Abstract
Nickel homeostasis is important for pathogenic and ureolytic bacteria, which use this metal ion as enzymatic cofactor. For example, in the human pathogen Helicobacter pylori an optimal balance between nickel uptake and incorporation in metallo-enzymes is fundamental for colonization of the host. Nickel is also used as cofactor to modulate DNA binding of the NikR regulator, which controls transcription of genes involved in nickel trafficking or infection in many bacteria. Accordingly, there is much interest in a systematic characterization of NikR regulation. Herein we use H. pylori as a model to integrate RNA-seq and ChIP-seq data demonstrating that NikR not only regulates metal-ion transporters but also virulence factors, non-coding RNAs, as well as toxin-antitoxin systems in response to nickel stimulation. Altogether, results provide new insights into the pathobiology of H. pylori and contribute to understand the responses to nickel in other bacteria.
Nickel homeostasis is of primary importance for many organisms and especially for pathogenic and ureolytic bacteria, which use this metal ion as enzymatic cofactor for the catalysis of redox reactions and Lewis acid-like functions, with important medical, agricultural and biotechnological implications1. The human pathogen H. pylori is a paradigmatic example, since its survival in the stomach relies on the catalytic activity of the two nickel-dependent metalloenzymes urease and hydrogenase, respectively involved in acid acclimation and energy metabolism of the bacterium2,3. Both activities are important for the colonization of the gastric epithelium, leading to long-term infections that correlate with many gastric diseases, including gastritis, peptic ulcers, gastric carcinoma and MALT lymphoma4. On the other hand, an excess of nickel ions can be noxious, poisoning other metallo-enzymes or producing reactive oxygen species (ROS)5. Nickel-utilizing bacteria must therefore maintain an optimal homeostasis of nickel ions, tightly controlling the balance between their uptake and incorporation in metallo-enzymes or storage proteins. One of the main regulatory factors of nickel homeostasis is the NikR protein, a ribbon-helix-helix (RHH) transcriptional regulator, whose orthologues are present in almost all the main bacterial and archeal clades6,7. Despite its widespread conservation, NikR regulation has been approached principally in H. pylori and E. coli, mainly through transcriptional analysis8,9 and in vitro protein-DNA binding studies10,11,12,13, leading to the characterization of several bona-fide regulatory targets14. While the EcNikR protein functions strictly as a nickel-dependent transcriptional repressor, HpNikR has been proposed to be a more versatile regulator, either inducing or repressing the transcription of a larger cohort of nickel-responsive genes9,11,15. However many studies focused on the regulation of already characterized metal-binding proteins, leaving a systematic characterization on NikR regulation unexplored13,16,17. In this work we sought to fill this gap, combining RNA-sequencing and ChIP-sequencing approaches to provide for the first time the comprehensive mapping of a bacterial nickel-responsive regulon.
Results
RNA–seq analysis determines NikR-dependent and nickel–responsive transcriptomes
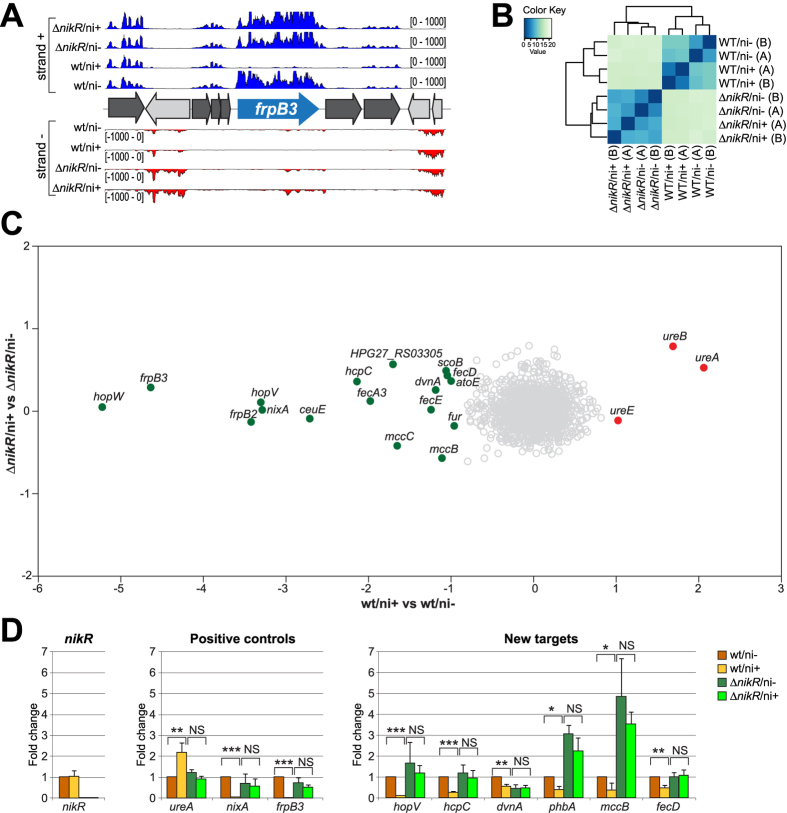
To elucidate the involvement of NikR in the nickel response of H. pylori, whole transcriptome analyses were performed by strand-specific RNA sequencing (Fig. S1). To this purpose, a set of nickel treated (ni+) or untreated (ni−) cultures of the wild type G27 strain and of a ΔnikR mutant grown to OD600 1.0–1.1 were used as starting material to produce strand specific RNA sequencing libraries. A minimum of 3 Millions of reads were obtained for each sample and for each of the two replicates (Bioproject: PRJNA313048), providing an optimal coverage of the transcripts (Supplementary Table S1). The reliability of the RNA-seq experiment is exemplified by the frpB3 genomic locus (Fig. 1A), which evidences a clear match between the strand-specificity of the RNA-seq tracks and the annotated CDS, along with the reduction of the signal in the wt/ni+ tracks, corresponding to the reported repression of the frpB3 gene in response to nickel excess17. Samples clustering clearly shows the reproducibility of replicates (average Pearson correlation value on normalized counts: 0.98) and their grouping according to genotypes and/or treatments (Fig. 1B).
Figure 1. Results of the RNA-sequencing experiment.
(A) RNA-seq profiles of the frpB3 genomic locus: wt/ni+, wt/ni−, ∆nikR/ni+ and ∆nikR/ni− strand specific coverages are shown in blue (strand+) and red (strand−). (B) Heatmap showing the Euclidean distances between samples and replicas as calculated from the regularized log transformation performed by DESeq2 on raw counts. (C) Effect of nickel treatment on H. pylori gene expression: log2FC values of nickel treated (ni+) vs untreated (ni−) samples are reported for wt (x-axis) and ∆nikR (y-axis) genotypes. DEGs induced and repressed after nickel addition are represented as filled red and green circles respectively (log2FC ≥ |1|; adj p < 0.01); empty grey circles correspond to non differential genes. (D) RT-qPCR validation of H. pylori nickel responsive genes expression. nikR (control), ureA, nixA, frpB3 (positive controls) and hopV, hcpC, dvnA, phbA, mccB and fecD (new targets) gene expression levels are reported for wt and ∆nikR strain in both ni+ and ni− conditions. Results show fold changes (mean ± SD) relative to the wt/ni− condition. ΔCts were obtained after normalization on 16S rRNA levels; at least three biological replicates were used for the analysis. SD = standard deviation. Ct = threshold cycle. Statistical significance is calculated using the t-test; *p < 0.05; **p < 0.01; ***p < 0.001.
Nickel treatment in a wild type (wt) background elicited a total of 20 differentially expressed genes (DEGs; log2FC ≥ |1|, adj p < 0.01), mapping to 14 transcriptional units (Supplementary Table S2 and Fig. 1C x-axis). We observed a transcriptional down-regulation for previously characterized targets of negative NikR regulation such as nixA, frpB2, frpB3, fecA3, fur and ceuE. We also recorded the significant induction of only three genes, all belonging to the nickel-responsive urease (Fig. 1C). In addition, the RNA–seq analysis pinpointed 11 novel nickel-repressed genes, including several interesting genes coding for membrane-associated proteins and transporters (Supplementary Table S2, Fig. 1C and D), and also a polycistronic operon carried on the pHPG27 plasmid (HPG27_RS07995/HPG27_RS08000; mccC/mccB). Oddly, we were not able to detect significant variations in the transcript levels of HPG27_RS07055* (hpn), HPG27_RS07080* (hpn2), HPG27_RS03075 (hydB), HPG27_RS00075 (groES) and HPG27_RS06735 (exbB), which were previously reported to be regulated by nickel or NikR9, nor the auto-repression of nikR in response to nickel excess8,9,18. When the same comparative analysis (ni+ vs ni−) was performed in the ΔnikR strain no genes were differentially expressed upon nickel treatment (Fig. 1C, y-axis), strongly suggesting that the nickel-dependent responses observed in the wt strain are mediated by the nikR gene product. These results were independently validated by qRT–PCR on a panel of 9 nickel–regulated genes (Fig. 1D), measuring their expression levels with and without nickel stimulation both in wt and in ΔnikR strains. For all these genes, nickel dependent regulation was lost in the ΔnikR mutant. In some cases we observed a de-repression corresponding to transcript levels measured in the wt/ni− condition (ureA, nixA, frpB3, hopV, hcpC and fecD), while in other cases a general decrease (dvnA) or increase (phbA and mccB) in mRNA levels was recorded, suggesting the integration with other layers of transcriptional control (e.g. Fur- or growth phase- dependent regulation, see below). The putative NikR gene targets hpn and hpn2, which did not change expression levels in all the four conditions analyzed in RNA-seq, were also included in the qRT-PCR analysis (Fig. S4A). Their lack of responsiveness to nickel and nikR deletion was confirmed, provisionally indicating that these two genes are not transcriptionally regulated by NikR under the tested conditions.
On the other hand, the comparative analysis between the wt and ΔnikR strains under similar nickel stimulation (ΔnikR/ni+ vs wt/ni+) outlined 194 DEGs (Supplementary Table S2 and Fig. S2), suggesting that in the ΔnikR strain a very large number of genes is deregulated, including many genes not responsive to nickel, likely due to indirect effects. The same comparative analysis in the absence of nickel treatment (ΔnikR/ni− vs wt/ni−) supported this interpretation. In fact, out of the 261 DEGs identified, only a handful of nickel-responsive genes pinpointed in the wt strain were spotted, while most differentially expressed transcripts belonged to stationary phase- and/or to the regulon of the Ferric uptake repressor Fur, which is itself part of the NikR regulon10 (Supplementary Table S2 and Fig. S2). Thus, the nikR deletion has a profound impact on the cell, indirectly affecting the transcription of many genes beyond the relatively tight cohort of nickel-responsive cistrons predicted to belong to its regulon.
Genome-wide analysis of NikR targets by ChIP-seq
To identify genomic regions bound in vivo by NikR, we performed Chromatin Immunoprecipitation assays with a specific NikR polyclonal antiserum (Fig. S3A), followed by deep sequencing (ChIP-seq) in wt and ΔnikR strains under nickel-replete (ni+) or untreated conditions (ni−) (Fig. S1). Two sets of biological replicates were employed for the IP analysis, obtaining at least 3 Millions raw reads for each replicate (Bioproject: PRJNA313048). On average more than 98.5% of the reads mapped with good quality on the H. pylori G27 reference genome (Supplementary Table 1). The ChIP-seq profiles of the wt strain showed NikR-specific and nickel-dependent enrichments that were absent from the ChIP-seq profiles of the ΔnikR mutant and from the control samples (INPUT) obtained by sequencing the sheared chromatin before immunoprecipitation (Fig. S2C; showing the enrichment profiles at the frpB2 locus). The lack of comparable profiles deriving from immunoprecipitations with antisera specific for other transcriptional regulators demonstrates that the ChIP-enrichment was specific for NikR. Since it is well established that NikR binds specifically to its operator sequences only in presence of the nickel cofactor19, we first identified the core of high fidelity NikR target binding sites in the wt strain treated with nickel, and then we analyzed their differential binding respect to wt untreated samples.
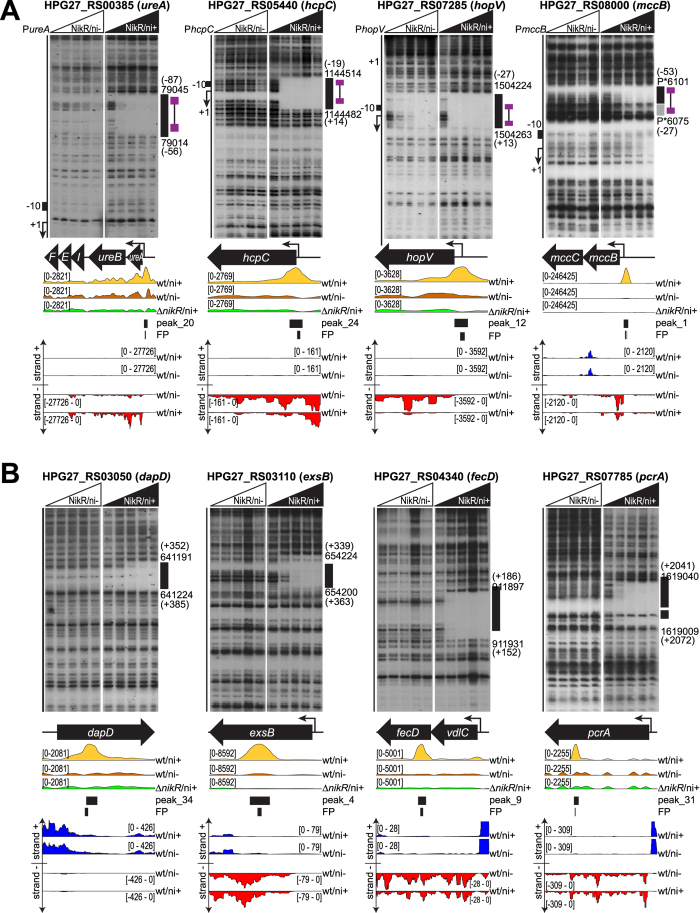
The peak regions were identified by comparing the ChIP-seq profiles of nickel-treated wt strain (wt/ni+) with those of the nickel-treated ΔnikR mutant (ΔnikR/ni+), setting the latter as negative control (background) of the whole experiment. Irreproducible Discovery Rate (IDR) analysis outlined the good reproducibility of the replicates (Supplementary Table S1). Consequently, an optimal set of 72 high-quality peaks was defined (Supplementary Table S3). These were mapped with respect to the list of putative H. pylori TSSs, obtained by remapping the H. pylori 26695 primary transcriptome annotation20 onto the H. pylori G27 reference genome, or by de novo 5′-end mapping primer extension analysis. 23 peaks were classified as bona-fide “promoter peaks” because they were centered between position −150/+30 with respect to a TSS. The remaining peaks were subdivided into “intragenic peaks” (41 peaks) and “intergenic peaks” (8 peaks) according to the position of their center respectively within or outside the annotated genes. Consistently, many “promoter peaks” overlap the promoters of known NikR–regulated operons. Moreover, we identified peaks on the promoters of the newly identified nickel–responsive operons: hopV, hopW, hcpC, dvnA and mccB. No peaks mapping to the promoters of hpn, hpn2, hydA and groES genes were detected (Fig. 2A, Figs S2 and S3).
Figure 2. Validation of new NikR promoters and internal peaks by DNase I footprinting.
(A) Radiolabeled PureA, PhcpC, PmccB and PhopV DNA probes were mixed with 0, 9.7, 29, 97 and 290 nM of the NikR tetramer, without nickel (left side of each panel) or with the addition of 150 μM NiSO4 (right side of each panel), before DNase I cleavage. On the right of each autoradiographic film, the G27 genomic coordinates of DNase I protected regions (black boxes) are reported, with position in brackets with respect to the transcriptional start site (TSS). Low affinity binding sites, if present, are shown as grey boxes surrounded by the same information. On the left, a schematic representation of the promoter is provided, with the TSS (+1, bent arrow) and the −10 region (black box). The position of the consensus sequence is reported with violet boxes, corresponding to the two conserved hemi-operator pentamers linked by a black line (15 nt spacer). In the middle panels a scheme of the corresponding transcriptional unit is shown, together with the normalized tag densities obtained from the ChIP-seq experiments (wt/ni+ in yellow, wt/ni− in orange and ∆nikR/ni+, negative control in green), the predicted peak extension by Homer2 and the DNaseI protected regions. Representation scales of ChIP-seq tracks are indicated on the left in brackets. In the bottom panels, the RNA-seq strand specific tracks of the corresponding genomic locus are visualized for wt/ni+ and wt/ni− samples (plus strand in blue, minus strand in red). P* indicates coordinates mapping on the pHPG27 plasmid. (B) Radiolabeled dapD, exsB, fecD and pcrA DNA probes were mixed with 0, 9.7, 29, 97 and 290 nM of NikR tetramers, without nickel (left side of each panel) or with the addition of 150 μM NiSO4 (right side of each panel). The same elements and information are reported as in panel A.
After the definition of high fidelity NikR binding sites, we set out to determine their nickel-dependence by comparing the read coverage of the 72 high-quality binding peaks in the nickel-treated wt strain (wt/ni+) with the read coverage at the same positions of the untreated culture (wt/ni−). 46 out of the 72 peaks resulted significantly enriched (log2FC > |1|, adj p–value < 0.01), suggesting a differential increase of NikR binding affinity to these targets in the presence of nickel (Supplementary Table S3). The remaining 26 peaks (36%) proved to be not differentially bound, suggesting that NikR binding to these positions is already saturated at the low nickel concentrations provided by the culture medium (Brucella Broth contains 0.2 μM nickel ions)15, or that these targets represent nickel-independent binding sites and/or derive from local flaws in ChIP-seq sensitivity.
Systematic definition of the NikR regulon by RNA-seq and ChIP-seq data integration
RNA-seq analysis identified 20 DEGs responding to nickel in a NikR-dependent fashion. These 20 genes belong to 14 different operons and among them, the urease operon was the only one to be up-regulated. On the other hand, ChIP-seq analysis revealed the presence of 46 NikR binding sites specifically enriched upon nickel stimulation. 19 of these binding sites are located at promoter regions, 10 of them map inside the promoters controlling operons which contain 14 of the 20 previously described DEGs, and one falls between nikR and exbB2 promoters (Supplementary Table S3). The peak on the fur promoter was detected but not differentially bound by NikR, suggesting that the regulator binds at this promoter in both high (ni+) and low nickel conditions (ni−). The remaining 5 genes deregulated in the RNA-seq experiment, but not directly associated to a called peak, belong to the operons led by the nixA, phbA and vdlC genes. Eventually, their ChIP-seq profiles were manually inspected, revealing three detectable peaks (all slightly below the threshold imposed for statistical significance) on the promoter regions. Furthermore for one of them, located on the nixA promoter, the direct NikR binding was previously demonstrated11. To confirm NikR binding on the nickel responsive operons identified by RNA-seq analysis, we performed in vitro DNaseI footprinting assays with purified recombinant NikR protein (Fig. 2A and Fig. S3). Remarkably, all the probes exhibited a detectable footprint of 32 bp (average size) starting at the minimal concentration of NikR used. Protection appeared only in presence of nickel, with the exception of hopV and hopW promoter regions for which in vitro binding was observed also in the absence of nickel, although the protection was remarkably stronger in the presence of the metal co-factor. For hopV, hopW, hcpC, vdlC, dvnA and mccB the NikR element overlapped the TSS and/or the core promoter region, while binding sites farther upstream (>50 bp from TSS) were verified only for ureA and phbA. Finally, 9 out of 19 promoter peaks individuated by ChIP-seq analysis that were differentially enriched after nickel stimulation, were not associated with differential nickel regulation in RNA-seq, suggesting that nickel-responsive binding of NikR to these regions does not result in measurable transcriptional effects, at the conditions used.
Extensive intragenic binding of NikR
A remarkable number of predicted NikR binding sites mapped in intragenic positions (41/72) and 23 of them were differentially enriched upon nickel stimulation. To exclude the possibility that this result derived from a bias in the ChIP experiment, footprinting assays were performed on a panel of intragenic differentially enriched binding sites (Fig. 2B). In all cases NikR exerted a clear and nickel-dependent protection centered within the called peak, validating ChIP-seq results and revealing extensive intragenic NikR binding not previously reported. Similarly to the promoter peaks not associated with nickel dependent transcriptional regulation, also the intragenic NikR targets were not affected by transcript levels variations (Fig. 2B), suggesting that more than 70% of NikR in vivo targets are orphan in terms of regulatory control. As such, the role of the transcriptionally orphan NikR binding sites may envisage also putative nucleoid-associated functions and/or (long-range) effects on gene expression, which certainly deserve further investigation in the future.
Three ncRNAs belong to the NikR regulon
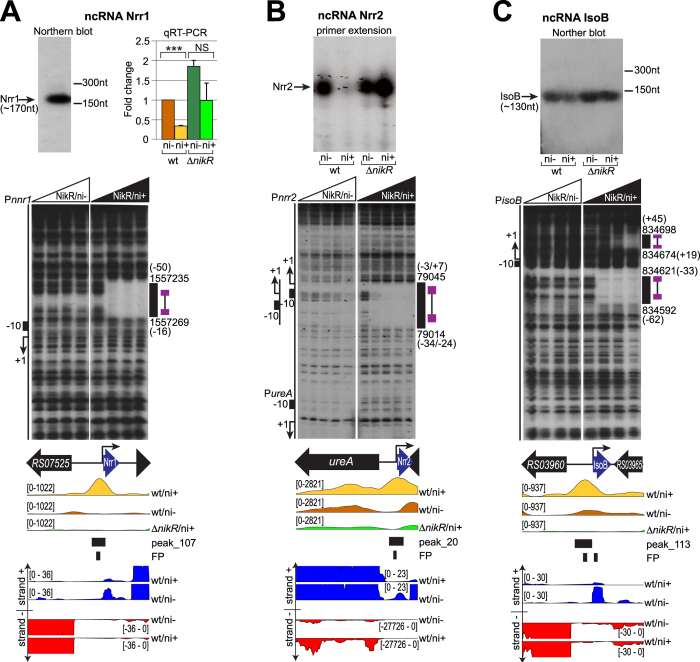
Interestingly, the ChIP-seq analysis mapped a NikR binding site to the promoter of a transcript corresponding to aapA6 (HPnc8050) in strain 2669520. This gene encodes a small ORF-encoding component of a putative class I toxin-antitoxin system. Analysis of the RNA-seq tracks showed a transcript downstream of this putative promoter also in strain G27: the TSS was validated by primer extension analysis, while RNA-blotting showed a single product of 170 nt (Fig. 3A), with only partial sequence similarity to the 5′ region of the aapA6 transcript. In fact, the transcript encoded by the G27 strain lacked any obvious ORF and no antisense transcript was detected on the opposite strand (Fig. 3A). These observations suggest that in strain G27 this locus encodes a non-coding RNA rather than a toxin-antitoxin system. The transcript was accordingly renamed Nrr1, for NikR-regulated sRNA1. The binding of NikR to the nrr1 promoter was confirmed in vitro by DNase I footprinting, with a strong protection occurring at the minimal concentration of protein tested only with the presence of nickel (Fig. 3A). Nrr1 expression levels were assayed by qRT-PCR, with results showing reduced transcript levels in response to nickel, and the loss of regulation in a ΔnikR genetic background, suggesting that NikR represses Nrr1 under nickel-replete conditions (Fig. 3A).
Figure 3. Validation of the new NikR-dependent nickel-regulated ncRNAs.
Top panels: transcriptional analysis of ncRNAs in wt/ni−, wt/ni+, ∆nikR/ni− and ∆nikR/ni+ conditions. (A) Northern blot of Nrr1 (left) and quantitative RT-qPCR of its transcript levels (right) (see legend 2D for details). (B) Primer extension analysis of the Nrr2 transcript. (C) Northern blot of the IsoB transcript. Middle panels: DNase I footprinting of radiolabeled Nrr1 (A), Nrr2 (B) and IsoB (C) DNA probes, mixed with 0, 9.7 (only panel B), 29, 97 and 290 nM of the NikR tetramer, without nickel (left side of each panel) or with the addition of 150 μM NiSO4 (right side of each panel). Uncropped blots and gels are provided in the Supplementary Information. Legends and symbols as in Fig. 2.
Another interesting case is represented by the intergenic region upstream of ureA, encompassing a divergent TSS, conserved in strain G27 and 2669520. RNA-seq data showed strong nickel and NikR-dependent down-regulation of this transcript (Fig. 3B), that corresponds to the conserved ncRNA HPnc0260. Direct NikR binding within the core promoter region was further validated by DNase I footprinting, demonstrating that a single NikR operator oppositely regulates the divergent ureA-HPnc0260 promoters (Fig. 3B), repressing the HPnc0260 ncRNA (renamed Nrr2: NikR-regulated sRNA2) while inducing the transcription of the urease operon.
Refinement of the ncRNA annotation also showed that a peak on the HPG27_RS03960 gene encompassed the promoter of a divergent transcript, homologous to the small ORF-encoding mRNA/antisense RNA family aapB/IsoB (HPnc4170 and HPnc4160). AapB-IsoB together were proposed to form one of several class I toxin–antitoxin loci, in which the Iso transcript functions as asRNA antitoxin to modulate the expression of the (sense) aap transcript20. While the IsoB antisense transcript was confirmed by primer extension analysis (not shown) and Northern blot analysis (Fig. 3C), the aapB transcript was barely detectable in the G27 transcriptome (Fig. 3C), and we were not able to confirm its expression with alternative techniques nor in different growth conditions (data not shown). Thus, IsoB may be the unique transcript arising from this locus, embodying a ncRNA that could target other mRNAs or Iso-Aap families in trans. Direct binding of NikR to the IsoB promoter was validated by footprinting analysis (Fig. 3C). Moreover, the IsoB transcript was down-regulated by nickel, while in the ΔnikR mutant the response to nickel vanished (Fig. 3C), strongly suggesting that, similarly to Nrr1 and Nrr2, IsoB is repressed by NikR. In conclusion, the integration of different genome-wide approaches permitted the identification of three new non coding transcripts repressed by NikR in nickel-replete conditions, suggesting that the responses to nickel may include extensive post-transcriptional regulatory events, which could explain some of the pleiotropic effects observed in the nikR knockout mutant (see also Fig. S2).
NikR consensus sequence
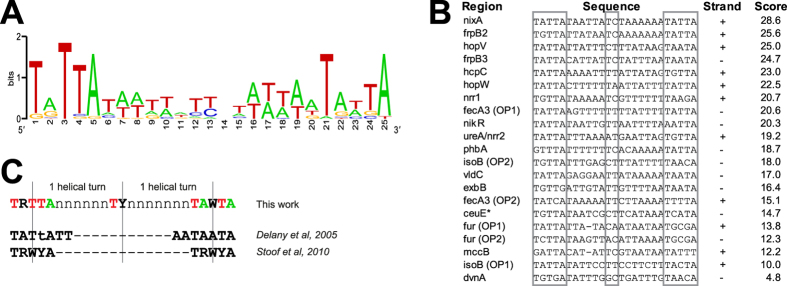
Finally, we used GLAM221, to investigate the consensus for NikR binding (Fig. 4A). Employing the promoter sequences protected by NikR in the footprinting experiments (Fig. 4B, this work and previously reported sequences10,11,13,16), we obtained the conserved pentamers TRTTA and TAWTA, positioned 15 nt apart from each other, with a relevant but less conserved TY element in between (Fig. 4A). This consensus sequence closely matches a TRWYA dyad motif predicted by bioinformatic analyses22. The 20 nt distance between the center of the 2 pentamers is in accordance with binding of one NikR tetramer to two hemi-operator regions separated by exactly two DNA helix turns23,24 (Fig. 4C). Moreover, the two half-sites of the consensus motif appear to be almost completely conserved among different H. pylori strains, hinting at a conserved operator readout mechanism (Supplementary Table S5). Interestingly, the first thymine of the second repeat is highly conserved among the NikR-bound promoters, since all the sequences used to generate the consensus have a T in that position, with the exception of PnikR, bound by NikR with lower affinity12 (see also Supplemenatry Table S6). Previous analysis of the sequence determinants for a tight DNA-protein binding has identified this position as an essential element for a low KD25. Coherently, the operators encompassing a thymine in this position, exhibit high binding affinity of NikR in our footprinting experiments (Supplementary Table S6). On the contrary, the presence of a cytosine characterizing one of the hemi-operator motifs of the 26695 ureA operator25, appears not to be a pre-requisite for high affinity binding of NikR.
Figure 4. NikR consensus sequence.
(A) Weblogo of the NikR consensus sequence elaborated by GLAM2 considering all validated NikR operators within gene promoters. (B) List of the DNA sequences aligned by GLAM2 to generate the consensus sequence, with the strand used for the alignment and the scores resulting from re-alignement of each sequence to the consensus. Homologous regions in G27 were used if the operator was originally characterized in a different strain. *The NikR operator on PceuE was re-mapped accordingly to the Maxam–Gilbert G + A reaction reported in ref. 14. (C) Proposed NikR consensus sequence and comparison with the published ones.
NikR regulates two hop paralogues encoding predicted outer membrane transporters
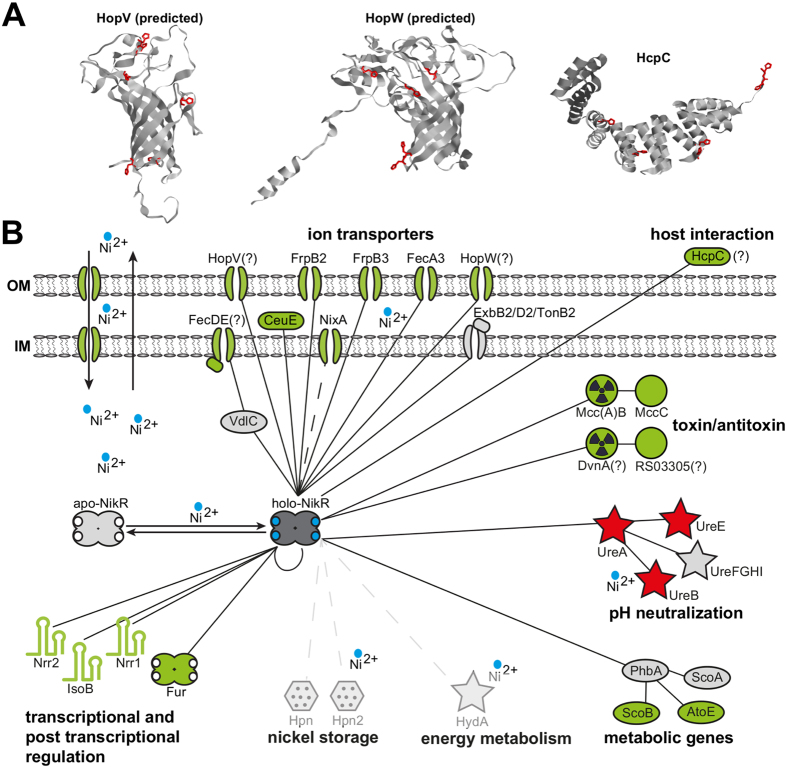
Our analysis also indicated that the hopV and hopW genes, encoding Helicobacter-specific outer membrane proteins, predicted to function as low-permeable broad range porins or as specific transporters for yet undetermined ions26, are directly regulated by NikR (Supplementary Table S3 and Fig. 5). Comparative genomic analysis with Ortholuge, a computational method that can generate precise orthologue predictions between species on a genome-wide scale27, indicated that hopV and hopW are conserved in gastric Helicobacter species, e.g. H. acinonychis and H. cetorum, while they are not conserved in non-gastric Helicobacter lineages. This prompts the hypothesis that these genes may have a role in nickel homeostasis, contributing to the adaptation to the gastric niche. Tertiary structure prediction indicated a transmembrane β-barrel fold for both H. pylori HopV and HopW, with histidine residues exposed on both surfaces as well as internal to the channel, tentatively suggesting a function as metal ion channels across the outer membrane (Fig. 5A). Interestingly, a hopV knockout mutant, showed a significant derepression of NikR regulatory targets under physiological growth conditions (Fig. S5), indicating lower intracellular concentration of nickel ions. This indirect evidence suggests that HopV may be involved in the trafficking of nickel ions.
Figure 5. Model of the NikR regulon.
(A) Predicted structures of HopV and HopW proteins, computed by the online tool Phyre2 and deposited crystallographic structure of the HcpC protein (PDB 1OUV). Histidine residues are highlighted in red. (B) The network connects NikR to the first gene of each transcriptional unit (TU) under direct (validated by footprinting) transcriptional control. Filled lines indicate interactions verified in this study, while dotted lines denote uncertain interactions. Genes part of the same TU are linked to the first gene of the cistron, with red, green and grey colours symbolizing induction, repression or absence of NikR-dependent regulation, respectively. Symbols are related to the predicted biological function.
Discussion
Much of our knowledge on nickel-dependent transcriptional responses derives from the detailed study of single genes or gene clusters controlled by the NikR family of regulators in different bacteria. These studies were generally biased towards nickel- or metal-binding proteins and transporters, providing a patchy view on NikR regulation. On the other hand, HpNikR is the only member of the NikR transcription factor family that has been addressed by genome-wide studies to date8,13. Our results indicate that in H. pylori NikR regulates 17 nickel-responsive transcriptional units, mainly repressed by the regulator, representing a fairly compact regulon with clearly defined functional roles (Fig. 5). This sets NikR as specific transcription factor dedicated to nickel-dependent responses rather than a pleiotropic regulator, despite other RHH-type regulators have been shown to act as global transcriptional regulators (e.g. Pseudomonas AmrZ28). In fact, considering the hundreds of DEGs indirectly affected by the nikR deletion, we can infer that some predicted targets validated by nikR mutational analysis reflect a pleiotropic adaptation to stresses deriving from the altered physiology and/or pervasive changes in the transcriptome of ΔnikR strains. One likely explanation is that together with the deregulation of the Fur regulon this effect arises from the deregulation of the NikR-dependent small ncRNAs, which may post-transcriptionally control the output of the nickel regulatory responses.
Interestingly, the NikR regulon includes both genes specific to the Helicobacter lineage as well as genes that are conserved in other bacteria. For example, the fecA, frpB and tonB-exbBD metal transporter systems, together with urease and urease chaperones, are widespread in Gram-negative organisms and have been repeatedly reported to respond to nickel29,30. In addition our analysis has included the vdlC-fecD-fecE operon in the NikR regulon. This operon comprises the fecD and fecE genes, that are annotated as putative components of an ABC transporter for iron (III) dicitrate. Nevertheless, their expression proved independent from iron stimulation31 and resulted to be regulated by NikR in response to nickel availability (Fig. S3). Moreover, the inactivation of fecD in H. mustelae showed reduced uptake of nickel and cobalt ions, along with reduced urease activity, indicative of a low availability of intracellular nickel, while no effect on iron trafficking was detected22. These observations indicate that FecD orthologues may act as nickel trafficking systems.
NikR also appears to regulate toxin-antoxin systems that are conserved in other bacterial species. For example, the mccB and mccC genes carried by the pHPG27 plasmid share similarity with the E. coli mccB and mccC genes, involved in the production of microcin C (McC). McC is a 1.2 kDa antibiotic peptide produced by many strains of the Enterobacteriaceae family, which inhibits the growth of phylogenetically-related species by interfering with the translation machinery, if the corresponding antitoxin is not expressed32. In E. coli the MccA pre-toxin is adenylated by MccB into the mildly active McC toxin intermediate33, that is eventually exported by the MccC protein, protecting the producer organism from their toxic effect. Recent findings indicate that the H. pylori strains containing this system are able to produce and export at least the intermediate form of McC34. These results suggest a strategy for rebuffing other microorganisms or cognate H. pylori strains lacking the episomal element, in order to succeed in the struggle for nickel acquisition. Similarly, the HPG27_RS03310-HPG27_RS03305 bicistronic operon appears to encode a nickel-responsive chromosomal antimicrobial system. In fact, HPG27_RS03310 exhibits 69% similarity with the dvnA gene of Carnobacterium divergens, encoding a class IIc Sec-secreted bacteriocin Divergicin A precursor35. It also shares high similarity to enterocin-B and enterocin-Q, the other two components of the class IIc bacteriocins. In C. divergens the dvnA cistron is followed by the dviA gene, encoding a transmembrane protein protecting the cell from the negative effect of the DvnA toxin36. In H. pylori the downstream gene in the operon (HPG27_RS03305) also encodes a membrane protein, that clusters together with HPG27_RS03310 in more than 50 different strains of H. pylori and H. acinonychis. Together, these findings represent the first evidence of nickel-responsive toxin-antitoxin systems, pointing at a possible selective antimicrobial weapon induced by nickel starvation, which may confer a selective advantage in conditions in which nickel represents a limiting source.
On the other hand, several of the NikR regulon members coding for membrane-associated or exposed proteins seem to be specific to Helicobacter. For example, in addition to the hopV and hopW OMP paralogues described before, the hcpC gene encodes for a secreted and OM-absorbed protein, which belongs to the family of Helicobacter cysteine-rich proteins37. HcpC is predicted to fold into a soluble α-helix-rich structure, with few histidines and a high number of exposed cysteine residues, hinting at a possible role in binding soluble metal ions (Fig. 5A). Members of the Hcp family have a poorly characterized putative β-lactamase function. Moreover, their expression associates with chronic atrophic gastritis38, and they have been shown to interact with host cells, inducing the production of IFNγ and other proinflammatory cytokines39. HcpC in particular fulfills different roles in the H. pylori virulence arsenal, ranging from immunoevasion to control of host invasion40. Evidence that hcpC is repressed by NikR and nickel, suggests that under nickel starvation the bacterium may boost its virulence, through one or more of the HcpC functions.
Our analysis also demonstrates that HpNikR, similarly to EcNikR, functions principally as repressor of transcription, with the only exception represented by the apparently positive regulation of the urease operon (Supplementary Table S2; see also below). Some of the down-regulated targets identified are well-known genes of the NikR regulon involved in metal trafficking: nixA, frpB2, frpB3, ceuE, fecA3 and fur. Hence, our analysis is in accordance with a significant portion of previous literature8,9,11,13,15,16,18,41. Moreover, our analysis pinpointed hopV and hopW to the NikR regulon, validating previous indications8, and identified new targets (hcpC, [vdlC]-fecD-fecE, mccB-mccC, [phbA]-scoB-atoE, mccB-mccC, dvnA-HPG27_RS03305) and non-coding transcripts (nrr1, nrr2, isoB; see Supplementary Table S6). Other genes that were previously shown to be under the control of NikR were not confirmed in our analysis: hpn, hpn2, nikR, exbB-exbD-tonB, hydA amongst others8,9. These discrepancies possibly arise from the different experimental set-up: we used a medium supplemented with FBS, which has a higher complexity compared to the more neutral ß-cyclodextrin used in the study of Muller et al.9. Moreover, while most of the previous investigators treated with low concentrations of nickel for prolonged times (see Supplementary Table S6), we chose a brief impulse of high nickel excess to focus on the nickel- and NikR-dependent responses. This approach likely minimizes the collateral effects of a prolonged exposure to nickel, which could elicit slow occurring adaptive responses through the activation of other pathways: the stress response circuit (hrcA, hspR, groELS8), the Fur regulon, growth-phase dependent effects and post-transcriptional regulation by non-coding RNAs. Nevertheless, we cannot exclude that our set-up underestimates the cohort of genes transcriptionally controlled by NikR, since the short time frame could be insufficient to detect significant transcriptional variations, in case of highly stable mRNAs and low affinity binding sites.
One particularly intriguing aspect emerging from this study is the absence of regulation of nikR in response to nickel, while this gene is widely considered as autorepressed. In fact, the NikR protein is able to bind its own promoter in presence of nickel ions8,10,12,25,42. Moreover, the expression levels (both mRNA and protein) decrease after prolonged exposure to nickel8,9,18. The divergent exbB-exbD-tonB operon is reported to be regulated by NikR similarly to the nikR gene: NikR binds to the exbB promoter, sharing the PnikR operators10, and the mRNA levels of the downstream genes are repressed in response to prolonged nickel treatment9. In the ChIP-seq experiment we were able to pinpoint a nickel-dependent peak on the nikR-exbB intergenic region, which indicates in vivo binding of NikR to this locus. However, we were unable to detect a trascriptional response of nikR and exbB-exbD-tonB operons under conditions promoting the response of the other genes belonging to the NikR regulon. In this respect, it is important to recall that PnikR is a weak binding target of NikR, with a 100-fold higher KD compared to other promoters12. Moreover, nickel-dependent variations of nikR mRNA or NikR protein levels were observed only after prolonged exposure to the metal (5 h with 10 μM nickel for mRNA and 10 h for protein)9, or were not detectable at all41. Similarly, variations of exbB transcript levels (and also variations of hpn, hpn2, and hydA mRNA) were observed only after prolonged treatment with nickel9. Because of the low-affinity binding of NikR on PnikR and PexbB promoters and the slow kinetics of transcriptional response of these genes to nickel treatment, it is possible that NikR binds and/or regulates these promoters only after prolonged nickel exposure. Another possibility is that regulation at these loci occurs through a more complex regualtory mechanism. Interestingly, Fur binds these promoters on two operators which partailly overlap those of NikR10. In vitro, Fur binding to the latter occurs in absence of metal ions, but increases when metal ions, including nickel, are added10. Competitive footprinting experiments also demonstrated that Fur is able to compete for NikR binding on the nikR promoter, suggesting that a competition between these two transcriptional factors could occur in vivo10. Moreover, since Fur is under the transcriptional control of NikR, the effect exerted by NikR on the nikR and exbB promoters could in principle be also mediated by Fur, resulting in an indirect and slower response kinetics at those promoters. Finally, it is worth noticing that Fur is able to use also nickel as co-factor to bind DNA10. As such it may contribute to transduce nickel-dependent responses, but only after prolonged exposure to the metal, since a 20 min nickel pulse is not sufficient to activate the Fur regulon (Fig. 1C). Taken together, our results suggest that the regulation of nikR, exbB, hpn, hpn2 and other genes with a slow response to nickel may be under the control of a different regulatory mechanism or circuit with respect to the more responsive members of the NikR regulon. The interplay between Fur and NikR seems to have a paramount role in these responses and clearly deserves further investigations.
Another paradigmatic example of the complexity of nickel regulation in H. pylori is represented by hpn and hpn2, which code for two highly expressed histidine-rich paralogues, fundamental for the colonization of the host43. Nevertheless, hpn and hpn2 respond to nickel very differently in distinct H. pylori strains (Fig. S4). For hpn this is likely imputable to significant sequence divergence within the promoter region (Fig. S4C). This observation appears striking, given the evolutionary importance of hpn and hpn2 in the adaptation to the gastric habitat and their essential role for colonization. For example, hpn2 was induced in response to the short (20 min) stimulation with 500 μM nickel in strain 26695, while we were not able to record any variation of the transcript in strain G27 (Fig. S4A), even though the low ChIP-seq enrichment values and the presence of a low affinity NikR binding site verified by in vitro footprinting, are compatible with a weak interaction of NikR with the promoter (Fig. S4B and S4D). Moreover, when the treatment with lower concentrations of nickel (20 μM) was prolonged for 5 hours, a sheer down-regulation of the hpn2 transcript was detected in strain G27 (Fig. S4A). Notably, these responses were maintained in the nikR knockout mutant, suggesting that another regulator participates to the transcriptional response of hpn2. Currently, the best candidate appears to be the ferric uptake regulator Fur, which is able to use nickel as co-repressor and bind to the Phpn2 promoter, repressing its transcription44.
Another possible explanation is that the positive effect of NikR may be postranscriptionally mediated by ncRNAs. The observation that positive responses on hpn, groESL, and hydA were recorded hierarchically after prolonged growth in nickel-replete media9, and do not respond promptly to nickel stimulation as all the other repressed targets, supports this hypothesis. Moreover, the hpn transcript is subjected to post-transcriptional control by aconitase, a metal-sensor contributing to bacterial pathogenesis45. Together with s-SodF, a recently identified sRNA derving form the 3′UTR of a nickel regulated transcript in Streptomyces coelicolor46, the three ncRNA regulated by NikR reported in this work represent the first examples of nickel-responsive sRNAs. Thus, in analogy to the conservation of iron-regulated small RNAs pioneered by the discovery of E. coli RyhB47, also nickel-responsive sRNAs maybe widespread and extend their function beyond metal homeostasis, involving the expression of virulence-associated factors in pathogenic bacteria. In this light, the possibility of a nickel-responsive post-transcriptional regulation of urease deserves further investigation, since other sRNA regulating urease have been reported both in H. pylori48 as well as in enterohemorrhagic E. coli49. In fact, the evidence that the unique NikR binding site upstream of the urease promoter is clearly associated with nickel-responsive repression of the divergent Nrr2 transcript, poses the question whether NikR binding to this operator also directly activates the ure promoter. A recent study suggested that NikR has acquired a RNA polymerase (RNAP) interacting domain50 and could therefore act as transcriptional activator. In this scenario, it is surprising to notice that upon the acquisition of a RNAP interacting domain a key transcription factor like NikR is engaged in the control of only one (urease) operon.
Finally, the integration of RNA-seq and ChIP-seq datasets allowed the identification of a large number of genomic loci bound by NikR in response to nickel excess, which are not related to transcriptional regulation. This is not surprising, since for many regulator targets identified by genome-wide location analyses, including those of HpFur44, a detectable transcriptional effect on the neighbouring genes is absent when the regulator is depleted51. This may be due to several reasons. NikR may be bound upstream of a gene where it has little impact on levels of transcription, because of the overriding influence of another regulatory mechanism or because its role is only to to fine-tune the levels of transcription. While this may be true for complex promoters, e.g. the arsR promoter were the interaction of Fur and NikR governs a sofisticated signal integration logic52, this possibility does not explain the role of intracistronic binding sites. Tentatively, intragenically bound NikR could hint at nucleoid-associated functions important for chromosome organization53. In this respect, the intragenic binding sites could also represent dedicated ‘parking bays’ for the regulator, hold in place by nucleoid topology in order to maximize regulator concentration at a spacially proximal regulatory element. Finally, it has not to be excluded that the intragenic targets may have no direct functional relevance, since bacterial genomes are permissive to transcription factor binding per se54.
In conclusion, the first comprehensive insight into bacterial nickel-dependent responses discloses a regulon specifically devoted to nickel-trafficking and metallo-enzyme genes, with the potential of extensive post-transcriptional regulatory capabilities, and an intimate interlink with the Fur regulatory circuit. Interestingly the NikR regulon also includes antibiotic and virulence factors not immediately associable with nickel homeostasis, which may contribute to shape responses in a bacterial community. As such the insights on the H. pylori NikR regulatory network contribute to a broader understanding of the nickel metabolism in other ureolytic microorganisms colonizing soil or the human oral, gastrointestinal, urinary and respiratory tracts, with evident biotechnological and biomedical implications.
Materials and Methods
Bacterial strains and growth conditions
All H. pylori strains used are listed in Supplementary Table 4. Bacteria were recovered from frozen glycerol stocks and propagated on BBL Brucella (BD) agar plates containing 5% fetal bovine serum FBS. Bacteria were grown at 37 °C in jars using CampyGen™ (Oxoid) gas–packs or in a water–jacketed thermal incubator (9% CO2, 91% air atmosphere, and 95% humidity) for 24–48 hr. Liquid cultures were grown in BBL Brucella Broth (Sigma–Aldrich) supplemented with 5% FBS and Dent’s or Skirrow’s antibiotic supplement at 37 °C in glass flasks with gentle agitation (125 rpm). In order to faithfully record the transcriptional responses to nickel transduced by NikR (DNA-binding) activity, and avoid the bias deriving from slow adaptation to growth in nickel-rich conditions instead, the bacteria were treated for short times with relatively high concentrations of nickel (500 μM Ni2+ for 20 min). E. coli strains were grown in Luria–Bertani (LB) agar or in LB broth. When required, 100 μg/ml ampicillin was added.
DNA manipulations
DNA amplification, restriction digestions and ligations were carried out with standard molecular techniques, with enzymes purchased from New England Biolabs.
RNA preparation, qRT assays, primer extension and Northern blot
Bacterial cultures of wild type and ΔnikR strains were grown to OD600 of 1.0–1.1 and split into 2 sub–cultures of 5 ml each that were treated either with 500 μM nickel (ni+) or with the same volume of sterile water (ni−) for 20 minutes. Samples were stopped by addition of 625 μl RNA stop solution (95% ethanol, 5% acid phenol) and RNAs were purified using 1 ml of Tri–reagent (Sigma-Aldrich) for each sample, following the manufacturer instructions. RNAs were treated with DNaseI prior to cDNA synthesis55. Two μl of 1:10 diluted cDNAs were added to 5 μl of PowerUp™ SYBR® Green Master Mix (TermoFisher) and 400 nM of forward and reverse oligonucleotides in a 10 μl final volume. The qRT–PCR program was performed as previously described55. Primer extension analysis was performed using 15 μg of total RNA and 0.1 pmol of radiolabeled probe. Northern blot assay was performed using 17 μg of total RNA and 1.25 pmol of radiolabeled oligo probe.
Overexpression and purification of recombinant NikR protein
Recombinant NikR protein was overexpressed and purified under native conditions as previously described19. The purified protein was dialyzed overnight against NikR footprinting buffer (20 mM Hepes pH 7.85, 50 mM KCl, 0.02% Igepal, 0.1 mM DTT and 10% glycerol) prior to the DNA binding experiments. A Bradford colorimetric assay (Bio–Rad) was used to quantify the protein fractions with bovine serum albumin as standard.
Preparation of a polyclonal α-NikR antibody
The α–NikR antisera were generated by immunizing rabbits with affinity–purified recombinant NikR protein (without tag) by Biotem Custom Antibodies and Services. After the final bleed, the antisera were purified by 3 sequential precipitations with 35% saturated (NH4)2SO4 and subsequent dissolution in water. The partially chemical purified α–NikR antibody was assayed by western blot analysis on H. pylori total extracts, showing a single major band corresponding to the expect molecular weight of NikR.
Chromatin Immunoprecipitation with a polyclonal α–NikR antibody
Bacterial cultures of wild type and ΔnikR strains were grown to OD600 of 1.0–1.1 and split into 2 sub–cultures of 50 ml each that were treated with 500 μM nickel (ni+) or with the same volume of sterile water (ni−) for 20 minutes. Samples were crosslinked with 1% formaldehyde for 10 min at room temperature, then the reaction was stopped by treatment with 125 mM glycine for 10 min at room temperature, centrifuged 3900 g for 10 min at 4 °C and washed twice in 50 ml of cold 1X PBS followed by same steps of centrifugation. Samples were resuspended in 500 μl of TE (10 mM Tris, 1 mM EDTA; pH 8) with 2 mg/ml lysozyme solution, incubated 1 hr at 4 °C on a rotation wheel, diluted with 500 μl of 2X sonication buffer (10 mM Tris pH 8, 1 mM EDTA pH 8, 200 mM NaCl, 0.2% Sodium deoxycholate, 0.2% Igepal, 0.2% SDS, 1.5% Triton X–100), sonicated with Bioruptor (Diagenode) at high power for 80 min (30 sec ON–30 sec OFF) and diluted with 200 μl of dilution buffer (10 mM Tris pH 8, 1 mM EDTA pH 8, 340 mM NaCl, 0.1% Sodium deoxycholate, 0.1% Igepal, 2.25% Triton X–100). After a centrifugation of 3400 g for 8 min at 4 °C, 50 μl of the surnatant were used for Input preparation, while 1.1 ml was incubated with 50 μl of recombinant protein G – sepharose (Life Technologies) pre–equilibrated in RIPA buffer (10 mM Tris pH 8, 1 mM EDTA pH 8, 140 mM NaCl, 0.1% Sodium deoxycholate, 0.1% Igepal, 1% Triton X–100) for 1 hr a 4 °C. The NikR antibody was added to the pre–cleared surnatants at a 1:200 dilution; after incubation at 4 °C for 16 hr on a rotation wheel 50 μl of protein G – sepharose beads pre–equilibrated in RIPA buffer were added and binding reaction was carried out for 3 hr at 4 °C on rotation. The beads were washed four times in cold RIPA buffer, twice in cold TE, resuspended in 100 μl of TE solution with 20 μg/mL RNase A, and incubated at 37 °C for 30 min. SDS was added at a final concentration of 0.5% and proteinase K at a final 50 μg/mL concentration, followed by 16 hr incubation at 37 °C and addition of the same amount of proteinase K. Surnatants were phenol–chloroform extracted twice, chloroform extracted twice and ethanol precipitated with the addition of 20 μg of glycogen (Sigma–Aldrich). Input samples were obtained from 50 μl of sonicated material, following the same procedures described for the IPs, starting from RNase incubation step.
DNase I footprinting
The DNA probes were prepared as follows: 1 pmol of pGEM–PureA, pGEM–cpdB, pGEM–vdlC, pGEM–PfecD, pGEM–PhopW, pGEM–PmccB, pGEM–Phpn and pGEM–Phpn2 vectors were linearized with NcoI, while pGEM–PhcpC, pGEM–PhopV, pGEM–PdvnA, pGEM–exsB, pGEM–dapD, pGEM–pcrA, pGEM–Pnnr1, pGEM–PisoB and pGEM–PphbA vectors were linearized with NdeI, dephosphorylated with calf intestinal phosphatase and labeled at the 5′ ends with 2 pmol of [γ–32P] ATP (6000 Ci/mmol; PerkinElmer) by using T4 polynucleotide kinase. The labeled DNA probe was further digested either with NdeI or NcoI and the products were separated by native polyacrylamide 4% gel electrophoresis, eluted and purified as previously described55. The binding reactions were carried out by using approximately 20 fmol of labeled probe and increasing concentrations of NikR protein (from 9.7 to 290 nM of the NikR tetramer) at room temperature for 15 min in a final volume of 50 μl in footprinting buffer with 300 ng of salmon sperm DNA (Invitrogen) as a nonspecific competitor. Afterwards, DNase I (0.075 U), diluted in footprinting buffer containing 10 mM CaCl2 and 2.5 mM MgCl2 was added to the reaction mixture (2 μl) and digestion was allowed to occur for 90 s. The reaction was stopped, purified and resuspended in formamide loading buffer; samples were denatured at 100 °C for 3 min, separated on 8 M urea –6% acrylamide sequencing gels in TBE buffer and autoradiographed; a modified G + A sequencing ladder protocol was employed to map the binding sites, all according to ref. 55.
ChIP-sequencing
Illumina libraries were prepared, for each of the conditions and strains analysed either from 5 ng of the two biological replicates of immunoprecipitated-DNA (IPs) or from 5 ng of the two biological replicates of the Input-DNA (INPUT) following the Illumina TruSeq ChIP-seq DNA sample preparation protocol; then each library was sequenced on a GAIIx or MiSeq Illumina sequencer and 51 bp single stranded reads were produced.
RNA-sequencing
Ribosomal RNAs were depleted starting from 1 μg of total RNA from each of the conditions analyzed by using the RiboZero Gram negative kit (Epicentre, Illumina). Strand specific RNA-seq libraries were prepared by using the ScriptSeqTM v2 RNAseq library preparation kit (Epicentre, Illumina) starting from 50 ng of previously rRNA depleted RNA from each biological replicate and for all the conditions analyzed. Then each library was sequenced on a GAIIx or MiSeq Illumina sequencer and 76 bp reads were produced. Bam files are publicly available at Sequence Reads Archive (SRA) under accession number BioProject PRJNA313048.
Reads mapping quality assessment
Bowtie 2 (v2.2.6) was used to align raw reads, produced from both ChIP and RNA sequencing experiments, to H. pylori G27 genome (RefSeq GCF_000021165.1). End-to-end mapping was performed and non-deterministic option was specified to force a single assignment of multi-mapping reads to the best scoring region (if present) or a random attribution in the case of regions with identical scores. High quality reads were then selected requiring: for uniquely mapping reads MAPQ (mapping quality) greater than 30 and alignment score greater than −10 in ChIP-seq or −15 in RNA-seq samples; for multi-mapping reads alignment score were set equal or greater than −10 for ChIP-seq or −15 for RNA-seq.
The quality of ChIP-Seq data was evaluated following ENCODE quality metrics56 and the numerical values obtained are provided in Supplementary Table S1. The cross-correlation analysis resulted in good NSC and RSC values. Moreover we obtained average PBC scores. For RNA-seq samples rRNA depletion, strand specificity and gene coverage were evaluated using BEDTools (v2.20.1) and SAMtools (v0.1.19) (see Supplementary Table S1).
ChIP-seq analysis
Irreproducible Discovery Rate procedure (IDR v 2.0.2) following ENCODE guidelines, and using Homer (v4.7.2) as peak caller, was performed to measure sample reproducibility and to identify consistent peaks. Homer parameters were set according to the authors’ indication for IDR calculation (-P 1 -LP 1 -poisson 1), -L was set to three and the fragment length was manually specified according to the median length of the sequencing library distribution. The “Fold Change vs Control” column was selected as ranking column for IDR calculations. R package DiffBind (v1.12.3) was adopted, without background reads subtraction, to determine differential bindings among the tested conditions. ΔnikR-nickel pooled samples were used as input/background for all the other experimental conditions. Peaks were manually classified as “promoter peaks” if centered −150/+30 from a TSS, as “intragenic peaks” if centered inside annotated genes and more than 30 nt apart from a TSS, and “intergenic peaks” if centered in unannotated regions and located farther than 150 nt form a TSS. TSS annotation was obtained cross-mapping onto G27 genome the 50 nt sequence upstream the 26695 published list of TSS20 and manually verifying the correspondence of the loci.
RNA-seq analysis
Strand specific reads overlapping to coding sequences for at least 50% of their length were considered to produce the raw-counts of each sample. The R package DESeq2 (v1.4.5) was then used to normalize the counts and to individuate differentially expressed features showing log2FC ≥ |1| and BH adjusted p-value lower than 0.01. Genes were annotated to the current version of H. pylori G27 RefSeq annotation (GCF_000021165.1) and Inparanoid v4.1 was adopted to obtain protein ortologues in the reference strain H. pylori 26695. Old H. pylori G27 annotation gene names, H. pylori 26695 ones and common gene names (if available) are reported in parallel to the last H. pylori G27 annotation to facilitate results comparison.
Consensus sequence analysis
The newly validated NikR promotorial binding sites as well as the previously individuated ones (or their homologous regions in G27 strain if the original strain was different from HPG27) were used as input for the consensus analysis. Considering the previous reports that highlighted an A/T-rich pseudo-palindromic recognition sequence for HpNikR targeted promoters10,22,25, we adopted GLAM2 which is specialized in finding gapped motifs to individuate NikR binding sequence, using the default parameters of the tool.
Availabiliy of data and materials
Raw data supporting the conclusions of this article are available in the Sequence Reads Archive under accession number BioProject PRJNA313048; additional datasets are available within the Supplementary Tables 1,2,3,4,5,6 associated with this article.
Additional Information
How to cite this article: Vannini, A. et al. Comprehensive mapping of the Helicobacter pylori NikR regulon provides new insights in bacterial nickel responses. Sci. Rep. 7, 45458; doi: 10.1038/srep45458 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
The authors wish to thank Vincenzo Scarlato for advice and support. We would like to thank Giada Caredda and Maria Vurchio (Institute of Biomedical Technologies, National Research Council, Milan) for technical and administrative support. This work was supported by Grants from the Italian Ministry of Education and University (2010P3S8BR_003, and 2010P3S8BR_002 to CP) and from grants by the University of Bologna to AD.
Footnotes
The authors declare no competing financial interests.
Author Contributions A.V., P.E.C., S.Pe., D.R. performed molecular and in vivo experiments; E.P., S.Pu. and C.P. carried out library preparation and sequencing; E.P. and S.Pu. performed bioinformatic analyses; A.V., C.P. and A.D. conceived experiments with the support of all authors. A.V., E.P., C.P. and A.D. wrote the manuscript; all authors reviewed the manuscript; G.D.B., C.P. and A.D. provided funding or resources.
References
- Sigel A., Sigel H. & Sigel R. Nickel and Its Surprising Impact in Nature. 2, (John Wiley & Sons, Ltd, 2007). [Google Scholar]
- Olson J. W. & Maier R. J. Molecular hydrogen as an energy source for Helicobacter pylori. Science 298, 1788–1790 (2002). [DOI] [PubMed] [Google Scholar]
- de Reuse H., Vinella D. & Cavazza C. Common themes and unique proteins for the uptake and trafficking of nickel, a metal essential for the virulence of Helicobacter pylori. Front. Cell. Infect. Microbiol. 3, 94 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salama N. R., Hartung M. L. & Müller A. Life in the human stomach: persistence strategies of the bacterial pathogen Helicobacter pylori. Nat. Rev. Microbiol. 11, 385–399 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macomber L. & Hausinger R. P. Mechanisms of nickel toxicity in microorganisms. Metallomics. 3, 1153–1162 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiter E. R. et al. Crystal structure of the nickel-responsive transcription factor NikR. Nat. Struct. Mol. Biol. 10, 794–799 (2003). [DOI] [PubMed] [Google Scholar]
- Rodionov D. A., Hebbeln P., Gelfand M. S. & Eitinger T. Comparative and functional genomic analysis of prokaryotic nickel and cobalt uptake transporters: evidence for a novel group of ATP-binding cassette transporters. J. Bacteriol. 188, 317–327 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras M., Thiberge J.-M., Mandrand-Berthelot M.-A. & Labigne A. Characterization of the roles of NikR, a nickel-responsive pleiotropic autoregulator of Helicobacter pylori. Mol. Microbiol. 49, 947–963 (2003). [DOI] [PubMed] [Google Scholar]
- Muller C. et al. Hierarchical regulation of the NikR-mediated nickel response in Helicobacter pylori. Nucleic Acids Res. 39, 7564–7575 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delany I. et al. In vitro analysis of protein-operator interactions of the NikR and fur metal-responsive regulators of coregulated genes in Helicobacter pylori. J. Bacteriol. 187, 7703–7715 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst F. D. et al. The nickel-responsive regulator NikR controls activation and repression of gene transcription in Helicobacter pylori. Infect. Immun. 73, 7252–7258 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosanjh N. S., West A. L. & Michel S. L. J. Helicobacter pylori NikR’s interaction with DNA: a two-tiered mode of recognition. Biochemistry 48, 527–536 (2009). [DOI] [PubMed] [Google Scholar]
- Jones M. D., Ademi I., Yin X., Gong Y. & Zamble D. B. Nickel-responsive regulation of two novel Helicobacter pylori NikR-targeted genes. Metallomics 7, 662–673 (2015). [DOI] [PubMed] [Google Scholar]
- Dosanjh N. S. & Michel S. L. J. Microbial nickel metalloregulation: NikRs for nickel ions. Curr. Opin. Chem. Biol. 10, 123–130 (2006). [DOI] [PubMed] [Google Scholar]
- Wolfram L., Haas E. & Bauerfeind P. Nickel represses the synthesis of the nickel permease NixA of Helicobacter pylori. J. Bacteriol. 188, 1245–1250 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielli A. et al. Growth phase and metal-dependent transcriptional regulation of the fecA genes in Helicobacter pylori. J. Bacteriol. 191, 3717–3725 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst F. D. et al. NikR mediates nickel-responsive transcriptional repression of the Helicobacter pylori outer membrane proteins FecA3 (HP1400) and FrpB4 (HP1512). Infect. Immun. 74, 6821–6828 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahlawane C. et al. Structural and mechanistic insights into Helicobacter pylori NikR activation. Nucleic Acids Res. 38, 3106–3118 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambelli B. et al. High-affinity Ni2+ binding selectively promotes binding of Helicobacter pylori NikR to its target urease promoter. J. Mol. Biol. 383, 1129–1143 (2008). [DOI] [PubMed] [Google Scholar]
- Sharma C. M. et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 464, 250–255 (2010). [DOI] [PubMed] [Google Scholar]
- Bailey T. L. et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–208 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoof J., Kuipers E. J. & van Vliet A. H. M. Characterization of NikR-responsive promoters of urease and metal transport genes of Helicobacter mustelae. Biometals 23, 145–159 (2010). [DOI] [PubMed] [Google Scholar]
- Mazzei L., Dobrovolska O., Musiani F., Zambelli B. & Ciurli S. On the interaction of Helicobacter pylori NikR, a Ni(II)-responsive transcription factor, with the urease operator: in solution and in silico studies. J. Biol. Inorg. Chem. 20, 1021–1037 (2015). [DOI] [PubMed] [Google Scholar]
- Schreiter E. R., Wang S. C., Zamble D. B. & Drennan C. L. NikR-operator complex structure and the mechanism of repressor activation by metal ions. Proc. Natl. Acad. Sci. USA 103, 13676–13681 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans S. E. & Michel S. L. J. Dissecting the role of DNA sequence in Helicobacter pylori NikR/DNA recognition. Dalton Trans. 41, 7946–7951 (2012). [DOI] [PubMed] [Google Scholar]
- Peck B. et al. Characterization of four members of a multigene family encoding outer membrane proteins of Helicobacter pylori and their potential for vaccination. Microbes Infect. 3, 171–179 (2001). [DOI] [PubMed] [Google Scholar]
- Fulton D. L. et al. Improving the specificity of high-throughput ortholog prediction. BMC Bioinformatics 7, 270 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Granero F., Redondo-Nieto M., Vesga P., Martín M. & Rivilla R. AmrZ is a global transcriptional regulator implicated in iron uptake and environmental adaption in P. fluorescens F113. BMC Genomics 15, 237 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora D. & Arioli S. Microbial Urease in Health and Disease. PLOS Pathog 10, e1004472 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noinaj N., Guillier M., Barnard T. J. & Buchanan S. K. TonB-dependent transporters: regulation, structure, and function. Annu. Rev. Microbiol. 64, 43–60 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gancz H., Censini S. & Merrell D. S. Iron and pH homeostasis intersect at the level of Fur regulation in the gastric pathogen Helicobacter pylori. Infect. Immun. 74, 602–614 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metlitskaya A. et al. Aspartyl-tRNA synthetase is the target of peptide nucleotide antibiotic Microcin C. J. Biol. Chem. 281, 18033–18042 (2006). [DOI] [PubMed] [Google Scholar]
- Regni C. A. et al. How the MccB bacterial ancestor of ubiquitin E1 initiates biosynthesis of the microcin C7 antibiotic. EMBO J. 28, 1953–1964 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantysh O. et al. Enzymatic synthesis of bioinformatically predicted microcin C-like compounds encoded by diverse bacteria. mBio 5, e01059–01014 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resende T., Correia D. M., Rocha M. & Rocha I. Re-annotation of the genome sequence of Helicobacter pylori 26695. J. Integr. Bioinforma. 10, 233 (2013). [DOI] [PubMed] [Google Scholar]
- Worobo R. W. et al. A signal peptide secretion-dependent bacteriocin from Carnobacterium divergens. J. Bacteriol. 177, 3143–3149 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumrese C. et al. The secreted Helicobacter cysteine-rich protein A causes adherence of human monocytes and differentiation into a macrophage-like phenotype. FEBS Lett. 583, 1637–1643 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L., Weck M. N., Michel A., Pawlita M. & Brenner H. Association between chronic atrophic gastritis and serum antibodies to 15 Helicobacter pylori proteins measured by multiplex serology. Cancer Res. 69, 2973–2980 (2009). [DOI] [PubMed] [Google Scholar]
- Deml L. et al. Characterization of the Helicobacter pylori cysteine-rich protein A as a T-helper cell type 1 polarizing agent. Infect. Immun. 73, 4732–4742 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roschitzki B., Schauer S. & Mittl P. R. E. Recognition of host proteins by Helicobacter cysteine-rich protein C. Curr. Microbiol. 63, 239–249 (2011). [DOI] [PubMed] [Google Scholar]
- Davis G. S., Flannery E. L. & Mobley H. L. T. Helicobacter pylori HP1512 is a nickel-responsive NikR-regulated outer membrane protein. Infect. Immun. 74, 6811–6820 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. & Zamble D. B. pH-responsive DNA-binding activity of Helicobacter pylori NikR. Biochemistry 48, 2486–2496 (2009). [DOI] [PubMed] [Google Scholar]
- Vinella D. et al. Evolution of Helicobacter: Acquisition by Gastric Species of Two Histidine-Rich Proteins Essential for Colonization. PLoS Pathog. 11, e1005312 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielli A. et al. In vivo dissection of the Helicobacter pylori Fur regulatory circuit by genome-wide location analysis. J. Bacteriol. 188, 4654–4662 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller H. K. & Auerbuch V. Bacterial iron-sulfur cluster sensors in mammalian pathogens. Metallomics 7, 943–956 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. M., Shin J.-H., Cho Y.-B. & Roe J.-H. Inverse regulation of Fe- and Ni-containing SOD genes by a Fur family regulator Nur through small RNA processed from 3′UTR of the sodF mRNA. Nucleic Acids Res. 42, 2003–2014 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massé E. & Gottesman S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. USA 99, 4620–4625 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Y., Feng J. & Sachs G. Helicobacter pylori 5′ureB-sRNA, a cis-encoded antisense small RNA, negatively regulates ureAB expression by transcription termination. J. Bacteriol. 195, 444–452 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber C. C. & Sperandio V. Global analysis of posttranscriptional regulation by GlmY and GlmZ in enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 83, 1286–1295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borin B. N., Tang W. & Krezel A. M. Helicobacter pylori RNA polymerase α-subunit C-terminal domain shows features unique to ɛ-proteobacteria and binds NikR/DNA complexes. Protein Sci. 23, 454–463 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade J. T., Struhl K., Busby S. J. W. & Grainger D. C. Genomic analysis of protein-DNA interactions in bacteria: insights into transcription and chromosome organization. Mol. Microbiol. 65, 21–26 (2007). [DOI] [PubMed] [Google Scholar]
- Roncarati D. et al. Metal-responsive promoter DNA compaction by the ferric uptake regulator. Nat. Commun. 7, 12593 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visweswariah S. S. & Busby S. J. W. Evolution of bacterial transcription factors: how proteins take on new tasks, but do not always stop doing the old ones. Trends Microbiol. 23, 463–467 (2015). [DOI] [PubMed] [Google Scholar]
- Wade J. T., Reppas N. B., Church G. M. & Struhl K. Genomic analysis of LexA binding reveals the permissive nature of the Escherichia coli genome and identifies unconventional target sites. Genes Dev. 19, 2619–2630 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelliciari S., Vannini A., Roncarati D. & Danielli A. The allosteric behavior of Fur mediates oxidative stress signal transduction in Helicobacter pylori. Front. Microbiol. 6, 840 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landt S. G. et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 22, 1813–1831 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.