Abstract
The large hepatitis delta antigen (HDAg-L) mediates hepatitis delta virus (HDV) assembly and inhibits HDV RNA replication. Farnesylation of the cysteine residue within the HDAg-L carboxyl terminus is required for both functions. Here, HDAg-L proteins from different HDV genotypes and genotype chimeric proteins were analyzed for their ability to incorporate into virus-like particles (VLPs). Observed differences in efficiency of VLP incorporation could be attributed to genotype-specific differences within the HDAg-L carboxyl terminus. Using a novel assay to quantify the extent of HDAg-L farnesylation, we found that genotype 3 HDAg-L was inefficiently farnesylated when expressed in the absence of the small hepatitis delta antigen (HDAg-S). However, as the intracellular ratio of HDAg-S to HDAg-L was increased, so too was the extent of HDAg-L farnesylation for all three genotypes. Single point mutations within the carboxyl terminus of HDAg-L were screened, and three mutants that severely inhibited assembly without affecting farnesylation were identified. The observed assembly defects persisted under conditions where the mutants were known to have access to the site of VLP assembly. Therefore, the corresponding residues within the wild-type protein are likely required for direct interaction with viral envelope proteins. Finally, it was observed that when HDAg-S was artificially myristoylated, it could efficiently inhibit HDV RNA replication. Hence, a general association with membranes enables HDAg to inhibit replication. In contrast, although myristoylated HDAg-S was incorporated into VLPs far more efficiently than HDAg-S or nonfarnesylated HDAg-L, it was incorporated far less efficiently than wild-type HDAg-L; thus, farnesylation was required for efficient assembly.
Hepatitis delta virus (HDV) is a subviral agent that possesses a closed circular single-stranded RNA genome of approximately 1.7 kilobases (27, 28). Due to extensive internal complementarity, the HDV genome assumes a partially double-stranded unbranched rod-like structure (5, 16). HDV is a satellite of hepatitis B virus (HBV) and requires the HBV envelope proteins, or surface antigens (HBsAg), for virion assembly (28). HDV virions possess an inner core composed of HDV RNPs and an outer envelope composed of host lipids and HBsAg proteins. HDV encodes two core proteins, the small and large hepatitis delta antigens (HDAg-S and HDAg-L), from a single coding sequence (33). HDAg-S is expressed early during infection and is required for HDV RNA replication (15). HDAg-L is produced later during infection, inhibits HDV RNA replication in a potent trans-dominant manner, and is required for HDV assembly through its interaction with HBsAg (8, 9, 12). In addition, HDAg-L can also be incorporated into the so-called empty particles derived from HBsAg-S, giving rise to virus-like particles (VLPs) devoid of HDV or HBV nucleic acids (32).
HDAg-L expression occurs after a host-mediated RNA editing event causes mutation of the HDAg-S UAG (amber) stop codon to a UGG (tryptophan) codon, thereby allowing translation of an additional carboxyl-terminal extension in the HDAg open reading frame (23). The resulting protein contains the entire amino acid sequence of HDAg-S plus a 19-amino-acid proline-rich carboxyl terminus. This extension contains the only cysteine residue of HDAg-L, which resides within a CXXQ farnesylation motif. Farnesylation has been shown to be important for both the virion assembly and replication inhibition functions of HDAg-L (11, 21, 26, 29). The role of HDAg-L in HDV assembly has been well characterized. Mutation of the CXXQ cysteine residue at position 211 (C211) abolishes HDV virion assembly (11), as does treatment of cells with farnesyl transferase (FTase) inhibitors (3, 4).
It is possible that the role of HDAg-L in HDV assembly is not limited to its direct interaction with HBsAg. One study suggests that a region within the HDAg-L carboxyl terminus spanning amino acids 197 to 210 functions as a nuclear export signal (NES), thereby facilitating trafficking of HDV RNPs out of the nucleus (20). The aforementioned study shows that mutation of the proline residue at position 205 to alanine (P205A) abolishes HDAg-L incorporation into VLPs. In addition, after microinjection into Xenopus oocytes, rabbit immunoglobulin G (IgG) molecules fused to a wild-type NES synthetic peptide can be found in both nuclear and cytoplasmic fractions of cell lysate, while IgG molecules bearing the P205A mutant NES peptide are confined to the nuclear fraction. This result suggests that the P205A mutant might be unable to support HDV assembly due to its defect in nuclear export and its inability to traffic to the site of envelope particle assembly.
The ability of HDAg-L to inhibit replication has been a point of some controversy; however, recent data clearly indicate that, even when expressed as a consequence of amber/W editing during HDV replication, HDAg-L is capable of inhibiting that process (29). It should also be noted that certain deletion mutants of HDAg that include a functional coiled-coil domain but that lack the carboxyl terminus of HDAg-L can inhibit replication even though these proteins are not farnesylated (19). In contrast, although farnesylated HDAg-L inhibits replication, nonfarnesylated HDAg-L does not (29). Hence, the mechanism by which deletion mutants of HDAg inhibit replication must be different from that used by HDAg-L. Since HDAg-L is the only viral protein naturally expressed that is known to be able to inhibit replication, it is the mechanism of action of this protein that is of interest in the present study.
HDV isolates are generally classified as genotype 1, 2, or 3 on the basis of differences in the nucleotide sequences of their genomes. The amino acid sequence of the carboxyl-terminal extension of HDAg-L, although highly conserved within HDV genotypes, is highly divergent among genotypes (6) (see also Fig. 1A). Previously, it was shown that assembly of genotype 2 HDV particles proceeds less efficiently that that of genotype 1 HDV particles, and it was hypothesized that the relatively low assembly efficiency associated with genotype 2 could account for its milder disease phenotype (13).
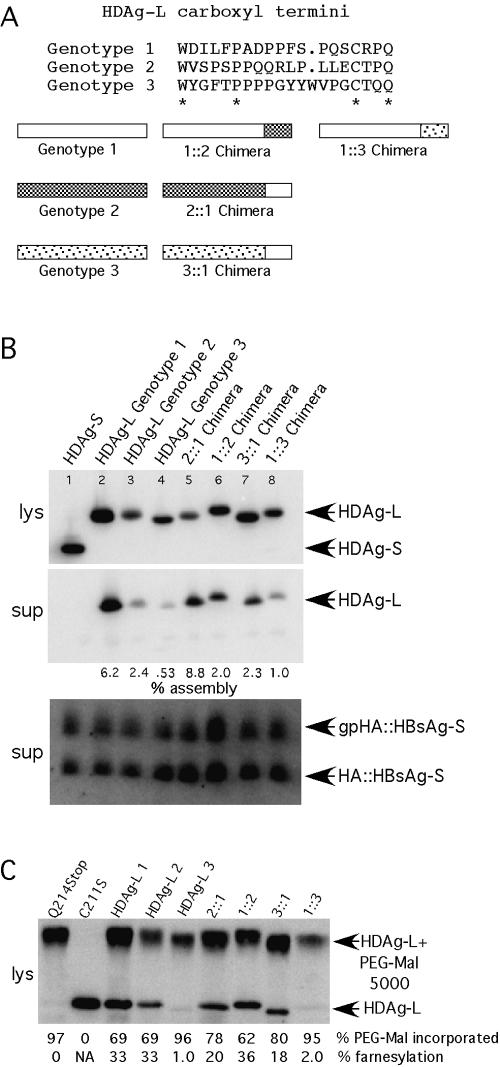
FIG. 1.
The sequence of the carboxyl-terminal region of HDAg-L is responsible for differences in assembly across HDV genotypes. (A) Schematic representations of mixed-genotype chimeric HDAg-L molecules and alignment of the carboxyl-terminal sequences used in this study. (B) Immunoblots for HDAg-L and HBsAg-S::HA. Percent assembly was calculated as HDAg-Lsup/(HDAg-Lsup+ HDAg-Llys) × 100 and was normalized to take into account sample-to-sample variation in HBsAg-S secretion. (C) The percentage of PEG-mal incorporation was calculated as HDAg-L-PEG-mal/(HDAg-L-PEG-mal + HDAg-L) × 100, using lysate from HuH7 cells transfected with the indicated HDAg expression constructs, harvested 3 days posttransfection. The percentage of farnesylation was calculated from PEG-mal incorporation values after normalization to the value obtained for HDAg-L(Q214Stop), which was taken to represent 0% farnesylation. Wild-type genotype 1, 2, and 3 HDAg-L samples appear in lanes 3, 4,and 5, respectively. Samples in which the 2::1 and 1::2 chimeras were expressed appear in lanes 6 and 7, respectively, and samples in which the 3::1 and 1::3 chimeras were expressed appear in lanes 8 and 9, respectively. NA, not applicable.
In this study, the contributions to assembly of carboxyl-terminal sequences from all three HDV genotypes were explored, as were the effects of various amino acid substitutions in the carboxyl-terminal region of genotype 1 HDAg-L. We developed a novel assay for quantifying the extent of HDAg-L farnesylation and used this assay to determine whether an assembly defect associated with a particular mutant resulted from an associated defect in farnesylation. In this way, we identified positions within the HDAg-L carboxyl terminus that were likely required for interaction with HBsAg and discovered that the presence of HDAg-S could stimulate farnesylation of HDAg-L.
While the carboxyl-terminal region unique to HDAg-L plays an important role in HDV assembly, the role, if any, of sequences in the amino-terminal portion of HDAg-L (amino acids 1 to 195) has not been carefully examined. One study shows that phosphorylation of the serine residue at position 177 in HDAg-S is required for HDV RNA replication and that this position is not phosphorylated in the context of HDAg-L (25). Interestingly, S177 resides within or immediately adjacent to a region of HDAg that has been shown to be conformationally distinct in HDAg-L versus HDAg-S (14). In the present study, we observed that HDAg-S bearing an amino-terminal human immunodeficiency virus type 1 (HIV-1) Gag myristoylation signal (MGARAS) could inhibit HDV RNA replication. Such inhibition was found to be dependent on myristoylation. Although myristoylation occurs at the amino terminus while farnesylation occurs at the carboxyl terminus, both modifications increase the affinity of the protein for membranes. Hence, membrane association per se was sufficient to enable HDAg to inhibit replication. Myristoylation also enabled HDAg-S to be incorporated into VLPs; however, in this case the phenotype was intermediate and the efficiency of incorporation was far less than that of HDAg-L. Hence, in addition to increasing the affinity of HDAg-L for membranes, farnesylation likely facilitates the direct interaction between HDAg-L and HBsAg.
MATERIALS AND METHODS
Construction of plasmids that express chimeric HDAg-L proteins.
For all plasmids used in this study that were generated by PCR, direct sequencing across the entire PCR insert was used to confirm that the plasmids did not contain any inadvertent mutations. The chimeric HDAg-L (1::2, 2::1, 1::3, and 3::1) expression plasmids were constructed as follows, where A::B indicates the amino-terminal 195 amino acids from genotype A HDAg-L and the carboxyl-terminal 19 or 20 amino acids from genotype B HDAg-L. With the exception of the 3::1 fusion, the chimeras were made via a two-round PCR strategy, where two partially overlapping oligonucleotides were used to generate the A::B fusion junction, and upstream and downstream oligonucleotides complementary to the parental vectors were used to generate a longer fragment, which could then be inserted into an appropriate vector backbone. For the 1::2 and 1::3 chimeras, the entire genotype 2 or genotype 3 carboxyl-terminal sequence was synthesized from the fusion junction oligonucleotides. The 3::1 chimera was made without using PCR, by ligating the 3,619-bp NdeI-NcoI fragment from pKW42 to the 126-bp NdeI-NcoI fragment from pcmvAg-L-Peru. Fusion junction oligonucleotides for the chimeric HDAg sequences were as follows: pBOM072 (1::2 chimera), OLI533 (5′GTACGCCCATCTCCCCCCCAACAACGCCTTCCACTCCTCGAGTGTACCCCCCAATGATAAAGCGGGTT3′) and OLI537 (5′GGGAGATGGGCGTACCCATGGAAATCCCTG3′); pBOM073 (1::3 chimera), OLI534 (5′TATGGGTTTACCCCGCCTCCCCCCGGGTATTACTGGGTCCCAGGGTGCACCCAACAATGAATAAAGCGGGTT3′) and OLI538 (5′CGGGGTAAACCCATACCATGGAAATCCCTG3′); and pBOM147 (2::1 chimera), OLI822 (5′CTTACCCATGGGAATCCCTGGGTCC3′) and OLI823 (5′AGACCCAAGCTTTCCGAGATTTCCT3′).
Construction of expression plasmids for single-amino-acid substitution mutants of genotype 1 HDAg-L.
Single-amino-acid substitutions in HDAg-L were constructed by PCR with the oligonucleotides listed below. For pBOM076, -077, -078, -079, -080, -081, -082, -083, and -091, the 3,227-bp SfiI-XmaI fragment from pDL444 and the 2,282-bp SalI-SfiI fragment from pKW42 were ligated with the appropriate XmaI-SalI-digested PCR product. For pBOM093, the PCR fragment was digested with XmaI and XhoI and ligated to the pDL444 fragment and the pKW42 fragment mentioned above. For pBOM094, the 2,173-bp SfiI-SalI fragment from pDL445 and the 2,097-bp EcoO109I-SfiI fragment from pKW42 were ligated with the SalI-EcoO109I-digested PCR product. The template for the PCR in all cases was pDL445. The vectors pBOM084 and pBOM085 express wild-type HDAg-S and HDAg-L, respectively, and were made by ligating the 1,708-bp SfiI-EcoRI fragment from pDL444 to the 2,747-bp EcoRI-SfiI fragment from pKW42 and pKW43, respectively. These vectors were made in order to take advantage of the polylinker sequence, which resides in pKW42 and pKW43. Mutagenic oligonucleotides for constructing genotype 1 HDAg-L single-amino-acid substitution mutations were as follows: pBOM076 (P201A), OLI544 (5′ACTGGGGTCGACAACTCTGGGGAGAAAAGGGCGGATCGGCTGCGAAGAGTAT3′); pBOM077 (P208A), OLI545 (5′ACTGGGGTCGACAACTCTGGGCAGAAAAGGGC3′); pBOM078 (L199A), OLI546 (5′ACTGGGGTCGACAACTCTGGGGAGAAAAGGGCGGATCGGCTGGGAAGGCTATATCCCAT3′); pBOM079 (I198A), OLI547 (5′CTGGGGTCGACAACTCTGGGGAGAAAAGGGCGGATCGGCTGGGAAGAGTGCATCCCATGGA3′); pBOM080 (D197A), OLI548 (5′ACTGGGGTCGACAACTCTGGGGAGAAAAGGGCGGATCGGCTGGGAGAGTATAGCCCATGGAAAT3′); pBOM081 (W196A), OLI549 (5′ACTGGGGTCGACAACTCTGGGGAGAAAAGGGCGGATCGGCTGGGAGAGTATATCCGCTGGAAATCCC3′); pBOM082 (P205A), OLI550 (5′ACTGGGGTCGACAACTCTGGGGAGAAAAGGCCGGATCGGCT3′); pBOM083 (C211S), OLI551 (5′ACTGGGGTCGACTACTCTGGGG A3′); pBOM091 (D203A), OLI569 (5′ACTGGGGTCGACAACTCTGGGGAGAAAAGGGCGGAGCGGCTGGGAAGAGTAT3′); pBOM092 (P204A), OLI570 (5′ACTGGGGTCGACAACTCTGGGGAGAAAAGGGCGCATCGGCTGGGAGAG3′); pBOM093 (P213A), OLI571 (5′AGAGTTCTCGAGCCCAGTGAATAAAGC3′); and pBOM094 (Q214Stop), OLI572 (5′GAGTTGTCGACCCTAGTGAATAAAGCGGG3′).
Construction of expression plasmids for myristoylated HDAgs.
Expression vectors for HDAg-L proteins bearing the amino-terminal HIV-1 Gag myristoylation signal, MGARAS, were constructed from pBOM084 (HDAg-S), pBOM085 (HDAg-L), and pBOM083 (C211S) templates, generating pBOM088 [expresses MGARAS:HDAg-L(C211S)], pBOM089 (expresses MGARAS:HDAg-S), pBOM090 (expresses MGARAS:HDAg-L), pBOM126 [expresses MAARAS:HDAg-L(C211S)], pBOM127 (expresses MAARAS:HDAg-S), and pBOM128 (expresses MAARAS:HDAg-L). For MGARAS constructs, OLI563 (5′CTCGAGTCTAGACCCACTGAGGACTGCCGCCTCTAGCCGAGATGGGTGCGAGAGCGTCAAGCCGGTCCGAGTCG3′), containing the entire MGARAS sequence and including an XbaI site, was used as the upstream oligonucleotide for PCR, and OLI038 (5′CCGAGAGAAGGGGGACTCCGGGAC3′) was used as the downstream oligonucleotide. PCR products were digested with XbaI and EcoRI and ligated into the 4,254-bp XbaI-EcoRI fragment from the appropriate parental vector (HDAg-S, -L, or C211S). For MAARAS constructs, overlapping OLI703 (5′GAGATGGCTGCGAGAGCGTCAAG C3′) and OLI704 (5′TCTCGCAGCCATCTCGGCTAGAGG3′) oligonucleotides were used to generate the Gly-to-Ala substitution, and OLI530 and OLI038 were used as upstream and downstream oligonucleotides. PCR products were again cut with XbaI and EcoRI and inserted into the appropriate parental vector.
Culture and transfection of HuH7 cells.
HuH7 cells were grown in 100-mm-diameter dishes at 37°C with 5% CO2 in Dulbecco's modified Eagle's medium with 4.5 mg of glucose/liter and without l-glutamine (Cellgro), supplemented with 10% fetal bovine serum (Sigma), 100 U of penicillin-streptomycin (GibcoBRL) per ml, 584 mg of l-glutamine (GibcoBRL) per liter, and nonessential amino acids (GibcoBRL). Cells were transfected by using the Lipofectamine PLUS (GibcoBRL) method. Briefly, 16 μg of CsCl-purified DNA was transfected with 20 μl of PLUS reagent and 35 μl of Lipofectamine. Cells were incubated with liposome-DNA complexes in the absence of serum for 3 to 4 h, after which time the serum-free medium was replaced with complete Dulbecco's modified Eagle's medium prepared as described above. In all transfections, the expression vector pSS15, which encodes secreted alkaline phosphatase driven by a cytomegalovirus immediate-early promoter, was included to provide a measure of overall transfection efficiency (1). This vector represented 10% of the total amount of transfected DNA.
Harvesting of cells and pelleting of HDV virions and HDAg-L virus-like particles.
Transfected cells were harvested at 5 days posttransfection for HDAg-L virus-like particle experiments. Cells were lysed in Laemmli buffer containing 2% sodium dodecyl sulfate (SDS) but lacking β-mercaptoethanol (β-ME), which was added at a final concentration of 2% prior to electrophoresis when appropriate. Lysate samples were passed 8 to 10 times through a 26-gauge tuberculin syringe in order to shear chromosomal DNA. Culture supernatant was clarified at 5,000 × g for 10 min, and virus-like particles were pelleted in the absence of a sucrose cushion by centrifugation of the clarified supernatant at 35,000 rpm in an SW41 (Beckman) rotor for 4 h at 4°C. Protein pellets were resuspended overnight at 4°C in 200 μl of Laemmli protein lysis buffer, and samples were boiled prior to loading.
Electrophoresis and immunoblotting.
Protein samples were heated at 95°C for 5 min and subjected to SDS-polyacrylamide gel electrophoresis (SDS-PAGE) through 12% polyacrylamide gels. β-ME was present at a final concentration of 2% for all experiments. After electrophoresis, proteins were transferred to nitrocellulose membranes by electrophoresis at 100 V for 1 h. Membranes were then stained with Ponceau S for 5 min for detection of transferred proteins. Subsequently, Ponceau stain was removed by brief washing with transfer buffer, and membranes were blocked for 30 min by incubation with 1% nonfat dry milk (NFDM) in 1× phosphate-buffered saline (PBS). Primary antibodies were incubated as follows. For detection of HDAg, rabbit anti-HDAg (α-HDAg) (30) was diluted 1:1,000 in 1% NFDM-1× PBS. For detection of hemagglutinin (HA)-tagged proteins, mouse monoclonal α-HA antibody (Babco) was diluted 1:400 in 1% NFDM-1× PBS. Blots were incubated with primary antibody overnight (16 to 18 h) in all cases. Incubation with primary antibody was followed by one wash for 15 min and three washes for 5 min each with 25 to 50 ml of 1× PBS. Secondary antibodies were incubated as follows. For blots incubated with α-HA primary antibody, 125I-labeled α-mouse IgG (80 μCi/ml; Perkin Elmer/NEN) was diluted 1:400 in 1% NFDM-1× PBS. For blots incubated with α-HDAg primary antibody, 125I-protein A (100 μCi/ml; Perkin Elmer/NEN) was diluted 1:1,000 in 1% NFDM-1× PBS. Membranes were incubated with secondary antibody or 125I-protein A for 1 to 2 h, washed as described above, and exposed to a Molecular Dynamics phosphorimager screen or to Kodak BioMax MS film for several hours or overnight. Quantitation of results was performed with a Molecular Dynamics Storm phosphorimager and the Molecular Dynamics ImageQuant software package.
Electrophoresis and Northern blotting.
Total cellular RNA was harvested from the same samples used for Western blotting when appropriate. RNA was isolated as follows. A 1/2 volume of a solution of 5 M potassium acetate-3 M acetic acid was added to an aliquot of the lysate, which was then vortexed and spun at full speed in a microcentrifuge for 5 min. The supernatant was placed in a clean microcentrifuge tube, and 1 volume of phenol was added. After mixing to obtain a single phase, 1/6 volume of chloroform was added. After vortexing and centrifugation as described above, the resulting supernatant was transferred to a new microcentrifuge tube, and RNA was precipitated with 1 volume of 2-propanol. RNA was pelleted by spinning in a microcentrifuge at full speed for 10 min, and pellets were dissolved in RNase-free H2O. In order to remove chromosomal DNA, the samples were treated with DNase RQ1 (Promega) for 30 min at 37°C. Loading buffer containing 96% formamide, 10 mM EDTA, and 0.1 mg each of xylene cyanol and bromphenol blue per ml was added to the RNA samples, which were loaded on 1.5% agarose gels made with 0.5× Tris-borate-EDTA containing 0.5 mg of ethidium bromide per ml. Electrophoresis was performed in 0.5× Tris-borate-EDTA-ethidium bromide at 100 V until the dye front reached the end of the gel. RNA was then transferred electrophoretically to Zeta-Probe membrane (Bio-Rad) for 1.5 h at 100 V and was subsequently cross-linked to the membrane by UV irradiation in a UV cross-linker (Fisher Scientific model FB-UVXL-1000), using the optimal cross-link setting. Prehybridization was performed with shaking at 55°C in NorthernMax PreHyb/Hyb buffer (Ambion) for approximately 2 h, and hybridization was performed overnight (16 to 18 h) with shaking, under the same temperature and buffer conditions. The riboprobe was made by in vitro transcription with the Ecl136II-linearized template pDL538 (18) in the presence of [32P]UTP (Perkin-Elmer/NEN).
Dominant negative inhibition assays.
The ability of HDAg-L variants to inhibit HDV replication was measured as previously described (19). Briefly, equal amounts of HDAg-L variant- and HDAg-S-expressing vectors were cotransfected together with a vector that expresses 1.1-unit-length HDV antigenomic RNA that contains a frameshift in the delta antigen open reading frame (17). At 6 days posttransfection, samples were harvested and subjected to Northern analysis with a radiolabeled probe complementary to the HDV genome.
Farnesylation assay.
At 3 days after transfection of HDAg expression vectors into HuH7 cells in six-well tissue culture dishes, cells were harvested in 300 μl of Laemmli buffer lacking β-ME. Samples were heated to 95°C for 5 min and were then passed 15 times through a 1-ml tuberculin syringe to shear chromosomal DNA. Sheared samples were then spun in a microcentrifuge for 5 min at maximum speed to pellet chromosomal DNA. A 10-μl aliquot of the cleared lysate was placed in a clean 1.5-ml microcentrifuge tube and was treated with 2.5 μl of 100 mM dithiothreitol for 5 min at room temperature to reduce disulfide bonds, which may have formed in the oxidative environment encountered during lysis. Following dithiothreitol treatment, 80 μl of 30 mM polyethylene glycol 5000-maleimide (PEG-mal) (Nektar Therapeutics) was added, and the reaction was allowed to proceed at 37°C for 1 h. Prior to use, the 30 mM PEG-mal solution was dialyzed overnight at 4°C in 3,500-kDa-cutoff dialysis tubing against 500 volumes of pH 6.5 PO4 buffer in order to remove unincorporated maleimide. The pegylation reaction (i.e., the covalent attachment of PEG-mal to free cysteine sulfhydryls [22]) was stopped by addition of 1 μl of β-ME, and 9 volumes of ice-cold 100% acetone were added. Total cellular protein was precipitated overnight at −80°C. Precipitated protein was pelleted by centrifugation for 10 min at full speed in a microcentrifuge, and the pellets were washed with ice-cold 85% acetone at −20°C for 1 h to remove any precipitated SDS. Protein pellets were dried under vacuum and then dissolved in 100 μl of Laemmli buffer containing 2% β-ME. Samples were run on SDS-12% polyacrylamide gels and then subjected to immunoblot analysis.
RESULTS
The carboxyl-terminal region of HDAg-L is responsible for differences in the efficiency of assembly among HDV genotypes. The carboxyl terminus of HDAg-L is thought play a critical role in assembly and incorporation of the associated protein into VLPs, yet the carboxyl-terminal sequences of HDAg-L from the three HDV genotypes share surprisingly little similarity (Fig. 1A) (6). We therefore investigated the importance of the HDAg-L carboxyl-terminal sequence in relation to VLP formation by comparing the efficiencies of incorporation of HDAg-L derived from each of the three HDV genotypes. As is shown in Fig. 1B (lanes 2 to 4), genotype 2 HDAg-L was incorporated into HBsAg-S particles at an efficiency which was 2.5-fold less than that of genotype 1 HDAg-L, while genotype 3 HDAg-L was incorporated at an efficiency which was 12-fold less that of genotype 1 HDAg-L.
Although a carboxyl-terminal extension containing a terminal CXXQ farnesylation motif is present in HDAg-L from each HDV genotype, there is little additional sequence similarity in this region across HDV genotypes. We therefore hypothesized that differences in the amino acid sequence in this region might account for the observed differences in assembly. In order to address this question, vectors that express chimeric HDAg-L proteins were constructed as indicated in Fig. 1B, where the carboxyl-terminal region of genotype 1 HDAg-L was fused with the HDAg-S region of genotype 2 or genotype 3 HDAg-L (2::1 and 3::1) and the genotype 2 and genotype 3 carboxyl termini were fused with the genotype 1 HDAg-S region (1::2 and 1::3). As is shown in Fig. 1C (lanes 5 to 8), the assembly efficiency of each chimeric protein correlated well with the assembly efficiency of its carboxyl-terminal parent. The genotype 1 carboxyl terminus enhanced assembly of genotype 2 and genotype 3 HDAgs, while the genotype 2 and genotype 3 carboxyl-termini inhibited assembly of genotype 1 HDAg. From this result we concluded that genotype-specific differences in the efficiency of VLP incorporation were specified by differences in the 19- to 20-amino-acid sequence present at the carboxyl terminus of HDAg-L.
Genotype 3 HDAg-L is farnesylated at very low levels in HuH7 cells.
Since farnesylation of HDAg-L is required for its incorporation into VLPs, we wondered whether the observed genotype-specific differences in VLP incorporation could be attributed to differences in levels of farnesylation. In order to quantify HDAg-L farnesylation, we developed a novel farnesylation assay, which exploits the ability of a polyethylene glycol-maleimide conjugate to covalently attach to, or pegylate, free cysteine sulfhydryls (22). Since HDAg-L contains only one cysteine, we measured the extent of farnesylation of HDAg-L, as the fraction of HDAg-L unable to react with PEG-mal. Since HDAg-L modified with PEG 5000-mal has a proportionately higher molecular weight, we could quantify the amounts of pegylated and unmodified HDAg-L by performing SDS-PAGE and immunoblotting.
Mutation of the terminal glutamine residue of HDAg-L to a stop codon (Q214Stop) abolishes the HDAg-L CXXQ farnesylation motif, and thus the resulting protein should not be farnesylated (11, 12). HDAg-L(Q214Stop) served as a positive control for PEG-mal incorporation, since 100% of the protein was expected to contain one free cysteine sulfhydryl and therefore all of the protein should become pegylated. Initially, using this protein, we observed variable levels of pegylation that ranged from 85 to 99%. Furthermore, we observed that the extent of HDAg-L(Q214Stop) pegylation varied with the different lots of PEG-mal reagent that were used. Since the PEG-mal reagent can be contaminated with small amounts of maleimide not conjugated to PEG, we hypothesized that when less than 100% pegylation of HDAg-L(Q214Stop) was observed, it resulted from reaction of that protein with unconjugated maleimide. Consistent with this interpretation, when the PEG-mal reagent was first dialyzed to remove free maleimide, nearly 100% of the HDAg-L(Q214Stop) was pegylated (for example, compare the Q214Stop lane in Fig. 2, where undialyzed PEG-mal reagent was used, with the same lanes in Fig. 1C, 3D, and 4C, where dialyzed reagent was used). For this reason, we calculated the extent of farnesylation by normalizing values obtained from different samples in each experiment to the observed amount of HDAg-L(Q214Stop) pegylation in the same experiment, where the value obtained for HDAg-L(Q214Stop) was assumed to represent 0% farnesylation.
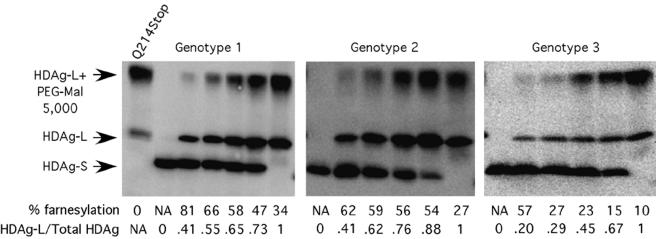
FIG. 2.
Farnesylation of HDAg-L is substantially increased in the presence of HDAg-S. HuH7 cells were transfected with various proportions of HDAg-L and HDAg-S from each of the three HDV genotypes. The HDAg-S/HDAg-L ratio reported was quantified from the actual levels of each protein as determined by immunoblotting and phosphorimager analysis. Farnesylation was then calculated as described for Fig. 1. NA, not applicable.
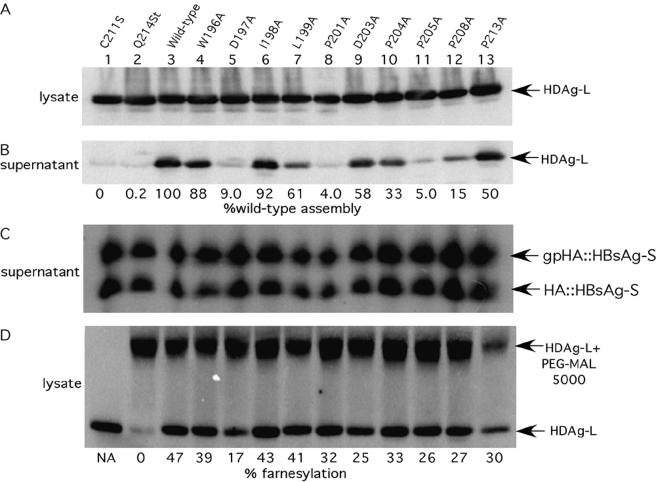
FIG. 3.
Some alanine substitution mutations in the C-terminal region of HDAg-L impair HDAg-L VLP assembly. Shown are immunoblots of proteins from the lysate and supernatant fractions of HuH7 cells transfected with various HDAg-expressing constructs and HBsAg-S::HA. Assembly was quantified as described for Fig. 1, and values were again normalized to take into account sample-to-sample variation in the level of HBsAg-S secretion. Assembly is expressed as the percentage of assembly of wild-type HDAg-L. Supernatant samples were 10 times more concentrated than lysate samples in all assembly experiments unless otherwise noted. Values for HDAg-Lsup are therefore calculated as total signal × 0.1, while values for HDAg-Llys are uncorrected. NA, not applicable.
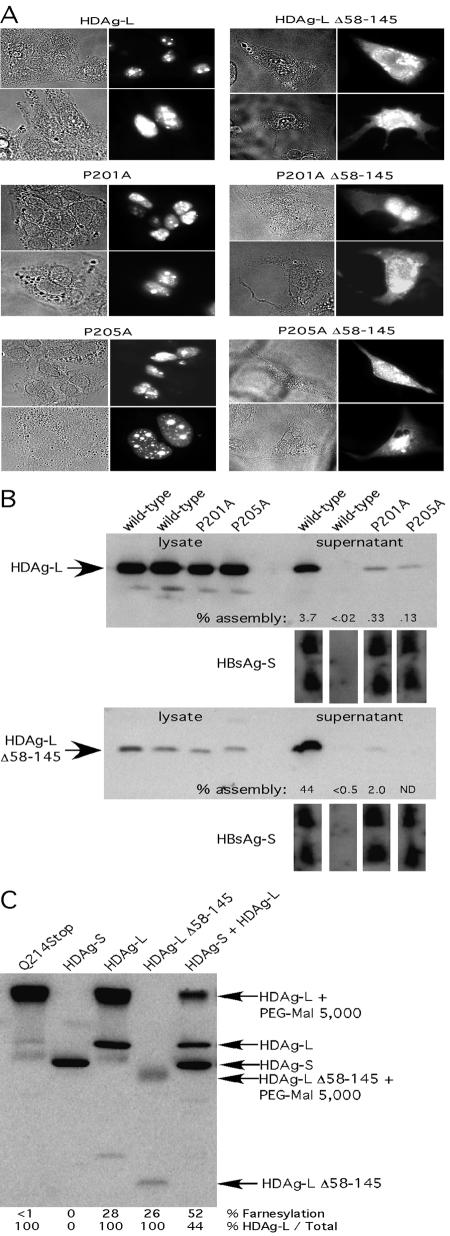
FIG. 4.
VLP formation by assembly-defective mutants is not restored by localization to the cytoplasm. (A) HuH7 cells were transfected with the indicated HDAg construct, and HDAg localization was analyzed by immunofluorescence microscopy. Monolayers were probed with rabbit anti-HDAg antiserum, followed by anti-rabbit tetramethyl rhodamine isocyanate-conjugated antibody. (B) HuH7 cells weretransfected with HDAg-L expression constructs and HBsAg-S::HA as indicated. The top panels show immunoblotting of samples transferred from an SDS-12% polyacrylamide gel, while the bottom panels show immunoblotting of samples from an SDS-15% polyacrylamide gel. Percent assembly was calculated as HDAg-Lsup/(HDAg-Lsup + HDAg-Llys) × 100 and was normalized to take into account sample-to-sample variation in HBsAg-S secretion. (C) HuH7 cells were transfected with HDAg-L expression constructs as indicated, harvested, and then subjected to Western analysis. Farnesylation was calculated as described for Fig. 1.
Mutation of the sole cysteine residue in HDAg-L to serine (C211S) produces a protein that, like HDAg-L(Q214Stop), cannot be modified by farnesylation, but in contrast, HDAg-L(C211S) contains neither cysteines nor free sulfhydryls (11, 12). HDAg-L(C211S) therefore was used as a control to monitor the specificity with which the PEG-mal reagent reacts with cysteines relative to other residues. As expected, no nonspecific reaction of PEG-mal with HDAg-L(C211S) was observed (Fig. 1C, lane 2).
Since the two control proteins behaved as expected, we next determined the extent of pegylation for wild-type HDAg-L for each of the three genotypes. Interestingly, genotype 3 HDAg-L was farnesylated at a much lower level (more than 10-fold lower) than were HDAg-Ls from both genotypes 1 and 2 (Fig. 1C, lanes 3 to 5). A similarly low level of farnesylation was observed with the 1::3 chimera but not with the 3::1 chimera, suggesting that the carboxyl-terminal region of genotype 3 HDAg-L was responsible for the hypofarnesylated phenotype. Since genotype 2 HDAg-L, which incorporates into VLPs poorly compared with genotype 1 HDAg-L, was farnesylated at a level similar to that of genotype 1, it seems unlikely that differences in farnesylation status alone can account for differences in assembly. However, it is likely that the remarkably low level of farnesylation of genotype 3 HDAg-L contributed to the relative inability of this protein to incorporate into HBsAg particles.
Farnesylation of HDAg-L is substantially increased in the presence of HDAg-S.
During early rounds of HDV RNA replication, HDAg-S is present in vast excess relative to HDAg-L, and the HDAg-L/HDAg-S ratio increases as replication progresses and edited HDV antigenomes accumulate within the cell. Since HDAg-L is never present in the absence of HDAg-S during a natural infection, we thought it prudent to ask whether HDAg-S exerts any effect on the farnesylation of HDAg-L. To this end, HuH7 cells were transfected with various proportions of vectors that express HDAg-S and HDAg-L, and farnesylation of HDAg-L was measured for each sample (Fig. 2). Somewhat surprisingly, the level of farnesylation of HDAg-L was significantly higher in the presence of excess HDAg-S for all three genotypes, and farnesylation decreased as the proportion of HDAg-L increased. We concluded that HDAg-S modulates HDAg-L farnesylation, perhaps through a direct interaction with that protein. It is possible that when HDAg-L forms homo-octamers, the carboxyl termini of two proximal HDAg-L subunits interact with each other and thereby hinder the access of the farnesyl transferase. Consistent with this possibility, we recently observed that two unprenylated HDAg-L carboxyl termini within a homo-octamer are in very close proximity and can form a disulfide bond when exposed to a mild oxidizing agent (10). Hence, it is likely that as the proportion of HDAg-S/HDAg-L within an octamer increases, the probability that one HDAg-L carboxyl terminus will be proximal to an HDAg-S rather than to a second HDAg-L also increases. In this model, HDAg-S stimulates farnesylation of HDAg-L by preventing HDAg-L self-association and thereby allowing the farnesyl transferase access to the carboxyl-terminal cysteine.
We next tested whether genotype 3 HDAg-L that was farnesylated at 57% due to the presence of genotype 3 HDAg-S could be incorporated into VLPs more efficiently than genotype 3 HDAg-L expressed in the absence of HDAg-S, where the level of farnesylation was less than 10%. Somewhat surprisingly, even though coexpression of genotype 3 HDAg-S led to increased farnesylation of genotype 3 HDAg-L, it did not stimulate VLP incorporation (data not shown), suggesting that there are impediments in addition to inefficient farnesylation that prevent the efficient incorporation of genotype 3 HDAg-L into VLPs.
Some alanine substitutions in the carboxyl-terminal region of HDAg-L impair HDAg-L VLP assembly but do not impair farnesylation.
In order to study the role in assembly of specific amino acid residues within the HDAg-L carboxyl terminus, we constructed single-amino-acid substitution mutations in 12 of the 19 carboxyl-terminal residues (Fig. 3). Mutation of the seven residues omitted from our analysis was previously shown to have no effect on VLP assembly (20), while the previously described P205A, C211S, and Q214Stop mutants were included in our study because they have been shown to inhibit incorporation into VLPs (11, 12, 20, 26). Furthermore, the P205A mutation was proposed to reside within a putative NES, spanning residues 197 to 210 of HDAg-L (20). Those authors hypothesized that the assembly defect associated with the P205A mutation results from the inability of the mutant protein to be exported from the nucleus and thereby restricts the access of the protein from the site of viral assembly (the endoplasmic reticulum membrane).
Of the 12 mutants tested here, the D197A, P201A, P205A, C211S, and Q214Stop mutants were severely impaired for incorporation into VLPs (Fig. 3B). As seen in Fig. 3D, some mutants were farnesylated at levels somewhat lower than that seen for wild-type HDAg-L; however, the efficiency of farnesylation did not correlate with the efficiency of assembly. For instance, only 25% of HDAg-L(D203A) was farnesylated, yet this protein was incorporated into VLPs with near-wild-type efficiency. In contrast, although both HDAg-L(P201A) and HDAg-L(P205A) were farnesylated more efficiently than was HDAg-L(D203A), the proline mutants were incorporated into VLPs 11- to 14-fold less efficiently than was the HDAg-L(D203A) mutant. We concluded that the assembly defects observed for P201A and P205A did not result from defects in HDAg-L farnesylation.
The same 12 mutants were also tested for their ability to inhibit HDV RNA replication. All of the mutants except Q214Stop and C211S were inhibitors of HDV RNA replication (data not shown). Since proteins bearing Q214Stop and C211S mutations are not farnesylated, this supported the notion that in order to inhibit replication, HDAg-L must be prenylated. Since both the P201A and P205A mutants were able to inhibit replication at wild-type efficiency but were incorporated into VLPs at greatly reduced efficiency, we concluded that the structural requirements for the assembly and replication inhibition functions of HDAg-L are distinct.
The assembly defects of the P201A and P205A mutants were not restored when the proteins were localized to the cytoplasm.
Both P201 and P205 reside in a putative HDAg-L NES, and we reasoned that if defective nuclear export was responsible for the observed assembly defect, relocalization of the mutant protein from the nucleus to the cytoplasm should restore VLP assembly by allowing access to the site of particle formation. In order to test this hypothesis, we utilized a mutant HDAg-L that contained a deletion of residues 58 to 145. This protein lacks both the HDAg nuclear localization signals and RNA binding domains and was previously observed to have localized predominantly to the cytoplasm (D. W. Lazinski, unpublished observation). Both the P201A and P205A mutations were placed in the Δ58-145 background, and in order to test whether the resulting proteins were actually localized to the cytoplasm, they were expressed in HuH7 cells and their intracellular localization was observed via immunofluorescence microscopy. As seen in Fig. 4A, although the full-length proteins (wild-type, P201A, and P205A) were localized almost exclusively to the nuclei of HuH7 cells, a significant fraction of the cognate Δ58-145 proteins were found in the cytoplasm.
We then tested the ability of the Δ58-145 mutants to assemble into HBsAg-S VLPs. As seen in Fig. 4B, while relocalization to the cytoplasm markedly enhanced incorporation of wild-type HDAg-L into VLPs, the assembly defects associated with the P201A and P205A mutants were not ameliorated by such relocalization. Since the assembly defects of the P201A and P205A mutants remained under conditions in which the requirement for an NES was eliminated, we concluded that in addition to any proposed role in nuclear export, these residues likely play a more direct role in HBsAg interaction.
Cytoplasmically localized HDAg-L was not farnesylated to a greater extent than wild-type HDAg-L.
Previous work has shown that HDAg-L shuttles between the nucleus and the cytoplasm (20). We hypothesized that if interaction with HDAg-S were to exert an effect on the kinetics with which HDAg-L shuttles, the amount of time spent by any given HDAg-L molecule in the cytoplasm might be increased. Since the host cell's FTase is located in the cytoplasm, increased time in the cytoplasm could result in increased farnesylation. If increased time in the cytoplasm indeed results in increased farnesylation, then cytoplasmically localized HDAg-L should be farnesylated to a higher level than HDAg-L that localizes to the nucleus. This possibility was tested by comparing the extent of farnesylation of wild-type HDAg-L with that of the Δ58-145 HDAg-L mutant. As shown in Fig. 4C, although a significant fraction of this mutant protein was localized to the cytoplasm, and even though its assembly into VLPs was enhanced about 20-fold versus wild-type HDAg-L, it was farnesylated to a similar extent as wild-type HDAg-L. We concluded that the stimulatory effect of HDAg-S on farnesylation seen in previous experiments did not occur as a consequence of a change in HDAg-L shuttling kinetics.
Myristoylated HDAg-S could be inefficiently assembled into HBsAg-S particles and could efficiently inhibit HDV RNA replication.
Our results here were fully consistent with prior work in that we found that the carboxyl-terminal region unique to HDAg-L played a critical structural role required for the protein's ability to both incorporate into VLPs and inhibit replication. Our results also underscored the importance of farnesylation in relation to both of these abilities. However, neither this work nor past efforts have ruled out the possibility that in addition to its carboxyl terminus, additional residues in the region shared with HDAg-S might also enable HDAg-L to interact with HBsAg. Furthermore, we were intrigued by the requirement for farnesylation. Is farnesylation required merely to increase the affinity of HDAg-L for membranes? If so, then any modification that increases membrane affinity might compensate for a loss of farnesylation. Alternatively, farnesylation of HDAg-L might induce very specific conformational changes in the protein or might enable very precise interactions with another protein. In such a case, other modifications that increase membrane affinity would not compensate for a loss of farnesylation. To address these issues we fused the membrane-targeting myristoylation signal MGARAS, from the HIV-1 matrix protein, to the amino termini of both HDAg-L(C211S) and HDAg-S. The glycine residue of this motif is modified cotranslationally by the covalent addition of a 14-carbon myristoyl moiety. A mutated myristoylation signal, MAARAS, lacking the myristoylated glycine residue, served to control for effects unrelated to myristoylation that resulted from the addition of six amino acids at the HDAg amino terminus.
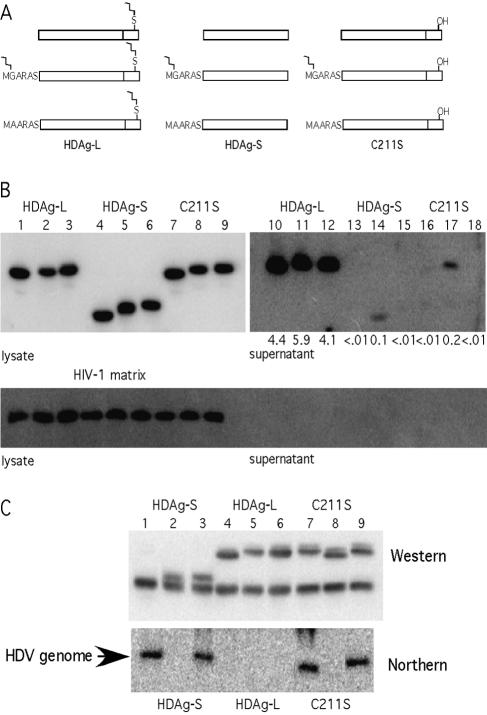
Figure 5A shows schematic diagrams of the proteins expressed in this experiment. To test the ability of these proteins to assemble into HBsAg-S particles, appropriate expression vectors were cotransfected into HuH7 cells with an HBsAg-S expression vector. As an internal control for the specificity of VLP incorporation, each transfection also contained a vector that expressed the myristoylated HIV-1 matrix protein. As shown in Fig. 5B, lanes 14 and 17, myristoylated HDAg-S and myristoylated HDAg-L(C211S) were assembled at low levels into HBsAg-S particles, while nonmyristoylated HDAg-S (lanes 13 and 15), nonmyristoylated HDAg-L(C211S) (lanes 16 and 18), and myristoylated HIV-1 matrix protein (Fig. 5B, lower panel) were not. Since myristoylated HDAg-S could be assembled into VLPs while myristoylated HIV-1 matrix protein could not be, we concluded that residues within the first 195 amino acids of HDAg could interact with HBsAg and thereby facilitate incorporation.
FIG. 5.
Myristoylated HDAg-S and HDAg-L C211S can be assembled into HBsAg-S particles and can inhibit HDV RNA replication. (A) Representations of myristoylated HDAg constructs. HDAg-L, -S, and -L(C211S) were N-terminally modified with either MGARAS (HIV-1 Gag minimal myristoylation signal) or MAARAS (mutated HIV-1 Gag minimal myristoylation signal). (B) Myristoylated HDAg-S and HDAg-L(C211S) can be assembled into HBsAg particles, while their nonmyristoylated counterparts and an unrelated myristoylated protein (HIV-1 p17 MA) cannot be. (C) Myristoylated HDAg-S and HDAg-L(C211S) can inhibit HDV RNA replication.
Although the efficiency of VLP incorporation for myristoylated HDAg-S was more than an order of magnitude lower than that for HDAg-L, it was still more than an order of magnitude higher than that for nonmyristoylated HDAg-S. Hence, when myristoylation was used to provide HDAg-S with a general affinity for membranes, its incorporation into VLPs was stimulated, but not nearly to the extent afforded by farnesylation. Thus, myristoylation could only partially compensate for the absence of farnesylation. We concluded that enhancing membrane association was just one of at least two distinct functions that farnesylation provides for HDAg-L.
Note also that the efficiency of VLP incorporation for myristoylated HDAg-S was nearly the same as that for myristoylated HDAg-L(C211S) even though the latter protein contained the carboxyl-terminal 19 amino acids thought to interact with HBsAg. Apparently, in this nonfarnesylated protein, the carboxyl-terminal 19 amino acids could not function to enhance the interaction with HBsAg. Based on these results, we hypothesized that with respect to assembly, farnesylation of HDAg-L provided a second role beyond increasing the protein's affinity for membranes. It induced a conformational change that activated the carboxyl-terminal 19 amino acids to a form competent for interaction with HBsAg.
We next evaluated the same set of proteins for their ability to inhibit HDV RNA replication. The corresponding expression vectors were cotransfected with an equivalent amount of a vector that expresses HDAg-S together with a vector that initiated HDV replication. At 6 days posttransfection, total RNA samples were harvested and subjected to Northern analysis. When the myristoylated (MGARAS) HDAg proteins were expressed (Fig. 5C), HDV RNA replication was inhibited, while no such inhibition was observed upon expression of the nonmyristoylated (MAARAS) HDAg-S and HDAg-L(C211S) proteins. Since a profound phenotype was observed when MGARAS but not MAARAS was appended to the amino terminus of HDAg-S, we concluded that the appended MGARAS signal was fully functional at directing efficient myristoylation. Consistent with the idea that the MGARAS tag was fully myristoylated, protein with it ran slightly faster than those with the MAARAS tag in SDS-PAGE (compare lane 5 with 6 and lane 8 with 9 in Fig. 5B and lane 2 with 3 and lane 8 with 9 in Fig. 5C). Note also that myristoylated HDAg-S was just as effective as wild-type HDAg-L at inhibiting replication; i.e., myristoylation could fully substitute for farnesylation in this assay. From this we concluded that with respect to the inhibition of replication, farnesylation of HDAg-L provides a single role; it increases the general affinity of the protein for membranes.
DISCUSSION
In this study we devised a novel assay to monitor the farnesylation status of HDAg-L. Moraleda et al. previously developed an alternate assay aimed at the same purpose (24). Both Moraleda et al. and we in the present study used gel electrophoresis to resolve prenylated from unprenylated forms of HDAg-L, so in this sense, both methods are equally direct. The difference between the two methods is that the one used by Moraleda et al. did not involve chemical treatment, while the present study used PEG 5000-maleimide, a reagent that reacts with the sole cysteine residue in unprenylated but not in prenylated HDAg-L. The method used by Moraleda et al. suffers from two main shortcomings. The first problem is that the difference in mobility between the prenylated and unprenylated forms that was observed was quite small. Rather than two well-separated bands, a doublet was observed, and this poor resolution hampered accurate quantification; for example, in Fig. 1C of the earlier paper, signal peaks from the prenylated and unprenylated proteins largely overlap (24). Furthermore, it is not known whether the method of Moraleda et al. will work with forms or variants of HDAg-L that travel through gels with altered mobility compared with that of wild-type HDAg-L, for example, the Δ58-145 mutant. In contrast, with the method developed for the present study, by virtue of the PEG 5000 moiety that is covalently attached to the unprenylated protein, a very substantial difference in the mobility of the unprenylated species was observed. This improved resolution facilitated accurate quantification and enabled the separation of prenylated and unprenylated species for a wide array of HDAg-L variants that differed greatly in their electrophoretic mobility.
A second and more significant problem with the method of Moraleda et al. is that the mechanism that accounts for the slightly slower mobility of prenylated HDAg-L relative to the unprenylated form is not understood. Although the process of farnesylation covalently joins a 15-carbon group to HDAg-L, it also removes the three carboxyl-terminal amino acids, and the net effect is a loss in molecular weight. Hence, farnesylated HDAg-L is expected to run faster, rather than slower, than the unfarnesylated form. In their discussion, Moraleda et al. concede this point, stating “This was in contrast to our expectation based on studies with other modified proteins, where farnesylated proteins migrate somewhat faster than the unmodified protein” (24). Bichko et al. showed that farnesylated HDAg-L is hyperphosphorylated relative to HDAg-L(Cys211Ala), a protein that is not farnesylated (2). Perhaps farnesylation of HDAg-L can target that protein to membranes, where it is subsequently phosphorylated, and it is this altered phosphorylation state that accounts for the slightly slower mobility observed. Moraleda et al. conceded this point as well, stating “It is possible that this discrepancy for Li is due to a difference in the levels of phosphorylation between L and Li” (24). If this is correct, then the method of Moraleda et al. represents an indirect measurement of farnesylation, as it would directly measure the status of phosphorylation. This would be a serious problem because it is not known whether farnesylated HDAg-L is hyperphosphorylated in the same manner and to the same extent under all conditions, for example, early versus late during replication or in the presence versus absence of HDAg-S. Furthermore, deletion mutants of HDAg-L that lack some of the phosphorylation sites but still contain the farnesylation site would likely not be resolved as a doublet. Since 1999, no other publication has used the method of Moraleda et al., and there has been no additional work to resolve the issue of just what change in HDAg-L accounts for the slower mobility of the farnesylated form.
In contrast to the method of Moraleda et al., the assay developed in the present study behaves entirely as expected on theoretical grounds. Maleimide is known to react only with free sulfhydryls, and consistent with this, HDAg-L(Cys211Ser), which lacks sulfhydryls, was not modified; the unprenylated protein HDAg-L(Gln214Stop) was completely modified; and HDAg-L, which is partially farnesylated, was partially modified. Furthermore, since the attachment of PEG 5000 greatly retards the mobility of the associated protein, there is no possibility that subtle changes in mobility that might result from other forms of modification could be misinterpreted as resulting from prenylation. Therefore, the method developed for this study directly accesses whether the Cys211 residue is modified or is not modified.
A cautionary note concerning the interpretation of HDAg-L species that are resolved as doublets is exemplified in Fig. 5. All of the HDAg-L bands in Fig. 5C, lanes 4 to 9, were resolved as doublets with fainter slower-migrating portions and darker faster-migrating portions. This included HDAg-L that was neither myristoylated nor farnesylated, HDAg-L that was myristoylated but not farnesylated, HDAg-L that was not myristoylated but was farnesylated, and HDAg-L that was both myristoylated and farnesylated. Hence, the appearance of doublets on this gel did not reflect the farnesylation status of the associated protein. We are not sure of the significance of the doublets, although we suspect that they result from different degrees of phosphorylation in HDAg-L. The issue has been difficult to resolve because we have had difficulty in reproducibly resolving the doublets. Nevertheless, these doublets illustrate another potential danger of attempting to quantify the extent of farnesylation by the method of Moraleda et al. and provide a third argument in support of the use of the PEG 5000-maleimide method developed in this study.
Among the three distinct HDV genotypes that have been identified, significant differences in the severity of disease observed in infected patients have been reported (6, 7, 13). In general, genotype 3 appears to be associated with the most severe disease course, while genotype 2 seems to be associated the least severe outcomes (6, 7, 13). A previous study comparing the efficiency of HDAg-L incorporation into VLPs for genotypes 1 and 2, but not genotype 3, proposed a direct correlation between the low assembly efficiency of HDV genotype 2 and its low virulence relative to genotype 1 (13). Here were compared the efficiency of assembly for all three HDV genotypes. In agreement with the prior report, we observed that genotype 2 HDAg-L was incorporated into VLPs 2.5-fold less efficiently than was genotype 1 HDAg-L. However, we also observed that genotype 3 HDAg-L was incorporated into VLPs 12-fold less efficiently than was genotype 1 HDAg-L. Given that genotype 3 is thought to be the most virulent HDV genotype, there was no direct correlation observed between the efficiency of VLP assembly and the reported severity of the associated disease.
We observed that genotypic differences in the efficiency of HDAg-L incorporation into VLPs were specified by the carboxyl-terminal sequences of these proteins. Hence, chimeras composed of the first 195 amino acids from genotype 2 or 3 but having the final 19 amino acids from genotype 1 (2::1 and 3::1) were incorporated with an efficiency similar to that of genotype 1 HDAg-L. Similarly, a chimera composed of the first 195 amino acids from genotype 1 but having the final 20 amino acids from genotype 3 (1::3) was incorporated with an efficiency similar to that of genotype 3 HDAg-L.
We wondered whether differences in the extent of farnesylation could account for the observed genotypic differences in the efficiency of HDAg-L incorporation. We found that genotype 2 HDAg-L is farnesylated to a similar extent as genotype 1 HDAg-L. In contrast, we observed that genotype 3 HDAg-L was farnesylated to a much lower extent than were HDAg-Ls from the other two genotypes. The local carboxyl-terminal sequence or structure was responsible for hypofarnesylation, as the 1::3 chimera was also hypofarnesylated.
Since in nature HDAg-L is never expressed in the absence of HDAg-S, we investigated whether coexpression of the latter protein could stimulate farnesylation of the former. Indeed, with all three genotypes we observed that HDAg-S could stimulate farnesylation of HDAg-L. We also observed that wild-type genotype 1 HDAg-L, which localizes to the nucleus, and a mutant genotype 1 HDAg-L that localizes to the cytoplasm were farnesylated to the same extent. Since the localization of HDAg-L does not affect its extent of farnesylation, HDAg-S must stimulate farnesylation via some mechanism unrelated to localization. One possible model is that HDAg-L might exist in dual conformations. In the functional conformation the CXXQ motif is exposed and accessible to the FTase, while in the nonfunctional conformation the CXXQ motif is buried and inaccessible to the FTase. Such a model would account for the partial farnesylation phenotype that we observed with HDAg-Ls from all three genotypes. Perhaps the genotype 3 HDAg-L carboxyl terminus has a greater propensity for being “buried” compared with that of genotype 1 protein. Furthermore, since HDAg-L expression is constrained by the mechanics of the HDV replication cycle to occur only in the presence of HDAg-S, it is possible that as an evolutionary consequence, HDAg-L functions optimally only when HDAg-S is also present. Such a scenario would account for the dramatic increase in farnesylation that was observed when HDAg-S and HDAg-L were coexpressed. By this model, HDAg-L would be stabilized in its functional conformation by interaction with HDAg-S and would function at a suboptimal level when HDAg-S is absent.
Even in the presence of genotype 3 HDAg-S, where the farnesylation of genotype 3 HDAg-L was efficient, the latter protein could still not be efficiently incorporated into VLPs. However, the inability of genotype 3 HDAg-L to be incorporated into VLPs does not reflect a general defect in assembly associated with this protein, as Bordier et al. observed efficient assembly of replicating genotype 3 RNPs into the same HBsAg particles used in this study (3). Perhaps the carboxyl-terminal residues of genotype 3 HDAg-L are properly exposed only when this protein is incorporated into an RNP. There is precedence for such a concept. Like genotype 3 HDAg-L, the hepatitis B virus core protein is unable to incorporate into enveloped particles in the absence of viral nucleic acid (31).
We also screened a panel of 12 alanine substitutions within the genotype 1 HDAg-L carboxyl terminus for their ability to abolish incorporation into VLPs. While some mutations, such as C211S and Q214Stop, impaired HDAg-L function by preventing farnesylation, others, such as D197A, P201A, and P205A, affected HDAg-L function differently. The assembly defects associated with mutation of D197 and P201 had not been previously described, and it is notable that P201 is the only residue in the carboxyl-terminal region, aside from those with clearly defined functions (i.e., W196 is required for amber/W editing, C211 is required for farnesyl addition, and Q214 is required for specifying farnesylation), that is evolutionarily conserved in every HDV isolate. Here, we observed that the dramatic assembly defects associated with the P201A and P205A mutations did not result from impaired farnesylation and persisted even when the mutants were localized to the cytoplasm. Although these two mutations nearly abolished the ability of HDAg-L to incorporate into VLPs, they had no affect on the protein's ability to inhibit HDV RNA replication. Hence, there are different structural requirements related to the two functions of HDAg-L.
It was somewhat surprising to find that myristoylated (MGARAS) HDAg-S could inhibit HDV RNA replication as potently as wild-type HDAg-L while the nonmyristoylated (MAARAS) derivative of this protein could not. Apparently, any HDAg protein capable of both incorporating into HDV RNPs and associating with membranes can potently interfere with RNA replication. This requirement for membrane association is consistent with the recent finding that small molecules that specifically inhibit farnesyl transferase activity also abolish the ability of HDAg-L to inhibit replication (29). We hypothesize that when one or several HDAg molecules competent for membrane association are incorporated into an HDV RNP, the resulting RNP then associates with the nuclear membrane and is sequestered from the site(s) where active transcription and replication occur.
We also observed that myristoylated (MGARAS) HDAg-S could be incorporated into VLPs while myristoylated HIV-1 matrix protein could not be, despite the fact that as a cytoplasmic protein, the HIV matrix protein should have preferential access to the site of VLP formation. From this result we concluded that residues present in HDAg-S facilitate VLP incorporation, most likely through a direct interaction with HBsAg. Furthermore, myristoylated HDAg-S could be incorporated into VLPs far more efficiently (at least 10-fold) than could nonmyristoylated (MAARAS) HDAg-S. From this result we concluded that the ability to associate with membranes enhances HDAg incorporation into VLPs. However, with respect to VLP incorporation, myristoylation could not fully compensate for the absence of farnesylation, and myristoylated HDAg-S was incorporated more than 10-fold less efficiently than was HDAg-L. This result suggests that while membrane anchoring is necessary and sufficient to enable the inhibition of HDV RNA replication, it is not sufficient to promote wild-type levels of assembly.
It is interesting that the phenotypes associated with myristoylated HDAg-S were indistinguishable from those associated with the HDAg-L(P201A) and HDAg-L(P205A) mutants. All three proteins could potently inhibit HDV RNA replication, yet all were incorporated into VLPs 10- to 25-fold less efficiently than was wild-type HDAg-L. This observation can be explained by a model in which farnesylation stabilizes a protein conformation necessary for presentation of important contact points in the carboxyl-terminal region to HBsAg-L that are needed for the efficient interaction with HBsAg. Such a model would explain why the incorporation of myristoylated HDAg-L(C211S) into VLPs was not significantly different from that of myristoylated HDAg-S. Although the former protein contains an intact carboxyl-terminal domain (with the exception of a very conservative cysteine-to-serine substitution), if farnesylation is required for correct presentation of the carboxyl-terminal domain, then the HDAg-L(C211S) protein would be incapable of presenting this domain and would behave like myristoylated HDAg-S, a protein that lacks the domain altogether. Such a model is also consistent with the observation that alanine substitution at positions D197, P201, and P205 inhibited VLP assembly even though farnesylation was not significantly affected. If these residues either directly contact HBsAg or are required to attain the proper structure that presents the HBsAg-interacting domain, then their conversion to alanine would inhibit HBsAg interaction even in the context of a properly farnesylated protein. We therefore conclude that farnesylation of HDAg-L stimulates HBsAg interaction both by targeting HDAg-L to the membrane and by inducing a conformational change that enables presentation of the HBsAg-interacting domain.
Acknowledgments
We thank John Coffin (Tufts University), Ralph Isberg (Tufts University), and Naomi Rosenberg (Tufts University) for helpful discussions and John Casey (Georgetown University) for providing constructs that contained genotype 2 and 3 HDAgs.
This work was supported by grant R01-AI40472 from the National Institutes of Health and by the Raymond and Beverly Sackler Research Foundation.
REFERENCES
- 1.Berger, J., J. Hauber, R. Hauber, R. Geiger, and B. R. Cullen. 1988. Secreted placental alkaline phosphatase: a powerful new quantitative indicator of gene expression in eukaryotic cells. Gene 66:1-10. [DOI] [PubMed] [Google Scholar]
- 2.Bichko, V., S. Barik, and J. Taylor. 1997. Phosphorylation of the hepatitis delta virus antigens. J. Virol. 71:512-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bordier, B. B., P. L. Marion, K. Ohashi, M. A. Kay, H. B. Greenberg, J. L. Casey, and J. S. Glenn. 2002. A prenylation inhibitor prevents production of infectious hepatitis delta virus particles. J. Virol. 76:10465-10472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bordier, B. B., J. Ohkanda, P. Liu, S. Y. Lee, F. H. Salazar, P. L. Marion, K. Ohashi, L. Meuse, M. A. Kay, J. L. Casey, S. M. Sebti, A. D. Hamilton, and J. S. Glenn. 2003. In vivo antiviral efficacy of prenylation inhibitors against hepatitis delta virus. J. Clin. Investig. 112:407-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casey, J. L. 2002. RNA editing in hepatitis delta virus genotype III requires a branched double-hairpin RNA structure. J. Virol. 76:7385-7397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casey, J. L., T. L. Brown, E. J. Colan, F. S. Wignall, and J. L. Gerin. 1993. A genotype of hepatitis D virus that occurs in northern South America. Proc. Natl. Acad. Sci. USA 90:9016-9020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casey, J. L., G. A. Niro, R. E. Engle, A. Vega, H. Gomez, M. McCarthy, D. M. Watts, K. C. Hyams, and J. L. Gerin. 1996. Hepatitis B virus (HBV)/hepatitis D virus (HDV) coinfection in outbreaks of acute hepatitis in the Peruvian Amazon basin: the roles of HDV genotype III and HBV genotype F. J. Infect. Dis. 174:920-926. [DOI] [PubMed] [Google Scholar]
- 8.Chang, F. L., P. J. Chen, S. J. Tu, C. J. Wang, and D. S. Chen. 1991. The large form of hepatitis delta antigen is crucial for assembly of hepatitis delta virus. Proc. Natl. Acad. Sci. USA 88:8490-8494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chao, M., S. Y. Hsieh, and J. Taylor. 1990. Role of two forms of hepatitis delta virus antigen: evidence for a mechanism of self-limiting genome replication. J. Virol. 64:5066-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cornillez-Ty, C. T., and D. W. Lazinski. 2003. Determination of the multimerization state of the hepatitis delta virus antigens in vivo. J. Virol. 77:10314-10326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glenn, J. S., J. A. Watson, C. M. Havel, and J. M. White. 1992. Identification of a prenylation site in delta virus large antigen. Science 256:1331-1333. [DOI] [PubMed] [Google Scholar]
- 12.Glenn, J. S., and J. M. White. 1991. trans-dominant inhibition of human hepatitis delta virus genome replication. J. Virol. 65:2357-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsu, S. C., W. J. Syu, I. J. Sheen, H. T. Liu, K. S. Jeng, and J. C. Wu. 2002. Varied assembly and RNA editing efficiencies between genotypes I and II hepatitis D virus and their implications. Hepatology 35:665-672. [DOI] [PubMed] [Google Scholar]
- 14.Hwang, S. B., and M. M. Lai. 1994. Isoprenylation masks a conformational epitope and enhances trans-dominant inhibitory function of the large hepatitis delta antigen. J. Virol. 68:2958-2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuo, M. Y., M. Chao, and J. Taylor. 1989. Initiation of replication of the human hepatitis delta virus genome from cloned DNA: role of delta antigen. J. Virol. 63:1945-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuo, M. Y., J. Goldberg, L. Coates, W. Mason, J. Gerin, and J. Taylor. 1988. Molecular cloning of hepatitis delta virus RNA from an infected woodchuck liver: sequence, structure, and applications. J. Virol. 62:1855-1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lazinski, D. W., and J. M. Taylor. 1994. Expression of hepatitis delta virus RNA deletions: cis and trans requirements for self-cleavage, ligation, and RNA packaging. J. Virol. 68:2879-2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lazinski, D. W., and J. M. Taylor. 1994. Recent developments in hepatitis delta virus research. Adv. Virus Res. 43:187-231. [DOI] [PubMed] [Google Scholar]
- 19.Lazinski, D. W., and J. M. Taylor. 1993. Relating structure to function in the hepatitis delta virus antigen. J. Virol. 67:2672-2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee, C. H., S. C. Chang, C. H. Wu, and M. F. Chang. 2001. A novel chromosome region maintenance 1-independent nuclear export signal of the large form of hepatitis delta antigen that is required for the viral assembly. J. Biol. Chem. 276:8142-8148. [DOI] [PubMed] [Google Scholar]
- 21.Lee, C. Z., P. J. Chen, M. M. Lai, and D. S. Chen. 1994. Isoprenylation of large hepatitis delta antigen is necessary but not sufficient for hepatitis delta virus assembly. Virology 199:169-175. [DOI] [PubMed] [Google Scholar]
- 22.Lu, J., and C. Deutsch. 2001. Pegylation: a method for assessing topological accessibilities in Kv1.3. Biochemistry 40:13288-13301. [DOI] [PubMed] [Google Scholar]
- 23.Luo, G. X., M. Chao, S. Y. Hsieh, C. Sureau, K. Nishikura, and J. Taylor. 1990. A specific base transition occurs on replicating hepatitis delta virus RNA. J. Virol. 64:1021-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moraleda, G., S. Seeholzer, V. Bichko, R. Dunbrack, J. Otto, and J. Taylor. 1999. Unique properties of the large antigen of hepatitis delta virus. J. Virol. 73:7147-7152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mu, J. J., D. S. Chen, and P. J. Chen. 2001. The conserved serine 177 in the delta antigen of hepatitis delta virus is one putative phosphorylation site and is required for efficient viral RNA replication. J. Virol. 75:9087-9095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Otto, J. C., and P. J. Casey. 1996. The hepatitis delta virus large antigen is farnesylated both in vitro and in animal cells. J. Biol. Chem. 271:4569-4572. [DOI] [PubMed] [Google Scholar]
- 27.Rizzetto, M., M. G. Canese, S. Arico, O. Crivelli, C. Trepo, F. Bonino, and G. Verme. 1977. Immunofluorescence detection of new antigen-antibody system (delta/anti-delta) associated to hepatitis B virus in liver and in serum of HBsAg carriers. Gut 18:997-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rizzetto, M., B. Hoyer, M. G. Canese, J. W. Shih, R. H. Purcell, and J. L. Gerin. 1980. Delta agent: association of delta antigen with hepatitis B surface antigen and RNA in serum of delta-infected chimpanzees. Proc. Natl. Acad. Sci. USA 77:6124-6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sato, S., C. T. Cornillez-Ty, and D. W. Lazinski. 2004. By inhibiting replication, the large hepatitis delta antigen can indirectly regulate amber/W editing and its own expression. J. Virol. 78:8120-8134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sato, S., S. K. Wong, and D. W. Lazinski. 2001. Hepatitis delta virus minimal substrates competent for editing by ADAR1 and ADAR2. J. Virol. 75:8547-8555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seeger, C., and W. S. Mason. 2000. Hepatitis B virus biology. Microbiol. Mol. Biol. Rev. 64:51-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang, C. J., P. J. Chen, J. C. Wu, D. Patel, and D. S. Chen. 1991. Small-form hepatitis B surface antigen is sufficient to help in the assembly of hepatitis delta virus-like particles. J. Virol. 65:6630-6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weiner, A. J., Q. L. Choo, K. S. Wang, S. Govindarajan, A. G. Redeker, J. L. Gerin, and M. Houghton. 1988. A single antigenomic open reading frame of the hepatitis delta virus encodes the epitope(s) of both hepatitis delta antigen polypeptides p24 delta and p27 delta. J. Virol. 62:594-599. [DOI] [PMC free article] [PubMed] [Google Scholar]