of teeth, especially in patients with gingivitis, are subjected to a protein environment that differs significantly from the typical salivary milieu, and includes the presence of both saliva and serum/blood-derived proteins and peptides (Carlén et al. 1998; Carlén et al. 2003; Rüdiger et al. 2012). It is likely that the serumnal constituents close to the gingival sulcus promote an acquired enamel pellicle different in composition to the pellicle that forms strictly in a salivary environment (Kajisa et al. 1990; Abbas et al. 1991). An important biological consequence of the changing protein environment near the gingival sulcus is that it may impact bacterial attachment and thus change the characteristics of the early biofilm. Indeed, some evidence points to the preferential adsorption of periodontitis-associated proteolytic Gram-negative species, such as Porphyromonas gingivalis, to these areas (Olczak et al. 2001; Biyikoğlu et al. 2012). Colonization with such species may further predispose the subject to progression from gingivitis to periodontitis (Lourenco et al. 2014).
Although much is known about pellicle composition on tooth enamel surfaces and, more recently, its impact on early biofilm formation, little knowledge is available on these events that occur near the gingival margin. The scientific question pertinent to this is whether the mixing of gingival fluid proteins with saliva proteins affects pellicle composition in this clinically critical region. This study represents the first hydroxyapatite adsorption analysis of saliva and serum proteins at different ratios using proteomic strategies to simultaneously quantify multiple constituents of complex protein mixtures. Furthermore, in an exploratory in vivo study, we investigated the incipient pellicle differences formed on serum- or saliva-coated tooth surfaces.
Materials and Methods
Saliva and Serum Sample Collection and Processing
This study was approved by the Institutional Review Board of Boston University Medical Center. Before enrollment, individuals provided informed consent to participate. For in vitro studies, human serum (AB blood type) from a male donor was obtained from a commercial source (Mediatech). For the in vivo studies, fresh human serum was used to coat the tooth surfaces. Whole saliva was collected from 3 subjects under masticatory stimulation, as described previously, and whole saliva supernatant (prepared by centrifugation) was used for all in vitro experiments (Thomadaki et al. 2011).
In Vitro Pellicle Formation by Saliva and Serum Proteins on Hydroxyapatite Surfaces
Protein concentration in whole saliva supernatant was 1.01 mg/mL. Serum protein concentration was 70 mg/mL and was diluted 70-fold. Aliquots of 1.0 mL undiluted whole saliva supernatant and 1.0 mL diluted human serum (each containing 1.0 mg/mL protein) were mixed with 800 μL of binding buffer containing 40 mM Tris and 50 mM NaCl, pH 6.8. The 1.8-mL mixture was then added to 5 mg of non-sintered hydroxyapatite (200–300 µm; 5.1741 m2/g; 3.15 (0.1) g/cm3) (Hitemco Medical). The suspensions were incubated while rotating end-over-end at 37 °C for 0, 5, 30, 60 or 240 min. After centrifugation, the 1.8-mL supernatants containing the unadsorbed proteins were removed and kept on ice. Hydroxyapatite sediments with the adsorbed salivary or serum proteins were washed 3 times in 1.0 mL of binding buffer and then dissolved in 1.8 mL of 0.2 M ethylenediaminetetraacetic acid (EDTA), pH 7.5, at room temperature for 1 h. Adsorbed and unadsorbed proteins were concentrated and desalted to a final volume of 300 μL in deionized water using an Amicon Ultra filter unit with a 3 kDa molecular weight cut-off.
SDS-PAGE and In-Gel Protein Digestion
Proteins in 20 µL aliquots of the unadsorbed and adsorbed protein fractions were mixed with 5 μL of 5× sample buffer, separated by 4% to 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and silver-stained (Chevallet et al. 2006). Selected bands were excised and subjected to a modified in-gel trypsin digestion procedure (Shevchenko et al. 1996). In brief, gel pieces were washed and dehydrated with pure acetonitrile, dried in a vacuum concentrator, and rehydrated in 50 mM ammonium bicarbonate solution containing 12.5 ng/µL modified sequencing-grade trypsin (Promega). After incubation at 37 °C for 16 h, the extracts were dried in a vacuum concentrator and stored at 4 °C until analysis.
Peptide Sequencing and Protein Identification by Liquid Chromatography Tandem Mass Spectrometry
Peptide samples dissolved in 0.1% formic acid were loaded onto a nano-scale C18 reverse-phase HPLC capillary column (Peng and Gygi 2001) via a Famos auto sampler (LC Packings), eluted with increasing concentrations of solvent B (97.5% acetonitrile, 0.1% formic acid), and subjected to mass spectrometric analysis using an LTQ Orbitrap Velos Pro ion-trap mass spectrometer (Thermo Fisher Scientific). Peptide sequences were determined from the acquired fragmentation patterns identifiable in the human proteome database using the Sequest program (Thermo Fisher Scientific) (Eng et al. 1994). The database search included a reversed version of all the sequences and the data were filtered by applying a DeltaCN value of more than 0.1 and Xcorr values over 2.0 and 3.0 for ions with Z = 2+ and 3+ charges, respectively; this ensured a less than 1% to 2% peptide false discovery rate.
Hydroxyapatite Adsorption and Visualization of CyDye-Labeled Saliva and Serum Protein
Whole saliva supernatant or diluted human serum (each 1.0 mg/mL; 1 mL aliquots) were adjusted to pH 8.5 with NaOH, and then combined with 20 μL of 0.4 mm CyDye working solution, using Cy3 for whole saliva and Cy5 for serum. Samples were processed as per the manufacturer’s instructions (Lumiprobe). Labeled proteins were mixed with 800 μL binding buffer and incubated with 5 mg hydroxyapatite, either in pure form or at various ratios (v/v) of whole saliva supernatant to human serum (1:9, 1:1, and 9:1). The bound and unbound proteins were recovered and desalted as described above. Electrophoresis was carried out for 160-µL aliquots in sample buffer without bromophenol tracking dye, as this interferes with Cy5. Protein gels were scanned with a Luminescent Imager Analyzer (LAS-4000, Fujifilm) using a laser/emission filter set of 520/575 nm for Cy3 and 630/670 nm for Cy5. The images were overlaid with FIJI imaging software (Schindelin et al. 2012). After CyDye imaging, the gels were silver-stained as described above.
In Vivo Pellicle Formation on Serum-Coated Tooth Surfaces
The buccal tooth surfaces in both arches, excluding the second and third molars in one subject, were thoroughly cleaned using a prophylaxis hand piece with a rubber cup, as described elsewhere (Heller et al. 2016). In a split-mouth study design, two-thirds of the cleaned coronal surfaces of incisors, canines, and premolars of the upper and lower right arch were covered immediately after tooth cleaning with 5 μL of undiluted serum derived from the same subject. The left arch was left uncoated. Exposures to the oral environment following prophylaxis were 0 min (subject closed the mouth and immediately thereafter the sample was collected), 30 min and 120 min. During this pellicle maturation phase, the subject was asked to refrain from eating, drinking (except water) or tooth brushing. A collection strip (electrode wick filter paper, Bio-Rad), presoaked in 3% citric acid, was then used to gently swipe the coronal buccal surfaces of incisors, canines and premolars as described (Siqueira and Oppenheim 2009), to collect the acquired enamel pellicle.
Protein Recovery from Collection Strips
Proteins were recovered from the collection strips by electroelution as described (Siqueira et al. 2007). Briefly, 30 μL of SDS sample buffer was added to the strips, and after boiling for 5 min, the strips and sample buffer were placed directly into 1 of the 10 wells of precast 4% to 12% SDS-PAGE gel (Bio-Rad). Gel electrophoresis was carried out as described above.
Results
Protein Identification of In Vitro–Formed Pellicles
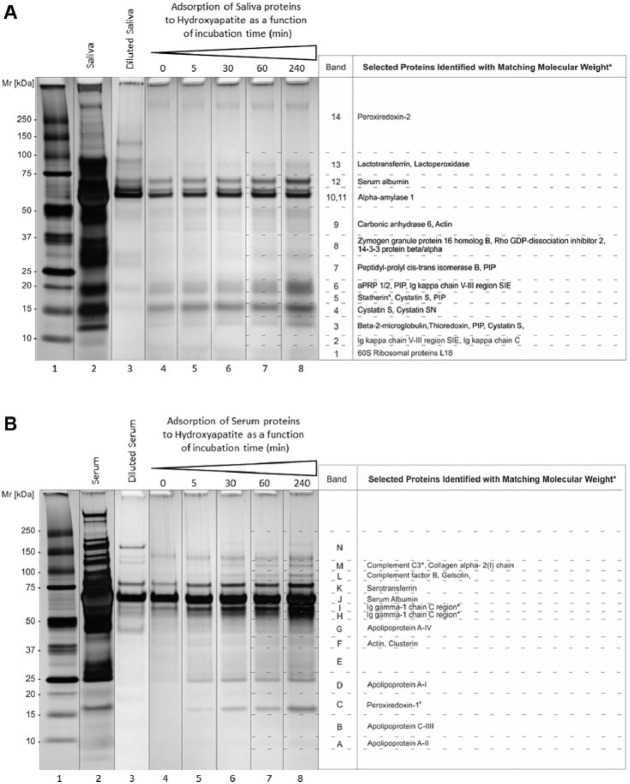
The first aim was to define the kinetic patterns of saliva and serum protein adsorption to HA, and to identify the adsorbed proteins by mass spectrometry. Experimental pellicles on hydroxyapatite powder were formed for 0 to 240 min with whole saliva supernatant or human serum (1 mg/mL) as the protein sources. Proteins adsorbed to hydroxyapatite (Fig. 1A) were separated by SDS-PAGE and visualized by silver staining. The selectivity of salivary protein adsorption to hydroxyapatite can be readily observed, as the hydroxyapatite-adsorbed protein patterns (Fig. 1, lanes 4–8) differed significantly to protein patterns of diluted whole saliva supernatant (Fig. 1A, lane 3). Whole saliva supernatant proteins with molecular weights of ~70 kDa and below 25 kDa rapidly adsorbed, most within the first 5 min of incubation. Selectivity of adsorption was also demonstrated for serum protein adsorption to hydroxyapatite (Fig. 1B, lane 3 vs. lanes 4–8). The prominent adsorbing proteins exhibited MW values of ~67 kDa and ~77 kDa. In contrast to saliva proteins, serum protein adsorption occurred at a slower but distinctly incremental rate, except for the highly predominant serum protein albumin (~67 kDa), which adsorbed rapidly.
Figure 1.
Identification of proteins in saliva and serum adsorbing to hydroxyapatite. (A) Saliva pellicles and (B) serum pellicles each formed after various incubation times with hydroxyapatite. Gels were silver-stained. Lane 1, Molecular weight standard; lane 2, original saliva and serum samples (20 µL); lane 3, diluted saliva and serum samples; lanes 4 to 8, adsorbed saliva and serum fractions harvested after 0, 5, 30, 60 and 240 min incubation with hydroxyapatite (20 µL). Proteins in the excised bands were identified by mass spectrometry. Shown are proteins identified by more than 2 peptides, with theoretical molecular weights matching their apparent molecular weights in the gels (± 10 kDa; exceptions are indicated with asterisks). These proteins were among the 95% most abundant proteins identified. The proteins were excised from gels and identified by mass spectrometric analysis in 2 independent experiments.
To identify the specific protein components in whole saliva- and serum-derived pellicles, protein bands were in-gel digested and analyzed by mass spectrometry. The excised protein bands of saliva pellicles were given numbers and the excised protein bands of the serum pellicles were given letters (Fig. 1), in ascending order from low- to high molecular weight. All proteins identified in the saliva and serum pellicles are listed in Appendix Table 1 (saliva: Table 1A; serum: Table 1B). They are ordered according to abundance, which was based on the percent contribution of the proteins to the total proteins in the sample, determined from the ion peak area in the first mass spectrum. In total, 82 proteins were identified in the saliva-derived pellicles and 84 proteins in the serum-derived pellicles. The proteins listed in Figure 1A and 1B were selected based on 3 criteria indicated in the legend. Salivary proteins identified in the whole saliva supernatant pellicles included statherin, acidic proline-rich proteins, cystatin S, cystatin SN, α-amylase 1, carbonic anhydrase 6 and lactoperoxidase (Fig. 1A), and this is consistent with the known affinity of these proteins for hydroxyapatite (Siqueira et al. 2012). Adsorbed serum proteins included albumin, apolipoprotein A-I, serotransferrin and peroxiredoxin 1 (Fig. 1B), some of which have previously been shown to adsorb to hydroxyapatite (Carlén et al. 1998) and be present in in vivo pellicle formation (Siqueira et al. 2007).
Competition between Saliva and Serum Proteins for Adsorption Sites on Hydroxyapatite
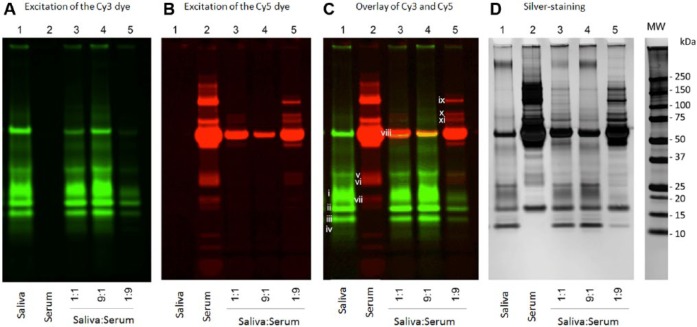
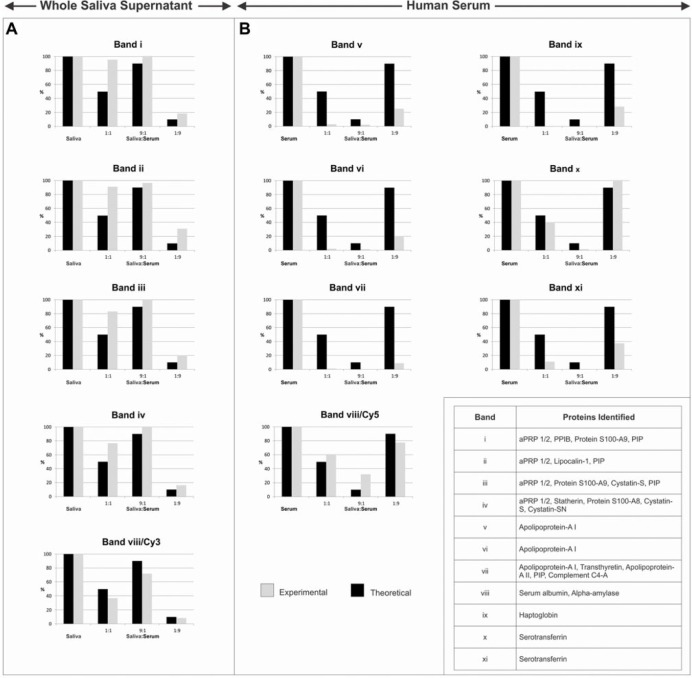
We next performed a competition assay to investigate relative affinities of saliva and serum proteins for hydroxyapatite. Proteins were labeled with Cy3 (green) and Cy5 (red) dyes to differentiate saliva and serum proteins, respectively, in the mixture (Unlü et al. 1997) (Fig. 2). The Cy3 and Cy5 labels did not generate any cross-talk when excited in a mixture at the Cy5 and Cy3 wavelengths, respectively (Fig. 2A, lane 2 and Fig. 2B, lane 1). Hydroxyapatite was incubated with Cy3-labeled saliva proteins, Cy5-labeled serum proteins, or both in a 1:1, 9:1 or 1:9 ratio (v/v). Labeling with Cy dyes did not affect the binding of the proteins to hydroxyapatite, since the adsorption patterns in Figure 2D (lanes 1 and 2; silver-stained CyDye gels) mimicked the adsorption patterns of unlabeled saliva and serum proteins in Figure 1. As expected, fractions containing only saliva or serum proteins (and mixed with hydroxyapatite) showed exclusively green- or red-labeled proteins. In the 1:1, 9:1 and 1:9 mixtures of saliva and serum proteins, a clear competition for binding to hydroxyapatite can be observed. For instance, lower (below 50 kDa) and higher (above 75 kDa) molecular weight serum proteins that adsorbed to hydroxyapatite in the absence of saliva proteins (Fig. 2C, lane 2) did not adsorb when saliva was present. This is evident in Figure 2C (lanes 3 to 5), containing various saliva:serum protein mixtures, where all lanes showed a significant presence of saliva proteins even when the saliva protein comprised just 10% of the total mixture. The 11 bands that showed adsorption/displacement are numbered with Roman numerals (i to xi) in Figure 2C. Bands i to iv in saliva, and band viii in serum strongly adsorbed in the saliva:serum mixture, whereas bands v to vii, and ix to xi in serum were rapidly displaced. The displacement was quantified by densitometric analysis of the black and white images of the Cy3 and Cy5 gels, and the proteins in the indicated bands were identified by mass spectrometric analysis (Fig. 3). The most dramatic increases in binding were observed for acidic proline-rich proteins 1 and 2 (aPRP 1/2), statherin, protein S100-A8/A9, and cystatin in serum, and the most rapid displacement was noted for the apolipoproteins, complement C4-A, haptoglobin, transthyretin and serotransferrin. Furthermore, the major saliva and serum proteins, amylase and albumin, in band viii/Cy3 and band viii/Cy5, respectively, showed less and more binding, respectively, compared with their calculated ratios in the mixtures.
Figure 2.
Electrophoretic separation and visualization of CyDye-labeled proteins from human whole saliva (Cy3) or serum (Cy5) adsorbed to hydroxyapatite. In each lane, 160 µL was applied. (A) Proteins visualized by excitation of the Cy3 dye. (B) Proteins visualized by excitation of the Cy5 dye. (C) Overlay of images in A and B. (D) Silver staining of the CyDye gel. Lane 1, whole saliva supernatant; lane 2, human serum; lanes 3 to 5: saliva:serum protein mixtures at 1:1, 9:1 and 1:9 (v/v), respectively. The excised gel bands are numbered i to xi. The experiment was conducted in duplicate.
Figure 3.
Densitometric analysis of selected proteins in experimental pellicles from saliva, serum and saliva:serum mixtures formed on hydroxyapatite. Quantitation was performed using black and white images of the Cy3- and Cy5-stained gels. The protein bands selected are indicated in Figure 2. Mass spectrometry was used to identify the proteins. Plotted are the theoretical percentage (black bars) of the various proteins and their experimental percentages (gray bars) relative to the band intensity in pure saliva and serum pellicle samples. The most abundant proteins detected in bands i to xi are indicated.
In Vivo Pellicle Formation on Saliva and Serum-Coated Enamel Surfaces
To gain preliminary insight into the effect of serum on the acquired enamel pellicle composition in vivo, we employed a split-mouth design in a single subject, where half the dentition immediately after prophylaxis was not coated (Fig. 4, left panel) while the other half was coated with serum (Fig. 4, right panel). The pellicle samples were collected after 0, 30 and 120 min of pellicle maturation by exposure to the oral environment. In the pellicle sample collected immediately after prophylaxis from the serum-coated surfaces, a clear presence of serum proteins was observed. In the 30 min (data not shown) and 120 min pellicle samples, many of the serum proteins (labeled “a”) had disappeared and were replaced by salivary proteins (labeled “b”), with the patterns of adsorbed proteins reminiscent of natural, acquired enamel pellicles. Based on the protein identifications shown in Figure 1, the displaced proteins labeled “a” represented peroxiredoxin-1, serotransferrin, complement factor B, complement factor C3, gelsolin and the collagen α-2(I) chain, whereas the primary adsorbing salivary proteins “b” represented acidic proline-rich proteins, statherin, cystatins and lactotransferrin. Several of these proteins were consistent with those identified in the in vitro competition assay.
Figure 4.
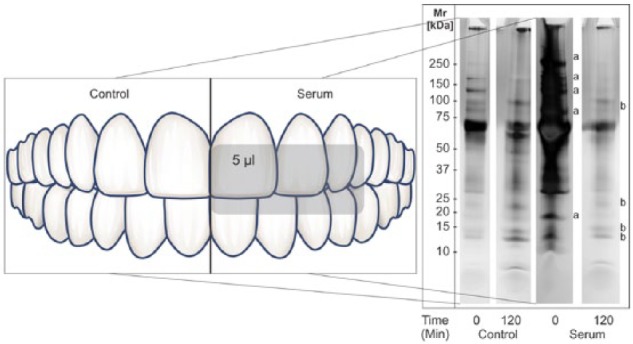
Composition of in vivo–formed pellicles on uncoated and serum-coated tooth surfaces in a split-mouth design of one subject. Proteins were harvested after 0, 30 (data not shown) and 120 min, subjected to SDS-PAGE, and visualized by silver staining. Note that the serum banding pattern in the 0-min sample (lane 3) had completely disappeared in the 120-min sample (lane 4), and resembled that of the saliva pellicle patterns in lane 2 from uncoated tooth surfaces. a, serum-derived proteins showing desorption; b, saliva-derived proteins showing adsorption.
Discussion
The biological importance of the acquired enamel pellicle and its relationship to biofilm formation has been established by several investigators (Kolenbrander et al. 1993; Scannapieco 1994). Therefore, it is important to elucidate the formation and functions of acquired enamel pellicles under conditions that mimic the environment of the gingival margin where the consequences of plaque accumulation are clinically very significant. The hydroxyapatite adsorption data with mixtures of saliva and serum proteins revealed that there is competition for adsorption among certain proteins. The in vivo implication is that there is an exchange of proteins in the gingival zone of the tooth surface. Consequently, the theoretical reciprocal concentration gradient of saliva and serum proteins between the gingival sulcus and the coronal portions of enamel may not reflect the actual ratios of salivary and serum proteins adsorbing to the tooth surface. The salivary proteins in the acquired enamel pellicle are therefore the major determinants of the functional and biological properties of this integument.
The ultimate composition of acquired enamel pellicle formed at the gingival margin in vivo is dependent on several physicochemical parameters dictated both by the protein structure and hydroxyapatite characteristics. These parameters are (a) the affinity constant of a protein for the hydroxyapatite surface (K), (b) the number of binding sites (N), (c) the nature of the binding site (e.g., exposed calcium or phosphate groups in the crystal lattice), and (d) the concentration of the protein. In general, a low isoelectric point and the presence of phosphorylated moieties in the protein structure increase the affinity of the protein for hydroxyapatite (Bernardi 1973). In saliva, phosphorylated proteins are abundant, and are represented by acidic proline-rich proteins, statherin, histatin 1, and cystatin S (Oppenheim et al. 2007). Moreno and coworkers (Moreno et al. 1982) determined that the K values for the 4 most abundant acidic proline-rich proteins range from 18.1 to 26.0 × 10-3 mL/µmol, with N values from 0.16 to 0.20 µmol/m2. Overall, the affinity constants for a limited set of the major salivary proteins are known (Moreno et al. 1982, 1984; Tabak et al. 1985; Johnsson et al. 1991), whereas much less information is available on serum protein adsorption parameters (Hlady and Füredi-Milhofer, 1977). Although apolipoproteins are acidic in nature, they do not contain any of the highly negatively charged phosphate groups (Gursky 2015). This may explain in part why their affinity for hydroxyapatite is lower than that of the phosphorylated salivary proteins. The molecular basis for these differences could be established by determining the N and K values of the identified serum proteins and assessing whether these saliva and serum proteins compete directly for the same binding sites on HA.
Data obtained by Carlén and coworkers (Carlén et al. 1998; Carlén et al. 2003; Rüdiger et al. 2012) on in vivo-formed acquired enamel pellicles showed the presence of plasma proteins fibrinogen, fibronectin, albumin, and IgG. However, these serum proteins were more readily detected on tooth surfaces near the gingival margin. It is possible that serum proteins will, to some extent, contribute to the in vivo pellicle, as the serum protein concentration in gingival crevicular fluid is 70-fold that of saliva. Such a high serum contribution to the protein environment surrounding the enamel surface is only expected in the zone immediately adjacent to the gingival sulcus. It is likely that serum protein adsorption to hydroxyapatite is primarily transient, perhaps except for albumin, and this is confirmed with our preliminary in vivo studies of the acquired enamel pellicle. This in vivo observation would have to be substantiated in a larger clinical study. Serum protein displacement at the gingival margin exposed to the oral environment is less likely to occur within the gingival sulcus or the periodontal pocket, which are hardly accessible to whole saliva.
The adsorption of phosphorylated salivary proteins to tooth surfaces is critically important for enamel protection, and the replacement of weakly adsorbing proteins from serum with less protective functions would be a significant benefit to the host. From a clinical perspective, shorter peptides from phosphorylated proteins with enamel-protective properties, such as statherin (Xiao et al. 2015) or casein (Reynolds 1997), are actively being pursued in the quest to protect against tooth erosion and dental caries. Our results suggest that such peptides, when applied locally, could rapidly replace proteins of lower protective value already adsorbed to the tooth surface at the gingival margin. Acquired enamel pellicle protein displacement thus represents a promising future direction to achieve pellicles with high protective functions, with the goal to diminish tooth demineralization and the formation of caries lesions. Furthermore, displacement therapies can also alter biofilm characteristics in this critical area of host vulnerabilities, affecting the initiation of gingivitis and periodontitis.
Author Contributions
D. Heller, contributed to conception, design, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript; E.J. Helmerhorst and F.G. Oppenheim, contributed to conception, design, data analysis, and interpretation, drafted and critically revised the manuscript. All authors gave final approval and agreed to be accountable for all aspects of the work.
Supplementary Material
Acknowledgments
The authors thank Dr. Olga Gursky for editing the manuscript and Renato Douek for his help with graphic design. We would like to pay tribute to Donald Ian Hay, who passed away on July 10, 2016. His pioneering work in the field of salivary protein biochemistry, particularly on the characterization of salivary protein adsorption and bacterial adhesion to oral surfaces, have laid the foundation for understanding the earliest phases of pellicle formation.
Footnotes
A supplemental appendix to this article is available online.
This work was supported by National Institutes of Health grants DE05672 (F.G.O.), DE07652 (F.G.O.), AI087803 (E.J.H.) and AI101067 (E.J.H.).
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- Abbas DK, Albandar JM, Messelt EB, Gjermo P. 1991. An in vivo model for the identification of serum proteins in the acquired subgingival pellicle. J Clin Periodontol. 18(5):341–345. [DOI] [PubMed] [Google Scholar]
- Bernardi G. 1973. Chromatography of proteins on hydroxyapatite. Methods Enzymol. 27:471–479. [DOI] [PubMed] [Google Scholar]
- Biyikoğlu B, Ricker A, Diaz PI. 2012. Strain-specific colonization patterns and serum modulation of multi-species oral biofilm development. Anaerobe. 18(4):459–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlén A, Börjesson AC, Nikdel K, Olsson J. 1998. Composition of pellicles formed in vivo on tooth surfaces in different parts of the dentition, and in vitro on hydroxyapatite. Caries Res. 32(6):447–455. [DOI] [PubMed] [Google Scholar]
- Carlén A, Rüdiger SG, Loggner I, Olsson J. 2003. Bacteria-binding plasma proteins in pellicles formed on hydroxyapatite in vitro and on teeth in vivo. Oral Microbiol Immunol. 18(4):203–207. [DOI] [PubMed] [Google Scholar]
- Chevallet M, Luche S, Rabilloud T. 2006. Silver staining of proteins in polyacrylamide gels. Nat Protoc. 1(4):1852–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawes C, Jenkins GN, Tonge CH. 1963. The nomenclature of the integuments of the enamel surface of teeth. Br Dent J. 115:65–68. [Google Scholar]
- Eng JK, McCormack AL, Yates JR. 1994. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 5(11):976–989. [DOI] [PubMed] [Google Scholar]
- Gursky O. 2015. Structural stability and functional remodeling of high-density lipoproteins. FEBS Lett. 589(19 Pt A): 2627–2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller D, Helmerhorst EJ, Gower AC, Siqueira WL, Paster BJ, Oppenheim FG. 2016. Microbial diversity in the early in vivo-formed dental biofilm. Appl Environ Microbiol. 82(6):1881–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hlady V, Füredi-Milhofer H. 1977. Adsorption of human serum albumin on precipitated hydroxyapatite. J Colloid Interface Sci. 69(3):460–468. [Google Scholar]
- Johnsson M, Richardson CF, Bergey EJ, Levine MJ, Nancollas GH. 1991. The effects of human salivary cystatins and statherin on hydroxyapatite crystallization. Arch Oral Biol. 36(9):631–636. [DOI] [PubMed] [Google Scholar]
- Kajisa L, Prakobphol A, Schiödt M, Fisher SJ. 1990. Effect of plasma on composition of human enamel and cementum pellicle. Scand J Dent Res. 98(6):461–471. [DOI] [PubMed] [Google Scholar]
- Kolenbrander PE, Ganeshkumar N, Cassels FJ, Hughes CV. 1993. Coaggregation: specific adherence among human oral plaque bacteria. FASEB J. 7(5):406–413. [DOI] [PubMed] [Google Scholar]
- Kolenbrander PE, London J. 1993. Adhere today, here tomorrow: oral bacterial adherence. J Bacteriol. 175(11):3247–3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamster IB, Ahlo JK. 2007. Analysis of gingival crevicular fluid as applied to the diagnosis of oral and systemic diseases. Ann N Y Acad Sci. 1098: 216–229. [DOI] [PubMed] [Google Scholar]
- Loe H, Holm-Pedersen P. 1965. Absence and presence of fluid from normal and inflamed gingivae. Periodontics. 3:171–177. [PubMed] [Google Scholar]
- Lourenco TG, Heller D, Silva-Boghossian CM, Cotton SL, Paster BJ, Colombo AP. 2014. Microbial signature profiles of periodontally healthy and diseased patients. J Clin Periodontol. 41(11):1027–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno EC, Kresak M, Hay DI. 1982. Adsorption thermodynamics of acidic proline-rich human salivary proteins onto calcium apatites. J Biol Chem. 257(6):2981–2989. [PubMed] [Google Scholar]
- Moreno EC, Kresak M, Hay DI. 1984. Adsorption of molecules of biological interest onto hydroxyapatite. Calcif Tissue Int. 36(1):48–59. [DOI] [PubMed] [Google Scholar]
- Olczak T, Dixon DW, Genco CA. 2001. Binding specificity of the Porphyromonas gingivalis heme and hemoglobin receptor HmuR, gingipain K, and gingipain R1 for heme, porphyrins, and metalloporphyrins. J Bacteriol. 183(19):5599–5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheim FG. 1970. Preliminary observations on the presence and origin of serum albumin in human saliva. Helv Odontol Acta. 14(1):10–17. [PubMed] [Google Scholar]
- Oppenheim FG, Salih E, Siqueira WL, Zhang W, Helmerhorst EJ. 2007. The salivary proteome and its genetic polymorphisms. Ann N Y Acad Sci. 1098:22–50. [DOI] [PubMed] [Google Scholar]
- Peng J, Gygi SP. 2001. Proteomics: the move to mixtures. J Mass Spectrom. 36(10):1083–1091. [DOI] [PubMed] [Google Scholar]
- Reynolds EC. 1997. Remineralization of enamel subsurface lesions by casein phosphopeptide-stabilized calcium phosphate solutions. J Dent Res. 76(9):1587–1595. [DOI] [PubMed] [Google Scholar]
- Rüdiger SG, Dahlén G, Carlén A. 2012. Pellicle and early dental plaque in periodontitis patients before and after surgical pocket elimination. Acta Odontol Scand. 70(6):615–621. [DOI] [PubMed] [Google Scholar]
- Scannapieco FA. 1994. Saliva-bacterium interactions in oral microbial ecology. Crit Rev Oral Biol Med. 5(3-4): 203–248. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods. 9(7):676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 68(5):850–858. [DOI] [PubMed] [Google Scholar]
- Siqueira WL, Custodio W, McDonald EE. 2012. New insights into the composition and functions of the acquired enamel pellicle. J Dent Res. 91(12):1110–1118. [DOI] [PubMed] [Google Scholar]
- Siqueira WL, Oppenheim FG. 2009. Small molecular weight proteins/peptides present in the in vivo formed human acquired enamel pellicle. Arch Oral Biol. 54(5):437–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siqueira WL, Zhang W, Helmerhorst EJ, Gygi SP, Oppenheim FG. 2007. Identification of protein components in in vivo human acquired enamel pellicle using LC-ESI-MS/MS. J Proteome Res. 6(6):2152–2160. [DOI] [PubMed] [Google Scholar]
- Tabak LA, Mirels L, Monte LD, Ridall AL, Levine MJ, Loomis RE, Lindauer F, Reddy MS, Baum BJ. 1985. Isolation and characterization of a mucin-glycoprotein from rat submandibular glands. Arch Biochem Biophys. 242(2):383–392. [DOI] [PubMed] [Google Scholar]
- Thomadaki K, Helmerhorst EJ, Tian N, Sun X, Siqueira WL, Walt DR, Oppenheim FG. 2011. Whole-saliva proteolysis and its impact on salivary diagnostics. J Dent Res. 90(11):1325–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unlü M, Morgan ME, Minden JS. 1997. Difference gel electrophoresis: a single gel method for detecting changes in protein extracts. Electrophoresis. 18(11):2071–2077. [DOI] [PubMed] [Google Scholar]
- Xiao Y, Karttunen M, Jalkanen J, Mussi MC, Liao Y, Grohe B, Lagugné-Labarthet F, Siqueira WL. 2015. Hydroxyapatite growth inhibition effect of pellicle statherin peptides. J Dent Res. 94(8):1106–1112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.