Abstract
Efficient and cost-effective fuel ethanol production from lignocellulosic materials requires simultaneous cofermentation of all hydrolyzed sugars, mainly including D-glucose, D-xylose, and L-arabinose. Saccharomyces cerevisiae is a traditional D-glucose fermenting strain and could utilize D-xylose and L-arabinose after introducing the initial metabolic pathways. The efficiency and simultaneous coutilization of the two pentoses and D-glucose for ethanol production in S. cerevisiae still need to be optimized. Previously, we constructed an L-arabinose-utilizing S. cerevisiae BSW3AP. In this study, we further introduced the XI and XR-XDH metabolic pathways of D-xylose into BSW3AP to obtain D-glucose, D-xylose, and L-arabinose cofermenting strain. Benefits of evolutionary engineering: the resulting strain BSW4XA3 displayed a simultaneous coutilization of D-xylose and L-arabinose with similar consumption rates, and the D-glucose metabolic capacity was not decreased. After 120 h of fermentation on mixed D-glucose, D-xylose, and L-arabinose, BSW4XA3 consumed 24% more amounts of pentoses and the ethanol yield of mixed sugars was increased by 30% than that of BSW3AP. The resulting strain BSW4XA3 was a useful chassis for further enhancing the coutilization efficiency of mixed sugars for bioethanol production.
1. Introduction
For the decreasing of fossil energy resources, fuel ethanol was proposed as an important renewable energy and the requirement of bioethanol production is more than ever [1, 2]. Future large-scale production of fuel ethanol will most certainly be based on lignocellulosic materials, which are the most abundant stock in the world [3]. In industry, economical and efficient fuel ethanol production process from lignocellulosic materials requires cofermentation of all hydrolyzed hexoses and pentoses [4]. Moreover, D-xylose and L-arabinose are the most abundant pentoses, which should be utilized [5].
Constructing microorganisms which are capable of fermenting mixed sugars simultaneously is the main challenge in optimizing the production and application of biofuels [6]. Saccharomyces cerevisiae is a traditional strain for fermenting ethanol, which is robust, safe, and suitable for fermenting lignocellulosic hydrolysates. Wild-type S. cerevisiae cannot utilize pentose because of the lack of initial metabolic pathways [7]. By expressing heterologous D-xylose and L-arabinose metabolic pathways, S. cerevisiae could obtain the metabolic capacity [8, 9]; however the efficiency still needs to be further improved.
The main strategies for constructing D-xylose-utilizing S. cerevisiae include two pathways [10, 11]. One is XR-XDH pathway, which contains D-xylose reductase (XR) and xylitol dehydrogenase (XDH), and converts D-xylose to xylulose in S. cerevisiae [12, 13]. Because of the cofactor imbalance in this pathway, the accumulation of byproduct xylitol was the main bottleneck to be solved [14]. Another is XI pathway, which only needs to introduce one D-xylose isomerase (XI), that directly converts D-xylose to xylulose [15, 16]. In this strategy, the activity of XI still needs to be increased [17, 18]. The xylulose from both pathways could then be phosphorylated to xylulose-5-P by endogenous xylulokinase. Xylulose-5-P was further entered into the endogenous pentose phosphate pathway (PPP) to produce ethanol. There are also two main L-arabinose metabolic pathways from fungi or bacteria, which are both candidates for constructing L-arabinose-metabolic yeasts. L-Arabinose could be converted to D-xylulose-5-phosphate through either fungal or bacterial pathway, and then D-xylulose-5-phosphate enters into PPP. The fungal pathway needs five enzymes including aldose reductase (AR), L-arabinitol-4-dehydrogenase (LAD), L-xylulose reductase (LXR), D-xylulose reductase (XDH), and xylulokinase (XK) [19–21]. In addition, this pathway contained two reduction reactions which utilize NADPH, two oxidation reactions which generate NADH, and a kinase reaction. The bacterial pathway contains only three initial enzymes including L-arabinose isomerase (AI), L-ribulokinase (RK), and L-ribulose-5-P-4-epimerase (RPE), which do not require any cofactor [22–24].
Coutilization of D-xylose and L-arabinose could be obtained by combining expression of their metabolic pathways. The first attempt is coexpression of the XR-XDH pathway of D-xylose and the bacterial pathway of L-arabinose [25, 26]. The resulting strains could successfully coutilize of D-xylose and L-arabinose with D-glucose; however, a large amount of L-arabinose was converted to byproduct L-arabitol due to the aldose reductase activity of XR. The D-xylose XI pathway and the bacterial pathway of L-arabinose were then carried out to ignore the effect of XR [24, 27]. The resulting strains could coutilize D-xylose and L-arabinose with D-glucose to produce ethanol well and significantly decreased the byproducts. In this case, the simultaneous utilization capacity of strains to convert D-glucose, D-xylose, and L-arabinose to bioethanol should be further improved. Due to the fungal pathway of L-arabinose metabolism containing the metabolic enzymes for D-xylose utilization, the resulting strains were also able to utilize D-xylose to produce ethanol. The capacity to metabolize mixed sugars of this pathway was also studied in S. cerevisiae [20, 28]. In summary, the studies for simultaneous utilization of pentoses with D-glucose still need to be further performed for high efficiency.
In the present study, we report a different strategy to construct D-glucose, D-xylose, and L-arabinose cofermenting S. cerevisiae. The XI and XR-XDH metabolic pathways were both introduced into the L-arabinose utilizing S. cerevisiae BSW3AP to obtain D-xylose and L-arabinose coutilization capacity. Evolutionary engineering was further used to improve the pentose metabolic efficiency. The sugar utilization of the resulting strain was tested on mixed D-glucose, D-xylose, and L-arabinose and the advantages of this strain were also discussed.
2. Materials and Methods
2.1. Plasmid and Strain Construction
The Ru-xylA fragments of D-xylose isomerase (XI) were cloned from plasmid pJX7 [15] and then inserted into pYX242-TEF1araA [29] between sites EcoR I and Nco I using Gibson assembly [30] to obtain pYX242-XIA (Figure 1). Escherichia coli DH5α was used for plasmid amplification, subcloning, and gene sequencing.
Figure 1.
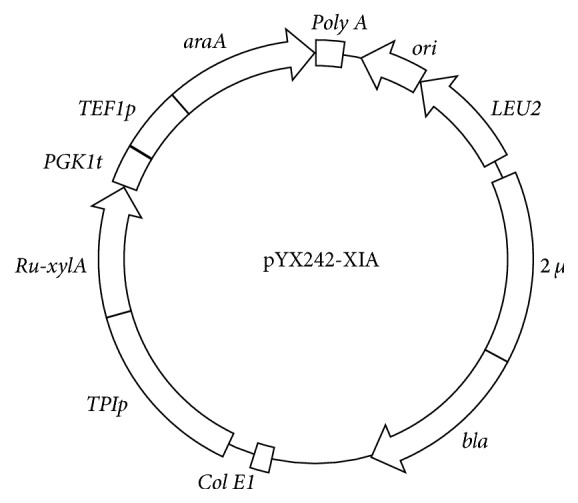
The physical map of plasmid pYX242-XIA.
The formerly obtained L-arabinose fermentation strain BSW3AP [29] was used as the chassis. The yeast transformation was conducted by the conventional lithium acetate transformation method [31]. The episomal plasmid pYX242-TEF1araA of BSW3AP was firstly lost by long time cultivation adding enough leucine and then resulting strain BSW4AP. The plasmid pYX242-XIA was then transferred into BSW4AP to obtain strain BSW4XA1. The plasmid pYMIK-xy127 [32] containing the XR-XDH pathway of D-xylose was digested by restriction endonuclease Hpa I and then integrated into the chromosome of BSW4XA1 to obtain BSW4XA2. After an extensive evolutionary engineering on D-xylose, the evolved strain BSW4XA3 exhibited better sugar coutilizing capacity.
S. cerevisiae strains and plasmids used in this study are listed in Table 1. The primers used in this study are summarized in Table 2.
Table 1.
S. cerevisiae strains and plasmids used in this work.
| Genotype/properties | Source/reference | |
|---|---|---|
| Strain | ||
| BSW3AP | CEN.PK102-3A derivative, gre3(−241, +338):: TPI1p–RKI1–RKI1t–PGK1p–TAL1–TAL1t–FBA1p–TKL1–TKL1t–ADH1p–RPE1–RPE1t–loxP, {YIp5-ara, pYX2422-TEF1araA}, selected for growth on L-arabinose | [29] |
| BSW4AP | BSW3AP derivative, discarding plasmid pYX2422-TEF1araA | Present work |
| BSW4XA1 | BSW4AP derivative, {pYX242-XIA} | Present work |
| BSW4XA2 | BSW4XA1 derivative, {pYX242-XIA, pYMIK-xy127} | Present work |
| BSW4XA3 | BSW4XA2 derivative, selected for growth on D-xylose | Present work |
| Plasmid | ||
| pYX2422-TEF1araA | pYX242-PGK1t-TEF1p-araA | [29] |
| pYX242-XIA | pYX242-Ru-xylA-PGK1t-TEF1p-araA | Present work |
| pYMIK-xy127 | Integration plasmid, KanMX4, ADH1p-XYL1-ADH1t, PGK1p-XYL2-PGK1t, PGK1p-XKS1-PGK1t | [32] |
Table 2.
The main DNA primers used in this work.
| Primers | Sequence (5′-3′) | Purpose |
|---|---|---|
| One-XI up | GCTTAAATCTATAACTACAAAAAACACATACAGGAATTCATGGCAAAAGAATATTTTCC | Cloning Ru-xylA genes |
| One-XI down | TAGAGACATGGGAGATCCTAGCTAGCTAGATCCATGGTTATTTGCAGTGGAGGGCGACG | |
|
| ||
| pYX242-ce-F | GGAGTTTAGTGAACTTGCAAC | To validate or sequence the plasmid pYX242 |
| pYX242-ce-R | CGACTCACTATAGGGCGAATTG | |
| PGKt-pYX2422-R | ATACGCTGAACCCGAACATAG | |
2.2. Media and Incubation
The yeast synthetic complete (SC) medium contains 1.7 g L−1 yeast nitrogen base (YNB, Sangon, China), 5 g L−1 ammonium sulfate (Sangon, China), and the complete supplement mixture 0.77 g L−1 CSM-URA or 0.67 g L−1 CSM-LEU-URA (MP Biomedicals, Solon, OH), and the needed carbon sources (D-glucose, D-xylose or L-arabinose) were used for cultivation of constructed yeasts and maintaining the required plasmids. About 200 μg mL−1 G418 was supplied in the culture when needed. Plasmids were amplified in E. coli strain DH5α (TransGen Biotech, China) growing on Luria-Bertani (LB) medium with 200 μg mL−1 ampicillin.
To cultivate BSW3AP derived strains by batch cultivation, the single colonies were preincubated two times in SC medium containing 20 g L−1 D-glucose for 24 h and 12 h, respectively. After that, cells were collected and used for batch cultivation in the same SC medium containing the needed carbon sources (D-glucose, D-xylose, or L-arabinose). All the batch incubations of yeasts were performed in 40 mL culture for aerobiotic cultivation or oxygen-limited fermentation at 30°C, 200 r min−1. All the cultivations of E. coli strains were performed at 37°C, 200 r min−1.
2.3. Growth Measurement
The culture optical density (OD600) was measured by a BioPhotometer plus (Eppendorf, Germany) to obtain the growth curves. The growth capacities of strains were determined by the exponential growth rates [33], which were analyzed by the linear regression coefficients of lnOD600 versus growth hours from the growth curves [34]. The dry cell weight (DCW) of the strains was calculated using the formula of dry weight (mg mL−1) which is equal to 0.266 × OD600 − 0.0762 [29].
2.4. The Analysis of Metabolites
The concentrations of D-glucose, D-xylose, xylitol, L-arabinose, L-arabitol, and ethanol were determined in the supernatant of filtered samples from oxygen-limited batch cultivation. The high performance liquid chromatography (HPLC) prominence LC-20A (Shimadzu, Japan) with a refractive index detector RID-10A (Shimadzu, Japan) and an Aminex HPX-87P ion exchange column (Bio-Rad, USA) was used to determine the concentration of the above chemicals at 80°C with a mobile phase of water at a flow rate of 0.6 mL min−1, as reported [25, 29].
3. Results
3.1. Coexpressing of D-Xylose Metabolic Pathways in BSW3AP
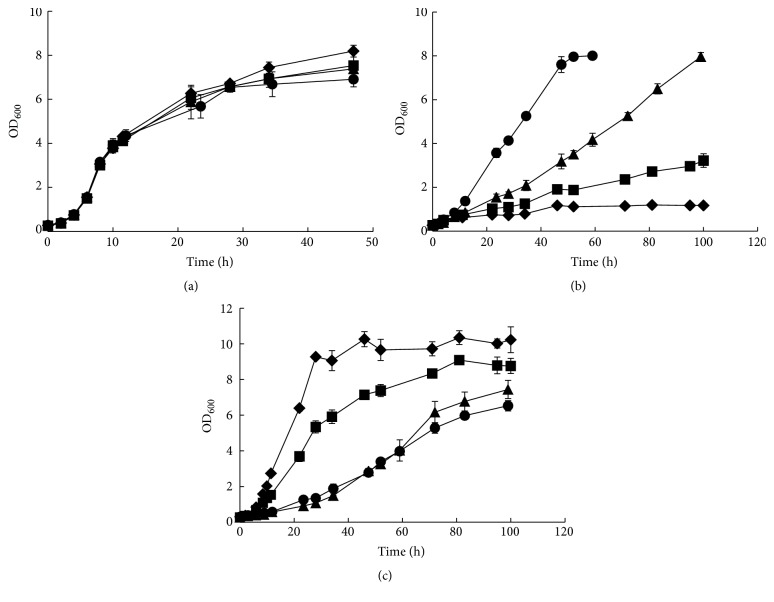
The D-xylose isomerase (XI) Ru-xylA fragments were formerly cloned from bovine rumen metagenome and tested to have higher enzyme activities than other tested ones in S. cerevisiae by our group [15]. So, this gene was overexpressed into the episomal plasmid pYX242-TEF1araA to construct plasmid pYX242-XIA (Figure 1). The formerly obtained L-arabinose fermentation strain BSW3AP [29] was firstly losing the plasmid by long time cultivation with enough leucine to obtain strain BSW4AP. The plasmid pYX242-XIA was then transferred into strain BSW4AP to obtain strain BSW4XA1, which obtained the D-xylose metabolic capacity and could slowly grow on D-xylose as sole carbon source with the maximum specific growth rate (μmax) 0.035 h−1 (Figure 2(b)). However, the D-xylose metabolic capacity of BSW4XA1 was not high enough and not improved after long time evolutionary engineering (data not shown). Furthermore, the D-xylose reductase (XR), xylitol dehydrogenase (XDH), and xylulokinase (XK) of XR-XDH pathway in the plasmid pYMIK-xy127 [32] were then integrated into the chromosome of BSW4XA1 to obtain BSW4XA2 and μmax on D-xylose was further increased up to 0.047 h−1 (Figure 2(b)). The growth capacities of BSW4XA1 and BSW4XA2 on D-glucose were not affected (Figure 2(a)), but μmax on L-arabinose was otherwise decreased from 0.2 h−1 to 0.158 h−1 and 0.047 h−1, respectively (Figure 2(c)).
Figure 2.
The growth curves of strains BSW3AP (◆), BSW4XA1 (■), BSW4XA2 (▲), and BSW4XA3 (●) on 20 g L−1 D-glucose (a), D-xylose (b), and L-arabinose (c), respectively. The strains were preincubated in SC-Leu-Ura medium containing 20 g L−1 D-glucose for 24 h and then transferred using 10 g L−1 D-glucose and 20 g L−1 L-arabinose as the carbon sources to incubate for 24 h. The cells were then collected and used for aerobiotic batch cultivation in 40 mL SC-Leu-Ura medium with 20 g L−1 D-glucose, D-xylose, or L-arabinose at 30°C, 200 r min−1, and the initial OD600 was 0.3. The data are the averages of three independent tests.
3.2. Improving D-Xylose Metabolic Capacity by Evolutionary Engineering
Evolutionary engineering was a useful tool for improving recombinant strains to growth on sugars [27, 35]. The strategy of evolutionary engineering was used here to evolve BSW4XA2 on D-xylose batch culture in air-limited condition. After approximately eight times of transfer, the estimated doubling time was stable at about 11 h (Figure 3). Selected on several D-glucose or D-xylose plates, a big colony was selected out and the ability of D-xylose efficient utilization remained after three times transfer on D-glucose. The selected strain was named BSW4XA3 and μmax on D-xylose was further increased to 0.062 h−1 (Figure 2(b)). It is noteworthy that the growth capacity of it on L-arabinose was not distinctly decreased (Figure 2(c)) and μmax was 0.046 h−1. The growth capacity of it on D-glucose was also not decreased (Figure 2(a)).
Figure 3.
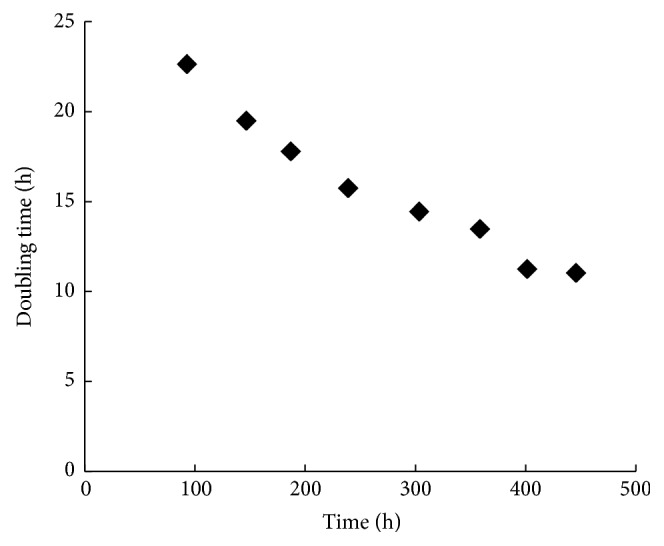
Adaptive cultivation of strain BSW4XA2 on D-xylose. The precultured BSW4XA2 was conducted in a series of batch cultures on 20 g L−1 D-xylose in air-limited condition until the doubling time is stable.
3.3. Coutilization of D-Glucose, D-Xylose, and L-Arabinose for Ethanol Production
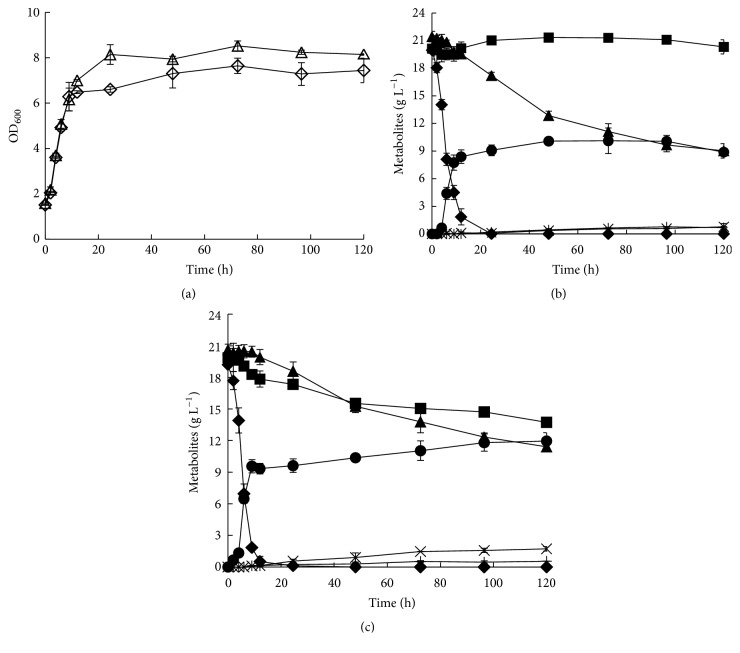
To investigate the capability of strain BSW4XA3 to utilize the mixed sugars of D-glucose, D-xylose, and L-arabinose, an oxygen-limited fermentation of BSW4XA3 and BSW3AP on 20 g L−1 D-glucose, 20 g L−1 D-xylose, and 20 g L−1 L-arabinose was performed (Figure 4 and Table 3). The strains BSW4XA3 and BSW3AP well grow on the mixed sugars with the maximum specific growth rates (μmax) of 0.17 h−1 and 0.16 h−1, respectively. The maximum OD600 of BSW4XA3 is lower than BSW3AP by about 10% (Figure 4(a)), which might indicate that more carbon sources can flow into the ethanol biosynthesis pathway instead of biomass synthesis in BSW4XA3. The strains BSW4XA3 and BSW3AP have similar D-glucose metabolic capacity, with the consumption rates 1.89 g h−1 g−1 DCW and 1.84 g h−1 g−1 DCW, respectively. After 6 h of fermentation, the strains started to prominently consume L-arabinose or D-xylose. BSW3AP could consume L-arabinose with a consumption rate of 0.1 g h−1 g−1 DCW and 12.42 g L−1 L-arabinose was consumed after 120 h of fermentation. During the same time, the L-arabinose consumption rate of BSW4XA3 was decreased to 0.059 g h−1 g−1 DCW and 9.22 g L−1 L-arabinose was consumed. BSW4XA3 successfully obtained the D-xylose metabolic capacity with a consumption rate of 0.055 g h−1 g−1 DCW to consume 6.14 g L−1 D-xylose after 120 h of fermentation. The consumption rates of L-arabinose and D-xylose were similar for BSW4XA3, which could utilize L-arabinose and D-xylose simultaneously. For ethanol productivity, the ethanol yield of BSW4XA3 from mixed sugars was higher than that of BSW3AP. After 120 h of fermentation, BSW4XA3 totally produced 12 g ethanol at an ethanol yield of 0.35 gethanol gconsumed sugars−1; however, BSW3AP only produced 8.9 g ethanol at an ethanol yield of 0.27 gethanol gconsumed sugars−1. In addition, the byproducts L-arabitol or xylitol of the two strains were little accumulated. The L-arabitol productivities of BSW3AP and BSW4XA3 were 0.78 g L−1 and 1.72 g L−1, respectively. Meanwhile, the xylitol productivities of BSW3AP and BSW4XA3 were 0.69 g L−1 and 0.55 g L−1, respectively.
Figure 4.
The metabolic capacities of BSW4XA3 and BSW3AP on mixed sugars of D-glucose, D-xylose, and L-arabinose. (a) The growth curves of strains BSW3AP (△) and BSW4XA3 (⋄). (b) The sugar utilization and sugar alcohols production of BSW3AP. (c) The sugar utilization and sugar alcohols production of BSW4XA3. D-Glucose (◆), D-xylose (■), L-arabinose (▲), ethanol (●), L-arabitol (×), and xylitol (+). The strains were preincubated in SC-Leu-Ura medium containing 20 g L−1 D-glucose for 24 h and then transferred using 20 g L−1 D-glucose, 20 g L−1 D-xylose, and 20 g L−1 L-arabinose as the carbon sources to incubate for 24 h. The cells were then collected and used for fermentation in 40 mL SC-Leu-Ura medium with 20 g L−1 D-glucose, 20 g L−1 D-xylose, and 20 g L−1 L-arabinose at 30°C, 200 r min−1, and the initial OD600 was 1.5. The data are the averages of three independent tests.
Table 3.
Physiological parameters of BSW4XA3 and BSW3AP fermentation on mixed sugars of 20 g L−1 D-glucose, 20 g L−1 D-xylose, and 20 g L−1 L-arabinose.
| Strain | μ max a (h−1) | Consumed sugars in 120 h (g L−1) | Sugar consumption rate (g h−1 g−1 DCW) | Ethanol production in 120 h (g L−1) | Ethanol yieldb (gethanol gconsumed sugars−1) | |||
|---|---|---|---|---|---|---|---|---|
| D-Xylose | L-Arabinose | D-Glucose | D-Xylose | L-Arabinose | ||||
| BSW3AP | 0.16 | 0.00 | 12.42 | 1.84 | 0.00 | 0.10 | 8.89 | 0.27 |
| BSW4XA3 | 0.17 | 6.14 | 9.22 | 1.89 | 0.055 | 0.059 | 11.95 | 0.35 |
The data are the averages of three independent tests.
aThe maximum specific growth rate.
bEthanol yield on all consumed sugars.
4. Discussion
The complete and simultaneous conversion of total sugars from lignocellulosic materials is important for cost-effective bioethanol production [6]. Except D-glucose, the main contents are pentoses D-xylose and L-arabinose. Some studies had been focused on pentose metabolism to obtain S. cerevisiae strains owing to pentoses metabolic capacity, but the efficiencies remain suboptimal [11].
Our group formerly obtained an efficient L-arabinose fermentation strain BSW3AP [29], and in this study we used it as a chassis to introduce two D-xylose metabolic pathways. The D-xylose isomerase (XI) Ru-xylA fragments, which were formerly cloned from bovine rumen metagenome by our group [15], were firstly introduced. After further coexpression of the XR-XDH pathway in the chromosome and evolutionary engineering on D-xylose, the D-xylose metabolic capacities were gradually increased. We finally selected a strain BSW4XA3, which could coutilize mixed sugars.
The former coutilization studies of D-xylose and L-arabinose paid more attention to metabolizing D-xylose first, and the resulting strains somehow presented the gradual utilization of D-glucose, D-xylose, and L-arabinose [20, 28]. The coutilization efficiencies were not high enough. Now, we converse the strategy, introducing two D-xylose metabolic pathways to an L-arabinose fermenting strain. The resulting strain BSW4XA3 well obtained the D-xylose metabolic capacity. BSW4XA3 also presented good simultaneous conversion of D-xylose and L-arabinose to bioethanol with similar consumption rates, and the D-glucose metabolic capacity was not affected. By introducing two D-xylose metabolic pathways and evolutionary engineering, the L-arabinose metabolic capacity of BSW4XA3 was decreased which might be due to the same downstream pathway of the two pentoses from intermediate xylulose 5-phosphate [20]. In strain BSW3AP, the metabolic flux in the pentose downstream pathway was only contributed by L-arabinose. However, a part of the metabolic flux was occupied by D-xylose in BSW4XA3, which might reduce the L-arabinose metabolic efficiency to some extent. Although the metabolic flux of L-arabinose was decreased, the total metabolic flux of pentoses was increased by 24%. It was reported that XR could convert a large amount of L-arabinose to L-arabitol [25, 26], but our results showed that BSW4XA3 metabolized 9.22 g L−1 L-arabinose to only produce 0.94 g L−1 more L-arabitol than BSW3AP, which meant that only 18% L-arabinose was metabolized to L-arabitol. The successful decrease of L-arabitol might be the reason that we used the L-arabinose metabolic strain as the host cell to keep a high L-arabinose metabolic capacity. Meanwhile, the xylitol production was less because the XI pathway played a key regulation function and also might be the expression level of XR which was within an appropriate scope. More D-xylose led to producing ethanol and the ethanol yield was increased by 30% compared with BSW3AP. Former reported results and our results all showed that the pentose metabolic capacity was prominently lower than that of D-glucose due to D-glucose-inhibition effect. To alleviate the phenomenon, the pentose metabolic flux could be further improved and a pentose specific transporter without inhibition by D-glucose might also be further needed. The resulting strain BSW4XA3 in this study was an important basis for further improving the coutilization efficiency of mixed sugars.
5. Conclusions
The complete and simultaneous conversion of total sugars from lignocellulosic materials is important for cost-effective bioethanol production. Using a formally obtained and efficient L-arabinose fermentation strain as the chassis cell, we successfully introduced two D-xylose metabolic pathways. The resulting strain presented good ability for simultaneous conversion of D-xylose and L-arabinose to bioethanol; meanwhile, the D-glucose metabolic capacity was not affected. Furthermore, the total pentose fermentation amounts and bioethanol productivity were also significantly increased. Our present work provides a basis for further improving the coutilization efficiency of mixed sugars in actual bioethanol production process of lignocellulosic materials.
Acknowledgments
This work was supported by the Natural Science Foundation of Shandong Province (ZR2014CL003) and the China Postdoctoral Science Foundation (2015M582121).
Conflicts of Interest
All the authors declare that there are no conflicts of interest.
Authors' Contributions
Chengqiang Wang and Jianzhi Zhao contributed equally to this paper.
References
- 1.Farrell A. E., Plevin R. J., Turner B. T., Jones A. D., O'Hare M., Kammen D. M. Ethanol can contribute to energy and environmental goals. Science. 2006;311(5760):506–508. doi: 10.1126/science.1121416. [DOI] [PubMed] [Google Scholar]
- 2.Mabee W. E. Policy options to support biofuel production. In: Olsson L., editor. Biofuels. Berlin, Germany: Springer; 2007. pp. 329–357. [DOI] [PubMed] [Google Scholar]
- 3.Hahn-Hägerdal B., Galbe M., Gorwa-Grauslund M. F., Lidén G., Zacchi G. Bio-ethanol—the fuel of tomorrow from the residues of today. Trends in Biotechnology. 2006;24(12):549–556. doi: 10.1016/j.tibtech.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 4.Galbe M., Zacchi G. A review of the production of ethanol from softwood. Applied Microbiology and Biotechnology. 2002;59(6):618–628. doi: 10.1007/s00253-002-1058-9. [DOI] [PubMed] [Google Scholar]
- 5.Seiboth B., Metz B. Fungal arabinan and L-arabinose metabolism. Applied Microbiology and Biotechnology. 2011;89(6):1665–1673. doi: 10.1007/s00253-010-3071-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim S. R., Ha S.-J., Wei N., Oh E. J., Jin Y.-S. Simultaneous co-fermentation of mixed sugars: a promising strategy for producing cellulosic ethanol. Trends in Biotechnology. 2012;30(5):274–282. doi: 10.1016/j.tibtech.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 7.Fonseca C., Romão R., Rodrigues De Sousa H., Hahn-Hägerdal B., Spencer-Martins I. l-Arabinose transport and catabolism in yeast. The FEBS Journal. 2007;274(14):3589–3600. doi: 10.1111/j.1742-4658.2007.05892.x. [DOI] [PubMed] [Google Scholar]
- 8.Wiedemann B., Boles E. Codon-optimized bacterial genes improve L-arabinose fermentation in recombinant Saccharomyces cerevisiae. Applied and Environmental Microbiology. 2008;74(7):2043–2050. doi: 10.1128/aem.02395-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim S. R., Park Y.-C., Jin Y.-S., Seo J.-H. Strain engineering of Saccharomyces cerevisiae for enhanced xylose metabolism. Biotechnology Advances. 2013;31(6):851–861. doi: 10.1016/j.biotechadv.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 10.Karhumaa K., Sanchez R. G., Hahn-Hägerdal B., Gorwa-Grauslund M.-F. Comparison of the xylose reductase-xylitol dehydrogenase and the xylose isomerase pathways for xylose fermentation by recombinant Saccharomyces cerevisiae. Microbial Cell Factories. 2007;6, article 5 doi: 10.1186/1475-2859-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee S.-M., Jellison T., Alper H. S. Bioprospecting and evolving alternative xylose and arabinose pathway enzymes for use in Saccharomyces cerevisiae. Applied Microbiology and Biotechnology. 2016;100(5):2487–2498. doi: 10.1007/s00253-015-7211-z. [DOI] [PubMed] [Google Scholar]
- 12.Jeffries T. W., Jin Y.-S. Metabolic engineering for improved fermentation of pentoses by yeasts. Applied Microbiology and Biotechnology. 2004;63(5):495–509. doi: 10.1007/s00253-003-1450-0. [DOI] [PubMed] [Google Scholar]
- 13.Jin Y.-S., Laplaza J. M., Jeffries T. W. Saccharomyces cerevisiae engineered for xylose metabolism exhibits a respiratory response. Applied and Environmental Microbiology. 2004;70(11):6816–6825. doi: 10.1128/aem.70.11.6816-6825.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krahulec S., Klimacek M., Nidetzky B. Analysis and prediction of the physiological effects of altered coenzyme specificity in xylose reductase and xylitol dehydrogenase during xylose fermentation by Saccharomyces cerevisiae. Journal of Biotechnology. 2012;158(4):192–202. doi: 10.1016/j.jbiotec.2011.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hou J., Shen Y., Jiao C., Ge R., Zhang X., Bao X. Characterization and evolution of xylose isomerase screened from the bovine rumen metagenome in Saccharomyces cerevisiae. Journal of Bioscience and Bioengineering. 2016;121(2):160–165. doi: 10.1016/j.jbiosc.2015.05.014. [DOI] [PubMed] [Google Scholar]
- 16.Kuyper M., Hartog M. M. P., Toirkens M. J., et al. Metabolic engineering of a xylose-isomerase-expressing Saccharomyces cerevisiae strain for rapid anaerobic xylose fermentation. FEMS Yeast Research. 2005;5(4-5):399–409. doi: 10.1016/j.femsyr.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 17.Lee S.-M., Jellison T., Alper H. S. Directed evolution of xylose isomerase for improved xylose catabolism and fermentation in the yeast Saccharomyces cerevisiae. Applied and Environmental Microbiology. 2012;78(16):5708–5716. doi: 10.1128/aem.01419-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hou J., Jiao C., Peng B., Shen Y., Bao X. Mutation of a regulator Ask10p improves xylose isomerase activity through up-regulation of molecular chaperones in Saccharomyces cerevisiae. Metabolic Engineering. 2016;38:241–250. doi: 10.1016/j.ymben.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 19.Hahn-Hägerdal B., Karhumaa K., Jeppsson M., Gorwa-Grauslund M. Metabolic engineering for pentose utilization in Saccharomyces cerevisiae. In: Olsson L., editor. Biofuels. Berlin, Germany: Springer; 2007. pp. 147–177. [DOI] [PubMed] [Google Scholar]
- 20.Bettiga M., Bengtsson O., Hahn-Hägerdal B., Gorwa-Grauslund M. F. Arabinose and xylose fermentation by recombinant Saccharomyces cerevisiae expressing a fungal pentose utilization pathway. Microbial Cell Factories. 2009;8, article 40 doi: 10.1186/1475-2859-8-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richard P., Verho R., Putkonen M., Londesborough J., Penttilä M. Production of ethanol from L-arabinose by Saccharomyces cerevisiae containing a fungal L-arabinose pathway. FEMS Yeast Research. 2003;3(2):185–189. doi: 10.1016/s1567-1356(02)00184-8. [DOI] [PubMed] [Google Scholar]
- 22.Schleif R. Regulation of the L-arabinose operon of Escherichia coli. Trends in Genetics. 2000;16(12):559–565. doi: 10.1016/s0168-9525(00)02153-3. [DOI] [PubMed] [Google Scholar]
- 23.Becker J., Boles E. A modified Saccharomyces cerevisiae strain that consumes L-arabinose and produces ethanol. Applied and Environmental Microbiology. 2003;69(7):4144–4150. doi: 10.1128/aem.69.7.4144-4150.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wisselink H. W., Toirkens M. J., Berriel M. D. R. F., et al. Engineering of Saccharomyces cerevisiae for efficient anaerobic alcoholic fermentation of L-arabinose. Applied and Environmental Microbiology. 2007;73(15):4881–4891. doi: 10.1128/aem.00177-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garcia Sanchez R., Karhumaa K., Fonseca C., et al. Improved xylose and arabinose utilization by an industrial recombinant Saccharomyces cerevisiae strain using evolutionary engineering. Biotechnology for Biofuels. 2010;3, article 13 doi: 10.1186/1754-6834-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karhumaa K., Wiedemann B., Hahn-Hägerdal B., Boles E., Gorwa-Grausland M.-F. Co-utilization of L-arabinose and D-xylose by laboratory and industrial Saccharomyces cerevisiae strains. Microbial Cell Factories. 2006;5, article 18 doi: 10.1186/1475-2859-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wouter Wisselink H., Toirkens M. J., Wu Q., Pronk J. T., Van Maris A. J. A. Novel evolutionary engineering approach for accelerated utilization of glucose, xylose, and arabinose mixtures by engineered Saccharomyces cerevisiae strains. Applied and Environmental Microbiology. 2009;75(4):907–914. doi: 10.1128/aem.02268-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bera A. K., Sedlak M., Khan A., Ho N. W. Y. Establishment of l-arabinose fermentation in glucose/xylose co-fermenting recombinant Saccharomyces cerevisiae 424A(LNH-ST) by genetic engineering. Applied Microbiology and Biotechnology. 2010;87(5):1803–1811. doi: 10.1007/s00253-010-2609-0. [DOI] [PubMed] [Google Scholar]
- 29.Wang C., Shen Y., Zhang Y., Suo F., Hou J., Bao X. Improvement of L-arabinose fermentation by modifying the metabolic pathway and transport in Saccharomyces cerevisiae. BioMed Research International. 2013;2013:9. doi: 10.1155/2013/461204.461204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gibson D. G. Enzymatic assembly of overlapping DNA fragments. Methods in Enzymology. 2011;498:349–361. doi: 10.1016/B978-0-12-385120-8.00015-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gietz R. D., Schiestl R. H., Willems A. R., Woods R. A. Studies on the transformation of intact yeast cells by the LiAc/SS‐DNA/PEG procedure. Yeast. 1995;11(4):355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 32.Wang Y., Shi W.-L., Liu X.-Y., et al. Establishment of a xylose metabolic pathway in an industrial strain of Saccharomyces cerevisiae. Biotechnology Letters. 2004;26(11):885–890. doi: 10.1023/b:bile.0000025897.21106.92. [DOI] [PubMed] [Google Scholar]
- 33.Young E. M., Tong A., Bui H., Spofford C., Alper H. S. Rewiring yeast sugar transporter preference through modifying a conserved protein motif. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(1):131–136. doi: 10.1073/pnas.1311970111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang C., Bao X., Li Y., et al. Cloning and characterization of heterologous transporters in Saccharomyces cerevisiae and identification of important amino acids for xylose utilization. Metabolic Engineering. 2015;30:79–88. doi: 10.1016/j.ymben.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 35.Peng B., Shen Y., Li X., Chen X., Hou J., Bao X. Improvement of xylose fermentation in respiratory-deficient xylose-fermenting Saccharomyces cerevisiae. Metabolic Engineering. 2012;14(1):9–18. doi: 10.1016/j.ymben.2011.12.001. [DOI] [PubMed] [Google Scholar]