Abstract:
Previous studies of resting state functional connectivity have demonstrated that the default‐mode network (DMN) is negatively correlated with a set of brain regions commonly activated during goal‐directed tasks. However, the location and extent of anti‐correlations are inconsistent across different studies, which has been posited to result largely from differences in whether or not global signal regression (GSR) was applied as a pre‐processing step. Notably, coordinates of seed regions‐of‐interest defined within the posterior cingulate cortex (PCC)/precuneus, an area often employed to study functional connectivity of the DMN, have been inconsistent across studies. Taken together with recent observations that the DMN contains functionally heterogeneous subdivisions, it is presently unclear whether these seeds map to different DMN subnetworks, whose patterns of anti‐correlation may differ. If so, then seed location may be a non‐negligible factor that, in addition to differences in preprocessing steps, contributes to the inconsistencies reported among published studies regarding DMN correlations/anti‐correlations. In this study, they examined anti‐correlations of different subnetworks within the DMN during rest using both seed‐based and point process analyses, and discovered that: (1) the ventral branch of the DMN (vDMN) yielded significantly weaker anti‐correlations than that associated with the dorsal branch of the DMN (dDMN); (2) vDMN anti‐correlations introduced by GSR were distinct from dDMN anti‐correlations; (3) PCC/precuneus seeds employed by earlier studies mapped to different DMN subnetworks, which may explain some of the inconsistency (in addition to preprocessing steps) in the reported DMN anti‐correlations. Hum Brain Mapp 38:2454–2465, 2017. © 2017 Wiley Periodicals, Inc.
Keywords: DMN, anti‐correlation, seed‐dependence, resting state
INTRODUCTION
Functional magnetic resonance imaging (fMRI) has been widely applied to investigate the intrinsic architecture of the brain in the past two decades [for reviews, see Biswal, 2012; Chen and Glover, 2015; Fox and Raichle, 2007; Lee et al., 2013; Van den Heuvel and Pol, 2010]. Multiple resting state networks (RSNs), that is, sets of cortical regions having correlated spontaneous fluctuations in the absence of a task, have been consistently identified across different studies [Damoiseaux et al., 2006; Smith et al., 2009]. The default‐mode network (DMN), also referred to as the task‐negative network, is one such RSN that has received substantial attention [Andrews‐Hanna, 2012; Greicius et al., 2003]. Core regions of the DMN—including posterior cingulate cortex (PCC)/precuneus, medial prefrontal cortex (MPFC), ventral anterior cingulate, lateral parietal cortex, inferior temporal cortex, and parahippocampal cortex—are robustly deactivated during goal‐oriented tasks, while they are actively engaged when subjects are in a passive state of internal mentation [see Buckner et al., 2008; Raichle and Hyman, 2015 for reviews].
It has been reported that in the resting state, fluctuations in the DMN may be negatively correlated (anti‐correlated) with a set of regions commonly recruited in attention‐demanding tasks, for example, dorsolateral prefrontal cortex (DLPFC), supramarginal gyrus (SMG), posterior parietal cortex, and supplementary motor area (SMA) [Carbonell et al., 2014; Chai et al., 2012; Chang and Glover, 2009; Fox et al., 2005, 2009; Fransson, 2005; Greicius et al., 2003; Kelly et al., 2008; Uddin et al., 2009; Wong et al., 2012]. Despite much study, the functional significance of this observed anti‐correlation is not well understood. It has been conjectured to reflect the antagonistic or competitive relationship between an introspective, self‐oriented process and an extrospectively‐oriented, attentive mode [Buckner et al., 2008; Fox et al., 2005; Fransson, 2005; Greicius et al., 2003], echoing the opposed neural fluctuations in the two networks during attention‐demanding tasks. This hypothesis is supported by subsequent studies reporting that anti‐correlation between the two networks is linked with behavioral variability in goal‐directed tasks [Hampson et al., 2010; Keller et al., 2015; Kelly et al., 2008] and subjects' vigilance states—it decreases in states of reduced consciousness, such as sedation [Barttfeld et al., 2015; Boveroux et al., 2010], sleep deprivation [De Havas et al., 2012; Yeo et al., 2015], or non‐dual awareness (equal awareness of internal and external experience, [Josipovic et al., 2011]), but increases in focused‐attention meditation, which increases the competition between intrinsic and extrinsic systems [Josipovic et al., 2011].
However, anti‐correlated regions, as well as the extent of anti‐correlations, are not consistent across different studies, and such inconsistency has frequently been attributed to the distinct preprocessing step of whether or not Global Signal Regression (GSR) was employed [Anderson et al., 2011; Chai et al., 2012; Chang and Glover, 2009; Fox et al., 2005, 2009; Fransson, 2005; Jo et al., 2010; Murphy et al., 2009; Uddin et al., 2009; Weissenbacher et al., 2009]. Functional MRI signals are modulated by cardiac and respiratory processes, which may induce non‐neural signal synchrony extensively across cortex [Birn et al., 2006, 2008; Chang et al., 2009; De Munck et al., 2008; Shmueli et al., 2007; Wise et al., 2004]. Thus, correction techniques that can effectively reduce artificial positive correlations induced by physiological noise (or other non‐neural sources) may be critical for revealing true anti‐correlations with respect to DMN. As an alternative to physiological denoising, global signal nuisance regression has been employed to reduce fluctuations that affect large areas of the brain, with the assumption that global signals are largely of non‐neural origin and should therefore be eliminated [Fox et al., 2005, 2009; Fransson, 2005; Weissenbacher et al., 2009]. However, the validity of this correction technique is questionable because the global signal also contains spontaneous fluctuations from true neural mechanisms; furthermore, it forces the summation of each brain voxel's correlation value with a given seed to be non‐positive, introducing spurious negative correlations [Murphy et al., 2009; Saad et al., 2012]. In line with the discussions above, studies not performing rigorous noise correction or GSR may fail to observe robust anti‐correlations.
Interestingly, if we revisit the coordinates of posterior cingulate cortex (PCC)/precuneus—the most popular seed region employed in previous studies to demonstrate the correlations/anti‐correlations of the DMN—we observe that the seed locations are rather inconsistent across literature, independent of their different preprocessing steps. Such observations motivate consideration of whether the seeds employed by earlier studies map to distinct functional subdivisions of the DMN whose anti‐correlation patterns differ substantially, given the growing evidence that the DMN is functionally heterogeneous. With independent component analysis (ICA), several groups have revealed that the DMN splits into dorsal and ventral branches [Damoiseaux et al., 2008; Shirer et al., 2012], or more subdivisions depending on the specified component numbers [Damoiseaux et al., 2008, 2012; Littow et al., 2010; Shirer et al., 2012; Westlye et al., 2011]. Uddin et al. [2009] also observed heterogeneous subdivisions within the DMN by placing seeds in MPFC and PCC, respectively. Using graph analysis combined with hierarchical clustering, [Andrews‐Hanna, 2012; Andrews‐Hanna et al., 2010] further identified the structure of DMN as a central hub (consisting of the anterior MPFC and PCC) that mediates personally‐significant affective information and two subsystems—a dorsal MPFC subsystem and a medial temporal lobe subsystem, which are involved in introspection about mental states and memory‐based reconstruction, respectively.
This topic may be of critical importance in that if divergent subnetworks of the DMN exhibit distinct anti‐correlation patterns, the PCC/precuneus seed coordinate is therefore a key factor (in addition to de‐noising steps) determining the observation of DMN anti‐correlations. Prior conclusions regarding anti‐correlation between the DMN and task‐positive regions and its dependence on preprocessing steps may thus need to be reassessed in this light. Furthermore, given that those early studies examining the impact of GSR may have been pinpointing different functional subnetworks, this factor ought to be considered in future studies wishing to examine DMN anti‐correlations using a PCC/precuneus seed coordinate from previous studies.
The aim of the present study is, therefore, to examine the anti‐correlated patterns associated with DMN subnetworks during resting state, so as to better understand how disparate anti‐correlation findings can be driven by the seed location as well as other common pre‐processing choices. Specifically, we first parcellated the PCC/precuneus region into separate units based on each voxel's functional connectivity with the rest of the brain, then focused on the seeds associated with the DMN to investigate: (1) whether the anti‐correlation patterns differ significantly across DMN subnetworks both with and without GSR; (2) whether inconsistent reports pertaining to anti‐correlations can be partly explained by the disparate PCC/precuneus seed locations and/or by the different preprocessing steps employed.
METHODS
Subjects and Experiments
Twenty healthy subjects (10 females), aged 31 ± 10 years and recruited from the Stanford community, participated in this study. All subjects provided written informed consent, using a protocol approved by the Stanford Institutional Review Board.
Each subject underwent an 8‐min rest scan for which they were instructed to remain awake, devoid of systematic thinking, and keep eyes closed. Data from 17 out of the 20 subjects were previously analyzed and reported in Chen et al. [2015], a study exploring a different scientific question.
FMRI data were collected at a 3T scanner with an 8‐channel radio frequency coil (GE Discovery 750, Milwaukee, WI). Thirty‐one oblique axial slices were acquired with 4‐mm slice thickness, no gap between slices. T2‐weighted fast spin echo structural images (TR = 3,000 ms, TE = 68 ms, ETL = 12, FOV = 22 cm, matrix 192 × 256) were acquired for anatomical reference. A gradient echo spiral‐in/out pulse sequence [Glover and Law, 2001] was used for T2*‐weighted functional imaging (TR = 2,000 ms, TE = 30 ms, flip angle = 77°, matrix 64 × 64, FOV = 22 cm, same slice prescription as the anatomical image, 240 time frames). Subjects' motions were minimized with a bite bar. Respiration and cardiac (pulse oximetry) data were recorded using the scanner's built‐in photoplethysmograph placed on a finger of the left hand (with the sampling rate of 100 Hz) and a pneumatic belt strapped around the upper abdomen (with the sampling rate of 25 Hz), respectively.
Data Preprocessing
Datasets were preprocessed using custom C and Matlab routines. Standard preprocessing included slice‐time correction, physiological noise correction with both RETROICOR [Glover et al., 2000] and RVHRCOR [Chang et al., 2009], and nuisance regression of scan drifts (linear and quadratic trends), six head motion parameters (see below). White matter and cerebrospinal fluid (CSF) signals in 3‐mm radius spheres centered at MNI (26, −12, 35) and (19, −33, 18) as used in Chang and Glover [2009] were extracted and renormalized to each subject's native space using SPM8 (Wellcome Department of Cognitive Neurology, London). Datasets were further spatially smoothed using a 3D Gaussian kernel (FWHM = 4 mm) and normalized to the MNI standard space (extrapolated to voxel dimension 2 mm × 2 mm ×2 mm) using SPM8. To avoid removing potentially meaningful fluctuations greater than 0.1 Hz, no low‐pass filtering was performed.
Analysis of Motion and Physiological Recordings
Motion parameters were estimated using methods described in Friston et al. [1996]. The maximum peak‐to‐peak excursion and root‐mean‐square (RMS) fluctuation for six motion parameters (3 translational and 3 rotational axes) were calculated. Rotations were converted to worst‐case translations by multiplying with an average head radius 68 mm [Thomason and Glover, 2008]. Maximum values (peak‐to peak and RMS excursions) over the 6 axes of motion are reported.
Although physiological nuisance regressors (derived from RETROICOR and RVHRCOR) were projected out of the data, residual effects may still remain due to imperfection of the models; moreover, physiological fluctuations may also link with BOLD signals in neural‐activity‐related manners [Bernardi et al., 1990; Chang et al., 2013b; Critchley et al., 2003; Force, 1996; Napadow et al., 2008]. Thus physiological variables were also estimated and are reported, including averaged heart rate (beat/min) and respiratory rate (cycle/min), the standard deviation of heart rates (estimated across sequences of 6 s long sliding windows centered at each 2‐s TR time point), and the mean of “respiratory variation” time series (the RMS amplitude of the respiration waveform across a 6 s sliding window centered at each TR time point, normalized to fractions of the maximum abdominal belt expansion).
Extraction of PCC/Precuneus Seeds Associated with the DMN
As PCC/precuneus encompasses a considerable mass of voxels, with connections to several neural circuitries other than the DMN, for example, the visual or cognitive networks [Bzdok et al., 2015; Cauda et al., 2010; Leech et al., 2012, 2011; Margulies et al., 2009; Vogt et al., 2006; Yang et al., 2014; Zhang et al., 2014], we first identified candidate DMN related PCC/precuneus sub‐regions via clustering analysis and then focused the ensuing analyses on these sub‐units only.
Voxels within the left/right PCC and precuneus delineated by the AAL atlas (∼7,500 voxels in total at 2 mm isotropic resolution) were parcellated into four functional units based on their functional connectivity with the averaged time series within each of the remaining gray matter ROIs (112 ROIs derived from AAL atlas excluding PCC/precuneus). The parcellation process was first performed at the individual‐subject level using K‐means clustering and then combined to form a group‐level result via normalized‐cut method (http://www.cis.upenn.edu/~jshi/software/).
Cluster centroids of two of the four resolved functional units were considered to be DMN related and chosen as the seeds for further comparison of PCC/precuneus positive/negative correlations. The MNI coordinates of the selected two seeds (denoted as SEED1 and SEED2, 6 mm radius spheres) are [0, −60, 46] and [0, −50, 26], respectively. See Supporting Information S1 for more details regarding clustering and seed selections.
Functional Connectivity with Respect to SEED1/SEED2
The spatial patterns of functional connectivity with SEED1 and SEED2 derived above were generated by computing their linear Pearson correlation with voxels across the brain. The analysis of functional connectivity was also performed on the dataset (1) without physiological correction (i.e., excluding RETROICOR and RVHRCOR), and (2) using the global signal (averaged signal across all brain voxels) regression (instead of RETROICOR and RVHRCOR for physiological noise correction) to evaluate the dependence of network patterns on different preprocessing steps. Other de‐noising steps described in section “Data Preprocessing” remain unchanged.
Of note, although SEED1 and SEED2 can be dissociated by virtue of belonging to different spatial clusters, their time series still exhibited substantial synchrony (Pearson correlation coefficients r = 0.60 ± 0.19 across the subjects). Thus, we performed an additional point‐process analysis [Tagliazucchi et al., 2012] on the dataset preprocessed by steps described in section “Data Preprocessing” to investigate the co‐activation patterns (CAPs, [Liu and Duyn, 2013]) specifically associated with SEED1 or SEED2 (i.e., at time frames when the two regions were unsynchronized with each other). See Supporting Information S2 for descriptions of detailed analysis and statistical testing.
Functional Connectivity of PCC/Precuneus Seeds Reported by Previous Studies
We selected several highly cited studies that had focused on DMN anti‐correlations and the effects of GSR upon them. We divided these studies into two groups: the “NonNeg” group [Carbonell et al., 2014; Fox et al., 2009; Murphy et al., 2009] only observed anti‐correlations when applying GSR, while the “Neg” group [Fox et al., 2009; Fransson, 2005; Uddin et al., 2009] observed DMN anti‐correlations even without global normalization or GSR.
To examine whether inconsistencies in the anti‐correlations reported by these studies can be partially attributable to different seed locations, we employed identical seed‐based correlation analysis (see section “Functional Connectivity with Respect to SEED1/SEED2” above) in our datasets.
RESULTS
Motion and Physiological Data
The peak‐to‐peak and RMS motion excursions across subjects were 0.39 ± 0.19 mm, 0.14 ± 0.08 mm (mean ± SD), respectively. The mean and standard deviation of heart rate were 62.8 ± 9.7 beat/min and 2.99 ± 1.16 beat/min. The averaged respiratory rate was 17.4 ± 2.3 cycle/min. The variation of respiratory volume was 15.6% ± 5.2%. See Supporting Information Table S1 for detailed information for each subject.
Positive/Negative Correlations with SEED1/SEED2
Figure 1 shows the t‐score map results of a group‐level analysis of functional connectivity with respect to SEED1/SEED2 and three types of preprocessing.
Figure 1.
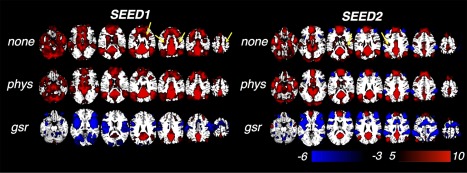
Regions positively/negatively correlated with two PCC seeds (group‐level t map). “none”: preprocessing without correction of physiological noise (standard preprocessing in section “Data Preprocessing” without model‐based physiological noise correction (RETROICOR, RVHRCOR)); “phys”: standard preprocessing in section “Data Preprocessing”; “gsr”: global signal regression without any additional physiological noise correction (“none” + global signal regression). Yellow arrows highlight reduced positive correlations after “phys” compared with “none”. [Color figure can be viewed at http://wileyonlinelibrary.com]
Focusing first on the results with model‐based physiological noise correction (RETROICOR and RVHRCOR, “phys”), regions positively correlated with SEED1/SEED2 both encompassed the central nodes of the DMN, including PCC/precuneus, MPFC, bilateral parietal cortex, hippocampus, parahippocampus, inferior temporal cortex, but varied in the correlation strength and spatial extents. SEED1‐DMN (regions significantly correlated with SEED1) exhibited stronger correlation with thalamus and DLPFC, while SEED2‐DMN (regions significantly correlated with SEED2) was more connected with the superior MPFC. Contrasting thresholded spatial patterns of SEED1‐/SEED2‐DMN (focusing on positive correlation only) to the functional network atlas reported by the Stanford FINDlab (http://findlab.stanford.edu/functional_ROIs.html, derived by ICA without GSR, [Shirer et al., 2012]), we note the strong correspondence between SEED1‐DMN and the ventral DMN (vDMN), and the correspondence between SEED2‐DMN and the dorsal DMN (dDMN), respectively (see Fig. 2).
Figure 2.
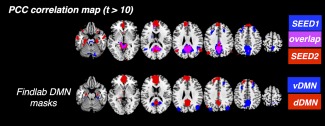
Correspondence between the spatial patterns of SEED1‐/SEED2‐DMN and the v/dDMN reported by Stanford FINDlab. [Color figure can be viewed at http://wileyonlinelibrary.com]
The spatial extents of regions negatively correlated with SEED2 were significantly larger than those correlated with SEED1 regardless of the preprocessing steps, which is quantitatively reflected in Figure 3 and qualitatively in Figure 1. After physiological noise correction, negative correlation was only barely observable with SEED1; by contrast, with SEED2 as the seed, typical task‐positive regions (including bilateral insula, DLPFC, SMG) were present even without any physiological noise correction (FDR corrected, P < 0.05).
Figure 3.
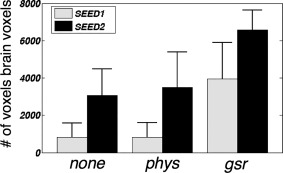
Number of voxels negatively correlated with SEED1/SEED2 (correlation coefficient r < −0.2) under different preprocessing conditions. P values from paired‐t tests (# of voxels negatively correlated with SEED1 vs. SEED2) under “none/phys/gsr” are 1.8 * 10−6, 1.6 * 10−6, and 1.3 * 10−5, respectively.
In line with previous literature, different preprocessing procedures also significantly altered connectivity patterns. Compared with “none,” model‐based physiological noise correction (“phys”) diminished the spatial extent of positive correlations (highlighted by yellow arrows in Fig. 1) while the spatial extent of negative correlations was slightly magnified. Similar trends became more prominent after GSR (“gsr”). Significantly reduced positive correlations were observed with both seeds. Bilateral temporal cortex and parahippocampal cortex were not correlated with either seed in the displayed t‐score range, and MPFC—a core node within the DMN—was no longer strongly correlated with SEED1.
GSR also produced extensive negative correlations with both seeds. Of note, the regions showing anti‐correlated activity with SEED1/SEED2 were not identical: negative correlation associated with SEED1 was stronger in the occipital cortex, rolandic operculum, superior temporal lobes, and pre‐/post‐central gyrus relative to that associated with SEED2, but was weaker in DLPFC, SMG, mid‐cingulate cortex, and superior parietal cortex.
CAPs associated with SEED1 or SEED2 (termed below as SEED1/SEED2‐CAP respectively) mirrored the correlation patterns shown in Figure 1 to a certain extent. Comparing regions with significant (FDR corrected, P < 0.01) positive amplitudes in SEED1‐CAP to SEED1‐DMN, the MPFC decoupled with PCC/precuneus, DLPFC and executive control network appeared in SEED1‐CAP. By contrast, SEED2‐CAP closely resembled SEED2‐DMN. Regions exhibiting significant negative amplitudes in SEED2‐CAP were akin to the anti‐correlated areas generated by linear correlation with the whole scan time points. No regions with clear structures survived the statistical threshold (FDR corrected, P < 0.05) for SEED2‐CAP. See Supporting Information S2 for results and figures.
Previous Studies: Seed‐Dependence of the Correlation/Anti‐Correlation Patterns with PCC/Precuneus
Figure 4 shows regions exhibiting strong positive and negative correlations using seed coordinates reported in previous literature (Table 1) applied to our data, for which only model‐based physiological noise correction (section “Data Preprocessing”) was applied. In general, seed locations from the “NonNeg” group, that is, studies that did not observe significant DMN anti‐correlations without GSR (a, b), yielded diminished anti‐correlations (Fig. 4A blue regions) compared with those from the “Neg” group (c–e), that is, studies reporting significant anti‐correlations (Table 1, quantitatively reflected in Fig. 4C). Spatial similarities (calculated as the linear Pearson correlation between gray matter intensities) between the network patterns generated by different seeds and SEED1‐/SEED2‐DMN (the “phys” result in Fig. 1) are shown in Fig. 4B. DMNs from the “NonNeg” group generally displayed lower spatial correlations with the SEED2‐DMN (dDMN) compared with the “Neg” group.
Figure 4.
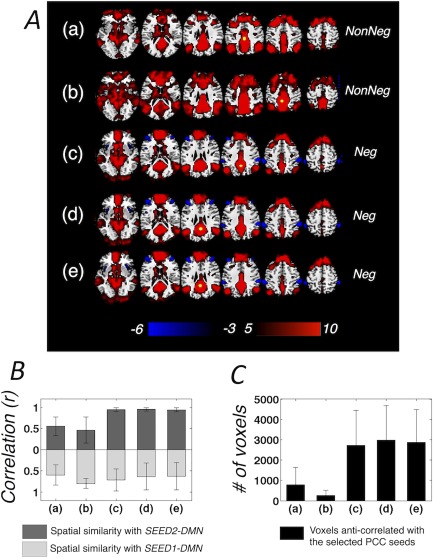
(A) Regions positively/negatively correlated with PCC seeds reported by previous literature (Table I, group t‐score map), seed locations are highlighted in yellow, only model‐based physiological de‐noising is applied; (B) Spatial similarity (linear Pearson correlation between gray matter voxel intensity) between the DMN patterns derived in (A) and SEED1‐/SEED2‐DMN, mean and standard deviation estimated across all the subjects (correlation values are positive moving away from 0 in both directions); (C) Number of voxels anti‐correlated with PCC (correlation coefficient r < −0.2), mean and standard deviation estimated across all the subjects. [Color figure can be viewed at http://wileyonlinelibrary.com]
Table 1.
PCC coordinates reported in previous literature and examined in the present study
| Group | ID | Study | Cluster centroid (mm) | |||||
|---|---|---|---|---|---|---|---|---|
| Talairach | MNI | |||||||
| x | y | z | x | y | z | |||
| NonNeg | (a)* | Fox et al., 2009 1 | −2 | −36 | 37 | 0 | −33 | 40 |
| (b)** | Murphy et al., 2009 | −5 | −49 | 40 | −4 | −47 | 45 | |
| Neg | (c) | Fransson, 2005 | 0 | −56 | 30 | 2 | −55 | 34 |
| (d) | Fox et al., 2009 2 | 0 | −52 | 27 | 2 | −51 | 31 | |
| (e) | Uddin et al., 2009 | −2 | −51 | 27 | −1 | −50 | 31 | |
“NonNeg” denotes studies not reporting significant DMN anti‐correlations without GSR; while “Neg” denotes studies reporting significant anti‐correlations even without GSR. Conversion between Talairach coordinate system and the MNI coordinate system was implemented with the transformation proposed in [Lancaster et al., 2007], which was demonstrated to effectively reduce the bias between Talairach/MNI coordinate disparity for analyses using the ICBM‐152 template).
*(a) Talairach ([−2 −36 37]) was first reported in [Fox et al., 2005] as the peak locus for intrinsically defined anti‐correlated networks [identified by a conjunction analysis of the correlation maps with respect to three seeds within the task negative network and three seeds within the task‐positive network (TPN)]. This coordinate was examined later in Fox et al. [2009] and Carbonell et al. [2014], both studies were not able to resolve significant anti‐correlations without GSR [Fox et al., 2009] or post‐hoc corrections that eliminate spurious correlations [Carbonell et al., 2014].
**(b) Talairach ([−5 −49 40]) was used in Fox et al. [2005] to yield significant anti‐correlations between the TPN and task negative network after GSR. The seed was tested in Murphy et al. [2009], and failed to observe significant anti‐correlations without GSR. Anderson et al. [2011] also used the seed reported in Fox et al. [2005], and was not able to yield significant anti‐correlations. However, the MNI coordinate of the PCC/precuneus seed reported by this study was [−5, −52, 40], differing from both (a)* and (b)** here. We posit this distinction to result from inconsistent “MNI—Talairach” conversion algorithms.
In line with the model‐based physiological corrections, after GSR (Fig. 5), the spatial patterns of anti‐correlations in the “NonNeg” group exhibited closer resemblance to anti‐correlated regions with respect to SEED1 (Fig. 1); whereas those of the “Neg” group are more similar to the negative networks of SEED2 (Fig. 1).
Figure 5.
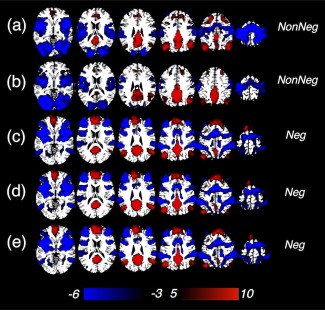
Regions positively/negatively correlated with PCC seeds reported by previous literature (Table I, group t‐score map), with GSR. [Color figure can be viewed at http://wileyonlinelibrary.com]
Considering that the analyses in the examined studies were mainly conducted in the Talairach coordinate system, one can ask whether our findings (normalized to the MNI template) can be readily compared with results in previous literature, as the conversion of seed coordinates from Talairach to MNI system is not definitive due to the significantly different brain shapes. To address these concerns, identical analyses were also performed in the Talairach coordinate system, and the conclusions were unaltered (see Supporting Information S3 for results and detailed analyses).
DISCUSSION
Positive/Negative Correlations with DMN Subnetworks
Comparing the spatial patterns of SEED1‐/SEED2‐DMN in our data with the DMN subnetworks resolved by other studies using ICA, it appears that the SEED1‐DMN corresponds to the ventral DMN reported by [Damoiseaux et al., 2012; Shirer et al., 2012], and posterior DMN in [Damoiseaux et al., 2008]; whereas SEED2‐DMN corresponds to the dorsal DMN reported by [Shirer et al., 2012], and resembles more closely the anterior component reported in [Damoiseaux et al., 2008, 2012]. The resolved network patterns of the two DMN branches partially overlapped with the subnetworks reported in Andrews‐Hanna et al. [2010] (ventral DMN vs. dorsal MPFC subnetwork, and dorsal DMN vs. medial temporal lobe subnetwork, respectively) but were not identical, which may be attributable to disparate scan conditions and computational techniques (instead of using seed‐based correlation or spatial ICA, graph‐analytic techniques on rest data and explicit task activation were employed to delineate DMN subnetworks in Andrews‐Hanna et al. [2010]).
SEED1 and SEED2 yielded disparate patterns of negative correlations. As reflected in Figure 1, and quantified in Figure 3, the spatial extents of negative correlations with SEED2 were significantly larger than with SEED1, regardless of the preprocessing step. Linking the correspondence between seed locations and the subnetworks of DMN, it appears that, during rest, the dDMN exhibits more robust anti‐correlations than the vDMN. Such heterogeneous anti‐correlations within the DMN have also been reported by a very recent study (Dixon et al., [2017]) that examines DMN subnetworks alternatively identified by Andrews‐Hanna et al. [2010]: negative correlations between the central core (PCC and MPFC) and dorsal attention network are stronger than both the dorsal MPFC and medial temporal lobe subsystems.
In line with the findings of previous studies, DMN anti‐correlations were sensitive to the use of GSR [Anderson et al., 2011; Carbonell et al., 2014; Chang and Glover, 2009; Fox et al., 2009; Murphy et al., 2009; Weissenbacher et al., 2009]. In contrast to cases of no correction or model‐based physiological noise correction, a wide range of negative correlations were present after GSR (Figs. 1, 3, and 5), with partially overlapping but distinct spatial patterns in SEED1 and SEED2 anti‐correlations. The influence of GSR on anti‐correlations and broader functional connectivity observations has been among the most controversial topics in functional connectivity data analysis. The advantages and pitfalls of GSR, as well as its alternatives, have been carefully reviewed in a recent consensus article [Murphy and Fox, 2016]. Thus, the present investigation does not aim to debate the effects of GSR on anti‐correlations, but rather, to emphasize a point largely omitted by earlier discussion: that the DMN is heterogeneous with respect to its anti‐correlated patterns, both with and without GSR.
A distinction between our study and earlier studies on DMN anti‐correlations is that we have utilized the full spectrum of time series without performing additional temporal filtering, to avoid eliminating potentially meaningful fluctuations greater than 0.1 Hz. However, we have also re‐analyzed the data with the inclusion of less than 0.1 Hz low‐pass filtering, and the results are presented in Supporting Information Figure S4. In concordance with observations in [Chang and Glover, 2009], low‐pass filtering slightly diminished the extent and magnitude of positive correlations but enhanced the extent and magnitude of negative correlations when preprocessed both with and without model based physiological noise correction, and similar trends appeared in most brain regions following GSR. However, the additional effects exerted by temporal filtering were very minor compared with those resulting from seed position and de‐noising steps.
Results of the point‐process analyses corroborated functional connectivity observations that SEED1/SEED2 have distinct DMN correlations/anti‐correlations. Particularly, regions showing significant negative (FDR corrected, P < 0.05) amplitudes in SEED2‐CAP well resembled SEED2‐DMN and common TPN regions reported in the literature while no regions demonstrating significant negative amplitudes in SEED1‐CAP under identical statistical threshold. These results, taken together, suggest the importance of seed locations in examining correlations and anti‐correlations with the DMN.
Seed Dependence of the DMN Anti‐Correlations in Previous Literature
As has been hypothesized, the spatial patterns of anti‐correlations produced by PCC/precuneus coordinates reported in previous studies were not consistent with each other (see Figs. 4 and 5). If we overlay the examined seeds on top of regions significantly correlated with SEED1 or SEED2 (Fig. 6), we can observe that the “Neg” group seeds lie inside the SEED2 network; while the “NonNeg” group seeds are located within the transition zone of SEED1/SEED2 network. Results presented here suggest that seed coordinates used in these studies may have been placed in different functional divisions of PCC/precuneus and the DMN.
Figure 6.
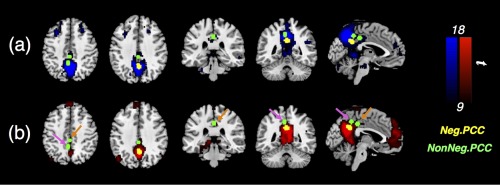
Different PCC seeds overlaid on regions significantly correlated with SEED1 [(a), blue]/SEED2 [(b), red]. The seeds used in [Murphy et al., 2009] and [Fox et al., 2009]1 are highlighted with magenta and orange arrows, respectively. [Color figure can be viewed at http://wileyonlinelibrary.com]
Of course, it may be possible that our findings cannot replicate those of prior studies due to differences in acquisition, recruited participants, and methods for analysis of correlation. To mitigate such concerns, we performed identical analyses on the resting‐state scans from a separate cohort of subjects (same acquisition environment as the present datasets) as a replication analysis and also examined functional connectivity with respect to different PCC/precuneus coordinates using the online platform Neurosynth (http://www.neurosynth.org/locations/), which computes and displays resting‐state functional connectivity for an arbitrary seed region using a sample of 1,000 subjects [Buckner et al., 2011; Choi et al., 2012; Yeo et al., 2011]. The seed‐dependence of the network patterns resulting from various preprocessing steps were remarkably similar, implying that choosing accurate seed locations is an important consideration when attempting to resolve the DMN anti‐correlations and correlations (see Supporting Information S5,6 for data descriptions, analyses, and results).
We also compared the DMN anti‐correlations in the present datasets to those network patterns reported by the original literature (see Table 1 and the associated comments for related studies). Observations in Figures 5 and 6 (with less stringent thresholds) well replicated prior results (the “NonNeg” group: Figure 1C and Supporting Information Figure 5 of Fox et al. [2009]1, Figure 6A of Carbonell et al. [2014]; the “Neg” group: Figure 3 of Fransson [2005], Supporting Information Figure 6 of Fox et al. [2009]2, and Figure 2 of Uddin et al. [2009]). As one discrepancy from our findings, Fox et al. used Talairach seed [−5 −49 40] to generate PCC correlations/anti‐correlations, finding that the reported antagonistic networks (Figs. 1 and 3 of [Fox et al., 2005]) resemble SEED2‐DMN/dDMN and its anti‐correlated regions instead of SEED1‐DMN/vDMN as we find here (Figs. 1 and 6b). This discrepancy may be due to the fact that Figure 1 of Fox et al. [2005] shows the result of a single subject, which may circumvent problems associated with registration/normalization at the group level, and Figure 3 in Fox et al. is produced by a conjunction analysis of correlations with respect to six seeds instead of the chosen PCC/precuneus seed alone. Murphy et al. used the identical seed to generate DMN anti‐correlations. Visually inspecting regions positively correlated with this seed (Figure 6 of Murphy et al. [2009]), they appear closer to our SEED1‐DMN/vDMN and Figure 5b, but notably, the negative network they obtained after GSR is closer to that associated with SEED2‐DMN/dDMN.
While choosing proper seed locations is critical, it may not be the only factor in revealing significant DMN anti‐correlations without GSR. For example, in the supplementary dataset, none of the PCC/precuneus seeds revealed significant anti‐correlations at the group level (Supporting Information Fig. S5.2.1, FDR corrected, P < 0.05) although the extents of anti‐correlations exhibited apparent patterns of seed‐dependence (Supporting Information Figs. S5.2.2 and S5.2.5). Several studies have reported time‐varying correlation patterns between the DMN and TPN across the duration of a single resting‐state scan, and suggested that the observed correlations/anti‐correlations with respect to PCC may only reflect the averaged pattern of several functionally distinct processes [Chang and Glover, 2010; Chen et al., 2015; Liu et al., 2013; Liu and Duyn, 2013; Smith et al., 2012; Yang et al., 2014; Karahanoglu et al., 2015; Dixon et al., 2017]. Thus, the relative portion of a resting‐state scan that is spent engaging in cognitive processes that are accompanied by negative interactions between the two networks, compared with that spent in processes that do not result in anti‐correlations may be an additional factor (beyond preprocessing and seed location) in determining whether salient anti‐correlation can be observed across a single scan session. Moreover, since the antagonistic relationship between DMN and TPN weakens under increased drowsiness, as evidenced by research on sleep deprivation and vigilance fluctuation [Chang et al., 2013a; De Havas et al., 2012; Wang et al., 2016; Yeo et al., 2015], the overall levels of arousal across scans and subjects ought to be considered as well. As such, mechanisms underlying inconsistent reports on DMN anti‐correlations are not yet conclusive—subjects' on‐going cognitive process, arousal levels, as well as the de‐noising steps in data analyses, all affect DMN anti‐correlations, but we can conclude that different seed locations contribute at least partly to those observations.
CONCLUSIONS
The major conclusions from our study are three‐fold. First, as revealed by both seed‐based correlation and point process analyses, the dDMN resulted in more prominent anti‐correlations compared with the vDMN in the resting state, which to our knowledge has not been described previously. Second, GSR induced overlapping but distinct anti‐correlations with respect to dDMN and vDMN. Third, the meta‐analysis of earlier studies suggested that the PCC/precuneus seeds used in different studies fell within different functional branches of the DMN, which, in addition to differences in pre‐processing, can account for the inconsistent observations of DMN anti‐correlations.
Supporting information
Supporting Information
ACKNOWLEDGMENTS
The authors gratefully acknowledge Anna Milazzo, Collin Price, and William Shirer for assistance with the Supporting Information data.
REFERENCES
- Anderson JS, Druzgal TJ, Lopez‐Larson M, Jeong EK, Desai K, Yurgelun‐Todd D (2011): Network anticorrelations, global regression, and phase‐shifted soft tissue correction. Hum Brain Mapp 32:919–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews‐Hanna JR (2012): The brain's default network and its adaptive role in internal mentation. Neuroscientist 18:251–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews‐Hanna JR, Reidler JS, Sepulcre J, Poulin R, Buckner RL (2010): Functional‐anatomic fractionation of the brain's default network. Neuron 65:550–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barttfeld P, Uhrig L, Sitt J, Sigman M, Jarraya B, Dehaene S (2015): Signature of consciousness in the dynamics of resting‐state brain activity (vol 112, pg 887, 2015). Proc Natl Acad Sci U S A 112:E5219–E5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi L, Salvucci F, Suardi R, Solda PL, Calciati A, Perlini S, Falcone C, Ricciardi L (1990): Evidence for an intrinsic mechanism regulating heart rate variability in the transplanted and the intact heart during submaximal dynamic exercise?. Cardiovasc Res 24:969–981. [DOI] [PubMed] [Google Scholar]
- Birn RM, Diamond JB, Smith MA, Bandettini PA (2006): Separating respiratory‐variation‐related fluctuations from neuronal‐activity‐related fluctuations in fMRI. Neuroimage 31:1536–1548. [DOI] [PubMed] [Google Scholar]
- Birn RM, Smith MA, Jones TB, Bandettini PA (2008): The respiration response function: The temporal dynamics of fMRI signal fluctuations related to changes in respiration. Neuroimage 40:644–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal BB (2012): Resting state fMRI: A personal history. Neuroimage 62:938–944. [DOI] [PubMed] [Google Scholar]
- Boveroux P, Vanhaudenhuyse A, Bruno MA, Noirhomme Q, Lauwick S, Luxen A, Degueldre C, Plenevaux A, Schnakers C, Phillips C, Brichant JF, Bonhomme V, Maquet P, Greicius MD, Laureys S, Boly M (2010): Breakdown of within‐ and between‐network resting state functional magnetic resonance imaging connectivity during propofol‐induced loss of consciousness. Anesthesiology 113:1038–1053. [DOI] [PubMed] [Google Scholar]
- Buckner RL, Krienen FM, Castellanos A, Diaz JC, Yeo BT (2011): The organization of the human cerebellum estimated by intrinsic functional connectivity. J Neurophysiol 106:2322–2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner RL, Andrews‐Hanna JR, Schacter DL (2008): The brain's default network: Anatomy, function, and relevance to disease. Ann N Y Acad Sci 1124:1–38. [DOI] [PubMed] [Google Scholar]
- Bzdok D, Heeger A, Langner R, Laird A, Fox P, Palomero‐Gallagher N, Vogt B, Zilles K, Eickhoff S (2015): Subspecialization in the human posterior medial cortex. Neuroimage 106:55–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonell F, Bellec P, Shmuel A (2014): Quantification of the impact of a confounding variable on functional connectivity confirms anti‐correlated networks in the resting‐state. Neuroimage 86:343–353. [DOI] [PubMed] [Google Scholar]
- Cauda F, Geminiani G, D'Agata F, Sacco K, Duca S, Bagshaw AP, Cavanna AE (2010): Functional connectivity of the posteromedial cortex. PLoS One 5: e13107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai XJ, Castanon AN, Ongur D, Whitfield‐Gabrieli S (2012): Anticorrelations in resting state networks without global signal regression. Neuroimage 59:1420–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Glover GH (2009): Effects of model‐based physiological noise correction on default mode network anti‐correlations and correlations. Neuroimage 47:1448–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Glover GH (2010): Time‐frequency dynamics of resting‐state brain connectivity measured with fMRI. Neuroimage 50:81–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Cunningham JP, Glover GH (2009): Influence of heart rate on the BOLD signal: The cardiac response function. NeuroImage 44:857–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Liu Z, Chen M, Liu X, Duyn J (2013a): EEG correlates of time‐varying BOLD functional connectivity. Neuroimage 72:227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Metzger CD, Glover GH, Duyn JH, Heinze HJ, Walter M (2013b): Association between heart rate variability and fluctuations in resting‐state functional connectivity. NeuroImage 68:93–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Glover G (2015): Functional magnetic resonance imaging methods. Neuropsychol Rev 25:289–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Chang C, Greicius M, Glover G (2015): Introducing co‐activation pattern metrics to quantify spontaneous brain network dynamics. Neuroimage 111:476–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi EY, Yeo BT, Buckner RL (2012): The organization of the human striatum estimated by intrinsic functional connectivity. J Neurophysiol 108:2242–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Critchley HD, Mathias CJ, Josephs O, O'Doherty J, Zanini S, Dewar BK, Cipolotti L, Shallice T, Dolan RJ (2003): Human cingulate cortex and autonomic control: Converging neuroimaging and clinical evidence. Brain 126:2139–2152. [DOI] [PubMed] [Google Scholar]
- Damoiseaux JS, Rombouts SA, Barkhof F, Scheltens P, Stam CJ, Smith SM, Beckmann CF (2006): Consistent resting‐state networks across healthy subjects. Proc Natl Acad Sci U S A 103:13848–13853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damoiseaux JS, Beckmann CF, Arigita EJ, Barkhof F, Scheltens P, Stam CJ, Smith SM, Rombouts SA (2008): Reduced resting‐state brain activity in the “default network” in normal aging. Cereb Cortex 18:1856–1864. [DOI] [PubMed] [Google Scholar]
- Damoiseaux JS, Seeley WW, Zhou J, Shirer WR, Coppola G, Karydas A, Rosen HJ, Miller BL, Kramer JH, Greicius MD, Alzheimer's Disease Neuroimaging Investigators (2012): Gender modulates the APOE epsilon4 effect in healthy older adults: Convergent evidence from functional brain connectivity and spinal fluid tau levels. J Neurosci 32:8254–8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Havas JA, Parimal S, Soon CS, Chee MW (2012): Sleep deprivation reduces default mode network connectivity and anti‐correlation during rest and task performance. Neuroimage 59:1745–1751. [DOI] [PubMed] [Google Scholar]
- De Munck JC, Goncalves SI, Faes TJ, Kuijer JP, Pouwels PJ, Heethaar RM, Lopes da Silva FH (2008): A study of the brain's resting state based on alpha band power, heart rate and fMRI. Neuroimage 42:112–121. [DOI] [PubMed] [Google Scholar]
- Dixon ML, Andrews‐Hanna JR, Spreng RN, Irving ZC, Mills C, Girn M, Christoff K (2017): Interactions between the default network and dorsal attention network vary across default subsystems, time, and cognitive states. Neuroimage 147:632–649. [DOI] [PubMed] [Google Scholar]
- Force T (1996): Heart rate variability. Standards of measurement, physiological interpretation, and clinical use. Task Force of the European Society of Cardiology and the North American Society of Pacing and Electrophysiology. Eur Heart J 17:354–381. [PubMed] [Google Scholar]
- Fox MD, Raichle ME (2007): Spontaneous fluctuations in brain activity observed with functional magnetic resonance imaging. Nat Rev Neurosci 8:700–711. [DOI] [PubMed] [Google Scholar]
- Fox MD, Snyder AZ, Vincent JL, Corbetta M, Van Essen DC, Raichle ME (2005): The human brain is intrinsically organized into dynamic, anticorrelated functional networks. Proc Natl Acad Sci U S A 102:9673–9678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Zhang D, Snyder AZ, Raichle ME (2009): The global signal and observed anticorrelated resting state brain networks. J Neurophysiol 101:3270–3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransson P (2005): Spontaneous low‐frequency BOLD signal fluctuations: An fMRI investigation of the resting‐state default mode of brain function hypothesis. Hum Brain Mapp 26:15–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston KJ, Williams S, Howard R, Frackowiak RS, Turner R (1996): Movement‐related effects in fMRI time‐series. Magn Reson Med 35:346–355. [DOI] [PubMed] [Google Scholar]
- Glover GH, Law CS (2001): Spiral‐in/out BOLD fMRI for increased SNR and reduced susceptibility artifacts. Magn Reson Med 46:515–522. [DOI] [PubMed] [Google Scholar]
- Glover GH, Li TQ, Ress D (2000): Image‐based method for retrospective correction of physiological motion effects in fMRI: RETROICOR. Magn Reson Med 44:162–167. [DOI] [PubMed] [Google Scholar]
- Greicius MD, Krasnow B, Reiss AL, Menon V (2003): Functional connectivity in the resting brain: A network analysis of the default mode hypothesis. Proc Natl Acad Sci U S A 100:253–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampson M, Driesen N, Roth J, Gore J, Constable R (2010): Functional connectivity between task‐positive and task‐negative brain areas and its relation to working memory performance. Magn Reson Imaging 28:1051–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo HJ, Saad ZS, Simmons WK, Milbury LA, Cox RW (2010): Mapping sources of correlation in resting state FMRI, with artifact detection and removal. NeuroImage 52:571–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josipovic Z, Dinstein I, Weber J, Heeger DJ (2011): Influence of meditation on anti‐correlated networks in the brain. Front Hum Neurosci 5:183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karahanoglu F, Van De Ville D (2015): Transient brain activity disentangles fMRI resting‐state dynamics in terms of spatially and temporally overlapping networks. Nat Commun 6:7751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller J, Hedden T, Thompson T, Anteraper S, Gabrieli J, Whitfield‐Gabrieli S (2015): Resting‐state anticorrelations between medial and lateral prefrontal cortex: Association with working memory, aging, and individual differences. Cortex 64:271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly AMC, Uddin LQ, Biswal BB, Castellanos FX, Milham MP (2008): Competition between functional brain networks mediates behavioral variability. NeuroImage 39:527–537. [DOI] [PubMed] [Google Scholar]
- Lancaster JL, Tordesillas‐Gutierrez D, Martinez M, Salinas F, Evans A, Zilles K, Mazziotta JC, Fox PT (2007): Bias between MNI and Talairach coordinates analyzed using the ICBM‐152 brain template. Hum Brain Mapp 28:1194–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Smyser C, Shimony J (2013): Resting‐state fMRI: A review of methods and clinical applications. Am J Neuroradiol 34:1866–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leech R, Rodrigo Braga, Sharp DJ (2012): Echoes of the brain within the posterior cingulate cortex. J Neurosci 32:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leech R, Kamourieh S, Beckmann CF, Sharp DJ (2011): Fractionating the default mode network: Distinct contributions of the ventral and dorsal posterior cingulate cortex to cognitive control. J Neurosci 31:3217–3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littow H, Elseoud AA, Haapea M, Isohanni M, Moilanen I, Mankinen K, Nikkinen J, Rahko J, Rantala H, Remes J, Starck T, Tervonen O, Veijola J, Beckmann C, Kiviniemi VJ (2010): Age‐related differences in functional nodes of the brain cortex ‐ a high model order group ica study. Front Syst Neurosci 4:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Duyn J (2013): Time‐varying functional network information extracted from brief instances of spontaneous brain activity. Proc Natl Acad Sci U S A 110:4392–4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Chang C, Duyn JH (2013): Decomposition of spontaneous brain activity into distinct fMRI co‐activation patterns. Front Syst Neurosci 7:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies DS, Vincent JL, Kelly C, Lohmann G, Uddin LQ, Biswal BB, Villringer A, Castellanos FX, Milham MP, Petrides M (2009): Precuneus shares intrinsic functional architecture in humans and monkeys. Proc Natl Acad Sci U S A 106:20069–20074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K, Fox MD (2016): Towards a consensus regarding global signal regression for resting state functional connectivity MRI. Neuroimage. doi: 10.1016/j.neuroimage.2016.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K, Birn RM, Handwerker DA, Jones TB, Bandettini PA (2009): The impact of global signal regression on resting state correlations: Are anti‐correlated networks introduced?. Neuroimage 44:893–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napadow V, Dhond R, Conti G, Makris N, Brown EN, Barbieri R (2008): Brain correlates of autonomic modulation: Combining heart rate variability with fMRI. NeuroImage 42:169–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raichle M, Hyman S (2015): The brain's default mode network. Annu Rev Neurosci 38:433–447. [DOI] [PubMed] [Google Scholar]
- Saad ZS, Gotts SJ, Murphy K, Chen G, Jo HJ, Martin A, Cox RW (2012): Trouble at rest: How correlation patterns and group differences become distorted after global signal regression. Brain Connect 2:25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirer WR, Ryali S, Rykhlevskaia E, Menon V, Greicius MD (2012): Decoding subject‐driven cognitive states with whole‐brain connectivity patterns. Cereb Cortex 22:158–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmueli K, van Gelderen P, de Zwart JA, Horovitz SG, Fukunaga M, Jansma JM, Duyn JH (2007): Low‐frequency fluctuations in the cardiac rate as a source of variance in the resting‐state fMRI BOLD signal. Neuroimage 38:306–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Fox PT, Miller KL, Glahn DC, Fox PM, Mackay CE, Filippini N, Watkins KE, Toro R, Laird AR, Beckmann CF (2009): Correspondence of the brain's functional architecture during activation and rest. Proc Natl Acad Sci U S A 106:13040–13045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Miller KL, Moeller S, Xu J, Auerbach EJ, Woolrich MW, Beckmann CF, Jenkinson M, Andersson J, Glasser MF, Van Essen DC, Feinberg DA, Yacoub ES, Ugurbil K (2012): Temporally‐independent functional modes of spontaneous brain activity. Proc Natl Acad Sci U S A 109:3131–3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagliazucchi E, Balenzuela P, Fraiman D, Chialvo DR (2012): Criticality in large‐scale brain FMRI dynamics unveiled by a novel point process analysis. Front Physiol 3:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomason ME, Glover GH (2008): Controlled inspiration depth reduces variance in breath‐holding‐induced BOLD signal. NeuroImage 39:206–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin LQ, Kelly AM, Biswal BB, Castellanos FX, Milham MP (2009): Functional connectivity of default mode network components: Correlation, anticorrelation, and causality. Hum Brain Mapp 30:625–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Heuvel M, Pol H (2010): Exploring the brain network: A review on resting‐state fMRI functional connectivity. Eur Neuropsychopharmacol 20:519–534. [DOI] [PubMed] [Google Scholar]
- Vogt BA, Vogt L, Laureys S (2006): Cytology and functionally correlated circuits of human posterior cingulate areas. Neuroimage 29:452–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Ong J, Patanaik A, Zhou J, Chee M (2016): Spontaneous eyelid closures link vigilance fluctuation with fMRI dynamic connectivity states. Proc Natl Acad Sci U S A 113:9653–9658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissenbacher A, Kasess C, Gerstl F, Lanzenberger R, Moser E, Windischberger C (2009): Correlations and anticorrelations in resting‐state functional connectivity MRI: A quantitative comparison of preprocessing strategies. Neuroimage 47:1408–1416. [DOI] [PubMed] [Google Scholar]
- Westlye ET, Lundervold A, Rootwelt H, Lundervold AJ, Westlye LT (2011): Increased hippocampal default mode synchronization during rest in middle‐aged and elderly APOE epsilon4 carriers: Relationships with memory performance. J Neurosci 31:7775–7783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise RG, Ide K, Poulin MJ, Tracey I (2004): Resting fluctuations in arterial carbon dioxide induce significant low frequency variations in BOLD signal. Neuroimage 21:1652–1664. [DOI] [PubMed] [Google Scholar]
- Wong CW, Olafsson V, Tal O, Liu TT (2012): Anti‐correlated networks, global signal regression, and the effects of caffeine in resting‐state functional MRI. Neuroimage 63:356–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Craddock R, Margulies D, Yan C, Milham M (2014): Common intrinsic connectivity states among posteromedial cortex subdivisions: Insights from analysis of temporal dynamics. Neuroimage 93:124–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo B, Tandi J, Chee M (2015): Functional connectivity during rested wakefulness predicts vulnerability to sleep deprivation. Neuroimage 111:147–158. [DOI] [PubMed] [Google Scholar]
- Yeo BT, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollinshead M, Roffman JL, Smoller JW, Zollei L, Polimeni JR, Fischl B, Liu H, Buckner RL (2011): The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol 106:1125–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Fan L, Zhang Y, Wang J, Zhu M, Zhang Y, Yu C, Jiang T (2014): Connectivity‐based parcellation of the human posteromedial cortex. Cereb Cortex 24:719–727. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information


