Abstract
In contrast to most gammaretrovirus envelope proteins (Env), the Gibbon ape leukemia virus (GaLV) Env protein does not mediate the infectivity of human immunodeficiency virus type 1 (HIV-1) particles. We made use of this observation to set up a directed evolution system by creating a library of GaLV Env variants diversified at three critical amino acids, all located around the R-peptide cleavage site within the cytoplasmic tail. This library was screened for variants that were able to functionally pseudotype HIV-1 vector particles. All selected Env variants mediated the infectivity of HIV-1 vector particles and encoded novel cytoplasmic tail motifs. They were efficiently incorporated into HIV particles, and the R peptide was processed by the HIV protease. Interestingly, in some of the selected variants, the R-peptide cleavage site had shifted closer to the C terminus. These data demonstrate a valuable approach for the engineering of chimeric viruses and vector particles.
Human immunodeficiency virus type 1 (HIV-1) vectors are usually pseudotyped with the vesicular stomatitis virus G protein (VSV-G) or the envelope protein (Env) of amphotropic murine leukemia virus (MLV) to broaden their host range (11). However, in contrast to the closely related MLV Env protein, the wild-type (WT) gibbon ape leukemia virus (GaLV) Env protein turned out to be unable to functionally pseudotype HIV-1 vector particles and to mediate gene transfer (19). The GaLV Env protein, like all other retroviral Env proteins, is composed of two subunits, the extracellular surface unit (SU) and the transmembrane unit (TM). The TM protein is a type I transmembrane protein containing a membrane fusion regulatory sequence, the R peptide, at the C terminus of its cytoplasmic tail (C tail). The R peptide is a common feature of oncoretroviruses that inhibits the fusogenicity of Env proteins expressed on the surfaces of infected cells and thus excludes cytotoxic effects (3). Upon virus particle formation, the R peptide is cleaved off by the viral protease, thus activating the fusogenic function of Env (16, 17). While there is only a low level of homology between R peptides, the membrane-proximal parts are more conserved, allowing the identification of R-peptide cleavage sites in the C tails of different gammaretroviruses by sequence alignment (3).
Mutational studies revealed that modifications in the GaLV TM protein are sufficient to overcome the HIV-1 pseudotyping defect. Chimeric GaLV Env proteins in which, e.g., the C tail was replaced with that of MLV Env, are fully active in mediating the infectivity of HIV-1 vector particles (19). Alternatively, genetic truncation of the GaLV R peptide also results in functional HIV-1 vector particles. Fine mapping within the C tail identified three critical amino acid residues (K618, I619, and R623), all of which are located in close proximity to the R-peptide cleavage site. The so-called RTM Env variant, in which these critical residues were exchanged with the corresponding residues of MLV Env, mediated the infectivity of HIV-1 particles with the same efficiency as the MLV Env protein (8, 19).
In order to comprehensively identify the molecular determinants in retroviral TM C tails governing viral infectivity, we used the GaLV Env/HIV pseudotype phenomenon as a model system to set up a molecular evolution approach. A GaLV-Env library encoding variants that were combinatorially diversified at the three critical amino acid residues was generated and screened for the ability to transfer HIV-1 vector-packaged reporter genes into target cells. The selection resulted in the identification of novel motifs that are able to overcome the block in pseudotyping HIV-1 vector particles with GaLV Env.
MATERIALS AND METHODS
Cell lines.
HEK-293, HEK-293T, and HT1080 cells were obtained from the American Type Culture Collection and were maintained in high-glucose (4.5 g/liter) Dulbecco's modified Eagle's medium (Gibco, Eggenstein, Germany) supplemented with 10% fetal calf serum (Biochrom, Berlin, Germany), benzylpenicillin (60 μg/ml), and streptomycin (100 μg/ml) at 37°C in an atmosphere of 5% CO2.
Envelope expression constructs.
For the generation of HIV-packageable GaLV Env-encoding vectors, the plasmids pHEIN and pALF-GaLV-WT were used as starting constructs (10, 19). First, silent mutagenesis of pALF-GaLV-WT was performed to introduce a SacII restriction site at codon 615 of the GaLV Env open reading frame to facilitate subsequent cloning steps. For this purpose, a Quickchange mutagenesis kit (Stratagene, La Jolla, Calif.) was used in combination with the primers GaLV-SacII(+) (5′-CAATGATAGGATATCCGCGGTTAAAATTCTGGTC-3′) and GaLV-SacII(−) (5′-GACCAGAATTTTAACCGCGGATATCCTATCATTG-3′). The resulting plasmid was then applied to a standard fusion PCR to generate an N-terminally hemagglutinin (HA)-tagged GaLV Env-encoding sequence by use of the following primer pairs: GaLV BamHI(+) (5′-ATGAGGATCCGCCACCATGGTATTGCTGCCTGGGTCC-3′) together with GaLV-HA-SLQ(+) (5′-TATCCATATGATGTTCCAGATTATGCTAGTCTGCAAAATAAGAACCCC-3′) and GaLV BsrGI(−) (5′-AGCATGTACACTCGAGTTAAAGGTTACCTTCGTTCTCTAGGG-3′) together with GaLV-GGT-HA(−) (5′-AGCATAATCTGGAACATCATATGGATACGTCCCGCCGCCGCCGAATACGC-3′). In parallel, a BsrGI site downstream of the 3′ HIV long terminal repeat (LTR) within pHEIN was deleted by XbaI and NheI digestion and subsequent religation. Subsequently, the fragment obtained by fusion PCR was inserted into this vector via BamHI and BsrGI sites, resulting in the plasmid pHGIN-WT. All other envelope constructs, including the plasmid library, were generated by the introduction of double-stranded oligonucleotides via SacII and BstEII sites into pHGIN-WT. For cloning of the plasmid pHGIN-X3, the primers GaLV-X3 [5′-GATATCCGCGGTT(G/C/A)NN(G/C/A)NNCTGTGATAA(G/C/A)NNCAGAAATATCAGGCCCTAGAGAACGAAGGTAACCTTT-3′] and GaLV-X(−) (5′-GTTACCTTCGTTCTCTAGAGC-3′) were annealed and subjected to Klenow polymerase-mediated primer extension. The constructs pHGIN-RTM and pHGIN-ΔR were generated by the same approach with the universal primer GaLV-X(−) together with the primer GaLV-pRTM (5′-ATATCCGCGGTTCAGGCCCTGGTCCTTACCCAGAAATATCAGGCCCTAGAGAACGAAGGTAACCTTTA-3′) or GaLV-ΔR (5′-GATATCCGCGGTTAAAATTCTGTGATAAAGACAGAAATATCAGGCCCTAGAGAACGAAGGTAACCTTT-3′). Mutants of the selected clones, numbers 2 and 4, were generated by the insertion of double-stranded oligonucleotides with overhanging ends that directly allowed insertion into SacII- and BstEII-digested pHGIN-WT. For example, mutant 2-L622V was generated by annealing and cloning the oligonucleotide pair 2-L622V(+) (5′-GGTTTATGCGCTGCTCGTCCAGCAGAAATATCAGGCTCTAGAGAACGAAG-3′) and 2-L622V(−) (5′-GCGGTTTATGCGCTGCTCGTCCAGCAGAAATATCAGGCTCTAGAGAACGA-′3). All other mutants were generated with basically the same oligonucleotides, with differences only in their central regions according to the respective mutation.
The ligation and cloning conditions for the library were basically as previously described (5), with the exception that ElectroTen-Blue bacterial cells (Stratagene) were used in 0.1-cm-wide cuvettes at 1.7 kV, 200 Ω, and 25 μF. Electroporated cells were plated and subsequently grown up in liquid medium for purification of the plasmid DNA.
Selection of libraries.
Twenty-four hours prior to transfection, 3 × 107 293T cells were seeded into T175 culture flasks. For the generation of HIV-1(VSV-G) pseudotype vectors, transfection was performed with 20 μg of pHGIN-X3, 20 μg of pCMVd8.2 (12), 5 μg of pMDG (12), and Lipofectamine Plus (Gibco) according to the supplier's instructions. On day 2 after transfection, the supernatant was collected and filtered through a 0.45-μm-pore-size filter. Subsequently, the filtrate was used to transduce approximately 5 × 106 HEK-293 cells. Starting on day 2 posttransduction, these cells were treated with 1 mg of G418 (Gibco)/ml for 7 days. To apply selection pressure, we seeded 3 × 106 of the surviving cells onto the bottom of a transwell chamber (Corning, Schiphol-Rijkand, The Netherlands) and transfected them on the following day with 10 μg of pCMVd8.2. On day 2 posttransfection, a permeable membrane coated with 3 × 106 HT1080 cells was added to the chamber and incubated for 3 days. Subsequently, the transduced cells were removed from the membrane by trypsinization and then cultivated in the presence of 1 μg of G418/ml for another 7 days. For initiation of the next selection cycle, G418-resistant cells were then seeded onto the bottom of a transwell chamber and transfected with pCMVd8.2 as described above.
For sequence analysis, genomic DNAs were isolated from transduced cells. PCRs were performed with the primers Seq2(+) (5′-CCCCTATTACTCCTCCTTCTGTTGCTCATCCTC-3′) and RT4(−) (5′-TTGGCCGCTTTACTTGTACACTCGAG-3′). The resulting PCR products were cloned by use of a pGEM-TEasy kit (Promega, Madison, Wis.), and plasmid DNAs from single bacterial clones were sequenced by use of a standard T7 primer (MWG, Ebersberg, Germany).
Vector production and titer determination.
For vector particle production, 8 × 105 HEK-293T cells were transfected with 3 μg of the pHGIN envelope construct and 3 μg of pCMVd8.2 or, for β-galactosidase assays, with 2 μg of the pHGIN envelope construct, 2 μg of pCMVd8.2, and 2 μg of pHRCMVlacZ (12). To inhibit HIV protease activity, we supplemented the cell culture medium with 1 μM saquinavir (NIH AIDS Research and Referent Reagents Program, Germantown, Md.). At 2 days posttransfection, the supernatants were harvested and passaged through a 0.45-μm-pore-size filter. Subsequently, 1.25 × 105 HT1080 cells were incubated with the filtrate for 2 h. Starting on day 2 posttransduction, these cells were treated with 1 mg of G418 (Gibco)/ml for 5 days. Surviving colonies were counted under a light microscope. For β-galactosidase assays, transduced cells were fixed at 2 days posttransduction with 2% formaldehyde and 0.5% glutaraldehyde. After being washed with phosphate-buffered saline (PBS), the samples were incubated with a staining solution (4 mM potassium ferricyanide, 4 mM potassium ferrocyanide, 2 mM MgCl2, and 80 μg of X-Gal [5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; Sigma, Deisenhofen, Germany]/ml) at 37°C overnight. The next day, blue colonies were counted under a light microscope.
FACS.
For fluorescence-activated cell sorting (FACS) analysis, 5 × 105 transfected cells were washed with PBA (PBS with 2% fetal calf serum and 0.1% sodium azide) and incubated with an anti-HA antibody (Sigma) in a 1:200 dilution for 45 min at 4°C. After being washed with PBA, the cells were incubated with diluted (1:50) phycoerythrin-labeled secondary antibodies directed against mouse immunoglobulin G (Sigma) for 30 min. After being washed, the cells were fixed with PBS-1% paraformaldehyde and subjected to FACS analysis (FACScan; Becton Dickinson, San Jose, Calif.).
Western blot analysis.
Cell culture supernatants were harvested at 2 days posttransfection and passaged through a 0.45-μm-pore-size filter (Sartorius, Göttingen, Germany). Subsequently, the filtrates were concentrated by ultracentrifugation at 35,000 rpm at 4°C for 90 min in a Beckman SW41 rotor through a 30% sucrose cushion. The TM subunit of Env was detected by the use of hybridoma supernatants containing the rat monoclonal antibody 42/114 (15) at a 1:12.5 dilution. A rabbit anti-rat immunoglobulin G-horseradish peroxidase conjugate (Dako, Glostrup, Denmark) was used as a secondary antibody at a 1:2,000 dilution. The HIV-1 capsid protein was detected with a mouse anti-human HIV p24 monoclonal antibody (Chemicon) together with a sheep anti-mouse-horseradish peroxidase conjugate (Amersham, Little Chalfont, United Kingdom) diluted 1:2,000 as a secondary antibody. Specific proteins were visualized by use of a SuperSignal chemiluminescence kit (Pierce, Rockford, Ill.).
RESULTS
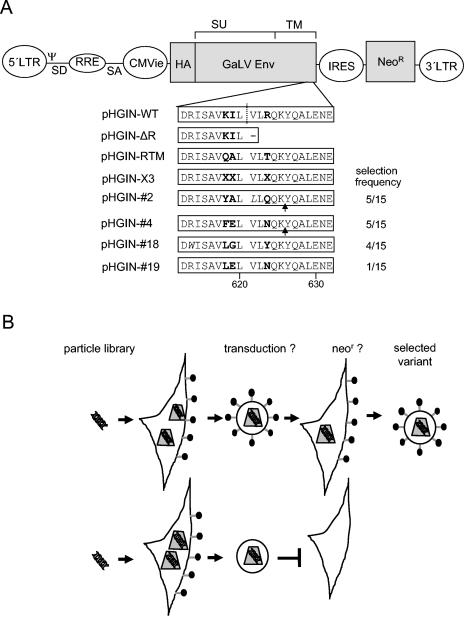
Starting with the bicistronic HIV-packageable vector pHEIN (10), we generated the plasmid pHGIN, which encodes an N-terminally HA-tagged GaLV envelope protein (Fig. 1A). The HA tag had no influence on the infectivity of MLV particles that were pseudotyped with the HA-GaLV Env protein, which therefore will be referred as the WT GaLV Env protein below (data not shown). Besides the Env protein, pHGIN encodes a neomycin resistance cassette preceded by an internal ribosome entry site. This construct was used as the parental plasmid for library construction. For construction of the library, we diversified the critical amino acids K618, I619, and R623 by the insertion of synthetic double-stranded oligonucleotides. In theory, the resulting pHGIN-X3 library encoded 4.1 × 103 different GaLV Env protein variants, which were well covered by the 106 bacterial clones obtained. Analyses of a representative number of single clones confirmed that there were diverse sequences with no obvious bias (not shown).
FIG. 1.
GaLV Env protein variants and experimental design. (A) Schematic representation of the plasmids used. The HIV-1-packageable pHGIN vector encodes an N-terminally HA-tagged GaLV Env protein under control of the cytomegalovirus immediate-early enhancer promoter (CMVie). The neomycin resistance gene (Neor) was driven by an internal ribosome entry site (IRES). Below the schematic, the amino acid sequences of GaLV TM protein C tails between positions 609 and 634 are shown for the control constructs, the library, and four selected clones (2, 4, 18, and 19). The numbers behind the selected clones indicate their frequencies after selection. Diversified amino acids are depicted in bold. The residues W613 in clone 19 and L621 in clone 2, shown in italics, changed during the selection process. The dotted line indicates the putative R-peptide cleavage site recognized by the GaLV protease (3). The arrows indicate the putative R-peptide cleavage site recognized by the HIV protease that resulted from the selection process. (B) Selection procedure. The coding regions of GaLV Env protein variants that were able to mediate the transduction of HIV particles were transferred to the target cell population. Transduced cells were amplified with G418 selection, and the coding sequences for functional GaLV Env proteins were mobilized by transfection with the HIV-1 Gag-Pol-encoding plasmid pHR-CMVΔ8.2 to initiate a second selection cycle.
To ensure the integration and expression of single library members per cell, we generated a VSV-G-pseudotyped HIV-1 particle library by cotransfection of HEK-293T cells with the plasmid pHGIN-X3 and plasmids encoding HIV-1 Gag-Pol (pHR-CMVΔ8.2) and VSV-G (pMDG) (12). Particles harvested from the supernatants were then incubated with HEK-293 cells, which were subsequently expanded and selected in the presence of G418, resulting in about 105 resistant colonies constituting the pool of library producer cells. For selection, the library producer cells were seeded onto the bottom of a transwell chamber and transfected with plasmid pHR-CMVΔ8.2 to initiate particle formation. From the next day on, these cells were cocultivated with the human fibrosarcoma cell line HT1080 and seeded on top of the permeable membrane for a further 3 days. Subsequently, the target cells were treated with G418 to select cells that were transduced by particles encoding infectious Env protein variants (Fig. 1B). Several hundred colonies were obtained which were then transfected with plasmid pHR-CMVΔ8.2 to initiate the next selection round. From the next day on, these cells were cocultivated with fresh HT1080 cells for another 3 days before selection for G418 resistance was conducted again.
Genomic DNAs isolated from G418-resistant cells obtained from both selection cycles were used for PCRs with GaLV Env-specific primers. PCR fragments were then cloned, and single bacterial colonies were picked for sequence analysis. Although after the first selection cycle no preference for certain amino acids was obvious, four predominant Env clones resulted from the second selection cycle (Fig. 1A). Among the four clones, three sequence motifs could be distinguished, with each appearing at a frequency of about 30%. There were no similarities to the parental WT or the RTM mutant. In general, a preference for aromatic or aliphatic amino acid residues at position 618 and for polar uncharged amino acids at position 623 was noted. Remarkably, for two of the four selected clones, two other amino acid changes within the C tail were observed, among which the exchange of arginine 613, which is highly conserved among the Env proteins of gammaretroviruses, for tryptophan was especially remarkable (Fig. 1A).
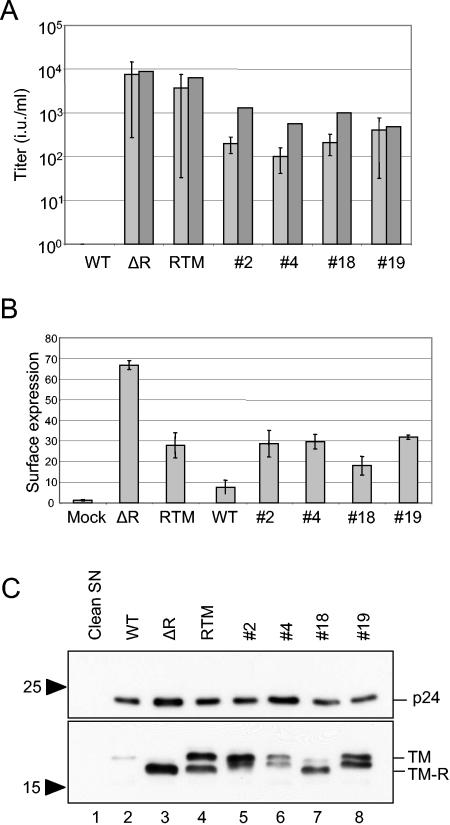
For a functional characterization of the selected Env proteins, pHGIN plasmids were reconstituted with the selected C-tail sequences to exclude any influence of other potential amino acid changes in the extracellular part of the Env reading frame. For this purpose, double-stranded oligonucleotides encoding the selected C tails were inserted into the SacII- and BstEII-restricted pHGIN-WT plasmid. Thus, for clone 18 the R613W exchange was not present in the reconstituted variant. Vector particles were produced in HEK-293T cells by cotransfection with the corresponding pHGIN-derived plasmids and pHR-CMVΔ8.2. At first, we addressed the question of whether the selected Env variants were able to mediate the infectivity of the HIV-1 particles. Titrations of the vector stocks were performed on HT1080 cells in the absence of Polybrene to precisely reflect the conditions of the selection procedure. Transduction events were monitored by counting G418-resistant colonies or, alternatively, after cross-packaging of the lacZ reporter gene, by a β-galactosidase assay. Data from both types of titrations were in good accordance and were clear-cut. While the WT GaLV Env protein did not mediate any infectivity, as expected, pseudotyping with all of the selected variants resulted in successful transduction (Fig. 2A). Overall, the titers of the different selected variants were similar, but they were about 1 log lower than those of the RTM and ΔR control variants.
FIG. 2.
Characterization of selected GaLV Env protein variants. HEK-293T cells were cotransfected with pHGIN plasmids encoding the indicated Env variants (see Fig. 1A) and with plasmid pHR-CMVΔ8.2 only. (A) Infectivity mediated by selected variants. Two days after transfection, cell culture supernatants were harvested for infectivity studies. Titers were determined on HT1080 cells in the absence of Polybrene by determining the numbers of neomycin-resistant colonies (light gray bars). Alternatively, the β-galactosidase reporter gene was cross-packaged into the vector particles, and the numbers of transduced cells were determined by a β-galactosidase assay (dark gray bars). (B) Cell surface expression. The cotransfected HEK-293T cells were analyzed by flow cytometry using anti-HA antibodies. The mean fluorescence values are shown. (C) Processing and HIV particle incorporation of GaLV TM protein variants. Virus particles were separated in sodium dodecyl sulfate-16% polyacrylamide gels, transferred to a nitrocellulose membrane, and stained with anti-HIV p24 (top) and anti-MLV TM (bottom) antibodies. The positions of the unprocessed (TM) and processed (TM-R) TM proteins are indicated.
Since a down regulation of GaLV Env protein expression in HIV-1 Gag-Pol-expressing cells has been reported (8), we determined the cell surface expression levels of the selected variants in pHR-CMVΔ8.2-transfected HEK-293T cells. At 2 days posttransfection, the cells were analyzed by FACS with an anti-HA tag antibody. Equally efficient transfections for all of the plasmids were verified by Western blot analyses of neomycin phosphotransferase expression (not shown). All selected variants showed about two- to threefold higher cell surface expression levels than the parental Env protein (Fig. 2B). Thus, these levels were in the same range as that in the RTM variant and about twofold lower than that in the ΔR variant.
To analyze HIV particle incorporation and to monitor the R-peptide cleavage of the selected Env protein variants, we concentrated the particles by ultracentrifugation and analyzed them by Western blotting with an anti-TM antibody. TM molecules were detectable in all of the different particles and, remarkably, also after expression of the parental WT Env protein (Fig. 2C). Although the signal intensity was by far the lowest for the WT Env protein, its detection was unexpected based on the results of previous reports (8). When the relative intensities of the bands derived from the TM proteins were normalized to those derived from the HIV p24 capsid protein, particle incorporation rates could be determined for the different Env variants. The incorporation of the parental TM molecule was <2% that of the ΔR or RTM-derived TM protein. All of the selected variants showed incorporation rates that were at least 8-fold (variant 4) or, in the case of variants 2 and 19, even 60- to 70-fold above that of the parental TM protein, thus being in the range of those of the RTM and ΔR variants.
The WT TM protein appeared as a single band corresponding to the uncleaved TM protein (Fig. 2C, lane 2). Notably, even after a long-time exposure of the blot, no TM cleavage product became detectable (not shown). In contrast, the RTM variant as well as all of the selected variants showed two distinct bands indicating that the R peptide was cleaved off. Processing was especially efficient for clone 18, which had a processing rate of almost 90%, while this rate was below or at about 50% in the case of the RTM variant and the other selected variants (Fig. 2C, lanes 4 to 8). Notably, the processed TM polypeptides of variants 2, 4, and 19 showed reduced electrophoretic mobilities compared to the ΔR TM protein. This may have been due to the amino acid changes in the membrane-proximal parts of their C tails and/or to a shift of the cleavage site towards the C terminus of the TM protein.
To further investigate the proteolytic process of the selected variants, we generated HIV particles with the different GaLV Env variants in the presence of the HIV protease inhibitor saquinavir. Western blot analysis of the HIV p24 protein revealed that HIV protease activity was completely blocked, as only the Gag precursor protein was detectable (not shown). Similarly, proteolytic processing of the TM proteins of all of the selected variants and of the RTM variant was completely inhibited (Fig. 3A). These data prove that the selected Env proteins were activated by HIV protease.
FIG. 3.
Molecular analysis of R-peptide processing in selected Env proteins. HEK-293T cells were cotransfected with pHGIN plasmids encoding the indicated Env variants (see Fig. 1A) and with plasmid pHR-CMVΔ8.2. Two days after transfection, cell culture supernatants were harvested and virus particles were concentrated by ultracentrifugation. Virus particles were separated in sodium dodecyl sulfate-16% polyacrylamide gels, transferred to a nitrocellulose membrane, and stained with anti-MLV TM antibodies. (A) Inhibition of R-peptide processing by saquinavir. Where indicated, transfected cells were cultivated in the presence of saquinavir (Saq.). (B) Western blot analysis of point mutants of clones 2 and 4. Stop codons are indicated by asterisks.
In order to identify the cleavage site in the C tails of variants 2 and 4, we generated a series of C-terminal truncation mutants. All mutations were introduced by the insertion of double-stranded oligonucleotides into the pHGIN-WT plasmid. Sequence analysis confirmed the identities of the constructs. All of the constructs were transfected into HEK-293T cells together with the plasmid pHR-CMVΔ8.2 to generate particles. To prevent any processing in the case of truncated variants, we generated the particles in the presence of saquinavir. Western blot analysis of the particles containing the clone 2- and 4-derived variants demonstrated that only truncations that were located C-terminally of residue Q624 resulted in retarded electrophoretic mobilities, similar to those observed for the cleavage products of clones 2, 4, and 19 (Fig. 3B, lanes 1 to 5; Table 1). The replacement of residue 627 with a stop codon still allowed syncytium formation by HEK-293T cells expressing the corresponding Env variants of clones 2 and 4, suggesting that proteolytic cleavage of the N-terminal peptide bond of this residue will activate membrane fusion (Table 1). Thus, based on the truncation mutants, K625 and Y626 were the most likely P1 residues of the HIV protease cleavage site.
TABLE 1.
Properties of truncation mutants of selected Env proteins
| Property | Result for indicated varianta
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| ΔR | 2-622* | 2-623* | 2-625* | 2-627* | 4-622* | 4-623* | 4-625* | 4-627* | |
| Gel migrationb | Unchanged | Unchanged | Unchanged | Retarded | Unchanged | Unchanged | Unchanged | Retarded | |
| Syncytium formationc | ++ | ++ | ++ | + | + | ++ | ++ | + | + |
Variants were named according to the position of the inserted stop codon.
Gel migration was compared to that of the ΔR variant.
Syncyitia included, on average, 5 to 10 (+) or >20 (++) nuclei.
We next exchanged residues L622, Q624, K625, and Y626 with valine residues, which strongly reduce or abolish HIV protease activity when introduced at the P1 position (14). For both selected variants, the exchange of residue K625 abolished proteolytic processing, while the processing of all other variants was unaltered or, rather, enhanced (Fig. 3B, lanes 6 to 15). In particular, the Q624V mutation enhanced the proteolytic activity of clone 2, while the L622V mutation shifted the cleavage site back to a position at or near that used by the RTM variant (Fig. 3B, lanes 6 and 7). Also, note the increased electrophoretic mobilities of the two K625V mutants, which demonstrate that the positive charge of this lysine residue reduces the electrophoretic mobility of the complete TM protein (compare lanes 8 and 9 for clones 2 and lanes 14 and 15 for clone 4). The unexpectedly slow mobilities of the clone 2, 4, and 19 TM processing products can therefore be explained by the presence of the K625 residue. Taken together, these data indicate that K625 is the most likely candidate for the P1 residue in the selected clones 2 and 4 and the closely related clone 19 (Fig. 1A).
DISCUSSION
In this report, we described an approach that allows us to monitor the molecular evolution of cytoplasmic tails of viral Env proteins when observed in the context of the structural proteins of other viruses. So far, directed evolution systems for retroviral Env proteins that have been diversified in their extracellular parts and screened for extension of the host range to include nonpermissive cells have been described (1, 5, 6, 18). The selection process established here is highly efficient. Within two rounds of selection, several GaLV Env variants mediating the infectivity of HIV particles were selected, starting from the completely nonfunctional parental GaLV Env WT protein. On the molecular level, the selected Env variants showed enhanced cell surface expression as well as strongly enhanced particle incorporation rates. Although it occurred at low rates, the parental Env protein was incorporated into HIV particles, as determined by the use of anti-TM as well as anti-HA antibodies detecting the SU subunit of Env (not shown). In contrast to incorporation, R-peptide cleavage was undetectable for the parental Env protein but was highly efficient for the selected Env proteins. Since 7 to 14 Env molecules per particle are sufficient to mediate retroviral infectivity, we concluded that R-peptide cleavage was the key issue for the selected variants to become functional and was the main driving force in our selection process (7). This extends and in part also adjusts results from a recent publication that identified expression and incorporation levels as blocking the GaLV/HIV-1 pseudotype phenomenon (8).
An interesting outcome of this study was the variability in the position of the R-peptide cleavage site that can be used by the HIV protease. At least two different sites could be distinguished. Selected variant 18 showed a stretch of hydrophobic residues within the diversified area. Based on the electrophoretic mobility of the cleavage product and the preference of the HIV protease for hydrophobic and aromatic residues in the vicinity of the cleaved peptide bond, this variant must be cleaved at or in close proximity to the R-peptide cleavage site that is used in the wild-type situation by GaLV. In contrast, the cleavage site(s) in the other three selected variants was shifted more toward the C terminus, most likely to residue K625. Site-directed mutagenesis excluded the neighboring residues Q624 and Y626 as P1 sites. The relatively long distance to the critical residues 618, 619, and 623 for functional incorporation of the GaLV Env into HIV particles makes residues located one or two positions further toward the C terminus from Y626 highly unlikely candidates for the P1 site. Although positively charged residues like K625 are rather unusual as P1 amino acids in HIV protease substrates, they have been previously described to be effective at some sites, e.g., the nucleocapsid-protease site in HIV-1 (9). It should also be kept in mind that the R peptide is membrane anchored via a so far unidentified palmitoylated residue, a modification that might alter HIV protease substrate recognition in this case (13). Yet variant 2-Q624V (VK↓YQ), which was most efficiently processed at this site, comes close to the HyHy↓HyQ consensus of class 3 substrate peptides of the HIV-1 protease (2).
Although a similar site of R-peptide processing has not been identified for other gammaretroviruses, cleavage at this site should activate the membrane fusion function of Env equally well. Previous studies of the related MLV R peptide (16 residues) demonstrated that syncytium formation by transfected cells, a correlate for the fusion capacity of Env, is possible with truncation mutants missing the C-terminal eight residues only (20, 21). Correspondingly, we observed that truncation at residue 626 in two of the selected clones still allowed syncytium formation. In this respect, it should also be mentioned that the R-peptide cleavage site in D-type retroviruses is in fact closer to the C terminus than that in MLV and is thus more similar to the site we identified here for some of the selected GaLV variants (4).
Overall, the selection procedure described here will be of high value for basic questions addressing retroviral particle assembly as well as for the engineering and optimization of chimeric viruses or vector particles.
Acknowledgments
This work was supported by grants BU1301/1-1 and BU1301/1-2 from the Deutsche Forschungsgemeinschaft to C.J.B. C.A.M. was supported by a Ph.D. scholarship of the Fonds der Chemischen Industrie and the Bundesministerium für Bildung und Forschung.
REFERENCES
- 1.Bahrami, S., T. Jespersen, F. S. Pedersen, and M. Duch. 2003. Mutational library analysis of selected amino acids in the receptor binding domain of envelope of Akv murine leukemia virus by conditionally replication competent bicistronic vectors. Gene 315:51-61. [DOI] [PubMed] [Google Scholar]
- 2.Beck, Z. Q., G. M. Morris, and J. H. Elder. 2002. Defining HIV-1 protease substrate selectivity. Curr. Drug Targets 2:37-50. [DOI] [PubMed] [Google Scholar]
- 3.Bobkova, M., J. Stitz, M. Engelstadter, K. Cichutek, and C. J. Buchholz. 2002. Identification of R-peptides in envelope proteins of C-type retroviruses. J. Gen. Virol. 83:2241-2246. [DOI] [PubMed] [Google Scholar]
- 4.Brody, B. A., S. S. Rhee, and E. Hunter. 1994. Postassembly cleavage of a retroviral glycoprotein cytoplasmic domain removes a necessary incorporation signal and activates fusion activity. J. Virol. 68:4620-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchholz, C. J., K. W. Peng, F. J. Morling, J. Zhang, F. L. Cosset, and S. J. Russell. 1998. In vivo selection of protease cleavage sites from retrovirus display libraries. Nat. Biotechnol. 16:951-954. [DOI] [PubMed] [Google Scholar]
- 6.Bupp, K., and M. J. Roth. 2003. Targeting a retroviral vector in the absence of a known cell-targeting ligand. Hum. Gene Ther. 14:1557-1564. [DOI] [PubMed] [Google Scholar]
- 7.Chertova, E., J. W. Bess, Jr., B. J. Crise, R. C. Sowder II, T. M. Schaden, J. M. Hilburn, J. A. Hoxie, R. E. Benveniste, J. D. Lifson, L. E. Henderson, and L. O. Arthur. 2002. Envelope glycoprotein incorporation, not shedding of surface envelope glycoprotein (gp120/SU), is the primary determinant of SU content of purified human immunodeficiency virus type 1 and simian immunodeficiency virus. J. Virol. 76:5315-5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christodoulopoulos, I., and P. M. Cannon. 2001. Sequences in the cytoplasmic tail of the gibbon ape leukemia virus envelope protein that prevent its incorporation into lentivirus vectors. J. Virol. 75:4129-4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grinde, B., C. E. Cameron, J. Leis, I. T. Weber, A. Wlodawer, H. Burstein, and A. M. Skalka. 1992. Analysis of substrate interactions of the Rous sarcoma virus wild type and mutant proteases and human immunodeficiency virus-1 protease using a set of systematically altered peptide substrates. J. Biol. Chem. 15:9491-9498. [PubMed] [Google Scholar]
- 10.Loewen, N., C. Bahler, W. Teo, T. Whitwam, M. Peretz, R. Xu, M. Fautsch, D. H. Johnson, and E. M. Poeschla. 2002. Preservation of aqueous outflow facility after second-generation FIV vector-mediated expression of marker genes in anterior segments of human eyes. Investig. Ophthalmol. Vis. Sci. 43:3686-3690. [PubMed] [Google Scholar]
- 11.Mochizuki, H., J. P. Schwartz, K. Tanaka, R. O. Brady, and J. Reiser. 1998. High-titer human immunodeficiency virus type 1-based vector systems for gene delivery into nondividing cells. J. Virol. 72:8873-8883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naldini, L., U. Blomer, P. Gallay, D. Ory, R. Mulligan, F. H. Gage, I. M. Verma, and D. Trono. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272:263-267. [DOI] [PubMed] [Google Scholar]
- 13.Olsen, K. E., and K. B. Andersen. 1999. Palmitoylation of the intracytoplasmic R peptide of the transmembrane envelope protein in Moloney murine leukemia virus. J. Virol. 73:8975-8981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petit, S. C., G. J. Hederson, C. A. Schiffer, and R. Swanstrom. 2002. Replacement of the P1 amino acid of human immunodeficiency virus type 1 Gag processing sites can inhibit or enhance the rate of cleavage by the viral protease. J. Virol. 76:10226-10233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pinter, A., W. J. Honnen, J. S. Tung, P. V. O'Donnel, and U. Hammerling. 1982. Structural domains of endogenous murine leukemia virus gp70s containing specific antigenic determinants defined by monoclonal antibodies. Virology 116:499-516. [DOI] [PubMed] [Google Scholar]
- 16.Ragheb, J. A., and W. F. Anderson. 1994. pH-independent murine leukemia virus ecotropic envelope-mediated cell fusion: implications for the role of the R peptide and p12E TM in viral entry. J. Virol. 68:3220-3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rein, A., J. Mirro, J. G. Haynes, S. M. Ernst, and K. Nagashima. 1994. Function of the cytoplasmic domain of a retroviral transmembrane protein: p15E-p2E cleavage activates the membrane fusion capability of the murine leukemia virus Env protein. J. Virol. 68:1773-1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schneider, R. M., Y. Medvedovska, I. Hartl, B. Voelker, M. P. Chadwick, S. J. Russell, K. Cichutek, and C. J. Buchholz. 2003. Directed evolution of retroviruses activatable by tumour-associated matrix metalloproteases. Gene Ther. 10:1370-1380. [DOI] [PubMed] [Google Scholar]
- 19.Stitz, J., C. J. Buchholz, M. Engelstadter, W. Uckert, U. Bloemer, I. Schmitt, and K. Cichutek. 2000. Lentiviral vectors pseudotyped with envelope glycoproteins derived from gibbon ape leukemia virus and murine leukemia virus 10A1. Virology 273:16-20. [DOI] [PubMed] [Google Scholar]
- 20.Yang, C., and R. W. Compans. 1997. Analysis of the murine leukemia virus R peptide: delineation of the molecular determinants which are important for its fusion inhibition activity. J. Virol. 71:8490-8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao, Y., L. Zhu, C. A. Benedict, D. Chen, W. F. Anderson, and P. M. Cannon. 1998. Functional domains in the retroviral transmembrane protein. J. Virol. 72:5392-5398. [DOI] [PMC free article] [PubMed] [Google Scholar]