Abstract
The conserved membrane-proximal external region (MPER) of human immunodeficiency virus type 1 (HIV-1) gp41 is a target of two broadly neutralizing human monoclonal antibodies, 2F5 and 4E10, and is an important lead for vaccine design. However, immunogens that bear MPER epitopes so far have not elicited neutralizing antibodies in laboratory animals. One explanation is that the immunogens fail to recreate the proper molecular environment in which the epitopes of 2F5 and 4E10 are presented on the virus. To explore this molecular environment, we used alanine-scanning mutagenesis across residues 660 to 680 in the MPER of a pseudotyped variant of HIV-1JR-FL, designated HIV-1JR2, and examined the ability of 2F5 and 4E10 to neutralize the Ala mutant viruses. The results show that the only changes to produce neutralization resistance to 2F5 occurred in residue D, K, or W of the core epitope (LELDKWANL). Likewise, 4E10 resistance arose by replacing one of three residues; two (W and F) were in the core epitope, and one (W) was seven residues C-terminal to these two (NWFDISNWLW). Importantly, no single substitution resulted in resistance of virus to both 2F5 and 4E10. Surprisingly, 8 out of 21 MPER Ala mutants were more sensitive than the parental pseudovirus to 2F5 and/or 4E10. At most, only small differences in neutralization sensitivity to anti-gp120 monoclonal antibody b12 and peptide T20 were observed with the MPER Ala mutant pseudoviruses. These data suggest that MPER substitutions can act locally and enhance the neutralizing activity of antibodies to this region and imply a distinct role of the MPER of gp41 during HIV-1 envelope-mediated fusion. Neutralization experiments showing synergy between and T20 and 4E10 against HIV-1 are also presented. The data presented may aid in the design of antigens that better present the MPER of gp41 to the immune system.
Eliciting broadly neutralizing antibodies against human immunodeficiency virus type 1 (HIV-1) by immunization is a major goal in HIV-1 vaccine development (9, 22a, 29, 34a). A few human monoclonal antibodies that neutralize a broad range of primary isolates of HIV-1 have been isolated (10, 20, 60, 77) and exemplify the antibodies it would be desirable to elicit in high titer with an HIV-1 vaccine (9). Monoclonal antibodies against the HIV-1 surface glycoprotein gp120 include b12, which binds to a discontinuous epitope overlapping the CD4-binding site, and 2G12, which binds to a glycan cluster on the outer face of gp120 (11, 53, 57, 66). In addition, there are two monoclonal antibodies against the transmembrane glycoprotein gp41, 2F5 and 4E10, which bind to neighboring linear epitopes in the membrane-proximal external region (MPER) of gp41 (8, 42, 60, 77).
The MPER of gp41 is highly conserved, and several studies have implicated it as an essential part of the cell fusion machinery (19, 40, 56). For vaccine development, linear neutralizing epitopes have a potential advantage over more complex ones in that relatively short peptides could be used to elicit a focused antibody response to the desired target so long as the elicited antibodies recapitulate the neutralizing activity of the original monoclonal antibodies. Owing to the linear nature of the neutralizing epitopes it bears and to its high amino acid conservation, the MPER of gp41 is an attractive target for HIV-1 vaccine development.
To date, immunization experiments involving the MPER of gp41 have generally focused on eliciting 2F5-like antibodies, primarily because 2F5 is so well characterized. Extensive epitope mapping studies of 2F5 with synthetic peptides (2, 5, 28, 65), phage-displayed peptide libraries (38, 42, 77), and protease protection assays (47) have revealed that peptides from gp41 of moderate length bind with high affinity to 2F5, e.g., ELLELDKWASLWN (2). At the core of the epitope, the residues D, K, and W were found to be most critical for recognition by 2F5 but by themselves were insufficient. A variety of immunogens bearing 2F5 epitope sequences in different contexts have failed to elicit neutralizing antisera against primary HIV-1, although high serum antibody titers against 2F5 epitope peptides have been achieved (16, 22, 28, 33, 34, 41).
These types of experiments have rested on the assumption that an antigen with high affinity to 2F5 should also be a good candidate for eliciting 2F5-like antibodies. This assumption is threatened, however, by the ability of the immune system to generate dominant antibody responses against irrelevant conformations of the 2F5 epitope peptides. The crystal structure of the 2F5 Fab in complex with the peptide ELDKWAS has been determined (45). In the crystal structure, ELDKWAS forms a β-turn, in which the core residues DKW are in contact with the paratope of 2F5. To increase the affinity of linear peptides to 2F5, a β-lactam constraint was used and shown to enhance the affinity of a 13-mer peptide to 2F5 (36). Immunizations of laboratory animals with this constrained peptide and other constrained 2F5 epitope mimics have not yet elicited HIV-1-neutralizing antibodies; however, a recent study suggests that some progress toward this goal may have been achieved (75).
The epitope of 4E10 has been somewhat less intensively studied but is also viewed as a potentially very valuable target for immunization studies. 4E10 is capable of neutralizing an extremely broad range of HIV-1 primary isolates, from virtually all subtypes, although it appears to have modest potency (77). The core of the 4E10 epitope, NWFDIT, maps just C-terminal to the 2F5 epitope on the gp41 ectodomain (60, 77). However, as for 2F5, there is evidence that more than just this core linear motif is important for 4E10 recognition. For example, the affinity of 4E10 for recombinant gp41 that has been denatured is less than its affinity for untreated gp41 (the reverse is true for 2F5) (4, 77). Moreover, 4E10 appears to bind with greater affinity to peptides with residues that flank the core epitope (77). Finally, 4E10 can neutralize HIV-1 isolates with sequences that differ from the subtype B consensus motif, NWFDIT, in the first, fourth, and sixth positions (77). Immunogenicity studies on 4E10 epitope peptides have not yet been reported. However, since 4E10 binds tightly to recombinant gp41 and yet recombinant gp41 does not elicit HIV-1-neutralizing antisera, it would appear that the 4E10 epitope is not highly immunogenic and that eliciting high titers of 4E10-like antibodies will be difficult.
It is not known exactly what antigenic form of gp41 elicited 2F5 and 4E10. gp41 on the virus or on HIV-1-infected cells undergoes extensive conformational changes during viral attachment in at least three states: the untriggered or native conformation, a receptor-activated and/or fusion-intermediate state, and the postfusion conformation. Structures of the six-helix bundle of gp41 have been determined and correspond to the postfusion state of gp41 (14, 63, 70, 78). The six-helix bundle consists of an inner core of parallel alpha-helices corresponding to the N-terminal heptad repeat (N-HR) region of gp41. Around this core are packed three alpha-helices, corresponding to the C-terminal heptad repeat (C-HR) region, in an antiparallel fashion. The 2F5 and 4E10 epitopes are not present in these six-helix bundle structures.
2F5 and 4E10 appear to bind to HIV-1-infected cells relatively poorly (58, 77), and there is evidence that these monoclonal antibodies have some postattachment neutralization activity (3, 24, 34), a mechanism of HIV-1 fusion inhibition that is also attributed to the peptide drug T20 (also called DP178, Fuzeon, and enfuvirtide) (37, 72). (T20 is a 36-mer peptide that corresponds to the C-HR region on the ectodomain of gp41 and inhibits HIV-1 envelope mediated fusion by inhibiting six-helix bundle formation [72].) Unfortunately, there is no detailed structural information on the native or fusion-intermediate states of gp41, making it extremely difficult to apply rational constraints to gp41 for vaccine purposes. Moreover, even if this structural information were available, there is no simple and reliable method to enhance the relative immunogenicity of neutralizing epitopes on a protein directly by using structural data. Thus, empirical approaches to vaccine development are essential, and more functional data involving the MPER of gp41 can aid immunogen design. Mutagenesis is one method for exploring protein function. Indeed, for 2F5 and 4E10, there has not been a systematic mutational analysis of their epitopes as they are presented on infectious HIV-1.
To determine the extent to which substitutions in the MPER of gp41 can modulate the exposure of the epitopes of 2F5 and 4E10 on intact virions, we performed alanine-scanning mutagenesis across residues 660 to 680 of an R5 HIV-1-pseudotyped virus and measured the ability of these monoclonal antibodies to neutralize the panel of Ala mutant viruses. Thus, unlike experiments designed to measure the affinity between an antibody and synthetic peptides or purified antigens, here, antibody neutralization titers against infectious mutants of HIV-1 are measured and compared. We identified substitutions in the MPER of gp41 that caused either resistance or increased sensitivity to 2F5 and to 4E10, and none of the substitutions significantly affected the sensitivity of the mutant pseudoviruses to T20 or other monoclonal antibodies. We also investigated the degree of synergy with which 2F5, 4E10, and T20 neutralize pseudotyped HIV-1 to gain further insight into the mechanism by which these reagents neutralize HIV-1. Significant synergy between 4E10 and T20 was observed, suggesting that their epitopes are non-mutually exclusive targets on gp41 that may be exploited for vaccine and drug design.
Taken as a whole, the present results show that only a few crucial residues in the MPER are indispensable for HIV-1 neutralization by 2F5 and 4E10. Moreover, the results show how the local sequence environment in which the epitopes of 2F5 and 4E10 are presented can profoundly influence their exposure on the virus without affecting other regions of HIV-1 Env. The enhancement of neutralizing epitopes on the MPER of gp41 may prove to be useful for HIV-1 vaccine design.
MATERIALS AND METHODS
Materials.
The following monoclonal antibodies were used in this study: b12 (53), b6 (53), 2F5 (42), 4E10 (8), Fab Z13 (77), 2G12 (66), D50 (21), D49 (21), 98-6 (73), hNMO1 (43), and single chain (sc) Fv X5 (39). HIV immunoglobulin was generously provided by J. Mascola (Vaccine Research Center, Washington, D.C.), 5-helix (54) was a gift from M. Root (Kimmel Cancer Center, Philadelphia, PA), and T20 was a gift from M. Franti and P. Poignard (TSRI, La Jolla, Calif.). Plasmid pSVIIIexE7pA−YU2 (62) was a gift from J. Sodroski; the envelope genes from JRFL and R2 (50) were cloned into pSVIIIexE7pA− as described previously (76). The following reagents were also obtained from the AIDS Research and Reference Reagent Program: pNL4-3.Luc.R−E− (17) (contributed by Nathaniel Landau) and U87.CD4.CCR5 cells (6) (contributed by H. Deng and D. Littman).
Engineering and production of HIV-1 mutant pseudovirions.
Plasmid pSVIIIexE7pA−JR-FL (62, 76) bears the KpnI fragment of the HIV-1JR-FL envelope gene, and if cotransfected into host cells with the luciferase reporter plasmid pNL4-3.Luc.R−E−, single-round infectious clones of HIV-1 pseudovirions are produced, the infectivity of which may be followed by luciferase activity in target cells (17). To engineer pSVIIIexE7pA−JR2, four point mutations were introduced into pSVIIIexE7pA−JR-FL with the Quikchange mutagenesis kit (Stratagene), which produces three substitutions in the MPER (i.e., S668N, T676S, and K677N) so that the sequence of the MPER of gp41JR2 is that of the HIV-1 R2 strain. DNA sequencing of the entire envelope gene (KpnI fragment) revealed no further mutations.
The Ala mutants were all generated with the pSVIIIexE7pA−JR2 template and the Quikchange kit according to the manufacturer's instructions. 293T cells grown in Dulbecco's modified Eagle's medium (Gibco) supplemented with penicillin, streptomycin, l-glutamine, and fetal bovine serum (10%) were transiently transfected with pSVIIIexE7pA−JR2 DNA and its mutants (2 μg) together with pNL4-3.Luc.R−E− (4 μg) with the calcium phosphate method. At 24 h posttransfection, the culture supernatants of all transfected cell cultures were replaced with fresh medium, and the cultures were incubated for a further 24 h. The supernatants containing pseudoviruses were harvested and either used fresh or, in some cases, frozen at −80°C until further use. We note that it was critical to clear the virus-containing supernatants from the transfected 293T cells for some mutants by centrifugation because significant background luciferase activity would otherwise be transferred to the target cells. We note here that the cells were washed twice carefully by centrifugation prior to seeding, since we found that even small amounts of the trypsin that is used to detach the cells can proteolyze and inactivate peptide T20. One hundred microliters of pseudovirions (roughly generating between 105 and 106 relative light units) were mixed with various concentrations of antibody, incubated for 1 h at 37°C, added to cells, and incubated for a further 3 days. The luciferase activity from triplicate wells was measured on a luminometer (EG&G Berthold LB 96V, Perkin Elmer, Gaithersburg, Md.), with the luciferase assay reagent (Promega), according to the manufacturer's instructions.
Synergy determination.
Antibodies were mixed at a constant ratio that was determined on the basis of their relative neutralization potency (90% inhibitory dose). Dose-response curves were then determined in the same assay for the antibody mixture and each of the antibodies in the mixture alone. The presence or absence of synergy was assessed with the computer program CalcuSyn (Biosoft, Ferguson, Mo.). The program automatically calculates the confidence interval, with values of less than 1, equal to 1, and greater than 1 indicating synergy, additive effect, and antagonism, respectively. To determine the confidence intervals, we ensured that all the basic requirements were met as recommended (15).
RESULTS
Generating a panel of MPER Ala mutants.
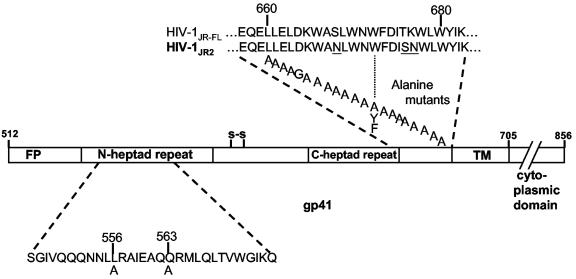
The alanine scan across the MPER of gp41 (i.e., residues 660 to 680 of gp160) was performed on a pseudovirus variant of HIV-1JR-FL (76) that contains three amino acid changes in gp41, designated HIV-1JR2 (Fig. 1). Baseline neutralization titers of 2F5, 4E10, T20, b12, 2G12, and purified immunoglobulin G from pooled sera from HIV-1-seropositive individuals were determined for HIV-1JR2, HIV-1JR-FL and HIV-1R2 (50) in a single-round infectivity assay with U87.CD4.CCR5 cells (6) and luciferase activity as a readout. As expected, JR2 and JR-FL showed very similar sensitivity to all of the antibodies and to T20, indicating that JR2 inherited the neutralization profile of JR-FL (Table 1). It is noteworthy that the R2 strain is ≈4-fold more sensitive than JR2 to 4E10 even though the sequence of the MPER is identical in R2 and JR2 and the two viruses are equally sensitive to 2F5.
FIG. 1.
Alanine scan of L660 to W680 of gp160JR2 (only gp41 is shown). The three amino acid differences between JR2 and JR-FL are shown underlined. Each A represents one Ala mutant. W672 was replaced with Phe (F) and Tyr (Y) in addition to Ala. The numbering scheme follows that of the HxB2 strain.
TABLE 1.
Neutralization of pseudotyped HIV-1JR-FL, HIV-1R2, and HIV-1JR2 by various antibodies and T20 peptide in the luciferase assay with U87.CD4.CCR5 cells
| Antibody or inhibitor | Inhibitory concn (μg/ml)
|
|||||
|---|---|---|---|---|---|---|
| R2
|
JR-FL
|
JR2
|
||||
| IC50 | IC90 | IC50 | IC90 | IC50 | IC90 | |
| 2F5 | 0.7 | 9 | 1.5 | 10.5 | 2 | 14.5 |
| 4E10 | 0.9 | 10 | 4 | 35 | 4.5 | 40 |
| b12 | 3.5 | 35 | 0.025 | 0.15 | 0.02 | 0.15 |
| 2G12 | >100 | >100 | 0.25 | 2 | 0.15 | 1.5 |
| Pooled HIV immu- noglobulin | 200 | 1,000 | 80 | 500 | 50 | 500 |
| T20 peptide | 0.025 | 0.5 | 0.05 | 0.5 | 0.05 | 0.5 |
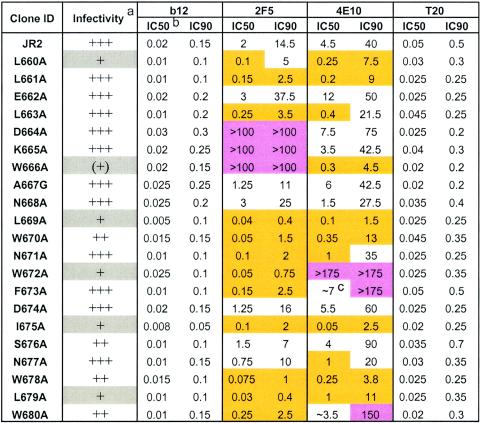
The Ala mutant pseudoviruses were harvested from supernatants of 293T cells cotransfected with the mutant Env vector and pNL4-3luc (17) DNA and used to infect U87.CD4.CCR5 target cells. Most of the 21 mutants produced the same relative luciferase activity as the parental virus, whereas the signal produced with six of them (L660A, W666A, L669A, W672A, I675A, and L679A) was severalfold to over an order of magnitude lower than that of the parental virus (Table 2). We note that the Ala substitutions that appeared to be most deleterious to viral titer were of hydrophobic residues and did not cluster together but rather were separated by two to five residues and dispersed along the MPER of gp41. We further note that the effect of freezing was generally to lower viral titer, typically by two- to threefold, such that it was necessary to use pseudovirus prepared from a fresh transfection in neutralization assays with the six attenuated pseudoviruses; the remaining mutants were used fresh or frozen with no observable difference in neutralization results.
TABLE 2.
Pseudotyped HIV-1JR2 and MPER Ala mutants in a single-round infection neutralization assay with U87.CD4.CCR5 cells against antibodies b12, 2F5, and 4E10 and peptide T20
Infectivity of Ala mutant pseudoviruses relative to parental HIV-1JR2 as measured by relative light units (RLU) produced in target cells by viral supernatants. +++, infectivity similar to that of wild-type HIV-1JR2; ++, >30% of wild-type infectivity; +, <30% of wild-type infectivity; (+), <10% of wild-type infectivity.
Concentrations are in micrograms per milliliter. IC50 and IC90 values that are >3-fold that of the parental (JR2) virus are highlighted in pink (mutant virus more neutralization resistant), whereas values that are less than one-third that of the IC50 or IC90 of JR2 are highlighted in gold (mutant virus more neutralization sensitive). Values represent the averages of three independent experiments performed in triplicate.
Values preceded by ≈ are approximate. Neutralization curves were shallow, so that there was greater error at the 50% infectivity level (see text).
MPER Ala substitutions affect HIV-1 neutralization sensitivity to 2F5 and 4E10 but not b12 or T20.
All of the gp41 MPER Ala mutant pseudoviruses were tested against monoclonal antibodies 2F5 and 4E10 in the single-round infectivity assay. In order to determine what effects these mutations might have on other regions of the virus, monoclonal antibody b12 and T20 were also included in the neutralization assays for comparison. As expected from previous epitope mapping studies, clones D664A, K665A, and W666A were resistant to 2F5, since DKW forms the core of the 2F5 epitope (Table 2). Surprisingly, the majority of the remaining mutants were more sensitive to 2F5, even for substitutions at positions that reportedly contribute binding affinity to 2F5 according to synthetic peptide mapping studies, an optimal sequence for which has been identified as ELLELDKWASLWN (2, 65). A similar scenario was observed with 4E10. Only three mutants, W672A, F673A, and W680 were resistant to neutralization by 4E10. The former two substitutions map to the center of the previously identified 4E10 core motif, NWFDI(T/S) (60, 77). Interestingly, the W680A substitution also led to significant resistance to 4E10 even though W680 is situated four residues C-terminal to NWFDIS. Thus, in the context of virus neutralization, the 4E10 epitope appears to require W680 (WFDISNWLW).
For 2F5, substitution of either D664, K665, or W666 in the core epitope produced resistance to 2F5, as measured at the level of 50% (IC50) and 90% (IC90) neutralization. Similarly, for 4E10, the substitution of W672A, in the core epitope, impacted IC50 and IC90 values. However, the substitutions of residues F673A and W680A, also in the 4E10 core epitope, impacted IC90 values but did not significantly affect the IC50 values. The reason for this surprising discrepancy is not clear at present but may relate to the way in which 4E10 recognizes its epitope on HIV-1 (see Discussion). Indeed, the inhibition curves for 4E10 and 2F5 typically appeared to be shallow rather than ideal sigmoidal curves, which was in contrast to the sigmoidal curves observed with some monoclonal antibodies (data not shown). As observed with 2F5, many of the Ala substitutions increased virus sensitivity to 4E10, and typically these were the same ones that enhanced neutralization by 2F5 (Table 2). We did not observe the reciprocal situation: no single substitution led to resistance to both 2F5 and 4E10.
Significantly, there were only very slight differences in neutralization by monoclonal antibody b12 and T20 within the panel of mutant pseudoviruses. These results indicate that the changes in the MPER of gp41 that affected virus sensitivity to 2F5 and 4E10 did not globally sensitize the viruses to other neutralizing agents. We note that a few isolates were also tested for neutralization by a preparation of polyclonal immunoglobulin G taken from HIV-1-seropositive donors as well as by other anti-gp120 monoclonal antibodies, including 2G12, hNMO1 (anti-V3 loop), and single-chain Fv X5. Again, no significant differences in neutralization titers between the selected mutants and the parental JR2 virus with these antibodies were observed (twofold or less; data not shown).
Substitutions in HIV-1JR2 W672.
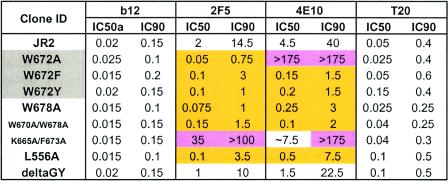
In the above analysis, W672 was found to be most crucial for the neutralizing activity of 4E10. Since W672 is absolutely conserved among all HIV-1 viruses, we decided to explore the effect of subtler substitutions at this position. Thus, mutants W672F and W672Y, in which the aromaticity at position 672 was conserved, were engineered and tested against the monoclonal antibody panel. Surprisingly, both the W672F and W672Y mutants were now more neutralization sensitive to 4E10 than wild-type JR2; these mutants were also more sensitive to 2F5 than JR2 (Table 3). Again, the sensitivity of W672F and W672Y mutants to monoclonal antibody b12 and T20 was unaffected.
TABLE 3.
Effects of conservative substitutions to W672, combining MPER Ala substitutions or substitutions in the N-terminal heptad repeat and cytoplasmic regions of gp41 on virus sensitivity to b12, 2F5, 4E10, and T20 in the single-round infection neutralization assay with U87.CD4.CCR5 cells
Concentrations are in micrograms per milliliter. IC50 and IC90 values that are >3-fold that of the parental (JR2) virus are highlighted in pink (mutant virus more neutralization resistant), whereas values that are less than one-third that of the IC50 or IC90 of JR2 are highlighted in gold (mutant virus more neutralization sensitive).
Mutant W672Y was further tested against monoclonal antibodies 2G12 and scFv X5 as well as the HIV-1 entry inhibitor 5-helix (54); these agents neutralized mutant W672Y at titers similar to those for the parental virus (W13F was not tested). Moreover, mutant W672Y remained neutralization resistant to several antibodies that bind tightly to JR2 envelope proteins but do not neutralize JR2, including 98-6 (anti-gp41), D50 (anti-gp41), and hNMO1 (data not shown). Therefore, W672Y is specifically more sensitive to the MPER-targeted antibodies 2F5 and 4E10. The results with W672F and W672Y indicate that there is not an absolute requirement for Trp at position 672 in order for 4E10 to neutralize virus. Thus, it appears that position 672 is pivotal for 4E10 activity: aromatic substitutions at this position enhance neutralization by 4E10, whereas an Ala substitution produces resistance.
We observed that the infectivity of mutants W672F and W672Y was attenuated, as it was for mutant W672A (data not shown). A previous report in a different system showed that substitutions at position W672 could diminish Env incorporation into nascent virions (56). This caused us to consider whether there might be an effect of the W672 substitutions on Env production. Thus, W672F, W672Y, and parental JR2 pseudoviruses were produced in 293T cells, pelleted, and lysed, and the relative p24 and Env contents were measured by a capture enzyme-linked immunosorbent assay. No significant differences in the relative amounts of p24 and Env were observed with JR2 and the W672F and W672Y mutants with this method (data not shown).
Combining substitutions in the MPER and comparison to substitutions in other regions of gp41.
We considered whether the Ala substitutions to the MPER that caused increased sensitivity to 2F5 and 4E10 might combine to produce pseudovirions with even greater sensitivity to these monoclonal antibodies. We avoided the substitutions that greatly reduced virus infectivity and chose to engineer the double mutants L661A/W678A and W670A/W678A. In addition, we made substitutions in the N-HR region of gp41, L556A and Q563A, which were previously shown to destabilize the six-helix bundle of gp41 and enhance the sensitivity of a replication-competent HIV-1HxB2 to 2F5 and the C-HR peptide, C34, in a fusion assay (24). Finally, we made a deletion mutant, Δ(G711,Y712), or deltaGY, which deletes an endocytic sorting signal in the cytoplasmic tail of gp41 (25, 71).
All of the above mutants showed severalfold-diminished viral titers which necessitated the use of freshly prepared pseudovirus for neutralization assays (data not shown); even so, the titers of mutants L661A/W678A, L556A/W678A, deltaGY/W678A, and Q563A were still too low to generate reliable neutralization data. The mutant W670A/W678A was found to be about as sensitive to 2F5 and 4E10 as the single mutant W678A (Table 3), so the effect of combining these substitutions on neutralization sensitivity to 2F5 and 4E10 did not appear to be additive. We found that mutant L556A was more sensitive than wild-type JR2 to 2F5 and 4E10, although not more sensitive than mutant W678A, but that mutant L556A and JR2 were equally sensitive to T20. The deltaGY mutant was only slightly more sensitive to 2F5 and 4E10.
We also engineered a double mutant to be resistant to both 2F5 and 4E10, K665A/F673A. As expected, this mutant was resistant to both 2F5 and 4E10, although the IC50 of 2F5 was 35 μg/ml, which is lower than observed with the single K665A mutant (IC50 > 200 μg/ml) (Table 3). This result might be explained by the increased sensitivity to 2F5 that is caused by F673A, as observed with the F673A single mutant (Table 3), which may permit the detection of some weak 2F5-neutralizing activity in the double mutant.
Neutralization of HIV-1 pseudoviruses with combinations of 2F5, 4E10, and T20.
We wished to investigate the level of synergy between 2F5, 4E10, and T20 in the single-round infectivity assay to potentially gain further insight into the mechanisms by which these agents inhibit HIV-1 infectivity. With the classical method of Chou et al. (15) for determining synergy, we performed neutralization assays on HIV-1JR2 pseudovirus against 2F5, 4E10, and T20, in which these agents were mixed in a fixed ratio that reflects their relative potency. Synergy in neutralization was observed between 4E10 and T20, whereas no synergy was observed between 2F5 and T20 or 4E10 (Table 4). In fact, some antagonism was observed in the 2F5 combinations. T20 contains the 2F5 epitope, so these two agents will interact in a mixture, and this can explain the antagonism that is observed. It has also been reported that 2F5 can interfere with the interaction between the N-HR and C-HR regions of gp41 (26) and that the N-HR/C-HR interaction in the six-helix bundle occludes the 2F5 epitope (23).
TABLE 4.
Neutralization of HIV-1JR2 by 2F5, 4E10, and T20 individually and in combinations in the pseudovirus assay in U87.CD4.CCR5 cellsa
| Antibody or peptide | ID90b (nM) | Dmc (nM) | rd | CIe at:
|
|
|---|---|---|---|---|---|
| ID50 | ID90 | ||||
| 2F5 | 160 | 23 | 0.97 | ||
| 4E10 | 240 | 56 | 0.99 | ||
| T20 | 270 | 29 | 0.98 | ||
| 2F5 + T20 (1:2.4) | 130 + 310 | 10.5 + 25 | 0.97 | 1.3 | 2.0 |
| 4E10 + T20 (1:0.84) | 100 + 84 | 12 + 10 | 0.98 | 0.70 | 0.37 |
| 4E10 + 2F5 (1:0.35) | 243 + 85 | 46 + 16 | 0.97 | 1.05 | 1.4 |
Neutralization synergy of antibody combinations for HIV-1JR2 was assessed by the classical approach in which dose-response curves were determined for each of the agents alone and in combinations mixed at a constant molar ratio (ratios shown in parentheses). The presence or absence of synergy was determined with the computer program CalcuSyn (15). Values represent the mean of two independent experiments for triplicate samples.
Ninety percent infections doses (ID90s) were calculated by estimating the 90% neutralization titer from the neutralization curves.
Dm, median effect dose; antibody concentration at half-maximal neutralization.
r, linear correlation coefficient.
According to Chou et al., combination indices (CIs) of 0.3 to 0.7 indicate synergism, 0.7 to 0.85 indicate moderate synergism, 0.85 to 0.9 indicate slight synergism, 0.9 to 1.1 indicate additivity, and above 1.1 indicate antagonism.
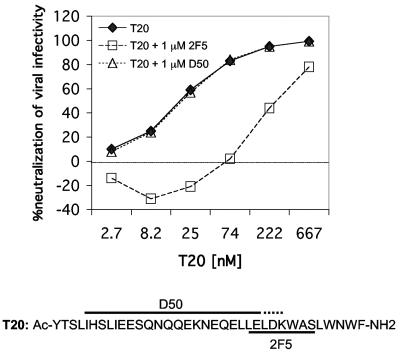
To further explore the antagonism that we observed between 2F5 and T20, we made use of the 2F5-resistant mutant, in this case, D664A, which is as sensitive to T20 as the parental virus (Table 2). In the presence of a molar excess of 2F5, T20 was much less potent against mutant D664A, suggesting that 2F5 inhibited the activity of T20 by preventing its binding to the target sequence during or leading up to fusion (Fig. 2). We included in this analysis a side-by-side comparison in which monoclonal antibody D50, which recognizes an epitope just N-terminal to the 2F5 epitope on gp41 (21), was substituted for 2F5. D50 also binds T20 (18), yet even in a molar excess over T20, D50 showed no effect whatsoever on the activity of T20 against the D664A mutant (Fig. 2). These results suggest that HIV-1 entry inhibition by T20 is completely indifferent to the presence of the nonneutralizing monoclonal antibody D50 but can be blocked by the neutralizing monoclonal antibody 2F5.
FIG. 2.
Neutralization of the 2F5-resistant HIV-1JR2 mutant pseudovirus D664A by T20 in the presence and absence of 2F5 or D50. Virus was preincubated with different concentrations of T20 in the presence or absence of a molar excess of either 2F5 or D50 (1 μM constant throughout) and then added to U87.CD4.CCR5 cells. Luciferase activity was measured after 72 h. The sequence of T20 is shown below the graph with the 2F5 epitope indicated, as well as the approximate region to which D50 binds, according to a previous study (21). Control experiments showed no effect of 1 μM D50 or 2F5 on the infectivity of the JR2 mutant D664A.
DISCUSSION
Neutralizing antibody selection pressures on HIV-1 appear to have resulted in circulating viruses in which conserved regions on the envelope spike of the virus are largely sequestered. One exception, at least under certain conditions, appears to be the MPER of gp41 (18, 58, 77). The MPER of gp41 is necessary for envelope-mediated fusion (19, 40, 56), which helps to explain its sequence conservation. Studies have variously suggested that the MPER is involved in membrane destabilization (55), recruiting additional gp41s to make a fusion pore (30), or that it simply provides a flexible tether to allow proper positioning of the fusion apparatus to facilitate membrane merger (20). Since its primary sequence is very highly conserved, the MPER does not evade neutralizing antibody by sequence variation, such as occurs with the variable loops of gp120. Nevertheless, antibodies such as 2F5 and 4E10 and the broad neutralizing activity that is associated with these two antibodies are infrequently observed in HIV-1-seropositive individuals (52, 69), suggesting that the epitopes of these monoclonal antibodies are very weakly immunogenic during natural infection.
One significant finding in this report was that resistance to 2F5 and 4E10 was only acquired by Ala substitutions to D664, K665, or W666, in the case of 2F5 and W672, F673, or W680 for 4E10. That so few of the residues within and very near the epitopes of these monoclonal antibodies can be changed to produce neutralization resistance to these monoclonal antibodies indicates that, in the context of virus, the sequence of the flanking residues is not nearly as important for monoclonal antibody neutralization as it is for monoclonal antibody affinity to short peptides. For example, it was shown that certain Ala substitutions near or within the 2F5 epitope sequence decrease the affinity of 2F5 to the peptide T20 (28). In particular, the affinity of 2F5 for T20 was diminished by greater than sixfold by the substitution L661A. In our present study, the same substitution made the JR2 virus more neutralization sensitive to 2F5 by a factor of about 6. There can be different explanations for this apparent paradox. For example, the L661A substitution that diminishes the affinity of 2F5 for the peptide may enhance the accessibility to the epitope on the functional Env trimer and therefore enhance neutralization. Alternatively, another unidentified property of the A661 virus, such as diminished fusion kinetics, may enhance the neutralizing activity of 2F5 compared to the L661 virus. Whatever the reason, it appears that HIV-1 provides a structural context for the epitope of 2F5 that permits considerable variability within or proximal to the epitope sequence that has been defined using peptides. Hence, for vaccine design, improving the structural context and immunogenicity of the 2F5 epitope may be more important than preserving an extended and correct primary amino acid sequence of the MPER of gp41. Likewise, for the epitope of 4E10, emphasis should be put into presenting the motif WFxxxxxxW in the correct structural environment and maximizing immunogenicity, and perhaps somewhat lesser emphasis on having a long MPER peptide with native sequence. This will almost certainly require an empirical approach in which numerous antigens are used to immunize animals and the sera are characterized for antibody titer and specificity as well as neutralizing activity.
Although escape mutants to 2F5 that contain substitutions in D664 and K665 have been described (49), escape mutants to 4E10 have not yet been reported. HIV-1 envelope mutations have been described that appear to cause resistance (7, 64, 68) or, reciprocally, enhanced sensitivity (24, 35, 48) simultaneously to a number of different antibody specificities, including those against gp41. Some such mutations are expected to affect virus sensitivity to 4E10 as well. Nevertheless, in our analysis, the only substitution to make the virus resistant to this monoclonal antibody at the IC50 level was W672A; F673A and W680A were reproducibly resistant to 4E10 at the IC90 level but not significantly so at the IC50 level. We found that the slope of the curves with 4E10 against F673A and W680A were shallow (nonsigmoidal), unlike the curves with another antibody, scFv X5, against the same mutants. Hence, the IC50s with 4E10 against these mutants were subject to greater error, and the shallow curves indicate that the dose response was not ideal. Nonsigmoidal binding curves have been observed previously in highly reproducible neutralization assays involving other neutralizing antibodies as well (M. B. Zwick, J. Binley, D. R. Burton, T. Wrin, and C. J. Petropoulos, unpublished observations). The basis for this behavior is not readily apparent but may reflect some heterogeneity in the mutant virus population or may be a consequence of 4E10's acting with different potencies at more than one step along the fusion pathway.
We show that 8 out of 21 Ala substitutions in the MPER produced enhanced neutralization sensitivity to both 2F5 and 4E10. Since the mutations that enhance sensitivity to either 4E10 or 2F5 occur at a number of different positions and often affect both antibodies at the same time and to a similar degree and combinations of these substitutions that produce infectious virus is nonadditive, we suggest that they are all producing the neutralization-hypersensitive phenotype by the same mechanism. Even very subtle changes to the MPER of gp41 (i.e., W672Y and W672F) greatly enhance viral sensitivity to 2F5 and 4E10, suggesting that they affect a very specific interaction.
The MPER of gp41 is likely to be somewhat flexible and exposed because of its susceptibility to proteases (70). Nevertheless, a peptide corresponding to the MPER was determined to be an alpha-helix in membrane-mimetic detergent micelles with nuclear magnetic resonance (59); an epitope peptide in complex with 4E10 has helical structure as well (13). The Trp-rich MPER peptide has been shown to bind and disrupt membranes (61). It seems unlikely that subtle changes such as W672Y or W672F could profoundly affect the helical secondary structure of the MPER in isolation or strongly affect the hydrophobic association between it and the lipid bilayer. However, the functional HIV-1 envelope spike is a putative trimer in which the MPER of one gp41 may be close to the MPER on a neighboring gp41. Moreover, MPER peptides have been shown to self-interact or oligomerize on membranes (1, 30, 55). We hypothesize that these substitutions weaken a specific interaction between the MPERs of neighboring gp41s. A weakening of the interaction between MPERs in the trimeric envelope spike could allow the MPERs to lie on the membrane. Indeed, the affinity of 2F5 and 4E10 for recombinant Env protein captured on proteoliposome beads has been shown to increase by pretreatment of the captured Env protein with lipids (27, 31). However, we cannot currently rule out other possibilities, such as a direct effect on an interaction between the MPER and coreceptor, gp120, the N-HR region of gp41, or the membrane.
The phenotype associated with the MPER substitutions that we studied may be influenced by additional factors, such as the type and strain of virus used, the kinetics of viral entry, and the type of target cell. For example, it was shown that certain Ala mutations in the MPER and W672F decreased envelope incorporation into nascent virions in a replication-competent HxB2 virus background (56). Although an extensive study of envelope incorporation was not performed in the present study with pseudoviruses, the Env-to-p24 ratios of mutants W672F and W672Y were identical to that of JR2 in a capture enzyme-linked immunosorbent assay format. In another report involving HIV-1HxB2 pseudoviruses, several substitutions in the MPER of gp41 had no effect on viral infectivity (12).
Another distinction between our results and those found in the literature involves a study in which HIV-1HxB2 mutants with reduced affinity to coreceptor were more sensitive to T20 because of slower fusion kinetics (51). Moreover, gp41 mutations that impair fusion by destabilizing the six-helix bundle and enhance viral sensitivity to 2F5 and the fusion-inhibiting peptide C34 have been described (24). In our analysis of the MPER of gp41, there was no significant increase in sensitivity of any of the Ala mutant viruses to T20 or to the CD4-induced antibody scFv X5, suggesting that a distinct mechanism is responsible for the increased sensitivity to 2F5 and 4E10. Nevertheless, it may be that the MPER Ala substitutions are affecting the kinetics of fusion, which is a complex, multistep process. Finally, the type of target cell may also affect the role of the MPER in HIV-1 entry into cells, and U87 cells differ from T cells in a number of ways. For example, U87 cells have high levels of heparan sulfates. Further studies are needed to address these issues and elucidate the fusion kinetics of these gp41 MPER mutants.
Cocktails of the broadly neutralizing immunoglobulin G1 anti-HIV-1 antibodies b12, 2G12, 2F5, and 4E10 have been argued to neutralize primary isolates of HIV-1 with a moderate degree of synergy (32, 77, 79). Although 2F5 and 4E10 compete with one another for binding to a synthetic peptide (residues 660 to 680 of gp41MN, LELDKWASLWNWFDITNWLW), and somewhat less efficiently for recombinant gp41, a study has suggested that combinations of 2F5 and 4E10 can neutralize HIV-1 synergistically (60). We did not observe synergy between 2F5 and 4E10, and 2F5 was found to antagonize T20, particularly for a 2F5-resistant mutant pseudovirus. In contrast, 4E10 and T20 reproducibly synergized with each other against HIV-1JR2. The 4E10 epitope may be a useful target that could be exploited for combination therapy with T20. Whether or not the MPER is amenable to small-molecule inhibitors is unknown, and 4E10 is only modestly potent. One possibility might be to enhance the neutralizing activity of 4E10 with in vitro affinity maturation techniques (74).
A recent report suggested that serum antibodies against T20 do not reduce its potency in vivo (67). Such antibodies may be more like D50 rather than 2F5, the latter of which we showed here to inhibit the activity of T20. 2F5 may antagonize T20 by impairing the access of T20 to the fusion intermediate. However, D50 also binds T20, albeit with lower affinity, so it too may be expected to sequester T20, but yet had no effect on its activity. This result may be explained by the inability of D50 to recognize the conformation(s) of T20 that 2F5 recognizes; the broadly cross-reactive antibody 2F5 appears to recognize a range of conformations of its epitope on peptides (28, 36, 38, 77). That D50 appears to bind with greater affinity to C-peptides when they are complexed with N-peptides of gp41 (18) suggests that D50 is indifferent to the equilibrium between T20 and the fusion intermediate on HIV-1. 2F5, in contrast, takes T20 “off pathway” in this equilibrium and likewise may neutralize HIV-1 by taking the C-HR region of gp41 off pathway in the fusion reaction.
The present results bear on HIV-1 vaccine strategies involving peptidomimetics of the MPER of gp41. Although high serum antibody titers against MPER peptides can be achieved (44), at least two potential hurdles remain in overcoming the lack of immunogenicity, specifically of the neutralizing epitopes on the MPER of gp41. One challenge is to constrain the MPER of gp41 in the correct conformation (i.e., minimize irrelevant conformations). The other challenge is to target antibodies to the correct face of the properly constrained antigen, which may involve attenuating the immunodominance of irrelevant antigenic faces or the imposed constraints. Additional monoclonal antibodies to the MPER, both neutralizing and nonneutralizing, may be useful as tools to help in the process of designing, and evaluating the antigenicity of candidate immunogens.
One potential solution to the immunodominance problem is to mask the unwanted determinants with carbohydrate, as is being pursued for gp120 (46), or by way of self-oligomerization or membrane association. Since 4E10 recognizes a very broad range of sequences, of which only a few residues are fixed, a polyvalent immunogen may prove to be more useful than a fixed sequence, as sequence-specific antibodies may be less likely to broadly neutralize. Such sophisticated approaches seem worthwhile for the MPER of gp41, given the difficulty of eliciting neutralizing antibodies against it but the extraordinary breadth with which 2F5 and 4E10 neutralize primary isolates of HIV-1.
ADDENDUM IN PROOF
Recently, an X-ray crystal structure of a complex of 2F5 with a more-extended (17-mer) peptide has been determined (G. Ofek, M. Tang, A. Sambor, H. Katinger, J. R. Mascola, R. Wyatt, and P. D. Kwong, J. Virol. 78:10724-10737, 2004). In this structure, the D, K, and W of the core epitope, LELDKWASL, are major contact residues. However, three other residue side chains have >50% surface area buried in the 2F5 paratope. In particular, L661 and L669 bury 68 and 52% of their surface, respectively, and yet, as shown in our study, their replacement by Ala enhances the sensitivity of the mutant viruses to 2F5 by severalfold.
Acknowledgments
We thank P. W. H. I. Parren, M. Franti, and P. Poignard for helpful suggestions.
We acknowledge support from the Elizabeth Glaser Pediatric AIDS Foundation (to M.B.Z.), the NIH, AI 058725 (to M.B.Z.) and AI 33292 (to D.R.B.), the Neutralizing Antibody Consortium of the International AIDS Vaccine Initiative, and the Pendleton Trust.
REFERENCES
- 1.Alfsen, A., and M. Bomsel. 2002. HIV-1 gp41 envelope residues 650-685 exposed on native virus act as a lectin to bind epithelial cell galactosyl ceramide. J. Biol. Chem. 277:25649-25659. [DOI] [PubMed] [Google Scholar]
- 2.Barbato, G., E. Bianchi, P. Ingallinella, W. H. Hurni, M. D. Miller, G. Ciliberto, R. Cortese, R. Bazzo, J. W. Shiver, and A. Pessi. 2003. Structural analysis of the epitope of the anti-HIV antibody 2F5 sheds light into its mechanism of neutralization and HIV fusion. J. Mol. Biol. 330:1101-1115. [DOI] [PubMed] [Google Scholar]
- 3.Binley, J. M., C. S. Cayanan, C. Wiley, N. Schulke, W. C. Olson, and D. R. Burton. 2003. Redox-triggered infection by disulfide-shackled human immunodeficiency virus type 1 pseudovirions. J. Virol. 77:5678-5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Binley, J. M., H. J. Ditzel, C. F. Barbas, 3rd, N. Sullivan, J. Sodroski, P. W. Parren, and D. R. Burton. 1996. Human antibody responses to HIV type 1 glycoprotein 41 cloned in phage display libraries suggest three major epitopes are recognized and give evidence for conserved antibody motifs in antigen binding. AIDS Res. Hum. Retroviruses 12:911-924. [DOI] [PubMed] [Google Scholar]
- 5.Biron, Z., S. Khare, A. O. Samson, Y. Hayek, F. Naider, and J. Anglister. 2002. A monomeric 3(10)-helix is formed in water by a 13-residue peptide representing the neutralizing determinant of HIV-1 on gp41. Biochemistry 41:12687-12696. [DOI] [PubMed] [Google Scholar]
- 6.Bjorndal, A., H. Deng, M. Jansson, J. R. Fiore, C. Colognesi, A. Karlsson, J. Albert, G. Scarlatti, D. R. Littman, and E. M. Fenyo. 1997. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J. Virol. 71:7478-7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouma, P., M. Leavitt, P. F. Zhang, I. A. Sidorov, D. S. Dimitrov, and G. V. Quinnan, Jr. 2003. Multiple interactions across the surface of the gp120 core structure determine the global neutralization resistance phenotype of human immunodeficiency virus type 1. J. Virol. 77:8061-8071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buchacher, A., R. Predl, K. Strutzenberger, W. Steinfellner, A. Trkola, M. Purtscher, G. Gruber, C. Tauer, F. Steindl, A. Jungbauer, et al. 1994. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res. Hum. Retroviruses 10:359-369. [DOI] [PubMed] [Google Scholar]
- 9.Burton, D. R., R. C. Desrosiers, R. W. Doms, W. C. Koff, P. D. Kwong, J. P. Moore, G. J. Nabel, J. Sodroski, I. A. Wilson, and R. T. Wyatt. 2004. HIV vaccine design and the neutralizing antibody problem. Nat. Immunol. 5:233-236. [DOI] [PubMed] [Google Scholar]
- 10.Burton, D. R., J. Pyati, R. Koduri, S. J. Sharp, G. B. Thornton, P. W. Parren, L. S. Sawyer, R. M. Hendry, N. Dunlop, P. L. Nara, et al. 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266:1024-1027. [DOI] [PubMed] [Google Scholar]
- 11.Calarese, D. A., C. N. Scanlan, M. B. Zwick, S. Deechongkit, Y. Mimura, R. Kunert, P. Zhu, M. R. Wormald, R. L. Stanfield, K. H. Roux, J. W. Kelly, P. M. Rudd, R. A. Dwek, H. Katinger, D. R. Burton, and I. A. Wilson. 2003. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science 300:2065-2071. [DOI] [PubMed] [Google Scholar]
- 12.Cao, J., L. Bergeron, E. Helseth, M. Thali, H. Repke, and J. Sodroski. 1993. Effects of amino acid changes in the extracellular domain of the human immunodeficiency virus type 1 gp41 envelope glycoprotein. J. Virol. 67:2747-2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cardoso, R., M. Zwick, R. Kunert, H. Katinger, D. R. Burton, and I. Wilson. 2003. Structural insights for 4E10 antibody neutralization on HIV-1. AIDS Vaccine 2003, 18-21 Sept. 2003, New York, N.Y. (abstract).
- 14.Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263-273. [DOI] [PubMed] [Google Scholar]
- 15.Chou, T. C., and M. P. Hayball. 1996. CalcuSyn, Windows software for dose effect analysis. BioSoft, Ferguson, Mo.
- 16.Coeffier, E., J. M. Clement, V. Cussac, N. Khodaei-Boorane, M. Jehanno, M. Rojas, A. Dridi, M. Latour, R. El Habib, F. Barre-Sinoussi, M. Hofnung, and C. Leclerc. 2000. Antigenicity and immunogenicity of the HIV-1 gp41 epitope ELDKWA inserted into permissive sites of the MalE protein. Vaccine 19:684-693. [DOI] [PubMed] [Google Scholar]
- 17.Connor, R. I., B. K. Chen, S. Choe, and N. R. Landau. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935-944. [DOI] [PubMed] [Google Scholar]
- 18.de Rosny, E., R. Vassell, S. Jiang, R. Kunert, and C. D. Weiss. 2004. Binding of the 2F5 monoclonal antibody to native and fusion-intermediate forms of human immunodeficiency virus type 1 gp41: implications for fusion-inducing conformational changes. J. Virol. 78:2627-2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dimitrov, A. S., S. S. Rawat, S. Jiang, and R. Blumenthal. 2003. Role of the fusion peptide and membrane-proximal domain in HIV-1 envelope glycoprotein-mediated membrane fusion. Biochemistry 42:14150-14158. [DOI] [PubMed] [Google Scholar]
- 20.D'Souza, M. P., D. Livnat, J. A. Bradac, and S. H. Bridges. 1997. Evaluation of monoclonal antibodies to human immunodeficiency virus type 1 primary isolates by neutralization assays: performance criteria for selecting candidate antibodies for clinical trials. AIDS Clinical Trials Group Antibody Selection Working Group. J. Infect. Dis. 175:1056-1062. [DOI] [PubMed] [Google Scholar]
- 21.Earl, P. L., C. C. Broder, R. W. Doms, and B. Moss. 1997. Epitope map of human immunodeficiency virus type 1 gp41 derived from 47 monoclonal antibodies produced by immunization with oligomeric envelope protein. J. Virol. 71:2674-2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eckhart, L., W. Raffelsberger, B. Ferko, A. Klima, M. Purtscher, H. Katinger, and F. Ruker. 1996. Immunogenic presentation of a conserved gp41 epitope of human immunodeficiency virus type 1 on recombinant surface antigen of hepatitis B virus. J. Gen. Virol. 77:2001-2008. [DOI] [PubMed] [Google Scholar]
- 22a.Ferrantelli, F., and R. M. Ruprecht. 2002. Neutralizing antibodies against HIV—back in the major leagues? Curr. Opin. Immunol. 14:495-502. [DOI] [PubMed] [Google Scholar]
- 23.Finnegan, C. M., W. Berg, G. K. Lewis, and A. L. DeVico. 2001. Antigenic properties of the human immunodeficiency virus envelope during cell-cell fusion. J. Virol. 75:11096-11105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Follis, K. E., S. J. Larson, M. Lu, and J. H. Nunberg. 2002. Genetic evidence that interhelical packing interactions in the gp41 core are critical for transition of the human immunodeficiency virus type 1 envelope glycoprotein to the fusion-active state. J. Virol. 76:7356-7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fultz, P. N., P. J. Vance, M. J. Endres, B. Tao, J. D. Dvorin, I. C. Davis, J. D. Lifson, D. C. Montefiori, M. Marsh, M. H. Malim, and J. A. Hoxie. 2001. In vivo attenuation of simian immunodeficiency virus by disruption of a tyrosine-dependent sorting signal in the envelope glycoprotein cytoplasmic tail. J. Virol. 75:278-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gorny, M. K., and S. Zolla-Pazner. 2000. Recognition by human monoclonal antibodies of free and complexed peptides representing the prefusogenic and fusogenic forms of human immunodeficiency virus type 1 gp41. J. Virol. 74:6186-6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grundner, C., T. Mirzabekov, J. Sodroski, and R. Wyatt. 2002. Solid-phase proteoliposomes containing human immunodeficiency virus envelope glycoproteins. J. Virol. 76:3511-3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joyce, J. G., W. M. Hurni, M. J. Bogusky, V. M. Garsky, X. Liang, M. P. Citron, R. C. Danzeisen, M. D. Miller, J. W. Shiver, and P. M. Keller. 2002. Enhancement of alpha-helicity in the HIV-1 inhibitory peptide DP178 leads to an increased affinity for human monoclonal antibody 2F5 but does not elicit neutralizing responses in vitro. Implications for vaccine design. J. Biol. Chem. 277:45811-45820. [DOI] [PubMed] [Google Scholar]
- 29.Klausner, R. D., A. S. Fauci, L. Corey, G. J. Nabel, H. Gayle, S. Berkley, B. F. Haynes, D. Baltimore, C. Collins, R. G. Douglas, J. Esparza, D. P. Francis, N. K. Ganguly, J. L. Gerberding, M. I. Johnston, M. D. Kazatchkine, A. J. McMichael, M. W. Makgoba, G. Pantaleo, P. Piot, Y. Shao, E. Tramont, H. Varmus, and J. N. Wasserheit. 2003. Medicine. The need for a global HIV vaccine enterprise. Science 300:2036-2039. [DOI] [PubMed] [Google Scholar]
- 30.Kliger, Y., S. A. Gallo, S. G. Peisajovich, I. Munoz-Barroso, S. Avkin, R. Blumenthal, and Y. Shai. 2001. Mode of action of an antiviral peptide from HIV-1. Inhibition at a post-lipid mixing stage. J. Biol. Chem. 276:1391-1397. [DOI] [PubMed] [Google Scholar]
- 31.Kwong, P., C. Huang, S. Majeed, G. Ofek, and T. Zhou. 2004. HIV-1 envelope structure. The 4th Frederick Workshop on the Cell Biology of Viral Entry, 4 May 2004 (abstract).
- 32.Li, A., H. Katinger, M. R. Posner, L. Cavacini, S. Zolla-Pazner, M. K. Gorny, J. Sodroski, T. C. Chou, T. W. Baba, and R. M. Ruprecht. 1998. Synergistic neutralization of simian-human immunodeficiency virus SHIV-vpu+ by triple and quadruple combinations of human monoclonal antibodies and high-titer anti-human immunodeficiency virus type 1 immunoglobulins. J. Virol. 72:3235-3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang, X., S. Munshi, J. Shendure, G. Mark, 3rd, M. E. Davies, D. C. Freed, D. C. Montefiori, and J. W. Shiver. 1999. Epitope insertion into variable loops of HIV-1 gp120 as a potential means to improve immunogenicity of viral envelope protein. Vaccine 17:2862-2872. [DOI] [PubMed] [Google Scholar]
- 34.Marusic, C., P. Rizza, L. Lattanzi, C. Mancini, M. Spada, F. Belardelli, E. Benvenuto, and I. Capone. 2001. Chimeric plant virus particles as immunogens for inducing murine and human immune responses against human immunodeficiency virus type 1. J. Virol. 75:8434-8439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34a.Mascola, J. R. 2003. Defining the protective antibody response for HIV-1. Curr. Mol. Med. 3:209-216. [DOI] [PubMed] [Google Scholar]
- 35.McCaffrey, R. A., C. Saunders, M. Hensel, and L. Stamatatos. 2004. N-linked glycosylation of the V3 loop and the immunologically silent face of gp120 protects human immunodeficiency virus type 1 SF162 from neutralization by anti-gp120 and anti-gp41 antibodies. J. Virol. 78:3279-3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McGaughey, G. B., M. Citron, R. C. Danzeisen, R. M. Freidinger, V. M. Garsky, W. M. Hurni, J. G. Joyce, X. Liang, M. Miller, J. Shiver, and M. J. Bogusky. 2003. HIV-1 vaccine development: constrained peptide immunogens show improved binding to the anti-HIV-1 gp41 MAb. Biochemistry 42:3214-3223. [DOI] [PubMed] [Google Scholar]
- 37.Melikyan, G. B., R. M. Markosyan, H. Hemmati, M. K. Delmedico, D. M. Lambert, and F. S. Cohen. 2000. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J. Cell Biol. 151:413-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Menendez, A., K. C. Chow, O. C. Pan, and J. K. Scott. 2004. Human immunodeficiency virus type 1-neutralizing monoclonal antibody 2F5 is multispecific for sequences flanking the DKW core epitope. J. Mol. Biol. 338:311-327. [DOI] [PubMed] [Google Scholar]
- 39.Moulard, M., S. K. Phogat, Y. Shu, A. F. Labrijn, X. Xiao, J. M. Binley, M. Y. Zhang, I. A. Sidorov, C. C. Broder, J. Robinson, P. W. Parren, D. R. Burton, and D. S. Dimitrov. 2002. Broadly cross-reactive HIV-1-neutralizing human monoclonal Fab selected for binding to gp120-CD4-CCR5 complexes. Proc. Natl. Acad. Sci. USA 99:6913-6918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Munoz-Barroso, I., K. Salzwedel, E. Hunter, and R. Blumenthal. 1999. Role of the membrane-proximal domain in the initial stages of human immunodeficiency virus type 1 envelope glycoprotein-mediated membrane fusion. J. Virol. 73:6089-6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muster, T., R. Guinea, A. Trkola, M. Purtscher, A. Klima, F. Steindl, P. Palese, and H. Katinger. 1994. Cross-neutralizing activity against divergent human immunodeficiency virus type 1 isolates induced by the gp41 sequence ELDKWAS. J. Virol. 68:4031-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muster, T., F. Steindl, M. Purtscher, A. Trkola, A. Klima, G. Himmler, F. Ruker, and H. Katinger. 1993. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J. Virol. 67:6642-6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ohno, T., M. Terada, Y. Yoneda, K. W. Shea, R. F. Chambers, D. M. Stroka, M. Nakamura, and D. W. Kufe. 1991. A broadly neutralizing monoclonal antibody that recognizes the V3 region of human immunodeficiency virus type 1 glycoprotein gp120. Proc. Natl. Acad. Sci. USA 88:10726-10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Opalka, D., A. Pessi, E. Bianchi, G. Ciliberto, W. Schleif, M. McElhaugh, R. Danzeisen, R. Geleziunas, M. Miller, D. M. Eckert, D. Bramhill, J. Joyce, J. Cook, W. Magilton, J. Shiver, E. Emini, and M. T. Esser. 2004. Analysis of the HIV-1 gp41 specific immune response using a multiplexed antibody detection assay. J. Immunol. Methods 287:49-65. [DOI] [PubMed] [Google Scholar]
- 45.Pai, E. F., M. H. Klein, P. Chong, and A. Pedyczak. October2000. World Intellectual Property Organization. Patent WO-00/61618.
- 46.Pantophlet, R., I. A. Wilson, and D. R. Burton. 2003. Hyperglycosylated mutants of human immunodeficiency virus (HIV) type 1 monomeric gp120 as novel antigens for HIV vaccine design. J. Virol. 77:5889-5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parker, C. E., L. J. Deterding, C. Hager-Braun, J. M. Binley, N. Schulke, H. Katinger, J. P. Moore, and K. B. Tomer. 2001. Fine definition of the epitope on the gp41 glycoprotein of human immunodeficiency virus type 1 for the neutralizing monoclonal antibody 2F5. J. Virol. 75:10906-10911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Polzer, S., M. T. Dittmar, H. Schmitz, and M. Schreiber. 2002. The N-linked glycan g15 within the V3 loop of the HIV-1 external glycoprotein gp120 affects coreceptor usage, cellular tropism, and neutralization. Virology 304:70-80. [DOI] [PubMed] [Google Scholar]
- 49.Purtscher, M., A. Trkola, A. Grassauer, P. M. Schulz, A. Klima, S. Dopper, G. Gruber, A. Buchacher, T. Muster, and H. Katinger. 1996. Restricted antigenic variability of the epitope recognized by the neutralizing gp41 antibody 2F5. AIDS 10:587-593. [DOI] [PubMed] [Google Scholar]
- 50.Quinnan, G. V., Jr., P. F. Zhang, D. W. Fu, M. Dong, and H. J. Alter. 1999. Expression and characterization of HIV type 1 envelope protein associated with a broadly reactive neutralizing antibody response. AIDS Res. Hum. Retroviruses 15:561-570. [DOI] [PubMed] [Google Scholar]
- 51.Reeves, J. D., S. A. Gallo, N. Ahmad, J. L. Miamidian, P. E. Harvey, M. Sharron, S. Pohlmann, J. N. Sfakianos, C. A. Derdeyn, R. Blumenthal, E. Hunter, and R. W. Doms. 2002. Sensitivity of HIV-1 to entry inhibitors correlates with envelope/coreceptor affinity, receptor density, and fusion kinetics. Proc. Natl. Acad. Sci. USA 99:16249-16254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Richman, D. D., T. Wrin, S. J. Little, and C. J. Petropoulos. 2003. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. USA 100:4144-4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roben, P., J. P. Moore, M. Thali, J. Sodroski, C. F. Barbas, 3rd, and D. R. Burton. 1994. Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding site of gp120 that show differing abilities to neutralize human immunodeficiency virus type 1. J. Virol. 68:4821-4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Root, M. J., M. S. Kay, and P. S. Kim. 2001. Protein design of an HIV-1 entry inhibitor. Science 291:884-888. [DOI] [PubMed] [Google Scholar]
- 55.Saez-Cirion, A., J. L. Arrondo, M. J. Gomara, M. Lorizate, I. Iloro, G. Melikyan, and J. L. Nieva. 2003. Structural and functional roles of HIV-1 gp41 pretransmembrane sequence segmentation. Biophys. J. 85:3769-3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Salzwedel, K., J. T. West, and E. Hunter. 1999. A conserved tryptophan-rich motif in the membrane-proximal region of the human immunodeficiency virus type 1 gp41 ectodomain is important for Env-mediated fusion and virus infectivity. J. Virol. 73:2469-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saphire, E. O., P. W. Parren, R. Pantophlet, M. B. Zwick, G. M. Morris, P. M. Rudd, R. A. Dwek, R. L. Stanfield, D. R. Burton, and I. A. Wilson. 2001. Crystal structure of a neutralizing human IgG against HIV-1: a template for vaccine design. Science 293:1155-1159. [DOI] [PubMed] [Google Scholar]
- 58.Sattentau, Q. J., S. Zolla-Pazner, and P. Poignard. 1995. Epitope exposure on functional, oligomeric HIV-1 gp41 molecules. Virology 206:713-717. [DOI] [PubMed] [Google Scholar]
- 59.Schibli, D. J., R. C. Montelaro, and H. J. Vogel. 2001. The membrane-proximal tryptophan-rich region of the HIV glycoprotein, gp41, forms a well-defined helix in dodecylphosphocholine micelles. Biochemistry 40:9570-9578. [DOI] [PubMed] [Google Scholar]
- 60.Stiegler, G., R. Kunert, M. Purtscher, S. Wolbank, R. Voglauer, F. Steindl, and H. Katinger. 2001. A potent cross-clade neutralizing human monoclonal antibody against a novel epitope on gp41 of human immunodeficiency virus type 1. AIDS Res. Hum. Retroviruses 17:1757-1765. [DOI] [PubMed] [Google Scholar]
- 61.Suarez, T., W. R. Gallaher, A. Agirre, F. M. Goni, and J. L. Nieva. 2000. Membrane interface-interacting sequences within the ectodomain of the human immunodeficiency virus type 1 envelope glycoprotein: putative role during viral fusion. J. Virol. 74:8038-8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sullivan, N., Y. Sun, Q. Sattentau, M. Thali, D. Wu, G. Denisova, J. Gershoni, J. Robinson, J. Moore, and J. Sodroski. 1998. CD4-Induced conformational changes in the human immunodeficiency virus type 1 gp120 glycoprotein: consequences for virus entry and neutralization. J. Virol. 72:4694-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tan, K., J. Liu, J. Wang, S. Shen, and M. Lu. 1997. Atomic structure of a thermostable subdomain of HIV-1 gp41. Proc. Natl. Acad. Sci. USA 94:12303-12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thali, M., M. Charles, C. Furman, L. Cavacini, M. Posner, J. Robinson, and J. Sodroski. 1994. Resistance to neutralization by broadly reactive antibodies to the human immunodeficiency virus type 1 gp120 glycoprotein conferred by a gp41 amino acid change. J. Virol. 68:674-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tian, Y., C. V. Ramesh, X. Ma, S. Naqvi, T. Patel, T. Cenizal, M. Tiscione, K. Diaz, T. Crea, E. Arnold, G. F. Arnold, and J. W. Taylor. 2002. Structure-affinity relationships in the gp41 ELDKWA epitope for the HIV-1 neutralizing monoclonal antibody 2F5: effects of side-chain and backbone modifications and conformational constraints. J. Peptide Res. 59:264-276. [DOI] [PubMed] [Google Scholar]
- 66.Trkola, A., M. Purtscher, T. Muster, C. Ballaun, A. Buchacher, N. Sullivan, K. Srinivasan, J. Sodroski, J. P. Moore, and H. Katinger. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 70:1100-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Walmsley, S., K. Henry, C. Katlama, M. Nelson, A. Castagna, J. Reynes, B. Clotet, J. Hui, M. Salgo, R. DeMasi, and J. Delehanty. 2003. Enfuvirtide (T-20) cross-reactive glycoprotein 41 antibody does not impair the efficacy or safety of enfuvirtide. J. Infect. Dis. 188:1827-1833. [DOI] [PubMed] [Google Scholar]
- 68.Watkins, B. A., S. Buge, K. Aldrich, A. E. Davis, J. Robinson, M. S. Reitz, Jr., and M. Robert-Guroff. 1996. Resistance of human immunodeficiency virus type 1 to neutralization by natural antisera occurs through single amino acid substitutions that cause changes in antibody binding at multiple sites. J. Virol. 70:8431-8437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307-312. [DOI] [PubMed] [Google Scholar]
- 70.Weissenhorn, W., A. Dessen, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387:426-430. [DOI] [PubMed] [Google Scholar]
- 71.West, J. T., S. K. Weldon, S. Wyss, X. Lin, Q. Yu, M. Thali, and E. Hunter. 2002. Mutation of the dominant endocytosis motif in human immunodeficiency virus type 1 gp41 can complement matrix mutations without increasing Env incorporation. J. Virol. 76:3338-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wild, C. T., D. C. Shugars, T. K. Greenwell, C. B. McDanal, and T. J. Matthews. 1994. Peptides corresponding to a predictive alpha-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc. Natl. Acad. Sci. USA 91:9770-9774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xu, J. Y., M. K. Gorny, T. Palker, S. Karwowska, and S. Zolla-Pazner. 1991. Epitope mapping of two immunodominant domains of gp41, the transmembrane protein of human immunodeficiency virus type 1, using ten human monoclonal antibodies. J. Virol. 65:4832-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang, W. P., K. Green, S. Pinz-Sweeney, A. T. Briones, D. R. Burton, and C. F. Barbas 3rd. 1995. CDR walking mutagenesis for the affinity maturation of a potent human anti-HIV-1 antibody into the picomolar range. J. Mol. Biol. 254:392-403. [DOI] [PubMed] [Google Scholar]
- 75.Zhang, H., Y. Huang, R. Fayad, G. T. Spear, and L. Qiao. 2004. Induction of mucosal and systemic neutralizing antibodies against human immunodeficiency virus type 1 (HIV-1) by oral immunization with bovine Papillomavirus-HIV-1 gp41 chimeric virus-like particles. J. Virol. 78:8342-8348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zwick, M. B., R. Kelleher, R. Jensen, A. F. Labrijn, M. Wang, G. V. Quinnan, Jr., P. W. Parren, and D. R. Burton. 2003. A novel human antibody against human immunodeficiency virus type 1 gp120 is V1, V2, and V3 loop dependent and helps delimit the epitope of the broadly neutralizing antibody immunoglobulin G1 b12. J. Virol. 77:6965-6978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zwick, M. B., A. F. Labrijn, M. Wang, C. Spenlehauer, E. O. Saphire, J. M. Binley, J. P. Moore, G. Stiegler, H. Katinger, D. R. Burton, and P. W. Parren. 2001. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J. Virol. 75:10892-10905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zwick, M. B., E. O. Saphire, and D. R. Burton. 2004. gp41: HIV's shy protein. Nat. Med. 10:133-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zwick, M. B., M. Wang, P. Poignard, G. Stiegler, H. Katinger, D. R. Burton, and P. W. Parren. 2001. Neutralization synergy of human immunodeficiency virus type 1 primary isolates by cocktails of broadly neutralizing antibodies. J. Virol. 75:12198-12208. [DOI] [PMC free article] [PubMed] [Google Scholar]