Abstract
Circling mouse (C57BL/6J-cir/cir) deleted the transmembrane inner ear (Tmie) gene is an animal model for human non-syndromic recessive deafness, DFNB6. In circling mouse, hair cells in the cochlea have degenerated and hair bundles have become irregularity as time goes on. Tmie protein carries out a function of the mechanoelectrical transduction channel in cochlear hair cells. Myosin7a (MYO7A) protein has key roles in development of the cochlear hair bundles as well as in the function of cochlear hair cells. To find whether Tmie protein interacts with MYO7A proteins in the cochlea postnatal developmental stage, we investigated expression of the MYO7A proteins in the cochlear hair cells of circling mice by western blot analysis and whole mount immunofluorescence at postnatal day 5 (P5). The expression of MYO7A showed statistically significant increase in the cochlea of C57BL/6J-+/cir and C57BL/6J-cir/cir mice than that of C57BL/6J-+/+ mice. The MYO7A intensity of the cochlear hair cells also increased in C57BL/6J-+/cir and C57BL/6J-cir/cir mice compared with those of C57BL/6J-+/+ mice. Taken together, the results indicate that Tmie protein may have an important role with MYO7A protein in the development and maintenance of the stereociliary bundles during postnatal developmental stage of the cochlea.
Keywords: Circling mice, cochlea, transmembrane inner ear (Tmie), myosin7a
Hearing loss is caused by environmental factors, genetic factors, or a combination of both. The hearing impairment caused by genetic factors accounts for about 50% of the total [1]. It has been reported that one in a thousand newborn babies and one in three hundred infants have congenital hearing loss. Besides, one in a thousand children suffers from profound hearing loss before adulthood [2,3]. Genes related to hearing loss have been determined rapidly for scores of years [4]. There are two types of hereditary hearing loss: syndromic hearing loss and non-syndromic hearing loss. Syndromic hearing loss is associated with other defects and occupies about 30% of hereditary hearing loss. Non-syndromic hearing loss is not associated with other diseases and occupies about 70% of hereditary hearing loss. Most non-syndromic hearing loss is sensorineural hearing loss (SNHL), which is caused by abnormality in the inner ear [5]. Non-syndromic hearing loss is classified into autosomal dominant deafness (DFNA), which accounts for about 15 to 20% of the total; autosomal recessive deafness (DFNB), which accounts for about 80%; X-linked deafness (DFN), which accounts for about 1%; and mitochondrial deafness, which accounts for at least 1% [6,7].
The circling mouse (C57BL/6J-cir/cir) is characterized by deafness, circling behavior, and head tossing. The mutated gene was transmitted by autosomal recessive inheritance with 100% penetrance [8,9]. The circling mice have a 40 Kbp genomic deletion that is involved in Tmie gene and Rn49018 on chromosome 9, which is homologous with human chromosome 3p21 [9]. Thus, the circling mice are reported as an animal model of DFNB6 in human [8,10]. According to previous research, stereociliary bundles of circling mice are disarranged and shorten at 10-day-old. Eventually, the organ of Corti degenerates completely by 21-day-old [10].
Recently, Tmie protein has been reported as being an essential element of the mechanoelectrical transduction channels in hair cells because it joins the tip-link to the mechanoelectrical transduction (MET) channels. Tmie protein is composed of a ternary complex with the protocadherin15 (PCDH15) protein of the tip-link and binding partner and tetraspan membrane protein of hair cell stereocilia (TMHS/LHFPL5) [11]. In cochlear hair cells, the motor protein of the stereocilia is composed of MYO7A, harmonin (a PDZ domain protein), and sans (a putative scaffolding protein), which exist on lateral links, ankle links, and upper tip-link density (UTLD) [12,13]. MYO7A is mostly located in stereocilia as well as in the cuticular plate [14,15] and plays an important role in the development and arrangement of hair bundles [16,17]. Furthermore, MYO7A is essential in the transduction of MET channels because it creates tension in the stereocilial links such as the tip link [18].
In this study, the expression levels of MYO7A protein were compared among C57BL/6J-+/+, C57BL/6J-+/cir, and C57BL/6J-cir/cir mice. The results suggest that Tmie protein associated with the expression of MYO7A protein during postnatal development of mechanosensory cells in circling mice.
Materials and Methods
Animals
Circling mouse was discovered from ICR outbred mice. After that, cir gene was maintained in the C57BL/6J genetic background (C57BL/6J-cir congenic mouse) [8]. We used littermate C57BL/6J-+/+, C57BL/6J-+/cir, and C57BL/6J-cir/cir mice without distinguish the sex. The animals were kept in specific-pathogen free animal care facility that maintained a regular environment: 22±2℃, 55±10% relative humidity, and a 12 hr light and 12 hr night routine cycle. Normal rodent pellet feeds (Cargill Agri Purina, Seongnam, Gyeonggi, Korea) and water were provided ad libitum. The animal study was conducted in accordance with the regulations of the Institutional Animal Care and Use Committees of Hallym University (Hallym 2009-58).
Genomic DNA preparation and genotyping
Genomic DNA was prepared from tails of mice. The tail tissues were incubated with a buffer (1X SSC: 439 µL, 1 M Tris-HCl: 5 µL, 0.5 M EDTA: 1 µL, 10% SDS: 50 µL, 20 mg/mL Proteinase K: 5 µL) at 53℃ for 5 hr. Then the genomic DNA was purified by phenol, phenol/chloroform, chloroform, and isopropanol reagents. Refined genomic DNA was washed with 70% ethanol and 100% ethanol and then dissolved in 50 µL of Tris-EDTA buffer (10 mM Tris-HCl, 1 mM EDTA). The concentration of DNA was measured at wavelengths of 260 and 280 nm. Genotyping was performed by polymerase chain reaction (PCR) of genomic DNA. In this study, we used 4 types of primers that are designed from the exon 6 of Tmie gene (5′GGCAGAAGTGGACCCACACACCA3′ and 5′TTCCTGAGGTGGCAGCCGGG3′) and the deletion breakpoint (5′GATTCCTGTGAGCCCCAGCACCA3′ and 5′ATCCATGTACAACATGGTAGAGACCTGGAAG3′) [9]. The PCR reaction buffer containing rTaq Plus 5x PCR Master Mix reagent (ELPiS, Daejeon, Korea), 50 ng genomic DNA, and 20 pmole of each primer was subjected to the conditions of initial denaturation at 95℃ for 5 min followed by 25 cycles of 95℃ for 30 sec, 69℃ for 30 sec, and 72℃ for 45 sec and then a final elongation period at 72℃ for 10 min. The PCR products, with total volumes of 30 µL, were separated by electrophoresis on 2% agarose gel. The PCR product band sizes of Tmie gene and the deletion breakpoint were 794 and 571 bp, respectively.
Western blot analysis
Two cochlea tissues were dissected from right and left temporal bone of P5 mice and homogenized in lysis buffer (iNtRON Biotechnology, Seongnam, Gyeonggi, Korea). The samples were centrifuged at 20,000 g for 30 min after sonication. Then 30 mg protein was boiled with 1x sample buffer for 5 min and loaded on 6–10% SDS polyacrylamide gel. After running at 100 V for 3 hr, the proteins were transferred to polyvinylidene fluoride transfer membrane (Millipore, Darmstadt, Hesse, Germany) at 250 mA for 2 hr. The membrane was blocked with 10% skim milk at room temperature for 1 hr and incubated at 4℃ overnight with primary antibody: anti-myosin7a (1:100, Abcam, Cambridge, Cambridgeshire, UK) or anti-Beta actin (1:10,000, Sigma, Saintlouis, Missouri, USA). Next it was incubated with horseradish peroxidase (HRP)-conjugated secondary antibody, anti-rabbit (1:2,500, Cell signaling technology, Danvers, Massachusetts, USA), and anti-mouse (1:50,000, Novus biologicals, Littleton, Colorado, USA) at room temperature for 1 hr and then proteins were detected by ECL (GE healthcare, Waukesha, Wisconsin, USA).
Whole mount immunofluorescence
For whole mount immunofluorescence of cochlea, temporal bones of littermate were obtained at 5 days after birth and fixed in 4% paraformaldehyde at 4℃ overnight. The organ of Corti were dissected from temporal bones. Samples were incubated in 5% Triton-X for 30 min and blocked with 1X PBS containing 5% horse serum and 3% BSA for 1 hr. Tissues were stained at 4℃ overnight by anti-myosin7a (1:250, Abcam, Cambridge, Cambridgeshire, UK). Next, the samples were washed in 1X PBS at 10 times for 15 min and incubated overnight at 4℃ with goat-anti-IgG Alexa 488 secondary antibody (1:300, Abcam, Cambridge, Cambridgeshire, UK) and Alexa 594 Phalloidin (1:250, Life Technologies, Carlsbad, California, USA). Finally, the tissues were washed with 1X PBS at 10 times for 15 min and mounted with antifade on glass slides. Pictures of outer hair cells (OHCs) and inner hair cells (IHCs) were taken from parts of the apex, middle, and base of the cochlea duct using confocal microscopy (LSM 710; Carl Zeiss, Oberkochen, Baden-Württemberg, Germany). Then the intensity of primary antibody was measured using a ZEN program.
Statistical analysis
For multiple comparisons, the one-way analysis of variance (ANOVA) was used. All data were analyzed by the SPSS statistics 22.0 (International Business Machines Corporation Software, USA). Statistically significant difference was defined at a P-value less than 0.05. Different alphabets represent a significant difference.
Results
Genotype analysis of circling mice
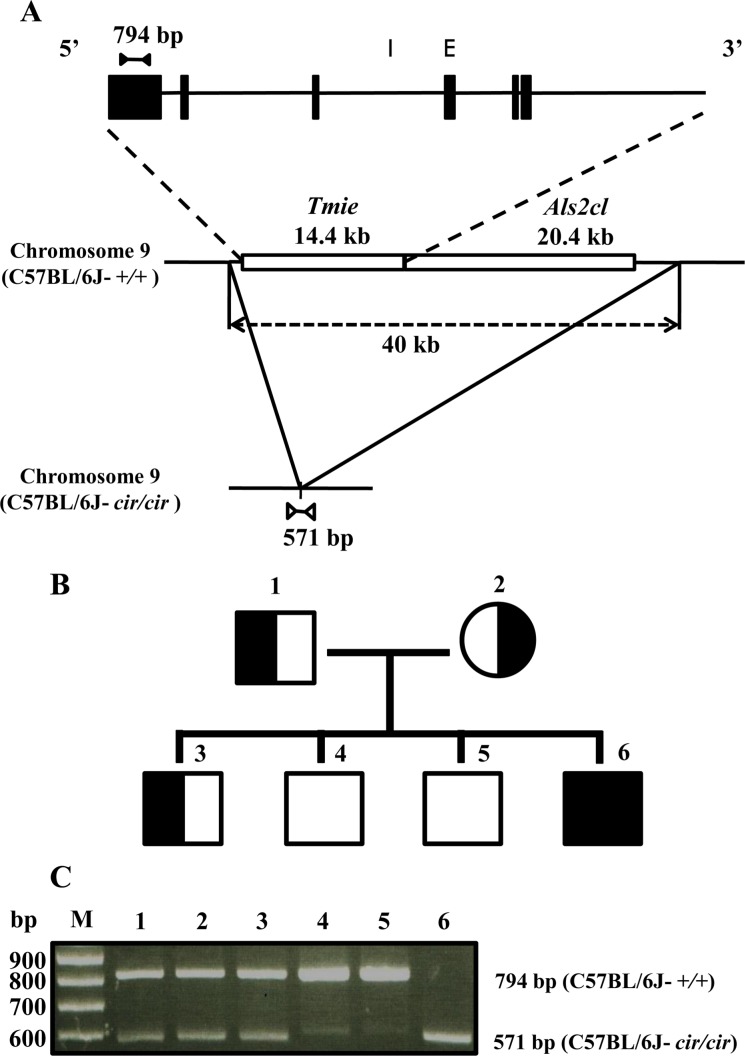
To identify the genotypes of C57BL/6J-+/+, C57BL/6J-+/cir, and C57BL/6J-cir/cir mice, we used two sets of primers: exon 6 primers and deletion point primers. The exon 6 primers were designed to amplify 794 bp products from normal mouse. The deletion point primers were designed to amplify 571 bp from circling homozygote mice (Figure 1A). In order to verify specificity of our primers, heterozygote (C57BL/6J-+/cir) male was crossed with heterozygote (C57BL/6J-+/cir) female. From this mating, four F1 progenies were obtained. These F1 progenies were used to identify genotypes (Figure 1B). As we expected, PCR products of 794 bp and 571 bp were detected from C57BL/6J-+/+ and C57BL/6J-cir/cir mice, respectively (Figure 1C). In our animal experiment, we used animals after genotyping by the same method.
Figure 1. Genotyping by genomic PCR in the circling mice. A. Location of the primers for genotyping. E: Exon. I: Intron. B. Pedigree of circling mice. Square and circle indicate each male and female. Also empty, compact, and half-and-half indicate C57BL/6J-+/+, C57BL/6J-cir/cir, and C57BL/6J-+/cir, respectively. C. C57BL/6J-+/+ and C57BL/6J-cir/cir mice had the 794 bp and 571 bp bands, respectively. M: Molecular size marker (100 bp DNA ladder).
Expression of MYO7A in the cochlea developmental stage of circling mice
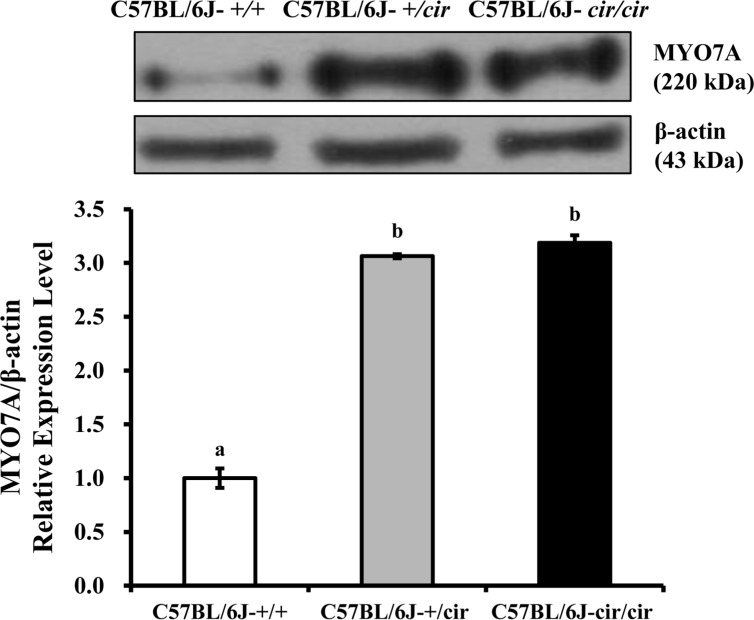
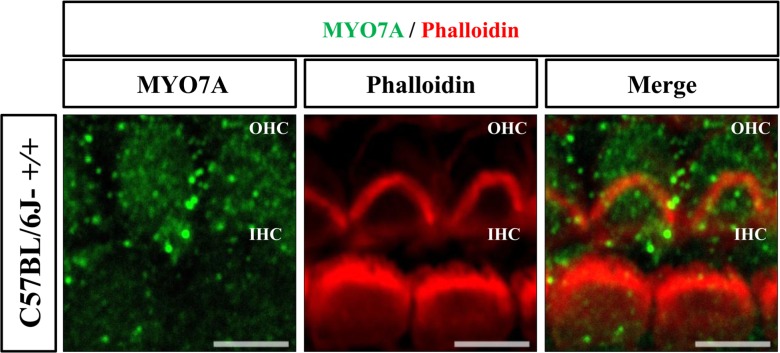
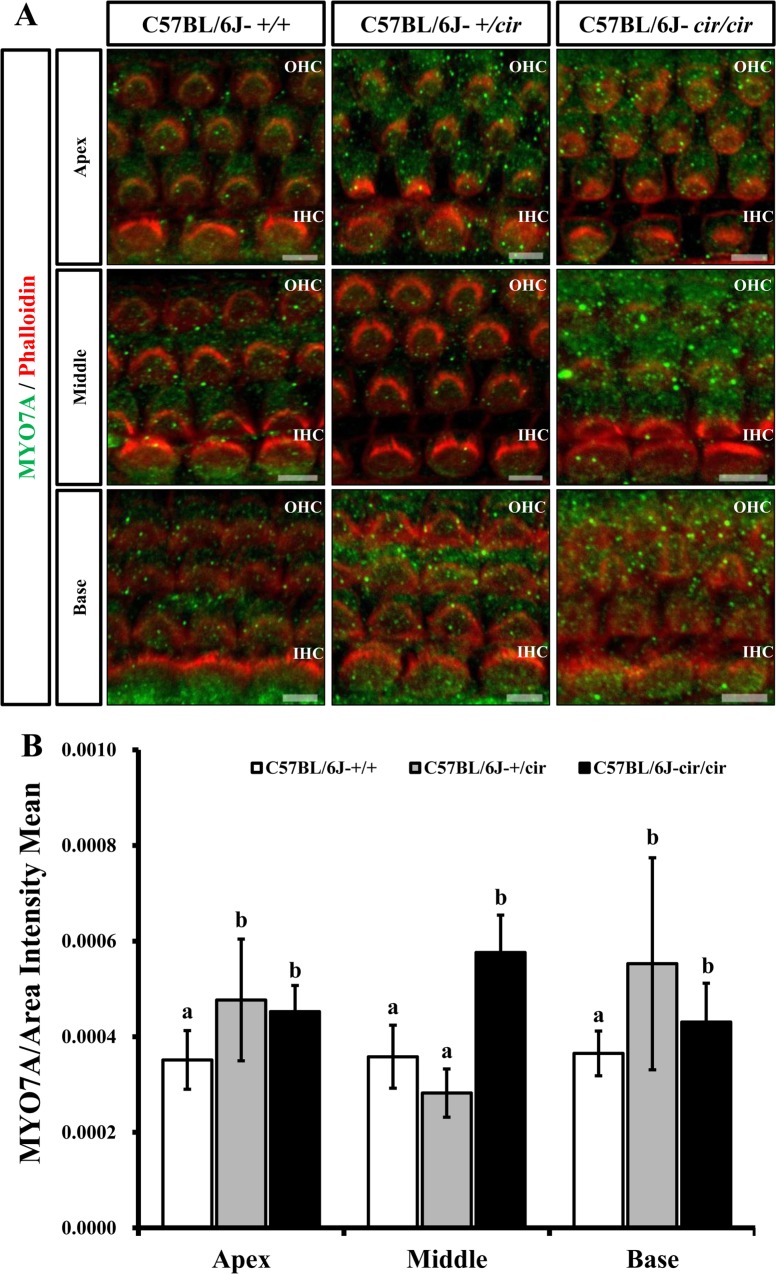
The expression of MYO7A protein in the cochlea was compared among C57BL/6J-+/+, C57BL/6J-+/cir, and C57BL/6J-cir/cir mice using western blot analysis and whole mount immunofluorescence. Expression levels of MYO7A in C57BL/6J-+/cir and C57BL/6J-cir/cir mice showed a statistically significant increase compared with C57BL/6J-+/+ mice at P5, but there was no difference in the expression level of MYO7A between C57BL/6J-+/cir and C57BL/6J-cir/cir mice (Figure 2). To confirm the western blot result, we measured the expression level of MYO7A in cochlear hair cells using whole mount immunofluorescence. In the middle part of the cochlear duct, stereociliary bundles of hair cells in C57BL/6J-+/+ mice were in a regular arrangement. MYO7A protein in C57BL/6J-+/+ mice was expressed at the cuticular plate and hair bundles (Figure 3). The MYO7A intensity of cochlear hair cells was upregulated at the apex and base parts of the cochlear duct in C57BL/6J-+/cir and C57BL/6J-cir/cir mice compared with C57BL/6J-+/+ mice. There was no difference in MYO7A intensity between C57BL/6J-+/cir and C57BL/6J-cir/cir mice at the part of apex and base cochlear turn (Figure 4).
Figure 2. Western blot analysis of MYO7A at P5. Proteins isolated from littermate cochlear tissues at P5 used for western blot analysis. Data were analyzed by one-way analysis of variance (ANOVA). Different alphabets indicate statistically significant difference (N=3).
Figure 3. Location of MYO7A in cochlear hair cells at P5 by whole mount immunofluorescence. The cochlear hair cells stained with MYO7A antibody (green) and phalloidin (red). Images acquired from the middle cochlear turn. OHC: outer hair cell. IHC: inner hair cell. Scale bar: 5 µm.
Figure 4. MYO7A expression in cochlear hair cells at P5 by whole mount immunofluorescence. The cochlear hair cells stained with MYO7A antibody (green) and phalloidin (red). A. Images acquired from parts of the apex, middle, and base cochlear turn. B. MYO7A intensity of the outer hair cells and inner hair cells was measured at parts of the apex, middle, and base of the cochlear duct. Data were analyzed by one-way analysis of variance (ANOVA). Different alphabets indicate statistically significant difference (N=3). OHC: outer hair cell. IHC: inner hair cell. Scale bar: 5 µm.
Discussion
Circling mice (C57BL/6J-cir/cir), with absence of Tmie gene, have been considered as an animal model for DFNB6 in human [19]. Tmie protein performs an essential function in the mechanoelectrical transduction channel as well as in development in cochlear hair bundles [11,20]. In this study, expression of MYO7A was investigated in C57BL/6J-+/+, C57BL/6J-+/cir, and C57BL/6J-cir/cir mice at postnatal day 5.
We have constructed two sets of primers to differentiate the genotypes of circling mice at P5. Three genotypes of uterine brothers; C57BL/6J-+/+, C57BL/6J-+/cir, and C57BL/6J-cir/cir identified by genotype analysis were used in this research (Figure 1). At P5, expression of MYO7A in the cochlea of C57BL/6J-+/cir and C57BL/6J-cir/cir mice was increased 3.0 fold compared with that of C57BL/6J-+/+ mice (Figure 2). Previous study reported that the cochlear hair cells in the circling mice were completely degenerated at postnatal day 21 [10]. Thus, expression of MYO7A protein from P6 to p21 should be investigated in further study. In addition, to know relationship between Tmie and other proteins which play key role in function of MET channels such as PCDH15, TMHS/LHFPL5, Transmembrane channel-like 1 (TMC1), and Transmembrane channel-like 2 (TMC2) also should be investigated [21,22,23].
The MYO7A is distributed at cuticular plates and stereocilia especially in the upper tip-link density (UTLD), lateral and ankle links [12,13,14,15]. In the present study, the MYO7A was expressed at the cuticular plate and hair bundles (Figure 3). The MYO7A intensity at base cochlear turn increased in C57BL/6J-+/cir and C57BL/6J-cir/cir mice compared with that of C57BL/6J-+/+ mice (Figure 4). The circling mice have a 40 Kbp genomic deletion including Tmie gene [9] and become deaf around P18 [24]. The Tmie protein is essential element of the MET channels [11]. The stereocilia of basal turn have more defect than that of the apical or middle turn [24]. Since the stereocilia of cochlear in circling mice have abnormal structure, there is possibility that the MYO7A protein were over-expressed to make up stereocilia abnormality. To confirm this hypothesis, the expression level of sans and harmonin should be measure in further study because the MYO7A forms complex with sans and harmonin at the UTLD [12]. The spiral ganglion neurons (SGN) in the basal turn decreased with age. The decreased number of SGN shift auditory brainstem response (ABR) thresholds [25]. Thus, the heterozygote of circling mice might be useful model for mechanism study of age-related hearing loss. Taken together, our results suggest that Tmie protein plays an important role in the maintenance and development of the stereocilia during the cochlea developmental stages by interacting with MYO7A protein.
Acknowledgments
This work was supported by the grants from Korea Ministry of Science, ICT and Future Planning to J. G. S. (KMPC:2014M3A9D5A01075129).
Footnotes
Conflict of interests: The authors declare that there is no financial conflict of interests to publish these results.
References
- 1.Schrijver I. Hereditary non-syndromic sensorineural hearing loss: transforming silence to sound. J Mol Diagn. 2004;6(4):275–284. doi: 10.1016/S1525-1578(10)60522-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mason JA, Herrmann KR. Universal infant hearing screening by automated auditory brainstem response measurement. Pediatrics. 1998;101(2):221–228. doi: 10.1542/peds.101.2.221. [DOI] [PubMed] [Google Scholar]
- 3.Parving A. The need for universal neonatal hearing screening--some aspects of epidemiology and identification. Acta Paediatr Suppl. 1999;88(432):69–72. doi: 10.1111/j.1651-2227.1999.tb01163.x. [DOI] [PubMed] [Google Scholar]
- 4.Stelma F, Bhutta MF. Non-syndromic hereditary sensorineural hearing loss: review of the genes involved. J Laryngol Otol. 2014;128(1):13–21. doi: 10.1017/S0022215113003265. [DOI] [PubMed] [Google Scholar]
- 5.Van Camp G, Willems PJ, Smith RJ. Nonsyndromic hearing impairment: unparalleled heterogeneity. Am J Hum Genet. 1997;60(4):758–764. [PMC free article] [PubMed] [Google Scholar]
- 6.ACMG (American College of Medical Genetics) Genetics Evaluation Guidelines for the Etiologic Diagnosis of Congenital Hearing Loss. Genetic Evaluation of Congenital Hearing Loss Expert Panel. ACMG statement. Genet Med. 2002;4(3):162–171. doi: 10.1097/00125817-200205000-00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hone SW, Smith RJ. Medical evaluation of pediatric hearing loss. Laboratory, radiographic, and genetic testing. Otolaryngol Clin North Am. 2002;35(4):751–764. doi: 10.1016/s0030-6665(02)00048-8. [DOI] [PubMed] [Google Scholar]
- 8.Lee JW, Ryoo ZY, Lee EJ, Hong SH, Chung WH, Lee HT, Chung KS, Kim TY, Oh YS, Suh JG. Circling mouse, a spontaneous mutant in the inner ear. Exp Anim. 2002;51(2):167–171. doi: 10.1538/expanim.51.167. [DOI] [PubMed] [Google Scholar]
- 9.Cho KI, Suh JG, Lee JW, Hong SH, Kang TC, Oh YS, Ryoo ZY. The circling mouse (C57BL/6J-cir) has a 40-kilobase genomic deletion that includes the transmembrane inner ear (tmie) gene. Comp Med. 2006;56(6):476–481. [PubMed] [Google Scholar]
- 10.Chung WH, Kim KR, Cho YS, Cho DY, Woo JH, Ryoo ZY, Cho KI, Hong SH. Cochlear pathology of the circling mouse: a new mouse model of DFNB6. Acta Otolaryngol. 2007;127(3):244–251. doi: 10.1080/00016480600827071. [DOI] [PubMed] [Google Scholar]
- 11.Zhao B, Wu Z, Grillet N, Yan L, Xiong W, Harkins-Perry S, Müller U. TMIE is an essential component of the mechanotransduction machinery of cochlear hair cells. Neuron. 2014;84(5):954–967. doi: 10.1016/j.neuron.2014.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grati M, Kachar B. Myosin VIIa and sans localization at stereocilia upper tip-link density implicates these Usher syndrome proteins in mechanotransduction. Proc Natl Acad Sci USA. 2011;108(28):11476–11481. doi: 10.1073/pnas.1104161108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maniak M. Cell adhesion: ushering in a new understanding of myosin VII. Curr Biol. 2001;11(8):R315–R317. doi: 10.1016/s0960-9822(01)00173-7. [DOI] [PubMed] [Google Scholar]
- 14.Hasson T, Heintzelman MB, Santos-Sacchi J, Corey DP, Mooseker MS. Expression in cochlea and retina of myosin VIIa, the gene product defective in Usher syndrome type 1B. Proc Natl Acad Sci USA. 1995;92(21):9815–9819. doi: 10.1073/pnas.92.21.9815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hasson T, Gillespie PG, Garcia JA, MacDonald RB, Zhao Y, Yee AG, Mooseker MS, Corey DP. Unconventional myosins in innerear sensory epithelia. J Cell Biol. 1997;137(6):1287–1307. doi: 10.1083/jcb.137.6.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lefèvre G, Michel V, Weil D, Lepelletier L, Bizard E, Wolfrum U, Hardelin JP, Petit C. A core cochlear phenotype in USH1 mouse mutants implicates fibrous links of the hair bundle in its cohesion, orientation and differential growth. Development. 2008;135(8):1427–1437. doi: 10.1242/dev.012922. [DOI] [PubMed] [Google Scholar]
- 17.Self T, Mahony M, Fleming J, Walsh J, Brown SD, Steel KP. Shaker-1 mutations reveal roles for myosin VIIA in both development and function of cochlear hair cells. Development. 1998;125(4):557–566. doi: 10.1242/dev.125.4.557. [DOI] [PubMed] [Google Scholar]
- 18.Kros CJ, Marcotti W, van Netten SM, Self TJ, Libby RT, Brown SD, Richardson GP, Steel KP. Reduced climbing and increased slipping adaptation in cochlear hair cells of mice with Myo7a mutations. Nat Neurosci. 2002;5(1):41–47. doi: 10.1038/nn784. [DOI] [PubMed] [Google Scholar]
- 19.Cho KI, Lee JW, Kim KS, Lee EJ, Suh JG, Lee HJ, Kim HT, Hong SH, Chung WH, Chang KT, Hyun BH, Oh YS, Ryoo ZY. Fine mapping of the circling (cir) gene on the distal portion of mouse chromosome 9. Comp Med. 2003;53(6):642–648. [PubMed] [Google Scholar]
- 20.Mitchem KL, Hibbard E, Beyer LA, Bosom K, Dootz GA, Dolan DF, Johnson KR, Raphael Y, Kohrman DC. Mutation of the novel gene Tmie results in sensory cell defects in the inner ear of spinner, a mouse model of human hearing loss DFNB6. Hum Mol Genet. 2002;11(16):1887–1898. doi: 10.1093/hmg/11.16.1887. [DOI] [PubMed] [Google Scholar]
- 21.Maeda R, Kindt KS, Mo W, Morgan CP, Erickson T, Zhao H, Clemens-Grisham R, Barr-Gillespie PG, Nicolson T. Tip-link protein protocadherin 15 interacts with transmembrane channel-like proteins TMC1 and TMC2. Proc Natl Acad Sci USA. 2014;111(35):12907–12912. doi: 10.1073/pnas.1402152111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiong W, Grillet N, Elledge HM, Wagner TF, Zhao B, Johnson KR, Kazmierczak P, Müller U. TMHS is an integral component of the mechanotransduction machinery of cochlear hair cells. Cell. 2012;151(6):1283–1295. doi: 10.1016/j.cell.2012.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fettiplace R. Is TMC1 the Hair Cell Mechanotransducer Channel? Biophys J. 2016;111(1):3–9. doi: 10.1016/j.bpj.2016.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee Y, Chang SY, Jung JY, Ahn SC. Reinvestigation of cochlear pathology in circling mice. Neurosci Lett. 2015;594:30–35. doi: 10.1016/j.neulet.2015.03.057. [DOI] [PubMed] [Google Scholar]
- 25.Tang X, Zhu X, Ding B, Walton JP, Frisina RD, Su J. Age-related hearing loss: GABA, nicotinic acetylcholine and NMDA receptor expression changes in spiral ganglion neurons of the mouse. Neuroscience. 2014;259:184–193. doi: 10.1016/j.neuroscience.2013.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]