Abstract
Dysbalance in gut microbiota has been linked to increased microbial translocation, leading to chronic inflammation in HIV-patients, even under effective HAART. Moreover, microbial translocation is associated with insufficient reconstitution of CD4+T cells, and contributes to the pathogenesis of immunologic non-response. In a double-blind, randomised, placebo-controlled trial, we recently showed that, compared to placebo, 12 weeks treatment with probiotic Saccharomyces boulardii significantly reduced plasma levels of bacterial translocation (Lipopolysaccharide-binding protein or LBP) and systemic inflammation (IL-6) in 44 HIV virologically suppressed patients, half of whom (n = 22) had immunologic non-response to antiretroviral therapy (<270 CD4+Tcells/μL despite long-term suppressed viral load). The aim of the present study was to investigate if this beneficial effect of the probiotic Saccharomyces boulardii is due to modified gut microbiome composition, with a decrease of some species associated with higher systemic levels of microbial translocation and inflammation. In this study, we used 16S rDNA gene amplification and parallel sequencing to analyze the probiotic impact on the composition of the gut microbiome (faecal samples) in these 44 patients randomized to receive oral supplementation with probiotic or placebo for 12 weeks. Compared to the placebo group, in individuals treated with probiotic we observed lower concentrations of some gut species, such as those of the Clostridiaceae family, which were correlated with systemic levels of bacterial translocation and inflammation markers. In a sub-study of these patients, we observed significantly higher parameters of microbial translocation (LBP, soluble CD14) and systemic inflammation in immunologic non-responders than in immunologic responders, which was correlated with a relative abundance of specific gut bacterial groups (Lachnospiraceae genus and Proteobacteria). Thus, in this work, we propose a new therapeutic strategy using the probiotic yeast S. boulardii to modify gut microbiome composition. Identifying pro-inflammatory species in the gut microbiome could also be a useful new marker of poor immune response and a new therapeutic target.
Introduction
Recent studies have shown that gut microbiota is impaired in HIV-patients, even after effective Highly Active Antirretroviral Therapy (HAART), and a large number of disease-associated bacteria have been identified. HIV-infection severely damages the gastrointestinal mucosal barrier resulting in microbial translocation [1–2], which in turn leads to continuous systemic inflammation and disease progression despite effective HAART [3–6]. Using high-resolution profiling of the bacterial community by 16S rDNA gene amplification and pyrosequencing, previous studies have identified a dysbiotic gut pattern in HIV-infected individuals, characterized by increased microbial translocation, chronic inflammation and hyperactivation of CD4+T cells, despite achieving long-term virologic suppression [7–11]. Moreover, microbial translocation is associated with insufficient reconstitution of CD4+T cells, and contributes to the pathogenesis of immunologic non-response [12–16]. A recent study have reported that HIV gut microbiome must be controlled for HIV risk factors, and after stratifying for sexual orientation, there was no solid evidence of an HIV-specific dysbiosis, but HIV-1 infection remained consistently associated with reduced bacterial richness and the lowest gut bacterial richness was observed in immunologic non-responders patients [17].
Recently the intestinal microbiome has been proposed as a novel therapeutic target for reducing chronic inflammation [18–19] and various interventions such as pre-probiotics have been proposed to improve the resident gut microbiome [20–22]. Since the specific effects of HIV infection on the particular gut bacterial taxa that contributes to chronic immunoactivation are unclear, it seems reasonable to propose treatments to improve the gut bacterial richness and HIV associated immune dysfunction. However, the beneficial effects of probiotics are strain-dependent and not all interventions are equally effective [23–25]. Saccharomyces boulardii is a probiotic whose clinical efficacy, anti-inflammatory and immunomodulatory effects are supported by extensive previous studies [26–30]. We recently demonstrated that treatment with S. boulardii significantly decreased plasma levels of microbial translocation (Lipopolysaccharide-binding protein or LBP) and inflammation parameters such as cytokine IL-6 in 44 HIV-treated patients, half of whom had an immunodiscordant response to antiretroviral therapy [31]. The aim of the present study was to investigate if this beneficial effect of the probiotic Saccharomyces boulardii is due to a modification in the gut microbiome composition of this patients, with a decrease in some species associated with higher systemic levels of microbial translocation and inflammation. In this study, we used 16S rDNA tagging to analyze the gut microbiome communities in these 44 patients, to assess the impact of probiotic treatment with S. boulardii compared to placebo.
In a sub-study of these patients, we explore if the immunological non-response was associated with higher systemic inflammation and microbial translocation parameters and a distinct gut microbiome pattern.
Subjects, material and methods
Study design
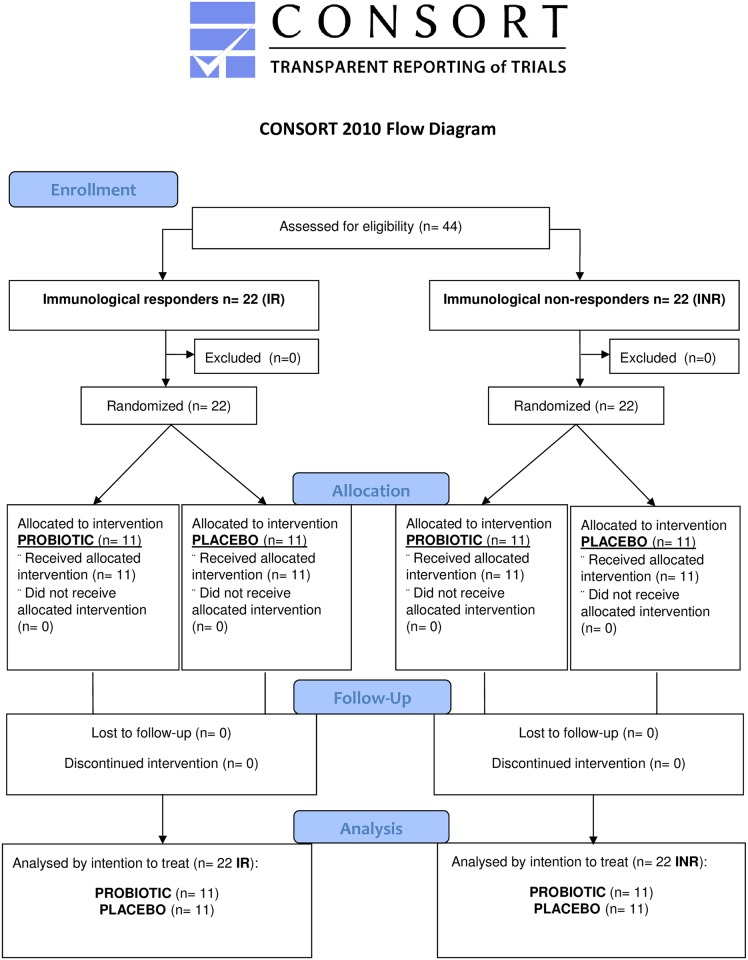
We performed a double-blind randomized, placebo-controlled trial to analyze the composition of the gut microbiota, microbial translocation, and inflammation parameters in 44 chronic HIV-infected patients with undetectable plasma viral load (<20 copies/mL) for at least 2 years and a stable HAART regimen. Half of the patients (n = 22) had already been previously selected as being immunodiscordant or immunological non-responders (INR), defined as individuals with an persistent unfavorable immunologic response (<270 CD4+Tcells/μL despite long-term suppressed viral load), while the other 22 had already been previously selected as immunologic responders (IR: CD4+T count >400 cells/μL). Once the patients had been included in the IR or INR group, they were double-blind randomized to oral supplementation with probiotic S. boulardii (capsules with 56.5 mg living yeasts, 2 capsules 3 times per day, n = 22), or placebo (2 capsules 3 times per day, n = 22) for 12 weeks. All 44 participants completed the study; none discontinued their participation because of side effects of the treatments or adverse events. (Fig 1. CONSORT Flow diagram). Markers of microbial translocation and inflammation, immunological and clinical data were collected before and after the intervention (Baseline and week 12), and follow-up visits were conducted until 48 weeks (See “S1 and S2 Protocols” Files). We recently reported the plasma findings in these patients [31]. All patients also provided 50gr of stool samples before and after the intervention, which were stored at -80°C within the first 24–48 hours; to allow us to compare gut microbiome composition before and after a 12-week treatment with probiotic or placebo. A computer-generated randomization list was used. Randomization was performed by a hospital pharmacist unrelated to the care of the patients, using sequentially numbered containers. The participants and the care providers were blinded after assignment to interventions.
Fig 1. CONSORT flow diagram.
The study was approved by the Clinical Research Ethics Committee of Hospital del Mar Medical Research Institute, number 2011/4508, on data 01/Feb/2012, according to the principles of the Declaration of Helsinki. Written informed consent was obtained from all participants before enrollment in the study. Patients were recruited between August 2012-July 2013. The study was registered during the enrollment because it had already been previously notified, registered and approved by the Hospital del Mar Medical Research Institute Ethics Committee. The authors confirm that related trial for this intervention is registered at ClinicalTrials.gov (PROB-VIH, Trial number: NCT01908049). “S1 Checklist”
Microbial translocation markers
Serum levels of soluble CD14 were quantified using a ELISA kit (Quantikine™, R&D Systems, Minneapolis, MN) on a Quanta-Lyser™ 160 robotic workstation (Inova Diagnostics, San Diego, CA). Serum LBP was measured using the Immulite™ chemiluminescent immunometric assay on its automated analyzer (Immulite One, Siemens Healthcare®, Llanberis, UK).
Inflammation markers
Milliplex MAP™ was used to analyze IL-6. Hs-CRP and β2 microglobulin were measured using the Immulite One™, and plasma fibrinogen with HemosIL™ reagents.
Microbiome taxonomy analyses & bioinformatics
DNA was extracted from frozen stool samples and amplified by PCR targeting 16S gene regions using primers described elsewhere [32]. We performed library preparation followed by Illumina sequencing according to standard protocols described previously [33]. The taxonomic composition of the intestinal microbiota was characterized by grouping sequences into OTUs (Organizational Taxonomic Units). We used the QIIME open source algorithm for handling and interpretation of the data from the raw sequences; following quality assessment of sequence data, we assigned taxonomic affiliations using the RDP_classifier from the Ribosomal Database Project [34].
Statistical analysis
No previous data were available to estimate the sample size; thus, for a Type I error rate of 0.05 and a 1-beta risk of 0.20 in a bilateral contrast, we computed that 22 patients were needed per study group to detect statistically significant differences between proportions. The assumed proportions for the power analysis for each group was 0.5. Continuous variables were expressed as the median and interquartile range, and discrete variables as percentages. Categorical variables were described as proportions. The Mann-Whitney U-test was used to compare medians, ANOVA to assess differences in continuous variables, and the Pearson's χ2 test to evaluate the association between categorical variables. We also conducted multivariate logistic regression analysis to study the correlation between microbial translocation and systemic inflammation parameters, and conditional backward stepwise regression analysis at baseline to identify parameters that were correlated with the immunodiscordance. The enter criteria was a p value<0.1. All the variables studied were selected based on the statistical significance. For the analysis of qualitative differences between responders and non-responders, we performed a median recoding of LBP, soluble CD4, β2 microglobuline and fibrinogen; the values obtained were defined as high or low. Statistical analyses were performed using SPSS software version 20.0 (SPSS, Inc., Chicago, IL) and several open source libraries in R (R Core Team 2014. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL http://www.R-project.org/). The glm (R) function has been used in the logistic regression analysis.
Results
Patient characteristics
This study included 44 patients. Half of each group were immunological non-responders (INR), the median age was 47.5 years, 84% were male, and the risk factors for acquiring HIV infection were Heterosexual (HTX) (46%), Men who Have Sex with Men (MHSM) (38%) and Injection drug users (IDU) (16%). The profiles of demographic, biological and clinical parameters were similar in the probiotic and placebo groups [31] (“S1 Table”). All patients had suppressed viral load for a median of 4.7 years, with NNRTI-based (75%) or PI-based HAART (25%). The median nadir CD4+T was 108 cells/μl, and 41% had AIDS diagnosis.
Microbial translocation is correlated with systemic inflammation
We tested for association between microbial translocation measurements and baseline parameters from 44 patients, and observed a statistically significant correlation between LBP levels and plasma levels of soluble CD14 (r = 0.48, p = 0.001), Hs-CRP (r = 0.63, p = 0.0001), β2 microglobulin (r = 0.59, p = 0.0001), Erythrocyte Sedimentation Rate-ESR (r = 0.57, p = 0.0001), Fibrinogen (r = 0.64, p = 0.0001), Cd4 nadir (r = -0.53, p = 0.0001), and IL-6 (r = 0.58, p = 0.0001). Using multivariate logistic regression analysis, we found that LBP was independently correlated with Hs-CRP, ESR and soluble CD14 (“S2 Table”).
Saccharomyces boulardii produces changes in some gut bacterial communities
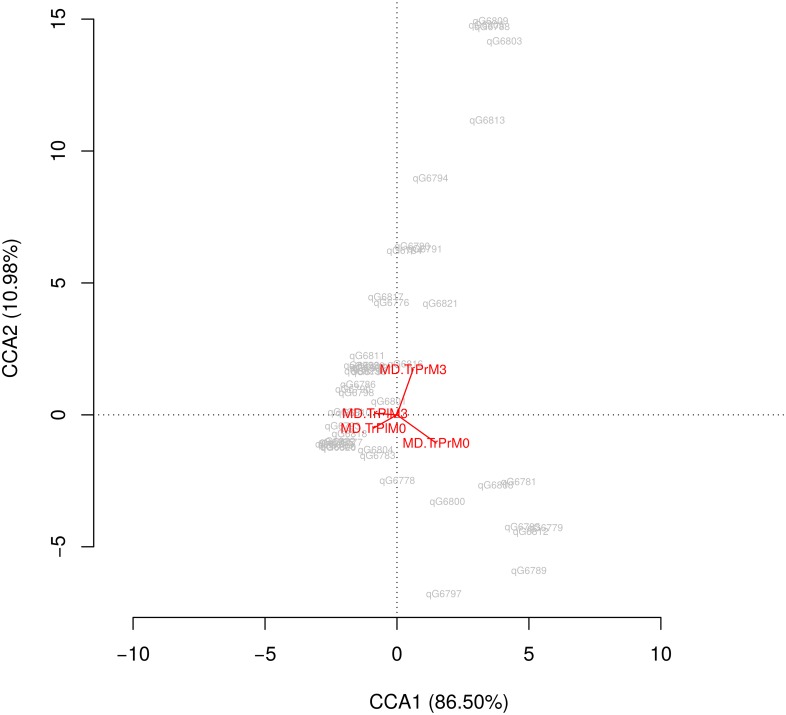
Canonical correspondance analysis shows the high influence of probiotic effect versus the time effect (vertical axis) (Fig 2). Changes in gut microbiome after the interventions are showed at “S1 Fig” (INRs group). After 12 weeks of probiotic treatment, we observed a significant decrease in levels of some Clostridiales species with respect to baseline compared with placebo (Fig 3). We also observed a significant decreased in Catenibacterium communities (2.65 to 0.04, p = 0.00003), and an increase in levels of Megamonas (p = 0.02) and Desulfovibrionales (Proteobacteria) (p = 0.05). These probiotic effects on gut microbiota composition were similar in the INR and IR groups.
Fig 2. Canonical correspondence analysis.
Fig 3. Changes in the proportion of Clostridiales after the interventions.
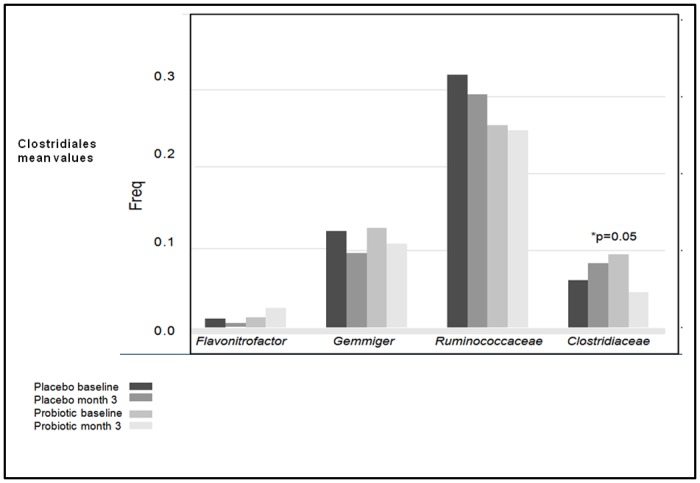
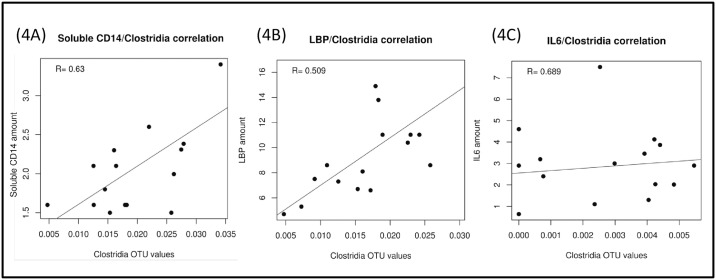
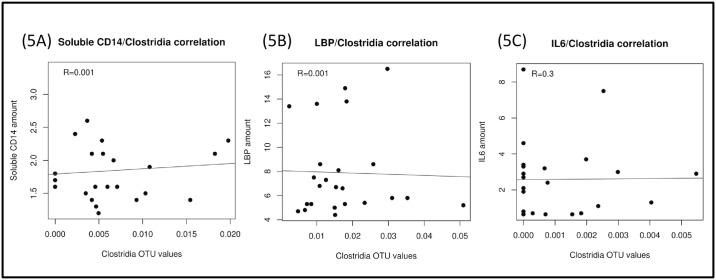
We then investigated the correlation between these different stool bacterial populations, parameters of microbial translocation and systemic inflammation, and observed a statistically significant correlation between the proportion of Clostridia genera and plasma concentrations of soluble CD14 (r = 0.63, p = 0.03), LBP (r = 0.71, p = 0.009), and IL-6 (r = 0.69, p = 0.0008; Fig 4). These correlations were present at baseline in INR group but not in the IRs probably because IRs have a lower relative abundance of Clostridiaceae at baseline. The correlations are not statistically significant in probiotic treated group, probably because of a decrease in these bacterial communities due to the probiotic effect (Fig 5).
Fig 4. Correlations between relative abundance of Clostridiales and soluble CD14 (4A), LBP (4B) and IL6 (4C) before the intervention (INR).
LBP, Lipopolisaccharide Binding-Proteine; IL6, interleukin-6.
Fig 5. Correlations between relative abundance of Clostridiales and soluble CD14 (5A), LBP (5B) and IL6 (5C) after probiotic treatment.
LBP, Lipopolisaccharide Binding-Proteine; IL6, interleukin-6.
Immunological non-response is associated with higher microbial translocation and the presence of some pro-inflammatory species
The baseline differences between the INR and IR groups are summarized in Table 1. Quantitative analysis of the baseline data indicates that the following variables showed statistically significant differences between the INR and IR group: age, current CD4+T count, CD4+T nadir, HCV coinfection, LBP and β2 microglobulin levels. The INR groups tended to have higher fibrinogen and soluble CD14 levels. Compared to the IR group, the INR group had a greater proportion of individuals with values greater than the median of LBP (68.2% vs 27.3%, p = 0.007); soluble CD14 (72.7% vs 40.9%, p = 0.03); β2 microglobuline (66.7% vs 33%, p = 0.022) and fibrinogen (65% vs 31.8%, p = 0.03).
Table 1. Differences between Immunological Responders (IR) and Non-Responders (INR).
| Responders (n = 22) |
Non–Responders (n = 22) |
p-value | |
|---|---|---|---|
| Demographics | |||
| Age (years) [SD] | 44 (37–49) | 52 (47–57) | 0.014 |
| Male [n (%)] | 21 (95.5) | 16 (72.7) | 0.042 |
| HIV infection | |||
| Risk factor [n (%)] | |||
| IDU | 0 (0) | 6 (100) | 0.03 |
| MHSM | 12 (70.6) | 5 (29.4) | 0.06 |
| HTX | 10 (50) | 10 (50) | 1 |
| Time with viral load <50 copies/ml (years) [median (IQR)] | 4.5 (3–9.25) | 5 (3.75–10) | 0.463 |
| Nadir CD4 cell count (/ml) [median (IQR)] | 244 (105–293) | 47.5 (9–113.75) | 0.000 |
| Zenith viral load (log10) [median (IQR)] | 4.88 (4.58–5.38) | 5.06 (4.39–5.41) | 0.770 |
| HCV co-infection [n (%)] | 0 (0) | 8 (100) | 0.002 |
| Current ART [n (%)] | |||
| NNRTI | 22 (68.8%) | 10 (31.3%) | 0.000 |
| PI | 0 | 11 (52.4%) | 0.000 |
| Absolute CD4T-count (cells/μl) [median (IQR)] | 483 (413–630) | 219 (169–266) | 0.000 |
| Absolute CD8T-count (cells/μl) [median (IQR)] | 699.5(472.5–860.5) | 594 (353–781) | 0.199 |
| Microbial translocation parameters [median (IQR)] | |||
| LBP (pg/mL) | 5.65 (5.2–6.5) | 7.4 (6.1–8.7) | 0.011 |
| Soluble CD14 (μg/mL) | 1.43 (1.31–1.93) | 1.69 (1.55–2.11) | 0.082 |
| Inflammation parameters[median (IQR)] | |||
| Hs-CRP(mg/dl) | 0.22 (0.07–0.35) | 0.16 (0.07–0.40) | 0.874 |
| IL -6(pg/mL) | 1.85 (0.7–2.85) | 2.7 (1.02–40.15) | 0.466 |
| Ig E (kU/L) | 48.9 (16.8–157) | 115 (21.6–360) | 0.467 |
| Fibrinogen (mg/dl) | 257.5 (219–279) | 293.5(259.5–324) | 0.062 |
| TNF-α(pg/mL) | 10.9 (8.7–12.3) | 12.7 (7.8–16.9) | 0.190 |
| ESR (mm/h) | 7 (2–10.5) | 10 (6–16.5) | 0.100 |
| Vit D 25 OH (ng/mL) | 30.97(20.07–0.33) | 37.04 (24.6–42.48) | 0.253 |
| β2microglobuline (μg/mL) | 1.7 (1.54–2.06) | 2.12 (1.93–2.86) | 0.002 |
| Gut microbiome composition (Relative abundance [%]) | |||
| Lachnospiraceae (Clostridia) | 0.017% | 0.093% | 0.03 |
| Proteobacteria | 0.053% | 0.248% | 0.06 |
ART, antiretroviral therapy; PI, protease inhibitor; NNRTI, non-nucleoside; reverse transcriptase inhibitor; LBP, Lipopolysaccharide-binding protein; sCD14, soluble CD14; hs-CRP, high sensitivity C-reactive protein; IL-6, Interleukin 6; ESR, Erythrocyte Sedimentation Rate; IQR, interquartile range.
High LBP was associated with being an INR (Relative Risk = 5.71; 95% CI = 1.56–20.93; p = 0.015) and in the conditional backward stepwise regression analysis, high LBP was the only variable that could explain immunological non-response (OR = 6.22, 95% CI = 1.63–23.75, p = 0.007) (“S3 Table”).
Analyzing gut microbiome composition at baseline, we observed a higher proportion of some Firmicutes species (Clostridia) in the INR group than in the IR group. The proportion of Proteobacteria was higher in immunodiscordant patients; the proportion of Lachnospiraceae genera was significantly higher in the INR group (OR = 3.4, p = 0.05) (“S4 Table”).
Discussion
This is the first clinical trial to use 16S rDNA sequencing to analyze changes in gut microbiome composition following treatment with Saccharomyces boulardii, and how these changes are correlated with microbial translocation and inflammation in HIV patients.
Microbial imbalance in the gut has been associated with increased microbial translocation, leading to chronic inflammation. In line with this, various microbial communities resident in the gut are correlated with plasma bacterial translocation markers (soluble CD14 and LBP) and pro-inflammatory cytokine interleukin-6 [10–11,35]. We recently reported a significant reduction in LBP and IL-6 levels in a small cohort of HAART-treated HIV patients supplemented with 12 weeks treatment with probiotic S. boulardii compared to placebo [31]. Here, we take a step further and use 16S rDNA tagging and high throughput sequencing to analyze microbiome profiles before and after the intervention, and to evaluate whether some predominant species are associated with increased bacterial translocation, systemic inflammation and poor immune response in these patients.
Following probiotic treatment, we observed a significant decrease in some Clostridiales, such as Clostridiaceae and Catenibacterium communities. Previous studies have used PCR or FISH techniques, rather than 16S rDNA sequencing, to evaluate the effects of supplementation with specific prebiotics in HAART-naïve infected HIV adults [21] and supplementation with probiotic S.boulardii in HIV-negative adults with enteritis [36]; these studies reported a consequent reduction in concentrations of pathogenic clostridia-related species, which are considered to be pro-inflammatory mucotropic bacteria, although these authors did not evaluate if this was correlated with a decrease in systemic inflammation. Recent studies have reported a higher proportion of Catenibacterium in HIV patients than in healthy individuals [9,11]. Catenibacterium is a Gram-positive, non-spore-forming and anaerobic genus from the family of Erysipelotrichidae. Taxa in this family were the most enriched [7], or a significant enrichment was reported [8–9], in the untreated HIV-infected subjects compared to HIV-uninfected individuals and was correlated with markers of microbial translocation and systemic inflammation in HIV patients [8]. In our study, the concentration of Catenibacterium, which have also been associated with other chronic diseases [37–39], also decreased following probiotic treatment.
We observed a relationship between bacterial translocation and various parameters of systemic inflammation. These data are consistent with previous studies demonstrating, in both treated and untreated HIV patients, a direct correlation between systemic parameters of bacterial translocation, and chronic immune activation and disease progression [40–48]. Furthermore, we found significant correlation between some bacterial taxa (Clostridia) and parameters of microbial translocation and systemic inflammation in immunological non-responders group (INR). Recent studies also have demonstrated a statistically significant correlation between some species of Clostridia, including Lachnospiraceae, and increased systemic immune activation parameters (TNF) in HIV-infected patients [11]. While the immunologic driving factors of gut dysbiosis related to HIV infection are likely to be complex interestingly, our study showed a significant reduction in these bacterial communities following probiotic treatment.
HIV-patients with deficient CD4+Tcell count recovery despite HAART have been defined as Immunological non-responders (INRs), and are estimated to account for up to 30% of all treated HIV-infected subjects. This is relevant because this subpopulation is associated with higher morbi-mortality. This phenomenon has been widely studied using different approaches, and its pathogenesis is known to be related to older age, nadir CD4 T-cell count, reduced thymic function, co-infection with hepatotropic viruses, and multiple genetic variants [12,13]. However, the ultimate cause of the discordant response is excessive CD4 T-cell destruction due to CD4+T hyperactivation [14], residual viral replication, and persistent antigenic stimulation due to microbial translocation. HAART-treated individuals have been reported to show an inverse correlation between levels of microbial translocation and sustained failure on CD4+T cell reconstitution [15,16]. A history of more advanced immunosuppression and the resulting damage to gut immunity and integrity are thought to be an important factor in the differences observed between groups in gut microbiome and markers of bacterial translocation and inflammation [49–51]. Our results are consistent with these findings, and extend them by comparing the composition of the gut microbial community in immunological responders and non-responders. Interestingly, the relative abundance of specific gut bacterial groups in INR patients is associated with greater bacterial translocation. Recent studies have shown that Proteobacteria, which are over-represented in our group of INRs, are responsible for much of the translocation after SIV-infection [52]; although the mechanisms remain unknown, Brenchley et al.[53] suggested their motility, high metabolic rate and possession of immune evasion genes as posibble reasons. Identifying gut species that contribute to the pathogenesis of immunologic non-response and persistent immune activation could be a new diagnostic and therapeutic tool in this group of HIV-patients with an increased risk of clinical progression and death.
The multiple anti-inflammatory mechanisms produced by S. boulardii provide molecular explanations to support its effectiveness in intestinal inflammatory states [30]. It has also been demonstrated that S. boulardii increases absolute numbers of the main habitual fermenting bacterial groups, and decreases concentrations of mucotropic occasional bacterial communities [54]. Several mechanisms of action have been identified directed against the host and pathogenic microorganisms, suppressing “bacteria overgrowth” and host cell adherence [30]. One limitation of this study is that, since S. boulardii is a yeast, we did not perform 16S rDNA gene amplification and parallel sequencing to demonstrate colonization by the probiotic, or changes in the gut mycobiome. A decrease in some dysbiotic bacterial genera, replacement of inflammogenic yeast and/or improved functional biostructure of colonic microbiota following S. boulardii treatment may prevent microbial translocation [54], although our study was not designed to determine the mechanism through which changes in gut microbiome composition alter its functional anatomy, metabolomics or mycobiome. Our study has some of the limitations of other studies in this topic. All are small works, and more extensive studies with better tools than 16s sequencing are needed before drawing definitive conclusions, including selection of the patients who could best respond to the probiotic treatment, as well as the doses and duration of the therapy. However, given the increasingly widespread use of metabolomic and metagenomic techniques, analysis of gut bacterial contents could be used in the future as a marker of a dysbiotic microbiome that contributes to chronic systemic inflammation and HIV progression. More importantly, the gut microbiome can be modified using certain strains of probiotics to generate a less pro-inflammatory profile; this new therapeutic target is worthy of further exploration.
Conclusions
We observed a change in gut microbiome composition following probiotic treatment (S. boulardii), with a decrease in some species which are directly correlated with systemic levels of microbial translocation and inflammation. The use of specific probiotics could be a new therapeutic strategy for HIV patients. In addition, our data suggest that identifying pro-inflammatory species in the gut microbiome could be a new marker of poor immune response.
Supporting information
(DOC)
(DOC)
(DOC)
INR, immunologic non-responders.
(TIF)
ART, antiretroviral therapy; PI, protease inhibitor; NNRTI, nonnucleoside; reverse transcriptase inhibitor; LBP, Lipopolisaccharide Binding-Proteine; sCD14, soluble CD14; hs-CRP, high sensitivity C-reactive protein; ESR, Erythrocyte Sedimentation Rate; IQR, interquartile range.
(DOCX)
LBP, Lipopolisaccharide Binding-Proteine; sCD14, soluble CD14, hs-CRP, high sensitivity C-reactive protein.
(DOCX)
LBP_basal, Lipopolisaccharide Binding-Proteine at baseline; sCD14, soluble CD14; β2microglob, Beta 2 microglobuline.
(DOCX)
(CSV)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work is supported by a Spanish Health Ministry “Grant for independent clinical research 2012” (EC-11-541) to Judit Villar, the principal investigator, www.msssi.gob.es. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Brenchley JM, Price DA, Schacker TW, Asher TE, Silvestri G, Douek DC, et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med 2006; 12(12): 1365–1371. 10.1038/nm1511 [DOI] [PubMed] [Google Scholar]
- 2.Nazli A, Chan O, Dobson-Belaire WN, Gray-Owen SD, Arsenault AL, Kaushic C, et al. Exposure to HIV-1 directly impairs mucosal epithelial barrier integrity allowing microbial translocation, PLoS Pathogens 2010; 6(4):e1000852 10.1371/journal.ppat.1000852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nowroozalizadeh S, Månsson F, da Silva Z, Norrgren H, Holmgren B, Jansson M, et al. Microbial translocation correlates with the severity of both HIV-1 and HIV-2 infections. J Infect Dis. 2010; 201(8):1150–4. [DOI] [PubMed] [Google Scholar]
- 4.Wallet MA, Rodriguez CA, Yin L, Saporta S, Chinratanapisit S, Hou W, et al. Microbial translocation induces persistent macrophage activation unrelated to HIV-1 levels or T-cell activation following therapy. AIDS2010; 24(9):1281–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tincati C, Biasin M, Bandera A, Violin M, Marchetti G, Gori A, et al. Early initiation of highly active antiretroviral therapy fails to reverse immunovirological abnormalities in gut-associated lymphoid tissue induced by acute HIV infection. Antivir Ther. 2009; 14(3):321–330. [PubMed] [Google Scholar]
- 6.Cassol E, Malfeld S, Mahasha P, Alfano M, Poli G, Rossouw T, et al. Persistent microbial translocation and immune activation in HIV-1-infected South Africans receiving combination antiretroviral therapy. J Infect Dis. 2010; 202(5):723–33. 10.1086/655229 [DOI] [PubMed] [Google Scholar]
- 7.Vujkovic-Cvijin I, Dunham RM, Iwai S, Hunt PW, Lynch SV, McCune JM, et al. Dysbiosis of the gut microbiota is associated with HIV disease progression and tryptophan catabolism, Science Translational Medicine 2013; 5(193):193ra91 10.1126/scitranslmed.3006438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dinh DM, Volpe GE, Duffalo C,Tai AK, Kane AV, Ward HD, et al. Intestinal microbiota, microbial translocation, and systemic inflammation in chronic HIV infection. J Infect Dis. 2015; 211(1):19–27. 10.1093/infdis/jiu409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lozupone CA, Rhodes ME, Neff CP, Fontenot AP, Campbell TB, and Palmer BE. HIV-induced alteration in gut microbiota: driving factors, consequences, and effects of antiretroviral therapy. Gut Microbes 2014; 5(4):562–570. 10.4161/gmic.32132 [DOI] [PubMed] [Google Scholar]
- 10.Ellis CL, Ma ZM, Mann SK, Miller CJ, Pollard RB, Asmuth DM, et al. Molecular Characterization of Stool Microbiota in HIV-Infected Subjects by Panbacterial and Order-Level 16S Ribosomal DNA (rDNA) Quantification and Correlations with Immune Activation. J Acquir Immune Defic Syndr. 2011; 57(5):363–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mutlu EA, Keshavarzian A, Losurdo J, Gorenz A, Burns C, Landay A, et al. A compositional look at the human gastrointestinal microbiome and immune activation parameters in HIV infected subjects, PLoSPathogens 2014; 10(2):IDe1003829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benveniste O, Flahault A, Sterkers G, Simon A, Ameisen JC, Leport C, et al. Mechanisms involved in the low-level regeneration of CD4+ cells in HIV-1-infected patients receiving highly active antiretroviral therapy who have prolonged undetectable plasma viral loads. J Infect Dis. 2005; 191 (10):1670–1679. 10.1086/429670 [DOI] [PubMed] [Google Scholar]
- 13.Gazzola L, Tincati C, Bellistri GM, Monforte A, Marchetti G. The absence of CD4R T cell count recovery despite receipt of virologically suppressive highly active antiretroviral therapy: clinical risk, immunological gaps, and therapeutic options. Clin Infect Dis 2009; 48(3):328–337. [DOI] [PubMed] [Google Scholar]
- 14.Massanella M, Negredo E, Perez-Alvarez N, Puig J, Ruiz-Hernandez R, Bofill M, et al. CD4 T-cell hyperactivation and susceptibility to cell death determine poor CD4 T-cell recovery during suppressive HAART. AIDS 2010; 24(7):959–68. 10.1097/QAD.0b013e328337b957 [DOI] [PubMed] [Google Scholar]
- 15.Marchetti G, Bellistri GM, Borghi E, Tincati C, Ferramosca S, La Francesca M, et al. Microbial translocation is associated with sustained failure in CD4+ T-cell reconstitution in HIV-infected patients on long-term highly active antiretroviral therapy. AIDS 2010; 22(15):2035–8. [DOI] [PubMed] [Google Scholar]
- 16.Merlini E, Bai F, Bellistri GM, Tincati C, d’Arminio Monforte A, Marchetti G. Evidence for polymicrobic flora translocating in peripheral blood of HIV-infected patients with poor immune response to antiretroviral therapy. PLoS One 2011; 6(4):e18580 10.1371/journal.pone.0018580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Noguera-Julian M, Rocafort M, Guillén Y, Rivera J, Casadellà M, Nowak P, et al. Gut Microbiota Linked to Sexual Preference and HIV Infection. EBioMedicine. 2016. January 28;5:135–46. 10.1016/j.ebiom.2016.01.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rajasuriar R, Khoury G, Kamarulzaman A, French MA, Cameron PU, Lewin SR. Persistent immune activation in chronic HIV infection: do any interventions work? AIDS 2013; 27 (8):1199–208. 10.1097/QAD.0b013e32835ecb8b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Assimakopoulos SF, Dimitropoulou D, Marangos M and Gogos CA. Intestinal barrier dysfunction in HIV infection: pathophysiology, clinical implications and potential therapies. Infection 2014; 42(6):951–9. 10.1007/s15010-014-0666-5 [DOI] [PubMed] [Google Scholar]
- 20.Hummelen R, Changalucha J, Butamanya NL, Cook A, Habbema JD, Reid G, et al. Effect of 25 weeks probiotic supplementation on immune function of HIV patients. Gut Microbes 2011; 2(2):80–85. [DOI] [PubMed] [Google Scholar]
- 21.Gori A, Rizzardini G, Van't Land B, Welling G, Garssen J, Clerici M, et al. Specific prebiotics modulate gut microbiota and immune activation in HAART-naïve HIV-infected adults: results of the “COPA” pilot randomized trial. NATURE 2011; 4(5): 554–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klatt NR, Canary LA, Sun X, Haddad EK, Estes JD, Brenchley JM, et al. Probiotic/prebiotic supplementation of antiretrovirals improves gastrointestinal immunity in SIV-infected macaques, The Journal of Clinical Investigation 2013; 123(2):903–907. 10.1172/JCI66227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cunningham-Rundles S, Ahrné S, Johann-Liang R, Grassey C, Bengmark S, Cervia JS, et al. Effect of probiotic bacteria on microbial host defense, growth and immune function in human immunodeficiency virus type- 1 infection. Nutrients 2011; 3(12):1042–1070. 10.3390/nu3121042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sandler NG, Zhang X, Bosch RJ, Deeks SG, Lederman MM, Gandhi RT et al. AIDS Clinical Trials Group A5296 Team. Sevelamer does not decrease lipopolysaccharide or soluble CD14 levels but decreases soluble tissue factor, low-density lipoprotein (LDL) cholesterol, and oxidized LDL cholesterol levels in individuals with untreated HIV infection. The Journal of Infectious Diseases 2014; 210(10):1549–1554. 10.1093/infdis/jiu305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stiksrud B, Nowak P, Nwosu FC, Hov JR, Dyrhol-Riise AM, Trøseid M, et al. Reduced Levels of D-dimer and Changes in Gut Microbiota Composition After Probiotic Intervention in HIV-Infected Individuals on Stable ART. J Acquir Immune Defic Syndr. 2015; 70 (4): 329–37. 10.1097/QAI.0000000000000784 [DOI] [PubMed] [Google Scholar]
- 26.Thomas S, Przesdzing I, Metzke D, Schmitz J, Radbruch A, Baumgart DC, et al. Saccharomyces boulardii inhibits lipopolysaccharide-induced activation of human dendritic cells and T cell proliferation. Clin Exp Immunol. 2009; 156 (1):78–87. 10.1111/j.1365-2249.2009.03878.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Im E, Pothoulakis C. Recent advances in Saccharomyces boulardii research. Gastroenterol Clin Biol. 2010; 34 Suppl 1:S62–S70. [DOI] [PubMed] [Google Scholar]
- 28.McFarland LV. Systematic review and meta-analysis of Saccharomyces boulardii in adult patients. World. J. Gastroenterol. 2010; 16(18):2202–22. 10.3748/wjg.v16.i18.2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Canonici A, Pellegrino E, Siret C, Terciolo C, Czerucka D, Bastonero S, et al. Saccharomyces boulardii Improves Intestinal Epithelial Cell Restitution by Inhibiting αvβ5 Integrin Activation State. PLoS ONE 2012; 7(9):p. e45047 10.1371/journal.pone.0045047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pothoulakis C. Review article: anti-inflammatory mechanisms of action of Saccharomyces boulardii. Aliment Pharmacol Ther. 2009; 30(8):26–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Villar-García J, Hernández JJ, Güerri-Fernández R, González A, Lerma E, Knobel H, et al. Effect of probiotics (Saccharomyces boulardii) on microbial translocation and inflammation in HIV-treated patients: a double-blind, randomized, placebo-controlled trial. J Acquir Immune Defic Syndr. 2015; 68(3):256–63. 10.1097/QAI.0000000000000468 [DOI] [PubMed] [Google Scholar]
- 32.Klindworth A, Pruesse E, Schweer T, Peplies J, Horn M, Glöckner FO, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013; 41(1):e1 10.1093/nar/gks808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caporaso JG, Lauber CL, Walters WA, Gilbert JA, Smith G, Knight R, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012; 6(8):1621–4. 10.1038/ismej.2012.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Q, Garrity GM, Tiedje JM, and Cole JR. Naïve Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl Environ Microbiol. 2007; 73(16):5261–5267. 10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perez-Santiago J, Gianella S, Massanella M, Little SJ, Richman DD, Smith DM, et al. Gut Lactobacillales are associated with higher CD4 and less microbial translocation during HIV infection, AIDS 2013; (27)12:1921–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swidsinski A, Loening-Baucke V, Kirsch S and Doerffel Y. Functional biostructure of colonic microbiota (central fermenting area, germinal stock area and separating mucus layer) in healthy subjects and patients with diarrhea treated with Saccharomyces boulardii. Gastroenterol Clin Biol. 2010; 34 Suppl 1:S79–92. [DOI] [PubMed] [Google Scholar]
- 37.Vaziri ND, Wong J, Pahl M, Piceno YM, Yuan J, DeSantis TZ, et al. Chronic kidney disease alters intestinal microbial flora. Kidney Int. 2013. February;83(2):308–15. 10.1038/ki.2012.345 [DOI] [PubMed] [Google Scholar]
- 38.Ciubotaru I, Green SJ, Kukreja S, Barengolts E. Significant differences in fecal microbiota are associated with various stages of glucose tolerance in African American male veterans. Transl Res. 2015. November;166(5):401–11. 10.1016/j.trsl.2015.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moreno-Indias I, Sánchez-Alcoholado L, García-Fuentes E, Cardona F, Queipo-Ortuño MI, Tinahones FJ. Insulin resistance is associated with specific gut microbiota in appendix samples from morbidly obese patients. Am J Transl Res. 2016. December 15;8(12):5672–5684. [PMC free article] [PubMed] [Google Scholar]
- 40.Ancuta P, Kamat A, Kunstman KJ, Kim EY, Autissier P, Wurcel A, et al. Microbial translocation is associated with increased monocyte activation and dementia in AIDS patients. PLoS One 2008; 3:e2516 10.1371/journal.pone.0002516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marchetti G, Cozzi-Lepri A, Merlini E, Bellistri GM, Castagna A, Galli M,et al. Microbial translocation predicts disease progression of HIV-infected antiretroviral-naive patients with high CD4+ cell count. AIDS 2011; 25(11):1385–94. 10.1097/QAD.0b013e3283471d10 [DOI] [PubMed] [Google Scholar]
- 42.Troseid M, Nowak P, Nyström J, Lindkvist A, Abdurahman S and Sönnerborg A, et al. Elevated plasma levels of lipopolysaccharide and high mobility group box-1 protein are associated with high viral load in HIV-1 infection: reduction by 2-year antiretroviral therapy. AIDS 2010; 24 (11):1733–1737. 10.1097/QAD.0b013e32833b254d [DOI] [PubMed] [Google Scholar]
- 43.Sandler NG, Wand H, Roque A, Law M, Nason MC, Emery S, et al. Plasma levels of soluble CD14 independently predict mortality in HIV Infection. J Infect Dis. 2011; 203 (6):780–790. 10.1093/infdis/jiq118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dillon SM, Lee EJ, Kotter CV, Robertson CE, Frank DN, Wilson CC, et al. An altered intestinal mucosal microbiome in HIV-1 infection is associated with mucosal and systemic immune activation and endotoxemia.Mucosal Immunol. 2014; 7(4):983–94. 10.1038/mi.2013.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marksa MA, Rabkin CS, Engels EA, Rager H, Goedert JJ, Chaturvedi AK, et al. Markers of microbial translocation and risk of AIDS-related lymphoma. AIDS 2013; 27(3)469–474. 10.1097/QAD.0b013e32835c1333 [DOI] [PubMed] [Google Scholar]
- 46.Kelesidis T, Kendall MA, Yang OO, Hodis HN, Currier JS. Biomarkers of microbial translocation and macrophage activation: association with progression of subclinical atherosclerosis in HIV-1 infection. J Infect Dis. 2012; 206 (10):1558–1567. 10.1093/infdis/jis545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blodget E, Shen C, Aldrovandi G, Gupta SK, Stein JH, Dubé MP, et al. Relationship between Microbial Translocation and Endothelial Function in HIV Infected Patients. PLoSONE 2012; 7(8): e42624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vázquez-Castellanos JF, Serrano-Villar S, Latorre A, Artacho A, Gosalbes MJ, Moya A, et al. Altered metabolism of gut microbiota contributes to chronic immune activation in HIV-infected individuals. Mucosal Immunol. 2015; 8(4):760–72. 10.1038/mi.2014.107 [DOI] [PubMed] [Google Scholar]
- 49.Jiang W, Lederman MM, Hunt P, Deeks SG, Douek DC, Brenchley JM, et al. Plasma levels of bacterial DNA correlate with immune activation and the magnitude of immune restoration in persons with antiretroviral-treated HIV infection. J Infect Dis. 2009; 199 (8): 1177–1185. 10.1086/597476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Piconi S, Trabattoni D, Gori A, Parisotto S, Capetti A, Rizzardini G, Clerici M, et al. Immune activation, apoptosis, and Treg activity are associated with persistently reduced CD4+ T-cell counts during antiretroviral therapy. AIDS 2010; 24 (13):1991–2000. 10.1097/QAD.0b013e32833c93ce [DOI] [PubMed] [Google Scholar]
- 51.Klatt NR, Funderburg NT and Brenchley JM. Microbial translocation, immune activation and HIV disease. Trends Microbiol. 2013; 21(1): 6–13. 10.1016/j.tim.2012.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Klase Z, Ortiz A, Deleage C, Mudd JC, Quiñones M, Schwartzman E, et al. Dysbiotic bacteria translocate in progressive SIV infection. Mucosal Immunol. September 2015;8(5):1009–1020. 10.1038/mi.2014.128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Williams B, Mirmonsef P, Boucher CA, Bushman F, Carrington-Lawrence S, Collman RG et al. A Summary of The First HIV Microbiome Workshop 2015. AIDS Res Hum Retroviruses. 2016. Oct-Nov;32(10–11):935–941. 10.1089/AID.2016.0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Swidsinski A, Loening-Baucke V, Schulz S, Manowsky J, Verstraelen H, Swidsinski S, et al. Functional anatomy of the colonic bioreactor: Impact of antibiotics and Saccharomyces boulardii on bacterial composition in human fecal cylinders. Syst Appl Microbiol. 2016. February;39(1):67–75 10.1016/j.syapm.2015.11.002 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
(DOC)
(DOC)
INR, immunologic non-responders.
(TIF)
ART, antiretroviral therapy; PI, protease inhibitor; NNRTI, nonnucleoside; reverse transcriptase inhibitor; LBP, Lipopolisaccharide Binding-Proteine; sCD14, soluble CD14; hs-CRP, high sensitivity C-reactive protein; ESR, Erythrocyte Sedimentation Rate; IQR, interquartile range.
(DOCX)
LBP, Lipopolisaccharide Binding-Proteine; sCD14, soluble CD14, hs-CRP, high sensitivity C-reactive protein.
(DOCX)
LBP_basal, Lipopolisaccharide Binding-Proteine at baseline; sCD14, soluble CD14; β2microglob, Beta 2 microglobuline.
(DOCX)
(CSV)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.