Abstract
Background
Pustular skin disorders are a category of difficult-to-treat and potentially life threatening conditions that involve the appearance of neutrophil rich pustules. The molecular basis of most pustular skin conditions has remained unknown.
Objective
We sought to investigate the molecular basis of three pustular skin disorders, generalized pustular psoriasis (GPP), palmoplantar pustulosis (PPP), and acute generalized exanthematous pustulosis (AGEP).
Methods
Microarray analyses were performed to profile genome-wide gene expression of skin biopsies obtained from GPP, PPP, AGEP patients and normal controls. Functional enrichment, gene network and k-means clustering analyses were used to identify molecular pathways dysregulated in the disorders. Immunohistochemistry and immunofluorescence were used to determine protein localization. qRT-PCR and ELISA were used to determine transcript and secreted cytokine levels. siRNA was employed to decrease transcript levels.
Results
Molecules and pathways related to neutrophil chemotaxis emerged as common alterations in GPP, PPP, and AGEP, consistent with the pustular phenotypes. Expression of two STEAP proteins, STEAP1 and 4, were elevated in patient skin, and co-localized with IL-36γ around neutrophilic pustules. STEAP1/4 expression clustered with and positively correlated with that of the IL-1, IL-36 family proteins, and CXCL1/8. STEAP4 expression was activated by cytokines and suppressed by inhibition of MEK1/2, whereas STEAP1 expression appeared less prone to such dynamic regulation. Importantly, STEAP1/4 knockdown resulted in impaired induction of a broad spectrum of pro-inflammatory cytokines including IL-1, IL-36, and neutrophil chemotaxins; CXCL1 and CXCL8. STEAP1/4 knockdown also reduced the ability of keratinocytes to induce neutrophil chemotaxis.
Conclusion
Transcriptomic changes in three pustular skin disorders GPP, PPP, and AGEP converged on neutrophil chemotaxis and diapedesis and cytokines known to drive neutrophil rich inflammatory processes including IL-1 and members of the IL-36 family. STEAP1 and STEAP4 positively regulate the induction of proinflammatory, neutrophil-activating cytokines.
Keywords: Pustular skin disorders, neutrophils, inflammation, transcriptomic profiling, STEAP, IL-1, IL-36, CXCL1, CXCL8
Introduction
Generalized pustular psoriasis (GPP), palmoplantar pustulosis (PPP), and acute generalized exanthematous pustulosis (AGEP) are pustular skin disorders whose symptoms include the appearance of pustules, circumscribed collections of fluids and neutrophils within the epidermis. GPP is characterized by sudden, often repeated, episodes of high-grade fever and generalized neutrophilic pustules. In contrast PPP is characterized by chronic, localized pustules on the palms and soles, and AGEP by eruptions classically as an adverse drug reaction. Despite different etiologies these reactions are all characterized by massive influx of neutrophils into the epidermis and these conditions can be difficult to treat and be life threatening (1–3).
GPP has been considered as a distinct form of psoriasis that is associated with IL36RN mutations and consequently over-stimulation of the IL-36 pathways (1). The same mutations were detected in PPP and AGEP, leading to speculation that similar molecular alterations may underlie these disorders (4, 5). Interestingly, gene expression study on PPP provided evidence that select neural genes were increased, raising the possibility that unique pathways are activated in this disease(6). However, the molecular abnormalities associated with GPP, PPP and AGEP have not been systematically compared on a transcriptome-wide level, and common and/or distinct mechanisms driving these inflammatory conditions have thereby remained largely elusive.
To this end, we have performed transcriptional profiling of skin biopsies from GPP/PPP/AGEP patients and healthy subjects. We report common molecular alterations including IL36RN, IL8, and other genes affecting neutrophil chemo-attraction as well as unique pathways enriched in each disease. A novel shared feature of GPP/PPP/AGEP was found to be upregulation of the STEAP family of proteins, which correlated with overexpression of a panel of pro-inflammatory cytokines. Both STEAP1 and STEAP4 were required for the crosstalk between these cytokines under inflammatory conditions. These findings open up possibilities in targeting the STEAP- pathways for the treatment of IL-36-associated, pustular skin conditions and other disorders that share similar inflammatory profiles.
Results
Transcriptional profiling of GPP, PPP and AGEP
To characterize the transcriptomic profiles in these pustular skin disorders, we performed microarrays on archived paraffin embedded skin tissues (FFPE) obtained from 30 GPP, 17 PPP, 14 AGEP patients, and 20 healthy subjects (NN) as controls. We also included normal palm and sole skin specimens from 10 healthy subjects (NNN) as controls for PPP. Histology staining confirmed the presence of intra-epidermal pustules (Figure 1A). In addition, we compared our PPP results with published transciptome profiles obtained from fresh tissues(6). The majority of genes altered in PPP (82% of PPP-increased genes and 76% of PPP-decreased genes) were confirmed by our study (see Figure E1 in the Online Repository). As negative controls, same number of randomly selected genes did not show consistent up- or down-regulation in PPP (see Figure E1 in the Online Repository). Collectively, reported PPP-increased genes exhibited significantly higher expression in PPP samples versus controls from our dataset (p = 0.0158) (see Figure E1 in the Online Repository), and reported PPP-decreased genes showed decreased expression in our PPP samples (p = 0.0002) (see Figure E1 in the Online Repository).
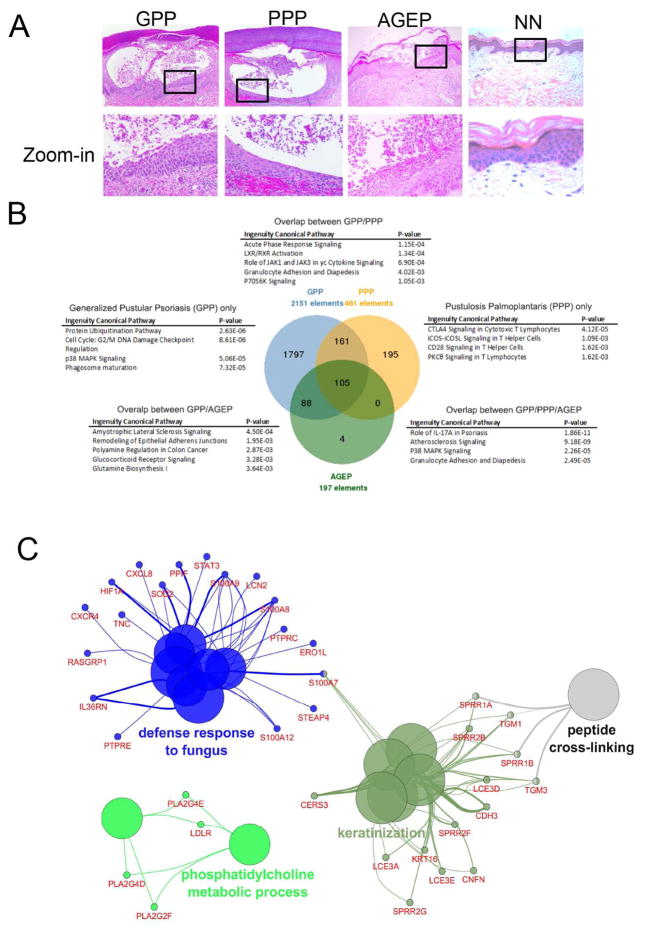
Figure 1. Transcriptional profiling of GPP, PPP, and AGEP.
A) H&E staining of GPP, PPP, and AGEP skin lesions and normal skin (NN). B) Functional enrichment of signaling pathways in DEGs common and specific to GPP, PPP, and AGEP. C) Biological processes linked to genes commonly altered in GPP, PPP and AGEP.
From our microarray experiments, GPP skin exhibited the most number of genes altered, with a total of 2151 differentially expressed genes (DEGs) identified compared to normal skin (FDR<0.05). Similarly, 461 DEGs were identified in PPP skin and 197 DEGs were identified in AGEP (Figure 1B). GPP showed the most unique mRNA-profile, with 83.5% of DEGs (1797 out of 2151) being GPP-specific, compared to 42.3% for PPP and 0.5% for AGEP. The top GPP-specific biological pathway enriched was protein ubiquitination that is known to be dysregulated in inflammatory and autoimmune diseases (Figure 1B) (7). It is also worth noting that we detected significant enrichment of the caspase signaling pathway in GPP-specific genes (p=7.31×10−5, 39 genes), including caspases-1, -4, and -11 that mediate inflammatory responses (8, 9). Caspase-1 activates IL-1β, an additional GPP-specific DEG, as part of an activated inflammasome. Importantly, autoinflammatory disorders such as Muckle Wells syndrome, Familial Cold Autoinflammatory Syndrome and CINCA (Chronic Infantile Neurological Cutaneous and Articular Syndrome), which share the periodic fever and systemic skin inflammation symptoms with GPP, are triggered by a hyperactivated inflammasome involving excessive caspase-1 activity and overproduction of IL-1β (10). These GPP-specific DEGs together with the extensive attribute of gene expression alterations might contribute to the systematic and severe features associated with GPP symptoms.
PPP-specific genes showed major enrichment in T-cell functions (Figure 1B). In addition, consistent with the localized lesions on palms and soles, we detected BBS1 as a PPP-only gene, whose mutation underlies human limb malformation, specifically abnormalities of the hands and feet (11, 12). We also observed enrichment of genes known to link with Twist1 (p=5.31×10−3, 7 genes), a crucial player in limb formation and patterning (13, 14), in the PPP-specific gene set only. Promoters of PPP-specific genes were significantly enriched with binding motifs for the Twist1 family of transcription factors (Z-score = 11.95; see Figure E2 in the Online Repository). The interactions between these limb-genes and inflammatory pathways could serve as one possible explanation for the localized PPP lesions on hands and feet.
Common processes enriched in GPP/PPP/AGEP included IL-17A signaling and granulocyte adhesion and diapedesis, consistent with a shared inflammation phenotype involving neutrophil infiltration (Figure 1B) (15, 16).
STEAPs are elevated in pustular skin disorders
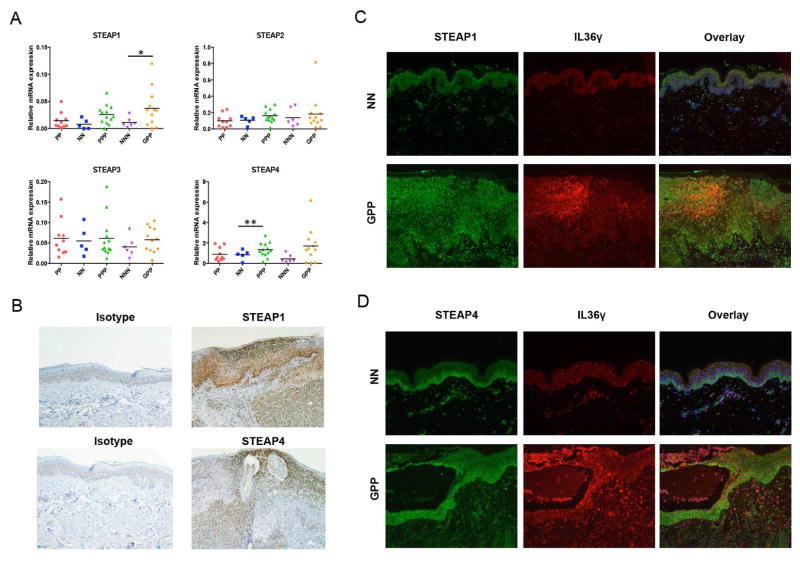
STEAP4 was one of the genes commonly altered in all three disease phenotypes (GPP, PPP, and AGEP) from the microarray analyses, with a potential function in defense response (Figure 1C). Thus, we decided to focus our subsequent studies on this family of proteins with the attempt to delineate a novel inflammatory pathway. Six-transmembrane epithelial antigens of prostate (STEAP) family of proteins are comprised of STEAP1–4, and share metalloreductases activities. STEAP1 and 2 are overexpressed in various types of cancers, and the cellular functions of STEAP1–4 have been linked to molecular trafficking, cell proliferation and death (17, 18). From qRT-PCR analyses, STEAP1 and STEAP4 levels were increased in PPP/GPP, but not in plaque psoriasis (PP) (Figure 2A). By contrast, STEAP2 and STEAP3 were not elevated in PPP, GPP, or PP. Immunostaining of STEAP1 and STEAP4 revealed their epithelial localization in normal skin and concentration in or around neutrophil infiltration of the epidermis, in a manner that partially overlaps with IL-36γ (Figure 2B, C). We also examined the mRNA levels in various cell types, including keratinocytes, fibroblasts, T-cells, B-cells, monocytes, macrophages and dendritic cells (see Figure E3 in the Online Repository). STEAP4, whose elevation was identified in GPP, PPP, and AGEP in microarray analyses (Figure 1C), was expressed the highest in keratinocytes. In addition, only in keratinocytes were both STEAP1 and STEAP4 expressed at relatively high levels. We therefore focused our subsequently analyses on keratinocytes.
Figure 2. Characterization of STEAPs in pustular psoriasis.
A) qRT-PCR of STEAP1–4 mRNA levels in PP, NN, PPP, NNN, and GPP skin. Mann-Whitney test. *, p < 0.05. **, p < 0.01. B) Immunohistochemistry of STEAP1 and STEAP4 in GPP skin. C, D) Immunofluorescence co-staining of STEAP1 (C), or STEAP4 (D), and IL36γ in NN and GPP skin.
STEAP1 and STEAP4 co-cluster with a group of inflammatory cytokines including IL-1, IL-36, and CXCL1/8
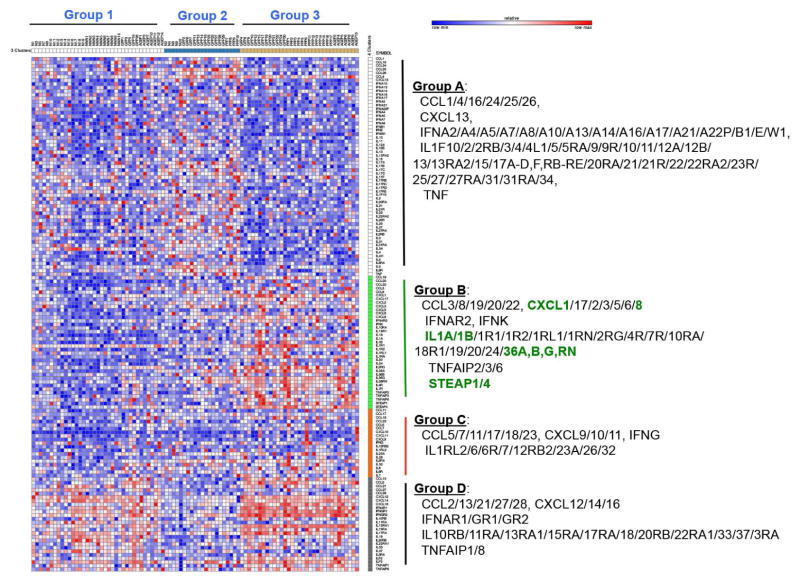
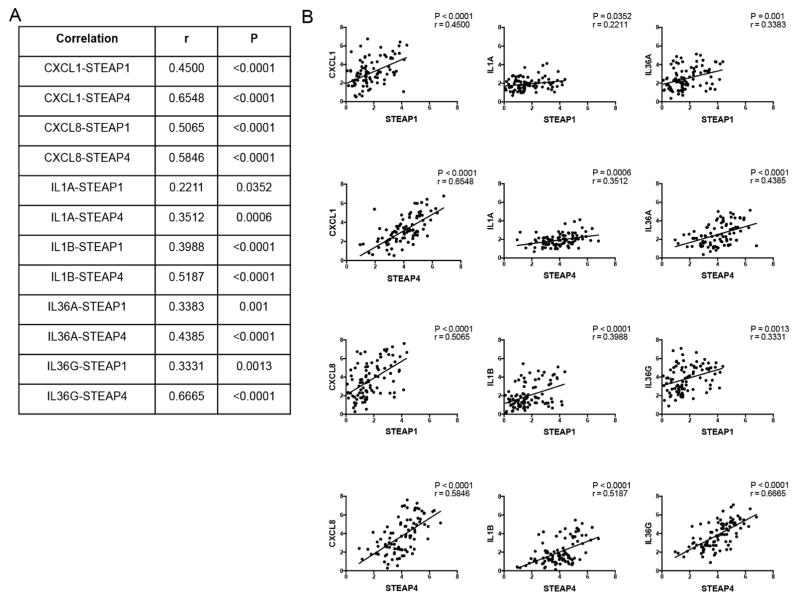
To investigate the cytokine activities in GPP, PPP, and AGEP, we performed clustering based on expression changes of various cytokines (ILs, IFNs, CCLs, CXCLs, TNF) and their related genes (receptors and induced genes). This led to the separation of cytokines into 4 distinct groups (A–D), and patients were subsequently stratified into 3 groups (subgroups 1–3) based on their distinct patterns of cytokine profiles (Figure 3). IL1A/B, IL36A/B/G/RN, and the neutrophil chemokines CXCL1 and CXCL8, were found to be clustered together with STEAP1 and STEAP4. This grouping of STEAP1/4 with neutrophil activators was a prominent feature of the patients in group 3 possibly related to a more severe sub-category of disease (Figure 3), and their expression levels correlated positively with IL1A, IL1B, IL36A, IL36G, CXCL1 and IL8 in all samples (GPP/PPP/AGEP/NN) (Figure 4), and was most prominent in patients with the most robust inflammatory phenotype (subgroup 3, see Figure E4 in the Online Repository), suggesting that the STEAP proteins function in the same pathway as IL-1, IL-36, and the neutrophil chemokines CXCL1 and IL8.
Figure 3. Cytokine profiling of pustular skin diseases and co-clustering with STEAP1 and STEAP4.
Expression changes of cytokine and related genes were clustered by K-means clustering. The resultant groups of samples were shown on top and genes were shown on the right.
Figure 4. Correlation between STEAP and cytokine expression shown as table (A) or graphs (B).
Correlation analyses are shown between STEAPs (STEAP1 or STEAP4) with selected Group B cytokines (CXCL1, CXCL8, IL1A, IL1B, IL36A, and IL36G).
For both GPP and PPP, patients in subgroup 3 exhibited significantly enriched expression of cell cycle checkpoint genes compared with patients in subgroup 2. For both GPP and AGEP, there was significantly increased expression of inflammatory genes including TLR and CCL2- pathways when comparing patients in subgroup 3 compared to subgroup 2 (see Figure E5 in the Online Repository). Thus, in the pustular skin disorders analyzed, stronger expression of the neutrophil activators correlated with dysregulated cell cycle checkpoints and more robust inflammatory profile.
Despite the enrichment of common molecular pathways across GPP, PPP, and AGEP, these patients showed limited overlap in terms of the specific genes altered (see Figure E6 in the Online Repository). Focusing on the checkpoint pathway that was highlighted in both GPP and PPP, 19 DEGs in the pathways were common to both diseases. 24 DEGs were PPP-specific and 16 were GPP-specific (see Figure E6 in the Online Repository). This indicates that a shared cytokine profile resulted in disease-specific gene expression changes, although they converged into similar manifestations on a higher order (e.g. checkpoint regulation).
STEAP4 expression is induced by pro-inflammatory cytokines
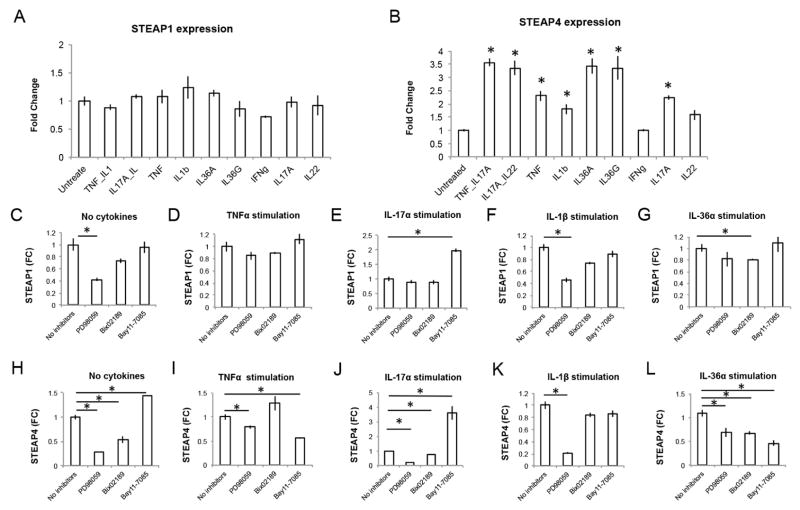
To investigate a potential role of STEAP1 and STEAP4 in cytokine responses, we first examined if the expression of STEAPs could be induced by inflammatory cytokines. In primary human keratinocytes, STEAP4 expression was induced by TNF-α, IL-1β, IL-36α, IL-36γ, IL-17A, as well as IL-17A combined with TNF-α or IL-22. By contrast, STEAP1 expression was not induced by these cytokine stimulations (Figure 5, A and B). We then analyzed the signaling pathways regulating STEAP expression using inhibitors of MEK1/2, MEK5, and the NF-κB pathway, respectively (Figure 5, C–L). The MEK1/2 inhibitor PD98059 inhibited STEAP1 expression both in baseline conditions and under IL-1β stimulation (Figure 5, C and F). However, the requirement for MEK1/2 activity was diminished under TNF-α and IL-17A treatment, indicating altered regulatory mechanisms (Figure 5, D and E). By contrast, STEAP4 remained sensitive to MEK1/2 inhibition under all conditions tested (Figure 5, H–L). In the GPP/PPP/AGEP/NN skin biopsies, expression of the MEK1/2 targets FOSL1, HIF1A, CRABP2 positively correlated with STEAP4 (see Figure E7 in the Online Repository) (19), supporting the possibility that MEK1/2 and STEAP4 function in the same pathway in vivo. The MEK5 inhibitor Bix02189 and the IκBα phosphorylation inhibitor Bay 11–7085 also had a greater effect on STEAP4 expression compared with STEAP1, suggesting STEAP4 expression is more susceptible to complex regulation by several cell signaling pathways.
Figure 5. Regulation of STEAP1 and STEAP4 expression.
A, B) Relative expression of STEAP1 (A) and STEAP4 (B) by qRT-PCR in untreated and cytokine treated keratinocytes. C–L) Relative expression of STEAP1 (C–G) and STEAP4 (H–L) by qRT-PCR under various conditions (no cytokines, TNF-α stimulation, IL-17A stimulation, IL-1β stimulation or IL-36α stimulation) with or without inhibitor co-treatment. * p<0.05, Student’s t-test.
STEAPs are required for optimal cytokine response and neutrophil chemotaxis
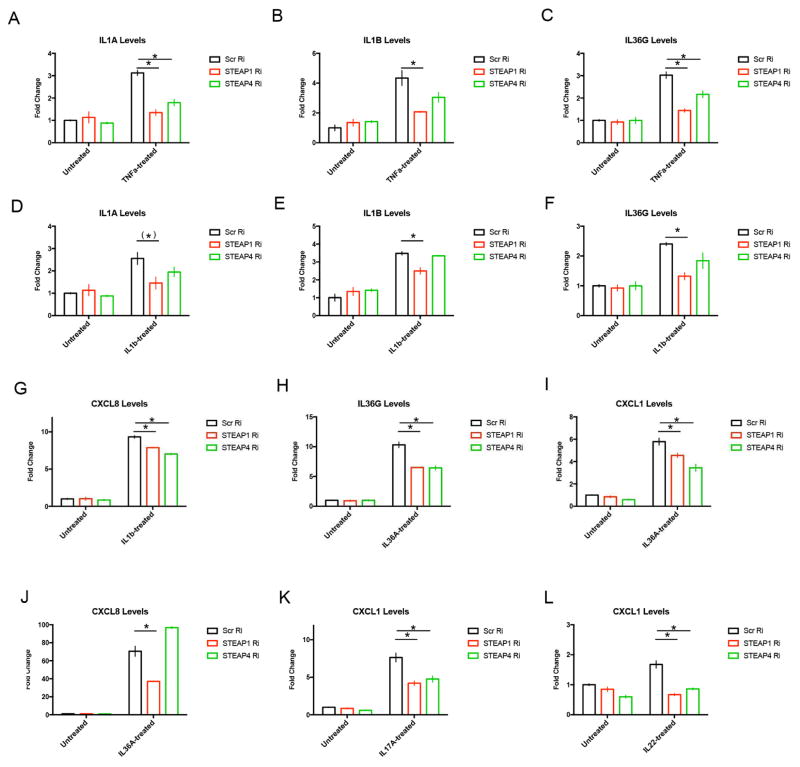
We analyzed the roles of STEAP1 and STEAP4 in mediating cytokine response by repressing STEAP expression in primary human keratinocytes (see Figure E8 in the Online Repository). We focused on induced expression of IL1A, IL1B, IL36A, IL36G, CXCL1, and IL8 because this group of cytokines co-clustered with STEAP1 and STEAP4 in GPP/PPP/AGEP patients (Figure 3). STEAP1 RNAi resulted in impaired induction of pro-inflammatory cytokines: induction of IL1A and IL1B by TNF-α and IL-1β; induction of IL36G by TNF-α, IL-1β and IL-36α; induction of CXCL1 by IL36-α, IL-17A and IL-22; as well as induction of CXCL8 by IL-1β and IL-36α; were all weakened (Figure 6 and see Figures E8–12 in the Online Repository). STEAP4 RNAi resulted in impaired induction of IL1A by TNF-α, induction of IL36G by TNF-α and IL-36α, induction of CXCL1 by IL-36α, IL-17A and IL-22, as well as induction of CXCL8 by IL-1β (Figure 6 and see Figures E10–11 in the Online Repository). The pattern of STEAP1 and STEAP4 function was found to be similar, although STEAP1 knockdown resulted in a broader suppression (see Figure E13 in the Online Repository).
Figure 6. Regulation of cytokine induction by STEAP1 and STEAP4.
Relative expression of various cytokines under the indicated cytokine treatments upon STEAP1/4 RNAi (Ri) or scrambed RNAi (Scr Ri) by qRT-PCR. Expressions were shown for IL1A (A, D), IL1B (B, E), IL36G (C, F, H), CXCL1 (I, K, L) and CXCL8 (G, J). * p<0.05, (*) p<0.1, Student’s t-test.
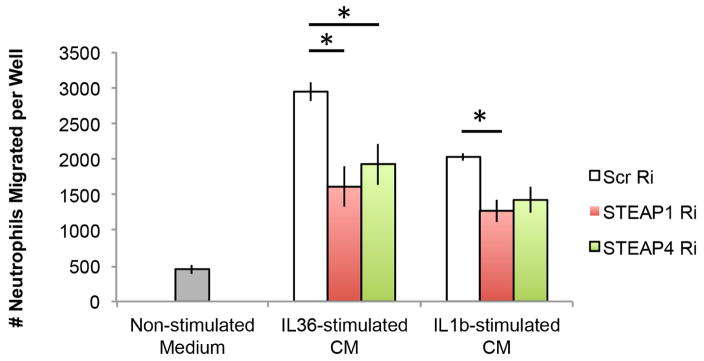
We then tested if the STEAP proteins regulate neutrophil chemotaxis towards the inflammatory environment conditioned by keratinocytes. We knocked down STEAP1 or STEAP4 in primary keratinocytes, and stimulated keratinocytes with IL36-α or IL-1β, which we had previously found to induce CXCL1/8 in a STEAP-dependent manner (Figure 6). We then collected conditioned medium from the stimulated keratinocytes and analyzed its ability to attract primary neutrophils by transwell assays. We found that conditioned medium from STEAP-reduced keratinocytes was less efficient in inducing neutrophil migration, compared to wild-type keratinocytes (Figure 7).
Figure 7. Regulation of neutrophil chemotaxis by STEAP1 and STEAP4.
Chemotaxis of neutrophils towards non-stimulated medium as negative control, and conditioned medium from IL-36α- or IL-1β-stimulated keratinocytes with scrambled RNAi (Scr Ri), STEAP1 RNAi (STEAP1 Ri) or STEAP4 RNAi (STEAP4 Ri). * p<0.05, Student’s t-test.
Collectively these data suggest that STEAPs are required for the maximal induction of pro-inflammatory neutrophil-attracting and activating cytokines as well as neutrophil chemotaxis, contributing to the shaping of inflammatory environments in various neutrophil driven inflammatory responses.
Discussion
We have performed transcriptional profiling of generalized pustular psoriasis (GPP), palmoplantar pustulosis (PPP), and acute generalized exanthematous pustulosis (AGEP) from FFPE tissues. Quantitative gene expression studies from FFPE tissues can be complicated by limitations including nucleic acid integrity and amplification efficiency(20). Comparison of our results with published PPP studies from fresh tissue has revealed 70%–80% overlap (see Figure E1 in the Online Repository). Therefore, profiling from FFPE clinical tissue samples remains a valuable method for analysis of archived specimens.
We have provided evidence that STEAP1 and STEAP4 are relevant in the inflammatory responses observed in GPP, PPP, and AGEP. In vivo, STEAP1 and STEAP4 are upregulated, and co-expressed with IL-1, IL-36, CXCL1, CXCL8 and additional pro-inflammatory cytokines in GPP/PPP/AGEP patients, delineating the inflammatory milieu for a subgroup of patients featuring dysregulated checkpoint signaling and excessive activation of inflammatory pathways involving TLRs and CCL2 (Figures 1–4). STEAP4 is inducible by various stimuli including IL-1β, IL-36α, IL-36γ, IL-17A, and TNF-α, amplifying the inflammatory reactions on top of that supported by STEAP1, whose expression is not cytokine induced (Figure 5). STEAP1 and 4 are required for the optimal induction of IL-1 and IL-36 cytokines as well as expression of the neutrophil chemokines: CXCL1 and IL-8 (Figure 6). Importantly, keratinocytes deficient in STEAP1/4 become less efficient in inducing neutrophil chemotaxis (Figure 7). Therefore, STEAP1 and STEAP4 support neutrophil-rich, pro-inflammatory responses in the skin as observed in pustular psoriasis.
In contrast to pustular psoriasis, STEAP1 and STEAP4 are not upregulated in plaque psoriasis (Figure 2). One important pathological difference between the two conditions is the distinctive appearance of neutrophil-rich pustules in pustular, but not plaque, psoriasis. We found that STEAP1/4 promote neutrophil chemotaxis towards the keratinocyte-conditioned environment (Figure 7), possibly via stimulating the production of various neutrophil-acting cytokines including the IL-36 proteins, CXCL1 and CXCL8 (Figure 6, see Figure E13 in the Online Repository). Therefore, the distinct neutrophil-activating activities of STEAP1 and STEAP4 may underlie the pathological alterations in pustular psoriasis, but not non-pustular psoriasis.
The identified molecular abnormalities may underlie the common pustular phenotypes observed in GPP, PPP, and AGEP. Aberrant IL-36 signaling is thought to drive inflammation in GPP, PPP, and AGEP patients (1, 4, 5, 21–23), and neutrophils play an important role in this pathway for their post-translational processing and activation of the IL-36 cytokines (24). IL-8 and CXCL1 are among the most potent chemoattractant molecules for the recruitment of neutrophils (25–28). TLR ligation stimulates neutrophil function, and CCL2 has a critical role in neutrophil recruitment (29–32). In addition, there is an emerging understanding of the effects of neutrophil activation on cell cycle checkpoints such as DNA damage response, G2/M checkpoint activation and subsequent replication errors, as well as how their dysregulation and genomic abnormalities produced in these processes may in turn amplify inflammation (33–36). The checkpoint pathway proteins overexpressed in the GPP/PPP/AGEP patients included BUB1B and MAD2L1, whose over-expression has been associated with chromosomal instability (37–39). There was also increased CDC6, CCNA1, and CCNB1 that may directly promote keratinocyte proliferation and allow for increased inflammatory gene expression mediated by CDK6 (40). The capture of these molecular alterations in our patient datasets has provided formal support for their involvement in human inflammatory diseases.
We have found that the above-mentioned alterations in cytokine responses are functionally linked by STEAP1 and STEAP4. The STEAP proteins were first identified as surface antigens overexpressed in prostate cancer, and later found to be overexpressed in many different cancers such as breast and bladder carcinoma and Ewing’s sarcoma (17, 18). They share a six-transmembrane domain and a C-terminal domain with homology to the F420: NADPH-oxidoreductase in archaea and bacteria, as well as a role in ion metabolism including iron and copper uptake (18, 41–43). STEAP1 and STEAP2 promote cancer cell proliferation and prevent apoptosis, whereas STEAP4 regulates adipocyte differentiation and modulates the effect of TNF-α on insulin resistance in mice. (44–47). However, the function of STEAP4 in humans remains unclear, particularly the role of STEAP4 in pre-adipocyte differentiation (48). Our studies have established a positive link between human STEAP1 and STEAP4 and pro-inflammatory cytokines in vivo in several neutrophil-driven disorders.
It is not yet known whether the inflammatory activities of STEAPs are in direct causal relationship with their regulation of ion metabolism. In GPP, PPP, and AGEP patients, we observed significant alterations in genes involved in iron biology including HEPHL1, LCN2, ALAS1, SOD2, and EROL, concomitant with STEAP upregulation (data not shown). HEPHL1 is a homologue of the ferroxidase hephaestin and may exhibit iron oxidation activity (49). Regulation of labile iron levels by LCN2 protects mice against sepsis (50). ALAS1 catalyzes the rate-limiting step in the biosynthesis of heme (51). SOD2 gains peroxidase activity when bound to iron, generating oxygen radicals (52). ERO1L expression is induced by iron deficiency (53). Therefore, the role of STEAPs in linking iron metabolism and inflammation would be of interest to address in future studies.
In conclusion, we have delineated transcriptomic changes in three pustular skin disorders GPP, PPP, and AGEP, which converged on neutrophil chemotaxis and diapedesis and cytokines known to drive neutrophil rich inflammatory processes including IL-1 and members of the IL-36 family. We identify a novel inflammatory pathway regulated by two STEAP proteins; STEAP1 and STEAP4 as positive regulators controlling expression of neutrophils activators in skin, and potential therapeutic targets in neutrophil driven pustular diseases.
Methods
Patient cohort
Archived formalin fixed paraffin embedded cases were identified by search of the University of Michigan Department of Pathology database. Cases with reported diagnosis of generalized pustular psoriasis (GPP), palmoplantar pustulosis (PPP), and acute generalized exanthematous pustulosis (AGEP) were identified, and the diagnosis was verified by slide review by a board-certified dermatopathologist (PWH) and chart review. Healthy controls and patients with chronic plaque psoriasis were identified in our clinic and biopsy was obtained for formalin fixation and paraffin embedding prior to processing and analyses. Healthy volunteers were recruited for blood draws for neutrophil isolation after providing written informed consent. All protocols were approved by the institutional review board of the University of Michigan, Ann Arbor, and the study was carried out in accordance with the Declaration of Helsinki principles.
Microarray
RNA extraction was performed with an E.Z.N.A. FFPE RNA isolation kit (Omega Bio-tek) using the xylene-based extraction method as specified by manufacturer and five 20μm thick FFPE sections of biopsy. RNA was eluted into water and stored at −80°C until analyzed. Affymetrix Human Gene ST 2.1 microarrays (Affymetrix) were processed at the University of Michigan Microarray Core Facility according to the manufacturer’s protocol. The raw microarray data (.CEL files) were processed in the publicly available software R (www.r-project.org) using a modified version of the “affy” package and Human Entrez Gene custom CDF annotation version 19 (54) (http://brainarray.mbni.med.umich.edu/Brainarray/Database/CustomCDF/genomic_curated_CDF.asp) using the Robust Multichip Average (RMA) method (55). Post-hybridization quality control checks were performed using RNA degradation score, relative log expression (RLE), and normalized unscaled standard errors (NUSE). Data were batch corrected using an implementation of ComBat v3(56)within the GenePattern pipeline (http://www.GenePattern.org). To remove background, we calculated the median values of all probe sets and removed those probes with expression values below the lowest median value + 1 standard deviation using a custom Perl script. Gene list comparisons, generation of Venn Diagrams, and enrichment of transcription factor motifs were performed on the adjusted expression data in Genomatix (www.genomatix.de) using the default settings (57). Biological processes linked to genes commonly altered in the three disorders were generated by GeneGO using DEGs common to GPP, PPP and AGEP as input and default settings. K-means clustering was performed using GENE-E (www.broadinstitute.org/cancer/software/GENE-E/). For K-means clustering, gene clustering was performed with default cluster number of 2–4. For each number of clustering, STEAP1 and STEAP4 co-clustered with CXCL1, CXCL8, IL-1, and IL-36 family proteins (Fig. 3, see Figures E14, 15 in the Online Repository). This was further confirmed with cluster number of 5 (see Figure E16 in the Online Repository). Patient clustering was performed with default cluster number of 2–3. For each number of clustering, the group of patient-only-samples (i.e. without healthy subjects) correlated with high expression of the STEAP1/4-CXCL1/8-IL1-IL36 group of genes (Fig. 3, see Figure E17 in the Online Repository). This was further confirmed with cluster number of 4 (see Figure E18 in the Online Repository). Therefore, representative image for gene cluster number of 4 and patient cluster number of 3 was shown in the main figure (Fig. 3). The clustering was also reiterated for 20 times and each time the above conclusion was validated, suggesting the robustness of the method (data not shown). Functional enrichment analyses and pathway generation were performed using Genomatix, Ingenuity IPA (www.qiagen.com/ingenuity), and ClueGO (57, 58). For correlation analyses, expression values (normalized relative expression from microarray analyses) were fitted by linear regression, and slope was tested for significant deviation from 0 (shown as P-values). The correlation coefficients for the pairwise comparisons are shown by r (Pearson correlation coefficients).
Statistics
Statistical analyses of the microarrays were performed using Significance Analysis of Microarrays method implemented in the MultiExperiment Viewer application (59). Mann-Whitney and Student’s t-test were used in gene expression comparisons, as specified in the figure legends. For Figure 2, STEAP mRNA levels were normalized to those of RPLPO in each sample. Each data point represents the sample from one individual. Lines show the mean of the different samples in the specified group. PP and PPP samples were compared to NN samples. GPP samples were compared to NNN samples. In Figures 5 and 6, each bar shows the mean from three experiments. Error bars are the standard error of the mean from the three experiments. Each treated condition was compared to the non-treated condition. In Figure 7, for the non-stimulated and IL1 β-stimulated experiments, each bar shows the mean from three experiments. Error bars are the standard error of the mean from the three experiments. For the IL-36 α - stimulated experiments, each bar shows the mean from the four experiments. Error bars are the standard error of the mean from the four experiments.
Keratinocyte culture, cytokine stimulations, inhibitors, RNAi, and gene expression analyses
Normal human keratinocytes (NHKs) were established from healthy adults as previously described(60) and grown in medium 154 CF (Thermofisher M154CF500) with human keratinocyte growth supplement (Thermofisher S0015). Inhibitors used included the MEK1/2 inhibitor PD98059 (Tocris 1213), the MEK5 inhibitor Bix02189 (Tocris 4842) and the IκBα phosphorylation inhibitor Bay 11–7085 (Tocris 1743) according to the manufecturer’s specifications. siRNA was introduced by electroporation using Lonza 4D-nucleofector following manufacturer’s instructions. Cytokine stimulations were performed as previously reported(61). qRT-PCR was performed on a 7900HT Fast Real-time PCR system (Applied Biosystems) with TaqMan Universal PCR Master Mix (ThermoFisher 4304437).
Neutrophil chemotaxis
Primary human neutrophils isolated from peripheral blood were directly used for chemotaxis towards keratinocyte-conditioned medium. To collect the conditioned medium, siRNA was introduced into keratinocytes and keratinocytes were stimulated with the indicated cytokines. Conditioned medium from stimulated keratinocytes were used in as chemoattractant for neutrophils. Chemotaxis assays were performed with 6.5 mm transwells with 3 μm pores (Corning). 2 × 104 freshly isolated neutrophils were plated on the top well of each transwell, and number of neutrophils migrated to the bottom wells was counted after incubation at 37°C for 1 hour.
H&E and Immunostaining
For H&E staining, slides were de-paraffinized and stained with hematoxylin (1:10 diluted) for 2 min, rinsed in running tap water for 5min, stained with eosin for 2 min, rinsed with water, dehydrated and mounted for microscopy. For immunohistochemistry, formalin fixed, paraffin-embedded human skin biopsies on slides were heated for 30min at 55°C, rehydrated, epitope retrieved, blocked, and incubated with primary antibody overnight at 4°C. Slides were washed, incubated with secondary antibody, developed with DAB and counterstained using hematoxylin for immunohistochemistry, or DNA-stained with DAPI for immunofluorescence. Antibodies were anti-STEAP1 (Lifespan Biosciences, LS-B1260), anti-IL36G (Santa Cruz Biotechnology sc-80056), anti-STEAP4 (Proteintech, 11994-1-AP).
Supplementary Material
Key Messages.
GPP, PPP, and AGEP are characterized by shared molecular alterations associated with neutrophilic inflammation
STEAP1 and STEAP4 were identified as a novel nexus that links the inflammatory responses shared among GPP/PPP/AGEP
Targeting STEAPs or their associated pathways may enable management of GPP, PPP, AGEP by controlling down-stream responses of IL-1 and IL-36
Acknowledgments
The work was in part supported by the University of Michigan Babcock Endowment Fund (A.J., Y.L, X.X, M.K.S., J.J.V., J.E.G.), NIH awards K08-AR060802 and R01-AR069071 (J.E.G.), the A. Alfred Taubman Medical Research Institute Kenneth and Frances Eisenberg Emerging Scholar Award (J.E.G.) and Doris Duke Charitable Foundation Grant #2013106 (J.E.G), and Novartis (J.E.G). AJ is supported by NIH K01 AR064765, and the National Psoriasis Foundation USA. JMK was partially supported by National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) of the National Institutes of Health under Award Number K08AR063668. We also thank Dr. Stefan W. Stoll for kindly providing primary keratinocyte cultures. We acknowledge Drs. Celine Berthier, Viji Nair, Philip Stuart and Dr. Lam C. Tsoi for assistance with biostatistical analyses. We apologize to those researchers whose work was not cited or discussed due to the space limitation.
Abbreviations
- GPP
generalized pustular psoriasis
- PPP
palmoplantar pustulosis
- AGEP
acute generalized exanthematous pustulosis
- DEG
differentially expressed genes
- STEAP
six-transmembrane epithelial antigens of prostate
- IL-1
interleukin-1
- IL-36
interleukin-36
- CXCL-1
chemokine (C-X-C motif) ligand 1
- IL-8/CXCL-8
interleukin-8
- IL-17
interleukin-17
- TNF
tumor necrosis factor
Footnotes
Author Contributions. Y.L., J.J.V., J.M.K., A.J., P.H., and J.E.G. designed the study and wrote the manuscript. Y.L., X.X., M.K.S, L.W., and A.J. collected and analyzed data.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Marrakchi S, Guigue P, Renshaw BR, Puel A, Pei XY, Fraitag S, Zribi J, Bal E, Cluzeau C, Chrabieh M, et al. Interleukin-36-receptor antagonist deficiency and generalized pustular psoriasis. N Engl J Med. 2011;365(7):620–8. doi: 10.1056/NEJMoa1013068. [DOI] [PubMed] [Google Scholar]
- 2.Langley RG, Krueger GG, Griffiths CE. Psoriasis: epidemiology, clinical features, and quality of life. Ann Rheum Dis. 2005;64(Suppl 2):ii18–23. doi: 10.1136/ard.2004.033217. discussion ii4–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mengesha YM, Bennett ML. Pustular skin disorders: diagnosis and treatment. Am J Clin Dermatol. 2002;3(6):389–400. doi: 10.2165/00128071-200203060-00003. [DOI] [PubMed] [Google Scholar]
- 4.Setta-Kaffetzi N, Navarini AA, Patel VM, Pullabhatla V, Pink AE, Choon SE, Allen MA, Burden AD, Griffiths CEM, Seyger MMB, et al. Rare Pathogenic Variants in IL36RN Underlie a Spectrum of Psoriasis-Associated Pustular Phenotypes. Journal of Investigative Dermatology. 2013;133(5):1366–9. doi: 10.1038/jid.2012.490. [DOI] [PubMed] [Google Scholar]
- 5.Navarini AA, Valeyrie-Allanore L, Setta-Kaffetzi N, Barker JN, Capon F, Creamer D, Roujeau JC, Sekula P, Simpson MA, Trembath RC, et al. Rare variations in IL36RN in severe adverse drug reactions manifesting as acute generalized exanthematous pustulosis. J Invest Dermatol. 2013;133(7):1904–7. doi: 10.1038/jid.2013.44. [DOI] [PubMed] [Google Scholar]
- 6.Bissonnette R, Suarez-Farinas M, Li X, Bonifacio KM, Brodmerkel C, Fuentes-Duculan J, Krueger JG. Based on Molecular Profiling of Gene Expression, Palmoplantar Pustulosis and Palmoplantar Pustular Psoriasis Are Highly Related Diseases that Appear to Be Distinct from Psoriasis Vulgaris. Plos One. 2016;11(5) doi: 10.1371/journal.pone.0155215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang J, Maldonado MA. The ubiquitin-proteasome system and its role in inflammatory and autoimmune diseases. Cell Mol Immunol. 2006;3(4):255–61. [PubMed] [Google Scholar]
- 8.Martinon F, Tschopp J. Inflammatory caspases: linking an intracellular innate immune system to autoinflammatory diseases. Cell. 2004;117(5):561–74. doi: 10.1016/j.cell.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 9.Shi J, Zhao Y, Wang Y, Gao W, Ding J, Li P, Hu L, Shao F. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature. 2014;514(7521):187–92. doi: 10.1038/nature13683. [DOI] [PubMed] [Google Scholar]
- 10.Scott AM, Saleh M. The inflammatory caspases: guardians against infections and sepsis. Cell Death Differ. 2007;14(1):23–31. doi: 10.1038/sj.cdd.4402026. [DOI] [PubMed] [Google Scholar]
- 11.Forsythe E, Beales PL. Bardet-Biedl syndrome. Eur J Hum Genet. 2013;21(1):8–13. doi: 10.1038/ejhg.2012.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tayeh MK, Yen HJ, Beck JS, Searby CC, Westfall TA, Griesbach H, Sheffield VC, Slusarski DC. Genetic interaction between Bardet-Biedl syndrome genes and implications for limb patterning. Human Molecular Genetics. 2008;17(13):1956–67. doi: 10.1093/hmg/ddn093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krawchuk D, Weiner SJ, Chen YT, Lu BC, Costantini F, Behringer RR, Laufer E. Twist1 activity thresholds define multiple functions in limb development. Dev Biol. 2010;347(1):133–46. doi: 10.1016/j.ydbio.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O’Rourke MP, Soo K, Behringer RR, Hui CC, Tam PP. Twist plays an essential role in FGF and SHH signal transduction during mouse limb development. Dev Biol. 2002;248(1):143–56. doi: 10.1006/dbio.2002.0730. [DOI] [PubMed] [Google Scholar]
- 15.Taylor PR, Roy S, Leal SM, Sun Y, Howell SJ, Cobb BA, Li XX, Pearlman E. Activation of neutrophils by autocrine IL-17A-IL-17RC interactions during fungal infection is regulated by IL-6, IL-23, ROR gamma t and dectin-2 (vol 14, pg 143, 2014) Nature Immunology. 2015;16(2) doi: 10.1038/ni.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pelletier M, Maggi L, Micheletti A, Lazzeri E, Tamassia N, Costantini C, Cosmi L, Lunardi C, Annunziato F, Romagnani S, et al. Evidence for a cross-talk between human neutrophils and Th17 cells. Blood. 2010;115(2):335–43. doi: 10.1182/blood-2009-04-216085. [DOI] [PubMed] [Google Scholar]
- 17.Hubert RS, Vivanco I, Chen E, Rastegar S, Leong K, Mitchell SC, Madraswala R, Zhou Y, Kuo J, Raitano AB, et al. STEAP: a prostate-specific cell-surface antigen highly expressed in human prostate tumors. Proc Natl Acad Sci U S A. 1999;96(25):14523–8. doi: 10.1073/pnas.96.25.14523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gomes IM, Maia CJ, Santos CR. STEAP proteins: from structure to applications in cancer therapy. Mol Cancer Res. 2012;10(5):573–87. doi: 10.1158/1541-7786.MCR-11-0281. [DOI] [PubMed] [Google Scholar]
- 19.Komatsu K, Buchanan FG, Otaka M, Jin M, Odashima M, Horikawa Y, Watanabe S, DuBois RN. Gene expression profiling following constitutive activation of MEK 1 and transformation of rat intestinal epithelial cells. Mol Cancer. 2006;5 doi: 10.1186/1476-4598-5-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fedorowicz G, Guerrero S, Wu TD, Modrusan Z. Microarray analysis of RNA extracted from formalin-fixed, paraffin-embedded and matched fresh-frozen ovarian adenocarcinomas. Bmc Med Genomics. 2009;2 doi: 10.1186/1755-8794-2-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakai N, Sugiura K, Akiyama M, Katoh N. Acute generalized exanthematous pustulosis caused by dihydrocodeine phosphate in a patient with psoriasis vulgaris and a heterozygous IL36RN mutation. JAMA Dermatol. 2015;151(3):311–5. doi: 10.1001/jamadermatol.2014.3002. [DOI] [PubMed] [Google Scholar]
- 22.Capon F. IL36RN Mutations in Generalized Pustular Psoriasis: Just the Tip of the Iceberg? Journal of Investigative Dermatology. 2013;133(11):2503–4. doi: 10.1038/jid.2013.361. [DOI] [PubMed] [Google Scholar]
- 23.Korber A, Mossner R, Renner R, Sticht H, Wilsmann-Theis D, Schulz P, Sticherling M, Traupe H, Huffmeier U. Mutations in IL36RN in Patients with Generalized Pustular Psoriasis. Journal of Investigative Dermatology. 2013;133(11):2634–7. doi: 10.1038/jid.2013.214. [DOI] [PubMed] [Google Scholar]
- 24.Henry CM, Sullivan GP, Clancy DM, Afonina IS, Kulms D, Martin SJ. Neutrophil-Derived Proteases Escalate Inflammation through Activation of IL-36 Family Cytokines. Cell Rep. 2016;14(4):708–22. doi: 10.1016/j.celrep.2015.12.072. [DOI] [PubMed] [Google Scholar]
- 25.Moser B, Clark-Lewis I, Zwahlen R, Baggiolini M. Neutrophil-activating properties of the melanoma growth-stimulatory activity. J Exp Med. 1990;171(5):1797–802. doi: 10.1084/jem.171.5.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schumacher C, Clark-Lewis I, Baggiolini M, Moser B. High- and low-affinity binding of GRO alpha and neutrophil-activating peptide 2 to interleukin 8 receptors on human neutrophils. Proc Natl Acad Sci U S A. 1992;89(21):10542–6. doi: 10.1073/pnas.89.21.10542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoffmann E, Dittrich-Breiholz O, Holtmann H, Kracht M. Multiple control of interleukin-8 gene expression. J Leukocyte Biol. 2002;72(5):847–55. [PubMed] [Google Scholar]
- 28.Lindley I, Aschauer H, Seifert JM, Lam C, Brunowsky W, Kownatzki E, Thelen M, Peveri P, Dewald B, Vontscharner V, et al. Synthesis and Expression in Escherichia-Coli of the Gene Encoding Monocyte-Derived Neutrophil-Activating Factor - Biological Equivalence between Natural and Recombinant Neutrophil-Activating Factor. P Natl Acad Sci USA. 1988;85(23):9199–203. doi: 10.1073/pnas.85.23.9199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hayashi F, Means TK, Luster AD. Toll-like receptors stimulate human neutrophil function. Blood. 2003;102(7):2660–9. doi: 10.1182/blood-2003-04-1078. [DOI] [PubMed] [Google Scholar]
- 30.Prince LR, Whyte MK, Sabroe I, Parker LC. The role of TLRs in neutrophil activation. Curr Opin Pharmacol. 2011;11(4):397–403. doi: 10.1016/j.coph.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 31.Xue ML, Thakur A, Cole N, Lloyd A, Stapleton F, Wakefield D, Willcox MD. A critical role for CCL2 and CCL3 chemokines in the regulation of polymorphonuclear neutrophils recruitment during corneal infection in mice. Immunol Cell Biol. 2007;85(7):525–31. doi: 10.1038/sj.icb.7100082. [DOI] [PubMed] [Google Scholar]
- 32.Reichel CA, Rehberg M, Lerchenberger M, Berberich N, Bihari P, Zahler S, Krombach F. Chemokines Ccl2 and Ccl3 mediate neutrophil recruitment via induction of protein synthesis and generation of lipid mediators. Eur J Clin Invest. 2009;39:35. doi: 10.1161/ATVBAHA.109.193268. [DOI] [PubMed] [Google Scholar]
- 33.Campregher C, Luciani MG, Gasche C. Activated neutrophils induce an hMSH2-dependent G2/M checkpoint arrest and replication errors at a (CA) 13-repeat in colon epithelial cells. Gut. 2008;57(6):780–7. doi: 10.1136/gut.2007.141556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harbort CJ, Soeiro-Pereira PV, von Bernuth H, Kaindl AM, Costa-Carvalho BT, Condino-Neto A, Reichenbach J, Roesler J, Zychlinsky A, Amulic B. Neutrophil oxidative burst activates ATM to regulate cytokine production and apoptosis. Blood. 2015;126(26):2842–51. doi: 10.1182/blood-2015-05-645424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bornstein S, White R, Malkoski S, Oka M, Han G, Cleaver T, Reh D, Andersen P, Gross N, Olson S, et al. Smad4 loss in mice causes spontaneous head and neck cancer with increased genomic instability and inflammation. J Clin Invest. 2009;119(11):3408–19. doi: 10.1172/JCI38854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hartlova A, Erttmann SF, Raffi FA, Schmalz AM, Resch U, Anugula S, Lienenklaus S, Nilsson LM, Kroger A, Nilsson JA, et al. DNA damage primes the type I interferon system via the cytosolic DNA sensor STING to promote antimicrobial innate immunity. Immunity. 2015;42(2):332–43. doi: 10.1016/j.immuni.2015.01.012. [DOI] [PubMed] [Google Scholar]
- 37.Schvartzman JM, Sotillo R, Benezra R. Mitotic chromosomal instability and cancer: mouse modelling of the human disease. Nat Rev Cancer. 2010;10(2):102–15. doi: 10.1038/nrc2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sotillo R, Hernando E, Diaz-Rodriguez E, Teruya-Feldstein J, Cordon-Cardo C, Lowe SW, Benezra R. Mad2 overexpression promotes aneuploidy and tumorigenesis in mice. Cancer Cell. 2007;11(1):9–23. doi: 10.1016/j.ccr.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sotillo R, Schvartzman JM, Socci ND, Benezra R. Mad2-induced chromosome instability leads to lung tumour relapse after oncogene withdrawal. Nature. 2010;464(7287):436–U138. doi: 10.1038/nature08803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmitz ML, Kracht M. Cyclin-Dependent Kinases as Coregulators of Inflammatory Gene Expression. Trends Pharmacol Sci. 2016;37(2):101–13. doi: 10.1016/j.tips.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 41.Ohgami RS, Campagna DR, McDonald A, Fleming MD. The Steap proteins are metalloreductases. Blood. 2006;108(4):1388–94. doi: 10.1182/blood-2006-02-003681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Finegold AA, Shatwell KP, Segal AW, Klausner RD, Dancis A. Intramembrane bis-heme motif for transmembrane electron transport conserved in a yeast iron reductase and the human NADPH oxidase. J Biol Chem. 1996;271(49):31021–4. doi: 10.1074/jbc.271.49.31021. [DOI] [PubMed] [Google Scholar]
- 43.Ohgami RS, Campagna DR, Greer EL, Antiochos B, McDonald A, Chen J, Sharp JJ, Fujiwara Y, Barker JE, Fleming MD. Identification of a ferrireductase required for efficient transferrin-dependent iron uptake in erythroid cells. Nat Genet. 2005;37(11):1264–9. doi: 10.1038/ng1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Challita-Eid PM, Morrison K, Etessami S, An Z, Morrison KJ, Perez-Villar JJ, Raitano AB, Jia XC, Gudas JM, Kanner SB, et al. Monoclonal antibodies to six-transmembrane epithelial antigen of the prostate-1 inhibit intercellular communication in vitro and growth of human tumor xenografts in vivo. Cancer Res. 2007;67(12):5798–805. doi: 10.1158/0008-5472.CAN-06-3849. [DOI] [PubMed] [Google Scholar]
- 45.Wang L, Jin Y, Arnoldussen YJ, Jonson I, Qu S, Maelandsmo GM, Kristian A, Risberg B, Waehre H, Danielsen HE, et al. STAMP1 Is Both a Proliferative and an Antiapoptotic Factor in Prostate Cancer. Cancer Research. 2010;70(14):5818–28. doi: 10.1158/0008-5472.CAN-09-4697. [DOI] [PubMed] [Google Scholar]
- 46.Wellen KE, Fucho R, Gregor MF, Furuhashi M, Morgan C, Lindstad T, Vaillancourt E, Gorgun CZ, Saatcioglu F, Hotamisligil GS. Coordinated regulation of nutrient and inflammatory responses by STAMP2 is essential for metabolic homeostasis. Cell. 2007;129(3):537–48. doi: 10.1016/j.cell.2007.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moldes M, Lasnier F, Gauthereau X, Klein C, Pairault J, Feve B, Chambaut-Guerin AM. Tumor necrosis factor-alpha-induced adipose-related protein (TIARP), a cell-surface protein that is highly induced by tumor necrosis factor-alpha and adipose conversion. Journal of Biological Chemistry. 2001;276(36):33938–46. doi: 10.1074/jbc.M105726200. [DOI] [PubMed] [Google Scholar]
- 48.Chen XH, Zhu C, Ji CB, Zhao YP, Zhang CM, Chen FK, Gao CL, Zhu JG, Qian LM, Guo XR. STEAP4, a gene associated with insulin sensitivity, is regulated by several adipokines in human adipocytes. Int J Mol Med. 2010;25(3):361–7. doi: 10.3892/ijmm_00000353. [DOI] [PubMed] [Google Scholar]
- 49.Petrak J, Vyoral D. Hephaestin - a ferroxidase of cellular iron export. Int J Biochem Cell B. 2005;37(6):1173–8. doi: 10.1016/j.biocel.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 50.Srinivasan G, Aitken JD, Zhang B, Carvalho FA, Chassaing B, Shashidharamurthy R, Borregaard N, Jones DP, Gewirtz AT, Vijay-Kumar M. Lipocalin 2 deficiency dysregulates iron homeostasis and exacerbates endotoxin-induced sepsis. J Immunol. 2012;189(4):1911–9. doi: 10.4049/jimmunol.1200892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Richardson DR, Lane DJR, Becker EM, Huang MLH, Whitnall M, Rahmanto YS, Sheftel AD, Ponka P. Mitochondrial iron trafficking and the integration of iron metabolism between the mitochondrion and cytosol. P Natl Acad Sci USA. 2010;107(24):10775–82. doi: 10.1073/pnas.0912925107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aguirre JD, Culotta VC. Battles with iron: manganese in oxidative stress protection. J Biol Chem. 2012;287(17):13541–8. doi: 10.1074/jbc.R111.312181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saletta F, Rahmanto YS, Noulsri E, Richardson DR. Iron Chelator-Mediated Alterations in Gene Expression: Identification of Novel Iron-Regulated Molecules That Are Molecular Targets of Hypoxia-Inducible Factor-1 alpha and p53. Mol Pharmacol. 2010;77(3):443–58. doi: 10.1124/mol.109.061028. [DOI] [PubMed] [Google Scholar]
- 54.Dai MH, Wang PL, Boyd AD, Kostov G, Athey B, Jones EG, Bunney WE, Myers RM, Speed TP, Akil H, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Research. 2005;33(20) doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 56.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118–27. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 57.Hodgin JB, Borczuk AC, Nasr SH, Markowitz GS, Nair V, Martini S, Eichinger F, Vining C, Berthier CC, Kretzler M, et al. A molecular profile of focal segmental glomerulosclerosis from formalin-fixed, paraffin-embedded tissue. Am J Pathol. 2010;177(4):1674–86. doi: 10.2353/ajpath.2010.090746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pages F, Trajanoski Z, Galon J. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25(8):1091–3. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34(2):374–8. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 60.Elder JT, Fisher GJ, Zhang QY, Eisen D, Krust A, Kastner P, Chambon P, Voorhees JJ. Retinoic acid receptor gene expression in human skin. J Invest Dermatol. 1991;96(4):425–33. doi: 10.1111/1523-1747.ep12469889. [DOI] [PubMed] [Google Scholar]
- 61.Johnston A, Gudjonsson JE, Aphale A, Guzman AM, Stoll SW, Elder JT. EGFR and IL-1 signaling synergistically promote keratinocyte antimicrobial defenses in a differentiation-dependent manner. J Invest Dermatol. 2011;131(2):329–37. doi: 10.1038/jid.2010.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.