Abstract
The proper function of neural circuits requires spatially and temporally balanced development of excitatory and inhibitory synapses. However, the molecular mechanisms coordinating excitatory and inhibitory synaptogenesis remain unknown. Here we demonstrate that SRGAP2A and its human-specific paralog SRGAP2C co-regulate the development of excitatory and inhibitory synapses in cortical pyramidal neurons in vivo. SRGAP2A promotes synaptic maturation, and ultimately the synaptic accumulation of AMPA and GABAA receptors, by interacting with key components of both excitatory and inhibitory postsynaptic scaffolds, Homer and Gephyrin. Furthermore, SRGAP2A limits the density of both types of synapses via its Rac1-GAP activity. SRGAP2C inhibits all identified functions of SRGAP2A, protracting the maturation and increasing the density of excitatory and inhibitory synapses. Our results uncover a molecular mechanism coordinating critical features of synaptic development and suggest that human-specific duplication of SRGAP2 might have contributed to the emergence of unique traits of human neurons while preserving the excitation/inhibition balance.
INTRODUCTION
The assembly and function of neural circuits requires synaptic excitation and inhibition to be tightly balanced (Nelson and Valakh, 2015). The balanced ratio of excitatory and inhibitory synapses (E/I ratio) and their relative distribution are attained early during development, before the density of synapses reaches its mature level (Benson and Cohen, 1996; Soto et al., 2011; Zhao et al., 2005). Remarkably, the E/I ratio is conserved between rodents and humans, even though the density of synapses is significantly higher in human cortical neurons (Defelipe et al., 2011). At least in the visual system, the E/I ratio in individual cells is reached before the onset of sensory experience (Soto et al., 2011), suggesting that developmental mechanisms set the equilibrium between the number of excitatory and inhibitory synapses and coordinate their formation. Very little is known about these mechanisms. At the molecular level, the assembly of excitatory and inhibitory synapses involves almost exclusive sets of proteins and the scarcity of genes known to co-regulate the density or the maturation of both types of synapses has obscured the identification of general principles for this critical aspect of cortical circuit development.
We have previously shown that Slit-Robo Rho-GTPase Activating Protein 2 (SRGAP2) regulates different aspects of cortical development from the migration and differentiation of pyramidal neurons (Guerrier et al., 2009) to the maturation and density of dendritic spines, small protrusions distributed along dendrites that receive the majority of excitatory synaptic inputs (Charrier et al., 2012). SRGAP2 encodes a protein highly conserved in mammals and is one of the few genes specifically duplicated in the human lineage (Dennis et al., 2012; Sudmant et al., 2010). The ancestral gene SRGAP2A has undergone two major partial duplications, which generated a human-specific gene, SRGAP2C, approximately 2–3 million years ago, at a critical time during evolution coinciding with the emergence of the Homo lineage (Dennis et al., 2012). SRGAP2C copy number is remarkably fixed in the human population and SRGAP2C is largely co-expressed with SRGAP2A in the developing and adult human brain (Charrier et al., 2012; Dennis et al., 2012). At the protein level, SRGAP2A contains three functional domains: a N-terminal F-BAR (Bin, Amphiphysin, Rvs) domain; a central Rho-GAP (Rho GTPase-Activating Protein) domain specific for the small GTPase Rac1; and a C-terminal SH3 (Src Homology 3) domain (Guerrier et al., 2009). SRGAP2A is primarily expressed in the neocortex during synaptogenesis and accumulates at excitatory synapses (Charrier et al., 2012; Guerrier et al., 2009), where it promotes the maturation of dendritic spines and limits their density in vivo (Charrier et al., 2012). SRGAP2C is a truncated form of SRGAP2A corresponding to the F-BAR domain lacking its last 49 amino acids (Charrier et al., 2012; Dennis et al., 2012). We have previously demonstrated that SRGAP2C physically interacts with SRGAP2A through its F-BAR domain and inhibits SRGAP2A function (Charrier et al., 2012). Inactivation of SRGAP2A, as well as heterologous expression of human-specific SRGAP2C in mouse cortical pyramidal neurons in vivo, induces the emergence of human traits of pyramidal neurons, such as neoteny during spine maturation and higher morphological complexity due to increased spine density and longer spine neck (Benavides-Piccione et al., 2002; Charrier et al., 2012; Elston et al., 2001; Huttenlocher and Dabholkar, 1997; Petanjek et al., 2011).
While the increase in the size and density of dendritic spines, as well as the delay of their maturation, might have been permissive for the emergence of human cognitive abilities during evolution (Defelipe, 2011; Geschwind and Rakic, 2013), these modifications raise fundamental questions about the impact of human-specific SRGAP2 duplication on the E/I equilibrium. Increasing excitation in cortical pyramidal neurons can induce hyperexcitability and has been implicated in cognitive, social and behavioral defects observed in neurodevelopmental disorders ranging from epilepsy to intellectual disability and autism spectrum disorders (Clement et al., 2012; Ramocki and Zoghbi, 2008; Yizhar et al., 2011). The evolutionary recent genomic fixation of SRGAP2C in modern human population implies the existence of developmental mechanisms that supported the coordinated evolution of excitatory and inhibitory synapses and preserved the E/I ratio.
In the present study, we tested the hypothesis that SRGAP2 paralogs co-regulate the development of excitatory and inhibitory synapses. We demonstrate that SRGAP2A co-regulates the maturation and the density of excitatory and inhibitory synapses in mouse cortical pyramidal neurons in vivo. Using an in utero gene replacement strategy in sparse cortical pyramidal neurons, we uncover the cellular and molecular mechanisms underlying SRGAP2A function in excitatory and inhibitory synapse development. We show that SRGAP2A has the unique property to couple small GTPase signaling to postsynaptic scaffolding proteins at both excitatory and inhibitory synapses through physical interactions with Homer and Gephyrin, respectively. Our results indicate that expressing SRGAP2C in mouse neurons inhibits all identified functions of SRGAP2A at synapses, from spine morphogenesis and inhibitory synapse formation to the accumulation of postsynaptic AMPA and GABAA receptors. Finally, we show that SRGAP2A inactivation co-regulates excitatory and inhibitory synaptic transmission. Our results identify a molecular mechanism coordinating excitatory and inhibitory synapse development in vivo and suggest that the human-specific duplication of SRGAP2 has modified the cell biology of cortical pyramidal neurons while preserving their E/I integrity.
RESULTS
Inhibition of SRGAP2A by SRGAP2C protracts the maturation and increases the density of inhibitory synapses
In the forebrain, most inhibitory synapses contain type A γ-Aminobutyric Acid Receptors (GABAAR) that are stabilized by a lattice of Gephyrin molecules underneath the plasma membrane (Tretter et al., 2012). Gephyrin is the core component of inhibitory postsynaptic scaffolds and can be used as a marker to label inhibitory synapses (Tretter et al., 2012). To address the potential role of SRGAP2 paralogs in inhibitory synaptogenesis, we chose an approach allowing the comparison of inhibitory and excitatory synapses at different developmental stages (i.e. juvenile and adult) with a single cell resolution in vivo. The expression of SRGAP2 paralogs was manipulated throughout development using sparse, cortex-specific, in utero electroporation (IUE). Neural progenitors generating layer 2/3 cortical pyramidal neurons were electroporated at embryonic day 15.5 (E15.5) with short hairpin RNAs (shRNA) against Srgap2a (Charrier et al., 2012) or a cDNA encoding SRGAP2C. The co-electroporation of a GFP-tagged Gephyrin and a cytosolic fluorescent protein (TdTomato) allowed the visualization of inhibitory synapses and dendritic spine morphology in neurons optically isolated by confocal microscopy (Figure 1A). This approach was recently shown to reliably label inhibitory synaptic contacts without affecting synaptic development or inhibitory neurotransmission (Chen et al., 2012; van Versendaal et al., 2012).
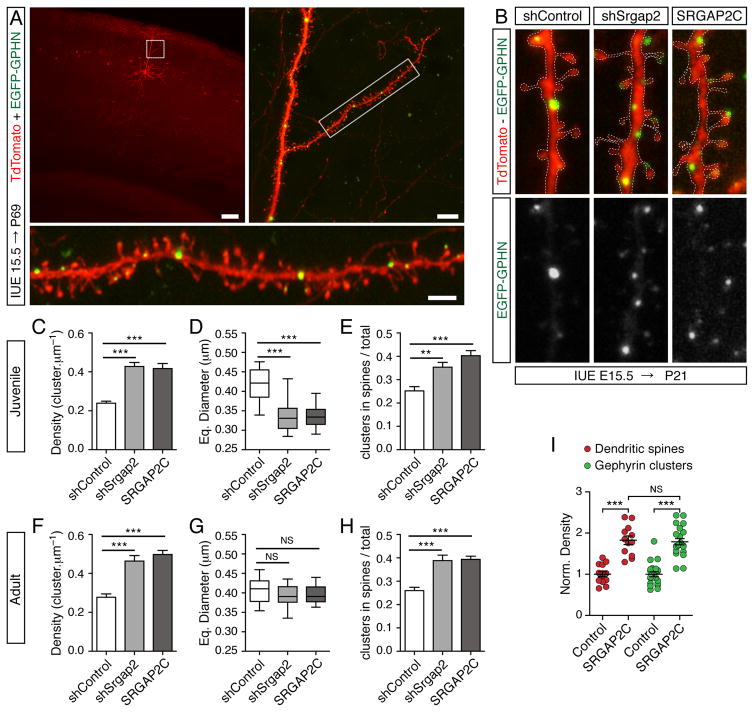
Figure 1. SRGAP2A and its human-specific paralog SRGAP2C regulate the maturation and the density of inhibitory synapses in vivo.
(A) Visualization of inhibitory synapses in sparse layer 2/3 cortical pyramidal neurons after in utero electroporation (IUE) with soluble TdTomato (red) and EGFP-Gephyrin (EGFP-GPHN, green). Inhibitory synapses in oblique apical dendrites (insets) are located in the dendritic shaft (large arrowheads) or directly in dendritic spines (small arrowheads). P69: postnatal day 69, shSrgap2: shRNA targeting mouse Srgap2a. Scale bars: 100 μm (top left), 10 μm (top right and bottom panels).
(B) Segments of dendrites from layer 2/3 pyramidal neurons expressing a control shRNA (shControl), shSrgap2 or SRGAP2C along with EGFP-Gephyrin in juvenile mice (P21). The dashed lines indicate the contour of dendrites (from TdTomato). Arrowheads point to examples of Gephyrin clusters in spines. Scale bar: 1 μm.
(C–H) Quantifications of Gephyrin cluster density (C, F), equivalent (Eq.) diameter (D, G), and proportion of Gephyrin clusters located in spines (E, H) in juveniles (C–E) and adult mice (F–H). Juveniles: nshControl = 32, nshSrgap2 = 36, nSRGAP2C = 31. Adults: nshControl = 21, nshSrgap2 = 23, nSRGAP2C = 22. Mean ± SEM in C, E, F, H. Box plot showing the distribution of the mean value per cell in D and G. *** p < 0.001, ** p < 0.01, NS (Not Significant): p > 0.05, Kruskal-Wallis test followed by Dunn’s Multiple comparison test. (I) Similar effects of SRGAP2C expression on the density of dendritic spines (red) and Gephyrin clusters (green) in adult cortical neurons (P>65). Norm. density: normalized density. Spines (data from Charrier et al., 2012): nControl = 14 and nSRGAP2C = 12. Gephyrin: same as in h. *** p < 0.001, NS: p > 0.05, Mann Whitney test. See also Figure S1 and S2.
We examined the density and the size of inhibitory synapses in oblique apical dendrites of the somatosensory cortex in juvenile (Postnatal day 21, P21) and adult mice (> P65). As previously described (Chen et al., 2012; Knott et al., 2002; Kubota et al., 2007; van Versendaal et al., 2012), Gephyrin clusters were distributed along the dendritic shaft (large arrowheads in Figure 1A), and some were located in dendritic spines (small arrowheads in Figure 1A). In juvenile mice (Figure 1B–E), Gephyrin cluster density was 0.24 ± 0.01 cluster/μm in control pyramidal neurons (Figure 1C), which is consistent with previous reports (Chen et al., 2012). Srgap2a knock-down using shRNA (shSrgap2) and SRGAP2C expression had similar effects: at P21, in both conditions, Gephyrin cluster density increased by ~75% (Figures 1C and S1) while Gephyrin cluster size decreased by ~20% (Figures 1D, S2). Unexpectedly, these effects were associated with a ~50% increase in the proportion of Gephyrin clusters located in dendritic spines (Figure 1E). In adult control neurons, the density (Figure 1F), the size (Figure 1G) and the subcellular distribution (Figure 1H) of Gephyrin clusters were similar to juvenile control neurons (Figure 1C–E), indicating that the morphological development of inhibitory synapses was largely complete by P21. By contrast, in SRGAP2A-deficient or SRGAP2C-expressing neurons, Gephyrin clusters underwent a substantial growth between juvenile and adult stages and reached a size undistinguishable from control (Figure 1D, G), suggesting that inhibitory synapses mature at a slower rate when SRGAP2A is inactivated. Furthermore, SRGAP2A-deficient and SRGAP2C-expressing neurons maintained a higher density of Gephyrin clusters (Figure 1F) and an enrichment of Gephyrin clusters in dendritic spines (Figure 1H) into adulthood. We found that 26 ± 1% of Gephyrin clusters were located in spines in control pyramidal neurons, which is in agreement with previous studies (Chen et al., 2012; Knott et al., 2002; Kubota et al., 2007). This percentage rose to 39 ± 2% following Srgap2a knock-down and 39 ± 1% following SRGAP2C expression, suggesting that local inhibition at the level of the spine is favored at higher synaptic density (see also Charrier et al., 2012). Together, these results demonstrate that (1) SRGAP2A accelerates the growth of inhibitory synapses and limits their density in a cell-autonomous manner; and (2) that SRGAP2C antagonizes the function of ancestral SRGAP2A during inhibitory synapse development. Strikingly, the protracted maturation and increased density of inhibitory synapses resulting from SRGAP2A loss of function or heterologous SRGAP2C expression paralleled the consequences on dendritic spines (Figures 1I, S1 and Charrier et al., 2012). Indeed, we have previously shown that in SRGAP2A-deficient neurons and in SRGAP2C-expressing neurons, spine maturation is delayed compared to control neurons and spine density is markedly increased in both juvenile and adult mice (Charrier et al., 2012). We conclude that SRGAP2 paralogs co-regulate the development of excitatory and inhibitory synapses in vivo and that SRGAP2C inhibits all identified functions of SRGAP2A.
SRGAP2A interacts with major components of excitatory and inhibitory postsynaptic scaffolds
The molecular mechanisms underlying the function of SRGAP2A at excitatory and inhibitory synapses are currently unknown. Although both types of synapses are assembled from dynamic networks of protein-protein interactions (Choquet and Triller, 2013) the sets of proteins that compose excitatory and inhibitory postsynaptic machineries are largely exclusive. SRGAP2A has a well-characterized Rac1-GAP activity (Guerrier et al., 2009) but few interacting partners have been identified, especially in neurons and at synapses. In order to identify other signaling pathways and protein-protein interaction networks that might be controlled by SRGAP2, we search for putative interaction motifs using sequence analysis (http://elm.eu.org). We identified a conserved proline-rich motif corresponding to a class II EVH1 binding site (PPMKF, residues P339–F343) in a predicted loop bridging two α-helixes in the F-BAR domain of SRGAP2 (see Guerrier et al., 2009 for the predicted secondary structure of SRGAP2 F-BAR domain). This motif is a canonical binding site for Homer family proteins (Tu et al., 1998), which are abundant components of excitatory postsynaptic densities (PSD) (Sheng and Hoogenraad, 2007). Homer plays a key role in the assembly of excitatory postsynaptic scaffolds via its interaction with Shank (Hayashi et al., 2009), and in the regulation of spine signaling via its binding to mGluR5, IP3 receptors and Ryanodine receptors (Bockaert et al., 2010; Ting et al., 2012). More specifically, Homer recruits into spines the Shank-GKAP-PSD95 complex, which ultimately stabilizes AMPA and NMDA glutamate receptors at the PSD (Hayashi et al., 2009; Sheng and Hoogenraad, 2007; Tu et al., 1999).
To assess a biochemical interaction between SRGAP2A and Homer, we isolated protein complexes associated with synaptic membranes from P15 mouse brains using subcellular fractionation and performed co-immunoprecipitation. Following Homer1 immunoprecipitation, SRGAP2A was detected in Western blots using two specific antibodies directed against its N-terminal domain or its C-terminal domain (Figure 2A). We noticed that the form of SRGAP2A immunoprecipitated in complex with Homer1 in brain lysates ran at a slightly higher apparent molecular weight than the majority of SRGAP2A present in the input. Although we do not know the basis for this, it might be due to differences in protein amounts between the input and immunoprecipitation lanes or to post-translational modifications of SRGAP2A such as phosphorylation, which is extensive based on publicly available mass spectrometry data (http://www.phosphosite.org). We then co-expressed GFP-Homer1c with hemagglutinin (HA)-tagged SRGAP2A in human embryonic kidney 293 (HEK) cells (Figure 2B). GFP and SRGAP2A-GFP were used as negative and positive controls, respectively. Using HA immunoprecipitation, we confirmed that SRGAP2A binds to Homer1c. Importantly, two point mutations within the class II EVH1 binding motif (P340L/F343C, EVH1dead mutant) were sufficient to abolish the interaction between SRGAP2A and Homer1c (Figure 2C), validating its specificity and providing a molecular tool to dissect its role in synaptic development in vivo (see below).
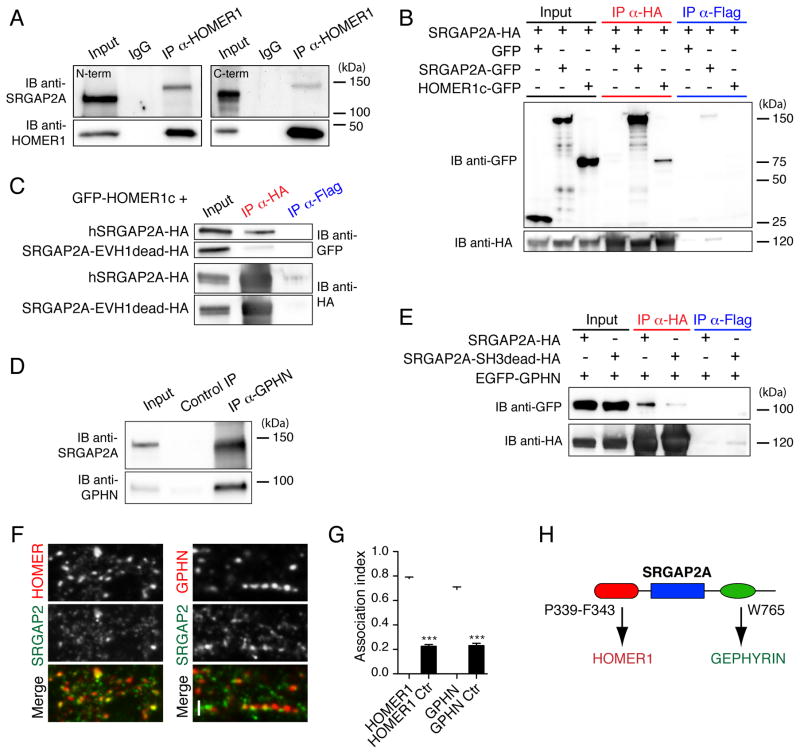
Figure 2. SRGAP2A interacts with Homer1 and Gephyrin.
(A) Co-immunoprecipitation (Co-IP) of endogenous SRGAP2A and Homer1 in synaptic fractions from P15 mouse brains. SRGAP2A was co-immunoprecipitated using anti-Homer1 antibody and detected in Western blot using two specific antibodies directed against its N-terminal domain (left) and its C-terminal domain (right). IB: immunoblot, α: anti.
(B) Co-IP of SRGAP2A-HA and GFP-HOMER1c in HEK cells.
(C) Mutation of the class 2 EVH1 binding motif of SRGAP2A (P340L/F343C, EVH1dead mutant) disrupts its interaction with HOMER1c in HEK cells.
(D) Co-IP of endogenous SRGAP2A and Gephyrin (GPHN) in synaptic fractions isolated from P15 mouse brains using GPHN.FingR.
(E) SRGAP2A interacts with EGFP-GPHN via its SH3 domain in HEK cells. SRGAP2A SH3dead mutant contains a W765A point mutation.
(F) Single section confocal images of endogenous SRGAP2A with Homer1 or GPHN in dissociated mouse cortical neurons after 17 days in vitro reveal close association (arrowheads) between SRGAP2A and both excitatory and inhibitory synapses. Scale bar: 2 μm.
(G) Histogram showing the fraction of Homer1 and gephyrin clusters associated with SRGAP2A immunoreactivity (association index). The control (Ctr) values correspond to the random fraction of Homer1 and gephyrin clusters associated with SRGAP2A in mismatched images. HOMER1 and HOMER1 Ctr: n = 21, GPHN and GPHN Ctr: n = 23. Mean ± SEM. *** p < 0.001, Mann-Whitney test.
(H) Schematic of SRGAP2A interactions with Homer and Gephyrin.
See also Figures S3 and S4.
While SRGAP2A interacts with Homer via a EVH1-binding site embedded in its F-BAR domain, Gephyrin was recently identified as a partner of SRGAP2A using an SH3-based photo-trapping assay (Okada et al., 2011). As mentioned above, Gephyrin forms high-order oligomers, which provide binding sites for most of the identified components of inhibitory synapses, including GABAARs (Fritschy et al., 2008; Tretter et al., 2012). To substantiate this interaction, we modified a Fibronectin intrabody generated with mRNA display directed against aa 1–113 of Gephyrin (GPHN.FingR, Gross et al., 2013) and showed that purified GPHN.FingR efficiently immunoprecipitates Gephyrin (Figure S3). Using this GPHN.FingR, we confirmed that SRGAP2A and Gephyrin interact in synaptic fractions from P15 mouse brains (Figure 2D). We then introduced a point mutation in the SH3 domain of SRGAP2A (W765A, SH3dead mutant) and performed co-immunoprecipitation experiments in HEK cells. EGFP-Gephyrin co-immunoprecipitated with HA-SRGAP2A but not with the SH3dead mutant (Figure 2E). We then wondered whether SRGAP2A colocalizes with Gephyrin in cortical neurons. We have previously shown that SRGAP2A accumulates postsynaptically and colocalizes with Homer1 clusters at excitatory synapses (Charrier et al., 2012). Using immunocytochemistry in dissociated cortical neurons after 17 days in vitro, we found that SRGAP2A associates with the majority of both Homer1 and Gephyrin clusters (Figure 2F–G). Remarkably, the EVH1dead and SH3dead mutants of SRGAP2A showed a decreased association with dendritic spines and Gephyrin clusters, respectively (Figure S4). Together, these results demonstrate that SRGAP2A has the unique property to interact, via two distinct domains, with two major components of excitatory and inhibitory postsynaptic scaffolds (Figure 2H).
Molecular determinants of SRGAP2-dependent spine development
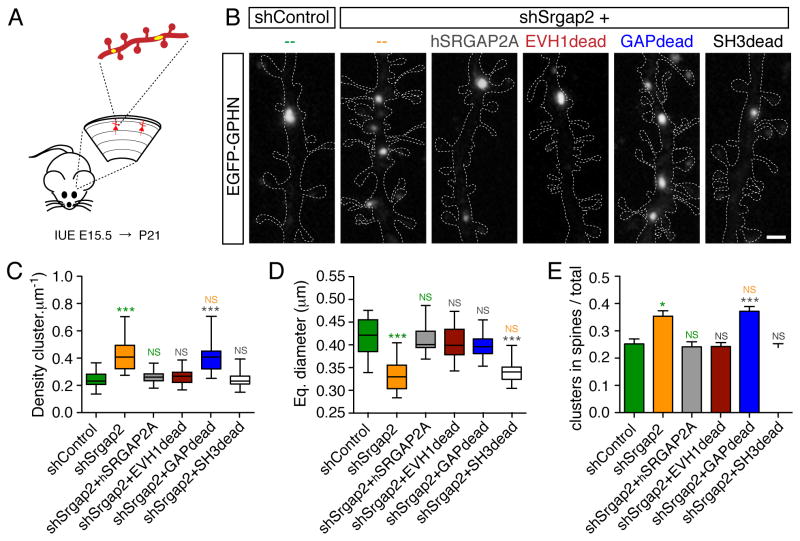
To determine the mechanisms underlying this co-regulation, we first investigated the molecular determinants of SRGAP2A function in the development of dendritic spines. To this end, we developed an in utero gene replacement strategy, which allows the characterization of cell-autonomous mechanisms of synaptic development in vivo (Figure 3A–E). This approach consists in knocking down endogenous Srgap2a in isolated layer 2/3 cortical pyramidal neurons and to replace it with mutant forms of SRGAP2A. We used an shRNA vector which also drives the expression of mVenus to visualize neuronal morphology. The co-electroporation at E15.5 of shSrgap2, which targets mouse Srgap2a but not human SRGAP2A (hSRGAP2A, Charrier et al., 2012), with hSRGAP2A cDNA (1μg/μl each), fully rescued spine density and morphology at P21 (Figure 3A–E), validating this strategy for the molecular dissection of SRGAP2A function in synaptic development in vivo.
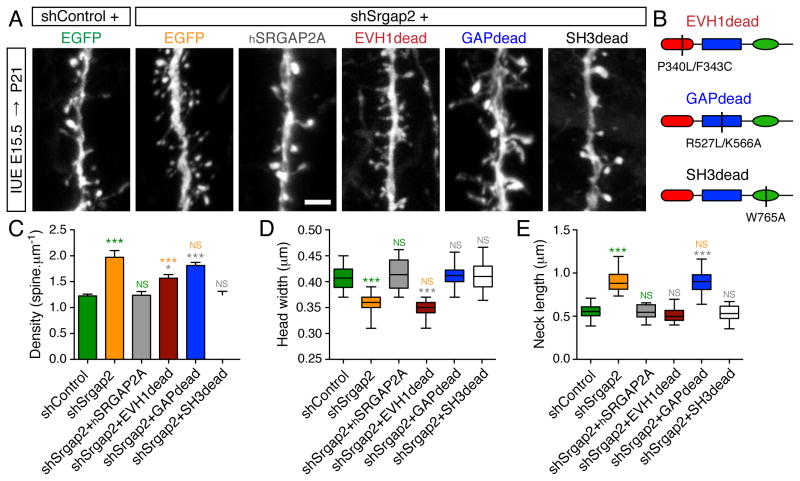
Figure 3. Molecular dissection of SRGAP2A function in spine development in vivo.
(A) Representative segments of oblique dendrites expressing mVenus from P21 mice in control condition (shControl), after knock-down of mouse Srgap2a (shSrgap2), replacement with hSRGAP2A or replacement with the indicated mutants in layer 2/3 cortical neurons using IUE. Scale bar: 2 μm.
(B) Schematic of hSRGAP2A mutants used in (A).
(C) Mean spine density (± SEM).
(D) Quantification of spine head widths.
(E) Quantification of spine neck lengths.
nshControl = 19, nshSrgap2 = 20, nshSrgap2+hSRGAP2A = 18, nshSrgap2+EVH1dead = 24, nshSrgap2+GAPdead = 25, nshSrgap2+SH3dead = 23. *** p < 0.001, * p < 0.05, NS: p > 0.05, Kruskal-Wallis test followed by Dunn’s Multiple comparison test. Green, orange, grey and blue symbols: comparison with shControl, shSrgap2, shSrgap2+hSRGAP2A, shSrgap2+GAPdead, respectively. See also Figures S2, S5 and S6.
To test the function of SRGAP2A-Homer interaction, we replaced endogenous SRGAP2A with the EVH1dead mutant (Figure 3B) in sparse cortical pyramidal neurons. We then analyzed spine morphology and density in juvenile mice. At that stage, the size of the spine head can be used as an indicator of spine maturation (Charrier et al., 2012; Harris and Stevens, 1989), while spine density and spine neck length contribute to dendritic and synaptic compartmentalization in cortical pyramidal neurons (Higley and Sabatini, 2008; Yuste, 2013). The EVH1dead mutant did not rescue spine head size, which was similar to shSrgap2 condition (Figure 3A, D and Figure S2), indicating a deficit in spine maturation. The EVH1dead mutant, however, completely rescued spine neck length and partially rescued spine density (Figure 3A, C and E). These effects mimicked shRNA-mediated Homer1 knock-down in pyramidal neurons in vivo (Figure S5, see also Hayashi et al., 2009), supporting the notion that SRGAP2A helps the formation of the Homer-based postsynaptic scaffold during spine maturation (see also Figure 6A–F).
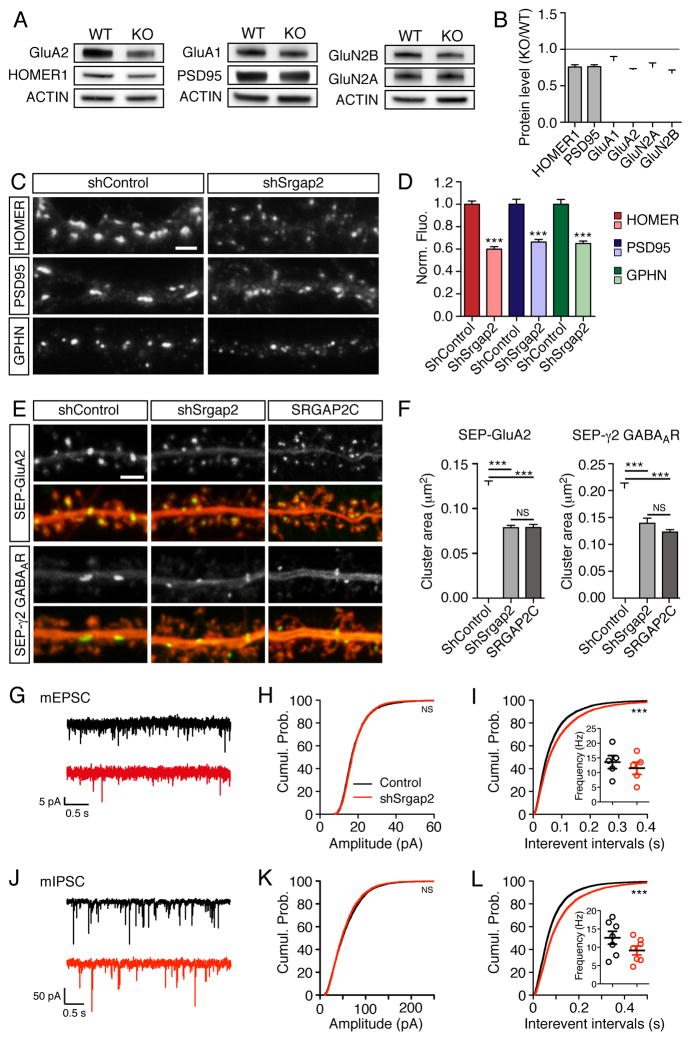
Figure 6. SRGAP2A regulates the assembly of excitatory and inhibitory postsynaptic scaffolds.
(A–B) Representative Western blots (A) and quantifications
(B) of the synaptic abundance of the indicated proteins in P15 wild-type (WT) and Srgap2a KO brains (n=4).
(C) Immunofluorescence of endogenous proteins in cortical neurons cultured for 17–18 days in control condition (shControl) or after Srgap2a knock-down (shSrgap2). Scale bar 2 μm.
(D) Normalized fluorescence intensity (Norm. Fluo.) associated with clusters of the indicated proteins in shControl and shSrgap2 conditions (n=40–42 cells).
(E) Live imaging of SEP-GluA2 and SEP-γ2 GABAA receptors in control pyramidal neurons, after Srgap2a knock-kown or after SRGAP2C expression (20–22 days in vitro). Scale bar: 2 μm.
(F) Mean receptor cluster size in the conditions described above. SEP-GluA2: nshControl = 34, nshSrgap2 = 35, nSRGAP2C = 32 ; SEP-γ2: nshControl = 36, nshSrgap2 = 25, nSRGAP2C = 35. Mean ± SEM.
(G) Representative traces of mEPSCs in control (black) and shSrgap2-electroporated (red) neurons.
(H–I) Quantification of mEPSC amplitude (nControl = 2883 from 5 cells, nshSrgap2 = 2811 from 5 cells) and interevent intervals (nControl = 7232, nshSrgap2 = 6587). Cumul. Prob.: cumulative probability. Inset indicates the mean frequency per cell.
(J) Representative traces of mIPSCs in control (black) and shSrgap2-electroporated (red) neurons.
(K–L) Quantification of mIPSC amplitude (nControl = 4370 from 7 cells, nshSrgap2 = 4125 from 7 cells) and interevent intervals (nControl = 11520, nshSrgap2 = 8262). Inset indicates the mean frequency per cell.
*** p < 0.001, NS: p > 0.05, Mann-Whitney test or Kruskal-Wallis test followed by Dunn’s multiple comparison test (panel F).
To assess the contribution of the Rac1-GAP activity of SRGAP2A, we inserted two point mutations within hSRGAP2A (R527L/K566A; GAPdead mutant) to generate a protein, which is no longer able to promote the GTPase activity of Rac1 and cannot bind to GTP-Rac1 (Guerrier et al., 2009) (Figure 3B). When expressed in replacement of endogenous SRGAP2A, the GAPdead mutant did not rescue spine density and spine neck length (Figure 3A, C and E). However, spine head size was similar to control condition (Figure 3A and D). These results are consistent with the function of Rac1 in dendritic spines (e.g. Cerri et al., 2011; Luo et al., 1996) and demonstrate that the Rac1-GAP activity of SRGAP2A limits spine neck length and spine density but is not required for the morphological maturation of dendritic spines.
We then tested the role of the SH3 domain of SRGAP2A. In addition to Gephyrin, the SH3 domain was previously shown to interact with Actin-regulating proteins (Okada et al., 2011), suggesting that it might regulate spine morphology. Unexpectedly, the SH3dead mutant behaved as wild-type hSRGAP2A (Figure 3A–E), suggesting that the SH3 domain of SRGAP2A is dispensable for dendritic spine development. Together, these results demonstrate that SRGAP2A couples Rac1-dependent signaling to excitatory PSDs via Homer1 and coordinates the regulation of spine density with the rate of their maturation in vivo.
Molecular determinants of SRGAP2-dependent inhibitory synapse development
To dissect the molecular mechanisms underlying SRGAP2A function in inhibitory synapse development, we used the in vivo approach used above for dendritic spines and selectively replaced endogenous SRGAP2A with mutated forms of hSRGAP2A in sparse layer 2/3 cortical pyramidal neurons using IUE (Figure 4A–B). The increased density and reduced size of Gephyrin clusters induced by mouse Srgap2a knock-down were fully rescued by the expression of hSRGAP2A in P21 cortical pyramidal neurons (Figure 4B–E). We then replaced SRGAP2A with the EVH1dead mutant. Surprisingly, despite significant effects on spine maturation and spine density (Figure 3A–D), expression of the EVH1dead mutant did not affect the density, the size or the distribution of Gephyrin clusters (Figure 4B–E, see also Figure S6). These results indicate that SRGAP2A binding to Homer does not affect the development of inhibitory synapses. They also suggest that SRGAP2-dependent regulations of excitatory synaptic development do not elicit indirect or homeostatic adaptations of inhibitory synaptic development.
Figure 4. Molecular dissection of SRGAP2A function in inhibitory synaptic development in vivo.
(A) Schematic: segments of oblique dendrites of layer 2/3 cortical pyramidal neurons were imaged from P21 mice following sparse IUE. EGFP-Gephyrin (yellow) labels inhibitory synapses, TdTomato (red) allows the visualization of dendritic morphology.
(B) EGFP-Gephyrin clusters in representative segments of dendrites in control condition (shControl) or after in utero replacement of mouse Srgap2a with indicated mutants of hSRGAP2A. For clarity, dashed lines define the contours of TdTomato fluorescence in dendrites. Scale bar: 1 μm.
(C–E) Quantification of Gephyrin cluster density (C), Gephyrin cluster equivalent (Eq.) diameter (D) and mean proportion of Gephyrin clusters located in dendritic spines (± SEM) (E). nshControl = 32, nshSrgap2 = 36, nshSrgap2+hSRGAP2A = 31, nshSrgap2+EVH1dead = 30, nshSrgap2+GAPdead = 32, nshSrgap2+SH3dead = 31. *** p < 0.001, * p < 0.05, NS: p > 0.05, Kruskal-Wallis test followed by Dunn’s Multiple comparison test. Green, Orange, grey: comparison with shControl, shSrgap2 and shSrgap2+hSRGAP2A, respectively. See also Figure S6
We then analyzed the role of the Rac1-GAP activity and of the SH3 domain of SRGAP2A. Replacement of SRGAP2A by the GAPdead mutant increased the density of Gephyrin clusters to the same extent as shSrgap2-mediated knockdown. However, the GAPdead mutant rescued the size of Gephyrin clusters, which was similar to control (Figure 4B–D). This uncovers the requirement of the Rac1-GAP activity of SRGAP2A for setting the density of inhibitory synapses but not for their maturation. Replacement of SRGAP2A with the SH3dead mutant had complementary effects: it rescued the effect of shSrgap2 on Gephyrin cluster density but not on Gephyrin cluster size (Figure 4B–D), indicating that SRGAP2A promotes inhibitory synaptic growth through its direct interaction with Gephyrin. Interestingly, the ratio of Gephyrin clusters located on dendritic spines increased with higher density, independently of Gephyrin cluster size (Figure 4C–E).
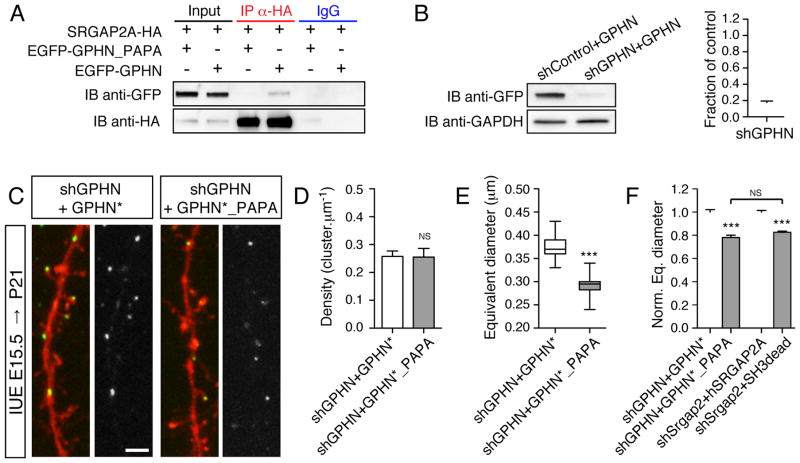
To further substantiate the importance of Gephyrin interaction with the SH3 domain of SRGAP2A in inhibitory synapse development, we performed the reverse experiment and analyzed the consequences of altering the SRGAP2A binding site on Gephyrin. The SRGAP2A binding site on Gephyrin was previously identified as a PGLP motif in the C-terminal E domain of Gephyrin (Okada et al., 2011). Disruption of the PGLP motif by introduction of two point mutations (P603A/P606A, GPHN_PAPA mutant) abolished Gephyrin interaction with SRGAP2A (Figure 5A). We then designed shRNA directed against Gephyrin (shGPHN, Figure 5B) and an shGPHN-resistant Gephyrin construct (GPHN*). Using in utero electroporation, we replaced endogenous Gephyrin with either GPHN* or GPHN*_PAPA mutant in sparse cortical pyramidal neurons (Figure 5C). In juvenile mice, GPHN* and GPHN*_PAPA clusters were present at the same density (Figure 5D). However, GPHN*_PAPA formed smaller clusters than GPHN* (Figure 5E). These results phenocopied what we previously observed using SRGAP2A SH3dead mutant (Figure 5F) and demonstrate that SRGAP2A specifically requires interaction with Gephyrin to regulate inhibitory synapse maturation. Altogether, these results show that the density and subcellular distribution of inhibitory synapses depend on the Rac1-GAP activity of SRGAP2A while direct binding of SRGAP2A to Gephyrin promotes their maturation.
Figure 5. Specific interaction between SRGAP2A and Gephyrin promotes inhibitory synapse maturation.
(A) Mutation of the binding site of SRGAP2A on Gephyrin (P603A/P606A, named GPHN_PAPA mutant) disrupted the interaction between SRGAP2A and Gephyrin in co-IP experiment in HEK cells.
(B) Validation of shGPHN in HEK cells using Western blot (left) and quantification. n = 3, mean ± SEM.
(C) Representative segments of dendrites from P21 mice after in utero replacement of endogenous Gephyrin with EGFP-tagged GPHN* or GPHN*_PAPA (green), and co-expression of soluble TdTomato (red). Scale bar 2 μm.
(D–E) Quantification of gephyrin cluster density (D) and equivalent diameter (E). nshGPHN+GPHN* = 15 (from 3 mice), n shGPHN+GPHN*_PAPA = 15 (from 4 mices), *** p < 0.001, NS: p > 0.05, Mann-Whitney test.
(F) Replacement of endogenous GPHN with GPHN*_PAPA mutant and replacement of endogenous SRGAP2A with hSRGAP2A SH3dead mutant have similar effects. The equivalent (eq.) diameter was normalized to the average value in shGPHN+GPHN* and shSrgap2+hSRGAP2A, respectively. *** p < 0.001, NS: p > 0.05, Kruskal-Wallis test, followed by Dunn’s Multiple comparison test.
Regulation of excitatory and inhibitory postsynaptic scaffold assembly by SRGAP2
We next tested if and how SRGAP2A inactivation impacts the formation of molecular scaffolds and the recruitment of ionotropic Glutamate and GABAA receptors during synaptic maturation. To address this question, we used several independent approaches. First, we isolated excitatory synaptic membranes using subcellular fractionation from juvenile (P15) wild-type and Srgap2a knock-out (KO) brains and compared the abundance of major synaptic proteins using quantitative Western blot analysis (Figure 6A). We observed a homogenous decrease (~25%) in the amount of the postsynaptic scaffolding proteins Homer1 and PSD-95, as well as in the level of AMPA (GluA1, GluA2) and NMDA (GluN2B, GluN2A) receptor subunits in synaptic fractions from the SRGAP2A-deficient brains (Figure 6A–B; comparison of the effect between proteins: p > 0.05, ANOVA). Because inhibitory synapses cannot be isolated using subcellular fractionation, we compared the consequences of SRGAP2A inactivation on glutamatergic and GABAergic synapses using immunocytochemistry and live cell imaging in cultured cortical neurons. In agreement with our biochemical and in vivo morphological data, Srgap2a knock-down in dissociated cortical neurons following lentiviral infection decreased the fluorescence associated with endogenous Homer1, PSD95 and Gephyrin clusters to a similar extent (Homer1: 60 ± 2%, PSD95: 66 ± 2%, Gephyrin: 66 ± 2% of the control value, Figure 6C–D). To estimate the postsynaptic accumulation of excitatory and inhibitory neurotransmitter receptors, we expressed low levels of super-ecliptic (SEP)-tagged receptor subunits, namely GluA2 for AMPA receptors (Lu et al., 2009) and γ2 for GABAARs (Tretter et al., 2012). SEP is a pH-sensitive variant of GFP, which enables the specific visualization of receptors present at the plasma membrane in live neurons (Miesenbock et al., 1998). SEP-tagged receptor subunits and shRNA vectors were introduced into cortical pyramidal neurons using IUE at E15.5, followed by dissociation and culture at E18.5, and live imaging after 20–22 days in vitro (Figure 6E). SRGAP2A knockdown significantly decreased the size of both GluA2-containing glutamate receptor clusters and γ2-containing GABAAR clusters to 62 ± 3% and 68 ± 5% of the control value, respectively (Figure 6E–F). Remarkably, SRGAP2C expression induced a similar decrease of receptor cluster size (GluA2: 62 ± 3% of the control value, γ2-GABAAR: 60 ± 2% of the control value). Together, these results further support the notion that SRGAP2 paralogs co-regulate excitatory and inhibitory synaptic maturation through the recruitment of structural components and neurotransmitter receptors during the assembly of postsynaptic molecular machineries.
Finally, we assessed how SRGAP2A inactivation might affect synaptic transmission. It is difficult to predict how an increase in synaptic density associated with decreased neurotransmitter receptor accumulation and increased morphological complexity (i.e. longer spine neck and more inhibitory synapses in spines) might impact synaptic currents. To address this issue, we performed whole-cell patch-clamp recording of miniature excitatory and inhibitory postsynaptic currents (mEPSCs and mIPSCs, respectively) in brain slices from juvenile (P20-P25) mouse brains. We compared layer 2/3 cortical pyramidal neurons in utero electroporated with shSrgap2 (identified based on their fluorescence) and neighboring non-electroporated control neurons (Figure 6G–L). Although we did not detect any difference in the amplitude of mEPSCs (Average amplitude: 19 ± 1 pA in both control and shSrgap2 neurons, Figure 6H) and mIPSCs (Average amplitude: 58 ± 1 pA in control, 56 ± 1 pA in shSrgap2, Figure 6K), we found that SRGAP2A inactivation induced a mild decrease in the frequency of both mEPSCs (Mean interevent interval: 70 ± 1 ms in control, 89 ± 1 ms in shSrgap2 neurons, Figure 6I) and mIPSCs (Mean interevent interval: 87 ± 1 ms in control, 116 ± 1 ms in shSrgap2 neurons, Figure 6L). Similar changes were observed in Srgap2a KO mice (data not shown). Our results suggest that SRGAP2A-deficient synapses are more frequently electrically “quiet” when recorded in the soma of juvenile neurons.
This is consistent with the delayed maturation of postsynaptic scaffolds and an increased compartmentalization of synaptic inputs (see Araya et al., 2006). We should be cautious however that synaptic currents originating form dendritic synapses (which represent the vast majority of synapses) are usually poorly detected in somatic voltage-clamp recordings due to space-clamp issues in pyramidal neurons (e.g. Williams and Mitchell, 2008; Spruston and Johnson, 2008). Therefore, it is possible that our recordings do not fully account for subtle changes in synaptic transmission that might happen in dendrites. Nonetheless, these results reinforce our main finding that SRGAP2A co-regulates the development of excitatory and inhibitory synapses and indicates that it maintains the equilibrium between excitatory and inhibitory synaptic transmission.
DISCUSSION
In the present study, we report that SRGAP2A, which is expressed in all mammals, co-regulates the development of excitatory and inhibitory synapses in cortical pyramidal neurons in vivo. Our results characterize a mechanism - the first to our knowledge - coupling the regulation of synapse number and synapse maturation at both excitatory and inhibitory synapses (Figure 7). They uncover molecular determinants by which SRGAP2A links major excitatory and inhibitory scaffolding molecules to its Rac1-GAP activity. Using point mutagenesis and an in utero gene replacement strategy in sparse cortical neurons, we demonstrate that SRGAP2A binding to Homer1 and Gephyrin promotes the maturation of excitatory and inhibitory synapses respectively, while its Rac1-GAP activity limits the density of both excitatory and inhibitory synapses. We show that the human-specific copy SRGAP2C inhibits all functions of SRGAP2A, which introduces cellular and molecular innovations during synaptic development that are characteristic features of human neurons, such as their protracted maturation and their increased density of synapses (Benavides-Piccione et al., 2002; Charrier et al., 2012; Elston et al., 2001; Huttenlocher and Dabholkar, 1997; Petanjek et al., 2011). Finally, we show that the co-regulation of the cell biology of excitatory and inhibitory synaptic development by SRGAP2A also maintains the proper equilibrium between excitatory and inhibitory synaptic transmission. We postulate that the unique properties of SRGAP2A in the coordination of excitatory and inhibitory synaptic development allowed the preservation of neuronal integrity following its partial human-specific duplications and may have been fundamental for the fixation of SRGAP2C in the human population.
Figure 7. Summary of the mechanism underlying the coordination of excitatory and inhibitory synaptic development by SRGAP2 paralogs.
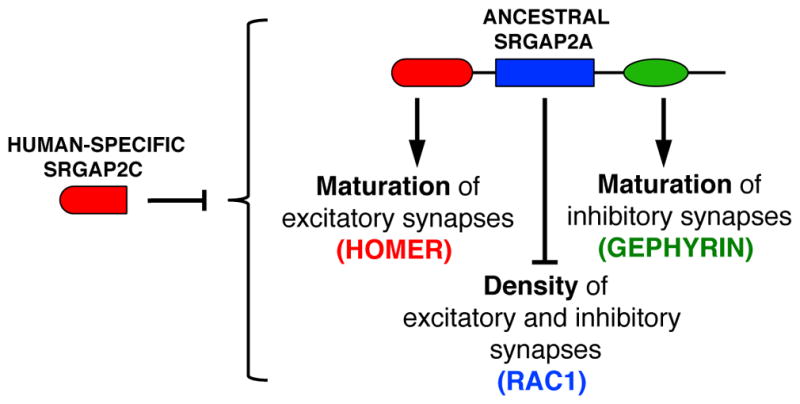
See text for details.
Mechanism coordinating the development of excitatory and inhibitory synapses
So far, the mechanisms underlying the development of excitatory and inhibitory synapses have been studied separately and little is known on the development of inhibitory synapses. This might be due to the great diversity of inhibitory synapses, the fact that they represent a small percentage (10 to 15%) of the total number of synapses in the neocortex and the difficulty to isolate them biochemically and morphologically compared with excitatory synapses associated with dendritic spines. Furthermore, although numerous molecules have been implicated in excitatory synaptic development (McAllister, 2007; Shen and Scheiffele, 2010), we still lack an integrated view of the mechanisms orchestrating synaptic maturation and determining synaptic density and distribution in vivo. Our results uncover shared molecular determinants of excitatory and inhibitory synaptic development in cortical pyramidal neurons and identify SRGAP2A as a major architect regulating the density, the geometry, the subcellular distribution and the timing of maturation of both excitatory and inhibitory synapses. By combining biochemical, cell biological and structure-function analyses in vivo, we characterized molecular interactions of SRGAP2A with binding partners and their functional relevance to synaptic development. We found that SRGAP2A promotes the maturation of both excitatory and inhibitory synapses via a class II EVH1 binding motif embedded in its F-BAR domain and via its SH3 domain, which interact with two key structural components of excitatory and inhibitory postsynaptic scaffolds: Homer and Gephyrin, respectively. It is not clear at this stage how SRGAP2A interactions with Homer and Gephyrin promote the assembly of excitatory and inhibitory postsynaptic scaffolds. One possibility is that SRGAP2A promotes the local recruitment of these scaffolding proteins to nascent synapses by interacting with highly curved membrane structures such as filopodia and spines (see Guerrier et al., 2009) or by coupling them with other as-yet-unidentified proteins involved in synaptic adhesion or serving as synaptic organizers. Another possibility is that SRGAP2A induces conformational changes favoring the oligomerization or the scaffolding properties of Homer and Gephyrin (Hayashi et al., 2009; Tretter et al., 2012). Nonetheless, SRGAP2A inactivation decreased the local accumulation of scaffolding molecules and neurotransmitter receptors at both excitatory and inhibitory synapses, which were less frequently detected in somatic voltage-clamp experiments in juvenile layer 2/3 cortical pyramidal neurons. Importantly, the possibility to dissociate the regulation of excitatory and inhibitory synaptic maturation in individual neurons by specifically disrupting SRGAP2A interaction with Homer or Gephyrin clearly indicates that SRGAP2A and SRGAP2C control a cell-autonomous, developmental mechanism rather than a homeostatic or adaptive response.
Our in vivo molecular dissection of SRGAP2 function in excitatory and inhibitory synaptic development also demonstrates that the Rac1-GAP activity of SRGAP2A limits the density of both excitatory and inhibitory synapses and concomitantly restricts their compartmentalization by controlling the length of the spine neck and the occurrence of inhibitory synapses in spines (Alvarez and Sabatini, 2007; Chiu et al., 2013; Yuste, 2011). Remarkably, these effects did not require direct binding of SRGAP2A to Homer or Gephyrin. In the neocortex, previous studies have implicated Rac1 signaling in the regulation of synaptic density, functional plasticity, critical periods and neurodevelopmental disorders (Cahill et al., 2009; Cerri et al., 2011; Luo et al., 1996). Our present results highlight SRGAP2 as a key determinant of the morphological complexity of cortical pyramidal neurons and suggest major functional implications of the inhibition of Rac1 GTPase signaling modulation by SRGAP2A in the evolution of human synapses.
Molecular regulation of synaptic development by human-specific SRGAP2C
Our results indicate that human-specific duplications of SRGAP2A resulted in a loss-of-function phenotype since SRGAP2C inhibits all identified functions of SRGAP2A. SRGAP2C expression mimics the disruption of SRGAP2A binding to Homer and Gephyrin and the inactivation of its Rac1-GAP activity, indicating that inhibition of the ancestral protein by its human-specific copy alleviates both structural and signaling constraints on synaptic development. Mechanistically, SRGAP2C heterodimerizes with SRGAP2A (Charrier et al., 2012) but it is unclear how it inhibits its function in neurons. Indeed, SRGAP2C might prevent the proper targeting of SRGAP2A to synapses, it might also interfere with SRGAP2A interactions with its partners by steric hindrance, by direct competition or by modifying SRGAP2A conformation, as was shown for other proteins containing both a F-BAR domain and an SH3 domain (Rao et al., 2010).
The unique property of SRGAP2A to bind both Homer and Gephyrin, two important scaffolding molecules during synaptogenesis expressed in virtually all excitatory and inhibitory synapses, suggests that SRGAP2C may universally modify synaptic development in human cortical pyramidal neurons, and maybe in other neuronal subtypes where it is expressed. The oligomerization properties of Homer and Gephyrin provide structural platforms for the assembly of excitatory and inhibitory synapses (Hayashi et al., 2009; Okabe, 2007; Tretter et al., 2012). Therefore, we postulate that SRGAP2C decelerates the assembly of postsynaptic scaffolds without altering the general principles of synaptic development. This hypothesis is supported by the homogenous decrease in the local accumulation of postsynaptic markers such as Homer1, PSD-95, NMDA receptors, AMPA receptors, Gephyrin and GABAA receptors in our biochemical and cell biological experiments. This is also consistent with mass spectrometry analyses of human versus rodent PSDs indicating that the synaptic proteome and interactome are evolutionary conserved (Bayes et al., 2012; Shanks et al., 2012). We should emphasize here that assembly of the Homer- and Gephyrin-based postsynaptic scaffolds might also be the target of other evolutionary mechanisms. Recently, the great-ape microRNA mir-1271, which is robustly expressed in the human forebrain, was shown to down-regulate both Homer1 and Gephyrin expression (Jensen and Covault, 2011).
Potential implications for the human brain
Based on transcriptome analysis of the developing human brain (http://www.brainspan.org) and on our previously published results (Charrier et al., 2012), it is likely that SRGAP2A is partially inhibited by SRGAP2C in human neurons. We have previously shown that the consequences of SRGAP2A inactivation is dose-dependent in juvenile mice but that SRGAP2A haploinsufficency ultimately leads in adults to an increase in spine size and density similar to its constitutive genetic inactivation (Charrier et al., 2012). Different levels of SRGAP2A inhibition would therefore mainly affect early cortical circuits and the timing of their maturation so that spatiotemporal modulations of SRGAP2A and SRGAP2C expression may contribute to the higher level of neoteny characterizing prefontal neocortical areas (Geschwind and Rakic, 2013).
Recent studies have emphasized the importance of protracting synaptic maturation and increasing the morphological complexity of human cortical pyramidal neurons in synaptic physiology and cognitive development. Although heterochrony may contribute to the high vulnerability of human synapses to perturbations, it is thought to extend the period of developmental plasticity and reinforce the role of the environment in the assembly of human cortical circuits (Geschwind and Rakic, 2013; Varki et al., 2008). Hence, accelerated timing of neuronal and synaptic maturation, as caused by haploinsufficiency in the synaptic gene SYNGAP1, has been associated with autism spectrum disorders and intellectual disability (Clement et al., 2012; Courchesne et al., 2007; Pinto et al., 2010). Furthermore, the higher morphological complexity of human cortical pyramidal neurons and the new geometry of dendritic spines may require more local routes for the trafficking of synaptic proteins, affect the propagation and the integration of spine signaling, modify the computational properties of cortical neurons and enforce the rearrangements of cortical circuits (Alvarez and Sabatini, 2007; Chiu et al., 2013; Cui-Wang et al., 2012; Geschwind and Rakic, 2013; Spruston, 2008; Yuste, 2013). As a consequence, the phenotypic outcomes of gene mutations affecting synaptic development and function (Krumm et al., 2014; Parikshak et al., 2013; Ting et al., 2012; Zoghbi and Bear, 2012) might differ between species. In the future, characterizing the cell biology of human neurons and the specificities of human synaptic development should provide new insights into the etiology of neurodevelopmental and psychiatric disorders.
EXPERIMENTAL PROCEDURES
Animals
All animals were handled according to protocols approved by French authorities and the Institutional Animal Care and Use Committee at Columbia University, New York. Timed-pregnant female mice were maintained in a 12-hour light/dark cycle and obtained by overnight breeding with males of the same strain. For timed-pregnant mating, noon after mating is considered embryonic day 0.5 (E0.5). Juveniles correspond to mice between postnatal day (P) 21 and P23. Adults correspond to mice between P65 and P75. The Srgap2 gene trapped allele (B6;129P2-Srgap2Gt(XH102)Byg/Mmcd) has been described previously (Charrier et al., 2012). Mice homozygous for this allele were referred to as Srgap2a KO.
In utero cortical electroporation and slice preparation
In utero cortical electroporation was performed at E15.5. See Supplemental experimental procedures for detailed methods.
Primary neuronal culture, lentiviral infection, live imaging and immunocytochemistry
Primary cultures were performed as described previously (Charrier et al., 2012) with few modifications. Please refer to Supplemental experimental procedures for details.
Cell line culture, transfection and lysis
Subcellular fractionation
Subcellular fractionation was performed from P15 mouse brains as described in Perez-Otano et al., 2006 with minor modifications. See Supplemental experimental procedures for details.
Co-immunoprecipitation and Western Blotting
Co-immunoprecipitation, Western blotting and antibodies used in this study are described in Supplemental experimental procedures.
Confocal image acquisition and analysis
Confocal images of isolated electroporated neurons in slices were blindly acquired and analyzed as detailed in Supplemental experimental procedures. Numbers of animals and dendrites analyzed per condition are indicated in Table S1.
Live cell imaging and analysis
Live cell imaging was performed at 37°C on a spinning disc confocal microscope as detailed in Supplemental experimental procedures.
Constructs and shRNAs
Plasmids are described in Supplemental experimental procedures.
Electrophysiology
Electrophysiology recordings are described in Supplemental experimental procedures.
Statistics
Details on statistical procedures and data representation can be found in Supplemental experimental procedures.
Supplementary Material
Acknowledgments
We thank Virginie Courchet for excellent management of mouse colonies and technical assistance, Antoine Triller and Serge Marty for comments on the manuscript, members of the Polleux and Triller laboratories for discussions, H. Cline, A. Maximov, A. Triller, R. Malinow (Addgene 24001), D. Arnold (Addgene 46296) and W.L. Jin for reagents. This work was supported by grants from the Agence Nationale de la Recherche (ANR-13-PDOC-0003, SYNHUMA to CC), Inserm (to CC), the National Institute for Neurological Disorders and Stroke (RO1NS067557 to FP), an award from the Fondation Roger de Spoelberch (to FP), the Netherlands Organization for Scientific Research (Rubicon 825.14.017 to ERS) and the European Molecular Biology Organization (EMBO ALTF 1055–2014 to ERS). The IBENS Protein facility and Imaging Facility received support implemented by the ANR under the program Investissements d’Avenir (ANR-10-LABX-54 MEMO LIFE, ANR-11-IDEX-0001-02 PSL* Research University and ANR- 10- INSB- 04- 01 France- BioImaging infrastructure). The IBENS Imaging facility is also supported by grants from Région Ile-de-France (NERF N°2009-44 and NERF N°2011-45), Fondation pour la Recherche Médicale (N° DGE 20111123023) and Fédération pour la Recherche sur le Cerveau - Rotary International France (2011).
Footnotes
AUTHOR CONTRIBUTIONS
CC and MF conducted experiments and analysis. RP performed electrophysiology experiments and analysis. ERS and JVK performed experiments and analysis in Figure S4. DS produced and purified GPHN.FingR. CC designed the study. CC made the figures. CC and FP interpreted the results and wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alvarez VA, Sabatini BL. Anatomical and physiological plasticity of dendritic spines. Annu Rev Neurosci. 2007;30:79–97. doi: 10.1146/annurev.neuro.30.051606.094222. [DOI] [PubMed] [Google Scholar]
- Araya R, Vogels TP, Yuste R. Activity-dependent dendritic spine neck changes are correlated with synaptic strength. Proc Natl Acad Sci U S A. 2006;111:2895–2904. doi: 10.1073/pnas.1321869111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayes A, Collins MO, Croning MD, van de Lagemaat LN, Choudhary JS, Grant SG. Comparative study of human and mouse postsynaptic proteomes finds high compositional conservation and abundance differences for key synaptic proteins. PLoS One. 2012;7:e46683. doi: 10.1371/journal.pone.0046683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benavides-Piccione R, Ballesteros-Yanez I, DeFelipe J, Yuste R. Cortical area and species differences in dendritic spine morphology. J Neurocytol. 2002;31:337–346. doi: 10.1023/a:1024134312173. [DOI] [PubMed] [Google Scholar]
- Benson DL, Cohen PA. Activity-independent segregation of excitatory and inhibitory synaptic terminals in cultured hippocampal neurons. J Neurosci. 1996;16:6424–6432. doi: 10.1523/JNEUROSCI.16-20-06424.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bockaert J, Perroy J, Becamel C, Marin P, Fagni L. GPCR interacting proteins (GIPs) in the nervous system: Roles in physiology and pathologies. Annu Rev Pharmacol Toxicol. 2010;50:89–109. doi: 10.1146/annurev.pharmtox.010909.105705. [DOI] [PubMed] [Google Scholar]
- Cahill ME, Xie Z, Day M, Photowala H, Barbolina MV, Miller CA, Weiss C, Radulovic J, Sweatt JD, Disterhoft JF, et al. Kalirin regulates cortical spine morphogenesis and disease-related behavioral phenotypes. Proc Natl Acad Sci U S A. 2009;106:13058–13063. doi: 10.1073/pnas.0904636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerri C, Fabbri A, Vannini E, Spolidoro M, Costa M, Maffei L, Fiorentini C, Caleo M. Activation of Rho GTPases triggers structural remodeling and functional plasticity in the adult rat visual cortex. J Neurosci. 2011;31:15163–15172. doi: 10.1523/JNEUROSCI.2617-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrier C, Joshi K, Coutinho-Budd J, Kim JE, Lambert N, de Marchena J, Jin WL, Vanderhaeghen P, Ghosh A, Sassa T, et al. Inhibition of SRGAP2 function by its human-specific paralogs induces neoteny during spine maturation. Cell. 2012;149:923–935. doi: 10.1016/j.cell.2012.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JL, Villa KL, Cha JW, So PT, Kubota Y, Nedivi E. Clustered dynamics of inhibitory synapses and dendritic spines in the adult neocortex. Neuron. 2012;74:361–373. doi: 10.1016/j.neuron.2012.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu CQ, Lur G, Morse TM, Carnevale NT, Ellis-Davies GC, Higley MJ. Compartmentalization of GABAergic inhibition by dendritic spines. Science. 2013;340:759–762. doi: 10.1126/science.1234274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet D, Triller A. The dynamic synapse. Neuron. 2013;80:691–703. doi: 10.1016/j.neuron.2013.10.013. [DOI] [PubMed] [Google Scholar]
- Clement JP, Aceti M, Creson TK, Ozkan ED, Shi Y, Reish NJ, Almonte AG, Miller BH, Wiltgen BJ, Miller CA, et al. Pathogenic SYNGAP1 mutations impair cognitive development by disrupting maturation of dendritic spine synapses. Cell. 2012;151:709–723. doi: 10.1016/j.cell.2012.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courchesne E, Pierce K, Schumann CM, Redcay E, Buckwalter JA, Kennedy DP, Morgan J. Mapping early brain development in autism. Neuron. 2007;56:399–413. doi: 10.1016/j.neuron.2007.10.016. [DOI] [PubMed] [Google Scholar]
- Cui-Wang T, Hanus C, Cui T, Helton T, Bourne J, Watson D, Harris KM, Ehlers MD. Local zones of endoplasmic reticulum complexity confine cargo in neuronal dendrites. Cell. 2012;148:309–321. doi: 10.1016/j.cell.2011.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defelipe J. The evolution of the brain, the human nature of cortical circuits, and intellectual creativity. Front Neuroanat. 2011;5:29. doi: 10.3389/fnana.2011.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis MY, Nuttle X, Sudmant PH, Antonacci F, Graves TA, Nefedov M, Rosenfeld JA, Sajjadian S, Malig M, Kotkiewicz H, et al. Evolution of human-specific neural SRGAP2 genes by incomplete segmental duplication. Cell. 2012;149:912–922. doi: 10.1016/j.cell.2012.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elston GN, Benavides-Piccione R, DeFelipe J. The pyramidal cell in cognition: a comparative study in human and monkey. J Neurosci. 2001;21:RC163. doi: 10.1523/JNEUROSCI.21-17-j0002.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritschy JM, Harvey RJ, Schwarz G. Gephyrin: where do we stand, where do we go? Trends Neurosci. 2008;31:257–264. doi: 10.1016/j.tins.2008.02.006. [DOI] [PubMed] [Google Scholar]
- Geschwind DH, Rakic P. Cortical evolution: judge the brain by its cover. Neuron. 2013;80:633–647. doi: 10.1016/j.neuron.2013.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross GG, Junge JA, Mora RJ, Kwon HB, Olson CA, Takahashi TT, Liman ER, Ellis-Davies GC, McGee AW, Sabatini BL, Roberts RW, Arnold DB. Recombinant probes for visualizing endogenous synaptic proteins in living neurons. Neuron. 2013;78:971–985. doi: 10.1016/j.neuron.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrier S, Coutinho-Budd J, Sassa T, Gresset A, Jordan NV, Chen K, Jin WL, Frost A, Polleux F. The F-BAR domain of srGAP2 induces membrane protrusions required for neuronal migration and morphogenesis. Cell. 2009;138:990–1004. doi: 10.1016/j.cell.2009.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand R, Polleux F. Neurogenin2 regulates the initial axon guidance of cortical pyramidal neurons projecting medially to the corpus callosum. Neural Dev. 2011;6:30. doi: 10.1186/1749-8104-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KM, Stevens JK. Dendritic spines of CA 1 pyramidal cells in the rat hippocampus: serial electron microscopy with reference to their biophysical characteristics. J Neurosci. 1989;9:2982–2997. doi: 10.1523/JNEUROSCI.09-08-02982.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi MK, Tang C, Verpelli C, Narayanan R, Stearns MH, Xu RM, Li H, Sala C, Hayashi Y. The postsynaptic density proteins Homer and Shank form a polymeric network structure. Cell. 2009;137:159–171. doi: 10.1016/j.cell.2009.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higley MJ, Sabatini BL. Calcium signaling in dendrites and spines: practical and functional considerations. Neuron. 2008;59:902–913. doi: 10.1016/j.neuron.2008.08.020. [DOI] [PubMed] [Google Scholar]
- Huttenlocher PR, Dabholkar AS. Regional differences in synaptogenesis in human cerebral cortex. J Comp Neurol. 1997;387:167–178. doi: 10.1002/(sici)1096-9861(19971020)387:2<167::aid-cne1>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Jensen KP, Covault J. Human miR-1271 is a miR-96 paralog with distinct nonconserved brain expression pattern. Nucleic Acids Res. 2011;39:701–711. doi: 10.1093/nar/gkq798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott GW, Quairiaux C, Genoud C, Welker E. Formation of dendritic spines with GABAergic synapses induced by whisker stimulation in adult mice. Neuron. 2002;34:265–273. doi: 10.1016/s0896-6273(02)00663-3. [DOI] [PubMed] [Google Scholar]
- Krumm N, O’Roak BJ, Shendure J, Eichler EE. A de novo convergence of autism genetics and molecular neuroscience. Trends Neurosci. 2014;37:95–105. doi: 10.1016/j.tins.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota Y, Hatada S, Kondo S, Karube F, Kawaguchi Y. Neocortical inhibitory terminals innervate dendritic spines targeted by thalamocortical afferents. J Neurosci. 2007;27:1139–1150. doi: 10.1523/JNEUROSCI.3846-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Shi Y, Jackson AC, Bjorgan K, During MJ, Sprengel R, Seeburg PH, Nicoll RA. Subunit composition of synaptic AMPA receptors revealed by a single-cell genetic approach. Neuron. 2009;62:254–268. doi: 10.1016/j.neuron.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Hensch TK, Ackerman L, Barbel S, Jan LY, Jan YN. Differential effects of the Rac GTPase on Purkinje cell axons and dendritic trunks and spines. Nature. 1996;379:837–840. doi: 10.1038/379837a0. [DOI] [PubMed] [Google Scholar]
- McAllister AK. Dynamic aspects of CNS synapse formation. Annu Rev Neurosci. 2007;30:425–450. doi: 10.1146/annurev.neuro.29.051605.112830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miesenbock G, De Angelis DA, Rothman JE. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature. 1998;394:192–195. doi: 10.1038/28190. [DOI] [PubMed] [Google Scholar]
- Nelson SB, Valakh V. Excitatory/Inhibitory Balance and Circuit Homeostasis in Autism Spectrum Disorders. Neuron. 2015;87:684–98. doi: 10.1016/j.neuron.2015.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe S. Molecular anatomy of the postsynaptic density. Mol Cell Neurosci. 2007;34:503–518. doi: 10.1016/j.mcn.2007.01.006. [DOI] [PubMed] [Google Scholar]
- Okada H, Uezu A, Mason FM, Soderblom EJ, Moseley MA, 3rd, Soderling SH. SH3 domain-based phototrapping in living cells reveals Rho family GAP signaling complexes. Sci Signal. 2011;4:rs13. doi: 10.1126/scisignal.2002189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikshak NN, Luo R, Zhang A, Won H, Lowe JK, Chandran V, Horvath S, Geschwind DH. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell. 2013;155:1008–1021. doi: 10.1016/j.cell.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Otano I, Lujan R, Tavalin SJ, Plomann M, Modregger J, Liu XB, Jones EG, Heinemann SF, Lo DC, Ehlers MD. Endocytosis and synaptic removal of NR3A-containing NMDA receptors by PACSIN1/syndapin1. Nat Neurosci. 2006;9:611–621. doi: 10.1038/nn1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petanjek Z, Judas M, Simic G, Rasin MR, Uylings HB, Rakic P, Kostovic I. Extraordinary neoteny of synaptic spines in the human prefrontal cortex. Proc Natl Acad Sci U S A. 2011;108:13281–13286. doi: 10.1073/pnas.1105108108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, Conroy J, Magalhaes TR, Correia C, Abrahams BS, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramocki MB, Zoghbi HY. Failure of neuronal homeostasis results in common neuropsychiatric phenotypes. Nature. 2008;455:912–918. doi: 10.1038/nature07457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao Y, Ma Q, Vahedi-Faridi A, Sundborger A, Pechstein A, Puchkov D, Luo L, Shupliakov O, Saenger W, Haucke V. Molecular basis for SH3 domain regulation of F-BAR-mediated membrane deformation. Proc Natl Acad Sci U S A. 2010;107:8213–8218. doi: 10.1073/pnas.1003478107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks NF, Savas JN, Maruo T, Cais O, Hirao A, Oe S, Ghosh A, Noda Y, Greger IH, Yates JR, 3rd, et al. Differences in AMPA and kainate receptor interactomes facilitate identification of AMPA receptor auxiliary subunit GSG1L. Cell Rep. 2012;1:590–598. doi: 10.1016/j.celrep.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen K, Scheiffele P. Genetics and cell biology of building specific synaptic connectivity. Annu Rev Neurosci. 2010;33:473–507. doi: 10.1146/annurev.neuro.051508.135302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng M, Hoogenraad CC. The postsynaptic architecture of excitatory synapses: a more quantitative view. Annu Rev Biochem. 2007;76:823–847. doi: 10.1146/annurev.biochem.76.060805.160029. [DOI] [PubMed] [Google Scholar]
- Soto F, Bleckert A, Lewis R, Kang Y, Kerschensteiner D, Craig AM, Wong RO. Coordinated increase in inhibitory and excitatory synapses onto retinal ganglion cells during development. Neural Dev. 2011;6:31. doi: 10.1186/1749-8104-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruston N. Pyramidal neurons: dendritic structure and synaptic integration. Nat Rev Neurosci. 2008;9:206–221. doi: 10.1038/nrn2286. [DOI] [PubMed] [Google Scholar]
- Spruston N, Johnston D. Out of control in the dendrites. Nat Neurosci. 2008;11:733–734. doi: 10.1038/nn0708-733. [DOI] [PubMed] [Google Scholar]
- Sudmant PH, Kitzman JO, Antonacci F, Alkan C, Malig M, Tsalenko A, Sampas N, Bruhn L, Shendure J, Genomes P, et al. Diversity of human copy number variation and multicopy genes. Science. 2010;330:641–646. doi: 10.1126/science.1197005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting JT, Peca J, Feng G. Functional consequences of mutations in postsynaptic scaffolding proteins and relevance to psychiatric disorders. Annu Rev Neurosci. 2012;35:49–71. doi: 10.1146/annurev-neuro-062111-150442. [DOI] [PubMed] [Google Scholar]
- Tretter V, Mukherjee J, Maric HM, Schindelin H, Sieghart W, Moss SJ. Gephyrin, the enigmatic organizer at GABAergic synapses. Front Cell Neurosci. 2012;6:23. doi: 10.3389/fncel.2012.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu JC, Xiao B, Naisbitt S, Yuan JP, Petralia RS, Brakeman P, Doan A, Aakalu VK, Lanahan AA, Sheng M, et al. Coupling of mGluR/Homer and PSD-95 complexes by the Shank family of postsynaptic density proteins. Neuron. 1999;23:583–592. doi: 10.1016/s0896-6273(00)80810-7. [DOI] [PubMed] [Google Scholar]
- Tu JC, Xiao B, Yuan JP, Lanahan AA, Leoffert K, Li M, Linden DJ, Worley PF. Homer binds a novel proline-rich motif and links group 1 metabotropic glutamate receptors with IP3 receptors. Neuron. 1998;21:717–726. doi: 10.1016/s0896-6273(00)80589-9. [DOI] [PubMed] [Google Scholar]
- van Versendaal D, Rajendran R, Saiepour MH, Klooster J, Smit-Rigter L, Sommeijer JP, De Zeeuw CI, Hofer SB, Heimel JA, Levelt CN. Elimination of inhibitory synapses is a major component of adult ocular dominance plasticity. Neuron. 2012;74:374–383. doi: 10.1016/j.neuron.2012.03.015. [DOI] [PubMed] [Google Scholar]
- Varki A, Geschwind DH, Eichler EE. Explaining human uniqueness: genome interactions with environment, behaviour and culture. Nat Rev Genet. 2008;9:749–763. doi: 10.1038/nrg2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SR, Mitchell SJ. Direct measurement of somatic voltage clamp errors in central neurons. Nat Neurosci. 2008;11:790–798. doi: 10.1038/nn.2137. [DOI] [PubMed] [Google Scholar]
- Yizhar O, Fenno LE, Prigge M, Schneider F, Davidson TJ, O’Shea DJ, Sohal VS, Goshen I, Finkelstein J, Paz JT, et al. Neocortical excitation/inhibition balance in information processing and social dysfunction. Nature. 2011;477:171–178. doi: 10.1038/nature10360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuste R. Dendritic spines and distributed circuits. Neuron. 2011;71:772–781. doi: 10.1016/j.neuron.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuste R. Electrical compartmentalization in dendritic spines. Annu Rev Neurosci. 2013;36:429–449. doi: 10.1146/annurev-neuro-062111-150455. [DOI] [PubMed] [Google Scholar]
- Zhao X, Shoji S, Lau P. Balanced GABAergic and glutamatergic synapse development in hippocampal neurons. Biochem Biophys Res Commun. 2005;330:1110–1115. doi: 10.1016/j.bbrc.2005.03.083. [DOI] [PubMed] [Google Scholar]
- Zoghbi HY, Bear MF. Synaptic dysfunction in neurodevelopmental disorders associated with autism and intellectual disabilities. Cold Spring Harb Perspect Biol. 2012:4. doi: 10.1101/cshperspect.a009886. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.