ABSTRACT
Although IGF1 is important for the proliferation and differentiation of chondrocytes, its underlying molecular mechanism is still unknown. Here we addressed the physiologic function of IGF1 in antler cartilage and explored the interplay of IGF1, IRS1/2 and RUNX1 in chondrocyte differentiation. The results showed that IGF1 was highly expressed in antler chondrocytes. Exogenous rIGF1 could increase the proliferation of chondrocytes and cell proportion in the S phase, whereas IGF1R inhibitor PQ401 abrogated the induction by rIGF1. Simultaneously, IGF1 could stimulate the expression of IHH which was a well-known marker for prehypertrophic chondrocytes. Further analysis evidenced that IGF1 regulated the expression of IRS1/2 whose silencing resulted in a rise of IHH mRNA levels, but the regulation was impeded by PQ401. Knockdown of IRS1 or IRS2 with specific siRNA could greatly enhance rIGF1-induced chondrocyte differentiation and reduce the expression of RUNX1. Extraneous rRUNX1 might rescue the effects of IRS1 or IRS2 siRNA on the differentiation. In antler chondrocytes, IGF1 played a role in modulating the expression of RUNX1 through IGF1R. Moreover, attenuation of RUNX1 expression advanced the differentiation elicited by rIGF1, while administration of rRUNX1 to chondrocytes treated with IGF1 siRNA or PQ401 reduced their differentiation. Additionally, siRNA-mediated downregulation of IRS1 or IRS2 in the chondrocytes impaired the interaction between IGF1 and RUNX1. Collectively, IGF1 could promote the proliferation and differentiation of antler chondrocytes. Furthermore, IRS1/2 might act downstream of IGF1 to regulate chondrocyte differentiation through targeting RUNX1.
KEYWORDS: antler, chondrocyte, differentiation, IGF1, IRS1/2, RUNX1
Introduction
Insulin-like growth factor 1 (IGF1) is an important growth factor in the regulation of cell proliferation and differentiation.1-3 Previous studies have found that IGF1 was highly expressed in deer antler which was the only bony organ capable of complete re-growth after shedding in mammals.4,5 Antler growth involves the fast proliferation of chondrocytes without becoming cancerous and differentiation which is a crucial step in the process of endochondral bone formation, and thus provides a valuable model for investigating the mechanisms implicated in cartilage development, endochondral ossification and rapid tissue growth.5,6 Although there is evidence that IGF1 expression was visualized in antler cartilage,4 its effects on the proliferation and differentiation of antler chondrocytes were still unknown.
Insulin receptor substrate 1 (IRS1) and IRS2 are cytoplasmic adaptor molecules that function as signaling intermediates downstream of activated cell surface receptors.7,8 Accumulating evidence has demonstrated that IRS1 and/or IRS2 might modulate the differentiation of murine embryonic stem (mES) cells, skin keratinocytes, adipocytes, myoblasts, osteoblasts, hemopoietic cells, etc.9-15 In epiphyseal cartilage, IRS1 protein was localized in the chondrocytes.16 But it is unclear whether IRS1 and IRS2 may regulate the differentiation of chondrocytes in response to IGF1.
Runt-related transcription factor 1 (RUNX1) is a member of the Runt family of transcription factors and plays an important role in skeletal development and cellular differentiation.17 In the mesenchymal cell specific RUNX1-deficient mice, differentiation of mesenchymal cells into chondrocytes was impaired.18 Simultaneously, there is a considerable amount of evidence to support functional roles of RUNX1 in osteoclast and chondrocyte differentiation.19-22 However, the function of RUNX1 in antler chondrocyte differentiation has never been addressed. Furthermore, whether RUNX1 may mediate the effects of IRS1/2 on chondrocyte differentiation responsiveness to IGF1 remains to be clarified.
In this study, we found that IGF1 could promote the proliferation and differentiation of antler chondrocytes. Further evidence showed that IRS1/2 might act downstream of IGF1 to regulate chondrocyte differentiation through targeting RUNX1.
Results
IGF1 mRNA expression in antler cartilage
In situ hybridization was used to localize the distribution of IGF1 mRNA in antler cartilage. The results showed that a high level of IGF1 mRNA signal was observed in the chondrocytes (Fig. 1A). However, once the DIG-labeled IGF1 antisense probe was replaced with DIG-labeled IGF1 sense probe, there was no corresponding signal in antler chondrocytes (Fig. 1A).
Figure 1.
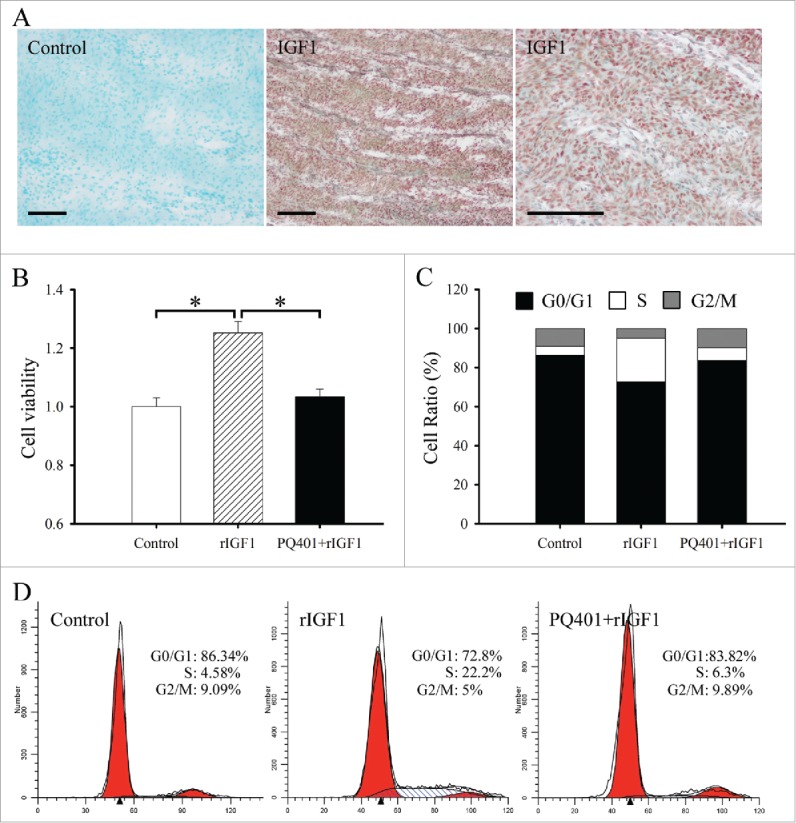
Effects of IGF1 on the proliferation and cell cycle of antler chondrocytes. (A) In situ hybridization of IGF1 expression in the cartilage of sika deer antler. Bar = 60μm. (B) Effects of IGF1 on the proliferation of antler chondrocytes. After antler chondrocytes were treated with rIGF1, or both rIGF1 and PQ401 for 24 h, MTS assay was performed. Data are shown mean ± SEM. *P < 0.05. (C and D), Effects of IGF1 on the cell cycle of antler chondrocytes. After antler chondrocytes were treated with rIGF1, or both rIGF1 and PQ401 for 24 h, flow cytometry was performed.
Effects of IGF1 on the proliferation and cell cycle of antler chondrocytes
MTS assay was performed to determine the effects of IGF1 on proliferation of antler chondrocytes. After treatment with recombinant human IGF1 protein (rIGF1), proliferation activity of chondrocytes was obviously enhanced compared with control (Fig. 1B). However, the enhancement was abrogated by a pretreatment with IGF1 receptor (IGF1R) inhibitor PQ401 (Fig. 1B).
To analyze the effects of IGF1 on cell cycle, flow cytometry was preformed. In antler chondrocytes, rIGF1 treatment led to a substantial accumulation of cells in the S phase with the simultaneous reduction in the proportion of cells in the G0/G1 and G2/M phases, whereas PQ401 pretreatment could reverse the effects of rIGF on cell cycle (Fig. 1C and D).
Effects of IGF1 on the differentiation of antler chondrocytes
To explore the role of IGF1 in the differentiation of antler chondrocytes, we analyzed its effects on the expression of Indian hedgehog (IHH) which was a well-known marker for prehypertrophic chondrocytes.23,24 The results showed that administration of rIGF1 to antler chondrocytes resulted in an obvious increase of IHH mRNA levels, which reached the peak at 24 h, whereas PQ401 pretreatment could dramatically attenuate the rIGF1-induced expression of IHH (Fig. 2A and B).
Figure 2.
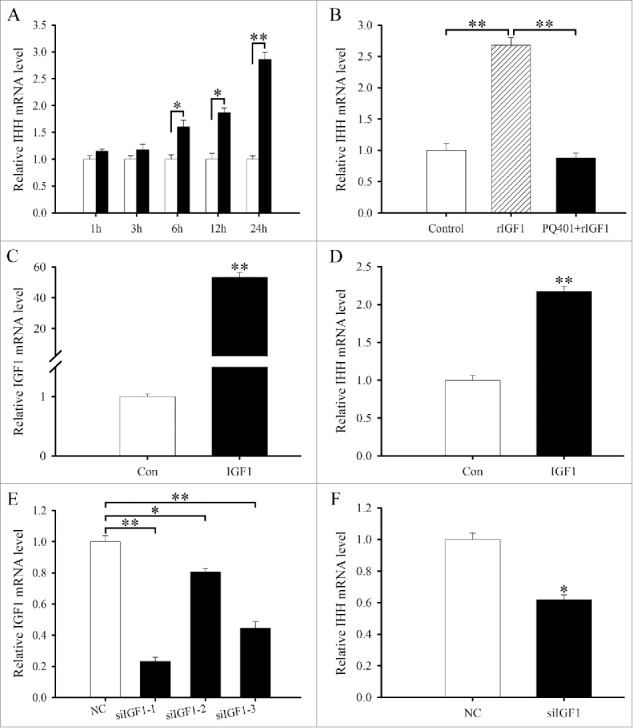
Effects of IGF1 on the differentiation of antler chondrocytes. (A) IHH expression in antler chondrocytes treated with rIGF1 for 1, 3, 6, 12 and 24 h. (B) IHH expression after antler chondrocytes were treated with rIGF1, or both rIGF1 and PQ401. (C) IGF1 expression after antler chondrocytes were transfected with IGF1 overexpression plasmid. (D) Effects of IGF1 overexpression on IHH expression. (E) IGF1 expression after antler chondrocytes were transfected with control siRNA, IGF1 siRNA 1, siRNA 2 or siRNA 3. (F) Effects of IGF1 siRNA on IHH expression. Con, empty pcDNA3.1 vector; IGF1, IGF1 overexpression plasmid; NC, negative control; siIGF1, IGF1 siRNA. *P < 0.05; **P < 0.01.
To further unveil the role of IGF1 in chondrocyte differentiation, we constructed the overexpression plasmid, transfected antler chondrocytes and examined the expression of IGF1 gene by real-time PCR. The result manifested that IGF1 expression was significantly elevated compared with control (Fig. 2C), indicating that IGF1 overexpression plasmid was available. Constitutive expression of IGF1 could stimulate the expression of IHH in antler chondrocytes (Fig. 2D). We next examined the effects of IGF1 siRNA on the differentiation of antler chondrocytes. After 3 different IGF1 siRNA duplexes were introduced into the chondrocytes, IGF1 siRNA 1 was the most effective one among them although IGF1 siRNA 1, 2 and 3 could noticeably down-regulate the expression of IGF1 compared with scrambled siRNA (Fig. 2E). Therefore, IGF1 siRNA 1 was selected to knock down IGF1 gene in the following study. Knockdown of IGF1 with specific siRNA could decrease the expression of IHH in antler chondrocytes (Fig. 2F).
IRS1/2 mediate the effects of IGF1 on the differentiation of antler chondrocytes
IRS1 and IRS2 are essential for intracellular signals and play an important role in cell differentiation.9-15 In antler cartilage, IRS1 and IRS2 were highly expressed in the chondrocytes (Fig. 3A–D). Treatment with siRNA targeted to IRS1 or IRS2, which efficiently suppressed the corresponding mRNA level, led to a remarkable increase in the expression of IHH (Fig. 4A and B).
Figure 3.
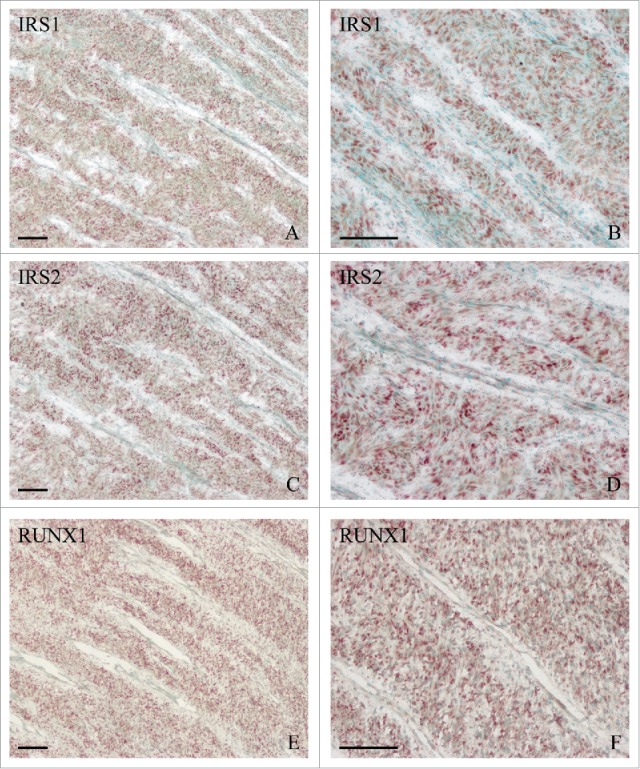
In situ hybridization of IRS1 (A, B), IRS2 (C, D) and RUNX1 (E, F) expression in the cartilage of sika deer antler. Bar = 60 μm.
Figure 4.
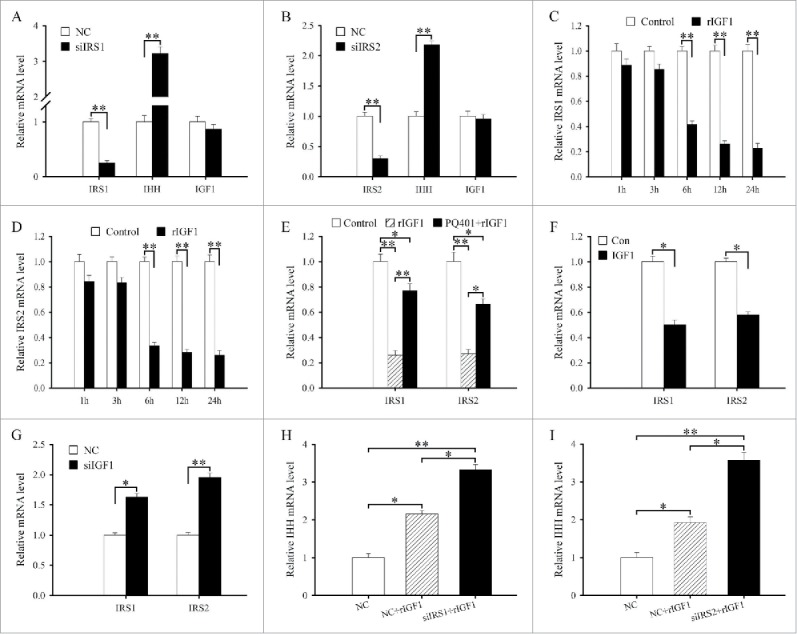
IRS1/2 mediate the effects of IGF1 on the differentiation of antler chondrocytes. (A) Effects of IRS1 siRNA on the expression of IRS1, IHH and IGF1. (B) Effects of IRS2 siRNA on the expression of IRS2, IHH and IGF1. (C) IRS1 expression in antler chondrocytes treated with rIGF1 for 1, 3, 6, 12 and 24 h. (D) IRS2 expression in antler chondrocytes treated with rIGF1 for 1, 3, 6, 12 and 24 h. (E) Expression of IRS1 and IRS2 after antler chondrocytes were treated with rIGF1, or rIGF1 and PQ401. (F) Effects of IGF1 overexpression on the expression of IRS1 and IRS2. (G) Effects of IGF1 siRNA on the expression of IRS1 and IRS2. (H) IRS1 mediated the effects of IGF1 on the expression of IHH. After transfection with IRS1 siRNA and addition of rIGF1, the expression of IHH was determined by real-time PCR. (I) IRS2 mediated the effects of IGF1 on the expression of IHH. siIRS1, IRS1 siRNA; siIRS2, IRS2 siRNA.
To elucidate the relationship between IGF1 and IRS1/2, we first examined the regulation of IGF1 on IRS1 and IRS2. The results evidenced that a dramatic reduction in the levels of IRS1 or IRS2 mRNA was observed at 6, 12 and 24 h after rIGF1 treatment (Fig. 4C and D). But PQ401 could reverse the downregulation of IRS1 and IRS2 expression elicited by rIGF1 (Fig. 4E). Overexpression of IGF1 could reduce the expression of IRS1 and IRS2 in antler chondrocytes, while silencing of IGF1 with specific siRNA augmented their expression (Fig. 4F and G). Meanwhile, IRS1 or IRS2 siRNA did not affect the expression of IGF1 in antler chondrocytes (Fig. 4A and B).
To ascertain whether IRS1 and IRS2 could mediate the effects of IGF1 on the differentiation of antler chondrocytes, we transfected the chondrocytes with IRS1 or IRS2 siRNA along with rIGF1 addition and then analyzed the expression of IHH. As expected, siRNA-mediated attenuation of IRS1 or IRS2 expression in the chondrocytes could further raise the induction of rIGF1 on IHH expression (Fig. 4H and I).
RUNX1 mediates the effects of IRS1/2 on the differentiation of antler chondrocytes
RUNX1 is important for skeletal development and chondrocyte differentiation.17-20 In antler cartilage, RUNX1 mRNA was localized in the chondrocytes (Fig. 3E and F). Addition of recombinant human RUNX1 protein (rRUNX1, 25 ng/ml) to chondrocytes led to a time-dependent decline in the expression of IHH (Fig. 5A). On the contrary, chondrocytes transfected with RUNX1 siRNA exhibited a reduction in RUNX1 expression and an increase in IHH expression compared with cells transfected with control siRNA (Fig. 5B).
Figure 5.
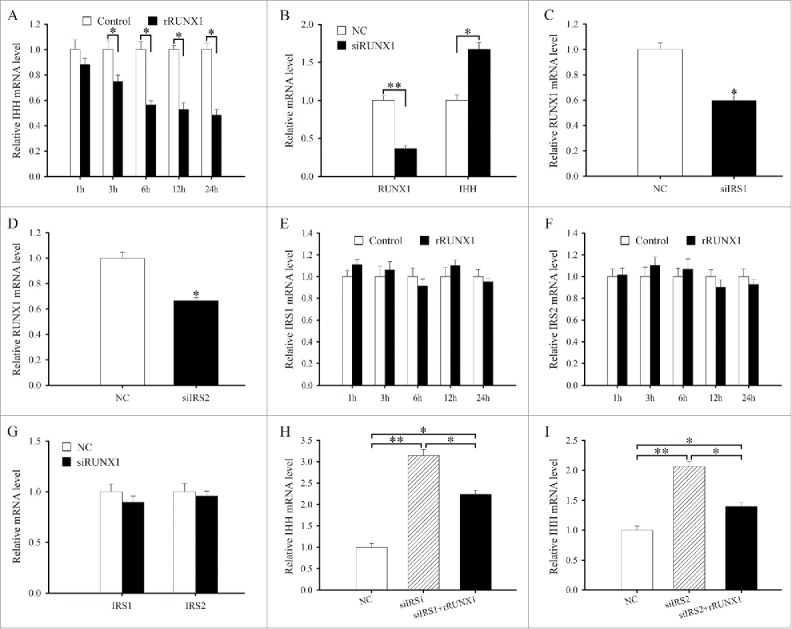
RUNX1 mediates the effects of IRS1/2 on the differentiation of antler chondrocytes. (A) IHH expression in antler chondrocytes treated with rRUNX1 for 1, 3, 6, 12 and 24 h. (B) Effects of RUNX1 siRNA on the expression of RUNX1 and IHH. (C) Effects of IRS1 siRNA on the expression of RUNX1. (D) Effects of IRS2 siRNA on the expression of RUNX1. (E) IRS1 expression in antler chondrocytes treated with rRUNX1 for 1, 3, 6, 12 and 24 h. (F) IRS2 expression in antler chondrocytes treated with rRUNX1 for 1, 3, 6, 12 and 24 h. (G) Effects of RUNX1 siRNA on the expression of IRS1 and IRS2. (H), rRUNX1 rescued the effects of IRS1 siRNA on the expression of IHH. After transfection with IRS1 siRNA and addition of rRUNX1, the expression of IHH was determined by real-time PCR. (I), rRUNX1 rescued the effects of IRS2 siRNA on the expression of IHH. siRUNX1, RUNX1 siRNA.
To dissect the crosstalk between IRS1/2 and RUNX1, we first investigated the effects of IRS1 or IRS2 on RUNX1 expression. Inhibition of IRS1 or IRS2 expression in antler chondrocytes with specific siRNA apparently reduced the expression of RUNX1 (Fig. 5C and D). In the meantime, after antler chondrocytes were treated with rRUNX1 or RUNX1 siRNA, we did not observe any statistically significant difference in the expression levels of IRS1 and IRS2 (Fig. 5E–G).
To assess whether the effects of IRS1 or IRS2 siRNA on chondrocyte differentiation could be rescued by rRUNX1, we treated antler chondrocytes with IRS1 or IRS2 siRNA and then stimulated the cells with rRUNX1 for 24 h. The result found that rRUNX1 could prevent the IRS1 or IRS2 siRNA-induced upregulation of IHH in the chondrocytes (Fig. 5H and I).
RUNX1 mediates the effects of IGF1 on the differentiation of antler chondrocytes
As stated above, the effects of IGF1 on the differentiation of antler chondrocytes were mediated by IRS1 and IRS2 which governed the expression of RUNX1. Based on the observation, we hypothesized that RUNX1 might mediate the role of IGF1 in chondrocyte differentiation. To confirm the hypothesis, we first studied the interaction between IGF1 and RUNX1. In antler chondrocytes, rIGF suppressed the expression of RUNX1 at 12 and 24 h, while PQ401 eliminated its inhibitory effects (Fig. 6A and B). Simultaneously, RUNX1 expression was downregulated after transfection with IGF1 overexpression plasmid and upregulated after treatment with IGF1 siRNA (Fig. 6C and D). However, neither rRUNX1 nor RUNX1 siRNA had any distinct impact on the expression of IGF1 in antler chondrocytes (Fig. 6E and F).
Figure 6.
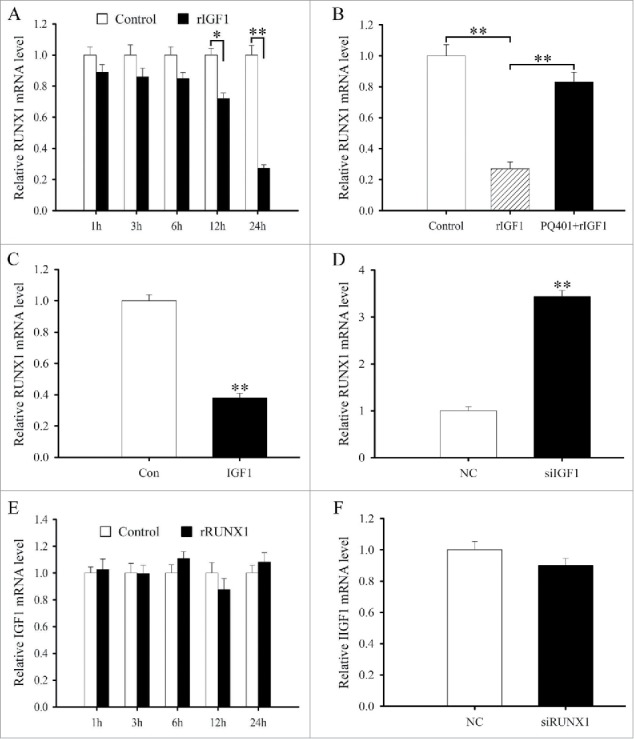
The interaction between IGF1 and RUNX1. (A) RUNX1 expression in antler chondrocytes treated with rIGF1 for 1, 3, 6, 12 and 24 h. (B) Expression of RUNX1 after antler chondrocytes were treated with rIGF1, or rIGF1 and PQ401. (C) Effects of IGF1 overexpression on the expression of RUNX1. (D) Effects of IGF1 siRNA on the expression of RUNX1. (E) IGF1 expression after antler chondrocytes were treated with rRUNX1. (F) Effects of RUNX1 siRNA on the expression of IGF1.
We next sought to determine whether RUNX1 expression might mediate the effectiveness of IGF1 on the differentiation of antler chondrocytes. After transfection with RUNX1 siRNA and then addition of rIGF1, we found that attenuation of RUNX1 expression could noticeably elevate the induction of IHH by rIGF1 (Fig. 7A). Conversely, administration of rRUNX1 to antler chondrocytes transfected with IGF1 siRNA caused a robust decline in the expression of IHH (Fig. 7B). Similarly, rRUNX1 was able to obviously enhance the inhibitory effects of PQ401 on the expression of IHH (Fig. 7C).
Figure 7.
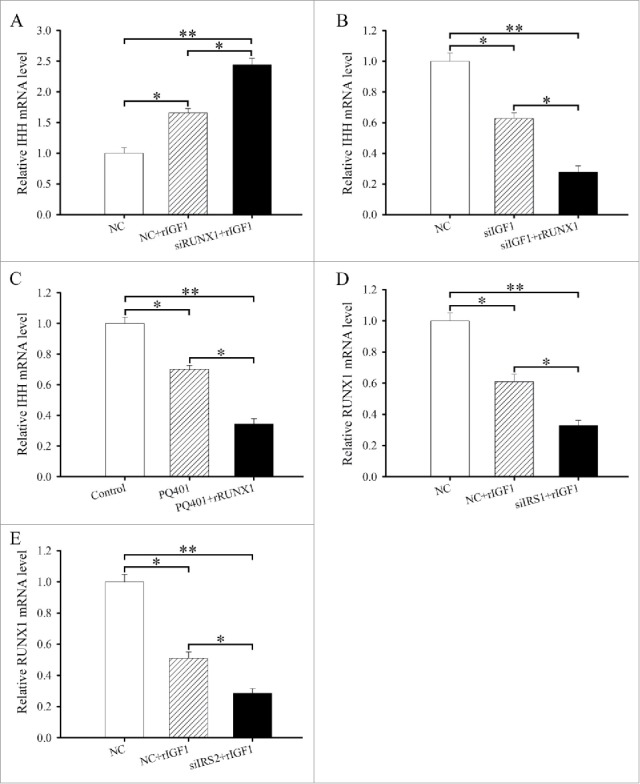
RUNX1 acts downstream of IRS1/2 to mediate the effects of IGF1 on the differentiation of antler chondrocytes. (A) RUNX1 siRNA enhanced the effects of IGF1 on the expression of IHH. After transfection with RUNX1 siRNA and addition of rIGF1, the expression of IHH was determined by real-time PCR. (B) rRUNX1 attenuated the effects of IGF1 siRNA on the expression of IHH. After transfection with IGF1 siRNA and addition of rRUNX1, the expression of IHH was determined by real-time PCR. (C) rRUNX1 attenuated the effects of PQ401 on the expression of IHH. (D) IRS1 mediated the regulation of IGF1 on the expression of RUNX1. (E) IRS2 mediated the regulation of IGF1 on the expression of RUNX1.
IGF1 regulates RUNX1 expression through IRS1/2 in antler chondrocytes
As described above, we have established that IGF1 might regulate the expression of IRS1 and IRS2 which directed RUNX1 expression. We next asked whether IRS1 and IRS2 might mediate the regulation of IGF1 on RUNX1 expression. To address this question, we administrated rIGF1 to antler chondrocytes treated with IRS1 or IRS2 siRNA and tested the expression of RUNX1. The result displayed that siRNA-mediated downregulation of IRS1 expression could evidently attenuate the expression of RUNX1 in the rIGF1-treated chondrocytes (Fig. 7D). Similarly, a drastic reduction of RUNX1 mRNA expression was also noted in the IRS2 siRNA-transfected chondrocytes following rIGF1 addition (Fig. 7E).
Discussion
Although IGF1 regulates the cellular proliferation and differentiation, its physiologic function in antler cartilage and underlying molecular mechanisms remains unresolved. In this study, we found that IGF1 was highly expressed in the chondrocytes and promoted the proliferation of antler chondrocytes. It is well established that the proliferation of animal cells proceeds through various stages of the cell cycle, G1, S, G2 and M phases.25 Administration of rIGF1 to antler chondrocytes led to an increased proportion of cells in the S phase, indicating that IGF1 controls the proliferation of antler chondrocytes by regulating the cell cycle progression at the G1-S phase transition. Previous studies have evidenced that IGF1 exerted its biologic role through binding its cognate cell membrane receptor IGF1R which was activated by autophosphorylation.26 PQ401, which suppressed the autophosphorylation of IGF1R, could abrogate the stimulatory effects of rIGF1 on the proliferation of antler chondrocytes and the accumulation of cells in the S phase, suggesting IGF1R requirement for the IGF1 action on chondrocyte proliferation.
Following this marked proliferation, chondrocytes begin to differentiate into prehypertrophic chondrocytes which are characterized by the elevated expression of IHH, a well-known marker for prehypertrophic chondrocytes.6,23,24 The present results demonstrated that both rIGF1 treatment and constitutive expression of IGF1 could induce the expression of IHH in antler chondrocytes, whereas inhibition of IGF1 with specific siRNA exhibited the opposite effects. PQ401 pretreatment could efficiently block the induction of rIGF1 on the expression of prehypertrophic chondrocyte marker IHH. Together these data reveal that IGF1 can advance the differentiation of antler chondrocytes into prehypertrophic chondrocytes through its receptor IGF1R. Further study found that IGF1 could down-regulate the expression of IRS1 and IRS2 in the chondrocytes, implying that IRS1 and IRS2 play a major negative role in cell differentiation. Indeed, IRS1 and IRS2 could also inhibit the differentiation of antler chondrocytes as indicated by the fact that attenuation of IRS1 or IRS2 obviously enhanced the expression of IHH. Consistent with our findings, the role of IRS1 and IRS2 in differentiation was also shown in the mES cells, myoblasts, osteoblasts and hemopoietic cells.12-15
It has previously reported that IRS1 and IRS2 served as intermediates to coordinate numerous key extracellular signals and mediate their biologic effects.27 In the present study, knockdown of IRS1 or IRS2 accelerated the rIGF1-induced differentiation of antler chondrocytes as evidenced by the increased expression of prehypertrophic chondrocyte marker IHH, suggesting a role of IRS1 and IRS2 in IGF1-mediated cell differentiation. This notion was supported by the finding that overexpression of IRS1 could abolish the IGF1-induced differentiation of hemopoietic cells and hippocampal neuronal cells, whereas constitutive expression of IRS2 prevented the stimulatory effects of IGF1R deficiency on skin keratinocyte differentiation.26,28,29
It has long been recognized that IGF1 binding to its specific receptor on plasma membrane activates the IGF1R intrinsic tyrosine kinase activity and regulates the expression of numerous genes.26,30-32 In antler chondrocytes, IGF1 could restrain the expression of RUNX1 which was important for sternum development, chondrogenesis and chondrocyte differentiation.18-20 The present study has established the importance of RUNX1 in the differentiation of antler chondrocytes. Moreover, knockdown of RUNX1 could enhance the induction in chondrocyte differentiation elicited by rIGF1, while addition of rRUNX1 elevated the repressive effects of IGF1 siRNA or PQ401 on chondrocyte differentiation. Taken together, these observations offer convincing proof that RUNX1 is a critical mediator of IGF1 function in the differentiation of antler chondrocyte into prehypertrophic chondrocytes. Simultaneously, rRUNX1 also rescued the effects of IRS1 or IRS2 siRNA on the expression of IHH, suggesting that RUNX1 is a downstream target of IRS1 and IRS2 in chondrocyte differentiation. Further analysis revealed that IRS1/2 could link and integrate the signaling from upstream activators to multiple downstream effectors to modulate various cellular activities.27 Attenuation of IRS1 or IRS2 expression in antler chondrocytes reinforced the regulation of rIGF1 on RUNX1 expression, implying that IRS1/2 may play an important role in crosstalk between IGF1 and RUNX1.
In summary, IGF1 may play an important role in the proliferation and differentiation of antler chondrocytes. IRS1/2 might act downstream of IGF1 to regulate chondrocyte differentiation through targeting RUNX1 (Fig. 8).
Figure 8.
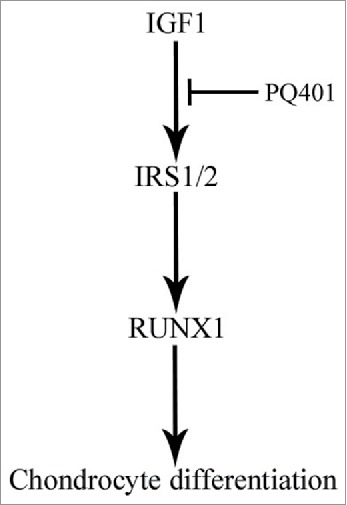
Schematic diagram shows the involvement of IGF1 in the differentiation of antler chondrocytes. IRS1/2 may act downstream of IGF1 to regulate chondrocyte differentiation through targeting RUNX1.
Materials and methods
Tissue collection
Antlers were harvested from anaesthetized sika deer at Changchun City Academy of Agricultural Sciences (Changchun, China). The antler removal procedures were approved by the Institutional Animal Care and Use Committee of Jilin University. Antler tissues were collected from 3-year-old health sika deer as growing for about 60 d after casting of the previous hard antler, and performed as described previously.33 Briefly, the distal 5 cm of growing tip was removed and sectioned sagittally along the longitudinal axis. A part of the tip was then cut into 4–6 mm pieces, flash frozen in liquid nitrogen and stored at −70°C for in situ hybridization, and the remaining tip was used for isolation of antler chondrocytes.
Isolation and culture of antler chondrocytes
The cartilaginous layer was dissected as described previously.33 The separated cartilage tissue was further cut into fine pieces (about 1 × 1 mm). After washing thoroughly with DMEM, the fine pieces were incubated with 0.1% hyaluronidase in D-Hanks for 1h at 38°C. The cartilaginous pieces were then washed twice with Hanks’ balanced salt solution (HBSS) and further digested with 0.1% type II collagenase for 6 h at 38°C with shaking. The digested cartilage tissue was filtered through a nylon mesh (pore size 125μm) and the cell suspension was centrifuged (200 g) for 10 min. Cells were washed twice in HBSS and then the cell pellet was resuspended in culture medium (DMEM medium supplemented with 10% fetal bovine serum, 100U/ml Penicillin and 100 μg/ml Streptomycin) and the viable cells were counted by trypan blue staining using a hemocytometer. 2 × 105 cells were cultured at 38°C with 5% CO2. When the cultured cells grew to 80–90% confluence, the adherent cells were digested with trypsase.
In situ hybridization
Total RNA from the antler cartilage was reverse-transcribed and amplified with the corresponding primers listed in Table 1. The amplified fragment of each gene was cloned into pGEM-T plasmid (pGEM-T Vector System 1, Promega, Madison, WI) and verified by sequencing. These plasmids were amplified with the primers for T7 and SP6 to prepare templates for labeling. Digoxigenin (DIG)-labeled antisense and sense cRNA probes were transcribed in vitro using a DIG RNA labeling kit (Roche Diagnostics GmbH, Mannheim, Germany).
Table 1.
Primers used in this study.
| Gene | Primer sequence | Size (bp) | Application |
|---|---|---|---|
| IGF1 | CTCCTCGCATCTCTTCTACC | 205 | In situ hybridization |
| GCAGCACTCATCCACTATTC | |||
| IRS1 | CAGTGGGGACTACATGCC | 307 | In situ hybridization |
| ACCTGTACAAGACAGGAGTTTG | |||
| IRS2 | GACCTGCGGGTCTACTG | 208 | In situ hybridization |
| GCGAGTAAGTCCGCTTC | |||
| RUNX1 | CCTTGAACCACTCCACTGCC | 268 | In situ hybridization |
| GACGGCAGAGTAG GGAACTG | |||
| IGF1 | CCTCGCATCTCTTCTACCTGG | 191 | Real time PCR |
| CACTATTCCCGTCTGGGGC | |||
| IHH | AGGAGTCGCTGCATTACGAA | 152 | Real time PCR |
| CTTGACGGAGCAATGCACG | |||
| IRS1 | CGGGTGAACCTCAGTCCCAA | 212 | Real time PCR |
| GAGCTGTGGCGTTTGACATC | |||
| IRS2 | CCAACGGGGACTACCTCAAC | 146 | Real time PCR |
| GTGAGGGGCCTTGTAGGAG | |||
| RUNX1 | CTTCACAAACCCACCGCAAG | 132 | Real time PCR |
| TCGGAAAAGGACAAGCTCCC | |||
| Gapdh | GAAGGGTGGCGCCAAGAGGG | 142 | Real time PCR |
| GGGGGCCAAGCAGTTGGTGG |
Frozen sections (10 μm) were mounted on 3-aminopropyltriethoxy-silane (Sigma)-coated slides and fixed in 4% paraformaldehyde solution in phosphate buffered saline (PBS). Hybridization was performed as described previously.34 Sections were counterstained with 1% methyl green. The positive signal was visualized as a dark brown color. The sense probe was also hybridized and served as a negative control. There was no detectable signal from sense probes.
MTS assay
Proliferation assays were performed using MTS reagent (Promega) according to the manufacturer's directions. Antler chondrocytes were seeded in 96-well plates and then treated for 24 h with recombinant human IGF1 protein (rIGF1, 100 nM). For further studies, chondrocytes were pretreated with IGF1 receptor (IGF1R) inhibitor PQ401 (10 μM) for 2 h before the addition of rIGF1. Finally, 20 μl MTS reagent was added to each well and incubated for 3 h. Absorbance was measured at 490 nm using a 96-well plate reader. The rIGF1 and PQ401 were dissolved in PBS and DMSO, respectively. Controls received the vehicle only.
Flow cytometry
Antler chondrocytes were starved in serum-free DMEM medium and treated with rIGF1 for 24 h in the absence or presence of PQ401 after synchronization. At the end of treatment, cells were harvested by trypsinization, centrifuged, washed with PBS and then fixed overnight at 4°C in 70% ethanol. The fixed cells were washed with PBS and stained with 0.5 mL PI/RNase staining buffer (BD Biosciences) for 15 min at room temperature. The stained cells were analyzed by flow cytometry.
Plasmid construction and transfection
Full-length IGF1 cDNA fragment was amplified by PCR from antler cartilage using the following primers: 5′-GAATTC (EcoRI) ATGGGAAAAATCAGCAGTC and 5′-GATATC (EcoRV) GCATGTCATTCTTCACTCTTT. The amplified products were purified and cloned into pGEM-T vector. Both pGEM-T-IGF1 and pcDNA3.1 vector were cut by EcoRI/EcoRV (TaKaRa, Dalian, China) at 37°C for 1 h, and then the fragments were ligated into pcDNA3.1 with T4 ligase (Promega) at 4°C overnight to construct pcDNA3.1-IGF1. An empty pcDNA3.1 expression vector was served as control.
Transfection of antler chondrocytes was performed according to the manufacturer's protocol for lipofectamine 2000 (Invitrogen). After transfection with control plasmid (empty pcDNA3.1 vector) or IGF1 overexpression plasmid, chondrocytes were collected at 24 h.
RNA interference
The small-interfering RNA (siRNA) duplexes for targeting IGF1, IRS1, IRS2 and RUNX1 as well as a scrambled sequence (control siRNA duplex, negative control) were designed and synthesized by GenePharma. The sequences were shown as follows: 5′-GCAGUCUUCCAACCCAAUUTT and 5′-AAUUGGGUUGGAAGACUGCTT (IGF1 siRNA 1); 5′-CCUCCUCGCAUCUCUUCUATT and 5′-UAGAAGAGAUGCGAGGAGGTT (IGF1 siRNA 2); 5′-GGAGUGCAGGAAACAAGAATT and 5′-UUCUUGUUUCCUGCACUCCTT (IGF1 siRNA 3); 5′-CCAGCAAGACCAUCAGCUUTT and 5′-AAGCUGAUGGUCUU GCUGGTT (IRS1 siRNA); 5′-UCGGCUUCGUGAAGCUCAATT and 5′-UUGAGCUUCAC GAAGCCGATT (IRS2 siRNA); 5′-GAACCACUCCACUGCCUUUTT and 5′-AAAGGCAGU GGAGUGGUUCTT (RUNX1 siRNA); 5′-UUCUCCGAACGUGUCACGUTT and 5′-ACGUG ACACGUUCGGAGAATT (nonspecific scrambled siRNA, negative control). Transfections for siRNA were performed according to Lipofectamine 2000 protocol. After transfection with IGF1, IRS1, IRS2 or RUNX1 siRNAs, antler chondrocytes were collected at 24 h.
Real-time PCR
Total RNAs from cultured chondrocytes were isolated using TRIPURE reagent according to the manufacturer's instructions (Roche) and reverse-transcribed into cDNA using MMLV reverse-transcriptase (Promega). Reverse transcriptase was performed at 42°C for 60 min with 2 μg total RNA in 25 μl volume. For real-time PCR, cDNA was amplified using FS Universal SYBR Green Real Master (Roche) on BIO-RAD CFX96™ Real Time Detection System. The conditions used for real-time PCR were as follows: 95°C for 3 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. All reactions were run in triplicate. The result was analyzed using CFX Manager Software. After analysis using the 2−ΔΔCt method, data were normalized to Gapdh expression. Primer sequences for real-time PCR were listed in Table 1.
Statistics
All the experiments were independently repeated at least 3 times. Significance of difference between 2 groups was compared by Independent-Samples T Test. The multiple comparisons were tested with one-way analysis of variance. The differences were considered significant at P < 0.05.
Disclosure of potential conflict of interest
No potential conflicts of interest were disclosed.
Funding
This work was financially supported by National Natural Science Foundation of China (31372390, 31472158 and 31672503) and Special Funds for Scientific Research on Public Causes (201303119).
References
- [1].Wang J, Zhou J, Bondy CA. Igf1 promotes longitudinal bone growth by insulin-like actions augmenting chondrocyte hypertrophy. FASEB J 1999; 13:1985-90; PMID:10544181 [DOI] [PubMed] [Google Scholar]
- [2].Wang Y, Cheng Z, Elalieh HZ, Nakamura E, Nguyen MT, Mackem S, Clemens TL, Bikle DD, Chang W. IGF-1R signaling in chondrocytes modulates growth plate development by interacting with the PTHrP/Ihh pathway. J Bone Miner Res 2011; 26:1437-46; PMID:21312270; http://dx.doi.org/ 10.1002/jbmr.359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Chen Y, Ke J, Long X, Meng Q, Deng M, Fang W, Li J, Cai H, Chen S. Insulin-like growth factor-1 boosts the developing process of condylar hyperplasia by stimulating chondrocytes proliferation. Osteoarthritis Cartilage 2012; 20:279-87; PMID:22281262; http://dx.doi.org/ 10.1016/j.joca.2011.12.013 [DOI] [PubMed] [Google Scholar]
- [4].Gu L, Mo E, Yang Z, Zhu X, Fang Z, Sun B, Wang C, Bao J, Sung C. Expression and localization of insulin-like growth factor-I in four parts of the red deer antler. Growth Factors 2007; 25:264-79; PMID:18092234; http://dx.doi.org/ 10.1080/08977190701773187 [DOI] [PubMed] [Google Scholar]
- [5].Li C, Zhao H, Liu Z, McMahon C. Deer antler–a novel model for studying organ regeneration in mammals. Int J Biochem Cell Biol 2014; 56:111-22; PMID:25046387; http://dx.doi.org/ 10.1016/j.biocel.2014.07.007 [DOI] [PubMed] [Google Scholar]
- [6].Park NR, Lim KE, Han MS, Che X, Park CY, Kim JE, Taniuchi I, Bae SC, Choi JY. Core Binding Factor β Plays a Critical Role During Chondrocyte Differentiation. J Cell Physiol 2016; 231:162-71; PMID:26058470; http://dx.doi.org/ 10.1002/jcp.25068 [DOI] [PubMed] [Google Scholar]
- [7].Mardilovich K, Pankratz SL, Shaw LM. Expression and function of the insulin receptor substrate proteins in cancer. Cell Commun Signal 2009; 7:14; PMID:19534786; http://dx.doi.org/ 10.1186/1478-811X-7-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Pankratz SL, Tan EY, Fine Y, Mercurio AM, Shaw LM. Insulin receptor substrate-2 regulates aerobic glycolysis in mouse mammary tumor cells via glucose transporter 1. J Biol Chem 2009; 284:2031-7; PMID:19056742; http://dx.doi.org/ 10.1074/jbc.M804776200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Fasshauer M, Klein J, Kriauciunas KM, Ueki K, Benito M, Kahn CR. Essential role of insulin receptor substrate 1 in differentiation of brown adipocytes. Mol Cell Biol 2001; 21:319-29; PMID:11113206; http://dx.doi.org/ 10.1128/MCB.21.1.319-329.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Miki H, Yamauchi T, Suzuki R, Komeda K, Tsuchida A, Kubota N, Terauchi Y, Kamon J, Kaburagi Y, Matsui J, Akanuma Y, Nagai R, Kimura S, Tobe K, Kadowaki T. Essential role of insulin receptor substrate 1 (IRS-1) and IRS-2 in adipocyte differentiation. Mol Cell Biol 2001; 21:2521-32; PMID:11259600; http://dx.doi.org/ 10.1128/MCB.21.7.2521-2532.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Sadagurski M, Nofech-Mozes S, Weingarten G, White MF, Kadowaki T, Wertheimer E. Insulin receptor substrate 1 (IRS-1) plays a unique role in normal epidermal physiology. J Cell Physiol 2007; 213:519-27; PMID:17508357; http://dx.doi.org/ 10.1002/jcp.21131 [DOI] [PubMed] [Google Scholar]
- [12].Rubin R, Arzumanyan A, Soliera AR, Ross B, Peruzzi F, Prisco M. Insulin receptor substrate (IRS)-1 regulates murine embryonic stem (mES) cells self-renewal. J Cell Physiol 2007; 213:445-53; PMID:17620314; http://dx.doi.org/ 10.1002/jcp.21185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wu S, Zhou B, Xu L, Sun H. IRS-2, but not IRS-1, can sustain proliferation and rescue UBF stabilization in InR or InR defective signaling of 32D myeloid cells. Cell Cycle 2009; 8:3218-26; PMID:19738441; http://dx.doi.org/ 10.4161/cc.8.19.9759 [DOI] [PubMed] [Google Scholar]
- [14].Hakuno F, Yamauchi Y, Kaneko G, Yoneyama Y, Nakae J, Chida K, Kadowaki T, Yamanouchi K, Nishihara M, Takahashi S. Constitutive expression of insulin receptor substrate (IRS)-1 inhibits myogenic differentiation through nuclear exclusion of Foxo1 in L6 myoblasts. PLoS One 2011; 6:e25655; PMID:21991327; http://dx.doi.org/ 10.1371/journal.pone.0025655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Contaldo C, Myers TJ, Zucchini C, Manara MC, Chiodoni C, Colombo MP, Nicoletti G, Lollini PL, Li T, Longobardi L, Scotlandi K, Spagnoli A. Expression levels of insulin receptor substrate-1 modulate the osteoblastic differentiation of mesenchymal stem cells and osteosarcoma cells. Growth Factors 2014; 32:41-52; PMID:24438070; http://dx.doi.org/ 10.3109/08977194.2013.870168 [DOI] [PubMed] [Google Scholar]
- [16].Hoshi K, Ogata N, Shimoaka T, Terauchi Y, Kadowaki T, Kenmotsu S, Chung UI, Ozawa H, Nakamura K, Kawaguchi H. Deficiency of insulin receptor substrate-1 impairs skeletal growth through early closure of epiphyseal cartilage. J Bone Miner Res 2004; 19:214-23; PMID:14969391; http://dx.doi.org/ 10.1359/JBMR.0301221 [DOI] [PubMed] [Google Scholar]
- [17].Smith N, Dong Y, Lian JB, Pratap J, Kingsley PD, van Wijnen AJ, Stein JL, Schwarz EM, O'Keefe RJ, Stein GS, Drissi MH. Overlapping expression of Runx1(Cbfa2) and Runx2(Cbfa1) transcription factors supports cooperative induction of skeletal development. J Cell Physiol 2005; 203:133-43; PMID:15389629; http://dx.doi.org/ 10.1002/jcp.20210 [DOI] [PubMed] [Google Scholar]
- [18].Kimura A, Inose H, Yano F, Fujita K, Ikeda T, Sato S, Iwasaki M, Jinno T, Ae K, Fukumoto S, Takeuchi Y, Itoh H, Imamura T, Kawaguchi H, Chung UI, Martin JF, Iseki S, Shinomiya K, Takeda S. Runx1 and Runx2 cooperate during sternal morphogenesis. Development 2010; 137:1159-67; PMID:20181744; http://dx.doi.org/ 10.1242/dev.045005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Wang Y, Belflower RM, Dong YF, Schwarz EM, O'Keefe RJ, Drissi H. Runx1/AML1/Cbfa2 mediates onset of mesenchymal cell differentiation toward chondrogenesis. J Bone Miner Res 2005; 20:1624-36; PMID:16059634; http://dx.doi.org/ 10.1359/JBMR.050516 [DOI] [PubMed] [Google Scholar]
- [20].Liakhovitskaia A, Lana-Elola E, Stamateris E, Rice DP, van 't Hof RJ, Medvinsky A. The essential requirement for Runx1 in the development of the sternum. Dev Biol 2010; 340:539-46; PMID:20152828; http://dx.doi.org/ 10.1016/j.ydbio.2010.02.005 [DOI] [PubMed] [Google Scholar]
- [21].Soung do Y, Kalinowski J, Baniwal SK, Jacome-Galarza CE, Frenkel B, Lorenzo J, Drissi H. Runx1-mediated regulation of osteoclast differentiation and function. Mol Endocrinol 2014; 28:546-53; PMID:24606124; http://dx.doi.org/ 10.1210/me.2013-1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Paglia DN, Yang X, Kalinowski J, Jastrzebski S, Drissi H, Lorenzo J. Runx1 Regulates Myeloid Precursor Differentiation Into Osteoclasts Without Affecting Differentiation Into Antigen Presenting or Phagocytic Cells in Both Males and Females. Endocrinology 2016; 157:3058-69; PMID:27267711; http://dx.doi.org/ 10.1210/en.2015-2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Day TF, Guo X, Garrett-Beal L, Yang Y. Wnt/beta-catenin signaling in mesenchymal progenitors controls osteoblast and chondrocyte differentiation during vertebrate skeletogenesis. Dev Cell 2005; 8:739-50; PMID:15866164; http://dx.doi.org/ 10.1016/j.devcel.2005.03.016 [DOI] [PubMed] [Google Scholar]
- [24].Correa D, Hesse E, Seriwatanachai D, Kiviranta R, Saito H, Yamana K, Neff L, Atfi A, Coillard L, Sitara D, Maeda Y, Warming S, Jenkins NA, Copeland NG, Horne WC, Lanske B, Baron R. Zfp521 is a target gene and key effector of parathyroid hormone-related peptide signaling in growth plate chondrocytes. Dev Cell 2010; 19:533-46; PMID:20951345; http://dx.doi.org/ 10.1016/j.devcel.2010.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Li DD, Yang ZQ, Guo CH, Yue L, Duan CC, Cao H, Guo B, Yue ZP. Hmgn1 acts downstream of C/EBPβ to regulate the decidualization of uterine stromal cells in mice. Cell Cycle 2015; 14:3461-74; PMID:26566865; http://dx.doi.org/ 10.1080/15384101.2015.1093704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Sadagurski M, Yakar S, Weingarten G, Holzenberger M, Rhodes CJ, Breitkreutz D, Leroith D, Wertheimer E. Insulin-like growth factor 1 receptor signaling regulates skin development and inhibits skin keratinocyte differentiation. Mol Cell Biol 2006; 26:2675-87; PMID:16537911; http://dx.doi.org/ 10.1128/MCB.26.7.2675-2687.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Dearth RK, Cui X, Kim HJ, Hadsell DL, Lee AV. Oncogenic transformation by the signaling adaptor proteins insulin receptor substrate (IRS)-1 and IRS-2. Cell Cycle 2007; 6:705-13; PMID:17374994; http://dx.doi.org/ 10.4161/cc.6.6.4035 [DOI] [PubMed] [Google Scholar]
- [28].Valentinis B, Romano G, Peruzzi F, Morrione A, Prisco M, Soddu S, Cristofanelli B, Sacchi A, Baserga R. Growth and differentiation signals by the insulin-like growth factor 1 receptor in hemopoietic cells are mediated through different pathways. J Biol Chem 1999; 274:12423-30; PMID:10212216; http://dx.doi.org/ 10.1074/jbc.274.18.12423 [DOI] [PubMed] [Google Scholar]
- [29].Morrione A, Navarro M, Romano G, Dews M, Reiss K, Valentinis B, Belletti B, Baserga R. The role of the insulin receptor substrate-1 in the differentiation of rat hippocampal neuronal cells. Oncogene 2001; 20:4842-52; PMID:11521195; http://dx.doi.org/ 10.1038/sj.onc.1204649 [DOI] [PubMed] [Google Scholar]
- [30].Dupont J, Khan J, Qu BH, Metzler P, Helman L, LeRoith D. Insulin and IGF-1 induce different patterns of gene expression in mouse fibroblast NIH-3T3 cells: identification by cDNA microarray analysis. Endocrinology 2001; 142:4969-75; PMID:11606465; http://dx.doi.org/ 10.1210/endo.142.11.8476 [DOI] [PubMed] [Google Scholar]
- [31].De Bruyne E, Bos TJ, Schuit F, Van Valckenborgh E, Menu E, Thorrez L, Atadja P, Jernberg-Wiklund H, Vanderkerken K. IGF-1 suppresses Bim expression in multiple myeloma via epigenetic and posttranslational mechanisms. Blood 2010; 115:2430-40; PMID:20086250; http://dx.doi.org/ 10.1182/blood-2009-07-232801 [DOI] [PubMed] [Google Scholar]
- [32].Latres E, Amini AR, Amini AA, Griffiths J, Martin FJ, Wei Y, Lin HC, Yancopoulos GD, Glass DJ. Insulin-like growth factor-1 (IGF-1) inversely regulates atrophy-induced genes via the phosphatidylinositol 3-kinase/Akt/mammalian target of rapamycin (PI3K/Akt/mTOR) pathway. J Biol Chem 2005; 280:2737-44; PMID:15550386; http://dx.doi.org/ 10.1074/jbc.M407517200 [DOI] [PubMed] [Google Scholar]
- [33].Wang ST, Gao YJ, Duan CC, Li DD, Tian XC, Zhang QL, Guo B, Yue ZP. Effects of PTHrP on expression of MMP9 and MMP13 in sika deer antler chondrocytes. Cell Biol Int 2013; 37:1300-7; PMID:23956041; http://dx.doi.org/ 10.1002/cbin.10168 [DOI] [PubMed] [Google Scholar]
- [34].Li DD, Zhao SY, Yang ZQ, Duan CC, Guo CH, Zhang HL, Geng S, Yue ZP, Guo B. Hmgn5 functions downstream of Hoxa10 to regulate uterine decidualization in mice. Cell Cycle 2016; 15:2792-805; PMID:27579887; http://dx.doi.org/ 10.1080/15384101.2016.1220459 [DOI] [PMC free article] [PubMed] [Google Scholar]


