Abstract
Over a decade passed between Friedman’s discovery of the mammalian leptin gene (1) and its cloning in fish (2) and amphibians (3). Since 2005, the concept of gene synteny conservation (vs. gene sequence homology) was instrumental in identifying leptin genes in dozens of species, and we now have leptin genes from all major classes of vertebrates. This database of LEP (leptin), LEPR (leptin receptor), and LEPROT (endospanin) genes has allowed protein structure modeling, stoichiometry predictions, and even functional predictions of leptin function for most vertebrate classes. Here, we apply functional genomics to model hundreds of LEP, LEPR, and LEPROT proteins from both vertebrates and invertebrates. We identify conserved structural motifs in each of the three leptin signaling proteins and demonstrate Drosophila Dome protein’s conservation with vertebrate leptin receptors. We model endospanin structure for the first time and identify endospanin paralogs in invertebrate genomes. Finally, we argue that leptin is not an adipostat in fishes and discuss emerging knockout models in fishes.
Keywords: leptin, leptin receptor, endospanin, in silico modeling, molecular evolution, obesity, adipostat, fish models
Introduction
In 1994, Friedman’s laboratory described leptin as a peptide hormone that is synthesized by adipose tissue (1) and soon after it was proposed to regulate appetite and metabolic rate by communicating energy stores to the central nervous system (4–6). In mammals, leptin is synthesized by adipose tissue and released into the blood; there it travels to the hypothalamus and binds to the leptin receptor, which stimulates reduction of appetite and increased mobilization of lipid for metabolism. Through this feedback loop, the brain regulates energy stores to remain relatively constant [“adipostat control” (4–6)]. Control of energy stores is central to an organism’s life history, and as such, it is a research focus for comparative biologists. Migratory birds fuel long-distance migration by dramatic changes in lipid stores (7), hibernating mammals accumulate lipid stores to survive long periods of torpor (8), snakes dramatically rework organs to process large and infrequent meals (9), amphibian survival after metamorphosis is tied to adipose stores (10), and fish routinely go months without eating during winter (11). Agricultural scientists also have a great interest in leptin, because manipulating energy acquisition and deposition has potential to influence production of commercially important species (12–14). Therefore, there has been great interest and effort expended toward cloning and characterizing leptin orthologs throughout vertebrates. Recent reviews thoroughly document the progress of the comparative leptin community (15–17). This review will focus on three areas: evolution of genes in the leptin signaling pathway, the status of leptin as an adipostat, and emerging non-mammal models for studying leptin signaling. These research topics have made significant progress in recent years, and they provide examples of how a comparative approach can inform the study of human leptin (hLEP) endocrinology.
Evolution of Leptin Signaling: Leptin and Leptin Receptor Genes among Vertebrates
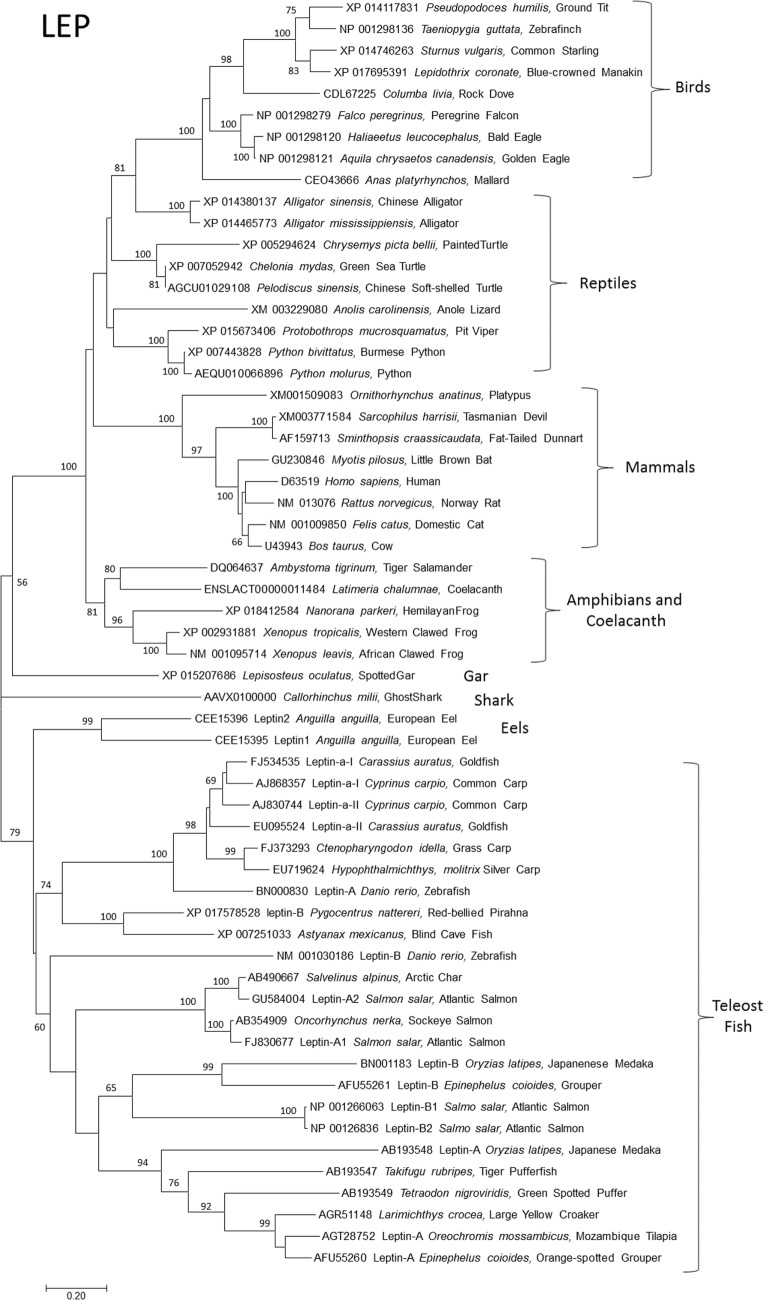
Although leptins in domestic mammals were identified soon after leptin in mice (18, 19), the first non-mammal leptin gene took over a decade to discover (2). This was due to the false assumption of sequence conservation among orthologs and was overcome by Kurokawa’s insight of gene order conservation or synteny (2). This major advance, along with progress on genome projects, has allowed identification of LEPs and LEPRs in all classes of vertebrates (Figures 1 and 2). It is now clear that the ancestral leptin that gave rise to leptins in tetrapods (birds, reptiles, amphibians, and mammals) is more closely related to coelacanth and shark (Callorhinchus milii) leptins vs. leptins from bony fish. In other words, bony fish leptins diverged along their own lineage independent of leptins in higher mammals (Figures 1 and 2). After the bony fish and tetrapods diverged, multiple paralogs of fish lep evolved. Tetrapods and their closest living relatives for which we have data (gar, coelacanth, Dipnoi not determined) express a single ortholog of leptin (Figure 1), with the exception of Anolis lizard, which has two lep, one of which is not expressed (15).
Figure 1.
Evolutionary relationships of vertebrate leptins (LEPs). Relationships of 59 amino acid sequences using the neighbor-joining method conducted in MEGA7. Numbers at nodes represent percentage of 500 bootstrap replicates. Nodes with no number indicate bootstrap support of less than 50%. Leptin amino acid sequences were manually aligned in MEGA7 informed by protein structural homologies. GenBank accession numbers represent protein accession.
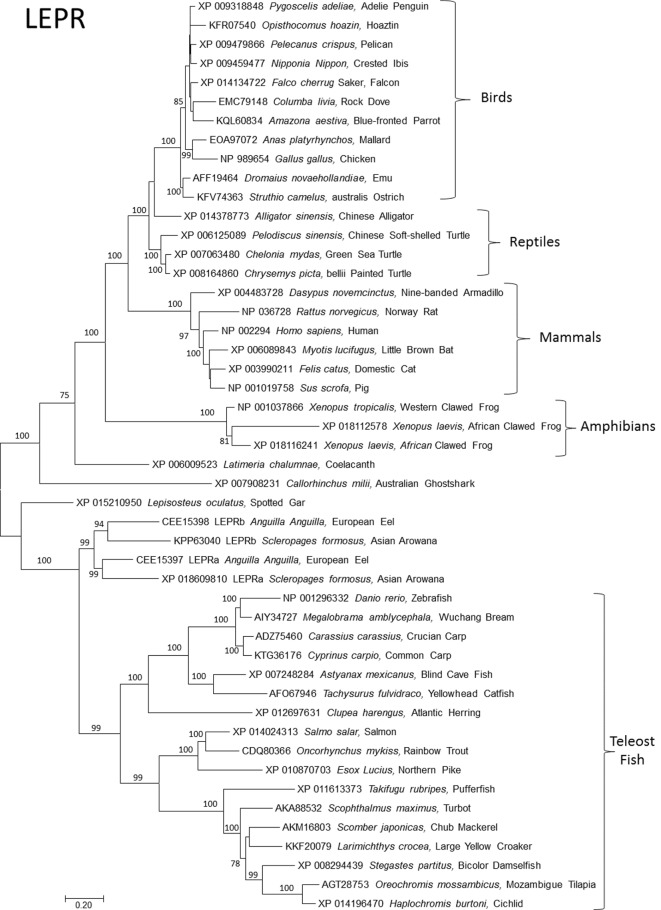
Figure 2.
Evolutionary relationships of vertebrate leptin receptor (LEPR). Relationships of 48 amino acid sequences using the neighbor-joining method conducted in MEGA7. Numbers at nodes represent percentage of 500 bootstrap replicates. Nodes with no number indicate bootstrap support of less than 50%. GenBank accession numbers represent protein accessions.
Bony fish typically expresses two paralogs of leptin, referred to as “A” and “B.” These are interpreted as arising during the whole genome duplication event in the Teleost fish lineage; more recent duplications (in salmonids and carps) are subtypes of A and B [see the study by Morini et al. (20) for an insightful discussion of leptin paralog evolution]. Leptin receptors are typically present as single orthologs per species, with the exception of recently identified duplicate lepr paralogs in European eel (20) and Asian arowana (Scleropages formosus) (acc# XP 018609810 and KPP63040). This duplication event appears to be ancient, but it is unresolved if the duplication of lepr was present in the ancestor of teleost fishes and then lost, or if loss of lepr predates teleosts (Figure 2).
Amphibians express a single paralog of lep and lepr, with lep expressed in multiple tissues, including adipose (3, 21). Xenopus responds to homologous recombinant leptin as an anorexigen, but not at all life stages (3). Xenopus leptin stimulates hind limb (3) and lung (22) development and may influence life history decisions in spadefoot toad (23). Xenopus lepr binds homologous and non-homologous leptins (3) and stimulates phosphorylation of intracellular signal transducer and activator of transcription (STAT) 3 and 5 (24). Less is known about reptile leptins. Several reports indicate that reptiles respond to non-homologous leptin treatment consistent with the mammalian model of leptin function [e.g., reduced appetite (25), reproductive effects (25, 26)]. In addition, studies using non-homologous leptin antibodies have documented leptin-like proteins that respond to seasonal changes in lipid (27–29), which are consistent with mammalian models. Denver et al. reported 2 lep (one which may be non-functional) and 1 lepr in the genome of the green anole (15). In general, amphibian lep and lepr expression and in vitro and in vivo function are more consistent with mammalian models than are similar data for fishes and birds.
What is the significance of multiple leptin paralogs? We assert that leptin-A and -B paralogs have distinct functions in teleosts. The fact that both paralogs (in multiple species of teleosts) are maintained throughout the teleost lineage (Figure 1) argues that each paralog has a distinct function. Where expression has been measured, A-type leps are typically expressed at higher message copies and with a more narrow tissue distribution than B-type (16, 30–33), but not in all species (2). If leptin-B paralogs are functional (and not pseudogenes), why is their expression lower and less tissue specific than A? Perhaps leptin-Bs are acting in an autocrine/paracrine manner, similar to that proposed for bird leptin (see below). Supporting this hypothesis is the observation that leptin-B is dramatically upregulated during regeneration of fin and heart in zebrafish (34), and after retinal injury (35), perhaps indicating local vs. circulating action. In addition, leptin-A knockdown in zebrafish embryos (via morpholino oligonucleotide) does not elicit a change in expression of leptin-B (36). If the A and B paralogs overlap functionally, one would expect a compensatory increase in B with decreased expression of A. Finally, in silico binding simulation of both paralogs predicts significantly lower binding energy of B vs. A to the leptin receptor (37). This indicates that something about the ligand–receptor interaction is different for leptin-B; perhaps it requires a second ligand or a higher local concentration of ligand (as in autocrine/paracrine signaling). To our knowledge, there are no published data on leptin-B protein expression or in vivo function other than regeneration (34–35). A leptin receptor reporter assay to assess functional differences between leptin paralogs, similar to that developed for Xenopus (24), and specific antibodies to document expression would be useful in addressing these questions.
Evolution of Leptin Signaling: Is There Another Major Signaling System for Energy Stores in Birds?
Arguably, bird leptin was the most difficult to identify among vertebrates, with over a decade of significant effort from multiple laboratories. A purported chicken leptin gene was reported early on, but independent laboratories were unable to amplify the sequence from chicken tissues, and it was absent in early builds of the chicken genome, despite the presence of a leptin receptor (38–40). The missing bird leptin gene was eventually found within regions of genomes that were refractory to characterization due to their high GC content and repetitive sequence (41–44). The advent of new methods of whole genome sequencing allowed identification of bird leptin in most major lineages of birds. Recently (45), RNAseq experiments in chicken documented highest leptin transcript copy number in brain (hypothalamus and cerebrum) and pituitary, with moderate expression in pancreas and testis, and low expression in liver and adipose [typically high expressing leptin tissues in fish and mammals, respectively (16)]. Further, Friedman-Einat’s group speculated that the high correlation between leptin and leptin receptor transcripts indicated that leptin in birds may not circulate, but instead acts as an autocrine/paracrine factor (45). Several lines of evidence support this hypothesis: bird leptin expression is primarily in CNS (42, 45), bird blood did not activate a sensitive chicken leptin receptor assay, even in birds with extreme adiposity (46), and genes with high GC content (such as bird leptin genes) are associated with low transcription rates (47). One study that supported a circulating leptin in birds documented that chicken serum and crow blood caused translocation of GFP-labeled STAT 3 to the nucleus in an expressed chicken leptin receptor assay (48). However, potent leptin receptor antagonists tested in chickens effectively block chicken leptin receptor in vitro but not in vivo (49).
The primary sequences of bird leptins have typically low but recognizable homology with other vertebrate leptins (41, 45), and bird leptin primary structure folds in silico into the conserved tertiary structure seen in all leptins (44, 45). Despite this structural conservation (or homology), it otherwise appears that bird leptins do not function similar to leptins in other vertebrates (detailed above). We assert that leptin signaling in birds is fundamentally different than it is in other vertebrates, which suggests that there is a leptin-independent pathway to manipulate energy stores. Birds make large-magnitude changes in adipose stores routinely as a life history strategy. Red knots undergo massive changes in body composition during their 9,000-km migration flights (7), emperor penguin lose ~50% of their body mass during a 4-month fast while incubating eggs on ice (50), and ptarmigan accumulate up to 30% of body mass as lipid in anticipation of winter storms (51). If leptin signaling is reduced/altered in birds, what signals these dramatic changes in lipid mobilization? Other major mammalian adipokine/appetite genes are missing in chickens, including resistin, TNFα, serpine 1, and omentin (52), and ghrelin in falcons (53). Thus, the “usual suspects” for neuroendocrine control of energy stores are either absent or play a fundamentally different role (52).
Evolution of Leptin Signaling: Analysis of Tertiary Structures Determined In Silico
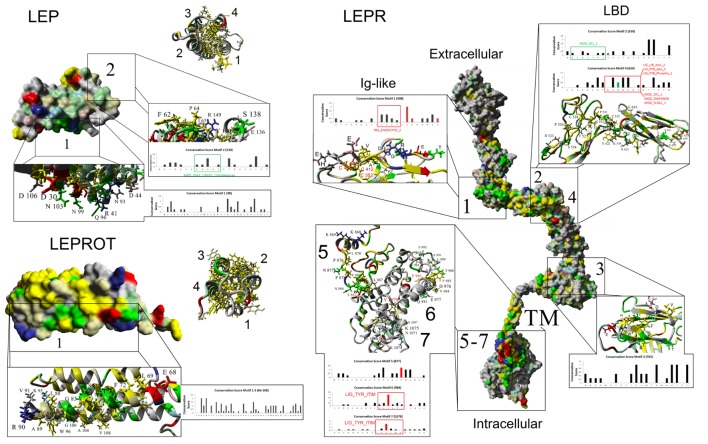
In the effort to understand the evolution of vertebrate leptin function, often the first data available are sequence data, and we have used these data to model leptins, leptin receptors, and their interaction. Comparing ~100 primary sequences per gene (Table 1), we can make some generalizations about structure. Vertebrate leptins demonstrate considerable primary amino acid sequence divergence, but despite this retain high tertiary structure conservation (predicted) when modeled with the hLEP structure (15, 37). We analyzed multiple tertiary structures (generated via in silico modeling) and proposed conservation of critical binding sites between leptin and the leptin receptor from fish to human (37). Combining our previous work (37) with our sequence-to-structure-to-function tools (63), we addressed the vertebrate evolution of LEP, LEPR, and the lesser-studied LEPROT. By using a total of 93 vertebrate LEP sequences and 89 vertebrate LEPR sequences (Table 1), we mapped conservation and linear motifs for each gene onto protein structures (Figure 3). Leptins contain a conserved disulfide bridge (Table 1) and several hydrophobic amino acids that are critical to maintaining the four-helix packing of the protein, even though sequence homology is low (~20%). On the surface of leptins, two linear motifs were identified, one for interaction with the Ig-like domain as suggested by Peelman et al. (64) and the other for the leptin-binding domain of LEPR. Utilizing molecular modeling and dynamics, we studied the structural integrity of the leptin protein among many taxa and determined that while sequence is highly divergent, the conservation of several hydrophobic amino acids and the disulfide bridge is sufficient to maintain protein folding in all classes of vertebrates. The leptin receptor conserves protein folding with seven highly conserved and selected linear motifs. There are also 16 conserved sites for posttranslational modification within the receptor (Table 1).
Table 1.
Vertebrate LEP, LEPR, LEPROT, and LEPROTL1 genes analyzed.
| Gene | Open reading frame sequences | AA start | AA end | Codons analyzed | Human model Z-score | Mouse–human homology (%) | Mouse–human alignment [root mean square deviation (RMSD), Å] | Molecular dynamic simulations carbon alpha (RMSD, Å) | Conserved posttranslational modifications |
|---|---|---|---|---|---|---|---|---|---|
| LEP | 93 | 22 | 167 | 13,578 | 0.28 | 84.93 | 0.342 | 1.33 | C117, C167 |
| LEPR | 89 | 29 | 1,158 | 100,570 | −3 | 75.35 | 0.457 | 2.06 | C196, N347, C352, C412, C413, C418, C447, C473, N624, N659, N688, N728, S882, Y986, Y1079, Y1141 |
| LEPROT | 150 | 1 | 131 | 19,650 | −0.56 | 94.66 | 0.346 | 2.85 | – |
Z-score is an indicator of how close (number of SDs from the mean) the predicted model fits chemical properties of all previously solved protein structures; RMSD is a measure of average distance between predicted models and native structures.
Figure 3.
Mapping protein conservation of leptin (LEP), leptin receptor (LEPR), and LEPROT/endospanin. Consurf analysis of LEP (top left), LEPR (right), and LEPROT/endospanin (bottom left) are shown as molecular surface plots of each structure. For LEP and LEPR, a picture of the four-helix bundle with conserved hydrophobic amino acids is shown as a ribbon diagram beside the surface plots of conservation. Top conserved motifs are magnified, identifying conserved amino acids that contribute to each motif. Amino acids are colored as followed: yellow, conserved hydrophobic; red, conserved polar acidic; blue, conserved polar basic; green, conserved hydrophilic; gray, not conserved. Amino acids with known posttranslational modifications are red (disulfide bonds of Cys-C or phosphorylation of Ser-S/Thr-T/Tyr-Y) and green (glycosylation of Asn-N) on the bar graphs of conservation. Predicted eukaryotic linear motifs are boxed and labeled on the bar graphs.
Concise Methods: Open reading frame (ORF) sequences were obtained for each gene from NCBI gene and aligned to the human ORF using ClustalW (54) in Mega (55). Codon selection was calculated using HyPhy (56) under a Muse-Gaut model (57) and standard Tamura-Nei model (58) for all sites in the LEP, LEPR, LEPROT, and LEPROTL1. Conservation scores were calculated using a combination of codon/amino acid fixation rates and dN-dS scores of selective pressure. A score of 2 at any site implies both a greater than 2 SDs above the mean for codon selection and a site that an amino acid is 100% conserved. A score of 0 implies no conservation of the amino acid and below the mean selective pressure (dN-dS). The scores were then put on a sliding window of 21 codons to calculate the top linear motifs within each gene. All numbering throughout the article is based on the full gene sequence of human.
Protein modeling was performed using our previously published LEP–LEPR interaction model (37) combined with I-TASSER- (59) generated extracellular and intracellular domains of LEPR joined by a single-pass transmembrane helix. The endospanin proteins were modeled with I-TASSER (59). Each structure was assessed for structural modeling reliability using a Z-score approach of a knowledge-based force field YASARA2 (60) relative to all solved structures of the PDB. Models were generated for both human and mouse and the structures aligned using MUSTANG to calculate sequence and atom alignments [in root mean square deviation (RMSD)]. Each protein was also run for 10 ns of molecular dynamic simulations (MDS) using the AMBER03 force field (61) to assess the average movement in RMSD of the carbon alpha positions throughout the proteins. For all four proteins, evolution was mapped onto protein structures using the sequence alignments above with the ConSurf tools (62). Homology modeling for the Drosophila UPD2 and Dome proteins was performed using YASARA (60) and structure scores calculated with the YASARA2 knowledge-based force field. BLAST analysis was performed for invertebrate genomes using all available sequences of ENSEMBL Metazoa BLAST (http://metazoa.ensembl.org/Multi/Tools/Blast?db=core) including Arthopoda, Nematoda, Lophotrochozoa, and Cnidaria. Sequences for metazoa, fungi, and plant endospanin orthologs (LEPROT and LEPROTL1 genes) were also pulled for ENSEMBL annotated orthologs.
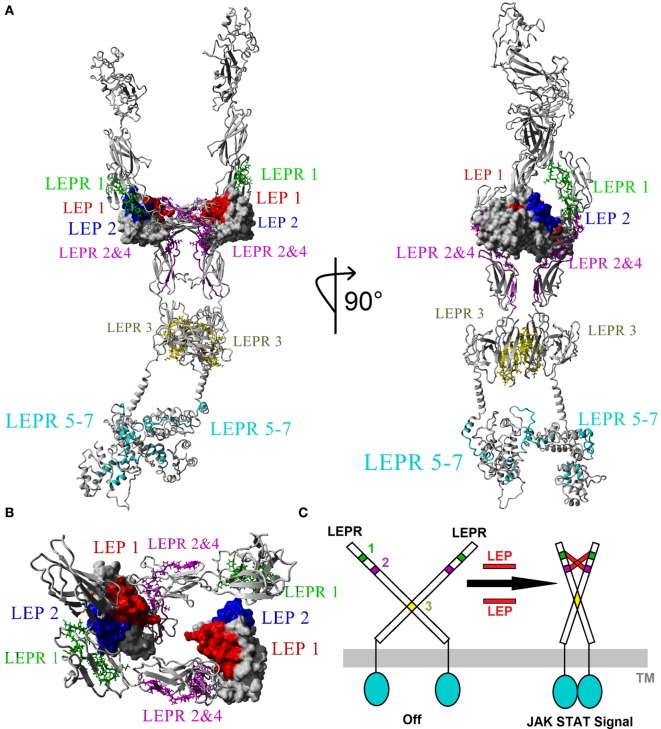
We hypothesize that the physiological effects of leptin are induced via binding with leptin receptor in a 2–2 molecular interaction, resulting in conformational stability to already dimerized receptors (37, 44, 65–67). There is evidence of higher order oligomerization states such as that of 4:4 stoichiometry (66, 68); however, very little is known about the structural basis for these states. Merging conserved motifs into the model of leptin–leptin receptor interaction, a 2–2 molecular interaction model was created (Figure 4A) using previous structures as a guide (66). When viewing the entire leptin receptor protein (Figure 4A), docking of leptin to leptin receptor accounted for all motifs. Motif 1 of leptin (red, Figure 4A) interacts with motifs 2 and 4 (magenta, Figure 4A) of leptin receptor, while motif 2 of leptin (blue, Figure 4A) interacts with motif 1 of LEPR (green, Figure 4A). Motif 3 of leptin receptor (yellow, Figure 4A) falls in the fibronectin type III 3 domain, known to control non-LEP-dependent dimerization of LEPR (67, 69, 70). Our models suggest with high probability that this motif contributes to dimerization of the receptor. In this dimer model, LEPR exists on the surface of cells as a dimer controlled by the conserved motif 3, such that the intracellular regions are not in close proximity to each other (Figure 4B). On two leptin molecules binding, the receptor is hinged by motif 3 (yellow, Figure 4C) to bring together motifs 1, 2, and 4 of LEPR to LEP motifs 1 and 2, resulting in intracellular domains of LEPR brought into close proximity for JAK and STAT activation (Figure 4C).
Figure 4.
Modeling the 2xLEP–2xLEPR interaction. (A) Each of the top motifs for leptin (LEP) and leptin receptor (LEPR) are colored in respective color coding. LEP: motif 1, red; motif 2, blue. LEPR: motif 1, green; motif 2 and 4, magenta; motif 3, yellow; motif 5–7, cyan. (B) Magnified view of the Ig-like and LBD of LEPR showing the 2–2 interaction model based on vertebrate evolution. (C) Model of endogenous dimerized LEPR being activated by LEP binding.
Evolution of Leptin Signaling: Invertebrate Leptin Signaling Genes
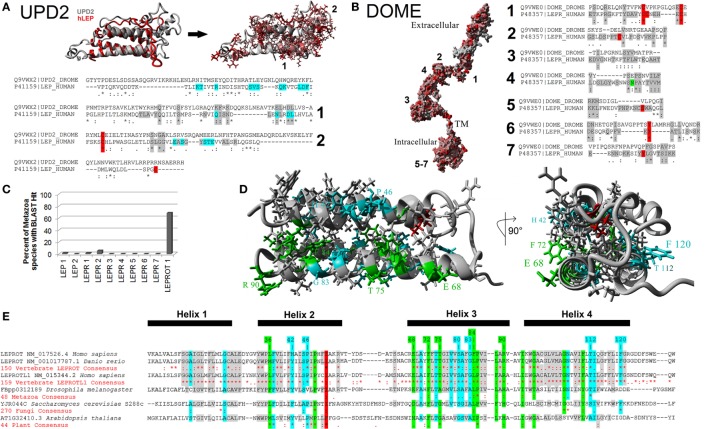
Rajan and Perrimon in 2012 described what they thought was a homologous leptin system in Drosophila melanogaster (71), through Unpaired 2 (Upd2) and Domeless (Dome) proteins. Similar to vertebrate LEPRs in fish (72), chicken (73), pig (74), cow (75), rat (76), and human (77), the Dome protein of Drosophila is critical for germline and follicle cell development through UPD signaling (78). Recent reports of a putative leptin signaling system in D. melanogaster through the UPD2 and Dome proteins (71), which are associated with phenotypes from tissue development (79), memory (80), and reproductive systems (78), proposes conserved leptin signaling components in invertebrates. Overlapping functions of vertebrate leptin receptor and Dome proteins suggest possible conserved tertiary structure.
To test homology between vertebrate and invertebrate systems, we modeled UPD2 (Figure 5A) and Dome proteins (Figure 5B) using our LEP:LEPR models and evaluated conservation of vertebrate motifs in the fly proteins. The UPD2 protein four-helix bundle was homologous to hLEP with some conserved amino acids contributing to packing and others that were surface exposed (Figure 5A). Structural alignments of hLEP to UPD2 had 8.24% homology and an alpha carbon average RMSD of 1.627 Å. The UPD2 model had a z-score of −1.375, which suggests that the model contains behaviors similar to most known protein structures. Aligning sequence of LEP to UPD2, 9 of 20 amino acids contributing to LEP–LEPR interaction were conserved in UPD2 (cyan, Figure 5A), fitting within our expectations based on zebrafish LEP modeling (37). Motif 1 of vertebrate LEP had 8 of 16 conserved amino acids, while motif 2 had 3 of 13 conserved amino acids. These data suggest a high probability of similar fold between leptin and UPD2 with a high number of amino acids conserved that are known to interact with leptin receptor, including motif 1 generated from our vertebrate evolutionary analysis.
Figure 5.
Defining the metazoan leptin system. (A) UPD2 model (gray) aligned with human LEP (hLEP, red). To the right of the structure overlay are identified amino acids in red that are conserved in the two proteins. Below the models are sequence alignments showing amino acids conserved in the top two motifs (gray), conserved posttranslational modifications (PTMs) (red), and known sites to interact with the receptor (cyan). (B) Dome model with amino acids in red conserved with human leptin receptor (LEPR). To the right of the model are sequence alignments showing amino acids conserved in the top seven motifs (gray) and conserved PTMs (red). (C) Using the top motifs of LEP, LEPR, and LEPROT, BLAST data for each in invertebrate genomes. (D,E) Models of LEPROT, now known as endospanin 1 (D), with conserved amino acids identified (E). Amino acids in red are conserved in all sequences, those in green conserved in at least four of the five groups, those in cyan conserved in at least three groups, and those in gray conserved in at least two groups.
The DOME and hLEPR models align with 22.94% homology and an average RMSD of alpha carbons of 0.823 Å (Figure 5B). To refine the functional conservation of DOME to hLEPR, we analyzed each of the top seven vertebrate motifs of LEPR. Each of the seven motifs of vertebrate LEP were found in the Dome sequence. Motif 1 had 6 of 14 critical amino acids conserved including two cysteine amino acids involved in disulfide bond formation. Motif 2 and 4 involved in the main interaction with LEP had hydrophobic and structural amino acids conserved with the vertebrate sequences. Motif 3 involved in non-LEP-dependent LEPR dimerization had three critical hydrophobic amino acids conserved. Of the intracellular three motifs 5–7, motif 6 was the most highly conserved including the known tyrosine phosphorylation site. To our knowledge, the combination of these seven motifs is not found in any other human protein, thus the high conservation of these motifs in Dome supports the assertion that this is indeed a homolog of vertebrate LEPR.
To probe the existence of the leptin signaling genes in other invertebrate genomes, a BLAST approach for the top motifs was used (Figure 5C). BLAST analysis of 54 invertebrate genomes was unable to identify invertebrate homologs. This is likely due to insertions and deletions seen in the motif alignments of Upd2 and Dome (Figures 5A,B), decreasing success of BLAST approaches. By using Ensembl Metazoa annotation tools (81), Upd2 homologs were only identified in the 12 sequenced Drosophila species, with no other invertebrates having annotated homologs. The Dome protein, however, has homologs found in 48 species of invertebrates according to ENSEMBL (http://metazoa.ensembl.org/Drosophila_melanogaster/Gene/Compara_Ortholog?db=core;g=FBgn0043903;r=X:19676061-19683518;t=FBtr0074756), with 22 being found as similar size of human LEPR and D. melanogaster Dome proteins. Outside of invertebrates, no homologs of Upd2 or Dome are yet reported. Contrary to LEP and LEPR, the LEPROT gene is found in many species from invertebrates to plants to fungi (Figures 5D,E).
Evolution of Leptin Signaling: Endospanin
Three years after the discovery of the leptin, Bailleul et al. established that the human LEPR transcribes a second, non-leptin receptor gene product (82). Initially named leptin receptor gene-related protein (OB-RGRP) or LEPROT (83), it is transcribed from an alternate AUG within the leptin receptor gene. The alternate start site is out of frame with the leptin receptor transcript, such that it produces a 131 amino acid protein that shares no primary sequence with LEPR.
LEPROT [recently renamed endospanin (84)] and its paralog LEPROTL1 (endospanin 2) are homologous with the yeast vesicle trafficking gene VPS55 (85). Knockout or disruption of VPS55 in yeast results in generally altered endosomal/vacuole trafficking (85, 86). In vertebrates, endospanin is proposed to specifically regulate endosomal trafficking and surface expression of the leptin receptor. Knockout LEPROT mice express more leptin receptors on the cell surface than wild-type, which makes them hyperresponsive to leptin and resistant to diet-induced obesity (87–89). Further, LEPR protein expression and LEPROT genomic copy number are negatively correlated in humans (90), and LEPROT may control tissue-specific expression of LEPR (91). Both endospanins 1 and 2 are known to interact with Rab13 and Rab8 (92), small G-proteins critical for trafficking between the trans-Golgi network and other cell compartments (93, 94). This suggests that endospanin1/2’s role is larger than just regulation of leptin receptor protein.
Is endospanin function conserved among vertebrates? While the BLAST approach did not identify invertebrate LEP and LEPR, the Ensembl Metazoa annotation (81) identified 48 invertebrate genomes as containing LEPROT homologous proteins. Further, 270 sequenced fungi and 44 sequenced plants contain a LEPROT homolog. We combined all of these sequences with 150 and 159 vertebrate LEPROT and LEPROTL1 sequences both to build the first tertiary structure prediction for endospanin and to determine critically conserved amino acids throughout eukaryote evolution (Figures 3 and 5D,E).
One amino acid is conserved in all 671 sequences studied (red), 13 amino acids are conserved in at least 4 of the 5 taxa (green), 16 in at least 3 of the 5 organism groupings (cyan), and 45 conserved in at least 2 of the 5 organism groupings (gray, Figures 5D,E). Using the total of 140 positions in the sequence alignment as shown, red represents 0.7%, green represents 9.3%, cyan represents 11.4%, and gray represents 32.1%, and thus 53.5% of the protein is identified to maintain conservation in at least one of the groupings. This value far exceeds that of LEPR and Dome proteins. Endospanin 1 protein contains a four-helix transmembrane bundle with high conservation of a hydrophobic core of the protein (Figure 5D). Noting conserved amino acids on our four-helix model (Figure 5D), 12 amino acids were conserved and surface exposed at positions 36, 42, 46, 68, 72, 75, 80, 83, 84, 90, 112, and 120 using the human LEPROT numbering (Figure 5E). These residues make up one side of the helix, suggesting possible interaction with another protein at this site.
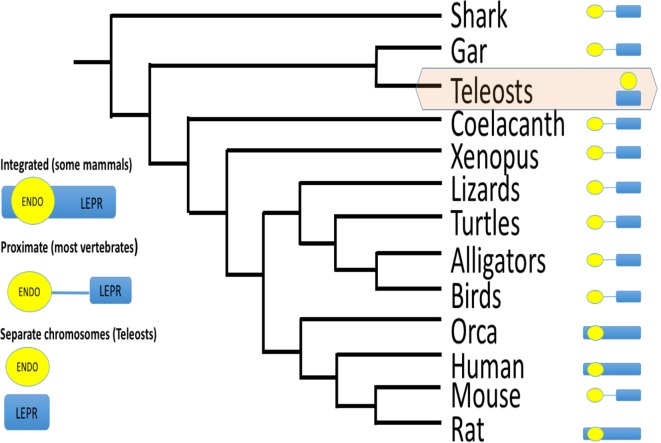
Another aspect of LEPROT genomics likely affects its influence on LEPR functional expression (i.e., on the surface of the cell). LEPROT’s original designation was as the “leptin receptor overlapping transcript” (82), indicating that LEPROT overlapped LEPR. Surveying Genbank for LEPROT and LEPR loci in all vertebrate classes, LEPROT overlaps or is adjacent to LEPR (within 150,000 bp and no intervening gene) in all cases. The one exception is teleost fishes, where LEPROT and LEPR are on different chromosomes (Figure 6). Gene proximity affects transcription rates (95). Given the high conservation of endospanin sequence, the conservation of its synteny with leptin receptor, and its effect on leptin receptor functional expression (87–89), it is likely that LEPROT and LEPR coevolved. We assert that because that synteny is broken in teleosts, it may be that control of leptin receptor expression is unique for teleosts among vertebrates.
Figure 6.
Schematic of gene order for endospanin and leptin receptor (LEPR) among vertebrates. For most vertebrate classes, endospanin (LEPROT) is either embedded within the LEPR gene, or within 150,000 bp, and without any gene between LEPROT and LEPR. For teleost fishes only, LEPROT and LEPR are on separate chromosomes. Data mined from Genbank queries. For example, LEPR search term returns chromosome 1, acc# NC_000001.11 for human LEPR, which also maps LEPROT within the human LEPR sequence, and chromosome 6, acc# NC_07117.6 for zebrafish LEPR but chromosome 2, acc# NC_007113.6 for zebrafish LEPROT.
Evolution of Leptin Signaling: What Does Gene Evolution Tell Us about Human Leptin Signaling?
Uncovering the evolutionary history of leptin signaling genes and modeling their structure is valuable as a self-contained enterprise, because it sets the stage for understanding functional differences among taxa. However, knowing how these genes are represented among vertebrates also has translational value. Modeling of Drosophila Dome as a leptin receptor and finding Dome homologs among other invertebrates provide an avenue for studying leptin signaling in other model systems. How changes in leptin signaling contribute to obesity is certainly complex, with interacting endocrine, neurological, epigenetic, and environmental variables. Added to this complexity is the interaction of multiple leptin receptor isoforms in transporting leptin across the blood–brain barrier. Decreased leptin signaling in the presence of high titers of circulating leptin, or leptin insensitivity/resistance, is often implicated as contributing to human obesity (96, 97). There is growing consensus that reduced blood–brain transport of leptin is a contributing factor to leptin insensitivity in the face of high leptin titers caused by obesity [reviewed in Ref. (98)]. Transport of leptin across the blood–brain barrier is facilitated by leptin receptors with short intracellular domains [commonly referred to as the ObRa paralog, as opposed to the ObRb paralog, which has a complete intracellular domain and is capable of mediating intracellular signaling (99, 100)]. This transport can be inhibited by a soluble form of the leptin receptor (ObRe), capable of binding leptin in serum (101, reviewed in 102). Sequencing cDNAs indicates that these isoforms are the result of alternate splicing of LEPR [e.g., Ref. (103)]; however, soluble receptors can also result from cleaving of membrane bound leptin receptors (102). All studies on leptin resistance (to our knowledge) are conducted in mammal models. Comparative study across model systems has the potential to illuminate how receptor paralog diversity contributes to leptin sensitivity. Given that endospanin controls surface expression of long-form LEPR [ObRb (82, 89)], it may also control expression of the other leptin receptor paralogs. Because endospanin is highly conserved, it represents an opportunity to study leptin sensitivity across models and an avenue to explore for human obesity treatment.
Leptin as an Adipostat: Lack of Evidence in Fishes
Recently, we made an argument that leptin in fishes does not fit the adipostat model proposed for mammals (16). Importantly, we are distinguishing between leptin’s proposed adipostat function and its anorexigen function. Leptin’s anorexigenic function is well documented among fishes (104, 105), amphibians (3), birds (although with non-homologous leptin) (106), lizards (25), and mammals (4). However, central to the adipostat model is the idea that serum leptin is proportional to total adipose stores, because adipose is the major producer of leptin in mammals (4–6). Kurokawa’s seminal study first noted that the primary tissue expressing leptin in fish was liver and not adipose (2). This was confirmed in many [e.g., Ref. (31–33)] but not all (107) fish species. Instead of decreasing as fat stores are depleted (as predicted by adipostat), plasma leptin consistently increases with fasting in salmonids (108–110) and flounder (111). Striberny et al. (112) found no evidence that this change in circulating leptin titer was mediated by the CNS. Further, Arctic charr will spontaneously stop feeding in winter even while leptin titers are falling and even if presented with food (11), but will resume eating during the time of year when leptin concentrations are rising (113, 114). The observation that leptin increases at the end of a long fast in fishes runs counter to leptin’s documented anorexigenic effects (above). It may be that plasma leptin titers in fasting fish are below the threshold that triggers an anorexigenic response. It is also possible that leptin injections result in supraphysiological concentrations of the hormone in serum, eliciting a response not seen with “normal” leptin signaling (115) and eliciting responses even with artifactual leptins (116). In fishes, increasing serum leptin commonly is interpreted as a signal to mobilize lipid stores in preparation for reproduction, rather than a response to fasting per se (113, 114).
Clearly, a decreasing leptin signal during winter and increase prior to reproduction is not consistent with the adipostat model proposed for mammals (2–4). The majority of leptin studies are done on rodents (16), and as such our view of leptin as an adipostat is likely biased by those studies. Rodents have high mass-specific metabolic rates and can only fast for hours, whereas hibernating mammals and ectothermic fishes routinely fast for months. Although leptin is thought to drive the prehibernation anorexia of some, but not all hibernating mammals [reviewed in Ref. (117)], organisms with life histories that are distinctly seasonal (but not necessarily hibernating) may change their set point for leptin sensitivity to accommodate different levels of activity and food availability between seasons (97, 112, 118, 119); thus, an adipostat as described for rodents may not be adaptive for fishes.
Total lipid stores (summing all tissues) clearly are not reflected in serum leptin (as evidenced by fasting fish that increase leptin titers above). However, many researchers (including us) have assumed that liver or gonad is the tissue that contributes to the bulk of serum leptin (because it gives the highest qPCR signal), but that may not be true. Salmonid adipocytes express detectable leptin (110, 120–123), and adipocytes cultured from food-restricted fish secrete significantly more leptin than those from fed fish, reflecting the response of the whole organism (110). Recent knockout models either affect adipose tissue [medaka lepr knockout (124)] or do not [zebrafish lep knockout (125)]. It may be that the bulk of leptin’s serum titer results from expression from liver, but that other tissues express leptin for autocrine/paracrine roles, as proposed for birds (see below). Well-controlled immunological studies, such as those for salmonids and tilapia (110, 126), are needed for a diversity of teleosts, along with how each tissue contributes to the serum/local pool under various physiological conditions.
How does the status of the adipostat model in fishes affect leptin as an adipostat in other vertebrates? We now know that human obesity is influenced by changes in food perception and metabolism after weight loss (127), and therefore a simplistic adipostat feedback loop does not adequately model human phenotypes. Documenting the response of appetite and leptin across vertebrates argues that it is possible to adjust leptin sensitivity, and even presents possible mechanisms for how sensitivity changes (e.g. endospanin).
Emerging Non-Mammal Models of Leptin Signaling
The obese (ob/ob) mouse, a long-standing model of human obesity (128), gained favor for leptin studies after Freidman’s laboratory cloned the truncated LEP gene (1). Together with the diabetic (db/db) mouse, a LEPR-deficient model, leptin administration effects have been demonstrated repeatedly. Intraperitoneal leptin injections in ob/ob mice causes 30% decrease in body mass, and db/db mice are similar to controls (5). Leptin’s pleiotropy was detailed using these models, and as a result, we now know that leptin affects reproduction, immune function, bone growth/resorption, and metabolic rate [reviewed in Ref. (129–132)]. The long-term normalizing effects of peripheral leptin injections on hLEP congenital deficiency reflect those in the ob/ob mouse [reviewed in Ref. (133)]. There are lines of fish (134) and birds (135) selected for high and low adiposity; however, few LEP and LEPR-null models are available for comparative (leptin) studies.
Our group used morpholino knockdown to generate zebrafish embryos with reduced leptin signaling (136, 137). We documented severe developmental defects in response to knockdown of LEPA or LEPR. Morphants were characterized by malformed sensory structures, bent notochord, poor yolk absorption, and low metabolic rate; these effects were rescued by coinjection of recombinant zebrafish leptin (136, 137). Microarray analysis of leptin-A “morphant” and “rescue” expression data identified differentially expressed genes that correspond to leptin signal transduction pathways [GnRH signaling, fatty acid metabolism, glycolysis/gluconeogenesis, MAP kinase, phosphoinositol signaling (138)]. The recent availability of CRISPR technology allowed direct comparison of zebrafish gene knockdown vs. gene knockout. “Morphant” and “mutant” phenotypes generally do not agree when targeting the same gene; typically morphants do not emulate mutant phenotypes (139, 140). Zebrafish morphants targeting (apparently) unrelated genes often share combinations of morphological markers ranging from disrupted eye, ear, and brain development; irregular body/tail curvature; or enlarged yolk (139, 141, 142). Non-specific MO off-target activity upregulates zebrafish tp53, which may induce apoptosis and global changes in gene expression (139, 143). For these reasons, we are hesitant to pursue antisense technologies. Similar to recent work by other laboratories, we are opting for knockout technologies as a means to generate comparative null models for many leptin signaling genes. Chisada et al. produced the first LEPR mutant fish, using the TILLING approach in medaka (124). Adult medaka LEPR mutants are hyperphagic, have elevated NPYa and AGRP, and decreased POMC mRNAs. Liver and muscle lipid does not increase in the LEPR mutants, but they accumulate visceral fat as adults (124). The medaka data are consistent with a mammalian adipostat model, but zebrafish are not. Michel et al. recently characterized an established (144) LEPR TILLING mutant in zebrafish and also generated CRISPR mutants for LEPA, LEPB, and LEPR (125). LEPR-null adults have no differences from wild-type in adiposity, body size, growth rate, mating success, or feeding behavior. However, LEPR mutants have altered glucose metabolism, and both LEPR and LEPA larvae have increased β-cell number (125).
Conclusion and Future Directions
Comparative leptin endocrinology has matured in the 11 years since the first non-mammal leptin was cloned. All major vertebrate classes are now represented in cloned leptins and leptin receptors, and investigation of invertebrate leptin signaling is beginning. Protein structures have been modeled for leptin, leptin receptor, and endospanins across an extensive evolutionary timescale, but models (although useful) are simply predictions to be tested. Now that we have identified conserved motifs and conserved sites of leptin–leptin receptor interaction, these predictions should be tested with in vitro functional assays.
The bird leptin problem has been solved in a genomic sense, but is just initiating physiologically. Now that the bird receptor assays (49) can be used with homologous ligands (hopefully soon), we should learn if birds are truly different among vertebrates in leptin signaling. We assert that understanding the endocrinology of how birds manipulate lipid stores will pay dividends in comparative endocrinology, agriculture, and human disease. Robust data from decades of research demonstrate that many species of birds perform large-scale manipulation of energy stores, and preliminary (but compelling) data indicate that they are doing so either without leptin (or other known adipokines) or by using leptin in a fundamentally different way (e.g., through an uncharacterized pathway). If leptin signaling in birds is truly different, it means that there is another way that vertebrates manipulate energy stores and thus potentially new avenues to pursue that will help us understand human obesity.
Amphibian leptin models are well developed with homologous recombinant leptin and receptor assays (15, 24), but relatively little is known for reptiles. These groups appear to adhere to the lipostat model, while birds and teleost fishes may not. As such, more species diversity in amphibian and reptile leptin studies could be very important in understanding leptin function as an adipostat.
In fishes, we are now past the point where one’s fish species of choice can be interpreted as representative of all fishes. Given that bony fishes have been on the planet ~370 MY longer than modern mammals (www.timetree.org), it is not surprising that they may be diverse in their leptin signaling. Phylogenetic analyses make it clear that teleost fishes are diverse in the structures of their leptin, leptin receptor, and LEPROT genes, and it is likely that reported differences among species represent true species divergence rather than methodological idiosyncrasies. Although we argue that there is a lack of evidence for adipostat function in fishes, the future may reveal that an “origin(s)” of that status within a fish clade, and we simply need to sample fish diversity more completely (e.g., non-teleost fishes need attention).
If we are to move forward, we must have comparable variables to assess species diversity. As such, reliance on relative qPCR for expression data does not allow quantitative comparisons among species; the community needs well-validated ELISAs (such as that developed for salmonids and tilapia) for multiple species. In this same light, the non-coding regions near leptin and leptin receptor need to be studied with more detail to gain an understanding of how expression is controlled throughout evolution. We need to pursue knockout models in non-mammals for laboratory approaches comparable to those using ob/ob and db/db mice. Finally, we need to measure leptin signaling responses of unmanipulated animals in the field and take advantage of the tremendous diversity of life histories that are well suited for leptin questions [The Krogh Principle (145)]. In doing so, the comparative community will contribute to understanding of human obesity similar to how Drosophila studies contributed to genetics, how shark-rectal gland contributed to kidney function, or how the squid giant axon contributed to neurobiology.
Ethics Statement
All procedures performed by the authors that used animals were approved by the institution’s IACUC.
Author Contributions
All authors contributed to drafting and editing of the manuscript. JWP completed all molecular modeling.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer RD and handling editor declared their shared affiliation, and the handling editor states that the process nevertheless met the standards of a fair and objective review.
Funding
RL and QL were funded by NIH 2R15DK079282-02 for some of the research highlighted in this review. RL was also supported by the University of Akron Faculty Research Committee. JWP was funded by NIH-K01ES025435 to perform all protein modeling and motif analysis.
Supplementary Material
The Supplementary Material for this article can be found online at http://journal.frontiersin.org/article/10.3389/fendo.2017.00058/full#supplementary-material.
References
- 1.Zhang Y, Proenca R, Maffei M, Barone M, Leopold L, Friedman JM. Positional cloning of the mouse obese gene and its human homologue. Nature (1994) 372:425–32. 10.1038/372425a0 [DOI] [PubMed] [Google Scholar]
- 2.Kurokawa T, Uji S, Suzuki T. Identification of cDNA coding for a homologue to mammalian leptin from pufferfish, Takifugu rubripes. Peptides (2005) 26:745–50. 10.1016/j.peptides.2004.12.017 [DOI] [PubMed] [Google Scholar]
- 3.Crespi EJ, Denver RJ. Leptin (ob gene) of the South African clawed frog Xenopus laevis. Proc Natl Acad Sci U S A (2006) 103:10092–7. 10.1073/pnas.0507519103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pelleymounter MA, Cullen MJ, Baker MB, Hecht R, Winters D, Boone T, et al. Effects of the obese gene product on body weight regulation in ob/ob mice. Science (1995) 269:540–3. 10.1126/science.7624776 [DOI] [PubMed] [Google Scholar]
- 5.Halaas JL, Gajiwala KS, Maffei M, Cohen SL, Chait BT, Rabinowitz D, et al. Weight-reducing effects of the plasma protein encoded by the obese gene. Science (1995) 269:543–6. 10.1126/science.7624777 [DOI] [PubMed] [Google Scholar]
- 6.Campfield LA, Smith FJ, Guisez Y, Devos R, Burn P. Recombinant mouse OB protein: evidence for a peripheral signal linking adiposity and central neural networks. Science (1995) 269:546–9. 10.1126/science.7624778 [DOI] [PubMed] [Google Scholar]
- 7.Hua N, Piersma T, Ma Z. Three-phase fuel deposition in a long-distance migrant, the red knot (Calidris canutus piersmai), before the flight to high Arctic breeding grounds. PLoS One (2013) 8:e62551. 10.1371/journal.pone.0062551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jastroch M, Giroud S, Barrett P, Geiser F, Heldmaier G, Herwig A. Seasonal control of mammalian energy balance: recent advances in the understanding of daily torpor and hibernation. J Neuroendocrinol (2016) 28(11). 10.1111/jne.12437 [DOI] [PubMed] [Google Scholar]
- 9.Secor SM, Carey HV. Integrative physiology of fasting. Compr Physiol (2016) 6:773–825. 10.1002/cphy.c150013 [DOI] [PubMed] [Google Scholar]
- 10.Scott DE, Casey ED, Donovan MF, Lynch TK. Amphibian lipid levels at metamorphosis correlate to post-metamorphic terrestrial survival. Oecologia (2007) 153:521–32. 10.1007/s00442-007-0755-6 [DOI] [PubMed] [Google Scholar]
- 11.Tveiten H, Johnsen HK, Jobling M. Influence of maturity status on the annual cycles of feeding and growth in Arctic charr reared at constant temperature. J Fish Biol (1996) 48:910–24. 10.1111/j.1095-8649.1996.tb01486.x [DOI] [Google Scholar]
- 12.Honig H, Ofer L, Elbaz M, Kaim M, Shinder D, Gershon E. Seasonal and parity effects on ghrelin levels throughout the estrous cycle in dairy cows. Gen Comp Endocrinol (2016) 235:64–9. 10.1016/j.ygcen.2016.06.006 [DOI] [PubMed] [Google Scholar]
- 13.Volkoff H, Estevan Sabioni R, Coutinho LL, Cyrino JEP. Appetite regulating factors in pacu (Piaractus mesopotamicus): tissue distribution and effects of food quantity and quality on gene expression. Comp Biochem Physiol A Mol Integr Physiol (2016) 203:241–54. 10.1016/j.cbpa.2016.09.022 [DOI] [PubMed] [Google Scholar]
- 14.McConn BR, Yi J, Gilbert ER, Siegel PB, Chowdhury VS, Furuse M, et al. Stimulation of food intake after central administration of gonadotropin-inhibitory hormone is similar in genetically selected low and high body weight lines of chickens. Gen Comp Endocrinol (2016) 232:96–100. 10.1016/j.ygcen.2016.01.004 [DOI] [PubMed] [Google Scholar]
- 15.Denver RJ, Bonett RM, Boorse GC. Evolution of leptin structure and function. Neuroendocrinology (2011) 94:21–38. 10.1159/000328435 [DOI] [PubMed] [Google Scholar]
- 16.Londraville RL, Macotela Y, Duff RJ, Easterling MR, Liu Q, Crespi EJ. Comparative endocrinology of leptin: assessing function in a phylogenetic context. Gen Comp Endocrinol (2014) 203:146–57. 10.1016/j.ygcen.2014.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gorissen M, Flik G. Leptin in teleostean fish, towards the origins of leptin physiology. J Chem Neuroanat (2014) 6(1–62):200–6. 10.1016/j.jchemneu.2014.06.005 [DOI] [PubMed] [Google Scholar]
- 18.Dyer CJ, Simmons JM, Matteri RL, Keisler DH. cDNA cloning and tissue-specific gene expression of ovine leptin, NPY-Y1 receptor, and NPY-Y2 receptor. Domest Anim Endocrinol (1997) 14:295–303. 10.1016/S0739-7240(97)00028-3 [DOI] [PubMed] [Google Scholar]
- 19.Pfister-Genskow M, Hayes H, Eggen A, Bishop MD. Chromosomal localization of the bovine obesity (OBS) gene. Mamm Genome (1996) 7:398–9. 10.1007/s003359900118 [DOI] [PubMed] [Google Scholar]
- 20.Morini M, Pasquier J, Dirks R, van den Thillart G, Tomkiewicz J, Rousseau K, et al. Duplicated leptin receptors in two species of eel bring new insights into the evolution of the leptin system in vertebrates. PLoS One (2015) 10:e0126008. 10.1371/journal.pone.0126008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boswell T, Dunn IC, Wilson PW, Joseph N, Burt DW, Sharp PJ. Identification of a non-mammalian leptin-like gene: characterization and expression in the tiger salamander (Ambystoma tigrinum). Gen Comp Endocrinol (2006) 146:157–66. 10.1016/j.ygcen.2005.08.001 [DOI] [PubMed] [Google Scholar]
- 22.Torday JS, Ihida-Stansbury K, Rehan VK. Leptin stimulates Xenopus lung development: evolution in a dish. Evol Dev (2009) 11:219–24. 10.1111/j.1525-142X.2009.00321.x [DOI] [PubMed] [Google Scholar]
- 23.Garcia NW, Pfennig KS, Burmeister SS. Leptin manipulation reduces appetite and causes a switch in mating preference in the plains spadefoot toad (Spea bombifrons). PLoS One (2015) 10:e0125981. 10.1371/journal.pone.0125981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cui MY, Hu CK, Pelletier C, Dziuba A, Slupski RH, Li C, et al. Ancient origins and evolutionary conservation of intracellular and neural signaling pathways engaged by the leptin receptor. Endocrinology (2014) 155:4202–14. 10.1210/en.2014-1301 [DOI] [PubMed] [Google Scholar]
- 25.Niewiarowski PH, Balk ML, Londraville RL. Phenotypic effects of leptin in an ectotherm: a new tool to study the evolution of life histories and endothermy? J Exp Biol (2000) 203(Pt 2):295–300. [DOI] [PubMed] [Google Scholar]
- 26.Putti R, Varricchio E, Gay F, Elena C, Paolucci M. Leptin effects on testis and epididymis in the lizard Podarcis sicula, during summer regression. Gen Comp Endocrinol (2009) 160:168–75. 10.1016/j.ygcen.2008.11.010 [DOI] [PubMed] [Google Scholar]
- 27.Goldberg DW, Leitão SAT, Godfrey MH, Lopez GG, Santos AJB, Neves FA, et al. Ghrelin and leptin modulate the feeding behaviour of the hawksbill turtle Eretmochelys imbricata during nesting season. Conserv Physiol (2013) 1:cot016. 10.1093/conphys/cot016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paolucci M, Rocco M, Varricchio E. Leptin presence in plasma, liver and fat bodies in the lizard Podarcis sicula: fluctuations throughout the reproductive cycle. Life Sci (2001) 69:2399–408. 10.1016/S0024-3205(01)01326-1 [DOI] [PubMed] [Google Scholar]
- 29.Spanovich S, Niewiarowski PH, Londraville RL. Seasonal effects on circulating leptin in the lizard Sceloporus undulatus from two populations. Comp Biochem Physiol B Biochem Mol Biol (2006) 143:507–13. 10.1016/j.cbpb.2006.01.001 [DOI] [PubMed] [Google Scholar]
- 30.Huang H, Wei Y, Meng Z, Zhang Y, Liu X, Guo L, et al. Polymorphisms of leptin-b gene associated with growth traits in orange-spotted grouper (Epinephelus coioides). Int J Mol Sci (2014) 15:11996–2006. 10.3390/ijms150711996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gorissen M, Bernier NJ, Nabuurs SB, Flik G, Huising MO. Two divergent leptin paralogues in zebrafish (Danio rerio) that originate early in teleostean evolution. J Endocrinol (2009) 201:329–39. 10.1677/JOE-09-0034 [DOI] [PubMed] [Google Scholar]
- 32.Chen T, Chen S, Ren C, Hu C, Tang D, Yan A. Two isoforms of leptin in the White-clouds Mountain minnow (Tanichthys albonubes): differential regulation by estrogen despite similar response to fasting. Gen Comp Endocrinol (2016) 225:174–84. 10.1016/j.ygcen.2015.08.002 [DOI] [PubMed] [Google Scholar]
- 33.Yuan X, Li A, Liang X-F, Huang W, Song Y, He S, et al. Leptin expression in mandarin fish Siniperca chuatsi (Basilewsky): regulation by postprandial and short-term fasting treatment. Comp Biochem Physiol A Mol Integr Physiol (2016) 194:8–18. 10.1016/j.cbpa.2016.01.014 [DOI] [PubMed] [Google Scholar]
- 34.Kang J, Hu J, Karra R, Dickson AL, Tornini VA, Nachtrab G, et al. Modulation of tissue repair by regeneration enhancer elements. Nature (2016) 532:201–6. 10.1038/nature17644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sifuentes CJ, Kim J-W, Swaroop A, Raymond PA. Rapid, dynamic activation of Müller glial stem cell responses in zebrafish. Invest Ophthalmol Vis Sci (2016) 57:5148–60. 10.1167/iovs.16-19973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dalman M. Characterization of Leptin Signaling in the Developing Zebrafish (Danio rerio) Using Molecular, Physiological, and Bioinformatic Approaches. Unpublished Ph.D. thesis, University of Akron; (2014). [Google Scholar]
- 37.Prokop JW, Duff RJ, Ball HC, Copeland DL, Londraville RL. Leptin and leptin receptor: analysis of a structure to function relationship in interaction and evolution from humans to fish. Peptides (2012) 38:326–36. 10.1016/j.peptides.2012.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pitel F, Faraut T, Bruneau G, Monget P. Is there a leptin gene in the chicken genome? Lessons from phylogenetics, bioinformatics and genomics. Gen Comp Endocrinol (2010) 167:1–5. 10.1016/j.ygcen.2009.10.006 [DOI] [PubMed] [Google Scholar]
- 39.Pitel F, Monbrun C, Gellin J, Vignal A. The chicken LEP (OB) gene has not been mapped. Anim Genet (2000) 31:281. 10.1111/j.1365-2052.2000.00610.pp.x [DOI] [PubMed] [Google Scholar]
- 40.Friedman-Einat M, Boswell T, Horev G, Girishvarma G, Dunn IC, Talbot RT, et al. The chicken leptin gene: has it been cloned? Gen Comp Endocrinol (1999) 115:354–63. 10.1006/gcen.1999.7322 [DOI] [PubMed] [Google Scholar]
- 41.Friedman-Einat M, Cogburn LA, Yosefi S, Hen G, Shinder D, Shirak A, et al. Discovery and characterization of the first genuine avian leptin gene in the rock dove (Columba livia). Endocrinology (2014) 155:3376–84. 10.1210/en.2014-1273 [DOI] [PubMed] [Google Scholar]
- 42.Wang D, Xu C, Wang T, Li H, Li Y, Ren J, et al. Discovery and functional characterization of leptin and its receptors in Japanese quail (Coturnix japonica). Gen Comp Endocrinol (2016) 225:1–12. 10.1016/j.ygcen.2015.09.003 [DOI] [PubMed] [Google Scholar]
- 43.Huang G, Li J, Wang H, Lan X, Wang Y. Discovery of a novel functional leptin protein (LEP) in zebra finches: evidence for the existence of an authentic avian leptin gene predominantly expressed in the brain and pituitary. Endocrinology (2014) 155:3385–96. 10.1210/en.2014-1084 [DOI] [PubMed] [Google Scholar]
- 44.Prokop JW, Schmidt C, Gasper D, Duff RJ, Milsted A, Ohkubo T, et al. Discovery of the elusive leptin in birds: identification of several “missing links” in the evolution of leptin and its receptor. PLoS One (2014) 9:e92751. 10.1371/journal.pone.0092751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seroussi E, Cinnamon Y, Yosefi S, Genin O, Smith JG, Rafati N, et al. Identification of the long-sought leptin in chicken and duck: expression pattern of the highly GC-rich avian leptin fits an autocrine/paracrine rather than endocrine function. Endocrinology (2016) 157:737–51. 10.1210/en.2015-1634 [DOI] [PubMed] [Google Scholar]
- 46.Yosefi S, Hen G, Rosenblum CI, Cerasale DJ, Beaulieu M, Criscuolo F, et al. Lack of leptin activity in blood samples of Adélie penguin and bar-tailed godwit. J Endocrinol (2010) 207:113–22. 10.1677/JOE-10-0177 [DOI] [PubMed] [Google Scholar]
- 47.Veloso A, Kirkconnell KS, Magnuson B, Biewen B, Paulsen MT, Wilson TE, et al. Rate of elongation by RNA polymerase II is associated with specific gene features and epigenetic modifications. Genome Res (2014) 24:896–905. 10.1101/gr.171405.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ohkubo T, Hirota K, Murase D, Adachi H, Nozawa-Takeda T, Sugita S. Avian blood induced intranuclear translocation of STAT3 via the chicken leptin receptor. Comp Biochem Physiol B Biochem Mol Biol (2014) 174:9–14. 10.1016/j.cbpb.2014.05.001 [DOI] [PubMed] [Google Scholar]
- 49.Gertler A, Shinder D, Yosefi S, Shpilman M, Rosenblum CI, Ruzal M, et al. Pegylated leptin antagonist with strong orexigenic activity in mice is not effective in chickens. J Exp Biol (2014) 217:180–4. 10.1242/jeb.095539 [DOI] [PubMed] [Google Scholar]
- 50.Robin JP, Frain M, Sardet C, Groscolas R, Le Maho Y. Protein and lipid utilization during long-term fasting in emperor penguins. Am J Physiol (1988) 254:R61–8. [DOI] [PubMed] [Google Scholar]
- 51.Mortensen A, Blix AS. Seasonal changes in the effects of starvation on metabolic rate and regulation of body weight in Svalbard Ptarmigan. Ornis Scand (1985) 16:20–4. 10.2307/3676570 [DOI] [Google Scholar]
- 52.Daković N, Térézol M, Pitel F, Maillard V, Elis S, Leroux S, et al. The loss of adipokine genes in the chicken genome and implications for insulin metabolism. Mol Biol Evol (2014) 31:2637–46. 10.1093/molbev/msu208 [DOI] [PubMed] [Google Scholar]
- 53.Seim I, Jeffery PL, Herington AC, Chopin LK. Comparative analysis reveals loss of the appetite-regulating peptide hormone ghrelin in falcons. Gen Comp Endocrinol (2015) 216:98–102. 10.1016/j.ygcen.2014.11.016 [DOI] [PubMed] [Google Scholar]
- 54.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics (2007) 23:2947–8. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- 55.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol (2011) 28:2731–9. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pond SLK, Frost SDW, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics (2005) 21:676–9. 10.1093/bioinformatics/bti079 [DOI] [PubMed] [Google Scholar]
- 57.Muse SV, Gaut BS. A likelihood approach for comparing synonymous and nonsynonymous nucleotide substitution rates, with application to the chloroplast genome. Mol Biol Evol (1994) 11:715–24. [DOI] [PubMed] [Google Scholar]
- 58.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol (1993) 10:512–26. [DOI] [PubMed] [Google Scholar]
- 59.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc (2010) 5:725–38. 10.1038/nprot.2010.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Krieger E, Joo K, Lee J, Lee J, Raman S, Thompson J, et al. Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: four approaches that performed well in CASP8. Proteins (2009) 77(Suppl 9):114–22. 10.1002/prot.22570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, et al. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem (2003) 24:1999–2012. 10.1002/jcc.10349 [DOI] [PubMed] [Google Scholar]
- 62.Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res (2010) 38:W529–33. 10.1093/nar/gkq399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Prokop JW, Leeper TC, Duan Z-H, Milsted A. Amino acid function and docking site prediction through combining disease variants, structure alignments, sequence alignments, and molecular dynamics: a study of the HMG domain. BMC Bioinformatics (2012) 13(Suppl 2):S3. 10.1186/1471-2105-13-S2-S3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Peelman F, Van Beneden K, Zabeau L, Iserentant H, Ulrichts P, Defeau D, et al. Mapping of the leptin binding sites and design of a leptin antagonist. J Biol Chem (2004) 279:41038–46. 10.1074/jbc.M404962200 [DOI] [PubMed] [Google Scholar]
- 65.Couturier C, Jockers R. Activation of the leptin receptor by a ligand-induced conformational change of constitutive receptor dimers. J Biol Chem (2003) 278:26604–11. 10.1074/jbc.M302002200 [DOI] [PubMed] [Google Scholar]
- 66.Moharana K, Zabeau L, Peelman F, Ringler P, Stahlberg H, Tavernier J, et al. Structural and mechanistic paradigm of leptin receptor activation revealed by complexes with wild-type and antagonist leptins. Structure (2014) 22:866–77. 10.1016/j.str.2014.04.012 [DOI] [PubMed] [Google Scholar]
- 67.Mancour LV, Daghestani HN, Dutta S, Westfield GH, Schilling J, Oleskie AN, et al. Ligand-induced architecture of the leptin receptor signaling complex. Mol Cell (2012) 48:655–61. 10.1016/j.molcel.2012.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zabeau L, Defeau D, Van der Heyden J, Iserentant H, Vandekerckhove J, Tavernier J. Functional analysis of leptin receptor activation using a Janus kinase/signal transducer and activator of transcription complementation assay. Mol Endocrinol (2004) 18:150–61. 10.1210/me.2003-0078 [DOI] [PubMed] [Google Scholar]
- 69.Maruyama IN. Activation of transmembrane cell-surface receptors via a common mechanism? The “rotation model”. Bioessays (2015) 37:959–67. 10.1002/bies.201500041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zabeau L, Defeau D, Iserentant H, Vandekerckhove J, Peelman F, Tavernier J. Leptin receptor activation depends on critical cysteine residues in its fibronectin type III subdomains. J Biol Chem (2005) 280:22632–40. 10.1074/jbc.M413308200 [DOI] [PubMed] [Google Scholar]
- 71.Rajan A, Perrimon N. Drosophila cytokine unpaired 2 regulates physiological homeostasis by remotely controlling insulin secretion. Cell (2012) 151:123–37. 10.1016/j.cell.2012.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Copeland DL, Duff RJ, Liu Q, Prokop J, Londraville RL. Leptin in teleost fishes: an argument for comparative study. Front Physiol (2011) 2:26. 10.3389/fphys.2011.00026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lei MM, Wu SQ, Li XW, Wang CL, Chen Z, Shi ZD. Leptin receptor signaling inhibits ovarian follicle development and egg laying in chicken hens. Reprod Biol Endocrinol (2014) 12:25. 10.1186/1477-7827-12-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Smolinska N, Kaminski T, Siawrys G, Przala J. Expression of leptin and its receptor genes in the ovarian follicles of cycling and early pregnant pigs. Animal (2013) 7:109–17. 10.1017/S1751731112001103 [DOI] [PubMed] [Google Scholar]
- 75.Sarkar M, Schilffarth S, Schams D, Meyer HHD, Berisha B. The expression of leptin and its receptor during different physiological stages in the bovine ovary. Mol Reprod Dev (2010) 77:174–81. 10.1002/mrd.21129 [DOI] [PubMed] [Google Scholar]
- 76.Ryan NK, Van der Hoek KH, Robertson SA, Norman RJ. Leptin and leptin receptor expression in the rat ovary. Endocrinology (2003) 144:5006–13. 10.1210/en.2003-0584 [DOI] [PubMed] [Google Scholar]
- 77.Dupuis L, Schuermann Y, Cohen T, Siddappa D, Kalaiselvanraja A, Pansera M, et al. Role of leptin receptors in granulosa cells during ovulation. Reproduction (2014) 147:221–9. 10.1530/REP-13-0356 [DOI] [PubMed] [Google Scholar]
- 78.Ghiglione C, Devergne O, Georgenthum E, Carballès F, Médioni C, Cerezo D, et al. The Drosophila cytokine receptor Domeless controls border cell migration and epithelial polarization during oogenesis. Development (2002) 129:5437–47. 10.1242/dev.00116 [DOI] [PubMed] [Google Scholar]
- 79.Brown S, Hu N, Hombría JC. Identification of the first invertebrate interleukin JAK/STAT receptor, the Drosophila gene domeless. Curr Biol (2001) 11:1700–5. 10.1016/S0960-9822(01)00524-3 [DOI] [PubMed] [Google Scholar]
- 80.Copf T, Goguel V, Lampin-Saint-Amaux A, Scaplehorn N, Preat T. Cytokine signaling through the JAK/STAT pathway is required for long-term memory in Drosophila. Proc Natl Acad Sci U S A (2011) 108:8059–64. 10.1073/pnas.1012919108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kersey PJ, Allen JE, Armean I, Boddu S, Bolt BJ, Carvalho-Silva D, et al. Ensembl Genomes 2016: more genomes, more complexity. Nucleic Acids Res (2016) 44:D574–80. 10.1093/nar/gkv1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bailleul B, Akerblom I, Strosberg AD. The leptin receptor promoter controls expression of a second distinct protein. Nucleic Acids Res (1997) 25:2752–8. 10.1093/nar/25.14.2752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Touvier T, Conte-Auriol F, Briand O, Cudejko C, Paumelle R, Caron S, et al. LEPROT and LEPROTL1 cooperatively decrease hepatic growth hormone action in mice. J Clin Invest (2009) 119:3830–8. 10.1172/JCI34997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Seron K, Couturier C, Belouzard S, Bacart J, Monte D, Corset L, et al. Endospanins regulate a postinternalization step of the leptin receptor endocytic pathway. J Biol Chem (2011) 286:17968–81. 10.1074/jbc.M111.224857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Belgareh-Touzé N, Avaro S, Rouillé Y, Hoflack B, Haguenauer-Tsapis R. Yeast Vps55p, a functional homolog of human obesity receptor gene-related protein, is involved in late endosome to vacuole trafficking. Mol Biol Cell (2002) 13:1694–708. 10.1091/mbc.01-12-0597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schluter C, Lam KKY, Brumm J, Wu BW, Saunders M, Stevens TH, et al. Global analysis of yeast endosomal transport identifies the vps55/68 sorting complex. Mol Biol Cell (2008) 19:1282–94. 10.1091/mbc.E07-07-0659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Séron K, Couturier C, Belouzard S, Bacart J, Monté D, Corset L, et al. Endospanins regulate a postinternalization step of the leptin receptor endocytic pathway. J Biol Chem (2011) 286:17968–81. 10.1074/jbc.M111.224857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vauthier V, Swartz TD, Chen P, Roujeau C, Pagnon M, Mallet J, et al. Endospanin 1 silencing in the hypothalamic arcuate nucleus contributes to sustained weight loss of high fat diet obese mice. Gene Ther (2014) 21:638–44. 10.1038/gt.2014.36 [DOI] [PubMed] [Google Scholar]
- 89.Couturier C, Sarkis C, Séron K, Belouzard S, Chen P, Lenain A, et al. Silencing of OB-RGRP in mouse hypothalamic arcuate nucleus increases leptin receptor signaling and prevents diet-induced obesity. Proc Natl Acad Sci U S A (2007) 104:19476–81. 10.1073/pnas.0706671104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jeon J-P, Shim S-M, Nam H-Y, Ryu G-M, Hong E-J, Kim H-L, et al. Copy number variation at leptin receptor gene locus associated with metabolic traits and the risk of type 2 diabetes mellitus. BMC Genomics (2010) 11:426. 10.1186/1471-2164-11-426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Satoh T, Yoshino S, Katano A, Ishizuka T, Tomaru T, Shibusawa N, et al. Isolation of a novel leptin receptor gene promoter preferentially functioning in neuronal cells. Biochem Biophys Res Commun (2009) 389:673–7. 10.1016/j.bbrc.2009.09.056 [DOI] [PubMed] [Google Scholar]
- 92.Hirvonen MJ, Büki KG, Sun Y, Mulari MTK, Härkönen PL, Väänänen KH. Novel interaction of Rab13 and Rab8 with endospanins. FEBS Open Bio (2013) 3:83–8. 10.1016/j.fob.2013.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nokes RL, Fields IC, Collins RN, Fölsch H. Rab13 regulates membrane trafficking between TGN and recycling endosomes in polarized epithelial cells. J Cell Biol (2008) 182:845–53. 10.1083/jcb.200802176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Huber LA, Pimplikar S, Parton RG, Virta H, Zerial M, Simons K. Rab8, a small GTPase involved in vesicular traffic between the TGN and the basolateral plasma membrane. J Cell Biol (1993) 123:35–45. 10.1083/jcb.123.1.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Akhtar W, de Jong J, Pindyurin AV, Pagie L, Meuleman W, de Ridder J, et al. Chromatin position effects assayed by thousands of reporters integrated in parallel. Cell (2013) 154:914–27. 10.1016/j.cell.2013.07.018 [DOI] [PubMed] [Google Scholar]
- 96.Myers MG, Cowley MA, Münzberg H. Mechanisms of leptin action and leptin resistance. Annu Rev Physiol (2008) 70:537–56. 10.1146/annurev.physiol.70.113006.100707 [DOI] [PubMed] [Google Scholar]
- 97.Scarpace PJ, Zhang Y. Leptin resistance: a prediposing factor for diet-induced obesity. Am J Physiol Regul Integr Comp Physiol (2009) 296:R493–500. 10.1152/ajpregu.90669.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Szczesna M, Zieba DA. Phenomenon of leptin resistance in seasonal animals: the failure of leptin action in the brain. Domest Anim Endocrinol (2015) 52:60–70. 10.1016/j.domaniend.2015.03.002 [DOI] [PubMed] [Google Scholar]
- 99.Hileman SM, Pierroz DD, Masuzaki H, Bjørbaek C, El-Haschimi K, Banks WA, et al. Characterizaton of short isoforms of the leptin receptor in rat cerebral microvessels and of brain uptake of leptin in mouse models of obesity. Endocrinology (2002) 143:775–83. 10.1210/endo.143.3.8669 [DOI] [PubMed] [Google Scholar]
- 100.Hileman SM, Tornøe J, Flier JS, Bjørbaek C. Transcellular transport of leptin by the short leptin receptor isoform ObRa in Madin-Darby Canine Kidney cells. Endocrinology (2000) 141:1955–61. 10.1210/endo.141.6.7450 [DOI] [PubMed] [Google Scholar]
- 101.Tu H, Kastin AJ, Hsuchou H, Pan W. Soluble receptor inhibits leptin transport. J Cell Physiol (2008) 214:301–5. 10.1002/jcp.21195 [DOI] [PubMed] [Google Scholar]
- 102.Schaab M, Kratzsch J. The soluble leptin receptor. Best Pract Res Clin Endocrinol Metab (2015) 29:661–70. 10.1016/j.beem.2015.08.002 [DOI] [PubMed] [Google Scholar]
- 103.Escobar S, Rocha A, Felip A, Carrillo M, Zanuy S, Kah O, et al. Leptin receptor gene in the European sea bass (Dicentrarchus labrax): cloning, phylogeny, tissue distribution and neuroanatomical organization. Gen Comp Endocrinol (2016) 229:100–11. 10.1016/j.ygcen.2016.03.017 [DOI] [PubMed] [Google Scholar]
- 104.Murashita K, Uji S, Yamamoto T, Rønnestad I, Kurokawa T. Production of recombinant leptin and its effects on food intake in rainbow trout (Oncorhynchus mykiss). Comp Biochem Physiol B Biochem Mol Biol (2008) 150:377–84. 10.1016/j.cbpb.2008.04.007 [DOI] [PubMed] [Google Scholar]
- 105.Li Z, Ceccarini G, Eisenstein M, Tan K, Friedman JM. Phenotypic effects of an induced mutation of the ObRa isoform of the leptin receptor. Mol Metab (2013) 2:364–75. 10.1016/j.molmet.2013.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kuo AY, Cline MA, Werner E, Siegel PB, Denbow DM. Leptin effects on food and water intake in lines of chickens selected for high or low body weight. Physiol Behav (2005) 84:459–64. 10.1016/j.physbeh.2005.01.014 [DOI] [PubMed] [Google Scholar]
- 107.Ohga H, Hirata D, Matsumori K, Kitano H, Nagano N, Yamaguchi A, et al. Possible role of the leptin system in controlling puberty in the male chub mackerel, Scomber japonicus. Comp Biochem Physiol A Mol Integr Physiol (2016) 203:159–66. 10.1016/j.cbpa.2016.09.009 [DOI] [PubMed] [Google Scholar]
- 108.Kling P, Rønnestad I, Stefansson SO, Murashita K, Kurokawa T, Björnsson BT. A homologous salmonid leptin radioimmunoassay indicates elevated plasma leptin levels during fasting of rainbow trout. Gen Comp Endocrinol (2009) 162:307–12. 10.1016/j.ygcen.2009.04.003 [DOI] [PubMed] [Google Scholar]
- 109.Trombley S, Maugars G, Kling P, Björnsson BT, Schmitz M. Effects of long-term restricted feeding on plasma leptin, hepatic leptin expression and leptin receptor expression in juvenile Atlantic salmon (Salmo salar L.). Gen Comp Endocrinol (2012) 175:92–9. 10.1016/j.ygcen.2011.10.001 [DOI] [PubMed] [Google Scholar]
- 110.Salmerón C, Johansson M, Angotzi AR, Rønnestad I, Jönsson E, Björnsson BT, et al. Effects of nutritional status on plasma leptin levels and in vitro regulation of adipocyte leptin expression and secretion in rainbow trout. Gen Comp Endocrinol (2015) 210:114–23. 10.1016/j.ygcen.2014.10.016 [DOI] [PubMed] [Google Scholar]
- 111.Fuentes EN, Safian D, Einarsdottir IE, Valdés JA, Elorza AA, Molina A, et al. Nutritional status modulates plasma leptin, AMPK and TOR activation, and mitochondrial biogenesis: implications for cell metabolism and growth in skeletal muscle of the fine flounder. Gen Comp Endocrinol (2013) 186:172–80. 10.1016/j.ygcen.2013.02.009 [DOI] [PubMed] [Google Scholar]
- 112.Striberny A, Ravuri CS, Jobling M, Jørgensen EH. Seasonal differences in relative gene expression of putative central appetite regulators in Arctic charr (Salvelinus alpinus) do not reflect its annual feeding cycle. PLoS One (2015) 10:e0138857. 10.1371/journal.pone.0138857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Frantzen M, Damsgård B, Tveiten H, Moriyama S, Iwata M, Johnsen HK. Effects of fasting on temporal changes in plasma concentrations of sex steroids, growth hormone and insulin-like growth factor I, and reproductive investment in Arctic charr. J Fish Biol (2004) 65:1526–42. 10.1111/j.1095-8649.2004.00564.x [DOI] [Google Scholar]
- 114.Frøiland E, Jobling M, Björnsson BT, Kling P, Ravuri CS, Jørgensen EH. Seasonal appetite regulation in the anadromous Arctic charr: evidence for a role of adiposity in the regulation of appetite but not for leptin in signalling adiposity. Gen Comp Endocrinol (2012) 178:330–7. 10.1016/j.ygcen.2012.06.017 [DOI] [PubMed] [Google Scholar]
- 115.Sahin-Efe A, Polyzos SA, Dincer F, Zaichenko L, McGovern R, Schneider B, et al. Intracellular leptin signaling following effective weight loss. Metabolism (2015) 64:888–95. 10.1016/j.metabol.2015.04.006 [DOI] [PubMed] [Google Scholar]
- 116.Lõhmus M, Olin M, Sundström LF, Troedsson MHT, Molitor TW, EL Halawani M. Leptin increases T-cell immune response in birds. Gen Comp Endocrinol (2004) 139:245–50. 10.1016/j.ygcen.2004.09.011 [DOI] [PubMed] [Google Scholar]
- 117.Florant GL, Healy JE. The regulation of food intake in mammalian hibernators: a review. J Comp Physiol B (2012) 182:451–67. 10.1007/s00360-011-0630-y [DOI] [PubMed] [Google Scholar]
- 118.Adam CL, Findlay PA. Decreased blood-brain leptin transfer in an ovine model of obesity and weight loss: resolving the cause of leptin resistance. Int J Obes (Lond) (2010) 34:980–8. 10.1038/ijo.2010.28 [DOI] [PubMed] [Google Scholar]
- 119.Ball HC, Londraville RL, Prokop JW, George JC, Suydam RS, Vinyard C, et al. Beyond thermoregulation: metabolic function of cetacean blubber in migrating bowhead and beluga whales. J Comp Physiol B (2017) 187(1):235–52. 10.1007/s00360-016-1029-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gong N, Einarsdottir IE, Johansson M, Björnsson BT. Alternative splice variants of the rainbow trout leptin receptor encode multiple circulating leptin-binding proteins. Endocrinology (2013) 154:2331–40. 10.1210/en.2012-2082 [DOI] [PubMed] [Google Scholar]
- 121.Pfundt B, Sauerwein H, Mielenz M. Leptin mRNA and protein immunoreactivity in adipose tissue and liver of rainbow trout (Oncorhynchus mykiss) and immunohistochemical localization in liver. Anat Histol Embryol (2009) 38:406–10. 10.1111/j.1439-0264.2009.00951.x [DOI] [PubMed] [Google Scholar]
- 122.Rønnestad I, Nilsen TO, Murashita K, Angotzi AR, Gamst Moen A-G, Stefansson SO, et al. Leptin and leptin receptor genes in Atlantic salmon: cloning, phylogeny, tissue distribution and expression correlated to long-term feeding status. Gen Comp Endocrinol (2010) 168:55–70. 10.1016/j.ygcen.2010.04.010 [DOI] [PubMed] [Google Scholar]
- 123.Salmerón C, Johansson M, Asaad M, Angotzi AR, Rønnestad I, Stefansson SO, et al. Roles of leptin and ghrelin in adipogenesis and lipid metabolism of rainbow trout adipocytes in vitro. Comp Biochem Physiol A Mol Integr Physiol (2015) 188:40–8. 10.1016/j.cbpa.2015.06.017 [DOI] [PubMed] [Google Scholar]
- 124.Chisada S, Kurokawa T, Murashita K, Rønnestad I, Taniguchi Y, Toyoda A, et al. Leptin receptor-deficient (knockout) medaka, Oryzias latipes, show chronical up-regulated levels of orexigenic neuropeptides, elevated food intake and stage specific effects on growth and fat allocation. Gen Comp Endocrinol (2014) 195:9–20. 10.1016/j.ygcen.2013.10.008 [DOI] [PubMed] [Google Scholar]
- 125.Michel M, Page-McCaw PS, Chen W, Cone RD. Leptin signaling regulates glucose homeostasis, but not adipostasis, in the zebrafish. Proc Natl Acad Sci U S A (2016) 113:3084–9. 10.1073/pnas.1513212113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Douros JD, Baltzegar DA, Breves JP, Lerner DT, Seale AP, Gordon Grau E, et al. Prolactin is a major inhibitor of hepatic leptin A synthesis and secretion: studies utilizing a homologous leptin A ELISA in the tilapia. Gen Comp Endocrinol (2014) 207:86–93. 10.1016/j.ygcen.2014.03.007 [DOI] [PubMed] [Google Scholar]
- 127.MacLean PS, Wing RR, Davidson T, Epstein L, Goodpaster B, Hall KD, et al. NIH working group report: innovative research to improve maintenance of weight loss. Obesity (Silver Spring) (2015) 23:7–15. 10.1002/oby.20967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Coleman DL. Obese and diabetes: two mutant genes causing diabetes-obesity syndromes in mice. Diabetologia (1978) 14:141–8. 10.1007/BF00429772 [DOI] [PubMed] [Google Scholar]
- 129.Wang B, Chandrasekera PC, Pippin JJ. Leptin- and leptin receptor-deficient rodent models: relevance for human type 2 diabetes. Curr Diabetes Rev (2014) 10:131–45. 10.2174/1573399810666140508121012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Procaccini C, La Rocca C, Carbone F, De Rosa V, Galgani M, Matarese G. Leptin as immune mediator: interaction between neuroendocrine and immune system. Dev Comp Immunol (2017) 66:120–9. 10.1016/j.dci.2016.06.006 [DOI] [PubMed] [Google Scholar]
- 131.Dupont J, Pollet-Villard X, Reverchon M, Mellouk N, Levy R. Adipokines in human reproduction. Horm Mol Biol Clin Investig (2015) 24:11–24. 10.1515/hmbci-2015-0034 [DOI] [PubMed] [Google Scholar]
- 132.Chen XX, Yang T. Roles of leptin in bone metabolism and bone diseases. J Bone Miner Metab (2015) 33:474–85. 10.1007/s00774-014-0569-7 [DOI] [PubMed] [Google Scholar]
- 133.Farooqi IS, Wangensteen T, Collins S, Kimber W, Matarese G, Keogh JM, et al. Clinical and molecular genetic spectrum of congenital deficiency of the leptin receptor. N Engl J Med (2007) 356:237–47. 10.1056/NEJMoa063988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gong N, Johansson M, Björnsson BT. Impaired central leptin signaling and sensitivity in rainbow trout with high muscle adiposity. Gen Comp Endocrinol (2016) 235:48–56. 10.1016/j.ygcen.2016.06.013 [DOI] [PubMed] [Google Scholar]
- 135.Yi J, Gilbert ER, Siegel PB, Cline MA. Fed and fasted chicks from lines divergently selected for low or high body weight have differential hypothalamic appetite-associated factor mRNA expression profiles. Behav Brain Res (2015) 286:58–63. 10.1016/j.bbr.2015.02.008 [DOI] [PubMed] [Google Scholar]
- 136.Liu Q, Dalman M, Chen Y, Akhter M, Brahmandam S, Patel Y, et al. Knockdown of leptin A expression dramatically alters zebrafish development. Gen Comp Endocrinol (2012) 178:562–72. 10.1016/j.ygcen.2012.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Dalman MR, Liu Q, King MD, Bagatto B, Londraville RL. Leptin expression affects metabolic rate in zebrafish embryos (D. rerio). Front Physiol (2013) 4:160. 10.3389/fphys.2013.00160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Tuttle M. In Silico Analysis of Zebrafish Leptin-A Knockdown Reveals Enrichment for Metabolism, Development, and Morpholino Artifacts. Unpublished MS thesis, University of Akron; (2016). [Google Scholar]
- 139.Kok FO, Shin M, Ni C-W, Gupta A, Grosse AS, van Impel A, et al. Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Dev Cell (2015) 32:97–108. 10.1016/j.devcel.2014.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lawson ND. Reverse genetics in zebrafish: mutants, morphants, and moving forward. Trends Cell Biol (2016) 26:77–9. 10.1016/j.tcb.2015.11.005 [DOI] [PubMed] [Google Scholar]
- 141.Danilova N, Sakamoto KM, Lin S. Ribosomal protein S19 deficiency in zebrafish leads to developmental abnormalities and defective erythropoiesis through activation of p53 protein family. Blood (2008) 112:5228–37. 10.1182/blood-2008-01-132290 [DOI] [PubMed] [Google Scholar]
- 142.Rai K, Chidester S, Zavala CV, Manos EJ, James SR, Karpf AR, et al. Dnmt2 functions in the cytoplasm to promote liver, brain, and retina development in zebrafish. Genes Dev (2007) 21:261–6. 10.1101/gad.1472907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Robu ME, Larson JD, Nasevicius A, Beiraghi S, Brenner C, Farber SA, et al. p53 activation by knockdown technologies. PLoS Genet (2007) 3:e78. 10.1371/journal.pgen.0030078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kettleborough RNW, Busch-Nentwich EM, Harvey SA, Dooley CM, de Bruijn E, van Eeden F, et al. A systematic genome-wide analysis of zebrafish protein-coding gene function. Nature (2013) 496:494–7. 10.1038/nature11992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lindstedt S. Krogh 1929 or “the Krogh principle”. J Exp Biol (2014) 217:1640–1. 10.1242/jeb.095505 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.