Abstract
Increased visceral fat, rather than subcutaneous fat, during the onset of obesity is associated with a higher risk of developing metabolic diseases. The inherent adipogenic properties of human adipose-derived stem cells (ASCs) from visceral depots are compromised compared with those of ASCs from subcutaneous depots, but little is known about the underlying mechanisms. Using ontological analysis of global gene expression studies, we demonstrate that many genes involved in retinoic acid (RA) synthesis or regulated by RA are differentially expressed in human tissues and ASCs from subcutaneous and visceral fat. The endogenous level of RA is higher in visceral ASCs; this is associated with upregulation of the RA synthesis gene through the visceral-specific developmental factor WT1. Excessive RA-mediated activity impedes the adipogenic capability of ASCs at early but not late stages of adipogenesis, which can be reversed by antagonism of RA receptors or knockdown of WT1. Our results reveal the developmental origin of adipocytic properties and the pathophysiological contributions of visceral fat depots.
Introduction
Obesity is defined as excess fat mass in the body and is generally associated with increased risk of developing metabolic diseases, such as cardiovascular diseases and type 2 diabetes (1). At least two main types of white adipose tissue (WAT) are present in human and animals—namely, subcutaneous (SC) fat and visceral (VS) fat. Body fat distribution is increasingly recognized as one of the key factors explaining the metabolic heterogeneity of obesity. Increased visceral adiposity is particularly associated with the risk of developing metabolic complications, whereas increased SC fat presents no or little risk and, rather, is considered to be protective (2–4). These two types of fat differ in their pathophysiological properties, including insulin sensitivity, adipokine secretion, lipolysis, and development of inflammation (5).
Adipose tissue expands not only through increased lipid storage in existing adipocytes (leading to hypertrophy) but also by the differentiation of new adipocytes from progenitor/stem cells (leading to hyperplasia). There are intrinsic differences in the properties of cells from different depots of WAT in vivo and in vitro. It is generally believed that when excess lipids systemically accumulate in the overnutrition state, cells from SC fat mainly undergo hyperplasia, whereas cells from VS fat tend to expand by hypertrophy in vivo (6). Although regulation of adipocyte differentiation has been extensively characterized (7,8), little is known about the molecular basis of regional differences in adipogenic differentiation capacities. Adipose-derived stem cells (ASCs) and adipose progenitor cells from SC and VS depots have intrinsic differences in vitro, such as proliferation and differentiation potentials (9–12). ASCs derived from SC fat differentiate easily into mature adipocytes, whereas VS-derived ASCs differentiate poorly in response to a standard induction cocktail (9). This explains the different expression levels of key adipogenic factors such as peroxisome proliferator–activated receptor (PPAR)-γ and C/EBPα in mature adipocytes and adipose tissue (13,14). As another example of inherent molecular differences, we recently demonstrated that distinct, selective cell surface markers are expressed in SC ASCs versus VS ASCs and reflect their adipogenic properties (15). In addition, previous reports showed that adipose tissue and cells from different depots have distinct patterns of gene expression, especially in the category of developmental genes (e.g., the Hox family), in humans and rodents (16–18). However, how these differences in developmental gene expression lead to functional differences of ASCs is not clear. We hypothesize that intrinsic differences in certain signaling pathways at the progenitor or stem cell level may account for depot-specific differences, with consequences in adipose cell properties and body fat distribution.
In this study, we found WT1-mediated upregulation of the retinoic acid (RA) signaling pathway in ASCs from VS fat, which leads to early, but not late-stage, inhibition of adipogenesis. Our data suggest a contribution of RA to controlling the depot-specific gene program during the functional development of adipocytes in human WAT.
Research Design and Methods
Isolation and Culture of ASCs
WAT was isolated from the SC depot of the abdominal region and the VS depot of the omental region of 10 human volunteers (S1–7 and S11–13) undergoing bariatric surgery, with approval by the National Healthcare Group Domain Specific Review Board, Singapore. The subjects S1–7, S11, and S12 have been described previously (15). S13 is a 47-year-old Chinese man. ASCs were isolated from WAT and cultured, as previously described (19). Only cells with a doubling time shorter than 36 h were used (up to p9), and cell samples with similar passage numbers were used for any comparative studies. Mesenchymal stem cell surface markers and the multipotency of ASCs used in this study were confirmed by flow cytometry and differentiation assays, respectively (15).
Adipogenesis and AdipoRed Staining
On day (D) 0, 2 days after reaching the confluent state, cells were induced with an adipogenic differentiation cocktail containing 1 μmol/L dexamethasone, 0.5 mmol/L isobutylmethylxanthine, and 167 nmol/L insulin plus 100 μmol/L indomethacin, 8 mg/L biotin, and 4 mg/L pantothenate. On D6, cells were switched to a medium with 1 μmol/L dexamethasone and 167 nmol/L insulin plus 100 μmol/L indomethacin and then maintained until at least D12.
The cells then were washed with PBS and stained with AdipoRed (Lonza) according to the manufacturer’s protocol. After 30 min, fluorescence readings were recorded at 485-nm excitation and 572-nm emission. Fluorescence images were captured using ImageXpress Micro (Molecular Devices).
Real-Time PCR
Total RNA was extracted using the TRIzol reagent (Invitrogen) and purified with the Column RNeasy Kit (Qiagen), according to the manufacturer’s instructions. cDNA was converted using the RevertAid H Minus First Strand cDNA Synthesis Kit (Fermentas). Quantitative PCR (qPCR) was performed using SYBR Green PCR Master Mix on a StepOnePlus Real-Time PCR System (Applied Biosystems) using the primer pairs shown in Supplementary Table 4. Relative mRNA were calculated and normalized to the level of GAPDH.
RA-Responsive Luciferase Assay
To determine relative intracellular levels of RA activity, a firefly luciferase under the control of an RA-responsive element (RARE) in the pGL3 vector and control plasmid pRL-Renilla luciferase under the control of the cytomegalovirus promoter were transfected together into 293T cells with Lipofectamine 2000 (Invitrogen) per the manufacturer’s protocol. Then, 24 h after transfection, the 293T cells were incubated with either fresh media (DMEM + 15% FBS) or human ASC–conditioned media. After another 24 h, RA activities were assessed by measuring the dual activities of firefly and Renilla luciferases per the manufacturer’s protocol (Promega) and normalizing firefly luciferase activities relative to the control (Renilla) luciferase.
Electrophoretic Mobility Shift Assay
Nuclear fractions from SC and VS ASCs were isolated using a previously described method (20). The oligonucleotides containing the WT1 binding site on the human ALDH1A2 promoter (5′-AGAACTCAGAGAGTGGGAGAGTGTTCCCT-3′) were hybridized to their respective complementary strands and labeled at the 3′ end with biotin tetraethylene glycol (Sigma-Aldrich). Nuclear extracts obtained from ASCs were incubated with the biotin-labeled probe and then subjected to electrophoresis, transferred to a nylon membrane, and ultraviolet cross-linked. The membrane then was probed with stabilized Streptavidin–horseradish peroxidase conjugate and developed using the LightShift Chemiluminescent electrophoretic mobility shift assay (EMSA) kit (Thermo Scientific/Pierce Biotechnology).
Chromatin Immunoprecipitation Assay
Chromatin immunoprecipitation assay was performed based on a previous protocol (21), with minor modifications. Briefly, ASCs were fixed with 1% formaldehyde, stopped by adding 125 mmol/L glycine, and then washed and collected in ice-cold PBS. The cell pellets were resuspended in lysis buffer, and the resulting pellet of crude nuclear extract was resuspended in a high-salt lysis buffer. The nuclear extracts were sonicated on ice using a Misonix 3000 to yield DNA fragments 200–500 base pairs in size. Immunoprecipitation was conducted overnight at 4°C with 2 μg of anti-WT1 antibody (sc-192x; Santa Cruz Biotechnology) or anti-IgG antibody. PureProteome preblocked protein A magnetic beads (100 μL; Millipore) were incubated with the samples at 4°C for 2 h with rotation and eluted for 2 h at 65°C. After the cross-linking was reversed overnight at 65°C, DNA was extracted and PCR performed using the following set of primers: WT1 forward: 5′-ATCCCATTCTGTTATCTACTCCC-3′; WT1 reverse: 5′-CCTGCACTGCTTTGTCTATATCT-3′.
Gene Knockdown Through Lentivirus
Short hairpin RNAs (shRNAs) targeting WT1 were designed using publicly available algorithms from GenScript. Nine different shRNAs were synthesized, and the one that provided the best knockdown efficiency was selected for further experiments. Stable knockdown of WT1 was achieved using lentivirus expressing the shRNA sequence GCCTCACTCCTTCATCAAACAACTCGAGATGTTTGATGAAGGAGTGAGGCTTTTT. Sense and antisense sequences are denoted in italics, whereas the loop sequence is underlined. A nontargeting shRNA was used as scrambled control. Lentiviral particles were produced by transient transfection of 293T cells with a transfer vector and pVSV-G, pMDL/pRRE, and pRSV-Rev packaging vectors. The viral supernatant was collected, filtered, and concentrated by ultracentrifugation. Stable cell lines were generated by transducing human ASCs at a multiplicity of infection of 4 in complete media containing polybrene.
Liquid Chromatography/Mass Spectrometry Analyses of Retinoids
To measure cellular retinol levels, highly sensitive ultraperformance liquid chromatography coupled with mass spectrometry (LC-MS) was used (22). For all-trans RA (atRA) extraction, samples were subjected to a two-step acid-base extraction as described before (23), with minor modifications. The details are described in the Supplementary Data.
Western Blot Analysis
Protein from ASCs was extracted in radioimmunoprecipitation assay buffer and subjected to Western blot analysis. The concentration of protein in lysates was determined using Bradford assay. Protein (20 μg) was separated on 4–20% Mini-PROTEAN gels (Bio-Rad) and then transferred to nitrocellulose membrane. The membranes were probed with primary antibodies, WT1 (1:1,000 dilution; Santa Cruz Biotechnology) and α-tubulin (1:5,000 dilution; Sigma). After subsequent washes and incubation with respective secondary antibodies, the horseradish peroxidase activity was detected using chemiluminescent reagents.
Data Analysis
All results are presented as means ± SEMs. Comparisons between groups were analyzed using two-sided, paired t tests. Differences with a P value <0.05 were considered significant.
Results
RNA-Seq and Microarray Analysis of SC and VS Fat Depots Highlight the Differential Regulation of Genes Involved in Retinoid Metabolism
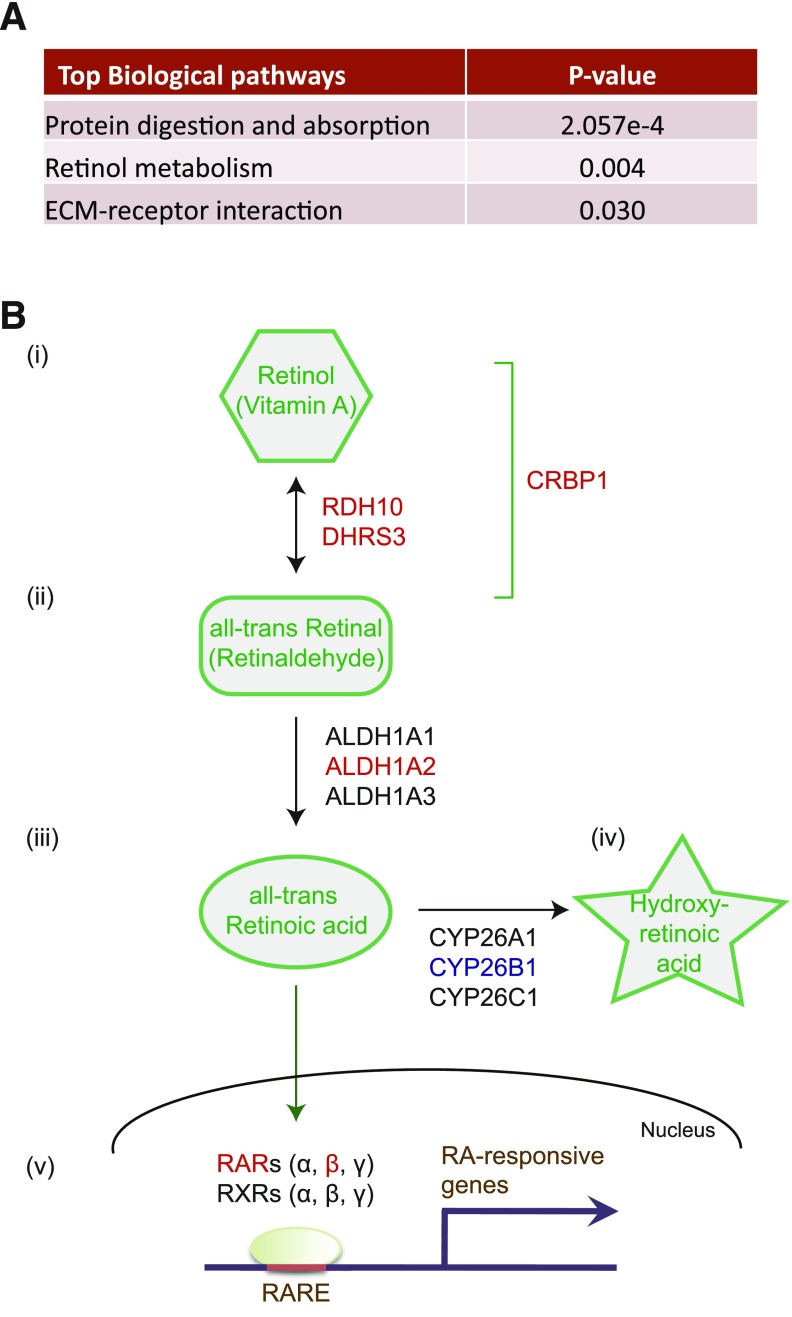
We performed next-generation sequencing analysis of eight paired SC and VS WAT depots from subjects without diabetes using the Illumina HiSeq 2000 system. Through the RNA-seq analysis, a total of 1,185 annotated genes exhibited significant differences between VS and SC fat depots (significance was defined as fold change >2.0 and a false discovery rate <0.05) (Supplementary Table 1). Of these genes, 792 had higher expression in VS fat, whereas 393 genes were predominant in SC fat. Ontological analysis of these differentially regulated genes highlighted just three categories that reached significance as assessed by P value <0.05 after Bonferroni correction (Fig. 1A). One of these was found to be “retinol metabolism,” which includes VS-enriched genes (set red in Supplementary Table 1) and SC-dominant genes (set blue in Supplementary Table 1).
Figure 1.
The metabolic pathway of retinoic acid. A: Gene ontology analysis of RNA-seq data from SC and VS adipose tissue identified retinol metabolism as the second most significant biological pathway that is differentially expressed between these two depots; this is defined by P value <0.05 after Bonferroni correction. ECM, extracellular matrix. B: Vitamin A/retinol is metabolized to atRA via two sequential enzymatic reactions. i: In the first reaction, RDH10 and DHRS3, two retinol dehydrogenases/reductases, reversibly oxidize retinol to retinal and vice versa. CRBP1 is a cellular retinol-binding protein that controls the availability of cellular retinol. ii: In the second reaction, ALDH1A1, ALDH1A2, and ALDH1A3 (retinaldehyde dehydrogenases) irreversibly oxidize retinal to atRA. iii: RA enters the nucleus and specifically binds the retinoic acid receptor family: RARs α, β, and γ or RXRs α, β, and γ. iv: atRA is metabolized to 4-hydroxy-retinoic acid by cytochrome P450 proteins (CYP26A1, CYP26B1, and CYP26C1). v: RARs are ligand-dependent transcription factors and, when bound to atRA, regulate the expression of many RA target genes through binding to the RARE. Proteins whose gene expression is upregulated in VS ASCs are highlighted in red, and those that are downregulated in VS-ASCs are blue.
Because different WAT regions contain a number of distinct cell types, especially infiltrating immune cells, it is plausible that many of the differentially regulated genes mentioned above are influenced by cell type differences in SC and VS depots. To determine what portion of these genes are influenced by intrinsic differences in the stem cell population, we performed global gene expression analysis of human ASCs from six paired SC and VS depots using an Illumina HumanHT-12 array. Of these, 171 genes showed a fold change >2.0 and a positive false discovery rate <0.05 (Supplementary Table 2). Among these genes, 85 were expressed predominantly in SC ASCs (Supplementary Table 2A) and 86 were expressed more in VS ASCs (Supplementary Table 2B). In our analysis, several HOX genes were differentially expressed in ASCs from different depots, as previously reported by others (16–18): HOXA2, HOXA4, and HOXA5 were upregulated in VS ASCs, whereas HOXC6, HOXC8, HOXC9, HOXA9, and HOXA10 were upregulated in SC ASCs. Consistent with our previous report (15), the cell surface protein MME/CD10 was selective for both SC ASCs and SC fat, whereas CD200 was higher in both VS ASCs and VS tissue at the gene expression level (Supplementary Tables 1 and 2).
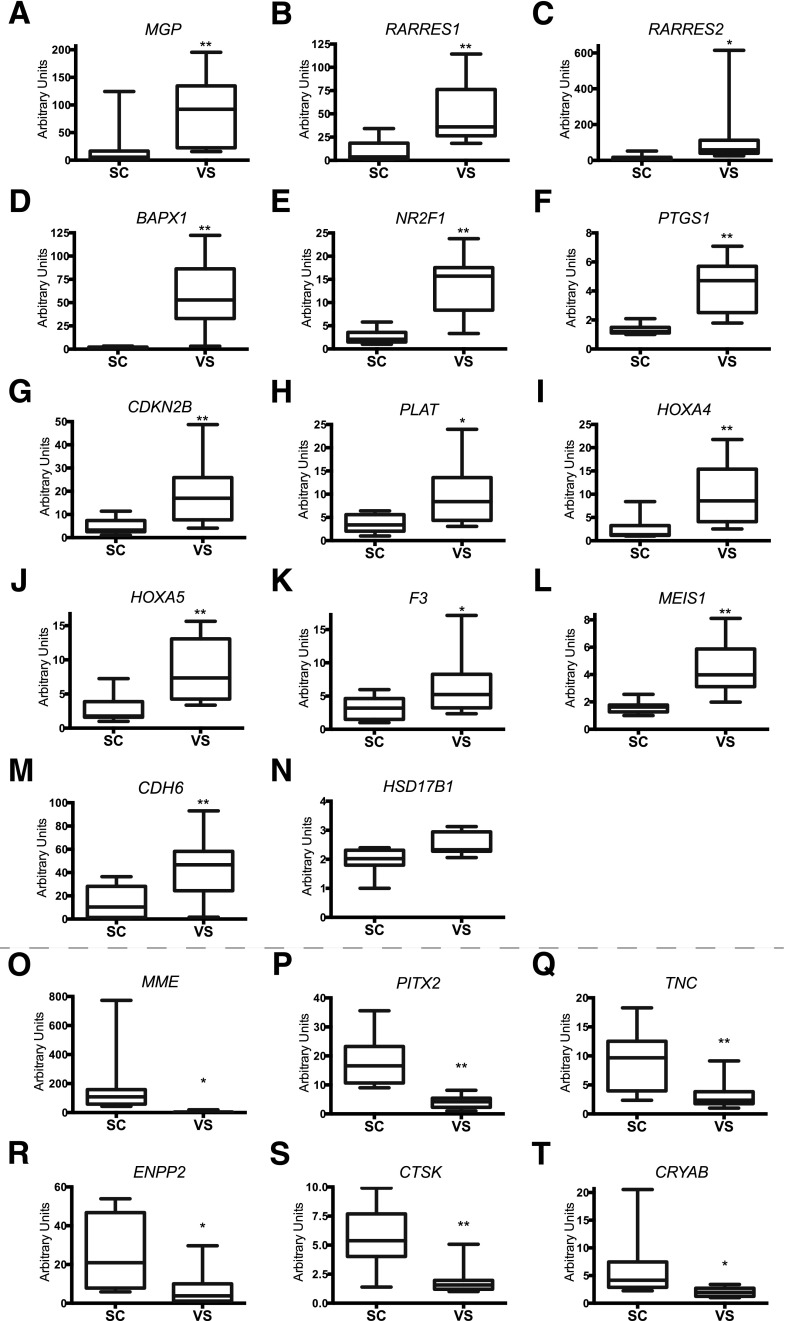
Similar to the RNA-seq analysis of tissues, ontological analysis of these differentially expressed genes in ASCs from different depots also revealed enrichment of genes categorized in the “retinoid metabolic process.” It was previously estimated that RA can modulate the expression of a few more than 530 genes (24). Of these, 15 RA-responsive genes among 86 of the upregulated genes in VS ASCs and 6 RA-responsive genes among 85 of the upregulated genes in SC ASCs were identified. We found that genes participating in RA synthesis (RDH10, CRBP1, and DHRS3) and reportedly modulated by RA (CRBP1, MGP, RARRES1, BAPX1, RARRES2, NR2F1, PTGS1, CDKN2B, PLAT, HOXA4, HOXA5, F3, HSD17B1, MEIS1, and CDH6) were upregulated in VS ASCs (Supplementary Table 3A). Also, six putative RA-responsive genes (MME, PITX2, TNC, ENPP2, CTSK, and CRYAB) were upregulated in SC ASCs (Supplementary Table 3B). The acquisition of RA-responsive genes in a depot-specific manner is highly significant: 15 of 86 VS-enriched genes (17.4%) and 6 of 85 SC-enriched genes (7.1%) versus 532 of the 31,000 total RA-responsive genes/all annotated genes from our microarray data (1.7%). These results indicate that a significant number of gene expression differences are present, even in stem cell populations between distinct fat depots, and that the retinoid/RA metabolism pathway may represent one of the most important categories underlying pathophysiological differences in VS versus SC fat.
Validation of Depot-Specific Expression of RA-Related Genes by qPCR
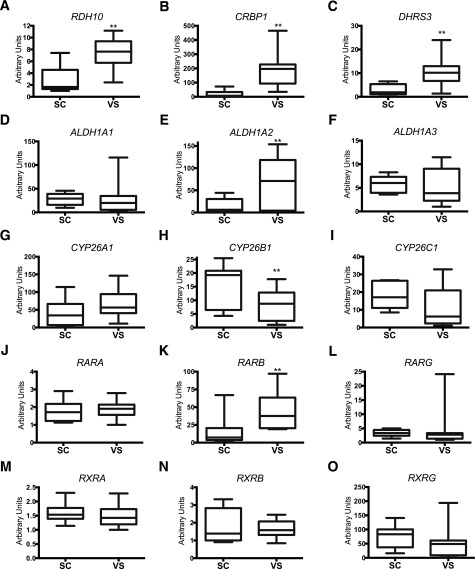
Our microarray results showed that the expression of genes involved in the first retinoid synthesis—RDH10, CRBP1, and DHRS3—was increased in VS ASCs when compared with that in SC ASCs. This observation led us to question whether other RA-related metabolic genes were differentially expressed in SC and VS ASCs. Retinol, a vitamin A species, is metabolized to RA through two sequential enzymatic reactions (25). In the first reaction, retinol dehydrogenase/reductase enzymes, such as RDH10 and DHRS3, reversibly oxidize or reduce retinol and retinal (also called retinaldehyde) (Fig. 1B). CRBP/RBP1, a cellular retinol-binding protein, controls the availability of cellular retinol. Reverse transcriptase qPCR confirmed that the expression of RDH10 (Fig. 2A), CRBP1 (Fig. 2B), and DHRS3 (Fig. 2C) was significantly upregulated in VS ASCs in almost all 10 subjects. In the second step, retinaldehyde dehydrogenases, which include ALDH1A1, ALDH1A2, and ALDH1A3, irreversibly oxidize retinal to RA. ALDH1A1 (Fig. 2D) and ALDH1A3 (Fig. 2F) did not show depot specificity, whereas ALDH1A2 expression (Fig. 2E) was significantly higher in VS ASCs than in SC ASCs. Finally, RA is metabolized to hydroxy-RA by cytochrome P450 proteins: CYP26A1, CYP26B1, and CYP26C1 (25). CYP26A1 (Fig. 2G) and CYP26C1 (Fig. 2I) did not show depot specificity, whereas CYP26B1 expression (Fig. 2H) was significantly lower in VS ASCs when compared with SC ASCs. In the downstream pathway, RA activates the RA receptor (RAR) family (RARs α, β, and γ or retinoid X receptors [RXRs] α, β, and γ), which contains ligand-dependent transcription factors of the nuclear receptor family, to regulate the expression of many RA target genes (Fig. 1B). Of the six genes measured, only RARB expression was upregulated in VS ASCs (Fig. 2K), whereas no change was observed in expression of the other five genes (Fig. 2J, L, M–O). Upregulation of RARB coincides with the results of an earlier report, which showed that gene expression of RARB, but not other RA receptors, is inducible by RA itself (26).
Figure 2.
Depot-specific expression profiles of genes involved in RA metabolism. qPCR was performed on 10 pairs of patient-derived SC and VS ASCs (S1–7 and S11–13). The graphs show differential mRNA expression of RDH10 (A), CRBP1 (B), DHRS3 (C), ALDH1A1 (D), ALDH1A2 (E), ALDH1A3 (F), CYP26A1 (G), CYP26B1 (H), CYP26C1 (I), RARA (J), RARB (K), RARG (L), RXRA (M), RXRB (N), and RXRG (O) in S1–7 and S11–13. The values are relative arbitrary units and are normalized to the housekeeping gene GAPDH. **P < 0.01 denotes significant differences in pairs of ASCs (n = 10).
In General, RA Induces VS ASC Genes and Suppresses SC ASC Genes
Using qPCR, we confirmed the depot specificity of 14 of the 15 genes that showed significantly higher expression in VS ASCs (Figs. 2B and 3A–M); only changes in HSD17B1 (Fig. 3N) did not reach statistical significance. All of the six genes were validated to exhibit significantly higher expression in SC ASCs (Fig. 3O–T).
Figure 3.
Differential expression of RA-regulated genes in SC and VS ASCs. The graphs show differential mRNA expression of genes upregulated in VS ASCs, including MGP (A), RARRES1 (B), RARRES2 (C), BAPX1 (D), NR2F1 (E), PTGS1 (F), CDKN2B (G), PLAT (H), HOXA4 (I), HOXA5 (J), F3 (K), MEIS1 (L), CDH6 (M), and HSD17B1 (N), and genes upregulated in SC ASCs, such as MME (O), PITX2 (P), TNC (Q), ENPP2 (R), CTSK (S), and CRYAB (T), in S1–7 and S11–13. The values are relative arbitrary units and are normalized to the housekeeping gene GAPDH. *P < 0.05 and **P < 0.01 denote significant differences in pairs of ASCs (n = 10). See also Supplementary Fig. 1.
To investigate the RA-mediated responses of these genes, we treated SC ASCs with different concentrations of exogenous RA for 48 h. Eleven of 14 VS ASC genes (CRBP1, MGP, RARRES1, RARRES2, NR2F1, CDKN2B, PLAT, HOXA4, HOXA5, F3, CDH6) showed increased expression upon RA treatment, generally in a concentration-dependent manner (Supplementary Fig. 1Ai–v). In contrast to the VS ASC genes listed above, four of six SC ASC genes (MME, TNC, ENPP2, CRYAB) showed decreased expression upon RA treatment in a dose-dependent manner (Supplementary Fig. 1Bi and ii). These data indicate that VS ASC–enriched genes are mostly induced by exogenous RA, whereas SC ASC genes are generally downregulated by RA treatment.
VS ASCs Exhibit Higher Endogenous RA Levels
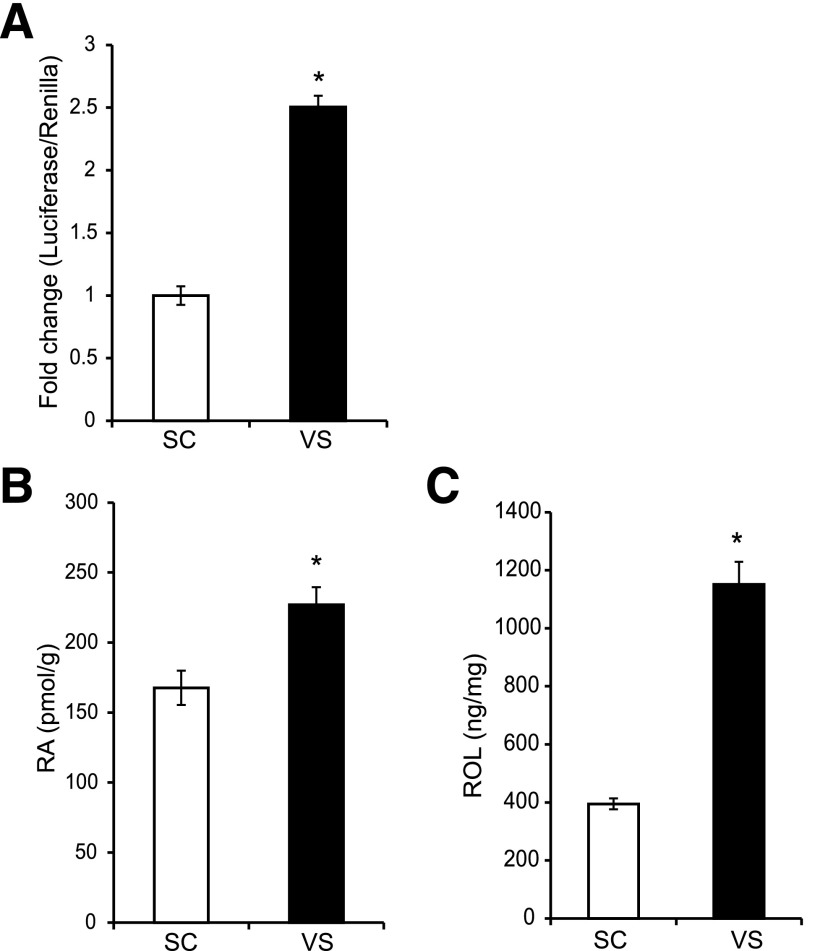
Since higher RA-mediated activity was observed in VS ASCs than in SC ASCs, we assessed endogenous RA levels using three different methods. First, a Luciferase reporter assay using the RARE indicated that VS ASCs rendered a significantly higher RA-responsive activity than SC ASCs (Fig. 4A). Second, endogenous levels of RA were assessed by ultrasensitive surface-enhanced Raman spectroscopy using the scheme and spectra shown in Supplementary Fig. 2A–C. The surface-enhanced Raman spectroscopy measurement indicated significant upregulation of RA in VS ASCs compared with SC ASCs (Supplementary Fig. 2D). A similar increase of endogenous RA in VS ASCs was observed when the conventional method using LC-MS was used (Fig. 4B). The level of the RA precursor retinol, as measured by LC-MS, also indicated a significant increase in VS ASCs compared with SC ASCs (Fig. 4C).
Figure 4.
Endogenous levels of RA are higher in VS ASCs. A: A representative graph showing increased levels of normalized RARE luciferase reporter activity in VS ASCs of S11 when compared with SC ASCs. B: A representative graph showing increased levels of RA in VS ASCs as measured by LC-MS. The data are averaged from cells of two subjects (S11 and S13). See also Supplementary Fig. 2. C: A representative graph showing increased levels of retinol (ROL) in VS ASCs of S11, measured by LC-MS. *P < 0.05 denotes significant change (n = 2).
RA Inhibits Adipocyte Differentiation of Human ASCs at Early Stages
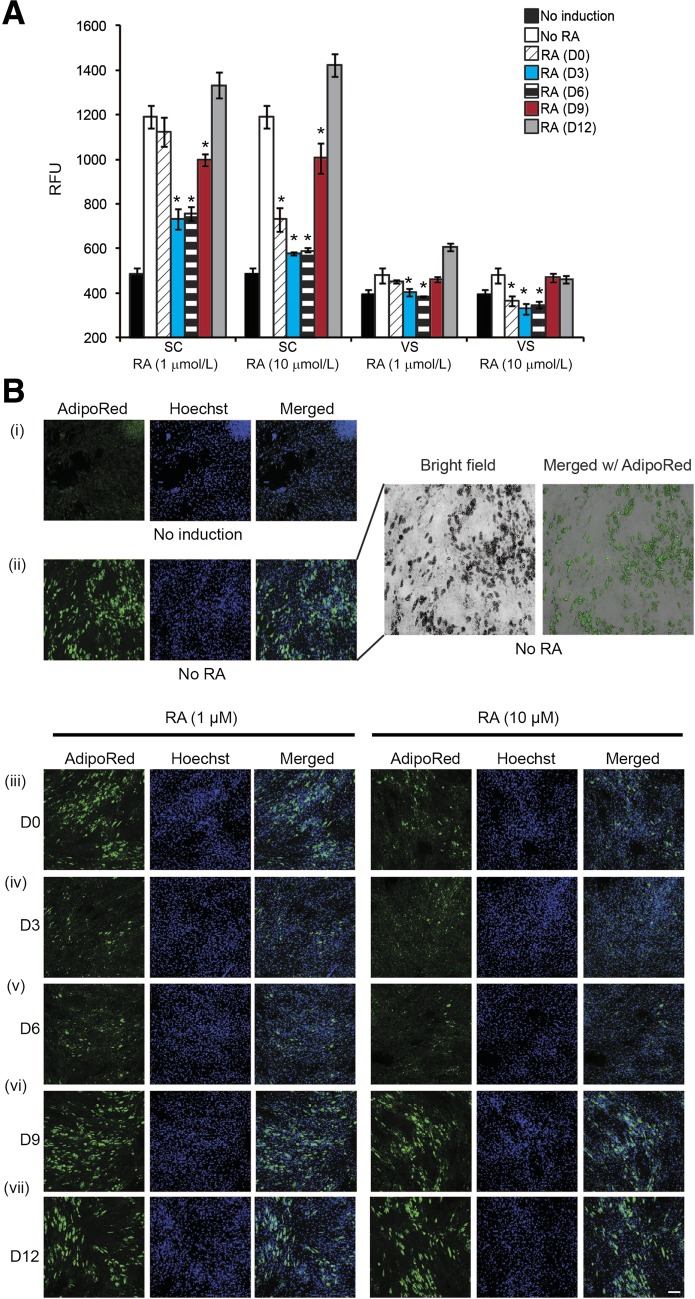
It was previously reported that RA is a potent inhibitor of adipocyte differentiation in mice (27,28). To determine whether RA affects the adipocyte differentiation of human ASCs and, if so, at which adipogenic stage, we treated ASCs with two different concentrations of RA (1 and 10 μmol/L) at different times during adipocyte differentiation. The results showed that the early addition of RA drastically inhibited adipocyte differentiation during the predifferentiation period and during the first 6 days after differentiation (D−2 to D0, D0 to D3, or D3 to D6) and did so to a greater extent in SC ASCs (Fig. 5A and Bi–v) than VS ASCs (Fig. 5A and Supplementary Fig. 3i–v). A higher concentration (10 μmol/L) of RA corresponded with a greater reduction in adipocyte formation, especially that of SC ASCs, when compared with a lower concentration (1 μmol/L) of RA. Importantly, the adipogenic level of SC ASCs treated with 10 μmol/L RA during D0 to D3 or D3 to D6 was similar to that of nontreated VS ASCs. Also, just 2 days of pretreatment with RA itself was sufficient to significantly reduce adipocyte formation in ASCs. RA treatment at later stages of differentiation—that is, D6 to D9 and D9 to D12—either minimally inhibited or failed to block the adipocyte formation in both SC and VS ASCs (Fig. 5A and Bvi–vii; Supplementary Fig. 3vi–vii). Furthermore, as shown in Supplementary Fig. 4, treatment of SC and VS ASCs with retinol (Supplementary Fig. 4Ai and ii) or retinal (Supplementary Fig. 4Bi and ii), intermediates in the RA synthesis pathway, significantly inhibited adipocyte differentiation at various time points during adipogenic induction, as observed by AdipoRed staining.
Figure 5.
RA inhibits the early stage of adipocyte differentiation of ASCs. Using a standard adipogenic cocktail, adipocyte differentiation was induced in ASCs from S11, with or without pretreatment/treatment with RA at various time points. A: A representative graph showing relative fluorescence units (RFUs) of AdipoRed staining on ASCs from S11. The ASCs were treated with 1 or 10 μmol/L of RA at different time points, as indicated by D0 (D−2 to D0), D3 (D0 to D3), D6 (D3 to D6), D9 (D6 to D9), and D12 (D9 to D12). *P < 0.05 denotes significant fold change (in RFUs) against respective “No RA” control samples, corresponding to the quantitation of lipid accumulation during adipocyte differentiation. B: Representative images (original magnification ×10) showing the lipid accumulation (AdipoRed, green) and the nuclei (Hoechst 33342, blue) in SC ASCs from S11 treated with 1 or 10 μmol/L of RA at different time points: no induction (i), without RA (ii), with RA treatment during D−2 to D0 (iii), D0 to D3 (iv), D3 to D6 (v), D6 to D9 (vi), or D9 to D12 (vii). The field in ii is magnified and merged with the bright-field image to show the overlap of AdipoRed staining and lipid droplet structures (top right). Similar results were obtained from experiments using cells from S12. For the purpose of presentation, the fluorescent intensities were enhanced to the same degree for all original images. Scale bar = 100 µm. See Supplementary Fig. 3 for representative images in VS ASCs.
Adipogenic Gene Expression Profiles in VS ASCs During Adipocyte Differentiation Are Similar to Those in RA-Treated SC ASCs
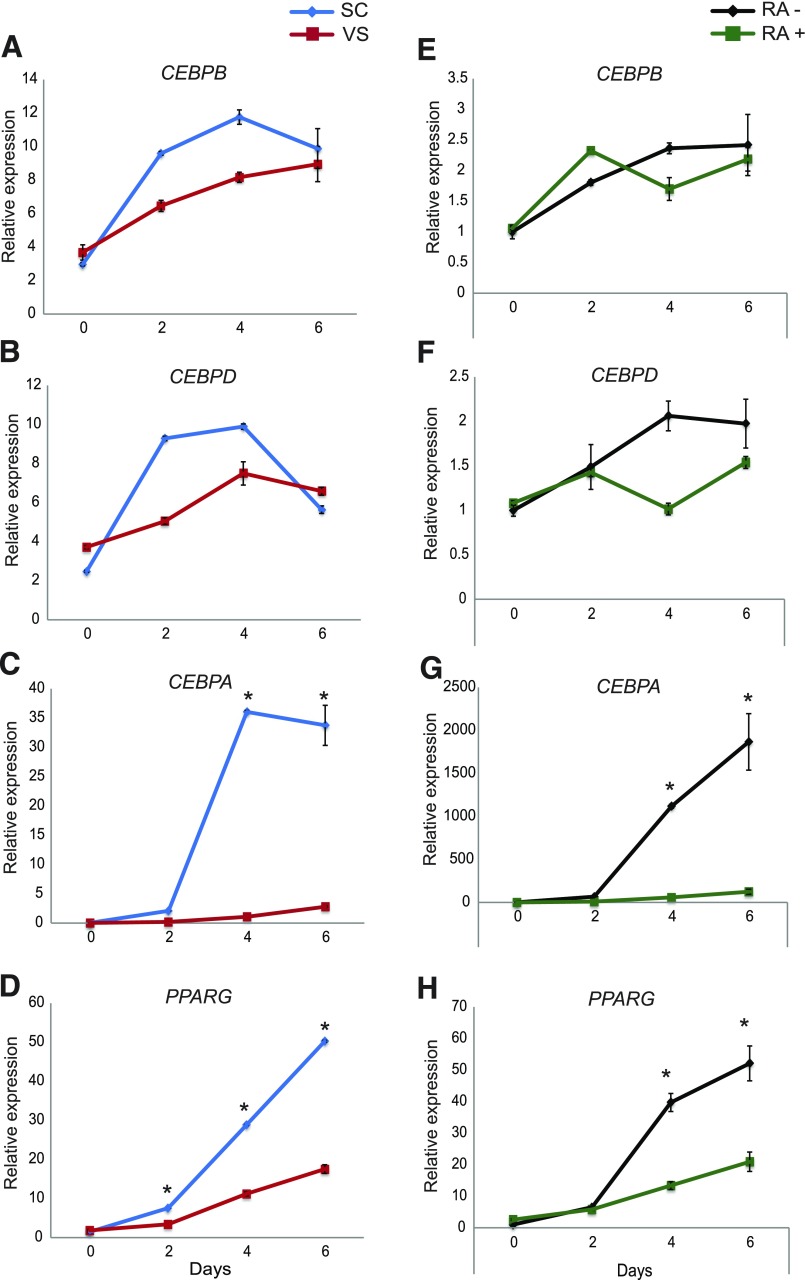
We examined changes in the expression of standard adipogenic genes during adipocyte differentiation in SC ASCs and VS ASCs. As previously reported (9,15,29), and as reflected in Fig. 5 and Supplementary Fig. 3, SC ASCs differentiated into mature adipocytes under a standard adipogenic cocktail substantially better than VS ASCs. The induction of early adipogenic markers (CEBPB and CEBPD) during adipogenesis was not significantly different between SC ASCs and VS ASCs (Fig. 6A and B). By contrast, the expression of late adipogenic markers (CEBPA and PPARG) was substantially increased after adipogenic induction in SC ASCs but was significantly defective in VS ASCs (Fig. 6C and D).
Figure 6.
RA-mediated differential expression of adipogenic genes during adipocyte differentiation of ASCs. Adipocyte differentiation was induced in SC and VS ASCs from S11 using a standard adipogenic cocktail. RNA was collected at various time points during differentiation (D0, D2, D4, and D6), and the expression of adipogenic genes was analyzed by qPCR. The representative graphs show gene expression of CEBPB (A), CEBPD (B), CEBPA (C), and PPARG (D) in SC and VS ASCs from S11 at the various time points indicated (n = 2). Adipocyte differentiation was induced in SC ASCs from S11 using a standard adipogenic cocktail, with or without RA (10 μmol/L) treatment. RNA was collected at various time points (D0, D2, D4, and D6). The representative graphs show gene expression of CEBPB (E), CEBPD (F), CEBPA (G), and PPARG (H) in SC ASCs treated with or without RA during differentiation at various time points (n = 2). *P < 0.05 denotes significance. Similar results were obtained in cells from S12.
Next, we determined the gene expression profiles of CEBPA, CEBPB, CEBPD, and PPARG with RA treatment during the early stages of adipocyte differentiation. The results showed no significant differences in the expression of early adipogenic genes CEBPB and CEBPD (Fig. 6E and F) in RA-treated SC ASCs compared with untreated control cells. By contrast, the expression of late adipogenic genes CEBPA and PPARG was dramatically reduced in RA-treated SC ASCs (Fig. 6G and H). These changes in the expression of early and late adipogenic markers in RA-treated SC ASCs (Fig. 6E–H) were very similar to those in VS ASCs (Fig. 6A–D). The results imply that the adipogenic defect of VS ASCs may be at least partially influenced by higher RA-mediated activities.
Visceral-Specific Developmental Factor May Act to Facilitate the RA Signaling Pathway by Modulating the Expression of the RA Synthesis Enzyme
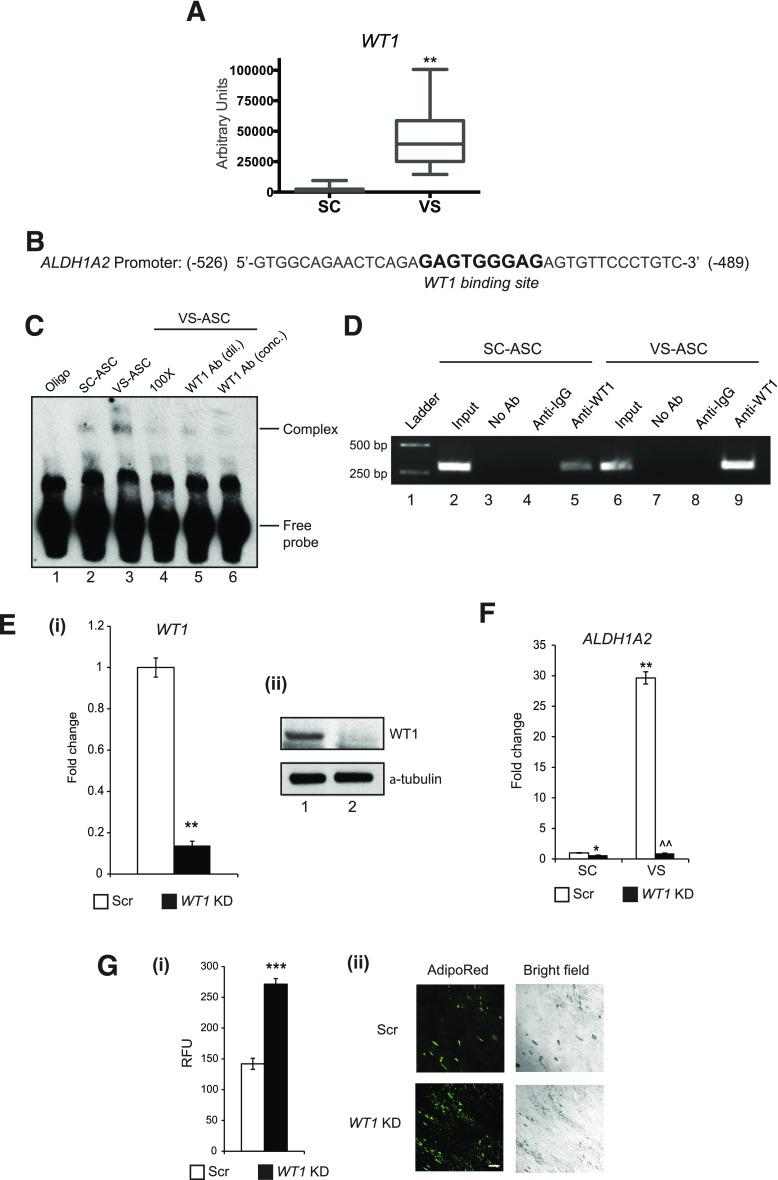
It was recently reported that the Wt1 gene, a mesothelial developmental marker, is specifically expressed in VS fat, but not in SC fat, in mice (30). Consistent with this result, we found that WT1 is selectively expressed 24-fold more, on average, in VS ASCs compared with SC ASCs, as assessed using qPCR; this reflects the presence of WT1 in the stem cell population of human VS fat (Fig. 7A). In the search for a developmental factor linking to the RA synthesis pathway, we found a WT1 binding site within the promoter region of the human ALDH1A2 gene (Fig. 7B) that is upregulated in VS ASCs and produces the dehydrogenase enzyme that oxidizes retinal into RA (Fig. 1B). Using EMSA, the antibody against WT1 was found to specifically bind the DNA probe spanning the ALDH1A2 promoter with higher affinity in VS ASCs than in SC ASCs (Fig. 7C). The specificity of WT1 was confirmed when a diminished band intensity or the disappearance of a shifted band is observed by (pre)incubating the VS ASC nuclear extracts with 100× the concentration of competitor oligos or WT1 antibodies. In addition, chromatin immunoprecipitation experiments showed that the anti-WT1 antibody, but not the control IgG antibody, pulled down the ALDH1A2 promoter region at a higher level in VS ASCs than SC ASCs (Fig. 7D). Finally, the WT1 gene was knocked down through the lentiviral construct in SC ASCs and VS ASCs. qPCR and Western blotting showed effective knockdown of WT1 in both gene and protein expression (Fig. 7Ei and ii). It was found that high expression of the ALDH1A2 gene in VS ASCs was dramatically suppressed by knockdown of WT1 compared with the scrambled control (Fig. 7F). Importantly, the knockdown of WT1 in VS ASCs significantly increased adipogenesis, as shown by the AdipoRed staining in Fig. 7Gi and ii. These results suggest that the developmental WT1 protein can upregulate the RA pathway directly through the induction of the ALDH1A2 gene in VS ASCs, and genetic ablation of WT1 improves their adipogenic capacity.
Figure 7.
WT1 mediates the upregulation of the RA synthesis enzyme in VS ASCs. A: A qPCR graph showing substantially higher WT1 expression in VS ASCs from S1–S7 and S11–13. **P < 0.01 denotes significant change in pairs of ASCs (n = 10). B: Consensus WT1 binding site was found within the promoter region of the human ALDH1A2 gene. C: EMSA was performed using nuclear extracts from SC and VS ASCs from S11. The representative gel shows increased WT1 binding in VS ASCs, as indicated by the shifted band in lane 3, when compared with that of SC ASCs (lane 2) (lane 1 represents oligonucleotides [oligo] only). Diminished band intensity or the disappearance of a shifted band is observed when VS ASC nuclear extracts were incubated/preincubated with 100× the concentration of competitor oligonucleotides (lane 4) or with diluted 1:1 (dil.; lane 5) or concentrated (conc.; lane 6) WT1 antibody (n = 2). D: A representative agarose gel image showing the increased binding of WT1 to the ALDH1A2 promoter in VS ASCs from S11 (lane 9) compared with SC ASCs (lane 5) as assessed by chromatin immunoprecipitation. The relative amounts of both SC and VS ASCs in the input were also assessed (lanes 2 and 6). Both no antibody (No Ab; lanes 3 and 7) and isotype-specific IgG (lanes 4 and 8) controls are shown (n = 2). E: (i) Graph showing mRNA expression of WT1 in WT1 knockdown (KD) ASCs from S11 (**P < 0.01) (n = 2). (ii) Western blot showing protein expression of WT1 in WT1 KD ASCs from S11; α-tubulin was used as the internal control on the gel (lane 1: scrambled control [Scr]; lane 2: WT1 KD) (n = 2). F: A representative graph showing the decrease in ALDH1A2 expression in WT1 KD cells in both SC and VS ASCs from S11. The values are expressed as fold change. Similar results were obtained from experiments using cells from S12. *P < 0.05 and **P < 0.01 denote significant fold change when compared with WT1 Scr SC ASCs; ^^P < 0.01 denotes significant fold change when compared with Scr VS ASCs (n = 2). G: (i) A representative graph showing relative fluorescence units (RFUs) of AdipoRed staining on Scr and WT1 KD VS ASCs from S11 (***P < 0.001) (n = 2). (ii) Representative images (original magnification ×10) showing AdipoRed staining and corresponding bright-field images in differentiated adipocytes of Scr and WT1 KD VS ASCs from S11 (n = 2). Scale bar = 100 µm. Similar results were obtained from experiments using cells from S12.
RA-Mediated Adipogenic Defects Are Reversed by Antagonism of the Downstream Target RAR
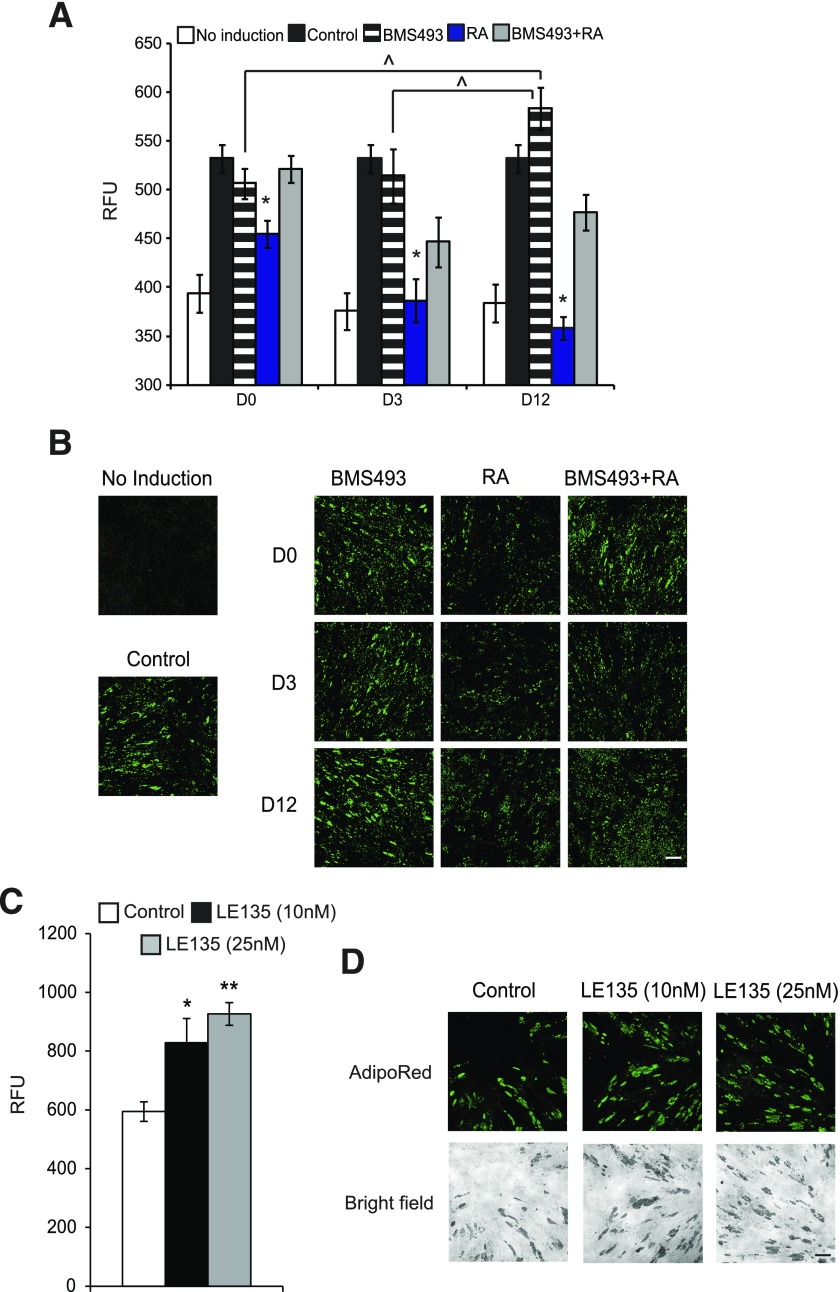
Finally, to address whether adipogenic defects caused by excessive RA can be reversed by modulating the RA pathway in VS ASCs, we used BMS493, an antagonist of RARs (α, β, γ), at different points during adipocyte differentiation. The result of AdipoRed staining and quantification demonstrated that treatment of BMS493 during D−2 and D12 significantly improved the adipogenic capacity of VS ASCs (Fig. 8A and B). BMS493 also reversed the adipogenic defect caused by excessive RA at all the times tested (Fig. 8A and B). We also treated VS ASCs with LE135, a RARβ-specific antagonist. AdipoRed staining revealed that treatment with LE135 significantly improved the adipogenic potential of VS ASCs, as shown in Fig. 8C and D. Together, these results propose a potential therapeutic strategy of RAR or RARβ antagonism that can relieve adipogenic dysfunction mediated by excessive RA signaling in VS fat.
Figure 8.
RA-mediated adipogenic defects are reversed by the antagonism of the downstream target RAR. A: A representative graph showing the relative fluorescence units (RFUs) of AdipoRed staining on VS ASCs from S11. Adipocyte differentiation was induced in ASCs using a standard adipogenic cocktail with or without pretreatment/treatment with BMS493 and/or RA at various time points. The ASCs were treated with 1 μmol/L of BMS493 and/or 10 μmol/L of RA at the time points indicated: D0 (D−2 to D0), D3 (D−2 to D3), and D12 (D−2 to D12). *P < 0.05 and ^P < 0.05 denote significant fold change in RFUs corresponding to the quantitation of lipid accumulation during adipocyte differentiation (n = 2). B: Representative images (original magnification ×10) showing lipid accumulation (AdipoRed, green) in VS ASCs from S11 treated with 1 μmol/L of BMS493 and/or 10 μmol/L of RA at the different time points indicated: D0 (D−2 to D0), D3 (D−2 to D3), and D12 (D−2 to D12). For the purpose of presentation, the fluorescent intensities were enhanced to the same degree for all images. Scale bar = 100 µm (n = 2). C: A representative graph showing the RFUs of AdipoRed staining on VS ASCs from S11. ASCs were treated with LE135 (10 or 25 nmol/L) for 3 days upon the induction of adipogenic differentiation with a standard adipogenic cocktail. *P < 0.05 and **P < 0.01 denote significant fold change in RFUs (n = 2). D: Representative images (original magnification ×10) showing lipid accumulation (AdipoRed, green) in VS ASCs from S11 treated with LE135 (10 or 25 nmol/L) for 3 days upon the induction of adipogenic differentiation. For the purpose of presentation, the fluorescent intensities were enhanced to the same degree for all images. Scale bar = 100 µm (n = 2). Similar results were obtained from experiments using cells from S12.
Discussion
In this study, through the ontological analysis of global gene expression, we found that a pathway regulated by RA is one of the major metabolic responses to developmental differences of stem cells from human fat depots. RA is a vitamin A metabolite that regulates many genes through specific binding to nuclear transcription factors, especially RARs (24,31). The endogenous concentration of RA is tightly regulated during embryonic and postnatal development and is essential for the proper functionality of adult tissues and cells. Several recent reports have indicated potential roles of retinoids (RA, retinol, retinal, and other vitamin A derivatives) in adipose tissue (32–34). Although the liver is the primary storage site of retinoids, adipose tissue is the second largest site and actively mediates retinoid metabolism (35,36). However, little is known about its fat depot–specific mechanisms and contributions, especially in the context of human stem/progenitor cells.
Our microarray and qPCR analysis indicated that genes that lead to RA synthesis, such as retinoid oxidoreductases (RDH10, DHRS3, and ALDH1A2), had higher expression in VS ASCs, whereas CYP26B1, a gene involved in the breakdown of RA, had higher expression in SC ASCs (Fig. 1B). RDH10 is a retinol dehydrogenase that has been shown to be essential for atRA biosynthesis in embryonic development (37), and DHRS3/retSDR1 seems to be a major retinal reductase that is also important for development in human and mice (38,39). ALDH1A2 is the primary isoform of retinal dehydrogenases that are critical for embryogenesis (40), whereas 1A1 and 1A3 isoforms are more important for postnatal and adult development and endocrine functions. In addition, the 1A2 isoform has a much lower Michaelis constant, Km (0.66 μmol/L), than that of 1A1 (11.6 μmol/L) and 1A3 (3.9 μmol/L) (25), indicating that, when all three isoforms are present, this isoform may be predominant in producing RA from retinal. Finally, for the catabolic enzymes, both CYP26A1 and CYP26B1 are important for embryonic development, but CYP26B1 is a major isoform in adult nonbrain tissues, at least in mice (25).
We found 14 genes that had higher expression in VS ASCs and were induced by RA. Of these, CDKN2B and CRBP1 are known to have an anti-adipogenic function (41,42). NR2F1 is expressed more in VS ASCs, and its paralog, NR2F2, has anti-adipogenic capacity as well (43). One VS ASC–specific and RA-inducible gene that is previously well studied is CRBP1. It was reported that overexpression of CRBP1 inhibits adipogenesis and knockdown of CRBP1 enhances adipogenesis in 3T3-L1 cells (41). Interestingly, CRBP1 knockout mice had increased adiposity, especially of epididymal fat (VS fat), but remained more glucose tolerant and insulin sensitive when fed a high-fat diet. Expression of the master adipogenic regulator PPAR-γ was significantly increased in WAT of CRBP1 knockout mice (41). Whether these VS-dominant genes are involved in the adipogenic defect or other VS-specific phenotypes in humans warrants further investigation.
Consistent with earlier studies showing a general inhibitory effect of RA in adipogenesis in vivo (27,44) or in vitro (28,45) in mice, we demonstrated that exogenous RA impaired adipocyte differentiation of human ASCs from different depots at the early stage of adipogenesis, although this occurred to a greater extent in SC ASCs. Interestingly, RA is known to enhance the commitment of embryonic stem cells into an adipocyte lineage when they were pretreated during embryoid body formation (46,47). It is possible that RA exerts distinct effects during different developmental stages or different stages of differentiation. Alternatively, only a higher concentration of RA, as used in our study (1 and 10 μmol/L) and observed in VS ASCs, may inhibit adipocyte differentiation by “overactivating” RAR-mediated signaling; the minimum concentration required for activating RAR is in the nanomolar range. However, the RA dose used for embryonic stem cells’ commitment into adipocytes is 0.1 to 1 μmol/L; we found that as little as 0.1 μmol/L RA is sufficient to inhibit ASC adipogenesis (data not shown). Further studies are necessary to delineate the mechanisms regulated by RA in different developmental/differentiation stages of cells. Together, our results suggest that RA, more of which is produced in VS ASCs than SC ASCs, modulates gene expression differences in ASCs from different depots, influences adipocyte differentiation, and may cause defects in the properties of adipose cells from visceral depots and in body fat distribution.
In addition, we found substantially higher expression of WT1, a developmental marker that marks all visceral organs within the abdominal and thoracic cavities, in human VS ASCs. We found that WT1 may control the expression of the VS-specific ALDH1A2 gene by directly binding to its promoter region in human VS ASCs. WT1 knockdown in VS ASCs led to the reduced expression of ALDH1A2 and improved adipogenesis. This result points to the developmental origin of accelerated RA production and signaling in the human VS depot.
Our finding of differential RA signaling may help to understand how cells from VS fat are compromised and lead to pathophysiological phenotypes of this depot, such as immune cell infiltration, inflammation, limited lipogenesis, altered adipokine secretion, and insulin resistance. It was recently reported that peritoneal macrophages receive the RA signal from the omental region and migrate to the peritoneal cavity in a manner dependent on RA-induced genes (48). Interestingly, RA synthesis enzymes, especially Raldh2 (the mouse ortholog of ALDH1A2), are abundantly expressed in peritoneum-associated tissues with mesothelial origins. It would be of interest to explore further how the higher level of RA in VS fat affects the functional polarization and migration of macrophages and other immune cells, compared with the SC fat depot, which is relatively free from proinflammatory immune infiltration and reactions. Taken together, our investigations pave the way for understanding the developmental basis of visceral adiposity and the potentially immune infiltrative milieu of this depot.
Supplementary Material
Article Information
Acknowledgments. The authors thank members of the Laboratory of Metabolic Medicine Bio-optical Imaging Group and the Singapore Bioimaging Consortium-Nikon Imaging Centre for helping with research activities.
Funding. This work is supported by intramural funding from the Biomedical Research Council of A*STAR to M.O., W.H., V.T., and S.Su. and by funding from the Singapore National Medical Research Council to S.-A.T.
Duality of Interest. S.Su. is a cofounder of Adigenics Pte Ltd, which has not had any financial or scientific influence on this study. No other potential conflicts of interest relevant to this article were reported.
Author Contributions. K.T. and S.Su. conceived and designed the study. K.T., S.Sr., X.H.D.C., and W.K.O. performed the experiments, analyzed data, and wrote the manuscript. C.R.Y., B.T., S.-A.L., K.V.K., and S.H. performed the experiments, analyzed data, and edited the manuscript. H.J., J.J.Y., J.P., M.A., and C.V. performed the experiments and analyzed data. J.S., A.S., and S.-A.T. handled human samples and advised on the clinical aspect of the study. W.S.B. and M.O. advised and supervised the experiments. W.H. supervised the experiments and edited the manuscript. V.T. and S.-A.T. supervised the experiments, analyzed data, and edited the manuscript. S.Su. supervised the experiments, analyzed data, and wrote the manuscript. S.Su. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db15-1315/-/DC1.
References
- 1.Lean ME. Pathophysiology of obesity. Proc Nutr Soc 2000;59:331–336 [DOI] [PubMed] [Google Scholar]
- 2.Misra A, Garg A, Abate N, Peshock RM, Stray-Gundersen J, Grundy SM. Relationship of anterior and posterior subcutaneous abdominal fat to insulin sensitivity in nondiabetic men. Obes Res 1997;5:93–99 [DOI] [PubMed] [Google Scholar]
- 3.Snijder MB, Dekker JM, Visser M, et al. . Associations of hip and thigh circumferences independent of waist circumference with the incidence of type 2 diabetes: the Hoorn Study. Am J Clin Nutr 2003;77:1192–1197 [DOI] [PubMed] [Google Scholar]
- 4.Tran TT, Yamamoto Y, Gesta S, Kahn CR. Beneficial effects of subcutaneous fat transplantation on metabolism. Cell Metab 2008;7:410–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wajchenberg BL, Giannella-Neto D, da Silva ME, Santos RF. Depot-specific hormonal characteristics of subcutaneous and visceral adipose tissue and their relation to the metabolic syndrome. Horm Metab Res 2002;34:616–621 [DOI] [PubMed] [Google Scholar]
- 6.Joe AW, Yi L, Even Y, Vogl AW, Rossi FM. Depot-specific differences in adipogenic progenitor abundance and proliferative response to high-fat diet. Stem Cells 2009;27:2563–2570 [DOI] [PubMed] [Google Scholar]
- 7.Koutnikova H, Auwerx J. Regulation of adipocyte differentiation. Ann Med 2001;33:556–561 [DOI] [PubMed] [Google Scholar]
- 8.Siersbæk R, Nielsen R, Mandrup S. Transcriptional networks and chromatin remodeling controlling adipogenesis. Trends Endocrinol Metab 2012;23:56–64 [DOI] [PubMed] [Google Scholar]
- 9.Macotela Y, Emanuelli B, Mori MA, et al. . Intrinsic differences in adipocyte precursor cells from different white fat depots. Diabetes 2012;61:1691–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tchkonia T, Tchoukalova YD, Giorgadze N, et al. . Abundance of two human preadipocyte subtypes with distinct capacities for replication, adipogenesis, and apoptosis varies among fat depots. Am J Physiol Endocrinol Metab 2005;288:E267–E277 [DOI] [PubMed] [Google Scholar]
- 11.Tchkonia T, Giorgadze N, Pirtskhalava T, et al. . Fat depot-specific characteristics are retained in strains derived from single human preadipocytes. Diabetes 2006;55:2571–2578 [DOI] [PubMed] [Google Scholar]
- 12.Ong WK, Sugii S. Adipose-derived stem cells: fatty potentials for therapy. Int J Biochem Cell Biol 2013;45:1083–1086 [DOI] [PubMed] [Google Scholar]
- 13.Gesta S, Tseng YH, Kahn CR. Developmental origin of fat: tracking obesity to its source. Cell 2007;131:242–256 [DOI] [PubMed] [Google Scholar]
- 14.Tchkonia T, Thomou T, Zhu Y, et al. . Mechanisms and metabolic implications of regional differences among fat depots. Cell Metab 2013;17:644–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ong WK, Tan CS, Chan KL, et al. . Identification of specific cell-surface markers of adipose-derived stem cells from subcutaneous and visceral fat depots. Stem Cell Rep 2014;2:171–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vohl MC, Sladek R, Robitaille J, et al. . A survey of genes differentially expressed in subcutaneous and visceral adipose tissue in men. Obes Res 2004;12:1217–1222 [DOI] [PubMed] [Google Scholar]
- 17.Gesta S, Blüher M, Yamamoto Y, et al. . Evidence for a role of developmental genes in the origin of obesity and body fat distribution. Proc Natl Acad Sci U S A 2006;103:6676–6681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tchkonia T, Lenburg M, Thomou T, et al. . Identification of depot-specific human fat cell progenitors through distinct expression profiles and developmental gene patterns. Am J Physiol Endocrinol Metab 2007;292:E298–E307 [DOI] [PubMed] [Google Scholar]
- 19.Sugii S, Kida Y, Berggren WT, Evans RM. Feeder-dependent and feeder-independent iPS cell derivation from human and mouse adipose stem cells. Nat Protoc 2011;6:346–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ye J, Cippitelli M, Dorman L, Ortaldo JR, Young HA. The nuclear factor YY1 suppresses the human gamma interferon promoter through two mechanisms: inhibition of AP1 binding and activation of a silencer element. Mol Cell Biol 1996;16:4744–4753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu YL, Peng XE, Wang D, Chen WN, Lin X. Human liver fatty acid binding protein (hFABP1) gene is regulated by liver-enriched transcription factors HNF3β and C/EBPα. Biochimie 2012;94:384–392 [DOI] [PubMed] [Google Scholar]
- 22.Wongsiriroj N, Jiang H, Piantedosi R, et al. . Genetic dissection of retinoid esterification and accumulation in the liver and adipose tissue. J Lipid Res 2014;55:104–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kane MA, Folias AE, Wang C, Napoli JL. Quantitative profiling of endogenous retinoic acid in vivo and in vitro by tandem mass spectrometry. Anal Chem 2008;80:1702–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balmer JE, Blomhoff R. Gene expression regulation by retinoic acid. J Lipid Res 2002;43:1773–1808 [DOI] [PubMed] [Google Scholar]
- 25.Kedishvili NY. Enzymology of retinoic acid biosynthesis and degradation. J Lipid Res 2013;54:1744–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prentice A, Matthews CJ, Thomas EJ, Redfern CP. The expression of retinoic acid receptors in cultured human endometrial stromal cells and effects of retinoic acid. Hum Reprod 1992;7:692–700 [DOI] [PubMed] [Google Scholar]
- 27.Berry DC, Noy N. All-trans-retinoic acid represses obesity and insulin resistance by activating both peroxisome proliferation-activated receptor beta/delta and retinoic acid receptor. Mol Cell Biol 2009;29:3286–3296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwarz EJ, Reginato MJ, Shao D, Krakow SL, Lazar MA. Retinoic acid blocks adipogenesis by inhibiting C/EBPbeta-mediated transcription. Mol Cell Biol 1997;17:1552–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tchkonia T, Giorgadze N, Pirtskhalava T, et al. . Fat depot origin affects adipogenesis in primary cultured and cloned human preadipocytes. Am J Physiol Regul Integr Comp Physiol 2002;282:R1286–R1296 [DOI] [PubMed] [Google Scholar]
- 30.Chau YY, Bandiera R, Serrels A, et al. . Visceral and subcutaneous fat have different origins and evidence supports a mesothelial source. Nat Cell Biol 2014;16:367–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.D’Ambrosio DN, Clugston RD, Blaner WS. Vitamin A metabolism: an update. Nutrients 2011;3:63–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frey SK, Vogel S. Vitamin A metabolism and adipose tissue biology. Nutrients 2011;3:27–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reichert B, Yasmeen R, Jeyakumar SM, et al. . Concerted action of aldehyde dehydrogenases influences depot-specific fat formation. Mol Endocrinol 2011;25:799–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sima A, Manolescu DC, Bhat P. Retinoids and retinoid-metabolic gene expression in mouse adipose tissues. Biochem Cell Biol 2011;89:578–584 [DOI] [PubMed] [Google Scholar]
- 35.O’Byrne SM, Wongsiriroj N, Libien J, et al. . Retinoid absorption and storage is impaired in mice lacking lecithin:retinol acyltransferase (LRAT). J Biol Chem 2005;280:35647–35657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bonet ML, Canas JA, Ribot J, Palou A. Carotenoids and their conversion products in the control of adipocyte function, adiposity and obesity. Arch Biochem Biophys 2015;572:112–125 [DOI] [PubMed] [Google Scholar]
- 37.Sandell LL, Sanderson BW, Moiseyev G, et al. . RDH10 is essential for synthesis of embryonic retinoic acid and is required for limb, craniofacial, and organ development. Genes Dev 2007;21:1113–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Billings SE, Pierzchalski K, Butler Tjaden NE, Pang XY, Trainor PA, Kane MA, Moise AR . The retinaldehyde reductase DHRS3 is essential for preventing the formation of excess retinoic acid during embryonic development. FASEB J 2013;27:4877–4889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haeseleer F, Huang J, Lebioda L, Saari JC, Palczewski K. Molecular characterization of a novel short-chain dehydrogenase/reductase that reduces all-trans-retinal. J Biol Chem 1998;273:21790–21799 [DOI] [PubMed] [Google Scholar]
- 40.Niederreither K, Subbarayan V, Dollé P, Chambon P. Embryonic retinoic acid synthesis is essential for early mouse post-implantation development. Nat Genet 1999;21:444–448 [DOI] [PubMed] [Google Scholar]
- 41.Zizola CF, Frey SK, Jitngarmkusol S, Kadereit B, Yan N, Vogel S. Cellular retinol-binding protein type I (CRBP-I) regulates adipogenesis. Mol Cell Biol 2010;30:3412–3420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Horswell SD, Fryer LG, Hutchison CE, et al. . CDKN2B expression in adipose tissue of familial combined hyperlipidemia patients. J Lipid Res 2013;54:3491–3505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Okamura M, Kudo H, Wakabayashi K, et al. . COUP-TFII acts downstream of Wnt/beta-catenin signal to silence PPARgamma gene expression and repress adipogenesis. Proc Natl Acad Sci U S A 2009;106:5819–5824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mercader J, Ribot J, Murano I, et al. . Remodeling of white adipose tissue after retinoic acid administration in mice. Endocrinology 2006;147:5325–5332 [DOI] [PubMed] [Google Scholar]
- 45.Shao D, Lazar MA. Peroxisome proliferator activated receptor gamma, CCAAT/enhancer-binding protein alpha, and cell cycle status regulate the commitment to adipocyte differentiation. J Biol Chem 1997;272:21473–21478 [DOI] [PubMed] [Google Scholar]
- 46.Monteiro MC, Wdziekonski B, Villageois P, et al. . Commitment of mouse embryonic stem cells to the adipocyte lineage requires retinoic acid receptor beta and active GSK3. Stem Cells Dev 2009;18:457–463 [DOI] [PubMed] [Google Scholar]
- 47.Phillips BW, Vernochet C, Dani C. Differentiation of embryonic stem cells for pharmacological studies on adipose cells. Pharmacol Res 2003;47:263–268 [DOI] [PubMed] [Google Scholar]
- 48.Okabe Y, Medzhitov R. Tissue-specific signals control reversible program of localization and functional polarization of macrophages. Cell 2014;157:832–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.