Abstract
As a master regulator involved in flower development, LEAFY-like gene has been demonstrated to play a key role in the flowering process regulation of angiosperms. Expression analysis of EjLFY-1, a LEAFY (LFY) homolog of loquat (Eriobotrya japonica Lindl.), indicated its participation in the regulation of flowering in loquat. To verify its function and potential value in the genetic engineering to shorten the juvenile phase, ectopic expression of EjLFY-1 in strawberry (Fragaria × ananassa) was achieved using Agrobacterium-mediated gene transfer of a plant expression vector with the loquat EjLFY-1 gene driven by the CaMV 35S promoter. Totally 59 plantlets were verified to be the transformants. The presence, expression and integration of EjLFY-1 in the transformants were assessed by PCR, quantitative real-time PCR and Southern blot, respectively. Constitutive expression of EjLFY-1 in strawberry accelerated the flowering process in strawberry with the shorten necessary period for flowering induction, development of flower and fruit set. While vegetative growth habits of the transformants in the first cropping season were consistent with the WT ones. Meanwhile, both the flowers and fruits of the transformants were also as same as those of the WT ones. Furthermore, the early-flowering habit was maintained in their asexual progeny, the runner plants. While with continuous asexual propagation, the clones showed a more strengthen early-flowering phenotype, such as the reduced vegetative growth and the abnormal floral organs in individual plantlets. These results demonstrated the function of this gene and at the same time provided us new insights into the utilization potential of such genes in the genetic engineering of perennial fruits.
Keywords: loquat (Eriobotrya japonica), LEAFY, transgenic strawberry, asexual progeny, flowering
Introduction
Flowering is one of the most important events in the life cycle of plants. Flowering process of plant is affected by both endogenous factors, such as hereditary characters or plant hormones levels, and environmental conditions, such as temperature or day length. Flowering at right time accommodating to the seasonal and endogenous signals is vital for the reproductive success of all plants. The flowering time determination in plants is controlled by complicated gene networks.
In the past decades, a great deal of progress has been accomplished in understanding the molecular basis of the regulatory mechanisms underlying flowering time in plants, especially studies in the molecular regulatory networks of Arabidopsis provides us a better understanding about it. Numerous genes involved in the flowering regulation were demonstrated. In Arabidopsis, for example, it has been reported that at least 180 genes are implicated in flowering-time control (Fornara et al., 2010), regardless of these unknown or undetected genes. One possible explanation for so many genes are involved in flowering regulation is that this process is affected by various factors. Normally, there are several genetically defined pathways which affect the flowering: the vernalization pathway, the photoperiod pathway, the gibberellin pathway, the autonomous pathway and the ambient temperature pathway (Fornara et al., 2010; Srikanth and Schmid, 2011). Such signaling pathways responding to endogenous and environmental signals converge on key regulators, such as FLOWERING LOCUS T (FT), which then activate other floral homeotic genes. For plant, although the organ where developmental decision leading to flowering occurs is shoot apex, leaf is the organ which sensing environmental signals. In Arabidopsis, the mobile signal FT has been demonstrated to be the long-distance signal moved from leaf to shoot apex and then induce the flowering (Corbesier et al., 2007). Expression of FT is affected by such pathway signals and its protein can promote flowering together with the meristem-specific bZIP transcriptional factor FD (Abe et al., 2005; Wigge et al., 2005). At SAM (shoot apical meristem), the FT-FD complex promotes the expression of the floral meristem identity genes such as LEAFY (LFY).
LEAFY and its homologs in other plants encode a kind of unique transcription factors that assign the floral fate of meristems, being thought as a meristem identity gene which determines floral identity (Moyroud et al., 2010). In the lfy mutants of Arabidopsis, homeotic transformations with leaf-like structures replacing the floral organs took place (Weigel et al., 1992). Expression of LFY is also regulated by many pathway signals. For example, GA (Eriksson et al., 2006) and auxin (Li et al., 2013) regulate the LFY expression in Arabidopsis and thereby affect the flowering habits. The master role of LFY as a meristem identity gene is reflected partly in activating its downstream flowering regulation factors, such as the directly activating of the expression of AP1, whose upregulation stands for the initiation of flower formation (Mandel and Yanofsky, 1995; Wagner et al., 1999; William et al., 2004; Liu et al., 2007). Coincided with its master role in flowering progress, over-expression of LFY or its homologs usually leads to an early-flowering phenotype not only in herbaceous plants such as Arabidopsis (Weigel and Nilsson, 1995) and rice (He et al., 2000), but also in woody plants such as hybrid aspen (Rottmann et al., 2000).
Interestingly, such precocious flowering phenotypes also could be found in the fruit trees such as citrus over-expressing AtLFY (Peña et al., 2001). Fruit trees are perennials with a juvenile phase lasting for years in which they are not competent to flower, despite whether in inductive environmental conditions or not. The existing of juvenile phase in fruit trees has been suggested to be a restricting factor for their genetic improvement or breeding process: most economic phenotypes associated with fruits of the hybrids cannot be detected in this phase. Such work mentioned above make it possible to break the juvenile stage of fruit trees with the gene modify technique. Actually, there have been several reports associated with the over-expression of flowering-related genes in fruit trees so as to shorten their juvenile phase. Duan et al. (2010) showed the expression of AtAP1 induced precocious flowering in transgenic kumquat. The juvenile period in apple has also been reduced with the silencing of the MdTFL1 gene (Kotoda et al., 2006) or over-expressing of FT or FT homologous gene (Trankner et al., 2010; Bergonzi and Albani, 2011).
Nevertheless, in perennial fruit species, new questions arouse regarding the stability of integration and expression of foreign genes, there are few reports reporting the stability of transgene integration and expression in plants growing in the field over years (Peña and Séguin, 2001; Rai and Shekhawat, 2014).
Loquat (Eriobotrya japonica Lindl.) is an evergreen fruit tree native to China and cultivated mainly in tropical and subtropical regions. Fruits of loquat can be consumed fresh or processed for jam, juice, wine, syrup, or as candied fruits (Lin et al., 1999). As a kind of woody fruit, loquat also has a long juvenile phase, which impedes both productivity and breeding efficiency. Esumi et al. (2005) first reported the existence of EjLFY-1, a LFY homolog from loquat, but its expression and function were not clarified until now for the insufficient transgenic system in loquat.
Most gene function analysis in fruits were carried out with ectopic expression in model plants, such as Arabidopsis or tobacco, for their easier transformation and shorter life cycles. But one significant defect is that the characters related to fruits cannot be found in the transformants originated from such model plants. Strawberry has been considered as a good candidate for the function analysis of such genes. Strawberry owns short reproductive cycle and produces fruits. More importantly, strawberry shares similar gene sequence and genomic microcolinearity with other members of the Rosaceae family, including a large amount of the fruit trees such as apple, peach, pear, plum, apricot, cherry, and other species (Slovin et al., 2009). In addition to sexual reproduction, marked by flower and seed formation, strawberry can also reproduce asexually. It sends out stolon (also called runners), which makes new plants by producing roots and forming leaf clusters at some of the nodes where they touch the ground (Figure 1).
FIGURE 1.
Vegetative reproduction of strawberry by its stolon. Arrows (from left to right) show the mother plant, stolon (runner) and the newly formed asexual plant.
Without the gene’s recombination and segregation, propagation with stolon buds in strawberry makes it feasible to sufficiently investigate the transgenic effects of the exogenous genes in their asexual progeny, which is especially important for fruit trees. In production, fruit species must be propagated in asexual ways, such as grafting or layering, to maintain their characters stable. The stability of the transgenic effect in the asexual propagated progeny is crucial for the application of genetic engineering in fruit trees.
The main objective of the this study was to evaluate the function of EjLFY-1 by investigating the ectopic expression effects of this gene on the flowering process in strawberry, and at the same time to investigate the stability of the effects of such transgene in the successive asexual propagated generations of the transformants.
Materials and Methods
Isolation of EjLFY-1 Sequences
Gene-specific forward and reverse primers, EjLFYfwd and EjLFYrev (Table 1), were designed based on the reported EjLFY-1 sequence (GenBank accession no. AB162033). Restriction enzyme sites were added additionally at the 5′-end and 3′-end of the primers for the subsequent vector construction. Flower bud and leaf of loquat cv. ‘Zaozhong No.6’ were used for the extraction of RNA and DNA, respectively. Full-length cDNA and DNA sequence of EjLFY-1 were obtained by PCR. PCR products were cloned into pGEM-T easy vector (Promega, Madison, WI, USA) and then sequenced.
Table 1.
Primers used in this study.
| Name | Primers sequences (5′–3′) | Cyclic amplification methods | Expected amplification sizes |
|---|---|---|---|
| EjLFYfwd EjLFYrev | GGCTCTAGAATGGATCCAGATGCCTTC ACAGAGCTCTCAATAGGGCAGGTGCTCGC | 32 cycles of 94° C for 1 min, 60°C for 1 min, 72°C for 1.5 min | 1.2 Kb |
| DIGUP DIGDW | GTAGACGACAAGGACGACGA GATGGAGAGACGGGGATGAG | 35 cycles of 94°C for 50 s, 53°C for 1 min, 72°C for 1 min | 498 bp |
| KANF KANR | GTTCTTTTTGTCAAGACCGACC CAAGCTCTTCAGCAATATCACG | 33 cycles of 94°C for 50 s, 61°C for 1 min, 72°C for 1 min | 560 bp |
| KANUP KANDP | TCCATCAGCAACGTGTCGGTGCT GTGGAAAGGCGGTGAGCGATGAT | 30 cycles of 94°C for 40 s, 51°C for 40 s, 72°C for 1 min | 500 bp |
| AGRIF AGRIR | TCCATCAGCAACGTGTCGGTGCT GTGGAAAGGCGGTGAGCGATGAT | 32 cycles of 94°C for 50 s, 59°C for 50 s, 72°C for 1 min | 1.0 Kb |
| EjLFY-F | AGGGAGCACCCGTTCATT | 40 cycles of 95°C for 10 s, 60°C for 20 s, | 223 bp |
| EjLFY-R | GCATCTTCGGCTTGTTGA | 72°C for 20 s | |
| FaAP1-F | AGAGGAAGGAGAAGGCAA | 40 cycles of 95°C for 10 s, 60°C for 20 s, | 230 bp |
| FaAP1-R | GAGAGTAAGGTCGAGCTGG | 72°C for 20 s | |
| FaTFL1-F | GTCACTGCCAAACCGAGAG | 40 cycles of 95°C for 10 s, 60°C for 20 s, | 231 bp |
| FaTFL1-R | GAGAACAAACACAAACCTG | 72°C for 20 s | |
Expression Analysis of EjLFY-1 in the Flowering Process of Loquat
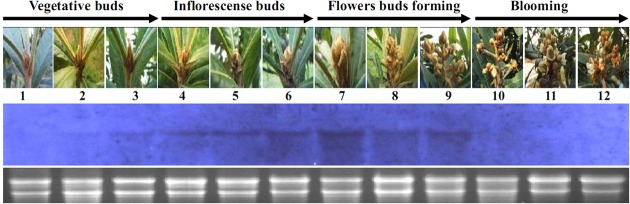
To determine the involvement of EjLFY-1 in floral development of loquat, we detected its expression levels in different floral differentiation stages, which were generally grouped into four groups: vegetative buds, inflorescence buds, flower buds forming and blooming. Totally 12 samples (buds or flowers) were collected with 10-day intervals. 10 μg RNA from each sample was electrophoresed in 1.2% agarose gel and transferred to Hybond N+ nylon membranes (Roche, Swiss). A gene-specific fragment (498 bp) amplified by a pair of primers (DIGUP and DIGDW, Table 1) at the conserved region of its 3′ end was used as the probe for hybridization. The specific methods of probe labeling and hybridization are as described in the instruction offered by Roche DIG High Prime DNA labeling and Detection Starter Kit I (Roche, Swiss).
Vector Construction and Strawberry Transformation
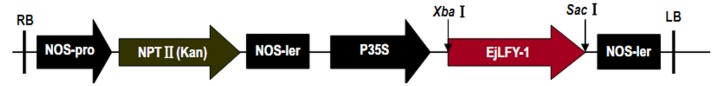
Full-length cDNA sequence of EjLFY-1 was subcloned into Xba I-Sac I sites of the binary vector pBI121 plasmid so that EjLFY-1 was under the control of CaMV 35S promoter (Figure 2). Recombinant plasmid pBI121-35S::EjLFY-1 was then introduced into the Agrobacterium tumefaciens strain EHA105.
FIGURE 2.
Vector construction. EjLFY-1 was inserted in sense orientation between the Xba I and Sac I sites of the binary vector pBI121. RB Right border, P35S cauliflower mosaic virus 35S promoter, Nos, nos terminator; LB, left border.
Strawberry (Fragaria × ananassa) cv. Tudla was used for Agrotransfection. A. tumefaciens-mediated transformation was performed with the leaf-disk procedure (Nehra et al., 1990). Leaf segments were excised from in vitro plantlets grown on MS basal medium (Murashige and Skoog, 1962). The leaf segments were then cocultivated with A. tumefaciens harboring the plasmid pBI121-35S::EjLFY-1, followed by regeneration under the selection medium (MS+1.5 mg L-1 TDZ+0.4 mg L-1 IBA) containing kanamycin (30 mg L-1). Regenerated buds were excised and transferred to 1/2MS medium containing kanamycin (50 mg L-1) for rooting. Resistant plants from separate leaf explants were defined as independent transgenic events.
Verification of Transgenic Plantlet
Genomic DNA were extracted from young leaves of kanamycin-resistant plants and the WT plants following a modified CTAB method (Ma et al., 2008), and then PCR analysis was carried out for detection of the insertion. Individual plants were tested for the presence of both EjLFY-1 and NPT II genes separately. To avoid the potential pollution caused by Agrobacterium may exist in the regenerated resistant plants, the existing of ChvA gene (Chromosomal virulence gene A) which specially belongs to Agrobacterium was also checked and excluded by PCR. Primers (AGRIF and AGRIR) and amplification methods are listed in Table 1.
For further verification of the integration of EjLFY-1, Southern blotting was performed using a randomly selected plantlet which had been confirmed by PCR. About 20 μg of genomic DNA was digested with EcoR I and then electrophoresed on a 0.8% (W/V) agarose gel. The digested DNA in the gel was then transferred to an Immobilon-Ny+ membrane (Millipore, USA). A fragment of NPT II gene obtained by PCR (primers: KANF and KANR, Table 1) was labeled as the probe using the DIG DNA Labeling and Detection Kit II (Roche, Diagnostics, USA). Hybridization was carried out according to the manufacturer’s instructions.
Non-transgenic strawberry (WT) was used as the control, except that the Agrobacterium strain containing recombined plasmid was used as a control for detection of the existence of ChvA gene.
Gene Expression Detection in the Transformants
Expression of EjLFY-1 in the first generation transgenic plantlet was detected by RT-PCR. Plantlets grown in greenhouse were used to extract the total RNA using an improved CTAB method as described by Chang et al. (2007). Before precipitation, the RNA was treated with RNase-free DNase I (Takara Biotechnology Co., Dalian, China) at 37°C for 4 h to eliminate the eventual genomic contamination. RNA integrity was assessed on a 1.0% agarose gel, and concentration was estimated using a DU800 spectrophotometer (Beckman Coulter, Fullerton, CA, USA). The first-stand cDNA synthesis was performed with 1 μg total RNA and Oligo d(T)18 primer using Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA, USA), according to the protocols provided by the manufacturers. Primers EjLFYfwd and EjLFYrev were used for the RT-PCR confirmation and 2 μl of the cDNA product was used as the template.
Quantitative real-time PCR (qRT-PCR) was performed to detect the expression of EjLFY-1 in the third-generation propagated clones originated from stolon buds of the transformants. Leaf was used as the material for the detection of EjLFY-1, shoot apex meristem was used to examine the expression level of FaAP1, a homolog of APETALA1 (AP1) in strawberry. Expression level of FaTFL1, homolog of the main floral repressor TERMINAL FLOWER 1 (TFL1) in strawberry, was also detected. Gene-specific primers used in qRT-PCR were also listed in Table 1. RNA extraction and cDNA synthesis were carried out following the method mentioned above. The qRT-PCR was performed using SYBR Green qPCR Kit (Takara Biotechnology Co., Dalian, China) following the manufacturer’s instructions. qPCR was repeated with three biological replicates, and each sample was assayed in triplicate by PCR. The 26S rRNA gene of strawberry was used as a internal control.
Field Trials
Independent transgenic lines and WT plants were rooted, transferred into pots and grown in a solar greenhouse in Shenyang Agricultural University (123°E, 41°N, Shenyang, Liaoning Province, China) at December of 2009. The agamic propagated plants from stolon buds of the transgenic plantlets were rooted and planted into new nutritional pots, and then separated from the donor plants.
At the end of each cropping seasons, old leaves and roots of the plantlets were partially removed and the plantlets were then transferred into a new nutritional pots separately.
Horticultural Traits Analysis
The plantlets were successively flowering after growing in the greenhouse for 3–4 months and their horticultural traits were recorded individually. Flower timing of each sample was measured as the time period from transferring into pots to formation of the first flower. Plant height, flower numbers were also recorded.
Data Analysis
The amino acid sequences were aligned and a phylogenetic tree was generated using the Clustal W method (Higgins et al., 1994). Boot strap values were derived from 1,000 replicate runs. The phylogenetic tree was constructed by MEGA 5.0 software (Tamura et al., 2011). Introns distribution diagram were drew with DNAMAN 6.0 software (Lynnon Biosoft, USA). The horticultural traits data were analyzed with EXCEL. Statistical analyses were performed using one-way ANOVA test, followed by Bonferroni’s test with SPSS software.
Results
Isolation and Sequence Characterization of EjLFY-1
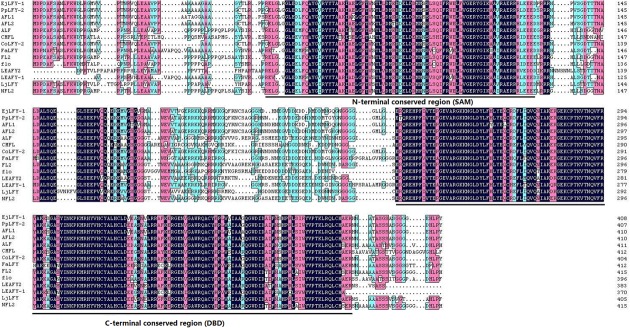
With a pair of gene-specific primers, a segment of 1,227 bp was isolated and sequenced. Sequence alignment showed that it is similar with the EjLFY-1 sequence which reported by Esumi et al. (2005). There are only three nucleotide acids and one amino acid diverse between the isolated sequence and the reported one, which may be used as molecular tool for cultivar differentiation. Alignment of the amino acids sequences encoded by EjLFY-1 and other LFY-like genes showed that they share the conserved domains with each other in the N-terminal and C-terminal, especially in the C-terminal conserve motif (Figure 3).
FIGURE 3.
Sequence alignment of EjLFY-1 and other LEAFY-like proteins. Dashes indicate gaps to maximize the alignment. DBD motif at the C-terminal and shoot apical meristem (SAM) motif at the N-terminal are shown by lines on bottom of the alignment.
A phylogenetic tree was constructed by using the deduced protein of EjLFY-1 and the other reported LFY-like proteins with Neighbor-Joining Method (Supplementary Figure S1). EjLFY-1 falls in a small group constructed by other homologs of Rosaceae fruit trees, including the PpLFY of pear and the AFL1 and AFL2 of apple. These data are consistent with the traditional taxonomy, indicating their closely relationship during evolution process.
Full-length DNA sequence of EjLFY-1 was also obtained. Sequencing result showed that gEjLFY-1 (GenBank accession no. AY551183) was 2,602 bp in length, indicating the existing of intron(s). Alignment of gEjLFY-1 with its cDNA sequence showed the existing of two introns. The first intron is 464 bp and the second one is 911 bp in length. The amount of introns in gEjLFY-1 is also same as those reported LFY-like genes of other Rosaseae plants such as strawberry, dog rose and apple (Supplementary Figure S2).
Expression of EjLFY-1 in the Flowering Process of Loquat
Northern blotting data suggested that the transcription of EjLFY-1 initiated in the buds that still in morphologic vegetative stages (Figure 4, lanes 2–3), except in the earliest detected vegetative bud (Figure 4, lane 1). Transcription level of EjLFY-1 was increased gradually during the swelling period of the buds when the inflorescence meristems were initiated (Figure 4, lanes 4–6). Then the transcription level reached the topmost point when the inflorescence meristems started to generate flower meristems (Figure 4, lane 7). Once the flowers came into being, expression of EjLFY-1 decreased (Figure 4, lanes 8–9). No transcripts were detected in little flowers or blooming flowers (Figure 4, lanes 10–12).
FIGURE 4.
Expression of EjLFY-1 during the flowering process of loquat. Materials were collected following the flowering development of loquat. For each lane, 10 μg of total RNA was loaded, blotted, and hybridized with an EjLFY-1 probe. rRNA stained with ethidium bromide showing the equivalence of RNA loading between the samples.
Generating and Verification of Transformants
An over-expression construct with EjLFY-1 CDS under the control of CaMV 35S promoter was introduced into strawberry cv. Tudla. Totally 59 resistant plantlets belonging to three independent lines (L1, L2, and L3) were got and rooted on MS medium containing kanamycin. The resistant plantlets and the WT ones were then transferred into pots and grown in greenhouse (Supplementary Figure S3).
To verify the genomic integration of the target gene, presence of EjLFY-1 and NPT II were confirmed by PCR separately. The expected specific fragments of both genes were detected in the randomly selected transformants (Figure 5A). While for the detection of ChvA gene, target fragment couldn’t be detected in any tested transformants (Figure 5A), indicating the detected fragments of EjLFY-1 or NPT II were caused by transformation event, excluded the possibility of Agrobacterium continuation.
FIGURE 5.
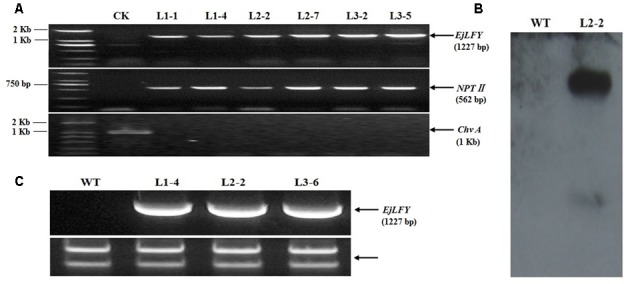
Verification of the transformants. (A) PCR detection for EjLFY-1, NPT II and Chv A genes, Marker: DL2000. (B) Southern blot analysis of L2-2. (C) Expression detection of EjLFY-1 in random selected plantlets of three transgenic lines.
L2-2, a random selected PCR-positive transgenic plantlet, was further tested by using Southern blotting detection for the integration of NPT II gene. Southern hybridization with the NPT II probe showed that two bands were visible in L2-2 while none was observed in the WT control (Figure 5B). This result provides a further evidence of the EjLFY-1 insertion in strawberry genotypes.
Expression level of EjLFY-1 in independent transgenic line was detected by RT-PCR (primers: EjLFYfwd and EjLFYrev, Table 1). Results showed that transcripts of EjLFY-1 could be detected in the tested transgenic lines (Figure 5C), but not in WT ones.
Over-expression of EjLFY-1 in Strawberry Promotes the Flowering Process
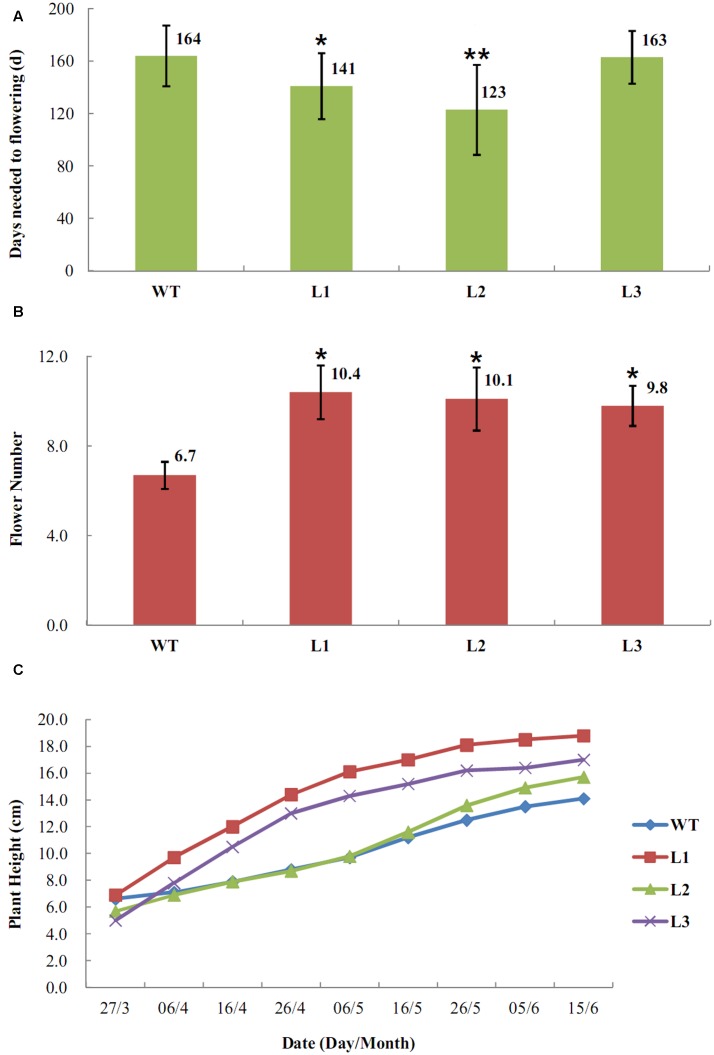
To evaluate the effect of over-expression of EjLFY-1 on the regulation of flowering process in strawberry, transgenic plantlets belonging to three independent lines (L1, L2, and L3) and the WT lines grown in small pots filled with nutrient soil were transferred into experimental greenhouse at December of 2009. It was found that plantlets belonging to L1 and L2 lines developed their inflorescences early and continuously at March of 2010, while the WT plants were still in vegetative growth at the meantime (Figures 6A–C). The early flowering habit of the transformants was very remarkable. Even at the April of 2010, when more than half plantlets of the L1 and L2 came into bloom, the WT ones didn’t develop inflorescences yet. Over-expression of EjLFY-1 promoted flowering process of the plants belongs to L1 and L2, their average flowering time were 23 and 41 days shorter than that of the WT, respectively (Figure 7). While for plants of line L3, the change of flowering formation period was negligible in compare with WT.
FIGURE 6.

Phenotypes of transformants and WT plants. (A) WT (right) and transgenic (left) plants. (C) Close-up of the inflorescence formed in transformants, which was not seen in the WT ones (B). (D) Morphology of the flowers and fruits of the transformants: Top left, flower of WT; top right, flower of one transgenic plantlet with doubled sepals; bottom, normal flower and fruit of transformants.
FIGURE 7.
Transgenic strawberry plants overexpressing EjLFY-1 showed early-flowering habits. (A) Average days needed for WT and transformants to flowering. Asterisks show significant difference comparing to WT (ANOVA, ∗P < 0.05, ∗∗P < 0.01). (B) Average flower numbers of the transformants and the WT ones. (C) Average plant height of the transformants and the WT ones.
Other Horticultural Traits in Transgenic Strawberry Plants
Other horticultural traits of the transformants were also tested at 2010. Over-expression of EjLFY-1 affected not only the flowering time of strawberry but also the flower quantities. Compared with WT plants, the transformants produced more flowers including the line L3 ones in which flowering time showed no significantly changes (Figure 7B). Even though, most flowers or fruits of the transformants showed no obviously changes compared with the WT ones, except that individual transgenic plantlets showing doubled sepals (Figure 6D).
Although over-expression of EjLFY-1 promoted the flowering process of the transgenic lines, it didn’t reduce the vegetative growth habit of those transformants at the first growing season. For example, three transgenic lines even showed a little higher average height phenotype than the WT ones at each investigated point except the beginning (Figure 7C).
Early-flowering Phenotype Could Be Maintained in Their Asexual Progeny
Almost all the fruit species in commercial production must be propagated by asexual ways so as to keep their stable traits. Normally the nursery stock of strawberry is runner plant which comes from stolon bud of the donor plant.
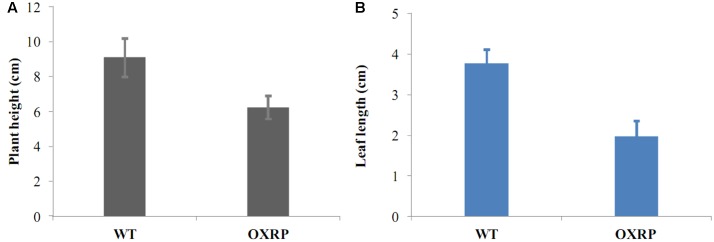
Early-flowering habit was also maintained in the runner plants originated from L1 and L2. And with the continuous asexual propagation, the clones showed a more strong early-flowering phenotype: the third-generation runner clones came into flowering at a very young stage (Figure 8A). And unlike the almost unchanged vegetative growth habits of the transformants in the first cropping season, runner clones showed an obviously reduction in vegetative growth. For example, plant showed a dwarf phenotype and their leaf were also extremely smaller than those of the WT ones (Figure 9).
FIGURE 8.

More strengthen phenotypes of the progeny propagated from stolon buds of the transgenic strawberry. (A) An very early flowering plant. (B) Abnormal flower in one plantlet with multiplied sepals formed at the incorrect position. (C) Flowering and fruiting of a runner plantlet (left) and the WT one which was in vegetative growth at June of 2014. Arrows showed the flower and fruit of the runner plantlet, and the stolon of the WT one.
FIGURE 9.
Runner transgenic plants were dwarf (A) with smaller leaves (B) than the normal ones.
Flowering of the runner clones were not only early but also continuously. Some runner plantlets of the transformants even could develop into flower in the hot summer, when the WT ones were still in their vegetative growth phase and only produced stolons (Figure 8C). In addition, variation of flower organs was also found in individual plantlets of the progeny, such as the multiplicated sepals (Figure 8B) in one runner plantlet originated from L1.
Gene Expression Detection in the Transgenic Asexual Progeny
To investigate whether the effect of ectopic expression of the transgene could be maintained in their asexual propagule, expression of EjLFY-1 was also detected in the runner plants originated from the transformants.
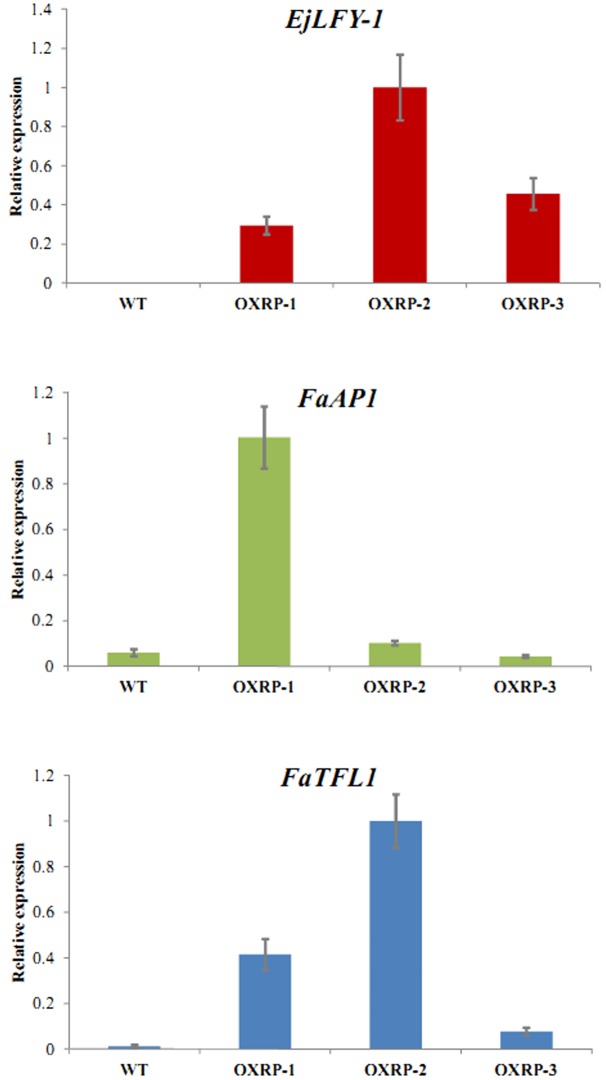
qRT-PCR detection showed that expression of EjLFY-1 also existed in the third-generation runner plants originated from the transformants, although the expression levels varies (Figure 10). As the downstream target gene of LFY homologs, expression of FaAP1, the AP1 homolog in strawberry, was also detected. Results showed that although the EjLFY-1 expression were higher in all the transgenic runner plants than that of the WT ones (in which were zero), expression of FaAP1 in those plants differed: its expression in OXRP-1 (Over-expression runner plant 1) was higher than not only the WT ones but also the OXRP-2 and OXRP-3 (Figure 10). The varied expression levels were also detected in FaTFL1, the main repressor in flowering process (Figure 10). Generally, the FaTFL1 expression in WT was lower than those in all the three tested transgenic runner plants.
FIGURE 10.
Expression detection of EjLFY-1, FaAP1, and FaTFL1 in third-generation runner plants originated from transgenic strawberry.
Discussion
Although the crop improvement of loquat is being carried out both by breeding programs and selection accessions from germplasm resources, the varieties grown mostly come from selections (Badenes et al., 2009). One reason for the slow breeding program in loquat is the existing of its long juvenile phase that last for several years. Genetic engineering technique provides a possible faster breeding method, but it is unavailable for loquat until now for its unclear molecular regulation mechanism of flowering and infeasible transformation system. Compared with other popular fruits, such as apple or citrus, whose genome had been sequenced, molecular mechanism of flowering in loquat is poorly understood and few flowering-related genes have been isolated and characterized.
LEAFY-like gene exists not only in angiosperms but also in plant species which don’t produce flowers (Moyroud et al., 2010). As a kind of meristem identity genes related to flowering in seed plants, LFY and its homologs have been demonstrated to not only distribute widely in plants but also function in distantly related species (Peña and Séguin, 2001; Moyroud et al., 2010). One possible reason is that LFY and its homologs are highly evolutionally conserved throughout the plant kingdom and show no apparent similarity to other proteins (Maizel et al., 2005). Two conserved domains of LFY protein were distinguished and, respectively, named as the DBD domain (DNA Binding Domain), which located in the N-terminal and leads it bind to the semi-palindromic 19-bp DNA cis-elements of the genes regulated by itself (Sayou et al., 2014), and the SAM domain (Sterile Alpha Motif), which located in the C-terminal and determines the genomic binding landscape (Sayou et al., 2016). Such characteristic motifs were also found in EjLFY-1 (Figure 3), suggesting its possible flowering regulator role in the flowering process of loquat.
Expression analysis revealed that EjLFY-1 started to increase its expression level in the buds following the transition from vegetative bud to inflorescence bud, decreased its expression level before the forming of flowers and eventually vanished after flowering (Figure 4). Comparing with expression of EjAP1, the AP1 homolog of loquat, EjLFY-1 started and ended its expression earlier (Liu et al., 2013), suggesting its possible key role in the regulation of flowering process of loquat.
To verify the function of EjLFY-1, a chimeric 35S::EjLFY-1 construct was introduced into strawberry. Most of the transgenic plantlets from lines 1 and 2 showed an early flowering phenotype in comparison with the wild-type ones (Figures 6, 7). Unlike the reduced vegetative development phenotype in the other reported transgenic plant species over-expressing LFY-like genes, strawberry over-expressing EjLFY-1 didn’t show obvious changes in the vegetative development habit at the first cropping season. Meanwhile the flowers and fruits of the transformants were almost as same as the wild ones (Figure 6D). Such results suggest the potential value of EjLFY-1 in genetic engineering for the fast breeding of loquat. For plantlets belonging to line 3, although the integration and expression were also verified by PCR, they didn’t show the similar early-flowering phenotype as the other two transformants lines. One possible reason for that is the insertion site effect of the transgene. However, it requires further examination.
The long juvenile phase of fruit trees limits their genetic improvement and prevents full domestication of them (Peña and Séguin, 2001). One of the major goals of fruits breeding programs is to reduce their juvenile phase and accelerate floral production. During the last decades, genetic engineering methods based on the use of transgenes have been successfully adopted to reduce the juvenile phase and accelerate the flowering process of some fruit species (Rai and Shekhawat, 2014). However, few reports mentioned the effectiveness and stabilization of the transgene over long periods or its asexual progeny, which was crucial for fruit trees because most of which are perennial and whose propagation in production are usually carried out through asexual reproduction ways in order to ensure reliability.
In this study, the observation results about the transgenic asexual clones lasting for three cropping seasons showed that the early-flowering habit induced by over-expression of EjLFY-1 could be maintained. Interestingly, with the continuous asexual propagation, the transgenic clones showed even a more consolidated early-flowering trait compared with their ancestor transformants of the first cropping season, such as the reduced vegetative growth habit and even abnormal floral organs (Figure 8). Reduction of the vegetative growth traits in those clones maybe caused by the continuous shorten of the vegetative growth, such as the earlier break of dormancy caused by accelerated flowering process. Such finding provides us new insights into the utilization potential of genetic engineering in the breeding of fruits.
The existing of abnormal floral organs in their clones suggests that continuously over-expression of EjLFY-1 may result in an excessive expression of the downstream floral organ related genes in strawberry, such as the AP1 homolog which may also control the outer two whorls of floral organs in strawberry just like what AP1 does in Arabidopsis (Bowman et al., 1993). Actually, the expression detection of EjLFY-1, FaAP1, and FaTFL1 (Figure 10) suggested that the ectopic expression of EjLFY-1 still exists in those runner clones, following a varied FaAP1 expression changes compared with the WT ones. TFL1 is a floral repressor which also binds to FD and suppresses the expression of floral promoters LEAFY and AP1 (Hanano and Goto, 2011). With the continuous expression of EjLFY-1 in strawberry and thereby the high expression level of FaAP1, expression of FaTFL1 was also activated in over-expression runner plants (OXRP) 1 and 2 (Figure 10), except the plant OXRP-3 in which the FaAP1 expression was not at a high level although the EjLFY-1 was also detected.
Although LFY has been demonstrated to function in distantly related species (Peña and Séguin, 2001; Moyroud et al., 2010), this is not the case for all the reported ones. It was shown that over-expression of AFL1 or AFL2, the LFY homologs in apple, can induce the early-flowering in Arabidopsis (Wada et al., 2002). In other reports, over-expression of AtLFY in apple didn’t show the same results instead of a columnar phenotype with shorten internodes (Flachowsky et al., 2010). Similar results were also reported in poplar (Rottmann et al., 2000). Perhaps it was caused by the divergent sequences of the LFY-like genes and their interacting factors among different species. In this study, we confirmed that the early-flowering habit existed in strawberry over-expressing EjLFY-1 and such habit could be maintained in their clones. For the closely evolutional relationship between strawberry and loquat, both belongs to Rosaceae family, it is promising that over-expression of EjLFY-1 in loquat could also accelerate its flowering process and break its juvenile stage.
Author Contributions
ZZ and SL conceived and designed research. YL, QZ, NM, HS, and CL conducted experiments. GH and JW contributed plant materials. YL and QZ analyzed data. YL wrote the manuscript. All authors read and approved the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We gratefully acknowledge help from Dr. Junhui Zhou, University of Maryland, College Park, with English editing.
Footnotes
Funding. This work was supported by the Education Administration Research Program of Liaoning Province (2008629 and L2013258) and the Natural Science Foundation of Liaoning Province (2014027015).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00496/full#supplementary-material
References
- Abe M., Kobayashi Y., Yamamoto S., Daimon Y., Yamaguchi A., Ikeda Y., et al. (2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309 1052–1056. 10.1126/science.1115983 [DOI] [PubMed] [Google Scholar]
- Badenes M. L., Lin S., Yang X., Liu C., Huang X. (2009). “Loquat (Eriobotrya Lindl.),” in Genetics and Genomics of Rosaceae, eds Folta K. M., Gardiner S. E. (New York, NY: Springer Science & Business Media; ) 525–538. 10.1007/978-0-387-77491-6_25 [DOI] [Google Scholar]
- Bergonzi S., Albani M. C. (2011). Reproductive competence from an annual and a perennial perspective. J. Exp. Bot. 62 4415–4422. 10.1093/jxb/err192 [DOI] [PubMed] [Google Scholar]
- Bowman J. L., Alvarez J., Weigel D., Meyerowizt E. M., Smyth D. R. (1993). Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development 119 721–743. [Google Scholar]
- Chang L., Zhang Z., Yang H., Li H., Dai H. (2007). Detection of strawberry RNA and DNA viruses by RT-PCR using total nucleic acid as a template. J. Phytopathol. 155 431–436. 10.1111/j.1439-0434.2007.01254.x [DOI] [Google Scholar]
- Corbesier L., Vincent C., Jang S., Fornara F., Fan Q., Searle I., et al. (2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316 1030–1033. 10.1126/science.1141752 [DOI] [PubMed] [Google Scholar]
- Duan Y. X., Fan J., Guo W. W. (2010). Regeneration and characterization of transgenic kumquat plants containing the Arabidopsis APETALA1 gene. Plant Cell Tiss. Organ. Cult. 100 273–281. 10.1007/s11240-009-9646-3 [DOI] [Google Scholar]
- Eriksson S., Böhlenius H., Moritz T., Nilsson O. (2006). GA4 is the active gibberellin in the regulation of LEAFY transcription and Arabidopsis floral initiation. Plant Cell 18 2172–2181. 10.1105/tpc.106.042317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esumi T., Tao R., Yonemori K. (2005). Isolation of LEAFY and TERMINAL FLOWER 1 homologues from six fruit tree species in the subfamily Maloideae of the Rosaceae. Sex Plant Reprod. 17 277–287. 10.1007/s00497-004-0239-3 [DOI] [Google Scholar]
- Flachowsky H., Hättasch C., Höfer M., Peil A., Hanke M. (2010). Overexpression of LEAFY in apple leads to a columnar phenotype with shorter internodes. Planta 231 251–263. 10.1007/s00425-009-1041-0 [DOI] [PubMed] [Google Scholar]
- Fornara F., de Montaigu A., Coupland G. (2010). Snapshot: control of flowering in Arabidopsis. Cell 141 550–550.e2. 10.1016/j.cell.2010.04.024 [DOI] [PubMed] [Google Scholar]
- Hanano S., Goto K. (2011). Arabidopsis TERMINAL FLOWER1 is involved in the regulation of flowering time and inflorescence development through transcriptional repression. Plant Cell 23 3172–3184. 10.1105/tpc.111.088641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z. H., Zhu Q., Dabi T., Li D. B., Weigel D., Lamb C. (2000). Transformation of rice with the Arabidopsis floral regulator LEAFY causes early heading. Transgenic. Res. 9 223–227. 10.1023/A:1008992719010 [DOI] [PubMed] [Google Scholar]
- Higgins D., Thompson J., Gibson T., Thompson J. D., Higgins D. G., Gibson T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4680. 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotoda N., Iwanami H., Takahashi S., Abe K. (2006). Antisense expression of MdTFL1, a TFL1-like gene, reduces the juvenile phase in apple. J. Am. Soc. Hortic. Sci. 131 74–81. [Google Scholar]
- Li W., Zhou Y., Liu X., Yu P., Cohen J. D., Meyerowitz E. M. (2013). LEAFY controls auxin response pathways in floral primordium formation. Sci. Signal. 6:ra23 10.1126/scisignal.2003937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S. Q., Sharpe R. H., Janick J. (1999). Loquat: botany and horticulture. Hortic. Rev, 23 233–276. [Google Scholar]
- Liu C., Zhou J., Bracha-Drori K., Yalovsky S., Ito T., Yu H. (2007). Specification of Arabidopsis floral meristem identity by repression of flowering time genes. Development 134 1901–1910. 10.1242/dev.003103 [DOI] [PubMed] [Google Scholar]
- Liu Y., Song H., Liu Z., Hu G., Lin S. (2013). Molecular characterization of loquat EjAP1 gene in relation to flowering. Plant Growth Regul. 70 287–296. 10.1007/s10725-013-9800-0 [DOI] [Google Scholar]
- Ma Y., Sun H., Zhao G., Dai H., Gao X., Li H., et al. (2008). Isolation and characterization of genomic retrotransposon sequences from octoploid strawberry (Fragaria × ananassa Duch.). Plant Cell Rep. 27 499–507. 10.1007/s00299-007-0476-7 [DOI] [PubMed] [Google Scholar]
- Maizel A., Busch M. A., Tanahashi T., Perkovic J., Kato M., Hasebe M., et al. (2005). The floral regulator LEAFY evolves by substitutions in the DNA binding domain. Science 308 260–263. 10.1126/science.1108229 [DOI] [PubMed] [Google Scholar]
- Mandel M. A., Yanofsky M. F. (1995). A gene triggering flower formation in Arabidopsis. Nature 377 522–524. 10.1038/377522a0 [DOI] [PubMed] [Google Scholar]
- Moyroud E., Kusters E., Monniaux M., Koes R., Parcy F. (2010). LEAFY blossoms. Trends Plant Sci. 15 346–352. 10.1016/j.tplants.2010.03.007 [DOI] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bioassay with tobacco tissue cultures. Physiol. Plant. 15 473–497. 10.1111/j.1399-3054.1962.tb08052.x [DOI] [Google Scholar]
- Nehra N. S., Chibbar R. N., Kartha K. K., Datla R. S., Crosby W. L., Stushnoff C. (1990). Genetic transformation of strawberry by Agrobacterium tumefaciens using a leaf disk regeneration system. Plant Cell Rep. 9 293–298. 10.1007/BF00232854 [DOI] [PubMed] [Google Scholar]
- Peña L., Martín-Trillo M., Juárez J., Pina J. A., Navarro L., Martínez-Zapater J. M. (2001). Constitutive expression of Arabidopsis LEAFY or APETALA1 genes in citrus reduces their generation time. Nat. Biotechnol. 19 263–267. 10.1038/85719 [DOI] [PubMed] [Google Scholar]
- Peña L., Séguin A. (2001). Recent advances in the genetic transformation of trees. Trends Biotechnol. 19 500–506. 10.1016/S0167-7799(01)01815-7 [DOI] [PubMed] [Google Scholar]
- Rai M. K., Shekhawat N. S. (2014). Recent advances in genetic engineering for improvement of fruit crops. Plant Cell Tiss. Organ. Cult. 116 1–15. 10.1007/s11240-013-0389-9 [DOI] [Google Scholar]
- Rottmann W. H., Meilan R., Sheppard L. A., Brunner A. M., Skinner J. S., Ma C., et al. (2000). Diverse effects of overexpression of LEAFY and PTLF, a poplar (Populus) homolog of LEAFY/FLORICAULA, in transgenic poplar and Arabidopsis. Plant J. 22 235–245. 10.1046/j.1365-313x.2000.00734.x [DOI] [PubMed] [Google Scholar]
- Sayou C., Monniaux M., Nanao M. H., Moyroud E., Brockington S. F., Thévenon E., et al. (2014). A promiscuous intermediate underlies the evolution of LEAFY DNA binding specificity. Science 343 645–648. 10.1126/science.1248229 [DOI] [PubMed] [Google Scholar]
- Sayou C., Nanao M. H., Jamin M., Posé D., Thévenon E., Grégoire L., et al. (2016). A SAM oligomerization domain shapes the genomic binding landscape of the LEAFY transcription factor. Nat. Commun. 7:11222 10.1038/ncomms11222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slovin J. P., Schmitt K., Folta K. M. (2009). An inbred line of the diploid strawberry Fragaria vesca f. semperflorens for genomic and molecular genetic studies in the Rosaceae. Plant methods 5 665–672. 10.1186/1746-4811-5-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikanth A., Schmid M. (2011). Regulation of flowering time: all roads lead to Rome. Cell Mol. Life Sci. 68 2013–2037. 10.1007/s00018-011-0673-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Steche G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trankner C., Lehmann S., Hoenicka H., Hanke M. V., Fladung M., Lenhardt D., et al. (2010). Over-expression of an FT-homologous gene of apple induces early flowering in annual and perennial plants. Planta 232 1309–1324. 10.1007/s00425-010-1254-2 [DOI] [PubMed] [Google Scholar]
- Wada M., Cao Q. F., Kotoda N., Soejima J. I., Masuda T. (2002). Apple has two orthologues of FLORICAULA/LEAFY involved in flowering. Plant Mol. Biol. 49 567–577. 10.1023/A:1015544207121 [DOI] [PubMed] [Google Scholar]
- Wagner D., Sablowski R. W., Meyerowitz E. M. (1999). Transcriptional activation of APETALA1 by LEAFY. Science 285 582–584. 10.1126/science.285.5427.582 [DOI] [PubMed] [Google Scholar]
- Weigel D., Alvarez J., Smyth D. R., Yanofsky M. F., Meyerowitz E. M. (1992). LEAFY controls floral meristem identity in Arabidopsis. Cell 69 843–859. 10.1016/0092-8674(92)90295-N [DOI] [PubMed] [Google Scholar]
- Weigel D., Nilsson O. (1995). A developmental switch sufficient for flower initiation in diverse plants. Nature 377 495–500. 10.1038/377495a0 [DOI] [PubMed] [Google Scholar]
- Wigge P. A., Kim M. C., Jaeger K. E., Busch W., Schmid M., Lohmann J. U., et al. (2005). Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309 1056–1059. 10.1126/science.1114358 [DOI] [PubMed] [Google Scholar]
- William D. A., Su Y., Smith M. R., Lu M., Baldwin D. A., Wagner D. (2004). Genomic identification of direct target genes of LEAFY. Proc. Natl. Acad. Sci. U.S.A. 101 1775–1780. 10.1073/pnas.0307842100 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.