Abstract
Hippocampal sclerosis of aging (HS-Aging) is a common neurodegenerative condition associated with dementia. To learn more about genetic risk of HS-Aging pathology, we tested gene-based associations of the GRN, TMEM106B, ABCC9, and KCNMB2 genes, which were reported to be associated with HS-Aging pathology in previous studies. Genetic data were obtained from the Alzheimer’s Disease Genetics Consortium (ADGC), linked to autopsy-derived neuropathological outcomes from the National Alzheimer’s Coordinating Center (NACC). Of the 3,251 subjects included in the study, 271 (8.3%) were identified as an HS-Aging case. The significant gene-based association between the ABCC9 gene and HS-Aging appeared to be driven by a region in which a significant haplotype-based association was found. We tested this haplotype as an expression Quantitative Trait Locus (eQTL) using two different public-access brain gene expression databases. The HS-Aging pathology protective ABCC9 haplotype was associated with decreased ABCC9 expression, indicating a possible toxic gain of function.
Keywords: CARTS, GWAS, PGRN, KATP, rs704180
1. Introduction
Hippocampal sclerosis of aging (HS-Aging) is a high-morbidity brain disease in people of advanced age (Corey-Bloom, et al., 1997). The prevalence of HS-Aging pathology ranges from 5 to 30% in older people in large autopsy series (Dickson, et al., 1994; Leverenz, et al., 2002; Nelson, et al., 2013; Zarow, et al., 2012). Clinical signs and symptoms of HS-Aging are similar to those of Alzheimer’s disease (AD) with amnestic memory deficits (Pao, et al., 2011; Zarow, et al., 2008). Because of the overlapping symptomology, HS-Aging is often clinically misdiagnosed as AD (Brenowitz, et al., 2014; Pao, et al., 2011; Zarow, et al., 2008). AD is characterized by the accumulation of amyloid plaques and neurofibrillary tangles (Hyman, et al., 2012), while HS-Aging is pathologically characterized by neuronal cell loss and gliosis in the hippocampus seen by hematoxylin and eosin (H&E) stain, which can occur unilaterally (~50%) or bilaterally (Nelson, et al., 2013; Zarow, et al., 2008). Whatever the laterality on H&E stain, the large majority of cases with HS-Aging show bilateral TAR DNA-binding protein 43 (TDP-43) pathology in limbic structures (Amador-Ortiz, et al., 2007; Nelson, et al., 2011b). Awareness of this common cause of dementia is rapidly increasing, and we recently recommended a revision of the terminology for describing this disease to cerebral age-related TDP-43 with sclerosis (CARTS) (Nelson, et al., 2016b). However, here we will maintain use of the term HS-Aging because the neuropathologic databases we assessed did not include TDP-43 pathologic information until quite recently.
Genetic risk factors for HS-Aging have been recently identified. Unlike AD, the apolipoprotein E (APOE) ε4 allele is not a risk factor for HS-Aging (Brenowitz, et al., 2014; Leverenz, et al., 2002; Nelson, et al., 2011b; Pao, et al., 2011; Troncoso, et al., 1996). By contrast, the following four genes (in the chronological order they were so identified) have been reported to harbor risk alleles associated with HS-Aging pathology: Granulin (GRN) on chromosome 17q, Transmembrane protein 106B (TMEM106B) on chromosome 7p, ATP-binding cassette subfamily member 9 (ABCC9) on chromosome 12p, and potassium channel subfamily M regulatory beta subunit 2 (KCNMB2) on chromosome 3q (Aoki, et al., 2015; Beecham, et al., 2014; Dickson, et al., 2010; Murray, et al., 2014; Nelson, et al., 2014; Nelson, et al., 2015b; Pao, et al., 2011).
Alleles near the coding portions of the GRN and TMEM106B genes were shown to have an association with HS-Aging using an allele test, following the known relationship of those two genes to frontotemporal lobar degeneration with TDP-43 inclusions (FTLD-TDP). Specifically, HS-Aging pathology was associated with the T-allele of the GRN single nucleotide polymorphism (SNP) rs5848 (Dickson, et al., 2010; Murray, et al., 2014; Pickering-Brown, et al., 2008; Rademakers, et al., 2008). For the other FTLD-related gene, TMEM106B, persons with eventual autopsy-proven HS-Aging pathology were more likely to have the T-allele than controls (Aoki, et al., 2015; Murray, et al., 2014; Rutherford, et al., 2012). We confirmed an increase in HS-Aging odds for each copy of the T-allele of TMEM106B rs1990622 (Nelson, et al., 2014).
The connections of the ABCC9 and KCNMB2 genes to HS-Aging risk were discovered via genome-wide association studies (GWAS), which are neither helped nor biased by prior mechanistic hypotheses. The association of ABCC9 SNP rs704178 with HS-Aging pathology was demonstrated in a GWAS using a recessive mode of inheritance (MOI) (Nelson, et al., 2014). The relationship of this locus with HS-Aging was subsequently tested in a different group of research subjects, and the association was replicated (Nelson, et al., 2015b). Beecham and colleagues reported the KCNMB2 SNP rs9637454 as the top SNP for HS pathology, although this association was not genome wide significant (Beecham, et al., 2014), and has not been replicated to date.
In the present study, we examined the associations of these four putative risk SNPs with HS-aging pathology, using genetic data obtained from Alzheimer’s Disease Genetics Consortium (ADGC) linked to neuropathological outcomes from the National Alzheimer’s Coordinating Center (NACC) (Nelson, et al., 2014; Nelson, et al., 2015b). Here we aggregated those data sets to attain greater statistical power for gene-wide association analyses, for the purpose of understanding better the association of multiple (often co-inherited) gene variants with disease development. Thus, we tested GRN, TMEM106B, ABCC9, and KCNMB2 for gene-based associations with HS-Aging pathology by aggregating SNPs and indels (small insertions or deletions) on each of those genes. In addition, we focused on the interesting region located around intronic SNP rs704178 on the ABCC9 gene that was identified in the previous work, and analyzed haplotype associations of the region with HS-Aging pathology and ABCC9 gene expression.
2. Material and methods
2.1 Study subjects
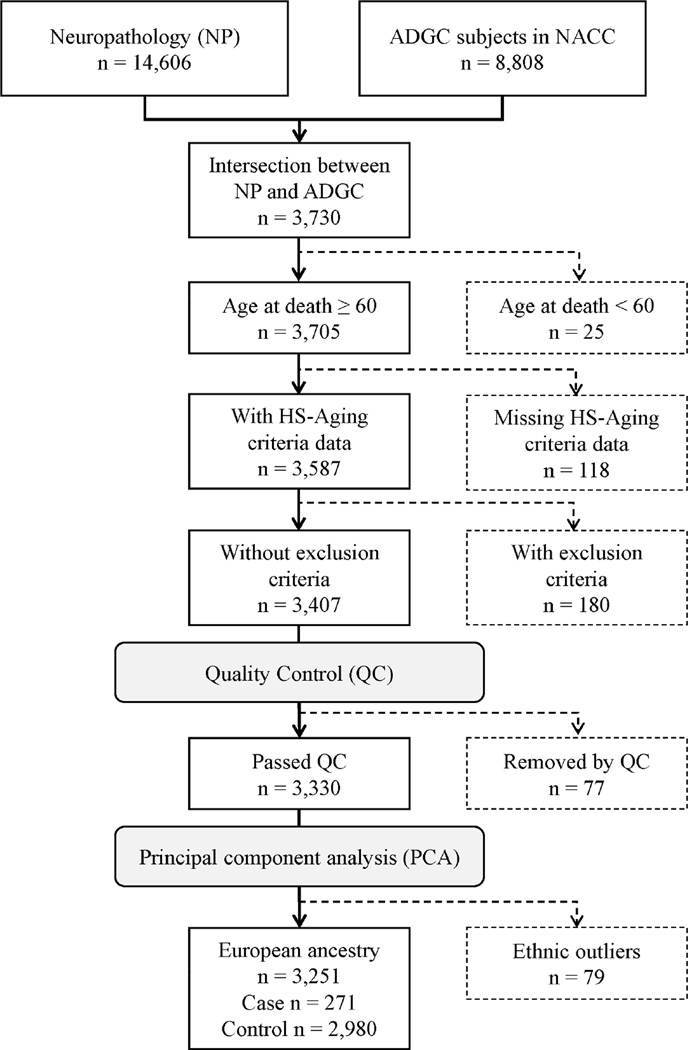
ADGC genotype data were linked to data from the National Institute on Aging (NIA)-funded 36 AD Centers (ADCs) and NACC registry phenotype information. Of 3,730 subjects with both genotype and autopsy information available to us, those who died at age 60 years or older were included in this study. Cases of HS-Aging were identified as patients who met at least one of the following criteria at autopsy; 1) the primary pathologic diagnosis was hippocampal sclerosis, 2) there was a contributing pathologic diagnosis of hippocampal sclerosis, or 3) medial temporal lobe sclerosis was present at autopsy. We then excluded 180 individuals who had FTLD with ubiquitin-positive inclusions, FTLD with no distinctive histopathology, FTLD-tau, or prion associated disease (Figure 1).
Figure 1.
Flow diagram of the subjects included in the analyses. Genetic data were obtained from subjects in ADGC who had the NACC individual IDs. Phenotype data were available from the neuropathological dataset in NACC. The inclusion/exclusion criteria, quality control and removal of ethnic outliers were applied in order.
Key: ADGC, Alzheimer’s Disease Genetics Consortium; NACC, National Alzheimer’s Coordinating Center; NP, neuropathological dataset; HS-Aging, hippocampal sclerosis of aging
2.2 Quality control of the ADGC genotype data
Standard quality control (QC) procedures were performed on the ADGC genotype data using PLINK v1.90a (Purcell, et al., 2007). Markers were excluded based on the following criteria: (1) minor allele frequency (MAF) < 1%; (2) call rate per variant (SNPs and indels) < 95%, (3) Hardy-Weinberg equilibrium test in controls < 10−5. (Supplemental Table 1). Samples were excluded based on the following criteria: (1) call rate per individual < 95%, (2) a high degree of relatedness per an estimated proportion of identical by descent (IBD) > 0.1875, (3) excess of ± 3.0 standard deviations of heterozygosity rate. Of the 3,407 individuals after the inclusion and exclusion criteria were applied, 3,330 passed the QC (Figure 1).
2.3 Identifying ethnic outliers
We performed principal component analysis (PCA) in EIGENSTRAT (Price, et al., 2006) using a linkage disequilibrium (LD) pruned subset of markers (pairwise r2 < 0.2) from our data merged to 1000 Genomes Project Phase 3 (1000 Genomes) (1000 Genomes Project Consortium, 2010) data after removing symmetric SNPs and flipping SNPs discordant for DNA strands between the two datasets. We then plotted the first and second principal components (PCs) for each individuals (n = 5,834: 2,504 from 1000 Genomes and 3,330 from the study) using the ggplot2 R package (version 2.2.0) (Wickham, 2009) in R (version 3.2; http://www.r-project.org). Based on the PC plot, 79 study subjects were removed as ethnic outliers (Figure 1 and Supplemental Figure 1). We reran the PCA for the remaining 3,251 European ancestries to derive orthogonal PCs which were used as covariates in the subsequent analyses.
2.4 Statistical analysis
2.4.1 Gene-based association analysis
Prior to gene-based association analyses, we performed the single variant association testing using logistic regression assuming each of the three most commonly used MOI (additive, dominant, and recessive) adjusted for age at death, sex and the top three PCs using PLINK v1.90a (Purcell, et al., 2007). Gene-based association analyses were conducted using GATES (Gene-Based Association Test Using Extended Simes Procedure) (Li, et al., 2011) as implemented in the open-source software Knowledge-Based Mining System for Genome-wide Genetic Studies (KGG; version 3.5) (Li, et al., 2011). GATES is a gene-based association test that combines the p-values of variants within a gene obtained from single variant association testing described above. We assigned variants to genes based on their physical positions at the UCSC Genome Browser GRCh37/hg19 human assembly (https://genome.ucsc.edu/) (Kent, et al., 2002), and defined gene boundaries as ± 5kb from 5’ and 3’ untranslated regions (UTRs). This gene-based association test adjusts for LD in European super population genotype data from the 1000 Genomes (1000 Genomes EUR) (1000 Genomes Project Consortium, 2010). The input data files to KGG contained four columns: chromosome number, marker ID, marker position, and single variant association p-value. We then obtained overall p-values for the associations of the target genes. Since those who live to advanced old age have a higher risk of HS-Aging pathology (Nelson, et al., 2011a; Nelson, et al., 2011b), there is a possibility that those who died earlier would be always identified as a control even if they have a genetic risk. Therefore, for sensitivity analysis against these possible misclassifications, we further performed these gene-based association tests in cases and controls who died at age 80 years or older. For the gene-based association test, statistical significance level was defined using the Bonferroni correction, yielding α = 0.05/(4 genes ×3 MOI ×2 age groups) = 0.0021 for the four examined genes and three MOI.
2.4.2 Haplotype-based association analysis for HS-Aging
After identifying the HS-Aging risk-associated region on the ABCC9 gene by generating a regional association plot using LocusZoom software (Pruim, et al., 2010), we performed additional post hoc haplotype analysis for the variants on the region. First, we selected tag variants using a pairwise SNP tagging approach with r2 ≥ 0.8 based on the 1000 Genomes EUR in Haploview version 4.2 (Barrett, et al., 2005). Maximum likelihood estimates of haplotype frequencies were computed using an expectation-maximization (EM) algorithm implemented in the functions haplo.em (for overall subjects) and haplo.group (for HS-Aging cases and controls) of the haplo.stats R package (version 1.7.7) (Sinnwell and Schaid, 2016) using R (version 3.2; http://www.r-project.org). The associations between common haplotypes (the estimated frequencies greater than 1% in entire subjects) and HS-Aging status assuming a recessive MOI were then tested with a haplotype score test adjusted for age at death, sex, and the top three PCs (Schaid, et al., 2002) implemented in the function haplo.score. The global and haplotype-specific empirical p-values were obtained via 107 Monte-Carlo simulations.
2.4.3 Haplotype-based expression Quantitative Trait Locus (eQTL) analysis for ABCC9 gene expression
We examined the association of the haplotypes with ABCC9 gene expression, focusing on the haplotypes that were identified in association analysis for HS-Aging pathology. We retrieved ABCC9 gene expression values in human brain and genotype data from two independent datasets: North American Brain Expression Consortium (NABEC) (Hernandez, et al., 2012) and United Kingdom Brain Expression Consortium (UKBEC) (Trabzuni, et al., 2011).
In the NABEC dataset, the expression data were available at Gene Expression Omnibus (GEO) public repository (http://www.ncbi.nlm.nih.gov/geo/) under the GEO accession GSE36192, consisting of two brain regions (cerebellum and frontal cortex) from 228 neurologically normal donors. The genotype data were obtained from the database of Genotypes and Phenotypes (dbGaP: http://www.ncbi.nlm.nih.gov/gap) under the dbGaP study accession phs000249.v2.p1. After the QC procedure with the same settings as we did for the ADGC genotype data was applied, the genotype data were imputed using Michigan Imputation Server (https://imputationserver.sph.umich.edu/start.html) (Das, et al., 2016) with the following parameters: 1000 Genome Phase 3 v5 reference panel, Eagle v2.3 phasing (Loh, et al., 2016), and EUR population. The imputed genotype with posterior probabilities < 0.9 were labeled as missing. Among the 228 NABEC subjects, 130 who died at age 30 years or older and passed the QC were included in the analysis (all of them were US Caucasians).
In the UKBEC dataset, gene expression for ten brain regions (cerebellar cortex, frontal cortex, hippocampus, medulla, occipital cortex, putamen, substantia nigra, thalamus, temporal cortex, and white matter) and genotype data from 134 “neuropathologically normal” individuals were obtained at BRAINEAC website (http://www.braineac.org/). The dosage files downloaded from the website (accessed 6/28/2016) were converted into PLINK file format using Genome-wide Complex Trait Analysis (GCTA) software version 1.24.4 (Yang, et al., 2011). The haplotype-based association analyses on ABCC9 gene expression were performed for the five haplotypes that were identified in the haplotype-based association analysis for HS-Aging assuming an additive MOI.
The analyses were carried out separately in the two datasets. We focused on ABCC9 gene expression through Illumina probe ID ILMN_1751453 in frontal cortex of the NABEC and through Affymetrix transcript ID t3446919 in the average of all ten regions of the UKBEC dataset. Expression data were quantile normalized and log2-transformed.
3. Results
Of the 3,251 included subjects from ADGC/NACC, 271 (8.3%) met at least one of the HS-Aging case criteria. Figure 2 shows the proportion of participants with HS-Aging pathology increased with age at death, from 3.1% (95% confidence interval (CI) is 1.6 to 5.4%) in those aged less than 70 years to 15.7% (95% CI is 12.8 to 19.0%) in those aged 90 years or older. The mean age at death in the cases was significantly higher than that in the controls (84.8 ± 8.4 years in the cases and 80.5 ± 8.8 years in the controls). No statistically significant differences were noted by case status and sex, APOE ε4 and microtubule-associated protein tau (MAPT) haplotype (H1 haplotype tagging rs8070723 A-allele and H2 tagging G-allele) frequencies (Table 1).
Figure 2.
Proportion and 95% confidence interval of hippocampal sclerosis of aging cases.
Table 1.
Comparison of selected characteristics between hippocampal sclerosis of aging cases and controls who died at age 60 years or older (n = 3,251)
| Variable | Cases n = 271 |
Controls n = 2,980 |
p-value |
|---|---|---|---|
| Age at death, mean (SD) | 84.8 (8.4) | 80.5 (8.8) | <0.001 |
| Sex, n (%) | |||
| Male | 124 (45.8) | 1,458 (48.9) | 0.349 |
| Female | 147 (54.2) | 1,522 (51.1) | |
| APOE, n (%)a | |||
| −/− | 114 (46.0) | 1,207 (44.1) | 0.749 |
| −/ε4 | 109 (43.9) | 1,216 (44.4) | |
| ε4/ε4 | 25 (10.1) | 314 (11.5) | |
| MAPT (rs8070723), n (%)b | |||
| H1/H1 | 176 (66.2) | 1,773 (60.1) | 0.146 |
| H1/H2 | 77 (28.9) | 1,022 (34.7) | |
| H2/H2 | 13 (4.9) | 154 (5.2) |
APOE genotype information was available for n = 2,985.
MAPT genotype information was available for n = 3,215.
Key: SD, standard deviation; APOE, apolipoprotein E; MAPT, microtubule-associated protein tau.
3.1 Single variant-based association
Table 2 shows the most associated variants on each of the four genes defined gene boundaries as ± 5kb from 5’ and 3’ UTRs. The highest association signals came from SNPs on the ABCC9 gene (rs7966849; p = 7.1 × 10−6 with an assumed recessive MOI and p = 4.4 × 10−5 with an assumed additive MOI) and on the KCNMB2 gene (rs73183328; p = 8.2 × 10−5 with an assumed additive MOI and p = 1.6 × 10−4 with an assumed dominant MOI). There was a series of small signals in high LD with the top SNP on the TMEM106B gene, and there was an associated region with small effects in low-to-moderate LD with the top SNP on the ABCC9 gene.
Table 2.
Most associated variant with hippocampal sclerosis of aging in four genes using a logistic regression model assuming a recessive/additive/dominant mode of inheritance in people who died at age 60 years or older (n = 3,251)
| Gene | MOI | Variant | Risk/protective alleles |
RAF in cases |
RAF in controls |
OR (95% CI)a | p-value | |
|---|---|---|---|---|---|---|---|---|
| GRN | ||||||||
| REC | rs72824731 | C/G | 9.5 | 8.4 | 3.88 (1.64 – 9.22) | 0.0021 | ||
| ADD | ] | rs2879096 | T/C | 28.6 | 24.4 | 1.25 (1.02 – 1.53) | 0.032 | |
| DOM | 1.38 (1.07 – 1.78) | 0.014 | ||||||
| TMEM106B | ||||||||
| REC | ] | rs3823612 | G/C | 64.6 | 56.5 | 1.53 (1.19 – 1.98) | 0.0011 | |
| ADD | 1.40 (1.16 – 1.68) | 3.6 × 10−4 | ||||||
| DOM | rs13229988 | A/G | 64.0 | 56.4 | 1.67 (1.16 – 2.40) | 0.0062 | ||
| ABCC9 | ||||||||
| REC | ] | rs7966849 | A/G | 60.3 | 51.2 | 1.84 (1.41 – 2.40) | 7.1 × 10−6 | |
| ADD | 1.46 (1.22 – 1.76) | 4.4 × 10−5 | ||||||
| DOM | rs829080 | C/T | 59.1 | 40.9 | 1.76 (1.18 – 2.62) | 0.0057 | ||
| KCNMB2 | ||||||||
| REC | rs13091964 | T/C | 96.1 | 92.9 | 1.84 (1.15 – 2.96) | 0.011 | ||
| ADD | ] | rs73183328 | A/G | 5.0 | 2.2 | 2.42 (1.56 – 3.76) | 8.2 × 10−5 | |
| DOM | 2.40 (1.52 – 3.78) | 1.6 × 10−4 |
Adjusted for age at death, sex and the top three principal components
Key: MOI, mode of inheritance; RAF, risk allele frequency; OR, odds ratio; CI, confidence interval; REC, recessive; ADD, additive; DOM, dominant.
3.2 Gene-based association
In the gene-based association analyses, 20, 222, 259 and 939 variants were mapped to the GRN, TMEM106B, ABCC9 and KCNMB2 genes, respectively. Table 3 shows the results of the gene-based association test in people aged 60 years. The ABCC9 gene had a significant gene-based association with HS-Aging assuming a recessive MOI when applying the Bonferroni correction (p = 2.4 × 10−4). There were nominally significant gene-based associations for the GRN gene assuming a recessive MOI, the TMEM106B gene assuming a recessive and an additive MOI, the ABCC9 gene assuming an additive MOI, and the KCNMB2 gene assuming an additive and a dominant MOI. For sensitivity analysis in people aged 80 years or older (n = 1,883: 203 in HS-Aging cases and 1,680 in controls), we confirmed the same results that the ABCC9 gene had a significant gene-based association with HS-Aging assuming a recessive MOI (p = 0.0017) (Supplemental Table 2).
Table 3.
Gene-based associations of the target four genes with hippocampal sclerosis of aging assuming a recessive/additive/dominant mode of inheritance in people who died at age 60 years or older (n = 3,251)
| Gene | # of variants |
Start position | End position | Gene-based p-value |
||
|---|---|---|---|---|---|---|
| REC | ADD | DOM | ||||
| GRN | 20 | 42,417,491 | 42,435,470 | 0.012 | 0.16 | 0.090 |
| TMEM106B | 222 | 12,245,848 | 12,281,890 | 0.028 | 0.0089 | 0.068 |
| ABCC9 | 259 | 21,945,324 | 22,094,628 | 2.4 × 10−4 | 0.0014 | 0.26 |
| KCNMB2 | 939 | 178,249,224 | 178,567,217 | 0.57 | 0.0079 | 0.016 |
Key: REC, recessive; ADD, additive; DOM, dominant
3.3 Haplotype-based association with HS-Aging
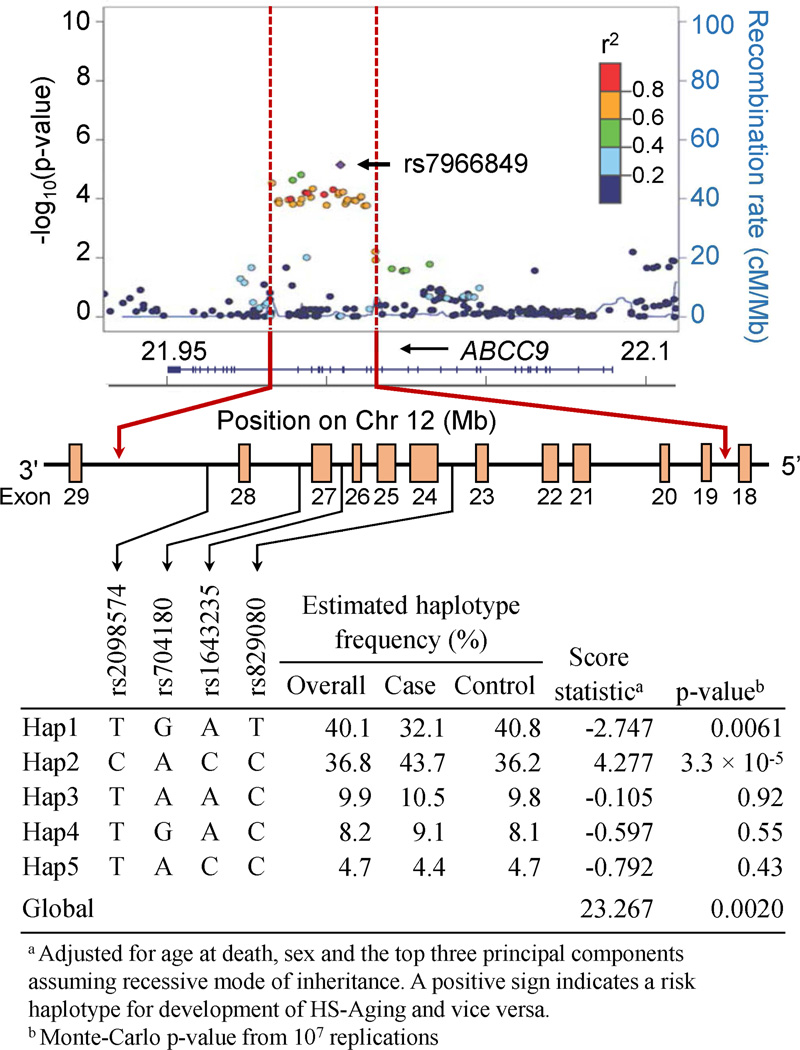
The single-variant-based association plots (Figure 3) imply that the significant gene-based association of the ABCC9 gene is driven by the region in which the most significant variants were located on the position 21,982,262 – 22,015,114 (all chromosomal positions we describe are referent to human assembly GRCh37/hg19). The top SNP (rs7966849) in this study is in high LD with rs704180 (r2 = 0.926) which was identified as the predominant risk SNP of HS-Aging (Nelson, et al., 2014; Nelson, et al., 2015b). Assuming a recessive MOI, there were 33 variants (30 SNPs and 3 indels) associated with HS-Aging pathology (each with p < 1.0 × 10−3) in this region, all of which are intronic. We selected four tag SNPs between exon 18 and 29 (Figure 3) of the ABCC9 gene when assuming a recessive MOI. The most frequent haplotypes were “Hap1” T-A-G-T (from 5’ to 3’) estimated to be present in 40.1% of observed chromosomes (32.1% in cases and 40.8% in controls), and “Hap2” C-C-A-C (36.8%; 43.7% in cases and 36.2% in controls). Hap1 was significantly associated with a lower risk of HS-Aging (score statistic = −2.747 and p = 0.0061) and Hap2 with a higher risk of HS-Aging (score statistic = 4.277 and p = 3.3 × 10−5).
Figure 3.
Estimation of haplotype frequencies and association using four tag single nucleotide polymorphisms on the ABCC9 gene when assuming a recessive mode of inheritance. Box indicates an exon.
3.4 Haplotype-based expression Quantitative Trait Locus (eQTL) association with ABCC9 gene expression
In haplotype-based association tests assuming an additive MOI, Hap1 was significantly associated with ABCC9 gene expression in both datasets (p = 0.0026 in the NABEC and p = 0.024 in the UKBEC). Compared with the association with rs704180 only, Hap1 had a stronger association with ABCC9 gene expression in the NABEC.
4. Discussion
In the large autopsy dataset derived from multiple research centers, we evaluated the genetic associations of four candidate genes (GRN, TMEM106B, ABCC9, and KCNMB2) for HS-Aging pathology. We found significant gene- and haplotype-based associations of the ABCC9 gene with HS-Aging, and these approaches provide new insights into the other candidate genes and variants that are associated with HS-Aging. The haplotype made up of the risk alleles at the region (Hap2: C-C-A-C) was significantly overrepresented in HS-Aging cases, and thus could be a risk haplotype, while the opposite haplotype (Hap1: T-A-G-T) was significantly overrepresented in controls, and thus could be a protective risk factor. We further revealed that the protective haplotype (i.e., Hap1) was associated with down-regulation of ABCC9 gene expression, and the results were consistent in two independent datasets.
Unlike the TMEM106B and GRN genes, the association between the ABCC9 gene and FTLD-TDP has never been reported. That is, the ABCC9 gene could potentially be a key gene on the distinction between FTLD-TDP and HS-Aging pathogenesis. The ABCC9 gene encodes a transmembrane protein, a part of an ATP-sensitive potassium (KATP) channel complex. KATP channel consists of two distinct subunits: an inwardly rectifying K+ channel (Kir6.x) and a regulatory sulfonylurea receptor (SURx) (Quast, et al., 2004). When the ATP levels drop due to hypoxia/ischemia or other stressor, vascular smooth muscle cell KATP channels open to increase K+ efflux, voltage-activated calcium channels close to block Ca2+ entry, and in turn, vasodilatation is induced (Sun and Feng, 2013; Sun and Hu, 2010). Given the critical roles in regulation of vascular tone, KATP channel dysfunction may be involved in cardio- and cerebrovascular diseases. In mouse experiments, knock-out Kir6.1 (encoded immediately downstream from ABCC9 on chromosome 12) and Abcc9 led to hypertension, coronary artery vasospasm, and sudden cardiac death (Chutkow, et al., 2002; Miki, et al., 2002). In addition, Leverenz and colleagues found in their community-based study that HS-Aging cases were more likely to have history of stroke, small vessel disease, and hypertension than AD cases (Leverenz, et al., 2002). Our group also reported that brains with HS-Aging pathology tended to have arteriolosclerosis in multiple cortical and subcortical regions (Neltner, et al., 2014). We note that known mutations in the human ABCC9 gene lead to a toxic gain of function (“Cantu syndrome”) also are associated with human cerebrovascular pathology - a phenotype of “tortuous cerebral vessels” detected on neuroimaging (Leon Guerrero, et al., 2016). These prior studies imply that cerebrovascular factors might be involved in developing HS-Aging via the KATP channel-dependent activity (Nelson, et al., 2015a). In addition, we recently reported that human brain gene expressions that are triiodothyronine (T3) responsive were correlated with the ABCC9 gene expression, and total T3 levels in cerebrospinal fluid (CSF) were significantly higher in HS-Aging cases than in controls (Nelson, et al., 2016a). Prior studies showed links between thyroid hormone (TH) levels and dementia (Annerbo and Lokk, 2013; Pasqualetti, et al., 2015; Rieben, et al., 2016), as well as TH levels and vascular diseases (Delitala, et al., 2015; Gao, et al., 2015; Sara, et al., 2015). Therefore it is possible that the ABCC9 gene variants may help mediate links between TH dysregulation, cerebrovascular disease, and HS-Aging pathology.
The TMEM106B gene did not have a significant gene-based association with HS-Aging when applying the Bonferroni correction, but nominal significance was found assuming a recessive and an additive MOI. Van Deerlin and colleagues identified rs1990622 T-allele as a risk factor for FTLD with TDP inclusions (FTLD-TDP) (Van Deerlin, et al., 2010). Here we report that rs3823612, which is in strong LD with rs1990622 (r2 = 0.975), is the variant on the TMEM106B gene that is most strongly associated with risk for HS-Aging pathology assuming a recessive and an additive MOI. However, there are 108 gene variants (96 SNPs and 12 indels) in near perfect LD with the top SNP rs3823612 over the gene (the range of r2 was from 0.930 to 0.996). Of the 108 variants, rs3173615 is a missense variant on exon 6, rs6460901 is a splice region variant, rs2302634 and rs2302633 are non-coding transcript exon variants, 19 variants are 5’ or 3’ UTR variants, 10 variants are upstream or downstream gene variants, and the remaining variants are intronic. Yu and colleagues reported that rs1990622 A-allele was associated with more advanced TDP-43 pathology which is the dominant feature of HS-Aging (Yu, et al., 2015). TDP-43 is also a major disease protein of other neurodegenerative diseases including FTLD and amyotrophic lateral sclerosis (ALS) (Neumann, et al., 2006). Nicholson and colleagues showed that rs3173615 (missense variant on exon 6), dictating the amino acid at codon 185 of threonine (ACC: T185) or serine (AGC: S185), was associated with higher TMEM106B protein levels in GRN mutation carriers (Nicholson, et al., 2013). Aberrant TDP-43 immunoreactivity is seen in both HS-Aging and FTLD-TDP, and rs1990622 A-allele is reported to be a risk allele of both HS-Aging and FTLD-TDP. However, these two diseases differ in clinical symptoms and pathological characteristics (Ighodaro, et al., 2015; Nelson, et al., 2011b).
The SNP on the KCNMB2 gene that was identified as a possible risk factor is rs9637454 (Beecham, et al., 2014), while in the current study we found that rs73183328 was the most strongly associated variant assuming an additive and a dominant MOI. Nominally significant gene-based association of the KCNMB2 gene with HS-Aging were found assuming an additive and a dominant MOI, although the gene-based associations were not significant when applying the Bonferroni correction. The KCNMB2 protein is the transmembrane β2 subunit of the large-conductance Ca2+- and voltage-activated K+ (BK) channel. The channel is formed by poreforming α-subunit encoded on the KCNMA1 gene (chromosome 10) and four β-subunits (β1 to β4) (Wu and Marx, 2010). The β2 subunit induces the BK channel inactivation with the coexpressed α-subunit leading to neuronal excitability by inhibiting K+ currents (Wallner, et al., 1999). Since inactivating BK channels are found in CA1 hippocampal neurons (Hicks and Marrion, 1998), HS-Aging may be related to the KCNMB2 gene via a process involving BK channel activation. It seems remarkable that both GWAS-identified putative HS-Aging risk genes (ABCC9 and KCNMB2) encode proteins that modify potassium channels.
There are limitations in this study. Since NACC data are derived from ADCs, the study design is not population-based. Also, HS pathologic diagnoses vary across calendar time and ADCs. Thus, there was probably some misclassification of HS-Aging diagnosis. However, neuropathologic evaluation is the gold standard for HS diagnosis, and thus the problem of misclassification, while ever-present, was minimized as much as possible. We did not obtain dense genetic information on the GRN gene. The previously identified SNP rs5848 as a HS-Aging risk SNP was removed in the process of the QC due to high missing rate. Therefore, we could not evaluate the GRN gene well in this study.
In summary, we confirmed that the ABCC9 gene had the significant gene-based association with HS-Aging when assuming a recessive MOI. The significant gene-based association of the ABCC9 gene is driven by the region in which a significant haplotype-based association was found. Although we did not find statistically significant gene-based associations of the other three genes (i.e., GRN, TMEM106B, and KCMNB2) with HS-Aging in this study, it does not mean that these genes are not associated with HS-Aging. Single variants may independently affect HS-Aging pathology rather than the entire gene, or there may be interactions between these genes conferring HS-Aging risk via other mechanisms, such as TDP-43 proteinopathies or ion channel dysfunction. In the future, we plan to examine what role the intronic region of the ABCC9 gene plays in developing HS-Aging pathology, and whether there are single variant-based and gene-based gene-gene interactions among these four genes to HS-Aging.
Supplementary Material
Table 4.
Haplotype association with ABCC9 gene expression in human brain assuming an additive mode of inheritance
| NABEC (Frontal cortex; n = 130 brains) |
UKBEC (10 brain regions; n = 134 brains) |
|||
|---|---|---|---|---|
| Score statistica | p-value | Score statistica | p-value | |
| Hap1 | −2.968 | 0.0026 | −2.250 | 0.024 |
| Hap2 | 1.450 | 0.15 | 1.740 | 0.081 |
| Hap3 | 1.686 | 0.091 | −0.048 | 0.96 |
| Hap4 | 0.214 | 0.83 | −0.822 | 0.41 |
| Hap5 | 0.878 | 0.38 | 1.952 | 0.051 |
| Global | 10.255 | 0.034 | 8.455 | 0.074 |
| rs704180 only | 0.010 | 0.011 | ||
A positive sign indicates up-regression of ABCC9 gene expression and vice versa.
Key: NABEC = North American Brain Expression Consortium (GEO accession: GSE36192); UKBEC = United Kingdom Brain Expression Consortium (http://www.braineac.org/).
Gene-based association of the ABCC9 gene with HS-Aging is significant
ABCC9 gene-based association is driven by a region with significant haplotypes.
Protective ABCC9 haplotype is associated with decreased ABCC9 expression.
Acknowledgments
The NACC database is funded by NIA/NIH Grant U01 AG016976. NACC data are contributed by the NIA funded ADCs: P30 AG019610 (PI Eric Reiman, MD), P30 AG013846 (PI Neil Kowall, MD), P50 AG008702 (PI Scott Small, MD), P50 AG025688 (PI Allan Levey, MD, PhD), P50 AG047266 (PI Todd Golde, MD, PhD), P30 AG010133 (PI Andrew Saykin, PsyD), P50 AG005146 (PI Marilyn Albert, PhD), P50 AG005134 (PI Bradley Hyman, MD, PhD), P50 AG016574 (PI Ronald Petersen, MD, PhD), P50 AG005138 (PI Mary Sano, PhD), P30 AG008051 (PI Steven Ferris, PhD), P30 AG013854 (PI M. Marsel Mesulam, MD), P30 AG008017 (PI Jeffrey Kaye, MD), P30 AG010161 (PI David Bennett, MD), P50 AG047366 (PI Victor Henderson, MD, MS), P30 AG010129 (PI Charles DeCarli, MD), P50 AG016573 (PI Frank LaFerla, PhD), P50 AG016570 (PI Marie-Francoise Chesselet, MD, PhD), P50 AG005131 (PI Douglas Galasko, MD), P50 AG023501 (PI Bruce Miller, MD), P30 AG035982 (PI Russell Swerdlow, MD) , P30 AG028383 (PI Linda Van Eldik, PhD), P30 AG010124 (PI John Trojanowski, MD, PhD), P50 AG005133 (PI Oscar Lopez, MD), P50 AG005142 (PI Helena Chui, MD), P30 AG012300 (PI Roger Rosenberg, MD), P50 AG005136 (PI Thomas Montine, MD, PhD), P50 AG033514 (PI Sanjay Asthana, MD, FRCP), P50 AG005681 (PI John Morris, MD), and P50 AG047270 (PI Stephen Strittmatter, MD, PhD). The National Institutes of Health, National Institute on Aging (NIH-NIA) supported this work through the following grants: ADGC, U01 AG032984, RC2 AG036528; NACC, U01 AG016976; NCRAD, U24 AG021886; NIA LOAD, U24 AG026395, R01AG041797; Data for this study were prepared, archived, and distributed by the National Institute on Aging Alzheimer’s Disease Data Storage Site (NIAGADS) at the University of Pennsylvania (U24-AG041689-01); Banner Sun Health Research Institute P30 AG019610; Boston University, P30 AG013846, U01 AG10483, R01 CA129769, R01 MH080295, R01 AG017173, R01 AG025259, R01AG33193; Columbia University, P50 AG008702, R37 AG015473; Duke University, P30 AG028377, AG05128; Emory University, AG025688; Group Health Research Institute, UO1 AG006781, UO1 HG004610, UO1 HG006375; Indiana University, P30 AG10133; Johns Hopkins University, P50 AG005146, R01 AG020688; Massachusetts General Hospital, P50 AG005134; Mayo Clinic, P50 AG016574; Mount Sinai School of Medicine, P50 AG005138, P01 AG002219; New York University, P30 AG08051, UL1 RR029893, 5R01AG012101, 5R01AG022374, 5R01AG013616, 1RC2AG036502, 1R01AG035137; Northwestern University, P30 AG013854; Oregon Health & Science University, P30 AG008017, R01 AG026916; Rush University, P30 AG010161, R01 AG019085, R01 AG15819, R01 AG17917, R01 AG30146; TGen, R01 NS059873; University of Alabama at Birmingham, P50 AG016582; University of Arizona, R01 AG031581; University of California, Davis, P30 AG010129; University of California, Irvine, P50 AG016573; University of California, Los Angeles, P50 AG016570; University of California, San Diego, P50 AG005131; University of California, San Francisco, P50 AG023501, P01 AG019724; University of Kentucky, P30 AG028383, AG05144; University of Michigan, P50 AG008671; University of Pennsylvania, P30 AG010124; University of Pittsburgh, P50 AG005133, AG030653, AG041718, AG07562, AG02365; University of Southern California, P50 AG005142; University of Texas Southwestern, P30 AG012300; University of Miami, R01 AG027944, AG010491, AG027944, AG021547, AG019757; University of Washington, P50 AG005136; University of Wisconsin, P50 AG033514; Vanderbilt University, R01 AG019085; and Washington University, P50 AG005681, P01 AG03991. The Kathleen Price Bryan Brain Bank at Duke University Medical Center is funded by NINDS grant # NS39764, NIMH MH60451 and by Glaxo Smith Kline. Genotyping of the TGEN2 cohort was supported by Kronos Science. The TGen series was also funded by NIA grant AG041232 to AJM and MJH, The Banner Alzheimer’s Foundation, The Johnnie B. Byrd Sr. Alzheimer’s Institute, the Medical Research Council, and the state of Arizona and also includes samples from the following sites: Newcastle Brain Tissue Resource (funding via the Medical Research Council, local NHS trusts and Newcastle University), MRC London Brain Bank for Neurodegenerative Diseases (funding via the Medical Research Council),South West Dementia Brain Bank (funding via numerous sources including the Higher Education Funding Council for England (HEFCE), Alzheimer’s Research Trust (ART), BRACE as well as North Bristol NHS Trust Research and Innovation Department and DeNDRoN), The Netherlands Brain Bank (funding via numerous sources including Stichting MS Research, Brain Net Europe, Hersenstichting Nederland Breinbrekend Werk, International Parkinson Fonds, Internationale Stiching Alzheimer Onderzoek), Institut de Neuropatologia, Servei Anatomia Patologica, Universitat de Barcelona. ADNI data collection and sharing was funded by the National Institutes of Health Grant U01 AG024904 and Department of Defense award number W81XWH-12-2-0012. ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California. We thank Drs. D. Stephen Snyder and Marilyn Miller from NIA who are ex-officio ADGC members. Support was also from the Alzheimer’s Association (LAF, IIRG-08-89720; MP-V, IIRG-05-14147) and the US Department of Veterans Affairs Administration, Office of Research and Development, Biomedical Laboratory Research Program. P.S.G.-H. is supported by Wellcome Trust, Howard Hughes Medical Institute, and the Canadian Institute of Health Research. Funding support for the “Brain eQTL (expression data) Study” was provided through the Division of Aging Biology and the Division of Geriatrics and Clinical Gerontology, NIA. The Brain eQTL (expression data) Study includes a genome-wide association study funded as part of the Intramural Research Program, NIA.
Funding
This work was supported by the National Cell Repository for Alzheimer’s Disease (U24 AG21886), and National Institute on Aging (K25 AG043546, UL1TR000117, and the UK-ADC P30 AG028383).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure statement
The authors declare no conflicts of interest.
This manuscript describes original work and is not under consideration by any other journal.
All authors approved the manuscript and this submission.
References
- 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amador-Ortiz C, Lin WL, Ahmed Z, Personett D, Davies P, Duara R, Graff-Radford NR, Hutton ML, Dickson DW. TDP-43 immunoreactivity in hippocampal sclerosis and Alzheimer’s disease. Ann Neurol. 2007;61(5):435–445. doi: 10.1002/ana.21154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annerbo S, Lokk J. A clinical review of the association of thyroid stimulating hormone and cognitive impairment. ISRN Endocrinol. 2013;2013:856017. doi: 10.1155/2013/856017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki N, Murray ME, Ogaki K, Fujioka S, Rutherford NJ, Rademakers R, Ross OA, Dickson DW. Hippocampal sclerosis in Lewy body disease is a TDP-43 proteinopathy similar to FTLD-TDP Type A. Acta Neuropathol. 2015;129(1):53–64. doi: 10.1007/s00401-014-1358-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Beecham GW, Hamilton K, Naj AC, Martin ER, Huentelman M, Myers AJ, Corneveaux JJ, Hardy J, Vonsattel JP, Younkin SG, Bennett DA, De Jager PL, Larson EB, Crane PK, Kamboh MI, Kofler JK, Mash DC, Duque L, Gilbert JR, Gwirtsman H, Buxbaum JD, Kramer P, Dickson DW, Farrer LA, Frosch MP, Ghetti B, Haines JL, Hyman BT, Kukull WA, Mayeux RP, Pericak-Vance MA, Schneider JA, Trojanowski JQ, Reiman EM, Alzheimer’s Disease Genetics C, Schellenberg GD, Montine TJ. Genome-wide association meta-analysis of neuropathologic features of Alzheimer’s disease and related dementias. PLoS Genet. 2014;10(9):e1004606. doi: 10.1371/journal.pgen.1004606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenowitz WD, Monsell SE, Schmitt FA, Kukull WA, Nelson PT. Hippocampal sclerosis of aging is a key Alzheimer’s disease mimic: clinical-pathologic correlations and comparisons with both alzheimer’s disease and non-tauopathic frontotemporal lobar degeneration. J Alzheimers Dis. 2014;39(3):691–702. doi: 10.3233/JAD-131880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chutkow WA, Pu J, Wheeler MT, Wada T, Makielski JC, Burant CF, McNally EM. Episodic coronary artery vasospasm and hypertension develop in the absence of Sur2 K(ATP) channels. J Clin Invest. 2002;110(2):203–208. doi: 10.1172/JCI15672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corey-Bloom J, Sabbagh MN, Bondi MW, Hansen L, Alford MF, Masliah E, Thal LJ. Hippocampal sclerosis contributes to dementia in the elderly. Neurology. 1997;48(1):154–160. doi: 10.1212/wnl.48.1.154. [DOI] [PubMed] [Google Scholar]
- Das S, Forer L, Schonherr S, Sidore C, Locke AE, Kwong A, Vrieze SI, Chew EY, Levy S, McGue M, Schlessinger D, Stambolian D, Loh PR, Iacono WG, Swaroop A, Scott LJ, Cucca F, Kronenberg F, Boehnke M, Abecasis GR, Fuchsberger C. Next-generation genotype imputation service and methods. Nat Genet. 2016;48(10):1284–1287. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delitala AP, Orru M, Filigheddu F, Pilia MG, Delitala G, Ganau A, Saba PS, Decandia F, Scuteri A, Marongiu M, Lakatta EG, Strait J, Cucca F. Serum free thyroxine levels are positively associated with arterial stiffness in the SardiNIA study. Clin Endocrinol (Oxf) 2015;82(4):592–597. doi: 10.1111/cen.12532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson DW, Baker M, Rademakers R. Common variant in GRN is a genetic risk factor for hippocampal sclerosis in the elderly. Neurodegener Dis. 2010;7(1–3):170–174. doi: 10.1159/000289231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson DW, Davies P, Bevona C, Van Hoeven KH, Factor SM, Grober E, Aronson MK, Crystal HA. Hippocampal sclerosis: a common pathological feature of dementia in very old (> or = 80 years of age) humans. Acta Neuropathol. 1994;88(3):212–221. doi: 10.1007/BF00293396. [DOI] [PubMed] [Google Scholar]
- Gao CX, Yang B, Guo Q, Wei LH, Tian LM. High thyroid-stimulating hormone level is associated with the risk of developing atherosclerosis in subclinical hypothyroidism. Horm Metab Res. 2015;47(3):220–224. doi: 10.1055/s-0034-1394370. [DOI] [PubMed] [Google Scholar]
- Hernandez DG, Nalls MA, Moore M, Chong S, Dillman A, Trabzuni D, Gibbs JR, Ryten M, Arepalli S, Weale ME, Zonderman AB, Troncoso J, O’Brien R, Walker R, Smith C, Bandinelli S, Traynor BJ, Hardy J, Singleton AB, Cookson MR. Integration of GWAS SNPs and tissue specific expression profiling reveal discrete eQTLs for human traits in blood and brain. Neurobiol Dis. 2012;47(1):20–28. doi: 10.1016/j.nbd.2012.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks GA, Marrion NV. Ca2+-dependent inactivation of large conductance Ca2+-activated K+ (BK) channels in rat hippocampal neurones produced by pore block from an associated particle. J Physiol. 1998;508(Pt 3):721–734. doi: 10.1111/j.1469-7793.1998.721bp.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman BT, Phelps CH, Beach TG, Bigio EH, Cairns NJ, Carrillo MC, Dickson DW, Duyckaerts C, Frosch MP, Masliah E, Mirra SS, Nelson PT, Schneider JA, Thal DR, Thies B, Trojanowski JQ, Vinters HV, Montine TJ. National Institute on Aging-Alzheimer’s Association guidelines for the neuropathologic assessment of Alzheimer’s disease. Alzheimers Dement. 2012;8(1):1–13. doi: 10.1016/j.jalz.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ighodaro ET, Jicha GA, Schmitt FA, Neltner JH, Abner EL, Kryscio RJ, Smith CD, Duplessis T, Anderson S, Patel E, Bachstetter A, Van Eldik LJ, Nelson PT. Hippocampal Sclerosis of Aging Can Be Segmental: Two Cases and Review of the Literature. J Neuropathol Exp Neurol. 2015;74(7):642–652. doi: 10.1097/NEN.0000000000000204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12(6):996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon Guerrero CR, Pathak S, Grange DK, Singh GK, Nichols CG, Lee JM, Vo KD. Neurologic and neuroimaging manifestations of Cantu syndrome: A case series. Neurology. 2016;87(3):270–276. doi: 10.1212/WNL.0000000000002861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leverenz JB, Agustin CM, Tsuang D, Peskind ER, Edland SD, Nochlin D, DiGiacomo L, Bowen JD, McCormick WC, Teri L, Raskind MA, Kukull WA, Larson EB. Clinical and neuropathological characteristics of hippocampal sclerosis: a community-based study. Arch Neurol. 2002;59(7):1099–1106. doi: 10.1001/archneur.59.7.1099. [DOI] [PubMed] [Google Scholar]
- Li MX, Gui HS, Kwan JS, Sham PC. GATES: a rapid and powerful gene-based association test using extended Simes procedure. Am J Hum Genet. 2011;88(3):283–293. doi: 10.1016/j.ajhg.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh PR, Danecek P, Palamara PF, Fuchsberger C, Y AR, H KF, Schoenherr S, Forer L, McCarthy S, Abecasis GR, Durbin R, A LP. Reference-based phasing using the Haplotype Reference Consortium panel. Nat Genet. 2016;48(11):1443–1448. doi: 10.1038/ng.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki T, Suzuki M, Shibasaki T, Uemura H, Sato T, Yamaguchi K, Koseki H, Iwanaga T, Nakaya H, Seino S. Mouse model of Prinzmetal angina by disruption of the inward rectifier Kir6.1. Nat Med. 2002;8(5):466–472. doi: 10.1038/nm0502-466. [DOI] [PubMed] [Google Scholar]
- Murray ME, Cannon A, Graff-Radford NR, Liesinger AM, Rutherford NJ, Ross OA, Duara R, Carrasquillo MM, Rademakers R, Dickson DW. Differential clinicopathologic and genetic features of late-onset amnestic dementias. Acta Neuropathol. 2014;128(3):411–421. doi: 10.1007/s00401-014-1302-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson PT, Estus S, Abner EL, Parikh I, Malik M, Neltner JH, Ighodaro E, Wang WX, Wilfred BR, Wang LS, Kukull WA, Nandakumar K, Farman ML, Poon WW, Corrada MM, Kawas CH, Cribbs DH, Bennett DA, Schneider JA, Larson EB, Crane PK, Valladares O, Schmitt FA, Kryscio RJ, Jicha GA, Smith CD, Scheff SW, Sonnen JA, Haines JL, Pericak-Vance MA, Mayeux R, Farrer LA, Van Eldik LJ, Horbinski C, Green RC, Gearing M, Poon LW, Kramer PL, Woltjer RL, Montine TJ, Partch AB, Rajic AJ, Richmire K, Monsell SE, Alzheimer’ Disease Genetic C, Schellenberg GD, Fardo DW. ABCC9 gene polymorphism is associated with hippocampal sclerosis of aging pathology. Acta Neuropathol. 2014;127(6):825–843. doi: 10.1007/s00401-014-1282-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson PT, Head E, Schmitt FA, Davis PR, Neltner JH, Jicha GA, Abner EL, Smith CD, Van Eldik LJ, Kryscio RJ, Scheff SW. Alzheimer’s disease is not “brain aging”: neuropathological, genetic, and epidemiological human studies. Acta Neuropathol. 2011a;121(5):571–587. doi: 10.1007/s00401-011-0826-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson PT, Jicha GA, Wang WX, Ighodaro E, Artiushin S, Nichols CG, Fardo DW. ABCC9/SUR2 in the brain: Implications for hippocampal sclerosis of aging and a potential therapeutic target. Ageing Res Rev. 2015a;24(Pt B):111–125. doi: 10.1016/j.arr.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson PT, Katsumata Y, Nho K, Artiushin SC, Jicha GA, Wang WX, Abner EL, Saykin AJ, Kukull WA, Alzheimer’s Disease Neuroimaging I, Fardo DW. Genomics and CSF analyses implicate thyroid hormone in hippocampal sclerosis of aging. Acta Neuropathol. 2016a;132(6):841–858. doi: 10.1007/s00401-016-1641-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson PT, Schmitt FA, Lin Y, Abner EL, Jicha GA, Patel E, Thomason PC, Neltner JH, Smith CD, Santacruz KS, Sonnen JA, Poon LW, Gearing M, Green RC, Woodard JL, Van Eldik LJ, Kryscio RJ. Hippocampal sclerosis in advanced age: clinical and pathological features. Brain. 2011b;134(Pt 5):1506–1518. doi: 10.1093/brain/awr053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson PT, Smith CD, Abner EL, Wilfred BJ, Wang WX, Neltner JH, Baker M, Fardo DW, Kryscio RJ, Scheff SW, Jicha GA, Jellinger KA, Van Eldik LJ, Schmitt FA. Hippocampal sclerosis of aging, a prevalent and high-morbidity brain disease. Acta Neuropathol. 2013;126(2):161–177. doi: 10.1007/s00401-013-1154-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson PT, Trojanowski JQ, Abner EL, Al-Janabi OM, Jicha GA, Schmitt FA, Smith CD, Fardo DW, Wang WX, Kryscio RJ, Neltner JH, Kukull WA, Cykowski MD, Van Eldik LJ, Ighodaro ET. “New Old Pathologies”: AD, PART, and Cerebral Age-Related TDP-43 With Sclerosis (CARTS) J Neuropathol Exp Neurol. 2016b;75(6):482–498. doi: 10.1093/jnen/nlw033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson PT, Wang WX, Partch AB, Monsell SE, Valladares O, Ellingson SR, Wilfred BR, Naj AC, Wang LS, Kukull WA, Fardo DW. Reassessment of risk genotypes (GRN, TMEM106B, and ABCC9 variants) associated with hippocampal sclerosis of aging pathology. J Neuropathol Exp Neurol. 2015b;74(1):75–84. doi: 10.1097/NEN.0000000000000151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neltner JH, Abner EL, Baker S, Schmitt FA, Kryscio RJ, Jicha GA, Smith CD, Hammack E, Kukull WA, Brenowitz WD, Van Eldik LJ, Nelson PT. Arteriolosclerosis that affects multiple brain regions is linked to hippocampal sclerosis of ageing. Brain. 2014;137(Pt 1):255–267. doi: 10.1093/brain/awt318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, McCluskey LF, Miller BL, Masliah E, Mackenzie IR, Feldman H, Feiden W, Kretzschmar HA, Trojanowski JQ, Lee VM. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314(5796):130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- Nicholson AM, Finch NA, Wojtas A, Baker MC, Perkerson RB, 3rd, Castanedes-Casey M, Rousseau L, Benussi L, Binetti G, Ghidoni R, Hsiung GY, Mackenzie IR, Finger E, Boeve BF, Ertekin-Taner N, Graff-Radford NR, Dickson DW, Rademakers R. TMEM106B p.T185S regulates TMEM106B protein levels: implications for frontotemporal dementia. J Neurochem. 2013;126(6):781–791. doi: 10.1111/jnc.12329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao WC, Dickson DW, Crook JE, Finch NA, Rademakers R, Graff-Radford NR. Hippocampal sclerosis in the elderly: genetic and pathologic findings, some mimicking Alzheimer disease clinically. Alzheimer Dis Assoc Disord. 2011;25(4):364–368. doi: 10.1097/WAD.0b013e31820f8f50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualetti G, Pagano G, Rengo G, Ferrara N, Monzani F. Subclinical Hypothyroidism and Cognitive Impairment: Systematic Review and Meta-Analysis. J Clin Endocrinol Metab. 2015;100(11):4240–4248. doi: 10.1210/jc.2015-2046. [DOI] [PubMed] [Google Scholar]
- Pickering-Brown SM, Rollinson S, Du Plessis D, Morrison KE, Varma A, Richardson AM, Neary D, Snowden JS, Mann DM. Frequency and clinical characteristics of progranulin mutation carriers in the Manchester frontotemporal lobar degeneration cohort: comparison with patients with MAPT and no known mutations. Brain. 2008;131(Pt 3):721–731. doi: 10.1093/brain/awm331. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, Boehnke M, Abecasis GR, Willer CJ. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast U, Stephan D, Bieger S, Russ U. The impact of ATP-sensitive K+ channel subtype selectivity of insulin secretagogues for the coronary vasculature and the myocardium. Diabetes 53 Suppl. 2004;3:S156–S164. doi: 10.2337/diabetes.53.suppl_3.s156. [DOI] [PubMed] [Google Scholar]
- Rademakers R, Eriksen JL, Baker M, Robinson T, Ahmed Z, Lincoln SJ, Finch N, Rutherford NJ, Crook RJ, Josephs KA, Boeve BF, Knopman DS, Petersen RC, Parisi JE, Caselli RJ, Wszolek ZK, Uitti RJ, Feldman H, Hutton ML, Mackenzie IR, Graff-Radford NR, Dickson DW. Common variation in the miR-659 binding-site of GRN is a major risk factor for TDP43-positive frontotemporal dementia. Hum Mol Genet. 2008;17(23):3631–3642. doi: 10.1093/hmg/ddn257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieben C, Segna D, da Costa BR, Collet TH, Chaker L, Aubert CE, Baumgartner C, Almeida OP, Hogervorst E, Trompet S, Masaki K, Mooijaart SP, Gussekloo J, Peeters RP, Bauer DC, Aujesky D, Rodondi N. Subclinical Thyroid Dysfunction and the Risk of Cognitive Decline: a Meta-Analysis of Prospective Cohort Studies. J Clin Endocrinol Metab. 2016;101(12):4945–4954. doi: 10.1210/jc.2016-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford NJ, Carrasquillo MM, Li M, Bisceglio G, Menke J, Josephs KA, Parisi JE, Petersen RC, Graff-Radford NR, Younkin SG, Dickson DW, Rademakers R. TMEM106B risk variant is implicated in the pathologic presentation of Alzheimer disease. Neurology. 2012;79(7):717–718. doi: 10.1212/WNL.0b013e318264e3ac. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sara JD, Zhang M, Gharib H, Lerman LO, Lerman A. Hypothyroidism Is Associated With Coronary Endothelial Dysfunction in Women. J Am Heart Assoc. 2015;4(8):e002225. doi: 10.1161/JAHA.115.002225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70(2):425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinnwell JP, Schaid DJ. haplo.stats: Statistical Analysis of Haplotypes with Traits and Covariates when Linkage Phase is Ambiguous. R package version 1.7.7. 2016 https://CRAN.R-project.org/package=haplo.stats.
- Sun HS, Feng ZP. Neuroprotective role of ATP-sensitive potassium channels in cerebral ischemia. Acta Pharmacol Sin. 2013;34(1):24–32. doi: 10.1038/aps.2012.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XL, Hu G. ATP-sensitive potassium channels: a promising target for protecting neurovascular unit function in stroke. Clin Exp Pharmacol Physiol. 2010;37(2):243–252. doi: 10.1111/j.1440-1681.2009.05190.x. [DOI] [PubMed] [Google Scholar]
- Trabzuni D, Ryten M, Walker R, Smith C, Imran S, Ramasamy A, Weale ME, Hardy J. Quality control parameters on a large dataset of regionally dissected human control brains for whole genome expression studies. J Neurochem. 2011;119(2):275–282. doi: 10.1111/j.1471-4159.2011.07432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troncoso JC, Kawas CH, Chang CK, Folstein MF, Hedreen JC. Lack of association of the apoE4 allele with hippocampal sclerosis dementia. Neurosci Lett. 1996;204(1–2):138–140. doi: 10.1016/0304-3940(96)12331-4. [DOI] [PubMed] [Google Scholar]
- Van Deerlin VM, Sleiman PM, Martinez-Lage M, Chen-Plotkin A, Wang LS, Graff-Radford NR, Dickson DW, Rademakers R, Boeve BF, Grossman M, Arnold SE, Mann DM, Pickering-Brown SM, Seelaar H, Heutink P, van Swieten JC, Murrell JR, Ghetti B, Spina S, Grafman J, Hodges J, Spillantini MG, Gilman S, Lieberman AP, Kaye JA, Woltjer RL, Bigio EH, Mesulam M, Al-Sarraj S, Troakes C, Rosenberg RN, White CL, 3rd, Ferrer I, Llado A, Neumann M, Kretzschmar HA, Hulette CM, Welsh-Bohmer KA, Miller BL, Alzualde A, Lopez de Munain A, McKee AC, Gearing M, Levey AI, Lah JJ, Hardy J, Rohrer JD, Lashley T, Mackenzie IR, Feldman HH, Hamilton RL, Dekosky ST, van der Zee J, Kumar-Singh S, Van Broeckhoven C, Mayeux R, Vonsattel JP, Troncoso JC, Kril JJ, Kwok JB, Halliday GM, Bird TD, Ince PG, Shaw PJ, Cairns NJ, Morris JC, McLean CA, DeCarli C, Ellis WG, Freeman SH, Frosch MP, Growdon JH, Perl DP, Sano M, Bennett DA, Schneider JA, Beach TG, Reiman EM, Woodruff BK, Cummings J, Vinters HV, Miller CA, Chui HC, Alafuzoff I, Hartikainen P, Seilhean D, Galasko D, Masliah E, Cotman CW, Tunon MT, Martinez MC, Munoz DG, Carroll SL, Marson D, Riederer PF, Bogdanovic N, Schellenberg GD, Hakonarson H, Trojanowski JQ, Lee VM. Common variants at 7p21 are associated with frontotemporal lobar degeneration with TDP-43 inclusions. Nat Genet. 2010;42(3):234–239. doi: 10.1038/ng.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallner M, Meera P, Toro L. Molecular basis of fast inactivation in voltage and Ca2+-activated K+ channels: a transmembrane beta-subunit homolog. Proc Natl Acad Sci U S A. 1999;96(7):4137–4142. doi: 10.1073/pnas.96.7.4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. ggplot2 : elegant graphics for data analysis. New York ; London: Springer Science + Business Media; 2009. [Google Scholar]
- Wu RS, Marx SO. The BK potassium channel in the vascular smooth muscle and kidney: alpha- and beta-subunits. Kidney Int. 2010;78(10):963–974. doi: 10.1038/ki.2010.325. [DOI] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88(1):76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, De Jager PL, Yang J, Trojanowski JQ, Bennett DA, Schneider JA. The TMEM106B locus and TDP-43 pathology in older persons without FTLD. Neurology. 2015;84(9):927–934. doi: 10.1212/WNL.0000000000001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarow C, Sitzer TE, Chui HC. Understanding hippocampal sclerosis in the elderly: epidemiology, characterization, and diagnostic issues. Curr Neurol Neurosci Rep. 2008;8(5):363–370. doi: 10.1007/s11910-008-0057-3. [DOI] [PubMed] [Google Scholar]
- Zarow C, Weiner MW, Ellis WG, Chui HC. Prevalence, laterality, and comorbidity of hippocampal sclerosis in an autopsy sample. Brain Behav. 2012;2(4):435–442. doi: 10.1002/brb3.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.