Abstract
Background
Mast cell (MC) progenitors leave the bone marrow, enter the circulation, and settle in the skin and other tissues. Their maturation in tissues is influenced by the surrounding microenvironment.
Objective
We tested the hypothesis that environmental factors play a role in MC maturation in the skin.
Methods
MCs were numerically, phenotypically, and functionally compared between germ-free (GF), SPF, and GF mice reconstituted with microbiota. Maturity of MCs was then correlated with skin levels of stem cell factor (SCF), a critical MC differentiation factor, and lipoteichoic acid (LTA), a TLR2 ligand. MCs were also evaluated in mice with keratinocyte-specific deletion of Scf.
Results
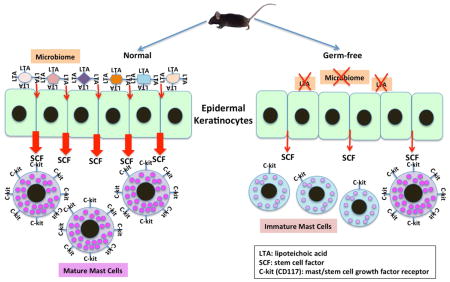
We found that GF mice express abnormally low amounts of stem cell factor (SCF), a critical MC differentiation factor, and contain MCs that are largely undifferentiated. Reconstituting the GF microbiota reverted this MC phenotype to normal, indicating that the phenotype is related to ongoing interactions of microbiota and skin. Consistent with the immaturity of GF MCs, degranulation-provoking compound 48/80 induced less edema in the skin of GF mice than in conventional mice. Our results show that the skin microbiome drives SCF production in keratinocytes, which triggers the differentiation of dermal MCs. Since the skin microbiome is a rich source of lipoteichoic acid (LTA), a TLR2 ligand, we mimicked the GF microbiome impact on MCs by applying LTA to the skin of GF mice. We also demonstrated that MC migration within the skin depends exclusively on keratinocyte-produced SCF.
Conclusion
This study has revealed a novel mechanism by which the skin microbiota signals the recruitment and maturation of MCs within the dermis via SCF production by LTA-stimulated keratinocytes.
Keywords: Mast cell, microbiome, germ free, SCF, keratinocyte, LTA
Graphical abstract
INTRODUCTION
Changes in the microbiome have been connected with different skin diseases such as atopic dermatitis, acne, and psoriasis 1. Specific commensal bacteria, such as Staphylococcus epidermidis, have been implicated in the modulation of skin inflammation 2 and recent studies suggest that the skin microbiota may play a role in the recruitment, accumulation, and function of various immune cells such as the modulation of T cell differentiation 1. Despite the essential role of mast cells (MCs) in several skin diseases 3, 4, little is known about whether or how the skin microbiome modulates the differentiation, survival, and function of MCs.
MCs are localized in close proximity to the environment. They originate from CD34+ bone marrow progenitor cells, migrate into the circulation and subsequently into peripheral tissues where their final differentiation takes place under the influence of cytokines in the surrounding tissue microenvironment 5–7. The tissue-specific influence is important for promoting MC accumulation and survival in the skin and gastrointestinal tract 8–10. Interestingly, a recent study demonstrated that the skin microbiota can extend into the dermis, thereby establishing physical contact between bacteria and various cells below the basement membrane. This contact allows normal commensal bacteria to directly communicate with dermal cells in a site previously thought to be sterile 11. The Gram-positive Actinobacteria phylum constitutes the majority of the skin microbiota 12. As the Gram-positive cell wall contains a high amount of lipoteichoic acid (LTA), this TLR2 ligand is one of the most abundant molecules on the surface of the skin. We have previously shown that LTA permeates the whole skin 13 and increases the anti-microbial capacity of skin MCs to defend against viral infections 14, 15. Thus, LTA derived from the skin microbiome is a candidate agonist for modulating MC biology.
Stem cell factor (SCF) is essential for the differentiation, survival, and migration of MCs 6, 16–19. SCF can be produced in a regulated fashion by various skin cells 16, 20, including keratinocytes 21, although the relevant stimuli remain to be explored. The microbiome has primary contact with the epidermis, particularly keratinocytes 11, but it is not known whether SCF production by keratinocytes can be modulated by the skin microbiome. Earlier studies have shown that murine MCs do not fully mature until 8 to 15 days after birth 22, 23, supporting the notion that environmental factors may drive the production of SCF or other factors that allow for MC differentiation in the skin. In this study, we have investigated this possibility by focusing on the role of commensal bacteria and their major product, LTA, in MC differentiation by stimulating keratinocytes to produce SCF in different conventional, gene-targeted, and gnotobiotic murine models.
RESULTS
Germ-free mice have immature mast cells in the dermis
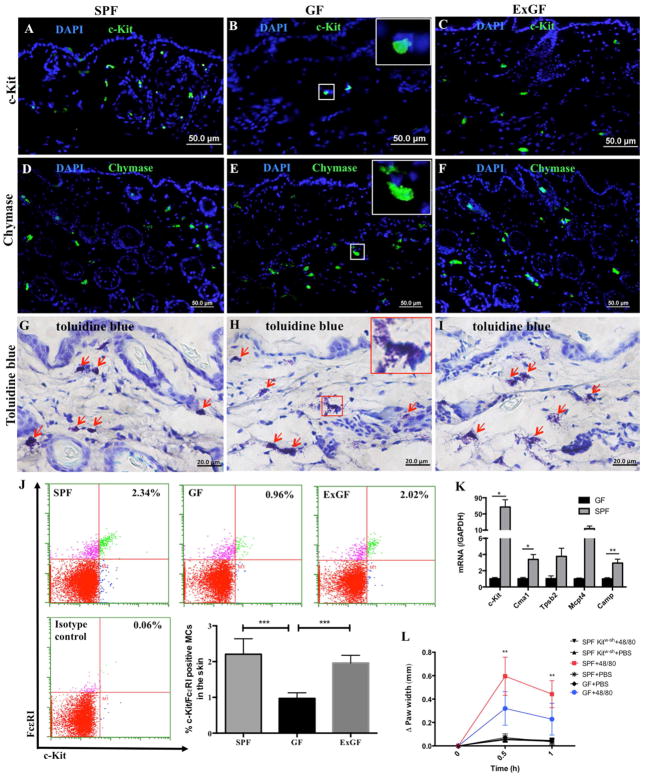
To determine the importance of the microbiome in MC maturation, we stained for the presence of c-Kit positive MCs in the skin of germ free (GF) mice, specific pathogen-free (SPF) conventional mice, and GF mice co-housed with SPF mice (ExGF) for 5 weeks to reconstitute their microbiome (as confirmed by bacterial plate cultures of gut microflora). We observed a significantly smaller population of c-Kit positive MCs in the GF mice (Figures 1A–C) that was normalized after bacterial reconstitution in the ExGF mice (isotype antibody control staining for all of the experiments are in Figure E1A–B). To extend this observation, dermal and epidermal cells were harvested from the skins of the three mouse populations and MCs were enumerated by FACS based on the presence of both the SCF receptor (c-Kit) and the high affinity IgE receptor (FcεRI) (Figure 1J). FACS confirmed that the skin of GF mice had markedly decreased numbers of c-Kit+ FcεRI+ MCs, while ExGF mice had normal MC numbers in the skin (Figure 1J).
Figure 1. Germ-free mice have immature mast cells in the dermis.
(A–C) Immunofluorescent staining for c-Kit (green) and DAPI (blue) in skin from SPF, GF, and ExGF (GF co-housed with SPF for 5 weeks) mice; (D–F) Immunofluorescent staining for chymase positive cells (green) and DAPI (blue) in skin from SPF, GF, and ExGF mice; (G–I) Toluidine blue staining of skin from SPF, GF, and ExGF mice (red arrows indicate MCs and the inset (in B, E and H) shows a magnification of the squared area in the image); (J) Flow cytometry plots and enumeration of mature MC numbers in samples of whole skin from SPF, GF, and ExGF mice; (K) qPCR analysis for MC markers in GF and SPF skin using the toluidine blue positive MCs collected from skin sections by laser capture microdissection; (L) Paw thickness in SPF, GF, and SPF MC-deficient Kitw-sh mice after injection of PBS (control) or compound 48/80. (*p<0.05, **p<0.01, ***p<0.001)
In parallel with these experiments, we stained skin sections for chymase and with toluidine blue to detect all mast cells regardless of functional state (Figure 1D–I). Surprisingly, these staining methods revealed similar numbers of skin MCs in GF, SPF and ExGF mice (Figure 1D–I), suggesting that MCs were present in GF mice, but that these cells were not positive for markers of differentiated cells such as c-Kit. To better characterize the phenotype of the MCs from GF, SPF, and ExGF mice, we isolated toluidine blue-positive cells from skin sections of these mice by laser capture microdissection (LCM). After RNA extraction, we assessed the mRNA expression profiles of several genes involved in and characteristic of MC maturation, including chymase (Cma1), tryptase beta 2 (Tpsb2), MC protease 4 (Mcpt4), SCF receptor (c-Kit), and cathelicidin antimicrobial peptide (Camp). As shown in Figure 1K, MCs from SPF mice expressed higher levels of all MC markers compared to MCs from GF mice, especially c-Kit, explaining the reduced numbers of c-Kit+ MCs in GF mice. Controls for the LCM tissue sections were negative for toluidine blue and in these sections, the genes of interest were undetectable (Figure E 1C–F). These results suggest that the skin microbiome markedly impacts MC gene expression and maturation.
To assess any functional differences between MCs in GF and SPF mice, we injected 0.5 μg (10 μl) of compound 48/80 (a substance known to induce MC degranulation) into the hind paws of three mice from each population. At 30 minutes and at 1 hour after injection, paw width was measured to assess for swelling due to 48/80-induced MC degranulation. The paws of SPF mice swelled to a greater extent than the paws of GF mice, suggesting that MCs in GF mice have altered functionality (Figure 1L).
Staphylococcal LTA promotes mast cell maturation in GF mice
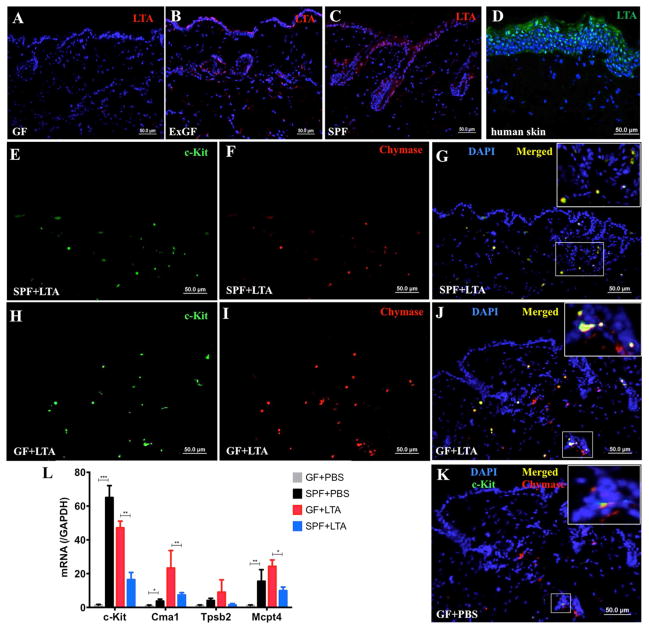
Given that LTA can have a strong influence on MC behavior in infections 14, 24 we hypothesized that the lack of microbiome, and thus the predicted lack of microbial-derived LTA in GF mice may account for the observed differences in MC maturation. To test this hypothesis, we first confirmed by immunostaining that LTA was indeed absent in the skin of GF mice (Figure 2A), whereas LTA could be clearly seen in the epidermis and hair follicles of ExGF and SPF mice (Figure 2B–C; isotype antibody control staining for these experiments are in Figure E 1G). As shown in Figure 2D, in human skin, LTA was not only present on the skin surface, but also found in the deeper layers of epidermal keratinocytes. As a staining control, we cultured normal human epidermal keratinocytes in vitro with and without LTA and stained with an anti-LTA monoclonal antibody. No LTA was detected in the keratinocytes unless they were treated with LTA (Figures S1H–J). Taken together, our data suggest that the skin is positive for LTA only in the presence of an intact microbiome.
Figure 2. Staphylococcal LTA promotes mast cell maturation in GF mice.
(A–C) Anti-LTA staining (red) and DAPI (blue) in skin from GF, ExGF, and SPF mice; (D) Anti-LTA staining (green) and DAPI (blue) at the human skin surface and in deeper epidermal layers; (E–J) Immunofluorescent staining for c-Kit (green), chymase (red), and DAPI (blue) in skin sections from SPF and GF mice, as indicated, after LTA injection. The inset (in Figure G, J and K) shows magnification of the squared area in the image; (K) Immunofluorescent staining for c-Kit (green), chymase (red), and DAPI (blue) in the skin of GF mice following PBS injection; (L) qPCR analysis for MC markers in GF and SPF skin using toluidine blue positive MCs collected from skin sections by laser capture microdissection after LTA injection. (*p<0.05, **p<0.01, ***p<0.001)
Next, to determine whether LTA can play a direct role in promoting MC maturation, we injected the backs of GF and SPF mice with 100 μl LTA (1 mg/ml). Immunofluorescent staining of the LTA-injected mouse skin 24 hours after injection revealed increased numbers of c-Kit positive MCs in both SPF and GF mice (Figure 2E–J); chymase expression also changed, compatible with qPCR expression in Figure 2J. For comparison, staining of skin from GF mice treated only with PBS is shown in Figure 2K. To confirm and extend the immunofluorescence studies, skin sections from the same mice were stained with toluidine blue, and toluidine blue-positive MCs were isolated by LCM and analyzed for changes in gene expression by qPCR. The dermal MCs from GF mice showed dramatically increased expressions of Cma1, Tpsb2, Mcpt4, and c-Kit (Figure 2L), demonstrating that the final maturation of MCs is influenced by bacterial products from the skin environment.
Staphylococcal LTA-induced MC maturation is not dependent on MC TLR2 expression
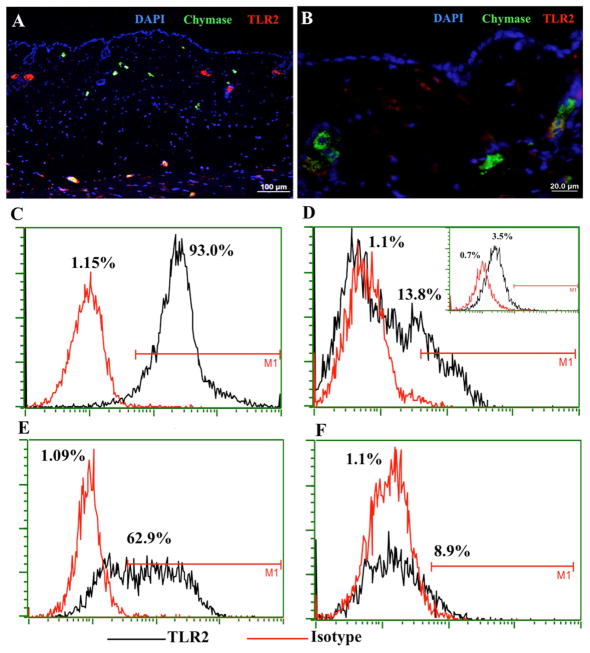
In order to determine the mechanism of LTA action, we first stained for the presence of TLR2, the receptor for LTA, on MCs in SPF mouse skin. Surprisingly, TLR2 was found to co-localize with MC chymase in the deeper layers of the dermis (Figure 3A), whereas in the upper dermal layer, MCs lacked TLR2 expression (Figure 3A–B). Isotype antibody control staining for this experiment is shown in Figure E 1K. This observation was also true for MCs in human skin (data not shown). The downregulation of Tlr2 in dermal mouse MCs was confirmed by qPCR on MCs collected by LCM (Figure E 1L).
Figure 3. TLR2 MC expression in the skin.
(A) SPF mouse skin was stained with anti-chymase (green), anti-TLR2 (red), and DAPI (blue) to define TLR2 presence on dermal MCs. TLR2 is visible in the deeper layer of dermis (yellow cells at the bottom) and green cells indicate MCs that do not express TLR2; (B) Magnification of chymase positive MCs that do not express TLR2. (C–D) FACS plots and enumeration of murine bone marrow-derived MCs that were differentiated in vitro without (C) or with (D) dermal fibroblasts for 7 days, stained with anti-TLR2 antibody, and then analyzed by FACS. The inset in (D) shows the expression of TLR2 in MCs derived from the bone marrow of Tlr2−/− mice; (E–F) Human CD34+ cord blood cell-derived MCs were differentiated in vitro without (E) or with (F) fibroblasts for 7 days, stained with anti-TLR2 antibody, and then analyzed by FACS.
Seeking to find an explanation for the observation that mast cells lose their TLR2 expression when entering the skin, we tried to co-culture MCs with different components of skin connective tissue, including fibroblasts (human fibroblasts with human MCs and mouse fibroblasts with mouse MCs), hyaluronic acid, collagen type I, fibronectin, or gelatin. After 7 days of co-culture, we performed FACS analysis on the MCs to determine the levels of TLR2 expression. Murine MCs differentiated from bone marrow and human MCs differentiated from CD34+ cord blood cells both showed significant expression of TLR2 when cultured alone (Figures 3C and E). Interestingly, after one week of co-culture with fibroblasts, a significant percentage of both murine and human MCs lost their expression of TLR2 (Figures 3D and F), resembling MCs cultured from the bone marrow of Tlr2 knockout mice that we used as a control (Figure 3D inset). In contrast, MCs that were cultured with extracellular matrix components did not lose their expression of TLR2 (Figure E 2). Therefore, we conclude that the direct contact of MC’s with fibroblasts promotes the downregulation of TLR2. Consequently, the effect of LTA on MC maturation is not via MC TLR2 expression, suggesting that LTA does not exert its effect directly on MCs, but is probably acting indirectly through TLR2 on other types of cells in the skin such as keratinocytes. Keratinocytes represent the interphase with the external environment and are definitely the cells that are most exposed to the skin microbiome. To verify Tlr2 expression in mouse keratinocytes, we isolated keratinocytes from mouse skin and showed by qPCR analysis that they were positive for Tlr2 (Figure E 2I) and that their expression of Tlr2 was increased by exposure to LTA.
Staphylococcal LTA promotes SCF expression in the epidermis
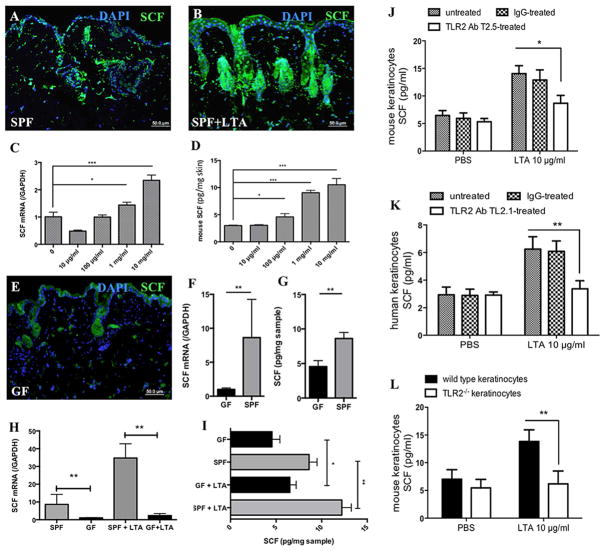
Given that SCF is the main inducer of MC differentiation 25, we hypothesized that SCF provides a possible link between LTA and MCs. To determine whether SCF is produced in the skin in response to commensal bacteria in vivo, we injected 100 μl of 1 mg/ml LTA into the skins of SPF mice. A significantly enhanced SCF signal was detected by immunofluorescent staining 24 hours after injection (Figure 4A–B). To extend this finding, we stimulated mice with different doses of LTA and after 24 h, dissected the skin and analyzed Scf (Kit ligand) mRNA expression by quantitative RT-PCR. As shown in Figure 4C, Scf expression at the site of injection increased significantly in a dose-dependent manner when compared to baseline. Furthermore, the observed increases in Scf mRNA resulted in corresponding increases in SCF protein levels (Figure 4D). To reveal the sources of SCF in response to LTA, the main resident cell types in the skin were also analyzed for SCF expression by qPCR and ELISA. We found that keratinocytes expressed high levels of SCF mRNA seven hours after LTA challenge (Figure E 3A), with an accompanying increase in SCF protein 16 hours after LTA stimulation (Figure E 3B). We also compared Scf expression in mouse fibroblasts versus keratinocytes in vitro. Both cell types upregulated Scf in response to LTA to similar degrees, but keratinocytes did so considerably faster than fibroblasts (Figure E 3C).
Figure 4. SCF expression in keratinocytes is low in the absence of microbiome.
(A–B) Anti-SCF (green) and DAPI (blue) immunostaining of SPF mouse skin after injection of PBS (A) or LTA (B); (C–D) Quantification of Scf expression in mouse skin by qPCR (C) and by ELISA (D) after LTA injection. (E) Anti-SCF (green) and DAPI (blue) immunostaining of GF mouse skin; (F) qPCR analysis of Scf in epidermis isolated from GF and SPF mice; (G) ELISA quantification of SCF in epidermis isolated from GF and SPF mice (normalized by tissue weight); (H) qPCR analysis of Scf in isolated epidermis of GF and SPF mice before and after injection of LTA; (I) ELISA quantification of SCF in isolated epidermis of GF and SPF mice (normalized by tissue weight) before and after injection of LTA; (J) ELISA quantification of SCF in mouse keratinocytes pre-treated for 1 h with either anti-TLR2 antibody Clone T2.5, or with the relevant isotype IgG control; (K) ELISA quantification of SCF in human keratinocytes pre-treated for 1 h with either anti-TLR2 antibody Clone TL2.1 or with the relevant isotype IgG control; (L) ELISA quantification of SCF in isolated mouse keratinocytes from Tlr2−/− and wild-type mice. (*p<0.05, **p<0.01, ***p<0.001)
To further investigate the role of LTA-induced expression of SCF in the skin, GF mice were analyzed for SCF expression (Figure 4E). Our results showed that GF mice exhibited lower expression levels of SCF protein in the skin when compared to SPF mice. To better evaluate the epidermal SCF levels in these mice, we separated epidermal cells from the dermis using dispase. SCF mRNA and protein levels in these isolated epidermal samples were much higher in SPF mice than in GF mice (Figure 4F–G). Moreover, when challenged with LTA, epidermal samples from SPF mice showed greater increases in SCF expression compared to samples from GF mice (Figure 4H–I).
It is known that keratinocytes express TLR2 in vivo and in vitro 26 and we confirmed by qPCR that keratinocytes increase Tlr2 expression in response to LTA (Figure E 2I); however, there are no reports that link TLR2 stimulation directly to SCF expression. Therefore, to investigate the mechanism of SCF induction by LTA in keratinocytes, we treated mouse keratinocytes and normal human keratinocytes with LTA for 24 h in the absence or presence of neutralizing anti-human TLR2 antibodies (for human keratinocytes) or anti-mouse TLR2 antibodies (for mouse keratinocytes), and measured SCF release in the medium by ELISA. LTA stimulation significantly increased SCF secretion in keratinocytes, which was blocked by anti-TLR2 antibodies (Figure 4J–K). In confirmation of these findings, keratinocytes from wild-type, but not Tlr2−/−, mice showed induction of SCF release upon LTA stimulation (Figure 4L). These data show that TLR2 is absolutely required for LTA-induced SCF expression in keratinocytes.
SCF from keratinocytes is essential for maintaining MCs in the skin
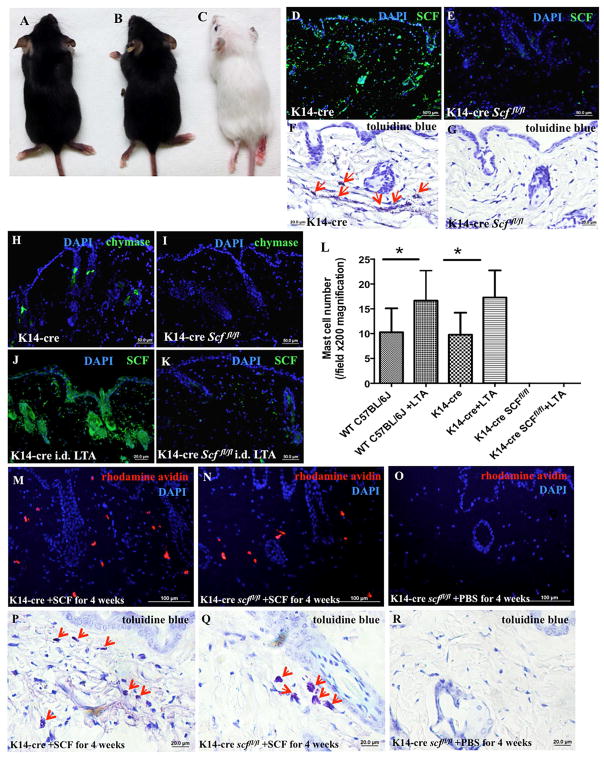
Based on our immunofluorescent staining, LTA induces the strongest expression of SCF in keratinocytes (Figure 4A–B); therefore, we sought to determine the relative importance of SCF in supporting MCs within the skin and in delivering signals from the microbiome. For these experiments, we bred K14-cre transgenic mice that express Cre recombinase under the control of the human keratin 14 promoter with mice containing floxed Scf alleles (Scffl/fl mice) to generate K14-cre Scffl/fl mice, which lack Scf in a keratinocyte-specific manner. These mice are albino, viable, fertile, normal in size, and do not display any gross physical or behavioral abnormalities (Figure 5A–C). As shown in Figure 5D–E, K14-cre Scffl/fl mice, unlike their K14-cre littermate controls, have no SCF protein in their keratinocytes, even as newborns (Figure E 5F and H). In contrast, skin fibroblasts from K14-cre Scffl/fl mice express Scf normally, as shown by anti-SCF antibody staining in vivo (Figure E 3E–F) and in vitro (Figure E 3G–H). Furthermore, these conditional knock-out mice have normal numbers of MCs and normal levels of SCF expression in the small intestinal mucosa (Figure E 5A–D) and bone marrow cells (Figure E 5E), and display no significant abnormalities in WBC counts in the blood (Table E 1).
Figure 5. K14-cre Scffl/fl mice lack SCF in the epidermis and MCs in the dermis.
(A–C) K14-cre (A) and Scffl/fl mice (B) exhibit a normal phenotype, while K14-cre Scffl/fl mice are albino (C); (D–E) K14-cre (D) and K14-cre Scffl/fl (E) mouse skin was immunostained with anti-SCF monoclonal antibody (green) and DAPI (blue); (F–G) Toluidine blue staining of the skins of K14-cre (F) and K14-cre Scffl/fl mice (G), red arrows indicate MCs; (H–I) Skins from K14-cre (H) and K14-cre Scffl/fl (I) mice were immunostained with anti-chymase monoclonal antibody (green) and DAPI (blue); (J–K) Following 100 μl LTA (1 mg/ml) injection, K14-cre (J) and K14-cre Scffl/fl (K) mouse skins were immunostained with anti-SCF monoclonal antibody (green) and DAPI (blue); (L) Quantification of MC numbers by positive toluidine blue staining shows that MC presence could not be rescued by LTA when SCF was conditionally deleted from skin epidermis (*p<0.05). (M–O) Rhodamine-avidin staining (red) and DAPI (blue) for MCs in the skins of K14-cre (M) and K14-cre Scffl/fl mice (N) subcutaneously injected with SCF (50 μg/kg/day) or PBS (O) for 4 weeks; (P–R) Toluidine blue staining for MCs (red arrows indicate MCs) in the skins of K14-cre (P) and K14-cre Scffl/fl mice (Q) subcutaneously injected with SCF (50 μg/kg/day) or PBS (R) for 4 weeks.
Toluidine blue staining (Figure 5F–G) and chymase immunofluorescent staining (Figure 5H–I) of mouse skin sections showed no evidence of MCs in the skin of K14-cre Scffl/fl mice. Notably, they lacked MCs in the dermis, even in newborn mouse pups (Figure E 5H and I). Furthermore, K14-cre Scffl/fl mice lacked melanocytes (Figure E 4A–D), which are known to be dependent on normal SCF expression 27, thus providing a likely explanation for the albino appearance. Together, these results demonstrate that keratinocyte-derived SCF is critical for maintaining normal numbers of MCs and melanocytes in the skin.
To evaluate if LTA stimulation could induce recruitment of MCs into the skin in the absence of keratinocyte-derived SCF, we injected LTA into control (K14-cre) and K14-cre Scffl/fl mice (Figure 5J–K). LTA did not induce SCF expression in K14-cre Scffl/fl mouse keratinocytes, and most importantly, it did not lead to MC recruitment (Figure 5L). In contrast, i.d. injection of LTA increased the number of MCs in the skin of K14-cre littermate control mice (Figure 5L). No inflammation was observed after LTA injection (data not shown), which is in accordance with our previously published data (Wang et al., 2010). Taken together, our data show that LTA signals to MCs by inducing the expression of SCF in keratinocytes. To directly demonstrate that skin MCs are dependent on high levels of SCF, as produced by keratinocytes, we injected SCF into the dermis of K14-cre Scffl/fl mice and examined MC numbers. SCF injection induced MC recruitment into the skin, reaching levels similar to wild-type mice (Figure 5M–R. For comparison, the melanocytes did not normalize upon acute injection of SCF (Figure E 4F), indicating that the melanocyte defect in the absence of SCF is likely to be developmental in nature
DISCUSSION
The importance of SCF in promoting MC differentiation and localization has long been established; however, the specific mechanisms by which MCs are recruited to the skin in order to become terminally differentiated remain unknown. Thus, we sought to determine whether the microbiota of the skin plays a role in dermal MC localization and development. Our studies prove that the microbiome influences MC maturation and functions. We have discovered an important system of communication between the skin microbiome and MC differentiation. The mechanism that we have now established works through the production of SCF in keratinocytes, which is essential for maintaining MCs within the dermis.
At first, we investigated the phenotype and maturation level of MCs in GF mice. When gene expression analysis was performed on MCs, cells from GF mice exhibited far lower gene expression levels than required for MC maturity. More specifically, their expression of c-Kit was greatly reduced. The most impressive and important observation was that when co-housing GF mice with SPF mice, the ExGF mice not only regained their microbiome, but also converted back to a normal mast cell phenotype in the skin. A striking and novel observation was the altered MC function in the absence of the microbiome. In fact, to further confirm that the microbiome is essential to MC maturation and function, we injected the paws of GF mice with 48/80 to induce in vivo MC degranulation and we noticed a significantly reduced inflammatory reaction in germ-free mice. This discovery may be important for a better understanding of the pathogenesis of skin diseases, such as atopic dermatitis, in which changes in the microbiome may be involved in disease flare-ups, and have even been proposed to be responsible for the diseases themselves 28.
The concept of c-Kit negative MCs was first proposed when assessing the MC profile of KitW-sh mice. Kitamura et al. demonstrated that the skin of KitW-sh mouse embryos maintained a high expression of c-Kit, while this expression was nearly abolished five days after birth 29. However, mRNA for MC-specific carboxypeptidase A was still detected in the five-day-old mice, confirming that c-Kit negative MCs may be present in the skin under certain circumstances. When we examined isolated skin MCs by FACS for the presence of c-Kit and the high affinity IgE receptor, a significantly lower number of MCs were detected in GF mice. The data collected by laser capture microdissection indicates that c-Kit negative MCs are present in the skin of GF mice. These same differences between GF and SPF recorded by FACS, were observed in the qPCR analysis of LMC of MCs in GF and SPF mice, indicating that the phenotype of the cells was different in the GF mice.
Our data shows that immature MCs in GF mice are capable of increasing their expression of c-Kit when a normal microbiome is reestablished. Moreover, they also mature upon stimulation with LTA. Immunofluorescent staining confirms that after LTA stimulation, the number of c-Kit positive MCs in the dermis of GF mice more closely resembles the MC composition in the skin of SPF mice. Additionally, when LTA is injected into the skin of GF mice, the expression of MC maturity markers increases drastically; however, when MCs are already exposed to a microbiome, like in SPF mice, the presence of additional LTA maintains these MC markers at a constant level. This confirms a heightened sensitivity of the GF MCs in responding to LTA. Thus, the skin microbiota, and more specifically, some of their byproducts, like LTA, are a key factor in priming MCs for proper development. The changes that we observed after exposing the GF skin with LTA is similar to what we observed with microbiome reconstitution. We chose to use LTA because we knew from our previous work that it had a positive influence on MCs during infections and because it is a defined molecule, as opposed to bacterial supernatant or living commensals that could have raised questions about the mechanism of activity on MCs.
Having established a clear association between the presence of microbiome, LTA, and MC maturation, we sought to determine how LTA exerts this effect on MCs. A recent paper 11 has demonstrated that the microbiota extends within the dermis, therefore enabling physical contact between bacteria and various cells below the basement membrane, establishing that normal commensal bacteria directly communicate with the host. However, we showed that MCs approaching the skin surface lose their TLR2 expression. Another paper has also suggested that MC TLR2 is internalized after MCs enter the skin 30 and we have further confirmed that MCs significantly downregulate their expression of TLR2 upon localization to the skin 30. Moreover we have proven that MCs need fibroblast contact to internalize TLR2. Based on the current knowledge, one might hypothesize that LTA may exert direct activity on MCs entering the skin; however, the fact that in the absence of SCF from keratinocytes MCs cannot re-enter the skin, even after injections of LTA, suggest to us that the effects of bacterial LTA signals must also be transmitted to MCs through a different ligand/receptor or through another cell type.
Because keratinocytes are the cells most exposed to the microbiome, we investigated keratinocyte expression of SCF. Our data suggests that LTA does influence MCs, not via direct interaction, but by triggering the production of SCF in neighboring keratinocytes. Both our mouse models and in vitro assays show that staphylococcal LTA can induce a significant upregulation of SCF in keratinocytes, indicating that a physical interaction between microbes and skin cells can maintain increased SCF levels in the epidermis. Based on these findings we used GF mice to assess whether LTA from the microbiota is necessary for SCF production and MC localization in the skin. Previous works 31 have shown that commensal bacteria play a key role in promoting the migration of MCs into the intestine, which, like the skin, is home to large communities of indigenous bacteria. A few studies have briefly discussed the presence of skin MCs in GF mice 31, 32, but none have provided specific information on their function and location in the skin. Our data demonstrates that GF keratinocytes produce SCF at lower levels compared to SPF mice, providing a connection to the incomplete skin MC maturation that is observed in GF mice.
Based on these results, we developed a mouse model that conditionally depletes SCF from the epidermis (K14-cre Scffl/fl). Analysis of these mice revealed a complete lack of dermal MCs, which not only suggests that SCF from keratinocytes is essential for maintaining MCs in the skin, but that it may be locally produced SCF that maintains MCs within their target organs. From these results, we have gained valuable information: first, that LTA requires keratinocyte-SCF production to transmit the signal to MCs; and second, that without keratinocyte-SCF production, MCs cannot be present in the skin. There is previous work that has shown that overexpressing SCF in keratinocytes leads to increased numbers of MCs in the skin 19; but never (until now) has it been shown that conditionally knocking out Scf from keratinocytes is sufficient to prevent MCs from entering the skin. In this sense, we have developed a useful ideal model for MC knock out studies, with potential application to additional organ systems, such as the gut and lung parenchyma.
Recently, other factors have been implicated in communications between the microbiome and resident immune cells of the skin. It has been shown that skin commensals modulate T-cell development and the immune response through a mechanism involving IL-1 and its receptor antagonist (IL-1ra) 33. According to this study, IL-1 signaling from keratinocytes is diminished in the absence of commensals. Indeed, IL-1α production by cutaneous cells is significantly reduced in GF, relative to SPF, mice and mono-association of GF mice with S. epidermidis restores the production of this cytokine. In contrast, keratinocytes from GF mice display increased levels of Il-1ra mRNA, relative to keratinocytes from SPF mice, and the addition of S. epidermidis to GF mice significantly reduces IL-1ra in keratinocytes 33. These results indicate that commensals control various aspects of functional IL-1 signaling. Our findings mimic this pattern of communication, starting with the microbiome, extending to keratinocytes, and ending with immune cell recruitment.
The findings of this study confirm the concept that both the microbiota and host cells work together to allow for normal skin MC physiology.
CONCLUSIONS
Our results show that the skin microbiome drives SCF production in keratinocytes, which triggers the development of dermal MCs. We further demonstrate that MC localization within the skin depends exclusively on keratinocyte-derived SCF. Thus, we present a novel mechanism by which the skin microbiota signals, through keratinocytes, recruitment of MCs within the dermis and conditions MC functions. This finding opens new perspectives on the possible modulation of the presence and function of mast cells under normal conditions and in different skin diseases.
METHODS
Mice
C57BL/6J wild-type control mice, K14-cre transgenic mice (a gift from Dr. Gallo at the University of California, San Diego), Scffl/fl mice (a gift from Dr. Morrison at the University of Texas Southwestern Medical Center) and Tlr2−/− mice (Jackson Laboratory) on a C57BL/6J background were housed at the University Research Center at the University of California, San Diego (UCSD). Germ-free mice on a C57BL/6 background (Taconic Biosciences) were house in the UCSD Gnotobiotic Mouse Facility. All animal experiments were approved by the UCSD Institutional Animal Care and Use Committee. K14-cre transgenic mice were bred with Scffl/fl mice for the generation of K14-cre Scffl/fl mice. K14-cre littermate controls were used in all experiments. At least three independent experiments were performed to assess reproducibility with at least four mice in each group.
Mast cell-deficient (C57BL/KitW-sh) mice were a donation from Dr. Peter Besmer’s laboratory (Developmental Biology Program, Memorial Sloan-Kettering Cancer Center at Cornell University, New York, NY). These mice have been extensively studied since they were generated 34. Adult KitW-sh mice had a profound deficiency in MCs in all tissues examined, but normal levels of major classes of other differentiated lymphoid cells.
For microbial reconstitution experiments, germ-free mice were co-housed in the same cage for 5 weeks with conventional, genetically matched, littermate mice under specific-pathogen-free (SPF) conditions. After 5 weeks of co-housing, bacterial reconstitution in GF mice was confirmed by CFU assays of fecal homogenates.
In vivo MC activation experiment
Intraplantar (i.pl.) injection was performed with compound 48/80 (0.5 μg/paw; 10 μl) or PBS alone in wild-type mice, germ-free mice, and KitW-sh mice. Change in hindpaw width was measured using digital calipers (±0.01 mm; Mitutoyo), and was calculated as the average of the left and right paw widths. Baseline paw widths for each mouse were taken before treatment and subtracted from post-treatment paw widths to estimate tissue edema 35.
Cell culture
Primary murine MCs were generated following our previously published protocol 24. Briefly, bone marrow cells were cultured in RPMI 1640 medium (Invitrogen) supplemented with recombinant murine IL-3 (1 ng/ml, R&D Systems) and recombinant murine stem cell factor (20 ng/ml, R&D Systems). After 4 weeks, MCs were consistently generated as confirmed by the expression of CD117 (c-Kit) and FcεRI and cell maturation was confirmed by metachromatic staining with toluidine blue. The purity of MCs was greater than 98%.
Primary human mast cells were derived from human cord blood CD34+CD45+ cells (Astarte Biologics) according to Kirshenbaum and Metcalfe 36. Briefly, CD34+CD45+ cells were cultured in serum-free medium (Stemline II, Sigma) containing recombinant human stem cell factor (100 ng/ml, R&D Systems), recombinant human IL-6 (100 ng/ml, R&D Systems), and recombinant human IL-3 (20 ng/ml, R&D Systems, first week only). After 10 weeks, hMCs were consistently generated as confirmed by the expression of CD117 and FcεRI. Cell maturation was confirmed by metachromatic staining with toluidine blue. The purity of hMCs was greater than 98%.
Dermal fibroblasts and epidermal keratinocytes
Primary normal human epidermal keratinocytes from Life Technologies were cultured in EpiLife medium supplemented with EpiLife Defined Growth Supplement and 60 μM CaCl2 (Invitrogen). Subconfluent cells were seeded in 6-, 12-, or 24-well plates and grown to 80% confluence before LTA was added at different concentrations.
Primary human and mouse dermal fibroblasts from iXCells Biotechnologies were cultured in DMEM (Invitrogen) with 10% fetal bovine serum (Hyclone). Subconfluent cells were seeded in 6-well plates and grown to 90% confluence before we added primary murine or human MCs to the well with mast cell growth factors (5×105 mast cells/well) for co-culture.
K14-cre Scffl/fl mouse dermal fibroblasts were isolated from the mouse skin as previously described 37 and cultured in 6-well plates.
Epidermal mouse keratinocytes were isolated from mice as previously described 38. Briefly, the skin was peeled off from neonates and the dermis was gently separated from the epidermis to a single cell solution. Cells were cultured in 6-well plates in supplemented EpiLife medium (Invitrogen). Cells were 80% confluent for all of the experiments.
Blocking experiments with anti-TLR2 antibodies
Isolated mouse epidermal keratinocytes and normal human epidermal keratinocytes (NHEKs, Life Technologies) were used. Cells were pre-incubated with anti-mouse TLR2 monoclonal antibody (Clone: T2.5, BioLegend), anti-human TLR2 monoclonal antibody (Clone: TL2.1, BioLegend) or its isotype control IgG (BioLegend). These antibodies are useful for blocking studies and have been reported to block TLR2-specific cell activation in vitro and in vivo 39–41.
Flow Cytometry (FACS)
Single-cell suspensions were prepared by mincing mouse skin tissue with scissors, followed by enzymatic digestion with collagenase Type II (Worthington), collagenase Type IV (Gibco), and 0.53mg/ml DNase I (Roche). Cells were stained with anti-CD117, anti-FcεRI, or anti-TLR2 monoclonal antibodies (BD Biosciences) according to the manufacturer’s instructions. Cells were analyzed with the Guava EasyCyte 8HT two laser, 6 color microcapillary-based benchtop flow cytometer (Millipore).
Laser capture microdissection for analysis of gene expression
After intradermal injection with 50 μl LTA (1 mg/ml, InvivoGen) or PBS, skin samples were collected and washed in RNAlater (QIAGEN) for 5 min. The samples were then immediately frozen in O.C.T. Compound (Sakura) with a dry ice/ethanol bath (−72 °C) and sectioned at a thickness of 8 μm in an RNase-free environment using RNaseZap spray (Thermo Fisher). Sections were treated with RNAlater for 4 min and then stained with 0.1% toluidine blue (Sigma, prepared in RNase-free water) using the LCM Frozen Section Staining Kit (Arcturus) according to the manufacturer’s instructions. We performed contamination-free isolation of 200 mast cells and 100 pieces of epidermis from each sample using the CellCut laser microdissection system (Molecular Machines & Industries). We extracted RNA from these samples using the PicoPure RNA Isolation Kit (Life Technologies), and the resulting total RNA was used for cDNA synthesis using the iSCRIPT cDNA Synthesis Kit (Bio-Rad). After performing pre-amplification PCR using pooled TaqMan Assays and TaqMan PreAmp Master Mix Kit (Life Technologies), we performed Real-Time Quantitative RT-PCR in an ABI 7300 Real-Time PCR system (Applied Biosystems). mRNA expression of type I Collagen alpha 1 (Col1a1) was undetectable in all samples, which confirmed that there was no contamination of skin fibroblasts in the laser-captured MCs and epidermis (data not shown).
Real-time quantitative RT-PCR
Total RNA was isolated using the RNeasy Mini Kit (QIAGEN) and 0.5μg of total RNA was used for cDNA synthesis using the iSCRIPT cDNA Synthesis Kit (Bio-Rad). Real-time quantitative RT-PCR was performed in an ABI 7300 Real-Time PCR system (Applied Biosystems) with Taqman primers and probes (Life Technologies). We used the comparative ΔΔCT method to quantify gene expression. Target gene expression levels in the test samples were normalized to endogenous GAPDH levels and reported as fold differences relative to GAPDH gene expression in untreated baseline controls 42. All assays were performed in triplicate and the experiments were repeated at least three times.
ELISA
ELISA kits (R&D systems) were used to determine SCF protein expression in mouse skin and normal human epidermal keratinocytes. All protein was detected from supernatants of tissue homogenate or cell culture media according to the manufacturer’s instructions. ELISA measurements were normalized to total weight or volume of each sample.
Histology and fluorescence staining
Animal skin samples were collected, fixed with buffered formalin, and embedded in paraffin. After deparaffinizing and rehydrating the sections, some sections were stained with toluidine blue (RICCA Chemical) or rhodamine-avidin 43 to identify MCs. Following antigen retrieval at 95–100°C for 30 minutes with citrate buffer (pH 6.0, Enzo Life Sciences), immunofluorescent staining of skin sections was performed using monoclonal antibodies against LTA (Abcam), mast cell chymase (Abcam), melanocyte combined markers HMB45+DT101+BC199 (Abcam), SCF (Santa Cruz Biotechnology), TLR2 (Abcam), c-Kit (Tonbo Biosciences) and their isotype controls (Santa Cruz Biotechnology). Alexa Fluor 488 and Alexa Fluor 594 conjugated secondary antibodies (Life Technologies) were used according to the manufacturer’s instructions. Slides were mounted in ProLong Anti-Fade reagent with DAPI (Molecular Probes). We imaged the sections using a Bx51 research microscope (Olympus) and the X-Cite 120 fluorescence illumination system (EXFO Photonic Solutions). All isotype controls showed negative staining (Figure E 1).
Statistical analyses
All data are presented as the mean±SD. At least three independent experiments were performed to assess reproducibility. Comparisons between groups were analyzed by Student’s t-test or ANOVA for multiple comparisons. For all statistical tests, p-values <0.05 were considered statistically significant (*p<0.05, **p<0.01, ***p<0.001).
Supplementary Material
Key Messages.
Skin microbiome influences mast cell maturation in the dermis; therefore, a normal microbiome is necessary for full maturation of mast cells and for their antimicrobial capacity
Gram positive bacterial products (LTA) enhance mast cell maturation by acting through epithelial cells
SCF produced by skin epithelial cells is necessary for correct mast cell localization in the dermis
The novel mechanism by which the skin microbiota signals the recruitment and maturation of MCs within the dermis via SCF production is important for understanding the pathogenesis of skin diseases involving MC function, such as atopic dermatitis or psoriasis, in which the role of the microbiome is still not clear.
Acknowledgments
We thank Dr. Richard Gallo and R.G. laboratory members for helpful discussions. Dr. Di Nardo’s lab is supported by the NIH – NAIAD with the NIH grant 5R01AI106874 to ADN.
Footnotes
Author contribution
A.D.N. conceived experiments, wrote the manuscript, and secured funding. Z.W., X.S., and N.M. performed experiments. L.E. and Y. M. generated and maintained the germ-free mice and performed the microbial reconstitution experiments. T.K. provided expertise and feedback.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nakamizo S, Egawa G, Honda T, Nakajima S, Belkaid Y, Kabashima K. Commensal bacteria and cutaneous immunity. Semin Immunopathol. 2015;37:73–80. doi: 10.1007/s00281-014-0452-6. [DOI] [PubMed] [Google Scholar]
- 2.Lai Y, Di Nardo A, Nakatsuji T, Leichtle A, Yang Y, Cogen AL, et al. Commensal bacteria regulate Toll-like receptor 3-dependent inflammation after skin injury. Nat Med. 2009;15:1377–82. doi: 10.1038/nm.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakamura Y, Oscherwitz J, Cease KB, Chan SM, Munoz-Planillo R, Hasegawa M, et al. Staphylococcus [dgr]-toxin induces allergic skin disease by activating mast cells. Nature. 2013;503:397–401. doi: 10.1038/nature12655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mashiko S, Bouguermouh S, Rubio M, Baba N, Bissonnette R, Sarfati M. Human mast cells are major IL-22 producers in patients with psoriasis and atopic dermatitis. Journal of Allergy and Clinical Immunology. 136:351–9.e1. doi: 10.1016/j.jaci.2015.01.033. [DOI] [PubMed] [Google Scholar]
- 5.Metcalfe DD, Baram D, Mekori YA. Mast cells. Physiol Rev. 1997;77:1033–79. doi: 10.1152/physrev.1997.77.4.1033. [DOI] [PubMed] [Google Scholar]
- 6.Halova I, Draberova L, Draber P. Mast cell chemotaxis - chemoattractants and signaling pathways. Front Immunol. 2012;3:119. doi: 10.3389/fimmu.2012.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu C, Liu Z, Li Z, Wu Y. Molecular regulation of mast cell development and maturation. Mol Biol Rep. 2009;37:1993–2001. doi: 10.1007/s11033-009-9650-z. [DOI] [PubMed] [Google Scholar]
- 8.Prussin C, Metcalfe DD. 4. IgE, mast cells, basophils, and eosinophils. J Allergy Clin Immunol. 2003;111:S486–94. doi: 10.1067/mai.2003.120. [DOI] [PubMed] [Google Scholar]
- 9.Galli SJ. Mast cells and basophils. Curr Opin Hematol. 2000;7:32–9. doi: 10.1097/00062752-200001000-00007. [DOI] [PubMed] [Google Scholar]
- 10.Galli SJ, Grimbaldeston M, Tsai M. Immunomodulatory mast cells: negative, as well as positive, regulators of immunity. Nat Rev Immunol. 2008;8:478–86. doi: 10.1038/nri2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakatsuji T, Chiang HI, Jiang SB, Nagarajan H, Zengler K, Gallo RL. The microbiome extends to subepidermal compartments of normal skin. Nat Commun. 2013;4:1431. doi: 10.1038/ncomms2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grice EA, Segre JA. The skin microbiome. Nature Rev Microbiol. 2011;9:244–53. doi: 10.1038/nrmicro2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Z, Macleod DT, Di Nardo A. Commensal Bacteria Lipoteichoic Acid Increases Skin Mast Cell Antimicrobial Activity against Vaccinia Viruses. J Immunol. 2012;189:1551–8. doi: 10.4049/jimmunol.1200471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Z, Lai Y, McLeod D, Cogen A, Di Nardo A. TLR2-activation turns mast cells in skin sentinels against viruses. JID. 2010:s124. [Google Scholar]
- 15.Aoki R, Kawamura T, Goshima F, Ogawa Y, Nakae S, Nakao A, et al. Mast cells play a key role in host defense against herpes simplex virus infection through TNF-alpha and IL-6 production. J Invest Dermatol. 2013;133:2170–9. doi: 10.1038/jid.2013.150. [DOI] [PubMed] [Google Scholar]
- 16.Ito M, Hiramatsu H, Kobayashi K, Suzue K, Kawahata M, Hioki K, et al. NOD/SCID/gamma(c)(null) mouse: an excellent recipient mouse model for engraftment of human cells. Blood. 2002;100:3175–82. doi: 10.1182/blood-2001-12-0207. [DOI] [PubMed] [Google Scholar]
- 17.Henz BM. Exploring the mast cell enigma: a personal reflection of what remains to be done. Exp Dermatol. 2008;17:91–9. doi: 10.1111/j.1600-0625.2007.00658.x. [DOI] [PubMed] [Google Scholar]
- 18.Okayama Y, Kawakami T. Development, migration, and survival of mast cells. Immunol Res. 2006;34:97–115. doi: 10.1385/IR:34:2:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kunisada T, Lu SZ, Yoshida H, Nishikawa S, Nishikawa S, Mizoguchi M, et al. Murine cutaneous mastocytosis and epidermal melanocytosis induced by keratinocyte expression of transgenic stem cell factor. J Exp Med. 1998;187:1565–73. doi: 10.1084/jem.187.10.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moller C, Karlberg M, Abrink M, Nakayama K, Motoyama N, Nilsson G. Bcl-2 and Bcl-XL are indispensable for the late phase of mast cell development from mouse embryonic stem cells. Experimental Hematology. 2007;35:385–93. doi: 10.1016/j.exphem.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 21.Welker P, Grabbe J, Czarnetzki BM. Human keratinocytes release mast cell differentiation factors other than stem cell factor. Int Arch Allergy Immunol. 1995;107:139–41. doi: 10.1159/000236956. [DOI] [PubMed] [Google Scholar]
- 22.Burton AL. Histochemical Studies on Developing Mast Cells. Anat Rec. 1964;150:265–9. doi: 10.1002/ar.1091500308. [DOI] [PubMed] [Google Scholar]
- 23.Kitamura Y, Kasugai T, Arizono N, Matsuda H. Development of mast cells and basophils: processes and regulation mechanisms. Am J Med Sci. 1993;306:185–91. doi: 10.1097/00000441-199309000-00011. [DOI] [PubMed] [Google Scholar]
- 24.Wang Z, Lai Y, Bernard JJ, Macleod DT, Cogen AL, Moss B, et al. Skin mast cells protect mice against vaccinia virus by triggering mast cell receptor S1PR2 and releasing antimicrobial peptides. J Immunol. 2012;188:345–57. doi: 10.4049/jimmunol.1101703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ito T, Smrz D, Jung MY, Bandara G, Desai A, Smrzova S, et al. Stem cell factor programs the mast cell activation phenotype. J Immunol. 2012;188:5428–37. doi: 10.4049/jimmunol.1103366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pivarcsi A, Kemeny L, Dobozy A. Innate immune functions of the keratinocytes. A review. Acta Microbiol Immunol Hung. 2004;51:303–10. doi: 10.1556/AMicr.51.2004.3.8. [DOI] [PubMed] [Google Scholar]
- 27.Yoshida H, Kunisada T, Grimm T, Nishimura EK, Nishioka E, Nishikawa SI. Review: melanocyte migration and survival controlled by SCF/c-kit expression. J Investig Dermatol Symp Proc. 2001;6:1–5. doi: 10.1046/j.0022-202x.2001.00006.x. [DOI] [PubMed] [Google Scholar]
- 28.Williams MR, Gallo RL. The role of the skin microbiome in atopic dermatitis. Curr Allergy Asthma Rep. 2015;15:65. doi: 10.1007/s11882-015-0567-4. [DOI] [PubMed] [Google Scholar]
- 29.Yamazaki M, Tsujimura T, Morii E, Isozaki K, Onoue H, Nomura S, et al. C-kit gene is expressed by skin mast cells in embryos but not in puppies of Wsh/Wsh mice: age-dependent abolishment of c-kit gene expression. Blood. 1994;83:3509–16. [PubMed] [Google Scholar]
- 30.Kurashima Y, Amiya T, Fujisawa K, Shibata N, Suzuki Y, Kogure Y, et al. The enzyme Cyp26b1 mediates inhibition of mast cell activation by fibroblasts to maintain skin-barrier homeostasis. Immunity. 2014;40:530–41. doi: 10.1016/j.immuni.2014.01.014. [DOI] [PubMed] [Google Scholar]
- 31.Kunii J, Takahashi K, Kasakura K, Tsuda M, Nakano K, Hosono A, et al. Commensal bacteria promote migration of mast cells into the intestine. Immunobiology. 2011;216:692–7. doi: 10.1016/j.imbio.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 32.Nahid MA, Satoh M, Chan EK. Mechanistic role of microRNA-146a in endotoxin-induced differential cross-regulation of TLR signaling. J Immunol. 2010;186:1723–34. doi: 10.4049/jimmunol.1002311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naik S, Bouladoux N, Wilhelm C, Molloy MJ, Salcedo R, Kastenmuller W, et al. Compartmentalized control of skin immunity by resident commensals. Science. 2012;337:1115–9. doi: 10.1126/science.1225152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Besmer P, Manova K, Duttlinger R, Huang EJ, Packer A, Gyssler C, et al. The kit-ligand (steel factor) and its receptor c-kit/W: pleiotropic roles in gametogenesis and melanogenesis. Dev Suppl. 1993:125–37. [PubMed] [Google Scholar]
- 35.Chatterjea D, Wetzel A, Mack M, Engblom C, Allen J, Mora-Solano C, et al. Mast cell degranulation mediates compound 48/80-induced hyperalgesia in mice. Biochem Biophys Res Commun. 2012;425:237–43. doi: 10.1016/j.bbrc.2012.07.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kirshenbaum AS, Metcalfe DD. Growth of human mast cells from bone marrow and peripheral blood-derived CD34+ pluripotent progenitor cells. Methods Mol Biol. 2006;315:105–12. doi: 10.1385/1-59259-967-2:105. [DOI] [PubMed] [Google Scholar]
- 37.Parrinello S, Samper E, Krtolica A, Goldstein J, Melov S, Campisi J. Oxygen sensitivity severely limits the replicative lifespan of murine fibroblasts. Nat Cell Biol. 2003;5:741–7. doi: 10.1038/ncb1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aasen T, Izpisua Belmonte JC. Isolation and cultivation of human keratinocytes from skin or plucked hair for the generation of induced pluripotent stem cells. Nat Protoc. 2010;5:371–82. doi: 10.1038/nprot.2009.241. [DOI] [PubMed] [Google Scholar]
- 39.Meng G, Rutz M, Schiemann M, Metzger J, Grabiec A, Schwandner R, et al. Antagonistic antibody prevents toll-like receptor 2-driven lethal shock-like syndromes. J Clin Invest. 2004;113:1473–81. doi: 10.1172/JCI20762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Flo TH, Halaas O, Lien E, Ryan L, Teti G, Golenbock DT, et al. Human toll-like receptor 2 mediates monocyte activation by Listeria monocytogenes, but not by group B streptococci or lipopolysaccharide. J Immunol. 2000;164:2064–9. doi: 10.4049/jimmunol.164.4.2064. [DOI] [PubMed] [Google Scholar]
- 41.Goo SY, Han YS, Kim WH, Lee KH, Park SJ. Vibrio vulnificus IlpA-induced cytokine production is mediated by Toll-like receptor 2. J Biol Chem. 2007;282:27647–58. doi: 10.1074/jbc.M701876200. [DOI] [PubMed] [Google Scholar]
- 42.Luu-The V, Paquet N, Calvo E, Cumps J. Improved real-time RT-PCR method for high-throughput measurements using second derivative calculation and double correction. Biotechniques. 2005;38:287–93. doi: 10.2144/05382RR05. [DOI] [PubMed] [Google Scholar]
- 43.McNeil BD, Pundir P, Meeker S, Han L, Undem BJ, Kulka M, et al. Identification of a mast-cell-specific receptor crucial for pseudo-allergic drug reactions. Nature. 2015;519:237–41. doi: 10.1038/nature14022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.