Abstract
Hereditary persistence of fetal hemoglobin deletion type-2 (HPFH-2) and Sicilian-δβ-thalassemia are conditions described as large deletions of the human β-like globin cluster, with absent β-globin chains and a compensatory variable increase in γ-globin. HPFH, in general, may be distinguished from DB-Thalassemia by higher fetal hemoglobin (HbF) levels, absence of anemia and hypochromic and microcytic erythrocytes. MicroRNAs (miRNAs) regulate a range of cellular processes including erythropoiesis and regulation of transcription factors such as the BCL11A and SOX6 genes, which are related to the regulation of γ-globin expression. In this report, a possible association among the overexpression of miRNAs and the expression of the γ-globin gene was analyzed in these two conditions. Forty-nine differentially expressed miRNAs were identified by microarrays in CD34+-derived erythroid cells of two subjects heterozygous for Sicilian-δβ-thalassemia, 2 for HPFH-2 and 3 for controls after 13 days of culture. Some of these miRNAs may participate in γ-globin gene regulation and red blood cell function. The BCL11A gene was found to be potentially targeted by 12 miRNAs that were up-regulated in HPFH-2 or in DB-Thal. A down-regulation of BCL11A gene expression in HPFH-2 was verified by quantitative polymerase chain reaction. These data suggest an important action for miRNA that may partially explain the phenotypic differences between HPFH-2 and Sicilian δβ-thalassemia and the increased expression of γ-globin in these conditions.
Keywords: Hereditary persistence of fetal hemoglobin deletion type-2, Sicilian-δβ-thalassemia, microRNAs, BCL11A, SOX6, hemoglobinopathies
Introduction
Delta-beta thalassemia (δβ-thalassemia) and hereditary persistence of fetal hemoglobin deletion type (HPFH) are conditions that are characterized by large deletions of variable extents in the human β-like globin cluster and a compensatory variable increase in γ-globin chains.1 DB-thalassemia is characterized by the maintenance of the expression of γ-globin genes in adulthood, with absence of δ- and β-globin gene expression and increase of fetal hemoglobin (HbF) concentrations, and the presence of normal or mild anemia with hypochromic and microcytic erythrocytes.1 Individuals that are heterozygous for DB-Thalassemia (δβ°) present HbF levels ranging from 4.0% to 18.6% distributed in a heterogeneous (heterocellular) subpopulation of so-called F cells.1,2 HPFH is distinguished from DB-Thalassemia by higher levels of HbF, balanced synthesis of α-like and β-like globin chains, absence of anemia and normocytic red-cell in heterozygotes, and relatively uniform HbF distribution (pancelular) among all red cells.1,3
A number of studies suggest various mechanisms to explain the increase in HbF levels in DB-Thalassemia and HPFH, such as the presence of one repressor element of gamma globin between the gamma and delta globin genes which is removed by deletion and also the presence of enhancer elements downstream from the β-globin gene which are brought closer to the beta cluster by the deletion. Moreover, deletions in HPFH are probably associated with the removal of silencer or enhancer elements in the proximity of the γ-globin genes. Recently, the discovery of a BCL11A binding site, characterized as a 3,5 kb intergenic region upstream from the δ-globin gene, seems to be required for efficient γ-globin blocking through binding with the BCL11A gene. This region is deleted in HPFH-2 and is preserved in Sicilian DB-thalassemia and may contribute to explain the differences between the phenotypes of these conditions.3
MicroRNAs (miRNAs) are small non-coding RNAs that are ∼22 nucleotides in length and that function as post-transcriptional regulators by hybridization between the miRNA and the targeted mRNA in its 3'UTR region, consequently repressing the translation of this mRNA.4–6 In vitro experiments show that miRNAs regulate a variety of cellular processes, including differentiation, development, apoptosis, oncogenesis, metabolism, erythropoiesis and regulation of transcription factors.7–10
A few studies have focused on the implication of miRNA expression for HbF induction. One study showed that human perinatal Hb switching is under the control of the kit receptor/miR 221-222 complex and that down modulation of these miRNAs stimulates erythropoietic proliferation through up modulation of the kit receptor.11 The over-expression of miRNAs-15a and 16-1 in patients with a trisomy of chromosome 13 led to high levels of HbF, due to the down-regulation of the MYB transcription factor, which control the γ-globin expression negatively.3 In another study, an over expression of miRNA-486-3p in erythroid cells was associated with reduced BCL11A protein levels and increased γ-globin expression.12 Finally, a study conducted on K-562 cells demonstrated an increase in γ-globin expression caused by miRNA-26b.13
To the best of our knowledge, there has been no comparative study of the profiles of miRNAs in DB-thalassemia and HPFH. Thus, we carried out analyses, comparing the miRNAs profiles of heterozygous individuals for Sicilian δβ-thalassemia (βA/βδβ) with heterozygous HPFH-2 individuals (βA/βHPFH). Forty-nine miRNAs were differentially expressed and identified in CD34+-derived erythroid cells after 13 days in culture with predominance of orthochromatic erythroblasts and erythroid enucleated cells. The BCL11A gene was found to be a potential target for several miRNAs that were up-regulated in HPFH-2 and Sicilian δβ-thalassemia, and the number of such miRNAs was significantly higher in HPFH-2 than in δβ-thalassemia.
Our findings suggest a novel pathway that may contribute to explain the differences in the expressions of γ-globin in Sicilian δβ-thalassemia and HPFH-2. Moreover, a deeper understanding of the interactions between miRNAs and γ-globin gene repressors, such as BCL11A, could lead to the discovery of new manners of activation of HbF in the adult erythroid cells of patients with hemoglobin disorders.
Materials and methods
Patients
Two Sicilian δβ-thalassemia patients and two heterozygotic HPFH-2 subjects were included in the study. Diagnosis was carried out by Hb electrophoresis, high-performance liquid chromatography (HPLC), and the deletion breakpoints were confirmed by DNA sequencing. The two δβ-thalassemia patients are brothers while the two HPFH-2 subjects are not related. Three healthy individuals were used as controls and one of these (CTRL3) is the uncle of the δβ-thalassemia patients (see Table 1 for clinical details). Informed, written consent was acquired from all patients and controls, and the Ethics Committee, UNICAMP, approved the study.
Table 1.
Clinical data of healthy controls, Sicilian δβ-thalassemia patients, and HPFH-2 subjects
| Controls |
HPFH-2 |
DB-Thal |
|||||
|---|---|---|---|---|---|---|---|
| Female (βA/βA) | Male (βA/βA) | Male (βA/βA) | Male (βA/βHPFH) | Male (βA/βHPFH) | Female (βA/βδβ) | Male (βA/βδβ) | |
| CTRL1 | CTRL2 | CTRL3 | P1 | P2 | DB1 | DB2 | |
| RBC (x106/µL) | 4.58 | 5.26 | 5.84 | 5.44 | 5.72 | 5.35 | 6.76 |
| HGB (g/dL) | 12 | 15.3 | 16.9 | 16.1 | 15.7 | 12.9 | 14.4 |
| MCV (fL) | 85.4 | 85.7 | 83.5 | 88 | 82.9 | 74.7 | 64.7 |
| MCH (pg) | 26.2 | 29.1 | 28.9 | 29.6 | 27.5 | 24.1 | 21.2 |
| RDW (%) | 14 | 14.1 | 14.1 | 14.1 | 16.4 | 19.1 | 18.8 |
| PLT (x103/µL) | 234 | 174 | 214 | 270 | 254 | 279 | 222 |
| HbA (%) | 86.7 | 87.1 | 86.6 | 65.3 | 64.8 | 70.9 | 81.8 |
| HbA2 (%) | 2.8 | 2.8 | 2.6 | 2.5 | 2.1 | 2.5 | 2.9 |
| HbF (%) | 0.2 | 0.5 | 0.2 | 25.5 | 25.3 | 18.8 | 6.2 |
Erythroid cell culture
Blood samples from normal volunteers, from two heterozygotic HPFH-2 subjects (βA/βHPFH) and from two heterozygotic Sicilian DB-Thalassemia patients (βA/βδβ) were cultured using a two-phase liquid culture procedure to examine the expression profile of miRNAs. One culture from each person was performed. Briefly, mononuclear cells were isolated from peripheral blood samples by centrifugation over a Ficoll-Hypaque gradient and after immunomagnetic separation, CD34+ cells were cultured in 5% CO2 at 37℃ for a period of 13 days in a suitable liquid medium for proliferation and differentiation—(Iscoves's Modified Dulbecco's Medium [IMDM; GIBCO™, Invitrogen Corporation, USA] prepared with bicarbonate sodium [Merck, Germany], glutamine [GIBCO™, Invitrogen Corporation, USA], alpha-thioglycerol [Sigma Aldrich, St. Louis, MO] penicillin/streptomycin [Sigma Aldrich], fungizone [Sigma Aldrich], and supplemented with bovine serum albumin [BSA] [USBiological, Swampscott, MA], 7.5% sodium bicarbonate [Sigma Aldrich], a solution of liposomes [cholesterol, oleic acid and phosphatidylcholine dipalmitol—Sigma Aldrich], apo-transferrin [Sigma Aldrich], and fetal bovine serum [FBS; 10%—days 0 through 7 and 30%—from 7 to 13; GIBCO™, Invitrogen Corporation, USA]). Cytokines were added to stimulate erythroid differentiation: 50 ng/mL SCF (R&D Systems, USA), 5 ng/mL interleukin-3 (IL-3; R&D Systems), and 2 units/mL of erythropoietin (Epo; Eprex3000, Vetter Pharma, Ravensburg, Germany). The viability and the cell numbers were determined by trypan blue staining. Samples of 1 × 107 cells were pelleted and resuspended in the lysis buffer of the mirVana™ miRNA Isolation Kit (Ambion, Life Technologies, Foster City, CA) and stored at −80℃ for total RNA extraction and complementary DNA (cDNA) synthesis.
Flow cytometry analysis
The erythroid differentiation process was monitored by flow cytometry using the following cell surface specific antibodies (Caltag Laboratories, Burlingame, CA): anti-transferrin receptor (FITC conjugated) (CD 71), anti-glycophorin A (PE-conjugated), and anti-fetal hemoglobin (FITC-conjugated). The cell concentration used was 1 x 105 cells in a final volume of 300 µL in phosphate-buffered saline. Data were quantified using a FACsCalibur instrument (Becton-Dickinson, USA) and 10,000 events were acquired for analysis using Cell Quest Software (Becton-Dickinson).
RNA extraction and cDNA synthesis
Total RNA was extracted from 1 x 107 cells from each culture using the mirVana™ miRNA Isolation Kit (Ambion, Life Technologies), according to the manufacturer’s guidelines. RNA samples were confirmed to be proteins and phenol free by spectrophotometry. RNA integrity was assessed by Agilent 6000 RNA Nano chips and an Agilent 2100 Bioanalyzer (Agilent Technologies, USA). cDNA was synthesized from total RNAs using 1 µg of RNA samples, and employing the RevertAid™ H Minus First Strand cDNA Synthesis Kit (Fermentas, Thermo Scientific, Lithuania).
miRNA expression profiling
The total RNA of CD34+-derived erythroid cells cultured for 13 days from two HPFH-2 individuals (βA/βHPFH), two Sicilian DB-Thalassemia patients (βA/βδβ), and three healthy controls (βA/βA) were Cy-3-labeled using the Agilent miRNA Complete Labeling and Hybridization Kit (Agilent Tech, Canada). Initially, 100 ng of each total RNA was dephosphorylated during incubation with calf intestinal phosphatase at 37℃ for 30 min and denatured in absolute dimethyl sulfoxide at 100℃ for 5 min. Each RNA sample was labeled with perCp-Cy3 using T4 ligase at 16℃ for 1 h before hybridizing on 8x 15K format Agilent Human miRNA array slides. The hybridizations were performed for 20 h at 55℃. The slides were washed according to Agilent’s instructions and scanned using a DNA microarray scanner (Agilent Technologies). The miRNA microarray data analysis was carried out by Agilent Feature Extraction program (v10.5). The microarray data were normalized to the quantile and analyzed using the GeneSpring GX 11.0 bioinformatics platform for hierarchical clustering of samples or miRNAs based on an ANOVA statistical analysis (P ≤ 0.05), fold change ≥2.0 and uncentered Pearson correlation metrics.
Real-time PCR
Quantitative real time polymerase chain reactions (PCRs) were performed using SYBR™ Green PCR Master Mix® (Applied Biosystems, Life Technologies) and primers were specifically designed to amplify mRNA from genes: BCL11A F-5′-CCCCAGCACTTAAGCAAACG-3′ and R-5′-GTGGTCTGGTTCATCATCTGTAAGA-3′; SOX6 F-5′-AATCTTAGGATCTCGCTGGAAAT-3′ and R-5′-AGTGGATCTTGCTTAGCCGG-3′. Gene expression levels were standardized to control levels from GAPDH primers: F-5′-GCACCGTCAAGGCTGAGAAC-3′ and R-5′-CCACTTGATTTTGGAGGGATCT-3′; ACTB F-5′-AGGCCAACCGCGAGAAG-3′, and R-5′-ACAGCCTGGATAGCAACGTACA-3′. Real-time PCR for miRNAs was performed using TaqMan® miRNA assay and the standard protocol (miR-146a: ID000468, Life Technologies). Normalization was performed using the RNU47 and RNU48 primer kit (ID 001223; ID 001006, Life Technologies). All reactions were run a StepOne Plus equipment (Applied Life Technologies) in experimental replicates and the relative expression was calculated using geNorm algorithms.
High-performance liquid chromatography
HPLC was used to quantify the hemoglobin variants and was carried out using the Variant II Hemoglobin Testing System (Bio-Rad, CA) at the Hematology Laboratory of the Department of Clinical Pathology—UNICAMP. The fractions of hemoglobins were quantified using 1 mL of whole blood from controls, Sicilian δβ-thalassemia patients, and from HPFH-2 subjects, before performing the CD34+-derived erythroid cell culture.
In silico analysis
The study of the interactions of differentially expressed miRNAs with some transcription factors as possible targets was carried out using miRanda–mirSVR (http://www.microrna.org). This database considers matches to human 3′ UTRs using an algorithm called mirSVR (support vector regression) for scoring the efficiency of miRanda-predicted miRNA target sites.14
DNA extraction and PCR
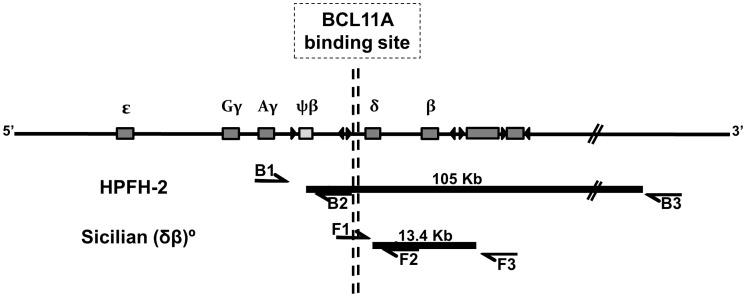
The genomic DNA (gDNA) from leukocytes of controls and patients were extracted from ethylenediaminetetraacetic acid-treated whole blood samples by the phenol–chloroform technique. The quantitation of the samples was performed in the NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, DE). Primers, previously established in other studies,15 were used to detect the regions of the deletion of each disease (breakpoints): B1 5′-GACATGGACTATTGTTCAATGA-3′, B2 5′-TGCTATGCCAACTCACTACC-3′, B3 5′-TTTATATATGAAATGCTACTGATT-3′, F1 5′-TTGGGTTTCTGATAGGCACTG-3′, F2 5′-GTGTCACCCATTAATGCCTTGTAC-3′ and F3 5′-TAGATCCCTTTGCCATTATG-3′. Pairs of primers were used to detect normal sequences, present in controls and heterozygotes (HPFH-2 = B1/B2 and Sicilian DB-thalassemia = F1/F2), and mutant sequences (HPFH-2 = B1/B3 and Sicilian DB-thalassemia = F1/F3), present only in heterozygotes, characterizing the deletions (Figure 1).
Sequencing
The sequencing for the two Sicilian DB-thalassemia patients was performed on an ABI 3500 Genetic Analyzer (Applied Biosystems, Hitachi) using the Big Dye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystem, CA, EUA). All steps were performed according to the manufacturers’ guidelines for the kits used in the experiment. The Chromas Lite v2.1.1 software (Technelysium, Australia) was used to analyze the sequences and to confirm the deletions. The HPFH-2 subjects were previously studied and deletions confirmed by sequencing.16–18
Results
High-performance liquid chromatography
The identification of hemoglobin variants was carried out using retention time (RT) windows and area percent and their relevant RBC parameters and HPLC chromatogramic findings are presented in Table 1. The subjects studied were previously diagnosed for HPFH-2 and Sicilian δβ-thalassemia, as described, using PCR and DNA sequencing to confirm the deletions.
miRNA expression profiling
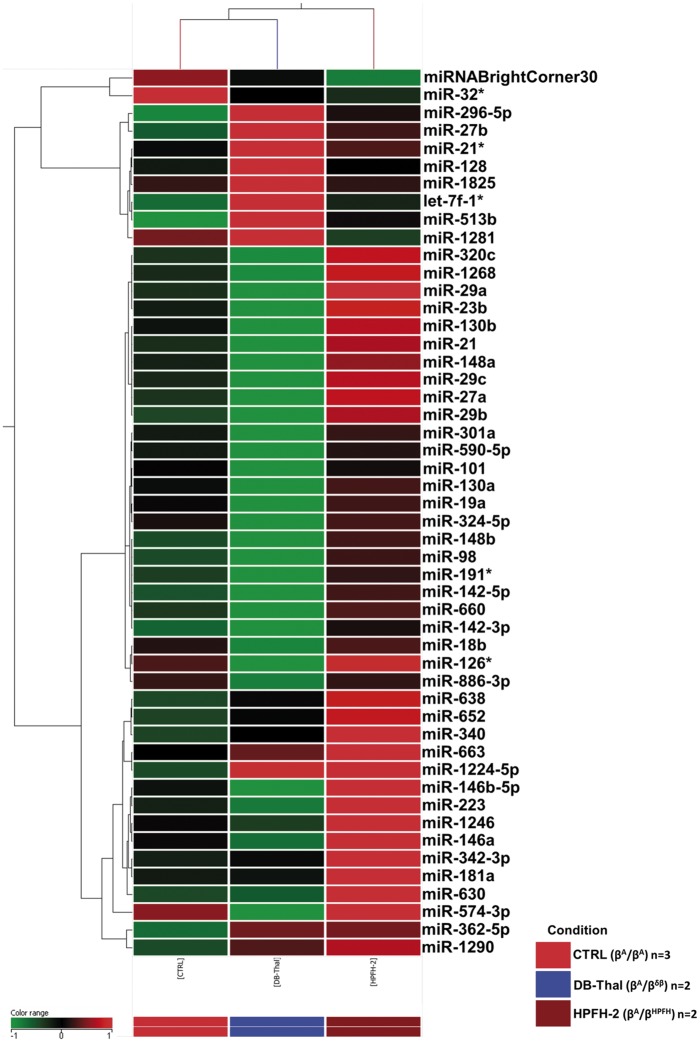
Forty-nine miRNAs that were differentially expressed in CD34+-derived erythroid cells after 13 days of culture were identified in a comparative analysis with two subjects that were heterozygous for Sicilian δβ-thalassemia (βA/βδβ), two subjects that were heterozygous for HPFH-2 (βA/βHPFH) and three healthy controls (βA/βA) (Figure 2 and Table S1).
Figure 1.
Representation of the human β-globin gene cluster with the deletions HPFH-2 and Sicilian DB-thalassemia. Approximate positions of the oligonucleotide primers used in the PCR for Breakpoint regions are shown. Details of the primers are provided in “Materials and methods” section. The BCL11A binding-site lies 1.5 kb 5' δ-globin gene (red vertical lines) and is removed in the HPFH-2 deletion, while it is maintained for Sicilian (δβ)0 deletion
Figure 2.
Mirnome (miRNA) expression profiles in CD34+-derived erythroid cells (after 13 days of culture) from two heterozygous for Sicilian δβ-thalassemia patients (βA/βδβ), two heterozygotic HPFH-2 subjects (βA/βHPFH) and three health controls (βA/βA). The dendrograms and heat map were acquired using the Agilent GeneSpring platform. Heat map legend: red = up-regulation, green = down-regulation, black = unmodulated (Pearson correlation metrics)
Of the 49 miRNAs that were differentially expressed in this comparative analysis, 36 miRNAs were up-regulated only in HPFH-2 (HPFH) (Figure 2 and Table S1). Another group of five different miRNAs was up-regulated only in δβ-thalassemia (DB-Thal) (Figure 2 and Table S1). Further one miRNA (hsa-miR-362-5p) were up-regulated in HPFH-2 and in δβ-thalassemia (DB-Thal) (Figure 2 and Table S1). Seven miRNAs were not considered in the analysis because they present at least one negative value in all the groups (Table S1).
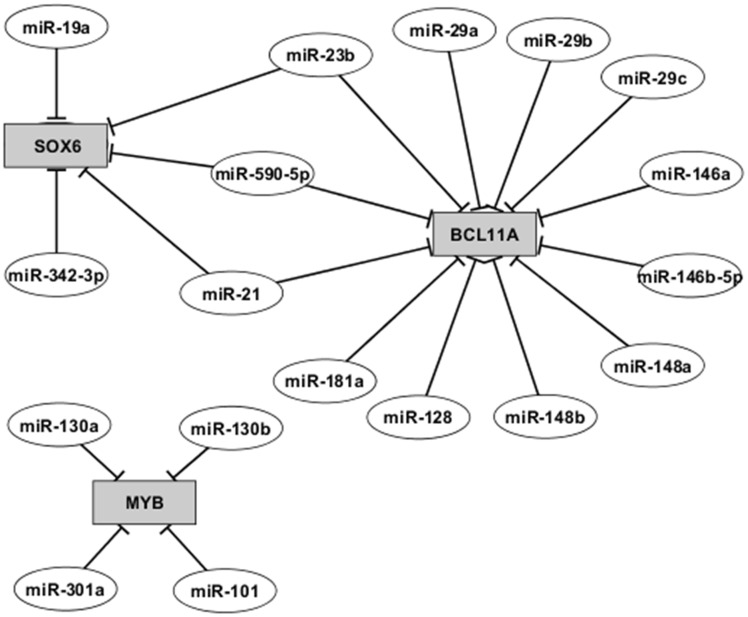
In silico analyses, using the 49 miRNAs differentially expressed in CD34+-derived erythroid cells, showed that 12 potentially target the BCL11A gene. According to MiRanda database, the BCL11A gene is potentially targeted by a total of 91 miRNAs that were checked against CD34+-derived erythroid cells. From these, 11 miRNAs that were up-regulated only in HPFH-2 (Figures 2 and 3; Table S1) and one miRNA (miR-128) up-regulated only in DB-Thal (Figures 2 and 3; Table S1) have one site in the BCL11A gene to which these miRNAs could bind and control the expression of this gene. Five miRNAs that were up-regulated only in HPFH-2 have SOX6 gene as a potential target. Three of these miRNAs also have the BCL11A gene as a potential target: miR-21, miR-23b and miR-590-5p (Figure 3). In silico analyses in the MiRanda database showed four miRNAs that were up-regulated only in HPFH-2 that have the MYB gene as a potential target: miR-101, miR-130a, miR-130b, and miR-301a (Figure 3; Table S1).
Figure 3.
Gene network of potential targets of the differentially expressed miRNAs found in the microarray data of CD34+-derived erythroid cells (after 13 days of culture) from two heterozygous for Sicilian δβ-thalassemia patients (βA/βδβ), two heterozygotic HPFH-2 subjects (βA/βHPFH) and three health controls (βA/βA)
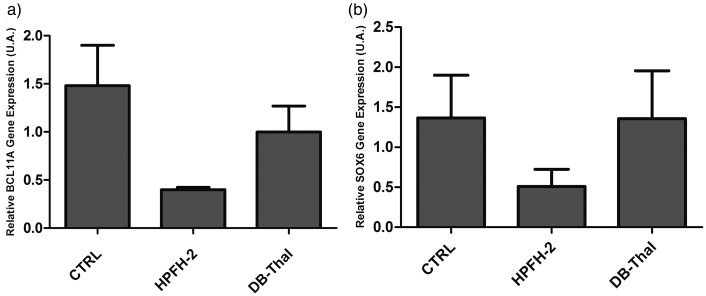
BCL11A and SOX6 mRNA gene expressions were determined by qPCR. Due to the small number of samples, it was not possible to compare them statistically but an apparent down-regulation of BCL11A gene expression in CD34+-derived erythroid cells (Figure 4(a)) was found in HPFH-2 and DB-Thal, when compared to controls. A down-regulation of SOX6 gene expression was found only in HPFH-2, when compared to controls and DB-Thal in CD34+-derived erythroid cells (Figure 4(b)).
Figure 4.
Relative transcriptional levels of BCL11A and SOX6 mRNA, as evaluated by quantitative reverse-transcription PCR (qRT-PCR). (a) BCL11A mRNA expression in CD34; (b) SOX6 mRNA expression in CD34; Controls = CTRL (n = 3); subjects heterozygous for HPFH-2 = HPFH-2 (n = 2); patients heterozygous for Sicilian δβ-thalassemia = DB-Thal (n = 2)
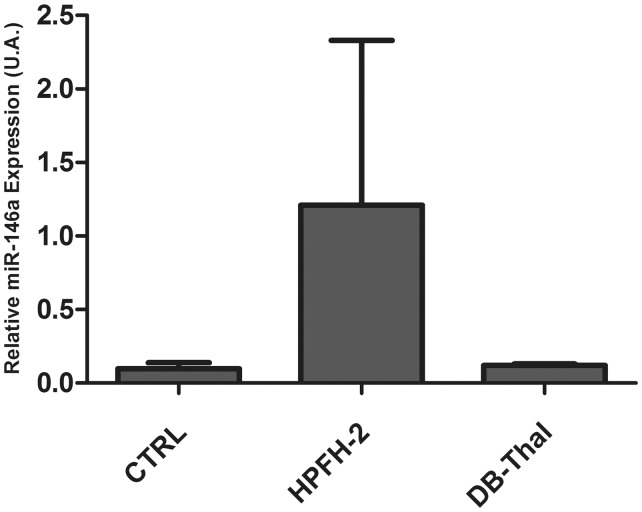
Five miRNAs were evaluated by qPCR and the results confirmed that the miRNA miR-146a is up-regulated in HPFH-2 compared to controls and to DB-Thal in CD34+-derived erythroid cells (Figure 5). The miRNAs 96, 210, 451 and 486-3p were also evaluated by qPCR in the seven samples (CTRL→ n = 3; HPFH-2→ n = 2; DB-Thal→ n = 2), and confirmed the data of the microarray analyses (Figure 2).
Figure 5.
Relative transcriptional levels of miR-146a in CD34+-derived erythroid cells, as evaluated by quantitative reverse-transcription PCR (qRT-PCR). Taqman qRT-PCR was used to evaluate the levels of miR-146a. Controls = CTRL (n = 3); subjects heterozygous for HPFH-2 = HPFH-2 (n = 2); patients heterozygous for Sicilian δβ-thalassemia = DB-Thal (n = 2)
Discussion
This study reports a comparative analysis of the miRprofile of CD34+-derived erythroid cells after 13 days in culture with predominance of orthochromatic erythroblasts from heterozygous for Sicilian DB-Thalassemia patients and heterozygous HPFH-2 subjects. Microarray data showed an up-regulation of 18 miRNAs in CD34+-derived erythroid cells for which in silico analysis designated target genes such as BCL11A, SOX6, and MYB. Of these, 12 miRNAs target BCL11A. The greater number of miRNAs in these interactions may suggest a down-regulation of BCL11A gene expression in HPFH-2, compared to δβ-thalassemia, and may indicate that this might be a mechanism by which of γ-globin gene expression is higher in HPFH-2 than in DB-thalassemia.
Genome-wide association studies have identified sequence variants in the genes, BCL11A and HBS1L-MYB, in independent non-anemic populations that influence HbF levels.19,20 Subsequently, several studies showed that HbF-regulating sequences have important effects on clinical course for sickle cell anemia and β-thalassemia.20,21 It was subsequently showed that BCL11A is a very important modulator of HbF switching and essential for blocking γ-globin gene expression in adult erythroid cells.22,23 An in vivo study with primary adult human erythroid cells showed that the BCL11A gene is probably associated with distal regulatory sequences and change the configuration of the cluster of β-globin. Furthermore, transcriptional blocking of γ-globin gene by BCL11A was shown to include long distance cooperation and reciprocal action with several transcription factors, including SOX6.24
The data presented herein strongly suggest a down-regulation of BCL11A and SOX6 genes in HPFH-2 on CD34+-derived erythroid cells as shown by qPCR analysis. These genes are considered to be possible targets of several miRNAs that were also found in this study to be up-regulated in HPFH-2 and DB-Thalassemia. Furthermore, the SOX6 gene is potentially targeted by five miRNAs that are up-regulated only in HPFH-2, compared to DB-Thal and CTRLs (Figure 3). Taken together, these data suggest that both BCL11A and SOX6 gene expressions could be, at least in part, modulated by these miRNAs and, consequently, influence the HbF levels.
Considering the differentially expressed miRNAs found in HPFH-2 and Sicilian δβ-thalassemia profiles, a few of these have been previously described as related to erythropoiesis or γ-globin gene regulation. miRNA-146a, which was found up-regulated in CD34+-derived erythroid cells possibly binds to a site in the 3′-UTR of the BCL11A gene, according to in silico analysis. Recently, this miRNA was evaluated in peripheral blood mononuclear cell of type 1 diabetes mellitus (T1D) patients and its effect on BCL11A gene expression was also determined.25 Data showed that the expression of miR-146a is lower in newly diagnosed T1D patients compared to controls and that BCL11A gene expression was significantly up-regulated in these patients. These data suggest that miR-146a may be active in the control of the expression of the BCL11A gene.
In another study, miRNA-146b-5p (miR-146b) from the family of miR-146a was transcriptionally activated by GATA-binding protein 1 (GATA-1), and promoted human erythroid and megakaryocytic differentiation via regulation of the PDGFRA signaling pathway.26 In our study, miR-146b-5p was found up-regulated in HPFH-2 in CD34+-derived erythroid cells, compared to DB-Thal and controls (Figure 5).
The miRNAs 21, 23b, and 590-5p were up-regulated in HPFH-2 and appear to have both BCL11A and SOX6 genes as potential targets. A study suggests that ETO2, an element of a protein complex containing regulators of hematopoiesis, including GATA-1 and SCL/TAL1, positively regulates the abundance of mature miRNAs, including miR-21.27 A recent study revealed that, during hypoxia, up-regulated miR-21 resulted in low catalase activity in young red blood cells.28 The miRNA-23b was studied in cytoskeleton remodeling during erythropoiesis. When miR-23b-3p level was overexpressed, the erythrocyte tropomodulin of 41 kDa (E-Tmod41) protein level was reduced during erythroid differentiation. The E-Tmod41 caps the pointed end of actin filament (F-actin) and is essential for the formation of erythrocyte membrane skeleton.29 There is no study associating miR-590-5p with erythroid cells.
The miRNA-19a that was up-regulated in HPFH-2 was associated to erythrocyte shapes and to the phospholipid and lipid profiles of the erythrocyte membrane.30
Furthermore, the miRNAs 130a and 130b which possible target MYB gene and were up-regulated in HPFH-2 have been associated with iron metabolism regulation.31
Although preliminary, due to small number of studied individuals, our results suggest that there are a number of miRNAs that are up-regulated in HPFH-2 and DB-thalassemia. Most of the miRNAs that have BCL11A and SOX6 as target genes are present only in HPFH-2 and this may indicate an important factor contributing to the difference in the γ-globin gene expression observed between these two entities.
These results contribute to confirm the potential therapeutic implications of the down-regulation of BCL11A in erythroid cells in sickle cell disease and β-thalassemia major patients.32,33
Author contributions
TAF contributed to the idea of the manuscript, performed the experimental work and analysis of the results and finally, in the writing of the manuscript; CL, DMA, and RF participated in the experimental work and in the analysis of the results; FFC was the supervisor of the study and participated in every step of the study. All authors have read and approved the final manuscript. The authors of this study are supported by FAPESP, Brazil. TAF was also supported by FAPESP (2013/06679-5). The authors thank Dr Nicola Conran, HEMOCENTRO-UNICAMP, for help with English correction and revision.
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- 1.Forget BG. Molecular basis of hereditary persistence of fetal hemoglobin. Ann NY Acad Sci 1998; 850: 39–44. [DOI] [PubMed] [Google Scholar]
- 2.Huisman HJ, Carver MFH, Baysal E, Efremov GD. HbVar: a database of human hemoglobin variants and thalassemias. Summaries of mutation categories. Pennsylvania University USA and McMaster University in Canada. Available at: http://globin.cse.psu.edu/ (accessed 17 August 2015).
- 3.Sankaran VG, Xu J, Byron R, Greisman HA, Fisher C, Weatherall DJ, Sabath DE, Groudine M, Orkin SH, Premawardhena A, Bender MA. A functional element necessary for fetal hemoglobin silencing. N Engl J Med 2011; 365: 807–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism and function. Cell 2004; 116: 281–97. [DOI] [PubMed] [Google Scholar]
- 5.Cai Y, Yu X, Hu S, Yu J. A brief review on the mechanisms of miRNA regulation. Genom Proteom Bioinformatics 2009; 7: 147–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 2010; 466: 835–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohneda K, Yamamoto M. Roles of hematopoietic transcription factors GATA-1 and GATA-2 in the development of red blood cell lineage. Acta Haematol 2002; 108: 237–45. [DOI] [PubMed] [Google Scholar]
- 8.Zhou Y, Ferguson J, Chang JT, Kluger Y. Inter.-and intra-combinatorial regulation by transcription factors and microRNAs. BMC Genom 2007; 8: 396–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Azzouzi I, Markus S, Oliver S. MicroRNAs as components of regulatory networks controlling erythropoiesis. Eur J Haematol 2012; 89: 1–9. [DOI] [PubMed] [Google Scholar]
- 10.Bauer DE, Kamran SC, Orkin SH. Reawakening fetal hemoglobin: prospects for new therapies for the b-globin disorders. Blood 2012; 120: 2945–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gabbianelli M, Testa U, Morsilli O, Pelosi E, Saulle E, Petrucci E, Castelli G, Giovinazzi S, Mariani G, Fiori ME, Bonanno G, Massa A, Croce CM, Fontana L, Peschle C. Mechanism of human Hb switching: a possible role of the kit receptor/miR 221–222 complex. Haematologica 2010; 95: 1253–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lulli V, Romania P, Morsilli O, Cianciulli P, Gabbianelli M, Testa U, Giuliani A, Marziali G. MicroRNA-486-3p regulates γ-globin expression in human erythroid cells by directly modulating BCL11A. PLoS One 2013; 8: e60436–e60436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alijani S, Alizadeh S, Kazemi A, Khatib ZK, Soleimani M, Rezvani M, Minayi N, Karami F, Tayebi B. Evaluation of the effect of miR-26b up-regulation on HbF expression in erythroleukemic K-562 cell line. Avicenna J Med Biotechnol 2014; 6: 53–6. [PMC free article] [PubMed] [Google Scholar]
- 14.Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol 2010; 11: R90–R90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Craig JE, Barnetson RA, Prior J, Raven JL, Thein SL. Rapid detection of deletions causing delta beta thalassemia and hereditary persistence of fetal hemoglobin by enzymatic amplification. Blood 1994; 83: 1673–82. [PubMed] [Google Scholar]
- 16.Gonçalves MS, Fahel S, Figueiredo MS, Kimura EJ, Nechtman F, Stoming TA, Arruda VR, Saad ST, Costa FF. Molecular identification of hereditary persistence of fetal hemoglobin type 2 (HPFH type 2) in patients from Brazil. Ann Hematol 1995; 70: 159–61. [DOI] [PubMed] [Google Scholar]
- 17.De Andrade TG, Fattori A, Saad ST, Sonati MF, Costa FF. Molecular identification of Sicilian (deltabeta) degrees-thalassemia associated with beta-thalassemia and hemoglobin S in Brazil. Braz J Med Biol Res 2002; 35: 873–6. [DOI] [PubMed] [Google Scholar]
- 18.De Andrade TG, Peterson KR, Cunha AF, Moreira LS, Fattori A, Saad ST, Costa FF. Identification of novel candidate genes for globin regulation in erythroid cells containing large deletions of the human beta-globin gene cluster. Blood Cells Mol Dis 2006; 37: 82–90. [DOI] [PubMed] [Google Scholar]
- 19.Menzel S, Garner C, Gut I, Matsuda F, Yamaguchi M, Heath S, Foglio M, Zelenika D, Boland A, Rooks H, Best S, Spector TD, Farrall M, Lathrop M, Thein SL. A QTL influencing F cell production maps to a gene encoding a zinc-finger protein on chromosome 2p15. Nat Genet 2007; 39: 1197–9.. [DOI] [PubMed] [Google Scholar]
- 20.Uda M, Galanello R, Sanna S, Lettre G, Sankaran VG, Chen W, Usala G, Busonero F, Maschio A, Albai G, Piras MG, Sestu N, Lai S, Dei M, Mulas A, Crisponi L, Naitza S, Asunis I, Deiana M, Nagaraja R, Perseu L, Satta S, Cipollina MD, Sollaino C, Moi P, Hirschhorn JN, Orkin S, Abecasis GR, Schlessinger D, Cao A. Genome-wide association study shows BCL11A associated with persistent fetal hemoglobin and amelioration of the phenotype of beta-thalassemia. Proc Natl Acad Sci USA 2008; 105: 1620–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lettre G, Sankaran VG, Bezerra MA, Araujo AS, Uda M, Sanna S, Cao A, Schlessinger D, Costa FF, Hirschhorn JN, Orkin SH. DNA polymorphisms at the BCL11A, HBS1L-MYB, and beta-globin loci associate with fetal hemoglobin levels and pain crises in sickle cell disease. Proc Natl Acad Sci U S A 2008; 105: 11869–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sankaran VG, Menne TF, Xu J, Akie TE, Lettre G, Van Handel B, Mikkola HK, Hirschhorn JN, Cantor AB, Orkin SH. Human fetal hemoglobin expression is regulated by the developmental stage-specific repressor BCL11A. Science 2008; 322: 1839–42. [DOI] [PubMed] [Google Scholar]
- 23.Sankaran VG, Xu J, Ragoczy T, Ippolito GC, Walkley CR, Maika SD, Fujiwara Y, Ito M, Groudine M, Bender MA, Tucker PW, Orkin SH. Developmental and species-divergent globin switching are driven by BCL11A. Nature 2009; 460: 1093–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu J, Sankaran VG, Ni M, Menne TF, Puram RV, Kim W, Orkin SH. Transcriptional silencing of {gamma}-globin by BCL11A involves long-range interactions and cooperation with SOX6. Genes Dev 2010; 24: 783–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang M, Ye L, Wang B, Gao J, Liu R, Hong J, Wang W, Gu W, Ning G. Decreased miR-146 expression in peripheral blood mononuclear cells is correlated with ongoing islet autoimmunity in type 1 diabetes patients. J Diabetes 2015; 7: 158–65. [DOI] [PubMed] [Google Scholar]
- 26.Zhai PF, Wang F, Su R, Lin HS, Jiang CL, Yang GH, Yu J, Zhang JW. The regulatory roles of microRNA-146b-5p and its target platelet-derived growth factor receptor α (PDGFRA) in erythropoiesis and megakaryocytopoiesis. J Biol Chem 2014; 289: 22600–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujiwara T, Okitsu Y, Katsuoka Y, Fukuhara N, Onishi Y, Ishizawa K, Harigae H. Expression profiling of ETO2-regulated miRNAs in erythroid cells: possible influence on miRNA abundance. FEBS Open Bio 2013; 3: 428–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song J, Yoon D, Christensen RD, Horvathova M, Thiagarajan P, Prchal JT. HIF-mediated increased ROS from reduced mitophagy and decreased catalase causes neocytolysis. J Mol Med (Berl) 2015; 93: 857–66. [DOI] [PubMed] [Google Scholar]
- 29.Mu W, Wang X, Zhang X, Zhu S, Sun D, Ka W, Sung LA, Yao W. Fluid shear stress upregulates E-Tmod41 via miR-23b-3p and contributes to F-actin cytoskeleton remodeling during erythropoiesis. PLoS One 2015; 10: e0136607–e0136607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ivanova SM, Iarlykova Iu V, Labetskaia OI, Karashtin VV, Levina AA, Shishkanova ZG, Tsybul'skaia MM, Kozinets GI. Influence of space flight factors on peripheral red blood cells in humans. Aviakosm Ekolog Med 1998; 32: 35–40. [PubMed] [Google Scholar]
- 31.Silva B, Faustino P. An overview of molecular basis of iron metabolism regulation and the associated pathologies. Biochim Biophys Acta 2015; 1852: 1347–59. [DOI] [PubMed] [Google Scholar]
- 32.Sankaran VG, Orkin SH. The switch from fetal to adult hemoglobin. Cold Spring Harb Perspect Med 2013; 3: a011643–a011643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Basak A, Sankaran VG. Regulation of the fetal hemoglobin silencing factor BCL11A. Ann N Y Acad Sci 2016; 1368: 25–30. [DOI] [PMC free article] [PubMed] [Google Scholar]