SUMMARY
Variability in induced pluripotent stem cell (iPSC) lines remains a concern for disease modeling and regenerative medicine. We have used RNA sequencing analysis and linear mixed models to examine the sources of gene expression variability in 317 human iPSC lines from 101 individuals. We found that ~50% of genome-wide expression variability is explained by variation across individuals and identified a set of expression quantitative trait loci that contribute to this variation. These analyses coupled with allele specific expression show that iPSCs retain a donor specific gene expression pattern. Network, pathway and key driver analyses showed that Polycomb targets contribute significantly to the non-genetic variability seen within and across individuals, highlighting this chromatin regulator as a likely source of reprogramming-based variability. Our findings therefore shed light on variation between iPSC lines and illustrate the potential for our dataset and other similar large-scale analyses to identify underlying drivers relevant to iPSC applications.
eTOC summary
Using large-scale analyses of over 300 iPSC lines, Chang, Quertermous, Lemischka and colleagues of the NHLBI NextGen consortium examine sources of gene expression variation between lines and illustrate how this approach can identify genetic and non-genetic drivers relevant to line variation with implications for iPSC characterization and disease modeling.
INTRODUCTION
Induced pluripotent stem cells (iPSC) and their differentiated progeny offer a unique platform to study developmental processes and could serve as an unlimited cell source for regenerative medicine. iPSC based models have also proven to be valuable for the study of Mendelian diseases and drug toxicity/efficacy, although their suitability for the study of complex human conditions has not been fully explored. An understanding of the variability in iPSC lines is necessary to develop homogenous iPSCs.
Recent steps towards addressing these questions are exemplified by studies showing that the source cell type (Bar-Nur et al., 2011; Kim et al., 2010), genetic abnormalities and aberrations in epigenetic reprogramming (Cahan and Daley, 2013; Liang and Zhang, 2013) or genetic background (Burrows et al., 2016; Kyttala et al., 2016; Rouhani et al., 2014) are putative contributors to variability in pluripotent lines. However, these studies have often been performed in mouse pluripotent stem cells or in a limited number of human ESC or iPSC lines, which limits the power and the ability to study transcriptional variability within lines derived from the same person or to detect variants associated with gene expression levels across the lines.
The NextGen Consortium was funded by the NHLBI with the mission of creating large-scale human iPSC libraries and generating accompanying genetic and genomic data for the study of genetically complex conditions and to serve as a resource for the greater scientific community. Using a non-integrative reprogramming approach, we have generated iPSC lines from 196 individuals with available genome-wide genotyping data. In the largest transcriptional profiling effort of human iPSCs to date, we have analyzed RNA-seq profiles of 317 iPSC clones from the first 101 (of 196) individuals to understand the landscape of transcriptional variability in human iPSC lines.
RESULTS
iPSC generation and quality control
We generated 3–7 iPSC lines from each of 196 individuals (Figure S1A and Table S1) and importantly, we established footprint-free iPSC lines through a non-integrative approach (Sendai virus). Reprogramming efficiency was assessed through Tra 1–60 live immuno-staining and Sendai virus clearance was detected as early as passage 9. Oct4 and Nanog protein expression showed no obvious differences in expression levels between individuals or between clones derived from the same individual (Figure S1B). So far, we have performed RNA-seq on 337 clones derived from the first 106 subjects recruited.
One of the main concerns in large-scale studies is cross-individual contamination of iPSC lines. To control for this, we used forensic genetics to compare the available genome wide genotyping data with the acquired RNA-seq data derived from the cultured iPSC lines. We excluded the samples where the genotype to RNA-seq matching rate was below 90%, which pointed to a possible cross-contamination between different individuals (20 lines from 5 individuals, 5.9% of the lines), rendering a total of 317 lines (101 individuals) with confirmed identity (Figure S1 and S2).
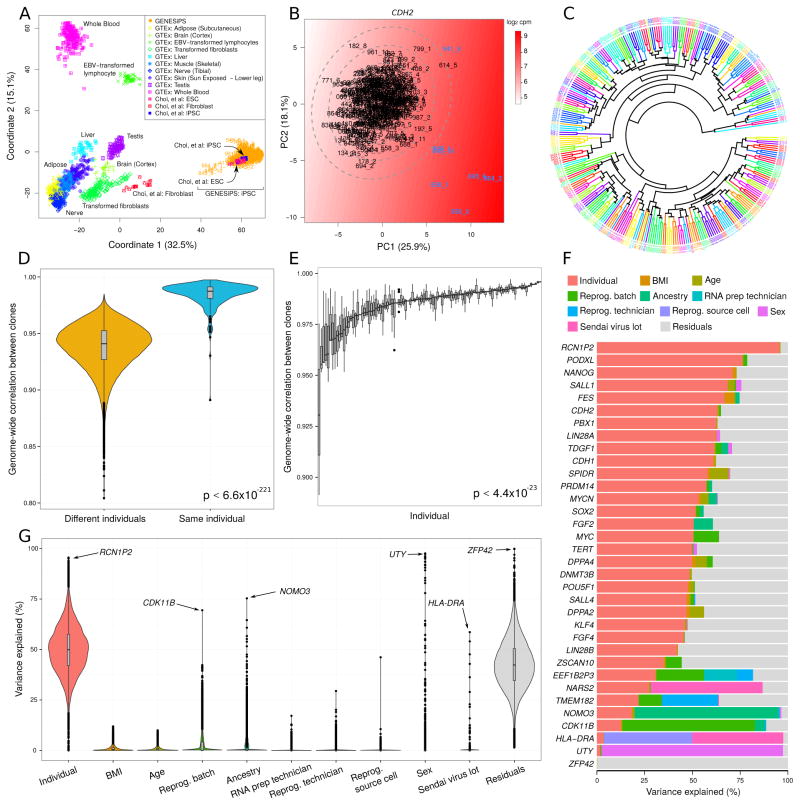
As the size of iPSC libraries becomes larger, the cost of performing teratoma assays to assess pluripotency becomes prohibitive and raises animal welfare concerns as well. Alternative biocomputational approaches based on gene expression data, exemplified by microarray-based Plurinet (Muller et al., 2011) and CellNet (Cahan et al., 2014), have recently demonstrated their utility. Thus, we sought to develop an affordable biocomputational RNA-seq based approach to assess the quality of our iPSC lines and to exclude partially reprogrammed or differentiated lines. We compared our RNA-seq dataset with a previously published iPSC and embryonic stem cell (ESC) dataset, where the quality of both cell types was assessed (Choi et al., 2015), and GTEx data for different mature tissues (GTEx Consortium, 2015). In a multidimensional scaling analysis of the combined RNA-seq data using 15,294 ENSEMBL genes passing a strict expression cutoff (STAR methods), our iPSC cohort clustered with iPSCs or ESCs and was distinct from mature tissues (Figure 1A). However, a small subset of our iPSC lines stood apart from the main trend, and to further examine the quality of our iPSC lines, we performed a principal component analysis using the expression of 23 established pluripotency markers and CDH2 (also known as NCAD), which is a well-known early marker of differentiation in iPSCs. We defined “bona fide” iPSC lines as those with variation in gene expression less than 3 standard deviations from the centroid (Figure 1B and S3). This analysis allowed us to identify 7 outliers, 6 of which showed low levels of core pluripotency factors OCT4, NANOG and LIN28A, among others, and high levels of the differentiation marker NCAD, pointing to an incomplete reprogramming or a partially differentiated phenotype. The remaining outlier clone expressed abnormally high levels of MYC, suggesting an abnormal transformation (Figure 1B and S3). These 7 clones were excluded from all subsequent analyses. Additionally, we analyzed chromosomal aberrations using gene expression data as described previously (Ben-David et al., 2011; Mayshar et al., 2010). Most of the clones with detectable chromosomal aberrations were excluded, with only two clones retained based on the pluripotency marker outlier analysis (data not shown).
Figure 1. Sources of iPSC Gene Expression Variability.
A) iPSCs from the current dataset cluster with previously characterized iPSCs and ESCs (Choi et al., 2015) and are distant from tissues studied in GTEx, based on multi-dimensional scaling. B) Outliers were identified with principal component analysis of 24 key stem cell genes. The color gradient represents smoothed expression of CDH2. Ellipses indicate 1, 2 and 3 standard deviations from the centroid. C) Hierarchical clustering of RNA-seq data indicates that multiple iPSC lines from the same individual cluster together (same color). D) Correlation of genome-wide gene expression profiles between multiple iPSC lines from the same individual are substantially higher than the correlation between profiles from different individuals. Violin plots represent the distribution of similarity scores with the width of the curve indicating the number of data points that fall in the region. E) The correlation between multiple lines from the same individual show substantial differences. Each bar represents an individual and shows the distribution of pairwise similarity values within the multiple iPSC lines from that individual. F) Expression variance is partitioned into fractions attributable to each experimental variable. Genes shown include 24 key stem cell genes, and genes for which one of the experimental variables explains a large fraction of total variance. G) Violin plots of the percentage of variance explained by each experimental variable over all the genes. For a small number of genes also shown in (F), the data point corresponding to the largest source of variation is indicated with an arrow. See also Figure S2, S3, S4 and Table S2
We examined the degree to which these “bona fide” iPSC lines retain a donor-specific genome-wide gene expression pattern. Hierarchical clustering indicated that iPSC lines derived from the same individual are more similar to each other than to iPSC lines from different individuals (Figure 1C). Supporting the clustering results, the correlation of genome-wide gene expression profiles between iPSC lines derived from the same individual was significantly higher than the correlation between lines derived from different individuals (one sided Mann-Whitney p<6.6×10−221) (Figure 1D). However, we did observe considerable heterogeneity in the degree of similarity between multiple iPSC lines from the same individual (Bartlett p < 4.4×10−23), implying that some individuals yielded consistent lines, while others yielded a more heterogeneous set (Figure 1E).
Characterizing sources of gene expression variability
Assessment of transcriptional variability in human iPSCs
We considered variability not only in terms of the magnitude of variance (the total amount a gene varies in expression) but also in terms of the contribution to variance (the percentage any single factor contributes to the variance for a given gene)(Figure S2). The magnitude of variance and the contribution to variance can be considered both across and within individuals. Across individual variability is defined as the variance between individuals after removing the technical effects. Within individual variability is defined as the variance within individuals after removing technical effects as well as individual effects, i.e. the variance within clones derived from the same individuals (Figure S2D). Therefore, a single gene may have both a large or small magnitude of variance and a large or small contribution to variance from a given source (Figure S2B and S2C).
Gene-level contributions to variance
We first characterized the contribution of diverse sources to the transcriptional variability at a gene-level resolution, using the variancePartition method (Hoffman and Schadt, 2016). This statistical and visualization framework fits a linear mixed model for each gene, and partitions the total variance into the contribution of each variable in the experimental design (e.g. donor, sex, reprogramming batch), plus the residual variance. Since the fractions sum to 1 for each gene, the variance fractions are easily interpretable across genes and sources of variation, and they are unrelated to the magnitude of variance. After removing variation across 8 sequencing batches and 2 RNA preparation methods (Figure 1G and S4), partitioning the variance of each gene into 10 components, plus residual variation, identified genes whose expression variation was attributable to multiple factors (Figure 1F). Variation across individuals explained a median of 49.9% of contribution to expression variance, with some genes showing substantial deviation from the genome-wide trend (Figure 1G). As expected, genes with over 2% contribution to variation explained by sex were highly enriched for being on the X or Y chromosome (Figure 1F and Table S2). Other attributes of the individuals, such as BMI, age and ancestry, are collinear with donor and therefore contributed to a very small fraction of the total variation (Figure 1F and Table S2). After removing the contribution of all other components, 42.3% of the total variance remained. This residual variation primarily represents the variation within the multiple iPSC lines derived from the same individual.
Cis-regulatory variants drive genetic background-associated changes in gene expression levels across individuals
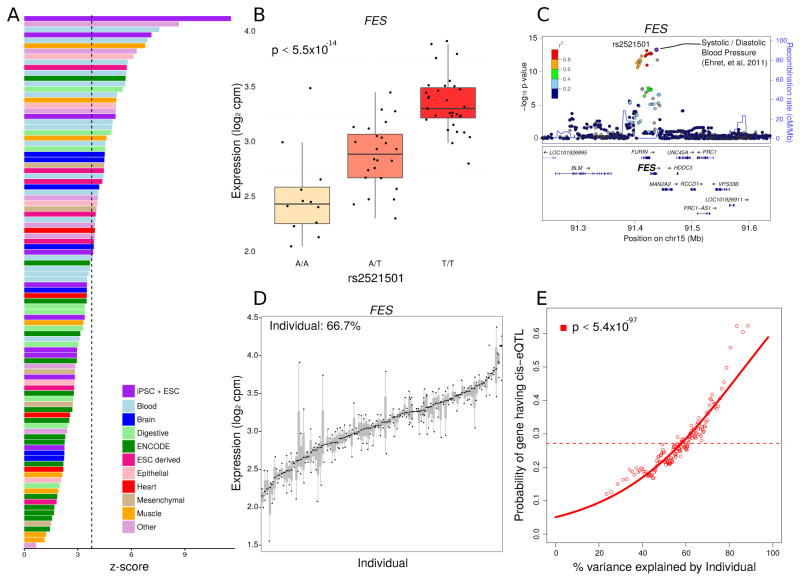
Genetic variation is known to be a major contributor to variation of gene expression (GTEx Consortium, 2015). We identified 4,150 cis-eQTLs at a false discovery rate of 5% when considering individuals of European ancestry (Figure S5A). These cis-eQTLs were generally located near the transcription start site of the corresponding gene (Figure S5B). Notably, these cis-eQTLs showed a degree of cell-type specificity as they were enriched in enhancers (Figure 2A) and promoters (Figure S5C) identified in iPSC and ESC cell lines (Roadmap Epigenomics Consortium, 2015). The cis-eQTLs detected here showed moderate overlap with eQTLs detected in multiple tissues by the GTEx Consortium (2015) (Figure S5D) and also moderate enrichment for proximity to GWAS hits from multiple phenotypes (Figure S5E). GWAS loci that are coincident with eQTLs have the potential to give a functional interpretation of GWAS hits. For example, the most significant marker associated with variation in FES expression (Figure 2B) is associated with variation in blood pressure (International Consortium for Blood Pressure Genome-Wide Association Studies, 2011) (Figure 2C), with the risk allele corresponding to an increase in gene expression. This suggests that iPSCs could be a good model to elucidate gene-SNP relationships and annotate complex GWAS results.
Figure 2. Function and Interpretation of eQTLs.
A) eQTLs show highest enrichment in enhancers in iPSCs and ESCs. Z-scores indicate the degree of enrichment in enhancers represented in cells and tissues samples from (Roadmap Epigenomics Consortium, 2015). Bars are colored based on tissue origin and the dashed line indicates the Bonferroni cutoff for multiple testing. B) rs2521501 is the most significant eQTL for the exemplary FES locus. Expression of FES is shown stratified by genotype at this SNP. C) LocusZoom plot shows −log10 p-values for variants in the FES locus. rs2521501 is an eQTL for FES and is also associated with systolic and diastolic blood pressure. D) FES shows high variation across individuals and low variation within individuals. Each bar represents an individual and the size of the bar represents the variation in FES expression within that individual. E) Probability of each gene having a cis-eQTL plotted against the percent variance explained by individual. Dashed lines indicate the genome-wide average probability, and curves indicate logistic regression smoothed probabilities as a function of the percent variance explained by individual. Points indicate a sliding window average of the probability of genes in each window having a cis-eQTL (window size is 200 genes with an overlap of 100 genes between windows). The p-value shown indicates the probability that an association as strong as between percent variance and eQTL probability occurs by chance according to the logistic regression smoothing. See also Figure S5
The variancePartition analysis illustrated that there are multiple components contributing to gene expression variation. For example, in FES, 66.7% of the contribution to variation was across individuals (Figure 2D). More generally, our genome-wide analysis indicated that genes with a higher across individual contribution to gene expression variation are significantly more likely to have a cis-eQTL detected in this dataset (logistic regression p < 5.4×10−97)(Figure 2E). This result is consistent with a model where cis-regulatory variants, rather than shared environment or technical processing, is a significant contributor to gene expression variation across individuals.
Consistency of allele specific expression within individuals
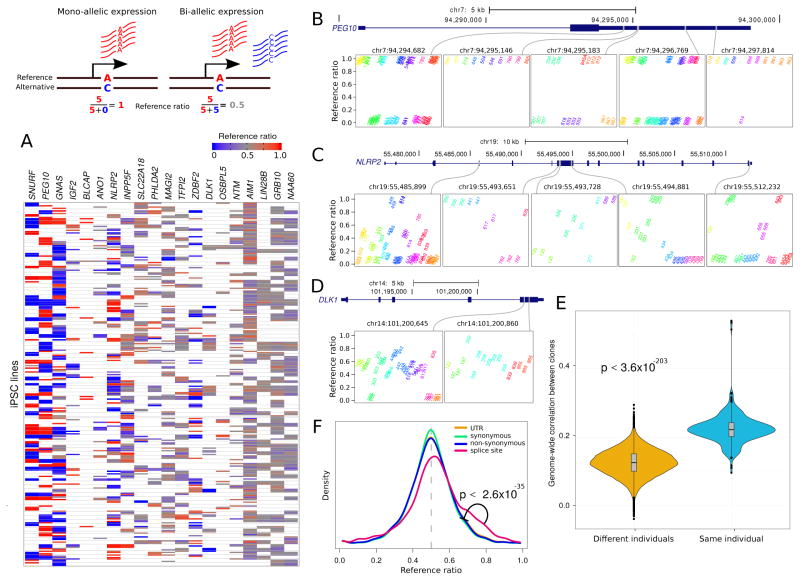
Allelic imbalance is a potential source of variability in iPSCs and we performed allele specific expression (ASE) analyses to gain insight into this process. Analysis of a subset of canonically imprinted genes (obtained from http://www.geneimprint.com following Rouhani et al., (2014)) showed that some genes had a marked allelic imbalance in most samples, while others had a reference ratio close to 0.5 in most samples (Figure 3A). We considered three of these genes as illustrative examples to demonstrate how genes with very different patterns of allelic imbalance still retain a donor-specific signature. A closer examination of PEG10 illustrated very strong allelic imbalance at 5 heterozygous sites (Figure 3B). While the direction of the imbalance varied likely due to a parent-of-origin (i.e. imprinting) rather than an allelic effect, multiple iPSC lines from the same individual showed a high degree of consistency in reference ratios. In contrast, NLRP2 showed more variability in the degree of allelic imbalance across individuals, but the multiple iPSC lines from the same individual showed remarkable consistency in reference ratios (Figure 3C). DLK1, as a representative member of the DIO3-DLK1 imprinted locus, showed balanced expression in most lines where allele specific expression can be detected (Figure 3D). This result agrees with previous reports that demonstrate a consistent deregulation of the DIO3-DLK1 locus in iPSCs (Nazor et al., 2012; Stadtfeld et al., 2010). Our results indicate that while some genes reported to be imprinted in other studies maintain strong allelic imbalance in iPSCs, other loci demonstrate consistent biallelic expression across lines derived from the same individual.
Figure 3. Allele-Specific Expression.
Diagram illustrates mono- and bi-allelic expression. A) Reference ratios for each set of canonically imprinted genes show the consistency of allele-specific expression (ASE) within multiple iPSC lines from the same individual. Red indicates expression of the reference allele, blue indicates expression of the alternative allele and grey indicates a mix. White indicates that ASE could not be assessed due to the lack of a heterozygous SNP with sufficient coverage. B) PEG10 exhibits strong allelic imbalance at 5 sites where the expressed allele is consistent in multiple iPSC lines from the same individual. Reference ratios are shown at 5 sites for individuals that are heterozygous at each site. Multiple iPSC lines from the same individual have the same color and labels indicate the individual identifier for each iPSC line. C) NLRP2 exhibits more variation in allele imbalance across individuals, but retains consistency in multiple iPSC lines from the same individual. D) DLK1 shows loss of imprinting but retains consistency within multiple iPSC lines from the same individual. E) Genome-wide correlation based on allelic imbalance at sites shared by each pair of individuals indicates that iPSC lines from the same individuals show higher similarity in ASE than iPSC lines from different individuals. F) Genome wide reference ratios for SNPs in splice site regions show increased expression of the reference allele, compared to SNPs in UTRs, or SNPs that cause synonymous or non-synonymous changes in coding regions.
This pattern was observed genome-wide, as the correlation of reference ratios between all pairs of samples indicated that iPSC lines were significantly more similar within the same individual than across individuals (one sided Mann-Whitney p<3.6×10−203) (Figure 3E). Furthermore, the distribution of reference ratios depended on the functional impact of the allele with imbalanced expression. Genome-wide reference ratios for SNPs in splice site regions show increased expression of the reference allele, compared to SNPs in UTRs, or SNPs that cause synonymous or non-synonymous changes in coding regions. Variants located in the UTR and synonymous or non-synonymous coding variants (156229, 78284 and 46382 variants respectively) had a similar distribution in reference ratios genome-wide (Figure 3F). However, the 3,547 variants affecting splice sites had significantly higher reference ratios (one sided Mann-Whitney p<2.6×10−35 compared to each of the other categories). This is consistent with transcripts containing variants that disrupt splice sites being subject to alternative splicing that affects the exon inclusion rate (Li et al., 2016) or being targeted by nonsense mediated decay so that the remaining transcripts disproportionately contain the reference allele (Rivas et al., 2015).
Novel biological insights from analysis of the magnitude of transcriptional variation in iPSCs
Highly transcriptionally varying genes associate with a range of pathways in human iPSC lines
To discover potential new drivers of iPSC variability and complement the variance partition analysis, we examined the magnitude of transcriptional variation in iPSC lines attributable to either across or within individual differences. Previous network analyses employing high variance gene expression filters, generally including more lowly expressed genes, have demonstrated the ability to identify biologically meaningful correlations that have elucidated complex traits (Zhang et al., 2013). Thus, we employed a relaxed set of filters (0.1 count per million, cpm, in 10 percent of our samples) on the expression data to include lowly expressed genes, given the increased power that is provided by network analysis in identifying drivers of transcriptional variability. We also performed a sensitivity analysis that confirmed that the covariance structure on which the networks rely was not significantly affected by the technical noise of genes with low expression levels (STAR methods).
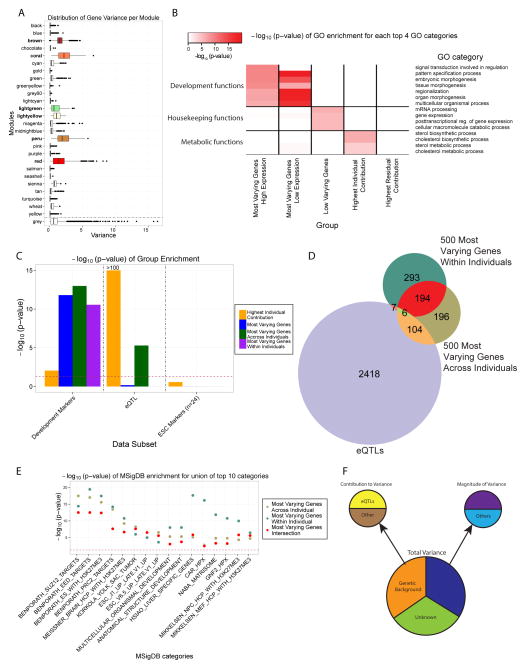
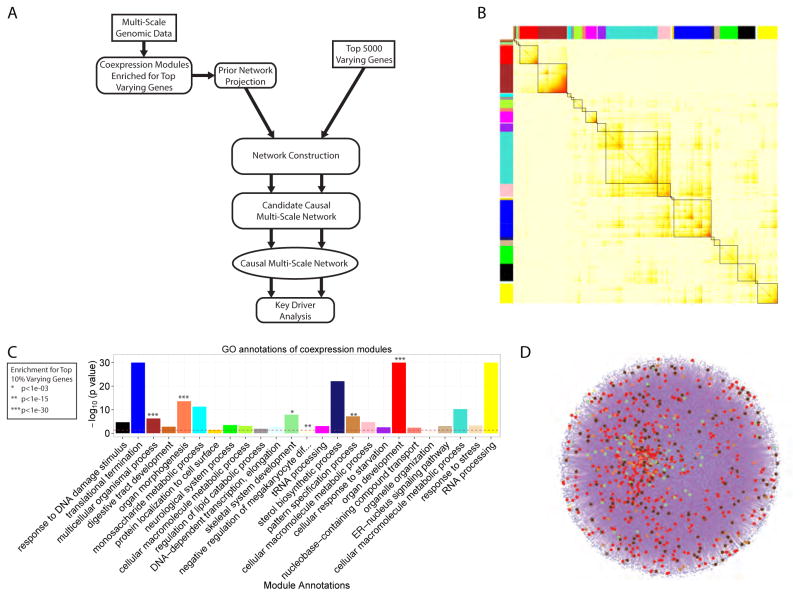
We applied standard co-expression network analysis (Figure 4A and 5B) to capture the robust gene-gene correlations in meaningful functional modules (Zhang et al., 2013). This analysis identified 10708 out of 25391 ENSEMBL genes passing the relaxed filter that were significantly co-expressed. Of the 25 modules of co-expressed genes identified in this analysis, 6 modules were enriched for the top 10% most varying genes (Figure 4A and 5C). These modules were comprised of 2204 genes, including 1127 of the top 10% most varying genes (OR = 16.1). To further investigate the covariance structure of the gene expression data and discover functions associated with co-expression modules, we identified the enriched GO terms for each of the 25 modules (Figure 4A, 5C and Table S3). All 6 of the modules significantly enriched for the most varying genes were also enriched for developmental functions such as organ development and morphogenesis.
Figure 4. Magnitude of Variance Defines High and Low Variable Genes and Pathways in Human iPSC Lines.
A) Distribution (boxplot) of the variance of all the genes in each module in the co-expression network. The grey module represents the ‘trash’ module (in which genes are not co-expressed). The 6 modules significantly enriched for the top 3000 most varying genes are colored according to the module name. B) Heatmap of the −log10 (p-value) for the top enriched Gene Ontology (GO) terms, grouped into general functional classes, for each category of genes considered. The categories are: (1) the 1000 most varying genes divided into 2 groups, the highly expressed ones (230 genes) and the nominally expressed ones (770 genes), (2) the 1000 least varying genes, (3) the 1000 genes with the highest individual contribution to variance, and (4) the 1000 genes with the highest residual contribution to variance. C) Distribution (bar-plot) of the −log10 (p-value) of the enrichment, assessed using the Fisher’s exact test, of the groups in the legend for development markers, eQTLs and ESC markers. D) Venn diagram of the top 500 most varying genes within individuals, across individuals and eQTL genes (1% FDR), E) −log10 (p-values) for the enrichment of the union of the 3 groups shown in (D) for top 10 MSigDB categories. F) Diagram recapitulating the different sources influencing the different types of gene expression variation in iPSCs. See also Figure S2B, S2C, S2D and Table S2, S3 and S4
Figure 5. Predictive Network Modeling Analysis Pipeline, co-Expression Network Results and Mapping onto Prior Network.
A) Diagram showing the different analysis steps from multi-scale data to predictive network modeling. B) The topological overlap matrix (TOM) of the iPSC co-expression network. Only genes included in co-expression modules are shown. C) Annotation of the modules with the most significantly enriched GO term. Modules significantly enriched for the top 3000 most varying genes are indicated. D) iPSC-specific prior network constructed from public databases (CPDB and MetaCore) and Roadmap Epigenomics Consortium iPSC data, with genes in the modules of interest mapped onto the network shown by dots colored according to the modules identity.
We also observed that genes with the highest magnitude of variance overall were strongly enriched for developmental functions and markers, regardless of expression levels (Figure 4B and 4C). The enrichment for these markers was conserved regardless of whether genes with the highest magnitude of variation were estimated across individuals or within individuals (Figure 4C), although genes with high magnitude of variance across individuals were also enriched for eQTLs (Figure 4C). Not surprisingly, genes with the lowest magnitude of variance were enriched for essential housekeeping processes (Figure 4B). Interestingly, while genes with the highest across individual contribution to variance were highly enriched for eQTLs and metabolic functions (Figure 2E, 4B and 4C), they were not enriched for developmental functions or markers (Figures 4B and 4C). Collectively these results reflect the complementary signals present in high magnitude of variance genes compared to genes whose variation can largely be explained by across individual contribution to variation.
Non-eQTL-related transcriptional variability is derived from Polycomb target genes
While variation in gene expression across individuals could be significantly attributed to eQTLs (Figure 4C and 4D), variability within lines derived from the same individual could not be as easily explained. However, we did find that developmental markers were overrepresented in genes with high magnitude of variance, both across and within individuals (Figure 4C). In fact, when restricting to the 500 most variable genes across and within individuals, corrected for technical confounders, we found 200 overlapping genes and 194 out of 200 that were not eQTL genes (Figure 4D and Table S4) (Fisher’s test p value=1.4e-218, OR=55.6). This indicates a common origin for a significant part of the magnitude of across and within individual variability, independent of eQTL effects.
Pathway enrichment analysis for the 500 genes with the highest within individual magnitude of variance showed that Polycomb repressive complex 2 (PRC2) and H3K27me3 mark-related targets were highly over-represented (Figure 4E). The same analysis for the 500 genes with the highest across individual magnitude of variance also showed enrichment for these pathways (Figure 4E). The same processes were also seen in the pathway enrichment of the 200 genes that overlap in terms of the within and across-individual magnitude of variance. However, while the 500 most varying genes across individuals were enriched for cis eQTLs (one sided Fisher’s test p value=4.0e-16, OR=2.6), the 200 genes in the overlap were under-represented (one sided Fisher’s test p value=1.4e-4, OR=0.28)(Figure 4D), suggesting that this set of variable genes may be independent of the genetic background.
Thus, we have described two main sources of variability in human iPSC lines. Through quantitation of the contribution of various sources to iPSC transcriptional variability using variancePartition analyses, eQTL-driven genetic background-associated variability was determined to be a significant contributor to variability across individuals. Further, analysis of the magnitude of variance uncovered molecular pathways contributing to iPSC transcriptional variability that are independent of the genetic background. Specifically, Polycomb targets were found to be an important factor explaining both across and within individual variability (Figure 4E and 4F).
Molecular networks for iPSC transcriptional variability
The availability of this large iPSC dataset made it possible to investigate the causal molecular mechanisms underlying the described variability. For this, we sought to build causal network models for iPSC transcriptional variability. The co-expression network constructed with our iPSC data characterizes the correlation structures among gene expression traits, reflecting sets (modules) of highly co-regulated genes operating in coherent biological pathways. However, such network modules do not reflect the probabilistic causal information needed to identify key driver genes of those network modules associated with within and across individual transcriptional variation. With the appropriate probabilistic causal network structure, we can predict which genes serve as key drivers that modulate the levels of gene expression in a significant proportion of genes comprising any given sub-network of interest (Zhang et al., 2013; Zhu et al., 2012). To achieve this, we developed a computational pipeline to reconstruct predictive network models. This pipeline integrates multiscale-omics data (including genotype), gene expression, and a prior network built from Roadmap Epigenomics Program histone modification data, and publicly available knowledge bases representing experimentally annotated and curated pathways, such as the ConsensusPathDB and MetaCore (Figure 5A and S2). The prior network was used to provide a comprehensive representation of iPSC biology and to compensate known shortcomings of RNA-seq data and co-expression networks (STAR methods).
To construct the predictive network model, we first created a gene list using a 2-step process to seed its construction. In the first step, we collected all genes in the 6 variation-associated co-expression modules (Figure 5B and 5C), as well as all genes in the GO and MSigDB terms related to pluripotency and development that were enriched in these 6 modules. In the second step, we expanded the set of defined genes by mapping them onto the prior iPSC-specific network we constructed separately to enhance the previous seeding gene list (Figure 5D). The gene list resulting from this 2-step process was then used to construct the predictive network model (STAR methods). The final network derived from the above seeding gene list was comprised of 13990 genes (13K network in Figures 6A, 7A and Table S5). This 13K network represents a probabilistic causal network model based on iPSC biology that captures causal relationships among the top varying genes. In this way, the network model serves to organize vast amounts of information captured in the iPSCs such that the information can be more directly queried in order to examine how the data may support existing hypotheses involving iPSCs or to generate novel hypotheses. As a result, the network model can help elucidate the mechanisms underlying across and within individual variation in the iPSCs. To establish the robustness of this network and select the most reliable key regulators, we also constructed a second network based on the top 5000 most varying genes (STAR methods, referred to as the 5K network in Figure S6A and S7A).
Figure 6. 13K Sub-Networks Downstream of Key Driver Genes of Interest Contribute to iPSC Variability.
A) Causal network covering the 13,990 genes comprising the co-expression modules enriched for the top 3000 most varying genes, the pathways related to development of these modules, and the mapping onto the prior network. The sub-networks 2 steps away from the key drivers of interest are shown in B) and C), with the key drivers shown in red and yellow respectively. See also Figure S6, S7 and Table S5 and S6
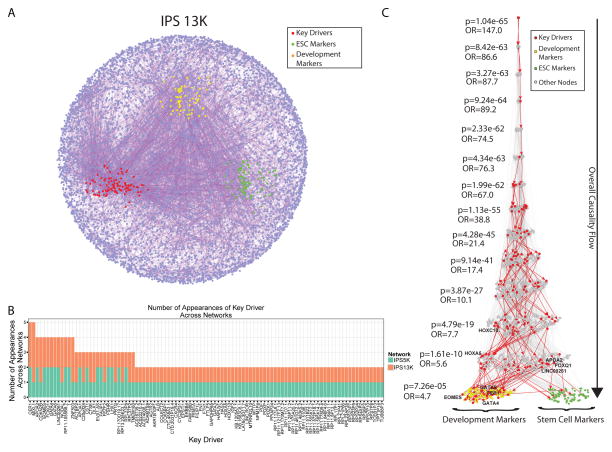
Figure 7. Bayesian Causal Gene Networks, Key Driver Gene Discovery and Network Validation with Prior Information.
A) Causal molecular networks covering the 13,990 genes comprising the co-expression modules enriched for the top 3000 most varying genes, the pathways related to development of these modules, and the mapping onto the prior network. The key drivers genes are highlighted in red, the stem cell markers in green and the development markers in orange. B) Distribution (histogram) of the number of appearances of any key driver gene in both networks, ranked by their total number of appearances. C) The Eiffel Tower plot shows the overall causality flow (top to bottom) from any stem cell (green) or development (yellow) markers to any upstream causal gene in the 13K network. It also shows the enrichment p-value of key driver genes (red) at every step upstream of the markers, assessed using a level-associated Fisher’s exact test. See also Figure S6, S7 and Table S5 and S7
Key drivers of iPSC transcriptional variability
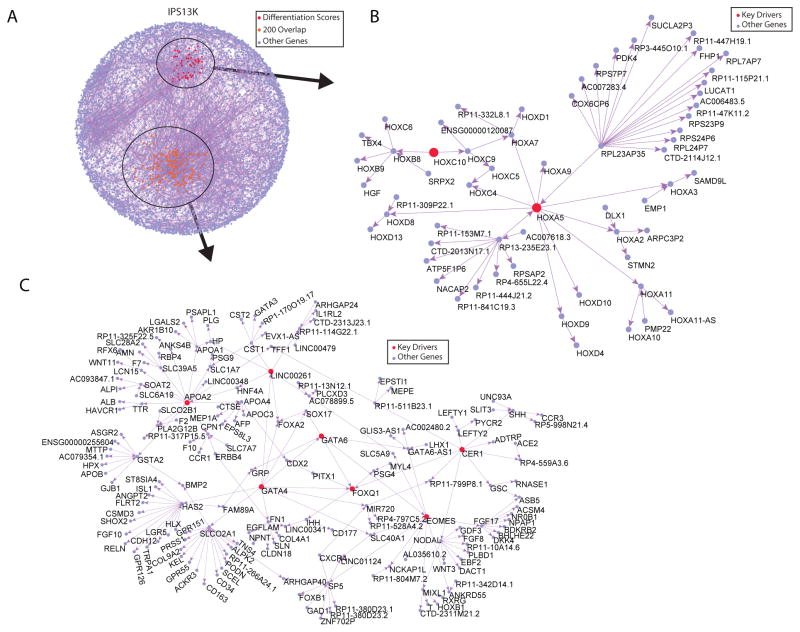
The organization of our iPSC data into causal network structures provides a way to identify regulators of biological processes of interest and derive novel hypotheses in an objective, data driven way. To illustrate the utility of the network model, we sought to identify regulators of functional and gene expression variation in human iPSCs by performing key driver analyses (KDA) (Zhang and Zhu, 2013) (Figures 6, 7, S6 and S7). To identify key drivers (KDs) of the developmental pathways that were enriched in the most varying genes, we mapped onto the 13K network the 200 most varying genes shared across and within individuals (Figure 4D and Table S4). After each of these mappings, we identified the largest connected sub-graph as the sub-network of interest on which to carry out the KDA. Interestingly, although the number of nodes in the 13K and 5K networks were substantially different, we found a high degree of overlap between the sets of KDs identified in each network, indicating that the topology across these networks at the level of key drivers was highly conserved (Figure 7B) and that the predicted KD genes present in both networks were highly robust to stochastic artifacts. Based on the KDAs from these 200 most varying genes, we considered only conserved and therefore robust KDs, and identified 7 key driver genes (GATA4, GATA6, EOMES, APOA2, LINC00261 (DEANR1), FOXQ1 and CER1) for the variability in iPSCs (Figure 6C and S6C).
Network analysis of iPSC differentiation efficiency
It is well known that iPSC differentiation to endothelial cells is strongly influenced by clone-to-clone variability. As another example of the potential utility of performing analyses based on the network structures we created, we sought to query whether transcriptional variation in the iPSCs could lend insight into the efficiency of iPSC differentiation to endothelial cells. These analyses were based on the endothelial cell differentiation scores from 73 lines derived from 23 subjects (Table S6). In this analysis we mapped onto the 13K network the top 500 differentially expressed genes for high versus low efficiency of differentiation to endothelial cells (Table S6 and Figure S7D) and identified the largest connected sub-graph as a sub-network of interest. Using KDA as described above we identified 2 potential key driver genes (HOXA5 and HOXC10) influencing the efficiency of differentiation of iPSCs to the endothelial lineage (Figure 6B and S6B).
Predictive causal networks capture iPSC biology through robust key drivers
To illustrate the central role the KDs play in iPSC biology, we examined the connectivity structure between the 9 KDs described above as well as of the 25 functional modules in the co-expression network and known iPSC/ESC and developmental marker genes to elucidate their causal relationships. Specifically, we extracted a sub-network consisting of all the nodes in the 13K network that was upstream of a list of 197 iPSC/ESC pluripotency and developmental markers (Table S7). We then examined the enrichment of this sub-network for KD genes and found significant enrichment of the KDs in every level upstream of the marker genes in both networks (Figure 7C and S7B). This same pattern of enrichment was not observed (Figure S7C) for KD genes downstream of the marker genes, corroborating that the KD genes are regulating the marker genes. This analysis emphasized that our predictive networks captures iPSC biology, allows us to better organize the data to discover meaningful KDs and enables the generation of new hypotheses.
DISCUSSION
Cis-eQTLs explain genetic background associated variability across individuals in iPSC lines
Using variancePartition (Hoffman and Schadt, 2016) we demonstrated that differences between individuals contribute the most (≈50%) to variation at a single gene level, when averaged genome-wide. Notably, some biological variables (donor age, BMI, sex and ancestry) and technical variables (reprogramming batch and technician, RNA preparation technician, Sendai virus lot and reprogramming cell source) affected the expression variation of only a small subset of genes. These observations support recent studies suggesting that iPSCs retain a donor-specific gene expression pattern (Burrows et al., 2016; Rouhani et al., 2014; Thomas et al., 2015). Furthermore, our analysis indicates that specific, detectable cis-eQTLs are responsible for a significant part of the across individuals variability in gene expression. Previous work has proposed the role of regulatory variants as major drivers of gene expression variation across iPSCs from different individuals (Burrows et al., 2016; Rouhani et al., 2014; Thomas et al., 2015), and the large size of our dataset strongly supports this model.
We show that cis-eQTLs identified in iPSCs are enriched for iPSC enhancers and promoters and that eQTLs in our iPSC cohort overlap those of other tissues. Our results are not definitive as to whether iPSCs will provide a more efficient mechanism for the identification of eQTLs compared to other cell types as suggested by (Thomas et al., 2015). Nevertheless, eQTLs detected in iPSCs can inform the interpretation of variants identified by GWAS, such as we show with FES. More importantly, the differentiation potential of iPSCs to distinct cell lineages may allow the study of tissue-specific eQTL effects.
Deregulation of allelic imbalance contributes to iPSC transcriptional variability
The retention of allelic imbalance at imprinted and other loci following the reprogramming and differentiation of iPSC lines has been a concern in terms of the genomic stability of iPSCs (Nazor et al., 2012; Stadtfeld et al., 2010). Our analysis using imputed genotype data was underpowered because exome or genome sequencing is required to increase the number of genes with heterozygous SNPs, and to detect rare, deleterious genetic variation that is more likely to impact allelic imbalance (GTEx Consortium, 2015; Lappalainen et al., 2013; Rivas et al., 2015). Nonetheless, we still observed robust allelic imbalance signals. Analyses of allelic imbalance of PEG10, NLRP2 and DKL1 illustrate how multiple iPSC lines from a single individual show consistent patterns, yet the variation across individuals is more complex. PEG10 retains the strong imbalance characteristic of imprinting. NLRP2 shows retention of allelic imbalance in some individuals but a larger range of variation in reference ratios across individuals. The DIO3-DLK1 imprinted locus harbors the protein coding genes DLK1, RTL1 and DIO3 and the long non-coding RNAs (lncRNAs) MEG3 and MEG8, in addition to the largest known microRNA (miRNA) cluster in the human genome (Benetatos et al., 2014). We have shown that DLK1 is affected by variation in allelic imbalance, which suggests that the whole locus is deregulated in iPSCs as previously shown (Nazor et al., 2012; Stadtfeld et al., 2010). Interestingly, all protein coding and lncRNA genes in the locus are among the overall top-500 most variable genes in our iPSC cohort (Table S2 and S4).
We observed consistency of genome-wide allelic imbalance patterns within iPSC lines from the same individual. A correlation metric based on allelic imbalance at shared sites (GTEx Consortium, 2015; Lappalainen et al., 2013) demonstrated a higher degree of similarity within iPSC lines from the same individuals compared to across individuals. Allelic imbalance also provided insight into the functional consequences of genetic variation, as variants affecting splice sites were significantly more likely to favor expression of the reference allele. Genes with aberrant splicing can be targets of non-sense mediated decay whereby the transcripts with the alternative allele are preferentially degraded (Rivas et al., 2015) or can affect the exon inclusion rate (Li et al., 2016).
Co-expression and predictive networks based key driver analysis allows novel insights into iPSC transcriptional and functional variability
Analysis of the overall magnitude of variance allowed us to identify highly variable and non-variable genes in our iPSC cohort. The latter were enriched for housekeeping pathways as described before (Kumar et al., 2014). However, highly variable genes showed a significant enrichment for pathways related to developmental processes such as pattern specification processes, regionalization and, organ and embryonic morphogenesis. In addition, the 6 co-expression modules enriched for the most varying genes were themselves enriched for developmental functions such as “organ development”, “skeletal system development”, “organ morphogenesis” and “central nervous system development”. Some of these functions relate to mesendodermal or ectodermal development. It is widely accepted that pluripotent circuitry maintenance is not based solely on the up-regulation of pluripotency-associated factors. The coordinated, simultaneous inhibition of both mesendodermal and ectodermal differentiation pathways through the action of core pluripotency factors in concert with Polycomb repressive complexes is also necessary (Cahan and Daley, 2013). Thus, our results suggest that developmental pathways contribute significantly to the overall variability in human iPSC lines and particularly to the within individual variability in iPSC lines.
Our predictive network and KDA found 7 possible regulators of variability (GATA4, GATA6, EOMES, APOA2, LINC00261 (DEANR1), FOXQ1 and CER1) in iPSCs and 2 possible regulators (HOXA5 and HOXC10) of endothelial differentiation efficiency (Bahrami et al., 2011; Rhoads et al., 2005). Surprisingly, many of the key drivers are strongly associated with early stages of mesendodermal development. A possible explanation for this finding is the alleged influence of the reprogramming cell source as has been previously proposed (Bar-Nur et al., 2011; Kim et al., 2010). Nevertheless, cell reprogramming requires the silencing of a set of differentiation-associated genes by the Polycomb repressor complex (Fragola et al., 2013). Thus, one can envision that in the case of erythroblasts, expressed genes associated with the mesodermal program will be the main targets of Polycomb mediated silencing during reprograming, as other programs, like ectoderm, will already be silent. Importantly, 7 out of 9 key drivers (HOXA5, HOXC10, GATA4, GATA6, EOMES, FOXQ1 and CER1) are known targets of Polycomb proteins (Bracken et al., 2006; Kim et al., 2006; Ku et al., 2008; Xie et al., 2013), and we have identified H3K27me3 and H3K4me3 histone marks on these genes in pluripotent embryonic stem cells in ENCODE data.
Pathway enrichment and KDA showed that Polycomb and H3K27me3 target genes accounted for a significant part of the variability found in genes with both the highest within or across individual variance. H3K27me3 is a repressive epigenetic mark associated with Polycomb complex recruitment to specific chromatin domains (Aloia et al., 2013) and the silencing of developmental regulators (Boyer et al., 2006) and differentiation associated genes (Surface et al., 2010). The overlap between the pathways associated with both across and within individual variability suggests that the reprogramming process itself is a primary determinant of both subsets of variability. However, we cannot exclude that dynamic fluctuations in chromatin state are associated with sporadic expression of certain Polycomb targets in iPSCs (Kumar et al., 2014).
Use of the iPSC resource to generate novel hypotheses
The application of our approaches to transcriptomic and network analysis to the large RNA-seq dataset has allowed us to generate novel hypotheses related to transcriptional variability in iPSCs: 1. Consistent deposition of silencing marks through PRC recruitment to differentiation-associated genes during reprogramming will help decrease the non-genetic background associated variability in iPSCs, 2. A subset of polycomb targets, i.e, the key drivers, may be central in the control of iPSC transcriptional variability and in the differentiation efficiency to the endothelial lineage. Although our data does not exclude a role for non-PRC mediated mechanisms, several reports support our hypotheses. For example, naive pluripotent cells have been shown to have less transcriptional variability than primed pluripotent stem cells and do not seem to rely on the Polycomb repressor complex (PRC) to silence developmental or differentiation associated genes (Gafni et al., 2013; Galonska et al., 2015).
Two lines of evidence describe different mechanisms of PRC regulation in pluripotent cells. First, there is evidence for a reciprocal regulation between Polycomb proteins and the DIO3-DLK1 locus. Several miRNAs in the locus have been postulated to target components of PRC2 (Liu et al., 2010) and the lncRNA MEG3 has been shown to direct PRC2 to specific target genes (Kaneko et al., 2014). Conversely, PRC2 is required for the proper expression of the DIO3-DLK1 locus in mESCs, preventing de novo DNA methylation (Das et al., 2015). The second possibility is based on the distinct metabolic profiles found in human naive versus primed ESCs (Sperber et al., 2015). This metabolic switch is regulated by Nicotinamide N-methyltransferase (NNMT) controlling S-adenosyl methionine (SAM) levels available for PRC2 mediated H3K27me3 histone methylation. Differences in SAM levels correlate with H3K27me3 mark changes found between naive and primed ESCs (Theunissen et al., 2014). Future experiments investigating the crosstalk between Polycomb proteins and the DIO3-DLK1 locus or metabolic regulation during the reprogramming process may lend insight into whether these processes will help to reduce the gene expression variability in iPSCs. Finally, experimental validation will help to dissect the direct and specific contribution of each of the key drivers. However, such validation will require an extensive effort to manipulate the action of the polycomb complex or the specific key drivers in the context of the different stages of reprogramming or differentiation to the endothelial lineage coupled with the generation and sequencing of a large number of iPSC lines.
Summary and implications for future studies
In summary, we have created a resource of well-characterized, “footprint-free” iPSC lines that will be available to the broader scientific community through WiCell. To the best of our knowledge, our results represent the most comprehensive attempt to define the variability in gene expression of human iPSCs, including technical variability, as well as across individual and within individual variation. Additional studies such as epigenetic profiling will help to draw a more complete canvas of the sources of gene expression variability in human iPSCs. Nonetheless, our analyses can serve as a roadmap to understand the variability in iPSCs and help improve iPSC-based model systems for human disease. Additionally, the eQTL characterization in our large iPSC library allows the directed selection of lines with haplotypes of interest to study the relationship between genetic variants and cellular function in iPSCs and their differentiated progeny. Finally, the co-expression, predictive network and key driver analyses offer the means to organize and directly query large amounts of information in iPSCs to examine how the data may support existing hypotheses or to generate novel hypotheses that help shed light into complex, unsolved questions.
STARS METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
The iPSC lines generated in this study are publicly available through WiCell (https://www.wicell.org/). Further information and requests for reagents may be directed to the corresponding author Thomas Quertermous (tomq1@stanford.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
The study included 201 subjects who had volunteered between October 2002 and October 2013 and were in general good health. Stanford Institutional Review Board approved the study protocol, and all subjects gave written informed consent for study participation. The study was performed at the Stanford Clinical and Translational Research Unit. Height and weight were measured while subjects were wearing light clothing and no shoes. Body mass index was calculated by dividing weight (in kilograms) by the square of height (in meters). See Table S1 for complete demographic data.
METHODS DETAILS
Peripheral Mononuclear Cell Isolation
After an overnight fast, blood was drawn in cell preparation tubes (BD Vacutainer CPT tube) with sodium citrate for the isolation of peripheral blood mononuclear cells. The tubes were centrifuged at 1800 g for 40 minutes at room temperature. The plasma and cells in the interphase were transferred to a new tube, resuspended in RPMI medium and centrifuged at 1500 rpm for 20 minutes at room temperature. The cells were frozen in RPMI medium supplemented with 12.5% of human serum albumin and 10% of DMSO and stored in liquid nitrogen for later use.
iPSC generation
T-cell activation protocol
PBMCs were thawed into 6-well plates pre-coated with hCD3 antibody (Ebioscience) and grown in LGM media (Lonza Clonetics) supplemented with 50 ng/ml IL-2 (R&D Systems), 0.4 μg/ml CD3 (Thermo Fisher Scientific) and CD28 (Thermo Fischer Scientific), 1 mM L-Glutamine (Thermo Fisher Scientific), 1% Penicillin-Streptomycin (Thermo Fisher Scientific) for 6 days until approximately 80% of cells express CD3 and CD28. Activated T-cells (5.0 E+05 cells) were transduced with the four Sendai viruses expressing Oct3/4, Sox2, Klf4, and c-Myc using the Cytotune 1.0 kit (Thermo Fisher Scientific) at a multiplicity of infection (MOI) 10 for all factors, and grown in LGM media supplemented with IL-2, CD3, L-Glutamine, and Penicillin-Streptomycin. After 24 hours, transduced cells were plated onto 0.1% Gelatin/PBS coated plates containing a monolayer of mouse embryonic fibroblasts (MEF)-irradiated cells (GlobalStem) and grown in iPSC A medium containing DMEM, 15% Fetal Calf Serum (FCS) (Gemini Biologicals), 1 mM L-Glutamine, 1% Penicillin-Streptomycin for 4 days. The cells were switched to iPSC B medium containing DMEM/F12, 20% Knockout Serum (Thermo Fisher Scientific), 1% Non-Essential Amino Acids (Thermo Fisher Scientific), 1mM L-Glutamine, 1% Penicillin-Streptomycin, 1 μg/mL Fungizone (Hyclone), 2-mercaptoethanol (Sigma), 20 ng/ml bFGF (R&D Systems), and 50 μg/mL Ascorbic Acid (Sigma) for the duration of reprogramming.
Erythroblast protocol
PBMCs were thawed into 6-well plates and expanded as erythroblasts in Expansion medium (EM) containing QBSF-60 (Quality Biologicals), 50 ng/ml SCF (R&D Systems), 10 ng/ml IL-3 (R&D Systems), 2 U/ml EPO (Amgen) 40 ng/ml IGF-1 (R&D Systems), 1 μM Dexamethasone (Sigma), 1% Penicillin- Streptomycin, 50 μg/ml Ascorbic Acid (Sigma), 1 μg/mL Fungizone for 9–12 days until approximately 90% of cells expressed CD36 and CD71. Erythroblasts (1.5–2.25 E+05 cells) were transduced with the four Sendai viruses expressing Oct3/4, Sox2, Klf4, and c-Myc using the Cytotune1.0 kit at an MOI of 10 for all factors. After 24 hours, transduced cells were washed and plated onto 0.1% Gelatin/PBS coated plates containing a monolayer MEF cells and grown in iPSC A medium containing DMEM/F12 10% FCS, 1% Non-Essential Amino Acids, 1 mM L-Glutamine, 1mM Penicillin-Streptomycin, 1 ug/mL Fungizone, 2- mercaptoethanol, 10 ng/ml bFGF, and 50 μg/mL Ascorbic Acid for 3 days. Cells were switched to medium containing 50% IPSC A medium and 50% iPSC Medium B containing DMEM/F12, 20% Knockout Serum, 1% Non-Essential Amino Acids, 1 mM L-Glutamine, 1mM Penicillin-Streptomycin, 1 ug/mL Fungizone, 2- mercaptoethanol, 10 ng/ml bFGF, and 50 μg/mL Ascorbic Acid for 2 days and switched to 100% IPSC B medium for the duration of reprogramming. Primary iPSC colonies emerged at day 9–12 post-transduction. To sub-clone passage 1 (P1) colonies iPSC cultures were subjected to live cell immunostaining with DAPI (Thermo Fisher Scientific) and anti-Human Tra-1-60 PE (Ebioscience) to identify pluripotent colonies. Tra-1-60+ colonies were manually picked using a fluorescent microscope housed in a biosafety cabinet and transferred with a micropipette to a 12-well plate coated with 0.1% Gelatin/PBS containing MEFs. iPSC P1 colonies were maintained in hESC medium containing DMEM/F12, 20% Knockout Serum, 1% Non-Essential Amino Acids, 1 mM L-Glutamine, 1mM Penicillin-Streptomycin, 1 μg/mL Fungizone, 2-mercaptoethanol, 10 ng/ml bFGF. IPSC cultures were routinely immunostained with human anti-Oct4 (Stemgent) and anti-Nanog (R&D Systems) to assess pluripotency.
Cell culture and expansion
iPSCs were expanded on 12-well plates coated with 0.1% Gelatin/PBS containing MEFs until passage 6–8 and were maintained in hESC medium as described. iPSCs were bulk passaged using either Accutase (Innovative Cell technologies) or 1 mM EDTA (Sigma). Cells were washed once with PBS and treated with pre-warmed Accutase or EDTA, incubated at 37 degrees for 1–5 minutes, washed once with PBS for Accutase passaging, and resuspended in fresh hESC medium containing either 10 μM Y-27632 Rock inhibitor (RI) (Stemgent) or 2 μM Thiazovivin (Millipore). Cells were detached with gentle pipetting and cell suspension was transferred to a new 12-well plate coated with 0.1% Gelatin/PBS containing a MEFs. After 12 hours incubation with RI or Thiazovivin, medium was changed daily with fresh hESC medium. iPSCs were passaged every 5–7 days and transitioned from MEF-feeder to feeder-free conditions at passage 6–8. To transition to feeder-free conditions, iPSCs were passaged as described and transferred to a 12-well or 6-well plate coated with 5% Matrigel (BD Bioscience) in DMEM/F-12. Cells were maintained in mTesr1 (Stem Cell Technologies) supplemented with 1 mM L-Glutamine, 1mM Penicillin-Streptomycin, 0.1 μg/ml Fungizone. To ensure Sendai virus was cleared from cells, it was tested by immunostaining cultures with anti-Sendai virus antibody (Abcam). Differentiating colonies were routinely eliminated from cultures.
Mycoplasma QC
PBMCs and iPSCs were grown in the absence of Penicillin-Streptomycin and Fungizone for 3 days, harvested for DNA extraction using either Easy-DNA kit (Thermo Fisher Scientific) or Purelink Genomic DNA kit (Thermo Fisher Scientific), and tested for mycoplasma contamination using a Mycoplasma PCR detection kit (BOCA scientific, e-Myco PLUS PCR Detection Kit).
Differentiation of iPSCs to endothelial cells
iPSCs were cultured in E8 medium (Thermo) on ESC-qualified Matrigel (Corning) until 80% confluency. The cells (1 to 24 dilution) were then plated onto a growth factor reduced Matrigel coated well in presence of 10 μM of Y-27632 (Selleckchem) in triplicate. 24 hours after plating, the medium was replaced with E8 medium (Thermo) for an additional day. The cells were then cultured for 3 days in DMEM/F12 supplemented with B27 and N2 (Thermo), 5μM CHIR-99021 (Selleckchem) and 25 ng/ml BMP-4 (Peprotech). For the final 2 days, the cells were grown in Stempro34 (Thermo) supplemented with 200ng/ml VEGF (Peprotech) and 5μM Forskolin (LC labs). Endothelial differentiation was performed in triplicates and calculated as CD31+/CD144+ cell percentage through flow cytometry. We differentiated 73 lines from 23 subjects and the cut-off for high versus low differentiation was set at 10% output of CD31+/CD144+ endothelial cells (range 0.99–35.34%)(Table S4)
RNA sequencing
iPSCs were grown under feeder-free conditions from passage 8–11 for RNA sequencing experiments. iPSCs were grown to 100% confluency, washed once with PBS, and harvested for RNA extraction using either miRNeasy Mini kit (Qiagen) or PureLInk RNA mini kit (Thermo Fisher Scientific). RNA was extracted as per manufacturers’ instruction. Total RNA was quantified using a Nanodrop (Thermo Scientific). RNA samples with a A260/280 ratio <1.8 or >2.3 were generally excluded from further processing.
RNA integrity was checked in Fragment Analyzer (Advanced Analytical) or 2100 Bioanalyzer using the RNA 6000 Nano assay (Agilent). All measured total RNA samples had RQN/RIN value of 7.0 or greater. The sequencing library was prepared with the standard TruSeq RNA Sample Prep Kit v2 protocol (Illumina). mRNA was isolated and fragmented. cDNA was synthesized using random hexamers, end-repaired and ligated with appropriate adaptors for sequencing. The library then underwent size selection and purification using AMPure XP beads (Beckman Coulter). The appropriate Illumina recommended 6 bp barcode bases are introduced at one end of the adaptors during the PCR amplification step. The size and concentration of the RNA-seq libraries were measured by Bioanalyzer and Qubit fluorometry (Thermo Fisher Scientific) before loading onto the sequencer. The mRNA libraries were sequenced on the Illumina HiSeq 2500 System with 100 nucleotide single-end reads, according to the standard manufacturer’s protocol (Illumina,).
RNA-seq pre-processing
RNA-seq reads were aligned to GRCh37 with STAR v2.4.0g1 (Dobin et al., 2013). Uniquely mapping reads overlapping genes were counted with featureCounts v1.4.4 (Liao et al., 2014) using annotations from ENSEMBL v70. All analysis used log2 counts per million (CPM) following TMM normalization (Robinson and Oshlack, 2010) implemented in edgeR (Robinson et al., 2010), unless stated otherwise. Genes with over 1 CPM in at least 30% of the experiments were retained for the strict cutoff and those with over 0.1 CPM in at least 10% of experiments were retained for the liberal cutoff. All analyses of RNA-seq data, with the exception of allele specific expression analysis, network analyses and functional analysis of variance, were performed on expression residuals after correcting for the effects of 8 sequencing batches and 2 RNA preparation kits. The mean expression value was added to residuals to preserve the scale of expression.
Visualization with violin-boxplots
The fraction of variation explained by each aspect of the study design is presented using a combined violin and boxplot (Wickham, 2009). The boxplots indicate the median, inner quartile range (IQR) and 1.5 times the IQR. Data beyond this are plotted as points. Violin plots indicate the density of data points based on their width.
Processing of genotype data
Genotype data were filtered to remove markers with over 5% missing entries, minor allele frequency below 1% and Hardy-Weinberg p-value < 1e-6. Genotypes were phased with SHAPEIT v2.r790 (Delaneau et al., 2012), and missing genotypes were imputed with Impute2 v2.3.2 (Howie et al., 2009) using the reference panel from the 1000 Genomes Project Phase 3 (The 1000 Genomes Project Consortium, 2015). Markers with high imputation quality (INFO>0.5; (Howie et al., 2009) and minor allele frequency over 1% were retained for downstream analysis.
Genotype and RNA-seq sample concordance
In order to ensure proper sample labeling, the concordance between array-based genotypes and RNA-seq variant calls was computed. This process ensured that multiple iPSC lines from the same individual had high concordance based on RNA-seq variants, and that array-based genotypes from each individual showed high concordance with the RNA-seq variants. RNA-seq variants were called with GATK v3.1.1 (DePristo et al., 2011) following GATK’s Best Practices. For each genotyped individual, only heterozygous sites were considered, and the concordance with each RNA-seq experiment was evaluated at only these sites. Only matches with more than 90% concordance rate were retained for further analysis. Mislabeled samples were relabeled only when its proper label could be determined unambiguously. Otherwise the RNA-seq experiment was excluded.
Integrating GTEx and Choi et al RNA-seq data
Gene expression values were quantile normalized, whereby for one sample at a time, the genes are ranked by RPKM magnitude and the ranks are transformed into the standard normal distribution. Visualization was preformed with multi-dimensional scaling of the RPKM values.
Identifying outliers in iPSC data
Principal components analysis was performed on expression data from 24 key stem cell genes: CDH1, CDH2, DNMT3B, DPPA2, DPPA4, FGF2, FGF4, KLF4, LIN28A, LIN28B, MYC, MYCN, NANOG, PBX1, PODXL, POU5F1, PRDM14, SALL1, SALL4, SOX2, TDGF1, TERT, ZFP42, ZSCAN10. iPSC lines greater than 3 standard deviations from the centroid, computed from the first 2 principal components using a robust covariance metric, were considered outliers.
variancePartition
The total variance was partitioned into the variance attributable to each experimental variable using a linear mixed model implemented in variancePartition v1.0.0 (Hoffman and Schadt, 2016) and the results visualized using the package’s functionalities. Continuous variables (i.e. Age and BMI) were modeled as fixed effects while the remaining categorical variables were modeled as random effects.
eQTL analysis
Following standard practice, only individuals of European ancestry were included in the eQTL analysis in order to avoid false positives due to the correlation between ancestry and gene expression. Principal components analysis based on genome-wide genotype data identified 81 individuals of European ancestry for eQTL analysis. eQTL analysis was performed with MatrixEQTL v2.1.1 (Shabalin, 2012) using the first 5 genotype principal components as covariates. Latent variables were identified in the gene expression data using PEER v1.0 (Stegle et al., 2010). Expression residuals were computed by removing the first 20 PEER components. Since multiple iPSC lines were assayed from each individual, the expression value for each individual was summarized as the mean expression residual value from the multiple lines for a given individual and gene. The mean values for each individual were subsequently quantile normalized for each gene. Cis-eQTL analysis considered markers within 1Mb of the transcription state site of each gene. False discovery rates were computed following Benjamini–Hochberg. Regions around eQTLs were visualized with locuszoom v1.3 (Pruim et al., 2010).
Allele-specific expression
This analysis exploits the fact that expression of a single gene can be separated into the transcripts originating from the maternal and paternal chromosomes when there is an expressed heterozygous genetic variant that distinguishes the two parental haplotypes. Instead of comparing gene expression across individuals, analysis of ASE (Lappalainen et al., 2013) uses an internal control by comparing the number of RNA-seq reads containing the reference allele to the total number of reads at a heterozygous site within the same experiment. Thus, a reference ratio of 0.5 indicates balanced expression of the two alleles while a significant deviation from 0.5 indicates allelic imbalance (Figure 3). ASE can only be detected in individuals with a heterozygous exonic SNP in the relevant gene. Despite these limitations, we illustrated this aspect of transcriptional variability. Allele-specific reads from RNA-seq were counted at heterozygous sites from imputed array-based genotype data. For a single individual, a site was considered heterozygous if the imputed dosage value was between 0.99 and 1.01 to retain genotyped sites, plus only imputed sites most likely to be true heterozygotes. In order to remove known biases in allele-specific expression (GTEx Consortium, 2015; Lappalainen et al., 2013), sites overlapping genomic regions with 50bp mapability score < 1 from the UCSC mapability track or showing excess bias in simulations were excluded. Only sites with over 30 reads were retained, and the reference fraction for each site was tested for a deviation from balanced expression using a binomial test. Sites with mono-allelic expression were only retained for canonically imprinted genes. Ideally, balanced expression would yield 50% reference alleles, but due to known reference bias, this fraction is slightly higher (Lappalainen et al., 2013). Instead, for each experiment, the genome-wide reference ratios were computed for each REF/ALT pair, and these values were used as empirical null values. These empirical null values were then used to compute a weighted reference ratio that was used in downstream analysis (Lappalainen et al., 2013). The reference ratio for each site was then tested against the matching REF/ALT genome-wide empirical null. False discovery rates were computed with qvalue. The genome-wide correlation between a pair of samples is the correlation between the weighted reference ratios for sites that pass the above cutoffs in both experiments (GTEx Consortium, 2015). The functional impact of variants was annotated with Variant Effect Predictor (McLaren et al., 2010).
Assessing eQTL overlap across datasets
For each gene with a genome-wide significant cis-eQTL in the current dataset, the most significant genetic marker was selected, and we tested whether there was evidence for that marker being an eQTL for the same gene in each tissue in the GTEx data (GTEx Consortium, 2015). We applied a widely used metric π1 that indicates the fraction of eQTL replicated in a second dataset (GTEx Consortium, 2015). This statistic avoids using an explicit p-value cutoff for identifying genome-wide significant eQTLs and is a better metric of eQTL sharing when studies have low power to replicate eQTLs at genome-wide significance. For a given tissue in GTEx, the p-values for markers corresponding to eQTLs in the current dataset were extracted and Storey’s q-value was used to estimate π0, the fraction of tests that come from the null model of no association between genotype and expression. Since π0 estimates the fraction of tests under the null model, π1 = 1−π0 is the estimated fraction of tests that are statistically significant. Thus π1 indicates estimated fraction of markers that are eQTLs in the current dataset as well as in the given GTEx tissue (GTEx Consortium, 2015). Reporting π1 for each tissue shows the degree of eQTL sharing between the current dataset and each GTEx tissue.
Enrichment of eQTLs near GWAS hits
We selected the most significant genetic marker for each gene with a genome-wide significant cis-eQTL in the current dataset. For each of these markers, we counted the genome-wide significant markers from the GWAS Catalog (Welter et al., 2014) associated with each phenotype that were within an r2 of 0.5 based on European individuals from the 1000 Genomes Project (The 1000 Genomes Project Consortium, 2015). In order to compute an odds ratio for the enrichment compared to the null model of no overlap between GWAS for each phenotype and eQTL signals, we used a permutation approach that generated 10,000 sets of markers with similar properties to the real set of eQTL markers. For each real eQTL marker, a corresponding marker was selected with minor allele frequency within 2%, distance to transcription start site within 1Kb, gene density within 30 genes per Mb. For each phenotype, the overlap between GWAS hits and real eQTLs from the current data was compared to the mean overlap under the empirical null distribution and this was used to compute odds ratios and 95% confidence intervals.
Integration of eQTLs with Epigenomics Roadmap
To assess how cis-eQTLs relate to known enhancer sequences, we tested for overlap between eQTLs and enhancer sequences from the (Roadmap Epigenomics Consortium, 2015). More specifically, we used chromatin states for enhancer sequences (active, genic, and weak enhancers), derived from a recent joint analysis that the Roadmap Epigenomics Consortium applied in different chromatin immunoprecipitation sequencing (ChIPseq) data across 98 human tissues and cell lines. We included tissues that were assayed for 6 different chromatin marks (H3K4me1, H3K4me3, H3K27ac, H3K36me3, H3K27me3, and H3K9me3). We tested for enrichment of significant eQTLs at FDR ≤ 5%, using as an “index” eQTL SNP (eSNP) the most significantly associated SNP per gene.
For each tissue or cell line, we counted the number of index eSNPs that lie within enhancer sequences respectively found in that tissue or cell line. To assess if this overlap is higher than expected by chance, we generated 1,000 sets of random SNPs matched with the index cis-eSNPs, in terms of allele frequency, gene density, distance from TSS, and density of tagSNPs arising from genomic variability of linkage disequilibrium. Z scores were estimated as:
Where observed is the number of index eSNPs that lie within enhancers, and meannull and SDnull are the mean and standard deviation of the null distribution of overlap, as estimated using the sets of permuted SNPs.
Co-expression Network Construction
Previous network analyses employing high variance gene expression filters, generally including more lowly expressed genes, have demonstrated the ability to identify biologically meaningful correlations that have elucidated complex traits (Zhang et al., 2013). For network construction purposes, we used the voom function from the limma R package (Ritchie et al., 2015) to normalize the gene counts and filtered out genes that did not have at least 0.1 counts per million (cpm) in 10% of the samples. We then adjusted the normalized counts for the following covariates: gender, reprogramming source cell, race, age, body mass index, RNA extraction method and insulin status. Co-expression networks were constructed using the coexpp R package(Langfelder and Horvath, 2008) (Michael Linderman and Bin Zhang (2011). coexpp: Large-scale Co-expression network creation and manipulation using WGCNA. R package version 0.1.0. https://bitbucket.org/multiscale/coexpp). Seeding gene lists to input in pathFinder were obtained by selecting genes in co-expression modules that were statistically enriched for the top 10% most varying genes.
Sensitivity Analysis
To demonstrate that the covariance structure was robust against technical effects (i.e., correlations driven by transcripts dominated by zero counts and coincident non-zero counts in a small number of samples) we constructed a similar co-expression network after adding a small amount of random noise to the zero reads of any gene in the raw RNA-seq count matrix. We then selected in this new network modules enriched for the top 10% most varying genes. The selected modules comprised of 2167 genes were homologous to the 6 modules described above, with an overlap of 2115 genes (p-value of Fisher’s exact test <10−300) and modular functional annotations (data not shown) and GO term enrichments that were virtually identical. These analyses confirmed that the covariance structure on which the networks rely were not significantly affected by the technical noise of genes with low expression levels.
Prior network construction
A prior network of gene and protein interactions was built by integrating two public databases: ConsensusPathDB, (CPDB) (Kamburov et al., 2013) and MetaCore (v6.24 from Thomson Reuters). While many of these interactions are at the protein level, the nodes in our network are gene IDs and we thereby implicitly assume equivalence between genes and proteins at the interaction level. We allow, however, two types of nodes that are not genes: protein complexes and protein families or classes (genes that code for proteins that are interchangeable for a given interaction). These two types of nodes were contained in the databases that we used to construct the network. They allow indirect interactions between genes through protein complexes or via a set of interchangeable proteins. The prior network contains interactions observed in a variety of human tissues. To make the network specific to iPSCs, we made use of Roadmap Epigenomics Project data (Roadmap Epigenomics Consortium, 2015) from iPSC lines to predict which genes are active, repressed or bivalent. Nodes predicted to be repressed in these iPSC lines were removed from the literature-curated prior network (complexes are removed if any component is repressed and families are removed if all family members are repressed) as well as all incoming and outgoing edges from these nodes. Thus any interactions that involve genes repressed in iPSC are removed and the resulting network should be iPSC-specific.
The epigenetic classification of genes into active, repressed and bivalent genes was done in three steps: first, peaks called from histone modification marks were used to predict active, repressed and bivalent sections of the genome. Individual transcripts were then classified based on the overlap of their promoter regions with these three classes of genome segments. Finally, genes were classified based on the classification of all of their associated transcripts.
To predict the state of genome segments we used the publicly available broad peak calls for histone modification marks H3K9me3 and H3K27me3 and gapped peak calls for H3K4me3, H3K9ac and H3K27ac. Using BEDOPS (Neph et al., 2012), active genome segments were defined as regions of the genome where H3K4me3 peaks overlapped with either H3K9ac or H3K27ac peaks and lacking H3K27me3 peaks. Repressed segments are characterized by either the absence of any histone modification marks or by the presence of either H3K9ac or H3K27ac peaks with no overlap with H3K4me3 peaks. Bivalent segments have overlapping H3K4me3 and H3K27me3 peaks.
Transcripts were classified into active/repressed/bivalent states by using a rule-based classifier. Specifically, a transcript was classified as active if its promoter region (defined as the transcription start site +- a certain window size, typically between 125bp and 1kb) overlapped with active segments. Transcripts not classified as active were considered bivalent if their promoters overlap with bivalent segments. Any unclassified transcripts that overlap with repressed marks were classified as repressed.
A gene was classified as active if any of its transcripts was active, as bivalent if none of its transcripts was active and at least one was bivalent, and as repressed if none of its transcripts was active or bivalent and at least one transcript was repressed.
There are several iPSC lines available in the Roadmap data. However for only two cell lines, iPS-18 and iPS-20b, ChIPseq data are available for all 5 histone modification marks (H3K4me3, H3K9ac, H3K9me3, H3K27ac and H3K27me3). After generating prior networks for each of these two cell-lines, the final iPSC-specific prior network was obtained by merging the two respective networks. The prior networks were built using RefSeq transcripts annotation downloaded from the UCSC Genome Browser in April 2015 and gene IDs were converted to ENSEMBL gene IDs using the biomaRt R package (Durinck et al., 2009). This final prior network contains 16,850 nodes and 246,139 edges.
pathFinder, a fast graphical algorithm
We developed pathFinder, an efficient graphical algorithm to extract neighborhood structures, given an initial gene set from a larger background network (in our case this will be the prior iPSC network). PathFinder is based on the classical Depth First Search (DFS) algorithm and allows users to expand an initial input gene set by including genes located in the paths connecting input genes in the background network. Since the background network contains directed and undirected edges, we transform the undirected edges into two directed edges with the same two end nodes but opposite directions. We do not allow these two edges to appear simultaneously in one path.
For every gene in the input list, the DFS explores all paths in the background network that start at that gene. The exploration of a path is stopped if it reaches length k (we used k=3), or arrives at a node with no valid child node(s). Only paths that start and end at genes included on the input list are retained. All nodes between the start and end genes on retained paths are included in the pathFinder output. When we apply pathFinder to the prior iPSC network, we only report genes along each path and not protein complexes or families. The final Bayesian network will not contain such nodes.
Bayesian network construction
We developed an integrative modeling pipeline to build causal and predictive network models by integrating multi-scale ‘–omics’ data, including genomic, transcriptomic, proteomic and epigenomic data, with the scientific literature and knowledgebase, specifically the ConsensusPathDB (Kamburov et al., 2013) and MetaCore (v6.24 from Thomson Reuters). Our pipeline constructed these networks in four steps: 1) We built a tissue-specific, multi-scale prior network from public databases (see section “Prior network construction” below). 2) We built co-expression modules from the gene expression data and extracted seeding gene lists from these modules to build Bayesian network (see section “Co-expression network construction and analysis” below). 3) To capture a comprehensive representation of the biology that is important in the context of pluripotency, we recruited the prior network from last step to compensate the limitations of using RNA-seq data and co-expression networks: i) RNA-seq data fails to capture protein-protein interaction; ii) linear Gaussian assumption in co-expression network may lead to missing non-linear correlations among genes, and iii) randomness in tuning the global parameters of co-expression network Our pipeline used this tissue-specific, multi-scale prior network to expand the set of seeding genes by using a graphical algorithm, pathFinder, that we developed for this purpose (see section “pathFinder, a fast graphical algorithm” above). The outcome of pathFinder was an expanded set of molecules which consists of the original seeding genes and genes, proteins and metabolites connecting to the original seeding genes in k steps (k=3 here) in the prior interaction network. 4) This expanded set of genes was used for constructing causal and predictive molecular interaction networks. We developed a Bayesian network component, which computes the Bayesian Dirichlet (BD) score at each step of the Markov chain Monte Carlo (MCMC) algorithm during the heuristic search for network structure. To avoid getting trapped at local maxima, we developed a hybrid search algorithm by integrating hill-climbing exploration for local neighbor structure with a global Hastings ratio for overall network structure update. For each move, we randomly selected a local node and calculate the BD score of all local neighbor structures reachable by a single move around the selected node. In this way, we segmented the global neighborhood of the current network structure into subsets of local neighbors, randomly selected a subset per step and explored all local structures within this subset. Then, we selected the local structure with maximal score in this subset as our candidate structure. Finally, we calculated the Hastings ratio between the BD scores of the current and candidate structures. This hybrid search algorithm made it feasible to efficiently search a very large structure space and ensured a probability for the MCMC chain to move out of a local maximal when the BD score of current structure was bigger than all its local neighbors. Our Bayesian network module integrated genetic data with gene expression data by translating the cis-eQTL genes into structural constraints during structure learning. Specifically, cis-eQTL genes were considered root nodes and other nodes weren’t allowed as their parents. The learned Bayesian network depicted causal molecular interactions.
Key driver analysis
To do Key Driver Analysis, we used the R package KDA (Zhang and Zhu, 2013)(KDA R package version 0.1, available at http://research.mssm.edu/multiscalenetwork/Resources.html). The package firstly defines a background sub-network by looking for a neighborhood K-step away from each node in the target gene list in the network. Then, stemming from each node in this sub-network, it assesses the enrichment in its k-step (k varies from 1 to K) downstream neighborhood for the target gene list. In this analysis, we used K=6.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis of gene expression data
Analysis was implemented in R (R Core Team, 2015). Hierarchical clustering used the complete-linkage algorithm, where each gene was centered and scaled to have a mean of 0 and a variance of 1. Results from clustering based on Euclidian distances between each pair of samples are shown, but results using a correlation-based distance (i.e. 1- correlation) show a very similar clustering of multiple iPSC lines from the same individual. Gene expression gradients were estimated by regressing the expression of a single gene on the first two principal components. The gradient corresponds to the predicted expression value based on this regression model. The relationship between the percentage of cross-individual variation for each gene and the probability that each gene has a genome-wide significant cis-eQTL was modeled with logistic regression in order to ensure a monotonically increasing smooth curve. A response variable was coded as 1 for genes with a detected cis-eQTL and 0 for genes without a cis-eQTL, and the percentage of variation explained by individual was used as a predictor. The standard Wald test for logistic regression was used to compute the p-value under the null hypothesis of no association between the response and predictor.
Additional statistical Analysis in R
For all enrichment tests, Fisher’s Exact Test was performed using R. All p values shown in the paper were FDR corrected (BH method). To test enrichment with GO annotations, the R packages goseq (Young et al., 2010), topGO (Alexa A and Rahnenfuhrer J (2010). topGO: topGO: Enrichment analysis for Gene Ontology. R package version 2.18.0.) and org.Hs.eg.db (Carlson M. org.Hs.eg.db: Genome wide annotation for Human. R package version 3.2.3.) were used. To test msigDB pathway enrichment, the R packages HTSanalyzeR (Wang et al., 2011), GSEABase (Morgan M, Falcon S and Gentleman R. GSEABase: Gene set enrichment data structures and methods. R package version 1.32.0., and gage (Luo et al., 2009) were used. All gene mapping was performed using the biomaRt R package (Durinck et al., 2009). Figures where generated using the R packages ggplot2 (Wickham, 2009) scales (Hadley Wickham (2012). scales: Scale functions for graphics. R package version 0.2.3. http://CRAN.R-project.org/package=scales), reshape2 (Wickham, 2007) and grid (Murrell, 2005). The lmFit function from the limma package was used for the differential expression analyses (Ritchie et al., 2015).
DATA AND SOFTWARE AVAILABILITY
Software
We used a number of previously published software resources as outlined in the individual method descriptions and key resources table. For the custom software:
variancePartition is a statistical and visualization framework fits a linear mixed model for each gene, and partitions the total variance into the contribution of each variable in the experimental design plus the residual variance: http://www.bioconductor.org/packages/release/bioc/html/variancePartition.html
Data resources
RNA-seq data is deposited at GEO: GSE79636 and dbGAP: phs001139.
Supplementary Material
Highlights.
Gene expression analysis characterizes 317 human iPSC lines from 101 individuals
eQTLs contribute significantly to across individual variation in iPSC lines
Polycomb target genes are a significant source of non-genetic variation
Predictive networks highlight candidate key drivers of differentiation efficiency
Acknowledgments
This work was supported by NIH grants U01HL107388 (TQ, EES, IL), R01GM114434 and IBM faculty award (GP), AHA (10FTF3360005, JWK), 1RF1AG051504-01, R01AG043076, R01MH097276 (RC)
Footnotes
Supplemental information for this article includes 7 figures and 7 supplemental items.
AUTHOR CONTRIBUTIONS
Conceptualization: T.Q., E.E.S., J.W.K., G.E.H., N.D.B., I.C-O, I.L., R.C. Methodology Development: G.E.H., K.Z., G.P., N.D.B., R.C. Software Programming: G.E.H., R.C., N.D.B., K.Z., G.P. Validation: D.P., S.W., G.P. Formal Analysis Application: G.E.H., S.W., K.Z., P.R., G.P., M.H., R.C., J.W.K. Investigation: I.C-O, P.C., F.A., M.S., A.P., S.D’S., P.L. Resources: P.C., F.A., G.M.R., A.P., S.D’S., J.W.K. Data Curation: G.E.H., F.A., I.C-O, S.D’S. Writing – Original Draft Preparation: I.C-O, P.C., S.D’S., R.C., J.W.K., G.E.H. Writing – Review & Editing Preparation: A.P., G.P., E.E.S.,I.L.,T.Q. Visualization: N.D.B., I.C-O, G.E.H., M.H. Supervision: G.M.R., G.P., S.D’S., E.E.S., I.L., T.Q., R.C., J.W.K. Project Administration: P.C., S.D’S., I.L., J.W.K. Funding Acquisition: E.E.S., T.Q., J.W.K.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aloia L, Di Stefano B, Di Croce L. Polycomb complexes in stem cells and embryonic development. Development. 2013;140:2525–2534. doi: 10.1242/dev.091553. [DOI] [PubMed] [Google Scholar]
- Bahrami SB, Veiseh M, Dunn AA, Boudreau NJ. Temporal changes in Hox gene expression accompany endothelial cell differentiation of embryonic stem cells. Cell adhesion & migration. 2011;5:133–141. doi: 10.4161/cam.5.2.14373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Nur O, Russ HA, Efrat S, Benvenisty N. Epigenetic memory and preferential lineage-specific differentiation in induced pluripotent stem cells derived from human pancreatic islet beta cells. Cell stem cell. 2011;9:17–23. doi: 10.1016/j.stem.2011.06.007. [DOI] [PubMed] [Google Scholar]
- Ben-David U, Mayshar Y, Benvenisty N. Large-scale analysis reveals acquisition of lineage-specific chromosomal aberrations in human adult stem cells. Cell stem cell. 2011;9:97–102. doi: 10.1016/j.stem.2011.06.013. [DOI] [PubMed] [Google Scholar]
- Benetatos L, Vartholomatos G, Hatzimichael E. DLK1-DIO3 imprinted cluster in induced pluripotency: landscape in the mist. Cellular and molecular life sciences: CMLS. 2014;71:4421–4430. doi: 10.1007/s00018-014-1698-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes & development. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows CK, Banovich NE, Pavlovic BJ, Patterson K, Gallego Romero I, Pritchard JK, Gilad Y. Genetic Variation, Not Cell Type of Origin, Underlies the Majority of Identifiable Regulatory Differences in iPSCs. PLoS genetics. 2016;12:e1005793. doi: 10.1371/journal.pgen.1005793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahan P, Daley GQ. Origins and implications of pluripotent stem cell variability and heterogeneity. Nature reviews Molecular cell biology. 2013;14:357–368. doi: 10.1038/nrm3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahan P, Li H, Morris SA, Lummertz da Rocha E, Daley GQ, Collins JJ. CellNet: network biology applied to stem cell engineering. Cell. 2014;158:903–915. doi: 10.1016/j.cell.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Lee S, Mallard W, Clement K, Tagliazucchi GM, Lim H, Choi IY, Ferrari F, Tsankov AM, Pop R, et al. A comparison of genetically matched cell lines reveals the equivalence of human iPSCs and ESCs. Nat Biotechnol. 2015;33:1173–1181. doi: 10.1038/nbt.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das PP, Hendrix DA, Apostolou E, Buchner AH, Canver MC, Beyaz S, Ljuboja D, Kuintzle R, Kim W, Karnik R, et al. PRC2 Is Required to Maintain Expression of the Maternal Gtl2-Rian-Mirg Locus by Preventing De Novo DNA Methylation in Mouse Embryonic Stem Cells. Cell reports. 2015;12:1456–1470. doi: 10.1016/j.celrep.2015.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nature methods. 2012;9:179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature genetics. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durinck S, Spellman PT, Birney E, Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nature protocols. 2009;4:1184–1191. doi: 10.1038/nprot.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragola G, Germain PL, Laise P, Cuomo A, Blasimme A, Gross F, Signaroldi E, Bucci G, Sommer C, Pruneri G, et al. Cell reprogramming requires silencing of a core subset of polycomb targets. PLoS genetics. 2013;9:e1003292. doi: 10.1371/journal.pgen.1003292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gafni O, Weinberger L, Mansour AA, Manor YS, Chomsky E, Ben-Yosef D, Kalma Y, Viukov S, Maza I, Zviran A, et al. Derivation of novel human ground state naive pluripotent stem cells. Nature. 2013;504:282–286. doi: 10.1038/nature12745. [DOI] [PubMed] [Google Scholar]
- Galonska C, Ziller MJ, Karnik R, Meissner A. Ground State Conditions Induce Rapid Reorganization of Core Pluripotency Factor Binding before Global Epigenetic Reprogramming. Cell stem cell. 2015;17:462–470. doi: 10.1016/j.stem.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman GE, Schadt EE. variancePartition: Quantifying and interpreting drivers of variation in complex gene expression studie. 2016 doi: 10.1186/s12859-016-1323-z. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS genetics. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Consortium for Blood Pressure Genome-Wide Association Studies. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamburov A, Stelzl U, Lehrach H, Herwig R. The ConsensusPathDB interaction database: 2013 update. Nucleic acids research. 2013;41:D793–800. doi: 10.1093/nar/gks1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S, Bonasio R, Saldana-Meyer R, Yoshida T, Son J, Nishino K, Umezawa A, Reinberg D. Interactions between JARID2 and noncoding RNAs regulate PRC2 recruitment to chromatin. Molecular cell. 2014;53:290–300. doi: 10.1016/j.molcel.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LI, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Paylor SW, Magnuson T, Schumacher A. Juxtaposed Polycomb complexes co-regulate vertebral identity. Development. 2006;133:4957–4968. doi: 10.1242/dev.02677. [DOI] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. Genome-wide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS genetics. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar RM, Cahan P, Shalek AK, Satija R, DaleyKeyser AJ, Li H, Zhang J, Pardee K, Gennert D, Trombetta JJ, et al. Deconstructing transcriptional heterogeneity in pluripotent stem cells. Nature. 2014;516:56–61. doi: 10.1038/nature13920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyttala A, Moraghebi R, Valensisi C, Kettunen J, Andrus C, Pasumarthy KK, Nakanishi M, Nishimura K, Ohtaka M, Weltner J, et al. Genetic Variability Overrides the Impact of Parental Cell Type and Determines iPSC Differentiation Potential. Stem cell reports. 2016;6:200–212. doi: 10.1016/j.stemcr.2015.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen T, Sammeth M, Friedlander MR, t Hoen PA, Monlong J, Rivas MA, Gonzalez-Porta M, Kurbatova N, Griebel T, Ferreira PG, et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–511. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YI, van de Geijn B, Raj A, Knowles DA, Petti AA, Golan D, Gilad Y, Pritchard JK. RNA splicing is a primary link between genetic variation and disease. Science. 2016;352:600–604. doi: 10.1126/science.aad9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G, Zhang Y. Genetic and epigenetic variations in iPSCs: potential causes and implications for application. Cell stem cell. 2013;13:149–159. doi: 10.1016/j.stem.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Liu L, Luo GZ, Yang W, Zhao X, Zheng Q, Lv Z, Li W, Wu HJ, Wang L, Wang XJ, et al. Activation of the imprinted Dlk1-Dio3 region correlates with pluripotency levels of mouse stem cells. The Journal of biological chemistry. 2010;285:19483–19490. doi: 10.1074/jbc.M110.131995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Friedman MS, Shedden K, Hankenson KD, Woolf PJ. GAGE: generally applicable gene set enrichment for pathway analysis. BMC bioinformatics. 2009;10:161. doi: 10.1186/1471-2105-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayshar Y, Ben-David U, Lavon N, Biancotti JC, Yakir B, Clark AT, Plath K, Lowry WE, Benvenisty N. Identification and classification of chromosomal aberrations in human induced pluripotent stem cells. Cell stem cell. 2010;7:521–531. doi: 10.1016/j.stem.2010.07.017. [DOI] [PubMed] [Google Scholar]
- McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller FJ, Schuldt BM, Williams R, Mason D, Altun G, Papapetrou EP, Danner S, Goldmann JE, Herbst A, Schmidt NO, et al. A bioinformatic assay for pluripotency in human cells. Nature methods. 2011;8:315–317. doi: 10.1038/nmeth.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell P. R Graphics. Boca Raton, Florida: Chapman and Hall/CRC; 2005. [Google Scholar]
- Nazor KL, Altun G, Lynch C, Tran H, Harness JV, Slavin I, Garitaonandia I, Muller FJ, Wang YC, Boscolo FS, et al. Recurrent variations in DNA methylation in human pluripotent stem cells and their differentiated derivatives. Cell stem cell. 2012;10:620–634. doi: 10.1016/j.stem.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neph S, Kuehn MS, Reynolds AP, Haugen E, Thurman RE, Johnson AK, Rynes E, Maurano MT, Vierstra J, Thomas S, et al. BEDOPS: high-performance genomic feature operations. Bioinformatics. 2012;28:1919–1920. doi: 10.1093/bioinformatics/bts277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, Boehnke M, Abecasis GR, Willer CJ. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; 2015. [Google Scholar]
- Rhoads K, Arderiu G, Charboneau A, Hansen SL, Hoffman W, Boudreau N. A role for Hox A5 in regulating angiogenesis and vascular patterning. Lymphatic research and biology. 2005;3:240–252. doi: 10.1089/lrb.2005.3.240. [DOI] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic acids research. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas MA, Pirinen M, Conrad DF, Lek M, Tsang EK, Karczewski KJ, Maller JB, Kukurba KR, DeLuca DS, Fromer M, et al. Effect of predicted protein-truncating genetic variants on the human transcriptome. Science. 2015;348:666–669. doi: 10.1126/science.1261877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roadmap Epigenomics Consortium. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhani F, Kumasaka N, de Brito MC, Bradley A, Vallier L, Gaffney D. Genetic background drives transcriptional variation in human induced pluripotent stem cells. PLoS genetics. 2014;10:e1004432. doi: 10.1371/journal.pgen.1004432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabalin AA. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics. 2012;28:1353–1358. doi: 10.1093/bioinformatics/bts163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperber H, Mathieu J, Wang Y, Ferreccio A, Hesson J, Xu Z, Fischer KA, Devi A, Detraux D, Gu H, et al. The metabolome regulates the epigenetic landscape during naive-to-primed human embryonic stem cell transition. Nature cell biology. 2015;17:1523–1535. doi: 10.1038/ncb3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Apostolou E, Akutsu H, Fukuda A, Follett P, Natesan S, Kono T, Shioda T, Hochedlinger K. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010;465:175–181. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegle O, Parts L, Durbin R, Winn J. A Bayesian framework to account for complex non-genetic factors in gene expression levels greatly increases power in eQTL studies. PLoS Comput Biol. 2010;6:e1000770. doi: 10.1371/journal.pcbi.1000770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surface LE, Thornton SR, Boyer LA. Polycomb group proteins set the stage for early lineage commitment. Cell stem cell. 2010;7:288–298. doi: 10.1016/j.stem.2010.08.004. [DOI] [PubMed] [Google Scholar]
- The 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theunissen TW, Powell BE, Wang H, Mitalipova M, Faddah DA, Reddy J, Fan ZP, Maetzel D, Ganz K, Shi L, et al. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell stem cell. 2014;15:471–487. doi: 10.1016/j.stem.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas SM, Kagan C, Pavlovic BJ, Burnett J, Patterson K, Pritchard JK, Gilad Y. Reprogramming LCLs to iPSCs Results in Recovery of Donor-Specific Gene Expression Signature. PLoS genetics. 2015;11:e1005216. doi: 10.1371/journal.pgen.1005216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Terfve C, Rose JC, Markowetz F. HTSanalyzeR: an R/Bioconductor package for integrated network analysis of high-throughput screens. Bioinformatics. 2011;27:879–880. doi: 10.1093/bioinformatics/btr028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, Klemm A, Flicek P, Manolio T, Hindorff L, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic acids research. 2014;42:D1001–1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. Reshaping Data with the reshape Package. Journal of Statistical Software. 2007;21:1–20. [Google Scholar]
- Wickham H. ggplot2: Elegant Graphics for Data Analysis. New York: Springer-Verlag; 2009. [Google Scholar]
- Xie R, Everett LJ, Lim HW, Patel NA, Schug J, Kroon E, Kelly OG, Wang A, D’Amour KA, Robins AJ, et al. Dynamic chromatin remodeling mediated by polycomb proteins orchestrates pancreatic differentiation of human embryonic stem cells. Cell stem cell. 2013;12:224–237. doi: 10.1016/j.stem.2012.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11:R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Gaiteri C, Bodea LG, Wang Z, McElwee J, Podtelezhnikov AA, Zhang C, Xie T, Tran L, Dobrin R, et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell. 2013;153:707–720. doi: 10.1016/j.cell.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Zhu J. Identification of Key Causal Regulators in Gene Networks. Proceedings of the World Congress on Engineering & Computer Science. 2013:2. [Google Scholar]
- Zhu J, Sova P, Xu Q, Dombek KM, Xu EY, Vu H, Tu Z, Brem RB, Bumgarner RE, Schadt EE. Stitching together multiple data dimensions reveals interacting metabolomic and transcriptomic networks that modulate cell regulation. PLoS biology. 2012;10:e1001301. doi: 10.1371/journal.pbio.1001301. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.