Abstract
Background
BRAFV600, NRAS, TP53 and BRAFNon-V600 are among the most common mutations detected in non-acral cutaneous melanoma (CM) patients. While several studies have identified clinical and pathologic features associated with BRAFV600 and NRAS mutations, limited data is available regarding the correlates and significance of TP53 and BRAFNon-V600 mutations.
Methods
We analyzed the patient demographics, primary tumor features, and clinical outcomes of a large cohort (n=926) of non-acral cutaneous melanoma patients that had undergone clinically-indicated molecular testing.
Results
The prevalence of BRAFV600, NRAS, TP53, and BRAFNon-V600 mutations were 43%, 21%, 19%, and 7%. The presence of a TP53 mutation was associated with older age (p=0.019), head and neck primary tumor site (p=0.0001), and longer overall survival (OS) from the diagnosis of stage IV disease on univariate (p=0.039) and multivariate (p=0.015) analyses. BRAFNon-V600 mutations were associated with older age (p=0.005), but no primary tumor features nor OS from stage IV. Neither TP53 nor BRAFNon-V600 mutations correlated significantly with OS with frontline ipilimumab treatment, and TP53 status was not significantly associated with outcomes with frontline BRAF inhibitor therapy. Eleven patients with BRAFNon-V600 mutations were treated with a BRAF inhibitor. Three patients were not evaluable for response due to treatment cessation for toxicities; the remaining patients had disease progression as the best response to therapy.
Conclusions
These results add to the understanding of the clinical features associated with TP53 and BRAFNon-V600 mutations in advanced CM patients, and they support the rationale to evaluate the prognostic significance of TP53 in other cohorts of melanoma patients.
Keywords: Melanoma, mutations, TP53, BRAFV600, BRAFNon-V600
Introduction
One of the most impactful discoveries in melanoma was the identification of missense mutations in the BRAF gene. Approximately 95% of these mutations result in substitutions for valine at position 600 of the BRAF protein (BRAFV600), most commonly with glutamic acid (BRAFV600E).1 These mutations markedly activate the RAS-RAF-MAPK signaling pathway. In addition to improving the understanding of the molecular pathogenesis of melanoma, this discovery led to the development of highly effective targeted therapies for patients with BRAFV600 mutations, including the mutant-selective BRAF inhibitors (BRAFi) vemurafenib and dabrafenib and the MEK inhibitors (MEKi) trametinib and cobimetinib. Together these developments strongly support the rationale to interrogate the significance of other somatic mutations in this disease to identify additional personalized therapeutic strategies.
Recently, we reported the results of a clinical next-generation sequencing (NGS) panel that encompassed commonly mutated regions in 46 genes in a large cohort of advanced melanoma patients.2 The most common mutations detected in the cutaneous melanoma patients were BRAFV600 (41%), NRAS (22%), TP53 (17%) and BRAFNon-V600 (7%). Previous studies have that interrogated the clinical and pathological features that correlate with BRAFV600 and NRAS mutations have identified a number of significant associations.3 However, very little is known about features of melanomas that are associated with TP53 mutations, which correlate with poor clinical outcomes in head and neck cancer4 and hematologic malignancies.5 Preclinical studies support that BRAFNon-V600 mutations are a potential therapeutic target,6 and clinical trials are ongoing to determine the efficacy of trametinib (MEKi) in melanoma patients with these mutations (NCT02296112). However, little is known at this time about the clinical features or outcomes associated with BRAFNon-V600 mutations in melanoma patients.
We have performed a retrospective analysis of a large single-institution cohort of advanced cutaneous melanoma patients with clinical NGS testing results, which encompassed regions of prevalent hotspot mutations in 50 genes. We have interrogated this cohort to identify clinical and pathological features that are associated with the presence of TP53 and BRAFNon-V600 mutations to improve our understanding of their significance in this disease.
Materials and Methods
Patient Selection and Clinical Data Collection
Under an Institutional Review Board-approved protocol, the results of clinically indicated molecular testing performed at The University of Texas MD Anderson Cancer Center (MDACC) from April of 2012 to November of 2014 for patients with non-acral cutaneous melanoma were reviewed. Patient demographics, primary tumor characteristics, treatments received, and overall survival were collected.
Mutation Testing
Molecular testing by a pan-cancer NGS panel of hotspot regions in 50 genes [Supplemental Table 1] was performed on DNA extracted from formalin-fixed, paraffin-embedded tissues from melanoma primary tumors or metastases using the AmpliSeq sequencing panel (CMS50; Life Technologies) as previously described.2, 7
Among detected mutations in the panel, TP53 mutations were further classified based on physicochemical or functional consequences including truncating mutations, missense mutations, DNA-binding domain mutations and UV signature mutations. Truncating mutations are mutations leading to a stop codon, frameshift and splice defect.8 In contrast, missense mutations are characterized by an amino acid change which results in a dominant-negative or a gain-of-function.9 DNA-binding domain mutations are mutations in codons 102 to 292 which induce inactivation of TP53 by eliminating DNA-binding contacts or altering structural stability of the core domain.9 Missense mutations are further stratified into high (≥75) and low risk (<75) by an evolutionary action score system (http://mammoth.bcm.tmc.edu/EAp53/) to predict highly deleterious TP53 functions which was validated in head and neck squamous cell carcinoma.10
In order to verify the coverage of CMS50 panel, potential hotspot mutations in cutaneous melanoma The Tumor Genome Atlas (TCGA) data11 were identified using the HotSpotter method as previously described which allows rapid and easy visualization of mutation data sets and identification of potential gene mutation hotspot sites and/or regions.12 The identified potential hotspot mutations by the HotSpotter analysis were compared to CMS50 panel.
Statistical Methods
The association between continuous parameters and mutational status was assessed by analysis of variance (ANOVA). Fisher's exact test was used to assess the association between categorical variables (primary tumor site, ulceration, elevated LDH) and mutational status. Wilcoxon rank sum tests were used to assess the association between continuous and ordinal variables (age, Breslow thickness, M stage) and mutational status. The method of Kaplan and Meier was used to estimate the distribution of overall survival, and log-rank testing was performed to determine the significance of observed differences. All statistical analyses were performed using R version 3.1.1. All statistical tests used a significance level of 5%. No adjustments for multiple testing were made.
Results
Patients Demographics and Mutation Frequency
A total of 926 patients with a non-acral cutaneous primary melanoma and clinical NGS testing results using the CMS50 panel were identified [Table 1]. The median age of the patients was 56 years (range 12-94) and 67.4% were male. Superficial spreading melanoma was the most common melanoma subtype, and the trunk was the most common primary tumor site. 531 patients (57.3%) were diagnosed with stage IV disease.
Table 1. Patients Demographics.
| Variable | All patients (N=926) |
|---|---|
| Median Age (range) | 56 (12-94) |
| Gender | |
| Male | 624 (67.4%) |
| Female | 302 (32.6%) |
| Subtype | |
| superficial spreading | 313 (33.8%) |
| nodular | 184 (19.9%) |
| lentigo maligna | 88 (9.5%) |
| desmoplastic | 7 (0.8%) |
| unclassified | 82 (8.9%) |
| not documented | 252 (27.2%) |
| Primary site | |
| Trunk | 339 (36.6%) |
| extremity | 256 (27.6%) |
| head and neck | 317 (34.2%) |
| other | 14 (1.5%) |
A BRAFV600 mutation was detected in 398 patients (43.0%), NRAS mutation in 195 (21.1%), TP53 mutation in 180 (19.4%), and BRAFNon-V600 mutation in 60 (6.5%) [Table 2]. The prevalence of the specific BRAFV600, NRAS, TP53 and BRAFNon-V600 substitutions detected in the cohort are presented in Supplemental Tables 2 – 5. TP53 mutations were detected in 11.3% of melanomas with a BRAFV600 mutation, 19.4% of tumors with an NRAS mutation, and 28.9% of tumors without a BRAFV600 or NRAS mutation [Supplemental Table 6 and Supplemental Fig 1A]. A BRAFNon-V600 mutation was detected in 0.5% of melanomas with a BRAFV600 mutation, 4.1% of tumors with an NRAS mutation, and 14.9% of tumors without a BRAFV600 or NRAS mutation [Supplemental Table 7 and Supplemental Fig 1B]. TP53 mutations were present in 8.9% of tumors with a BRAFNon-V600 mutation.
Table 2.
Prevalence of mutations in full cohort of non-acral cutaneous melanomas.
| Mutation | No. of pt (%) | Mutation | No. of pt (%) |
|---|---|---|---|
| BRAFV600 | 398 (43.0%) | HNF1A | 9 (1.0%) |
| NRAS | 195 (21.1%) | RET | 9 (1.0%) |
| TP53 | 180 (19.4%) | STK11 | 8 (0.9%) |
| BRAFNon-V600 | 60 (6.5%) | CSF1R | 7 (0.8%) |
| CDKN2A | 59 (6.4%) | GNAS | 7 (0.8%) |
| PTEN | 36 (3.9%) | EZH2 | 6 (0.6%) |
| KIT | 35 (3.8%) | MLH1 | 5 (0.5%) |
| ERBB4 | 34 (3.7%) | SMAD4 | 5 (0.5%) |
| EGFR | 31 (3.3%) | SMARCB1 | 5 (0.5%) |
| KDR | 30 (3.2%) | SMO | 5 (0.5%) |
| IDH1 | 27 (2.9%) | ABL1 | 4 (0.4%) |
| CTNNB | 24 (2.6%) | AKT1 | 4 (0.4%) |
| KRAS | 24 (2.6%) | FGFR1 | 4 (0.4%) |
| ATM | 18 (1.9%) | NOTCH1 | 4 (0.4%) |
| APC | 16 (1.7%) | GNAQ | 3 (0.3%) |
| PKI3CA | 16 (1.7%) | SRC | 3 (0.3%) |
| RB1 | 16 (1.7%) | JAK2 | 2 (0.2%) |
| FLT3 | 15 (1.6%) | JAK3 | 2 (0.2%) |
| MET | 15 (1.6%) | VHL | 2 (0.2%) |
| PTPN11 | 15 (1.6%) | ALK | 1 (0.1%) |
| FBXW7 | 13 (1.4%) | ERBB2 | 1 (0.1%) |
| HRAS | 13 (1.4%) | GNA11 | 1 (0.1%) |
| FGFR2 | 11 (1.2%) | MPL1 | 1 (0.1%) |
| PDGFRA | 11 (1.2%) | NPM1 | 1 (0.1%) |
| FGFR3 | 9 (1.0%) |
Table 5. Multivariate analysis of survival from diagnosis of stage IV melanoma.
| HR | 95% CI | P value | |
|---|---|---|---|
| Age | 1.01 | 1.00-1.02 | 0.05 |
| Gender | |||
| Female | |||
| Male | 0.82 | 059-1.14 | 0.23 |
| stage IV M1 classification | |||
| M1a | |||
| M1b | 0.91 | 0.42-1.97 | 0.8 |
| M1c | 1.92 | 0.92-4.00 | 0.08 |
| LDH | |||
| WNL | |||
| elevated | 1.01 | 0.70-1.47 | 0.94 |
| unknown | 0.8 | 0.11-5.93 | 0.83 |
| Mutation | |||
| BRAFV600 mutation (vs BRAFV600 ND) | 0.93 | 0.64-1.34 | 0.7 |
| NRAS mutation (vs NRAS ND) | 1.2 | 0.82-1.76 | 0.35 |
| BRAFNon-V600 mutation (vs BRAFNon-V600) | 0.74 | 0.39-1.76 | 0.37 |
| TP53 mutation (vs TP53 ND) | 0.62 | 0.42-0.91 | 0.015 |
CI: confidence interval, HR: hazard ratio, LDH: lactate dehydrogenase, WNL: within normal limit, ND: not detected
Clinical and pathologic characteristics associated with TP53 mutations
Analysis of the full patient cohort showed that the presence of a TP53 mutation that was detectable by the clinical CMS50 panel was associated with increased age at diagnosis (median 58 versus 56 years, p=0.019) [Table 3]. TP53 mutations were also significantly associated with primary tumor site (p=0.0001), as they were more common in tumors of the head and neck (27.4%) than the trunk (15.9%) or extremity (14.7%). TP53 mutations were not significantly associated with primary tumor thickness or ulceration status. Among the 531 patients that had developed stage IV disease, there was no significant association between TP53 mutation status and M stage or serum LDH at the diagnosis of stage IV disease.
Table 3. Demographics, primary tumor features, and clinical outcomes of melanoma patients with TP53 or BRAFNon-V600 mutations.
| Variable | All (N=926) | |||||
|---|---|---|---|---|---|---|
|
| ||||||
| TP53 mutant | ND TP53 | P value | BRAFNon-V600 mutant | ND BRAFNon-V600 | P value | |
| No. of Patients (%) | 180 (19.4%) | 746 (80.6%) | 60 (6.5%) | 866 (93.5%) | ||
| Median Age (range) | 58 (12-89) | 56 (13-94) | 0.019 | 60 (29-89) | 56 (12-94) | 0.005 |
| Gender (%) | 0.052 | 0.88 | ||||
| Male | 132 (21.2%) | 492 (78.8%) | 41 (6.6%) | 583 (93.4%) | ||
| Female | 48 (15.9%) | 254 (84.1%) | 19 (6.3%) | 283 (93.7%) | ||
| Primary site (%) | 0.0001 | 0.38 | ||||
| Trunk | 54 (15.9%) | 285 (84.1%) | 21 (6.2%) | 318 (93.8%) | ||
| Extremity | 38 (14.8%) | 218 (85.2%) | 12 (4.7%) | 244 (95.3%) | ||
| Head and neck | 87 (27.4%) | 230 (72.6%) | 26 (8.2%) | 291 (91.8%) | ||
| Other | 1 (7.1%) | 13 (92.9%) | 1 (7.1%) | 13 (92.9%) | ||
| Breslow thickness (mean) | 3.31mm | 3.35mm | 0.9 | 2.9mm | 3.38mm | 0.32 |
| Ulceration (%) | 0.9 | 0.63 | ||||
| Yes | 47 (18.7%) | 205 (81.3%) | 18 (7.1%) | 234 (92.9%) | ||
| No | 80 (19.2%) | 337 (80.8%) | 25 (6.0%) | 392 (94.0%) | ||
| Not available | 53 (20.6%) | 204 (79.4%) | 17 (6.6%) | 240 (93.4%) | ||
|
| ||||||
| Patients with stage IV | N=531 | |||||
|
| ||||||
| No. of Patients (%) | 107 (20.1%) | 424 (79.9%) | 41 (7.7%) | 490 (92.3%) | ||
| M stage (%) | 0.5 | 0.56 | ||||
| M1a | 5 (15.2%) | 28 (84.8%) | 1 (3.0%) | 32 (97.0%) | ||
| M1b | 22 (23.9%) | 70 (76.1%) | 8 (8.7%) | 84 (91.3%) | ||
| M1c | 80 (19.7%) | 326 (80.3%) | 32 (7.9%) | 374 (92.1%) | ||
| LDH at stage IV | 0.6 | 0.3 | ||||
| Elevated (%) | 57 (19.3%) | 239 (80.7%) | 26 (8.8%) | 270 (91.2%) | ||
| WNL (%) | 46 (20.4%) | 180 (79.6%) | 14 (5.9%) | 212 (94.1%) | ||
| Missing (%) | 4 (44.4%) | 5 (55.6%) | 1 (11.1%) | 8 (88.9%) | ||
|
| ||||||
| Patients with stage IV (tested within 12 mo) | N=417 | |||||
|
| ||||||
| Median OS from stage IV (Months, 95% CI) | 18.8 (14.0-23.6) | 15.3 (13.4-17.2) | 0.039 | 17.9 (14.7-17.9) | 16.1 (14.3-17.9) | 0.53 |
CI: confidence interval, LDH: lactate dehydrogenase, No.: Number, OS: overall survival, WNL: within normal limit, ND: not detected mutation
Similar analyses were performed to assess the characteristics associated with TP53 mutations separately in melanomas with a concurrent BRAFV600 mutation, in melanomas with a concurrent NRAS mutation, and in melanomas without a BRAFV600 or NRAS mutation [Supplemental Table 6]. There were no significant associations with TP53 mutation status in tumors with BRAFV600 mutations or with NRAS mutations. Significant associations with age at diagnosis (p=0.001) and primary tumor site (p=0.009) were observed in tumors without a BRAFV600 or NRAS mutation.
TP53 mutations and overall survival from stage IV
In order to avoid bias due to inclusion of patients with unusually long survival intervals prior to molecular testing, the analysis of outcomes after stage IV diagnosis were limited to patients with clinical NGS testing performed within 12 months of stage IV diagnosis (n=417). The prevalence of TP53 mutations detected by the CMS50 panel among these patients was 21.1%. Patients with a TP53 mutation had longer overall survival (OS) from the diagnosis of stage IV on univariate analysis (median 18.8 versus 15.3 months, p=0.039) [Fig. 1A]. OS from stage IV was very similar for patients with a BRAFV600 mutation with or without a TP53 mutation (median 17.5 versus 16.9 months, p=0.89) [Supplemental Fig. 2A]. In contrast, non-significant trends for longer OS with TP53 mutation were observed among patients with NRAS mutations (median 21.7 versus 13.0 months, p=0.07) and among patients without a BRAFV600 or NRAS mutation (median 19.3 versus 15.8 months, p=0.08) [Supplemental Fig. 2B/C].
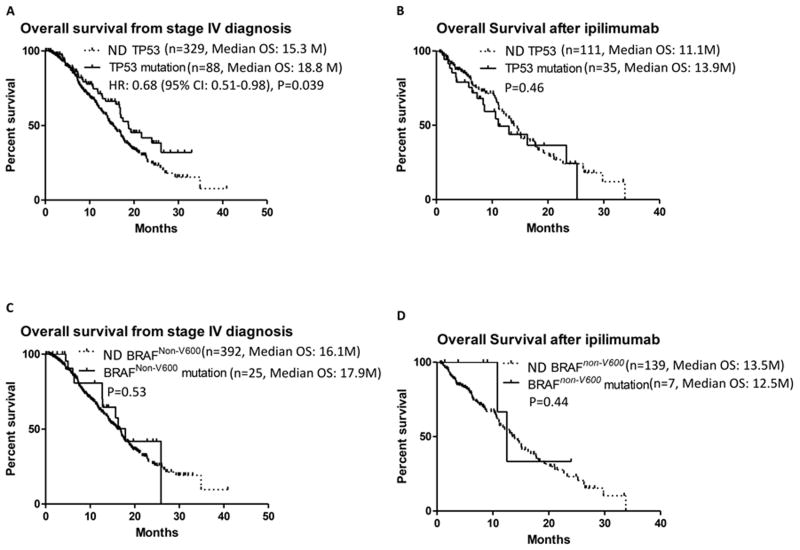
Figure 1.
Kaplan Meier analysis of survival after stage IV diagnosis (A) and after frontline ipilimumab treatment (B) based on TP53 mutation status. Kaplan Meier survival curve after stage IV diagnosis (C) and after frontline ipilimumab treatment (D) based on BRAFNon-V600 mutation status. Dashed line, not detected gene mutation (ND); solid line, gene mutation present.
To further evaluate the prognostic significance of TP53 mutations, outcomes with FDA-approved therapies among the stage IV cohort were reviewed. A total of 146 patients with clinical NGS testing performed within 12 months of stage IV diagnosis were treated with frontline ipilimumab, including 35 patients with a TP53 mutation. No significant correlation was observed between TP53 mutation status and OS from the start of ipilimumab treatment in the full cohort (median 11.1 months for TP53 mutant versus 13.9 months for TP53 mutation not detected, p=0.46) [Fig. 1B]. There was also no significant association with OS from the start of ipilimumab by TP53 mutation among the cohorts defined by the concurrent BRAFV600 and NRAS mutation status [Supplemental Fig. 3A, B and C].
Among the patients with a BRAFV600 mutation, 64 patients in the cohort were treated with an FDA-approved BRAF inhibitor as first-line therapy for stage IV disease [Supplemental Table 8]. The objective response rate (ORR) did not differ significantly between the patients with (n=9, ORR 55.6%) and without a TP53 mutation (n=55, ORR 47.3%, p=0.7). TP53 mutation status was also not significantly associated with progression-free survival (PFS) (p=0.42) or OS (p=0.49) with BRAF inhibitor treatment [Supplemental Fig. 4A and B].
The outcomes associated with TP53 mutations were further characterized by analyzing the nature of the observed mutations in the stage IV patients, as different types of mutations in TP53 may have different prognostic significance. Consistent with the strong association of non-acral cutaneous melanoma with UVR exposure, 58.2% of the detected TP53 mutations in the cohort were substitutions that are associated with UVR-related DNA damage. This rate is relatively similar to the rate observed in the cutaneous melanoma TCGA cohort (45.3%).11 Patients with UVR-related (n=55, median OS 17.5 months) and non-UVR related TP53 mutations (n=31, median OS 24.0 months) had trends for improved OS from stage IV compared to patients without a detectable TP53 mutation (median OS 15.3 months), but these differences did not reach statistical significance (p=0.09 and p=0.11, respectively) [Supplemental Fig. 5A]. Most TP53 mutations were missense mutations (n=61, median OS 19.3 months), and patients with such mutations had improved OS compared to patients without TP53 mutations detected by the CMS50 panel (p=0.002). In contract, the presence of a truncating mutation in TP53 was less common (n=21) and was not associated with a significant difference in survival from stage IV (median OS 12.9 months, p=0.93) [Supplemental Fig. 5B]. The majority (n=74; 84.1%) of the mutations detected in TP53 affected the DNA binding domain of the protein, and the presence of such mutations was associated with improved OS compared to patients without a mutation detected (median OS 18.6 months, p=0.05). Although mutations in other domains of TP53 were not significantly associated with survival from stage IV (p=0.39), there was very limited power for this analysis due to the small number of such patients in this cohort (n=11) [Supplemental Fig. 5C]. Finally, categorization of the TP53 mutations into two groups based on evolutionary score10 showed that high risk evolutionary score (N=51) correlated significantly with OS from stage IV (median OS 21.7 months, p=0.04) but not low risk (N=10, median OS 19.3 months, p=0.16) [Supplemental Fig. 5D].
Clinical and pathologic characteristics associated with BRAFNon-V600 mutations
Twenty seven different BRAFNon-V600 mutations were detected in a total of 60 patients in the cohort [Supplemental Table 5]. BRAFNon-V600 mutations were detected in 2 patients with BRAFV600 mutations (0.5%), 8 patients with NRAS mutations (4.1%), and 50 patients with neither BRAFV600 nor NRAS mutations (14.9%) [Supplemental Fig. 1B]. In the full cohort, the presence of a BRAFNon-V600 mutation (6.5%) was associated with significantly older age at diagnosis (median 60 versus 56, p=0.005) [Table 3]. No other significant correlations with patient demographics or primary tumor characteristics were identified. Among the 531 patients diagnosed with stage IV disease, the presence of a BRAFNon-V600 mutation (n=41, 7.7%) was not associated significantly with M1 stage or serum LDH levels.
Clinical and pathologic characteristics of BRAFNon-V600 mutations were examined separately in melanomas with a concurrent BRAFV600, a concurrent NRAS mutation, and in melanomas without a BRAFV600 or NRAS mutation (Supplemental Table 7). Non-significant trends for deeper Breslow thickness with BRAFNon-V600 mutation were observed in tumors without a BRAFV600 or NRAS mutation (p=0.08), but no statistically significant associations were detected.
BRAFNon-V600 mutations and overall survival from stage IV
A BRAFNon-V600 mutation was detected in 25 (6.0%) of patients with mutation testing performed within 12 months of the diagnosis of stage IV disease. There was no significant difference in OS from stage IV between patients with (median 17.9 months) and those without (median 16.1 months, p=0.53) a BRAFNon-V600 mutation [Figure 1C]. There was no patient with a BRAFV600 mutations who had mutation testing within 12 months of the diagnosis of stage IV diagnosis who had a concurrent BRAFNon-V600 mutation, and only 1 among 86 patients with an NRAS mutation. Analysis of patients without a concurrent BRAFV600 or NRAS mutation failed to detected a significant association between OS from stage IV and BRAFNon-V600 mutation status (p=0.95) [Supplemental Figure 6].
Twenty one (8.9%) of the patients treated with ipilimumab had a BRAFNon-V600 mutation. There was no significant correlation between BRAFNon-V600 mutation status and OS from the start of ipilimumab treatment was observed (median 12.5 versus 13.5 months, p=0.44) [Figure 1D].
Eleven patients with a BRAFNon-V600 mutation were treated with an FDA-approved BRAF inhibitor (vemurafenib, n=9; dabrafenib, n=1; dabrafenib + trametinib, n=1). All of the patients had stage 4 disease, and none of the patients had a concurrent BRAFV600 or NRAS mutation [Table 4]. Three patients treated with vemurafenib were not evaluable for response due to early severe drug related toxicities; the remaining 8 patients had disease progression as their best response to therapy. The median PFS and OS were 1.9 months (95% CI: 1.48-2.32) and 7.1 months (95% CI: 4.54-9.66), respectively [Supplemental Figure 7A and B]. Since BRAF K601 and L597 mutations are located in the activating kinase domain of BRAF, and a BRAF inhibitor demonstrated some antitumor activity in a patient with a BRAF L597 mutant melanoma,13 clinical outcomes of patients with BRAF K601 and L597 mutations and other BRAFNon-V600 mutations with BRAF inhibitor treatment. There was no difference in PFS and OS between patients with BRAF K601 and L597 mutations and the other BRAFNon-V600 mutations [supplemental Figure 7C and D]. No responses were observed among 3 patients with BRAFNon-V600 mutations (BRAF L597R, G466E, G469E) treated with trametinib (MEKi) monotherapy [Table 4].
Table 4. Clinical outcomes of patients with BRAFnon-V600 mutations treated with a selective BRAF inhibitor and/or a MEK inhibitor.
| Patient # | Age | Gender | BRAFNon-V600 mutation | Stage | Treatment | Response | PFS (Months) | OS (Months) |
|---|---|---|---|---|---|---|---|---|
| 1 | 84 | M | G469R | M1c | Vemurafenib | NE | NE | NE |
| 2 | 35 | M | K601E | M1c | Vemurafenib | NE | NE | NE |
| 3 | 85 | M | G469R | M1a | Vemurafenib | NE | NE | NE |
| 4 | 56 | F | L597R | M1c | Vemurafenib | PD | 4.3 | 11.2 |
| 5 | 51 | M | S467L | M1c | Vemurafenib | PD | 1.9 | 5.6 |
| 6 | 69 | F | G466V | M1c | Vemurafenib | PD | 0.9 | 3.7 |
| 7 | 44 | M | L597Q | M1c | Vemurafenib | PD | 2.0 | 5.4 |
| 8 | 70 | M | G469R | M1c | Vemurafenib | PD | 3.5 | 9.1 |
| 9 | 74 | M | G466E | M1c | Vemurafenib | PD | 1.9 | + 30.6 |
| 10 | 52 | M | K601E | M1c | Dabrafenib | PD | 0.7 | + 64.5 |
| 11 | 36 | M | K601E | M1c | Dabrafenib and trametinib | PD | 2.0 | + 4.5 |
| 12 | 57 | F | L597R | M1c | Trametinib | PD | 1.7 | 4.8 |
| 13 | 76 | M | G466E | M1c | Trametinib | PD | 1.5 | 5.2 |
| 14 | 66 | M | G469E | M1c | Trametinib | PD | 1.3 | 7.1 |
PFS, progression free survival; OS, overall survival; PD, progressive disease; NE, not evaluable.
Multivariate analysis
Multivariate Cox regression modeling was performed to identify significant predictors of OS from stage IV diagnosis, including gender, M1 stage, LDH, and BRAFV600, NRAS, TP53 and BRAFNon-V600 mutation status [Table 5]. Age and the presence of stage IV M1c disease were associated with non-significant trend of shorter OS from stage IV (p=0.05 and p=0.08). The presence of a TP53 mutation was associated with improved OS from stage IV (HR 0.62, 95% CI 0.42-0.91, p=0.015).
Discussion
A number of previous studies have described the clinical and pathologic features associated with BRAFV600 and NRAS mutations,3, 14, 15 which are the most common hotspot oncogenic mutations detected in cutaneous melanomas. While many other mutations have been identified in melanoma, the ability to assess their clinical associations has been limited by their relative scarcity. In this study, we have utilized a retrospective cohort of 926 non-acral cutaneous melanoma patients that underwent clinically indicated NGS testing to expand the integrated analysis of molecular and clinical features in this disease. This analysis focused on TP53 and BRAFNon-V600 mutations, as they are particularly common in cutaneous melanomas without BRAFV600 or NRAS mutations, and as preclinical data supports potential functional roles for both events in this disease.
TP53 encodes a tumor suppressor gene that is altered in many cancer types. Similar with the rate observed in the melanoma TCGA (15%),11 we identified TP53 mutations in 19% of the full cohort of cutaneous melanoma patients who had undergone molecular testing with the clinical CMS50 NGS panel. Compared to patients who did not have an alteration detected by this panel, TP53 mutations were associated with older age and increased head and neck primary tumor location. These clinical associations are consistent with the observed correlation between ultraviolet radiation (UVR)-induced DNA damage and TP53 mutations in cutaneous melanoma.16, 17 In this cohort, 58% of TP53 mutations were consistent with UV-induced DNA damage, which is again similar to the rate reported in the melanoma TCGA (45.3%).11 While preclinical studies have shown that TP53 mutations accelerate BRAFV600-driven melanomagenesis,17, 18 we observed that TP53 mutations were associated with a significantly improved survival from the diagnosis of stage IV disease on univariate and multivariate analyses. The observed association of improved outcomes in melanomas with TP53 mutations contrasts with associations with poor clinical outcomes in a number of other cancers, including breast cancer,19 head and neck cancer,4 and hematologic malignancies.5 However, these results are consistent with a previous study that identified improved clinical outcomes in melanomas with overexpression of the P53 protein,20 which is usually associated with mutations in the TP53 gene. Our analysis also showed that missense substitution mutations in TP53, mutations that affected the DNA binding domain of the P53 protein and high risk evolutionary score of TP53 mutations were associated with significant improvements in OS from stage IV, but truncating mutations, mutations affecting other domains and low risk evolutionary score were not. While these findings may further help to interpret the results of TP53 molecular testing results, the analysis of truncating mutations, mutations affecting other domains and low risk evolutionary score were limited by the small number of patients with such alterations in this cohort.
At this time it is unclear why TP53 mutations are associated with improved survival in stage IV melanoma patients. The presence of a somatic mutation in TP53 was also associated with a trend for improved OS from initial diagnosis in the full melanoma TCGA cohort, as patients with a TP53 mutation (n=64) had a median OS of 170 months, while patients without a TP53 mutation (n=350) had a median OS of 114 months (p=0.44). Analysis limited to the TCGA patients with stage III or IV metastases also showed a trend for improved OS with a TP53 mutation (median OS Not reached versus 72.8 months, p=0.25). While the observed differences are interesting, the TCGA cohort included stage II-IV melanomas, many tumors were collected after previous recurrences, the date of stage IV disease is unknown for the overwhelming majority of patients, and there is not sufficient data to evaluate associations with known stage-specific prognostic factors for the cohort. Thus, studies of additional cohorts with appropriate power and annotation will need to be evaluated in the future to further assess the prognostic significance of TP53 mutations in melanoma. Notably, our analysis of survival was limited to patients with molecular testing performed within 12 months of stage IV diagnosis to avoid biasing results by including patients who survived for a prolonged period of time before undergoing molecular testing, another factor to consider in the evaluation of molecular prognostic factors. Recent studies support that increased missense mutation burden correlates with improved survival in metastatic melanoma patients.21 As TP53 mutations are associated with increased mutation burden, including in the melanoma TCGA cohort (p<0.0001 by Mann-Whitney test), it is possible that the presence of this mutation signifies the likelihood of increased mutation burden, and therefore enhanced anti-tumor immune surveillance. While this is a reasonable hypothesis, we did not observe a significant association between the presence of a TP53 mutation and an improvement in overall survival with ipilimumab. As the relatively low response and long-term survival rates for ipilimumab are modest, this analysis was somewhat limited to detect a positive association. Additional studies may be performed in the future to examine outcomes in patients treated with more active immunotherapy regimens such as anti-PD-1 antibodies and adoptive T cell therapy.
BRAFNon-V600 mutations were detected in 6% of the full cohort of cutaneous melanomas in this study, also consistent with our previous report2 and the melanoma TCGA. BRAFNon-V600 mutations were extremely rare in cutaneous melanomas with a concurrent BRAFV600 mutation (0.5%), but they were more frequent in melanomas with an NRAS mutation (4%) and in melanomas with neither of those oncogenic driver mutations (15%). In contrast to the TP53 mutation analysis, there was no significant association observed between BRAFNon-V600 mutations and overall survival from stage IV diagnosis or with ipilimumab treatment.
Consistent with previous clinical data,22, 23 patients with BRAFNon-V600 mutations in this cohort that were treated with FDA-approved BRAF inhibitors had no clinical responses. The failure of a selective BRAF inhibitor in BRAFNon-V600 mutant melanoma can be explained by preclinical data which showed activation of endogenous CRAF in BRAFnon-V600 mutant melanoma.6 In contrast to other BRAFNon-V600 mutations, BRAF K601 and L597 mutations are located in the activating domain of BRAF, and a clinical response to a BRAF inhibitor has previously been reported in one metastatic melanoma patient with a BRAF L597 mutation.13 However, we did not observe any clinical responses in this study in patients with a BRAFNon-V600 mutation. While there is evidence that MEK inhibitors may be an effective strategy for melanomas with BRAFNon-V600 mutations, and objective response to a selective MEK inhibitor was reported in BRAF L597S mutant melanoma,6, 24 no responses were seen among 3 patients with BRAF L597R, G466E or G469E mutations in this cohort that were treated with trametinib. A prospective, open-label phase II study (NCT02296112), is currently ongoing to further characterize the safety and activity of trametinib in patients with BRAFNon-V600 mutations, and particularly to interrogate if clinical activity is enhanced with specific mutations. Notably the findings that BRAFNon-V600 mutations do not correlate significantly with overall survival from stage IV or with ipilimumab treatment is beneficial for the interpretation of the clinical results seen in that study.
As our study was conducted at a single tertiary cancer center, it is possible that different results may be detected in other melanoma patient populations. Indeed, we do note that serum LDH, which is part of the AJCC staging system for stage IV melanoma, was not prognostic in the cohort of patients with molecular testing with 12 months of the diagnosis of stage IV. The reason for this finding in this cohort is unknown. However, our cohort is quite large, and thus likely represents significant clinical heterogeneity that is seen at melanoma centers. Notably, the clinical NGS panel did not perform NGS of the entire encoding region of TP53 or BRAF [Supplemental Table 1]. Thus, the actual prevalence of TP53 and BRAFNon-V600 mutations could be higher than were detected using this clinical panel. However, the observed rates of TP53 and BRAFNon-V600 mutations are very similar to those reported in the initial report of the melanoma TCGA effort,11 which included whole exome sequencing (WES). This is likely due to the fact that the regions that were sequenced in the clinical panel were selected based on existing data about the location of common mutations. Indeed, the residues in TP53 that are assessed for mutations in the CMS50 panel largely overlap with the mutations that were detected in the melanoma TCGA [Supplemental Fig. 8]. However, we do note the limitation that the clinical CMS50 panel is not optimal to detect medium and long deletions, nor chromosomal copy number changes, affecting the TP53 gene. Based on our observations, there is a strong rationale for a comprehensive evaluation of the TP53 gene in other cohorts of clinically annotated stage IV melanoma patients.
In conclusion, this study reports the analysis of the largest single-institution cohort of clinically and molecularly characterized advanced cutaneous melanoma patients to date. The size of this cohort has allowed us to interrogate the clinical and pathological features that are significantly associated with TP53 and BRAFNon-V600 mutations. Our results suggest that in contrast to many other tumor types, TP53 mutations may correlate with a favorable prognosis in advanced cutaneous melanoma patients, at least for those who undergo molecular testing within 12 months of stage IV diagnosis. In addition, our studies show that BRAFNon-V600 mutations are not prognostic in stage IV melanoma patients, information which will augment the design and interpretation of current and future clinical trials in this patient population. Further studies including prospective and mechanistic analysis are needed to validate our findings.
Supplementary Material
Acknowledgments
This work was supported by the MDACC Melanoma SPORE (P50 CA093459), NIH National Cancer Institute grant (T32 CA163185 and CA009666), philanthropic contributions to the MDACC Melanoma Moon Shots Program, the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation, the Melanoma Research Foundation, the AIM at Melanoma Foundation, and the Miriam and Jim Mulva Research Funds.
Footnotes
Conflicts of interest: There are no conflicts of interest.
Author Contributions: Clinical and Pathological Data Collection: Lauren Haydu, Alan Siroy, Michael Tetzlaff, Rodabe Amaria, Jennifer Wargo, Jennifer McQuade, Jeff Gershenwald,
Molecular Data Annotation: Aron Joon, Mark Routbort, Jan Kemnade, Scott Woodman, Alexander Lazar
Data Analysis: Aron Joon, Roland Bassett, Jason Roszik
Manuscript Preparation: Dae Won Kim, Michael Davies, Patrick Hwu, Alexander Lazar, Jeff Gershenwald, Jennifer Wargo, Scott Woodman, Rodabe Amaria
Overall content guarantor: Michael Davies
References
- 1.Hodis E, Watson IR, Kryukov GV, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siroy AE, Boland GM, Milton DR, et al. Beyond BRAF(V600): clinical mutation panel testing by next-generation sequencing in advanced melanoma. J Invest Dermatol. 2015;135:508–515. doi: 10.1038/jid.2014.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ellerhorst JA, Greene VR, Ekmekcioglu S, et al. Clinical correlates of NRAS and BRAF mutations in primary human melanoma. Clin Cancer Res. 2011;17:229–235. doi: 10.1158/1078-0432.CCR-10-2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poeta ML, Manola J, Goldwasser MA, et al. TP53 mutations and survival in squamous-cell carcinoma of the head and neck. N Engl J Med. 2007;357:2552–2561. doi: 10.1056/NEJMoa073770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wattel E, Preudhomme C, Hecquet B, et al. p53 mutations are associated with resistance to chemotherapy and short survival in hematologic malignancies. Blood. 1994;84:3148–3157. [PubMed] [Google Scholar]
- 6.Wan PT, Garnett MJ, Roe SM, et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116:855–867. doi: 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 7.Singh RR, Patel KP, Routbort MJ, et al. Clinical validation of a next-generation sequencing screen for mutational hotspots in 46 cancer-related genes. J Mol Diagn. 2013;15:607–622. doi: 10.1016/j.jmoldx.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 8.Petitjean A, Mathe E, Kato S, et al. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum Mutat. 2007;28:622–629. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- 9.Petitjean A, Achatz MI, Borresen-Dale AL, Hainaut P, Olivier M. TP53 mutations in human cancers: functional selection and impact on cancer prognosis and outcomes. Oncogene. 2007;26:2157–2165. doi: 10.1038/sj.onc.1210302. [DOI] [PubMed] [Google Scholar]
- 10.Neskey DM, Osman AA, Ow TJ, et al. Evolutionary Action Score of TP53 Identifies High-Risk Mutations Associated with Decreased Survival and Increased Distant Metastases in Head and Neck Cancer. Cancer Res. 2015;75:1527–1536. doi: 10.1158/0008-5472.CAN-14-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cancer Genome Atlas N. Genomic Classification of Cutaneous Melanoma. Cell. 2015;161:1681–1696. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roszik J, Woodman SE. HotSpotter: efficient visualization of driver mutations. BMC Genomics. 2014;15:1044. doi: 10.1186/1471-2164-15-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bahadoran P, Allegra M, Le Duff F, et al. Major clinical response to a BRAF inhibitor in a patient with a BRAF L597R-mutated melanoma. J Clin Oncol. 2013;31:e324–326. doi: 10.1200/JCO.2012.46.1061. [DOI] [PubMed] [Google Scholar]
- 14.Jakob JA, Bassett RL, Jr, Ng CS, et al. NRAS mutation status is an independent prognostic factor in metastatic melanoma. Cancer. 2012;118:4014–4023. doi: 10.1002/cncr.26724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Long GV, Menzies AM, Nagrial AM, et al. Prognostic and clinicopathologic associations of oncogenic BRAF in metastatic melanoma. J Clin Oncol. 2011;29:1239–1246. doi: 10.1200/JCO.2010.32.4327. [DOI] [PubMed] [Google Scholar]
- 16.Zerp SF, van Elsas A, Peltenburg LT, Schrier PI. p53 mutations in human cutaneous melanoma correlate with sun exposure but are not always involved in melanomagenesis. Br J Cancer. 1999;79:921–926. doi: 10.1038/sj.bjc.6690147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Viros A, Sanchez-Laorden B, Pedersen M, et al. Ultraviolet radiation accelerates BRAF-driven melanomagenesis by targeting TP53. Nature. 2014;511:478–482. doi: 10.1038/nature13298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Terzian T, Torchia EC, Dai D, et al. p53 prevents progression of nevi to melanoma predominantly through cell cycle regulation. Pigment Cell Melanoma Res. 2010;23:781–794. doi: 10.1111/j.1755-148X.2010.00773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olivier M, Langerod A, Carrieri P, et al. The clinical value of somatic TP53 gene mutations in 1,794 patients with breast cancer. Clin Cancer Res. 2006;12:1157–1167. doi: 10.1158/1078-0432.CCR-05-1029. [DOI] [PubMed] [Google Scholar]
- 20.Essner R, Kuo CT, Wang H, et al. Prognostic implications of p53 overexpression in cutaneous melanoma from sun-exposed and nonexposed sites. Cancer. 1998;82:309–316. doi: 10.1002/(sici)1097-0142(19980115)82:2<317::aid-cncr10>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 21.Gupta S, Artomov M, Goggins W, Daly M, Tsao H. Gender Disparity and Mutation Burden in Metastatic Melanoma. J Natl Cancer Inst. 2015;107 doi: 10.1093/jnci/djv221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hong DS, Vence L, Falchook G, et al. BRAF(V600) inhibitor GSK2118436 targeted inhibition of mutant BRAF in cancer patients does not impair overall immune competency. Clin Cancer Res. 2012;18:2326–2335. doi: 10.1158/1078-0432.CCR-11-2515. [DOI] [PubMed] [Google Scholar]
- 23.Falchook GS, Long GV, Kurzrock R, et al. Dabrafenib in patients with melanoma, untreated brain metastases, and other solid tumours: a phase 1 dose-escalation trial. Lancet. 2012;379:1893–1901. doi: 10.1016/S0140-6736(12)60398-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dahlman KB, Xia J, Hutchinson K, et al. BRAF(L597) mutations in melanoma are associated with sensitivity to MEK inhibitors. Cancer Discov. 2012;2:791–797. doi: 10.1158/2159-8290.CD-12-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.