Abstract
Two acidic domains of the Potato leafroll virus (PLRV) coat protein, separated by 55 amino acids and predicted to be adjacent surface features on the virion, were the focus of a mutational analysis. Eleven site-directed mutants were generated from a cloned infectious cDNA of PLRV and delivered to plants by Agrobacterium-mediated mechanical inoculation. Alanine substitutions of any of the three amino acids of the sequence EWH (amino acids 170 to 172) or of D177 disrupted the ability of the coat protein to assemble stable particles and the ability of the viral RNA to move systemically in four host plant species. Alanine substitution of E109, D173, or E176 reduced the accumulation of virus in agrobacterium-infiltrated tissues, the efficiency of systemic infection, and the efficiency of aphid transmission relative to wild-type virus, but the mutations did not affect virion stability. A structural model of the PLRV capsid predicted that the amino acids critical for virion assembly were located within a depression at the center of a coat protein trimer. The other amino acids that affected plant infection and/or aphid transmission were predicted to be located around the perimeter of the depression. PLRV virions play key roles in phloem-limited virus movement in plant hosts as well as in transport and persistence in the aphid vectors. These results identified amino acid residues in a surface-oriented loop of the coat protein that are critical for virus assembly and stability, systemic infection of plants, and movement of virus through aphid vectors.
Members of the family Luteoviridae are icosahedral viruses with small (≈6-kb) RNA genomes that infect phloem-associated tissues of their plant hosts (29). They are transmitted between plants by aphids in a circulative, persistent manner. Although the virus can survive for extended periods in often hostile environments in the aphid, the virus does not replicate in the insect (11). Features of the virion regulate local and systemic movement of the virus in plant hosts and regulate the recognition, transport, and persistence of virus in aphid tissues, yet little is known about the specific properties and biologically active domains of the luteovirus particle.
The icosahedral virions of members of the Luteoviridae are composed of two structural proteins, the major 22- to 24-kDa capsid protein (CP) encoded by open reading frame (ORF) 3, and a minor species referred to as the readthrough protein (RT) (17). The RT, encoded by ORF 3 and the downstream adjacent ORF 5, is translated by occasional suppression of the CP termination codon (16). The virion structure has not been resolved for any member of the Luteoviridae, but the capsids are thought to be assembled from 180 subunits according to T = 3 quasi-symmetry. A variable but minor number of the 180 capsids are RT subunits incorporated into the virion via their CP moiety (16), although sequences in the RT domain regulate the incorporation (24). The ≈50-kDa RT domain encoded by ORF 5 is believed to protrude from the surface of the virion. Virions can be assembled from the CP alone, but virions without the RT are less efficient in systemic infection of plant hosts and cannot be transmitted by their aphid vectors (4, 6, 23).
Based on known structures and structural protein sequences of distantly related icosahedral RNA viruses, the CP can be divided into an N-terminal arginine-rich domain and the shell domain (7). The charged arginine-rich domain is likely found to be internal and interacting with the viral RNA. The shell domain would make up the core of the capsid. The shell domain three-dimensional structure is highly conserved among plant and animal viruses and consists of an eight-stranded β-barrel often referred to as a jelly roll configuration (7).
There is limited information on the structure of poleroviruses. Potato leafroll virus (PLRV)-specific monoclonal antibody binding mapped three surface-exposed domains on the virion structure (32). Based on sequence alignments with other icoahedral viruses whose structures were known, two of the surface domains were located on loops known to be important antigenic sites on structurally related viruses. More recently, independent structure models were developed for the CP of two members of the genus Polerovirus in the Luteoviridae, PLRV and Beet western yellows virus (BWYV) (2, 31). The models support the data from the epitope-mapping studies and locate the surface epitopes on the loops between β-strands G and H and between β-strands B and C. Although the best-fit structure models for PLRV and BWYV both locate epitope 10 (32) on the G-H loop, the structural features of the loop differed between these closely related viruses (2).
Partially contained within this epitope and extending a few residues upstream is one domain of an acidic patch region (31). This acidic patch domain contains four acidic amino acid residues within 12 conserved amino acid residues of the G-H loop (Fig. 1). E170 is highly conserved among most members of the Luteoviridae sequenced except for the RMV strain of Barley yellow dwarf virus (BYDV RMV), where there is an E-to-V substitution. Position 173 is a highly conserved acidic residue, usually D in poleroviruses and BYDV RMV and E in luteoviruses. E176 is highly conserved among the poleroviruses and BYDV RMV but replaced by V or I in other luteoviruses. D177 is highly conserved in most members of the Luteoviridae, although replaced by the amide N in some luteoviruses. A portion of this domain, residues HDSSEDQ, was defined as an epitope exposed at the surface of virions (32).
FIG. 1.
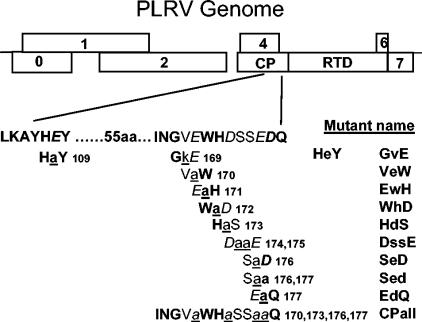
Genome organization of potato leafroll virus (PLRV) and description of the CP mutants. The wild-type amino acid sequence of the two domains of the acidic patch are provided in the single-letter code. The domains are separated by 55 amino acids. Bold letters indicate residues that are conserved among all poleroviruses whose sequence is known. The five acidic amino acids (D or E) are in italics. The 11 single and multiple amino acid mutants are defined under the wild-type sequence. The amino acid changes (all alanine substitutions except one) are underlined and are shown in the center of the three- or four-letter mutant designations. The numbers indicate the amino acid residue position within the PLRV CP. Note that the upstream domain overlaps the virus movement protein (P17) encoded by ORF 4, whereas the downstream domain does not.
A second acidic patch domain is located 55 amino acids upstream in the PLRV sequence. The upstream acidic residue (E109) lies within a stretch of seven amino acids (LKAYHEY) that is highly conserved in the CP among all members of the Luteoviridae with the exception of two residues. In many isolates of the Luteovirus genus there is an E-to-R substitution (acidic to basic) and a conserved A-to-L substitution. According to the PLRV model, these two domains would be brought into close proximity at the center of a CP trimer when the virion is assembled (31).
Here we report a comprehensive mutational analysis of this conformational acidic patch domain and define the amino acids that are critical to virion assembly and stability, host-dependent systemic infection, and aphid transmission. Our findings differ somewhat from those of a report on mutations within the downstream acidic patch domain of BWYV (2) and suggest that the two related poleroviruses are dissimilar in CP monomer structure and interactions.
MATERIALS AND METHODS
Generation of recombinant cDNA PLRV CP constructs.
Plasmid pBPY was described previously (15). Briefly, this plasmid contains a full-length, infectious cDNA clone of PLRV ligated into plasmid pBIN19 to produce pBNUP110 for use in agrobacterial infection and transformation (8, 21). A DNA fragment containing the yeast 2μm plasmid DNA plus the trp gene selectable marker was subsequently introduced into the transferred DNA of pBNUP110 close to the left border. This yielded plasmid pBPY, which was able to replicate in Saccharomyces cerevisiae (15).
Site-directed mutations were generated by primer extension mutagenesis with pBPY as a template and oligonucleotides PLRV5′p3438 (corresponding to PLRV nucleotides 3438 to 3461) and PLRV3′p4382 (complementary to PLRV nucleotides 4382 to 4402) as the external primers. The sequences of the partially overlapping forward and reverse mutagenic primers are available on request. The resulting PCR fragments were inserted into linearized pBPY by homologous recombination in S. cerevisiae (15). The 11 mutants constructed for this study are shown in Fig. 1. The CP of the various mutants was sequenced to verify that each contained only the desired changes. The resulting plasmids were introduced into Agrobacterium tumefaciens strain LBA4404 and used for plant infection.
Agrobacterium-mediated infection of plants.
Two different inoculation techniques were used depending on the assay. Plants were either inoculated with agrobacteria to produce a phloem-limited systemic infection or infiltrated with agrobacteria to produce a localized infection of mesophyll tissue. The latter method generates a nearly synchronous infection of large numbers of cells, allowing easier detection and analysis of transcription and translation products. For inoculation of agrobacteria, A. tumefaciens carrying the desired plasmid was grown in YEB broth with kanamycin (50 μg/ml) for 48 h at 25°C. The culture was centrifuged and resuspended in 1/10th volume of water and used for infection of Nicotiana benthamiana, Nicotiana clevelandii, Physalis floridana, and Solanum tuberosum plantlets that were generally at the five- to six-leaf stage (21). Approximately 40 μl of Agrobacterium suspension was injected with a Hamilton syringe into the midrib or petioles of leaves. Inoculated plants were placed in a growth room (20°C, continuous light) for 7 days prior to being transferred to a greenhouse. Systemically infected plants were identified 3 to 4 weeks postinoculation by double-antibody sandwich enzyme-linked immunosorbent assay (DAS-ELISA) (14).
For infiltration with agrobacteria, A. tumefaciens cultures were grown as above. The cells were resuspended to an optical density at 600 nm of 0.3 to 0.4 in water and infiltrated into fully expanded N. benthamiana leaves with a 5-ml syringe barrel. Plants were kept in a growth room (20°C, continuous light) for 5 to 6 days, and the infiltrated tissues were collected for analysis. Virus was rarely detected outside of the infiltrated areas, although occasionally a plant would develop a phloem-limited systemic infection.
Analysis of CP mutants.
The translation and accumulation of CP and RT were measured in samples of 0.1 to 0.2 g of agrobacterium-infiltrated tissue by Western blotting as described by Nurkiyanova et al. (21). Tissue was disrupted in 250 μl of 2X sodium dodecyl sulfate sample buffer (27) and heated to 95°C for 5 min.
To identify if the CP mutants were able to assemble stable virions, ELISA and an RNase sensitivity assay were used. For ELISA, 0.1 to 0.2 g of agrobacterium-infiltrated tissue was disrupted in 500 μl of 1X phosphate-buffered saline (pH 7.2) and tested by either DAS-ELISA (14) or triple antibody sandwich (TAS)-ELISA with monoclonal antibody SCR3 as described by Nurkiyanova et al. (21). The PLRV CP-specific monoclonal antibody SCR3 recognizes an epitope located near the N terminus of the CP (32).
The RNase sensitivity assay was similar to one described by Reutenauer et al. (24). Briefly, 0.1 to 0.2 g of agrobacterium-infiltrated tissue was frozen in liquid nitrogen, homogenized in 200 μl of piperazine ethanesulfonic acid (PIPES) buffer (50 mM PIPES, pH 6.5, and 0.1% Tween), and incubated at 37°C for 30 min. Total RNA was extracted with an RNeasy plant mini kit (Qiagen Inc., Valencia, Calif.). Another sample of the same agrobacterium-infiltrated tissue was frozen in liquid nitrogen, and total RNA was extracted immediately without PIPES buffer or incubation at 37°C. Viral RNA was detected by Northern blotting with a psoralen- and biotin-labeled RNA (BrightStar psoralen-biotin labeling kit; Ambion Inc., Austin, Tex.) probe complementary to the 3′-terminal 112 nucleotides of PLRV RNA and a nonisotopic biotin detection system (BrightStar BioDetect detection kit; Ambion Inc.).
The progeny viruses from systemically infected plants were analyzed by immunocapture reverse transcription-PCR and direct sequencing of the PCR products. For immunocapture of virus particles, plant sap from systemically infected plants was incubated overnight in thin-walled tubes precoated with polyclonal antibodies against PLRV CP (Agdia, Inc., Elkhart, Ind.). The tubes were washed three times with 1X phosphate-buffered saline and once with diethyl pyrocarbonate-treated water. Reverse transcription-PCR analysis was done with primers PLRV5′p3438 (corresponding to PLRV nucleotides 3438 to 3461) and PLRV3′p4382 (complementary to PLRV nucleotides 4382 to 4402) and the One Step reverse transcription-PCR kit (Invitrogen, Carlsbad, Calif.). The reverse transcription-PCR parameters used included a reverse transcription step at 53°C for 45 min and 95°C for 2 min, followed by PCR, 40 cycles of 95°C for 30 s, 53°C for 30 s, and 72°C for 1.5 min, followed by 72°C for 10 min. The amplified fragments were sequenced with an Applied Biosystems Division automated 3700 DNA analyzer (ABI 3700) with Big Dye Terminator chemistry and AmpliTaq-FS DNA polymerase (Applied Biosystems).
Aphid transmission assays.
Systemically infected P. floridana or N. clevelandii plants were used as a virus source in the aphid transmission test. Nonviruliferous Myzus persicae older instars and adults were allowed a 24- to 48-h acquisition access period on detached leaves of P. floridana or N. clevelandii. The aphids were transferred to healthy plants (10 aphids per plant) for a 72-h inoculation access period and then killed by fumigation. The plants were transferred to a greenhouse and tested by DAS-ELISA at 3 to 4 weeks postinoculation. The progeny viruses from systemically infected plants were analyzed by immunocapture reverse transcription-PCR and direct sequencing of the PCR products.
PLRV structural model development.
Based on the preliminary results with the threading program 3DPSSM (13) and the Structure Prediction Meta Server (SPMS) (5, 10), initial three-dimensional models for the PLRV capsid protein were generated by the sequence alignments to Rice yellow mottle virus (RYMV) CP obtained from the SPMS and generated by the ORFeus and FFAS03 (25) threading programs (two highest-score alignments from SPMS; see Table 4), and the X-ray crystallographic coordinates of RYMV CP (PDB code 1F2N) as input for the Modeller program (26). Modeller is a program for comparative protein modeling that generates three-dimensional coordinates through satisfaction of spatial restraints. The program generates a set of distance and dihedral-angle restraints from the structure used as the template and produces a model for the target sequence that minimizes the violations of the set of restraints.
TABLE 4.
Sequence and structure similaritiesa of the monomer of the capsid protein from rice yellow mottle virus (PDB code 1F2N) with structurally related proteins and with our model for PLRV
| Virusd | Sequence identity (%) | No. of Aligned/gap positions | Cα rmsdb | PDB code |
|---|---|---|---|---|
| CoMV | 33.5 | 185/6 | 1.1 | 1NG0 |
| SeMV | 23.8 | 185/12 | 1.6 | 1SMV |
| TNV | 18.5 | 178/6 | 1.8 | 1C8N |
| TBSV | 15.6 | 160/32 | 2.1 | 2TBV |
| PhMV | 10.1 | 148/44 | 2.8 | 1QJZ |
| BPMV | 8.8 | 136/69 | 3.7 | 1BMV |
| STNV | 5.7 | 140/68 | 3.5 | 2STV |
| PLRVc | 22.2 | 158/43 | 1.2 |
Computed by the CE method (28).
rmsd, root mean square deviations computed with the positions of the Cα atoms after optimal superposition.
Model of PLRV from this work.
CoMV, cocksfoot mottle virus; SeMV, sesbania mosiac virus; TNV, tobacco necrosis virus; TBSV, tomato bushy stunt virus; PhMV, physalis mottle virus; BPMV, bean pod mottle virus; STNV, satellite tobacco necrosis virus.
Generation of the final model was carried out by going through a repetitive process in which the current three-dimensional model was compared visually with the structures of the template and the proteins were identified as structurally related to the template. This led to the identification of regions in the target sequence that were improperly aligned. A new trial alignment was produced, and the three-dimensional model was updated. This process was particularly important in assigning the loop regions, where template and target differed substantially. In such cases, the structurally related proteins that provided the better template for that particular loop region were used. Based on the sequence identity of target and template sequences as well as on the similarities among the experimentally determined structures of the related capsid proteins used to produce the alignments, the Cα trace of the models for the PLRV are expected to be within 4 to 5 Å root mean square deviation from the actual structure.
RESULTS AND DISCUSSION
Mutations in the PLRV acidic patch domains.
Eleven site-directed mutants were generated (Fig. 1), and with one exception, all were alanine substitutions. V169 was substituted with K because an alanine substitution would have been a conservative replacement and K is present in other luteoviruses at this position. The E-to-A substitution at position 109 resulted in an amino acid substitution (S to L) in the P17 movement protein encoded by the overlapping ORF 4 (Fig. 1). The P17 coding sequence terminates prior to the downstream domain of the acid patch, and therefore the 10 downstream acidic patch domain mutants express wild-type P17. In addition, two other previously described mutants (15) were used as controls. The mutant CPRD translated only a full-length RT and not the 22-kDa CP. This mutant does not assemble virus particles. The second mutant, ΔRTD, translated CP but not RT. A similar BWYV mutant was shown to be able to assemble stable virus particles but was not transmissible by aphids (4).
Effect of the acidic patch mutations on virus infection in inoculated cells.
The infiltration of N. benthamiana leaf sections with agrobacteria expressing infectious cDNA clones of the various mutants led to a nearly synchronous infection of all cells and facilitated the evaluation of virus replication and virus protein translation. CP was visualized on stained polyacrylamide gels loaded with crude sap extracts from agrobacterium-infiltrated tissue, indicating a high density of infected cells. In the infiltrated tissues, virus infection was not confined to the phloem as in systemically infected leaves (15) (data not shown).
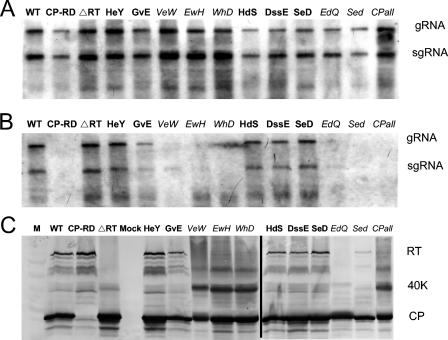
Northern blot hybridization analysis of total RNA extracted from agrobacterium-infiltrated tissues with a riboprobe complementary to the 3′-terminal region of the viral RNA indicated that all the mutants were replication competent and produced the expected genomic and subgenomic RNAs (Fig. 2A). Although the relative amounts of viral RNA detected varied among infiltrated tissue samples, repeated observations indicated that RNA yields from all mutants were similar to wild-type yields (Fig. 2A and data not shown).
FIG. 2.
Replication, encapsidation, and translation of PLRV RNA in agrobacterium-infiltrated Nicotiana benthamiana tissue. (A) Northern blot analysis of total RNA extracted from 0.1 to 0.2 g of tissue infiltrated with Agrobacterium tumefaciens carrying plasmid pBPY, encoding mutations in the CP of PLRV. (B) Detection of viral RNA from the same tissue used in panel A except that total RNA was extracted after exposing the RNA to conditions that allow only encapsidated RNA to remain intact. The positions of the genomic (g) and subgenomic (sg) RNAs are indicated in panels A and B. (C) Detection of PLRV structural proteins from total protein extracts of agrobacterium-infiltrated tissue that were separated by sodium dodecyl sulfate-12.5% polyacrylamide gel electrophoresis, blotted to nitrocellulose, and probed with antibodies prepared against purified PLRV. The positions of the 22-kDa CP, the full-length RT, and a 40-kDa intermediate protein are indicated. Several minor bands are apparent RT degradation products. ΔRTO (abbreviated ΔRTD on figure) does not produce the RT, and the minor bands are not present in this sample. Minor bands below the CP are assumed to be CP degradation products.
Two methods were used to examine the ability of the various mutated CPs to form virions. An RNase sensitivity assay as well as DAS-ELISA and TAS-ELISA were compared. The translation of CP and RT was confirmed by Western blot analysis. The RNase sensitivity assay revealed that several of the mutations interfered with either virion assembly or stability (Fig. 2B). Most notably, alanine substitutions of E170 (mutant VeW), W171 (mutant EwH), H172 (mutant WhD), and D177 (mutant EdQ) resulted in a consistent and dramatic reduction or loss of detection of protected RNA. Not surprisingly the double alanine substitution of E176 and D177 (Sed) and the substitution of all four acidic amino acids in the downstream domain (CPall) also resulted in a reduction of protected RNA. CPRD was a negative control in this RNase protection assay. CPRD was not able to assemble virions and the viral RNA was not protected (Fig. 2B, lane 2). The wild-type and ΔRTD viruses served as positive controls. ΔRTD could assemble stable particles and did protect the RNA, similar to the wild type (Fig. 2B, lane 3 versus lane 1).
Two BWYV CP mutants (2) were similar to our EwH and EdQ mutants. A substitution of W166 with arginine in the BWYV CP (analogous to W171 in the PLRV CP, in mutant EwH) also resulted in a packaging defect. On the other hand, a substitution of D172 with the amide form, asparagine, in the BWYV CP (analogous to D177 in the PLRV CP, in mutant EdQ) did not interfere with assembly, but the mutant did not accumulate to wild-type levels in systemically infected plants and was not aphid transmitted unless a second mutation was present. It is unknown if the contrasting result is influenced by a difference in charge, side chain, or relative position of the amino acid in the two virus CP structures (see below).
Repeated observations revealed that the PLRV CP with the lysine substitution of V169 (GvE) or the alanine substitution of E109 (HeY), D173 (HdS), S174 and S175 (DssE), or E176 (SeD) were consistently able to protect against RNA degradation similar to the wild-type or ΔRTD virus (Fig. 2 and data not shown). The RNase assay provides a qualitative measure of RNA protection and does not allow us to determine the exact level of protection relative to the wild-type virus. Clearly, some degree of RNA degradation occurs in all samples. Note that the GvE RNA in Fig. 2 appears to be less protected than the wild-type RNA. This observable difference was not consistent in repeated experiments, and similar to results for HeY, HdS, DssE, and SeD, a significant amount of RNA is protected from degradation, and the ELISA data indicate that virions are assembled. Substitution of D168 and E171 with their amide forms in the BWYV CP (analogous to D173 and E176 in the PLRV CP) also resulted in no observable differences in RNA packaging (2).
An unexpected observation in our experiments and a result in contrast to previous reports (2, 17) was the encapsidation of the subgenomic RNA. The high concentration of virus-encoded proteins and genomic and subgenomic RNAs in the agrobacterium-infiltrated tissues may be responsible for this phenomenon (see below). Poleroviruses do encapsidate RNAs other than genomic RNA. When the PLRV CP gene was expressed in insect cells, virus-like particles that did encapsidate random cellular RNAs were formed (9). Poleroviruses also encapsidate umbravirus RNA (30) and satellite or satellite-like RNAs (19, 22)
Viral CP was undetectable by ELISA from tissue infiltrated with any of the mutants that the RNase assay suggested did not assemble virions (Table 1). Unassembled CP or RT is not detected efficiently by either DAS-ELISA or TAS-ELISA. Mutants that did assemble virions (based on RNase assay results) were detected by ELISA at either wild-type levels (GvE and DssE) or 50 to 95% less than wild-type levels (HeY, HdS, and SeD). The latter three mutants were detected more efficiently, relative to the wild type, by TAS-ELISA than DAS-ELISA (Table 1). We interpret this to indicate that the surface features of these mutants were altered relative to the wild type but the alterations did not affect proper assembly of the virions.
TABLE 1.
Virus antigen titer in tissue infiltrated with wild-type or acidic patch mutants of PLRV
| PLRV | Virus antigen in tissuea (% of wild-type level)
|
|
|---|---|---|
| DAS-ELISA | TAS-ELISA | |
| Wild type | 100 | 100 |
| HeY | 15-50 | 40-100 |
| GvE | 100 | 100 |
| VeW | 0 | 0 |
| EwH | 0 | 0 |
| WhD | 0 | 0 |
| HdS | 5-20 | 40-100 |
| DssE | 100 | 100 |
| SeD | 10-50 | 50-100 |
| Sed | 0 | 0 |
| EdQ | 0 | 0 |
| CPall | 0 | 0 |
Range of virus antigen levels in tissue agroinfitrated with the various mutants determined in two or three independent experiments. A value of zero indicates that absorbance values were not above the background (<0.05). Double-antibody sandwich ELISA used the same PLRV-specific polyclonal antibodies for trapping and detection. The detection antibody was conjugated to alkaline phosphatase. The triple-antibody sandwich ELISA trapped virus with a PLRV-specific polyclonal antibody and detected it with a PLRV-specific monoclonal antibody.
Interestingly, while the levels of CP translation of all the acidic patch mutants in infiltrated tissues were comparable to that of the wild type based on Western blotting analysis (Fig. 2C), the translation or stability of RT was either reduced or below detection in all of the mutants that were assembly deficient. The alanine substitution of E176 (SeD) did not affect RT accumulation, whereas the substitution of D177 (EdQ) abolished RT accumulation, and the double substitution E176 and D177 (Sed) partially restored RT accumulation (Fig. 2C). Brault et al. (2) also reported a reduction in RT accumulation in the W166 assembly-deficient mutant of BWYV CP that was mentioned earlier. In contrast to our results, they observed a reduction in CP accumulation for the W166 mutant. We observed the appearance of another protein with an approximate size of 40 kDa in several of the mutants in which a drastic reduction of RT was observed (Fig. 2C, VeW, EwH, WhD, and CPall). This protein is recognized by a CP-specific monoclonal antibody, indicating that it is either a truncated RT or a dimeric form of the CP.
ORF 3 and ORF 5 were sequenced in their entirety in several of the mutants that accumulated the 40-kDa protein. There are no nucleotide changes other than the introduced changes in the CP. Sequences within ORF 5 regulate the translation efficiency of the RT protein, and considerable secondary structure in this region effects transcription and translation (3, 18, 20). However, no sequences in ORF 3 have been shown to regulate either translation efficiency or the size of the translation product. One possibility is that the RT protein is not stable unless incorporated into particles and is degraded rapidly.
Effects of acidic patch mutants on systemic infection of plants.
The GvE, DssE, and SeD mutants systemically infected both N. benthamiana and N. clevelandii as well as P. floridana and potato (S. tuberosum cv. Russet Burbank) with similar or slightly lower efficiencies than wild-type virus (Table 2). While inoculation of N. clevelandii with the mutant HeY resulted in wild-type efficiencies of infection, few N. benthamiana plants became infected, and we have been unable to systemically infect P. floridana or potato plants with this mutant by agroinoculation. However, P. floridana plants were infected by aphid transmission of the mutant virus HeY (see below). The mutant HdS infected N. clevelandii, P. floridana, and potato systemically, but repeated attempts to agroinoculate N. benthamiana failed (Table 2). None of the mutants that failed to assemble stable particles infected any plants systemically (Fig. 2 and Table 2). This observation is consistent with a previous report (33) that virion formation is essential for virus movement in N. clevelandii, and our data extend these observations to other hosts. The CP gene was sequenced from progeny virus in multiple plants infected with each mutant described above. The consensus sequences revealed only the expected mutation, with no other fixed changes.
TABLE 2.
Systemic infection of four host plant species agroinoculated with PLRV acidic patch mutants
| PLRV | No. of plants moculated no. infecteda
|
|||
|---|---|---|---|---|
| N. clevelandii | N. benthamiana | S. tuberosum | P. floridana | |
| None | 0/15 | 0/15 | 0/3 | 0/5 |
| Wild type | 14/15 | 13/15 | 5/5 | 3/5 |
| HeY | 11/15 | 3/15 | 0/3 | 0/5 |
| GvE | 11/15 | 9/15 | 2/3 | 3/5 |
| VeW | 0/13 | 0/15 | 0/3 | 0/5 |
| EwH | 0/15 | 0/15 | 0/3 | 0/5 |
| WhD | 0/15 | 0/15 | 0/3 | 0/5 |
| HdS | 13/15 | 0/15 | 1/3 | 1/5 |
| DssE | 15/15 | 13/15 | 3/3 | 2/5 |
| SeD | 12/15 | 8/15 | 2/3 | 1/5 |
| Sed | 0/15 | 0/15 | 0/3 | 0/5 |
| EdQ | 0/15 | 0/15 | 0/3 | 0/5 |
| CPall | 0/15 | 0/15 | 0/3 | 0/5 |
Number of plants agroinoculated/number of plants systemically infected.
Aphid transmission from agroinoculated plants.
N. clevelandii and P. floridana plants systemically infected with HeY, GvE, HdS, DssE, and SeD were used as virus sources for aphid transmission experiments. Mutant DssE was transmitted at wild-type efficiencies in all experiments (Table 3). In initial experiments with N. clevelandii as a source plant for mutant GvE, transmission to all three recipient hosts was reduced relative to that with the wild type. However, when P. floridana was used as a virus source and as a recipient plant, transmission efficiency was similar to that of wild-type virus. Mutant SeD was not transmitted in any experiments when N. clevelandii was used as a virus source and was transmitted inefficiently (3 of 15) when P. floridana was the virus source. HdS was transmitted inefficiently or not at all regardless of the source. Substitutions of the uncharged amide forms of E176 (SeD) and D173 (HdS) in equivalent positions in BWYV reduced transmission relative to wild-type virus but less so than the mutations in PLRV (2). HeY was transmitted inefficiently or not at all from N. clevelandii. The two P. floridana plants infected with HeY in an early experiment died prior to being used as source plants, and repeated attempts to infect additional P. floridana by either aphid transmission or agroinfection failed. The CP gene from PLRV in all of the aphid-inoculated plants described above was sequenced, and all the plants contained the expected mutation, with no other apparent changes.
TABLE 3.
Aphid transmission of PLRV mutants from systemically infected Nicotiana clevelandii to either N. benthamiana, N. clevelandii, or Physallis floridana or from P. floridana to P. floridanaa
| PLRV | No. of plants infested/no. infected with virus
|
|||
|---|---|---|---|---|
|
N. clevelandii source
|
P. floridana source, P. floridana recipient | |||
| N. benthamiana recipient | N. clevelandii recipient | P. floridana recipient | ||
| None | 0/9 | 0/9 | 0/9 | NTb |
| Wild type | 12/15 | 7/13 | 4/8 | 23/35 |
| HeY | 0/5 | 2/9 | 2/17 | NT |
| GvE | 3/10 | 1/8 | 2/5 | 26/35 |
| HdS | 0/10 | 1/10 | 0/5 | 0/15 |
| DssE | 2/2 | 5/5 | 5/5 | 5/5 |
| SeD | 0/10 | 0/10 | 0/5 | 3/15 |
Plants were tested by ELISA 3 weeks postinoculation. In general, five plants were used for each construct for each experiment.
NT, not tested.
Mapping the acidic patch mutants on a model of the PLRV capsid protein.
The model presented here was derived from a three-dimensional structure-based comparison of the PLRV CP sequence with the CP of other icosahedral plant viruses whose structures have been determined. An initial BLAST search of the Brookhaven protein databank (1) for the PLRV sequence did not provide reliable pairwise alignments, i.e., no homologous sequence was detected. A search for the most probable three-dimensional structure with the threading program 3DPSSM (13) identified several plant viruses CP whose structures have been solved as possible templates. In spite of the relatively low sequence identity between PLRV and these templates (10 to 14%), the level of certainty assigned by 3DPSSM to these structures is high (>50%).
The 3DPSSM program also provides sequence alignments of the PLRV CP with each template. These CP sequences included monomer A of physalis mottle virus, satellite tobacco necrosis virus, and monomer 1 of the middle component of bean pod mottle virus. A second structural search with the Structure Prediction Meta Server (SPMS) (5, 10) assigned the highest (3D-Jury) score to the alignment of the PLRV sequence with that corresponding to the structure of the capsid protein from RYMV. SPMS assigned slightly lower scores to alignments with sequences corresponding to the structures of monomer A of tomato bushy stunt virus, monomer A of cocksfoot mottle virus, monomer A of sesbania mosaic virus, and monomer C of tobacco necrosis virus. When the structures of these proteins are optimally superimposed (28), the root mean squared deviation of the Cα atoms used for optimal superposition in the pairwise alignments range from 1.1 to 3.7 Å (Table 4), indicating that the structures are quite similar. The level of sequence identity between RYMV and PLRV, tomato bushy stunt virus, cocksfoot mottle virus, sesbania mosaic virus, and tobacco necrosis virus ranges from ≈6% to ≈34% (Table 4). Information on the structural alignment of these proteins is available on request.
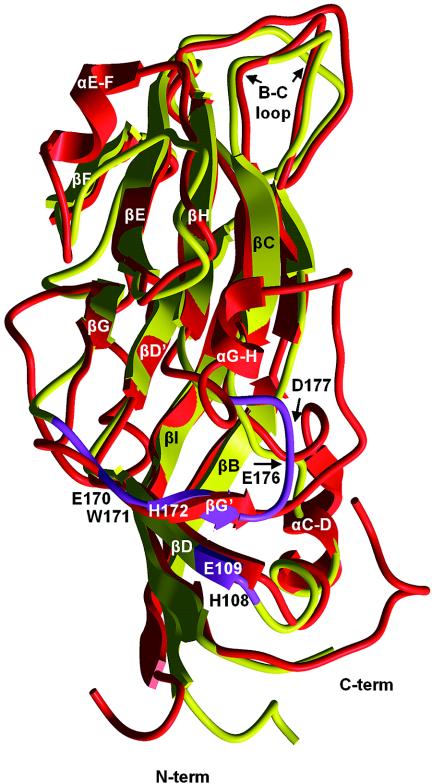
Our model is expected to be similar to that predicted by Terradot et al. (31) with a few changes in the region that includes strands E and F, none of which should have a large effect on the predicted structure of the acidic patch within a subunit (Fig. 3) or the interaction of the acidic patch at the center of a CP trimer (Fig. 4). Figure 3 shows a ribbon representation of the structural superimposition of the PLRV CP (yellow) with the RYMV P (red). The eight β-sheets form the barrel configuration of the CP protein monomer. The regions containing amino acid residues in the acidic patch domains (Fig. 3, purple) are reported to be in loops C and D and G and H in the previous polerovirus CP models (2, 31); however, βD and βG are split into two domains, and the acidic patch residues are partially contained in loops and partially in β sheets (Fig. 3). The two domains are predicted to be in close proximity in the CP monomer (Fig. 3) and oriented into the threefold axis of symmetry of the virion (Fig. 4).
FIG. 3.
Superposition of ribbon diagrams of the RYMV capsid protein (in red), whose structure has been determined by X-ray crystallographic data, and predicted structure of the PLRV capsid protein (in yellow). The purple fragments in the PLRV model indicate the positions of residues that form the acidic patch regions. Those residues lie in the loop regions between strands C and D and strands G and H. Notice that βD and βG are divided into two domains, designated βD and βD′ and βG and βG′. Positions of amino acid residues that are biologically active are noted.
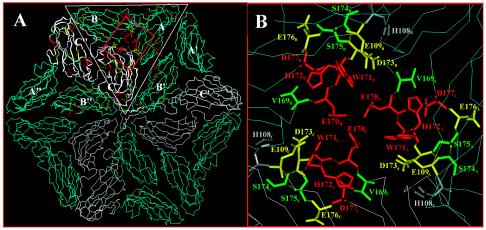
FIG. 4.
Structures of the PLRV capsid protein subunits predicted by a three-dimensional sequence comparison with RYMV. (A) Asymmetric trimer of three subunits within the triangle and their relationship to the larger hexamer face of the icosahedral PLRV particle. The region around the threefold axis of symmetry is boxed and enlarged in panel B. Amino acid residues that were mutated in the acid patch domains are color coded based on their biological phenotype. Red indicates amino acids that were required for stable virion formation. Yellow indicates amino acids that were not critical for virion assembly but did affect host range and aphid transmission. Green indicates amino acids that did not confer an observable phenotype. Histidine 108 was not mutated in this study but may interact with the acidic E109 or D173.
According to the model, W171 is oriented horizontally, forming the walls of a depression at the threefold axis of symmetry (Fig. 4B). E170 is projected into the center of the threefold axis of symmetry, forming a negatively charged depression. H172 surrounds the edge of a depression and is oriented toward D177 in a manner that may allow their opposite charges to interact and stabilize the structure (Fig. 4B). These four residues were important for the assembly and stability of virions (mutants VeW, EwH, WhD, and EdQ) (Fig. 2B).
Brault et al. (2) reported that the W residue in BWYV, corresponding to PLRV W171, was projecting into the center of the threefold axis of symmetry. Mutation of this residue to arginine (R) also resulted in an assembly-deficient virus. Mutational analysis of the corresponding adjacent E and H residues of BWYV was not done, but the features of the BWYV model would likely position these residues away from the threefold axis. A change in the BWYV residue corresponding to D177 to its amide form (asparagine) did not result in an assembly-deficient virus but did result in a virus that was not aphid transmissible unless there was a second substitution. This suggests that the negative charge is not important in assembly but may contribute to aphid-virus recognition; however, the alternative structural model proposed for BWYV locates this residue away from the threefold axis (2) (see below).
PLRV CP residues E109 and D173 (mutants HeY and HdS, respectively) are adjacent and located near the edge of the depression in our model (Fig. 4B). They may contribute further to the negative charge of the center of the threefold axis of symmetry, or they may interact with the positively charged H108. Neither of these mutations has an effect on the assembly or stability of virions, but they do reduce the transmission efficiency of the virus by aphids and have an effect on the host range. S174, S175, and E176 also surround the depression but are oriented away from the depression. None of these five residues has any detectable affect on virion assembly, but several affect plant-virus or aphid-virus interactions. The defined epitope 10, HDSSEDQ, is not totally accessible in this model, although H108, E109, S174, S175, and E176 appear to be in a somewhat linear arrangement (Fig. 4B) and may be the actual epitope located on the side of the depression. Mutations in the BWYV equivalents of D173 and E176 to their amide (uncharged) form had only slight effects on virus titer and aphid transmission and did not affect virion assembly. The alanine substitutions of D173 and E176 in PLRV were more significant in the reduction of virus titer and aphid transmission but, as in BWYV, did not affect virion assembly. These amino acid residues are located further away from the threefold axis of symmetry in the BWYV model (2), whereas they are adjacent to the threefold axis of symmetry in the PLRV model (Fig. 4). This positional difference, if accurate, may account for the observed differences in virus-aphid interactions between PLRV and BWYV.
As indicated by Brault et al. (2), until crystallographic data are available, we will only be able to simulate the structure of the polerovirus particle, but the models will be useful in predicting interactions. Such predictions can be tested by mutational analysis of the genome coupled with various biological assays. Whereas the available biological data for the BWYV CP sequence do not appear to fit the Terradot model (31), our model provides an independent confirmation of the main features of the Terradot model, and the biological data on the acidic patch mutants further support the model. Coupling the findings from this study and that of Brault et al. (2), it is apparent that the conserved downstream domain of the acidic patch is a biologically active region involved in CP subunit interactions, plant-virus interactions, and aphid-virus recognition.
Acknowledgments
We thank Dawn Smith and Tom Hammond for assistance with aphid transmission assays and greenhouse work.
This research was partially supported by USDA CSREES NRI grant 96-01120 and a USDA ARS postdoctoral research associate grant. P.P. is supported by a grant-in-aid from the Scottish Executive Environment and Rural Affairs Department to the SCRI. Part of this research was carried out by using resources of the Cornell Theory Center, which receives funding from Cornell University, New York state, federal agencies, foundations, and corporate partners.
REFERENCES
- 1.Berman, H. M., J. Westbrook, Z. Feng, G. Gilliland, T. N. Bhat, H. Weissig, I. N. Shindyalov, and P. E. Bourne. 2000. The protein data bank. Nucleic Acids Res. 28:235-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brault, V., M. Bergdoll, J. Mutterer, V. Prasad, S. Pfeffer, M. Erdinger, K. E. Richards, and V. Ziegler-Graff. 2003. Effects of point mutations in the major capsid protein of beet western yellows virus on capsid formation, virus accumulation, and aphid transmission. J. Virol. 77:3247-3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown, C. M., S. P. Dinesh Kumar, and W. A. Miller. 1996. Local and distant sequences are required for efficient readthrough of the barley yellow dwarf virus PAV coat protein gene stop codon. J. Virol. 70:5884-5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruyere, A., V. Brault, V. Ziegler-Graff, M. T. Simonis, J. F. J. M. van den Heuvel, K. Richards, H. Guilley, G. Jonard, and E. Herrbach. 1997. Effects of mutations in the beet western yellows virus readthrough protein on its expression and packaging and on virus accumulation, symptoms, and aphid transmission. Virology 230:323-334. [DOI] [PubMed] [Google Scholar]
- 5.Bujnicki, J. M., A. Elofsson, D. Fischer, and L. Rychlewski. 2001. Structure prediction meta server. Bioinformatics 17:750-751. [DOI] [PubMed] [Google Scholar]
- 6.Chay, C. A., U. B. Gunasinge, S. P. Dinesh Kumar, W. A. Miller, and S. M. Gray. 1996. Aphid transmission and systemic plant infection determinants of barley yellow dwarf luteovirus-PAV are contained in the coat protein readthrough domain and 17-kDa protein, respectively. Virology 219:57-65. [DOI] [PubMed] [Google Scholar]
- 7.Dolja, V. V., and E. V. Koonin. 1991. Phylogeny of capsid proteins of small icosahedral RNA plant viruses. J. Gen. Virol. 72:1481-1486. [DOI] [PubMed] [Google Scholar]
- 8.Franco-Lara, L. F., D. K. McGeachy, U. Commandeur, R. R. Martin, M. A. Mayo, and H. Barker. 1999. Transformation of tobacco and potato with cDNA encoding the full-length genome of potato leafroll virus: evidence for a novel virus distribution and host effects on virus multiplication. J. Gen. Virol. 80:2813-2822. [DOI] [PubMed] [Google Scholar]
- 9.Gildow, F. E., B. Reavy, M. A. Mayo, G. H. Duncan, J. A. T. Woodford, J. W. Lamb, and R. T. Hay. 2000. Aphid acquisition and cellular transport of potato leafroll virus-like particles lacking P5 readthrough protein. Phytopathology. 90:1153-1161. [DOI] [PubMed] [Google Scholar]
- 10.Ginalski, K., A. Elofsson, D. Fischer, and L. Rychlewski. 2003. 3D-Jury: a simple approach to improve protein structure predictions. Bioinformatics 19:1015-1018. [DOI] [PubMed] [Google Scholar]
- 11.Gray, S., and F. E. Gildow. 2003. Luteovirus-aphid interactions. Annu. Rev. Phytopathol. 41:539-566. [DOI] [PubMed] [Google Scholar]
- 12.Harrison, S. C., A. J. Olson, C. E. Schutt, F. K. Winkler, and G. Brigogne. 1978. Tomato bushy stunt virus at 2.9 A resolution. Nature 276:368-373. [DOI] [PubMed] [Google Scholar]
- 13.Kelley, L. A., R. M. MacCallum, and M. J. Sternberg. 2000. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol. 299:501-522. [DOI] [PubMed] [Google Scholar]
- 14.Lee, L., P. Palukaitis, and S. M. Gray. 2002. Host-dependent requirement for the potato leafroll virus 17-kDa protein in virus movement. Mol. Plant-Microbe Interact. 15:1086-1094. [DOI] [PubMed] [Google Scholar]
- 15.Liang, D., S. M. Gray, I. Kaplan, and P. Palukaitis. 2004. Site-directed mutagenesis and generation of chimeric viruses by homologous recombination in yeast to facilitate analysis of plant-virus interactions. Mol. Plant-Microbe Interact. 17:571-576. [DOI] [PubMed] [Google Scholar]
- 16.Mayo, M. A., and W. A. Miller. 1999. The structure and expression of luteovirus genomes, p. 23-42. In H. G. Smith and H. Barker (ed.), The luteoviridae. CABI Publishing, Wallingford, England.
- 17.Mayo, M. A., and V. Ziegler Graff. 1996. Molecular biology of luteoviruses. Adv. Virus Res. 46:413-460. [DOI] [PubMed] [Google Scholar]
- 18.Miller, W. A., C. M. Brown, and S. P. Wang. 1997. New punctuation for the genetic code: luteovirus gene expression. Semin. Virol. 8:3-13. [Google Scholar]
- 19.Miller, W. A., T. Hercus, P. M. Waterhouse, and W. L. Gerlach. 1991. A satellite RNA of barley yellow dwarf virus contains a novel hammerhead structure in the self-cleavage domain. Virology 183:711-720. [DOI] [PubMed] [Google Scholar]
- 20.Mohan, B. R., S. P. Dineshkumar, and W. A. Miller. 1995. Genes and cis-acting sequences involved in replication of barley yellow dwarf virus-PAV RNA. Virology 212:186-195. [DOI] [PubMed] [Google Scholar]
- 21.Nurkiyanova, K. M., E. V. Ryabov, U. Commandeur, G. H. Duncan, T. Canto, S. M. Gray, M. A. Mayo, and M. E. Taliansky. 2000. Tagging potato leafroll virus with the jellyfish green fluorescent protein gene. J. Gen. Virol. 81:617-626. [DOI] [PubMed] [Google Scholar]
- 22.Passmore, B. K., M. Sanger, L. S. Chin, B. W. Falk, and G. Bruening. 1993. Beet western yellows virus-associated RNA: an independently replicating RNA that stimulates virus accumulation. Proc. Natl. Acad. Sci. USA 90:10168-10172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reinbold, C., F. E. Gildow, E. Herrbach, V. Ziegler-Graff, M. C. Goncalves, J. F. J. M. van den Heuvel, and V. Brault. 2001. Studies on the role of the minor capsid protein in transport of beet western yellows virus through Myzus persicae. J. Gen. Virol. 82:1995-2007. [DOI] [PubMed] [Google Scholar]
- 24.Reutenauer, A., V. Ziegler-Graff, H. Lot, D. Scheidecker, H. Guilley, K. Richards, and G. Jonard. 1993. Identification of beet western yellows luteovirus genes implicated in viral replication and particle morphogenesis. Virology 195:692-699. [DOI] [PubMed] [Google Scholar]
- 25.Rychlewski, L., L. Jaroszewski, W. Li, and A. Godzik. 2000. Comparison of sequence profiles: strategies for structural predictions using sequence information. Protein Sci. 9:232-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sali, A., and T. L. Blundell. 1993. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234. [DOI] [PubMed]
- 27.Sambrook, J., E. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 28.Shindyalov, I. N., and P. E. Bourne. 1998. Protein structure alignment by incremental combinatorial extension (CE) or the optimal path. Protein Eng. 11:739-747. [DOI] [PubMed] [Google Scholar]
- 29.Smith, H. G., and H. Barker (ed.). 1999. The luteoviridae. CABI Publishing, Wallingford, England.
- 30.Taliansky, M. E., D. J. Robinson, and A. F. Murant. 2000. Groundnut rosette disease virus complex: biology and molecular biology. Adv. Virus Res. 55:357-400. [DOI] [PubMed] [Google Scholar]
- 31.Terradot, L., M. Souchet, V. Tran, and D. G. Ducray-Bourdin. 2001. Analysis of a three-dimensional structure of potato leafroll virus coat protein obtained by homology modeling. Virology 286:72-82. [DOI] [PubMed] [Google Scholar]
- 32.Torrance, L. 1992. Analysis of epitopes on potato leafroll virus capsid protein. Virology 191:485-489. [DOI] [PubMed] [Google Scholar]
- 33.Ziegler Graff, V., V. Brault, J. D. Mutterer, M. T. Simonis, E. Herrbach, H. Guilley, K. E. Richards, and G. Jonard. 1996. The coat protein of beet western yellows luteovirus is essential for systemic infection but the viral gene products P29 and P19 are dispensable systemic infection and aphid transmission. Mol. Plant-Microbe Interact. 9:501-510. [Google Scholar]