Abstract
Genetic variation of human immunodeficiency virus (HIV-1) represents a major obstacle for AIDS vaccine development. To decrease the genetic distances between candidate immunogens and field virus strains, we have designed and synthesized an artificial group M consensus env gene (CON6 gene) to be equidistant from contemporary HIV-1 subtypes and recombinants. This novel envelope gene expresses a glycoprotein that binds soluble CD4, utilizes CCR5 but not CXCR4 as a coreceptor, and mediates HIV-1 entry. Key linear, conformational, and glycan-dependent monoclonal antibody epitopes are preserved in CON6, and the glycoprotein is recognized equally well by sera from individuals infected with different HIV-1 subtypes. When used as a DNA vaccine followed by a recombinant vaccinia virus boost in BALB/c mice, CON6 env gp120 and gp140CF elicited gamma interferon-producing T-cell responses that recognized epitopes within overlapping peptide pools from three HIV-1 Env proteins, CON6, MN (subtype B), and Chn19 (subtype C). Sera from guinea pigs immunized with recombinant CON6 Env gp120 and gp140CF glycoproteins weakly neutralized selected HIV-1 primary isolates. Thus, the computer-generated “consensus” env genes are capable of expressing envelope glycoproteins that retain the structural, functional, and immunogenic properties of wild-type HIV-1 envelopes.
The high level of genetic variability of HIV-1 poses a major hurdle for AIDS vaccine development. Genetic differences among HIV-1 groups M, N, and O are extensive, ranging from 30 to 50% in the gag and env genes, respectively (14, 20, 33, 35). HIV-1 also frequently recombines among different subtypes to create circulating recombinant forms (CRFs) and novel recombinants (5, 27, 28). To overcome the challenge of HIV-1 diversity, “centralized” HIV-1 genes have been proposed to use in HIV-1 immunogen design. These strategies include using consensus sequences, the most frequent base found in a given position, or ancestral or center-of-the-tree sequences, both modeled from phylogenetic trees (9, 10, 12, 18, 23, 24). A sequence that is central to all HIV-1 epidemic strains within group M would increase amino acid similarities with contemporary field HIV-1 isolates relative to intersubtype distances and therefore might be useful in a setting where diverse subtypes and recombinants are cocirculating (12). However, because centralized genes are artificially made, it has been of great concern that these genes may not be able to fold into native conformations, perform biological functions of native Env, preserve Env antigenic epitopes, or induce salutary immune responses when used as immunogens. To address these concerns, we synthesized an artificial group M consensus env gene (CON6 gene) and studied its biological, antigenic, and immunological properties. Our studies demonstrated that CON6 proteins are biologically functional and are immunogenic for eliciting immune responses to wild-type HIV-1 strains.
MATERIALS AND METHODS
Expression of CON6 gp120 and gp140CF proteins by using rVVs.
To generate secreted forms of group M consensus env gene (CON6) envelope glycoproteins, CON6 gp120 and gp140CF plasmids were constructed by introducing stop codons after the gp120 cleavage site (REKR) and before the membrane-spanning domain (YIKIFIMIVGGLIGLRIVFAVLSIVN), respectively. The gp120/gp41 cleavage site and fusion domain of gp41 were deleted in the gp140CF protein. Recombinant vaccinia viruses (rVVs) containing CON6 env genes were generated as described previously (21) and confirmed by PCR and nucleotide sequence analysis. Recombinant CON6 gp120 and gp140CF glycoproteins were purified with agarose Galanthus nivalis lectin beads (Vector Labs, Burlingame, Calif.) and stored at −70°C until use.
MAbs and gp120 wild-type envelopes.
Human monoclonal antibodies (MAbs) known to bind conformational epitopes on gp120 (A32), the gp120 V3 loop (F39F), and the CCR5 binding site (17b) were kindly provided by James Robinson (Tulane Medical School, New Orleans, La.) (37, 38). MAbs 2F5, 447-52D, IgG1b12, and 2G12 and soluble CD4 (sCD4) were obtained from the National Institutes of Health (NIH) AIDS Research and Reference Reagent Program (Bethesda, Md.) (13, 25, 26, 34). T8 is a murine MAb that maps to the gp120 C1 region (a gift from P. Earl, NIH). BaL (subtype B), 96ZM651 (subtype C), and 93TH975 (subtype E) gp120s were provided by QBI, Inc., and the Division of AIDS, NIH. 92U037 (subtype A) and 93BR029 (subtype F) gp140 proteins (secreted and uncleaved) were purified from CHO cell lines (obtained from the Centralised Facility for AIDS Reagents, National Institute for Biological Standards and Control [NIBSC], Hertfordshire, United Kingdom) by using agarose Galanthus nivalis lectin beads (Vector Labs).
BN-PAGE analysis.
Blue native polyacrylamide gel electrophoresis (BN-PAGE) analysis of CON6 gp120 and gp140CF proteins was carried out according to the methods described by others (30, 31), with minor modifications due to the highly basic pIs of HIV-1 Env proteins. Lectin column-purified proteins were diluted in a buffer containing 50 mM MOPS (morpholinepropanesulfonic acid), 50 mM Tris-HCl (pH 7.7), 20% glycerol, and 0.05% Coomassie blue. Protein samples were loaded onto a 3 to 8% Tris-acetate NuPAGE gel (Invitrogen, Carlsbad, Calif.), and electrophoresis was carried out for 1.5 h at 150 V with 50 mM MOPS-50 mM Tris-HCl (pH 7.7)-0.03% Coomassie blue as the cathode running buffer and 50 mM MOPS-50 mM Tris HCl (pH 7.7) as the anode buffer. Thyroglobulin, ferritin, catalase, and alcohol dehydrogenase (Amersham Biosciences) were included in each run, as was HIV-1 89.6 gp120 Env monomer.
Surface plasmon resonance biosensor measurements and ELISA.
Surface plasmon resonance biosensor measurements were determined on a BIAcore 3000 instrument, and data analysis was performed with BIAevaluation 3.0 software (BIAcore Inc, Upsaala, Sweden). Anti-gp120 MAbs (T8, A32, 17b, and 2G12) or sCD4 in 10 mM Na-acetate buffer (pH 4.5) were directly immobilized to a CM5 sensor chip, using a standard amine coupling protocol for protein immobilization. Fast protein liquid chromatography-purified CON6 gp120 monomer or gp140CF oligomer recombinant proteins were flowed over CM5 sensor chips at concentrations of 100 and 300 μg/ml, respectively. Binding of CON6 envelope proteins was monitored in real time at 25°C with a continuous flow of phosphate-buffered saline (150 mM NaCl, 0.005% surfactant P20 [pH 7.4]) at 10 to 30 μl/min. Enzyme-linked immunosorbent assay (ELISA) was performed to determine the reactivities of various MAbs to CON6 gp120 and gp140CF proteins as described previously (16). For assay of human MAb binding to gp120 or gp140 proteins, end point titers were defined as the highest titer of any MAb (beginning at 20 μg/ml) at which the MAb bound CON6 gp120 and gp140CF Env proteins ≥3-fold over the background control (nonbinding human MAb).
Measurement of kinetics of binding of CON6 Env proteins to sCD4 and anti-HIV-1 MAbs.
Soluble CD4 and MAbs (A32 and T8) were immobilized on individual flow cells of the CM5 sensor chip. Association (on) and dissociation (off) rates were measured by injecting serial dilutions of Env proteins (1.3 to 83 nM for 89.6 gp120, 34 to 1100 nM for CON6 gp120, and 62.5 to 1000 nM for CON6 gp140CF) over each ligand surface at a flow rate of 20 μl/min. For kinetic analysis, nonspecific signal was subtracted from a control immunoglobulin G (IgG) surface (mouse IgG). Rate constants were derived from global curve-fitting analysis, using a Langmuir 1:1 binding model.
Infectivity and coreceptor usage.
HIV-1/SG3Δenv and CON6 or control env plasmids were cotransfected into human 293T cells. Pseudotyped virion preparations were harvested, filtered, analyzed for p24 content (Bechman Coulter, Inc., Miami, Fla.), and then used to infect JC53-BL cells (using serial fivefold dilutions) (8, 36). JC53-BL cells express CD4, CCR5, and CXCR4 receptors and contain a β-galactosidase gene and a firefly luciferase gene stably integrated under the transcriptional control of an HIV-1 long terminal repeat. To determine the coreceptor usage of the CON6 env gene, JC53-BL cells were treated with 1.2 μM AMD3100 and 4 μM TAK799 for 1 h at 37°C and then infected with equal amounts of p24 (5 ng) of each Env-pseudotyped virus. Infectivity in the absence of the blocking agent was set as 100%. Blockage efficiency was expressed as the percentage of the infectivity of the virus in the absence of blocking agent.
Immunizations.
All animals were housed in the Duke University Animal Facility under Association for Assessment and Accreditation of Laboratory Animal Care guidelines with animal use protocols approved by the Duke University Animal Use and Care Committee. For induction of antienvelope antibodies, each of four outbred guinea pigs (Harlan Sprague, Inc., Chicago, Ill.) was given 100 μg of either purified CON6 gp120 or gp140CF subcutaneously in RIBI-CWS adjuvant every 3 weeks (total of five immunizations). Serum samples were heat inactivated (56°C, 1 h) and stored at −20°C until use. For induction of antienvelope T-cell responses, 6- to 8-week-old female BALB/c mice (Frederick Cancer Research and Developmental Center, National Cancer Institute, Frederick, Md.) were immunized intramuscularly (five mice per group) with 50 μg of plasmid DNA three times at 3-week intervals. Three weeks after the last DNA immunization, mice were boosted with 107 PFU of rVV expressing either gp120 or gp140CF glycoprotein.
Neutralization assays.
Neutralization assays were performed with either an MT-2 assay as described previously (3) or a luciferase-based multiple-replication-cycle HIV-1 infectivity assay in 5.25.GFP.Luc.M7 cells with a panel of HIV-1 primary isolates (3, 4). In the luciferase-based assay, neutralizing antibodies were measured as a function of a reduction in luciferase activity in 5.25.EGFP.Luc.M7 cells (kindly provided by Nathaniel R. Landau, Salk Institute, La Jolla, Calif.) (2). Five hundred 50% tissue culture infectious doses of cell-free virus was incubated (1 h, 37°C) with the indicated serum dilutions in 150 μl in triplicate in 96-well flat-bottom culture plates. The 5.25.EGFP.Luc.M7 cells were suspended at a density of 5 × 105/ml in medium containing DEAE-dextran (10 μg/ml). Cells (100 μl) were added until 10% of cells in control wells (no test serum sample) were positive for green fluorescent protein expression by fluorescence microscopy. The suspension of cells (50 μl) was transferred to 96-well white solid plates (Costar, Cambridge, Mass.) for measurement of luciferase activity by using Bright-Glo substrate (Promega, Madison, Wis.) on a Wallac 1420 Multilabel Counter (Perkin-Elmer Life Sciences, Boston, Mass.). Neutralization titers in the luciferase assays were those where relative luciferase units were reduced by ≥50% compared to corresponding preimmune serum. Neutralization titers in the MT-2 assay were determined by a 50% reduction in virus-induced cell killing, which corresponds to a ∼90% reduction in infectivity (3). Only samples with titers beyond 1:20 (i.e., >1:30) and neutralization titers three times over prebleed levels were considered positive. Absorption of sera with HIV-1 V3 peptides was performed at a saturating final peptide concentration of 50 μg/ml.
Enzyme-linked immune spot assay.
Spleens were harvested 2 weeks after the rVV boost, and single-cell suspensions of splenocytes from individual immunized mice were prepared by mincing and forcing through a 70-μm nylon cell strainer (BD Labware, Franklin Lakes, N.J.). Overlapping Env peptides of CON6 gp140 (159 peptides, 15-mers overlapping by 11) were purchased from Boston Bioscience, Inc. (Royal Oak, Mich.). Overlapping Env peptides of MN gp140 (subtype B; 170 peptides, 15-mers overlapping by 11) and Chn19 gp140 (subtype C; 69 peptides, 20-mers overlapping by 10) were obtained from the NIH AIDS Research and Reference Reagent Program. Splenocytes from each mouse (five mice per group) were stimulated in vitro with CON6 overlapping Env peptides pools. Ninety-six-well polyvinylidene difluoride plates (MultiScreen-IP, Millipore, Billerica, Mass.) were coated with an anti-gamma interferon (anti-IFN-γ) MAb (5 μg/ml) (AN18; Mabtech, Stockholm, Sweden). After the plates were blocked at 37°C for 2 h with complete HEPES-buffered RPMI medium, 50 μl of the pooled overlapping envelope peptides (two CON6 pools with 79 or 80 peptides in each pool, two MN pools with 85 peptides in each pool, and one Chn19 pool with 69 peptides) at a final concentration of 5 μg of each peptide per ml was added to the plate, and then 50 μl of splenocytes at a concentration of 107/ml was added to the wells in duplicate and incubated for 16 h at 37°C with 5% CO2. The plates were incubated with 100 μl of a 1:1,000 dilution of streptavidin-alkaline phosphatase (Mabtech), and purple spots were developed with 100 μl of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside-nitroblue tetrazolium (Plus) alkaline phosphatase substrate (Moss, Pasadena, Md.). Spot-forming cells (SFCs) were measured with an Immunospot counting system (CTL Analyzers, Cleveland, Ohio). Total responses for each envelope peptide pool are expressed as SFCs per 106 splenocytes.
RESULTS
CON6 envelope gene design, construction, and expression.
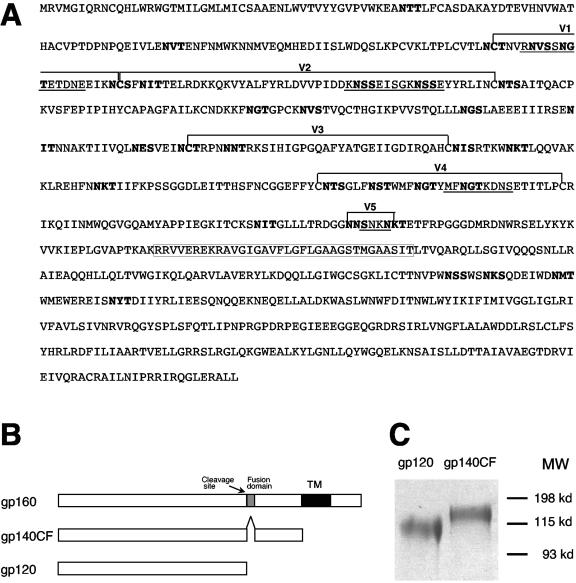
A synthetic group M consensus env gene (CON6 gene) was constructed by generating consensus sequences of env genes for each HIV-1 subtype from sequences in the Los Alamos HIV sequence database and then generating a consensus sequence of all subtype consensuses, thus avoided biasing the group M consensus towards heavily sequenced subtypes (12, 19). The hypervariable regions (V1, V2, V4, and V5) in the env gene evolve by rapid insertion and deletion and not by point mutation. Consensus sequences were not generated for these regions. Instead, five highly variable regions (V1, V2, V4, V5, and a region in the cytoplasmic domain of gp41) from a CRF08_BC recombinant strain (98CN006) were then used to fill in the nonconsensus variable regions in the CON6 sequence, with the thought that these regions might be able to act coordinately to recreate a functional, properly folded envelope when embedded in the context of the M group consensus sequence. The V3 region generally evolves by point mutation with relatively few insertions and deletions, and so in CON6 Env it is based on the group M consensus and not on 98CN006 (Fig. 1A). For high levels of expression, the codons of the CON6 env gene were optimized based on codon usage of highly expressed human genes (1, 15). The codon-optimized CON6 env gene (gp160) was constructed and subcloned into pcDNA3.1 DNA at EcoRI and BamHI sites (11). High levels of protein expression were confirmed by Western blot analysis after transfection into 293T cells (not shown). To obtain soluble recombinant CON6 Env proteins for biological characterization and use as immunogens, rVVs were generated to express secreted gp120 and gp140CF (Fig. 1B). Both proteins were purified from the supernatants of rVV-infected 293T cells by using agarose Galanthus nivalis lectin beads. The purity of each protein was >95% as determined by sodium dodecyl sulfate(SDS)-PAGE analysis (Fig. 1C).
FIG. 1.
Generation and expression of the group M consensus env gene (CON6). (A) The complete amino acid sequence of CON6 gp160 is shown. The five regions from the wild-type CRF08_BC (98CN006) env gene are indicated by underlined letters. Variable regions are indicated by brackets above the sequences. Potential N-liked glycosylation sites are highlighted with boldface letters. The deletion in gp140CF construct is shown in a box. (B) Constructs of CON6 gp120 and gp140CF. (C) Expression of CON6 gp120 and gp140CF. CON6 gp120 and gp140CF were purified from the cell culture supernatants of rVV-infected 293T cells with agarose Galanthus nivalis lectin columns. Both gp120 and gp140CF were separated on a 6% SDS-polyacrylamide gel under reducing conditions and stained with Coomassie blue.
CON6 gp160 is biologically active and uses CCR5 as its coreceptor.
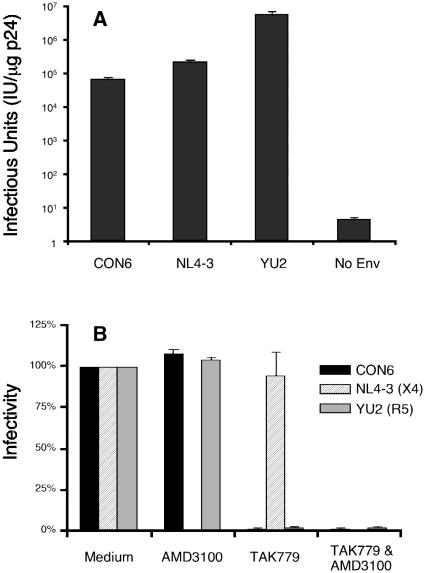
To examine whether the synthetic group M consensus env gene expressed a glycoprotein that could pseudotype an env-deficient HIV-1 provirus, CON6 gp160 was cotransfected with HIV-1/SG3Δenv (along with the NL4-3 and YU2 gp160 genes for controls) into human 293T cells, and the resulting supernatants were analyzed for the presence of infectious viruses in the JC53-BL cell assay (8, 36). JC53-BL cells express CD4, CCR5, and CXCR4 receptors and contain a β-galactosidase gene stably integrated under the transcriptional control of an HIV-1 long terminal repeat. These cells can thus be used to quantify the infectious titers of pseudotyped viral stocks by staining for β-galactosidase expression and counting the number of blue cells (infectious units) per microgram of p24 of input virus. As shown in Fig. 2A, CON6-derived glycoprotein conferred infectivity to HIV-1/SG3Δenv when complemented in trans; however, in three independent experiments, the resulting CON6 Env pseudovirion stocks were reduced in their infectivity by 1 to 2 log units compared to wild-type NL4-3 or YU2 Env pseudovirions.
FIG. 2.
Infectivity and coreceptor usage of CON6 envelope. (A) CON6 and control env plasmids were cotransfected with an HIV-1/SG3Δenv backbone into human 293T cells to generate Env pseudovirions. Culture supernatants were analyzed for p24 content and then used to infect JC53-BL cells (using serial fivefold dilutions). The infectivity was determined by counting the number of blue cells (infectious units) per microgram of p24 of pseudovirions after staining the infected cells for β-galactosidase expression. (B) Coreceptor usage of the CON6 env gene was determined on JC53-BL cells treated with AMD3100 and/or TAK799 for 1 h (37°C) and then infected with equal amounts of p24 (5 ng) of each Env pseudovirion. Infectivity in the control group (no blocking agent) was set as 100%. The blockage efficiency was expressed as the percentage of the infectivity of the virus in the absence of the blocking agent. Data shown are means ± standard deviations from three independent experiments.
Experiments were next performed to determined the coreceptor usage of pseudovirions containing the CON6 gp160 envelope gene. When treated with the CXCR4-blocking agent AMD3100, the infectivity of NL4-3 Env pseudovirions was blocked, while the infectivity of YU2 or CON6 Env pseudovirions was not inhibited (Fig. 2B). In contrast, when treated with the CCR5-blocking agent TAK779, the infectivity of NL4-3 Env pseudovirions was not affected, while the infectivity of YU2 or CON6 Env pseudovirions was inhibited. When treated with both blocking agents, the infectivity of all pseudovirions was inhibited. Taken together, these data showed that the CON6 env gene expressed a glycoprotein that was incorporated into virus particles, mediated virus entry (albeit inefficiently) into appropriate target cells, and used the CCR5 coreceptor for its entry into target cells.
Oligomeric status of CON6 gp120 and gp140CF proteins.
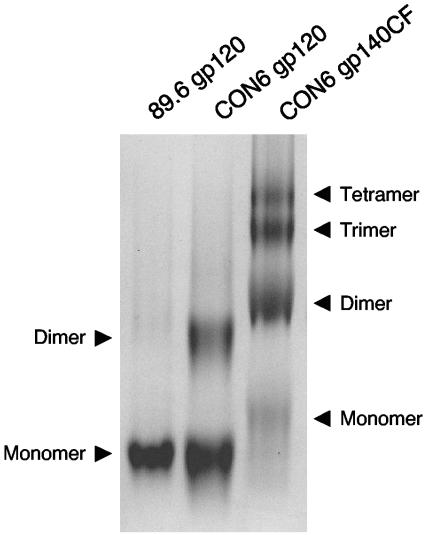
Upon size exclusion chromatography, dimer and higher oligomeric forms of CON6 gp120 and gp140CF, respectively, were detected (data not shown). To more precisely determine the oligomeric status of CON6 Env proteins, we analyzed both CON6 gp120 and gp140CF by BN-PAGE analysis as previously reported (29, 31). A representative result of three determinations is presented in Fig. 3. In the blue native gel, CON6 gp120 proteins migrated as monomers and dimers. However, formation of dimers is not unique to this consensus Env protein, as Center et al. have previously shown that the wild-type HIV-1 Env V2 domain mediates disulfide-linked gp120 dimer formation (6). Dimer formation of CON6 gp120 and gp140CF Env was also disulfide linkage mediated (not shown). On BN-PAGE, the size of CON6 gp120 monomers was very similar to that of 89.6 gp120 monomers. Dimers and oligomeric species (trimers and tetramers) were observed for CON6 gp140CF proteins in the BN-PAGE analysis (Fig. 3). Dimers and trimers of CON6 gp140CF were the most abundant multimer forms, while a lesser gp140CF tetrameric band was detected.
FIG. 3.
BN-PAGE analysis of CON6 gp120 and gp140CF proteins. Affinity-purified 89.6 gp120, CON6 gp120, and CON6 gp140CF proteins were loaded onto a 3 to 8% Tris-acetate NuPAGE gel, and electrophoresis was carried out for 1.5 h at 150 V with 50 mM MOPS-50 mM Tris-HCl (pH 7.7)-0.03% Coomassie blue as the cathode running buffer and 50 mM MOPS-50 mM Tris-HCl (pH 7.7) as the anode buffer.
The CD4 binding domain and other HIV-1 epitopes are preserved on CON6 proteins.
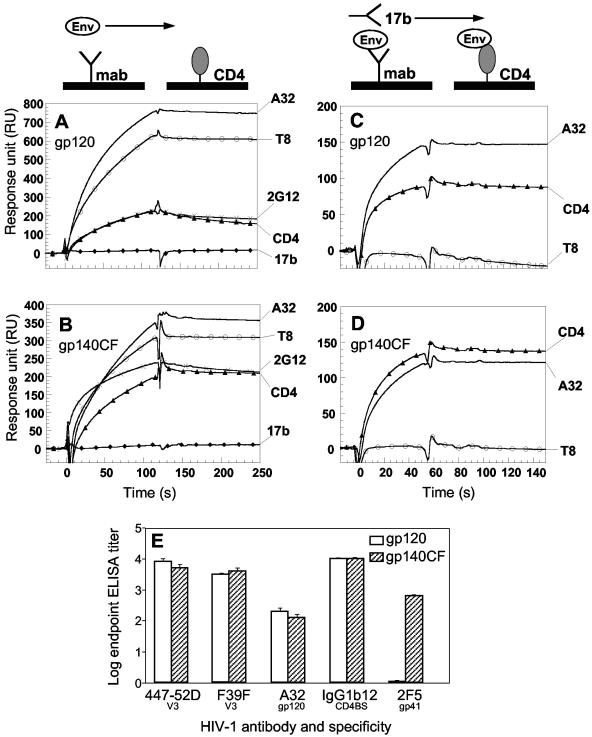
To examine the folding of CON6 env gene-derived gp120 and gp140CF glycoproteins, CON6 proteins were assayed for their ability to bind to sCD4 as well as to several well-characterized anti-gp120 MAbs and to undergo CD4-induced conformational changes. First, BIAcore CM5 sensor chips were coated with either sCD4 or MAbs to monitor their binding to CON6 Env proteins. Both monomeric CON6 gp120 and trimeric gp140CF efficiently bound sCD4 and anti-gp120 MAbs T8, 2G12, and A32, but not MAb 17b (Fig. 4A and B). MAb 17b binds to a CD4-inducible epitope located near the CCR5 binding site of gp120; the binding of either sCD4 or A32 to wild-type gp120 can cause the conformational change that exposes the 17b epitope (37, 38). To determine whether the 17b epitope could also be induced on purified CON6 Envs, sCD4, A32, or T8 was applied to sensor chips to capture CON6 gp120 or gp140CF, and then 17b binding activity was monitored. As shown in Fig. 4C and D, both CON6 gp120 and gp140CF were induced to undergo conformational changes and bound 17b only following binding to sCD4 and A32 but not T8.
FIG. 4.
Binding of CON6 gp120 and gp140CF to sCD4 and anti-Env MAbs. (A and B) Each of the indicated MAbs and sCD4 were covalently immobilized to a CM5 sensor chip (BIAcore), and CON6 gp120 (A) or gp140CF (B) was injected over each surface (100 and 300 μg/ml, respectively). (C and D) To determine induction of 17b MAb binding to CON6 gp120 and gp140CF, CON6 gp120 (C) or gp140CF (D) proteins were captured (400 to 580 response units) on individual flow cells immobilized with sCD4 or MAb A32 or T8. Following stabilization of each of the surfaces, MAb 17b was injected and allowed to flow over each of the immobilized flow cells. (E) To determine binding of CON6 gp120 and gp140CF to human MAbs in ELISA, titers of stock solutions of 20 μg of MAbs 447-52D, F39F, A32, IgG1b12, and 2F5 were determined with CON6 gp120 and gp140CF glycoproteins. MAbs 447-52D (V3), F39F (V3) A32 (gp120), and IgG1b12 (CD4 binding site) each bound to both CON6 gp120 and gp140CF well, while 2F5 (anti-gp41 ELDKWAS) bound only CON6 gp140CF. The concentrations at the end point titer (end titer where the experimental versus the control value was ≥3.0) with gp120 for MAb 447-52D and F39F binding were <0.003 and 0.006 μg/ml, respectively; that for MAb A32 was <0.125 μg/ml; that for IgG1b12 was <0.002 μg/ml; and that for 2F5 with gp140CF was 0.016 μg/ml.
Both CON6 gp120 and gp140CF bound to sCD4, T8, and A32 with high affinity (Kd in the nanomolar range) as determined by measuring the on and off rates of binding in BIAcore analysis (Table 1). The bindings of CON6 gp120 and gp140CF to immobilized sCD4 were similar to each other (34.7 and 13.8 nM, respectively) but lower than that observed with 89.6 gp120 (5.7 nM). The differences were largely in the on rates (ka), with CON6 Env proteins binding with lower on rates. Similar binding activities of CON6 Env proteins in on rates were also observed when rates of binding to A32 and T8 were compared. However, it is important to note that the reported affinity of HIV-1 gp120 proteins binding to sCD4 spans a relatively wider range, from about 1 to 2 nM (89.6, YU2, and ADA) to 20 to 28 nM (JRFL and BH10) (17, 22, 39, 40). Thus, the affinities for CON6 gp120 and gp140CF fall within this range.
TABLE 1.
Kinetics of sCD4 and anti-HIV-1 MAb binding to CON6 Env proteinsa
| Ligand | Analyte | ka (M−1 s−1) | kd (s−1) | Kd (nM) |
|---|---|---|---|---|
| sCD4 | 89.6 gp120 | 9.8 × 104 | 5.6 × 10−4 | 5.7 |
| CON6 gp120 | 1.7 × 104 | 5.9 × 10−4 | 34.7 | |
| CON6 gp140 | 1.6 × 104 | 2.2 × 10−4 | 13.8 | |
| T8 | 89.6 gp120 | 3.9 × 104 | 9.5 × 10−5 | 2.4 |
| CON6 gp120 | 1.2 × 104 | 3.0 × 10−4 | 25.0 | |
| CON6 gp140 | 1.1 × 104 | 2.0 × 10−4 | 19.1 | |
| A32 | 89.6 gp120 | 1.6 × 105 | 5.3 × 10−5 | 0.3 |
| CON6 gp120 | 1.3 × 104 | 4.2 × 10−5 | 3.2 | |
| CON6 gp140 | 9.7 × 103 | 1.1 × 10−5 | 1.2 |
ka, on rate; kd, off rate; Kd, affinity.
In an ELISA, both CON6 gp120 and gp140CF bound well to neutralizing V3 MAbs 447-52D and F39F and to the potent neutralizing CD4 binding site MAb IgG1b12. MAb 2F5, which neutralizes HIV-1 primary isolates by binding to a C-terminal gp41 epitope, also bound well to CON6 gp140CF (Fig. 4E).
Reaction of CON6 gp120 with sera of different subtypes.
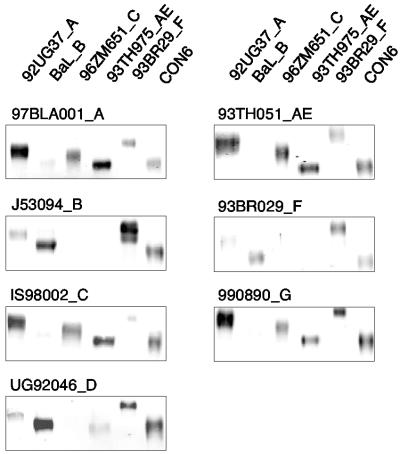
To determine whether multiple subtype linear epitopes are preserved on CON6 gp120, a panel of recombinant Env proteins (one each for subtypes A, B, C, F, and CRF01_AE gp120 or gp140 protein) was assembled. Equal amounts of each Env protein (100 ng) were loaded on SDS-polyacrylamide gels, transferred to nitrocellulose, and reacted with subtype A through G patient sera (1:1,000 dilution) in Western blot assays. For each HIV-1 subtype, four to six patient sera were tested. One serum representative of each subtype is shown in Fig. 5. Whereas sera of all subtypes tested showed variable reactivities among Envs of different subtypes in the panel, patient sera of all subtypes reacted equally well with CON6 gp120 Env protein, demonstrating that cross-reactive HIV-1 Env epitopes recognized by patient sera were well preserved on the CON6 Env protein.
FIG. 5.
Western blot analysis of Env proteins of multiple subtypes against antisera of multiple subtypes. Equal amounts of Env proteins (100 ng) were separated on SDS-10% polyacrylamide gels (gp120: Bal, 96ZM651, 93TH975, and CON6; gp140: 92UG37, and 93BR029). Following electrophoresis, proteins were transferred to nitrocellulose membranes and reacted with sera from HIV-1-infected patients (1:1,000). Protein-bound antibody was probed with fluorescence-labeled secondary antibodies, and the images were scanned and recorded on an Odyssey infrared imager (Li-Cor, Lincoln, Nebr.). Subtypes are indicated by single letters after Env protein and serum designations. Four to six sera were tested for each subtype, and reaction patterns were similar among all sera from the same subtype. One representative result for each subtype serum is shown.
Induction of T-cell responses to CON6 envelope overlapping peptides.
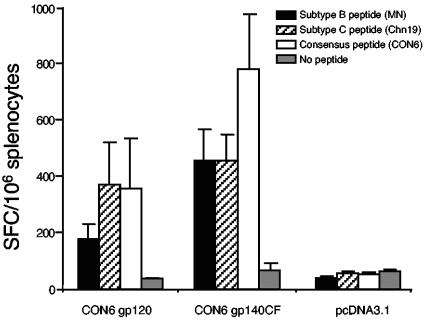
T-cell immune responses induced by CON6 envelope immunogens were assayed against overlapping CON6 Env peptides as well as subtype B or C peptide pools (MN and Chn19, respectively). Three groups of mice (five mice per group) were primed with CON6 gp120, CON6 gp140CF, or empty vector DNA control and boosted with rVV expressing the corresponding envelope glycoproteins or a control vaccinia virus strain (WR). IFN-γ SFCs were detected when splenocytes from mice immunized with either CON6 gp120 or gp140CF proteins were stimulated with CON6, MN, or Chn19 peptide pools (Fig. 6). The T-cell immune responses induced by CON6 gp140CF appeared to be more robust than those induced by CON6 gp120 for any peptide pool. These data demonstrated that CON6 gp120 and gp140CF immunogens were capable of inducing T-cell responses to both CON6 epitopes and to subtype B and subtype C wild-type epitopes.
FIG. 6.
T-cell immune responses induced by CON6 Env immunogens in mice. Splenocytes were isolated from individual immunized mice (five mice per group). After splenocytes were stimulated in vitro with overlapping Env peptide pools of CON6, Chn19_C, MN_B, and medium, IFN-γ-producing cells were determined by the enzyme-linked immune spot assay. Total responses for each immunogen and peptide pool are expressed as SFCs per million splenocytes. The values are the means ± standard errors of the means of IFN-γ SFCs (n = 5 mice per group).
Induction of antibodies that neutralize selected HIV-1 primary isolates.
To determine whether CON6 envelope immunogens induced antibodies that could neutralize HIV-1 primary isolates, guinea pigs were immunized with either CON6 gp120 or gp140CF protein. Sera collected after four or five immunizations were used for neutralization assays and compared to the corresponding prebleed sera (Table 2). All gp120 and gp140CF sera showed neutralization of two of eight subtype B primary isolates (BXO8 and SF162), and most gp120 and gp140CF sera showed weak neutralization of subtype B isolate QH0692 and SS1196. Sporadic weak neutralization or no neutralization was detected against SHIV SF162P3 and SHIV89.6P, as well as against the other subtype A, B, C, D, and CRF01_AE primary isolates tested. Thus, these data showed that while the group M consensus CON6 envelope was indeed capable of inducing antibodies that could neutralize select subtype B HIV-1 primary isolates, the level of neutralization was weak for all isolates neutralized except HIV-1 SF162.
TABLE 2.
Ability of group M consensus HIV-1 envelope CON6 gp120 and gp140CF proteins to induce antibodies that neutralize HIV-1 primary isolates
| HIV isolate (subtype)a | Titerb with the indicated guinea pig or control
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CON6 gp120 protein
|
CON6 gp140CF protein
|
Positive controlc
|
|||||||||
| 646 | 647 | 648 | 649 | 650 | 651 | 652 | 653 | TriMab2 | CD4-IgG2 | HIV-positive serum | |
| 92RW020 (A) | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | NTd | NT | 693 |
| BAL (B) | <20 | <20 | <20 | <20 | <20 | 200 | <20 | <20 | NT | NT | 3307 |
| BX08 (B) | 438 | 326 | 364 | 417 | 481 | 480 | 652 | 246 | 0.7 | NT | 2384 |
| BG1168 (B) | <20 | <20 | <20 | <20 | 40 | <20 | <20 | 25 | 2.7 | NT | NT |
| 6101 (B) | <20 | <20 | <20 | <20 | <20 | <20 | 72 | <20 | 1.1 | NT | NT |
| SF162 (B) | 2,146 | 388 | 110 | 302 | 235 | 5,502 | 15,098 | 199 | NT | NT | >540 |
| SS1196 (B) | 246 | >20 | 185 | 86 | 381 | 449 | 333 | 117 | NT | NT | 301 |
| QH0692 (B) | 31 | 32 | 34 | <20 | <20 | 33 | 30 | 45 | 0.8 | NT | 769 |
| PAVO (B) | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | 209 | NT | NT |
| SHIV 89.6P (B) | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | NT | NT | NT |
| SHIV SF162P3 (B) | <20 | 30 | 48 | <20 | 27 | <20 | <20 | <20 | NT | 0.2 | NT |
| DU368 (C) | 25 | 35 | 62 | <20 | <20 | <20 | <20 | 23 | NT | 2.3 | NT |
| S021 (C) | <20 | <20 | 33 | <20 | <20 | <20 | <20 | <20 | NT | 803 | NT |
| S080 (C) | 24 | 37 | 70 | 41 | <20 | <20 | <20 | 52 | NT | 3.4 | NT |
| DU179 (C) | <20 | <20 | <20 | <20 | <20 | <20 | <20 | 540 | NT | 0.8 | NT |
| S007 (C) | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | NT | NT | NT |
| S017 (C) | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | NT | NT | NT |
| TV-1 (C) | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | NT | NT | NT |
| 93ZR001 (D) | 314 | 170 | 156 | <20 | 339 | 339 | 181 | 262 | NT | NT | 693 |
| CM244 (CRF01_AE) | 35 | <20 | 85 | <20 | 31 | <20 | <20 | 25 | NT | NT | 693 |
SHIV 89.6P was tested in the MT-2 assay; all other HIV isolates were tested in the M7-luciferase assay. HIV-1 isolates QH0692, SS1196, SF162, 6101, BX08, BG1168, and BAL were assayed with post-injection 5 serum; other HIV-1 isolates were assayed with post-injection 4 serum.
Prebleed values for sera with positive neutralization titers ranged from <20 to 69. All positive neutralization titers were ≥3-fold over background (boldface) and were >30.
HIV-positive serum was either a HIV-1-positive human serum (LEH03) or an anti-gp120 guinea pig serum (used only with isolate SS1196) with known neutralizing activity for HIV-1 isolate SS1196. TriMab2 is a mixture of human MAbs 2F5, b12, and 2G12. Values for TriMab2 and CD4-IgG2 are in micrograms per milliliter.
NT, not tested.
Ability of group M consensus V3 peptide to absorb some neutralizing activity of CON6 gp120 and gp140CF-induced antibodies.
To determine the contribution of the group M consensus V3 loop antibodies to the ability to neutralize HIV-1 primary isolates and to determine the specificity of epitopes of gp120- and gp140CF-induced antibodies for both T-cell-line-adapted and primary HIV-1 strains, we have tested anti-gp120 and -gp140CF sera in the presence of 50 μg of group M consensus peptide (TRPNNNTRKSIHIGPGQAFYATGEIIGDIRQAH) per ml (final concentration) for the ability to neutralize HIV-1 (Tables 3 and 4). For anti-gp120 guinea pig sera, the V3 peptide absorbed a mean of 77% of the neutralizing activity for the TCLA strain HIV-1 MN and 53% of the activity for the primary isolate HIV-1 SS1196, while the group M consensus V3 peptide absorbed only 30% of the neutralizing activity for primary isolate HIV-1 BX08 (P < 0.03 for BX08 versus MN) (Table 3). Similarly, for anti-gp140CF sera, the V3 peptide absorbed 87% of anti-MN activity, 66% of anti-SS1196 activity, and only 19% of anti-BX08 activity (P < 0.01 for BX08 versus MN) (Table 4). Taken together, these data demonstrated that the neutralizing activity for group M consensus antisera for HIV MN and to a lesser extent for SS1196 is predominantly against the V3 loop, and specificities of antibodies distinct from the V3 antibodies absorbed by the V3 peptide are responsible for the majority of CON6 Env-induced neutralization activity for the HIV-1 primary isolate BX08.
TABLE 3.
Ability of group M consensus V3 peptides to absorb CON6 gp120- induced neutralizing activity to T-cell line-adapted HIV-1 MN and to HIV-1 primary isolates SS1196 and BX08
| Guinea pig no. | HIV-1 Isolate | Neutralizing antibody titera
|
||
|---|---|---|---|---|
| Before absorption | After absorption | % Absorption | ||
| 646 | MN | 6,166 | 2,309 | 63 |
| 647 | MN | 1,089 | 292 | 73 |
| 648 | MN | 1,364 | 195 | 86 |
| 649 | MN | 388 | 57 | 85 |
| Mean ± SEM | 77 ± 5 | |||
| 646 | SS1196 | 1,408 | 406 | 68 |
| 647 | SS1196 | 380 | 251 | 34 |
| 648 | SS1196 | 469 | 195 | 58 |
| 649 | SS1196 | 113 | 57 | 50 |
| Mean ± SEM | 53 ± 7 | |||
| 646 | BX08 | 510 | 333 | 35 |
| 647 | BX08 | 454 | 173 | 62 |
| 648 | BX08 | 388 | 258 | 34 |
| 649 | BX08 | 124 | 137 | −10 |
| Mean ± SEM | 30 ± 15 | |||
Neutralizing titers are from the multiple-round luciferase assay as described in Materials and Methods.
TABLE 4.
Ability of group M consensus V3 peptides to absorb CON6 gp140CF-induced neutralizing activity to T-cell line-adapted HIV-1 MN and to HIV-1 primary isolates SS1196 and BX08
| Guinea pig no. | HIV-1 Isolate | Neutralizing antibody titera
|
||
|---|---|---|---|---|
| Before absorption | After absorption | % Absorption | ||
| 650 | MN | 2,179 | 168 | 94 |
| 651 | MN | 7,729 | 942 | 88 |
| 652 | MN | 3,400 | 954 | 72 |
| 653 | MN | 860 | 57 | 93 |
| Mean ± SEM | 87 ± 5 | |||
| 650 | SS1196 | 644 | 46 | 93 |
| 651 | SS1196 | 547 | 223 | 59 |
| 652 | SS1196 | 506 | 256 | 49 |
| 653 | SS1196 | 368 | 133 | 64 |
| Mean ± SEM | 66 ± 9 | |||
| 650 | BX08 | 409 | 196 | 52 |
| 651 | BX08 | 267 | 137 | 49 |
| 652 | BX08 | 178 | 223 | −28 |
| 653 | BX08 | 254 | 248 | 2 |
| Mean ± SEM | 19 ± 20 | |||
Neutralizing titers are from the multiple-round luciferase assay as described in Materials and Methods.
DISCUSSION
In this study, we have tested a new strategy for HIV-1 immunogen design by generating a group M consensus env gene (CON6 gene) with the purpose of decreasing the genetic distance between this candidate immunogen and circulating virus strains. A critical question that this study has answered is whether such a synthetic gene can encode an envelope glycoprotein with antigenicity and immunogenicity properties resembling those of wild-type HIV-1 envelope glycoproteins. We have demonstrated this important proof of concept that the synthetic group M consensus Env protein was biologically functional, preserved many well-studied antigen epitopes, cross-reacted with patient sera of multiple subtypes, and were immunogenic for inducing both T- and B-cell anti-HIV-1 responses in mice or guinea pigs.
The genetic distance between the group M consensus Env sequence and other subtype Env sequences is only about 15%, which is half of that seen between wild-type subtypes (30%) (12). The CON6 Env was generated by choosing the most common amino acid at most positions in all subtypes in constant and V3 regions (12, 19). Since only the most common amino acids were selected, it was anticipated that the majority of antibody and T-cell epitopes would be well preserved. We postulate that polyclonal T- and B-cell responses to a consensus immunogen would be likely to include a greater number of shared epitopes with improved potential to cross-react with circulating strains than any one natural strain. Cross-reactivity of the CON6 Env protein to patient sera of multiple subtypes indicated that cross-reactive antibody binding epitopes were preserved on this artificial group M consensus Env protein. These data suggest that the group M consensus Env protein would be an ideal candidate for use in diagnostic test kits for HIV-1 antibodies.
Infectivity of CON6-Env pseudovirions was demonstrated by using a single-round infection system, although the ability to mediate viral entry was reduced, indicating a suboptimal envelope conformation affecting entry and/or uncoating events. Nonetheless, BIAcore data showed that both CON6 gp120 and gp140CF bound sCD4 and a number of conformationally sensitive MAbs that also are reactive with wild-type HIV-1 Env proteins. The group M consensus envelope utilized the CCR5 coreceptor, which is an important consideration for a vaccine candidate since primary HIV infections tend to be with viruses with CCR5 usage (7, 32).
CON6 Env proteins were immunogenic for inducing both T- and B-cell immune responses, including neutralizing antibodies, albeit of limited strength and breadth for the latter. When used for immunization in mice, both CON6 gp120 and gp140CF proteins induced T-cell immune responses to subtype B and subtype C wild-type Envs (Fig. 6). However, future studies are needed to test the breadth of T-cell immune responses in mice and nonhuman primates by expanding the number of strains of mice and native Env overlapping peptide sets tested and to directly determine whether CON6 envelope genes are better than wild-type env genes for induction of cross-subtype T-cell responses. These studies with multiple strains of mice and with nonhuman primates are under way.
Many contemporary envelope glycoproteins express epitopes to which potent neutralizing human MAbs bind, yet when used as immunogens themselves, they do not induce broadly neutralizing anti-HIV-1 antibodies. Our neutralizing antibody studies did demonstrate as proof of concept the ability of CON6 Envs to induce antibodies that weakly neutralized selected HIV-1 primary isolates. However, it is also clear that the primary isolates that are most difficult to neutralize (e.g., PAVO, 6101, BG1168, 92RW020, and CM244) were either only weakly or not neutralized by anti-CON6 gp120 or gp140CF sera (Table 2). For the TCLA strain MN and one of two primary isolates tested (SS1196), antibodies to the group M consensus V3 loop were responsible for the majority of the neutralizing activity in both gp120 and gp140CF sera (Tables 3 and 4). Thus, for centralized envelopes such as CON6 to be candidates as immunogens, significant improvements in consensus Env immunogenicity will need to be made.
BN-PAGE analysis of CON6 gp120 and gp140CF showed oligomer formation of CON6 gp140CF in the form of trimers and tetramers. The similarity of neutralizing antibody responses induced by CON6 gp120 and gp140CF proteins indicated that with the CON6 group M consensus protein, the oligomeric form did not add enhanced immunogenicity. It will be important to compare the neutralizing capacity of antibodies induced by newer consensus Env proteins in which all variable loops are consensus to that of the spectrum of neutralizing antibodies induced by CON6 Envs.
Three computer models (consensus, ancestor, and center of the tree) have been proposed to generate centralized HIV-1 genes (9, 10, 12, 19, 23, 24). The biology of HIV-1 gives rise to star-like phylogenies, and as a consequence of this, the three kinds of sequences are very similar to each other (10). Any of the three will reduce the protein distances between immunogens and field virus strains. In the case of the M group central sequence studied here, distances to all subtypes and recombinants are essentially reduced to intrasubtype levels (12, 24). Within subtypes, distances are roughly halved (12). However, given the fact that HIV-1 is diversifying under host immune pressure, the small number of differences between the three model sequences may be enriched for immunologically important positions.
Consensus, ancestral, and center-of-the-tree sequences, despite their similarities, have theoretical advantages and disadvantages (10, 12, 23). Global sequencing is generally conducted with viruses sampled during chronic infections that have been subjected to within-host immune pressure, not with transmitted viruses sampled during acute infection. While consensus sequences are arguably the most representative of current circulating viral populations, ancestral and center-of-the-tree sequences hypothetically may have an advantage of recreating potent epitopes that have tended to escape over time during chronic infections but, for reasons of viral fitness and transmission, tend to revert to a more ancestral form in a new host. However, even phylogenetic reconstructions of center-of-the-tree and ancestor sequences may miss such epitopes if they are inadequately represented in the sequences sampled and used to reconstruct the trees. Furthermore, focusing a vaccine response on epitopes that for the most part have escaped and are rare in a contemporary population may be a disadvantage, no matter how potent the response to a particular epitope is. Another potentially useful strategy would be to derive central sequences by using only samples obtained during acute infection, but at this time such samples are inadequately represented in the database.
The CON6 env gene was our first prototype gene designed in 2000 from 1999 Los Alamos HIV sequence database sequences. A next-generation group M consensus env (CON-S) has been generated based on 2000 database sequences. The CON-S env gene contains minimum consensus variable regions and is currently being tested for antigenicity and immunogenicity. The data in our present study demonstrate that our initial prototype consensus Env shares antigenic, functional, and immunogenic properties with wild-type Env. Future studies with newer centralized Env sequences will determine the full extent of breadth of T- and B-cell responses that can be generated by centralized artificial genes.
Acknowledgments
We acknowledge Maria Rhein and Casey Paleos for expert technical assistance; Marcia Kalish, Clement Zeh, and Guido Ferrari for HIV-1-infected patient sera; and the Centralised Facility for AIDS Reagents, NIBSC, Hertfordshire, United Kingdom, for HIV-1-infected patient sera and CHO cell lines that express 92UG037 and 93BR029 gp140 proteins.
This work was supported by NIAID grants AI85338, AI54497, AI55386, PO-1 AI52816, and IPCAVD AI35351; Proteomics Core of the Duke Center for Translational Research grant PO-1 AI51445; and the NIAID AIDS Research and Reference Reagent Program. E.A.W. was supported by NIH training grant 5T32 AI07392. B.T.K. was funded through an NIH-DOE interagency agreement.
REFERENCES
- 1.Andre, S., B. Seed, J. Eberle, W. Schraut, A. Bultmann, and J. Haas. 1998. Increased immune response elicited by DNA vaccination with a synthetic gp120 sequence with optimized codon usage. J. Virol. 72:1497-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brandt, S. M., R. Mariani, A. U. Holland, T. J. Hope, and N. R. Landau. 2002. Association of chemokine-mediated block to HIV entry with coreceptor internalization. J. Biol. Chem. 277:17291-17299. [DOI] [PubMed] [Google Scholar]
- 3.Bures, R., A. Gaitan, T. Zhu, C. Graziosi, K. M. McGrath, J. Tartaglia, P. Caudrelier, R. El Habib, M. Klein, A. Lazzarin, D. M. Stablein, M. Deers, L. Corey, M. L. Greenberg, D. H. Schwartz, and D. C. Montefiori. 2000. Immunization with recombinant canarypox vectors expressing membrane-anchored glycoprotein 120 followed by glycoprotein 160 boosting fails to generate antibodies that neutralize R5 primary isolates of human immunodeficiency virus type 1. AIDS Res. Hum. Retroviruses 16:2019-2035. [DOI] [PubMed] [Google Scholar]
- 4.Bures, R., L. Morris, C. Williamson, G. Ramjee, M. Deers, S. A. Fiscus, S. Abdool-Karim, and D. C. Montefiori. 2002. Regional clustering of shared neutralization determinants on primary isolates of clade C human immunodeficiency virus type 1 from South Africa. J. Virol. 76:2233-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carr, J. K., B. T. Foley, T. Leitner, M. O. Salminen, B. T. Korber, and F. McCutchan. 1998. Reference sequences representing the principal genetic diversity of HIV-1 in the pandemic, p. III-10-III-19. In B. Korber, C. Kuiken, B. Foley, B. H. Hahn, F. McCutchan, J. W. Mellors, and J. Sodrosky (ed.), Human retroviruses and AIDS 1998: a compilation and analysis of nucleic acid and amino acid sequence. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, N.Mex.
- 6.Center, R. J., P. L. Earl, J. Lebowitz, P. Schuck, and B. Moss. 2000. The human immunodeficiency virus type 1 gp120 V2 domain mediates gp41-independent intersubunit contacts. J. Virol. 74:4448-4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Connor, R. I., K. E. Sheridan, D. Ceradini, S. Choe, and N. R. Landau. 1997. Change in coreceptor use correlates with disease progression in HIV-1-infected individuals. J. Exp. Med. 185:621-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Derdeyn, C. A., J. M. Decker, J. N. Sfakianos, X. Wu, W. A. O'Brien, L. Ratner, J. C. Kappes, G. M. Shaw, and E. Hunter. 2000. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J. Virol. 74:8358-8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellenberger, D. L., B. Li, L. D. Lupo, S. M. Owen, J. Nkengasong, M. S. Kadio-Morokro, J. Smith, H. Robinson, M. Ackers, A. Greenberg, T. Folks, and S. Butera. 2002. Generation of a consensus sequence from prevalent and incident HIV-1 infections in West Africa to guide AIDS vaccine development. Virology 302:155-163. [DOI] [PubMed] [Google Scholar]
- 10.Gao, F., T. Bhattacharya, B. Gaschen, J. Taylor, J. P. Moore, V. Novitsky, K. Yusim, D. Lang, B. Foley, S. Beddows, M. Alam, B. Haynes, B. H. Hahn, and B. Korber. 2003. Consensus and ancestral state HIV vaccines. Science 299:1517-1518. [Google Scholar]
- 11.Gao, F., Y. Li, J. Decker, F. W. Peyerl, F. Bibollet-Ruche, C. M. Rodenburg, Y. Chen, D. S. Shaw, S. Allen, R. Musonda, G. M. Shaw, J. Z. Allan, N. L. Letvin, and B. H. Hahn. 2003. Codon usage optimization of HIV-1 subtype C gag, pol, env and nef genes: in vitro expression and immune responses in DNA vaccinated mice. AIDS Res. Hum. Retroviruses 9:817-823. [DOI] [PubMed] [Google Scholar]
- 12.Gaschen, B., J. Taylor, K. Yusim, B. Foley, F. Gao, D. Lang, V. Novitsky, B. Haynes, B. H. Hahn, T. Bhattacharya, and B. Korber. 2002. Diversity considerations in HIV-1 vaccine selection. Science 296:2354-2360. [DOI] [PubMed] [Google Scholar]
- 13.Gorny, M. K., T. C. VanCott, C. Hioe, Z. R. Israel, N. L. Michael, A. J. Conley, C. Williams, J. A. Kessler II, P. Chigurupati, S. Burda, and S. Zolla-Pazner. 1997. Human monoclonal antibodies to the V3 loop of HIV-1 with intra- and interclade cross-reactivity. J. Immunol. 159:5114-5122. [PubMed] [Google Scholar]
- 14.Gurtler, L. G., P. H. Hauser, J. Eberle, A. von Brunn, S. Knapp, L. Zekeng, J. M. Tsague, and L. Kaptue. 1994. A new subtype of human immunodeficiency virus type 1 (MVP-5180) from Cameroon. J. Virol. 68:1581-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haas, J., E. C. Park, and B. Seed. 1996. Codon usage limitation in the expression of HIV-1 envelope glycoprotein. Curr. Biol. 6:315-324. [DOI] [PubMed] [Google Scholar]
- 16.Haynes, B. F., M. A. Moody, C. S. Heinley, B. Korber, W. A. Millard, and R. M. Scearce. 1995. HIV type 1 V3 region primer-induced antibody suppression is overcome by administration of C4-V3 peptides as a polyvalent immunogen. AIDS Res. Hum. Retroviruses 11:211-221. [DOI] [PubMed] [Google Scholar]
- 17.Kim, M., B. Chen, R. E. Hussey, Y. Chishti, D. Montefiori, J. A. Hoxie, O. Byron, G. Campbell, S. C. Harrison, and E. L. Reinherz. 2001. The stoichiometry of trimeric SIV glycoprotein interaction with CD4 differs from that of anti-envelope antibody Fab fragments. J. Biol. Chem. 276:42667-42676. [DOI] [PubMed] [Google Scholar]
- 18.Korber, B., B. Gaschen, K. Yusim, R. Thakallapally, C. Kesmir, and V. Detours. 2001. Evolutionary and immunological implications of contemporary HIV-1 variation. Br. Med. Bull. 58:19-42. [DOI] [PubMed] [Google Scholar]
- 19.Korber, B., M. Muldoon, J. Theiler, F. Gao, R. Gupta, A. Lapedes, B. H. Hahn, S. Wolinsky, and T. Bhattacharya. 2000. Timing the ancestor of the HIV-1 pandemic strains. Science 288:1789-1796. [DOI] [PubMed] [Google Scholar]
- 20.Kuiken, C. L., B. Foley, E. Freed, B. H. Hahn, P. A. Marx, F. McCutchan, J. W. Mellors, S. Wolinksy, and B. Korber. 2002. HIV sequence compendium 2002. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, N.Mex.
- 21.Moss, B., and P. Earl. 1998. Expression of proteins in mammalian cells using vaccinia viral vectors, p. 16.15.1-16.19. 9. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley & Sons, Inc., Indianapolis, Ind.
- 22.Myszka, D. G., R. W. Sweet, P. Hensley, M. Brigham-Burke, P. D. Kwong, W. A. Hendrickson, R. Wyatt, J. Sodroski, and M. L. Doyle. 2000. Energetics of the HIV gp120-CD4 binding reaction. Proc. Natl. Acad. Sci. USA 97:9026-9031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nickle, D. C., M. A. Jensen, G. S. Gottlieb, D. Shriner, G. H. Learn, A. G. Rodrigo, and J. I. Mullins. 2003. Consensus and ancestral state HIV vaccines. Science 299:1515-1517. [DOI] [PubMed] [Google Scholar]
- 24.Novitsky, V., U. R. Smith, P. Gilbert, M. F. McLane, P. Chigwedere, C. Williamson, T. Ndung'u, I. Klein, S. Y. Chang, T. Peter, I. Thior, B. T. Foley, S. Gaolekwe, N. Rybak, S. Gaseitsiwe, F. Vannberg, R. Marlink, T. H. Lee, and M. Essex. 2002. Human immunodeficiency virus type 1 subtype C molecular phylogeny: consensus sequence for an AIDS vaccine design? J. Virol. 76:5435-5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nyambi, P. N., J. Nkengasong, P. Lewi, K. Andries, W. Janssens, K. Fransen, L. Heyndrickx, P. Piot, and G. van der Groen. 1996. Multivariate analysis of human immunodeficiency virus type 1 neutralization data. J. Virol. 70:6235-6243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Purtscher, M., A. Trkola, G. Gruber, A. Buchacher, R. Predl, F. Steindl, C. Tauer, R. Berger, N. Barrett, A. Jungbauer, et al. 1994. A broadly neutralizing human monoclonal antibody against gp41 of human immunodeficiency virus type 1. AIDS Res. Hum. Retroviruses 10:1651-1658. [DOI] [PubMed] [Google Scholar]
- 27.Robertson, D. L., J. P. Anderson, J. A. Bradac, J. K. Carr, B. Foley, R. K. Funkhouser, F. Gao, B. H. Hahn, M. L. Kalish, C. Kuiken, G. H. Learn, T. Leitner, F. McCutchan, S. Osmanov, M. Peeters, D. Pieniazek, M. Salminen, P. M. Sharp, S. Wolinsky, and B. Korber. 2000. HIV-1 nomenclature proposal. Science 288:55-56. [DOI] [PubMed] [Google Scholar]
- 28.Robertson, D. L., J. P. Anderson, J. A. Bradac, J. K. Carr, B. Foley, R. K. Funkhouser, F. Gao, B. H. Hahn, M. L. Kalish, C. Kuiken, G. H. Learn, T. Leitner, F. McCutchan, S. Osmanov, M. Peeters, D. Pieniazek, M. Salminen, P. M. Sharp, S. Wolinsky, and B. Korber. 2000. HIV-1 nomenclature proposal: a reference guide to HIV-1 classification, p. 492-505. In C. L. Kuiken, B. Foley, B. H. Hahn, P. A. Marx, F. McCutchan, J. W. Mellors, J. I. Mullins, S. Wolinksy, and B. Korber (ed.), Human retroviruses and AIDS 1999: a compilation and analysis of nucleic acid and amino acid sequences. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, N.Mex.
- 29.Sanders, R. W., M. Vesanen, N. Schuelke, A. Master, L. Schiffner, R. Kalyanaraman, M. Paluch, B. Berkhout, P. J. Maddon, W. C. Olson, M. Lu, and J. P. Moore. 2002. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J. Virol. 76:8875-8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schagger, H., W. A. Cramer, and G. von Jagow. 1994. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 217:220-230. [DOI] [PubMed] [Google Scholar]
- 31.Schulke, N., M. S. Vesanen, R. W. Sanders, P. Zhu, M. Lu, D. J. Anselma, A. R. Villa, P. W. Parren, J. M. Binley, K. H. Roux, P. J. Maddon, J. P. Moore, and W. C. Olson. 2002. Oligomeric and conformational properties of a proteolytically mature, disulfide-stabilized human immunodeficiency virus type 1 gp140 envelope glycoprotein. J. Virol. 76:7760-7776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shankarappa, R., J. B. Margolick, S. J. Gange, A. G. Rodrigo, D. Upchurch, H. Farzadegan, P. Gupta, C. R. Rinaldo, G. H. Learn, X. He, X. L. Huang, and J. I. Mullins. 1999. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J. Virol. 73:10489-10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simon, F., P. Mauclere, P. Roques, I. Loussert-Ajaka, M. C. Muller-Trutwin, S. Saragosti, M. C. Georges-Courbot, F. Barre-Sinoussi, and F. Brun-Vezinet. 1998. Identification of a new human immunodeficiency virus type 1 distinct from group M and group O. Nat. Med. 4:1032-1037. [DOI] [PubMed] [Google Scholar]
- 34.Trkola, A., M. Purtscher, T. Muster, C. Ballaun, A. Buchacher, N. Sullivan, K. Srinivasan, J. Sodroski, J. P. Moore, and H. Katinger. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 70:1100-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vanden Haesevelde, M., J. L. Decourt, R. J. De Leys, B. Vanderborght, G. van der Groen, H. van Heuverswijn, and E. Saman. 1994. Genomic cloning and complete sequence analysis of a highly divergent African human immunodeficiency virus isolate. J. Virol. 68:1586-1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wei, X., J. M. Decker, H. Liu, Z. Zhang, R. B. Arani, J. M. Kilby, M. S. Saag, X. Wu, G. M. Shaw, and J. C. Kappes. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46:1896-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wyatt, R., P. D. Kwong, E. Desjardins, R. W. Sweet, J. Robinson, W. A. Hendrickson, and J. G. Sodroski. 1998. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393:705-711. [DOI] [PubMed] [Google Scholar]
- 38.Wyatt, R., J. Moore, M. Accola, E. Desjardin, J. Robinson, and J. Sodroski. 1995. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J. Virol. 69:5723-5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang, C. W., Y. Chishti, R. E. Hussey, and E. L. Reinherz. 2001. Expression, purification, and characterization of recombinant HIV gp140. The gp41 ectodomain of HIV or simian immunodeficiency virus is sufficient to maintain the retroviral envelope glycoprotein as a trimer. J. Biol. Chem. 276:39577-39585. [DOI] [PubMed] [Google Scholar]
- 40.Zhang, W., A. P. Godillot, R. Wyatt, J. Sodroski, and I. Chaiken. 2001. Antibody 17b binding at the coreceptor site weakens the kinetics of the interaction of envelope glycoprotein gp120 with CD4. Biochemistry 40:1662-1670. [DOI] [PubMed] [Google Scholar]