Abstract
Primary effusion lymphomas (PELs) are specifically associated with Kaposi's sarcoma-associated herpesvirus (KSHV) infection and most frequently occur in human immunodeficiency virus (HIV)-positive individuals as lymphomatous effusions in the serous cavities without a detectable solid tumor mass. Most PELs have concomitant Epstein-Barr virus (EBV) infection, suggesting that EBV is an important pathogenetic cofactor, although other as yet unidentified cofactors, such as cellular genetic alterations, are also likely to play a role. Lymphomatous effusions that lack KSHV also occur; these are frequently EBV associated in the setting of HIV infection. Here we used gene expression profile analysis to determine the viral impact on cellular gene expression and the pathogenesis of these lymphomatous effusions. Our results show that many genes, including cell cycle and signal transduction regulators, are differentially expressed between KSHV-positive PELs and KSHV-negative lymphomatous effusions and also between KSHV-positive, EBV-positive and KSHV-positive, EBV-negative PELs. Our results confirm that KSHV plays an important role in the pathogenesis of PELs, as its presence selects for a very distinct cellular gene expression category and a clearly different lymphoma type. Within the KSHV-positive PELs, the effect of EBV is more subtle but nevertheless clear.
Primary effusion lymphoma (PEL) is a rare and distinct subtype of non-Hodgkin's lymphoma which is preferentially associated with infection by Kaposi's sarcoma-associated herpesvirus (KSHV) (3-5). It most frequently occurs in human immunodeficiency virus (HIV)-positive patients as lymphomatous effusions in the serous cavities without a detectable solid tumor. The majority of PELs exhibit indeterminate immunophenotypes, i.e., express CD45, CD138, and activation-associated antigens (CD30, CD38, and HLA-DR), lack surface expression of B-cell markers (CD19, CD20, CD79a, and immunoglobulin), but exhibit clonal rearrangements and somatic hypermutation of the immunoglobulin genes, suggesting that they originate from postgerminal center B cells (9, 17). While the term PEL is currently employed for the above-described distinct clinicopathologic category of lymphoma, other lymphomatous effusions, including Burkitt's lymphoma and diffuse large B-cell lymphomas, which are frequently Epstein-Barr virus (EBV) positive but KSHV negative (13), may involve the body cavities.
KSHV is a member of the Gammaherpesvirinae family, which includes Epstein-Barr virus (6). KSHV does not transform B cells in vitro (18) and therefore, although it appears to be essential, probably is not sufficient by itself for the development of PEL. In addition, most PELs exhibit concomitant EBV infection, suggesting that EBV is an important cofactor in PEL development, although little or no expression of EBV transforming genes is found in PEL (10, 21). Other cofactors, such as unidentified cellular genetic alterations, are likely to play a role in PEL development as well.
Gene expression profiling has made it possible to simultaneously analyze the expression of thousands of genes pertinent to various biologic functions (14). With this method, Klein et al. (12) and more recently Jenner et al. (11) have demonstrated that gene expression patterns can distinguish KSHV-positive PELs from KSHV-negative malignant lymphomas and have documented that PELs are more closely related to neoplastic plasma cells and EBV-positive immunoblasts than to other malignant B cells. However, this comparison did not include KSHV-negative lymphomatous effusions and did not address the difference between KSHV-positive, EBV-positive PELs and KSHV-positive, EBV-negative PELs. The present study is aimed at understanding whether the viral status confers biological differences among lymphomatous effusions and to identify transforming cofactors or other cellular pathways involved in the development of PEL.
We used the Affymetrix HG-U133A microarray that contains approximately 33,000 genes to analyze the gene expression profile of nine cell lines (three from each virologic group) and three PEL patient samples. Our results suggest that KSHV-positive PELs are very different from other lymphomatous effusions and that the genes that are differentially expressed include apoptosis regulators, cell cycle regulators, transcriptional factors, and signal transduction regulators. KSHV clearly plays a dominant role in the phenotype of PEL. Within the KSHV-positive PELs, two subgroups can be identified, which were correlated with their EBV status. Among these genes, four were regulators of the mitogen-activated protein kinase pathway that were upregulated in the KSHV-positive, EBV-negative PELs, suggesting that in the absence of EBV, events that lead to activation of the mitogen-activated protein kinase pathway may act as a cofactor for the development of PEL.
MATERIALS AND METHODS
Cell collection and RNA preparation.
Four KSHV-positive, EBV-positive PEL cell lines (BC-1, BC-2, BC-5, BCS-6, and JCS-1), five KSHV-positive, EBV-negative PEL cell lines (BC-3, PEL-5, BCBL-1, BCP-1, and VG-1), and three KSHV-negative, EBV-positive cell lines (BCKN-1, IBL-4, and SM-1) were derived from lymphomatous effusions and grown in RPMI plus 20% heat-inactivated fetal calf serum, penicillin, streptomycin, and glutamine. Of these cell lines, three from each virology category were used for microarray analysis, and all were used for reverse transcription-PCR and protein analysis. The VG-1, BCP-1, and JSC-1 cell lines were kindly provided by Christian Brander, Shou-Jiang Gao, and Richard Ambinder, respectively.
The three PEL cases were received for diagnostic evaluation at the New York-Presbyterian Hospital/Weill Medical College of Cornell University. The diagnosis of PEL was made based on infection by KSHV and on the specific morphological, immunophenotypic, and molecular features. All three primary cases were from HIV-positive patients. Two of the three primary cases are EBV positive (patients 1 and 2) and one is EBV negative (patient 3). Mononuclear cells were isolated from the serous fluids by Ficoll-Hypaque (Pharmacia, Piscataway, N.J.) density gradient centrifugation and cryopreserved with standard techniques. Total RNA was isolated with RNeasy mini kit (Qiagen, Valencia, Calif.). Microarray analysis of the same three PEL cases with a smaller chip array (U95A) has been reported previously (12).
Immunohistochemistry and in situ hybridization.
Immunohistochemical staining was performed on formalin-fixed, paraffin-embedded cell blocks from PEL patients 1 to 3. Staining was performed with a TechMate 500 automated immunostainer (Ventana Medical Systems Inc., Tucson, Ariz.) and ChemMate ABC peroxidase secondary detection kit (Ventana Medical Systems Inc.). For demonstration of KSHV antigens, we used a rat monoclonal antibody to latent nuclear antigen (LANA; ORF-73) and a rabbit polyclonal antibody to viral interleukin-6 (IL-6) made in our laboratory against the PDVTPDVHDK peptide as described (19). Immunostaining was performed according to a modified monoimmunoperoxidase (MIP) protocol (Ventana Medical Systems Inc.). For demonstration of KSHV LANA, biotinylated polyclonal goat anti-rat immunoglobulin (multiple adsorption) (BD Biosciences Pharmingen, San Diego, Calif.) was added to secondary antibodies. Prior to staining, paraffin tissue sections were pretreated for 40 min in a water bath at 95°C with antigen retrieval Citra Plus (BioGenex, San Ramon, Calif.). The peroxidase reaction was developed with liquid diaminobenzidine (DAB) substrate chromogen included in the detection kit. Negative controls with rat, mouse, or rabbit immunoglobulin G were run in parallel. Sections were counterstained with hematoxylin and mounted in Permount (Fisher Scientific).
In situ hybridization for EBV-encoded RNA (EBER) transcripts was performed with the Epstein-Barr probe in situ hybridization kit (Novocastra, Newcastle upon Tyne, United Kingdom) according to the manufacturer's protocol.
Preparation of labeled cRNA and hybridization to high-density oligonucleotide microarray.
Double-stranded cDNA was synthesized from 10 mg of total RNA with oligo(dT)24 T7 primer with the SuperScript Choice system kit (Invitrogen). Biotinylated cRNA was synthesized with a high yield transcription kit (Affymetrix, Santa Clara, Calif.); 20 μg of fragmented cRNA was hybridized to the HG-U133A microarray (Affymetrix) according to the manufacturer's instructions. After scanning, the expression data were obtained with Affymetrix software. The expression data were then analyzed by Genespring 6.0 software (Silicon Genetics, Redwood City, Calif.). The results have been submitted to NCBI Gene Expression Omnibus.
Statistical analysis.
Unsupervised hierarchical clustering of all nine effusion lymphoma cell lines was performed with Pearson correlation, based on 11,024 genes that had raw data greater than 150 in at least one of the 18 samples. To identify genes that are differentially expressed between lymphomatous effusions according to viral status, parametric Welch t test and Benjamini and Hochberg false discovery rate for multiple testing correction were used, where the P value was <0.01 between KSHV-positive PELs and KSHV-negative lymphomatous effusions, and ≤0.05 between KSHV-positive, EBV-positive and KSHV-positive, EBV-negative PELS. All the gene trees and condition trees for supervised hierarchical clustering were done with Pearson and Spearman correlation. The analysis was done with Genespring 6.0 software (Silicon Genetic, Redwood city, Calif.).
Western blotting.
Whole-cell extracts were prepared from 5 × 106 cells. The cells were washed in phosphate-buffered saline and lysed in 500 μl of radioimmunoprecipitation assay (RIPA) lysis buffer (50 mmol of Tris-HClper liter, 1% NP-40, 0.25% sodium deoxycholate, 150 mmol of NaCl per liter, 1 mmol of EDTA per liter, 1 mmol of phenylmethylsulfonyl fluoride per liter, 0.5 μg of aprotinin per liter, 0.5 μg of leupeptin per liter, 1 mmol of Na3VO4 per liter, 1 mmol of NaF per liter) for 15 min at 4°C. The extracted proteins were quantified with a protein bioassay (Bio-Rad, Hercules, Calif.) with a bovine serum albumin standard; 40 μg of protein was resolved on a 10% acrylamide gel and transferred to nitrocellulose. The membranes were blocked, incubated with p38delta antibody (Santa Cruz Biotechnology, Santa Cruz, Calif.), washed and incubated with horseradish peroxidase-conjugated rabbit immunoglobulin (Amersham, Piscataway, N.J.). Detection was performed with chemiluminescence (Amersham). The membranes were stripped and reprobed with actin antibody (Sigma, St. Louis, Mo.). Detection was performed as for p38delta antibody.
Real-time quantitative PCR analysis.
Real-time quantitative reverse transcription-PCR analysis was performed on an ABI Prism 7000 sequence detection system with the SYBR Green dye. DNase-treated total RNA (1.0 μg) was reverse transcribed with the reverse transcription system (Promega) and resuspended in 100 μl of sterile distilled H2O. The real-time PCR contained 1 μl of cDNA, 5 pmol of both the forward and reverse primers, and 12.5 μl of SYBR Green dye master mix (Applied Biosystems, San Francisco, Calif.). Sequence-specific primers were designed by means of Primer Express 2.0 software (Applied Biosystems, San Francisco, Calif.). The primers used were GADD45β forward, 5′-TTGCAACATGACGCTGGAA-3′, and reverse,5′-GGTCACCGTCTGCATCTTCTG-3′; caspase1 forward, 5′-CCGCAAGGTTCGATTTTCA-3′, and reverse, 5′ACTCTTTCAGTGGTGGGCATCT 3′; SCAP 2 forward, 5′-TCTGATGGAGCCCAGTTTCC, and reverse, 5′-CAAGGTAGCCAGCCTTTAGAACA; follicular lymphoma translocation variant 1 (FVT1) forward, 5′-GGGCGCATGTGGTGGTTA-3′, and reverse, 5′-ATAGCACTCGATAGCAATGCACTT-3′; and β-actin forward, 5′-CCTGGCACCCAGCACAA-3′, and reverse, 5′-GCCGATCCACACGGAGTACT-3′. No amplification was observed in the no-template controls for each primer set.
PCR conditions were as follows: one cycle at 50°C for 2 min, one cycle at 95°C for 10 min, and 40 cycles at 95°C for 15 s and 60°C for 1 min. All reactions were done in triplicate, and CT value, the threshold cycle number, for each cell line or primary sample was calculated. A CT value is a quantitative measurement of the mRNA levels of the genes tested. The higher the value, the lower the mRNA content (16). CT values for β-actin were used for normalization purposes. The averages of the normalized CT values from KSHV-positive, EBV-positive and KSHV-positive, EBV-negative cell lines and primary samples were used in order to determine the increases in gene expression of the KSHV-positive, EBV-negative group by the ΔΔCT method (16).
RESULTS
Presence of KSHV determines the distinct cellular gene expression profile of lymphomatous effusions.
To confirm the impact of KSHV in lymphomatous effusions, we first compared the gene expression profile of KSHV-positive PELs and KSHV-negative, EBV-positive lymphomatous effusions with an unsupervised clustering method. All cell lines used were derived from patients with lymphomatous effusions. The EBV-positive but KSHV-negative cell lines were classified by morphology and immunophenotype as large B-cell lymphoma (BCKN-1), immunoblastic large B-cell lymphoma (IBL-4), and small noncleaved cell lymphoma (SM-1). Cell line SM-1 had an 8;14 balanced translocation, and IBL-4 had a 2;8 translocation, both involving the c-myc locus and cytogenetically consistent with Burkitt's lymphoma. While these three cases are heterogeneous with respect to their morphological and molecular classification, they all presented as primary lymphomatous effusions.
Previous studies have compared larger numbers of well-defined categories of HIV-associated lymphomas, including Burkitt's lymphoma, with PELs (11, 12), but such studies have not specifically evaluated Burkitt's and other KSHV-negative lymphomas presenting as effusions. Six cell lines were KSHV positive. They were further divided into two subtypes according to their EBV status. RNA extracted from these cell lines was converted into labeled cRNA and hybridized to HG-U133A Affymetrix Gene Chips containing approximately 33,000 genes. To show the reproducibility of the technique, all extractions and analyses were done in duplicate. Samples from the same cell line were found to cluster under the smallest branches, indicating reproducibility.
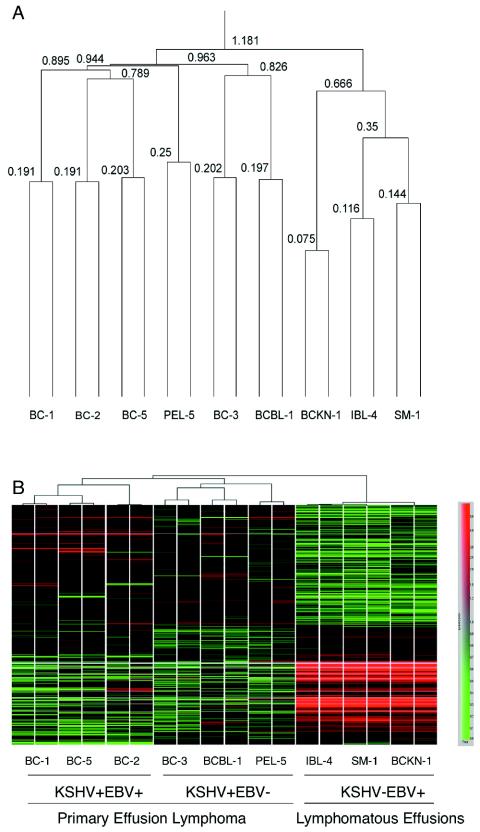
Two-dimensional unsupervised hierarchical clustering was performed on 11,024 genes whose expression levels were greater than 150 on raw data in at least 1 of 18 samples with Pearson correlation as the similarity measure. As shown in Fig. 1A, the sample dendrogram derived two major branches. One branch contained all three KSHV-negative cell lines, whereas the second branch contained all six KSHV-positive PEL cell lines. Clearly, PEL displays a profile that is distinguishable from that of KSHV-negative, EBV-positive lymphomatous effusions.
FIG. 1.
Clustering of gene expression in all PELs and lymphomatous effusions. (A) Unsupervised hierarchical clustering of gene expression analysis was performed on 11,024 genes when raw expression data were greater than 150 in at least one of the 18 samples with Pearson correlation. The number above each line indicates the vertical length of each line as the correlation of relatedness. The sample dendrogram shows that all six KSHV-positive PEL cell lines (BC-1, BC-2, BC-5, PEL-5, BC-3, and BCBL) are clustered under one branch, showing the high degree of similarity in gene expression pattern. Three KSHV-negative cell lines obtained from lymphomatous effusions (BCKN-1, IBL-4, and SM-1) clustered under the other branch. This figure shows that KSHV-positive PEL are clearly different from KSHV-negative lymphomatous effusions. (B) Hierarchical clustering of statistically differentially expressed genes among KSHV-positive, EBV-positive PELs, KSHV-positive, EBV-negative PELs, and KSHV-negative, EBV-positive lymphomatous effusions is shown. Each column represents an individual sample. Those with the most similar gene expression patterns are clustered under one branch. Each row represents an individual gene. The color of each cell represents the relative gene expression level according to the color scale shown at the right side. KSHV-positive PELs have a distinct gene expression profile compared to KSHV-negative, EBV-positive lymphomatous effusions. Half of the genes are upregulated and the other half are downregulated in the PEL group compared to the KSHV-negative, EBV-positive lymphomatous effusions. Among 514 differentially expressed genes are apoptosis regulators, cell cycle regulators, transcriptional factors, and signal transduction regulators.
To determine the genes with significant differential expression between these two groups, parametric Welch t test with P value (<0.01) and Benjamini and Hochberg false discovery rate for multiple testing correction were used. About 500 genes are differentially expressed. Pearson correlation analyses of these genes was used for hierarchical clustering, which is shown in Fig. 1B. About half of the genes are upregulated and the other half are downregulated in the PEL group compared to the KSHV-negative, EBV-positive lymphomatous effusions. Among these genes are transcriptional factors and cell cycle signal transduction and apoptosis regulators. The gene expression profiles demonstrate that PEL has a characteristic gene expression profile that is clearly distinguishable from that of KSHV-negative, EBV-positive lymphomatous effusions. These results also confirms previous studies (11, 12) indicating that PELs have a homogeneous phenotype independent of the presence of EBV. This phenotype is defined by the common pattern of expression of approximately 500 genes.
Two subtypes of PELs can be identified by unsupervised clustering according to their EBV status.
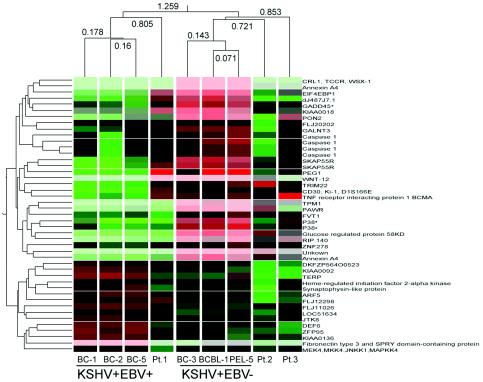
Although PELs display a more homogenous expression profile regardless of their EBV status compared to other lymphomatous effusions, we wanted to know if there are subtle differences in the gene expression profile between EBV-positive and EBV-negative PELs. The unsupervised hierarchical clustering analysis shown in Fig. 1A revealed that among the KSHV-positive cell lines, two subtypes can be identified based on EBV status. The sample dendrogram (Fig. 1A) shows that all three KSHV-positive, EBV-positive cell lines (BC-1, BC-2, and BC-5) are clustered under one branch (the correlation of relatedness is 0.895, indicated by the vertical length of the line), thus showing a high degree of similarity in their gene expression patterns. Two KSHV-positive, EBV-negative cell lines (BCBL-1 and BC-3) were clustered under one branch (the correlation of relatedness is 0.826), and a third one, PEL-5, appeared distinct. This shows that among KSHV-positive PELs, two main subtypes can be distinguished, those containing and those lacking EBV coinfection.
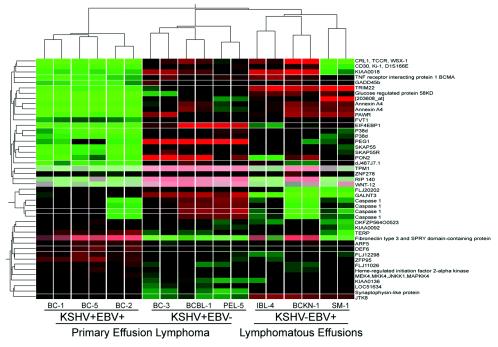
To determine which genes are differentially expressed between these two PEL subtypes, we compared three KSHV-positive, EBV-negative PEL cell lines with three KSHV-positive, EBV-negative PEL cell lines. The parametric Welch t test with P value (< = 0.05) and Benjamini and Hochberg false discovery rate for multiple testing correction were used for statistical analysis. About 45 probes showed differences between the two subtypes representing a total of 40 different genes, as several probes contained sequences from the same gene. Although the KSHV-negative, EBV-positive group was not included in the statistical analysis, we evaluated the expression levels of these 40 genes in this group, which was included for hierarchical clustering. Figure 2 shows hierarchical clustering of these 45 probes among nine cell lines with Spearman correlation. These results show that although PELs have a common phenotype defined by the expression of 500 genes, two subtypes of PELs can still be identified with 40 genes being differentially expressed. Interestingly, the KSHV-negative, EBV-positive group had an expression pattern most similar to that of the KSHV-single-positive PELs.
FIG. 2.
Hierarchical clustering of 40 statistically differentially expressed genes between KSHV-positive, EBV-positive PELs (three cell lines), KSHV-positive, EBV-negative PELs (three cell lines), and KSHV-negative, EBV-positive lymphomatous effusions (3 cell lines). Of 45 probes representing 40 different genes, SKAP55R (Src family-associated phosphoprotein 2 or SKAP55-related protein), p38δ (mitogen-activated protein kinase 13, SAPK4), GADD45β, and caspase 1 (ICE) are involved in the mitogen-activated protein kinase pathway. All these genes are upregulated in the KSHV-positive, EBV-negative PEL group. MKK4 is also a component of the JNK/p38 mitogen-activated protein kinase pathway and is upregulated in the KSHV-positive, EBV-positive PELs.
Differential gene expression between EBV-positive and EBV-negative PELs can be confirmed in additional cell lines and by different methodology.
Of the 40 genes, SKAP55R (Src family-associated phosphoprotein 2 or SCAP2), p38delta (p38δ, mitogen-activated protein kinase 13, SAPK4), GADD45β, ΜΚΚ4 (ΜΕΚ4, mitogen-activated protein kinase 4), and caspase I (ICE) are directly or indirectly involved in the JNK/p38 mitogen-activated protein kinase pathway (7, 8, 15, 22). MKK4 is upregulated in the KSHV-positive, EBV-positive PEL cell lines. The other four genes (SKAP55R, p38δ, GADD45β, and caspase I) are upregulated in the KSHV-positive, EBV-negative PEL group. The sixth gene, follicular lymphoma variant translocation 1 (FVT1) (20), is also upregulated in the KSHV-positive, EBV-negative PELs.
The expression of these molecules on PEL cell lines and patient samples was confirmed by real-time PCR (Table 1). Expression is presented as the ΔCT, which is the value that represents the normalized measure of the number of PCR cycles that it takes to see an amplified PCR product (normalized threshold cycle number). The lower the ΔCT, the higher the transcript copy number for a particular gene. We also included additional cell lines, two EBV-positive (BCS-6 and JSC-1) and two EBV-negative (BCP-1 and VG-1), that had not been tested in the microarray analysis. As shown in Table 1, the expression level of mRNAs for p38δ, GADD45β, caspase 1, SKAP55R, and FVT-1 is higher in all KSHV-positive PEL cell lines lacking EBV than in those containing the two viruses, with the notable exception of the JSC-1 cell line, which had lower expression of all these genes. As the JSC-1 cell line behaves more like those lacking EBV, it was excluded from the general comparison. When the overall difference in expression levels was calculated, we found that expression was increased 54.6-fold (p38δ), 3.8-fold (GADD45β), 9.3-fold (caspase 1), 45-fold (SKAP55R), and 6.8-fold (FVT-1) more in single-positive cell lines (KSHV only) than in double-positive cell lines (with EBV and KSHV).
TABLE 1.
Quantitative RT-PCR analysis for selected genes that are differentially expressed in EBV-positive and EBV-negative categories of KSHV-positive PELsa
| Gene | Value | Cell lines
|
Patient specimens
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EBV+
|
EBV−
|
EBV+
|
||||||||||||
| BC-1 | BC-2 | BC-5 | BCS6 | BC-3 | BCBL-1 | PEL-5 | BCP-1 | VG1 | JSC-1 | Pt1 | Pt2 | Pt3b | ||
| P38δ | ΔCT | 12.3 | 17.5 | 17.8 | 17.8 | 11.0 | 10.28 | 9.69 | 11.1 | 11.0 | 9.44 | 15.39 | 10.45 | 9.22 |
| Avg ΔCT ± SD | 16.38 ± 2.70 | 10.61 ± 0.61 | ||||||||||||
| Fold increase in EBV-PEL | 54.6 | |||||||||||||
| GADD45β | ΔCT | 10.0 | 10.6 | 11.7 | 9.12 | 7.57 | 8.62 | 7.79 | 8.73 | 9.66 | 8.78 | 6.63 | 6.01 | 6.95 |
| Avg ΔCT ± SD | 10.38 ± 1.1 | 8.47 ± 0.83 | ||||||||||||
| Fold increase in EBV-PEL | 3.8 | |||||||||||||
| Caspase 1 | ΔCT | 5.19 | 5.53 | 5.15 | 9.84 | 4.88 | 3.79 | 1.74 | 3.89 | 1.82 | 3.68 | 3.53 | 2.96 | 3.36 |
| Avg ΔCT ± SD | 6.43 ± 2.28 | 3.22 ± 1.39 | ||||||||||||
| Fold increase in EBV-PEL | 9.3 | |||||||||||||
| SKAP55R | ΔCT | 15.3 | 18.4 | 11.5 | 10.5 | 7.66 | 9.49 | 8.38 | 8.68 | 7.95 | 7.7 | 7.29 | 4.69 | 5.26 |
| Avg ΔCT ± SD | 13.92 ± 3.63 | 8.43 ± 0.71 | ||||||||||||
| Fold increase in EBV-PEL | 45 | |||||||||||||
| FVT-1 | ΔCT | 12.5 | 13.2 | 10.9 | 12.16 | 9.48 | 11.39 | 8.59 | 8.95 | 8.65 | 8.60 | 11.78 | 7.07 | 7.28 |
| Avg ΔCT ± SD | 12.17 ± 0.97 | 9.41 ± 1.16 | ||||||||||||
| Fold increase in EBV-PEL | 6.8 | |||||||||||||
For each cell line and primary sample, a real-time quantitative RT-PCR was performed in triplicate for the genes shown. The standard deviation (SD) for the triplicates was ≤0.5. The mean CT values for each cell line and primary sample were calculated (avg CT), and the normalized values (ΔCT) were determined from average β-actin CT values. The average ΔCT values were calculated for the KSHV+ EBV+ cell line group (BC-1, BC-2, BC-5, and BCS-6) and for the KSHV+ EBV− cell line group (BC-3, BCBL-1, PEL-5, BCP-1, and VG-1). The difference between the two groups for each gene (ΔΔCT) was used to determine the increase in gene expression of the KSHV+ EBV− group. Although data for the JCS-1 cell line are presented, they were not included in the calculations because the CT values for all four genes were consistently atypical for the KSHV+ EBV+ group and more similar to those of the KSHV+ EBV− group.
In this case a small proportion of cells were positive for EBV but the majority of KSHV-infected cells were negative (Fig. 4).
The expression levels of mRNAs for p38δ, caspase 1, SKAP55R, and FVT-1 were higher in a single-positive patient sample (patient 3) than the double-positive patient 1, so these two primary samples behaved like the cell lines for these four genes. However, patient 2 had an expression pattern for these specific genes that most closely resembled an EBV-negative specimen. Notably, only approximately 10% of the cells in this specimen were positive for EBER (see Fig. 4), suggesting in fact an intermediate pattern and perhaps an in vivo situation reminiscent of the JSC-1 cell line.
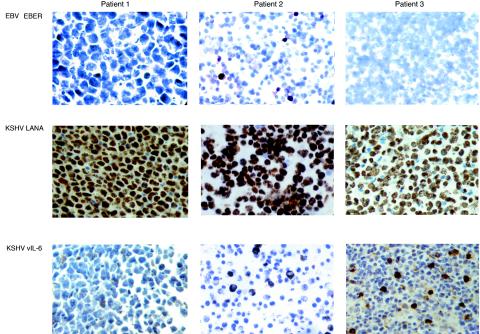
FIG. 4.
Pattern of EBV and KSHV positivity in PEL in vivo. In situ hybridization for EBER was performed in three cases of PEL without previous culture. Patient 1 showed strung positivity in all the cells, mostly nuclear but also spilling into the cytoplasm. Patient 2 showed only a small proportion of EBER-positive cells. Patient 3 was negative. Immunohistochemistry for KSHV in the specimens from the three patients showed that the vast majority of cells were positive for LANA, and a proportion also expressed viral IL-6. Magnification, 400×.
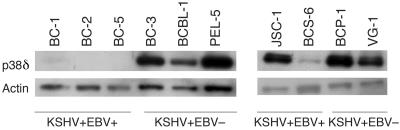
An antibody for P38δ antibody is available for Western blot analysis. Figure 3 shows that the KSHV-positive, EBV-positive cell lines BC-1, BC-2, BC-5, and BCS-6 have no expression of p38δ, while the KSHV-positive, EBV-negative cell lines BC-3, BCBL-1, PEL-5, BCP-1, and VG-1 have very high levels of p38δ. Except for JSC-1, which is a KSHV-positive, EBV-positive cell line, the newly included cell lines showed an expression pattern similar to those used for microarray analysis.
FIG. 3.
Western blot confirming the expression of p38δ/mitogen-activated protein kinase 13/SAPK4 in KSHV-positive PEL cell lines. Expression of p38δ protein in all PEL cell lines is shown in the upper row. Expression of actin protein is shown in the lower row. All KSHV-positive, EBV-negative PEL cell lines have high expression of p38δ. All KSHV-positive, EBV-positive PEL cell lines except JSC-1 have low or no expression of p38δ.
Cellular gene expression in primary PEL cases.
To validate the data obtained from cell lines, we evaluated three PEL primary specimens that had not been cultured prior to RNA extraction. By PCR analysis, the specimens from patients 1 and 2 were positive for EBV, while that from patient 3 was negative. However, by in situ hybridization for EBERs, we found that EBV was only identifiable in a small proportion of the cells in patient 2 (Fig. 4). In contrast, all the cells from patient 1 were clearly EBER positive, and those from patient 3 were all negative. This illustrates that EBV positivity may be heterogeneous in vivo. In contrast to EBV, the vast majority of the cells from all three patients were positive for KSHV, as demonstrated by detection of LANA, and a significant proportion of the cells also expressed viral IL-6.
We determined whether we could predict the viral status of the three primary cases of PEL based on the 45 gene probes that were differentially expressed in the KSHV-positive cells lines according to EBV status. Hierarchical clustering of six cell lines and three primary patient samples was performed with Pearson correlation (Fig. 5). The dendrogram shows that patient 1 (KSHV-positive, EBV-positive) was correctly clustered under a branch with the KSHV-positive, EBV-positive PELs, patient 2 (intermediate between the two classes, perhaps due to a low proportion of EBER-positive cells), and patient 3 (KSHV-positive, EBV-negative) were clustered under the other branch with the KSHV-positive, EBV-negative PELs. Thus, the 45 gene probes differentially expressed in KSHV-positive, EBV-positive PELs versus KSHV-positive, EBV-negative PELs represent a consistent phenotypic difference between the two subtypes.
FIG. 5.
Viral status of three primary patient samples can be predicted with the 45 gene probes generated from supervised clustering. Hierarchical clustering of nine cell lines and three primary patient samples was based on 45 gene probes. The number above or below each horizontal line indicates the vertical distance of each line as the correlation of relatedness. The dendrogram shows that patient 1 was correctly clustered under one branch with the KSHV-positive, EBV-positive PELs, and patients 2 and 3 was correctly clustered under one branch with the KSHV-positive, EBV-negative PELs.
DISCUSSION
Previous studies have shown that the gene expression profile of PEL is distinct from that of other non-Hodgkin's B-cell lymphomas, confirming their unique phenotype (11, 12). These studies have included larger panels of AIDS lymphomas, but they did not focus specifically on lymphomatous effusions. In this study we confirmed that KSHV positivity is associated with a very distinct gene expression pattern also among lymphomatous effusions. About 500 genes, including apoptosis regulators, cell cycle regulators, transcriptional factors, and signal transduction regulators, are differentially expressed between KSHV-positive PEL and KSHV-negative lymphomatous effusions.
We also found that among PELs, two subgroups corresponding to viral status (KSHV-positive, EBV-positive versus KSHV-positive, EBV-negative) can be identified by unsupervised hierarchical clustering methods. Subsequently, we identified 40 genes that are differentially expressed between these two subgroups. Among these 40 genes, expression of four genes (p38δ, GADD45, caspase 1, and SKAP55R), which are regulators of the mitogen-activated protein kinase pathway, is significantly higher in the KSHV-positive, EBV-negative PELs. MKK4 is a regulator of the JNK/p38 mitogen-activated protein kinase pathway but is lower in the KSHV-positive, EBV-negative PELs. A negative feedback loop by p38α has been reported for MKK6 (1), so our findings could be explained by hypothesizing that expression of active p38δ similarly leads to downregulation of MKK4. While total p38δ protein is clearly more abundant in EBV-negative than in double positive PELs, it is unclear if it is phosphorylated and active (at the time of this writing there is no available antibody to the phosphorylated form of this protein), and its natural substrate in these cells is not yet known. Nevertheless, the apparent overrepresentation of proteins related to mitogen-activated protein kinase signaling in the genes that were identified as being differentially expressed between the two categories of PEL suggests the involvement of this pathway.
KSHV and EBV are gammaherpesviruses, and they are more closely related to each other than to other known human viruses. It is interesting that lymphomas containing KSHV, regardless of EBV coinfection, form a distinct cluster, i.e., that lymphomas containing both viruses have a gene expression profile that more closely resembles that of KSHV alone than that of EBV alone, suggesting that KSHV infection plays a dominant role in their phenotype. This can be partially explained by the previous observations that in the presence of both viruses, the expression of latent EBV genes is restricted (10, 21). In contrast, in all KSHV-infected cells, at least four viral proteins are produced that can affect proliferation and/or survival of the infected cells. A recent study showed that in vitro infection of KSHV-positive PEL cells with EBV results in cell lines that express a limited number of EBV-encoded genes but that EBV confers higher tumorigenicity in nude mice to these cell lines (23).
Since EBV immortalizes B cells in vitro but KSHV does not, and the majority of PELs are infected by both viruses, EBV must play an important role as a cofactor in PEL development, but once these lymphomas develop, KSHV appears to be the driving force. It is also interesting that the KSHV-negative, EBV-positive cell lines appear to more closely resemble the KSHV-positive, EBV-negative cell lines with respect to the 40 genes that are differentially expressed in the two KSHV-positive subgroups. Some of these genes appear to distinguish the cases infected by both viruses from those infected by a single virus, regardless of whether it is EBV or KSHV.
To validate the differences found in the microarray analysis, we tested four additional KSHV-positive cell lines by quantitative reverse transcription-PCR for a subset of genes. The overexpression of these genes was consistently higher in all cell lines lacking EBV with one exception. The JSC-1 cell line, which contains both EBV and KSHV genomic sequences, has a pattern for all of the differentially expressed genes that is most consistent with the EBV-negative cases. We confirmed the genomic positivity for EBV (not shown), so loss of this virus did not explain the gene expression pattern. Data recently published by Jenner et al. are consistent with our findings (11). This study used a cDNA microarray of ≈5,700 genes to examine gene expression. While not described in their text, the cluster analysis shows that the KSHV-positive cell lines examined could be separated according to EBV status with the notable exception of JSC-1. PEL-SY and HBL-6 are double positive, and they clustered together, while the single-positive cell lines included, BCP-1, BC-3, and BCBL-1, clustered more tightly together. JSC-1 had a gene expression profile like that of the EBV-negative lines examined. Consistent with the hypothesis that EBV is defective in the JSC-1 cell line, Cannon et al. have reported that this cell line produces KSHV viral particles but not EBV when induced to undergo lytic reactivation, and only low levels of EBV latent gene expression were detected (2). The argument can be made that as yet uncharacterized defects in the EBV genome or more restricted latency account for the finding that this particular cell line has the gene expression profile of a KSHV-positive, EBV-negative cell line.
The number of primary patient samples was too small to draw firm conclusions. Another study showing gene expression profiling has been published, which included the same three primary cases reported here and six additional primary PEL specimens (12). However, a different array was used, so direct comparison was not possible. Nevertheless, an important observation emerged from evaluating these three cases. One of the cases had less than 10% EBER-positive cells (patient 2) and it clustered more closely with the EBV-negative samples. This case also had a pattern of expression of selected genes examined by quantitative reverse transcription-PCR that more closely resembled the EBV-negative case. Thus, EBV can be heterogeneous in vivo, and cases exist in which EBV is present in only a small proportion of cells. It is also possible that in patient 2 all the neoplastic cells are positive for EBV but that these EBV-positive cells have little or no EBV gene transcription (including of EBERs). Regardless, lack of EBER expression in the majority of cells appears to correlate with a cellular gene expression profile most similar to that of the EBV-negative PELs.
In summary, our results demonstrate that lymphomatous effusions containing KSHV have a unique gene expression profile distinct from that of those lacking this virus, further confirming the distinct immunophenotype of this disease. We show for the first time that among the KSHV-associated primary effusion lymphomas, the presence of EBV is associated with a gene expression signature that is different from that of EBV-negative cases. Finally, we demonstrate that a set of genes are more highly expressed in KSHV-positive PELs lacking EBV and that four of these genes are either directly or indirectly involved in the mitogen-activated protein kinase pathway, suggesting that alterations in this pathway may compensate for the lack of this virus. Based on these data we hypothesize that in the absence of EBV infection, alterations of the mitogen-activated protein kinase pathway may act as a cofactor for PEL development.
REFERENCES
- 1.Ambrosino, C., G. Mace, S. Galban, C. Fritsch, K. Vintersten, E. Black, M. Gorospe, and A. R. Nebreda. 2003. Negative feedback regulation of MKK6 mRNA stability by p38α mitogen-activated protein kinase. Mol. Cell, Biol. 23:370-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cannon, J. S., D. Ciufo, A. L. Hawkins, C. A. Griffin, M. J. Borowitz, G. S. Hayward, and R. F. Ambinder. 2000. A new primary effusion lymphoma-derived cell line yields a highly infectious Kaposi's sarcoma herpesvirus-containing supernatant. J. Virol. 74:10187-10193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cesarman, E., Y. Chang, P. S. Moore, J. W. Said, and D. M. Knowles. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body cavity-based lymphomas. N. Engl. J. Med. 332:1186-1191. [DOI] [PubMed] [Google Scholar]
- 4.Cesarman, E., and D. M. Knowles. 1997. Kaposi's sarcoma-associated herpesvirus: A lymphotropic human herpesvirus associated with Kaposi's sarcoma, primary effusion lymphoma, and multicentric Castleman's disease. Semin. Diagn. Pathol. 14:54-66. [PubMed] [Google Scholar]
- 5.Cesarman, E., and D. M. Knowles. 1999. The role of Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) in lymphoproliferative diseases. Semin. Cancer Biol. 9:165-174. [DOI] [PubMed] [Google Scholar]
- 6.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 7.Curtis, D. J., S. M. Jane, D. J. Hilton, L. Dougherty, D. M. Bodine, and C. G. Begley. 2000. Adaptor protein SKAP55R is associated with myeloid differentiation and growth arrest. Exp. Hematol. 28:1250-1259. [DOI] [PubMed] [Google Scholar]
- 8.Dhanasekaran, N., and E. Premkumar Reddy. 1998. Signaling by dual specificity kinases. Oncogene 17:1447-1455. [DOI] [PubMed] [Google Scholar]
- 9.Gaidano, G., A. Gloghini, V. Gattei, M. F. Rossi, A. M. Cilia, C. Godeas, M. Degan, T. Perin, V. Canzonieri, D. Aldinucci, G. Saglio, A. Carbone, and A. Pinto. 1997. Association of Kaposi's sarcoma-associated herpesvirus-positive primary effusion lymphoma with expression of the CD138/syndecan-1 antigen. Blood 90:4894-4900. [PubMed] [Google Scholar]
- 10.Horenstein, M. G., R. G. Nador, A. Chadburn, E. M. Hyjek, G. Inghirami, D. M. Knowles, and E. Cesarman. 1997. Epstein-Barr virus latent gene expression in primary effusion lymphomas containing Kaposi's sarcoma-associated herpesvirus human herpesvirus-8. Blood 90:1186-1191. [PubMed] [Google Scholar]
- 11.Jenner, R. G., K. Maillard, N. Cattini, R. A. Weiss, C. Boshoff, R. Wooster, and P. Kellam. 2003. Kaposi's sarcoma-associated herpesvirus-infected primary effusion lymphoma has a plasma cell gene expression profile. Proc. Natl. Acad. Sci. USA 100:10399-10404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klein, U., A. Gloghini, G. Gaidano, A. Chadburn, E. Cesarman, R. Dalla-Favera, and A. Carbone. 2003. Gene expression profile analysis of AIDS-related primary effusion lymphoma (PEL) suggests a plasmablastic derivation and identifies PEL-specific transcripts. Blood 101:4115-4121. [DOI] [PubMed] [Google Scholar]
- 13.Knowles, D. M., G. A. Chamulak, M. Subar, J. S. Burke, M. Dugan, J. Wernz, C. Slywitzky, P.-G. Pelicci, R. Dalla-Favera, and B. Raphael. 1988. Lymphoid neoplasia associated with the acquired immunodeficiency syndrome (AIDS). Ann. Intern. Med. 108:744-753. [DOI] [PubMed] [Google Scholar]
- 14.Lipshutz, R. J., S. P. Fodor, T. R. Gingeras, and D. J. Lockhart. 1999. High density synthetic oligonucleotide arrays. Nat. Genet. 21:20-24. [DOI] [PubMed] [Google Scholar]
- 15.Liu, J., H. Kang, M. Raab, A. J. da Silva, S. K. Kraeft, and C. E. Rudd. 1998. FYB (FYN binding protein) serves as a binding partner for lymphoid protein and FYN kinase substrate SKAP55 and a SKAP55-related protein in T cells. Proc. Natl. Acad. Sci. USA 95:8779-8784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 17.Matolcsy, A., R. G. Nador, E. Cesarman, and D. M. Knowles. 1998. Immunoglobulin VH gene mutational analysis suggests that primary effusion lymphomas derive from different stages of B cell maturation. Am. J. Pathol. 153:1609-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mesri, E. A., E. Cesarman, L. Arvanitakis, S. Rafii, M. A. S. Moore, D. N. Posnett, D. M. Knowles, and A. S. Asch. 1996. Human herpesvirus-8/Kaposi's sarcoma-associated herpesvirus is a new transmissible virus that infects B cells. J. Exp. Med. 183:2385-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moore, P. S., C. Boschoff, R. A. Weiss, and Y. Chang. 1996. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science 274:1739-1744. [DOI] [PubMed] [Google Scholar]
- 20.Rimokh, R., M. Gadoux, M. F. Bertheas, F. Berger, M. Garoscio, G. Deleage, D. Germain, and J. P. Magaud. 1993. FVT-1, a novel human transcription unit affected by variant translocation t(2;18)(p11;q21) of follicular lymphoma. Blood 81:136-142. [PubMed] [Google Scholar]
- 21.Szekely, L., F. Chen, N. Teramoto, B. Ehlin-Henriksson, K. Pokrovskaja, A. Szeles, A. Manneborg-Sandlund, M. Lowbeer, E. T. Lennette, and G. Klein. 1998. Restricted expression of Epstein-Barr virus (EBV)-encoded, growth transformation-associated antigens in an EBV- and human herpesvirus type 8-carrying body cavity lymphoma line. Gen. Virol. 79:1445-1452. [DOI] [PubMed] [Google Scholar]
- 22.Takekawa, M., and H. Saito. 1998. A family of stress-inducible GADD45-like proteins mediate activation of the stress-responsive MTK1/MEKK4 MAPKKK. Cell 95:521-530. [DOI] [PubMed] [Google Scholar]
- 23.Trivedi, P., K. Takazawa, C. Zompetta, L. Cuomo, E. Anastasiadou, A. Carbone, S. Uccini, F. Belardelli, K. Takada, L. Frati, and A. Faggioni. 2004. Infection of HHV-8+ primary effusion lymphoma cells with a recombinant Epstein-Barr virus leads to restricted EBV latency, altered phenotype, and increased tumorigenicity without affecting TCL1 expression. Blood 103:313-316. [DOI] [PubMed] [Google Scholar]