Abstract
Epstein-Barr virus (EBV) resides as a persistent infection in human leukocyte antigen (HLA) class II+ B lymphocytes and is associated with a number of malignancies. The EBV lytic-phase protein gp42 serves at least two functions: gp42 acts as the coreceptor for viral entry into B cells and hampers T-cell recognition via HLA class II molecules through steric hindrance of T-cell receptor-class II-peptide interactions. Here, we show that gp42 associates with class II molecules at their various stages of maturation, including immature αβIi heterotrimers and mature αβ-peptide complexes. When analyzing the biosynthesis and maturation of gp42 in cells stably expressing the viral protein, we found that gp42 occurs in two forms: a full-length type II membrane protein and a truncated soluble form. Soluble gp42 is generated by proteolytic cleavage in the endoplasmic reticulum and is secreted. Soluble gp42 is sufficient to inhibit HLA class II-restricted antigen presentation to T cells. In an almost pure population of Burkitt's lymphoma cells in the EBV lytic cycle, both transmembrane and soluble forms of gp42 are detected. These results imply that soluble gp42 is generated during EBV lytic infection and could contribute to undetected virus production by mediating evasion from T-cell immunity.
Epstein-Barr virus (EBV) is a large DNA virus belonging to the family Herpesviridae, subfamily Gammaherpesviriniae, genus Lymphocryptovirus. Like other herpesviruses, EBV persists for life, establishing a latent infection in B lymphocytes with occasional viral reactivation (29). Approximately 90% of the adult population carries EBV DNA. EBV is the causative agent of infectious mononucleosis and is associated with malignancies that originate from lymphoid cells (e.g., Burkitt's lymphoma and Hodgkin's lymphoma) and epithelial cells (e.g., nasopharyngeal carcinoma). In immunosuppressed or immunocompromised individuals, EBV can cause (fatal) lymphoproliferative disease. In contrast, in healthy individuals, EBV is well controlled by the immune system. The widespread and mostly asymptomatic persistent EBV infections in adults reflect the balance between viral replication and host immune control.
EBV dedicates part of its genome to immune evasion functions (15). Modulation of T-cell recognition is an important target for EBV, as the virus resides intracellularly for most of its life cycle. Fragments of viral proteins can be displayed at the cell surface by human leukocyte antigen (HLA) class I and class II molecules for the activation of T cells carrying receptors of the appropriate specificity. Activated T cells may then lead to elimination of the pathogen, among other ways through destruction of the infeced cell. To escape from antiviral immunity, EBV should interfere with both CD8+ and CD4+ T-cell responses, particularly as the virus infects B lymphocytes expressing both classes of HLA molecules. The EBV nuclear protein EBNA1 contains a glycine-alanine repeat domain that renders the protein resistant to proteasomal degradation and inhibits the translation of its own mRNA in cis (14, 43). In this way, blocking the generation of antigenic peptides to be presented to CD8+ T cells in the context of HLA class I could account for the limited EBNA1-specific cytolytic activity towards EBV-infected B cells (13). Although this block is not complete, as shown by the recent generation of gamma interferon-secreting cytotoxic T-cell clones against EBNA1 from the blood of EBV-positive donors (12, 37, 40), it is conceivable that evasion from HLA class I-restricted cytolysis contributes to undetected expression of a viral protein, EBNA1, that is indispensable for maintaining the EBV episome in latently infected cells. Recently, Ressing et al. demonstrated that the gp42 protein functions as an immune evasion molecule for HLA class II-restricted T-cell responses (27). Mature HLA class II molecules, expressed at the cell surface, are composed of α- and β-chain heterodimers loaded with peptide (4, 26). Newly synthesized major histocompatibility complex (MHC) class II α and β chains associate with the invariant chain (Ii chain) in the endoplasmic reticulum (ER). Signals in the cytoplasmic tail of the Ii chain direct the αβIi complex from the trans-Golgi network to the acidic MHC class II-loading compartment. Proteases within this compartment cleave the Ii chain to enable peptide binding within the MHC class II binding groove. Stable MHC class II αβ-peptide complexes proceed from the MHC class II-loading compartment to the cell surface, where they can be recognized by CD4+ T-helper cells. EBV gp42 binds to HLA class II-peptide complexes, thereby creating a new conformation at the cell surface that can no longer engage T-cell receptors (TCRs). Thus, cells expressing gp42 have impaired recognition by T-helper cells (27).
The gp42 protein is expressed late in the EBV replication cycle and is incorporated into the viral envelope. Herpesvirus envelope proteins play an essential role during various stages of infection, including viral attachment, entry, cell-to-cell spread, and envelopment and maturation of nascent virus particles. In particular the viral proteins that mediate attachment and entry contribute to determining the cellular tropism of the virus. In this respect, EBV displays refined adaptation to infection of B lymphocytes that is mediated by viral glycoproteins binding to cellular receptors with a restricted distribution. Attachment of EBV particles to B cells occurs through interaction of the main envelope protein, gp350, with complement receptor type 2 (23). The subsequent fusion events critical to EBV entry into host cells involve gH-gL-gp42 complexes, in which gp42 binds to HLA class II molecules on B cells (1, 16, 35, 41, 42). Apart from B cells, EBV infects cells of epithelial origins. The gp42 protein appears to be a molecular switch for this cellular tropism. Whereas the presence of gp42 in the viral envelope is essential to infection of B lymphocytes, virions that predominantly carry gH-gL dimers are better suited for infection of class II− epithelial cells (1).
The gp42 open reading frame (ORF), BZLF2, encodes a 223-amino-acid type II membrane protein, comprised of a short cytoplasmic tail, a single transmembrane domain, and an ER lumenal or extracellular portion (Fig. 1A). Within the extracellular gp42 region, residues 34 to 58 are thought to be required for gH-gL binding (42), whereas residues 100 to 223 are involved HLA class II β-chain association (18, 20, 35). Here, we studied the biosynthesis, posttranslational modifications, and intracellular trafficking of EBV gp42. Interactions of gp42 with HLA class II complexes were evaluated at various maturation stages. We show that EBV gp42 is posttranslationally modified to produce truncated soluble gp42 (s-gp42) that can mediate HLA class II immune evasion.
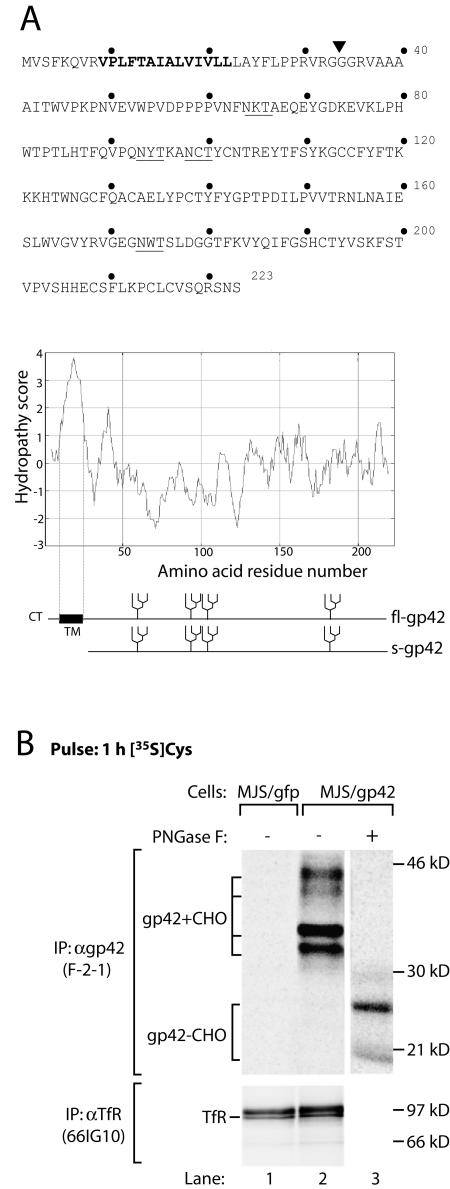
FIG. 1.
EBV gp42 occurs in two forms. (A) Amino acid sequence (single-letter code) and Kyte-Doolittle hydropathy plot of EBV gp42. Bold type highlights the putative transmembrane domain. The N-linked glycosylation sites are underlined. A signal sequence cleavage site is predicted between residues 33 and 34 (inverted triangle); fl-gp42 and the truncated protein potentially generated upon cleavage (s-gp42) are depicted. CT, cytoplasmic tail; TM, transmembrane domain. (B) MJS/gp42 cells (lanes 2 and 3) and control MJS/gfp cells (lane 1) were metabolically labeled for 1 h with [35S]Cys (gp42) or with[35S]Met (TfR). After lysis of the cells, MAb F-2-1 was used to isolate EBV gp42 molecules. Portions of the immunoprecipitates were treated with PNGase F to remove N-linked glycans. Glycosylated (gp42+CHO) and nonglycosylated (gp42−CHO) gp42 polypeptides are indicated. As a control protein, TfR was precipitated with MAb 66IG10. Samples were analyzed by reducing SDS-12% PAGE. IP, immunoprecipitation; α, anti-.
MATERIALS AND METHODS
Cells and isolation of EBV lytic cycle cells.
Cell lines were maintained in complete culture medium consisting of RPMI 1640 medium (Gibco BRL) supplemented with 8% fetal calf serum (27).
Human melanoma-derived cells, Mel JuSo (MJS) cells, were transduced with retroviruses carrying the gene of interest and a marker gene, that for green fluorescent protein (GFP), separated by an internal ribosomal entry site to achieve coexpression of both genes; control cells were transduced with an internal ribosomal entry site-GFP-carrying control retrovirus (MJS/gfp cells) (27). Genes of interest included the EBV BZLF2 ORF for gp42 in the wild-type configuration (MJS/gp42 cells) or with the predicted signal sequence replaced by that of the murine MHC class I H-2Kb molecule (MJS/Kbss-gp42 cells).
T2 and .221 are related EBV-transformed B-cell lines that are HLA class II− and HLA class II+, respectively (30, 32).
Akata is a human EBV-positive cell line, which was derived from a Burkitt's lymphoma and displays a strictly nonproductive infection but can be induced into the lytic cycle by ligation of cell surface immunoglobulin G (IgG); 10 to 40% of the cells then express lytic cycle antigens (36). To obtain more pure populations of cells in the lytic cycle, Akata cells were stably transfected with a reporter plasmid (pHEBO-prBMRF1-rat CD2-GFP) in which the EBV lytic cycle promoter region for the BMRF1 gene was inserted upstream of a chimeric gene encoding the extracellular and transmembrane domains of rat CD2 with GFP replacing the functional cytosolic domain. The resulting cells were called AKBM cells (M. E. Ressing et al., unpublished data). Upon ligation of AKBM cells with goat anti-human IgG antibody (Cappel), the expression of rat CD2-GFP was induced only in cells that had entered the lytic cycle. To isolate lytic cycle-positive cells, the cells were stained with rat CD2-specific antibody OX34 (9) and were positively selected by magnetic cell sorting with anti-mouse IgG2a/b Microbeads and MS columns (Miltenyi Biotech) according to the manufacturer's guidelines; routinely, more than 90% sort purity was achieved.
Antibodies and gp42.Fc protein.
Mouse monoclonal antibody (MAb) F-2-1 is specific for EBV gp42 (17). Polyclonal antiserum #32 against amino acids 61 to 77 of EBV gp42 was raised in a rabbit by using synthetic peptide VNFNKTAEQEYGDKEVK conjugated to tetanus toxoid (6).
The following HLA-DR-specific antibodies were used: MAb Tü36 (31), kindly provided by A. Ziegler; MAb L243 (11); and MAb HB10A against isolated β chains (3), kindly provided by P. Cresswell. MAb 66IG10 recognizing the human transferrin receptor (TfR) (38) was kindly provided by J. Neefjes.
gp42.Fc is a soluble recombinant protein containing the extracellular portion of gp42 (amino acids 34 to 223) linked to the Fc portion of human IgG1; the fusion protein was expressed from mammalian cells (35).
Biochemical analysis, immunoprecipitation, SDS-PAGE, and Western blotting.
For pulse-chase experiments, cells were cultured in methionine (Met)- and cysteine (Cys)-free RPMI medium (BioWhittaker) for 60 min at 37°C (starvation) prior to metabolic labeling with [35S]Met (250 μCi/ml; 35S Redivue Promix, a mixture of >70% [35S]Met and <30% [35S]Cys; Amersham) or [35S]Cys (300 μCi/ml; Amersham) (pulse). Incorporation of the label was terminated by replacing the medium with complete culture medium supplemented with 1 mM Met and 0.1 mM Cys (chase). In some experiments, 5 nM concanamycin B (ConB), an inhibitor of vacuolar H+ ATPases, was included in the starvation, pulse, and chase media to inhibit endolysosomal proteolysis. A 10 μM stock solution of ConB (obtained from Ajinomoto Co., Kanagawa, Japan) was prepared in ethanol. Precleared NP-40 cell lysates (or culture supernatants) were subjected to immunoprecipition with various specific antibodies for 2 h at 4°C. Immune complexes were washed with NET buffer (0.5% NP-40, 5 mM EDTA, 50 mM Tris-Hcl [pH 7.4], 150 mM NaCl) and were prepared for sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) in three ways: (i) boiled for 5 min in nonreducing sample buffer, (ii) incubated for 1 h at 37°C in nonreducing sample buffer (final concentrations: 2% SDS, 30 mM Tris-HCl [pH 6.8], 5% glycerol, 0.05% bromophenol blue), or (iii) boiled for 5 min in reducing sample buffer (final concentrations: 2% SDS, 50 mM Tris-HCl [pH 8.0], 10% glycerol, 5% 2-mercaptoethanol, 0.05% bromophenol blue).
Western blot analysis was performed with boiled immunoprecipitates to identify the nature of coprecipitating proteins (27); total cell lysates were analyzed in parallel. Western blots were stained with HLA-DR β-chain-specific MAb HB10A.
To analyze oligosaccharide modifications, peptide N-glycanase F (PNGase F) and endoglycosidase H (endo H) treatments were performed according to the manufacturer's instructions (New England Biolabs).
Gels were dried and exposed to a phosphorimaging screen, which was scanned with Personal Molecular Imager FX (Bio-Rad) and analyzed with Quantity One software.
In vitro transcription and translation.
For in vitro transcription and translation, plasmid pcDNA3 with the BZLF2 ORF followed by a sequence encoding four additional C-terminal methionines (gp42-4Met) was linearized and used for in vitro transcription with T7 polymerase (Promega). Transcripts were translated for 30 min at 30°C in the presence of [35S]Met (translation grade; Amersham) in rabbit reticulocyte lysate to which microsomes were or were not added (39). Samples were boiled in sample buffer and separated by SDS-PAGE (see above).
Edman degradation.
To determine the proteolytic cleavage site, s-gp42 was isolated from culture supernatants of MJS/gp42 cells by affinity chromatography with gp42-specific MAb F-2-1. Purified s-gp42 was subjected to 20 cycles of Edman degradation, and the resulting amino acids were compared to the EBV gp42 sequence to identify the N-terminal residues of truncated s-gp42.
Flow cytometry.
The binding of s-gp42 to HLA class II molecules was analyzed by flow cytometry. Supernatants of MJS/gp42 cells and control MJS/gfp cells were harvested after 3 days of culturing and were incubated with HLA class II+ gp42− cells for 2 h at 37°C to permit the binding of s-gp42; gp42.Fc was included as a positive control and standard. After extensive washing, HLA class II-bound gp42 was visualized by indirect immunofluorescence with anti-gp42 antiserum #32 as the primary antibody, followed by goat anti-rabbit immunoglobulin conjugated to phycoerythrin (PE) (Jackson). Cells were analyzed by using a FACSCalibur (Becton Dickinson) with CellQuest software.
T-helper-cell assays.
In 96-well round-bottom microtiter plates, 10,000 responder T cells were incubated with 100,000 irradiated (2,500 rads) HLA-DR3+ peripheral blood mononuclear cells (PBMC) in the presence or absence of antigenic peptide or protein in RPMI 1640 medium containing 2 mM glutamine, 100 IU of penicillin/ml, and 100 μg of streptomycin/ml (Gibco BRL) and supplemented with 10% pooled human serum. In some experiments, 10 μg of soluble gp42.Fc/ml was added to the cultures. After 3 days of incubation, [3H]thymidine was added for an additional 16 h to measure T-cell proliferation. As described elsewhere (27), we used T-cell clone Rp15.1.1, which is specific for the mycobacterial hsp65-derived epitope p3-15 in the context of HLA-DR3. Purified protein derivative (PPD; 10 μg/ml; Statens Serum Institute, Copenhagen, Denmark), which contains mycobacterial hsp65, was used as a specific antigen. The mitogen phytohemagglutinin (PHA; 2 μg/ml; HA16; Wellcome, Dartford, United Kingdom) was used as a potent antigen-independent stimulus for T-cell activation.
Allophycocyanine-labeled HLA-DR4 tetramers were generated as descibed previously (27) and contained a peptide of residues 307 to 319 of the influenza virus hemagglutinin (HA307-319). Polyclonal influenza virus-specific T cells directed against the HLA-DRB*0401-restricted HA307-319 epitope were generated as described previously (27) and were labeled with 5 μM 5 (and 6)-carboxyfluorescein diacetate succinimidyl ester (CFSE; Molecular Probes). In some experiments, EBV gp42.Fc was added during the 2-week culture period (recall T-cell response). At day 15, responding T cells were harvested and were stained with the HLA-DR4-HA307-319 tetramers in combination with a PE-conjugated anti-CD4 antibody (Becton Dickinson); stained cells were analyzed by flow cytometry. Living cells were gated on the basis of propidium iodide (PI) exclusion, and proliferating cells were selected by loss of CFSE staining.
RESULTS
EBV gp42 undergoes various posttranslational modifications.
Biosynthesis and posttranslational modifications of the EBV lytic cycle protein gp42 were studied, using a cell line stably expressing gp42 (MJS/gp42) (27). The gp42 gene, BZLF2, encodes a type II membrane protein with a transmembrane region spanning amino acids 9 to 22 (Fig. 1A). This topology is supported by the detection of gp42 at the cell surface with a MAb directed against the C-terminal part of gp42 (F-2-1) (data not shown). The protein sequence of gp42 lacks methionine residues, apart from the starting amino acid that is removed in the cytoplasm. For this reason, we used [35S]Cys to visualize the viral protein by metabolic labeling, as gp42 contains 11 cysteine residues. Immunoprecipitation of gp42 from metabolically labeled MJS/gp42 cells with MAb F-2-1 yielded, in addition to the 42-kDa band representing fully mature gp42, other protein bands (with molecular masses of approximately 30, 34, and 38 kDa) that were absent from control MJS/gfp cells (Fig. 1B). Since four potential N-linked glycosylation sites are contained within the extracellular portion of gp42 (Fig. 1A), we examined whether gp42 was differentially glycosylated. Unexpectedly, when N-linked glycans were removed by digestion of F-2-1 immunoprecipitates with PNGase F, two bands of approximately 25 and 21 kDa remained (Fig. 1B, lane 3). These results indicate that differential glycosylation accounts only in part for the higher-molecular-mass forms of gp42 and that additional variations result from the generation of two BZLF2-encoded protein backbones.
Two protein backbones are generated early in EBV gp42 biosynthesis.
To dissect intracellular modifications of gp42 in more detail, we performed a pulse-chase experiment with MJS/gp42 cells (Fig. 2A and B). Different forms of gp42 were detectable over time. First, glycosylated intermediates with apparent molecular masses of 30 to 34 kDa were formed (Fig. 2A, lanes 1 to 3) and matured into fully modified 38- and 42-kDa forms from 4 h onward (lanes 4 and 5). Treatment with endo H removes high-mannose N-linked glycans but not complex-type glycans that arise after the transport of glycoproteins beyond the ER and cis-Golgi compartments. At early time points (<2 h of chase time), the majority of gp42 molecules were sensitive to endo H digestion, but from 2 h of chase onward, gp42 became endo H resistant; this feature coincided with a large shift in the apparent molecular mass, which is indicative of the acquisition of complex-type oligosaccharides (Fig. 2B). Like PNGase F digestion (Fig. 2A, lane 7), treatment with endo H revealed two gp42 protein backbones with sizes that correspond to the predicted molecular masses of deglycosylated full-length gp42 (fl-gp42) (25 kDa) and a shorter polypeptide (21 kDa) (Fig. 2B, lanes 1 to 3). Since both forms of gp42 are observed after pulses as short as 10 min (Fig. 2B, lane 2), the shorter form is likely to be generated within the ER during or shortly after translation, a scenario which is reminiscent of proteolytic cleavage by signal peptidases.
FIG. 2.
A truncated form of EBV gp42 is secreted over time. (A) MJS/gp42 cells were labeled for 30 min with [35S]Cys and chased for the times indicated. EBV gp42 molecules were recovered from cell lysates (lanes 1 to 7) and from supernatants (lanes 8 to 13) with MAb F-2-1. To reveal the protein backbones, portions of the immunoprecipitates were treated with PNGase F (lanes 7 and 13). (B) Digestions with endo H were performed to examine protein transport beyond the ER and cis-Golgi compartments; endo H-sensitive (endo HS) and endo H-resistant (endo HR) gp42 forms are indicated. All samples were analyzed by SDS-12% PAGE. Untreated F-2-1 precipitates were boiled in nonreducing sample buffer, whereas PNGase F- and endo H-digested samples were denatured under reducing conditions. (C) EBV gp42 was analyzed after in vitro translation of gp42-4Met mRNA in the presence (lanes 2 to 4) or in the absence (lane 1) of microsomes. Portions of the samples were digested with saturating (lane 3) or suboptimal (lane 4) amounts of PNGase F. fl-gp42 molecules bearing one to four N-linked glycans are indicated (+1 through +4); fl-gp42+CHO and s-gp42+CHO overlap such that, for instance, fl-gp42 plus one N-linked glycan runs with a mobility comparable to that of s-gp42 plus two N-linked glycans. See the legend to Fig. 1 for definitions of abbreviations.
The gp42 protein contains a potential signal sequence cleavage site between amino acids 33 and 34 (Fig. 1A) (24) and, in the absence of cleavage, the N-terminal hydrophobic domain can serve as a signal anchor. Upon removal of the N-terminal 33 amino acids from gp42, its predicted molecular mass is approximately 21 kDa. Since this size corresponds to the molecular mass observed for the shorter form of gp42, the combined data suggest that gp42 may be proteolytically cleaved by signal peptidase activity.
EBV A soluble form of gp42 is secreted after proteolytic cleavage.
The C-terminal part of gp42 may be secreted when the N-terminal membrane anchor domain is removed. Indeed, in a pulse-chase experiment comparing cells and supernatants, a shorter form of gp42, s-gp42 (with an apparent molecular mass of 38 kDa under reducing conditions), accumulated in supernatants over time (Fig. 2A, lanes 9 to 12). In contrast, fl-gp42 remained cell associated (Fig. 2A, lanes 1 to 5). Upon PNGase F digestion, the respective longer and shorter protein backbones of the transmembrane and secreted forms of gp42 were observed (Fig. 2A, lanes 7 and 13).
To determine the contribution of ER-associated factors to gp42 modification, in vitro translation was performed in the presence or absence of microsomal membranes. Four methionine residues were added to the C-terminal end of gp42 to permit visualization of the viral protein translated in the presence of [35S]Met (Fig. 2C). Upon translation in the absence of microsomes, gp42 cannot undergo signal peptidase cleavage or glycosylation and, accordingly, the protein migrated at the rate of backbone fl-gp42 (fl-gp42−CHO; Fig. 2C, lane 1). In contrast, when microsomal membranes were provided, gp42 acquired high-mannose glycans, as reflected by distinct bands with retarded migration on SDS-PAGE (Fig. 2C, lane 2). Partial PNGase F digestion indicated that all four putative N-linked glycosylation sites were used (Fig. 2C, lane 4). Two protein bands were observed following complete PNGase F digestion (Fig. 2C, lane 3), consistent with the possibility that gp42 is posttranslationally cleaved in the N-terminal region within the ER.
Next, we identified the cleavage site by N-terminal sequencing of s-gp42 purified from culture supernatants of MJS/gp42 cells. Amino acids corresponding to positions 40, 41, and 42 of the gp42 sequence were detected in the first cycle of Edman degradation, indicating that proteolytic cleavage occurs at these positions. Therefore, s-gp42 appears to be truncated at a region different from the predicted signal peptide cleavage site.
We conclude that the EBV BZLF2 ORF encodes a 25-kDa protein that is in part cleaved posttranslationally, yielding a truncated protein backbone of 21 kDa. Both forms are heavily glycosylated, which results in cell surface expression of fl-gp42 and secretion of a soluble form of gp42 with a molecular mass of 38 kDa.
s-gp42 binds to immature αβIi and mature, SDS-stable αβ HLA class II complexes.
Since HLA class II molecules serve as interaction partners for gp42, we determined whether the soluble form can associate with HLA-DR molecules. MAb F-2-1 cannot be used to detect gp42-class II complexes because it recognizes (and blocks) a region of gp42 involved in class II binding (16). Therefore, we generated rabbit polyclonal antiserum #32 against amino acids 61 to 77 of gp42, a region that can be deleted without abolishing HLA class II binding (35).
To investigate gp42 binding to cell surface-exposed HLA class II molecules, culture supernatants from MJS/gp42 cells and control MJS/gfp cells were incubated with class II+ (MJS and .221) or class II− (T2) cells. HLA class II-s-gp42 complexes were detected with serum #32 and were analyzed by flow cytometry. Complex formation was observed only for culture supernatants from MJS/gp42 cells in combination with class II+ cells (Fig. 3A). Similarly, a recombinant fusion protein consisting of amino acids 34 to 223 of gp42 fused to the Fc portion of human IgG1 (gp42.Fc) was capable of associating with class II molecules at the surface of MJS cells. Together, these results indicate that the truncated form of EBV gp42 binds to mature HLA class II complexes.
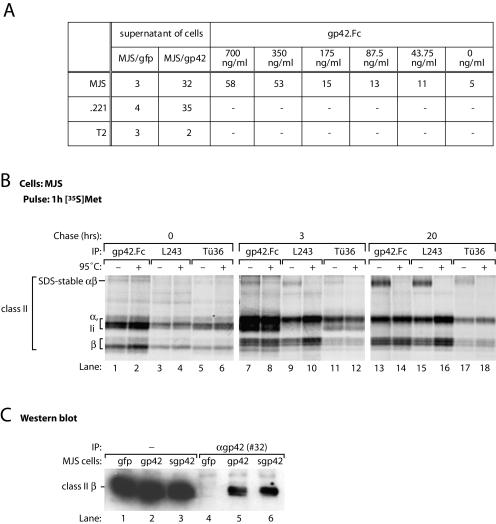
FIG. 3.
s-gp42 is capable of binding to both immature and mature HLA-DR molecules. (A) Culture supernatants of MJS/gp42 cells and control MJS/gfp cells were incubated with HLA class II+ MJS and .221 cells or with HLA class II− T2 cells; for comparison, the binding of gp42.Fc was included. Cell-associated gp42 was detected with antiserum #32 and analyzed by flow cytometry. Mean fluorescence values are depicted; −, not tested. (B) MJS cells were metabolically labeled with [35S]Met for 1 h and chased for 3 or 20 h before lysis. Immunoprecipitation (IP) was performed with s-gp42.Fc or HLA-DR-specific MAbs L243 and Tü36. Immune complexes were incubated in nonreducing sample buffer for 3 h at 37°C (−) or for 5 min at 95°C (+) prior to separation by SDS-12% PAGE. (C) Total lysates of MJS/gp42 cells (lanes 2 and 5), MJS/Kbss-gp42 cells (lanes 3 and 6), and control MJS/gfp cells (lanes 1 and 4) were used directly (lanes 1 to 3) or were subjected to immunoprecipitation with rabbit antiserum #32 against gp42 (lanes 4 to 6). Total lysates and immune complexes were boiled in nonreducing sample buffer, separated by SDS-12% PAGE, and blotted onto polyvinylidene difluoride membranes. Western blots were stained with a MAb specific for HLA-DR β chains (HB10A) and visualized by enhanced chemiluminescence.
Interactions of EBV gp42 with HLA class II at different stages of maturation were studied with gp42.Fc, which allows the direct precipitation of interaction partners of s-gp42 with protein A/G-Sepharose beads. Biochemically, mature stages of class II-peptide complexes can be discriminated from immature class II-Ii heterotrimers by the acquisition of an SDS-stable conformation, which dissociates upon heating to 95°C (22). In lysates from (nontransduced) MJS cells, exogenously added truncated gp42 bound to HLA class II αβIi (in particular at early chase times) (Fig. 3B, lanes 1, 2, 7, and 8) as well as SDS-stable αβ-peptide complexes (most prominently after overnight chase) (lane 13). These results imply that gp42 can associate with class II at different stages of maturation.
To examine whether s-gp42, when produced in MJS cells, complexes with HLA class II, we generated a mutant in which the predicted signal sequence cleavage site of gp42 (between amino acids 33 and 34) was replaced by the fully cleavable signal sequence of the murine MHC class I molecule H-2Kb (MJS/Kbss-gp42 cells), which resulted in complete cleavage and secretion of mutant gp42 molecules (data not shown). By Western blotting, we investigated whether HLA class II molecules are coprecipitated with s-gp42 in MJS/Kbss-gp42 cells following immunoprecipitation with serum #32 (Fig. 3C). In wild-type gp42-expressing cells, class II β chains were detected in gp42 precipitates (Fig. 3C, lane 5). Kbss-gp42 interacted with HLA-DR β chains to a similar extent (Fig. 3C, lane 6) (the same results were obtained for class II α chains; data not shown). From these experiments, we conclude that s-gp42 interacts with all configurations of HLA-DR molecules.
EBV gp42 associates intracellularly with HLA class II at various stages of maturation.
We next analyzed the intracellular association of gp42 and HLA class II molecules in a temporal fashion. EBV gp42 was immunoprecipitated from lysates of [35S]Cys-labeled MJS/gp42 cells with serum #32. The maturation of gp42 displayed a pattern identical to that observed with MAb F-2-1 (compare Fig. 2A and 4A, lanes 1 to 6). In addition, a protein band at a molecular mass of 25 to 28 kDa was revealed by serum #32, possibly representing the HLA class II β chain. As opposed to gp42, HLA class II molecules are visualized more efficiently with [35S]Met radiolabeling than with [35S]Cys radiolabeling. In [35S]Met-labeled MJS/gp42 cell lysates, immunoprecipitations with serum #32 and MAb Tü36 (against HLA-DR) displayed comparable patterns (Fig. 4A, compare lanes 7 to 12 and lanes 13 to 18), indicating that class II α, β, and Ii chains coprecipitated with gp42 (lanes 7 to 11). In particular the maturation of β chains over time (from 25 to 28 kDa) clearly reflects the association between gp42 and HLA class II.
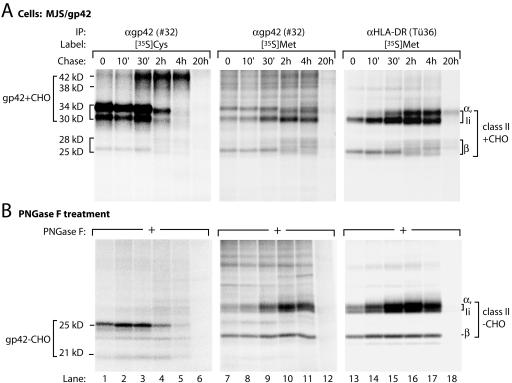
FIG. 4.
EBV gp42 associates with HLA-DR molecules intracellularly shortly after biosynthesis. (A) MJS/gp42 cells were metabolically labeled with [35S]Cys (lanes 1 to 6) or with [35S]Met (lanes 7 to 18) for 30 min and chased for the times indicated. For immunoprecipitation (IP), rabbit antiserum #32 against gp42 and MAb Tü36 against HLA-DR were used. (B) Half of the immunoprecipitates were treated with PNGase F to remove N-linked glycans. Immune complexes were denatured in nonreducing (A) or reducing (B) sample buffer and analyzed by SDS-12% PAGE.
Class II α and Ii chains and the immature forms of gp42 all migrate at approximately 30 to 35 kDa. To discriminate among these proteins, the samples were subjected to digestion with PNGase F (Fig. 4B). In this way, the BZLF2 gene products (gp42−CHO, 21 and 25 kDa; Fig. 4B, lanes 1 to 6) could be distinguished clearly from the coprecipitating class II chains (class II−CHO, lanes 7 to 12 and lanes 13 to 18). These data support the early binding of gp42 to class II molecules in MJS/gp42 cells, and this association must be relatively strong, since it was preserved in NP-40 lysates and persisted through washes with 0.1% SDS.
gp42 interacts with αβIi complexes shortly after synthesis, because coprecipitation was already detectable after a 30-min labeling period (Fig. 4, lanes 1 and 7) and remained apparent for at least 4 h after translation (lanes 5 and 11). All components of the gp42-class II complex traveled beyond the ER, since their N-linked glycans underwent further maturation, as can be inferred from the more diffuse bands that migrated at a higher molecular mass (Fig. 4A, lanes 10 and 11). Indeed, as was reported earlier, complexes of gp42 and αβ-peptide were detected at the cell membrane by using surface iodination of MJS/gp42 cells (27). At the cell surface, both free gp42 and gp42-class II αβ complexes were found.
The combined data show that endogenously expressed EBV gp42 associates with HLA class II molecules upon synthesis in the ER. The resulting complexes are detectable at all stages of transport and maturation.
EBV gp42 is degraded intracellularly by ConB-sensitive proteases.
Lytic infection of HLA class II-expressing cells results in the release of EBV particles with reduced amounts of gp42 incorporated into gH-gL complexes (1). The amounts of gp42 increased when EBV-producing cells were treated with inhibitors of endolysosomal proteolysis (1). These data indicate that, possibly as a result of associating with HLA class II, gp42 molecules are targeted to endolysosomal compartments, leading to their degradation.
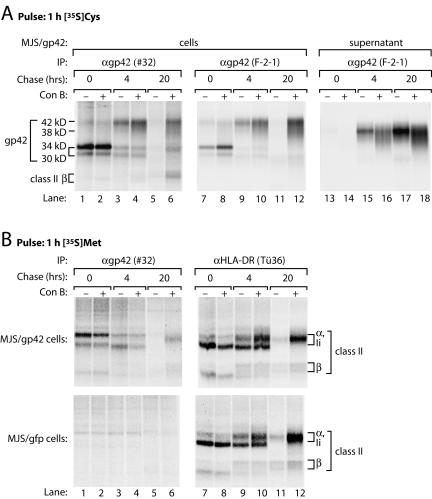
In MJS/gp42 cells, s-gp42 was still present in supernatants, but neither cell-associated gp42 protein nor gp42-class II interactions were detectable after a chase period of 20 h (Fig. 2A and B, lanes 6, and Fig. 4, lanes 6 and 12). This result could have been due to protein turnover occurring in a post-Golgi compartment, as gp42 acquired complex-type oligosaccharide modifications that were endo H resistant (Fig. 2B). Therefore, we investigated whether the degradation of gp42 was inhibited by ConB. ConB disturbs the activity of vacuolar H+ ATPases, which are necessary to create the acidic environment required for the functioning of endolysosomal proteases. In MJS/gp42 cells labeled with [35S]Cys, fl-gp42 and s-gp42 were still detectable after 20 h of chase at a more neutral vacuolar pH (Fig. 5A, lanes 6, 12, and 18). Moreover, coprecipitation of HLA class II molecules (visualized by [35S]Met labeling) with gp42 remained apparent after 20 h of chase only in the presence of ConB (Fig. 5B, compare lanes 5 and 6). Thus, inhibition of endolysosomal proteolysis leads to stabilization of gp42 and gp42-class II complexes.
FIG. 5.
Treatment with ConB prevents the degradation of EBV gp42 and stabilizes gp42-HLA-DR complexes. MJS/gp42 cells (A and B, upper panels) and control MJS/gfp cells (B, lower panels) were mock treated (−) or treated (+) with ConB during starvation, labeling, and chase to prevent endolysosomal proteolysis. Pulse-labeling was performed for 1 h with [35S]Cys (A) or [35S]Met (B); chase times were 4 and 20 h. EBV gp42 molecules were isolated from cell lysates (lanes 1 to 12) and from supernatants (lanes 13 to 18) with antiserum #32 (A and B, lanes 1 to 6) or MAb F-2-1, which is specific for free gp42 (A, lanes 7 to 18); HLA-DR complexes were precipitated with MAb Tü36 (B, lanes 7 to 12). All samples were analyzed by nonreducing SDS-12% PAGE. IP, immunoprecipitation.
s-gp42 is sufficient to mediate HLA class II immune escape.
Previously it was shown that endogenous expression of gp42 by antigen-presenting cells (MJS/gp42) interferes with the activation of HLA class II-restricted T cells (27). Since we observed that a truncated form of gp42 is generated and secreted, we next analyzed whether the interaction of s-gp42 with HLA-DR molecules also interferes with antigen recognition by T-helper cells.
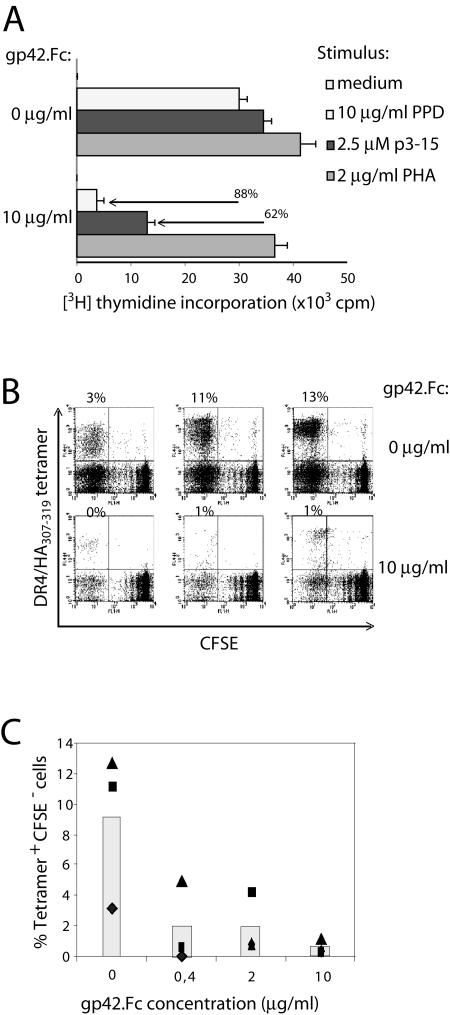
In functional T-cell assays, HLA-DR3+ PBMC were used to present antigen to the DR3-restricted CD4+ T-cell clone Rp15.1.1, which recognizes the p3-15 epitope from hsp65 of Mycobacterium tuberculosis (27). Antigen was provided either as synthetic p3-15 peptide or as PPD (containing hsp65), which requires endogenous processing by antigen-presenting cells to generate the antigenic epitope. T-cell proliferation in response to antigen was strongly diminished by the addition of gp42.Fc to the cultures (62 and 88% inhibition for p3-15 peptide and PPD, respectively) (Fig. 6A); the levels of inhibition were similar for exogenous and endogenous antigen presentation. Mitogenic TCR stimulation by PHA was not markedly influenced by the presence of gp42.Fc. These data demonstrate that antigen-specific T-helper-cell recognition is hampered by the addition of s-gp42.
FIG. 6.
s-gp42 is sufficient to inhibit antigen-specific HLA-DR-restricted T-cell recognition. (A) Effects of gp42.Fc on activation of M. tuberculosis-specific Rp15.1.1 T cells were examined upon coculturing with HLA-DR3+ PBMC in the absence or in the presence of PPD as a source of hsp65, the related p3-15 peptide, or the mitogen PHA. [3H]Thymidine incorporation is shown with error bars for triplicate samples. (B) TCR-HLA-DR-peptide interactions in the presence of various concentrations of gp42.Fc were evaluated with PBMC that were obtained from an HLA-DR4+ donor, labeled with CFSE, and stimulated in vitro with HA307-319 peptides. After 2 weeks, responding T cells were incubated with specific allophycocyanine-conjugated tetramers and were stained with PE-conjugated MAbs to CD4. To study the influence of gp42.Fc on recall responses induced by TCR-HLA-DR-peptide, we used the same approach, except that gp42.Fc was provided during the 2-week in vitro stimulation. Responding T cells were incubated with HLA-DR4/HA307-319 tetramers and stained for CD4. Each flow cytometry dot plot represents approximately 30,000 CD4+ PI− cells; percent values represent the percentages of tetramer-positive CFSE− cells among the total CD4+ PI− T cells (upper leftquadrant). (C) The percentages of tetramer-positive CFSE− T cells (among the CD4+ PI− cells) in the experiment shown in panel B are plotted against the concentration of gp42.Fc. The symbols (▴, ▪, and ♦) represent the three wells of the triplicates.
Using HLA-DR4 tetramers, we evaluated the effect of s-gp42 on T-cell expansion induced by TCR triggering in vitro (Fig. 6B and C). Polyclonal influenza virus-specific T cells were generated from a healthy HLA-DR4+ donor by stimulation of CFSE-labeled PBMC with synthetic peptides representing the viral HA307-319 epitope. After 2 weeks, less than 1% tetramer-positive CD4+ T cells were detected when PBMC were left unstimulated (data not shown). Triplicate cultures of HA307-319 peptide-stimulated PBMC yielded, on average, 9% tetramer-positive responding CD4+ T cells that had divided and had thus become CFSE− (3 to 13% CFSE− tetramer-positive CD4+ cells) (Fig. 6B, upper panels). Addition of 10 μg of gp42.Fc/ml during in vitro restimulation largely abolished the expansion of HLA-DR4/HA307-319-specific T-helper cells in the recall response (reduction to <1%) (Fig. 6B, lower panels), and this effect was dose dependent (Fig. 6C).
Taken together, these results show that s-gp42 interferes with the activation of antigen-specific CD4+ T cells.
The two forms of gp42 are produced in B cells during EBV lytic cycle.
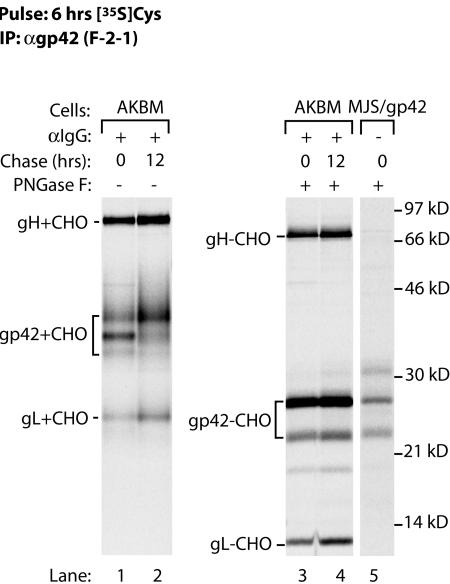
Having observed the generation of both fl-gp42 and truncated gp42 in transduced MJS/gp42 cells, we investigated whether the same also occurs in EBV-infected B lymphocytes. So far, it has been difficult to study lytic-phase protein expression in EBV-positive cells, because no fully permissive culture system for EBV is currently available. At best, up to 40% productively infected B cells can be obtained by cross-linking surface IgG on Akata Burkitt's lymphoma cells (17). Here, we used a novel strategy to isolate pure populations of cells that entered the lytic cycle (Ressing et al., unpublished). We generated an Akata Burkitt's lymphoma cell line, AKBM, in which a reporter protein, rat CD2-GFP, is expressed under the control of an early lytic cycle promoter of EBV. AKBM cells were cultured for 20 h with anti-human IgG antibody to induce viral replication (17). Cells in the lytic cycle were positively selected for surface expression of rat CD2-GFP, pulse-labeled for 6 h with [35S]Cys, and chased for 0 or 12 h in the absence of radiolabel. EBV gp42 was recovered from NP-40 cell lysates with MAb F-2-1. The cells expressed gp42 complexed with gH-gL (Fig. 7, lanes 1 and 2). Removal of N-linked glycans by PNGase F induced a shift in the mobility of gp42 from induced AKBM cells (Fig. 7, lanes 3 and 4) such that the gp42 backbones migrated at positions comparable to those of the two bands observed in MJS/gp42 cells (lane 5). These data demonstrate that, as in MJS/gp42 cells, gp42 is expressed as a full-length protein and as a truncated form during replicative EBV infection of B cells.
FIG. 7.
During EBV lytic infection of B cells, both forms of gp42 are generated. AKBM cells were cultured with anti-IgG antibody to induce EBV reactivation. At 20 h postinduction, AKBM cells in the lytic cycle were positively selected for the expression of the inducible rat CD2-GFP reporter protein, pulse-labeled for 6 h with [35S]Cys, and chased for up to 12 h prior to NP-40 cell lysis. SDS-12% PAGE analysis is shown for proteins immunoprecipitated with MAb F-2-1. Portions of the samples were subjected to PNGase F treatment to reveal protein backbones (lanes 3 and 4); for comparison, gp42−CHO from MJS/gp42 cells is also shown (lane 5). See the legend to Fig. 1 for definitions of abbreviations.
DISCUSSION
In the present study, we showed that EBV gp42 occurs in two alternatively processed forms: a full-length membrane protein and a shorter, soluble protein. s-gp42 is generated by proteolytic cleavage of fl-gp42 at about residue 41. Both forms of gp42 bind to HLA class II either as immature αβIi or mature αβ-peptide complexes. Moreover, s-gp42 is sufficient to mediate antigen-specific evasion from T-helper-cell recognition by blocking the TCR-HLA class II-peptide association.
The processing of EBV fl-gp42 to generate truncated s-gp42 is a posttranslational event, as indicated by the observation that in vitro translation of BZLF2 mRNA in the absence of microsomes yielded a single 25-kDa product (Fig. 2C), excluding the possibility of differential mRNA splicing. Microsomal membranes are required to produce the 21- and 25-kDa protein backbones of gp42, indicating that an ER-resident enzyme is responsible for proteolytic cleavage. Consistent with ER localization of the protease involved, the inhibition of endolysosomal proteases by ConB did not abolish the generation of s-gp42 in MJS/gp42 cells (Fig. 5). Since gp42 processing occurs irrespective of the expression of other viral proteins, gp42 is cleaved by a cellular factor rather than a virus-encoded protease. The ER-resident serine endopeptidase, signal peptidase, can remove signal peptides from nascent chains of membrane and secreted proteins (5). Signal peptidase recognizes a motif comprised of amino acids with small side chains in the −1 and −3 positions relative to the cleavage site. For EBV gp42, a signal peptidase cleavage site is predicted between residues 33 and 34 (24), and cleavage at this location could result in secretion of the C-terminal part of gp42. We found by N-terminal sequencing of secreted s-gp42 that proteolytic cleavage most likely occurs near amino acids 40 to 42. In the absence of a specific motif in this region, it is at present unclear whether s-gp42 is generated through cleavage (i) by signal peptidase, followed by trimming of the N terminus of the protein, or (ii) by another endoproteinase.
The nature of the two alternative forms of EBV gp42 was elucidated by combining different culture systems to express the viral protein. First, cells stably expressing gp42 in isolation (MJS/gp42 cells) facilitated the analysis of protein biosynthesis and maturation. In these cells, two gp42 backbones were found to result from cleavage of a proportion of ER-resident fl-gp42, causing the removal of the N-terminal membrane anchor (Fig. 1B and 2A and B). As a consequence, mature gp42 occurred both as a full-length transmembrane protein of 42 kDa and as a truncated soluble polypeptide of 38 kDa (Fig. 2A and B). Second, B cells productively infected with EBV were used to assess whether gp42 was similarly modified in the presence of its viral interaction partners, gH and gL. To obtain pure populations of productively infected B cells, we used a novel approach that is based on the inducible expression of a reporter protein, rat CD2-GFP, which is controlled by an early EBV promoter. Following induction of the lytic cycle with anti-IgG antibody, the population of productively infected Akata Burkitt's lymphoma cells expressing the reporter protein is positively selected. Whereas cross-linking of surface IgG generally results in 10 to 40% of cells entering the lytic cycle, the reporter-based isolation procedure routinely yields 90% purity (Ressing et al., unpublished). This novel selection protocol allowed us to study gp42 in the context of productive EBV infection. Thus, from PNGase F digestion of gp42 immune complexes, we deduced that both fl-gp42 and s-gp42 were generated in B cells during the EBV lytic cycle (Fig. 7). In future experiments, we will exploit this approach to further explore gp42-class II interactions biochemically and functionally in the context of a natural EBV infection. Since all EBV proteins are coexpressed, s-gp42 generated during productive EBV infection could associate with gH and gL. Both forms of gp42 are present in gH-gL-gp42 complexes from lytically induced AKBM cells (Fig. 7 and data not shown), indicating that s-gp42 has retained the capacity to bind gH and gL. Therefore, the region required for interactions with gH and gL (42) can be narrowed down to gp42 residues 40 to 58.
In earlier studies detecting gH-gL-gp42 heterotrimers on the membranes of virus-producing cells and in the viral envelope, gp42 was represented by polypeptides with molecular masses of 42 and 38 kDa (17, 42). Here, we show that these alternative forms of gp42 arise as a consequence of protelolytic cleavage of a proportion of newly synthesized gp42 proteins within the ER. The combined data imply that fl-gp42 and truncated gp42 are generated during the course of a natural EBV infection.
Recently, gH-gL complexes of two betaherpesviruses, HCMV and human herpesvirus 6 (HHV-6), were found to also associate with a third viral glycoprotein, gO or gQ, respectively (7, 19); these arrangements mirror the EBV gH-gL-gp42 interactions. Comparison of the sequences for EBV gp42 with those for gO and gQ does not reveal any obvious similarities. It remains to be established whether the HCMV gO and HHV-6 gQ proteins have functions in viral entry and/or immune evasion comparable to those of EBV gp42. For alphaherpesviruses, no additional viral interaction partners have been found to complex with gH and gL (34). In herpes simplex virus (HSV), another viral protein, gD, is involved in host cell penetration. HSV gD interacts with entry receptors, including HveA (reviewed in reference 2). Like the situation for EBV gp42 (16), viral entry of susceptible cells can be blocked by providing HSV gD-specific antibodies or soluble forms of gD (10). Interestingly, gD is secreted from cells infected with HSV type 2. The soluble form of gD is fully glycosylated and is thought to be derived from the full-length protein by posttranslational cleavage (21). These findings resemble our data for EBV gp42.
EBV gp42 serves at least two functions: gp42 is the coreceptor for viral entry into B cells (16) and acts as an HLA class II immune evasion molecule (27). With respect to the physiological significance of s-gp42, we suggest that it could contribute to EBV propagation in the following ways. The binding of s-p42 could block EBV entry receptors on lytic B cells, thereby preventing their reinfection. In support of this notion, gp42.Fc was previously shown to be effective in blocking the entry of EBV into B cells (16). At the time of virus spread, s-gp42 could serve as a decoy for antiviral humoral immunity by neutralizing gp42-specific antibodies. During viral reactivation, s-gp42 could inhibit HLA class II-restricted T-cell recognition of EBV-producing B cells, thereby creating a window for undetected virus production.
At the cell surface, EBV gp42 has been demonstrated to bind HLA class II molecules (1, 27), and the interaction sites have been identified through a combination of X-ray structure analysis of gp42-HLA-DR1 αβ complexes (20, 33) and viral infection experiments with cells expressing mutated HLA class II β1 chains (18). Here, we report for the first time an intracellular association between immature HLA-DR αβIi heterotrimers and endogenously synthesized gp42 in the ER. The resulting complexes mature and migrate to the cell surface as gp42-class II αβ-peptide complexes (Fig. 4 and 5) (27). Likewise, s-gp42 can bind to both immature and mature HLA class II molecules (Fig. 3). Finally, fl-gp42 proteins are degraded in endolysosomal compartments (Fig. 5). Experiments with virus-producing cells have indicated that the degradation of EBV gp42 is dependent on the coexpression of class II proteins (1). Gene products of other herpesviruses, e.g., murine cytomegalovirus m06, HHV-7 U21, and human cytomegalovirus pp65, divert MHC molecules toward lysosomes for destruction as a strategy to prevent antigen presentation to T cells (8, 25, 28). It is tempting to speculate that an association with EBV gp42 might predispose HLA class II molecules to degradation, but surface levels of class II molecules were not downregulated on MJS/gp42 cells (27). Irrespective of these data, we have found that MJS/gp42 cells have a significantly reduced capacity to activate antigen-specific T-helper-cell responses (27). Immune escape from HLA class II-restricted T-cell recognition by gp42 relies on steric interference with TCR-class II-peptide interactions upon gp42-class II association. Here, we have shown that exogenously added s-gp42 can also inhibit T-helper-cell activation in an antigen-specific manner, suggesting that naturally produced EBV s-gp42 may perform the same function in vivo.
Acknowledgments
We acknowledge the assistance of Lieke van der Velden with experiments. We thank Hidde Ploegh for helpful advice and Danijela Koppers-Lalic for critically reading the manuscript.
This study was financially supported by the Dutch Cancer Society (grant KWF/RUL 1998-179) and by The Netherlands Organization for Scientific Research. S.K. and M.R. were supported by the Medical Research Council, London, United Kingdom, and S.K. was also supported by a short-term EMBO fellowship.
REFERENCES
- 1.Borza, C. M., and L. M. Hutt-Fletcher. 2002. Alternate replication in B cells and epithelial cells switches tropism of Epstein-Barr virus. Nat. Med. 8:594-599. [DOI] [PubMed] [Google Scholar]
- 2.Campadelli-Fiume, G., F. Cocchi, L. Menotti, and M. Lopez. 2000. The novel receptors that mediate the entry of herpes simplex viruses and animal alphaherpesviruses into cells. Rev. Med. Virol. 10:305-319. [DOI] [PubMed] [Google Scholar]
- 3.Clark, A. E., and T. Yakoshi. 1984. HB10A, p. 195. In A. Bernard, L. Baunsell, J. Dausset, and S. Schlossman (ed.), Leukocyte typing. Springer-Verlag KG, Heidelberg, Germany.
- 4.Cresswell, P. 1994. Assembly, transport, and function of MHC class II molecules. Annu. Rev. Immunol. 12:259-293. [DOI] [PubMed] [Google Scholar]
- 5.Dalbey, R. E., and G. von Heijne. 1992. Signal peptidases in prokaryotes and eukaryotes—a new protease family. Trends Biochem. Sci. 17:474-478. [DOI] [PubMed] [Google Scholar]
- 6.Drijfhout, J. W., W. Bloemhoff, J. T. Poolman, and P. Hoogerhout. 1990. Solid-phase synthesis and applications of N-(S-acetylmercaptoacetyl) peptides. Anal. Biochem. 187:349-354. [DOI] [PubMed] [Google Scholar]
- 7.Huber, M. T., and T. Compton. 1998. The human cytomegalovirus UL74 gene encodes the third component of the glycoprotein H-glycoprotein L-containing envelope complex. J. Virol. 72:8191-8197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hudson, A. W., P. M. Howley, and H. L. Ploegh. 2001. A human herpesvirus 7 glycoprotein, U21, diverts major histocompatibility complex class I molecules to lysosomes. J. Virol. 75:12347-12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jefferies, W. A., J. R. Green, and A. F. Williams. 1985. Authentic T helper CD4 (W3/25) antigen on rat peritoneal macrophages. J. Exp. Med. 162:117-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson, D. C., R. L. Burke, and T. Gregory. 1990. Soluble forms of herpes simplex virus glycoprotein D bind to a limited number of cell surface receptors and inhibit virus entry into cells. J. Virol. 64:2569-2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lampson, L. A., and R. Levy. 1980. Two populations of Ia-like molecules on a human B cell line. J. Immunol. 125:293-299. [PubMed] [Google Scholar]
- 12.Lee, S. P., J. M. Brooks, H. Al Jarrah, W. A. Thomas, T. A. Haigh, G. S. Taylor, S. Humme, A. Schepers, W. Hammerschmidt, J. L. Yates, A. B. Rickinson, and N. W. Blake. 2004. CD8 T cell recognition of endogenously expressed Epstein-Barr virus nuclear antigen 1. J. Exp. Med. 199:1409-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levitskaya, J., M. Coram, V. Levitsky, S. Imreh, P. M. Steigerwald-Mullen, G. Klein, M. G. Kurilla, and M. G. Masucci. 1995. Inhibition of antigen processing by the internal repeat region of the Epstein-Barr virus nuclear antigen-1. Nature 375:685-688. [DOI] [PubMed] [Google Scholar]
- 14.Levitskaya, J., A. Sharipo, A. Leonchiks, A. Ciechanover, and M. G. Masucci. 1997. Inhibition of ubiquitin/proteasome-dependent protein degradation by the Gly-Ala repeat domain of the Epstein-Barr virus nuclear antigen 1. Proc. Natl. Acad. Sci. USA 94:12616-12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levitsky, V., and M. G. Masucci. 2002. Manipulation of immune responses by Epstein-Barr virus. Virus Res. 88:71-86. [DOI] [PubMed] [Google Scholar]
- 16.Li, Q., M. K. Spriggs, S. Kovats, S. M. Turk, M. R. Comeau, B. Nepom, and L. M. Hutt-Fletcher. 1997. Epstein-Barr virus uses HLA class II as a cofactor for infection of B lymphocytes. J. Virol. 71:4657-4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li, Q., S. M. Turk, and L. M. Hutt-Fletcher. 1995. The Epstein-Barr virus (EBV) BZLF2 gene product associates with the gH and gL homologs of EBV and carries an epitope critical to infection of B cells but not of epithelial cells. J. Virol. 69:3987-3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McShane, M. P., M. M. Mullen, K. M. Haan, T. S. Jardetzky, and R. Longnecker. 2003. Mutational analysis of the HLA class II interaction with Epstein-Barr virus glycoprotein 42. J. Virol. 77:7655-7662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mori, Y., P. Akkapaiboon, X. Yang, and K. Yamanishi. 2003. The human herpesvirus 6 U100 gene product is the third component of the gH-gL glycoprotein complex on the viral envelope. J. Virol. 77:2452-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mullen, M. M., K. M. Haan, R. Longnecker, and T. S. Jardetzky. 2002. Structure of the Epstein-Barr virus gp42 protein bound to the MHC class II receptor HLA-DR1. Mol. Cell 9:375-385. [DOI] [PubMed] [Google Scholar]
- 21.Murata, T., F. Goshima, H. Takakuwa, and Y. Nishiyama. 2002. Excretion of herpes simplex virus type 2 glycoprotein D into the culture medium. J. Gen. Virol. 83:2791-2795. [DOI] [PubMed] [Google Scholar]
- 22.Neefjes, J. J., and H. L. Ploegh. 1992. Inhibition of endosomal proteolytic activity by leupeptin blocks surface expression of MHC class II molecules and their conversion to SDS resistance alpha beta heterodimers in endosomes. EMBO J. 11:411-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nemerow, G. R., C. Mold, V. K. Schwend, V. Tollefson, and N. R. Cooper. 1987. Identification of gp350 as the viral glycoprotein mediating attachment of Epstein-Barr virus (EBV) to the EBV/C3d receptor of B cells: sequence homology of gp350 and C3 complement fragment C3d. J. Virol. 61:1416-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 25.Odeberg, J., B. Plachter, L. Branden, and C. Soderberg-Naucler. 2003. Human cytomegalovirus protein pp65 mediates accumulation of HLA-DR in lysosomes and destruction of the HLA-DR alpha-chain. Blood 101:4870-4877. [DOI] [PubMed] [Google Scholar]
- 26.Pieters, J. 1997. MHC class II restricted antigen presentation. Curr. Opin. Immunol. 9:89-96. [DOI] [PubMed] [Google Scholar]
- 27.Ressing, M. E., D. van Leeuwen, F. A. Verreck, R. Gomez, B. Heemskerk, M. Toebes, M. M. Mullen, T. S. Jardetzky, R. Longnecker, M. W. Schilham, T. H. Ottenhoff, J. Neefjes, T. N. Schumacher, L. M. Hutt-Fletcher, and E. J. Wiertz. 2003. Interference with T cell receptor-HLA-DR interactions by Epstein-Barr virus gp42 results in reduced T helper cell recognition. Proc. Natl. Acad. Sci. USA 100:11583-11588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reusch, U., W. Muranyi, P. Lucin, H. G. Burgert, H. Hengel, and U. H. Koszinowski. 1999. A cytomegalovirus glycoprotein re-routes MHC class I complexes to lysosomes for degradation. EMBO J. 18:1081-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rickinson, A., and E. Kieff. 1996. Epstein-Barr virus, p. 2397-2446. In B. Fields, D. Knipe, P. Howley, R. Chanock, J. Melnick, T. Monath, B. Roizman, and S. Straus (ed.), Virology. Lippincott-Raven, Philadelphia, Pa.
- 30.Salter, R. D., and P. Cresswell. 1986. Impaired assembly and transport of HLA-A and -B antigens in a mutant TxB cell hybrid. EMBO J. 5:943-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shaw, A. R., J. K. Chan, S. Reid, and J. Seehafer. 1985. HLA-DR synthesis induction and expression in HLA-DR-negative carcinoma cell lines of diverse origins by interferon-gamma but not by interferon-beta. JNCI 74:1261-1268. [PubMed] [Google Scholar]
- 32.Shimizu, Y., and R. DeMars. 1989. Production of human cells expressing individual transferred HLA-A,-B,-C genes using an HLA-A,-B,-C null human cell line. J. Immunol. 142:3320-3328. [PubMed] [Google Scholar]
- 33.Silva, A. L., J. Omerovic, T. S. Jardetzky, and R. Longnecker. 2004. Mutational analyses of Epstein-Barr virus glycoprotein 42 reveal functional domains not involved in receptor binding but required for membrane fusion. J. Virol. 78:5946-5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spear, P. G., and R. Longnecker. 2003. Herpesvirus entry: an update. J. Virol. 77:10179-10185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spriggs, M. K., R. J. Armitage, M. R. Comeau, L. Strockbine, T. Farrah, B. Macduff, D. Ulrich, M. R. Alderson, J. Mullberg, and J. I. Cohen. 1996. The extracellular domain of the Epstein-Barr virus BZLF2 protein binds the HLA-DR beta chain and inhibits antigen presentation. J. Virol. 70:5557-5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takada, K., and Y. Ono. 1989. Synchronous and sequential activation of latently infected Epstein-Barr virus genomes. J. Virol. 63:445-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tellam, J., G. Connolly, K. J. Green, J. J. Miles, D. J. Moss, S. R. Burrows, and R. Khanna. 2004. Endogenous presentation of CD8+ T cell epitopes from Epstein-Barr virus-encoded nuclear antigen 1. J. Exp. Med. 199:1421-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van de Rijn, M., A. H. Geurts van Kessel, V. Kroezen, A. J. van Agthoven, K. Verstijnen, C. Terhorst, and J. Hilgers. 1983. Localization of a gene controlling the expression of the human transferrin receptor to the region q12 leads to qter of chromosome 3. Cytogenet. Cell Genet. 36:525-531. [DOI] [PubMed] [Google Scholar]
- 39.van der Wal, F. J., J. D. Oliver, and S. High. 1998. The transient association of ERp57 with N-glycosylated proteins is regulated by glucose trimming. Eur. J. Biochem. 256:51-59. [DOI] [PubMed] [Google Scholar]
- 40.Voo, K. S., T. Fu, H. Y. Wang, J. Tellam, H. E. Heslop, M. K. Brenner, C. M. Rooney, and R. F. Wang. 2004. Evidence for the presentation of major histocompatibility complex class I-restricted Epstein-Barr virus nuclear antigen 1 peptides to CD8+ T lymphocytes. J. Exp. Med. 199:459-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang, X., and L. M. Hutt-Fletcher. 1998. Epstein-Barr virus lacking glycoprotein gp42 can bind to B cells but is not able to infect. J. Virol. 72:158-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang, X., W. J. Kenyon, Q. Li, J. Mullberg, and L. M. Hutt-Fletcher. 1998. Epstein-Barr virus uses different complexes of glycoproteins gH and gL to infect B lymphocytes and epithelial cells. J. Virol. 72:5552-5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yin, Y., B. Manoury, and R. Fahraeus. 2003. Self-inhibition of synthesis and antigen presentation by Epstein-Barr virus-encoded EBNA1. Science 301:1371-1374. [DOI] [PubMed] [Google Scholar]