Abstract
The ability of DNA Double Strand Breaks (DSBs) to cluster in mammalian cells has been subjected to intense debate over the past few years. Here we used a high throughput chromosome conformation capture assay (Capture Hi-C) to investigate clustering of DSBs induced at defined loci in the human genome. We unambiguously found that DSBs do cluster but only when induced in transcriptionally active genes. Clustering of damaged genes mainly occurs during the G1 cell cycle phase and coincides with delayed repair. Moreover DSB clustering depends on the MRN complex, as well as the Formin 2 (FMN2) nuclear actin organizer and the LINC (LInker of Nuclear and Cytoplasmic skeleton) complex, suggesting that active mechanisms promote DSB clustering. This work reveals that when damaged, active genes exhibit a very peculiar behavior compared to the rest of the genome, being mostly left unrepaired and clustered in G1 while being repaired by homologous recombination in post-replicative cells.
Keywords: DSB repair, clustering, translocation, transcription, Capture Hi-C
Introduction
Translocations, that occur when two DNA Double Strand breaks (DSBs) are abnormally rejoined, represent highly deleterious genome rearrangements favoring cancer initiation and progression. However, the mechanisms that drive their formation are yet poorly deciphered. One prerequisite for translocation is the juxtaposition of two distant DSBs, an event that would be favored if DSBs cluster, i.e. are brought together in close spatial proximity within the nucleus. In budding yeast, a lacO or TetO-tagged genomic locus exhibits increased motion when damaged using the I-SceI endonuclease. Intriguingly, this phenomena, referred to as “local mobility”, is also accompanied by a “global mobility” process whereby the other chromosomes also explore wider volumes in the nuclear space although to a lesser extent than the damaged locus itself1–3. Moreover, inducing two DSBs by the HO and I-SceI endonucleases triggers the formation of a single Rad52 focus, suggesting that DSBs can indeed coalesce in yeast4.
However, whether DSBs cluster in higher eukaryotes has been subjected to a strong controversy over the past decade, due to conflicting results obtained using microscopy based methods5–13 (for review14). On one hand, DSBs induced on a linear track by α particles, etoposide or high dose of radiation can move over long distances and cluster over time5–8. In addition, DSBs induced at subtelomeric regions in cells that use alternative lengthening of telomeres (ALT) for telomere maintenance or induced by a restriction enzyme (AsiSI) undergo clustering15,16. Finally acquisition of thousands of time-lapse movies demonstrated that two damaged loci can frequently be juxtaposed and lead to translocation17. However, in other studies X-ray or heavy ion induced DSBs did not exhibit this peculiar behavior and dynamic analysis of a single broken chromosome end showed only a small scale local motion9–13, arguing against clustering.
DSBs are mainly repaired by Homologous Recombination (HR) and Non Homologous End Joining (NHEJ) that rely on profoundly different mechanisms. NHEJ directly reseals the two DNA ends and is available throughout the cell cycle. In contrast, HR uses a homologous copy of the broken locus as a template. It depends on the generation of single strand DNA (a process called resection) which is initiated by the MRN complex and followed by nucleoprotein filament assembly, homology search, strand invasion and DNA synthesis (reviewed in18). Importantly this pathway is strongly suppressed during the G1 cell cycle phase, most likely to minimize the use of an illegitimate copy as a template19. Besides the tight control exerted by the cell cycle stage on repair pathway usage, a growing body of evidence suggests that repair is also regulated by chromatin and varies across the genome (reviewed in20). High resolution profiles of RAD51 and XRCC4 (involved respectively in HR and NHEJ), on a genome wide scale, following induction of multiple, sequence-specific, DSBs by a restriction enzyme (AsiSI) revealed that, in post-replicative cells, DSBs lying in intergenic regions or silent genes are mostly repaired by NHEJ, while those occurring in active genes are channeled to HR21. The trimethylation of lysine 36 of histone H3 (H3K36me3), a histone mark previously linked to transcriptional elongation, contributes to target HR at transcriptionally active loci21–24.
Since the initial chromatin context controls the repair pathway choice and because the ability of a locus to roam in the nucleus has been proposed to be regulated by the chromatin compaction state (for example25), we hypothesized that the propensity of DSBs to cluster could also depend on the properties of the broken loci, i.e., their genomic position. Here, in order to assess this hypothesis, we studied clustering between DSBs induced by AsiSI, scattered throughout the genome and located both in transcriptionally active and inactive regions, using a chromosome conformation capture (3C) derivative technique (Capture Hi-C26). We unveiled that DSB clustering mainly occurs between breaks induced in active genes, during G1, in a manner that depends on the nuclear actin organizer Formin 2 (FMN2) and the LINC (LInker of Nuclear and Cytoplasmic skeleton) complex. Furthermore, clustering correlates with delayed repair in G1, which may indicate that instead of promoting repair, clustering may rather contribute to “sequester” persistent DSBs.
Results
Capture Hi-C experimental design in DIvA cells
The DIvA (DSB inducible via AsiSI) human cell line allows the induction of roughly a hundred DSBs located in euchromatin (since AsiSI activity is inhibited by DNA methylation) upon 4 hydroxytamoxifen (4OHT) treatment21,27. Using time lapse microscopy we recently reported that AsiSI-induced DSBs exhibit clustering in an ATM-dependent manner16. However, the identity of DSBs able to coalesce, among the 100 breaks induced by AsiSI, was unknown. In order to identify those DSBs we performed Hi-C, followed by sequence capture and high throughput sequencing (Capture Hi-C)26 (Fig. 1a). Capture probes were designed throughout γH2AX domains established following DSB induction in DIvA cells21 surrounding the hundred most cleaved AsiSI sites in vivo (see details in Supplementary Note). 3 control regions of equivalent size (2Mb) but devoid of DSB were included. An average of 50 probes were designed per domain with a high coverage at the immediate vicinity of the DSB on a +/- 10kb window (one probe per HindIII fragment i.e. 1 probe /5kb) and a lower coverage on the rest of the domain (1 probe for 10 HindIII fragments, i.e approximately 1 probe/ 50kb) (Supplementary Tables 1-2). Capture Hi-C was performed in undamaged or damaged cells, in two biological, independent, replicates. Di-tags analyses behaved as expected indicating that both 3C libraries and capture steps worked efficiently (Fig. S1a-c). Both replicates exhibited a strong correlation (Fig. S1d-e).
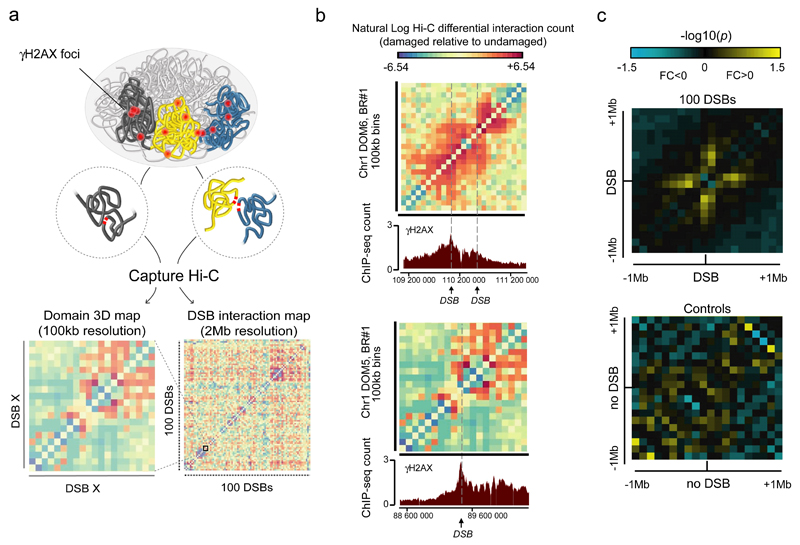
Figure 1. Capture Hi-C reveals DSB-induced local changes in chromosome folding.
a. General experimental pipeline: DSBs are introduced at known positions throughout the human genome following 4-hydroxytamoxifen (4-OHT) addition. Capture Hi-C experiments are next conducted both before and after DSB induction in order to identify whether and which DSBs interact within the nucleus following damage (Supplementary Note). 100kb resolution contact maps help to determine changes in cis interaction within γH2AX domains, while 2 megabase (2Mb) resolution interaction maps, allow to identify domains able to interact (i.e. cluster) following damage.
b. The differential (damaged versus undamaged), 100kb resolution, interaction map is shown for two domains captured on the chromosome 1. Data are expressed as natural log of differential interaction count (normalized, see Online Methods). Arrows indicate DSBs positions. γH2AX profile obtained by ChIP-seq at the same chromosomal locations21 following damage are shown (bottom panel).
c. Averaged interaction heatmap between damaged versus undamaged cells at 100kb resolution within 2Mb captured domains, around the 100 DSBs (top panel) or around the 3 control domains devoid of DSB (bottom panel). Both biological replicates are taken into account. -log10(p) are indicated, negative fold changes (damaged<undamaged) in blue, positive fold change (damaged>undamaged) in yellow.
Cis interactions are increased within γH2AX domains following damage
In order to visualize the interaction data, differential contact matrices between untreated and treated samples were calculated at 100kb resolution on captured regions. Interestingly, we could observe changes in cis interactions within γH2AX domains after damage, in both independent biological replicates (Fig. 1b, Fig. S2). On average, cis interactions were reproducibly enhanced around DSBs following damage, in contrast to control regions (Fig. 1c). Increased contact frequency was observed on both sides of the break, over a region spanning roughly one megabase (500kb on each side), which is reminiscent of the average size of γH2AX domains. The interaction pattern, observed as a cross centered on the DSB (Fig. 1c), indicates that the regions surrounding the break frequently come into contact with the DSB and further support our previous hypothesis that H2AX phosphorylation is sustained thanks to direct contact of loci in cis with the DSB28.
Clustering occurs between DSBs induced on the same or different chromosomes
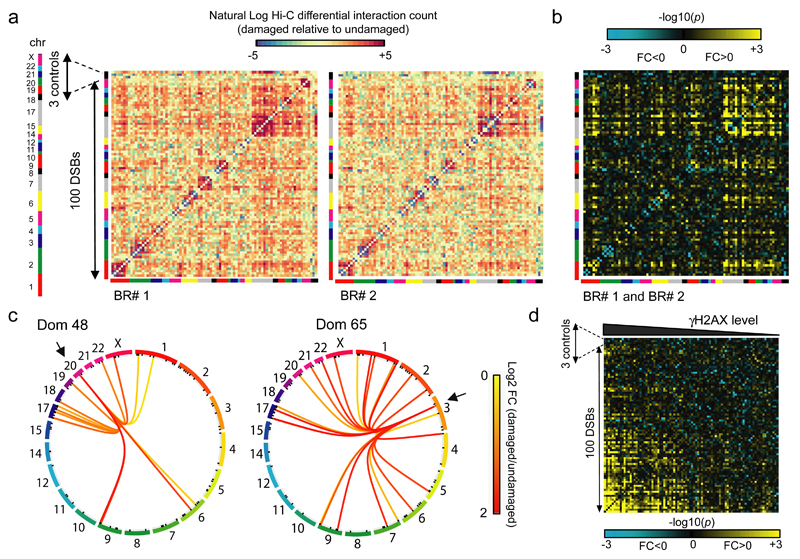
In order to identify DSBs capable of interacting (i.e. clustering), we next created differential contact matrices (damaged vs undamaged) between all 2 megabases (Mb) domains for each replicates. Individual captured domains were first sorted by their order of appearance on the genome (from chromosome 1 to chromosome X, colored lines) with the 3 control regions placed at the end of the matrix (black line). The heatmaps for the pairs of independent replicates showed strong similarity (Fig. 2a). The statistical contact matrix, based on both replicates, clearly indicated that some, but not all, genomic loci tend to interact when damaged (Fig. 2b). Although smaller chromosomes (chr17-22), which tend to be located in the center of the nucleus29, presented more clustered DSBs than longer ones, we did not find any correlation between the position of the DSBs in the nucleus and the clustering ability (Fig. S3a). Visualization of Hi-C data using Circos Plots clearly indicated that DSB-induced long range interactions were not restricted to the same chromosome and that clustering could also occur between DSBs induced on different chromosomes (Fig. 2c, Fig. S3b). In addition, DSBs clustering was favored between loci that are initially in a close proximity (Fig. S3a-c) but DSBs induced on physically close domains (belonging to the same chromosome, Fig. S3c left panels) did not necessarily cluster, indicating that spatial proximity is likely required but not sufficient for DSB clustering. Finally, when DSBs were sorted based on damage induced γH2AX levels obtained by ChIP-seq in DIvA cells21, the DSBs exhibiting higher γH2AX levels demonstrated a higher ability to cluster together (Fig. 2d). Altogether, these data indicate that DSBs on the human genome can undergo clustering even when induced on different chromosomes and in a manner correlated to the level of γH2AX.
Figure 2. Clustering can occur between DSBs induced on different chromosomes and correlates with γH2AX.
a. Differential heatmaps (damaged versus undamaged) between each of the 2Mb domain captured on the human genome, for both biological replicates. Data are expressed as natural log of differential count. DSBs are ordered according to their positions on the genome and chromosomes are indicated by colors bars. The 3 control regions (devoid in DSB) are positioned at the end (black line). Each square represents a 2Mb domain captured, surrounding a DSB.
b. The number of interactions between each domain were measured and p values between damaged and undamaged samples were computed based on both replicates (Online Methods). –log10(p) are indicated, with negative fold changes (FC<0, damaged<undamaged) in blue, and positive fold change (FC>0, damaged>undamaged) in yellow. DSB are ordered according to their positions on the genome and chromosomes are indicated by colors bars. The 3 control regions (devoid in DSB) are positioned at the end as indicated.
c. Circos plots showing the statistically significant (p<0.05) interactions induced after 4OHT treatment for two selected DSB-containing captured domains. Connecting lines are colored according to the log2 fold change between damaged and undamaged cells. p-values were computed from damaged and undamaged samples using biological replicates (Online Methods).
d. Interaction heatmap between damaged versus undamaged cells at a 2Mb domain resolution, based on both biological replicates (Online Methods). -log10(p) are indicated, with negative fold changes (damaged<undamaged) in blue, positive fold change (damaged>undamaged) in yellow. DSBs are sorted based on their level of γH2AX analyzed by ChIP-seq21. Controls have been placed on the right side of the matrix as indicated.
Clustering is enhanced between DSBs induced in transcriptionally active regions
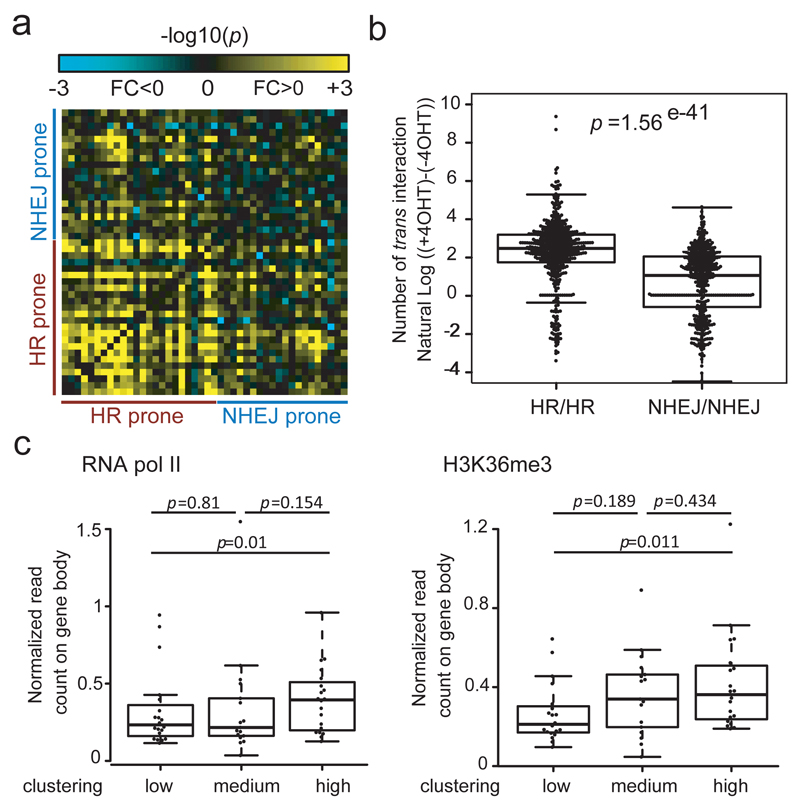
In a previous study, we discovered that DSBs induced on the genome are not necessarily repaired by homologous recombination (HR) in post-replicative cells, even though the sister chromatid is available. Indeed, using RAD51 and XRCC4 ChIP-seq data we determined the repair pathway used at each AsiSI-induced DSB and found that in G2, DSBs induced in active genes bodies or promoters are mostly repaired by HR (hereafter designated as HR-prone) while, in contrast, DSBs induced in silenced regions are unable to recruit RAD51 and undergo NHEJ (NHEJ-prone)21. We thus wondered whether DSB clustering could depend on the pathway used for repair. Strikingly, sorting the captured domains based on RAD51 ChIP-seq signal revealed that DSBs showing high levels of RAD51 recruitment also showed statistically significant increased clustering (Fig. S4a). Enhanced clustering was observed at HR-prone compared to NHEJ-prone DSBs (Fig. 3a) and interactions engaged following damage between each of the HR-prone loci were significantly higher than interactions engaged between NHEJ-prone DSBs (Fig. 3b). Importantly, analysis of DSBs levels in DIvA cells confirmed that cleavage efficiency was similar between both categories (Fig. S4b) indicating that the differential behavior of HR-prone and NHEJ-prone DSBs toward clustering was not due to DSB induction efficiency.
Figure 3. DSBs induced in transcriptionally active genes and repaired by homologous recombination in post-replicative cells undergo clustering.
a. Interaction heatmap between damaged versus undamaged cells at a 2Mb domain resolution for DSBs defined as HR-prone or NHEJ-prone (21, Online Methods). -log10(p) are indicated, with negative fold changes (damaged<undamaged) in blue, positive fold change (damaged>undamaged) in yellow.
b. Box-plot showing the distribution of DSB-induced interactions between each HR-prone DSB with any other HR- prone DSB (HR/HR) or each NHEJ-prone DSB with any other NHEJ-prone DSB (NHEJ/NHEJ). The difference between the two classes is highly significant (Wilcoxon Mann-Whitney test). Center line: median; Box limits: 1st and 3rd quartiles; Whiskers: Maximum and minimum without outliers; Points: outliers (Online Methods).
c. DSBs were classified based on their ability to cluster (high, medium, low). RNA polymerase II occupancy (left panel) and H3K36me3 enrichment (right panel) analyzed in DIvA cells by ChIP-Seq (21, Online Methods) were averaged over the closest genes. Highly clustered DSBs also exhibited the highest RNA PolII and H3K36me3 levels (Wilcoxon Mann-Whitney test). Center line: median; Box limits: 1st and 3rd quartiles; Whiskers: Maximum and minimum without outliers; Points: outliers.
Since HR-prone DSBs were found to mostly occur within active genes, we further investigated the correlation between clustering ability and the transcriptional status of the damaged locus. RNA polymerase II occupancy and H3K36me3 enrichment (indicators of transcriptional activity) previously analyzed by ChIP-seq in DIvA cells21 were calculated at DSB regions exhibiting low, medium or high clustering ability (Online Methods). DSBs that exhibited high clustering were those lying in regions that showed the highest transcriptional activity (Fig. 3c). Altogether these data indicate that in human cells, DSBs are able to cluster but this peculiar behavior is favored at breaks induced in transcriptionally active chromatin and repaired by HR in post replicative cells.
Clustering is enhanced during G1 cell cycle phase
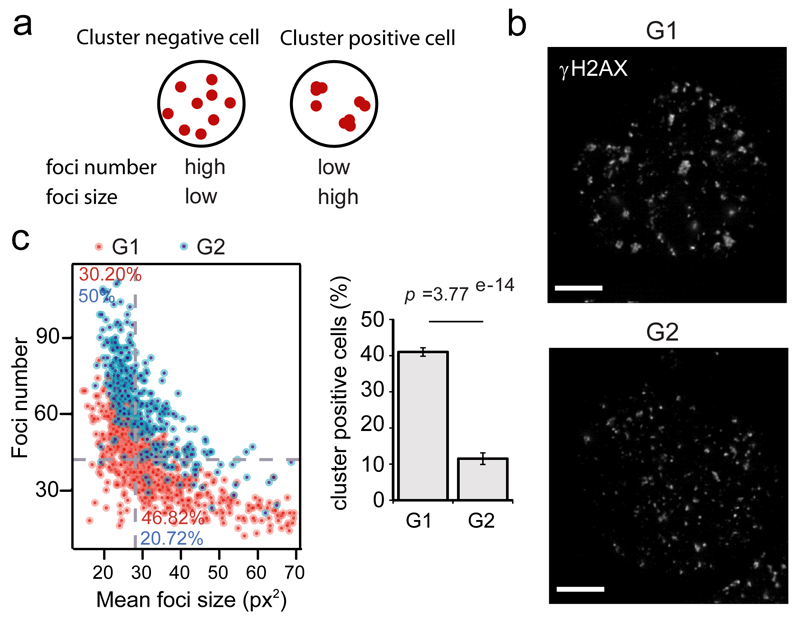
The fact that clustering occurs preferentially at DSBs repaired by HR could indicate that clustering is somehow linked to homology search, a process required for HR and that likely involves enhanced chromatin mobility2. However, previous studies suggested that clustering was enhanced in the G1 phase of the cell cycle5, during which HR is strongly inhibited19. This seeming contradiction prompted us to investigate whether DSB clustering was also enhanced in G1 in DIvA cells. Importantly, we previously reported by ChIP-chip that both the number of DSBs induced by AsiSI and the extent of γH2AX spreading were identical between G1 and G227. Hence, clustering can also be inferred by measuring the number and size of γH2AX foci detected within the nuclei (Fig. 4a). Larger foci were observed in G1 compared to G2 cells (Fig. 4b). In order to systematically measure γH2AX foci number and size in a large number of G1 and G2 damaged nuclei we performed high throughput microscopy (Online Methods). Cell cycle stage was determined using Hoechst staining (Fig. S5a). For each cells, the number of γH2AX foci was plotted against the average foci size and “clustering-positive cells” were defined as cells exhibiting a low number of large foci (corresponding of the bottom-right gate in Fig. 4c, left panel, see Online Methods). This analysis revealed a robust and significant decrease of the number of cluster-positive cells in G2 compared to G1 (Fig. 4c, right panel). In agreement, in G2, foci were in average smaller, while their number were in average higher, compared to G1 cells (Fig. S5b). Altogether these data indicate enhanced DSB clustering in G1.
Figure 4. DSB clustering is favored during the G1 cell cycle phase.
a. AsiSI induces a known number of DSBs in a homogeneous fashion in the cell population. Measure of γH2AX foci number and foci size in each nuclei can thus be used to infer clustering. Low clustering ability leads to more foci of smaller size (left, “cluster negative” cells) while high clustering ability leads to fewer and bigger foci (right, “cluster positive” cells).
b. G1 and G2 DIvA nuclei, synchronized by a double thymidine block followed by a release (respectively 15h and 8h), stained for γH2AX, following damage induction. Scale bar: 5µM.
c. DIvA cells were treated with 4OHT (4h) and subjected to high throughput microscopy following γH2AX staining. Left panel: Scatterplot obtained for a representative experiment, showing the number of γH2AX foci (y axis) and the average foci size (x axis) for each cells in G1 or in G2 (based on Hoechst staining, Fig. S5a). The plot is divided in four area based on the medians of foci number and foci size (dotted grey lines). Percent of cells from G1 population (in red) or G2 population (in blue) that are “cluster positive” (bottom right) or “cluster negative” (top left) are indicated. Right panel: Quantification showing the percent of “cluster positive” cells in G1 and in G2, analyzed from independent experiments. Mean, s.e.m and p value (paired t test) are shown (n=14, independent experiments).
Clustered, HR-prone, DSBs exhibit delayed repair
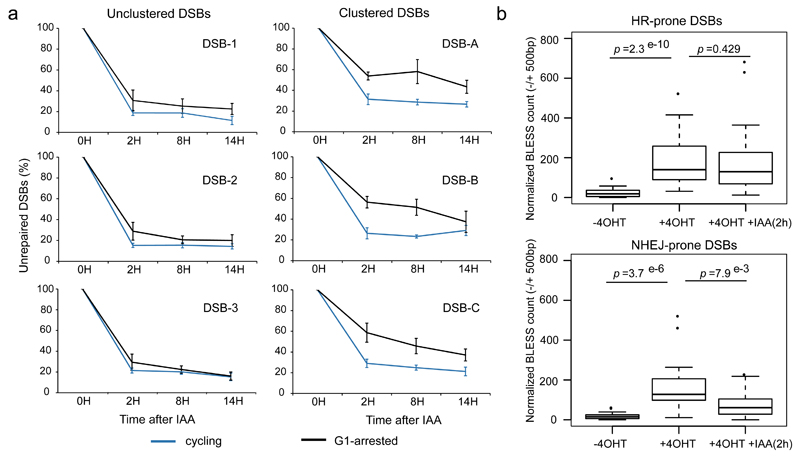
Of note, long range mobility of DSBs has often been associated with persisting break or delayed repair. For example, unrepairable DSBs in yeast move toward the nuclear periphery30,31. Similarly, in higher eukaryotes, DSBs induced in heterochromatin or in the ribosomal DNA, also exhibit relocalization to the periphery of the heterochromatin focus32,33 or the nucleolus respectively34,35, two sub nuclear compartments suggested to be refractory to efficient repair. Therefore, clustering of damaged active genes observed in G1 could reflect delayed repair. Hence, we measured repair kinetics at clustered and unclustered DSBs in G1-arrested compared to cycling cells. Briefly, we used an assay previously developed21,36 (Online Methods) that allows DSBs purification in damaged cells (4OHT) or at different time point during repair (after auxin addition) followed by qPCR at selected DSBs (Fig. S5c, Supplementary Table 3). Importantly, lovastatin treatment performed to arrest the cells in G1 (Fig. S5d) did not alter the auxin-induced degradation of AsiSI (Fig. S5e). Remarkably, repair at three selected DSBs that undergo clustering in G1 (Fig. 5a right panels) exhibited delayed repair in G1-arrested cells, even 8h post auxin addition, compared to normally cycling cells. In contrast, unclustered DSBs were rapidly repaired in both cycling or G1 arrested cells (Fig. 5a left panels).
Figure 5. Clustered, HR-prone, DSBs exhibit delayed repair in G1.
a. Following DSB induction by 4OHT, repair kinetics was measured after auxin (IAA) addition (that triggers AsiSI degradation and subsequent DSB repair), at 3 unclustered (left panels) or 3 clustered (right panels) DSBs either in cycling cells (blue), or G1 arrested cells (following a lovastatin treatment, black). Briefly, repair kinetics was assessed by collecting cells at different time points after auxin addition and performing a ligation and purification mediated quantitative PCR (described in21,36, see Fig. S5c). DSB level before auxin addition (time point 0h) is set to 100%.
b. BLESS was performed in untreated, 4OHT treated (4h) and 4OHT+IAA (2h) treated DIvA cells (Fig. S6), synchronized in G1. BLESS reads count (normalized) is shown on a -/+ 500bp window surrounding DSBs HR-prone (top panel) or NHEJ-prone (bottom panel) for each condition as indicated. p values are indicated (Wilcoxon Mann-Whitney test). Center line: median; Box limits: 1st and 3rd quartiles; Whiskers: Maximum and minimum without outliers; Points: outliers.
In order to assess repair kinetics in G1 on a global scale we further performed direct in situ Breaks Labeling, Enrichment on Streptavidin and next generation Sequencing (BLESS), a technique developed to capture and identify DSBs at a genome wide level37,38(Fig. S6a-b). Analyses of BLESS data indicated that DSBs were induced in G1 following 4OHT addition and partially repaired after 2h of auxin treatment (see an example Fig. S6c and averaged signal at AsiSI sites Fig. S6d). We found that while NHEJ-prone DSBs exhibited almost complete repair two hours after auxin addition, in contrast HR-prone DSBs remained largely unrepaired (Fig. 5b). Altogether, these data indicate that DSBs induced during G1 in transcriptionally active genes are not fully proficient for repair and prone to cluster.
MRN, FMN2 and the SUN2 component of the LINC complex sustain clustering
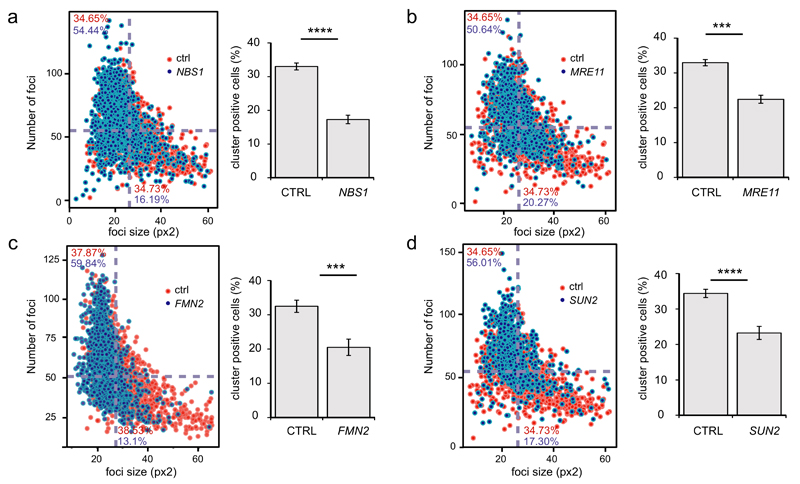
In order to get insights into the mechanisms that could mediate clustering, we further used high throughput microscopy combined with siRNA mediated depletion of selected candidates (Fig. S7, Supplementary Table 4). Previous studies implicated the MRN complex in DSB mobility/clustering and in favouring translocations5,17. Interestingly, depletion of both MRE11 and NBS1 led to a drop in cluster–positive cells and clustering index (Fig. 6a-b, Fig. S8), further confirming the role of the MRN complex in clustering. Depletion of known components of the DDR (RNF8, 53BP1, XRCC4) did not significantly alter clustering (Fig. S8a-b). In addition, two recent studies have pointed towards the role of cytoskeleton proteins in DSB mobility and/or repair. On one hand, disruption of the LINC complex, a complex embedded in the nuclear envelope that connects the cytoskeleton to the nucleoskeleton, decreased mobility of DSBs and unprotected telomeres within the nucleus39. On another hand, actin has been found to polymerize within the nucleus shortly following DNA damage induction in a formin2 (FMN2) actin organizer dependent manner40. We found that both pathways are contributing to DSB clustering. Indeed, depletion of SUN2 (but not of SUN1) and FMN2 led to a decrease in cluster-positive cells and clustering index (Fig. 6c-d, Fig. S8a-b). These data indicate that DSBs cluster in G1 in a manner that depends on the MRN complex, but also on microtubule-and actin-related networks, suggesting that active mechanisms may be at work in this process. Finally, since clustering mostly involves DSBs induced in transcriptionally active regions, we investigated whether transcription inhibition would affect clustering. A pretreatment with 5,6-dichloro-1-β-D-ribofuranosylbenzimidazole (DRB), a well-known transcription inhibitor, indeed led to decreased clustering (Fig. S8c-d). However, such a reduction of clustering was not observed using another transcription inhibitor (triptolide), suggesting that transcription per se is not responsible for induced clustering of damaged active genes (see discussion).
Figure 6. DSB clustering depends on the MRN complex as well as on the LINC complex and the FMN2 actin organizer.
a. DIvA cells were transfected with control siRNA (in red) or a siRNA against NBS1 (in blue), treated with 4OHT (4h) and subjected to high throughput microscopy following γH2AX staining. Left panel: Average foci size (x axis) and number of foci (y axis) were determined in each cells and plotted against each other as in Fig. 4c. Right panel shows the quantification (mean and s.e.m) of cluster positive cells obtained from independent experiments (n=5). (**** p<0.001, paired t test).
b. Same as in a, except that a siRNA against MRE11 was used (n=3) (*** p<0.005, paired t test).
c. Same as in a, except that a siRNA against FMN2 was used (n=7) (*** p<0.005, paired t test).
d. Same as in a, except that a siRNA against SUN2 was used (n=6) (**** p<0.001, paired t test).
Discussion
In summary, thanks to the analysis of long range contact following DSB induction, we uncovered in this study of number of important features of the DDR.
First, damaged induced changes within γH2AX domains revealed a cross-shaped pattern indicating that regions located on -/+ 500kb surrounding the DSB frequently come into contact with the break. We previously reported that ATM, the main H2AX kinase, is recruited on restricted regions (~10kb) while γH2AX can span over 1-2Mb windows16. In addition, in yeast, Tel1 and Mec1 (ATM and ATR homologs) are able to phosphorylate H2A in trans41. Therefore we previously proposed that γH2AX spreading likely occurs through the dynamics of the chromatin fibre around DSBs, which would bring distant H2AX containing nucleosomes into the physical proximity of ATM, bound to the vicinity of DNA ends14,28. The fact that the increased contacts between loci in cis and the break itself occur on a region reminiscent of γH2AX domain (1Mb in average, Fig. 1c) sustains this hypothesis.
Moreover, we unambiguously established that DSBs can undergo clustering even between different chromosomes. Furthermore, we show that this is not an intrinsic property of DSBs but it depends on the nature of the damaged locus since DSBs cluster only when induced in transcriptionally active genes. This likely accounts for the discrepancies previously observed since various DNA damaging agents do not necessarily induce DSBs at equivalent genomic locations. Of note, it has previously been shown that translocations largely occur in active genes42–45. While this was rather attributed to an increased susceptibility of active genes to breakage, our results suggest that increased clustering of damaged active genes may also contribute to the translocation pattern.
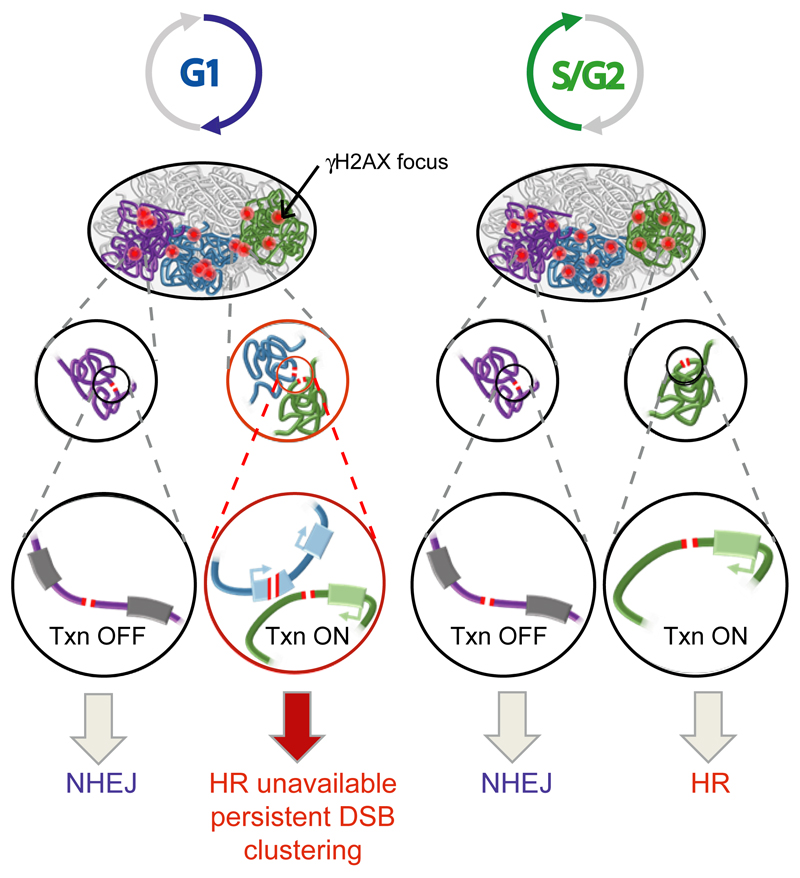
Importantly, our study also revealed that DSBs induced in active genes, albeit repaired by HR in post replicative cells, are refractory for repair in G1. Since clustering was found to be enhanced in G1 compared to G2, it suggests that these “persisting breaks” display large scale motion to further coalesce within foci (Fig. 7). Interestingly, this behavior is reminiscent of other large scale DSB mobility events that have been almost systematically associated with persistent or “difficult” DSBs (30,32,35,46, reviewed in 14). The reasons underlying repair deficiency at active genes in G1 remain unknown. Although clustering was altered by DRB-mediated transcriptional inhibition (Fig. S8), it was not reduced upon treatment with other inhibitors (such as triptolide, data not shown). This suggests that the delayed repair observed at active genes should be attributed to chromatin/secondary DNA structures assembled at transcribed regions rather than to transcription itself. RNA pol II machinery occupancy, supercoiling or RNA: DNA hybrids that form at transcribing loci47 may delay broken ends resealing sufficiently to allow minimal resection, which would subsequently impede classical NHEJ. In post replicative cells, such processed ends would be further handled by the HR machinery. In contrast, in G1, these DSBs would persist due to the down regulation of HR activity (Fig. 7). The fact that MRN, whose activity sustains the initiation of resection, is involved in clustering (Fig. 6a-b) further supports this hypothesis, although it remains to be established whether MRN contributes to clustering through promoting resection.
Figure 7. Model for DSB clustering.
While DSBs in intergenic regions and inactive genes are mostly repaired by NHEJ throughout the cell cycle, the repair of DSB induced in active genes is dependent on the cell cycle phase. In post-replicative cells, DSBs in transcribed regions are prone to HR repair, whereas in G1, those breaks persist longer and coalesce with each other, forming clusters of unrepaired active genes.
Given the increased risk of translocation associated with DSB clustering, one may wonder why this potentially deleterious mechanism has been selected throughout evolution. Studies in yeast suggested that DSBs could cluster in order to promote “repair center” assembly to concentrate and favor repair machineries recruitment. However, we found here that DSB clustering coincides with delayed repair, standing against the idea of “repair center” in human cells. Rather, we favour the hypothesis that DSB clustering occurs to sequester and/or prepare DSBs for faithful repair. Indeed, using the sole available pathway that can accommodate resected/processed ends in G1, i.e. alternative non homologous end joining, at damaged active genes, would be too detrimental for the cell given the high mutation rate associated with this pathway. Clustering may help to inhibit such error prone repair pathway and/or to sequester DSBs from the rest of the genome while waiting for a more appropriated cell cycle phase.
Of note, a mild replication stress leads to the formation of 53BP1/γH2AX bodies (also known as OPT bodies) following chromosome breakage during mitosis, that will persist throughout the G1 phase. These bodies have been proposed to contain common fragile sites (CFS)48,49 previously shown to correlate with long genes50,51. Hence, similarly to the DSBs clusters described here, OPT bodies may contain multiple clustered DSBs induced at transcriptionally active long genes, under replicated and damaged during the division, that would be refractory to C-NHEJ repair in G1.
Finally we also provide insights into the mechanisms that promote DSB clustering. We found that clustering relies on nucleo- and cytoskeleton networks, since it depends on the LINC complex and FMN2 actin organizer (Fig. 6c-d). On one hand, a function of the LINC complex has been recently identified in promoting DSBs and uncapped telomeres mobility in higher eukaryotes39. Moreover, both in yeast and in Drosophila, LINC subunits homologs contribute to the localization of unrepaired and heterochromatic DSBs to the nuclear periphery31,52,53. Therefore, the LINC complex might exert an evolutionary conserved role in promoting DSB mobility and clustering/relocalization of persistent DSBs. On another hand, nuclear actin was found to polymerize within the nucleus following DNA damage in a FMN2 dependent manner40, an actin nucleator protein known to be phosphorylated and upregulated upon DNA damage54,55. Whether FMN2 contributes to DSB clustering through its ability to promote nuclear actin filament that would alter chromatin mobility thus represents an exciting possibility.
Since, most of the cells in multicellular organisms are non-proliferative and arrested in G1, it will be of utmost importance to integrate these novel concepts not only in our understanding of translocation processes, but also in our view of adverse effects during cancer chemo and radio-therapy. The possibility that active mechanisms could be involved in translocations will deserve close attention as the cytoskeleton is a well-known and robust target for cancer therapy.
Online Methods
Biotinylated RNA bait library design
A genomic restriction map was created to generate a list of all HindIII fragments in the genome, and this was subsequently filtered against 2 megabase (Mb) around the 100 best cleaved AsiSI sites21 and on 3 control regions of 2Mb devoid of the AsiSI recognition site (chr1:185500000- 187500000; chr17:70000000-72000000; chr5:111000000-113000000). Probes were designed on every HindIII fragment in a +/-10 kb region around the AsiSI and the center of control regions. When 2 windows of 20kbp overlapped, each probe on the overlapping region were synthesized only once. Over the rest of the 2Mb window, baits were designed every 10 HindIII fragments, i.e. ~50kb. When two 2Mb windows overlapped leading to a HindIII fragment captured twice, the fragment just downstream was selected in order to capture each fragment only once. For each selected restriction fragment, two 120bp capture probes were designed, one to each end of the fragment. Because of the size selection step following sonication, the probes had to fall entirely within a region no more than 500bp from the end of the fragment. Each probe was required to have no more than 3 consecutive bases masked by repeat masker, and they had to have a GC content of 25 to 65% to match the efficient capture range of the SureSelect target enrichment system (Agilent Technologies). Where multiple probes passed these criteria, the probe nearest the end of the restriction fragment was chosen. The full list of HindIII sites near which RNA capture probes were designed is available in Supplementary Table 1. Coordinates and names of captured domains as well as the size of the gaps between captured regions are available Supplemental Table 2.
Capture Hi-C
Capture Hi-C library generation was carried out as described previously56. Briefly, 30.106 cells were fixed in 2% formaldehyde for 10 minutes. After overnight digestion with HindIII, DNA ends were labeled with biotin-14–dATP using the klenow fragment of DNA pol I and religated in preserved nuclei. 50 µg of DNA was sheared to an average size of 450 bp (Covaris). Sheared DNA was then end repaired, adenine tailed and double size selected from 250 to 600 bp (AMPure XP beads). Chimerae marked by biotin were pulled down using MyOne Streptavidin C1 DynaBeads (Invitrogen) and ligated to PE adaptors (Illumina). The Hi-C libraries (retained on the beads) were amplified using PE primers (Illumina) with 10 PCR amplification cycles. Capture was next carried out on Hi-C library, with SureSelect target enrichment, using the custom-designed biotinylated RNA bait library and custom paired-end blockers according to the manufacturer's instructions (Agilent Technologies). After library enrichment by capture, a post-capture PCR amplification step was carried out using PE PCR 1.0 and PE PCR 2.0 primers with 7 PCR amplification cycles. Captured-Hi-C libraries were performed on damaged (4OHT treated) or undamaged (untreated) samples in biological duplicates (i.e. independent experiments) (BR#1 and BR#2) and were sequenced on the Illumina HiSeq 1000 platform. High throughput sequencing data have been deposited to Array Express under accession number E-MTAB-4846.
Samples sequencing, mapping and quality control
Samples were sequenced using an HiSeq 1000, generating 51 bp paired-end reads, at the Babraham Institute in Cambridge. The quality of each raw sequencing file (fastq) was checked using FastQC. After quality control, fastq files were aligned against the reference human genome hg19 using the HiCUP pipeline57: this pipeline identifies HiC junction (hicup_truncater) in the fastq file, generates a paired-end read alignment file in .BAM format (hicup_mapper) by mapping each read pair corresponding to a Hi-C di-tag (purified Hi-C fragment) and filters out artefactual read pairs (hicup_filter and hicup_deduplicator). The pipeline also provides a report summarizing quality control during the pipeline: number of too short and truncated reads, number of unmapped reads, number of filtered wrong pairs (invalid, same fragment, religation…), di-tag length and duplication summary (Fig. S1b). Capture efficiency was shown to be reproducible for each sample, and efficient (Fig. S1c-d).
Interaction counts
Seqmonk (http://www.bioinformatics.babraham.ac.uk/projects/seqmonk), was used to visualize aligned bam files and to compute the number of interactions between our regions of interest. Briefly, the number of interactions were calculated over the 2Mb windows, where probes were designed, around the 100 best-cleaved AsiSI sites and 3 control regions. When sites were less than 1Mb apart, their 2Mb regions were merged, leading to 88 domains (85 sites and 3 control). The number of di-tags that correspond to any possible pair of domains were calculated with Seqmonk and interaction reports containing the 3228 pairs of domains were used for further visualizations and analyses. When indicated (Fig.1b, Fig. S2), each domain was subdivided into 100kbp windows, leading to a Seqmonk interaction report containing the number of interactions between 1,574,425 pairs of sub-domains.
R Analysis and Normalization
Each count matrix was then loaded in R-software for further analysis. Manipulation and analyses were performed using the Biocondutor :GenomicRanges R package58 available from the Bioconductor software project (http://www.bioconductor.org/). To compare different samples, the number of interactions were normalized by the total number of aligned read pairs captured on at least one end. The number of interactions between 2Mb or 100kb domains were further normalized against the total number of probes used to capture those specific windows, to account for the variation in probe density across each domain. In other words, for each pair of domains (100kb or 2Mb), the number of interactions was normalized against the sum of probes designed in each of the two domains.
Differential heatmaps of damaged versus undamaged cells
The number of interactions between each pair of domains (2Mb or 100kb) measured in the +4OHT (damaged) and in -4OHT (undamaged) samples were independently subtracted for both pairs of replicate samples. Differential heat maps were drawn with a natural log (ln) scale (on absolute values, then multiplied by direction of change), as follows: sign (+4OHT - -4OHT)*log|+4OHT - -4OHT|. When indicated, domains were ordered based on RAD51 or γH2AX levels (respectively averaged over +/-2kb or +/-40kb) analyzed by ChIP-seq21.
Statistical heatmaps
To evaluate the significance of the differences observed between +4OHT and -4OHT, p-values were computed for each pair of domains, taking into account both replicates, using EdgeR (Empirical analysis of Digital Gene Expression data in R) available through Bioconductor :EdgeR R package59. For a symmetrical visualization that would take into account the direction of the fold change (where up and down represent enhanced and decreased interaction levels between samples), p-values were next transformed as follow: plotted Value = sign(logFoldChange) x -log10(Pvalue), and represented as heatmaps. Significant negative changes (decreased interaction following damage) are plotted in blue, while significant positive changes (increased interaction following damage) are plotted in yellow. Plotted values are statistically significant when value is > 1.3 and < -1.3 (~ p-value = 0.05).
Determination of HR-prone and NHEJ-prone DSBs
HR/NHEJ categories were computed based on BLESS data in DIvA cells (Clouaire T, Aguirrebengoa M, Rocher V, Biernacka A, Skrzypczak M, Aymard F, Iacovoni JS, Dojer N, Fongang B, Rowicka M, Ginalski K, and Legube G, manuscript in preparation) and ChIP-Seq against XRCC4 and Rad5121. Sites were ordered by their average level of BLESS in a window of 1 kb centered on the AsiSI site. The 80 most cleaved sites were selected. A ratio between the level of RAD51 on a 4kb window and the level of XRCC4 in a window of 500bp around each DSB was computed (as performed in21). We discriminated the 30 sites with the highest ratio as HR-prone site and the 30 sites with the lowest ratio as NHEJ-prone sites.
Average interactions maps
For each 2Mb domain, vectors containing the number of interactions between pairs of 100kb windows were generated for each replicate damaged and undamaged samples. A mean vector was computed based on all vectors for all 100 DSBs and the 3 control regions. EdgeR was then used on these mean vectors in order to identify significant differences in the number of interactions between 100kb windows. Results were shown as heatmaps, after pvalue transformation, as described above.
Circos plot
Circos plots represent interactions between a captured domain and all other domains on the genome. Only significantly increased interactions (EdgeR p-value <0.05; logFoldChange > 0) between undamaged and damaged condition were drawn, using the Bioconductor OmicCircos R package 60. Connecting lines are coloured based on the log2 fold change.
Correlation with RNA polII and H3K36me3 occupancy
The domains were separated into 3 equal categories, based on the number of differential interactions each domain possesses with the others (low, medium or high clustering ability). Transcriptional activity around AsiSI sites was determined using PolIIS2P and H3K36me3 ChIP seq average levels from the closest gene (from -500bp of TSS to the TTS) neighboring each AsiSI sites21.
Cell lines and cell culture
DIvA (AsiSI-ER-U20S) and AID-DIvA (AID-AsiSI-ER-U20S) cells were previously developed21,27 using U2-OS from ATCC. They were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with antibiotics, 10% FCS (Invitrogen) and either 1 μg/mL puromycin (DIvA cells) or 800 μg/mL G418 (AID-DIvA cells) at 37 °C under a humidified atmosphere with 5% CO2. The cell lines were regularly checked for mycoplasma contamination.
Treatment and siRNA transfection
For AsiSI-dependent DSB induction, cells were treated with 300 nM 4OHT (Sigma, H7904) for 4 h. For transcriptional inhibition, cells were pre-treated with DRB (20µM or 100µM as indicated) for 1h prior 4h of 4OHT, and left during the entire 4OHT treatment (Fig. S8d). For cell synchronization in G1, cells were incubated with 2 mM thymidine for 16 h, released for 14 h and subjected to the second thymidine treatment for 19 h. G1 and G2 cells were treated with 4OHT respectively after 11h and 4 hours and fixed 4 hours later (Fig.4b). For BLESS experiment (Fig. 5b) cells were treated with 4OHT (4h) 12h after release, followed by an auxin treatment of 2h before harvesting. For cell arrest in G1 (Fig. 5a), cells were treated with 40µM Lovastatin for 48 hours. siRNA transfections were performed with the Cell Line Nucleofactor kit V (Amaxa) according to the manufacturer's instructions or using interferin (PolyPlus). Sequences for siRNAs are shown in Supplemental table 4. To assess the efficiency of siRNAs, mRNA were extracted using the Qiagen RNeasy kit (#69504) and reverse transcribed using the AMV reverse transcriptase (Promega, M510F). cDNA were quantified by RT-qPCR (primer sequences are shown in Supplementary Table 3) and normalized to P0 cDNA levels.
Immunofluorescence
Cells grown on glass coverslips were fixed with 4% Paraformaldehyde and immunofluorescence was carried out following a standard protocol (described in 27). A mouse monoclonal Upstate Biotechnology antibody (JBW 301) was used to stain γH2AX. Secondary antibodies were purchased from Molecular Probes. Image acquisition was performed using MetaMorph on a wide field microscope equipped with a with a cooled charge-coupled device camera (CoolSNAP HQ2), using a 100X objective. Deconvolution was performed using Richardson lucy algorithm with total variation (TV) regularization61. The algorithm is implemented in the image J software deconvolution lab (http://bigwww.epfl.ch/algorithms/deconvolutionlab/). For Fig. S3a, the 3D coordinates and the volume of each foci was obtained using the 3D foci picker plugin from ImageJ with the following setting: background was set on automatic, minimum pixel number in the focus on 20 and tolerance was variable depending on the experiment to detect a similar amount of foci (between 100 and 150 per cells). Using these positions and an estimation of the 3D coordinates of the cell center, the 3D Euclidian distance to the cell center was computed for each foci within each cell, with the formula: and correlated to volume (x_foci * y_foci * z_foci ) of each foci. Nineteen cells, from 3 independent experiments, were analyzed.
Repair Assay
DIvA cells were treated with 4OHT for 4 hours to induce damage and were then were washed three times in pre-warmed PBS and further incubated with 500 μg/mL auxin (Sigma, I5148) to allow the repair. Cells were collected by trypsinisation at different time after auxin treatment (0h, 2h, 8 and 14h), their DNA was then extracted in order to perform repair assay. The full procedure for the repair assay has been previously described21,36. A biotinylated double-stranded oligonucleotide, cohesive with AsiSI sites, was ligated in vitro to genomic DNA after break induction. DNA was extracted, then fragmented by EcoRI digestion at 37 °C for 2 h. DNA was pulled down with streptavidin beads (Sigma) at 4 °C overnight. Beads were resuspended in 100 μL of water and digested with HindIII at 37 °C for 4 h. After phenol/chloroform purification and precipitation, DNA was resuspended in 100 μL water. Proportion of unrepaired DSBs is then quantified by qPCR (Syber premix, TaKaRa) in triplicates (primer sequences are provided in Supplementary Table 3).
Direct in situ Breaks Labeling, Enrichment on Streptavidin and next generation Sequencing (BLESS)
After 4OHT and or auxin treatment in G1 synchronized cells, DSBs labelling was carried out using BLESS method as described previously37. Briefly, cells were fixed with formaldehyde, lysed and mildly digested with proteinase K at 37ºC in order to purify intact nuclei, then DSBs were blunted and 5'phosphorylated using Quick Blunting Kit (NEB). Subsequently, a biotinylated proximal linker (Sigma) was ligated to DSBs using T4 ligase (NEB). Next, DNA was extracted by precipitation with isopropanol and sonicated using Covaris S220 ultrasonicator to create fragments approximately 400bp long. Labeled, biotinylated fragments were captured on streptavidin beads (Invitrogen) and ligated to a distal linker (Sigma). Resulting DNA fragments were then linearized by I-SceI (NEB) digestion and amplified by PCR. Libraries were prepared using TruSeq DNA LT Sample Prep Kit (Illumina), followed by 2x61 bp and 2x70 bp next generation sequencing on Illumina HiSeq 2500. Raw data were filtered using BLESS adapter in order to keep only the end proximal to the cut from the paired read. These proximal reads were aligned using bwa-aln and PCR-duplicates were cleaned using samtools. The coverage and the normalization by the total number of reads was then generated using R. For each site we computed the average level of normalized counts in a window of +/-500bp around sites.
High throughput microscopy
DIvA cells were plated in 96 well plates (2500 cell per plates) after transfection with siRNAs using the Nucleofactor Kit V, Amaxa. 48h after, cells were treated with 4OHT (300nM) for 4h. After fixation, permeabilisation and saturation steps, γH2AX was stained overnight with the mouse anti-γH2AX monoclonal antibody (Millipore) and a secondary antibody anti mouse Alexa 647 (Molecular Probes). Nuclei were labeled with Hoechst 33342 (Sigma) at the final concentration of 1µg/ml during 30 minutes. γH2AX foci clustering were further analyzed using Operetta automated high-content screening microscope (PerkinElmer). For the quantitative image analysis, multiple fields per well were acquired with a 40X objective lens to visualize about 2000 cells/well in triplicate. Each picture was analyzed through the integrated software Harmony. Briefly, the Hoechst-nuclei were selected following the B method applying appropriate parameters such as size and intensity of fluorescent objects to eliminate false positive compounds.
Then the Alexa 647-γH2AX foci were detected using the D method with the following parameters: detection sensitivity=1, splitting coefficient=1, background correction> 0.5 to 0.9. We retained the selection of γH2AX foci intensity, foci number and foci area as parameters to measure variation of clustering for each nucleus in each well. G1 and G2 nuclei were selected based on the Hoechst intensity, following visualization of Hoechst distribution in all cells.
In each nuclei, and for each condition, the clustering index (Fig.S5b, FigS.8b) was calculated by dividing the average foci size with the number of foci. Median clustering index for the whole nuclei population (at least 1000 nuclei for each condition) was next calculated. G1 (Fig. S5b) or Ctrl siRNA treated cells (Fig. S8b) were next set to 1. A minimum of 4 independent experiments were done for each conditions and averaged clustering index together with s.e.m were plotted. One sample t-test was performed to assess statistical relevance across all biological replicates.
The scatter plots were drawn using the average foci size and the number of foci for each nuclei. Medians of the foci size and number of foci were used to define subpopulations (gates) corresponding to cluster positive and cluster negative cells. Outliers, defined as any point that lies over 1.5 x Interquantile Range (Q3-Q1) below the first quartile (Q1) or above the third quartile (Q3) in a data set, were excluded from graphical representations and further analysis. Number of nucleus in the top-left gate (cluster-negative) and the bottom right gate (cluster-positive cells) were then counted and the proportion of nuclei in these two gates for each condition is indicated on the scatter plot for representative experiments (Fig. 4cc, Fig. 6a-d, Fig. S8). The proportion of cluster positive cells was calculated for at least 3 biological replicates in each condition and average was plotted together with s.e.m (Fig. 4c, Fig. 6a-d, Fig. S8).
Statistics
For all box-plot representations, outliers were defined as data below 1stQuart - 1.5x(3rdQuart - 1stQuart) and above 3rdQuart + 1.5x(3rdQuart - 1stQuart). Center line represents the median, box limits the 1st-3rd quartiles, whiskers the maximum and minimum values without outliers, and points shows outliers.
For each figure statistical test of hypothesis were chosen regarding data distribution: For interaction counts of Hi-C data (Fig. 1c, Fig. 2b, Fig. 2d, Fig. 3a and Fig. S4a), the EdgeR methods was applied to compute P-values on number of Hi-C interactions for each couple of domains. Indeed EdgeR implements statistical methodology based on the negative binomial distributions that can be applied to differential signal analysis of genomic data that produce counts. The Fisher test (var.test function in R) was also applied to verify the homoscedasticity within groups: BR1 vs BR2 -4OHT pval = 0,745, BR1 vs BR2 +4OHT pval = 0,969, and between groups BR1-4OHT vs BR1+4OHT : pval = 0,694, BR1-4OHT vs BR2+4OHT : pval = 0,666, BR2-4OHT vs BR1+4OHT : pval = 0,946, BR2-4OHT vs BR2+4OHT : pval = 0,915. So the variances are homogeneous within and between groups. For figures Fig. 3b, Fig. 3c, Fig. 5b, Fig. S2d and Fig. S6d the Shapiro-Wilk Test (shapiro.test function in R) was applied to each sample in order to check whether samples came from a normally distributed population or not. As the hypothesis that data are normally distributed could be rejected with more than 95 % of confidence, the non-parametric unpaired Mann–Whitney–Wilcoxon Test (also called Wilcoxon Sum Rank Test, wilcoxon.test function in R) was applied in order to test distribution differences between 2 samples.
Supplementary Material
Acknowledgments
We thank Simon Andrews, Kristina Tabbada and Steven Wingett (Babraham Institute) for probes design and quality control of Hi-C data. Funding was provided by Polish National Science Centre (2011/02/A/NZ2/00014 to K.G. and 2015/17/D/NZ2/03711 to M.S.) and Foundation for Polish Science (TEAM to K.G.). Funding to M. R. was provided by NIH (NIH 5 R01 GM 112131). M.A. and E.G. were supported by the Fondation pour la Recherche Médicale (FRM). Funding in GL laboratory was provided by grants from the European Research Council (ERC-2014-CoG 647344), Agence Nationale pour la Recherche (ANR-14-CE10-0002-01and ANR-13-BSV8-0013), the Institut National contre le Cancer (INCA PLBIO15-199) and the Ligue Nationale contre le Cancer (LNCC).
Footnotes
Accession Code
High throughput sequencing data have been deposited to Array Express under accession number E-MTAB-4846.
Data Availability Statement
Source data for Fig. 3b-c, Fig. 4c, Fig. 5a-b and Fig. 6 a-d are available with the paper online. Other data and source codes are available upon request.
Author contributions
F.A, E.G, B.M.J, B.B and C.A performed experiments. M.A, V.R and J.S.I performed bioinformatic analyses of Hi-C and BLESS datasets. A.B, K.G, M.S, and M.R performed BLESS experiments. P.F contributed to Capture Hi-C experimental design, experiments and analyses. G.L conceived and analyzed experiments. F.A and G.L wrote the manuscript. All authors commented and edited the manuscript.
Competing Financial Interests Statement
The authors declare no competing financial interests.
References
- 1.Dion V, Kalck V, Horigome C, Towbin BD, Gasser SM. Increased mobility of double-strand breaks requires Mec1, Rad9 and the homologous recombination machinery. Nature cell biology. 2012;14:502–509. doi: 10.1038/ncb2465. [DOI] [PubMed] [Google Scholar]
- 2.Mine-Hattab J, Rothstein R. Increased chromosome mobility facilitates homology search during recombination. Nature cell biology. 2012;14:510–517. doi: 10.1038/ncb2472. [DOI] [PubMed] [Google Scholar]
- 3.Neumann FR, et al. Targeted INO80 enhances subnuclear chromatin movement and ectopic homologous recombination. Genes & development. 2012;26:369–383. doi: 10.1101/gad.176156.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lisby M, Mortensen UH, Rothstein R. Colocalization of multiple DNA double-strand breaks at a single Rad52 repair centre. Nature cell biology. 2003;5:572–577. doi: 10.1038/ncb997. [DOI] [PubMed] [Google Scholar]
- 5.Aten JA, et al. Dynamics of DNA double-strand breaks revealed by clustering of damaged chromosome domains. Science. 2004;303:92–95. doi: 10.1126/science.1088845. [DOI] [PubMed] [Google Scholar]
- 6.Krawczyk PM, et al. Chromatin mobility is increased at sites of DNA double-strand breaks. Journal of cell science. 2012;125:2127–2133. doi: 10.1242/jcs.089847. [DOI] [PubMed] [Google Scholar]
- 7.Krawczyk PM, Stap J, van Oven C, Hoebe R, Aten JA. Clustering of double strand break-containing chromosome domains is not inhibited by inactivation of major repair proteins. Radiation protection dosimetry. 2006;122:150–153. doi: 10.1093/rpd/ncl479. [DOI] [PubMed] [Google Scholar]
- 8.Neumaier T, et al. Evidence for formation of DNA repair centers and dose-response nonlinearity in human cells. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:443–448. doi: 10.1073/pnas.1117849108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Becker A, Durante M, Taucher-Scholz G, Jakob B. ATM alters the otherwise robust chromatin mobility at sites of DNA double-strand breaks (DSBs) in human cells. PloS one. 2014;9:e92640. doi: 10.1371/journal.pone.0092640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jakob B, Splinter J, Taucher-Scholz G. Positional stability of damaged chromatin domains along radiation tracks in mammalian cells. Radiation research. 2009;171:405–418. doi: 10.1667/RR1520.1. [DOI] [PubMed] [Google Scholar]
- 11.Kruhlak MJ, et al. Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks. The Journal of cell biology. 2006;172:823–834. doi: 10.1083/jcb.200510015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kruhlak MJ, Celeste A, Nussenzweig A. Spatio-temporal dynamics of chromatin containing DNA breaks. Cell Cycle. 2006;5:1910–1912. doi: 10.4161/cc.5.17.3169. [DOI] [PubMed] [Google Scholar]
- 13.Soutoglou E, et al. Positional stability of single double-strand breaks in mammalian cells. Nature cell biology. 2007;9:675–682. doi: 10.1038/ncb1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marnef A, Legube G. Organizing DNA repair in the nucleus: DSBs hit the road. Current opinion in cell biology. 2017;46:1–8. doi: 10.1016/j.ceb.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 15.Cho NW, Dilley RL, Lampson MA, Greenberg RA. Interchromosomal homology searches drive directional ALT telomere movement and synapsis. Cell. 2014;159:108–121. doi: 10.1016/j.cell.2014.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caron P, et al. Non-redundant Functions of ATM and DNA-PKcs in Response to DNA Double-Strand Breaks. Cell reports. 2015;13:1598–1609. doi: 10.1016/j.celrep.2015.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roukos V, et al. Spatial dynamics of chromosome translocations in living cells. Science. 2013;341:660–664. doi: 10.1126/science.1237150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mladenov E, Magin S, Soni A, Iliakis G. DNA double-strand-break repair in higher eukaryotes and its role in genomic instability and cancer: Cell cycle and proliferation-dependent regulation. Seminars in cancer biology. 2016;37-38:51–64. doi: 10.1016/j.semcancer.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 19.Orthwein A, et al. A mechanism for the suppression of homologous recombination in G1 cells. Nature. 2015;528:422–426. doi: 10.1016/j.semcancer.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clouaire T, Legube G. DNA double strand break repair pathway choice: a chromatin based decision? Nucleus. 2015;6:107–113. doi: 10.1080/19491034.2015.1010946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aymard F, et al. Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nature structural & molecular biology. 2014;21:366–374. doi: 10.1038/nsmb.2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfister SX, et al. SETD2-dependent histone H3K36 trimethylation is required for homologous recombination repair and genome stability. Cell reports. 2014;7:2006–2018. doi: 10.1016/j.celrep.2014.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carvalho S, et al. SETD2 is required for DNA double-strand break repair and activation of the p53-mediated checkpoint. eLife. 2014;3:e02482. doi: 10.7554/eLife.02482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daugaard M, et al. LEDGF (p75) promotes DNA-end resection and homologous recombination. Nature structural & molecular biology. 2012;19:803–810. doi: 10.1038/nsmb.2314. [DOI] [PubMed] [Google Scholar]
- 25.Hajjoul H, et al. High-throughput chromatin motion tracking in living yeast reveals the flexibility of the fiber throughout the genome. Genome research. 2013;23:1829–1838. doi: 10.1101/gr.157008.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schoenfelder S, et al. The pluripotent regulatory circuitry connecting promoters to their long-range interacting elements. Genome research. 2015;25:582–597. doi: 10.1101/gr.185272.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iacovoni JS, et al. High-resolution profiling of gammaH2AX around DNA double strand breaks in the mammalian genome. The EMBO journal. 2010;29:1446–1457. doi: 10.1038/emboj.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aymard F, Legube G. A TAD closer to ATM. Molecular & cellular oncology. 2016;3:e1134411. doi: 10.1080/23723556.2015.1134411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bolzer A, et al. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS biology. 2005;3:e157. doi: 10.1371/journal.pbio.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagai S, et al. Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science. 2008;322:597–602. doi: 10.1126/science.1162790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oza P, Jaspersen SL, Miele A, Dekker J, Peterson CL. Mechanisms that regulate localization of a DNA double-strand break to the nuclear periphery. Genes & development. 2009;23:912–927. doi: 10.1101/gad.1782209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chiolo I, et al. Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell. 2011;144:732–744. doi: 10.1016/j.cell.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jakob B, et al. DNA double-strand breaks in heterochromatin elicit fast repair protein recruitment, histone H2AX phosphorylation and relocation to euchromatin. Nucleic acids research. 2011;39:6489–6499. doi: 10.1093/nar/gkr230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harding SM, Boiarsky JA, Greenberg RA. ATM Dependent Silencing Links Nucleolar Chromatin Reorganization to DNA Damage Recognition. Cell reports. 2015;13:251–259. doi: 10.1016/j.celrep.2015.08.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Sluis M, McStay B. A localized nucleolar DNA damage response facilitates recruitment of the homology-directed repair machinery independent of cell cycle stage. Genes & development. 2015;29:1151–1163. doi: 10.1101/gad.260703.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chailleux C, et al. Quantifying DNA double-strand breaks induced by site-specific endonucleases in living cells by ligation-mediated purification. Nature protocols. 2014;9:517–528. doi: 10.1038/nprot.2014.031. [DOI] [PubMed] [Google Scholar]
- 37.Crosetto N, et al. Nucleotide-resolution DNA double-strand break mapping by next-generation sequencing. Nature methods. 2013;10:361–365. doi: 10.1038/nmeth.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mitra A, Skrzypczak M, Ginalski K, Rowicka M. Strategies for achieving high sequencing accuracy for low diversity samples and avoiding sample bleeding using illumina platform. PloS one. 2015;10:e0120520. doi: 10.1371/journal.pone.0120520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lottersberger F, Karssemeijer RA, Dimitrova N, de Lange T. 53BP1 and the LINC Complex Promote Microtubule-Dependent DSB Mobility and DNA Repair. Cell. 2015;163:880–893. doi: 10.1016/j.cell.2015.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Belin BJ, Lee T, Mullins RD. DNA damage induces nuclear actin filament assembly by Formin -2 and Spire-(1/2) that promotes efficient DNA repair. [corrected] eLife. 2015;4:e07735. doi: 10.7554/eLife.07735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee CS, Lee K, Legube G, Haber JE. Dynamics of yeast histone H2A and H2B phosphorylation in response to a double-strand break. Nature structural & molecular biology. 2014;21:103–109. doi: 10.1038/nsmb.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chiarle R, et al. Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell. 2011;147:107–119. doi: 10.1016/j.cell.2011.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klein IA, et al. Translocation-capture sequencing reveals the extent and nature of chromosomal rearrangements in B lymphocytes. Cell. 2011;147:95–106. doi: 10.1016/j.cell.2011.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei PC, et al. Long Neural Genes Harbor Recurrent DNA Break Clusters in Neural Stem/Progenitor Cells. Cell. 2016;164:644–655. doi: 10.1016/j.cell.2015.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwer B, et al. Transcription-associated processes cause DNA double-strand breaks and translocations in neural stem/progenitor cells. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:2258–2263. doi: 10.1073/pnas.1525564113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsouroula K, et al. Temporal and Spatial Uncoupling of DNA Double Strand Break Repair Pathways within Mammalian Heterochromatin. Molecular cell. 2016;63:293–305. doi: 10.1016/j.molcel.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 47.Ginno PA, Lim YW, Lott PL, Korf I, Chedin F. GC skew at the 5' and 3' ends of human genes links R-loop formation to epigenetic regulation and transcription termination. Genome research. 2013;23:1590–1600. doi: 10.1101/gr.158436.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harrigan JA, et al. Replication stress induces 53BP1-containing OPT domains in G1 cells. The Journal of cell biology. 2011;193:97–108. doi: 10.1083/jcb.201011083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lukas C, et al. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nature cell biology. 2011;13:243–253. doi: 10.1038/ncb2201. [DOI] [PubMed] [Google Scholar]
- 50.Durkin SG, Glover TW. Chromosome fragile sites. Annual review of genetics. 2007;41:169–192. doi: 10.1146/annurev.genet.41.042007.165900. [DOI] [PubMed] [Google Scholar]
- 51.Le Tallec B, et al. Common fragile site profiling in epithelial and erythroid cells reveals that most recurrent cancer deletions lie in fragile sites hosting large genes. Cell reports. 2013;4:420–428. doi: 10.1016/j.celrep.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 52.Kalocsay M, Hiller NJ, Jentsch S. Chromosome-wide Rad51 spreading and SUMO-H2A.Z-dependent chromosome fixation in response to a persistent DNA double-strand break. Molecular cell. 2009;33:335–343. doi: 10.1016/j.molcel.2009.01.016. [DOI] [PubMed] [Google Scholar]
- 53.Ryu T, et al. Heterochromatic breaks move to the nuclear periphery to continue recombinational repair. Nature cell biology. 2015;17:1401–1411. doi: 10.1038/ncb3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Matsuoka S, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 55.Yamada K, Ono M, Perkins ND, Rocha S, Lamond AI. Identification and functional characterization of FMN2, a regulator of the cyclin-dependent kinase inhibitor p21. Molecular cell. 2013;49:922–933. doi: 10.1016/j.molcel.2012.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nagano T, et al. Comparison of Hi-C results using in-solution versus in-nucleus ligation. Genome biology. 2015;16:175. doi: 10.1186/s13059-015-0753-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wingett S, et al. HiCUP: pipeline for mapping and processing Hi-C data. F1000Research. 2015;4:1310. doi: 10.12688/f1000research.7334.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lawrence M, et al. Software for computing and annotating genomic ranges. PLoS computational biology. 2013;9:e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hu Y, et al. OmicCircos: A Simple-to-Use R Package for the Circular Visualization of Multidimensional Omics Data. Cancer informatics. 2014;13:13–20. doi: 10.4137/CIN.S13495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dey N, et al. Richardson-Lucy algorithm with total variation regularization for 3D confocal microscope deconvolution. Microscopy research and technique. 2006;69:260–266. doi: 10.1002/jemt.20294. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.