Abstract
The folding of genomic DNA from the beads-on-a-string like structure of nucleosomes into higher order assemblies is critically linked to nuclear processes. We have calculated the first 3D structures of entire mammalian genomes using data from a new chromosome conformation capture procedure that allows us to first image and then process single cells. This has allowed us to study genome folding down to a scale of <100 kb and to validate the structures. We show that the structures of individual topological-associated domains and loops vary very substantially from cell-to-cell. By contrast, A/B compartments, lamin-associated domains and active enhancers/promoters are organized in a consistent way on a genome-wide basis in every cell, suggesting that they could drive chromosome and genome folding. Through studying pluripotency factor- and NuRD-regulated genes, we illustrate how single cell genome structure determination provides a novel approach for investigating biological processes.
Introduction
Our understanding of nuclear architecture has been built on electron and light microscopy studies that suggest the existence of territories pervaded by an inter-chromosomal space through which molecules diffuse to and from their sites of action1. In parallel, biochemical studies, in particular chromosome conformation capture experiments (3C, Hi-C etc.) where DNA sequences in close spatial proximity in the nucleus are identified after restriction enzyme digestion and DNA ligation, have provided molecular information about chromosome folding2. At a mega-base scale, Hi-C experiments have partitioned the genome into two (A/B) compartments3. In addition, they have provided evidence for 0.5-1.0 Mb “topological-associated domains” (TADs)4–6, as well as smaller loops (hundreds of kilobases)7. 3C-type experiments have further shown that enhancers make direct physical interactions with promoters, and that these interactions are stabilized by a network of protein-protein interactions involving CTCF, cohesin and mediator8,9. Although probabilistic methods can be used to calculate ensembles of low-resolution models that are consistent with population Hi-C data10,11, understanding genome structure at higher resolution requires the development of single cell approaches.
In mitotic cells both A/B-compartments and TADs disappear12 and thus the structural complexity of interphase chromosomes is reestablished during G1 phase. To study interphase genome structure, we have combined imaging with an improved Hi-C protocol (Fig. 1a) to determine whole genome structures of single G1 phase haploid mouse embryonic stem cells (mESCs) at a 100 kb scale. The structures allow us to study TAD/loop structure genome-wide, to analyze the principles underlying genome folding, and to understand which factors may be important for driving chromosome/genome structure. We also illustrate how combining single-cell genome structures, with population-based ChIP- and RNA-seq data, provides new insight into the organization of pluripotency factor- and Nucleosome Remodeling Deacetylase (NuRD)-regulated genes.
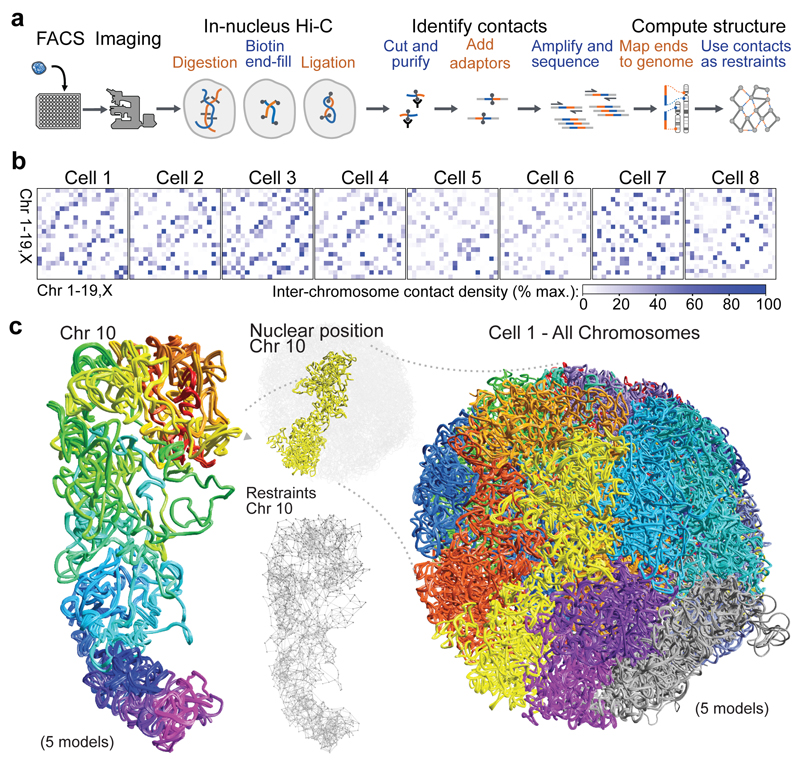
Fig. 1. Calculation of 3D genome structures from single cell Hi-C data.
a, Schematic of the protocol used to image and process single nuclei. b, Colour density matrices representing the relative number of contacts observed between different pairs of chromosomes. c, Five superimposed structures from a single cell, from repeat calculations using 100 kb particles and the same experimental data, with the chromosomes coloured differently. An expanded view of Chromosome 10 is shown, coloured from red through to purple (centromere to telomere), together with an illustration of the restraints determining its structure.
Calculation of intact genome structures from single-cell Hi-C data
We imaged haploid mESC nuclei, expressing fluorescently tagged CENP-A (the centromeric histone H3 variant) and histone H2B proteins, to select G1 phase cells (Extended Data Fig. 1a) and to later validate the structures. Hi-C processing of eight individual mESCs yielded 37,000-122,000 contacts (Extended Data Table 1), representing 1.2-4.1% recovery of the total possible ligation junctions. In single cells, unlike in population data, Hi-C contacts are observed between distinct and different sets of chromosomes (Fig. 1b and Extended Data Fig. 1b).
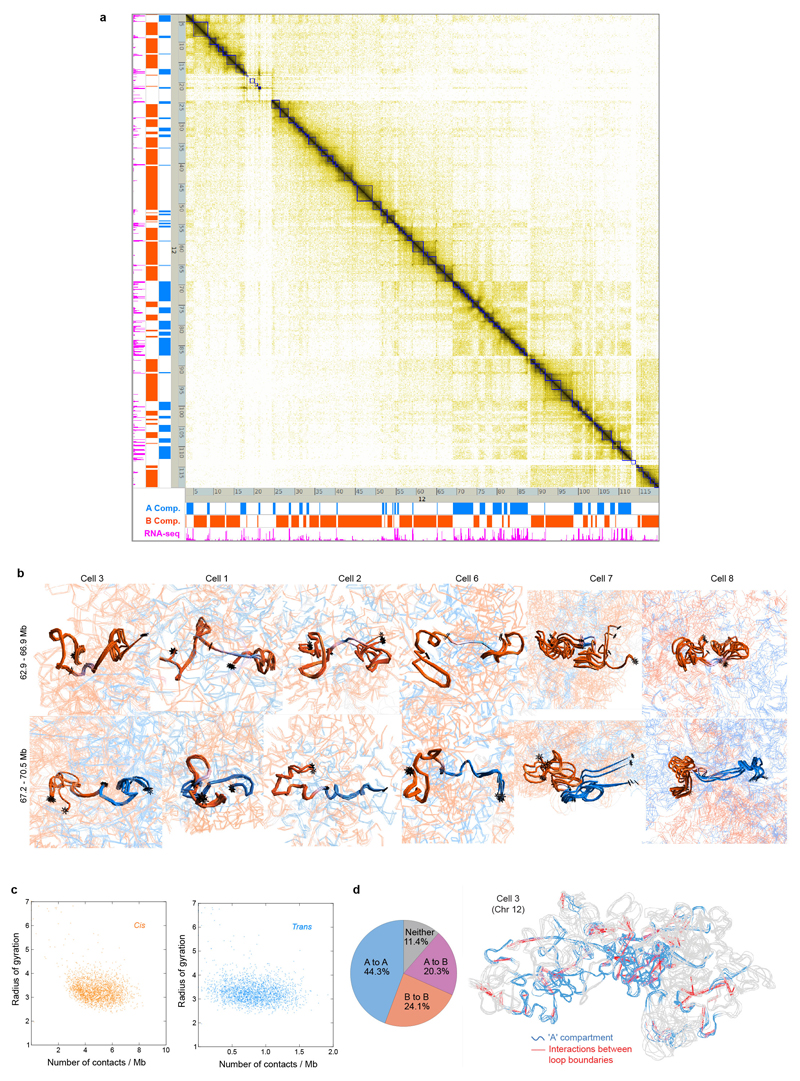
Using a particle-on-a-string representation and an extended simulated annealing protocol we calculated highly consistent 3D genome structures [ensemble root mean square deviations (RMSDs) < 1.75 particle radii] with discrete chromosome territories (Fig. 1c and Supplementary Videos 1, 2). The structures were calculated with an average of 1-3 Hi-C contact derived restraints for each 100 kb particle (with a total of 26,000-75,000 restraints, Extended Data Table 2 and Extended Data Fig. 1c). Recalculation after randomly omitting 10-70% of the data reliably generated the same folded conformation (RMSD < 2.5 particle radii). Moreover, structure calculations after randomly merging half the data from two different cells resulted in a vast increase in the number of violated experimental restraints (37.4 % have a distance >4 particle radii, compared to 5-6% for the separate data), and generated compacted, highly inconsistent structures (Extended Data Fig. 1d). Thus, single-cell Hi-C data cannot result from independent sampling of contacts from a single underlying conformation. In addition, cells with either a broken/recombined chromosome (Extended Data Fig. 1e) or with a duplicated chromosome (Extended Data Fig. 1f) can be immediately recognized from the data.
Validation of the structures and analysis of single-cell contacts
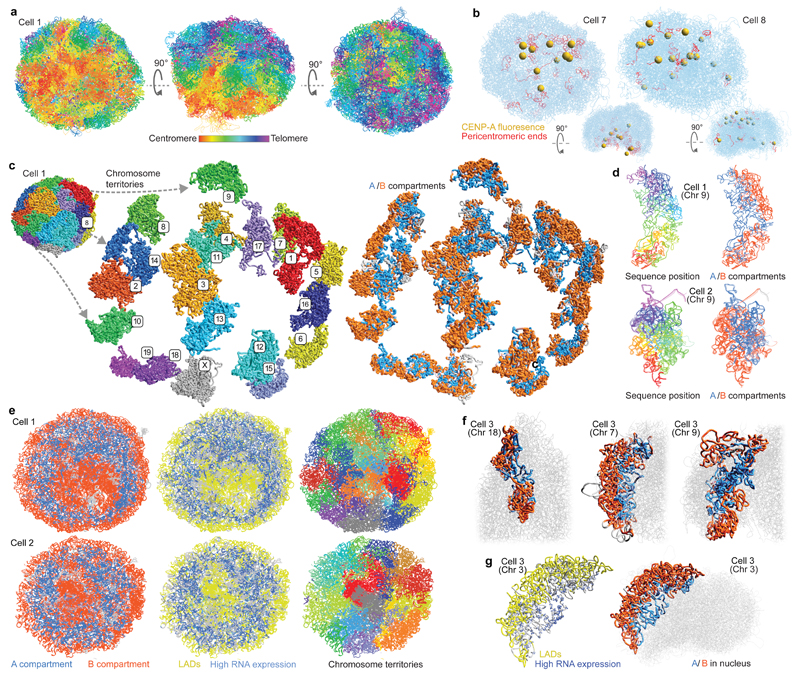
A consistent Rabl configuration (with centromeres and telomeres clustered on opposite sides of the nucleus) was observed in all G1 phase mESCs, strongly validating the structures (Fig. 2a, Extended Data Fig. 2a and Supplementary Video 3). Fig. 2b shows two examples of CENP-A image superposition with the corresponding genome structure from the same cell, providing independent evaluation of the reliability of the structure. Cell 7 shows typical clustering of the pericentromeric regions in a cavity on one side of the structure, which is clearly supported by the centromere positions in the CENP-A image. In Cell 8 the centromeres are more diffusely distributed in both the image and the structure. The structures were additionally validated through: 1) comparison with previous imaging studies, and both our own and previous DNA-FISH experiments; and 2) testing structural predictions using super-resolution microscopy (see below).
Fig. 2. Large-scale structure of the genome.
a, Five superimposed structures from a single cell in three different orientations with the chromosomes coloured from red through to purple (centromere to telomere). b, Superposition of two single cell structures with images of mEos3.2-tagged CENP-A recorded from the same single cells. The centromeres from the images are shown as yellow spheres and the centromeric ends of the chromosomes are coloured red. The same structures after rotation through 90º are shown below. c, 3D structure of a haploid mES genome with expanded views of the separate chromosome territories (left), and the spatial distribution of the A (blue) and B (red) compartments (right). d, Structure of chromosome 9 from two different cells coloured (left) from red through to purple (centromere to telomere), or (right) according to whether the sequence is found in either the A (blue) or B (red) compartments. e, Cross-sections through five superimposed 3D structures from two different cells, coloured according to whether: (left) the sequence is in the A or B compartment; (centre) is part of a constitutive lamin-associated domain (cLAD) (yellow) or contains highly expressed genes (blue); and (right) chromosome identity. f, Structures of selected chromosomes from a single cell illustrating the different ways chromosomes can contribute to the A/B compartments. g, Chromosome 3 from a single cell with the positions of highly expressed genes shown as blue circles (larger circles indicate higher expression) and lamin associated regions shown in yellow (left), and where the sequence is coloured according to whether it is in the A or B compartment (right).
The single cell Hi-C data shows fairly uniform coverage of long range contacts across both the A and B compartments, suggesting similar restriction enzyme/ligase accessibility in each (Extended Data Fig. 2b). Importantly, the contact probability is preserved for all nearby particles, showing that the entire structure is consistent with the Hi-C contact data (Extended Data Fig. 2c). We noticed an increase in contact density in some regions that coincided with sites of early DNA replication13, but after studying violated experimental restraints we were unable to identify any region that cannot be described by a single structural conformation, i.e. where replication appeared to have begun (Extended Data Fig. 2b).
Comparison of haploid and diploid mESCs using RNA-seq and ChIP experiments respectively showed that the levels of gene expression are highly correlated with each other (Spearman’s rho=0.97, P<10-15) (Extended Data Fig. 2d) and that protein-genome interactions are highly similar (Extended Data Fig. 2e). This allowed us to utilise published ChIP-seq data when analysing the haploid structures.
The large-scale 3D architecture of the genome is conserved in all cells
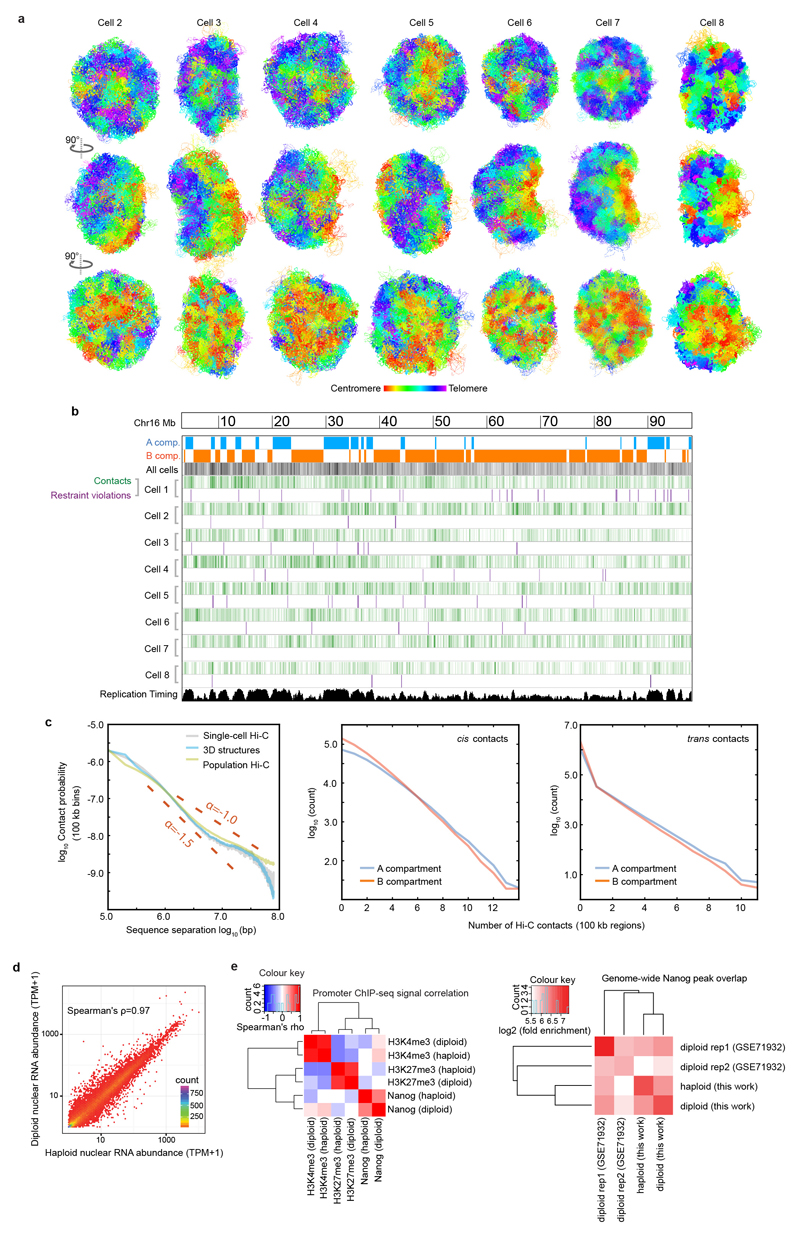
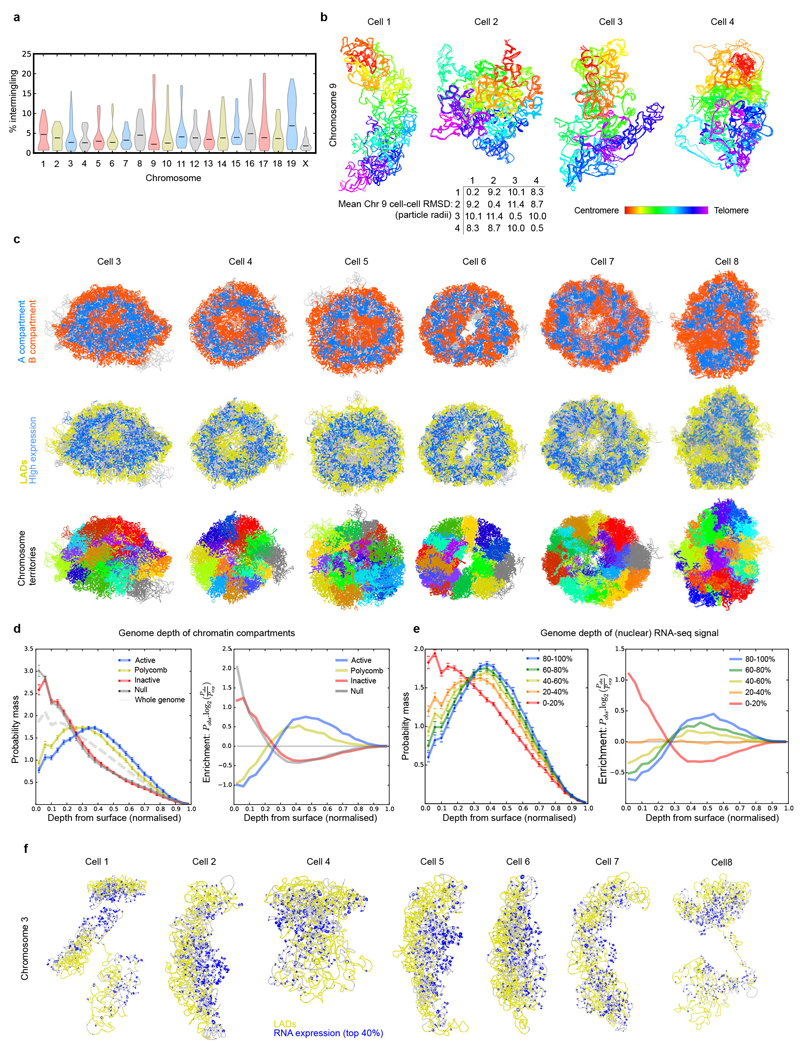
Discrete chromosome territories can be seen in all the intact genome structures (Fig. 2c and Supplementary Video 1), although there is a significant degree (5-10%) of chromosome intermingling (Extended Data Fig. 3a). Whilst chromosome structure varies dramatically from cell-to-cell, we find that regions belonging to the A or B compartments always cluster together and A segregates from B (Fig. 2d and Extended Data Fig. 3b). This is supported by recent imaging experiments showing that A and B compartment TADs are organized in a spatially polarized manner in single chromosomes14, providing further validation of our structures. In all cells the chromosomes then pack together to give an outer ring of B compartment, an inner ring of A compartment, and an internal region of B compartment around the hollow nucleoli (Fig. 2e, Extended Data Fig. 3c and Supplementary Video 4). The nucleolus is often close to the nuclear membrane with the A compartment forming a bowl-like structure. To achieve this organization chromosomes can fold in from the surface towards the nucleoli, or fold in and back out again, or go all the way through the nucleus (Fig. 2f and Supplementary Video 5). Chromatin states computed from the genome-wide association of post-translationally modified histones in mammalian cells15 (a completely independent method), also show a similar organization (Extended Data Fig. 3d). Likewise, regions that constitutively associate with Lamin B1 (cLAD’s)16,17 are confined to either the nuclear membrane or nucleolar periphery in every cell, consistent with reshuffling between these regions each cell cycle18,19. Highly expressed genes, however, mostly lie in the inner ring of A compartment (Figs. 2e,g, Extended Data Figs. 3c,e,f and Supplementary Videos 6, 7).
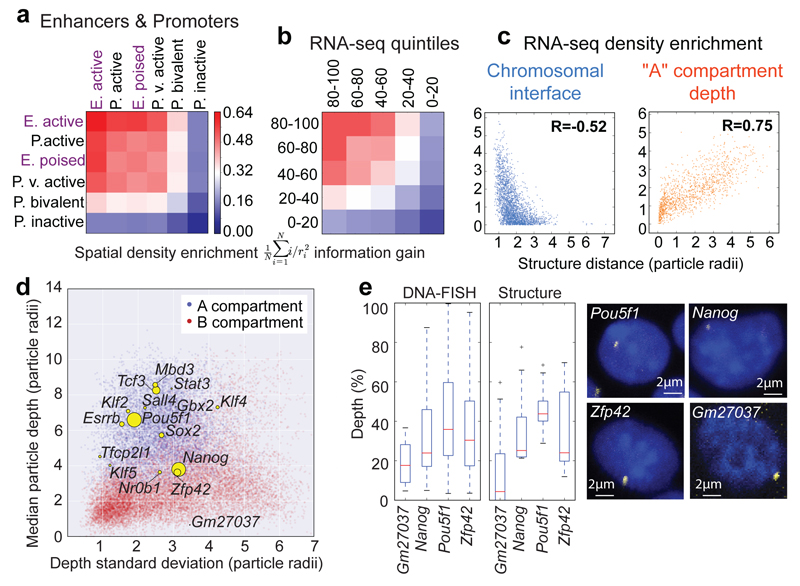
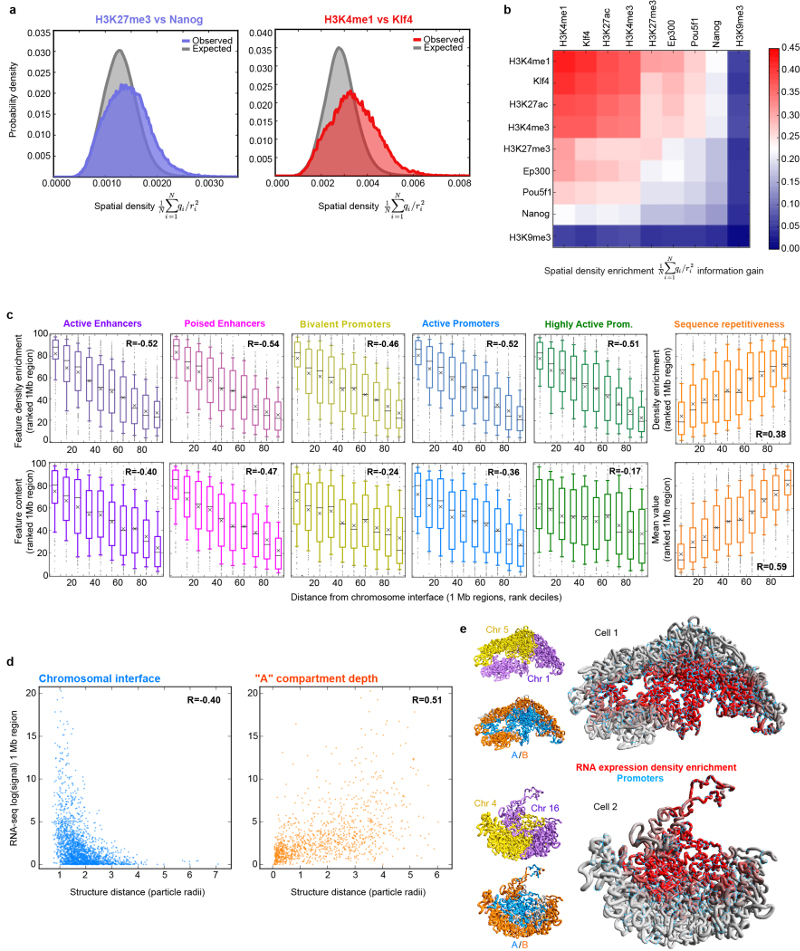
By mapping ChIP-seq data onto the single cell genome structures we observed 3D clustering of histones H3K4me1, H3K27ac, and H3K4me3, consistent with the presence of enhancer/promoter clusters or transcription factories (Extended Data Figs. 4a,b). Annotating enhancers and promoters for activity (see Supplementary Methods) showed that active enhancers spatially associate most strongly with each other, followed by active enhancers with active promoters (Fig. 3a). We also found a pronounced clustering of highly expressed genes, in single cells, after mapping nuclear RNA-seq data onto the structures (Fig. 2g), and the greater the level of gene expression the larger the effect (Fig. 3b). Genome-wide analysis also showed that active/poised enhancers and active/bivalent promoters have a clear preference for being located at chromosomal interfaces (Extended Data Fig. 4c). Interestingly, there are very clear correlations between a gene’s expression level, and both localization to a chromosomal interface and depth within the A compartment (Fig. 3c and Extended Data Figs. 4d,e). We also related the preferred positions of pluripotency genes20 to gene expression and found that two highly expressed genes, Zfp42/Rex1 and Nanog, have variable positions in our structures (Fig. 3d). They are either found near the nuclear membrane or buried. DNA-FISH experiments, where Pou5f1 is a typical highly expressed (and usually buried) gene control, verified these conclusions providing further validation of the structures (Fig. 3e).
Fig. 3. Relationship between genome folding and gene expression.
The enrichment in spatial density of: a, enhancers and promoters annotated using ChIP-seq data; b, gene expression determined from nuclear RNA-seq data, with genes separated according to their relative level of expression. In both (a) and (b) the data are presented in hierarchical order, grouping the most similar datasets together. c, The enrichment in the spatial density of gene expression vs distance from the nearest inter-chromosomal interface (left) and the outer surface of the A compartment (right). d, Median vs standard deviation of the depth from the nuclear periphery for particles in the A (blue) or B (red) compartments. Particles containing pluripotency genes are indicated by yellow circles – the sizes illustrate relative levels of expression. e, Comparison of nuclear depth in either the 3D structures (n=8) or DNA-FISH analysis of the Nanog (n=84 cells) and Zfp42 (n=142 cells) genes, with Pou5f1 (n=189 cells) as a control. Gm27037 (n=16 cells), a pseudo-gene, provided a non-pluripotency factor control.
Notably, the A/B compartments, cLAD, ChIP- and RNA-seq data were all determined from populations of cells. Their consistent organization in every cell suggests that overall chromosome/genome conformation may be driven by a combination of interactions of LADs with the nuclear membrane/nucleolus and the clustering of active enhancers/promoters, which can be modulated by chromatin remodeling21. That genome structure is driven by transcription is supported by live cell imaging of histone-GFP fusion proteins during C. elegans development, which shows that knock-down of RNA Pol II leads to a collapse of the chromatin to a ring inside the nuclear membrane22.
Folding of chromosomes into Topologically Associated Domains (TADs) or CTCF/Cohesin loops
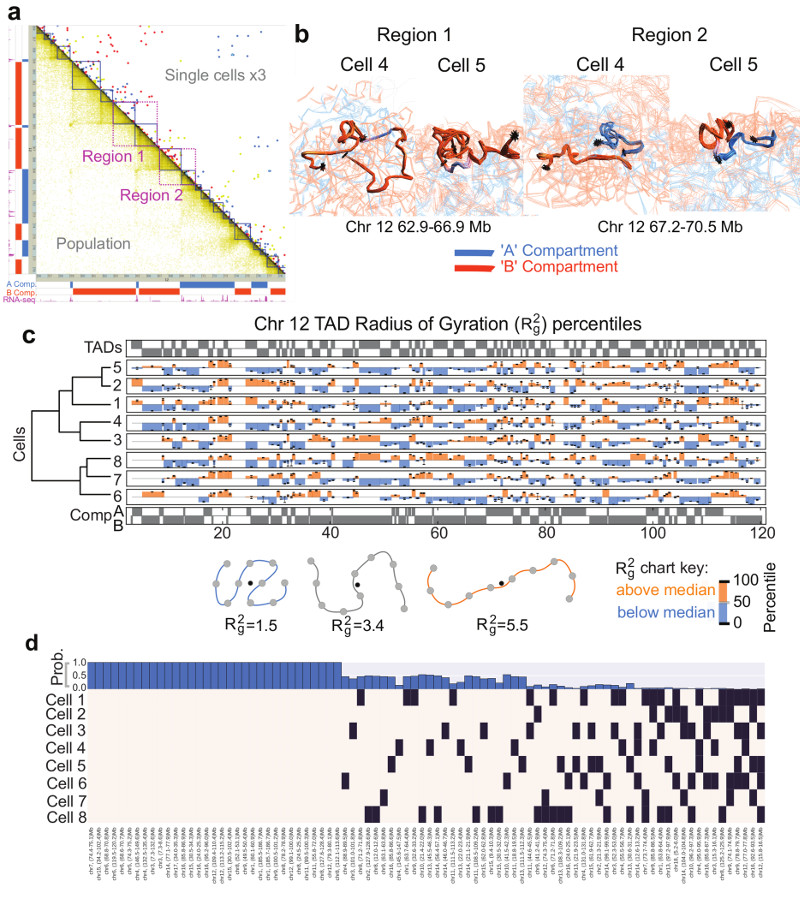
As in previous studies5,9,23, we observed an alignment between highly expressed genes and both A/B compartment and TAD boundaries (Fig. 4a and Extended Data Fig. 5a). Analysis of four TADs, either side of highly expressed genes (Regions 1 and 2 in Fig. 4a), illustrates that in some cells a particular TAD is compacted, often such that its two boundaries are close enough to interact, whilst in others it is completely extended. This difference is not due to a lack of data because the structures obtained from repeated calculations using identical experimental restraints are very well defined (Fig. 4b and Extended Data Fig. 5b).
Fig. 4. Structure of topological-associated domains (TADs) and CTCF/Cohesin loops.
a, Part of the Hi-C contact map from Chromosome 12 showing: (above the diagonal) contacts observed in three different single cells (coloured red, yellow and blue); (below the diagonal) the corresponding population Hi-C data. TADs identified by Dixon et al. (Ref. 5) are shown in dark blue, and the two regions analysed in panel b are shown in magenta. b, Ensembles of five superimposed structures showing: (left) two B compartment TADs (Region 1 in a); (right) TADs either side of an A/B compartment boundary (Region 2 in a). The TADs are coloured according to whether they are in the A (blue) or B (red) compartments, with white indicating a transitional segment (between A and B). Boundaries are marked by asterisks. c, The mean radius of gyration (ROG) of Chromosome 12 TADs ± the standard error of the mean. The data are scaled according to TAD size, and presented as quantile values for the chromosome. Values below the 50th percentile value are colored blue and above it red. The ROG values for multiple cells are presented in hierarchical cluster order, grouping the most similar cell traces together. A schematic illustrating the calculation of the ROG as a measure of the compaction of a particle chain is shown below. d, Analysis illustrating whether CTCF/Cohesin loops with sequence separation >600 kb identified by Rao, et al. (Ref. 7) could be formed in the different single cells. A black square indicates that a loop could be formed, whilst a white square indicates that the two relevant particles are too far apart in the structure. The bar chart across the top shows the probability, for each loop, of random particles (pairs with the same sequence separation) forming the same number of contacts, or better.
We systematically studied compaction in chromosome 12 TADs (Extended Data Fig. 5a), by computing the radius of gyration (ROG) after excluding possible sites of early DNA replication where TAD structure might be disrupted. As with previous studies of the Tsix TAD24 individual TAD compaction varies widely from highly extended to compacted states (Fig. 4c), consistent with ligation occurring between almost every site in population Hi-C data. The structures of both compact and extended TADs are well defined and there is little correlation between the ROG and Hi-C contact density (Extended Data Fig. 5c), further showing that extended TAD structures do not result from a lack of experimental contacts. Analysis of TAD structure in all the other chromosomes gave analogous results (Extended Data Fig. 6). It is noteworthy that compaction in the structures often appears to involve the formation of loops within a TAD (see Fig. 4b, Extended Data Fig. 5b and Supplementary Videos 8-11) and it will be interesting to investigate whether these structures are related to supercoiling25,26 or loop extrusion27–29.
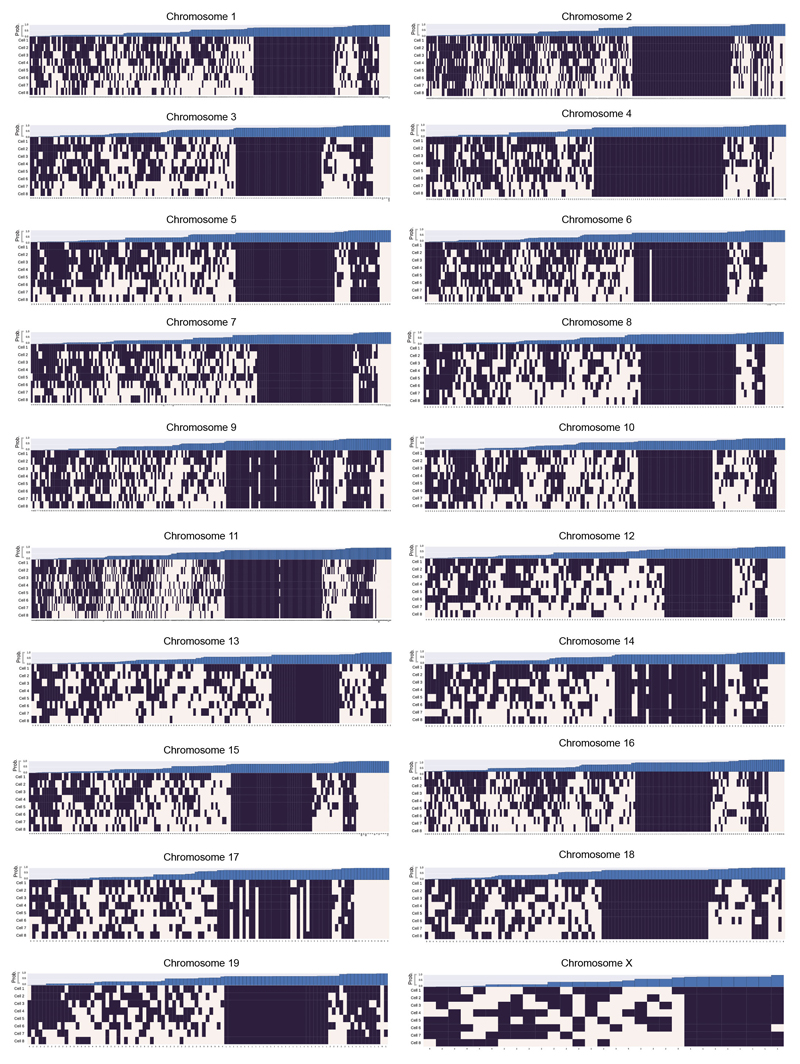
We found that CTCF/Cohesin loops identified in high-resolution Hi-C data from mouse B-lymphoblasts7 mostly involve interactions where at least one end of the loop is in (or very near to) the A compartment (Extended Data Fig. 5d). Considering the 88 largest loops from 2,823 in total (with sequence separation >600 kb), we found that 33% do not form in any of the cells whereas the boundaries of the remainder contact each other in 12-62% of the cells (Fig. 4d). Extending this analysis to all 2,823 loops in 8 cells showed that the boundaries interact in 62.1% (Extended Data Fig. 7). Our genome-wide results suggest that TADs and CTCF/Cohesin loops do not form in all cells, in agreement with previous DNA-FISH experiments by Rao et al. (Ref. 7) who showed that four representative loops form in only a proportion of cells.
Our structures provide snapshots of genome folding at a particular time in different cells, and thus do not provide information about dynamics. They are, however, strikingly consistent with what one would expect from recently proposed loop-extrusion models, where TADs and CTCF/Cohesin loops might be expected to have highly dynamic and variable structures as Cohesin rings are driven to stable binding sites7,27–29. It is not known what drives the movement of Cohesin rings in mammalian cells, but previous studies in yeast suggest that it might be RNA polymerase molecules and transcription30. This would be consistent with our observation that CTCF/Cohesin loops7 are mostly found in the A compartment (where transcription levels are higher), studies in Drosophila suggesting that TADs result from the compaction of chromatin due to transcription31,32, and recent studies of the inactive mouse X chromosome that show a global loss of TAD structure except at expressed genes33,34.
Understanding the nature of gene networks in single mESCs
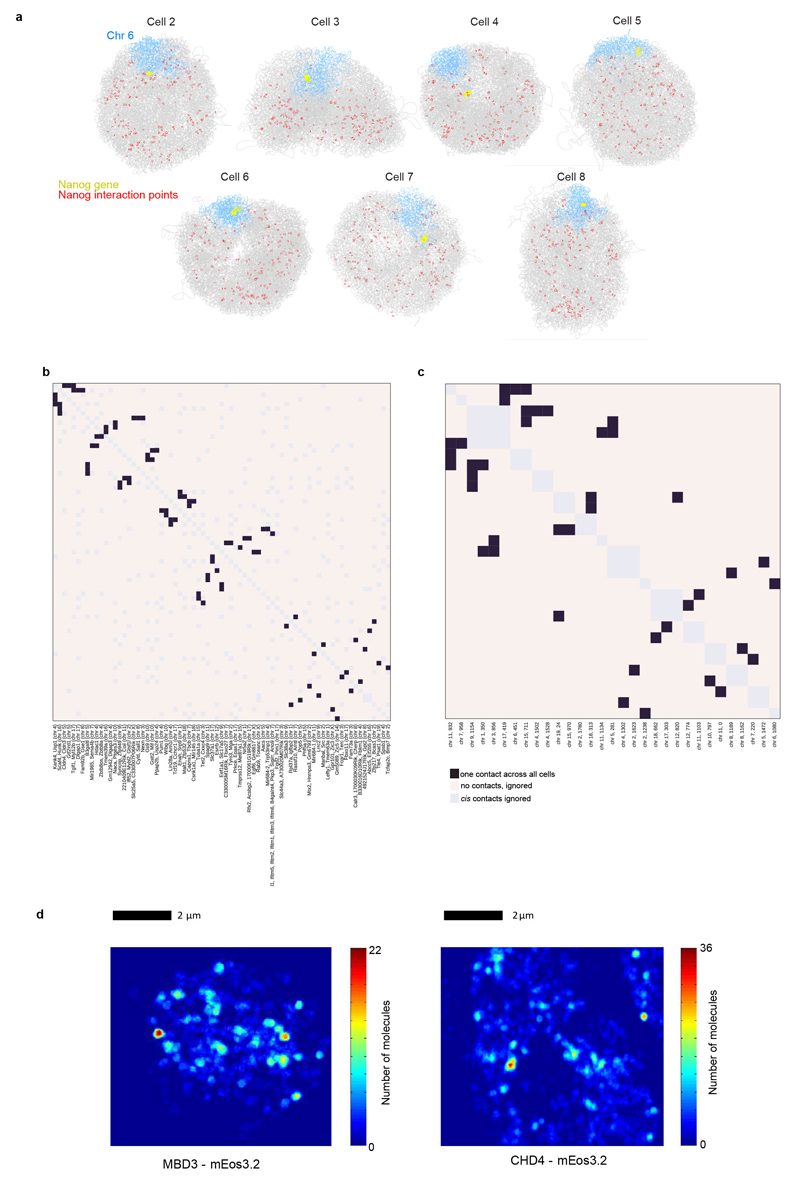
In addition to CTCF, Cohesin and Mediator, previous studies have implicated key pluripotency factors as well as the Polycomb complexes (PRC1 and PRC2) in organizing 3D genome structure in mESCs. Analysis of one of the published 4C Nanog-gene interaction networks35 showed that only one (or two) of the previously identified 4C contacts can be identified in each single cell structure, showing that the propensity for particular genes to interact is low (Fig. 5a and Extended Data Figs. 8a,b). Analysis of Pou5f1-gene interacting regions36 gave very similar results (Extended Data Fig. 8c).
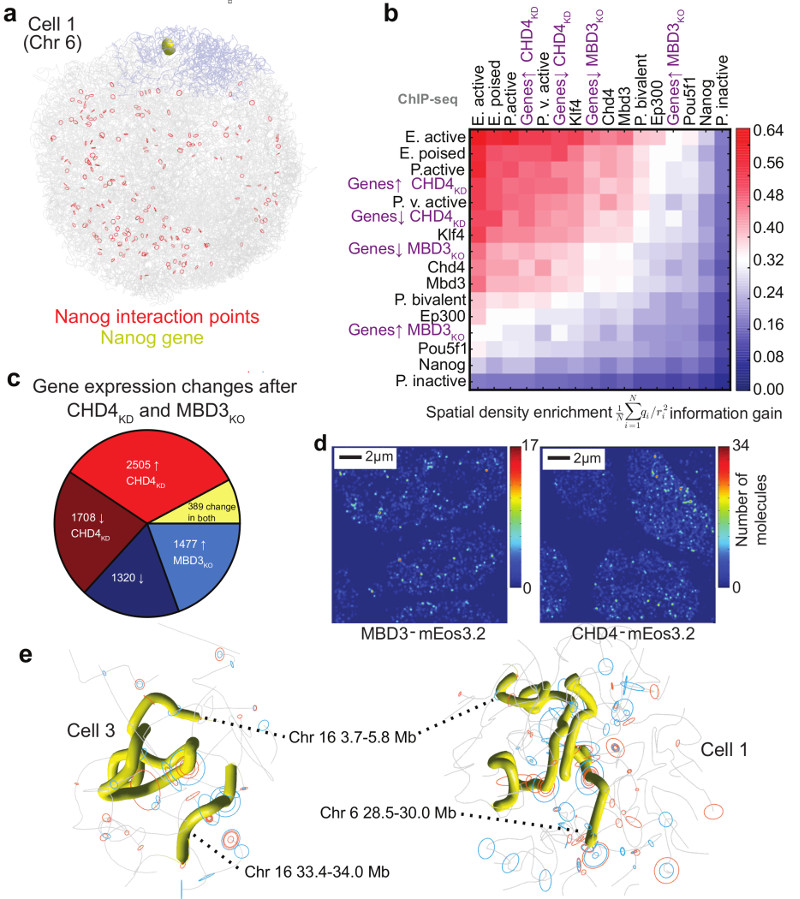
Fig. 5. Understanding the nature of gene networks in mouse ESCs.
a, Structure of an individual cell illustrating the interactions identified between the Nanog gene in Chromosome 6 (coloured yellow and blue, respectively) and other regions of the genome (red circles) in a population 4C experiment35. b, The spatial density enrichment of NuRD components (CHD4 and MBD3), pluripotency factors and NuRD regulated genes, as well as annotated enhancers and promoters defined using ChIP-seq data. c, Pie chart showing the numbers of NuRD regulated genes in different classes. d, A heat map showing clustering of CHD4 and MBD3 molecules in 2D super-resolution PALM in fixed mESCs. e, Structures of a region of chromosome 16 in two different cells, showing clustering of regions containing genes that are highly regulated by NuRD (highlighted in yellow). The positions of genes in either the CHD4-knockdown or MBD3-null cells that are down-regulated (red circles) or up-regulated (blue circles) are indicated by circles (larger for more highly regulated).
We mapped ChIP-seq data for different pluripotency factors onto the single cell genome structures and showed that, in single cells, Klf4 spatially clusters strongly with itself, H3K4me1, H3K27ac, and H3K4me3, i.e. with active enhancers/promoters (Extended Data Figs. 4a,b). This analysis also suggested 3D clustering of histone H3K27me3 (a marker for Polycomb complexes), but lower levels of 3D clustering of Nanog, both with itself and with H3K27me3. These results are consistent with previous mESC imaging experiments36,37, and strongly validate our single cell structures. They support the proposal that Klf4 organizes long-range chromosomal interactions36, and suggest that the observed large-scale 3D segregation of Nanog and H3K27me337 mostly results from Nanog and PRC complexes binding to separated sequences in chromosomes. However, whilst they suggest that Klf4-bound genes cluster, they also show that there is little propensity for “particular” Klf4-bound genes to interact with each other.
Next, we used the structures to study genes regulated by the NuRD complex, which plays a key role in controlling the earliest stages of differentiation of mESCs38. Whilst ChIP-seq experiments showed that CHD4 (the chromatin remodeling component) and MBD3 (part of the deacetylase core)39 are widely distributed (data not shown), we surprisingly found marked 3D clustering of NuRD-regulated genes (Figs. 5b,c). Super-resolution microscopy and single particle tracking using photo-activated light microscopy (PALM) in fixed and live cells, respectively, showed clustering of both the chromatin remodeling and deacetylase sub-modules (as illustrated by the mEos3.2 tagged CHD4 and MBD3 proteins, respectively), consistent with the 3D clustering of NuRD-regulated genes (Fig. 5d and Extended Data Fig. 8d). Interestingly, whilst our structures show that regions containing highly NuRD-regulated genes cluster, the actual regions that interact vary from cell-to-cell (Fig. 5e). In addition, we found that most genes are up/down regulated in either the CHD4 depletion experiment (CHD4-KD) or in the MBD3 knockout cells (MBD3-KO), but not both (Fig. 5c), suggesting that the chromatin remodeling and deacetylase sub-modules may function separately. However, despite regulating different sets of genes, it is notable that genes that are down-regulated in the MBD3-KO cells cluster more strongly than those that are up regulated, and that genes that are down-regulated in the MBD3-KO cluster more strongly with genes that are up-regulated in the CHD4-KD (and vice-versa) (Fig. 5b). Although further work is necessary to understand what drives the formation of NuRD clusters, the 3D clustering of CHD4 and MBD3 with active enhancers and promoters is noteworthy (Fig. 5b).
Conclusion
The structures allow the first genome-wide analysis of 3D interactions of individual regulatory elements/genes in single cells. In combination with 3D imaging they show that whilst Klf4- and NuRD-regulated genes interact and cluster to form foci, the genes they bring together are very variable. Our combination of imaging with genome structure determination will allow further studies of these and many other biological processes. In addition, the finding that chromosomes have a Rabl configuration in mammalian G1-phase cells may underlie slight preferences in long-range chromosomal interactions – e.g. those leading to translocation events involved in disease40.
Extended Data
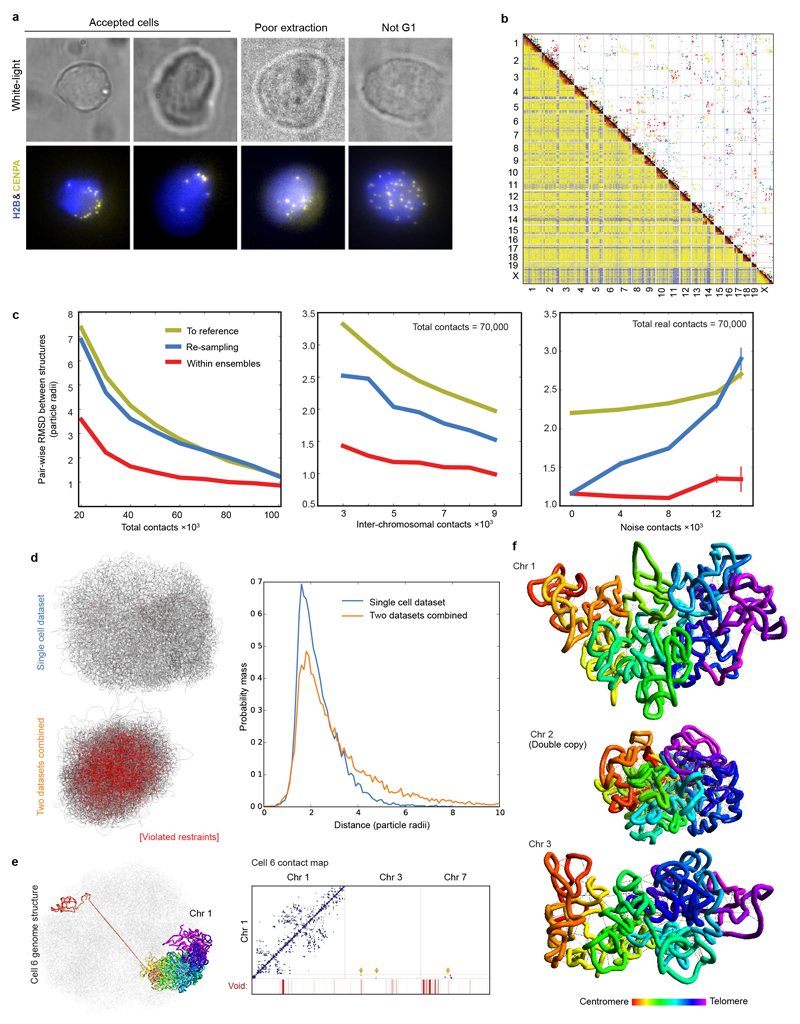
Extended Data Fig. 1. Quality control for Hi-C processing and 3D structure calculation.
a, Comparison of 3D images of CENP-A in haploid mES nuclei, expressing mEos3.2-tagged CENP-A and tandem iRFP-tagged histone H2B, with their corresponding white light images. b, Comparison of three single cell Hi-C contact maps (above the diagonal with contacts coloured red, yellow and blue), with the population Hi-C map (below the diagonal). c, An analysis of the accuracy and precision of the 100 kb structure calculation procedure for Cell 1. The graphs show how the global (dis)similarity of structures is affected by: the total number of contacts (left); the number of inter-chromosomal contacts (middle); and the number of random noise contacts (right). Mean RMSD values for all pairs of conformations ± the standard error of the mean are shown for: the precision within ensembles arising from ten re-calculations using the same contacts (red); the variation across ensembles arising from different random resampling (blue); and (as a measure of accuracy) the similarity to the best ensemble of structures (yellow). d, An example of a structure calculation carried out using either a single dataset, or after randomly merging 50% of the data from two different cells (lower). Strongly violated experimental restraints (>4 particle radii apart) are shown in red. The plot (right) shows the probability of any two particles connected by an experimental restraint being violated to different degrees. e, (left) The structure of chromosome 1 from Cell 6, where part of the chromosome lies at the opposite side of the genome structure, with no intermediate chromosome folding, illustrating the presence of a chromosomal break or recombination event. The contact map (right) shows that there are no contacts from the disconnected region to any other part of chromosome 1, but clear contacts to chromosomes 3 and 7. f, An example of an attempted calculation of the haploid genome structure for a cell containing a duplicated chromosome 2 shows many violations of the experimental restraints for that chromosome and a much more compacted structure (here compared with chromosomes 1 and 3). The structures are coloured according to position in the chromosome sequence from red through to purple (centromere to telomere).
Extended Data Fig. 2. Validation and analysis of single cell contacts.
a, Structure of the entire haploid mESC genome from cells 2 to 8. The structural ensemble is represented by five superimposed conformations from repeat calculations, and is shown in three different orientations (after rotation through 90º relative to each other) with the chromosomes coloured according to their position in the chromosome sequence from red through to purple (centromere to telomere). b, Correspondence between the distribution of Hi-C contacts (both cis and trans), violations of the distance restraints in the 3D structures, and DNA replication timing13 for a representative chromosome (Chromosome 12). c, (left) Log scale plots of contact probability (Pcont) against sequence separation (S). The slopes for a power law relationship (Pcont ∝ Sα) where α is either -1.0 or -1.5 are also indicated. Data is shown for the combined single cell Hi-C contact data, for all of the non-sequential particles that are close to each other in the structures (<2 particle radii apart), and for the population Hi-C data. (right) The distribution in the number of intra- (cis) or inter-chromosomal (trans) contacts between 100 kb regions in the single-cell Hi-C data is shown for both the A and B compartments. d, Correlation of gene expression levels (left), and hierarchically clustered heat maps showing the pairwise enrichment of ChIP-seq peak overlaps between haploid and diploid mES cells (centre), and between Nanog ChIP-seq peak overlaps between haploid and diploid ES cells used in this study, as well as that previously published from diploid ES cells (right)41.
Extended Data Fig. 3. Chromosome interactions.
a, Violin plot showing the proportion of each chromosome that intermingles with other chromosomes. b, Pair-wise comparison of the chromosome structure in different cells by root mean square deviation (RMSD) analysis. Four models of chromosome 9 from a selection of different cells are shown, coloured according to the chromosome sequence (from red through to purple, centromere to telomere), together with a table showing the RMSD between the chromosomal 3D coordinates for each cell (bottom). c, Further cross-sections from cells 3-8 through the structures of haploid genomes (see Fig. 2e), coloured according to: (top) whether the sequence is in the A or B compartment; (centre) whether the sequence is part of a constitutive lamin-associated domain (cLAD) or contains highly expressed genes (coloured yellow and blue, respectively); and (bottom) identity of the chromosomes. In each case the figures show an ensemble of five superimposed conformations arising from repeat calculations using different randomly generated sets of coordinates. d, An analysis of the genome depth of various chromatin class categories, determined by k-means clustering of 100 kb segments according to the presence of histone H3 ChIP-seq data15. The Active class is associated with H3K4me3, Polycomb with H3K27me3, Inactive with H3K9me3, and null the remainder. (left) The probability distribution for each of the categories at different normalized nucleus depths. (right) The divergence of the probability distribution for each category from the whole genome average. Data is shown for the genome structures of all cells. e, An analysis of the genome depth for regions with differing levels of gene expression, as measured by nuclear RNA-seq. Here RNA-seq signal peaks were ranked and split into five classes. As in panel d, the probability distribution for each class with regard to genome depth is shown (left), together with the divergence of each distribution from the genome as a whole (right). f, Further comparisons of the structure of chromosome 3 from different cells, coloured according to whether the sequence is part of the constitutive LAD domains (yellow), with the positions of highly expressed genes indicated by the presence of blue rings (larger circles indicate higher expression).
Extended Data Fig. 4. Relationship between genome folding and gene expression.
a, Calculation of 3D spatial clustering compared to a random hypothesis where the same data were circularly permuted around the sequence, and repeating the calculations, using the same structure. Two examples, showing strong (Klf4/H3K4me1) and weaker (Nanog/H3K27me3) spatial co-localisation, compared to random, are shown. b, The enrichment in spatial density (after removal of any clustering expected from their being located nearby in the same chromosome sequence), of histone H3 with various post-translational modifications and selected pluripotency factors as determined using ChIP-seq data. The enrichment is calculated over all cells as the Kullback-Liebler divergence of the normalized spatial density distribution from a random, circularly permuted, expectation (see Supplementary Methods for more details), and the data are presented in hierarchical order, grouping the most similar datasets together. c, Box and whisker plots showing enhancer, promoter and repetitive sequence content (lower row), and the enrichment in spatial density of different types of enhancer, promoter and repetitive sequence (upper row), after the data have been divided into ten groups based on increasing distance from the nearest inter-chromosomal interface. The whiskers represent the 10th and 90th percentiles, the boxes represent the range from the 25th to 75th percentile, and outliers are shown as dots. Mean and median values are shown with black crosses and bars, respectively. The R-values are the Pearson’s correlation coefficient on the underlying, unranked data. d, Plots of the level of gene expression as measured by the nuclear RNA-seq signal within 1 Mb regions against distance from the nearest inter-chromosomal interface (left) and the outer surface of the A compartment (right). e, Examples of inter-chromosomal interfaces from two different cells where the chromosomes are coloured increasingly brightly red for higher enrichment in the density of gene expression, compared to what would be expected for a given sequence separation. The remainder of the two chromosomes is coloured grey, and the positions of promoters are indicated by blue circles. The same views are shown with the two different chromosomes coloured yellow and blue (upper), or with their regions in the A and B compartments coloured blue and red (lower).
Extended Data Fig. 5. Chromosome folding into compartments, TADs and loops.
a, A contact map showing the population Hi-C data for chromosome 12 with TADs identified using the directionality index5 in blue. On the left hand side and below, data tracks are shown identifying the A and B compartments (in blue and red, respectively), and highly expressed genes (in magenta). b, Further comparisons (see Fig. 4b) showing the structures (and their variability) of two B compartment TADs either side of a highly expressed gene(s) in a short region of A compartment, or at a boundary between the A and B compartments (lower). Ensembles of five superimposed conformations, from repeat calculations using the same experimental data, are shown with pairs of TADs highlighted and coloured according to whether they are in the A or B compartments (blue and red, respectively), with white indicating a transitional segment (between A and B). TAD boundaries are marked by asterisks. c, Scatter plots of the mean radius of gyration for 1 Mb regions of genome structure compared to the average number of single-cell Hi-C contacts, within the same region, considering a 1 Mb sliding analysis window. Data is shown for all genome structures and split according to cis contacts (left) and trans contacts (right). d, Structure of Chromosome 12, with the A compartment coloured blue and positions of CTCF/Cohesin loops identified by Rao et al. (Ref. 7) indicated by dotted red lines. The pie chart shows the numbers of loops between sequences in the A and B compartments.
Extended Data Fig. 6. Chromosome folding into TADs.
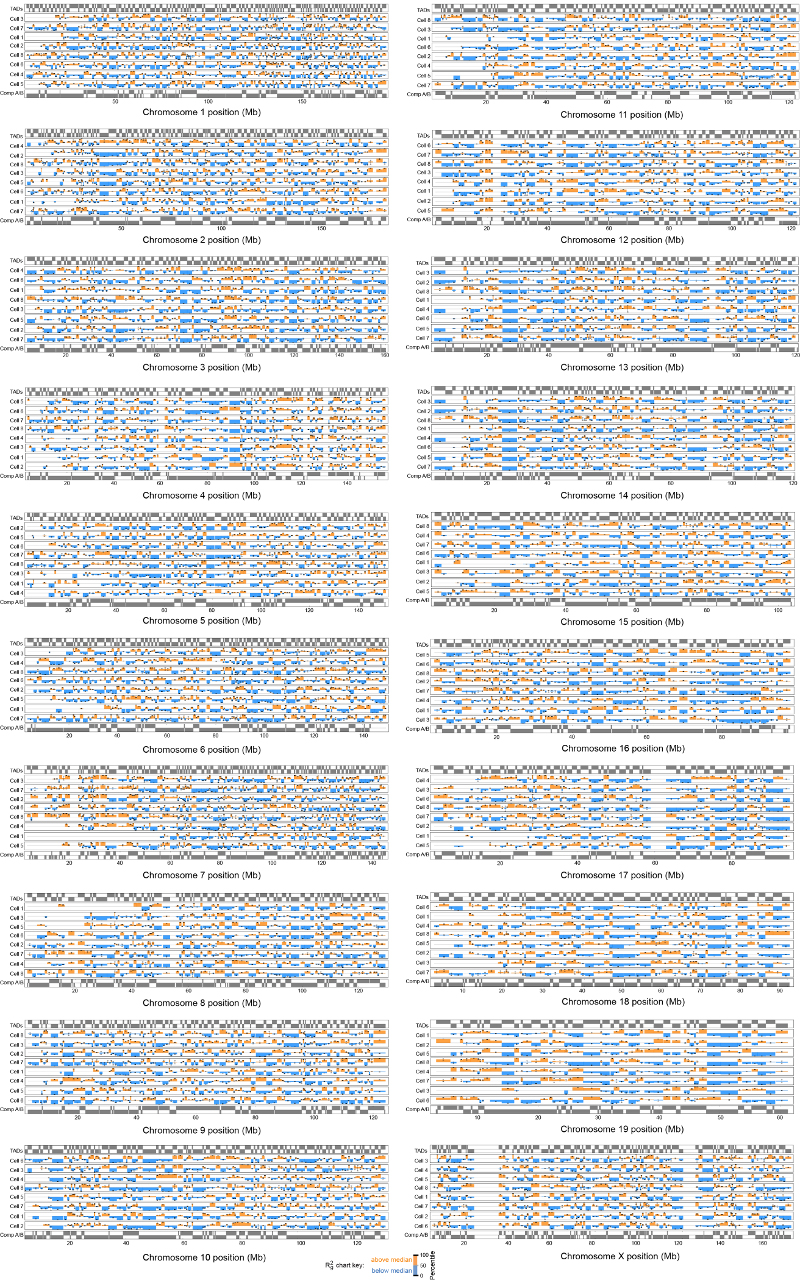
Bar charts of the mean radius of gyration (ROG) values of TADs identified using the directionality index5 for all the different chromosomes. The data are mean values over all structure conformations, scaled according to TAD size, and presented as quantile values for the chromosome. The 50th percentile value corresponds to the central grey line. Values below this are colored blue and above this are red. TADs that contain both regions of early replication timing (above 90th percentile) and moderate restraint violation (see Extended Data Fig. 2b) are excluded from the calculation. The errors in the ROG are the percentiles at ± the standard error of the mean. Values for multiple cells are presented in hierarchical cluster order, grouping the most similar cells together.
Extended Data Fig. 7. Chromosome folding into loops.
A genome-wide analysis illustrating whether CTCF/Cohesin loops7 could be formed in the different single cells, in each chromosome. A black square indicates that the two boundaries in the loop could interact, whilst a white square indicates that the two relevant particles are too far apart in the structure. The loop boundary separation, in particles, is shown along the x axis. The bar chart across the top shows the probability, for each loop, of random particles (pairs with the same sequence separation) forming the same number of contacts, or better. The probability of choosing a set of loop boundary points, which interact more frequently than we observed is 0.00072 (see Supplementary Methods).
Extended Data Fig. 8. Understanding the nature of gene networks in mouse embryonic stem cells.
a, Structures of Cells 2-8 illustrating the interactions identified between the Nanog gene and other regions of the genome by population 4C (Ref. 34). Chromosome 6 is coloured in blue, with the position of the Nanog gene highlighted in yellow, whilst the remainder of the chromosomes are coloured grey. Interacting positions in the genome are indicated by red circles. b, Heat map showing the number of times a particular interaction is detected between two of the 4C Nanog-interacting points35. c, Heat map showing the number of times a particular interaction is detected between two of the 4C Pou5f1-interacting points36. In both b,c the interaction points are presented in hierarchical order grouping the regions that show the most interactions together. d, 2D single molecule tracking using photo-activated light microscopy (PALM) in live mESCs shows clustering of CHD4 and MBD3. In both cases, a heat map of a single cell is shown where the pixels have been colour-coded according to the density of molecules detected in that region.
Extended Data Table 1.
Summary of the data collected for the eight cells analysed in the paper, and statistics for the sequence analysis.
| Cell | Input read pairs | Unique mapped pairs | Primary contacts | Final contacts* | Normal ligation % | Single read % | Trans % | Promiscuous ends | Mean redundancy |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 1,969,076 | 1,235,949 | 110,042 | 122,475 | 89.36 | 21.5 | 11.65 | 4342 | 7.88 |
| 2 | 1,621,648 | 944,140 | 65,636 | 84,129 | 92.29 | 21.7 | 6.30 | 2320 | 7.70 |
| 3§ | 1,937,061 | 1,326,834 | 32,534 | 37,604 | 68.67 | 14.0 | 22.66 | 1826 | 24.15 |
| 4 | 1,517,614 | 704,831 | 75,740 | 92,748 | 89.99 | 23.7 | 6.64 | 771 | 6.39 |
| 5 | 1,592,161 | 883,678 | 61,855 | 74,417 | 89.86 | 21.2 | 9.67 | 1833 | 10.12 |
| 6 | 1,493,430 | 643,721 | 60,334 | 75,352 | 92.21 | 21.7 | 5.21 | 699 | 7.70 |
| 7† | 4,796,232 | 1,191,086 | 46,438 | 55,792 | 80.60 | 16.5 | 5.51 | 1655 | 17.26 |
| 8† | 1,776,396 | 810,661 | 35,157 | 42,586 | 90.40 | 20.13 | 7.42 | 1423 | 16.00 |
Final contact counts include those with genome sequence mapping ambiguities that were resolved by initial 100 kb resolution genome structure calculations.
These cells have fluorescent image data.
Contacts in this cell were sequenced using the Nextera protocol.
Extended Data Table 2.
Summary of statistics from the genome structure calculation process for the eight cells analysed in the paper. The separate statistics for calculations involving 400 to 100 kb particles relate to the different stages of the hierarchical structure calculation protocol (see Supplementary Methods for details), with the last section representing the final, highest quality structures.
| Cell | 400 kb Particles | 200 kb Particles | 100 kb Particles (without ambiguous restraints) | 100 kb Particles (with ambiguous restraints) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RMSD* | NRest† | %V§ >3 | %V§ >4 | RMSD* | NRest† | %V§ >3 | %V§ >4 | RMSD* | NRest† | %V§ >3 | %V§ >4 | RMSD* | NRest† | %V§ >3 | %V§ >4 | |
| 1 | 0.13 | 26,453 | 2.94 | 0.76 | 2.32 | 43,150 | 1.29 | 0.08 | 0.56 | 71,729 | 2.81 | 0.57 | 0.41 | 74,945 | 2.29 | 0.22 |
| 2 | 0.29 | 22,838 | 0.16 | 0.00 | 0.50 | 35,244 | 0.29 | 0.00 | 0.88 | 47,598 | 1.04 | 0.15 | 0.63 | 56,159 | 0.74 | 0.13 |
| 3 | 0.24 | 17,944 | 0.81 | 0.28 | 0.51 | 21,049 | 1.66 | 0.08 | 0.89 | 22,822 | 4.82 | 1.36 | 0.70 | 25,952 | 2.87 | 0.42 |
| 4 | 0.33 | 25,126 | 2.35 | 0.68 | 0.58 | 37,594 | 1.26 | 0.14 | 1.03 | 50,516 | 2.49 | 0.94 | 0.74 | 60,420 | 1.14 | 0.17 |
| 5 | 0.32 | 23,062 | 1.18 | 0.30 | 0.65 | 34,103 | 0.86 | 0.10 | 1.08 | 44,298 | 2.12 | 0.73 | 0.86 | 51,824 | 1.24 | 0.21 |
| 6 | 0.38 | 25,084 | 0.60 | 0.10 | 0.69 | 35,526 | 0.70 | 0.06 | 1.09 | 44,687 | 0.99 | 0.22 | 0.91 | 53,710 | 0.74 | 0.17 |
| 7 | 0.81 | 18,301 | 0.07 | 0.04 | 1.26 | 26,585 | 0.03 | 0.00 | 1.91 | 33,588 | 0.11 | 0.01 | 1.75 | 39,257 | 0.13 | 0.02 |
| 8 | 0.49 | 15,877 | 0.00 | 0.00 | 0.93 | 22,956 | 0.00 | 0.00 | 1.52 | 28,083 | 0.02 | 0.02 | 1.26 | 32,482 | 0.03 | 0.02 |
The precision of structure ensembles in terms of mean pairwise all-particle RMSDs, in units of particle radii.
Restraint numbers include all experimental Hi-C contacts between non-adjacent particles, and thus increase as the particle size decreases. (NB – A single restraint may correspond to multiple Hi-C contacts, i.e. between the same two particle regions.)
The percentage of violated restraints was calculated for thresholds of 3.0 and 4.0 particle radii, respectively.
Supplementary Material
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Acknowledgements
We thank Andy Riddell for cell sorting, Peter Humphreys for confocal microscopy, Alex Peter Gunnarson for the density mapping software, the CRUK Cambridge Institute for DNA sequencing, Takashi Nagano and Peter Fraser for processing the initial haploid mES cells, and Wendy Dean, Stefan Schoenfelder and Stephen Wingett for helpful advice. We thank the Wellcome Trust (082010/Z/07/Z), the EC FP7 4DCellFate project (277899) and the MRC (MR/M010082/1) for financial support.
Footnotes
Data Availability Statement
The ChIP-seq, RNA-seq and Hi-C data, structures and images reported in this study have been made available at the Gene Expression Omnibus (GEO) repository under accession code GSE80280.
Author Contributions
DL, SB and YC developed the protocol and carried out imaging/Hi-C processing. TJS developed the software with assistance from LPA and KJW. AO’S-K, JC, MR and BH carried out the CHD4/MBD3 depletion experiments, associated RNA- and ChIP-seq, and created the mEos3.2-Halo tagged ES cell lines. ML and AW provided the initial samples of haploid mESCs. SFL, MP and DK designed and built the microscope. LM, MS and LDiC carried out ChIP- and RNA-Seq experiments, whilst AF, EB and BL carried out bioinformatics analysis. TJS and EDL designed experiments, analyzed the results and wrote the manuscript with contributions from all the other authors.
Author Information
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing interests.
References
- 1.Cremer T, et al. The 4D nucleome: Evidence for a dynamic nuclear landscape based on co-aligned active and inactive nuclear compartments. FEBS Lett. 2015;589:2931–2943. doi: 10.1016/j.febslet.2015.05.037. [DOI] [PubMed] [Google Scholar]
- 2.Bickmore WA, van Steensel B. Genome architecture: domain organization of interphase chromosomes. Cell. 2013;152:1270–1284. doi: 10.1016/j.cell.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nora EP, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sexton T, et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 7.Rao SS, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zuin J, et al. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc Natl Acad Sci U S A. 2014;111:996–1001. doi: 10.1073/pnas.1317788111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Phillips-Cremins JE, et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013;153:1281–1295. doi: 10.1016/j.cell.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalhor R, Tjong H, Jayathilaka N, Alber F, Chen L. Genome architectures revealed by tethered chromosome conformation capture and population-based modeling. Nature biotechnology. 2012;30:90–98. doi: 10.1038/nbt.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tjong H, et al. Population-based 3D genome structure analysis reveals driving forces in spatial genome organization. Proc Natl Acad Sci U S A. 2016;113:E1663–1672. doi: 10.1073/pnas.1512577113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naumova N, et al. Organization of the mitotic chromosome. Science. 2013;342:948–953. doi: 10.1126/science.1236083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foti R, et al. Nuclear Architecture Organized by Rif1 Underpins the Replication-Timing Program. Mol Cell. 2016;61:260–273. doi: 10.1016/j.molcel.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang S, et al. Spatial organization of chromatin domains and compartments in single chromosomes. Science. 2016;353:598–602. doi: 10.1126/science.aaf8084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Julienne H, Zoufir A, Audit B, Arneodo A. Human genome replication proceeds through four chromatin states. Plos Comput Biol. 2013;9:e1003233. doi: 10.1371/journal.pcbi.1003233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peric-Hupkes D, et al. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell. 2010;38:603–613. doi: 10.1016/j.molcel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meuleman W, et al. Constitutive nuclear lamina-genome interactions are highly conserved and associated with A/T-rich sequence. Genome research. 2013;23:270–280. doi: 10.1101/gr.141028.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Koningsbruggen S, et al. High-resolution whole-genome sequencing reveals that specific chromatin domains from most human chromosomes associate with nucleoli. Molecular biology of the cell. 2010;21:3735–3748. doi: 10.1091/mbc.E10-06-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kind J, et al. Single-cell dynamics of genome-nuclear lamina interactions. Cell. 2013;153:178–192. doi: 10.1016/j.cell.2013.02.028. [DOI] [PubMed] [Google Scholar]
- 20.Dunn SJ, Martello G, Yordanov B, Emmott S, Smith AG. Defining an essential transcription factor program for naive pluripotency. Science. 2014;344:1156–1160. doi: 10.1126/science.1248882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Therizols P, et al. Chromatin decondensation is sufficient to alter nuclear organization in embryonic stem cells. Science. 2014;346:1238–1242. doi: 10.1126/science.1259587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kruger AV, et al. Comprehensive single cell-resolution analysis of the role of chromatin regulators in early C. elegans embryogenesis. Dev Biol. 2015;398:153–162. doi: 10.1016/j.ydbio.2014.10.014. [DOI] [PubMed] [Google Scholar]
- 23.Sofueva S, et al. Cohesin-mediated interactions organize chromosomal domain architecture. EMBO J. 2013;32:3119–3129. doi: 10.1038/emboj.2013.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giorgetti L, et al. Predictive polymer modeling reveals coupled fluctuations in chromosome conformation and transcription. Cell. 2014;157:950–963. doi: 10.1016/j.cell.2014.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Naughton C, et al. Transcription forms and remodels supercoiling domains unfolding large-scale chromatin structures. Nat Struct Mol Biol. 2013;20:387–395. doi: 10.1038/nsmb.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kouzine F, et al. Transcription-dependent dynamic supercoiling is a short-range genomic force. Nat Struct Mol Biol. 2013;20:396–403. doi: 10.1038/nsmb.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo Y, et al. CRISPR Inversion of CTCF Sites Alters Genome Topology and Enhancer/Promoter Function. Cell. 2015;162:900–910. doi: 10.1016/j.cell.2015.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanborn AL, et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci U S A. 2015;112:E6456–6465. doi: 10.1073/pnas.1518552112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fudenberg G, et al. Formation of Chromosomal Domains by Loop Extrusion. Cell reports. 2016;15:2038–2049. doi: 10.1016/j.celrep.2016.04.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lengronne A, et al. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature. 2004;430:573–578. doi: 10.1038/nature02742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eagen KP, Hartl TA, Kornberg RD. Stable Chromosome Condensation Revealed by Chromosome Conformation Capture. Cell. 2015;163:934–946. doi: 10.1016/j.cell.2015.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhimulev IF, et al. Genetic organization of interphase chromosome bands and interbands in Drosophila melanogaster. PLoS One. 2014;9:e101631. doi: 10.1371/journal.pone.0101631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Minajigi A, et al. Chromosomes. A comprehensive Xist interactome reveals cohesin repulsion and an RNA-directed chromosome conformation. Science. 2015;349 doi: 10.1126/science.aab2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giorgetti L, et al. Structural organization of the inactive X chromosome in the mouse. Nature. 2016;535:575–579. doi: 10.1038/nature18589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Wit E, et al. The pluripotent genome in three dimensions is shaped around pluripotency factors. Nature. 2013;501:227–231. doi: 10.1038/nature12420. [DOI] [PubMed] [Google Scholar]
- 36.Wei Z, et al. Klf4 organizes long-range chromosomal interactions with the oct4 locus in reprogramming and pluripotency. Cell stem cell. 2013;13:36–47. doi: 10.1016/j.stem.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 37.Denholtz M, et al. Long-range chromatin contacts in embryonic stem cells reveal a role for pluripotency factors and polycomb proteins in genome organization. Cell stem cell. 2013;13:602–616. doi: 10.1016/j.stem.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reynolds N, et al. NuRD suppresses pluripotency gene expression to promote transcriptional heterogeneity and lineage commitment. Cell stem cell. 2012;10:583–594. doi: 10.1016/j.stem.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang W, et al. The Nucleosome Remodeling and Deacetylase Complex NuRD is built from preformed catalytically active sub-modules. J Mol Biol. 2016 doi: 10.1016/j.jmb.2016.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y, et al. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell. 2012;148:908–921. doi: 10.1016/j.cell.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murakami K, et al. NANOG alone induces germ cells in primed epiblast in vitro by activation of enhancers. Nature. 2016;529:403–407. doi: 10.1038/nature16480. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.