ABSTRACT
WRNIP1 interacts with WRN helicase, which is defective in the premature aging disease Werner syndrome. WRNIP1 belongs to the AAA+ ATPase family and is conserved from Escherichia coli to human. The protein contains an ubiquitin-binding zinc finger (UBZ) domain at the N terminus and an ATPase domain in the middle region. In addition to WRN, WRNIP1 interacts with proteins involved in multiple cellular pathways, including RAD18, monoubiquitylated PCNA, DNA polymerase δ, RAD51, and ATMIN. Mgs1, the yeast homolog of WRNIP1, may act downstream of ubiquitylation of PCNA to mobilize DNA polymerase δ. By contrast, the functions of WRNIP1 in higher eukaryotic cells remain obscure, although data regarding the roles of WRNIP1 in DNA transactions have emerged recently. Here, we first describe the functions of Mgs1 in DNA transaction. We then describe various features of WRNIP1 and discuss its possible roles based on recent studies of the function of WRNIP1.
KEYWORDS: ATMIN, DNA polymerase δ, DNA polymerase η, Mgs1, PCNA, RAD18, WRNIP1, WRN, ubiquitin
Introduction
Genome stability is essential for life. However, living cells are constantly exposed to environmental and endogenous DNA-damaging agents that cause the formation of DNA lesions, which block replication. In addition to DNA adducts, other factors such as topological stress, aberrant DNA structure, and depletion of nucleotide pools can induce replication block. Multiple mechanisms have evolved in prokaryotic and eukaryotic cells to remove DNA lesions, allowing cells to cope with replication block.
DNA helicases, enzymes that unwind structured DNA, play essential roles in preserving genome integrity. In particular, the helicases implicated in the DNA damage response play prominent roles in DNA damage sensing, fork maintenance, replication restart, and DNA repair. The RecQ family consists of highly conserved helicases with diverse roles in multiple DNA metabolic processes. One member of the RecQ family, WRN, is mutated in patients with Werner syndrome (WS), a genetic disease characterized by premature aging associated with genome instability and an elevated risk of cancer.1
WRNIP1 was originally identified as Werner helicase-interacting protein (WHIP) in a yeast 2-hybrid screen2 and subsequently renamed Werner helicase-interacting protein 1 according to HUGO nomenclature conventions. WRNIP1 belongs to the AAA+ ATPase family and is conserved from Escherichia coli to human. The homology of WRNIP1 to members of the replication factor C family and its ability to stimulate the DNA-synthesis activity of DNA polymerase δ3 suggest a possible role for WRNIP1 in DNA replication and/or replication-related DNA transactions. The Saccharomyces cerevisiae homolog of WRNIP1, Mgs1 (maintenance of genome stability 1), is an evolutionarily conserved AAA+ ATPase that is crucial for maintenance of genomic stability.4
In this review, we will provide an overview of the properties and functions of Mgs1, and then focus on WRNIP1. Specifically, we will address the protein's structure and interacting partners, and recent advances in understanding the roles of WRNIP1 in DNA replication and maintenance of stalled replication forks.
Function of Mgs1
MGS1 was identified by computational research for S. cerevisiae proteins homologous to RuvB.4 Accordingly, Mgs1 is similar to E. coli RuvB and the eukaryotic clamp loader protein RFC, and all 3 of these proteins share the Walker A/B and sensor I/II motifs characteristic of the AAA+ ATPase family. Mgs1 has DNA-dependent ATPase and single-strand DNA annealing activities. Deletion of MGS1 results in an elevated rate of mitotic recombination, and this phenotype is synergistic with mutation in SGS1, which encodes a RecQ family helicase considered to be the yeast homolog of WRN: the mgs1 sgs1 double mutant exhibits more severe growth defects and genome instability than either single-gene mutant.4,5
Mutation in MGS1 is synthetically lethal with mutation in RAD6, a gene involved in DNA damage tolerance, suggesting that Mgs1 participates in handling stalled replication forks.6 The Rad6-dependent DNA damage tolerance pathway allows stalled replication forks to bypass the causative damage, via either error-prone translesion DNA synthesis (TLS) or an error-free damage avoidance mechanism that may involve template switching to the undamaged sister chromatid. The Rad6–Rad18 complex, which conjugates ubiquitin to PCNA, participates in both branches of the DNA damage tolerance pathway.7 In the damage avoidance mechanism, the Mms2–Ubc13–Rad5 complex ubiquitylates monoubiquitylated PCNA to generate polyubiquitylated PCNA.8
Mgs1 physically associates with PCNA and is thought to suppress the Rad6-mediated DNA damage tolerance pathway in the absence of exogenous DNA damage.9 Like WRNIP1, Mgs1 has a ubiquitin-binding zinc finger (UBZ) domain (Fig. 1) and preferentially interacts with ubiquitylated PCNA, especially polyubiquitylated PCNA, both in vitro and in vivo.10 Overexpression of Mgs1 sensitizes cells to DNA-damaging agents such as MMS and HU, and also suppresses damage-induced mutagenesis.4,11 The increase in DNA damage sensitivity upon overexpression of Mgs1 is dependent on the UBZ domain and the presence of Rad18,10 implying that excess Mgs1 cripples the post-replication repair pathway by interacting with ubiquitylated PCNA. Overexpression of Mgs1 exacerbates the temperature sensitivity and DNA damage sensitivity of DNA polymerase δ mutants, whereas deletion of MGS1 reduces temperature and damage sensitivity.6,11,12 Mgs1 inhibits the interaction of Pol32, a subunit of DNA polymerase δ with ubiquitylated PCNA10; thus Mgs1 acts downstream of PCNA ubiquitylation to mobilize DNA polymerase δ. Because Pol32 is also a subunit of the error-prone DNA polymerase ζ,13 competition between Mgs1 and Pol32 for interaction with ubiquitylated PCNA could explain the suppression of damage-induced mutagenesis by Mgs1 overexpression.
Figure 1.
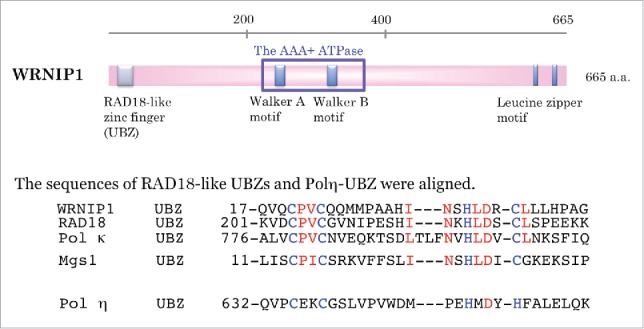
Schematic showing the functional domains of WRNIP1. The sequence of the Rad18-like ubiquitin-binding zinc finger (UBZ) within WRNIP1 is aligned with those of RAD18, Polκ, and Mgs1, and that of polη UBZ. Zinc ion-coordinating residues are denoted by blue letters. RAD18-like CCHC zinc fingers are denoted by red letters.
The synthetic lethality of the mgs1 and rad6 mutations can be rescued by expression of Mgs1 lacking the UBZ domain,10 suggesting that Mgs1 functions in other pathways in which ubiquitylation of PCNA is not required. In the context of DNA replication, Mgs1 may contribute to processing of Okazaki fragments by stimulating Fen1, an endonuclease required for the removal of the flap ends of these fragments.14
PCNA is modified with SUMO as well as ubiquitin; sumoylated PCNA then recruits Srs2 DNA helicase, which displaces Rad51 from the single-stranded DNA-Rad51 complex. Synthetic lethality seems to be rescued by the Rad51-dependent homologous recombination pathway since mutation of SRS2 or genes involved in sumoylation of PCNA suppresses synthetic lethality (Fig. 2A).9 Thus, when the Rad6-dependent DNA damage tolerance and HR pathways are blocked, an alternative pathway involving Mgs1 becomes indispensable for life, even in the absence of exogenous DNA damage.15 Physical association of Mgs1 with non-ubiquitylated PCNA, along with the genetic and physical interaction of Mgs1 with DNA polymerase δ, suggests that the role of Mgs1 in the alternative pathway is related to the regulation of DNA polymerase δ. This issue should be addressed in a future study.
Figure 2.
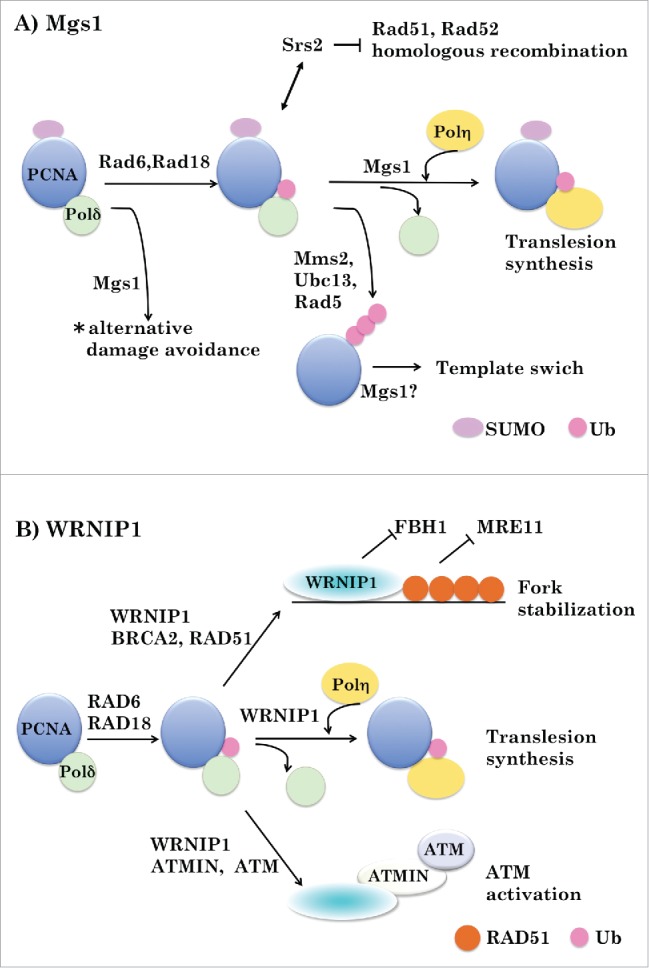
Possible roles of Mgs1 and WRNIP1 in DNA transaction. A. Role of Mgs1. PCNA plays the important role in regulating the DNA damage tolerance pathway via its modification with ubiquitin and SUMO in yeast. SUMOylated PCNA interacts with Srs2 helicase, which prevents homologous recombination by disrupting DNA-Rad51 filaments. The 2 types of ubiquitylated PCNA (monoubiquitylated PCNA and polyubiquitylated PCNA) channels the lesion bypass to translesion synthesis and template switch, respectively. Mgs1 induces polymerase switch from Polδ to Polη during TLS. Mgs1 may also act in the template switch. *This pathway is essential when the RAD6 pathway and homologous recombination are disabled. B. Role of WRNIP1. WRNIP1 stabilizes RAD51 nucleofilaments formed by BRCA2, by counteracting FBH1 and protects replication forks from nucleolytic degradation by MRE11. WRNIP1 induces polymerase switch from Polδ to Polη during TLS. WRNIP1 links ubiquitylated PCNA with ATMIN/ATM to activate ATM signaling in response to replication stress.
The observations described above suggest that Mgs1 acts to mobilize DNA polymerase δ at the replication fork by directly interfering with the PCNA–DNA polymerase δ interaction. Mgs1 may facilitate release of the polymerase by blocking the interaction of the enzyme with ubiquitylated PCNA during the polymerase switch associated with TLS and the events preceding strand exchange in preparation for template switching (Fig. 2A).
The WRNIP1 gene and structure of the WRNIP1 protein
The human WRNIP1 gene, which maps to chromosome 6 p25.2, encodes a protein similar to the replication factor C (RFC) family of clamp loader proteins; however, WRNIP1 does not form a stable complex with RFC proteins.3 Annotated domains in WRNIP1 include a Rad18-like zinc finger domain at the N terminus, an ATPase domain in the middle region, and 2 leucine zipper motifs in the C terminus (Fig. 1). Gel-filtration chromatography and glycerol gradient sedimentation revealed that WRNIP1 exists in a homo-oligomeric complex, most likely an octamer.3
The UBZ domain of WRNIP1
The UBZ domain, identified in members of the Y-family of translesion DNA polymerases (polymerase η and κ), is required for post-replicative DNA repair. UBZ domains, which chelate zinc ions via cysteine or histidine residues, are classified into 2 families: CCHH-type zinc fingers (found in polymerase η) and CCHC-type zinc fingers (found in DNA polymerase κ, WRNIP1, and RAD18) (Fig. 1).16 In addition, UBZ domains can be categorized into at least 2 types, DNA polymerase η and WRNIP1, based on their ubiquitin-binding surfaces.17 WRNIP1 can interact with monoubiquitin, as well as polyubiquitin chains linked via lysine 48 or 63.16,18 Moreover, WRNIP1 itself is modified by ubiquitylation and sumoylation, and the UBZ domain of WRNIP1 is involved in the ubiquitylation reaction.18 Polyubiquitin chains conjugated to WRNIP1 are linked through lysines 11, 48, or 63.19 The physiologic importance of these modifications remains unclear.
The localization of WRNIP1 to nuclear foci upon exposure to DNA-damaging agents such as UVC light is dependent on the UBZ domain. In addition, WRNIP1 accumulates very rapidly at laser-irradiated sites, and this repositioning also requires the UBZ domain.16,20 Mutation of the conserved aspartate in the UBZ domain to alanine (D37A) abolishes WRNIP1 binding to both mono- and polyubiquitin. In addition, WRNIP1 D37A cannot form nuclear foci after UVC irradiation nor accumulate at laser-irradiated sites. Because ubiquitylated PCNA is bound by WRNIP1,21 it may be one of the targets of the WRNIP1 UBZ domain upon UVC or laser light irradiation.
The ATPase domain of WRNIP1
The ATPase domain in the middle region of WRNIP1 contains the Walker A/B and sensor I/II motifs characteristic of AAA+ ATPase family proteins. WRNIP1 has DNA-dependent ATPase activity, preferring template-primer mimic DNA such as poly(dA)/oligo(dT), and binds DNA in an ATP-dependent manner.3,22 In addition, the interaction between WRNIP1 and WRN requires ATP, and mutation of the Walker A motif of WRNIP1 abolishes this interaction.23 However, the ATPase domain of WRNIP1 is dispensable for accumulation of WRNIP1 at sites of DNA damage.20
As in the WRNIP1-WRN interaction, p97 (also known as valosin-containing protein, VCP), another AAA+ ATPase, interacts with WRN in an ATP-dependent manner.24 p97/VCP is a ubiquitin-dependent protein segregase that separates ubiquitylated proteins from tightly bound partner proteins, enabling their selective degradation. For example, p97/VCP facilitates degradation of the largest subunit of RNA polymerase II, Rpb1, at sites of stalled transcription.25
Following DNA damage by UV light, p97/VCP is recruited to blocked replication forks by the adaptor protein DVC1 (also known as Spartan), where it facilitates the switch from replicative to TLS polymerase, as well as the removal of TLS polymerase.26,27 This function is similar to the activities of Mgs1, discussed in the previous section, and WRNIP1, discussed in a later section.
The leucine zipper domain of WRNIP1
WRNIP1 contains 2 leucine zipper motifs in its C-terminal region (leucine zipper domain), which is required for oligomerization of the protein.16 The oligomerization of WRNIP1 is important for its localization to nuclear foci following UV irradiation. Both the N-terminal region containing the UBZ domain and the C-terminal region containing the leucine zipper domain, but not the ATPase domain in the middle region, are required for accumulation of WRNIP1 at laser-irradiated sites.20 These observations indicate that oligomerization of WRNIP1, mediated by the leucine zipper domain, is important for proper subnuclear localization of WRNIP1.
WRNIP1-interacting proteins
WRNIP1 interacts with multiple proteins involved in various cellular pathways; these include FGFR1OP (fibroblast growth factor receptor 1 oncogene partner),28 Nup107,29 p97/VCP,30 RAD18,22 ubiquitylated PCNA,21 DNA polymerase δ,3 DNA polymerase η,31,32 RAD51, BRCA2,33 ATMIN,21 and WRN.2 In this section, we focus on the WRNIP1-interacting proteins involved in DNA replication and/or repair: WRN, RAD18, DNA polymerase δ, and DNA polymerase η.
WRN
WRN is mutated in patients with WS, a rare autosomal recessive disorder characterized by premature aging and early onset of age-related diseases, including arteriosclerosis, melituria, cataracts, and malignant neoplasms. Somatic cells derived from WS patients have reduced lifespans in vitro34 and elevated rates of chromosomal translocations, rearrangements, and deletions.35 WRN, a member of the RecQ DNA helicase family, has 3′-5′ DNA helicase and 3′-5′ exonuclease activities, and is involved in many DNA transactions such as DNA recombination, repair, transcription, and telomere maintenance.
A yeast 2-hybrid screen revealed that WRN interacts with WRNIP1, and that this interaction is mediated by the N-terminal region of WRN.2 Subsequently, we learned that the WRN–WRNIP interaction is dependent on ATP.23
The functional relationship between WRNIP1 and WRN was investigated by generating WRNIP1/WRN double-knockout (wrnip1/wrn) cells in the chicken DT40 cell line. In contrast to yeast mgs1/sgs1 mutants, wrnip1/wrn cells grow at a rate similar to that of wild-type cells; however, the frequency of spontaneously occurring sister chromatid exchange (SCE) is slightly higher in wrnip1/wrn cells than in the wild-type or either single knockout.23 In addition, wrnip1/wrn cells are more sensitive to camptothecin than either single mutant, suggesting that WRNIP1 acts independently of WRN in dealing with some types of DNA lesions.
RAD18
RAD18, which has single-stranded DNA-binding activity, is a ubiquitin ligase that participates in monoubiquitylation with the aid of the ubiquitin-conjugating enzyme RAD6. WRNIP1 and RAD18 interact both in vitro and in living cells.22 In contrast to the WRNIP1–WRN interaction, the WRNIP1–RAD18 interaction is not dependent on ATP. WRNIP1 binds DNA in an ATP-dependent manner, and the affinity of binding is increased by RAD18. By contrast, WRNIP1 inhibits DNA binding by RAD18, suggesting that WRNIP1 preferentially recognizes the RAD18–DNA complex via an interaction with RAD18 and binds the DNA, thereby displacing RAD18.22
The yeast mgs1 and rad18 mutations are synthetically lethal, whereas WRNIP1/RAD18 double-knockout (wrnip1/rad18) DT40 cells are viable but grow slightly slower than wild-type cells. The frequency of spontaneously occurring SCE is higher in wrnip1/rad18 cells than in either single mutant, suggesting that these 2 proteins function in different pathways in the absence of exogenous DNA damage.36 Moreover, the high sensitivity of rad18 cells to camptothecin is partially suppressed by disruption of WRNIP1, suggesting that WRNIP1 acts upstream of RAD18 in the process of dealing with DNA lesions induced by camptothecin.
DNA polymerase δ
DNA polymerase δ a replicative polymerase involved in lagging-strand synthesis, was recently suggested to play a role in leading-strand synthesis as well.37 DNA polymerase δ in higher eukaryotic cells has 4 subunits, of which WRNIP1 interacts with 3: p125, p50, and p12.3 On poly(dA)/oligo(dT) template, purified WRNIP1 stimulates the DNA-synthesis activity of DNA polymerase δ by increasing initiation frequency, but the mechanism underlying this increase remains unclear. One possibility is that WRNIP1 strengthens DNA polymerase δ binding to template primers. Alternatively, given that WRNIP1 (like Mgs1) acts as a mobilizer for DNA polymerase δ, it may promote detachment of DNA polymerase δ from the template after DNA synthesis, making DNA polymerase δ molecules available for a new round of initiation.
DNA polymerase η
DNA polymerase η, encoded by POLH (which is the gene responsible for the xeroderma pigmentosum variant), is a translesion DNA polymerase that can efficiently and accurately bypass UV-induced cyclobutane pyrimidine dimers (CPDs). The interaction between WRNIP1 and DNA polymerase η has been confirmed by proteomics31 and co-immunoprecipitation analyses.32
WRNIP1/POLH double-knockout (wrnip1/polh) DT40 cells grow slightly slower than the wild type or either single knockout, suggesting that the encoded proteins play non-overlapping roles under non-stressed conditions. wrnip1/polh cells exhibit suppression of the phenotypes observed in polh cells upon UV irradiation, including UV sensitivity, delayed repair of CPDs, elevated rates of mutation and SCE, and reduced rate of replication fork progression.32 Therefore, it is likely that WRNIP1 acts upstream of DNA polymerase η after UV irradiation.
Functions of WRNIP1 in DNA transactions
WRNIP1 was first identified 16 y ago, but its functions in the cell remained obscure. Recently, however, data have accumulated regarding the roles of WRNIP1 in DNA transactions. In this section, we discuss the contributions of WRNIP1 to TLS after UV irradiation, as well as to maintenance of replication forks and checkpoint activation following DNA replication stress.
Translesion DNA synthesis (TLS)
TLS enables DNA synthesis on a damaged template via insertion of correct or incorrect bases into the daughter strand, allowing bypass of DNA lesions. This process requires a polymerase switch in which the standard replicative polymerase is replaced by specialized translesion polymerase. In UV-irradiated cells, the RAD6–RAD18 complex is recruited to the irradiated sites, where it monoubiquitylates PCNA. This modification of PCNA triggers the exchange of a replicative polymerase (e.g., DNA polymerase δ) for the translesion polymerase DNA polymerase η. WRNIP1 accumulates at UV-irradiated sites in a UBZ domain-dependent manner,16,20 interacts with RAD18, and displaces it from the DNA.WRNIP1 also interacts directly with DNA polymerase δ to stimulate its DNA-synthesis activity by promoting recycling of DNA polymerase δ on template/primer DNA.3 Furthermore, WRNIP1 interacts with DNA polymerase η, and genetic data suggest that it functions upstream of DNA polymerase η32 Based on these observations, WRNIP1 may stimulate detachment of DNA polymerase δ from ubiquitylated PCNA and recruit DNA polymerase η to sites of DNA damage during TLS (Fig. 2B). The high affinity of WRNIP1 for polyubiquitin and its interaction with RAD51 suggest that WRNIP1 itself is involved in the template switching mechanism. This issue should be addressed in a future study.
Stabilization of replication forks
DNA replication can be stalled for many reasons, including topological stress, DNA lesions, and depletion of nucleotide pools. Proper handling of stalled replication forks is essential for the maintenance of genome stability. BRCA2 is involved in stabilization of stalled replication forks, independent of its double-strand break repair function, by stabilizing RAD51 filaments and blocking degradation of replication forks by MRE11.38,39
WRNIP1 is recruited to stalled replication forks, where it interacts with BRCA2 and RAD51.33 In WRNIP1-depleted cells, MRE11 generates more ssDNA than in control cells under both unperturbed and HU-treated conditions, and the amount of RAD51 on stalled replication forks is considerably lower. The aforementioned phenotypes and chromosome instability in WRNIP1-depleted cells are suppressed by depletion of FBH1, which can disrupt RAD51 filaments. Thus, WRNIP1 is directly involved in preventing MRE11-mediated degradation of stalled replication forks by stabilizing RAD51 on ssDNA, and this function is not dependent on the ATPase activity of WRNIP1. The ATPase activity is essential for recovery of perturbed replication forks, but the contribution of WRNIP1 to this process remains unclear.
Checkpoint activation
The ataxia telangiectasia mutated (ATM) kinase responds to the presence of DNA damage, especially damage induced by ionizing radiation (IR), by activating the cell cycle checkpoint and promoting DNA repair.40,41 However, the molecular mechanism by which ATM is activated by replication stress remains obscure. The contribution of ATMIN (ATM interactor) to ATM activation is complementary to that of NBS1, i.e., it is dispensable for IR-induced ATM activation.42 ATMIN is required for ATM-mediated signaling and recruitment of 53BP1 to DNA damage sites upon replication stress43 Moreover, WRNIP1 binds ATMIN and facilitates its ATM-activating function.21 Upon induction of replication stress by aphidicolin, WRNIP1 interacts with monoubiquitylated PCNA and forms foci, dependent upon ubiquitylation of PCNA by RAD18. Depletion of RAD18, WRNIP1, or ATMIN prevents activation of ATM upon replication stress, but not after IR. Thus, WRNIP1 recognizes monoubiquitylated PCNA and connects with ATMIN/ATM to activate ATM signaling in response to replication stress.
Conclusions and future perspectives
WRNIP1 and its yeast homolog, Mgs1, play important roles following ubiquitylation of PCNA, acting downstream of RAD18 to mobilize DNA polymerase δ. Although the mgs1 and rad18 mutations are synthetically lethal, wrnip1/rad18 double knockouts are viable and grow slightly slower than wild-type cells. Given that activation of the recombination pathway by inhibition of Srs2 function rescues the synthetic lethality of mgs1/rad18 cells, it seems likely that this pathway is active in wrnip1/rad18 cells. In fact, the frequency of spontaneous SCE is considerably higher in wrnip1/rad18 cells than in wild-type cells. Future studies should seek to determine the UBZ domain–independent function of Mgs1 in the alternative pathway, which becomes essential when the RAD6 and HR pathways are blocked, as well as whether WRNIP1 has a similar function. In addition, it would be interesting to elucidate the coordination of the 2 events in which WRNIP1 participates, i.e., checkpoint activation involving ATM and stabilization of replication forks. Recently, we obtained results suggesting that WRNIP1 regulates the function of PrimPol, which is thought to function in TLS and restart DNA synthesis by synthesising primers (to be published elsewhere).
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dr. Shusuke Tada for critical reading of the manuscript and for helpful comments.
Funding
The studies in our laboratory were supported by JSPS KAKENHI Grant Numbers JP26440065, JP23370065, and JP20390020.
References
- [1].Yu CE, Oshima J, Fu YH, Wijsman EM, Hisama F, Alisch R, Matthews S, Nakura J, Miki T, Ouais S, Martin GM, Mulligan J, Schellenberg GD. Positional cloning of the Werner's syndrome gene. Science 1996; 272:258-262; PMID:8602509; http://dx.doi.org/ 10.1126/science.272.5259.258 [DOI] [PubMed] [Google Scholar]
- [2].Kawabe Y, Branzei D, Hayashi T, Suzuki H, Masuko T, Onoda F, Heo SJ, Ikeda H, Shimamoto A, Furuichi Y, Seki M, Enomoto T. A novel protein interacts with the Werner's syndrome gene product physically and functionally. J Biol Chem 2001; 276:20364-20369; PMID:11301316; http://dx.doi.org/ 10.1074/jbc.C100035200 [DOI] [PubMed] [Google Scholar]
- [3].Tsurimoto T, Shinozaki A, Yano M, Seki M, Enomoto T. Human Werner helicase interacting protein 1 (WRNIP1) functions as a novel modulator for DNA polymerase δ. Genes Cells 2005; 10:13-22; PMID:15670210; http://dx.doi.org/ 10.1111/j.1365-2443.2004.00812.x [DOI] [PubMed] [Google Scholar]
- [4].Hishida T, Iwasaki H, Ohno T, Morishita T, Shinagawa H. A yeast gene, MGS1, encoding a DNA-dependent AAA+ ATPase is required to maintain genome stability. Proc Natl Acad Sci USA 2001; 98:8283-8289; PMID:11459965; http://dx.doi.org/ 10.1073/pnas.121009098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Branzei D, Seki M, Onoda F, Yagi H, Kawabe Y, Enomoto T. Characterization of the slow-growth phenotype of S. cerevisiae whip/mgs1 sgs1 double deletion mutants. DNA Repair 2002; 1:671-682; PMID:12509289; http://dx.doi.org/ 10.1016/S1568-7864(02)00073-3 [DOI] [PubMed] [Google Scholar]
- [6].Hishida T, Ohno T, Iwasaki H, Shinagawa H. Saccharomyces cerevisiae MGS1 is essential in strains deficient in the RAD6-dependent DNA damage tolerance pathway. EMBO J 2002; 21:2019-2029; PMID:11953321; http://dx.doi.org/ 10.1093/emboj/21.8.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 2002; 419:135-141; PMID:12226657; http://dx.doi.org/ 10.1038/nature00991 [DOI] [PubMed] [Google Scholar]
- [8].Parker JL, Ulrich HD. Mechanistic analysis of PCNA poly-ubiquilyation by the ubiquitin protein ligases Rad18 and Rad5. EMBO J 2009; 28:3657-3666; PMID:19851286; http://dx.doi.org/ 10.1038/emboj.2009.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hishida T, Ohya T, Kubota Y, Kamada Y, Shinagawa H. Functional and physical interaction of yeast Mgs1 with PCNA: impact on RAD6-dependent DNA damage tolerance. Mol Cell Biol 2006; 26:5509-5517; PMID:16809783; http://dx.doi.org/ 10.1128/MCB.00307-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Saugar I, Parker JL, Zhao S, Ulrich HD. The genome maintenance facror Mgs1 is targeted to sites of replication stress by ubiquitylated PCNA. Nucleic Acids Res 2012; 40:245-257; PMID:21911365; http://dx.doi.org/ 10.1093/nar/gkr738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Branzei D, Seki M, Onoda F, Enomoto T. The product of Saccharomyces cerevisiae WHIP/MGS1, a gene related to replication factor C genes, interacts functionally with DNA polymerase δ. Mol Genet Genomics 2002; 268:371-386; PMID:12436259; http://dx.doi.org/ 10.1007/s00438-002-0757-3 [DOI] [PubMed] [Google Scholar]
- [12].Vijeh Motlagh ND, Seki M, Branzei D, Enomoto T. Mgs1 and Rad18/Rad5/Mms2 are required for survival of Saccharomyces cerevisiae mutants with novel temperature/cold sensitive alleles of the DNA polymerase δ subunit, Pol31. DNA Repair 2006; 5:1459-1474; PMID:16949354; http://dx.doi.org/ 10.1016/j.dnarep.2006.07.006 [DOI] [PubMed] [Google Scholar]
- [13].Siebler HM, Lada AG, Baranovskiy AG, Tahirov TH, Pavlov YI. A novel variant of DNA polymerase ζ, Rev3ΔC, highlights differnetial regulation of Pol32 as a subunit of polymerase δ versus ζ in Saccharomyces cerevisiae. DNA Repair 2014; 24:138-149; PMID:24819597; http://dx.doi.org/ 10.1016/j.dnarep.2014.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kim JH, Kang YH, Kang HJ, Kim DH, Ryu GH, Kang MJ, Seo YS. In vivo and in vitro sutudies of Mgs1 suggest a link between genome instability and Okazaki fragment processing. Nucleic Acids Res 2005; 33:6137-6150; http://dx.doi.org/ 10.1093/nar/gki900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Barbour L, Xiao W. Regulation of altanative replication bypass pathways at stalled replication forks and its effects on genome stability : a yeast model. Mutat Res 2003; 532:137-155; PMID:14643434; http://dx.doi.org/ 10.1016/j.mrfmmm.2003.08.014 [DOI] [PubMed] [Google Scholar]
- [16].Crosetto N, Bienko M, Hibbert RG, Perica T, Ambrogio C, Kensche T, Hofmann K, Sixma TK, Dikic I. Human Wrnip1 is localized in replication factories in a ubiquitin-binding zinc figer-dependent manner. J Biol Chem 2008; 283:35173-35185; PMID:18842586; http://dx.doi.org/ 10.1074/jbc.M803219200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Suzuki N, Rohaim A, Kato R, Dikic I, Wakatsuki S, Kawasaki M. A novel mode of ubiquitin recognition by ubiquitin-binding zinc finger domain of WRNIP1. FEBS J 2016; 283:2004-2017; PMID:27062441; http://dx.doi.org/ 10.1111/febs.13734 [DOI] [PubMed] [Google Scholar]
- [18].Bish R, Myers MP. Werner helicase interacting protein1 binds polyubiquitin via its zinc finger domain. J Biol Chem 2007; 282:23184-23193; PMID:17550899; http://dx.doi.org/ 10.1074/jbc.M701042200 [DOI] [PubMed] [Google Scholar]
- [19].Bish RA, Fregoso OI, Piccini A, Myers MP. Conjugation of complex polyubiquitin chains to WRNIP1. J Proteome Res 2008; 7:3481-3489; PMID:18613717; http://dx.doi.org/ 10.1021/pr800217q [DOI] [PubMed] [Google Scholar]
- [20].Nomura H, Yashimura A, Edo T, Kanno S, Tada S, Seki M, Yasui A, Enomoto T. WRNIP1 accumulates at laser light irradiated sites rapidly via its ubiquitin-binding zinc finger domain and independently from its ATPase domain. Biochem Biophys Res Commun 2012; 417:1145-1150; PMID:22209848; http://dx.doi.org/ 10.1016/j.bbrc.2011.12.080 [DOI] [PubMed] [Google Scholar]
- [21].Kanu N, Zhang T, Burrell RA, Chakraborty A, Cronshaw J, DaCosta C, Grönroos E, Pemberton HN, Anderton E, Gonzalez L, Sabbioneda S, Ulrich HD, Swanton C, Behrens A. RAD18, WRNIP1 and ATMIN promore ATM signalling in response to replication stress. Oncogene 2016; 35:4009-4019; PMID:26549024; http://dx.doi.org/ 10.1038/onc.2015.427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Yoshimura A, Seki M, Kanamori M, Tateishi S, Tsurimoto T, Tada S, Enomoto T. Physical and functional interaction between WRNIP1 and RAD18. Genes Genet Syst 2009; 84:171-178; PMID:19556710; http://dx.doi.org/ 10.1266/ggs.84.171 [DOI] [PubMed] [Google Scholar]
- [23].Kawabe Y, Seki M, Yoshimura A, Nishino K, Hayashi T, Takeuchi T, Iguch S, Kusa Y, Ohtsuki M, Tsuyama T, Imamura O, Matsumoto T, Furuichi Y, Tada S, Enomoto T. Analyses of the interaction of WRNIP1 with Werner syndrome protein (WRN) in vitro and in the cell. DNA Repair 2006; 5:816-828; PMID:16769258; http://dx.doi.org/ 10.1016/j.dnarep.2006.04.006 [DOI] [PubMed] [Google Scholar]
- [24].Indig FE, Partridge JJ, von Kobbe C, Aladjem MI, Latterrich M, Bohr VA. Werner syndrome protein directly binds to the AAA ATPase p97/VCP in an ATP-dependent fashion. J Struct Biol 2004; 146:251-259; PMID:15037256; http://dx.doi.org/ 10.1016/j.jsb.2003.11.009 [DOI] [PubMed] [Google Scholar]
- [25].Verma R, Oania R, Fang R, Smith GT, Deshaies RJ. Cdc48/p97 mediates UV-dependent turnover of RNA pol II. Mol Cell 2011; 41:82-92; PMID:21211725; http://dx.doi.org/ 10.1016/j.molcel.2010.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ghosal G, Leung JW, Nair BC, Fong KW, Chen J. Proliferating cell nuclear antigen (PCNA)-binding protein C1orf124 is a regulator of transletion synthesis. J Biol Chem 2012; 287:34225-34233; PMID:22902628; http://dx.doi.org/ 10.1074/jbc.M112.400135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Mosbech A, Gibbs-Seymour I, Kagias K, Thorslund T, Beli P, Povlsen L, Nielsen S V, Smedegaard S, Sedgwick G, Lukas C, Hartmann-Petersen R, Lukas J, Choudhary C, Pocock R, Bekker-Jensen S, Mailand N. DVC1(C1orf124) is a DNA damage-targeting p97 adaptor that promotes ubiquitin-dependent responses to replication blocks. Nat Struct Mol Biol 2012; 19:1084-1092; PMID:23042605; http://dx.doi.org/ 10.1038/nsmb.2395 [DOI] [PubMed] [Google Scholar]
- [28].Mano Y, Takahashi K, Ishikawa N, Takano A, Yasui W, Inai K, Nishimura H, Tsuchiya E, Nakamura Y, Daigo Y. Fibroblast growth factor receptor 1 oncogene partner as a novel prognostic biomarker and therapeutic target for lung cancer. Cancer Sci 2007; 98:1902-1913; PMID:17888034; http://dx.doi.org/ 10.1111/j.1349-7006.2007.00610.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kaur S, White TE, DiGuilio AL, Glavy JS. The discovery of a Werner helicase interacting protein (WHIP) association with the nuclear pore complex. Cell Cycle 2010; 9:3106-3111; PMID:20676042; http://dx.doi.org/ 10.4161/cc.9.15.12524 [DOI] [PubMed] [Google Scholar]
- [30].Yu CC, Yang JC, Chang YC, Chuang JG, Lin CW, Wu MS, Chow LP. VCP phosphorylation-dependent interaction partners prevent apoptosis in Helicobacter pylori-infected gastric epithelial cells. PLoS One 2013; 8:e55724; PMID:23383273; http://dx.doi.org/ 10.1371/journal.pone.0055724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Yuasa MS, Masutani C, Hirano A, Cohn MA, Yamaizumi M, Nakatani Y, Hanaoka F. A human DNA polymerase η complex containing Rad18, Rad6 and Rev1; proteomic analysis and targeting of the complex to the chromatin-bound fraction of cells undergoing replication fork arrest. Genes Cells 2006; 11:731-744; PMID:16824193; http://dx.doi.org/ 10.1111/j.1365-2443.2006.00974.x [DOI] [PubMed] [Google Scholar]
- [32].Yoshimura A, Kobayashi Y, Tada S, Seki M, Enomoto T. WRNIP1 functions upstream of DNA polymerase η in the UV-induced DNA damage response. Biochem Biophys Res Commun 2014; 452:48-52; PMID:25139235; http://dx.doi.org/ 10.1016/j.bbrc.2014.08.043 [DOI] [PubMed] [Google Scholar]
- [33].Leuzzi G, Marabitti V, Pichierri P, Franchitto A. WRNIP1 protects stalled forks from degradation and promotes fork restart after replication stress. EMBO J 2016; 35:1437-1451; PMID:27242363; http://dx.doi.org/ 10.15252/embj.201593265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Martin GM. Cellular aging-clonal senescence. A review (Part I). Am J Pathol 1977; 89:484-512. [PMC free article] [PubMed] [Google Scholar]
- [35].Fukuchi K, Martin GM, Monnat RJ Jr.. Mutator phenotype of Werner syndrome is characterized by extensive deletions. Proc Natl Acad Sci USA 1989; 86:5893-5897; PMID:2762303; http://dx.doi.org/ 10.1073/pnas.86.15.5893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Yoshimura A, Seki M, Hayashi T, Kusa Y, Tada S, Ishii Y, Enomoto T. Functional Relationships between Rad18 and WRNIP1 in vertebrate cells. Biol Pharm Bull 2006; 29:2192-2196; PMID:17077513; http://dx.doi.org/ 10.1248/bpb.29.2192 [DOI] [PubMed] [Google Scholar]
- [37].Johnson RE, Klassen R, Prakash L, Prakash S. A major role of DNApolymerase δ in replication of both the leading and lagging DNA strands. Mol Cell 2015; 59:163-175; PMID:26145172; http://dx.doi.org/ 10.1016/j.molcel.2015.05.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell 2011; 145:529-542; PMID:21565612; http://dx.doi.org/ 10.1016/j.cell.2011.03.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Costanzo V. Brca2, Rad51 and Mre11: Performing balancing acts on replication forks. DNA Repair 2011; 10:1060-1065; PMID:21900052; http://dx.doi.org/ 10.1016/j.dnarep.2011.07.009 [DOI] [PubMed] [Google Scholar]
- [40].Derheimer FA, Kastan MB. Multiple roles of ATM in monitoring and maintaining DNA integrity. FEBS Lett 2010; 584:3675-3681; PMID:20580718; http://dx.doi.org/ 10.1016/j.febslet.2010.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Lee JH, Paull TT. Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene 2007; 26:7741-7748; PMID:18066086; http://dx.doi.org/ 10.1038/sj.onc.1210872 [DOI] [PubMed] [Google Scholar]
- [42].Kanu N, Behrens A. ATMIN defines an NBS-independent pathway of ATM signalling. EMBO J 2007; 26:2933-2941; PMID:17525732; http://dx.doi.org/ 10.1038/sj.emboj.7601733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Schmidt L, Wiedner M, Velimezi G, Prochazkova J, Owusu M, Bauer S, Loizou JI. ATMIN is required for the ATM-mediated signaling and recruitment of 53BP1 to DNA damage sites upon replication stress. DNA Repair 2014; 24:122-130; PMID:25262557; http://dx.doi.org/ 10.1016/j.dnarep.2014.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]


