Abstract
Activation of G protein-coupled estrogen receptor (GPER) by its agonist, G1, protects the heart from stressors such as pressure-overload, ischemia, a high-salt diet, estrogen loss, and aging, in various male and female animal models. Due to nonspecific effects of G1, the exact functions of cardiac GPER cannot be concluded from studies using systemic G1 administration. Moreover, global knockdown of GPER affects glucose homeostasis, blood pressure, and many other cardiovascular-related systems, thereby confounding interpretation of its direct cardiac actions. We generated a cardiomyocyte-specific GPER knockout (KO) mouse model to specifically investigate the functions of GPER in cardiomyocytes. Compared to wild type mice, cardiomyocyte-specific GPER KO mice exhibited adverse alterations in cardiac structure and impaired systolic and diastolic function, as measured by echocardiography. Gene deletion effects on left ventricular dimensions were more profound in male KO mice compared to female KO mice. Analysis of DNA microarray data from isolated cardiomyocytes of wild type and KO mice revealed sex-based differences in gene expression profiles affecting multiple transcriptional networks. Gene Set Enrichment Analysis (GSEA) revealed that mitochondrial genes are enriched in GPER KO females, whereas inflammatory response genes are enriched in GPER KO males, compared to their wild type counterparts of the same sex. The cardiomyocyte-specific GPER KO mouse model provides us with a powerful tool to study the functions of GPER in cardiomyocytes. The gene expression profiles of the GPER KO mice provide foundational information for further study of the mechanisms underlying sex-specific cardioprotection by GPER.
Keywords: GPER, knockout, cardiomyocyte, microarray
1. Introduction
Cardiovascular disease (CVD) remains the leading cause of death in women in the United States, accounting for almost 400 million female deaths in 2013 [1]. The CVD morbidity rate in premenopausal women is lower compared with age-matched men, while the rate exceeds that of men after menopause [2,3]. The female sex hormone estrogen is thought to contribute to the observed differences in CVD morbidity between men and women and between pre- and postmenopausal women [2,4]. However, there is no clear evidence of an association between estrogen-based hormone replacement therapy and lower rates of cardiovascular outcomes or all-cause mortality in the postmenopausal population [5]. Rather, the available evidence shows that the risk of CVD, as well as breast cancer, increases with estrogen-based hormone therapy [5–8]. A better understanding of the relationship between estrogen and CVD risk in postmenopausal women, as well as the mechanisms and specific receptors that drive the estrogen effect, is needed to inform the development of specific cardioprotective treatments with fewer side effects.
The G protein-coupled estrogen receptor (GPER), also called GPR30, is an estrogen receptor (ER) that is expressed in the heart. GPER was first cloned from human B-cell lymphoblasts in 1996 [9], and in the intervening decade, several studies have demonstrated that activation of GPER protects the heart from various stresses such as pressure-overload, ischemia, a high-salt diet, estrogen loss, and aging in both male and female animal models [10–13]. Both in vivo and in vitro studies have examined the cardioprotective effects of GPER activated by its agonist, G1; however, G1 also binds and activates ERα36 and has nonspecific cardiac effects that confound analysis of GPER function [14]. A whole-body GPER knockout (KO) mouse has also been generated, though global elimination of GPER was shown to influence glucose homeostasis and blood pressure via various actions on smooth muscle cells, endothelial cells, fibroblasts, and cardiac mast cells [15–21].
To determine the specific function of GPER in cardiomyocytes, we generated a cardiomyocyte-specific GPER KO mouse. We compared basal cardiac function and structure, systemic blood pressure, and exercise tolerance in male and female wild type (WT) and cardiomyocyte-specific GPER KO mice and performed DNA microarray analysis to identify differential gene expression profiles and transcriptional networks. We hypothesize that GPER deletion in cardiomyocytes induces the morphometric impairments and functional declines in the heart of both sexes compared to their intact counterparts. The microarray data from GPER KO vs. intact cardiomyocytes would be important for future studies on the exact roles of GPER in the heart and the underlying mechanisms of sex-specific cardioprotection.
2.0 Materials and Methods
All mice were housed in a facility approved by the Association for Assessment and Accreditation of Laboratory Animal Care with a 12-h light/dark cycle and constant temperature and humidity. Mice had ad libitum access to standard chow (Nestle Purina, St. Louis, MO) and tap water. All procedures were in performed in compliance with the Guide for the Care and Use of Laboratory Animals, published by the National Institutes of Health, and were reviewed and approved by the Wake Forest School of Medicine’s Animal Care and Use Committee before commencement of the study (Approved protocol #A13-178).
2.1. Generation of cardiomyocyte-specific GPER KO mice
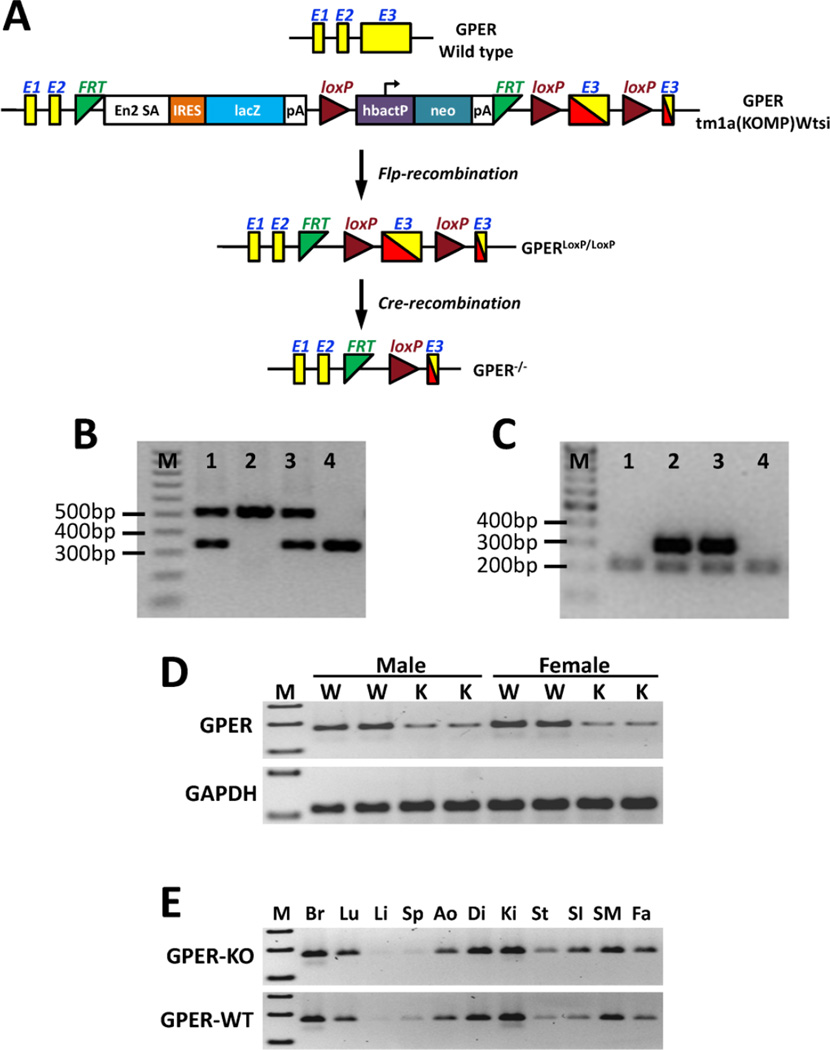
An embryonic stem (ES) cell clone (EPD0334_2_H06) carrying a GPER KO allele with conditional potential (GPERtm1a(KOMP)Wtsi) and flanking loxP sites within exon 3 was generated, expanded, and injected into C57BL/6 blastocysts as part of the Knockout Mouse Project (KOMP), as previously described [22] (Figure 1). Germ-line transmission was achieved by mating male chimeric founders with C57BL/6N female mice. The C57BL/6N mouse harboring a first mutation KO at the GPER locus was crossed with a Flp recombinase deleter mouse, C57BL/6N-Tg(CAG-Flpo)1Afst/Mmucd (Mutant Mouse Regional Resource Center, University of California, Davis, CA), to remove the selection cassette and generate a GPER allele with a floxed region in exon 3, which is the only coding exon of the GPER gene. Cardiomyocyte-specific GPER KO mice (GPERf/f/Cre) were generated by crossing homozygous GPER-floxed animals (GPERf/f) with transgenic mice expressing Myh6-Cre recombinase under the control of the cardiomyocyte-specific α myosin-heavy chain 6 promoter (Myh6) (The Jackson Laboratory, Bar Harbor, ME). Genotyping of the GPER floxed allele was performed using PCR with primers 1 (5′-GAA CCC ACA GCT CTC TTG TGT GC-3′) and 2 (5′-GAG TGT GTG GTG TGG GAA TTT GAG G-3′), which amplified a 530-bp fragment, whereas the GPER WT allele produced a 345-bp fragment. The α-Myh6-Cre transgene was detected by PCR using the primers 5′-ATG ACA GAC AGA TCC CTC CTA TCT CC-3′ and 5′-CTC ATC ACT CGT TGC ATC ATC GAC-3′, which amplified a 300-bp fragment.
Figure 1.
Development of the cardiomyocyte-specific GPER KO mouse. (A) Targeting strategy for the conditional disruption of the GPER gene. (B) A representative image showing genotyping results for the floxed GPER transgene from mouse tails. Sample 2: 530 bp band, homozygous floxed GPER transgene (GPERf/f); Sample 4: 345 bp band, WT GPER; Samples 1 and 3: 530 bp and 345 bp bands, heterozygous floxed GPER transgene. (C) A representative image showing genotyping results for the Cre transgenic mice. Samples 2 and 3: 300 bp band, Cre positive; Samples 1 and 4: 200 bp band, Cre negative (internal positive control for PCR). (D) RT-PCR results showing knockdown of GPER expression in cardiomyocytes isolated from adult (8 weeks old) GPERf/f/Cre mice (knockout, K) compared with GPERf/f mice (W). (E) RT-PCR results showing GPER expression in various tissues from GPERf/f/Cre (knockout, KO) vs. GPERf/f (WT) mice. M: marker; Br: brain; Lu: lung; Li: liver; Sp: spleen; Ao: aorta; Di: diaphragm; Ki: kidney; St: stomach; SI: small intestine; SM: skeletal muscle; Fa: fat.
2.2. Blood pressure (BP) measurement
Systolic blood pressure (SBP) was measured noninvasively using a volume-pressure recording tail-cuff method on conscious, restrained mice using the CODA 6 system (Kent Scientific Corp, Torrington, CT). Briefly, mice were gently placed in the restrainer and allowed to rest quietly for 10 min at 30°C prior to obtaining 5 acclimation cycles followed by 10 measurement cycles. SBP measurements were collected and averaged. All measurements were performed between 9:00–11:00 a.m. for all groups, to account for any diurnal variations.
2.3. Exercise capacity test
The maximal exercise capacity test (time to exhaustion during a standardized exercise protocol) was performed using a motorized treadmill (Scientific Instruments, Stoelting, Wood Dale, IL). Mice were familiarized with the motor-driven, one-lane rodent treadmill by walking at a speed of 20 cm/s, 10 min/d, for 1 week. Each exercise test was performed after at least 1 day of rest. The protocol for exercise capacity evaluation consisted of 3 min at 12 meters/min, with 1.2-meter/min increases in speed every 3 min until the mouse reached exhaustion. Time to exhaustion (in min) was determined when the mouse was unable to continue running and sat at the lower end of the treadmill for more than 5 s, despite gentle nudging.
2.4. Echocardiography
Left ventricular (LV) structure and function were determined in 15-week-old male and female cardiomyocyte-specific GPER KO mice (GPERf/f/Cre and WT mice (GPERf/f) using a commercially available echocardiograph equipped with both PureWave 12-4 MHz sector and 15-7 MHz linear transducers (CX50 CompactXtreme System; Philips Medical Systems) by the same investigator (L.G.), who was blinded to the experimental groups as previously described [23]. In brief, mice were anesthetized with an isoflurane (1.5%) oxygen mixture by nose cone and secured in the supine position to a warm (37.5°C) imaging platform. A 15 MHz linear probe was used to obtain 2D-guided, LV M-mode images using parasternal long and short axis views for measures of LV end diastolic and end systolic dimensions (LVEDD and LVESD) and posterior and anterior wall thicknesses (PWTed and AWTed) at end diastole. A 12 MHz phased array probe was used to obtain the apical 4-chamber view for transmitral inflow Doppler (early transmitral filling or maximum E-wave velocity) and septal tissue Doppler measurements (early mitral annular descent or e′) of diastolic function. Heart rate was determined from five consecutive RR intervals from pulsed-Doppler inflow tracings. The fractional shortening (FS) of the LV was expressed as %FS = (LVEDD-LVESD)/LVEDD × 100. The relative wall thickness (RWT) was calculated as (PWTed + AWTed)/LVEDD. The early mitral inflow filling velocity-to-early mitral annular descent velocity ratio, or E/e′, was used to estimate LV filling pressure. E/e′ is a useful measure to assess the severity of LV stiffness or diastolic dysfunction.
2.5. Cardiomyocyte isolation
Mice at 18–20 weeks of age were injected i.p. with 200 µl heparin (Sagent Pharmaceutical Inc., Schaumburg, IL, 100 IU/mouse) 10 min prior to anesthesia with pentobarbital (Akorn Inc., Lake Forest, IL, 100 mg/kg body weight) by i.p. injection. Upon verification of deep anesthesia by the absence of response to tail/toe pinches, the heart was quickly removed and trimmed in an ice-cold, calcium-free perfusion buffer (126 mM NaCl, 4.4 mM KCl, 1 mM MgCl2, 4 mM NaHCO3, 10 mM HEPES, 11 mM glucose, 30 mM 2,3-butanedine monoxime [Sigma, St. Louis, MO], 5 mM taurine [Sigma], pH 7.35). The heart was then cannulated through the aorta on an Easycell System for Cardiomyocyte Isolation (Harvard Apparatus, Holliston, MA) and perfused at 37°C with calcium-free perfusion buffer at a flow rate of 3 ml/min for 4–5 min until the effluent became clear. The heart was switched to digestion buffer (perfusion buffer plus 50 µM CaCl2 and 0.5 mg/ml collagenase II [Worthington Biochemical Corp., Freehold, NJ]), and perfused for 10–15 min at a flow rate of 4 ml/min until the heart was pale and flaccid. The heart was pulled from the cannula and the ventricles were transferred to a 60-mm sterile dish containing 5 ml of transfer buffer (perfusion buffer plus 0.1 mM CaCl2 and 2% bovine serum albumin [Sigma]) and cut into small pieces. The minced tissue was incubated in a 37°C water bath for 10 min. The cell suspension was filtered through a 100-µm mesh cell strainer (BD Biosciences, San Jose, CA) to remove tissue debris and spun at 420 rpm at room temperature for 2 min. After removing the supernatant, the cardiomyocytes were washed with 1 ml of PBS and centrifuged at 1500 rpm at 4°C for 3 min. The cells were suspended in 1 ml of QIAzol (Qiagen Inc, Valencia, CA), mixed, and homogenized before storing at −80°C.
2.6. DNA microarray assay
Total RNA was isolated from cardiomyocytes using the RNeasy Lipit Tissue Mini Kit (Qiagen Inc) and further purified using RNeasy MinElute Cleanup Kit (Qiagen Inc) followed by quality assessment on an Agilent 2100 bioanalyzer. Samples with RIN values >8.0 and a 260/280 ratio between 1.8 and 2.1 were carried forward for cRNA synthesis and hybridization to GeneAtlas MG-430 PM Array Strips (Affymetrix, Santa Clara, CA) following the manufacturer’s recommended protocol. Briefly, approximately 250 ng of purified total RNA was reverse transcribed and biotin labeled to produce biotinylated cRNA targets according to the standard Affymetrix GeneAtlas 3’-IVT Express labeling protocol (GeneAtlas 3’ IVT Expression Kit User Manual, P/N 702833 Rev. 4, Affymetrix). Following fragmentation, 6 µg of biotinylated cRNA was hybridized for 16 h at 45°C on the Affymetrix GeneAtlas Mouse MG-430 PM Array Strip. Strips were washed and stained using the GeneAtlas Fluidics Station according to standard Affymetrix operating procedures (GeneAtlas™ System User’s Guide, P/N 08-0306 Rev. A January 2010). Strips were subsequently scanned using the GeneAtlas Imager system according to the standard Affymetrix protocol. Fluidics control, scan control, and data collection were performed using the GeneAtlas Instrument Control Software version 1.0.5.267. All microarray analyses were performed by the Wake Forest School of Medicine Microarray Shared Resource Core.
2.7. Microarray data analysis
Gene expression profiles of cardiomyocytes detected by DNA microarray were estimated by RMA normalization and log2 converted probe set pixel intensity [24]. Paired analysis of differentially expressed genes between cardiomyocyte-specific GPER KO and WT cardiomyocytes from male and female mice were identified by Empirical Bayes method [25] implemented in the R Biocondoctor limma package, where the selection thresholds used were logFC (fold change) >1 and FDR (False Discovery rate) P<0.01.
EPIG (Extracting Patterns and Identifying differentially expressed Genes) analysis [26] was employed to evaluate gene expression data from WT and cardiomyocyte-specific GPER KO male and female mice. EPIG uses an un-supervised profile approach to extract patterns from the whole data set and then categorize genes to one of the patterns if they met all the following criteria: signal/noise ratio (SNR) >3 (i.e., the corresponding P-value is <0.05), fold change >1, and the maximum Pearson correlation r value to a given pattern of >0.64.
2.8. Ingenuity pathway analysis (IPA) and Gene set enrichment analysis (GSEA)
Functional annotation to gene ontology was performed using IPA software (Ingenuity Systems, Inc., Redwood City, CA). GSEA was performed to determine whether genes belonging to a biological pathway or a previously determined functional group were significantly overrepresented at the top or bottom of a ranked gene list compared to controls without a predefined cut-off value. This bioinformatic tool evaluates all significantly measured targets derived from a microarray experiment at the level of gene sets, which are defined based on prior biological knowledge. Thus, biologically relevant information is not missed by losing target genes due to an “arbitrarily” chosen cut-off value. In this study, expression data of all 21,782 genes were compared against functional gene sets to determine whether any of these sets were enriched in GPER KO cardiomyocytes vs. WT cardiomyocytes.
2.9. RT-PCR
Total RNA was extracted from tissues (brain, lung, liver, spleen, aorta, diaphragm, kidney, stomach, small intestine, skeletal muscle, and fat) or cardiomyocytes, and purified and qualified as described above for the microarray assay. Complimentary first strand DNA was synthesized from oligo (dT) primed total RNA using the Omniscript RT kit (Qiagen Inc). PCR amplification of GPER and the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were performed under the following PCR conditions: initial denaturation at 95°C for 10 min, followed by 30 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s. The size of the amplified GPER product was 172 bp and GAPDH was 110 bp. PCR products were visualized on 1% agarose gel (Sigma) stained with ethidium bromide and observed under ultraviolet light.
2.10. Real-time quantitative PCR
Relative quantification of mRNA levels by real-time qPCR was performed using a SYBR Green PCR kit (Qiagen Inc), as previously described [11]. Amplification and detection were performed with the ABI7500 Sequence Detection System (Applied Biosystems, Foster City, CA). Sequence-specific oligonucleotide primers were designed according to published GenBank sequences and confirmed with OligoAnalyzer 3.0. The primer sequences are listed in the supplemental table. The relative target mRNA levels in each sample were normalized to GAPDH. Expression levels are reported relative to the geometric mean of the control group.
2.11. Statistical analyses
For all experiments except the microarray analysis, values are reported as mean ± SEM. Two-way ANOVA was used to determine the effect of GPER status (KO vs. WT) and sex (male vs. female), followed by Tukey’s multiple comparisons test. Data were analyzed using the software GraphPad Prism Version 6 (GraphPad Software, Inc., La Jolla, CA). Differences for all tests were considered significant at P<0.05.
3.0 Results
3.1. Confirmation of GPER KO specifically in cardiomyocytes
Figure 1B is a representative image of the genotyping results for homozygous and heterozygous loxP-flanked GPER (GPERf/f) mice and WT GPER mice. The mice grew normally into adulthood without any health problems. Figure 1C shows a representative image of genotyping results for the Cre transgenic mice. RT-PCR confirmed a significant knockdown of GPER expression in cardiomyocytes isolated from GPERf/f/Cre mice (KO) compared with GPERf/f mice (WT) (Figure 1D). GPER expression in other tissues (brain, lung, liver, spleen, aorta, diaphragm, kidney, stomach, small intestine, skeletal muscle, and fat) was not different between the GPER KO and WT mice (Figure 1E).
Expression of ERα and ERβ was also determined by real-time RT-qPCR. As shown in Table 1, knockdown of GPER in the cardiomyocytes did not affect ERα or ERβ mRNA levels in cardiomyocytes in either male or female mice.
Table 1.
Body weight, systolic BP, treadmill time, selected gene mRNA levels in cardiomyocytes
| Male | Female | P Values | |||||
|---|---|---|---|---|---|---|---|
| WT | KO | WT | KO | Sex | KO | Sex×KO | |
| n | 4 | 4 | 4 | 4 | |||
| Body weight (g) | 31.1 ± 0.8 | 28.0 ± 0.5 | 22.4 ± 0.9 | 22.4 ± 0.8 | <0.0001 | 0.068 | 0.059 |
| Systolic BP (mmHg) | 108 ± 4 | 114 ± 3 | 109 ± 2 | 111 ± 2 | 0.813 | 0.223 | 0.544 |
| Treadmill time (min) | 18.8 ± 1.6 | 21.5 ± 1.6 | 33.6 ± 2.6 | 28.9 ± 1.9 | <0.0001 | 0.624 | 0.117 |
| Gene mRNA levels in cardiomyocytes | |||||||
| GPER | 1.00 ± 0.08 | 0.44 ± 0.04 | 1.16 ± 0.26 | 0.39 ± 0.04 | 0.394 | <0.001 | 0.081 |
| ERα | 1.00 ± 0.15 | 0.81 ± 0.07 | 0.87 ± 0.07 | 0.99 ± 0.24 | 0.869 | 0.814 | 0.320 |
| ERβ | 1.00 ± 0.14 | 0.81 ± 0.04 | 0.85 ± 0.12 | 0.79 ± 0.14 | 0.301 | 0.472 | 0.625 |
| ANF | 1.00 ± 0.27 | 3.38 ± 0.16 | 1.07 ± 0.18 | 3.73 ± 0.28 | 0.371 | <0.0001 | 0.555 |
| BNP | 1.00 ± 0.11 | 2.25 ± 0.45 | 1.59 ± 0.33 | 2.97 ± 0.72 | 0.179 | 0.014 | 0.880 |
WT: wild type (GPERf/f); KO: cardiomyocyte-specific GPER knockout (GPERf/f/Cre); BP: blood pressure; ER: estrogen receptor; ANF: atrial natriuretic factor; BNP: brain natriuretic peptide. Two-way ANOVA was used to determine the significant differences in respect to sex (male vs. female), GPER status (WT vs. KO), and interaction (gender × GPER status).
3.2. Body weight, blood pressure, and exercise capacity
Body weight, blood pressure, and exercise tolerance characteristics are shown in Table 1. Body weight was significantly higher in male vs. female mice [sex effect: F(1,17) = 86.4, P<0.0001], irrespective of GPER deletion in cardiomyocytes [KO effect: F(1,17) = 3.79, P=0.068]. There were no effects of sex or GPER deletion on systolic blood pressure. While exercise tolerance, as measured by treadmill stress test, was greater in female mice vs. male mice [sex effect: F(1,17) = 41.9, P<0.0001], GPER deletion in cardiomyocytes did not influence physical performance [KO effect: F(1,17) = 0.25, P=0.624].
3.3. Heart structure and function
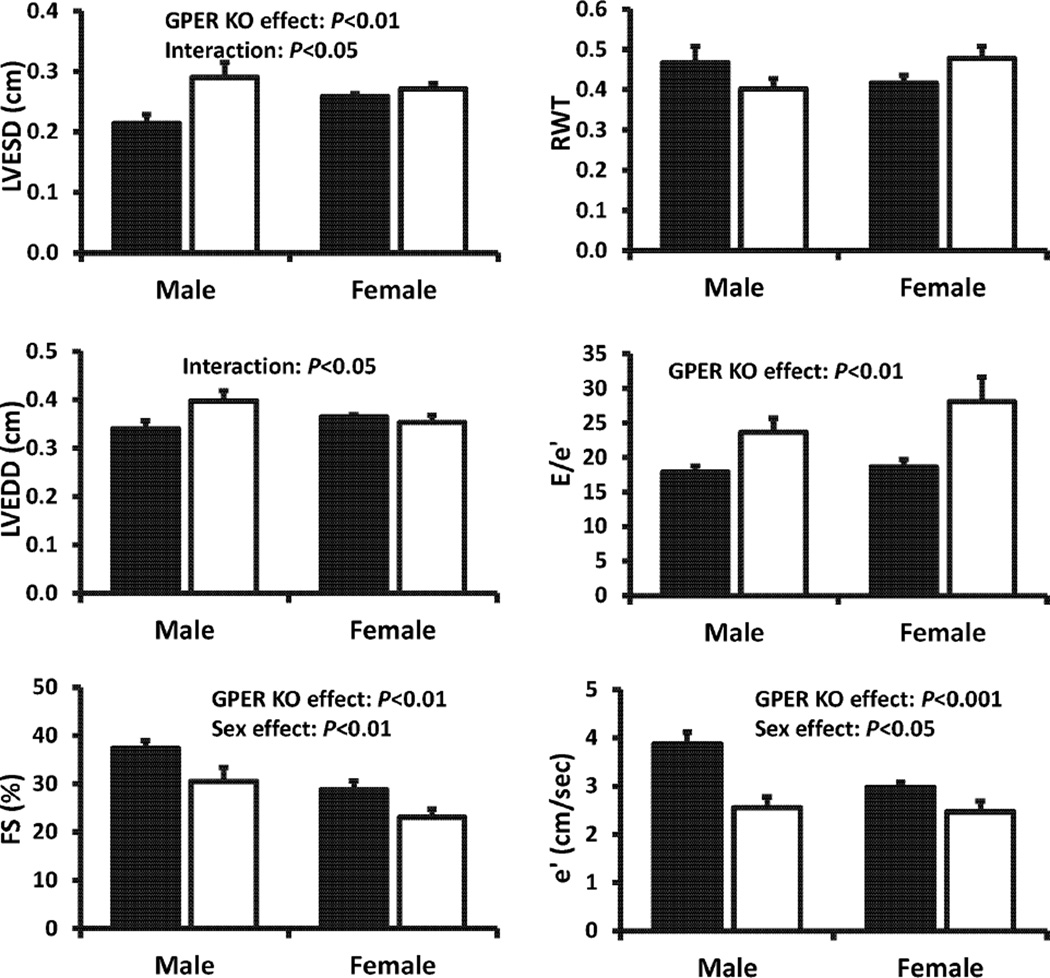
Echocardiographic indices of LV dimensions, wall thickness, and systolic and diastolic function are shown in Figure 2. Heart rates under isoflurane anesthesia were not significantly different between sexes (481 ± 22 and 401 ± 21 beats/min in WT males and females, respectively) [sex effect: F(1,18) = 2.24, 0.299], and the deletion of GPER in cardiomyocytes did not influence this variable [KO effect F(1,18) = .77, 0.39]. There was a significant interaction between sex and KO [sex × KO effect: F(1,18) = 7.51, P=0.014]; heart rate dropped by 15% in male rats with cardiomyocyte-specific deletion of GPER, whereas it increased by 8% in females. LVESD exhibited a marked increase in cardiomyocyte-specific GPER KO mice compared to WT mice [KO effect: F(1,18) = 11.58, P=0.003], and this effect was more profound among the males [sex × KO effect: F(1, 18) = 6.295, P=0.022]. For LVEDD and RWT, there were no overt sex or cardiomyocyte-specific GPER deletion effects. Nonetheless, among males, GPER deletion increased LVEDD by 13% compared to WT mice, whereas in females, this LV dimension was not affected by gene deletion [sex × KO effect: F(1,18) = 4.99, P=0.038]. An interaction effect between sex and cardiomyocyte-specific GPER deletion was also observed for RWT [sex × KO effect: F(1,18) = 4.34, P=0.052]. Specifically, among males, deletion of GPER in cardiomyocytes had a wall-thinning effect compared to WT males, whereas among females, GPER knockdown in cardiomyocytes had an overall wall-thickening effect compared to WT females. Global systolic function was affected by sex and deletion of GPER in cardiomyocytes, as the %FS was lower in both females vs. males [sex effect: F(1,18) = 8.47, P=0.009) and in the GPER KO vs. WT mice [KO effect: F(1,18) = 20.42, P=0.0003]. The tissue Doppler measure of myocardial relaxation, mitral annular descent (e′) was also significantly affected by both sex and GPER deletion. Specifically, e′ was lower in female vs. male mice [sex effect: F(1,17) = 4.99, P=0.039], and in cardiomyocyte-specific GPER KO vs. WT mice [KO effect: F(1,17) = 20.49, P=0.0003). The effect of GPER deletion on this parameter of diastolic function appeared to be more profound in males than in females [sex × KO effect: F(1,17) = 4.37, P=0.052]. Although there were no sex differences in LV filling pressures (E/e′), there was a significant effect of cardiomyocyte-specific GPER KO [F(1,17) = 16.74, P=0.0008], particularly between female KO and female WT mice (P<0.05).
Figure 2.
Echocardiographic measurements in cardiomyocyte-specific GPER KO (open bar) vs. WT (GPER intact, solid bar) mice. Values are means ± SEM; n=4/group. Two-way ANOVA was used to determine the significant differences with respect to sex (male vs. female), GPER status (WT vs. KO), and the interaction (sex × GPER status).
Atrial natriuretic factor (ANF) and brain natriuretic peptide (BNP) are biomarkers for cardiac hypertrophy and heart failure. Consistent with the echocardiographic findings, results from real-time qPCR showed that ANF and BNP levels in cardiomyocytes were significantly higher in cardiomyocyte-specific GPER KO mice compared with WT mice, irrespective of sex (ANF, KO effect: F(1,12) = 117.6, P<0.0001; BNP, F(1,12) = 8.203, P=0.014] (Table 1).
3.4. DNA microarray paired analysis results from isolated cardiomyocytes
45,141 probe sets in the DNA microarray assay detected 21,781 genes, with some probe sets corresponding to the same Entrez Gene ID. The gene expression changes in GPER KO vs. WT cardiomyocytes were different between female and male mice. Tables 2–5 list the top 50 most significantly upregulated and downregulated genes in cardiomyocytes of female and male cardiomyocyte-specific GPER KO vs. WT mice. Some genes, including ANF and BNP (Table 1), were selected for verification by real-time qPCR.
Table 2.
Top 50 most significantly upregulated genes in cardiomyocytes of female cardiomyocyte-specific GPER KO vs. WT mice
| Gene Symbol | Gene Description | Fold Change |
P Value |
|---|---|---|---|
| Myh7 | myosin, heavy polypeptide 7, cardiac muscle, beta | 6.87 | 0.00000 |
| 2810474O19Rik | RIKEN cDNA 2810474O19 gene | 3.32 | 0.00130 |
| Tmem 184c | transmembrane protein 184C | 2.73 | 0.00011 |
| Ndnf | neuron-derived neurotrophic factor | 2.72 | 0.00000 |
| Fgl2 | fibrinogen-like protein 2 | 2.63 | 0.00003 |
| Xist | inactive X specific transcripts | 2.62 | 0.00028 |
| Malat1 | metastasis associated lung adenocarcinoma transcript 1 | 2.46 | 0.01767 |
| Trpm7 | transient receptor potential cation channel, member M7 | 2.35 | 0.01411 |
| Acta1 | actin, alpha 1, skeletal muscle | 2.28 | 0.00318 |
| Nppa | natriuretic peptide type A | 2.26 | 0.00014 |
| Ankrd1 | ankyrin repeat domain 1 (cardiac muscle) | 2.22 | 0.00000 |
| Gck | glucokinase | 2.21 | 0.00001 |
| Phldb2 | pleckstrin homology-like domain, family B, member 2 | 2.17 | 0.00014 |
| Mybpc2 | myosin binding protein C, fast-type | 2.14 | 0.00000 |
| Zfp445 | zinc finger protein 445 | 2.11 | 0.00874 |
| Kdm7a | lysine (K)-specific demethylase 7A | 2.09 | 0.00391 |
| Dmxl2 | Dmx-like 2 | 2.06 | 0.00088 |
| Fam 198b | family with sequence similarity 198, member B | 2.06 | 0.00062 |
| Sorbs2 | sorbin and SH3 domain containing 2 | 2.05 | 0.01998 |
| Atrx | α thalassemia/mental retardation syndrome X-linked homolog | 2.04 | 0.02410 |
| Nbeal1 | neurobeachin like 1 | 2.04 | 0.01435 |
| Kif5b | kinesin family member 5B | 2.02 | 0.00769 |
| Gas5/Snord47 | growth arrest specific 5/small nucleolar RNA, C/D box 47 | 1.98 | 0.03187 |
| Rtn4 | reticulon 4 | 1.98 | 0.00000 |
| Rif1 | Rap 1 interacting factor 1 homolog (yeast) | 1.94 | 0.01003 |
| Akap9 | A kinase (PRKA) anchor protein (yotiao) 9 | 1.92 | 0.02423 |
| Tlr4 | toll-like receptor 4 | 1.92 | 0.00132 |
| Arid4b | AT rich interactive domain 4B (RBPl-like) | 1.91 | 0.00883 |
| Trove2 | TROVE domain family, member 2 | 1.90 | 0.00000 |
| Gdap10 | ganglioside-induced differentiation-associated-protein 10 | 1.90 | 0.02673 |
| Cfh | complement component factor h | 1.90 | 0.01042 |
| Ranbp2 | RAN binding protein 2 | 1.89 | 0.01033 |
| Kif5b | kinesin family member 5B | 1.87 | 0.01335 |
| Trip 11 | thyroid hormone receptor interactor 11 | 1.85 | 0.00204 |
| Cwf1912 | CWF19-like 2, cell cycle control (S. pombe) | 1.85 | 0.01437 |
| Acbd5 | acyl-Coenzyme A binding domain containing 5 | 1.84 | 0.02449 |
| H2-Aa | histocompatibility 2, class II antigen A, alpha | 1.84 | 0.00074 |
| Usp9x | ubiquitin specific peptidase 9, X chromosome | 1.81 | 0.00489 |
| Zfp367 | zinc finger protein 367 | 1.81 | 0.00794 |
| Ccnd2 | cyclin D2 | 1.80 | 0.00014 |
| Lin7c | lin-7 homolog C (C. elegans) | 1.80 | 0.04039 |
| Pcolce | procollagen C-endopeptidase enhancer protein | 1.79 | 0.00052 |
| Zfp62 | zinc finger protein 62 | 1.79 | 0.03314 |
| Phlda3 | pleckstrin homology-like domain, family A, member 3 | 1.79 | 0.00000 |
| Pnisr | PNN interacting serine/arginine-rich | 1.79 | 0.04485 |
| Lgals3 | lectin, galactose binding, soluble 3 | 1.79 | 0.00888 |
| Pde1c | phosphodiesterase 1C | 1.78 | 0.00067 |
| Cast | calpastatin | 1.78 | 0.01134 |
| Nabp1 | nucleic acid binding protein 1 | 1.78 | 0.00223 |
| Taok1 | TAO kinase 1 | 1.77 | 0.01954 |
Table 5.
Top 50 most significantly downregulated genes in cardiomyocytes of male cardiomyocyte-specific GPER KO vs. WT mice
| Gene Symbol | Gene Description | Fold Change |
P Value |
|---|---|---|---|
| Dmd | dystrophin, muscular dystrophy | 0.21 | 0.00012 |
| Amy1 | amylase 1, salivary | 0.26 | 0.00000 |
| Gpr116 | G protein-coupled receptor 116 | 0.39 | 0.00162 |
| Pah | phenylalanine hydroxylase | 0.43 | 0.00832 |
| Aldob | aldolase B, fructose-bisphosphate | 0.44 | 0.00000 |
| Gpcpd1 | glycerophosphocholine phosphodiesterase GDE1 homolog | 0.47 | 0.00307 |
| Ppp2r3a | protein phosphatase 2, regulatory subunit B”, alpha | 0.49 | 0.00002 |
| Pfkfb1 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 | 0.50 | 0.00003 |
| Pde7a | phosphodiesterase 7A | 0.50 | 0.01219 |
| Fbln1 | fibulin 1 | 0.53 | 0.00075 |
| Gdap10 | ganglioside-induced differentiation-associated-protein 10 | 0.54 | 0.03215 |
| Iigp1 | interferon inducible GTPase 1 | 0.55 | 0.00188 |
| Ankrd32 | ankyrin repeat domain 32 | 0.55 | 0.03600 |
| Pdgfd | platelet-derived growth factor, D polypeptide | 0.56 | 0.00006 |
| 2310040G07Rik | RIKEN cDNA 2310040G07 gene | 0.56 | 0.01438 |
| Exoc2 | exocyst complex component 2 | 0.56 | 0.00000 |
| Luc712 | LUC7-like 2 (S. cerevisiae) | 0.57 | 0.03289 |
| Kcnj3 | potassium inwardly-rectifying channel, subfamily J, member 3 | 0.57 | 0.00332 |
| Fabp4 | fatty acid binding protein 4, adipocyte | 0.58 | 0.00018 |
| Ano 10 | anoctamin 10 | 0.58 | 0.00000 |
| Prrg 1 | proline rich Gla (G-carboxyglutamic acid) 1 | 0.58 | 0.00031 |
| Dpyd | dihydropyrimidine dehydrogenase | 0.58 | 0.00003 |
| Gpr22 | G protein-coupled receptor 22 | 0.58 | 0.00771 |
| Btnl9 | butyrophilin-like 9 | 0.59 | 0.00825 |
| Ddx3y | DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked | 0.59 | 0.02995 |
| Ets1 | E26 avian leukemia oncogene 1,5’ domain | 0.59 | 0.03764 |
| B230214O09Rik | RIKEN cDNA B23 0214O09 gene | 0.59 | 0.00219 |
| Cyr61 | cysteine rich protein 61 | 0.59 | 0.01297 |
| D7Ertd715e | DNA segment, Chr 7, ERATO Doi 715, expressed | 0.59 | 0.01290 |
| Wif1 | Wnt inhibitory factor 1 | 0.59 | 0.01242 |
| Cav2 | caveolin 2 | 0.60 | 0.04897 |
| Kcnd2 | potassium voltage-gated channel, Shal-related family, member 2 | 0.60 | 0.00045 |
| Il15 | interleukin 15 | 0.61 | 0.03395 |
| Gbp6 | guanylate binding protein 6 | 0.61 | 0.00999 |
| Etl4 | enhancer trap locus 4 | 0.61 | 0.00752 |
| Acot1 | acyl-CoA thioesterase 1 | 0.61 | 0.02195 |
| Ttll1 | tubulin tyrosine ligase-like 1 | 0.61 | 0.00000 |
| Eltd1 | EGF, latrophilin seven transmembrane domain containing 1 | 0.61 | 0.01848 |
| Btnl9 | butyrophilin-like 9 | 0.61 | 0.02541 |
| Dcakd | dephospho-CoA kinase domain containing | 0.62 | 0.00000 |
| Cyyr1 | cysteine and tyrosine-rich protein 1 | 0.62 | 0.02812 |
| Zbtb20 | zinc finger and BTB domain containing 20 | 0.62 | 0.04300 |
| Hnrnpa3 | heterogeneous nuclear ribonucleoprotein A3 | 0.63 | 0.02435 |
| Cyr61 | cysteine rich protein 61 | 0.63 | 0.01999 |
| Ube2b | ubiquitin-conjugating enzyme E2B | 0.63 | 0.03951 |
| Trdn | triadin | 0.63 | 0.00371 |
| 1810011O10Rik | RIKEN cDNA 1810011O10 gene | 0.64 | 0.01249 |
| Ssbp2 | single-stranded DNA binding protein 2 | 0.64 | 0.00017 |
| Tmod3 | tropomodulin 3 | 0.64 | 0.04622 |
| PggtIb | protein geranylgeranyltransferase type I, beta subunit | 0.64 | 0.03485 |
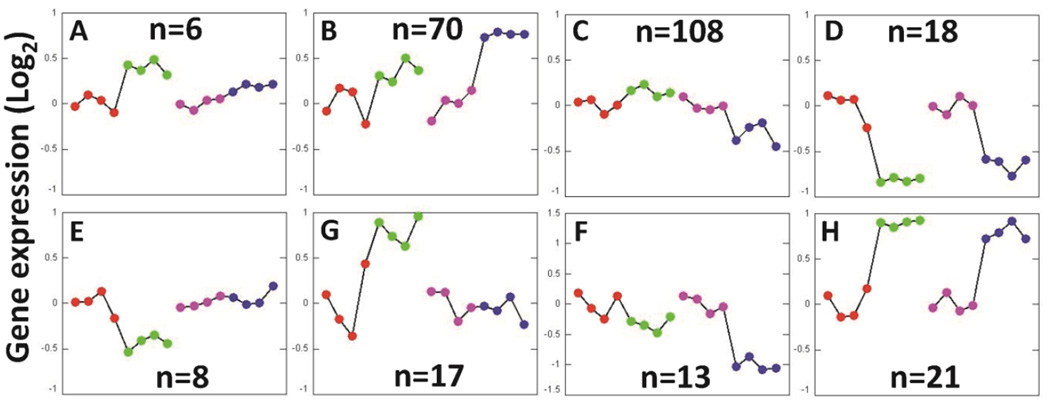
3.5. EPIG analysis based on sex and GPER expression
278 probes were found to be significantly responsive to the cardiomyocyte-specific knockout of GPER from male and female mice and categorized into eight different patterns, as summarized in Figure 3. Similar gene expression patterns between GPER KO and their WT female and male counterparts are displayed in panels 3D and 3H, while the other panels illustrate gene expression patterns with marked differences respect to sex and cardiomyocyte-specific GPER deletion. The names of the genes in each panel are available upon request.
Figure 3.
Gene expression patterns extracted by EPIG from microarray data from cardiomyocyte-specific GPER KO and WT (GPER intact) mice. The y-axis is the average log2 of the ratio values from the top 6 gene probe set profiles within the respective pattern. The average expression levels in one of the WT female mice were aligned around 0 (y-axis) in the EPIG analysis. Each point along the x-axis represents a mouse sample. Within a single pattern from left to right: WT female (red), GPER KO female (green), WT male (pink), and GPER KO male (blue). The numbers of significant genes for each pattern are shown in the figures.
3.6. Pathway analysis
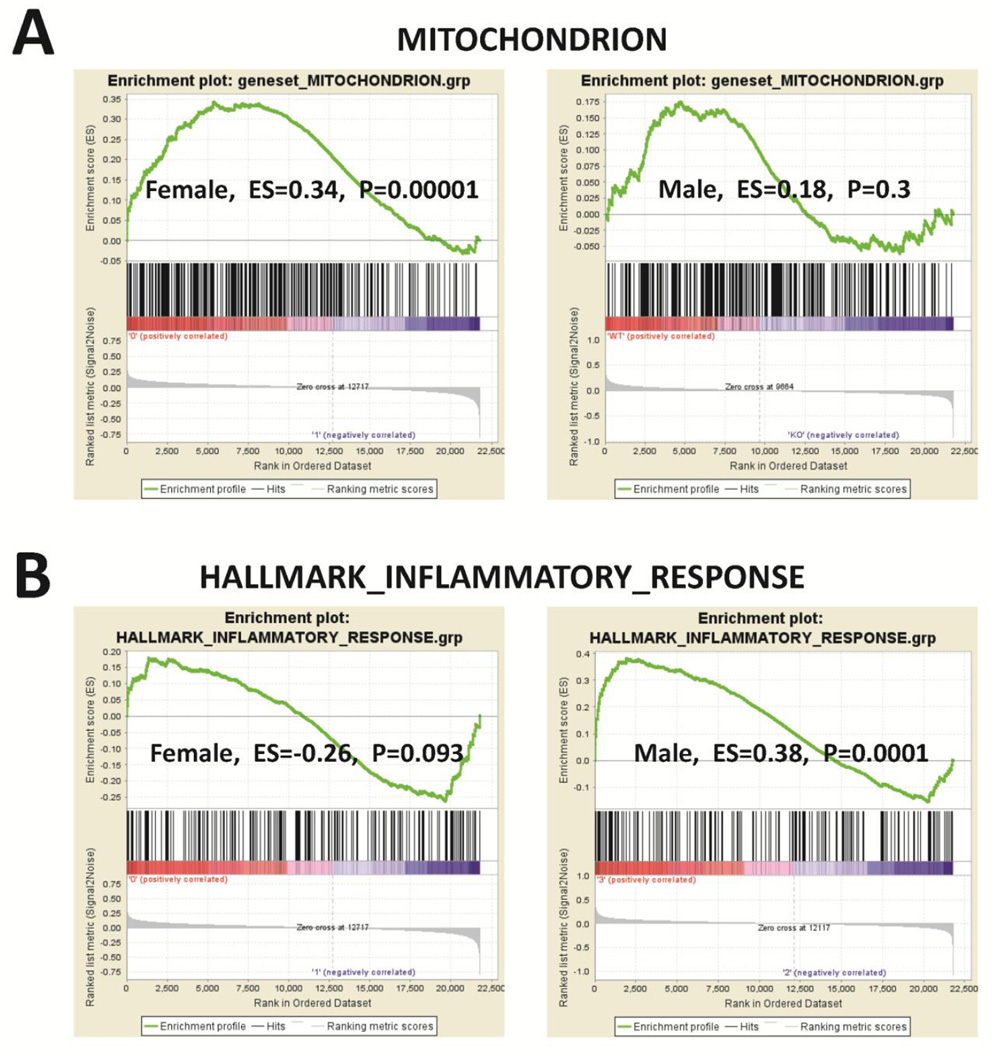
To examine the differences in the mitochondrial and inflammatory response gene expressions between GPER KO and WT cardiomyocytes, microarray data were loaded into GSEA 2.0.1 using GSEA gene sets “MITOCHONDRION (including 314 genes)” and “HALLMARK_INFLAMMATORY_RESPONSE (including 193 genes).” The results showed that the mitochondrial genes were enriched in GPER-KO vs. WT cardiomyocytes from female, but not male, mice. In contrast, the inflammatory response genes were enriched in GPER KO vs. WT cardiomyocytes from male, but not female, mice (Figure 4). The heat maps and the individual mitochondrial and inflammatory gene changes in GPER knockout versus intact cardiomyocytes from both sexes are presented in Data In Brief [27].
Figure 4.
GSEA analysis for the enrichment of “MITOCHONDRION (including 314 genes)” and “HALLMARK_INFLAMMATORY_RESPONSE (including 193 genes)” gene sets in cardiomyocyte-specific GPER KO vs. WT (GPER intact) cardiomyocytes. The bar-code plot indicates the positions of genes in each gene set; red and blue colors represent positive and negative Pearson correlations with GPER expression, respectively. The results show that mitochondrial genes were enriched in GPER KO vs. WT cardiomyocytes from female, but not male, mice. Inflammatory response genes were enriched in GPER KO vs. WT cardiomyocytes from male, but not female, mice. ES: enrichment score.
IPA revealed differences in canonical pathways and genes related to various diseases and disorders, molecular and cellular functions, physiological system development and functions, and cardiotoxicity between GPER KO vs. WT cardiomyocytes of male and female mice (Tables 6 and 7).
Table 6.
Summary of IPA in cardiomyocytes of female cardiomyocyte-specific GPER KO vs. WT mice
|
Top Canonical Pathways | ||
| Name | p-value | Overlap |
| Cardiomyocyte Differentiation via BMP Receptors | 2.38E-03 | 10.0 % (2/20) |
| Maturity Onset Diabetes of Young (MODY) Signaling | 2.62E-03 | 9.5% (2/21) |
| Epoxysqualene Biosynthesis | 7.26E-03 | 50.0 % (1/2) |
| Antigen Presentation Pathway | 8.00E-03 | 5.4% 2/37 |
| Trehalose Degradation II (Trehalase) | 1.09E-02 | 33.3% (1/3) |
|
Top Diseases and Disorders | ||
| Name | p-value | #Molecules |
| Psychological Disorders | 1.84E-02 - 1.30E-04 | 9 |
| Neurological Disease | 2.16E-02 - 1.94E-04 | 18 |
| Skeletal and Muscular Disorders | 1.84E-02 - 1.94E-04 | 15 |
| Hematological Disease | 2.16E-02 - 5.76E-04 | 8 |
| Immunological Disease | 2.16E-02 - 5.76E-04 | 7 |
|
Molecular and Cellular Functions | ||
| Name | p-value | #Molecules |
| Carbohydrate Metabolism | 1.71E-02 - 3.90E-05 | 6 |
| Molecular Transport | 1.81E-02 - 3.90E-05 | 11 |
| Small Molecule Biochemistry | 1.83E-02 - 3.90E-05 | 14 |
| Cell Death and Survival | 2.16E-02 - 9.80E-05 | 27 |
| Cellular Development | 1.81E-02 - 1.12E-04 | 19 |
|
Physiological System Development and Function | ||
| Name | p-value | #Molecules |
| Embryonic Development | 2.13E-02 - 9.80E-05 | 12 |
| Hematological System Development and Function | 1.81E-02 - 1.12E-04 | 8 |
| Behavior | 7.26E-03 - 1.30E-04 | 3 |
| Nervous System Development and Function | 1.81E-02 - 1.30E-04 | 15 |
| Tissue Development | 1.81E-02 - 1.30E-04 | 15 |
|
Cardiotoxicity | ||
| Name | p-value | #Molecules |
| Cardiac Arrhythmia | 1.97E-01 - 9.33E-04 | 4 |
| Cardiac Congestive Cardiac Failure | 3.64E-03 - 3.64E-03 | 1 |
| Heart Failure | 2.36E-01 - 3.64E-03 | 2 |
| Cardiac Transformation | 7.26E-03 - 7.26E-03 | 1 |
| Cardiac Hypertrophy | 4.09E-01 - 8.95E-03 | 5 |
Table 7.
Summary of IP A in cardiomyocytes of male cardiomyocyte-specific GPER KO vs. WT mice
|
Top Canonical Pathways | ||
| Name | p-value | Overlap |
| Dendritic Cell Maturation | 3.24E-04 | 3.4% 6/177 |
| B Cell Development | 6.61E-04 | 9.1% 3/33 |
| Antigen Presentation Pathway | 9.27E-04 | 8.1% 3/37 |
| Autoimmune Thyroid Disease Signaling | 1.86E-03 | 6.4 % 3/47 |
| Graft-versus-Host Disease Signaling | 1.98E-03 | 6.2 % 3/48 |
|
Top Diseases and Disorders | ||
| Name | p-value | #Molecules |
| Neurological Disease | 7.99E-03 - 6.59E-10 | 42 |
| Endocrine System Disorders | 5.67E-03 - 4.82E-09 | 37 |
| Gastrointestinal Disease | 7.77E-03 - 4.82E-09 | 44 |
| Metabolic Disease | 5.19E-03 - 4.82E-09 | 28 |
| Immunological Disease | 7.99E-03 - 6.33E-09 | 32 |
|
Molecular and Cellular Functions | ||
| Name | p-value | #Molecules |
| Cell Death and Survival | 7.99E-03 - 4.95E-07 | 49 |
| Cellular Movement | 7.52E-03 - 8.19E-07 | 31 |
| Cell-To-Cell Signaling and Interaction | 7.79E-03 - 3.12E-05 | 31 |
| Cellular Assembly and Organization | 7.99E-03 - 7.97E-05 | 30 |
| Cell Morphology | 7.79E-03 - 8.66E-05 | 29 |
|
Physiological System Development and Function | ||
| Name | p-value | #Molecules |
| Immune Cell Trafficking | 7.52E-03 - 8.19E-07 | 21 |
| Hematological System Development and Function | 7.79E-03 - 1.29E-06 | 27 |
| Endocrine System Development and Function | 5.19E-03 - 1.45E-04 | 7 |
| Cardiovascular System Development and Function | 5.75E-03 - 1.59E-04 | 19 |
| Organ Morphology | 5.19E-03 - 1.59E-04 | 5 |
|
Cardiotoxicity | ||
| Name | p-value | #Molecules |
| Cardiac Hypertrophy | 1.17E-01 - 2.08E-06 | 11 |
| Congenital Heart Anomaly | 1.42E-03 - 1.42E-03 | 2 |
| Cardiac Infarction | 3.43E-03 - 1.46E-03 | 5 |
| Cardiac Necrosis/Cell Death | 2.61E-01 - 1.83E-03 | 6 |
| Cardiac Arrhythmia | 2.68E-01 - 3.13E-03 | 5 |
3.7. Cardiac hypertrophy-related gene expression
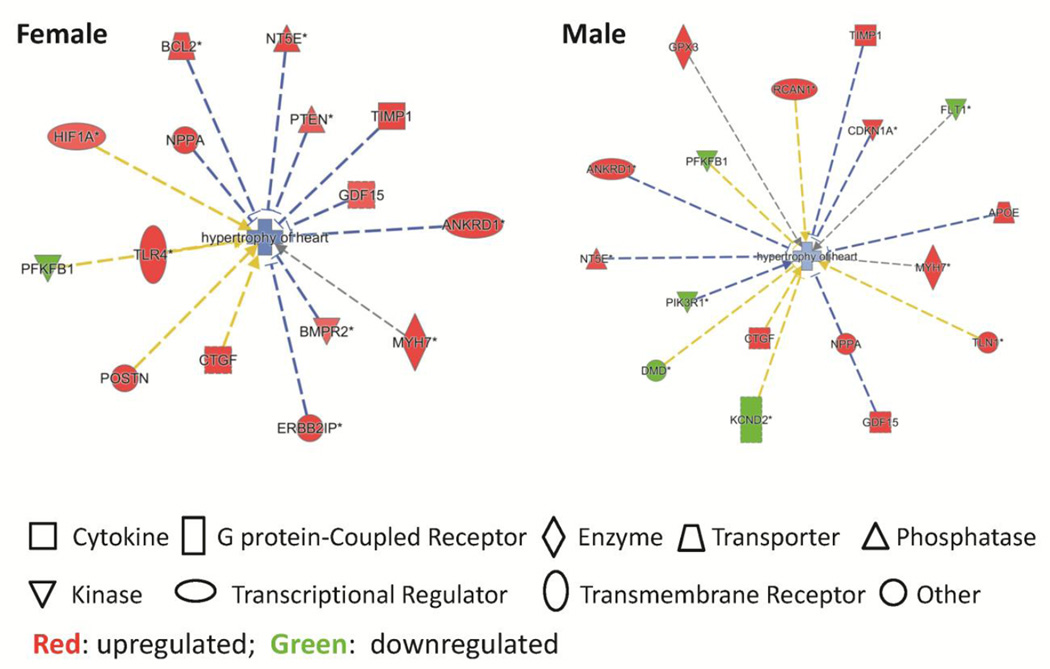
Although LV hypertrophy was not overtly apparent in vivo in the hearts of cardiomyocyte-specific GPER KO mice compared to WT mice (Table 1), IPA revealed significant sex-specific differences in cardiac hypertrophy-related gene expression between GPER KO and WT cardiomyocytes (Figure 5). Specifically, in female mice, the hypertrophy-related genes that were elevated in GPER KO vs. WT cardiomyocytes included ANKRD1, BCL-2, BMPR2, CTGF, ERBB2IP, GDF15, HIF1A, MYH7, NPPA, NT5E, POSTN, PTEN, TIMP1, and TLR4; while PFKFB1 was lower. In male mice, the hypertrophy-related genes that were elevated in GPER KO vs. WT cardiomyocytes included ANKRD1, APOE, CDKN1A, CTGF, GDF15, GPX3, MYH7, NPPA, NT5E, RCAN1, TIMP1, and TLN1; while DMD, FLT1, KCND2, PFKFB1, and PIK3R1 were lower (Figure 5).
Figure 5.
Cardiac hypertrophy-related gene expression analyzed by the IPA software.
4.0 Discussion
The present study is the first to describe the successful generation of a cardiomyocyte-specific GPER KO mouse. Compared to WT mice, echocardiography revealed the presence of adverse alterations in cardiac structure and impairments in systolic and diastolic function in cardiomyocyte-specific GPER KO mice of both sexes, with more profound increases in LV dimensions among male KO mice. Analysis of DNA microarray data from cardiomyocytes of GPER KO and WT mice showed differential expression profiles of genes affecting multiple transcriptional networks between male and female mice after cardiomyocyte-specific GPER deletion. GSEA revealed that mitochondrial genes were enriched in GPER-KO cardiomyocytes from females compared to WT males, while inflammatory response genes were enriched in GPER-KO cardiomyocytes from males compared to WT females. Taken together, our findings provide strong evidence confirming the cardioprotective effect of GPER that had been suggested by previous studies of non-specific activation of GPER with G1 and global GPER knockout techniques.
The discovery of GPER, especially its expression in the heart, has opened up a new direction for research into the cardioprotective effects of estrogen and the mechanisms underlying CVD risk in older women after the loss of ovarian estrogen function. Studies in animal models using an agonist of GPER, G1, have shown that both chronic and acute treatment with G1 limits LV dysfunction and improves cardiac remodeling induced by ischemia/reperfusion, hypertension, high-salt diet, or estrogen loss [10,11,13,28–31]. In male global GPER knockout mice, the LV is enlarged and LV contractility and relaxation capacities are reduced [32]. Despite these examples of GPER-mediated cardioprotection, the exact roles of cardiac GPER could not be concluded, as the specificity of G1 binding to GPER is questionable [14]. The systemic administration of G1 or global GPER knockout also have effects on other tissues, which could influence whole body metabolism, blood pressure, endothelial function and inflammatory responses [15,18,33,34], ultimately interfering with myocardial structure and function. Compared to global GPER KO mice [32], this study demonstrated that GPER deletion specifically in cardiomyocytes impaired heart structure and functions not only in male, but also in female mice.
The sex hormones, including the estrogens, play important roles in maintaining cardiac structure and function in both females and males [35]. Results from this study demonstrate that GPER expression levels in cardiomyocytes are the same in WT mice of both sexes; however, the effects of GPER deletion in cardiomyocytes on LV dimensions had different effects in males and females. Male mice with cardiomyocyte-specific GPER deletion exhibited profound increases in both LVEDD and LVESD compared to their female GPER KO mice. RWT was lower in male cardiomyocyte-specific GPER KO mice compared to WT male mice, whereas in female GPER KO mice, RWT was higher compared to WT mice. This interaction between sex and cardiomyocyte-specific GPER gene deletion accounted for nearly 19% of the total variance in RWT. Studies of global GPER KO mice also found that both systolic and diastolic function were impaired in males, but not females [32]. Consistent with the observed sex differences in cardiac structure and function with cardiomyocyte-specific GPER deletion, we found differential expression of cardiomyocyte genes in the male vs. female KO mice, providing a set of foundational data for future studies aimed at ascertaining the mechanisms underlying sex differences in cardioprotection by GPER.
The different effects of GPER deletion in LVESD, LVEDD, and the gene expression profiles between male and female mice might be due to the effects of endogenous estrogens in female mice through its other receptors, ERα and ERβ, which also play important roles in cardioprotection, in addition to GPER. In ovariectomized female rats, the ERβ agonist 8β-VE2 attenuates DOCA-induced hypertension and cardiac hypertrophy [36], while ERβ deletion augments the transverse aortic constriction (TAC)-induced cardiac hypertrophy in both male and female mice [37,38]. The functional roles of ERα in the heart are not consistent. While the activation of ERα by its agonist attenuates cardiac hypertrophy and improves heart function in TAC, spontaneous hypertensive, and DOCA animal models [36,39,40], ERα KO mice develop cardiac hypertrophy after TAC to the same extent as that in WT littermate [38]. More studies are needed to determine the exact roles of the individual estrogen receptors and their interactions in response to various stresses in the female versus the male heart.
A variety of pathways and mechanisms may mediate the cardioprotective effects of GPER. Using GSEA, we found that mitochondrial genes were enriched in GPER KO vs. WT cardiomyocytes from female, but not male, mice. Previous studies suggest that estrogens have significant regulatory roles in maintaining normal mitochondrial properties by stabilizing the structural assembly of mitochondria, attenuating mitochondrial ROS production, and modulating mitochondrial ATP synthesis in the heart [41]. The protective effects of estrogens might be mediated, at least partially, by GPER, since activation of GPER by G1 inhibits mitochondria permeability transition pore opening and protects the heart against ischemia-reperfusion injury [28]. More recently, studies in H9c2 cardiomyocytes have shown that G1 mimics the beneficial actions of estradiol on enhancing mitochondrial function and biogenesis, while the GPER antagonist G15 neutralizes the beneficial effects on bioenergetics [42]. Although confirmation of any alterations in mitochondrial structure and function between cardiomyocyte-specific GPER KO vs. WT mice will require further study, our microarray data and GSEA provided direct evidence that the cardioprotective effects of GPER in the female might be related to enhancements in mitochondrial function.
In addition to the mitochondria gene changes, GSEA revealed that inflammatory response genes were enriched in GPER KO vs. WT cardiomyocytes, specifically among males. Previous studies suggest that the anti-inflammatory effects of estrogens might mediate cardioprotection [43–46]. Moreover, the agonist to GPER, G1, has been shown to inhibit inflammatory changes induced by tumor necrosis factor (TNF) in human umbilical vein endothelial cells [47,48]. However, the exact role of GPER on mediating the inflammatory responses of cardiomyocytes to various stressors remains unclear. The gene expression profile in GPER KO vs. WT cardiomyocytes suggest that GPER might mediate the anti-inflammatory actions of estrogens, and that the anti-inflammatory effects of GPER in cardiomyocytes is sex-specific.
5.0 Limitations and future studies
The present study focused on the gene expression profiles in cardiomyocytes of GPER KO mice which provides foundational information for further studies on the mechanisms underlying sex-specific cardioprotection by GPER. In order to get a high yield of adult cardiomyocytes with high quality, the cardiomyocyte isolation had to be performed immediately after the hearts were removed from the chest. Therefore, heart weights and morphology were not determined in this study. Future studies are in progress emphasizing potential GPER-related alterations in whole heart morphometric and histopathology, as well as single cardiomyocyte functional dynamics.
We also determined ERβ mRNA in cardiomyocytes using real-time PCR. We realize that some investigators report that ERβ is not detectable in the heart [49,50], while its existence in both cardiomyocytes and cardiac fibroblasts has been demonstrated at mRNA and protein levels, and might mediate the cardioprotective effects of estrogen [51–57]. Even though ERβ expression is not the focus of the present study, future investigations using a more specific ERβ antibody and/or PCR product sequencing are needed to confirm whether ERβ is expressed in the heart.
In summary, cardiomyocyte-specific GPER KO in mice induced adverse alterations in cardiac morphology and abnormalities in systolic and diastolic function, with different effects in males and females. Moreover, cardiomyocyte gene expression profile changes with the loss of GPER may also be influenced by sex, particularly for the families of genes relating to mitochondrial and inflammatory responses. Along with the powerful tool of the cardiomyocyte-specific GPER KO mouse model, the microarray data generated in this study provides the necessary foundation for ascertaining the exact roles of GPER in the heart and the underlying mechanisms of sex-specific cardioprotection in future studies.
Supplementary Material
Table 3.
Top 50 most significantly upregulated genes in cardiomyocytes of male cardiomyocyte-specific GPER KO vs. WT mice
| Gene Symbol | Gene Description | Fold Change |
P Value |
|---|---|---|---|
| Myh7 | myosin, heavy polypeptide 7, cardiac muscle, beta | 7.26 | 0.00000 |
| Ndnf | neuron-derived neurotrophic factor | 3.48 | 0.00000 |
| Lgals3 | lectin, galactose binding, soluble 3 | 3.15 | 0.00002 |
| Timp1 | tissue inhibitor of metalloproteinase 1 | 2.83 | 0.00000 |
| Fgl2 | fibrinogen-like protein 2 | 2.80 | 0.00001 |
| H2-Aa | histocompatibility 2, class II antigen A, alpha | 2.72 | 0.00000 |
| Nppa | natriuretic peptide type A | 2.65 | 0.00002 |
| Ctss | cathepsin S | 2.47 | 0.00016 |
| Col3a1 | collagen, type III, alpha 1 | 2.26 | 0.00555 |
| Cdkn1a | cyclin-dependent kinase inhibitor 1A (P21) | 2.23 | 0.00261 |
| Phlda3 | pleckstrin homology-like domain, family A, member 3 | 2.17 | 0.00000 |
| Lyz1 | lysozyme 1 | 2.17 | 0.00017 |
| H2-Eb1 | histocompatibility 2, class II antigen E beta | 2.16 | 0.00013 |
| Mybpc2 | myosin binding protein C, fast-type | 2.15 | 0.00000 |
| Lcn2 | lipocalin 2 | 2.12 | 0.00002 |
| Lyz2 | lysozyme 2 | 2.08 | 0.00023 |
| Cd74 | CD74 antigen | 2.07 | 0.00000 |
| Fcgr2b | Fc receptor, IgG, low affinity IIb | 2.05 | 0.00044 |
| Slc38a4 | solute carrier family 38, member 4 | 2.04 | 0.00189 |
| Tubb2a | tubulin, beta 2A class IIA | 2.03 | 0.00025 |
| Ccl 6 | chemokine (C-C motif) ligand 6 | 2.01 | 0.00224 |
| Nppb | natriuretic peptide type B | 2.00 | 0.00017 |
| Rtn4 | reticulon 4 | 1.99 | 0.00000 |
| Fam 198b | family with sequence similarity 198, member B | 1.99 | 0.00095 |
| Dpysl3 | dihydropyrimidinase-like 3 | 1.98 | 0.00000 |
| Pcolce | procollagen C-endopeptidase enhancer protein | 1.93 | 0.00017 |
| Tmem 176a | transmembrane protein 176A | 1.91 | 0.00001 |
| Hn 1 | hematological and neurological expressed sequence 1 | 1.88 | 0.00000 |
| Comtd 1 | catechol-O-methyltransferase domain containing 1 | 1.86 | 0.00000 |
| Socs3 | suppressor of cytokine signaling 3 | 1.83 | 0.02286 |
| Ctgf | connective tissue growth factor | 1.83 | 0.00093 |
| Tmem 176b | transmembrane protein 176B | 1.83 | 0.00000 |
| Ephx1 | epoxide hydrolase 1, microsomal | 1.81 | 0.00363 |
| Mthfd2 | methylenetetrahydrofolate dehydrogenase (NAD+ dependent), methenyltetrahydrofolate cyclohydrolase |
1.80 | 0.00097 |
| Stbd1 | starch binding domain 1 | 1.80 | 0.00000 |
| Ccl9 | chemokine (C-C motif) ligand 9 | 1.80 | 0.00011 |
| Ctsz | cathepsin Z | 1.79 | 0.00021 |
| Tyrobp | TYRO protein tyrosine kinase binding protein | 1.78 | 0.00025 |
| Apoe | apolipoprotein E | 1.77 | 0.00090 |
| Mt2 | metallothionein 2 | 1.77 | 0.00207 |
| C1qc | complement component 1, q subcomponent, C chain | 1.77 | 0.00104 |
| Acta1 | actin, alpha 1, skeletal muscle | 1.76 | 0.02983 |
| H2-Aa | histocompatibility 2, class II antigen A, alpha | 1.75 | 0.00240 |
| Slc38a1 | solute carrier family 38, member 1 | 1.75 | 0.00097 |
| 2610002J02Rik | RIKEN cDNA 2610002J02 gene | 1.75 | 0.00069 |
| H2-Ab1 | histocompatibility 2, class II antigen A, beta 1 | 1.73 | 0.00001 |
| Cpxm2 | carboxypeptidase X 2 (M14 family) | 1.71 | 0.04565 |
| Tspo | translocator protein | 1.70 | 0.00000 |
| Scd2 | stearoyl-Coenzyme A desaturase 2 | 1.70 | 0.00147 |
| Grk5 | G protein-coupled receptor kinase 5 | 1.70 | 0.00183 |
Table 4.
Top 50 most significantly downregulated genes in cardiomyocytes of cardiomyocyte-specific GPER female KO vs. WT mice
| Gene Symbol | Gene Description | Fold Change |
P Value |
|---|---|---|---|
| Amy1 | amylase 1, salivary | 0.37 | 0.00002 |
| Acot1 | acyl-CoA thioesterase 1 | 0.41 | 0.00031 |
| Il15 | interleukin 15 | 0.50 | 0.00523 |
| Ssbp2 | single-stranded DNA binding protein 2 | 0.51 | 0.00000 |
| Ano 10 | anoctamin 10 | 0.52 | 0.00000 |
| Dcakd | dephospho-CoA kinase domain containing | 0.53 | 0.00000 |
| Pfkfb1 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 | 0.53 | 0.00010 |
| 2310040G07Rik | RIKEN cDNA 2310040G07 gene | 0.54 | 0.00915 |
| 1700040L02Rik | RIKEN cDNA 1700040L02 gene | 0.54 | 0.00006 |
| Exoc2 | exocyst complex component 2 | 0.55 | 0.00000 |
| Dmd | dystrophin, muscular dystrophy | 0.56 | 0.07599 |
| Maob | monoamine oxidase B | 0.56 | 0.00000 |
| Ucp3 | uncoupling protein 3 (mitochondrial, proton carrier) | 0.56 | 0.00810 |
| Ctnna3 | catenin (cadherin associated protein), alpha 3 | 0.56 | 0.00001 |
| Aldob | aldolase B, fructose-bisphosphate | 0.57 | 0.00022 |
| Wif1 | Wnt inhibitory factor 1 | 0.58 | 0.01025 |
| Egflam | EGF-like, fibronectin type III and laminin G domains | 0.58 | 0.00031 |
| Acot2 | acyl-CoA thioesterase 2 | 0.60 | 0.00067 |
| G0s2 | G0/G1 switch gene 2 | 0.60 | 0.00614 |
| Acot1/Acot2 | acyl-CoA thioesterase 1 / acyl-CoA thioesterase 2 | 0.60 | 0.00105 |
| Immp2l | IMP2 inner mitochondrial membrane peptidase-like | 0.60 | 0.00002 |
| Sox4 | SRY (sex determining region Y)-box 4 | 0.61 | 0.02574 |
| Fdft1 | farnesyl diphosphate farnesyl transferase 1 | 0.61 | 0.00068 |
| Btg1 | B cell translocation gene 1, anti-proliferative | 0.61 | 0.00854 |
| Csrp2 | cysteine and glycine-rich protein 2 | 0.61 | 0.01669 |
| Kcnj3 | potassium inwardly-rectifying channel, subfamily J, member 3 | 0.61 | 0.00813 |
| Fbp2 | fructose bisphosphatase 2 | 0.64 | 0.00494 |
| Tm4sf1 | transmembrane 4 superfamily member 1 | 0.64 | 0.02878 |
| Apbb1 | amyloid beta precursor protein-binding, family B, member 1 | 0.65 | 0.00003 |
| Tesc | tescalcin | 0.65 | 0.00412 |
| Ttll1 | tubulin tyrosine ligase-like 1 | 0.65 | 0.00001 |
| Leprel4 | leprecan-like 4 | 0.65 | 0.00178 |
| Adhfe1 | alcohol dehydrogenase, iron containing, 1 | 0.65 | 0.00118 |
| Fah | fumarylacetoacetate hydrolase | 0.65 | 0.00130 |
| Entpd4 | ectonucleoside triphosphate diphosphohydrolase 4 | 0.66 | 0.04073 |
| Dpyd | dihydropyrimidine dehydrogenase | 0.66 | 0.00043 |
| Fbln1 | fibulin 1 | 0.66 | 0.01377 |
| Efnb3 | ephrin B3 | 0.66 | 0.00088 |
| Stom | stomatin | 0.66 | 0.02360 |
| Lipe | lipase, hormone sensitive | 0.67 | 0.00956 |
| Cdh5 | cadherin 5 | 0.67 | 0.04511 |
| Bcl2l11 | BCL2-like 11 (apoptosis facilitator) | 0.67 | 0.00215 |
| Adra1b | adrenergic receptor, alpha 1b | 0.67 | 0.00369 |
| Slc22a3 | solute carrier family 22 (organic cation transporter), member 3 | 0.67 | 0.00014 |
| AI464131 | expressed sequence AI464131 | 0.68 | 0.00596 |
| Fgf9 | fibroblast growth factor 9 | 0.68 | 0.00002 |
| Snrpn/Snurf | small nuclear ribonucleoprotein N upstream reading frame | 0.68 | 0.00089 |
| Smco4 | single-pass membrane protein with coiled-coil domains 4 | 0.68 | 0.00035 |
| Khdrbs3 | KH domain containing, RNA binding, signal transduction associated 3 | 0.68 | 0.00147 |
| Tmem 164 | transmembrane protein 164 | 0.69 | 0.02876 |
Highlights.
Cardiomyocyte-specific GPER knockout leads to left ventricular dysfunction.
Effects of GPER knockout in the heart are sex-dependent.
GPER in the female heart is related to the enhancements in mitochondrial function.
Inflammatory response genes are enriched in male GPER knockout cardiomyocytes.
Acknowledgments
The GPER mouse strain used for this research project was created from ES cell clone (EPD0334_2_H06) generated by the Wellcome Trust Sanger Institute and made into mice by the KOMP Repository (WWW.KOMP.org) and the Mouse Biology Program (www.mousebiology.org) at the University of California Davis. The mouse strain used for this research project, C57BL/6N-Tg(CAG-Flpo)1Afst/Mmucd, identification number 036512-UCD, was obtained from the Mutant Mouse Regional Resource Center (MMRRC) at UC Davis, a NIH funded strain repository, and was originally donated to the MMRRC by Dr. Konstantinos Anastassiadis from Technische Universitaet Dresden. We appreciate the assistance of Ms. Lou Craddock at Wake Forest University Comprehensive Cancer Center Microarray facility in running the microarray. This work was funded by National Institutes of Health Grants AG-042758 (L.G.), AG-033727 (L.G.), and HL-051952 (C.M.F.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, Cushman M, et al. Heart disease and stroke statistics--2015 update: a report from the American Heart Association. Circulation. 2015;131:e29–e322. doi: 10.1161/CIR.0000000000000152. [DOI] [PubMed] [Google Scholar]
- 2.Mosca L, Benjamin EJ, Berra K, Bezanson JL, Dolor RJ, Lloyd-Jones DM, et al. Effectiveness-based guidelines for the prevention of cardiovascular disease in women--2011 update: a guideline from the American Heart Association. J. Am. Coll. Cardiol. 2011;57:1404–1423. doi: 10.1016/j.jacc.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kelsey SF, James M, Holubkov AL, Holubkov R, Cowley MJ, Detre KM. Results of percutaneous transluminal coronary angioplasty in women. 1985–1986 National Heart, Lung, and Blood Institute’s Coronary Angioplasty Registry. Circulation. 1993;87:720–727. doi: 10.1161/01.cir.87.3.720. [DOI] [PubMed] [Google Scholar]
- 4.Lloyd-Jones D, Adams RJ, Brown TM, Carnethon M, Dai S, De Simone G, et al. Heart disease and stroke statistics--2010 update: a report from the American Heart Association. Circulation. 2012;121:e46–e215. doi: 10.1161/CIRCULATIONAHA.109.192667. [DOI] [PubMed] [Google Scholar]
- 5.Boardman H, Hartley L, Eisinga A, Main C, Figuls MR. Cochrane corner: oral hormone therapy and cardiovascular outcomes in post-menopausal women. Heart. 2016;102:9–11. doi: 10.1136/heartjnl-2015-308287. [DOI] [PubMed] [Google Scholar]
- 6.Hale GE, Shufelt CL. Hormone therapy in menopause: An update on cardiovascular disease considerations. Trends. Cardiovasc. Med. 2015;25:540–549. doi: 10.1016/j.tcm.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 7.Hvidtfeldt UA, Tjønneland A, Keiding N, Lange T, Andersen I, Sørensen TI, et al. Risk of breast cancer in relation to combined effects of hormone therapy, body mass index, and alcohol use, by hormone-receptor status. Epidemiology. 2015;26:353–361. doi: 10.1097/EDE.0000000000000261. [DOI] [PubMed] [Google Scholar]
- 8.Bassuk SSJE. The timing hypothesis: Do coronary risks of menopausal hormone therapy vary by age or time since menopause onset? Metabolism. 2016;65:794–803. doi: 10.1016/j.metabol.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Owman C, Blay P, Nilsson C, Lolait SJ. Cloning of human cDNA encoding a novel heptahelix receptor expressed in Burkitt’s lymphoma and widely distributed in brain and peripheral tissues. Biochem. Biophys. Res. Commun. 1996;228:285–292. doi: 10.1006/bbrc.1996.1654. [DOI] [PubMed] [Google Scholar]
- 10.Deschamps AM, Murphy E. Activation of a novel estrogen receptor, GPER, is cardioprotective in male and female rats. Am. J. Physiol. Heart. Circ. Physiol. 2009;297:H1806–H1813. doi: 10.1152/ajpheart.00283.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang H, Jessup JA, Lin MS, Chagas C, Lindsey SH, Groban L. Activation of GPR30 attenuates diastolic dysfunction and left ventricle remodelling in oophorectomized mRen2. Lewis rats. Cardiovasc. Res. 2012;94:96–104. doi: 10.1093/cvr/cvs090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao Z, Wang H, Jessup JA, Lindsey SH, Chappell MC, Groban L. Role of estrogen in diastolic dysfunction. Am. J. Physiol. Heart. Circ. Physiol. 2004;306:H628–H640. doi: 10.1152/ajpheart.00859.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alencar AK, da Silva JS, Lin M, Silva AM, Sun X, Ferrario CM, et al. Effect of age, estrogen status, and late-Life GPER activation on cardiac structure and function in the Fischer344×Brown Norway female rat. J. Gerontol. A. Biol. Sci. Med. Sci. 2016 Mar 22; doi: 10.1093/gerona/glw045. pii: glw045. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang C, Lv X, Jiang CJS. The putative G-protein coupled estrogen receptor agonist G-1 suppresses proliferation of ovarian and breast cancer cells in a GPER-independent manner. Am. J. Transl. Res. 2012;4:390–402. [PMC free article] [PubMed] [Google Scholar]
- 15.Sharma G, Prossnitz ER. GPER/GPR30 Knockout Mice: Effects of GPER on metabolism. Methods. Mol. Biol. 2016;1366:489–502. doi: 10.1007/978-1-4939-3127-9_38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meoli L, Isensee J, Zazzu V, Nabzdyk CS, Soewarto D, Witt H, et al. Sex- and age-dependent effects of Gpr30 genetic deletion on the metabolic and cardiovascular profiles of diet-induced obese mice. Gene. 2014;540:210–216. doi: 10.1016/j.gene.2014.02.036. [DOI] [PubMed] [Google Scholar]
- 17.Ford J, Hajibeigi A, Long M, Hahner L, Gore C, Hsieh JT, et al. GPR30 deficiency causes increased bone mass, mineralization, and growth plate proliferative activity in male mice. J. Bone. Miner. Res. 2011;26:298–307. doi: 10.1002/jbmr.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Z, Cheng L, Liang H, Duan W, Hu J, Zhi W, et al. GPER inhibits diabetes-mediated RhoA activation to prevent vascular endothelial dysfunction. Eur. J. Cell. Biol. 2016;95:100–113. doi: 10.1016/j.ejcb.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 19.Gui Y, Shi Z, Wang Z, Li JJ, Xu C, Tian R, et al. The GPER agonist G-1 induces mitotic arrest and apoptosis in human vascular smooth muscle cells independent of GPER. J. Cell. Physiol. 2015;230:885–895. doi: 10.1002/jcp.24817. [DOI] [PubMed] [Google Scholar]
- 20.Wang H, Zhao Z, Lin M, Groban L. Activation of GPR30 inhibits cardiac fibroblast proliferation. Mol. Cell. Biochem. 2015;405:135–148. doi: 10.1007/s11010-015-2405-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao Z, Wang H, Lin M, Groban L. GPR30 decreases cardiac chymase/angiotensin II by inhibiting local mast cell number. Biochem. Biophys. Res. Commun. 2015;459:131–136. doi: 10.1016/j.bbrc.2015.02.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bharadwaj MS, Strawn WB, Groban L, Yamaleyeva LM, Chappell MC, Horta C, et al. Angiotensin-converting enzyme 2 deficiency is associated with impaired gestational weight gain and fetal growth restriction. Hypertension. 2011;58:852–858. doi: 10.1161/HYPERTENSIONAHA.111.179358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 25.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 26.Chou JW, Zhou T, Kaufmann WK, Paules RS, Bushel PR. Extracting gene expression patterns and identifying co-expressed genes from microarray data reveals biologically responsive processes. BMC. Bioinformatics. 2007;8:427. doi: 10.1186/1471-2105-8-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang H, Sun X, Chou J, Lin M, Ferrario CM, Zapata-Sudo G, Groban L. Inflammatory and mitochondrial gene expression data in GPER-deficient cardiomyocytes from male and female mice. Data In Brief (Submitted for publication) doi: 10.1016/j.dib.2016.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bopassa JC, Eghbali M, Toro L, Stefani E. A novel estrogen receptor GPER inhibits mitochondria permeability transition pore opening and protects the heart against ischemia-reperfusion injury. Am. J. Physiol. Heart. Circ. Physiol. 2010;298:H16–H23. doi: 10.1152/ajpheart.00588.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Filice E, Recchia AG, Pellegrino D, Angelone T, Maggiolini M, Cerra MC. A new membrane G protein-coupled receptor (GPR30) is involved in the cardiac effects of 17beta-estradiol in the male rat. J. Physiol. Pharmacol. 2009;60:3–10. [PubMed] [Google Scholar]
- 30.Weil BR, Manukyan MC, Herrmann JL, Wang Y, Abarbanell AM, Poynter JA, et al. Signaling via GPR30 protects the myocardium from ischemia/reperfusion injury. Surgery. 2010;148:436–443. doi: 10.1016/j.surg.2010.03.011. [DOI] [PubMed] [Google Scholar]
- 31.Jessup JA, Lindsey SH, Wang H, Chappell MC, Groban L. Attenuation of salt-induced cardiac remodeling and diastolic dysfunction by the GPER agonist G-1 in female mRen2.Lewis rats. PLoS. One. 2010;5:e15433. doi: 10.1371/journal.pone.0015433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Delbeck M, Golz S, Vonk R, Janssen W, Hucho T, Isensee J, et al. Impaired left-ventricular cardiac function in male GPR30-deficient mice. Mol. Med. Rep. 2011;4:37–40. doi: 10.3892/mmr.2010.402. [DOI] [PubMed] [Google Scholar]
- 33.Zimmerman MA, Budish RA, Kashyap S, Lindsey SH. GPER-novel membrane estrogen receptor. Clin. Sci. (Lond) 2016;130:1005–1016. doi: 10.1042/CS20160114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao TZ, Ding Q, Hu J, He SM, Shi F, Ma LT. GPER expressed on microglia mediates the anti-inflammatory effect of estradiol in ischemic stroke. Brain. Behav. 2016;6:e00449. doi: 10.1002/brb3.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Smet MA, Lapauw B, De Backer T. Sex steroids in relation to cardiac structure and function in men. Andrologia. 2016 May 2; doi: 10.1111/and.12610. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 36.Arias-Loza PA, Hu K, Dienesch C, Mehlich AM, König S, Jazbutyte V, et al. Both estrogen receptor subtypes, alpha and beta, attenuate cardiovascular remodeling in aldosterone salt-treated rats. Hypertension. 2007;50:432–438. doi: 10.1161/HYPERTENSIONAHA.106.084798. [DOI] [PubMed] [Google Scholar]
- 37.Fliegner D, Schubert C, Penkalla A, Witt H, Kararigas G, Dworatzek E, et al. Female sex and estrogen receptor-beta attenuate cardiac remodeling and apoptosis in pressure overload. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2010;298 doi: 10.1152/ajpregu.00825.2009. R1597-F1606. [DOI] [PubMed] [Google Scholar]
- 38.Skavdahl M, Steenbergen C, Clark J, Myers P, Demianenko T, Mao L, et al. Estrogen receptor-beta mediates male-female differences in the development of pressure overload hypertrophy. Am. J. Physiol. Heart. Circ. Physiol. 2005;288:H469–H476. doi: 10.1152/ajpheart.00723.2004. [DOI] [PubMed] [Google Scholar]
- 39.Pelzer T, Jazbutyte V, Hu K, Segerer S, Nahrendorf M, Nordbeck P, et al. The estrogen receptor-alpha agonist 16alpha-LE2 inhibits cardiac hypertrophy and improves hemodynamic function in estrogen-deficient spontaneously hypertensive rats. Cardiovasc. Res. 2005;67:604–612. doi: 10.1016/j.cardiores.2005.04.035. [DOI] [PubMed] [Google Scholar]
- 40.Westphal C, Schubert C, Prelle K, Penkalla A, Fliegner D, Petrov G, et al. Effects of estrogen, an ERα agonist and raloxifene on pressure overload induced cardiac hypertrophy. PLoS. One. 2012;7:e50802. doi: 10.1371/journal.pone.0050802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rattanasopa C, Phungphong S, Wattanapermpool J, Bupha-Intr T. Significant role of estrogen in maintaining cardiac mitochondrial functions. J. Steroid. Biochem. Mol. Biol. 2015;147:1–9. doi: 10.1016/j.jsbmb.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 42.Sbert-Roig M, Bauzá-Thorbrügge M, Galmés-Pascual BM, Capllonch-Amer G, García-Palmer FJ, Lladó I, et al. GPER mediates the effects of 17β-estradiol in cardiac mitochondrial biogenesis and function. Mol. Cell. Endocrinol. 2016;420:116–124. doi: 10.1016/j.mce.2015.11.027. [DOI] [PubMed] [Google Scholar]
- 43.McLarty JL, Li J, Levick SP, Janicki JS. Estrogen modulates the influence of cardiac inflammatory cells on function of cardiac fibroblasts. J. Inflamm. Res. 2013;6:99–108. doi: 10.2147/JIR.S48422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu X, Tang Z, Cong B, Du J, Wang C, Wang L, et al. Estrogens increase cystathionine-γ-lyase expression and decrease inflammation and oxidative stress in the myocardium of ovariectomized rats. Menopause. 2013;20:1084–1091. doi: 10.1097/GME.0b013e3182874732. [DOI] [PubMed] [Google Scholar]
- 45.Pechenino AS, Lin L, Mbai FN, Lee AR, He XM, Stallone JN, et al. Impact of aging vs. estrogen loss on cardiac gene expression: estrogen replacement and inflammation. Physiol. Genomics. 2011;43:1065–1073. doi: 10.1152/physiolgenomics.00228.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stice JP, Chen L, Kim SC, Jung JS, Tran AL, Liu TT, et al. 17β-Estradiol, aging, inflammation, and the stress response in the female heart. Endocrinology. 2011;152:1589–1598. doi: 10.1210/en.2010-0627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chakrabarti S, Davidge ST. Analysis of G-protein coupled receptor 30 (GPR30) on endothelial inflammation. Methods. Mol. Biol. 2016;1366:503–516. doi: 10.1007/978-1-4939-3127-9_39. [DOI] [PubMed] [Google Scholar]
- 48.Chakrabarti S, Davidge ST. G-protein coupled receptor 30 (GPR30): a novel regulator of endothelial inflammation. PLoS. One. 2012;7:e52357. doi: 10.1371/journal.pone.0052357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pugach EK, Blenck CL, Dragavon JM, Langer SJ, Leinwand LALA. Estrogen receptor profiling and activity in cardiac myocytes. Mol. Cell. Endocrinol. 2016;431:62–70. doi: 10.1016/j.mce.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Förster C, Kietz S, Hultenby K, Warner M, Gustafsson JA. Characterization of the ERbeta−/−mouse heart. Proc. Natl. Acad. Sci. U. S. A. 2004;101:14234–14239. doi: 10.1073/pnas.0405571101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Irsik DL, Carmines PK, Lane PH. Classical estrogen receptors and ERα splice variants in the mouse. PLoS. One. 2013;8:e70926. doi: 10.1371/journal.pone.0070926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schuster I, Mahmoodzadeh S, Dworatzek E, Jaisser F, Messaoudi S, Morano I, Regitz-Zagrosek V. Cardiomyocyte-specific overexpression of oestrogen receptor β improves survival and cardiac function after myocardial infarction in female and male mice. Clin. Sci. (Lond) 2016;130:365–376. doi: 10.1042/CS20150609. [DOI] [PubMed] [Google Scholar]
- 53.Gao J, Xu D, Sabat G, Valdivia H, Xu W, Shi NQ. Disrupting KATP channels diminishes the estrogen-mediated protection in female mutant mice during ischemia-reperfusion. Clin. Proteomics. 2014;11:19. doi: 10.1186/1559-0275-11-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pfaffl MW, Lange IG, Daxenberger A, Meyer HH. Tissue-specific expression pattern of estrogen receptors (ER): quantification of ER alpha and ER beta mRNA with real-time RT-PCR. APMIS. 2001;109:345–355. doi: 10.1034/j.1600-0463.2001.090503.x. [DOI] [PubMed] [Google Scholar]
- 55.Watanabe T, Akishita M, He H, Miyahara Y, Nagano K, Nakaoka T, et al. 17 beta-estradiol inhibits cardiac fibroblast growth through both subtypes of estrogen receptor. Biochem. Biophys. Res. Commun. 2003;311:454–459. doi: 10.1016/j.bbrc.2003.09.232. [DOI] [PubMed] [Google Scholar]
- 56.Saunders PT, Fisher JS, Sharpe RM, Millar MR. Expression of oestrogen receptor beta (ER beta) occurs in multiple cell types, including some germ cells, in the rat testis. J. Endocrinol. 1998;156:R13–R17. doi: 10.1677/joe.0.156r013. [DOI] [PubMed] [Google Scholar]
- 57.Grohé C, Kahlert S, Löbbert K, Stimpel M, Karas RH, Vetter H, et al. Cardiac myocytes and fibroblasts contain functional estrogen receptors. FEBS. Lett. 1997;416:107–112. doi: 10.1016/s0014-5793(97)01179-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.