Abstract
Viral infectivity factor (Vif) is one of the human immunodeficiency virus (HIV) accessory proteins and is conserved in the primate lentivirus group. This protein is essential for viral replication in vivo and for productive infection of nonpermissive cells, such as peripheral blood mononuclear cells (PBMC). Vif counteracts an antiretroviral cellular factor in nonpermissive cells named CEM15/APOBEC3G. Although HIV type 1 (HIV-1) Vif protein (Vif1) can be functionally replaced by HIV-2 Vif protein (Vif2), its identity is very small. Most of the functional studies have been carried out with Vif1. Characterization of functional domains of Vif2 may elucidate its function, as well as differences between HIV-1 and HIV-2 infectivity. Our aim was to identify the permissivity of different cell lines for HIV-2 vif-minus viruses. By mutagenesis specific conserved motifs of HIV-2 Vif protein were analyzed, as well as in conserved motifs between Vif1 and Vif2 proteins. Vif2 mutants were examined for their stability, expression, and cellular localization in order to characterize essential domains of Vif2 proteins. Viral replication in various target cells (PBMC and H9, A3.01, U38, and Jurkat cells) and infectivity in single cycle assays in the presence of APOBEC3G were also analyzed. Our results of viral replication show that only PBMC have a nonpermissive phenotype in the absence of Vif2. Moreover, the HIV-1 vif-minus nonpermissive cell line H9 does not show a similar phenotype for vif-negative HIV-2. We also report a limited effect of APOBEC3G in a single-cycle infectivity assay, where only conserved domains between HIV-1 and HIV-2 Vif proteins influence viral infectivity. Taken together, these results allow us to speculate that viral inhibition by APOBEC3G is not the sole and most important determinant of antiviral activity against HIV-2.
Human immunodeficiency virus type 1 (HIV-1) and HIV-2 are etiologic agents of AIDS (8, 17). The genomic organization of these viruses is very similar, resulting in an average protein homology of 35% (33). Structural differences of HIV-1 and HIV-2 reflect distinct biological and clinical behaviors, namely, in infectivity. Although HIV-1 is found worldwide, HIV-2 is still mainly localized in West Africa, suggesting that HIV-2 is less infectious (38, 54).
The viral infectivity of HIV is modulated by the viral infectivity factor (Vif) (15, 49). HIV-1 Vif (Vif1) is a 192-amino-acid protein conserved in all known lentiviruses with the exception of equine infectious anemia virus (35). Functional studies on Vif1 established that it acts during the late stage of viral production, in a cell-dependent manner, and its absence impairs the early stages of infection. Accordingly, it was demonstrated that Vif-deficient HIV-1 is impaired in endogenous reverse transcription to complete proviral DNA in newly infected cells (3, 9, 10, 15, 34, 49, 51, 55). Cells have been classified as permissive (Vif is dispensable for HIV replication) and nonpermissive (Vif is required for HIV replication) (1, 10, 40, 55). Vif is essential for viral replication in primary T lymphocytes, in macrophages, and in some T-cell lines such as H9 (16, 40). The intracellular localization of Vif1 was reported at the plasma membrane (18, 19, 48), the cytoplasm (18, 23, 32, 47), the cytoskeleton (21, 23), and the nucleus (7, 22), as well as in purified virus preparations (3, 23, 26).
Recently, it was demonstrated that Vif acts by interaction with an antiviral host factor (44). This cellular factor (CEM15) was identified in nonpermissive cells for HIV-1 and later called APOBEC3G. This protein belongs to a family of nucleic acid-editing enzymes related to APOBEC1, a cytidine deaminase that edits the apolipoprotein B mRNA. APOBEC3G acts as a DNA mutator of the minus strand of retroviral DNA, converting cytosine to uracil (20, 28, 29, 61). Vif protein promotes the degradation of APOBEC3G from virus-producing cells by inducing its ubiquitination and subsequent proteasome degradation (30, 31, 45, 60, 61).
The HIV-2 vif gene has only 25% identity with vif from HIV-1 (39). Analysis of the HIV-2 Vif protein (Vif2) has been less extensive than studies with Vif1. Differences in cytopathicity, host range, and susceptibility to neutralizing antibodies have been reported for various HIV-2 isolates (5, 43). Characterization of Vif2 functional domains may elucidate its function, as well as differences between HIV-1 and HIV-2 infectivity. Moreover, since Vif is essential for viral infection, its molecular characterization should provide information on potential antiretroviral strategies.
Functional studies of Vif1 have identified important sequence motifs and amino acid residues (19, 50, 58). Two cysteine residues (27) and a major conserved motif (144-SLQXLA-149) (59) are essential for Vif1 function. Mutations in the phosphorylation site of Vif (Ser144) cause a defect in viral infectivity (57-59). The C-terminal domain of Vif1 is required for membrane localization and interaction with membrane-associated proteins (19). It was also showed that Vif interacts with the Gag precursor (4). Deletion analysis showed that Vif is packaged into a ribonucleoprotein complex by an interaction of its central region with viral genomic RNA and suggested that its incorporation into virions is important for its function (25). More recently, amino acid residues Glu88 and Trp89 in the central hydrophilic region of Vif1 were demonstrated to be critical for viral infectivity by enhancing the steady-state expression of Vif (14).
The aim of the present study was to identify the permissivity of different cell lines for HIV-2 vif-minus viruses. We performed mutational analyses of Vif2 with deletions and substitutions introduced into specific conserved motifs of the protein (39), as well as in conserved motifs between Vif1 and Vif2 (35). Mutant proteins were also characterized for their stability, expression, and cellular localization. Virus replication in various target cells and infectivity in single-cycle assays in the presence of APOBEC3G was also analyzed.
MATERIALS AND METHODS
Vectors and DNA constructs.
The proviral expression vector pKP59 (HIV-2ROD), the vesicular stomatitis virus glycoprotein expression vector (pVSV-G), and plasmids coding for pHIV-1NL43 and pHIV-1NL43 vif-minus viruses were obtained from AIDS Research and Reference Reagent Program. A 2,268-bp fragment of HIV-2ROD containing the vif gene was cloned in plasmid pBK-CMV (Stratagene). This plasmid was used for PCR-mediated site-directed mutagenesis of vif gene. Vif2 defective plasmid (MDSTOP) was constructed by altering the vif sequence encoding amino acids 51 and 52 (TGGTGG) for two in-frame stop codons (TGATGA). MD1A and MD1B are deletion mutants in the conserved motif of Vif2 protein (amino acids 42 to 61). Vif MD1A has a deletion from amino acids 42 to 50, and Vif MD1B has a deletion from amino acids 51 to 61. MD5 (amino acids 24 to 31) and MD8 (amino acids 147 to 153) are deletion mutants of Vif2 protein in conserved regions between Vif2 and Vif1. M2 was altered in the conserved region of HIV-2 Vif protein between amino acids 120 and 130 by substitution of Arg123, Arg124, and Arg127 for alanine. Vif M6 and Vif M7 are substitution mutants in the conserved cysteine amino acids Cys116Arg and Cys134Arg/Cys135Gly. The integrity of Vif protein mutants was confirmed by sequencing. All Vif2 mutants were cloned into proviral HIV-2ROD by replacing the BclI/PmaCI fragment of pKP59 (amino acids 4665 to 6238) with the corresponding fragment from pBK-CMV Vif mutant clones. APOBEC3G was amplified from the H9 cell line cDNA by using the oligonucleotides 5′-GAATTCAAGGATGAAGCCTCACTTCAGA-3′ and 5′-GACTGCAGCCCATCCTTCAGTTTTCCTG-3′ and cloned in pcDNA3.1 (Invitrogen). The sequence of hemagglutinin (HA) tag was added at the C-terminal end of APOBEC3G. The expression plasmids of Vif wild-type and Vif mutant clones were constructed by PCR amplification and cloned into BamHI and EcoRI restriction sites of plasmid AS1B containing the first 15 codons of the HA sequence under control of cytomegalovirus (CMV) promoter. Expression plasmids NP and TE contain the open reading frames of a nuclear protein and the thioesterase cloned by PCR amplification in plasmid AS1B.
Immunolabeling.
HeLa cells were cultured in complete Dulbecco modified Eagle medium supplemented with 10% fetal calf serum. Cells (106) were grown on coverslips and transfected with the corresponding DNA by the CaCl2 method (41). Mouse and rat anti-HA monoclonal antibodies (Roche) were used as primary antibodies. Secondary antibodies conjugated with fluorescein isothiocyanate or Cy3 were obtained from Jackson Immunoresearch Laboratories (0.5 to 1 μg/ml; multiple-labeling grade). Immunolabeling was performed according to the method of Pombo et al. (36). Confocal microscopy was performed by using a hybrid Bio-Rad MRC1000/MRC1024 confocal laser-scanning microscope (running under Comos 7.0a) equipped with an argon-krypton laser and coupled to a Nikon Diaphot 200 inverted microscope (×60 PlanApo oil-immersion objective lens; numerical aperture, 1.4). Kalman-filtered images (n = 6 to 15) were collected sequentially with a minimum iris aperture (0.7 mm), and the minimum laser power that filled the whole gray scale in the low scan/low signal mode.
In vitro transcription and translation.
Mutant and wild-type vif genes cloned in plasmid pBK-CMV were expressed by using a transcription-translation reaction with a TNT Coupled Reticulocyte Lysate System (Promega) performed according to the manufacturer's instructions. Radiolabeled products were resolved by standard reducing sodium dodecyl sulfate-7.5% polyacrylamide gel electrophoresis (SDS-7.5% PAGE) and exposed to X-ray film.
Cell culture and virus.
293T, HeLa, HeLa CD4+, and HeLa P4 cells were propagated in Dulbecco modified Eagle medium containing 10% fetal bovine serum (FBS). T-cell lines SupT1, H9, Jurkat, and A3.01 and the monocyte/macrophage cell line U38 were grown in complete RPMI 1640 medium supplemented with 10% FBS. All cell cultures were maintained at 37°C in 5% CO2. Human peripheral blood mononuclear cells (PBMC) from healthy donors were treated for 72 h with phytohemagglutinin and maintained in RPMI 1640 medium supplemented with 15% FBS and 20 U of interleukin-2 (Roche)/ml. Virus stocks were prepared by cotransfection of 293T cells with HIV-2ROD or HIV-2ROD vif mutant proviruses plus pVSV-G with Fugene reagent according to the manufacturer's protocol (Roche). Virus-containing supernatants were harvested 48 h posttransfection, and cellular debris was removed by centrifugation at 3,000 rpm for 10 min at 4°C. Assay of p24 antigen was performed as recommended by the manufacturer (Virinostika HIV-1 Antigen; bioMérieux). The activity of virion-associated reverse transcriptase was measured according to the manufacturer's protocol (Lenti-RT Cavidi).
Viral infections.
PBMC, HeLa CD4+, and T-cell lines (106) were infected with p24 and p26 normalized HIV-1 and HIV-2 virus samples (30,000 pg/ml). At 24 h postinfection the cells were washed three times with phosphate-buffered saline. All p26 values were adjusted by a factor of 10 due to the low level of detection of HIV-2 capsid antigen by the HIV-1 assay kit used (data not shown). To monitor infections, aliquots were taken at the indicated time points, and HIV-2 p24/p26 antigen levels were determined.
Total RNA and Northern blot analysis.
Total RNAs were isolated from uninfected cells and prepared by using RNeasy minikits (Qiagen). RNA samples (20 μg of each cell line) were analyzed by electrophoresis on denatured 1.2% agarose gels and transferred to a nylon membrane. The filters were hybridized with [γ-32P]dATP (3,000 Ci/mmol; Amersham Pharmacia Biotech) end-labeled probes by using T4 polynucleotide kinase (USB-Amersham) and visualized by autoradiography.
To detect APOBEC3G mRNA, the oligonucleotide 5′-GACTGCAGCCCATCCTTCAGTTTTCCTG-3′ corresponding to the terminal end of the APOBEC3G gene was used. β-Actin mRNA was identified by using the primer 5′-GGACTCGTCATACTCCTGCTTGC-3′ complementary to the Homo sapiens β-actin gene terminal end.
Immunoprecipitation and Western blotting.
For Western blot analysis 293T cells were cotransfected by Fugene (Roche) with 1 μg of DNA from APOBEC3G containing plasmid and the proviral plasmids Vif2 STOP, Vif2 wild type, or Vif2 mutant cloned in ASIB. At 48 h posttransfection, cells were lysed in radioimmunoprecipitation buffer (0.15 M NaCl, 0.05 M Tris-HCl [pH 7.2], 1% Triton X-100, 0.1% SDS, and 1% sodium deoxycholate) in ice and immunoprecipitated with anti-HA affinity matrix (Roche). Proteins were separated by SDS-polyacrylamide gel electrophoresis, and protein bands were transferred onto nitrocellulose membrane (Amersham Biosciences). Proteins were detected by ECL (Amersham Biosciences) with horseradish peroxidase-conjugated rat anti-HA monoclonal antibody 3F10 (Roche).
One-cycle infectivity assay.
A one-cycle infectivity assay was used to measure the ability of wild-type or mutant Vif proteins to complement a single-round replication of HIV-2 vif-minus virus in trans. Briefly, 293T cells were cotransfected by Fugene (Roche) with 1 μg of DNA proviral plasmids encoding HIV-2ROD vif-minus or HIV-2ROD plus VSV-G envelope, with 1 μg of APOBEC3G and HA-Vif2 wild-type or HA-Vif2 mutant proteins. As controls, cells were transfected with HIV-1NL43 vif-minus complemented by HA-Vif wild type plus VSV-G envelope and APOBEC3G. For determinations of the titers of APOBEC3G necessary to inhibit the HIV-1 vif-minus and HIV-2 vif-minus mutants, 293T cells were transfected with proviral plasmids encoding Vif and without Vif plus VSV-G envelope, along with increasing concentrations of APOBEC3G. The plasmid ratios of provirus to APOBEC3G were 1:0, 1:1, 1:4, and 1:10. Viruses in the supernatant were collected at 48 h posttransfection, and virus titers were measured by using the p26 antigen. Normalized viral titers were used to infect P4 target cells. On day 2, infected cells were lysed and the ability of wild-type or mutant Vif proteins to complement a single round of infection was measured by a chemiluminescent β-galactosidase assay according to the manufacturer's protocol (Roche). Values shown are the percentage of infectivity relative to HIV vif-minus mutant complemented with Vif wild-type protein.
RESULTS
Characterization of cell line permissivity for Vif-minus HIV-2.
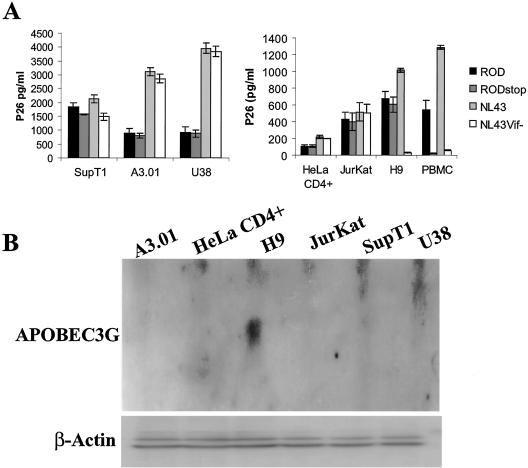
Several studies have identified the cellular nonpermissivity for HIV-1 vif-minus replication in which H9 constitutes the standard nonpermissive cell line for Vif function (15, 55). Nevertheless, studies carried out with HIV-2 vif-minus virus in different laboratories produced contradictory results regarding cellular nonpermissivity (24, 32, 37, 53). Therefore, we analyzed HIV-2 vif-minus replication in different cell lines for functional analysis of Vif protein. To define the cellular nonpermissivity to Vif-defective HIV-2, we constructed a HIV-2ROD vif-minus (ROD-STOP) where two in-frame stop codons were introduced at the N-terminal region of vif gene. Proviral plasmids HIV-2ROD and HIV-2RODSTOP were used to cotransfect 293T cells with pVSV-G. Pseudotyped viruses in the supernatant were harvested, quantified, and used to compare their abilities to replicate in several cell types, namely, PBMC and A3.01, Jurkat, H9, SupT1, HeLa CD4+, and U38 cells. Ten days after infection, the ability of viruses to replicate was measured by quantification of p26 antigen levels in culture supernatants. As a control, we carried out the same infection experiments with HIV-1NL43 and HIV-1NL43 vif-minus proviral plasmids (Fig. 1A). In general, we observed that infections by pseudotyped HIV-1 produce higher levels of p24/p26 than pseudotyped HIV-2 viruses and also that some cell lines are more productive (SupT1, A3.01, and U38) than others (HeLa CD4+ and Jurkat). Two patterns of viral replication were observed. First, PBMC did not support the spread of HIV-2ROD vif-minus virus, corresponding to the nonpermissive character, since it was also observed for HIV-1NL43 vif-minus virus, as previously described (15, 55). A second group that includes all other cell lines tested showed that the lack of Vif expression had no visible consequences in HIV-2 viral replication, corresponding to a permissive character. The monocyte/macrophage cell line U38, derived from U937 cell line, is permissive for HIV-2ROD vif-minus (Fig. 1A) as described previously with HIV-2 chimera strain La317 infection of U937 cells (24). In contrast to HIV-1 vif-minus, H9 cells show a permissive character to Vif-defective HIV-2ROD, confirming previous reports (24). These results show that HIV-2ROD vif-minus virus has different cell-type permissivities compared to HIV-1 vif-minus virus. To confirm cell line phenotypes, we measured APOBEC3G expression by Northern analysis (Fig. 1B), and we verified that APOBEC3G was only expressed in H9 cells.
FIG. 1.
Characterization of cell line permissivity for Vif-defective HIV-2. (A) The abilities of HIV-2ROD and HIV-2RODSTOP viruses to replicate were compared in several cell lines, namely, SupT1, A3.01, U38, and HeLaCD4+, Jurkat, and H9 cells and PBMC. HIV-1NL43 and HIV-1NL43 vif-minus viruses were used for control of cell line permissivity character. Quantification of p26 levels was performed in cell culture supernatants 10 days after infection. PBMC are the only cells corresponding to the nonpermissive character for HIV-2 replication in the absence of Vif. (B) Northern analysis of APOBEC3G expression in A3.01, Jurkat, H9 HeLa CD4+, SupT1, and U38 cells. RNAs extracted from uninfected cells were resolved by electrophoresis and subjected to Northern analysis with 32P-labeled APOBEC3G probe (top panel). For internal control, a β-actin specific probe was used (bottom panel).
In vitro and in vivo characterization of HIV-2 Vif mutants.
To characterize the functional domains of Vif2, mutants were generated in conserved motifs and specific amino acid residues of the protein. In Fig. 2 is shown the comparison of Vif2 and Vif1 proteins where the conserved motifs are indicated (39). An identity of 25% was observed between these two proteins. Conserved motifs in HIV-2 Vif proteins are boxed (continuous line), as well as conserved motifs between Vif proteins from HIV-1 and HIV-2 (broken line). Essential amino acid residues in Vif1 are also indicated.
FIG. 2.
Sequence alignment of Vif proteins from HIV-2 and HIV-1. Conserved motifs in Vif2 proteins are boxed (continuous line), as well as conserved motifs between Vif protein from HIV-1 and HIV-2 (broken line). Arrows indicate essential cysteines and phosphorylated amino acid residues in Vif1 protein.
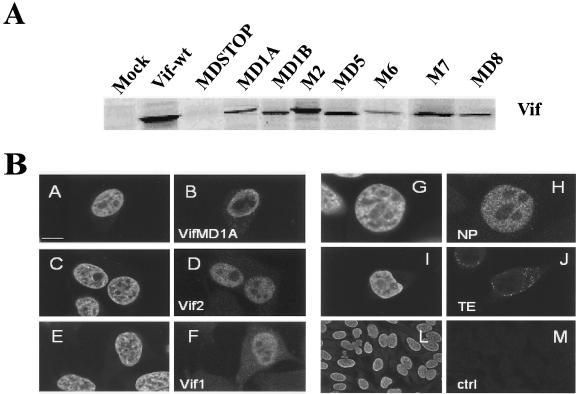
To assess the stability of Vif2 mutant proteins, we performed in vitro transcription-translation experiments. Mutants of vif genes were cloned into the eukaryotic expression plasmid pBK-CMV, and in vitro expression was analyzed. The results shown in Fig. 3A indicate that Vif2 mutant proteins are expressed with molecular weights similar to those of the wild-type protein. Differences in the molecular weight of Vif2 deletion mutants are visible. As expected. no protein was expressed when Vif2 mutant MDSTOP was used since two in-frame stop codons (TGATGA) were introduced in the N-terminal portion of vif gene. To confirm in vivo expression of Vif2 mutants as well as cellular localization, HeLa cells were transfected with plasmids encoding vif wild-type (Vif1 and Vif2) and vif mutant (VifMD1A) constructs cloned in fusion with HA epitope at the N terminus. As controls of nuclear and cytoplasmic localization, we used a nuclear protein (NP) and a cytoplasmic protein (TE-thioesterase) cloned in the same vector. At 18 h posttransfection, protein localization was detected with monoclonal antibody against HA epitope (Fig. 3B, subpanels B, D, F, H, and J) and TOTO-3 for nucleic acids staining (Fig. 3B, subpanels A, C, E, G, I, and L). The results show the expression of wild-type HA-Vif2 and HA-Vif1 (Fig. 3B, subpanels D and F) and of HA-Vif2 MD1A (Fig. 3B, subpanel B) in the nucleus, similar to that obtained with all other mutants (data not shown). Comparison of subpanels A and B, C and D, and E and F in Fig. 3B shows that HA-Vif2 and HA-Vif1 wild-type and HA-Vif2 MD1A present a nuclear localization pattern confirmed by TOTO-3 staining. These results were obtained under mild conditions of fixation (4% paraformaldehyde). Nevertheless, when methanol fixation was used, the morphology of cells was altered and Vif is seen in both the nucleus and the cytoplasm, probably indicating that internal cellular structures were disrupted (data not shown). Although Vif has been described predominantly as a cytoplasmic protein (11, 18, 19, 32, 48), several reports also show the presence of substantial Vif protein in the nucleus (7, 18, 22, 23, 48). For example, Vif from FIV is primarily in the nucleus (7), but in HIV-1-transfected cells Vif is less evident in nuclear fractions (18, 22, 23, 48). Therefore, our results of HIV-2 Vif protein localization in the nucleus show that its localization may depend on different viral settings. Further assays will have to be performed to analyze the importance of this cellular localization. Furthermore, our results demonstrate that Vif2 mutant constructs present a stable cellular expression.
FIG. 3.
(A) Analysis of in vitro transcription-translation products with different DNA constructs encoding Vif2 wild-type and Vif2 mutant proteins. The results show that Vif2 mutant proteins are expressed with molecular weights similar to those of the wild-type protein. Nevertheless, an increased mobility is noted for deletion mutants. As expected, when vif-containing stop codon (MDSTOP) was used no protein was expressed. (B) Cellular localization of Vif2 mutants by using immunolabeling detection. HeLa cells were transfected with Vif2 and Vif1 wild-type and Vif2 mutant constructs cloned in fusion with HA epitope at the N terminus. Cells were fixed 18 h posttransfection, and detection was performed by indirect immunofluorescence. Nucleic acids were counterstained with TOTO-3 (in subpanels A, C, E, G, I, and L). The results show the expression of HA-Vif2 MD1A (subpanel B) and wild-type HA-Vif2 and HA-Vif1 (subpanels D and F) in the nucleus. Positive controls for nuclear localization NP (subpanel H) and cytoplasmic localization TE (subpanel J) were used. Nontransfected cells (subpanels L and M) show no background staining or nonspecific reactivity from the antibodies. Equatorial optical sections were collected on a confocal laser-scanning microscope. Bars, 10 μm.
Conserved Vif motifs are essential for HIV-2 replication.
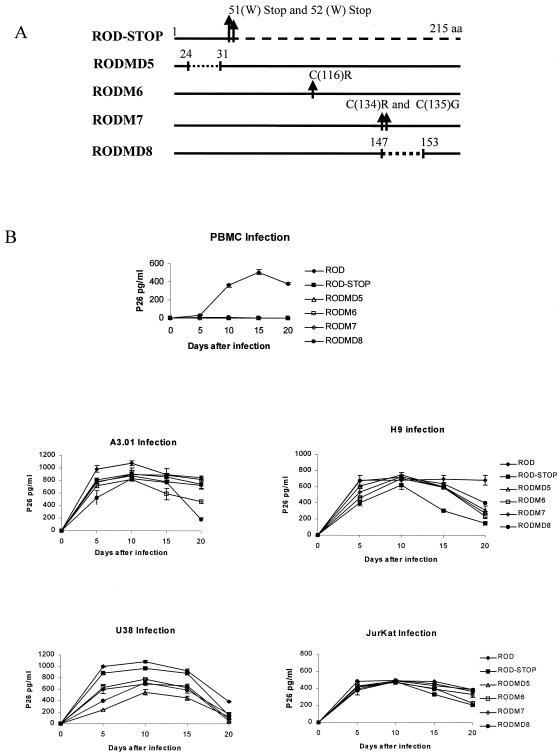
To evaluate which conserved motifs of Vif are essential for viral replication, protein mutants were introduced into HIV-2ROD proviral DNA as detailed in Materials and Methods. Viral stocks were produced by cotransfection of 293T cells with pVSV-G and proviral clones of HIV-2 vif mutants. At 48 h posttransfection, Vif mutant viruses present in supernatant were normalized for p26 antigen and used for infection of permissive and nonpermissive cells, as described above. The viral kinetics in cells infected with HIV-2ROD and the HIV-2ROD vif-minus and HIV-2ROD vif mutants are shown in Fig. 4B. The mutants shown in Fig. 4A are those with alterations in conserved regions between Vif1 and Vif2, namely, HIV-2RODM6 and HIV-2RODM7 with the substitution of essential cysteines and HIV-2RODMD5 and HIV-2RODMD8 with deletions in conserved motifs described previously for Vif1 (13, 27, 35, 50). PBMC infected by HIV-2ROD produce large amounts of viral particles, as detected by p26 levels. Nevertheless, when these cells are infected with HIV-2ROD Vif mutants MD5, M6, M7, and MD8, viral replication is inhibited, indicating that these motifs of Vif2 are essential for HIV-2 infectivity. In contrast, viral infection of A3.01, H9, Jurkat, and U38 cells showed no significant differences in p26 levels with HIV-2ROD and HIV-2ROD Vif mutants, as expected for permissive cells. Nevertheless, HIV-2ROD vif-minus and all HIV-2ROD Vif mutants show a pronounced decrease in p26 levels after 15 days of infection in H9 cells.
FIG. 4.
Conserved motifs between Vif proteins of HIV-1 and HIV-2. (A) Genetic structure of Vif2 mutant proteins constructed by site-direct mutagenesis by altering the conserved motifs between HIV-1 and HIV-2 Vif proteins. Mutant vif genes were introduced into the proviral clone HIV-2ROD. Mutant Vif MDSTOP was made by altering the sequence of amino acids 51 and 52 (TGGTGG) for stop codons (TGATGA). Mutants HIV-2RODMD5 (amino acids 24 to 31) and HIV-2RODMD8 (amino acids 147 to 153) are deletion mutants Vif2 in the first and second conserved regions between Vif1 and Vif2. Mutants HIV-2RODM6 and HIV-2RODM7 result from the substitution of Cys116Arg and Cys134Arg/Cys135Gly in Vif2, respectively. (B) Viral replication kinetics of HIV-2 wild-type, HIV-2RODSTOP, HIV-2RODMD5, HIV-2RODM6, HIV-2RODM7, and HIV-2RODMD8 viruses. PBMC and A3.01, H9, U38, and Jurkat cells were infected with recombinant viruses as described in Materials and Methods. Cell-free supernatants collected at different time points were subjected to p26 antigen assay. The data are expressed as the means of at least three independent experiments ± the standard deviations.
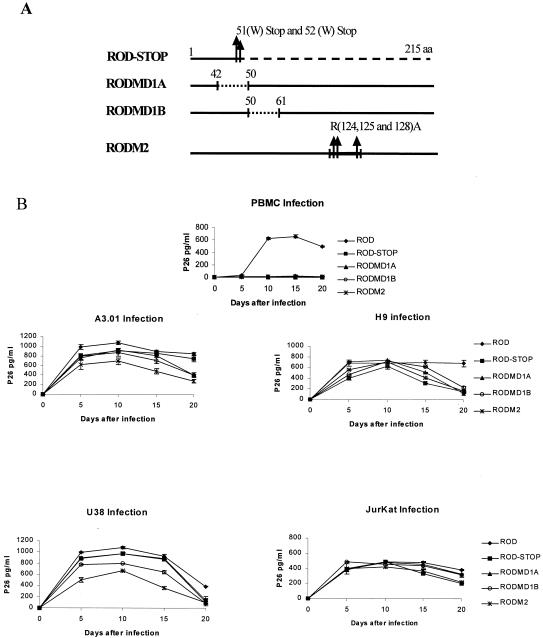
Similar replication analysis was performed with HIV-2 with deletions (HIV-2RODMD1A and HIV-2RODMD1B) and substitution (HIV-2RODM2) in conserved motifs of Vif2 protein (Fig. 5A). The results presented in Fig. 5B with nonpermissive PBMC show that Vif mutants failed to trigger detectable levels of virus replication as assayed by p26 antigen, whereas the same mutants reveal wild-type-like kinetics for infection in permissive cells. These patterns of infection suggest that conserved motifs 42-VPHHKVGWAWWTCSRVIFPL-61 (MD1A and MD1B) and 121-EVRRAIRGEK-130 (M2) are essential for Vif function in HIV-2 replication. To confirm that the absence of viral production with HIV-2 Vif mutants in nonpermissive cells is Vif dependent and not due to an abnormal viral expression, we analyzed the major viral proteins. Radioimmunoprecipitation analysis showed the same expression level of gp140 Env protein and p26 capsid proteins in HIV-2 wild-type and Vif mutant viruses, indicating a good integrity of the mutant viruses used for the infectivity studies (data not shown). These results show that all of the conserved regions in Vif2 are specifically important for the replication of HIV-2.
FIG. 5.
Conserved motifs between Vif proteins from HIV-2 are essential for viral infectivity. (A) Genetic structure of Vif2 mutant proteins constructed by site-direct mutagenesis by altering the conserved motifs of HIV-2 Vif proteins. Mutant vif genes were introduced into the proviral clone HIV-2ROD. Mutant Vif STOP was made by altering the sequence of amino acids 51 and 52 (TGGTGG) for stop codons (TGATGA). HIV-2RODMD1A and HIV-2RODMD1B are deletion mutants in the first conserved motif of Vif2 protein (amino acids 42 to 61). HIV-2RODMD1A has a deletion from amino acid residues 42 to 50 and HIV-2RODMD1B from residues 50 to 61 of Vif2. HIV-2RODM2 is a mutant generated in the second conserved region of Vif2 (amino acids 122 to 130) by substitution of Arg124, Arg125 and Arg128 to alanine. (B) Viral replication kinetics of HIV-2 wild-type, HIV-2RODSTOP, HIV-2RODMD1A, HIV-2RODMD1B and HIV-2RODM2. PBMC, A3.01, H9, U38 and Jurkat cells were infected with recombinant viruses as described in Materials and Methods. Cell-free supernatants collected at different time points were subjected to p26 antigen assay. The data are expressed as means of at least three independent experiments ± the standard deviations.
Functional relationship between HIV-2 Vif and APOBEC3G.
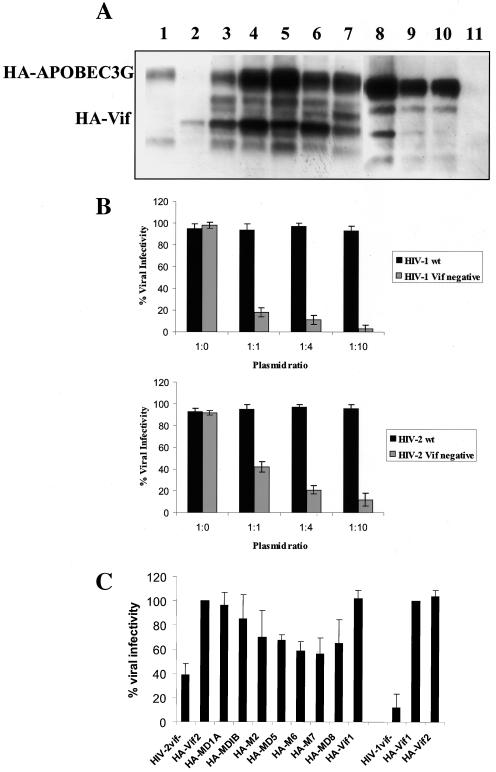
HIV-1 Vif function was associated with the inhibition of the antiviral cellular protein APOBEC3G (20, 28, 29, 46, 61). This link was confirmed by APOBEC3G expression in H9 cells and the lack of replication of HIV-1 vif-minus in those cells. Moreover, it was possible to change the cell permissivity to HIV-1 vif-minus when APOBEC3G was expressed (44). As shown above, our results with HIV-2 are in partial disagreement with the model that APOBEC3G completely inhibits HIV-1 replication in the absence of Vif protein. This stems from the finding that HIV-2 vif-minus replicates in nonpermissive H9 cells that express APOBEC3G (Fig. 1B). To confirm this hypothesis and determine the titer of APOBEC3G necessary to inhibit HIV-2 vif-minus, we performed a single-cycle infectivity assay with 293T cells cotransfected with APOBEC3G and HIV-2 vif-minus or HIV-1 vif-minus. Compared to HIV-1 vif-minus, the results in Fig. 6B show that infectivity of HIV-2 vif-minus is consistently higher for all amounts of APOBEC3G used. These results are in agreement with the lower level of restriction of HIV-2 replication in H9 cells that occurs in the absence of Vif, indicating that APOBEC3G does not affect similarly these viruses. To evaluate whether this difference is also reflected in Vif mutant phenotype, HIV-2 vif-minus transcomplemented with plasmids expressing HA-Vif2 and HA-Vif2 mutants were produced from 293T cells cotransfected with a Vif/APOBEC3G ratio of 1:1. By infection of P4 target cells, HIV-2 vif-minus showed a significant reduction of 60% in viral infectivity compared to the values obtained for HIV-2 vif-minus transcomplemented with a plasmid expressing HA-Vif2 or HA-Vif1 (Fig. 6C). We have used HIV-1NL43 vif-minus as a control and confirmed a strong reduction in viral infectivity (80%) (44). A small reduction of 30 to 40% was observed when HA-Vif2 mutants MD5, M6, M7, and MD8 were tested. In the case of M7 and MD8, the decrease in viral infectivity is associated with very low levels of Vif expression (Fig. 6A, lanes 7 to 10); this is probably due to the instability of these mutants in the presence of APOBEC3G. For mutants MD5 and M6, a similar reduction in infectivity was observed compared to that of M7 and MD8; expression is higher for mutants MDS and M6, suggesting a role for these domains in Vif function. In contrast, HA-Vif2 mutants MD1A, MD1B, and M2 show values of viral infectivity similar to those obtained for HIV-2 vif-minus transcomplemented with HA-Vif wild type, indicating the lack of a functional role (Fig. 6C). Therefore, these results suggest that the motifs 24-SLVKYLKY-31 (MD5), Cys116 (M6), Cys134-Cys135 (M7), and 147-SLQFLAL-153 (MD8) of Vif2 protein may have an important role for its function and stability when APOBEC3G is highly expressed in a transfection assay, which is not the case in H9 when the deaminase is expressed in a steady-state. Moreover, when 293T cells are cotransfected in a ratio of 1:1 with Vif2 mutants and APOBEC3G, a consistent expression of deaminase protein is detected, as measured by densitometry (Fig. 6A). These results show that with a decrease of ∼50% in viral infectivity when APOBEC3G is coexpressed with HIV-2 vif-minus, no alteration is observed in the steady state of deaminase. In addition, our results also show that the decrease in HIV-2 vif-minus infectivity using Vif2 mutants does not correlate with increase of APOBEC3G expression but may result from differences of Vif2 expression levels or other than APOBEC3G effect (Fig. 6A). These results are in agreement with the viral replication kinetics in H9 cells, where the presence of APOBEC3G does not strongly affect viral replication, suggesting a minor role of APOBEC3G on HIV-2 vif-minus replication.
FIG. 6.
Functional relationship between Vif2 proteins and APOBEC3G. (A) Expression analysis of HA-Vif wild-type or HA-Vif mutant proteins and APOBEC3G in cotransfected 293 cells. Cell lysates were immunoprecipitated, and HA fusion proteins were detected by Western blotting with horseradish peroxidase-conjugated anti-HA monoclonal antibody and by enhanced chemiluminescence. Cells were transfected with 1 μg of DNA plasmid as follows: lane 1, HA-APOBEC3G; lane 2, HA-Vif2 wild type (wt); lane 3, HA-APOBEC3G and HA-Vif2 wt; lane 4, HA-APOBEC3G and HA-MD1A; lane 5, HA-APOBEC3G and HA-MD1B; lane 6, HA-APOBEC3G and HA-M2; lane 7, HA-APOBEC3G and HA-MD5; lane 8, HA-APOBEC3G and HA-M6; lane 9, HA-APOBEC3G and HA-M7; lane 10, HA-APOBEC3G and MD8; and lane 11, HA-Vif2STOP. (B) Titers of APOBEC3G concentrations necessary for inhibition of HIV-1 vif-minus were determined by single-cycle infectivity assay. Briefly, 293T cells were transfected with proviral HIV-1NL43 vif-minus and HIV-2ROD vif-minus DNA plus increasing concentrationsAPOBEC3G, with plasmid ratios of 1:0, 1:1, 1:4, and 1:10. Viruses in the supernatant were collected, and its virus titers were normalized by using p26 antigen to infect P4 cells. Infectivity was measured based on the level of β-galactosidase expression as detailed in Materials and Methods. Values shown are percentages of infectivity relative to HIV vif-minus complemented with HA-Vif wild-type. The results are representative of three independent experiments. (C) Single-cycle infectivity assay was used to evaluate the effect of APOBEC3G in infectivity of HIV-2. 293T cells were cotransfected with proviral HIV-2ROD vif-minus DNA and plasmids expressing HA-Vif2 wild-type or HA-Vif2 mutant proteins together with APOBEC3G. The ratio of proviral DNA to APOBEC3G was 1:1. As controls, cells were transfected with HIV-1NL43 vif-minus transcomplemented by HA-Vif1 and HA-Vif2 wild type (wt; right side of the graphic). Viruses in the supernatant were collected, and its virus titers were normalized by using p26 antigen to infect P4 cells. Infectivity was measured based on the level of β-galactosidase expression as detailed in Materials and Methods. Values shown are percentages of infectivity relative to HIV vif-minus complemented with HA-Vif wild-type. The results are representative of four independent experiments.
DISCUSSION
HIV-2 and HIV-1 share ca. 50% of nucleotide sequence. Furthermore, Vif proteins from the two viruses have only 25% amino acid identity, which may explain some of the differences in infectivity seen between these two viruses (8). In the present study we determined the replication capacity of HIV-2 Vif-defective virus and HIV-2 Vif mutant viruses in various cell lines to identify its permissivity and the structure-function relationship of Vif protein. Previous reports show contradictory results about the nonpermissive phenotype for HIV-2 vif-minus (24, 32, 37). We confirm here that H9, SupT1, and A3.01 cells are permissive for HIV-2 replication in the absence of Vif, as observed for chimera LA 317 (HIV-2ROD+GH) and HIV-2Kr strains (24, 32, 37). Other cells, such as Jurkat, HeLa CD4+, and U38 cells, are also permissive for HIV-2ROD vif-minus replication. Although U937 cells have been previously classified as nonpermissive in infectious studies with HIV-2Kr and HIV-2Isy strains, our results confirm reports in which U38 cells derived from monocytic U937 have shown a permissive phenotype for HIV-2La317 (24). These apparently contradictory results may indicate that in some cell lines the permissive phenotype is strain dependent. Surprisingly, our results show that none of the cell lines used is nonpermissive for HIV-2 vif-minus replication. Only PBMC show a nonpermissive viral replication in absence of Vif, suggesting that other factors not expressed in the cell lines used may interfere with HIV-2 vif-minus replication. Moreover, HIV-1 vif-minus nonpermissive cell line H9 does not show a similar phenotype for Vif-defective HIV-2. These results may indicate that APOBEC3G does not affect HIV-2 vif-minus virus as it does for HIV-1.
Comparison of amino acid sequences from Vif proteins of HIV-1 and HIV-2 show the presence of two conserved motifs (35). The results shown in Fig. 4 confirm that the “SLQFLA” motif in Vif protein is essential for HIV-2 viral replication, as already described for HIV-1, SIV, and FIV (7, 12, 50). Nevertheless, expression of this deletion mutant is low, indicating a structural constraint of this region in Vif protein. Moreover, the conserved motif of Vif represented by amino acids 24 to 31 (SLVKYLKY) is also essential for HIV-2 replication, and its expression is similar to wild-type Vif protein. As with HIV-1 Vif, the conserved cysteines (M6 and M7) in Vif2 are important for its function, indicating a structural conformational role of these residues due to a lack of expression when some of them are eliminated (M7). In addition, Vif2 specific conserved motifs are also essential for viral infectivity (39). The presence of these motifs in HIV-2 Vif protein may explain the different infectivity phenotype of this virus compared to HIV-1 in the absence of Vif.
Previous studies of Vif1 wild type and Vif1 mutants have shown a predominant cytoplasmic localization (4, 18, 19, 22, 32) that was not observed with the HIV-2 Vif wild type or Vif mutants in our study. These reports show that cellular localization of Vif has been mainly observed in the cytoplasm, interacting with membranes, Gag, and intermediate filaments (18, 23, 32, 48). Nevertheless, a consistent small fraction of HIV-1 Vif is always present in the nucleus when these experiments are performed 48 h posttransfection. Our results show that Vif2 wild-type and Vif2 mutants are consistently present in the nucleus of HeLa cells at 18 h after transfection. By a similar approach, it has also been found that Vif1 in HeLa transfected cells is predominantly seen in the nucleus (48). It is conceivable that an initial steady-state of Vif expression is mainly localized in the nucleus and consequently released to the cytoplasm. Further studies are necessary to completely elucidate this hypothesis.
The function of Vif directly involves the degradation and inactivation of the deaminase APOBEC3G. Nevertheless, in H9 cells that stably express APOBEC3G our replication studies show no differences during 15 days of replication whether HIV-2ROD and HIV-2ROD vif-minus are used. In contrast, results of a single-cycle replication assay with viruses produced in 293T cells cotransfected with APOBEC3G showed differences in viral infectivity between the Vif2 mutants and the wild-type Vif2. HIV-2ROD vif-minus shows 40% viral infectivity in presence of APOBEC3G compared to HIV-2ROD. This small reduction in infectivity was obtained with a 1:1 plasmid ratio (Vif/APOBEC3G), but was reduced to nearly 10-fold with higher APOBEC3G concentrations. In our assay, HIV-1 vif-minus shows always a greater reduction in viral infectivity, with an ∼100-fold reduction at the highest APOBEC3G concentration. This relative comparison with HIV-2 and HIV-1 vif-minus in the presence of APOBEC3G indicates that this deaminase restricts less HIV-2 infectivity in the absence of Vif. We may hypothesize that other APOBEC family members may be involved in restricting HIV-2 infectivity (56), other than APOBEC3G, and that Vif2 is more specific for these factors. In addition, we may speculate that HIV-2 Gag proteins may also be involved in blocking APOBEC3G activity, reducing the role of Vif (2, 6, 52).
As shown in a single-cycle assay, the decrease in viral infectivity is dependent on the Vif2 mutant used. When mutations are localized in similar conserved regions of Vif1 and Vif2, the decrease in viral infectivity is more evident. In contrast, when mutations are introduced in conserved motifs specific for Vif2 protein, no effect on viral infectivity was detected. These results suggest that HIV-2 vif-minus inhibition due to APOBEC3G presence may follow a different mechanism of action.
Studies on the regulation of HIV-1 infectivity by APOBEC3G show that its activity is essential but not the sole determinant of antiviral activity (46). Our results show that only mutated domains conserved between Vif1 and Vif2 proteins influence HIV-2 infectivity when APOBEC3G is present. This fact supports the hypothesis that Vif2 targets the deaminase differently, and it is conceivable that the level of APOBEC3G expression may influence HIV-2 vif-minus infectivity. This is consistent with the result that the decrease of HIV-2 vif-minus infectivity is more evident only when 293T cells are transfected with APOBEC3G. Therefore, the steady-state expression of APOBEC3G in H9 cells may not be sufficient for the antiviral effect observed with HIV-1 vif-minus.
Our results allow us to speculate that viral inhibition by APOBEC3G is not the sole and most important determinant of antiviral activity against HIV-2. This hypothesis is compatible with our results of viral replication and single-cycle infectivity, in which a limited effect of APOBEC3G exists. Furthermore, the lack of viral replication in PBMC supports this conclusion. Further studies are in progress to define the relationship between APOBEC3G family members and HIV-2 Vif protein.
Acknowledgments
This study was supported by grant POCTI/SAU/1411/01 from the Fundação para a Ciência e Tecnologia to I.B. and grant POCTI/MGI/33096/01 to J.G. and by the Comissão Nacional de Luta Contra a Sida. A.C.R., A.M.S., and M.S.-M. are the recipients of doctoral fellowships from the Fundação para a Ciência e Tecnologia.
The 293T, HeLa, HeLa CD4+, HeLa P4, SupT1, H9, Jurkat, A3.01, and U38 cell lines were obtained from the AIDS Research and Reference Reagent Program. We thank L. Lang Xia and R. Bénarous for the plasmid containing the thioesterase gene. We are also grateful to M. João Gama and E. Rodrigues from UBM-CPM for assistance with Northern blotting. We thank Sofia Coelho, Elsa Anes, and Quirina Santos Costa for helpful assistance.
REFERENCES
- 1.Akari, H., J. Sakuragi, Y. Takebe, K. Tomonaga, M. Kawamura, M. Fukasawa, T. Miura, T. Shinjo, and M. Hayami. 1992. Biological characterization of human immunodeficiency virus type 1 and type 2 mutants in human peripheral blood mononuclear cells. Arch. Virol. 123:157-167. [DOI] [PubMed] [Google Scholar]
- 2.Alce, T. M., and W. Popik. 2004. APOBEC3G is incorporated into virus-like particles by a direct interaction with HIV-1 Gag nucleocapsid protein. J. Biol. Chem. 279:34083-34086. [DOI] [PubMed] [Google Scholar]
- 3.Borman, A. M., C. Quillent, P. Charneau, C. Dauguet, and F. Clavel. 1995. Human immunodeficiency virus type 1 Vif− mutant particles from restrictive cells: role of Vif in correct particle assembly and infectivity. J. Virol. 69:2058-2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouyac, M., M. Courcoul, G. Bertoia, Y. Baudat, D. Gabuzda, D. Blanc, N. Chazal, P. Boulanger, J. Sire, R. Vigne, and B. Spire. 1997. Human immunodeficiency virus type 1 Vif protein binds to the Pr55Gag precursor. J. Virol. 71:9358-9365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castro, B. A., S. W. Barnett, L. A. Evans, J. Moreau, K. Odehouri, and J. A. Levy. 1990. Biologic heterogeneity of human immunodeficiency virus type 2 (HIV-2) strains. Virology 178:527-534. [DOI] [PubMed] [Google Scholar]
- 6.Cen, S., F. Guo, M. Niu, J. Saadatmand, J. Deflassieux, and L. Kleiman. 2004. The Interaction between HIV-1 Gag and APOBEC3G. J. Biol. Chem. 279:33177-33184. [DOI] [PubMed] [Google Scholar]
- 7.Chatterji, U., C. K. Grant, and J. H. Elder. 2000. Feline immunodeficiency virus Vif localizes to the nucleus. J. Virol. 74:2533-2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clavel, F., K. Mansinho, S. Chamaret, D. Guetard, V. Favier, J. Nina, M. O. Santos-Ferreira, J. L. Champalimaud, and L. Montagnier. 1987. Human immunodeficiency virus type 2 infection associated with AIDS in West Africa. N. Engl. J. Med. 316:1180-1185. [DOI] [PubMed] [Google Scholar]
- 9.Courcoul, M., C. Patience, F. Rey, D. Blanc, A. Harmache, J. Sire, R. Vigne, and B. Spire. 1995. Peripheral blood mononuclear cells produce normal amounts of defective Vif− human immunodeficiency virus type 1 particles which are restricted for the preretrotranscription steps. J. Virol. 69:2068-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan, L., and K. Peden. 1992. Cell-free transmission of Vif mutants of HIV-1. Virology 190:19-29. [DOI] [PubMed] [Google Scholar]
- 11.Fouchier, R. A., J. H. Simon, A. B. Jaffe, and M. H. Malim. 1996. Human immunodeficiency virus type 1 Vif does not influence expression or virion incorporation of gag-, pol-, and env-encoded proteins. J. Virol. 70:8263-8269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujita, M., A. Sakurai, N. Doi, M. Miyaura, A. Yoshida, K. Sakai, and A. Adachi. 2001. Analysis of the cell-dependent replication potentials of human immunodeficiency virus type 1 vif mutants. Microbes Infect. 3:1093-1099. [DOI] [PubMed] [Google Scholar]
- 13.Fujita, M., A. Sakurai, A. Yoshida, S. Matsumoto, M. Miyaura, and A. Adachi. 2002. Subtle mutations in the cysteine region of HIV-1 Vif drastically alter the viral replication phenotype. Microbes Infect. 4:621-624. [DOI] [PubMed] [Google Scholar]
- 14.Fujita, M., A. Sakurai, A. Yoshida, M. Miyaura, A. H. Koyama, K. Sakai, and A. Adachi. 2003. Amino acid residues 88 and 89 in the central hydrophilic region of human immunodeficiency virus type 1 Vif are critical for viral infectivity by enhancing the steady-state expression of Vif. J. Virol. 77:1626-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gabuzda, D. H., K. Lawrence, E. Langhoff, E. Terwilliger, T. Dorfman, W. A. Haseltine, and J. Sodroski. 1992. Role of vif in replication of human immunodeficiency virus type 1 in CD4+ T lymphocytes. J. Virol. 66:6489-6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gabuzda, D. H., H. Li, K. Lawrence, B. S. Vasir, K. Crawford, and E. Langhoff. 1994. Essential role of vif in establishing productive HIV-1 infection in peripheral blood T lymphocytes and monocyte/macrophages. J. Acquir. Immune Defic. Syndr. 7:908-915. [PubMed] [Google Scholar]
- 17.Gallo, R. C., P. S. Sarin, E. P. Gelmann, M. Robert-Guroff, E. Richardson, V. S. Kalyanaraman, D. Mann, G. D. Sidhu, R. E. Stahl, S. Zolla-Pazner, J. Leibowitch, and M. Popovic. 1983. Isolation of human T-cell leukemia virus in acquired immune deficiency syndrome (AIDS). Science 220:865-867. [DOI] [PubMed] [Google Scholar]
- 18.Goncalves, J., P. Jallepalli, and D. H. Gabuzda. 1994. Subcellular localization of the Vif protein of human immunodeficiency virus type 1. J. Virol. 68:704-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goncalves, J., B. Shi, X. Yang, and D. Gabuzda. 1995. Biological activity of human immunodeficiency virus type 1 Vif requires membrane targeting by C-terminal basic domains. J. Virol. 69:7196-7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harris, R. S., K. N. Bishop, A. M. Sheehy, H. M. Craig, S. K. Petersen-Mahrt, I. N. Watt, M. S. Neuberger, and M. H. Malim. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803-809. [DOI] [PubMed] [Google Scholar]
- 21.Henzler, T., A. Harmache, H. Herrmann, H. Spring, M. Suzan, G. Audoly, T. Panek, and V. Bosch. 2001. Fully functional, naturally occurring and C-terminally truncated variant human immunodeficiency virus (HIV) Vif does not bind to HIV Gag but influences intermediate filament structure. J. Gen. Virol. 82:561-573. [DOI] [PubMed] [Google Scholar]
- 22.Huvent, I., S. S. Hong, C. Fournier, B. Gay, J. Tournier, C. Carriere, M. Courcoul, R. Vigne, B. Spire, and P. Boulanger. 1998. Interaction and co-encapsidation of human immunodeficiency virus type 1 Gag and Vif recombinant proteins. J. Gen. Virol. 79(Pt. 5):1069-1081. [DOI] [PubMed] [Google Scholar]
- 23.Karczewski, M. K., and K. Strebel. 1996. Cytoskeleton association and virion incorporation of the human immunodeficiency virus type 1 Vif protein. J. Virol. 70:494-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawamura, M., R. Shimano, T. Ogasawara, R. Inubushi, K. Amano, H. Akari, and A. Adachi. 1998. Mapping the genetic determinants of human immunodeficiency virus type 2 for cell tropism and replication efficiency. Arch. Virol. 143:513-521. [DOI] [PubMed] [Google Scholar]
- 25.Khan, M. A., C. Aberham, S. Kao, H. Akari, R. Gorelick, S. Bour, and K. Strebel. 2001. Human immunodeficiency virus type 1 Vif protein is packaged into the nucleoprotein complex through an interaction with viral genomic RNA. J. Virol. 75:7252-7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu, H., X. Wu, M. Newman, G. M. Shaw, B. H. Hahn, and J. C. Kappes. 1995. The Vif protein of human and simian immunodeficiency viruses is packaged into virions and associates with viral core structures. J. Virol. 69:7630-7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma, X. Y., P. Sova, W. Chao, and D. J. Volsky. 1994. Cysteine residues in the Vif protein of human immunodeficiency virus type 1 are essential for viral infectivity. J. Virol. 68:1714-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mangeat, B., P. Turelli, G. Caron, M. Friedli, L. Perrin, and D. Trono. 2003. Broad antiretroviral defense by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99-103. [DOI] [PubMed] [Google Scholar]
- 29.Mariani, R., D. Chen, B. Schrofelbauer, F. Navarro, R. Konig, B. Bollman, C. Munk, H. Nymark-McMahon, and N. R. Landau. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114:21-31. [DOI] [PubMed] [Google Scholar]
- 30.Marin, M., K. M. Rose, S. L. Kozak, and D. Kabat. 2003. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 9:1398-1403. [DOI] [PubMed] [Google Scholar]
- 31.Mehle, A., B. Strack, P. Ancuta, C. Zhang, M. McPike, and D. Gabuzda. 2003. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J. Biol. Chem. 279:7792-7798. [DOI] [PubMed] [Google Scholar]
- 32.Michaels, F. H., N. Hattori, R. C. Gallo, and G. Franchini. 1993. The human immunodeficiency virus type 1 (HIV-1) Vif protein is located in the cytoplasm of infected cells and its effect on viral replication is equivalent in HIV-2. AIDS Res. Hum. Retrovir. 9:1025-1030. [DOI] [PubMed] [Google Scholar]
- 33.Myers, G., B. Korber, S. Wain-Hobson, R. Smith, and G. Pavlakis. 1993. Human retrovirus and AIDS 1993. Los Alamos National Laboratory, Los Alamos, N.Mex.
- 34.Nascimbeni, M., M. Bouyac, F. Rey, B. Spire, and F. Clavel. 1998. The replicative impairment of Vif− mutants of human immunodeficiency virus type 1 correlates with an overall defect in viral DNA synthesis. J. Gen. Virol. 79(Pt. 8):1945-1950. [DOI] [PubMed] [Google Scholar]
- 35.Oberste, M. S., and M. A. Gonda. 1992. Conservation of amino-acid sequence motifs in lentivirus Vif proteins. Virus Genes 6:95-102. [DOI] [PubMed] [Google Scholar]
- 36.Pombo, A., P. Cuello, W. Schul, R. G. Roeder, P. R. Cook, and S. and Murphy. 1998. Regional and temporal specialization in the nucleus: a transcriptionally-active nuclear domain rich in PTF, Oct1, and PIKA antigens associates with specific chromosomes early in the cell cycle. EMBO J. 17:1768-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reddy, T. R., G. Kraus, O. Yamada, D. J. Looney, M. Suhasini, and F. Wong-Staal. 1995. Comparative analyses of human immunodeficiency virus type 1 (HIV-1) and HIV-2 Vif mutants. J. Virol. 69:3549-3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Remy, G. 2003. HIV-2 infection in the world: a geographical perspective. Sante 8:440-446. [PubMed] [Google Scholar]
- 39.Ribeiro, A. C., P. J. Moniz, and I. Barahona. 1998. HIV type 2 Vif proteins have specific conserved amino acid motifs. AIDS Res. Hum. Retrovir. 14:465-469. [DOI] [PubMed] [Google Scholar]
- 40.Sakai, H., R. Shibata, J. Sakuragi, S. Sakuragi, M. Kawamura, and A. Adachi. 1993. Cell-dependent requirement of human immunodeficiency virus type 1 Vif protein for maturation of virus particles. J. Virol. 67:1663-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sambrook, J., and D. W. Russel. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring, N.Y.
- 42.Santos Ferreira, M. O., J. Moniz Pereira, and J. L. Champalimaud. 1993. Biologic prospective, p. 113-117. In M.-M. Galteau, G. Siest, and J. Henry (ed.), Comptes rendus du 8e colloque de Pont-à-Mousson. John Libbey Eurotext, Montrouge, France.
- 43.Schuitemaker, H., M. Koot, N. A. Kootstra, M. W. Dercksen, R. E. de Goede, R. P. van Steenwijk, J. M. Lange, J. K. Schattenkerk, F. Miedema, and M. Tersmette. 1992. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus population. J. Virol. 66:1354-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646-650. [DOI] [PubMed] [Google Scholar]
- 45.Sheehy, A. M., N. C. Gaddis, and M. H. Malim. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 9:1404-1407. [DOI] [PubMed] [Google Scholar]
- 46.Shindo, K., A. Takaori-Kondo, M. Kobayashi, A. Abudu, K. Fukunaga, and T. Uchiyama. 2003. The enzymatic activity of CEM15/Apobec-3G Is essential for the regulation of the infectivity of HIV-1 Virion, but not a sole determinant of Its antiviral activity. J. Biol. Chem. 278:44412-44416. [DOI] [PubMed] [Google Scholar]
- 47.Simon, J. H., E. A. Carpenter, R. A. Fouchier, and M. H. Malim. 1999. Vif and the p55Gag polyprotein of human immunodeficiency virus type 1 are present in colocalizing membrane-free cytoplasmic complexes. J. Virol. 73:2667-2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simon, J. H., R. A. Fouchier, T. E. Southerling, C. B. Guerra, C. K. Grant, and M. H. Malim. 1997. The Vif and Gag proteins of human immunodeficiency virus type 1 colocalize in infected human T cells. J. Virol. 71:5259-5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simon, J. H., and M. H. Malim. 1996. The human immunodeficiency virus type 1 Vif protein modulates the postpenetration stability of viral nucleoprotein complexes. J. Virol. 70:5297-5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simon, J. H., A. M. Sheehy, E. A. Carpenter, R. A. Fouchier, and M. H. Malim. 1999. Mutational analysis of the human immunodeficiency virus type 1 Vif protein. J. Virol. 73:2675-2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sova, P., and D. J. Volsky. 1993. Efficiency of viral DNA synthesis during infection of permissive and non-permissive cells with vif-negative human immunodeficiency virus type 1. J. Virol. 67:6322-6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Svarovskaia, E. S., H. Xu, J. L. Mbisa, R. Barr, R. J. Gorelick, A. Ono, E. O. Freed, W. S. Hu, and V. K. Pathak. 2004. Human apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3G (APOBEC3G) is incorporated into HIV-1 virions through interactions with viral and nonviral RNAs. J. Biol. Chem. 279:35822-35828. [DOI] [PubMed] [Google Scholar]
- 53.Talbott, R., G. Kraus, D. Looney, and F. Wong-Staal. 1993. Mapping the determinants of human immunodeficiency virus 2 for infectivity, replication efficiency, and cytopathicity. Proc. Natl. Acad. Sci. USA 90:4226-4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.UNAIDS. 2000. Report on the global HIV/AIDS epidemic. UNAIDS, Geneva, Switzerland.
- 55.von Schwedler, U., J. Song, C. Aiken, and D. Trono. 1993. Vif is crucial for human immunodeficiency virus type 1 proviral DNA synthesis in infected cells. J. Virol. 67:4945-4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wiegand, H. L., B. P. Doehle, H. P. Bogerd, and B. R. Cullen. 2004. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 23:2451-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang, X., and D. Gabuzda. 1998. Mitogen-activated protein kinase phosphorylates and regulates the HIV-1 Vif protein. J. Biol. Chem. 273:29879-29887. [DOI] [PubMed] [Google Scholar]
- 58.Yang, X., and D. Gabuzda. 1999. Regulation of human immunodeficiency virus type 1 infectivity by the ERK mitogen-activated protein kinase signaling pathway. J. Virol. 73:3460-3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang, X., J. Goncalves, and D. Gabuzda. 1996. Phosphorylation of Vif and its role in HIV-1 replication. J. Biol. Chem. 271:10121-10129. [DOI] [PubMed] [Google Scholar]
- 60.Yu, X., Y. Yu, B. Liu, K. Luo, W. Kong, P. Mao, and X. F. Yu. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302:1056-1060. [DOI] [PubMed] [Google Scholar]
- 61.Zhang, H., B. Yang, R. J. Pomerantz, C. Zhang, S. C. Arunachalam, and L. Gao. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424:94-98. [DOI] [PMC free article] [PubMed] [Google Scholar]