Abstract
Obesity is associated with insulin resistance, but significant variability exists between similarly obese individuals, pointing to qualitative characteristics of body fat as potential mediators. To test the hypothesis that obese, insulin-sensitive (IS) individuals possess adaptive adipose cell/tissue responses, we measured subcutaneous adipose cell size, insulin suppression of lipolysis, and regional fat responses to short-term overfeeding in BMI-matched overweight/obese individuals classified as IS or insulin resistant (IR). At baseline, IR subjects exhibited significantly greater visceral adipose tissue (VAT), intrahepatic lipid (IHL), plasma free fatty acids, adipose cell diameter, and percentage of small adipose cells. With weight gain (3.1 ± 1.4 kg), IR subjects demonstrated no significant change in adipose cell size, VAT, or insulin suppression of lipolysis and only 8% worsening of insulin-mediated glucose uptake (IMGU). Alternatively, IS subjects demonstrated significant adipose cell enlargement; decrease in the percentage of small adipose cells; increase in VAT, IHL, and lipolysis; 45% worsening of IMGU; and decreased expression of lipid metabolism genes. Smaller baseline adipose cell size and greater enlargement with weight gain predicted decline in IMGU, as did increase in IHL and VAT and decrease in insulin suppression of lipolysis. Weight gain in IS humans causes maladaptive changes in adipose cells, regional fat distribution, and insulin resistance. The correlation between development of insulin resistance and changes in adipose cell size, VAT, IHL, and insulin suppression of lipolysis highlight these factors as potential mediators between obesity and insulin resistance.
Introduction
The prevalence of obesity has increased dramatically over the past three decades (1), contributing to increasing prevalence of type 2 diabetes (2). This relationship is largely mediated by insulin resistance, which affects ∼30% of the U.S. adult population (3) and is linearly related to adiposity (4). This association appears to be causal because dietary and surgical weight loss leads to sustained improvement in insulin sensitivity among IR humans (5). Overweight and moderately obese humans, who comprise 60% of U.S. adults, are metabolically heterogeneous, however, and insulin resistance can vary more than sixfold at any given BMI within this range (6). Why some individuals develop insulin resistance in the setting of excess body weight and others appear to be “protected” has not been fully elucidated. Given that obesity is not synonymous with insulin resistance, it is likely that biological characteristics of adipose tissue at the cellular and molecular level and/or regional distribution are contributors to insulin sensitivity beyond fat mass per se.
We previously demonstrated that insulin resistant (IR) humans, compared with BMI-matched insulin sensitive (IS) humans, demonstrated larger adipose cells, accumulation of small adipose cells, and decreased expression of adipocyte-differentiation genes (7,8). These findings were consistent with the hypothesis (9) that larger adipose cells reflect impaired differentiation and recruitment of new adipose cells as well as the observation that adipose cell size independently predicts insulin resistance and the development of type 2 diabetes (10). We hypothesized that during weight gain, impairment in adipose cell differentiation and recruitment, with a concomitant decrease in the capacity to store triglyceride (TG) in subcutaneous fat, would place affected individuals at risk for insulin resistance. This hypothesis derives support from several lines of data. First, in human and animal lipodystrophy models, absence of subcutaneous fat is associated with ectopic fat and severe insulin resistance (11,12), and in fatless mice, transplantation of subcutaneous fat restores IS (11). Second, thiazolidinediones, which promote adipocyte differentiation, redistribute fat from visceral to subcutaneous depots and reduce intrahepatic fat while dramatically improving insulin sensitivity (13–15). Finally, cross-sectional studies demonstrate decreased expression of adipogenic genes in adipose tissue of IR subjects or subjects with diabetes compared with healthy control subjects (7,16,17).
To test the hypothesis that the relative inability of adipocytes to store TG in subcutaneous adipose tissue (SAT) contributes to obesity-related insulin resistance, we designed an overfeeding study in overweight-to-moderately obese individuals classified as IS or IR, whose adipose cells would be near or at maximal storage capacity and who would likely exhibit differential adipose cell and tissue responses to additional TG storage demands. We hypothesized that in response to modest weight gain, the IR subjects would demonstrate characteristics of impaired adipocyte differentiation and TG storage, including adipocyte hypertrophy, lipolysis/increased circulating free fatty acid (FFA), fat deposition in visceral and intrahepatic depots, and worsening insulin resistance. IS subjects, we hypothesized, due to enhanced ability to differentiate and recruit new adipose cells, would be protected from adipocyte hypertrophy, ectopic fat deposition, lipolysis, and/or worsening of insulin resistance.
Research Design and Methods
Subjects
Healthy “overweight” adults, aged 30–60 years, from the San Francisco Bay Area were recruited by local newspaper advertisements. After an overnight fast, potential subjects were screened in the Stanford Clinical Translational Research Unit (CTRU), where written, informed consent was obtained, medical history was obtained, and physical examination and screening laboratory tests were performed. Eligibility requirements included BMI 25–35 kg/m2, stable body weight within 2 kg during the prior 3 months, fasting plasma glucose <126 mg/dL in the absence of medications known to alter blood glucose/insulin sensitivity, and no history of major organ disease, inflammatory condition, malignancy, uncontrolled hypertension (>160/90 mmHg), bariatric surgery or liposuction, active psychiatric condition, use of weight loss medication, or intense physical activity >7 h per week. Hematocrit, liver enzymes, and TG were required to be ≥30%, less than three times the upper limit of normal, and <400 mg/dL, respectively.
Measurements Performed at Baseline and at Peak Body Weight
Anthropometric Measures
Height was measured with a standardized CTRU stadiometer at screening. Weight was averaged from three separate visits after overnight fasting. BMI was calculated from height and weight (kg/m2), and percentage of body fat (%BF) was calculated using the Deurenberg formula (18,19). Waist circumference was measured, with arms raised at end expiration, between the iliac crest and the bottom of the rib cage. Morning systolic and diastolic blood pressures were averaged from three separate visits. Subjects who met the inclusion criteria underwent baseline metabolic testing for insulin sensitivity, meal tolerance test, SAT biopsy, and radiologic measures to quantify abdominal, intraabdominal, thigh, and intrahepatic fat, as described below. All baseline measures were repeated at peak weight.
Insulin-Mediated Glucose Uptake
Whole-body insulin-mediated glucose uptake (IMGU) was quantified using the modified (20) insulin-suppression test as originally described and validated for measurement of muscle (21,22) and adipose (23) insulin resistance. Briefly, after an overnight fast, subjects were infused for 180 min with octreotide (0.27 µg/m2/min), insulin (25 mU/m2/min), and glucose (240 mg/m2/min). Blood was drawn at 10-min intervals from 150 to 180 min of the infusion to measure plasma glucose (oximetric method) and insulin (radioimmunoassay) concentrations: the mean of these four values comprised the steady-state plasma glucose (SSPG) and insulin concentrations for each individual. At steady state, insulin concentrations (65 μU/mL) are similar in all subjects, and the SSPG provides a direct relative measure of IMGU: the higher the SSPG concentration, the more IR the individual. Although the SSPG is distributed continuously, for the purpose of this study, we defined IS as SSPG <120 mg/dL and IR as SSPG ≥150, largely to provide separation between the two groups. Individuals with SSPG between 120 and 150 mg/dL were excluded.
Insulin Suppression of Lipolysis
We have previously validated the test above for quantification of in vivo insulin suppression of lipolysis: higher FFA concentrations during steady state with fixed insulin concentrations reflect resistance to insulin suppression of lipolysis (23).
Meal Tolerance Test
After an overnight fast, subjects consumed a standardized test meal consisting of 43% carbohydrate, 42% fat, and 15% protein. The meal was prepared by the CTRU research kitchen and administered over 20 min. Blood was drawn from subjects before and hourly after the meal. The area under the curve (AUC), using the trapezoidal method, was calculated for plasma FFA and insulin concentrations.
Quantification of Regional Fat Mass and Intrahepatic Fat
The volume of SAT, visceral adipose tissue (VAT), and midthigh fat was quantified with CT scans (California Advanced Imaging, Atherton, CA), as previously described (24). Percent VAT was calculated as VAT/(VAT + SAT) × 100. Intrahepatic lipid (IHL) was quantified using 1H-MRS on a 3.0T GE Healthcare scanner. Briefly, scout images were obtained to identify three 15 × 15 × 20 mm3 voxels within the right lobe that is devoid of biliary or vascular structures. Intrahepatic TG content was determined using a point-resolved spectroscopy technique (PROBE/SV, GE Healthcare). Two sequential measurements were made from each voxel and results averaged. Peak areas from resulting spectra were quantified using Spectral Analysis software (GE Healthcare) with internal water referencing. Percentage of IHL content was calculated as the fat-to-water ratio (area under the resonance peaks) and expressed in decimal form.
Adipose Tissue Biopsy
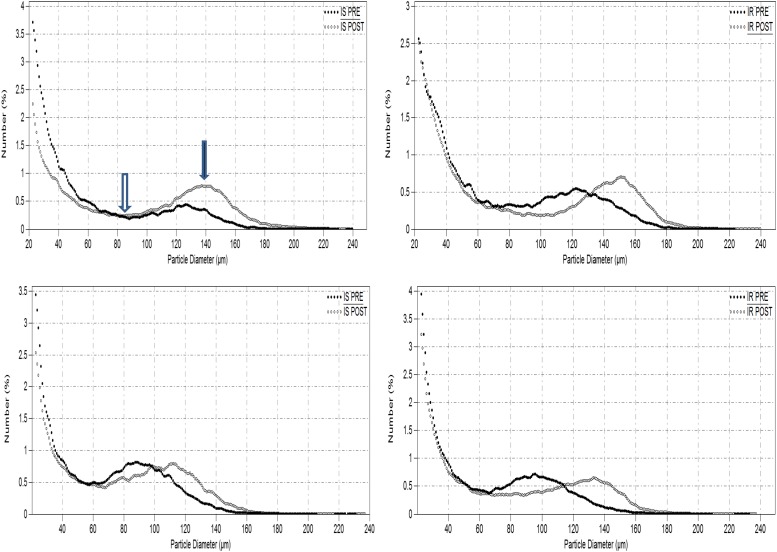
Adipose tissue was obtained under sterile conditions and local anesthesia. A 1-cm scalpel incision was made inferior to the umbilicus, from which 2 g of SAT was removed. Two samples of 20–30 mg of tissue were immediately fixed in osmium tetroxide and incubated in a water bath at 37°C for 48 h, as previously described (7), after which adipose cell size was determined using a Multisizer 3 Coulter Counter (Beckman Coulter, Miami, FL) with a 400-μm aperture. The effective cell-size range using this aperture is 20 to 240 μm. Data averaged from the two duplicate tissue samples were expressed as cell count at each cell diameter, yielding a frequency histogram.
Analysis of adipose cell-size distribution from Multisizer graphs (Fig. 1A) entailed, for each subject, identification of the “nadir,” defined as the midway point between which two cell populations were typically present in increased frequency. “Percent small cells” was defined as the percentage of adipose cells below the nadir, and “peak diameter” was defined as the diameter at which the frequency of the large adipose cell population peaked. We have previously shown that these measures are more descriptive than the mean or median cell size (7,8).
Figure 1.
Representative Beckman Multisizer profiles for adipose cell size distribution are shown in two IS and two IR subjects at baseline (PRE) and at peak weight (POST). Nadir, indicated by open arrow, separates two populations of small cells, distributed as a double exponential tail to the left of nadir, and large cells, distributed as a Gaussian curve. The black arrow indicates peak diameter (center of Gaussian curve).
Measurement of Adipose Tissue Gene Expression
From a larger nanostring analysis of 20 subjects in this cohort, we selected 10 lipogenic genes to analyze for insulin resistance and response to weight gain. RNA was isolated by the Hybrid-R RNA extraction kit (GeneAll BioTechnologies, Palo Alto, CA) according to the manufacturer’s instructions. RNA amount and quality were checked by Bioanalyzer analysis. RNA samples were sent to the Stanford Genomic Core Facility for analysis. Inter- and intracartridge replicates were included in the assay. Normalization of nCounter results was done using nSolver Analysis Software, version 2.5 (NanoString Technologies), according to the manufacturer’s guidelines. RNA counts were normalized using the expression of four reference genes (GAPDH, CLTC, GUSB, and HPRT1) and positive controls in each sample, as previously described (25). Baseline gene expression for each subject was expressed relative to a single IS control subject, and fold-change from baseline at peak weight was calculated for each individual relative to their baseline value.
Dietary Intervention
All subjects were given a controlled hypercaloric diet with calorie excess/day to induce 3.2-kg weight gain over 4 weeks, followed by 1 week of weight stabilization before repeat of metabolic, radiologic, and adipose measures. Before the baseline biopsy, subjects were instructed to avoid extreme macronutrient composition, dining out more than once weekly, and use of alcoholic beverages of more than one per day for women or more than two per day for men. Up to two fructose-sweetened beverages per day were allowed before and during weight gain. Three-day food diaries were obtained at baseline and at weekly visits during the study, and data were entered into the ESHA Food Processor for data analysis.
After the baseline biopsy, subjects began the weight gain period by adding an average of 880 calories/day in addition to their usual daily intake. Exact caloric excess, administered by a research dietitian as snacks and beverages, with fixed macronutrient composition of 50% carbohydrate, 35% fat (<7% saturated fat), and 15% protein, was calculated individually using the Harris Benedict equation (26) for each subject to attain weight gain of 0.8 kg/week (3.2 kg total). The usual diet was required to be the same as baseline. Subjects were prohibited from changing their physical activity pattern or starting new medications during the study. Weekly visits with a study dietitian for weight checks, return of food diary, dispensation of snacks, and caloric adjustment, if needed, ensured compliance. After 1 week of weight maintenance, subjects underwent peak weight biopsy, metabolic/radiologic tests. For ethical reasons, subjects then underwent supervised weight loss for 6–8 weeks so that they returned to their baseline weight. Per protocol design, subjects were not restudied after weight loss.
Statistical Analysis
Baseline comparison of IR versus IS subjects used unpaired Student t tests. Measures related to fat distribution, FFA concentrations, and adipose cell size were adjusted for %BF via ANCOVA to account for residual confounding. Student t tests were used to compare within-group change from baseline to peak weight for IR and IS groups. Comparison of between-group differences in variable changes with weight gain used ANCOVA with adjustment for Δ%BF. Independent predictors of ΔSSPG were identified via general linear regression models including the following variables: IS/IR group, predictor of interest, interaction between IS/IR group and the predictor of interest, and Δ%BF. For variables with significant “group” interactions with the predictor of interest, IS and IR groups were analyzed separately, using comparable models.
Results
Comparison of IS and IR Groups at Baseline
The study enrolled 35 subjects, and 15 IS and 16 IR completed the study. One IS and three IR subjects dropped out after 2 weeks of overfeeding due to physical discomfort or noncompliance with diet. Reported total dietary macronutrient composition was nearly identical between groups at baseline (protein, 17%; carbohydrate, 47%; fat, 35%; saturated fat, 13%), with little change at peak weight: for both groups, carbohydrate intake increased by 1–2% and saturated fat decreased by 2–3%. Subject characteristics are reported in Table 1. Age, sex, BMI, and %BF did not differ significantly between the groups. Although race did not differ statistically by group, because there were more Asians in the IR group, comparison of all baseline variables in Asians versus non-Asians within this group was undertaken, revealing virtually identical measures of all fat-related indices, including adipose cell size. By design, the IS group had a lower mean SSPG than the IR group. Other factors that differed between groups included fasting plasma glucose and TG, which were significantly higher in the IR group, and HDL cholesterol, which was significantly lower in the IR group. The FFA and insulin AUCs during the meal tolerance test were significantly higher in the IR group. Plasma FFAs during steady state of the infusion study were higher in the IR group, but the difference fell short of statistical significance.
Table 1.
Comparison of demographic and clinical characteristics
| Variable | Baseline |
Peak weight |
P ∆** | ||||
|---|---|---|---|---|---|---|---|
| IS (n = 15) | IR (n = 16) | P | IS | IR | P | ||
| Age (years) | 54 ± 8 | 57 ± 6 | 0.27 | — | — | — | — |
| Sex (n) | 0.42 | — | — | — | — | ||
| Male | 8 | 8 | |||||
| Female | 7 | 8 | |||||
| Race (n) | 0.06 | — | — | — | — | ||
| Caucasian | 12 | 9 | |||||
| Asian | 0 | 5 | |||||
| Black | 3 | 1 | |||||
| Hispanic | 0 | 1 | |||||
| BMI (kg/m2) | 29.3 ± 2.4 | 30.7 ± 2.7 | 0.11 | 30.5 ± 2.6§ | 31.5 ± 2.7§ | 0.31 | 0.22 |
| Weight (kg) | 86.2 ± 10.1 | 89.4 ± 11.2 | 0.42 | 89.6 ± 10.3§ | 92.1 ± 11.1§ | 0.52 | 0.23 |
| %BF | 37.0 ± 7.0 | 39.1 ± 7.7 | 0.43 | 38.3 ± 7.2§ | 40.2 ± 7.8§ | 0.48 | 0.38 |
| Waist (cm) | 100 ± 7 | 105 ± 6 | 0.049 | 107 ± 7‡ | 108 ± 7‡ | 0.41 | 0.48 |
| SSPG (mg/dL) | 82 ± 24 | 200 ± 40 | <0.001 | 118 ± 41§ | 216 ± 35† | <0.001 | 0.01 |
| Fasting glucose (mg/dL) | 94 ± 9 | 102 ± 11 | 0.02 | 98 ± 9 | 106 ± 15 | 0.10 | 0.80 |
| Blood pressure (mmHg) | |||||||
| Systolic | 122 ± 13 | 125 ± 7 | 0.47 | 127 ± 12 | 124 ± 14 | 0.56 | 0.62 |
| Diastolic | 77 ± 5 | 81 ± 6 | 0.05 | 80 ± 5† | 81 ± 5 | 0.66 | 0.20 |
| TG (mg/dL) | 80 ± 39 | 138 ± 82 | 0.007 | 100 ± 38 | 185 ± 115‡ | 0.54 | 0.13 |
| Cholesterol (mg/dL) | 190 ± 34 | 181 ± 27 | 0.78 | 194 ± 29† | 197 ± 26† | 0.74 | 0.79 |
| VLDL | 16.4 ± 8.2 | 20.0 ± 7.6 | 0.037 | 26.3 ± 12.4 | 37.0 ± 23.1‡ | 0.025 | 0.13 |
| LDL | 111 ± 36 | 108 ± 22 | 0.94 | 108 ± 30 | 114 ± 24 | 0.01 | 0.26 |
| HDL | 63 ± 18 | 50 ± 14 | 0.04 | 66 ± 22 | 50 ± 17 | 0.027 | 0.69 |
| SAT (cm3)* | 147 ± 54 | 140 ± 34 | 0.04 | 162 ± 51 | 148 ± 37† | 0.018 | 0.68 |
| VAT (cm3)* | 37 ± 22 | 64 ± 16 | <0.001 | 44 ± 28‡ | 73 ± 27 | 0.01 | 0.76 |
| %VAT* | 20 ± 12 | 32 ± 8 | <0.001 | 22 ± 13‡ | 33 ± 11 | 0.005 | 0.86 |
| Thigh (cm3)* | 61 ± 24 | 54 ± 22 | 0.02 | 68 ± 21 | 54 ± 21 | 0.001 | 0.86 |
| IHL (lipid/H2O)* | 0.03 ± 0.21 | 0.23 ± 0.31 | 0.02 | 0.07 ± 0.04‡ | 0.30 ± 0.22‡ | 0.002 | 0.03 |
| FFA (mmol/L) | |||||||
| Fasting* | 374 ± 80 | 374 ± 106 | 0.16 | 369 ± 169 | 348 ± 88 | 0.90 | 0.75 |
| AUC* | 958 ± 266 | 1,234 ± 237 | 0.01 | 919 ± 233 | 1,152 ± 202 | 0.01 | 0.77 |
| Insulin suppression of lipolysis* | 71 ± 50 | 123 ± 69 | 0.09 | 94 ± 56† | 124 ± 86 | 0.43 | 0.046 |
| Insulin AUC (μU/mL) | 92 ± 40 | 151 ± 89 | 0.01 | 110 ± 43§ | 199 ± 114‡ | 0.01 | 0.023 |
Data are shown as mean ± SD of IS vs. IR subjects at baseline, peak weight, and changes for each variable after weight gain, with change from baseline denoted by symbols in peak weight column for each group. AUC was calculated by the trapezoidal method.
*Regional fat depot mass was adjusted for sex and %BF (baseline and peak weight); IHL and all FFA comparisons were adjusted for %BF (baseline or peak weight) to minimize potential confounding;
†Paired t test comparing peak weight to baseline P < 0.05;
‡Paired t test comparing peak weight to baseline P < 0.01;
§Paired t test comparing peak weight to baseline P < 0.001;
**ANCOVA comparing ∆absolute value between IR and IS groups with adjustment for Δ%BF. Because baseline SSPG differed substantially (by design), comparison was for %change SSPG.
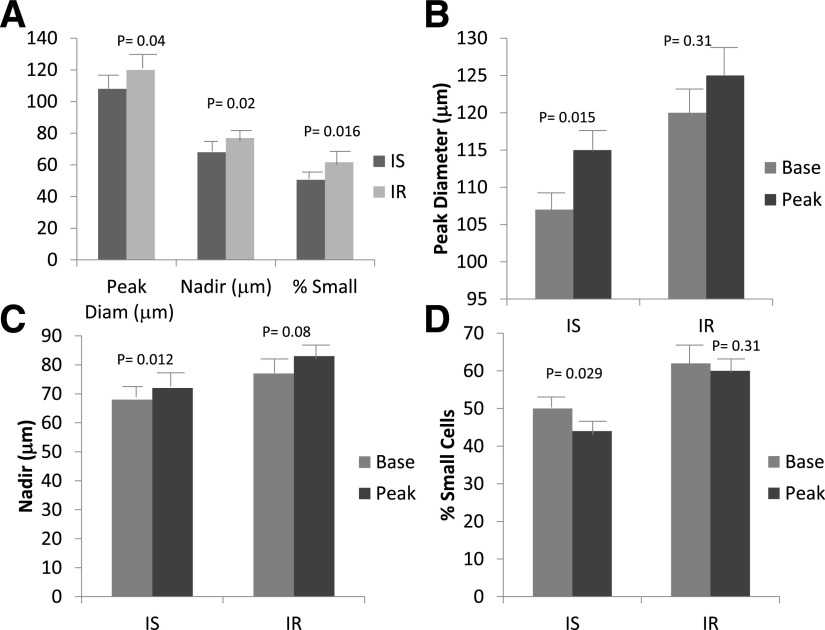
Comparisons of baseline regional and intrahepatic fat (IHL), adjusted for BMI and sex, are reported in Table 1. SAT and thigh fat were significantly greater in the IS group. VAT and %VAT were significantly greater in the IR subgroup, and IHL was nearly eightfold greater. Changes in these variables with weight gain and between-group differences at peak weight are discussed below. Baseline differences in adipose cell size and distribution are shown in Fig. 2A. The IR subjects had a significantly larger peak diameter, higher nadir, and greater percentage of small cells than the IS subjects.
Figure 2.
Measures of adipose cell size and distribution in IS and IR subjects at baseline (A) and changes with weight gain for peak diameter (B), nadir (C), and percentage of small cells (D). Mean ± SEM, analyzed via ANCOVA with adjustment for %BF (A) or paired Student t test (B, C, and D). Diam, diameter.
Comparisons of Changes in Clinical Variables From Baseline
Changes in variables from baseline to peak weight for IS and IR groups are reported in Table 1, with statistical significance indicated by symbols in the peak weight column. Weight gain was linear for all subjects. Body weight increased significantly, by 3.9% and 3.0% in the IS and IR groups, respectively, and %BF increased by 5.6% and 5.0%: the change between groups was not statistically significant. Waist circumference increased significantly, by 5.5% and 3.2% in IS and IR groups, respectively, also not statistically significantly different between groups. Insulin resistance, measured by SSPG, increased significantly in both groups (45% in the IS and 8% in the IR group), and to a significantly greater degree in the IS group than in the IR group. Fasting plasma glucose and systolic blood pressure did not change significantly in either group. Diastolic blood pressure increased significantly in the IS group alone. Fasting plasma cholesterol increased significantly in both groups, whereas fasting TG and VLDL concentrations increased significantly in the IR group only. Fasting and FFA-AUC concentrations decreased slightly but not statistically significantly in both groups. Insulin suppression of lipolysis worsened significantly in the IS subgroup alone, and the insulin AUC during the meal tolerance test increased significantly in both groups but to a significantly greater degree in the IR group.
Changes in regional fat distribution revealed an increase in SAT in the IS and IR groups, which reached statistical significance only in the IR group. However, VAT and %VAT, although higher at baseline in the IR group, increased significantly only in the IS subjects. Changes in thigh fat were not statistically significant in either group. Finally, IHL increased significantly in both groups with weight gain, with a larger absolute but smaller relative (30% vs. 133%) gain in the IR group.
Changes in adipose cell size and distribution after weight gain, shown in Fig. 2B–D, also differed according to IS/IR group. The peak diameter increased in both groups, but this change was only statistically significant in the IS group (108 ± 15 to 115 ± 14 μm [P = 0.015] vs. 120 ± 16 to 125 ± 13 μm [P = 0.31]). The nadir shifted significantly to the right in the IS group (P = 0.012) but did not change in the IR group, and the percentage of small cells decreased significantly in the IS (P = 0.029) but not in the IR group.
Gene Expression
Similar to clinical and cell-size data, genes reflecting metabolically active adipose cells were upregulated at baseline in the IS compared with the IR group, after adjustment for %BF (Table 2). These included lipid metabolism (lipogenic and lipolytic) and glucose uptake genes. Expressions of lipogenic and lipolytic genes were highly correlated (data not shown). Also similar to clinical and cell size data, expression of lipid metabolism genes decreased significantly with overfeeding in the IS group only, whereas GLUT4 increased significantly in the IR but not the IS group, thus demonstrating significantly divergent patterns of change in IS versus IR subjects.
Table 2.
Relative expression of genes related to glucose uptake and lipid metabolism in adipose tissue from IS and IR subjects at baseline and changes with weight gain
| Gene | IS baseline* (n = 10) | IR baseline* (n = 10) | P baseline | IS ∆ | IR ∆ | P ∆ |
|---|---|---|---|---|---|---|
| Glut4 | 1.1 ± 0.52 | 0.62 ± 0.41 | 0.02 | −0.08 ± 0.33 | 0.47 ± 0.50‡ | 0.01 |
| FABP4 | 1.08 ± 0.22 | 0.89 ± 0.28 | 0.08 | −0.21 ± 0.21‡ | 0.13 + 0.41 | 0.04 |
| PEPCK | 1.00 ± 0.22 | 0.67 ± 0.24 | 0.003 | −0.18 ± 0.24† | 0.16 ± 0.37 | 0.01 |
| ACC1 | 1.67 ± 1.1 | 1.13 ± 0.74 | 0.21 | 0.29 ± 0.99 | 0.49 ± 0.72 | 0.53 |
| CD36 | 1.21 ± 0.40 | 1.08 ± 0.31 | 0.37 | −0.03 ± 0.46 | 0.08 ± 0.52 | 0.52 |
| FATP1 | 1.00 ± 0.17 | 0.85 ± 0.22 | 0.06 | −0.14 ± 0.19† | 0.10 ± 0.36 | 0.08 |
| LPL | 1.44 ± 0.55 | 1.22 ± 0.40 | 0.29 | 0.02 ± 0.61 | 0.23 ± 0.53 | 0.39 |
| HSL | 1.35 ± 0.37 | 1.02 ± 0.35 | 0.049 | −0.19 ± 0.30 | 0.30 ± 0.56 | 0.03 |
| ATGL | 1.47 ± 0.32 | 1.07 ± 0.29 | 0.01 | −0.30 ± 0.39† | 0.17 ± 0.26 | 0.006 |
*Baseline comparison adjusted for baseline %BF; change comparison adjusted for ∆%BF. Relative expression at peak weight was normalized to baseline expression for each gene and change calculated as the difference between peak weight and baseline relative expression;
†Paired t test comparing peak weight to baseline P < 0.05;
‡Paired t test comparing peak weight to baseline P < 0.01.
Baseline Predictors of ΔSSPG
Of the baseline predictors of interest, including VAT, SAT, IHL, peak diameter, and percentage of small cells, after adjustment for Δ%BF, the only independent predictor of ΔSSPG was peak diameter of adipose cells, which was inversely associated with ΔSSPG: the larger the cells at baseline, the less the SSPG changed with weight gain (r = −0.62, P = 0.008).
Change in Variables as Predictors of ΔSSPG
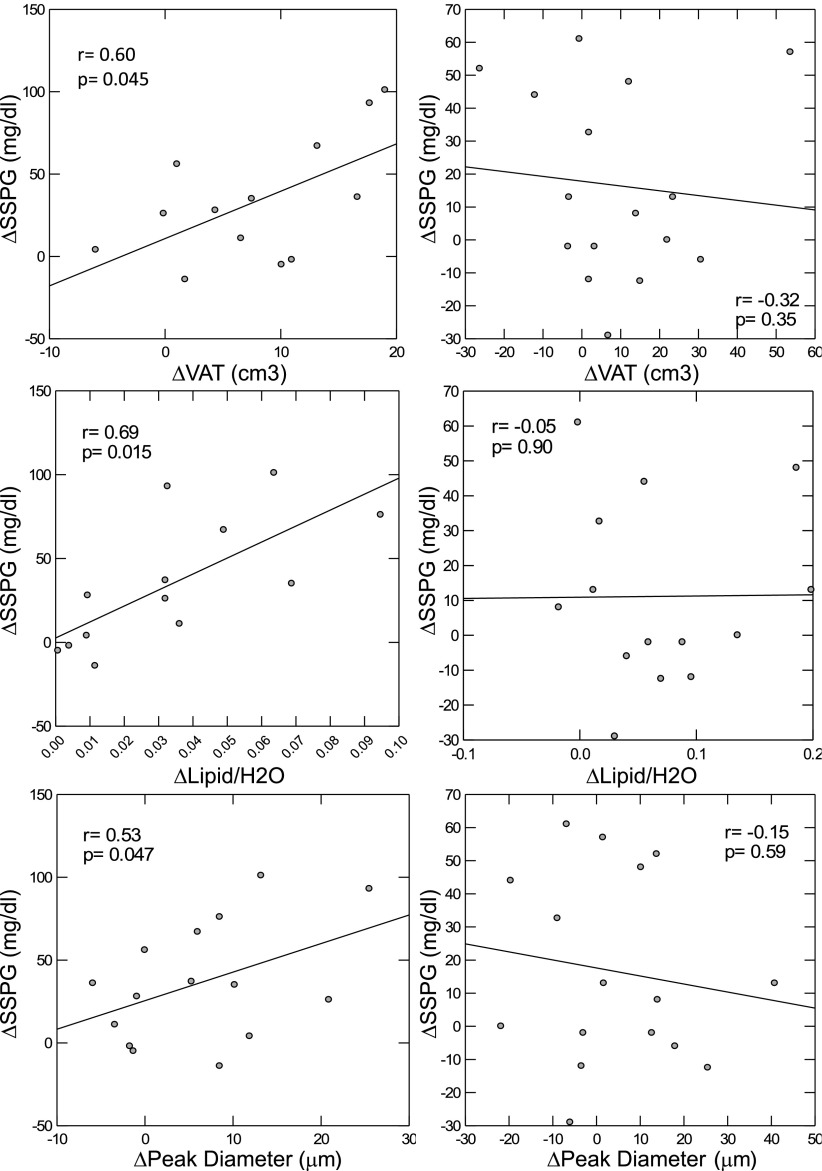
Weight gain–related changes in four variables were significantly associated with deterioration in insulin resistance after adjusting for Δ%BF and IR/IS group. In the group as a whole, ΔVAT, ΔIHL, and Δpeak diameter all significantly predicted ΔSSPG (P = 0.028, P = 0.004, and P = 0.04, respectively), and all three demonstrated an interaction with IS/IR group (P = 0.025, P = 0.006, and P = 0.04, respectively). Linear regression analyses by IR/IS group demonstrated that in the IS group alone, ΔVAT, ΔIHL, and Δpeak diameter were directly and significantly associated with ΔSSPG, as shown in Fig. 3. ΔInsulin suppression of lipolysis (FFA concentration at steady state) also independently predicted ΔSSPG, (r = 0.60, P = 0.04), with no interaction by group. Other adipocyte, regional fat, and FFA variables did not predict ΔSSPG.
Figure 3.
Change in insulin resistance, as measured by SSPG, as a function of ΔVAT, IHL (Lipid/H2O), and peak diameter of adipose cells in IS (left) and IR (right) humans. General linear regression with adjustment for Δ%BF. Standardized r and P values are reported.
Ancillary Analyses
Exploration of whether adipose cell characteristics predicted change in regional fat depots revealed that smaller baseline peak diameter and higher percentage of small cells both predicted ΔVAT independent of Δ%BF (r = 0.42, P = 0.03, and r = 0.40, P = 0.03, respectively). ΔPeak diameter predicted ΔIHL (r = 0.81, P = 0.029) with a significant group interaction (P = 0.017), but by-group analysis revealed a significant association in the IS group only (P = 0.026).
Discussion
In contrast to our hypothesis that IS subjects would demonstrate adaptive adipose tissue and metabolic responses to weight gain, we found the opposite: IS subjects exhibited maladaptive adipose tissue responses and developed clinically significant insulin resistance. Adipose mass expanded in the visceral and intrahepatic depots, and adipose cell hypertrophy was evident. FFA concentrations under steady-state insulin conditions increased by 133%, indicating resistance to insulin suppression of lipolysis, whereas AUC FFA concentrations after a standardized test meal were not increased, likely due to concomitant increases in the insulin AUC. Muscle insulin resistance, as measured by SSPG, worsened by 45% in the IS group compared with 8% in the IR group with similar weight gain. Interestingly, the magnitude of change in all of these variables, including VAT, IHL, adipose cell peak diameter, and insulin suppression of lipolysis, significantly predicted the degree to which SSPG worsened. Further, these associations were independent of weight gain per se, implying that differential cellular and regional fat distribution patterns of adipose tissue may contribute to the metabolic heterogeneity of obesity.
Interestingly, with the exception of IHL, which increased significantly in both groups, and SAT, which increased significantly only in the IR group, significant changes in adipose tissue variables were limited to the IS group. Further, with the exception of Δinsulin suppression of lipolysis, which correlated with ΔSSPG in both groups, correlations between the change in these variables (VAT, IHL, peak diameter) and ΔSSPG were also limited to the IS group.
Despite the relatively adverse adipose and metabolic responses to weight gain in the IS group, the IS subjects demonstrated a significantly healthier profile at baseline, including waist circumference, VAT, %VAT, IHL, FFA-AUC concentrations, adipose cell size, nadir, and percentage of small cells, as well as fasting plasma glucose, HDL cholesterol, TG, and diastolic blood pressure. Thus, our study reveals a paradox: although the IS subjects had a healthier adipose and metabolic profile at baseline, with the exception of TG and VLDL cholesterol, they decompensated to a greater degree with weight gain. Expression of lipid and glucose metabolism genes in SAT closely mirrored what was observed clinically: IS subjects demonstrated significantly higher expression of GLUT4 and multiple genes related to lipid metabolism at baseline, but these declined significantly with weight gain, whereas the IR group had lower expression at baseline and no change with weight gain, excepting an increase in GLUT4. As adipose cell gene expression and cell size in the SAT of IS subjects approached those seen in IR subjects, systemic insulin suppression of lipolysis deteriorated significantly, as did muscle insulin resistance and fat deposition in ectopic and visceral depots. Although gene expression does not always reflect function, together these observations make a strong case for disordered lipid and glucose metabolism in adipose cells as a mediator between weight gain and development of systemic insulin resistance.
These findings have important clinical implications because they demonstrate that overweight individuals who are metabolically healthy can quickly become metabolically unhealthy as a result of weight gain. Although it has been generally accepted that overweight-to-moderately obese individuals can be metabolically healthy or unhealthy, to what degree an individual can switch “categories” is not known. Indeed, it is perhaps best not to consider metabolic health in terms of categories but rather as a continuum along which all individuals can move with different slopes and/or threshold BMIs. Among our overweight IS subjects, the degree of adipose and metabolic decompensation varied, further highlighting this concept. The only predictor appeared to be smaller baseline adipose cell size. What BMI constitutes the “tipping point” for an individual to shift from metabolically healthy to unhealthy is not currently known. The current results suggest that BMI and weight gain per se are inadequate measures of this risk and that weight gain causes metabolic decompensation at different thresholds in different individuals. Identifying an individual’s tendency to respond poorly to weight gain and/or whether they have approached their metabolic “tipping point” would be clinically useful for diabetes prevention.
Results presented in this study also have important biological implications. The mechanisms linking excess body fat to insulin resistance are still not clear. One hypothesis contends that adipocyte hypertrophy causes insulin resistance. Prior support for this hypothesis is found in animal, in vitro, and human cross-sectional studies. The current results extend these data by demonstrating that the degree of adipose cell enlargement resulting from weight gain in healthy overweight humans is independently associated with the development of insulin resistance. By inference, adipocyte hypertrophy reflects impaired differentiation of new cells in the setting of increased TG storage demands. The scope of “impaired differentiation” includes decreased proliferation and/or commitment of new preadipocytes, differentiation of preadipocytes, and dysfunctional terminal maturation resulting in decreased TG storage capacity. An alternative explanation for enlarged adipose cells would be inherently increased capacity for TG storage, potentially due to differences in lipid metabolism, blood supply, or external restriction.
The current results suggest that hypertrophy of adipose cells is not due to increased capacity for TG storage capacity, because the lipid metabolism genes declined as cells enlarged, systemic metabolic health declined, and ectopic fat increased. Other insights to be gleaned from the current results include a significant decrease in the percentage of small cells in the IS group, indicating that perhaps early differentiation/recruitment from preadipocytes is impaired and a contributor to the observed adipocyte enlargement. Overfeeding studies in lean mice are consistent with this notion: early increase in adipose cell size is followed by stable cell size with increased number of small adipose cells (27). Furthermore, overfeeding Zucker fa/fa rats causes oscillations between adipose cell hypertrophy and hyperplasia, suggesting that when adipose cells reach their maximal size, hyperplasia is triggered to provide additional fat storage and prevent lipotoxicity (28). Data in humans are scant, but a small overfeeding study in obese Pima Indians showed no change in mean adipose cell size but rather an increase in small adipose cell number, presumably because the large adipose cells were already enlarged to capacity (29), perhaps similar to our IR subgroup that had larger adipose cells at baseline.
That our IR subjects did not experience significant adipose cell enlargement or worsening of IS is of interest, especially because they were not at the upper limit of the SSPG distribution. They similarly did not experience significant expansion of VAT, although SAT and IHL increased significantly. Indeed, the composite increase in measured fat (SAT, VAT, thigh) was 17 cm3 in the IR versus 29 cm3 in IS group, despite similar weight gain, implying potential deposition of fat in unmeasured depots in the IR group. The high capacity among IR subjects to store fat in the liver may reflect a tendency to store fat in other unmeasured ectopic sites, which could explain the lack of adipocyte hypertrophy and relatively low “measured” fat mass expansion. Alternatively, relative protection from further adipocyte hypertrophy and metabolic decompensation among IR individuals might have resulted from upregulation of adipocyte differentiation genes in chronically stressed adipocytes, consistent with significant SAT increase among IR subjects.
Our findings are partly supported by two publications (30,31) showing that individuals with smaller subcutaneous abdominal adipose cells at baseline experienced greater cellular enlargement (30) and greater reduction in IS (31). In these studies, respectively, subjects were leaner (22.1 ± 0.5 and 25.5 ± 2.3 kg/m2), were younger (27–29 years), and had a more prolonged weight gain period (4.6 and 7.6 kg over 8 weeks). Furthermore, like our findings, the second study showed that smaller baseline adipose cell size predicted a greater increase in %VAT but not IHL or SAT. Neither change in adipose cell size nor relationship to development of insulin resistance was reported. Another overfeeding study (32) in considerably heavier subjects (BMI 36.6 ± 4 kg/m2), classified as metabolically normal or metabolically abnormal obese (MAO), showed that MAO, similar to our IR group, exhibited greater increases in absolute IHL in response to weight gain, TG and VLDL concentrations, and hepatic insulin resistance. Unlike our results, adipose tissue and muscle insulin sensitivity both worsened in the MAO subjects. Calorie excess derived from fast food restaurants or heavier baseline BMI in these subjects may have contributed to differing results.
Study limitations include possible lack of generalizability to premenopausal women and African American or Hispanics. We found that Asians, when matched for insulin resistance, demonstrated baseline and weight-gain characteristics that were virtually identical to those in the Caucasians in this study. In addition, we were unable to biopsy multiple depots due to an already high subject burden, so whether changes in adipose cell size/distribution occurred in other body fat depots is unclear. Finally, establishing causality in human studies is difficult. In the current study, weight gain was manipulated, which led to changes in adipose tissue variables that were significantly associated with development of insulin resistance, which implies, but does not prove, causality.
In conclusion, the current results provide strong support for the hypothesis that disordered adipose cell function contributes to the metabolic heterogeneity of obesity. Results uniquely demonstrate the importance of adipose cell enlargement, in association with decreased expression of lipogenic genes, and expansion of VAT and IHL as potential mediators of obesity-related insulin resistance. Further, they demonstrate that IS subjects are not protected from metabolic deterioration during weight gain, likely undergoing an early stage of decompensation as they approach the phenotype exhibited by the IR subjects. Further research is needed to determine what drives or prevents adipocyte hypertrophy and what cellular and molecular processes are activated that link adipose cell enlargement and expansion of VAT and IHL to systemic insulin resistance.
Supplementary Material
Article Information
Funding. This work was supported by American Diabetes Association grant 1-11-CT-35.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. T.M.L. directed the study and conducted the metabolic tests and fat biopsies, prepared data, and contributed to the manuscript. C.C., D.P., and C.A. oversaw enrollment, clinical visits, data collection, and compliance with humans subjects’ ethics requirements. L.-F.L. oversaw all laboratory aspects. D.S. oversaw hepatic imaging studies. S.W.C. contributed to study design and data interpretation and analyzed adipose tissue samples for cell size data. T.M.L. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Prior Presentation. Parts of this study were presented in abstract form at the 74th Scientific Sessions of the American Diabetes Association, San Francisco, CA, 13–17 June 2014.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db15-1213/-/DC1.
References
- 1.Flegal KM, Carroll MD, Kit BK, Ogden CL. Prevalence of obesity and trends in the distribution of body mass index among US adults, 1999-2010. JAMA 2012;307:491–497 [DOI] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention (CDC) Increasing prevalence of diagnosed diabetes--United States and Puerto Rico, 1995-2010. MMWR Morb Mortal Wkly Rep 2012;61:918–921 [PubMed] [Google Scholar]
- 3.Ford ES, Giles WH, Dietz WH. Prevalence of the metabolic syndrome among US adults: findings from the third National Health and Nutrition Examination Survey. JAMA 2002;287:356–359 [DOI] [PubMed] [Google Scholar]
- 4.Bogardus C, Lillioja S, Mott DM, Hollenbeck C, Reaven GM. Relationship between degree of obesity and in vivo insulin action in man. Am J Physiol 1985;248:E286–E291 [DOI] [PubMed]
- 5.McLaughlin T, Schweitzer P, Carter S, et al. . Persistence of improvement in insulin sensitivity following a dietary weight loss programme. Diabetes Obes Metab 2008;10:1186–1194 [DOI] [PubMed] [Google Scholar]
- 6.McLaughlin T, Allison G, Abbasi F, Lamendola C, Reaven G. Prevalence of insulin resistance and associated cardiovascular disease risk factors among normal weight, overweight, and obese individuals. Metabolism 2004;53:495–499 [DOI] [PubMed] [Google Scholar]
- 7.McLaughlin T, Sherman A, Tsao P, et al. . Enhanced proportion of small adipose cells in insulin-resistant vs insulin-sensitive obese individuals implicates impaired adipogenesis. Diabetologia 2007;50:1707–1715 [DOI] [PubMed] [Google Scholar]
- 8.McLaughlin T, Lamendola C, Coghlan N, et al. . Subcutaneous adipose cell size and distribution: relationship to insulin resistance and body fat. Obesity (Silver Spring) 2014;22:673–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spalding KL, Arner E, Westermark PO, et al. . Dynamics of fat cell turnover in humans. Nature 2008;453:783–787 [DOI] [PubMed] [Google Scholar]
- 10.Weyer C, Foley JE, Bogardus C, Tataranni PA, Pratley RE. Enlarged subcutaneous abdominal adipocyte size, but not obesity itself, predicts type II diabetes independent of insulin resistance. Diabetologia 2000;43:1498–1506 [DOI] [PubMed] [Google Scholar]
- 11.Gavrilova O, Marcus-Samuels B, Graham D, et al. . Surgical implantation of adipose tissue reverses diabetes in lipoatrophic mice. J Clin Invest 2000;105:271–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robbins DC, Horton ES, Tulp O, Sims EA. Familial partial lipodystrophy: complications of obesity in the non-obese? Metabolism 1982;31:445–452 [DOI] [PubMed] [Google Scholar]
- 13.de Souza CJ, Eckhardt M, Gagen K, et al. . Effects of pioglitazone on adipose tissue remodeling within the setting of obesity and insulin resistance. Diabetes 2001;50:1863–1871 [DOI] [PubMed] [Google Scholar]
- 14.McLaughlin TM, Liu T, Yee G, et al. . Pioglitazone increases the proportion of small cells in human abdominal subcutaneous adipose tissue. Obesity (Silver Spring) 2010;18:926–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eliasson B, Smith U, Mullen S, Cushman SW, Sherman AS, Yang J. Amelioration of insulin resistance by rosiglitazone is associated with increased adipose cell size in obese type 2 diabetic patients. Adipocyte 2014;3:314–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang X, Jansson PA, Nagaev I, et al. . Evidence of impaired adipogenesis in insulin resistance. Biochem Biophys Res Commun 2004;317:1045–1051 [DOI] [PubMed] [Google Scholar]
- 17.Deurenberg P, Weststrate JA, Seidell JC. Body mass index as a measure of body fatness: age- and sex-specific prediction formulas. Br J Nutr 1991;65:105–114 [DOI] [PubMed] [Google Scholar]
- 18.Deurenberg P, Andreoli A, Borg P, et al. . The validity of predicted body fat percentage from body mass index and from impedance in samples of five European populations. Eur J Clin Nutr 2001;55:973–979 [DOI] [PubMed] [Google Scholar]
- 19.Dubois SG, Heilbronn LK, Smith SR, Albu JB, Kelley DE, Ravussin E; Look AHEAD Adipose Research Group . Decreased expression of adipogenic genes in obese subjects with type 2 diabetes. Obesity (Silver Spring) 2006;14:1543–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen SW, Reaven GM, Farquhar JW. Comparison of impedance to insulin-mediated glucose uptake in normal subjects and in subjects with latent diabetes. J Clin Invest 1970;49:2151–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pei D, Jones CN, Bhargava R, Chen YD, Reaven GM. Evaluation of octreotide to assess insulin-mediated glucose disposal by the insulin suppression test. Diabetologia 1994;37:843–845 [DOI] [PubMed] [Google Scholar]
- 22.Greenfield MS, Doberne L, Kraemer F, Tobey T, Reaven G. Assessment of insulin resistance with the insulin suppression test and the euglycemic clamp. Diabetes 1981;30:387–392 [DOI] [PubMed] [Google Scholar]
- 23.McLaughlin T, Yee G, Glassford A, Lamendola C, Reaven G. Use of a two-stage insulin infusion study to assess the relationship between insulin suppression of lipolysis and insulin-mediated glucose uptake in overweight/obese, nondiabetic women. Metabolism 2011;60:1741–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McLaughlin T, Lamendola C, Liu A, Abbasi F. Preferential fat deposition in subcutaneous versus visceral depots is associated with insulin sensitivity. J Clin Endocrinol Metab 2011;96:E1756–E1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davé V, Yousefi P, Huen K, Volberg V, Holland N. Relationship between expression and methylation of obesity-related genes in children. Mutagenesis 2015;30:411–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harris JA, Benedict FG. A Biometric Study of Basal Metabolism in Man (Publication 279). Washington, DC, Carnegie Institute of Washington, 1919 [Google Scholar]
- 27.Faust IM, Johnson PR, Stern JS, Hirsch J. Diet-induced adipocyte number increase in adult rats: a new model of obesity. Am J Physiol 1978;235:E279–E286 [DOI] [PubMed] [Google Scholar]
- 28.Jo J, Guo J, Liu T, et al. . Hypertrophy-driven adipocyte death overwhelms recruitment under prolonged weight gain. Biophys J 2010;99:3535–3544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kashiwagi A, Mott D, Bogardus C, Lillioja S, Reaven GM, Foley JE. The effects of short-term overfeeding on adipocyte metabolism in Pima Indians. Metabolism 1985;34:364–370 [DOI] [PubMed] [Google Scholar]
- 30.Tchoukalova YD, Votruba SB, Tchkonia T, Giorgadze N, Kirkland JL, Jensen MD. Regional differences in cellular mechanisms of adipose tissue gain with overfeeding. Proc Natl Acad Sci U S A 2010;107:18226–18231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johannsen DL, Tchoukalova Y, Tam CS, et al. . Effect of 8 weeks of overfeeding on ectopic fat deposition and insulin sensitivity: testing the “adipose tissue expandability” hypothesis. Diabetes Care 2014;37:2789–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fabbrini E, Yoshino J, Yoshino M, et al. Metabolically normal obese people are protected from adverse effects following weight gain. J Clin Invest 2015;125:787–795 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.